ABSTRACT
The spatial information of xenobiotics distribution, metabolism, and toxicity mechanisms in situ has drawn increasing attention in both pharmaceutical and environmental toxicology research to aid drug development and environmental risk assessments. Mass spectrometry imaging (MSI) provides a label‐free, multiplexed, and high‐throughput tool to characterize xenobiotics, their metabolites, and endogenous molecules in situ with spatial resolution, providing knowledge on spatially resolved absorption, distribution, metabolism, excretion, and toxicity on the molecular level. In this perspective, we briefly summarize applications of MSI in toxicology on xenobiotic distribution and metabolism, quantification, toxicity mechanisms, and biomarker discovery. We identified several challenges regarding how we can fully harness the power of MSI in both fundamental toxicology research and regulatory practices. First, how can we increase the coverage, sensitivity, and specificity in detecting xenobiotics and their metabolites in complex biological matrices? Second, how can we link the spatial molecular information of xenobiotics to toxicity consequences to understand toxicity mechanisms, predict exposure outcomes, and aid biomarker discovery? Finally, how can we standardize the MSI experiment and data analysis workflow to provide robust conclusions for regulation and drug development? With these questions in mind, we provide our perspectives on the future directions of MSI as a promising tool in spatial toxicology research.
Keywords: mass spectrometry imaging, spatial toxicology, xenobiotics, drug distribution, environmental pollutant, ADMET
1. Spatial Toxicology: Elucidating Xenobiotic‐Biological Interactions in Spatial Context
The absorption, distribution, metabolism, excretion, and toxicity (ADMET) of xenobiotics represent key research topics in pharmacology and toxicology. The concept of ADMET is mostly used in pharmaceutical toxicology to evaluate drugs, whereas it is also applicable to toxicology of environmental pollutants and toxins [1]. Measurements of xenobiotics and their metabolites, as well as endogenous biomolecules, have enabled in‐depth elucidation of ADMET mechanisms at the molecular level and facilitate drug development and risk assessment. In recent years, the spatial contexts of toxicity responses in tissues and cells have attracted increasing attentions. Biological organisms are highly heterogeneous across scales. Molecules, organelles, and cells of different functions form spatially organized compartments to carry out specialized biological functions and interact with each other in spatial context, such as ligand‐receptor interactions and cell‐to‐cell signaling. Xenobiotics entering the body are not uniformly distributed throughout the body but have preferential localizations in specific tissues and cells. The spatial heterogeneity of biological organisms also results in different xenobiotic metabolism and toxicity at different locations. We define the term “spatial toxicology,” derived from the term “spatial biology,” as the subfield of toxicology focusing on elucidating the ADMET mechanisms in biological organisms in spatial context. Compared to measuring bulk, homogenized samples, spatial toxicology investigates xenobiotic‐biological interactions in situ from samples where the tissue and cellular architectures are preserved. Spatial toxicology approaches can be used to reveal highly localized effects in specific regions of the tissues, offering unique and significant insights into how xenobiotics interact with the highly heterogeneous biological organisms.
2. Mass Spectrometry Imaging in Spatial Toxicology
Many tools are available to study the ADMET mechanisms in situ with spatial resolution, such as whole‐body autoradiography [2], positron emission tomography (PET) [3], spectroscopy [4], and recently, spatial transcriptomics [5]. Mass spectrometry imaging (MSI) provides label‐free, highly multiplexed, and high‐throughput measurements to characterize xenobiotics, their metabolites, and endogenous molecules in situ on various types of samples [6, 7, 8, 9, 10, 11]. Matrix‐assisted laser desorption/ionization MS (MALDI‐MS) imaging uses highly focused laser pulses to desorb and ionize chemical matrix and sample materials to achieve chemical imaging. It has been the most commonly applied MSI method for a wide range of molecules. Desorption electrospray ionization (DESI), an emerging MSI technique in recent years, achieves spatial resolution by directing a flow of electrospray onto the sample surface. Secondary ion mass spectrometry (SIMS) is a less commonly used MSI technique but provides unique capabilities in 3D depth profiling by using a focused primary ion beam onto the sample surface and generating secondary ions of samples for measurements. With a proper ion beam source, SIMS can achieve submicron spatial resolution, whereas the nanoSIMS further pushes the limit to as small as 50 nm of lateral resolution [12]. Finally, although not frequently mentioned in the field of MSI, laser ablation inductively coupled plasma MS (LA‐ICP‐MS) is a specialized tool in elemental imaging and plays key roles in studying heavy metal exposure and biological metal homeostasis [13].
Utilization of MSI for toxicology research started in the early 2000s, concurrent with the development and commercialization of MSI instrumentations, and has steadily increased since then, as shown by a keyword search query to the PubMed (Figure 1). A large fraction of MSI's application in toxicology is for drug development. Drug distribution is an indispensable part in the pharmaceutical R&D pipeline, as drugs needs to be distributed to their intended target site in the right form for desired effects. A handful of reviews have summarized in detail the application of MSI in pharmaceutical research and drug development [14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28]. On the other hand, the application of MSI in environmental toxicology is less common, and MSI has not been incorporated in the pipeline of toxicity risk assessment for environmental contaminants. Two recent reviews summarized examples of MSI application in environmental sciences, which included MSI analysis for environmental contaminants [29, 30]. LA‐ICP‐MS has been used to visualize the localization of nano/microsized particulates and heavy metals in rodent tissues and wheat grains [31, 32, 33, 34]. MSI has also been applied to study the spatial distribution and toxicity of pesticides in plants, honeybees, and zebrafish among other organisms [35, 36, 37, 38], and recently, to investigate the spatial distribution of per‐ and polyfluoroalkyl substances (PFAS), a category of emerging contaminants, in zebrafish and rodent models [39, 40, 41, 42, 43]. The application of MSI in the area of environmental toxicology is still rising.
FIGURE 1.
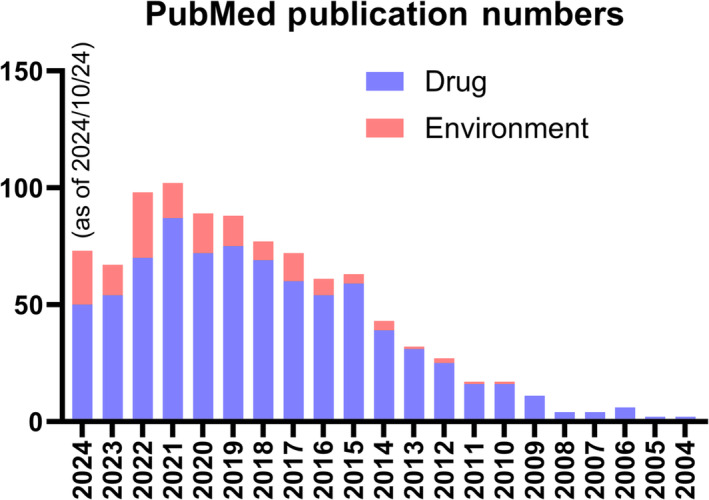
PubMed search results using searching query of (“mass spectrometry imaging”[Title/Abstract] OR “imaging mass spectrometry”[Title/Abstract]) AND (“drug”[Title/Abstract] OR “pharmaceutical”[Title/Abstract]) for drug and (“mass spectrometry imaging”[Title/Abstract] OR “imaging mass spectrometry”[Title/Abstract]) AND (“environmental”[Title/Abstract] OR “pollutant”[Title/Abstract] OR “contaminant”[Title/Abstract]) for environment.
To date, MSI has been an emerging and powerful tool in toxicology to study xenobiotic distribution and metabolism, quantification, toxicity mechanisms, and biomarker discovery, whereas the application is mainly limited in laboratory studies without being fully incorporated into the pipelines for drug discovery and risk assessment. In this perspective, we identify three challenges regarding how we can fully harness the power of MSI in fundamental toxicology research, drug discovery, and regulatory practices. The challenges are as follows: (1) How can we increase the coverage, sensitivity, and specificity in detection of xenobiotics and their metabolites in complex biological matrices? (2) How can we link the spatial distribution to toxicity consequences so that we can understand toxicity mechanisms, predict exposure outcomes, and aid biomarker discovery in spatial context? (3) How can we standardize the MSI experiment and data analysis workflow to provide robust conclusions for regulation and drug development? With these questions in mind, we provide our perspectives on the future directions of using MSI as a promising tool in toxicology research.
3. Comprehensive Spatial Mapping of Xenobiotics and Their Metabolism in Situ
One great advantage of MSI in toxicology is its capability to colocalize the xenobiotics and their metabolites in situ with high spatial resolution. Compared to quantitative whole‐body autoradiography (qWBA), a standard tool to study drug ADMET in pharmacology, MSI can unambiguously identify xenobiotics and their metabolites with high spatial resolution and is label‐free, eliminating the use of radioactively labeled compounds [44, 45]. As an example, using MALDI‐MS imaging, Sun et al. [46] showed differential distribution of the drug pirfenidone and its metabolites in mouse lung and kidney, providing valuable information on drug metabolism in situ in relation to histological features. However, it seems that MSI cannot fully replace qWBA yet due to challenges in quantification in complex tissue matrices (being discussed later) and in effective ionization and identification of xenobiotics and their metabolites.
Many xenobiotic molecules, particularly environmental pollutants, are hard to ionize by common MALDI‐ and ESI‐based MS imaging techniques, such as polychlorinated biphenyls (PCBs), polycyclic aromatic hydrocarbons (PAHs), polybrominated diphenyl ethers (PBDE), and other persistent organic pollutants (POPs). Due to their nonpolar or low‐polar properties and high volatility, these pollutants are commonly analyzed with gas chromatography (GC)‐MS with electron ionization (EI) and chemical ionization (CI) techniques [47, 48], whereas unfortunately, EI and CI are not compatible with imaging. Several reports showed that graphene or graphene oxide films can be used as a MALDI matrix for the detection of PAH [49], octachlorodibenzo‐p‐dioxin [50], and nitro‐PAH [51]. A recent work by Huang et al. [52] used tetraphenyl phosphonium chloride (Ph4PCl) as an additive to enhance the electrospray ionization of polyhalogenated compounds. The additive was applied to an air flow‐assisted ionization source to map the spatial distribution of chlorinated paraffins and hexabromocyclododecane in exposed zebrafish. Multiphoton laser desorption/ionization has also shown ionization of PCBs and PAHs [53, 54]. Overall, both laser‐based and electrospray‐based ionization have successfully shown MS imaging for nonpolar and low‐polar xenobiotics by developing novel matrices, manipulating laser configurations, and exploring new additives.
In addition, xenobiotics and their metabolism products may have distinct physiochemical properties, making it challenging to have comprehensive coverage for both precursor and products in one MSI run. For example, POPs like PCBs can be metabolized in vivo to hydroxylated forms like OH‐PCBs. These metabolites are more polar and commonly analyzed via liquid chromatography (LC)‐MS with ESI. Therefore, it is possible to use ESI‐based ionization such as DESI to image the POP hydroxylated metabolites in tissues. Recently, Zheng et al. [55] showed that atmospheric pressure chemical ionization (APCI) and atmospheric pressure photoionization (APPI) can efficiently ionize POPs such as PAHs and PCBs as well as their hydroxylated metabolites. Thus, although not commercially available, a UV laser ablation (LA)‐APCI‐MS setup [56] holds great potential to spatially map POPs and their metabolites simultaneously. Notably, the previously mentioned work using Ph4PCl as an ESI additive achieved simultaneous detection of polyhalogenated xenobiotics and endogenous metabolites of distinct properties, making it a highly promising method for mapping xenobiotics, their metabolites, and endogenous molecules on one sample [52]. It should be noted that the high volatility of many POPs will require careful design of sample preparation for imaging; for example, the common vacuum drying step in MALDI‐MSI sample preparation may cause the loss of volatile molecules. Compared to (halogenated) hydrocarbons, drug molecules are usually easier to ionize using MALDI and ESI. MSI of drugs and their metabolites have been reported using MALD‐MS imaging [44, 46, 57, 58, 59], although the differences in ionization efficiency should be considered to derive quantitative models of drug ADMET.
Without a pre‐ionization separation such as chromatography, MS imaging of xenobiotics also faces the challenges of ion suppression from abundant biological molecules, and such effect is sample‐dependent. As an example, Li et al. [43] showed detection of similar perfluorooctane sulfonate (PFOS) levels in the kidney, spleen, heart, and brain of dosed animals using LC‐MS/MS, but the MALDI‐MS imaging results only showed PFOS distribution in kidney, spleen, and heart, not in brain, indicating a tissue‐dependent ion suppression on PFOS detection in MALDI‐MS imaging. Possible solutions include increasing ionization efficiencies with secondary ionization, developing new MALDI matrices, and on‐tissue derivatization. In general, ionization of xenobiotics represents a challenge for comprehensively mapping and colocalizing the spatial distribution of xenobiotic and their metabolites, and future efforts are needed in this direction to enable more effective, efficient, and biological matrix‐tolerating ionization and sample preparation methods.
Finally, the isomerism of xenobiotics and their metabolites also poses challenges in the chromatography‐free MSI techniques, as mass analyzers cannot simply differentiate isomers with the same chemical formula. Isomers of xenobiotics and their metabolites widely exist. As an example, branched chain PFAS may take up to 60% in abiotic environmental samples and show differential accumulation patterns as well as health effects in biological organisms [60]. Ion mobility spectrometry (IMS), a gas‐phase separation technique, represents a powerful tool for isomer separation post‐ionization [61, 62]. Recently, Zheng et al. [55] demonstrated ion mobility separation of parent and metabolized PAH, PCB, and PBDE isomers using drift tube IMS, and our group demonstrated the separation of PFOS branched vs linear isomers using MALDI‐MS coupled with trapped ion mobility spectrometry (TIMS) [63]. With the development of IMS, we expect combination of MSI and IMS to be applied to resolve the isomerism of xenobiotics and their metabolites in situ.
4. Uncovering Toxicity Mechanisms and Biomarkers in Highly Heterogeneous Tissues
MS imaging is an emerging tool to understand spatially resolved toxicity mechanisms [37, 41, 64, 65, 66, 67, 68]. LC‐MS‐based proteomics, lipidomics, and metabolomics are widely used to understand the changes of endogenous biomolecules upon xenobiotic exposure; however, the spatial information was lost during sample preparation. MS imaging helps to identify features that changed their levels at different regions. By atmospheric pressure‐MALDI‐MS imaging, Zeng et al. [68] resolved changes of endogenous lipids in different regions of kidney (cortex, medulla, and juxtamedullary cortex) after acute cadmium exposure. Another work by Liu et al. [37] on MS imaging of zebrafish showed differential patterns of lipids in zebrafish eyes after fipronil exposure. Spatially resolved isotope tracing with MS imaging was also used to discover mechanisms of action of a central nervous system drug [69]. These reports showcase that MS imaging can pinpoint the molecular changes with spatial resolution in highly heterogeneous tissues. It should be noted that compared to LC‐MS, MS imaging is less quantitative due to the biological matrix interference, crystal heterogeneity (MALDI only), and analyte‐dependent variations in ionization. Therefore, researchers need to be careful when quantitatively comparing the signal intensities in two tissues sections. Conclusions should be confirmed with enough biological replicates. Several ways to minimize sample‐to‐sample variation include matching the sectioning plane of control and exposed tissues, mounting the control and exposed sections on the same slide, performing MS imaging in one instrumental run, and randomizing the order of imaging runs.
Compared to MS imaging, LC‐MS‐based omics approach provides more quantitative analysis and more comprehensive coverage of molecules. Thus, combination of both methods represents a strategy for comprehensive investigations on toxicity mechanisms. Using zebrafish as a model, Ma et al. [64] identified affected metabolites and molecular pathways after indoxacarb exposure with the LC‐MS metabolomics data, followed by MS imaging to confirm changes of the identified metabolites in situ in zebrafish livers. A similar approach was used for mouse models after cadmium exposure [67]. In both studies, MS imaging was used to confirm the results from LC‐MS metabolomics by showing decreased signal intensities in tissues. An interesting thinking of reversing their roles is to use MSI to identify spatially resolved features as potential biomarkers for xenobiotic exposure and then use (ideally spatial) LC‐MS to quantitatively confirm the identity of potential biomarkers with standards, retention time, and fragmentation patterns. This approach focuses on using MSI as the main tool for biomarker discovery, thus reducing the possibility of missing low‐abundant but highly localized metabolites using LC‐MS approaches. This approach requires researchers to ensure a good coverage of molecular profiles in MSI analysis. For example, multiple matrices and different polarities can be used to increase coverage for MALDI‐MSI [68]. In addition, spatial sampling approaches, such as laser capture microdissection (LCM) [70] or liquid microextraction [65], together with small‐volume LC‐MS analysis, may be considered to catch up with the spatial resolution of MSI when two approaches are integrated.
Another exciting development is the application of MS imaging in protein biomarker discovery for xenobiotic exposure. Meistermann et al. [65] demonstrated the application of MALDI‐MSI in the spatial profiling of proteins in kidneys and discovered a protein, transthyretin, as a biomarker for gentamicin nephrotoxicity. The protein signals found in MSI were also confirmed by liquid microextraction on tissue surface followed by LC‐MS. An intriguing recent study used MALDI‐MSI to monitor drug target engagement by measuring histone poly acetylation, identified by mass shifts, under histone deacetylase drug treatment [66]. Both studies focused on protein biomarkers. Protein MSI is challenging as tandem MS‐based sequencing is necessary for protein identification, posing challenges in ionization efficiency, instrument capabilities, and data processing [71]. Thus, to use it as a biomarker discovery tool, integration with LC‐MS‐based proteomics should be considered for protein identification.
The potential of MSI in understanding spatially resolved toxicity mechanisms has yet to be fully harnessed. In addition to what has been demonstrated, we provide our perspective on its potential future developments. First, MSI can be multiplexed and/or coupled with other imaging modalities to provide multidimensional pictures of xenobiotic toxicity in biological organisms. These developments of multimodal imaging include multiplexing different MSI techniques [72] and/or combining MSI with other imaging modalities such as histopathology and immunohistochemical staining [73], in situ fluorescence hybridization [74], and infrared spectroscopic imaging [75]. Such multimodal combinations will help to link the spatial molecular features detected by MSI to phenotypes (e.g., pathological changes and cell types) and biological endpoints (e.g., gene expression). Secondly, MSI can be applied in causative mechanistic studies, such as profiling xenobiotics and their metabolism after blocking xenobiotic receptors and/or genetically manipulating model organisms. Finally, MSI can be used to elucidate subcellular toxicity mechanisms by further pushing the resolution and sensitivity to resolve the spatial distribution of xenobiotics and endogenous molecules in subcellular compartments, which will be a significant milestone for spatial toxicology studies.
5. Standardizing MSI Practice for Regulatory and R&D Purposes
Although MSI has proven a powerful tool for spatial toxicology research, it has not yet been systematically incorporated into the pipelines of regulatory and R&D processes. Standardization of MSI practices is mentioned by several reviews as a necessity for MSI to be validated to achieve the metrology and standard needed for regulatory submissions [14, 18, 23]. Many variables exist in MSI workflow from sample preparation to instrumentation. For example, in MALDI‐MSI analysis, sample preparation includes tissue freezing, cryo‐sectioning, mounting, drying, and matrix application, and instrumental analysis can be done on mass spectrometers with different configurations with tunable laser intensity/profile and ion optics. Whereas these variations make MSI highly tunable and adaptable for various research questions, they also pose challenges in standardizing MSI practices for regulatory and R&D purposes. Guidelines of using LC‐MS/MS for measuring drugs and environmental contaminants in different sample types have been developed and validated by authorities such as US Food and Drug Administration and Environmental Protection Agency. MSI methods will need to be similarly standardized in all aspects including sample type, sample preparation, instrumental parameters, and data analysis. And their reproducibility, accuracy, precision, specificity, and sensitivity need to be validated across instruments and sites, in order to make MSI results acceptable for regulatory submissions. Studies on multiplatform and multisite comparisons are valuable in the efforts for standardization. Boskamp et al. [76] tested the site‐to‐site reproducibility of MALDI‐MSI by using a single human teratoma sample and a tissue microarray of tumor samples and comparing MALDI‐MSI results obtained in two independent labs with varying protocols. They found that a cross‐normalization strategy, which captured and matched the statistical distribution of spectral intensities, can significantly reduce intersample and interlab batch effects and also cross‐protocol variations. This cross‐normalization represents a promising data preprocessing step for MSI standardization to minimize site‐to‐site variations. Future multiplatform and multisite research will keep shedding light on the key factors in optimizing MSI standardization.
Achieving quantitative MSI (qMSI) also represents a challenge in standardization. Quantification of xenobiotics and endogenous metabolites is important for studies in xenobiotic ADMET and biomarker discovery. However, qMSI is intrinsically hard due to the spatial heterogeneity of biological matrices on tissue, which affect the analyte ionization efficiency (“matrix effects”) and extraction efficiency in situ. Internal standards can be used for relative quantification [77]. For absolute quantification in qMSI, several strategies have been developed to build calibration curves, including in‐solution, on‐tissue (under‐tissue as a variant), and in‐tissue [78, 79]. In‐solution calibration curves are collected from standards directly spotted onto the target plate/slide, whereas on‐tissue strategy spots the standards onto (or under) an untreated, blank tissue section. Compared to in‐solution, the on‐tissue strategy helps to minimize matrix effects. The in‐tissue strategy addresses both matrix effect and analyte extraction efficiency by creating tissue mimetics that are spiked and mixed with different concentrations of standards. However, it is the most time‐ and sample‐consuming strategy. Balancing the pros and cons, on‐tissue calibration curves are currently the most common qMSI strategy. Recently, an intriguing machine learning‐based virtual calibration qMSI strategy was reported to map the drug distribution in whole‐animal sections, which is highly heterogeneous and hard to perform on‐tissue strategy [80]. The authors implemented machine learning‐based regression models to predict calibration factors for correcting matrix effects and extractability based on endogenous metabolite signals. They successfully demonstrated pharmacokinetic evaluation of drugs in whole‐animal sections. This method is yet to be tested in more scenarios, whereas it holds high potential for quantification of xenobiotics and their metabolites in highly heterogeneous samples. Finally, the selection of mass spectrometer also affects the results of quantification. MSI experiments using DESI showed that a triple quadrupole MS provided overall best performance compared to other quadrupole time‐of‐flight instruments [81]. In general, qMSI is an important part in developing MSI standardization and should be systematically validated for regulatory and R&D purposes.
6. Outlook
Spatial toxicology represents a subfield of toxicology that investigates xenobiotic ADMET mechanisms in spatial context in cells and tissues. It provides significant insights into xenobiotic‐biological interactions in highly heterogeneous biological organisms. Compared to measurement on the ensemble averages of bulk samples, spatial toxicology approaches provide the opportunity to reveal highly localized effects in specific regions in tissues or even cells, aiding accurate assessments of drug safety/efficacy and pollutant risks. MSI, a label‐free, highly multiplexed, and high‐throughput analysis to measure xenobiotics, their metabolites, and endogenous biological molecules, is a powerful tool to study spatial toxicology. With the development of MSI methodology, instrumentation, and data analysis, together with the integration with other analytical modalities, MSI holds high potential to play a major role for future spatial toxicology research, and standardization of MSI practices will further help this methodology to be validated and integrated for regulatory and R&D practices.
Conflicts of Interest
The author declares no conflicts of interest.
Acknowledgements
The author would like to thank the start‐up support from Michigan State University and a Starter Grant from the Society for Analytical Chemists of Pittsburgh to T.A.Q.
Funding: This work was supported by the Society for Analytical Chemists of Pittsburgh and the Michigan State University.
Data Availability Statement
The dataset used to plot Figure 1 will be available upon request to the corresponding author. No other datasets were generated or analyzed in the current manuscript.
References
- 1. van Vugt‐Lussenburg B. M. A., Capinha L., Reinen J., et al., ““Commandeuring” Xenobiotic Metabolism: Advances in Understanding Xenobiotic Metabolism,” Chemical Research in Toxicology 35, no. 7 (2022): 1184–1201, 10.1021/acs.chemrestox.2c00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Solon E. G., “Autoradiography Techniques and Quantification of Drug Distribution,” Cell and Tissue Research 360, no. 1 (2015): 87–107, 10.1007/s00441-014-2093-4. [DOI] [PubMed] [Google Scholar]
- 3. Fischman A. J., Alpert N. M., and Rubin R. H., “Pharmacokinetic Imaging,” Clinical Pharmacokinetics 41, no. 8 (2002): 581–602, 10.2165/00003088-200241080-00003. [DOI] [PubMed] [Google Scholar]
- 4. El‐Mashtoly S. F., Petersen D., Yosef H. K., et al., “Label‐Free Imaging of Drug Distribution and Metabolism in Colon Cancer Cells by Raman Microscopy,” Analyst 139, no. 5 (2014): 1155–1161, 10.1039/C3AN01993D. [DOI] [PubMed] [Google Scholar]
- 5. Nault R., Saha S., Bhattacharya S., Sinha S., Maiti T., and Zacharewski T., “Single‐Cell Transcriptomics Shows Dose‐Dependent Disruption of Hepatic Zonation by TCDD in Mice,” Toxicological Sciences 191, no. 1 (2023): 135–148, 10.1093/toxsci/kfac109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xiao Y., Deng J., Yao Y., Fang L., Yang Y., and Luan T., “Recent Advances of Ambient Mass Spectrometry Imaging for Biological Tissues: A Review,” Analytica Chimica Acta 1117 (2020): 74–88, 10.1016/j.aca.2020.01.052. [DOI] [PubMed] [Google Scholar]
- 7. Spraker J. E., Luu G. T., and Sanchez L. M., “Imaging Mass Spectrometry for Natural Products Discovery: A Review of Ionization Methods,” Natural Product Reports 37, no. 2 (2020): 150–162, 10.1039/C9NP00038K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moore J. L. and Charkoftaki G., “A Guide to MALDI Imaging Mass Spectrometry for Tissues,” Journal of Proteome Research 22, no. 11 (2023): 3401–3417, 10.1021/acs.jproteome.3c00167. [DOI] [PubMed] [Google Scholar]
- 9. Buchberger A. R., DeLaney K., Johnson J., and Li L., “Mass Spectrometry Imaging: A Review of Emerging Advancements and Future Insights,” Analytical Chemistry 90, no. 1 (2018): 240–265, 10.1021/acs.analchem.7b04733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Prentice B. M., “Imaging With Mass Spectrometry: Which Ionization Technique Is Best?,” Journal of Mass Spectrometry 59, no. 5 (2024): e5016, 10.1002/jms.5016. [DOI] [PubMed] [Google Scholar]
- 11. Soudah T., Zoabi A., and Margulis K., “Desorption Electrospray Ionization Mass Spectrometry Imaging in Discovery and Development of Novel Therapies,” Mass Spectrometry Reviews 42, no. 2 (2023): 751–778, 10.1002/mas.21736. [DOI] [PubMed] [Google Scholar]
- 12. Nuñez J., Renslow R., J. B. Cliff, III , and Anderton C. R., “NanoSIMS for Biological Applications: Current Practices and Analyses,” Biointerphases 13, no. 3 (2017): 03B301, 10.1116/1.4993628. [DOI] [PubMed] [Google Scholar]
- 13. Sabine Becker J., Zoriy M., Wu B., Matusch A., and Susanne Becker J., “Imaging of Essential and Toxic Elements in Biological Tissues by LA‐ICP‐MS,” Journal of Analytical Atomic Spectrometry 23, no. 9 (2008): 1275–1280, 10.1039/B805228J. [DOI] [Google Scholar]
- 14. Schulz S., Becker M., Groseclose M. R., Schadt S., and Hopf C., “Advanced MALDI Mass Spectrometry Imaging in Pharmaceutical Research and Drug Development,” Current Opinion in Biotechnology 55 (2019): 51–59, 10.1016/j.copbio.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 15. Goodwin R. J. A., Bunch J., and McGinnity D. F., “Chapter Six—Mass Spectrometry Imaging in Oncology Drug Discovery,” in Advances in Cancer Research. Applications of Mass Spectrometry Imaging to Cancer, vol. 134, eds. Drake R. R. and McDonnell L. A. (Cambridge, MA: Academic Press, 2017): 133–171, 10.1016/bs.acr.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 16. Karlsson O. and Hanrieder J., “Imaging Mass Spectrometry in Drug Development and Toxicology,” Archives of Toxicology 91, no. 6 (2017): 2283–2294, 10.1007/s00204-016-1905-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Swales J. G., Hamm G., Clench M. R., and Goodwin R. J. A., “Mass Spectrometry Imaging and Its Application in Pharmaceutical Research and Development: A Concise Review,” International Journal of Mass Spectrometry 437 (2019): 99–112, 10.1016/j.ijms.2018.02.007. [DOI] [Google Scholar]
- 18. Nilsson A., Goodwin R. J. A., Shariatgorji M., Vallianatou T., Webborn P. J. H., and Andrén P. E., “Mass Spectrometry Imaging in Drug Development,” Analytical Chemistry 87, no. 3 (2015): 1437–1455, 10.1021/ac504734s. [DOI] [PubMed] [Google Scholar]
- 19. Chen Y., Xie Y., Li L., Wang Z., and Yang L., “Advances in Mass Spectrometry Imaging for Toxicological Analysis and Safety Evaluation of Pharmaceuticals,” Mass Spectrometry Reviews 42, no. 5 (2023): 2207–2233, 10.1002/mas.21807. [DOI] [PubMed] [Google Scholar]
- 20. Kumar B. S., “Recent Developments and Applications of Ambient Mass Spectrometry Imaging in Pharmaceutical Research: An Overview,” Analytical Methods 16, no. 1 (2023): 8–32, 10.1039/D3AY01267K. [DOI] [PubMed] [Google Scholar]
- 21. Spruill M. L., Maletic‐Savatic M., Martin H., Li F., and Liu X., “Spatial Analysis of Drug Absorption, Distribution, Metabolism, and Toxicology Using Mass Spectrometry Imaging,” Biochemical Pharmacology 201 (2022): 115080, 10.1016/j.bcp.2022.115080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sugiura Y. and Setou M., “Imaging Mass Spectrometry for Visualization of Drug and Endogenous Metabolite Distribution: Toward In Situ Pharmacometabolomes,” Journal of Neuroimmune Pharmacology 5, no. 1 (2010): 31–43, 10.1007/s11481-009-9162-6. [DOI] [PubMed] [Google Scholar]
- 23. Nishidate M., Hayashi M., Aikawa H., et al., “Applications of MALDI Mass Spectrometry Imaging for Pharmacokinetic Studies During Drug Development,” Drug Metabolism and Pharmacokinetics 34, no. 4 (2019): 209–216, 10.1016/j.dmpk.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 24. Lietz C. B., Gemperline E., and Li L., “Qualitative and Quantitative Mass Spectrometry Imaging of Drugs and Metabolites,” Advanced Drug Delivery Reviews 65, no. 8 (2013): 1074–1085, 10.1016/j.addr.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Holbrook J. H., Kemper G. E., and Hummon A. B., “Quantitative Mass Spectrometry Imaging: Therapeutics & Biomolecules,” Chemical Communications 60, no. 16 (2024): 2137–2151, 10.1039/D3CC05988J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen B., Vavrek M., and Cancilla M. T., “From Molecules to Visuals: Empowering Drug Discovery and Development With Mass Spectrometry Imaging,” Journal of Mass Spectrometry 59, no. 5 (2024): e5029, 10.1002/jms.5029. [DOI] [PubMed] [Google Scholar]
- 27. Phulara N. R. and Seneviratne H. K., “Mass Spectrometry Imaging‐Based Multi‐Omics Approaches to Understand Drug Metabolism and Disposition,” Journal of Mass Spectrometry 59, no. 7 (2024): e5042, 10.1002/jms.5042. [DOI] [PubMed] [Google Scholar]
- 28. Rajbhandari P., Neelakantan T. V., Hosny N., and Stockwell B. R., “Spatial Pharmacology Using Mass Spectrometry Imaging,” Trends in Pharmacological Sciences 45, no. 1 (2024): 67–80, 10.1016/j.tips.2023.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maloof K. A., Reinders A. N., and Tucker K. R., “Applications of Mass Spectrometry Imaging in the Environmental Sciences,” Current Opinion in Environmental Science & Health 18 (2020): 54–62, 10.1016/j.coesh.2020.07.005. [DOI] [Google Scholar]
- 30. Herruzo‐Ruiz A. M., Peralbo‐Molina Á., López C.‐M., Michán C., Alhama J., and Chicano‐Gálvez E., “Mass Spectrometry Imaging in Environmental Monitoring: From a Scarce Existing Past to a Promising Future,” Trends in Environmental Analytical Chemistry 42 (2024): e00228, 10.1016/j.teac.2024.e00228. [DOI] [Google Scholar]
- 31. Chen B., Lum J. T.‐S., Huang Y., Hu B., and Leung K. S.‐Y., “Integration of Sub‐Organ Quantitative Imaging LA‐ICP‐MS and Fractionation Reveals Differences in Translocation and Transformation of CeO2 and Ce3+ in Mice,” Analytica Chimica Acta 1082 (2019): 18–29, 10.1016/j.aca.2019.07.044. [DOI] [PubMed] [Google Scholar]
- 32. Kuraś R., Stępnik M., Domeradzka‐Gajda K., and Janasik B., “The Use of LA‐ICP‐MS as an Auxiliary Tool to Assess the Pulmonary Toxicity of Molybdenum (IV) Sulfide (MoS2) Nano‐ and Microparticles,” International Journal of Occupational Medicine and Environmental Health 37, no. 1 (2024): 18–33, 10.13075/ijomeh.1896.02305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu J., Cui J., Wei X., et al., “Investigation on Selenium and Mercury Interactions and the Distribution Patterns in Mice Organs With LA‐ICP‐MS Imaging,” Analytica Chimica Acta 1182 (2021): 338941, 10.1016/j.aca.2021.338941. [DOI] [PubMed] [Google Scholar]
- 34. Zhang X.‐Y., Geng L.‐P., Gao P.‐P., et al., “Bioimaging of Pb by LA‐ICP‐MS and Pb Isotopic Compositions Reveal Distributions and Origins of Pb in Wheat Grain,” Science of the Total Environment 802 (2022): 149729, 10.1016/j.scitotenv.2021.149729. [DOI] [PubMed] [Google Scholar]
- 35. Taira S., Tokai M., Kaneko D., Katano H., and Kawamura‐Konishi Y., “Mass Spectrometry Imaging Analysis of Location of Procymidone in Cucumber Samples,” Journal of Agricultural and Food Chemistry 63, no. 27 (2015): 6109–6112, 10.1021/acs.jafc.5b00957. [DOI] [PubMed] [Google Scholar]
- 36. Gerbig S., Brunn H. E., Spengler B., and Schulz S., “Spatially Resolved Investigation of Systemic and Contact Pesticides in Plant Material by Desorption Electrospray Ionization Mass Spectrometry Imaging (DESI‐MSI),” Analytical and Bioanalytical Chemistry 407, no. 24 (2015): 7379–7389, 10.1007/s00216-015-8900-2. [DOI] [PubMed] [Google Scholar]
- 37. Liu W., Nie H., Liang D., Bai Y., and Liu H., “Phospholipid Imaging of Zebrafish Exposed to Fipronil Using Atmospheric Pressure Matrix‐Assisted Laser Desorption Ionization Mass Spectrometry,” Talanta 209 (2020): 120357, 10.1016/j.talanta.2019.120357. [DOI] [PubMed] [Google Scholar]
- 38. Zhang Y., Chen D., Du M., et al., “Insights Into the Degradation and Toxicity Difference Mechanism of Neonicotinoid Pesticides in Honeybees by Mass Spectrometry Imaging,” Science of the Total Environment 774 (2021): 145170, 10.1016/j.scitotenv.2021.145170. [DOI] [PubMed] [Google Scholar]
- 39. Shi Q., Zhang X., Liu X., Yan C., and Lu S., “Visualization of PFOA Accumulation and Its Effects on Phospholipid in Zebrafish Liver by MALDI Imaging,” Analytical and Bioanalytical Chemistry 416 (2024): 2493–2501, 10.1007/s00216-024-05214-y. [DOI] [PubMed] [Google Scholar]
- 40. Bian Y., He M.‐Y., Ling Y., et al., “Tissue Distribution Study of Perfluorooctanoic Acid in Exposed Zebrafish Using MALDI Mass Spectrometry Imaging,” Environmental Pollution 293 (2022): 118505, 10.1016/j.envpol.2021.118505. [DOI] [PubMed] [Google Scholar]
- 41. Chen Y., Jiang L., Zhang R., et al., “Spatially Revealed Perfluorooctane Sulfonate‐Induced Nephrotoxicity in Mouse Kidney Using Atmospheric Pressure MALDI Mass Spectrometry Imaging,” Science of the Total Environment 838 (2022): 156380, 10.1016/j.scitotenv.2022.156380. [DOI] [PubMed] [Google Scholar]
- 42. Yang C., Lee H. K., Zhang Y., et al., “In Situ Detection and Imaging of PFOS in Mouse Kidney by Matrix‐Assisted Laser Desorption/Ionization Imaging Mass Spectrometry,” Analytical Chemistry 91, no. 14 (2019): 8783–8788, 10.1021/acs.analchem.9b00711. [DOI] [PubMed] [Google Scholar]
- 43. Li X., Li T., Wang Z., et al., “Distribution of Perfluorooctane Sulfonate in Mice and Its Effect on Liver Lipidomic,” Talanta 226 (2021): 122150, 10.1016/j.talanta.2021.122150. [DOI] [PubMed] [Google Scholar]
- 44. Stoeckli M., Staab D., and Schweitzer A., “Compound and Metabolite Distribution Measured by MALDI Mass Spectrometric Imaging in Whole‐Body Tissue Sections,” International Journal of Mass Spectrometry 260, no. 2 (2007): 195–202, 10.1016/j.ijms.2006.10.007. [DOI] [Google Scholar]
- 45. Perez C. J., Tata A., de Campos M. L., Peng C., and Ifa D. R., “Monitoring Toxic Ionic Liquids in Zebrafish (Danio Rerio) With Desorption Electrospray Ionization Mass Spectrometry Imaging (DESI‐MSI),” Journal of the American Society for Mass Spectrometry 28, no. 6 (2017): 1136–1148, 10.1007/s13361-016-1515-9. [DOI] [PubMed] [Google Scholar]
- 46. Sun N., Fernandez I. E., Wei M., et al., “Pharmacokinetic and Pharmacometabolomic Study of Pirfenidone in Normal Mouse Tissues Using High Mass Resolution MALDI‐FTICR‐Mass Spectrometry Imaging,” Histochemistry and Cell Biology 145, no. 2 (2016): 201–211, 10.1007/s00418-015-1382-7. [DOI] [PubMed] [Google Scholar]
- 47. Focant J.‐F., Pirard C., Eppe G., and de Pauw E., “Recent Advances in Mass Spectrometric Measurement of Dioxins,” Journal of Chromatography. A 1067, no. 1 (2005): 265–275, 10.1016/j.chroma.2004.10.095. [DOI] [PubMed] [Google Scholar]
- 48. Ayala‐Cabrera J. F., Santos F. J., and Moyano E., “Recent Advances in Analytical Methodologies Based on Mass Spectrometry for the Environmental Analysis of Halogenated Organic Contaminants,” Trends in Environmental Analytical Chemistry 30 (2021): e00122, 10.1016/j.teac.2021.e00122. [DOI] [Google Scholar]
- 49. Zhang J., Dong X., Cheng J., Li J., and Wang Y., “Efficient Analysis of Non‐Polar Environmental Contaminants by MALDI‐TOF MS With Graphene as Matrix,” Journal of the American Society for Mass Spectrometry 22, no. 7 (2011): 1294–1298, 10.1007/s13361-011-0143-7. [DOI] [PubMed] [Google Scholar]
- 50. Zhou X., Wei Y., He Q., Boey F., Zhang Q., and Zhang H., “Reduced Graphene Oxide Films Used as Matrix of MALDI‐TOF‐MS for Detection of Octachlorodibenzo‐p‐Dioxin,” Chemical Communications 46, no. 37 (2010): 6974–6976, 10.1039/C0CC01681K. [DOI] [PubMed] [Google Scholar]
- 51. Ma N., Bian W., Li R., et al., “Quantitative Analysis of Nitro‐Polycyclic Aromatic Hydrocarbons in PM 2.5 Samples With Graphene as a Matrix by MALDI‐TOF MS,” Analytical Methods 7, no. 9 (2015): 3967–3971, 10.1039/C5AY00341E. [DOI] [Google Scholar]
- 52. Huang Y., Shang H., Wang C., et al., “Spatially Resolved Co‐Imaging of Polyhalogenated Xenobiotics and Endogenous Metabolites Reveals Xenobiotic‐Induced Metabolic Alterations,” Environmental Science & Technology 57, no. 48 (2023): 19330–19340, 10.1021/acs.est.3c05817. [DOI] [PubMed] [Google Scholar]
- 53. Shitamichi O., Matsui T., Hui Y., Chen W., and Imasaka T., “Determination of Persistent Organic Pollutants by Gas Chromatography/Laser Multiphoton Ionization/Time‐of‐Flight Mass Spectrometry,” Frontiers of Environmental Science & Engineering 6, no. 1 (2012): 26–31, 10.1007/s11783-011-0374-7. [DOI] [Google Scholar]
- 54. Mahajan T. B., Ghosh U., Zare R. N., and Luthy R. G., “Microscale Detection of Polychlorinated Biphenyls Using Two‐Step Laser Mass Spectrometry,” International Journal of Mass Spectrometry 212, no. 1 (2001): 41–48, 10.1016/S1387-3806(01)00470-5. [DOI] [Google Scholar]
- 55. Zheng X., Dupuis K. T., Aly N. A., et al., “Utilizing ion Mobility Spectrometry and Mass Spectrometry for the Analysis of Polycyclic Aromatic Hydrocarbons, Polychlorinated Biphenyls, Polybrominated Diphenyl Ethers and Their Metabolites,” Analytica Chimica Acta 1037 (2018): 265–273, 10.1016/j.aca.2018.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Herdering C., Reifschneider O., Wehe C. A., Sperling M., and Karst U., “Ambient Molecular Imaging by Laser Ablation Atmospheric Pressure Chemical Ionization Mass Spectrometry,” Rapid Communications in Mass Spectrometry 27, no. 23 (2013): 2595–2600, 10.1002/rcm.6727. [DOI] [PubMed] [Google Scholar]
- 57. Marko‐Varga G., Fehniger T. E., Rezeli M., Döme B., Laurell T., and Végvári Á., “Drug Localization in Different Lung Cancer Phenotypes by MALDI Mass Spectrometry Imaging,” Journal of Proteomics 74, no. 7 (2011): 982–992, 10.1016/j.jprot.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 58. Tang W., Chen J., Zhou J., et al., “Quantitative MALDI Imaging of Spatial Distributions and Dynamic Changes of Tetrandrine in Multiple Organs of Rats,” Theranostics 9, no. 4 (2019): 932–944, 10.7150/thno.30408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kertesz V., van Berkel G. J., Vavrek M., Koeplinger K. A., Schneider B. B., and Covey T. R., “Comparison of Drug Distribution Images From Whole‐Body Thin Tissue Sections Obtained Using Desorption Electrospray Ionization Tandem Mass Spectrometry and Autoradiography,” Analytical Chemistry 80, no. 13 (2008): 5168–5177, 10.1021/ac800546a. [DOI] [PubMed] [Google Scholar]
- 60. Schulz K., Silva M. R., and Klaper R., “Distribution and Effects of Branched Versus Linear Isomers of PFOA, PFOS, and PFHxS: A Review of Recent Literature,” Science of the Total Environment 733 (2020): 139186, 10.1016/j.scitotenv.2020.139186. [DOI] [PubMed] [Google Scholar]
- 61. Dodds J. N., Alexander N. L. M., Kirkwood K. I., et al., “From Pesticides to Per‐ and Polyfluoroalkyl Substances: An Evaluation of Recent Targeted and Untargeted Mass Spectrometry Methods for Xenobiotics,” Analytical Chemistry 93, no. 1 (2021): 641–656, 10.1021/acs.analchem.0c04359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Foster M., Rainey M., Watson C., et al., “Uncovering PFAS and Other Xenobiotics in the Dark Metabolome Using Ion Mobility Spectrometry, Mass Defect Analysis, and Machine Learning,” Environmental Science & Technology 56, no. 12 (2022): 9133–9143, 10.1021/acs.est.2c00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Reynolds A. J., Smith A. M., and Qiu T. A., “Detection, Quantification, and Isomer Differentiation of Per‐ and Polyfluoroalkyl Substances (PFAS) Using MALDI‐TOF With Trapped Ion Mobility,” Journal of the American Society for Mass Spectrometry 35, no. 2 (2024): 317–325, 10.1021/jasms.3c00369. [DOI] [PubMed] [Google Scholar]
- 64. Ma L., Yin Z., Xie Q., et al., “Metabolomics and Mass Spectrometry Imaging Reveal the Chronic Toxicity of Indoxacarb to Adult Zebrafish (Danio Rerio) Livers,” Journal of Hazardous Materials 453 (2023): 131304, 10.1016/j.jhazmat.2023.131304. [DOI] [PubMed] [Google Scholar]
- 65. Meistermann H., Norris J. L., Aerni H.‐R., et al., “Biomarker Discovery by Imaging Mass Spectrometry: Transthyretin Is a Biomarker for Gentamicin‐Induced Nephrotoxicity in Rat,” Molecular & Cellular Proteomics 5, no. 10 (2006): 1876–1886, 10.1074/mcp.M500399-MCP200. [DOI] [PubMed] [Google Scholar]
- 66. Munteanu B., Meyer B., von Reitzenstein C., et al., “Label‐Free in Situ Monitoring of Histone Deacetylase Drug Target Engagement by Matrix‐Assisted Laser Desorption Ionization‐Mass Spectrometry Biotyping and Imaging,” Analytical Chemistry 86, no. 10 (2014): 4642–4647, 10.1021/ac500038j. [DOI] [PubMed] [Google Scholar]
- 67. Zeng T., Guo W., Jiang L., et al., “Integration of Omics Analysis and Atmospheric Pressure MALDI Mass Spectrometry Imaging Reveals the Cadmium Toxicity on Female ICR Mouse,” Science of the Total Environment 801 (2021): 149803, 10.1016/j.scitotenv.2021.149803. [DOI] [PubMed] [Google Scholar]
- 68. Zeng T., Zhang R., Chen Y., Guo W., Wang J., and Cai Z., “ In Situ Localization of Lipids on Mouse Kidney Tissues With Acute Cadmium Toxicity Using Atmospheric Pressure‐MALDI Mass Spectrometry Imaging,” Talanta 245 (2022): 123466, 10.1016/j.talanta.2022.123466. [DOI] [PubMed] [Google Scholar]
- 69. Jin B., Pang X., Zang Q., et al., “Spatiotemporally Resolved Metabolomics and Isotope Tracing Reveal CNS Drug Targets,” Acta Pharmaceutica Sinica B 13, no. 4 (2022): 1699–1710, 10.1016/j.apsb.2022.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dilillo M., Pellegrini D., Ait‐Belkacem R., de Graaf E. L., Caleo M., and McDonnell L. A., “Mass Spectrometry Imaging, Laser Capture Microdissection, and LC‐MS/MS of the Same Tissue Section,” Journal of Proteome Research 16, no. 8 (2017): 2993–3001, 10.1021/acs.jproteome.7b00284. [DOI] [PubMed] [Google Scholar]
- 71. Han J., Permentier H., Bischoff R., Groothuis G., Casini A., and Horvatovich P., “Imaging of Protein Distribution in Tissues Using Mass Spectrometry: An Interdisciplinary Challenge,” TrAC Trends in Analytical Chemistry 112 (2019): 13–28, 10.1016/j.trac.2018.12.016. [DOI] [Google Scholar]
- 72. Gorman B. L., Taylor M. J., Tesfay L., et al., “Applying Multimodal Mass Spectrometry to Image Tumors Undergoing Ferroptosis Following In Vivo Treatment With a Ferroptosis Inducer,” Journal of the American Society for Mass Spectrometry 35 (2023): 5–12, 10.1021/jasms.3c00193. [DOI] [PubMed] [Google Scholar]
- 73. Kaya I., Michno W., Brinet D., et al., “Histology‐Compatible MALDI Mass Spectrometry Based Imaging of Neuronal Lipids for Subsequent Immunofluorescent Staining,” Analytical Chemistry 89, no. 8 (2017): 4685–4694, 10.1021/acs.analchem.7b00313. [DOI] [PubMed] [Google Scholar]
- 74. Bourceau P., Geier B., Suerdieck V., et al., “Visualization of Metabolites and Microbes at High Spatial Resolution Using MALDI Mass Spectrometry Imaging and in Situ Fluorescence Labeling,” Nature Protocols 18 (2023): 1–30, 10.1038/s41596-023-00864-1. [DOI] [PubMed] [Google Scholar]
- 75. Neumann E. K., Comi T. J., Spegazzini N., et al., “Multimodal Chemical Analysis of the Brain by High Mass Resolution Mass Spectrometry and Infrared Spectroscopic Imaging,” Analytical Chemistry 90, no. 19 (2018): 11572–11580, 10.1021/acs.analchem.8b02913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Boskamp T., Casadonte R., Hauberg‐Lotte L., Deininger S., Kriegsmann J., and Maass P., “Cross‐Normalization of MALDI Mass Spectrometry Imaging Data Improves Site‐To‐Site Reproducibility,” Analytical Chemistry 93, no. 30 (2021): 10584–10592, 10.1021/acs.analchem.1c01792. [DOI] [PubMed] [Google Scholar]
- 77. Wang Y. and Hummon A. B., “Quantification of Irinotecan in Single Spheroids Using Internal Standards by MALDI Mass Spectrometry Imaging,” Analytical Chemistry 95, no. 24 (2023): 9227–9236, 10.1021/acs.analchem.3c00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Tobias F. and Hummon A. B., “Considerations for MALDI‐Based Quantitative Mass Spectrometry Imaging Studies,” Journal of Proteome Research 19, no. 9 (2020): 3620–3630, 10.1021/acs.jproteome.0c00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kibbe R. R. and Muddiman D. C., “Quantitative Mass Spectrometry Imaging (qMSI): A Tutorial,” Journal of Mass Spectrometry 59, no. 4 (2024): e5009, 10.1002/jms.5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Song X., He J., Pang X., et al., “Virtual Calibration Quantitative Mass Spectrometry Imaging for Accurately Mapping Analytes Across Heterogenous Biotissue,” Analytical Chemistry 91, no. 4 (2019): 2838–2846, 10.1021/acs.analchem.8b04762. [DOI] [PubMed] [Google Scholar]
- 81. Lamont L., Hadavi D., Viehmann B., et al., “Quantitative Mass Spectrometry Imaging of Drugs and Metabolites: A Multiplatform Comparison,” Analytical and Bioanalytical Chemistry 413, no. 10 (2021): 2779–2791, 10.1007/s00216-021-03210-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset used to plot Figure 1 will be available upon request to the corresponding author. No other datasets were generated or analyzed in the current manuscript.


