Summary
Tlx3, a master regulator of the fate specification of excitatory neurons, is primarily known to function in post-mitotic cells. Although we have previously identified TLX3 expression in the proliferating granule neuron progenitors (GNPs) of cerebellum, its primary role is unknown. Here, we demonstrate that the dysfunction of Tlx3 from the GNPs significantly reduced its proliferation through regulating anti-proliferative genes. Consequently, the altered generation of GNPs resulted in cerebellar hypoplasia, patterning defects, granule neuron-Purkinje ratio imbalance, and aberrant synaptic connections in the cerebellum. This altered cerebellar homeostasis manifested into a typical autism-like behavior in mice with motor, and social function disabilities. We also show the presence of TLX3 variants with uncharacterized mutations in human cases of autism spectrum disorder (ASD). Altogether, our study establishes Tlx3 as a critical gene involved in developing GNPs and that its deletion from the early developmental stage culminates in autism.
Subject areas: Neuroscience, Molecular Neuroscience, Cellular Neuroscience, Omics, Transcriptomics
Graphical abstract

Highlights
-
•
Tlx3 regulates proliferation and differentiation of cerebellar granule neurons
-
•
Tlx3 ablation leads to upregulation of anti-proliferative genes
-
•
Deletion of Tlx3 causes cerebellar patterning defects and cerebellar hypoplasia
-
•
Loss of Tlx3 induces autistic behavior in mouse
Neuroscience, Molecular Neuroscience, Cellular Neuroscience, Omics, Transcriptomics
Introduction
The homeobox family of transcription factors plays a significant role in vertebrate brain development, and the Hox family of genes, which is a subset of the homeobox family, is primarily involved in the patterning of the hindbrain.1 Tlx (T cell leukemia) genes belong to the Hox family of genes and are expressed in various regions of the developing embryo, including the spleen, spinal cord and branchial arches.2,3,4,5,6,7 Tlx1 (Hox11), Tlx2 (Hox11L1/Enx), and Tlx3 (Hox11L2/Rnx) are the three members of the Tlx family, and each of these genes has distinctive expression pattern and function during development. Tlx1/Hox11 controls the genesis of the spleen, and its deletion leads to asplenia,6,8 whereas Tlx2/Hox11L1 is essential for the intact positional specification and differentiated cell fate of enteric neurons, as noted in Tlx2-deficient mice which developed hyperganglionic megacolon.9 However, Tlx3 becomes quite distinct from its family members since it is predominantly expressed in the nervous system, and Tlx3-null mice die within one day after birth from central respiratory failure.10 Although the distinct expression of Tlx3 in the spinal cord, brain stem, and cerebellum has been well characterized, its functional role in cerebellum is still not fully understood.10,11,12,13 In the spinal cord, Tlx3 acts as a post-mitotic selector gene that promotes glutamatergic over GABAergic cell fates and also regulates dorsal horn pain modulatory peptidergic neurons of the spinal cord.11,14,15 Concurrently, Tlx3 expression is present in the brain stem and contributes explicitly only to the development of first-order relay visceral sensory neurons and noradrenergic centers.12
To date, there are no reports regarding the functional role of TLX3 in the cerebellum. We and others have shown that TLX3 is expressed explicitly in the posterior lobes of the cerebellum between the embryonic day (E) 15 and postnatal day (PN) 20 stages.10,13,16 Although TLX3 was known to be expressed exclusively in post-mitotic neurons, its expression was also evident in the outer proliferating external granular layer (EGL) and internal granular layer (IGL) of the posterior cerebellum.16 The EGL harbors the proliferating cerebellar granule neurons (cGN) and the IGL has the differentiated GN. The cerebellum development starts from early E9 and continues until PN21, to form a complete foliated structure with ten lobules (I–X) identified by its fissures. These lobules can be subdivided into four major lobes such as the anterior lobe (I–V), central lobe (VI–VII), posterior lobe (VIII–IX), and flocculonodular lobe (X) based on specific gene expression patterns.17 After completing the neurogenesis, adult mouse cerebellum contains four distinct layers, which are precisely arranged by diverse cell types generated from the embryonic germinal centers, such as the ventricular zone and upper rhombic lip. GABAergic cell types of the cerebellum, such as Purkinje cells (PC), Golgi, Lugaro, stellate, basket, and glial fibers, are originated from the ventricular zone and are derived by the bHLH transcription factor Ptf1a. Simultaneously, glutamatergic cell types, granule cells (GC), and unipolar brush cells (UBC) are derived from the embryonic structure upper rhombic lip, marked by the transcription factor Atoh1.18,19 Numerous transcription factors, cytokines, and signaling pathways regulate the proliferation, migration, differentiation, and maturation of these diverse cell types. Thus, it is a tightly regulated process, and any alterations could potentially induce developmental disorders, including autism spectrum disorder (ASD), attention deficit hyperactivity disorder (ADHD), and developmental dyslexia. In addition, the cerebellum is implicated in various functions, and disruptions to it during development can have long-lasting consequences for motor skills, learning, and cognition.20 In addition, we have also demonstrated that Pax6 can regulate the expression of Tlx3 in the posterior cerebellum, which in turn controls Chrnα3 expression and other genes associated with ASD.16 Hence, the restricted expression pattern of TLX3 during the critical stages of cerebellar development prompted us to assess its significance in cerebellar neurogenesis as well as its implications.
Here, we demonstrated the role of TLX3 in the early development of the cerebellum and also provide evidence that its misexpression could manifest into autism-like behavior in mouse model. To understand the role of Tlx3 in early cerebellum development and its association with ASD, we have used cGN-specific Tlx3-conditional knock-out mice. Here, the expression of Tlx3 was specifically disrupted in the proliferating cerebellar granule neuron precursors (cGNPs) of the cerebellum. We show that loss of TLX3 results in decreased proliferation of cerebellar GNPs by directly targeting the anti-proliferative genes, leading to cerebellar hypoplasia, altered cell-type ratio, and synaptic connections. The resulting altered cerebellar homeostasis during the early embryonic stages culminated to a typical ASD-like phenotype with observable impairments in motor, and social functions in adult mice. It was also intriguing to find that pathogenic variation of TLX3 is present in human populations, with reported mutations in a few ASD cases and developmental disorders.
Results
TLX3 expression is crucial for the proliferation of granule neuron progenitors and patterning of the developing cerebellum
Tlx3 is a homeobox gene known to express from the E15 to PN20 stage in the posterior lobe of the developing cerebellum.16 Although, Tlx3 has been designated as a post-mitotic selector gene, our previous studies showed that TLX3 expression is also confined to the proliferating cGNPs of EGL in the mouse posterior cerebellum other than the differentiated neurons.16 Here, we showed the co-expression of TLX3 with Pax6 and Ki67, confirming that TLX3 is expressed in the proliferating GN progenitors of EGL (Figures 1A and 1B). We also assessed the expression pattern of Tlx3 in the various cell types of cerebellum by analyzing scRNA-seq database21 and found that Tlx3 is only present in the clusters of granule neuron progenitors (GNP) and GN beginning from E15 to PN10 (Figure 1C). Therefore, we further sought to determine the function of TLX3 in actively dividing cGNPs of EGL and to understand the mechanism by which TLX3 contributes to the development of cerebellum. To elucidate this, we generated a cerebellar GNP-specific Tlx3 conditional knockout mice with floxed exon 2. The mice harboring floxed sites in Tlx3 alleles were crossed with Atoh1-Cre mice (Figure 1D), having Cre recombinase active under the GNP-specific Atoh1 promoter. Atoh1 marks all the early GNPs of rhombic lip(RL),22,23,24 and the synchronous activation of Tlx3 and Atoh1 promoters occurs only in cerebellar GNPs and not in other brain regions such as the spinal cord or brain stem. We first confirmed the knockout of exon2 of Tlx3 by real-time PCR at various stages of development (Figure 1E). Further, the effective deletion of Tlx3 only in the GNPs of the posterior cerebellum during E16 was confirmed whereas, it remained intact in the dorsal spinal cord and brain stem (Figures S1A–S1H and S2A–S2H). Moreover, IGV Snapshot showed the deletion of Tlx3 exon 2 regions in respective KOs (Figure S1I). Since Tlx3 gets knocked out only in the cGNPs, the progeny of the Tlx3 cKO (Tlx3fl/fl; Atoh1 Cre+/−) mouse was not embryonically lethal and was fertile. There were no observable morphological differences in Tlx3 cKO mice during the postnatal weeks. However, by the age of three months, most of the Tlx3 cKO mice had developed skin lesions in the neck, and hind limbs, near the tail due to excessive grooming (Figure S3A), and we did observe 20% mortality among adult littermates by the age of 6–9 months (Figure S3B). Body weight between control (Tlx3fl/fl; Atoh1 Cre−/−) and Tlx3 cKO (Tlx3fl/fl; Atoh1 Cre+/−) at three months of age showed no significant difference (Figure S3C).
Figure 1.
TLX3 regulates proliferation of granule neuron progenitors in cerebellum
(A) Immunofluorescence staining of TLX3 and Pax6 (marker of cerebellar granule neurons) in the sagittal section of cerebellum at PN7. Scale bar, 200μm.
(B) Immunofluorescence staining of TLX3 and Ki67 (proliferation marker) in the sagittal section of cerebellum at PN7. Scale bar, 200μm.
(C) Heatmap showing the expression of Tlx3 in various clusters of cerebellum identified from scRNA-seq published database beginning from E10 to PN10.
(D) Cartoon illustrating the generation of Tlx3 conditional knockout mice (Tlx3fl/fl; Atoh1 Cre).
(E) Bar graph depicts real-time PCR analysis of Tlx3 confirming deletion of exon 2 at different stages of development in Tlx3 cKO.
(F and G) Immunofluorescence staining of BrdU (12 h BrdU pulsing) and Pax6 (marker of cerebellar granule neurons) in the sagittal section of E16 control cerebellum (Tlx3fl/fl; Atoh1 Cre−/−) and Tlx3 conditional knockout (Tlx3fl/fl; Atoh1 Cre+/−). Scale bar, 100μm. (f’ and g’) Enlarged image of the boxed region from (F and G), respectively.
(H and I) Immunofluorescence staining of BrdU (12 h BrdU pulsing) and Pax6 (marker of cerebellar granule neurons) in the sagittal section of PN7 control cerebellum (Tlx3fl/fl; Atoh1 Cre−/−) and Tlx3 conditional knockout (Tlx3fl/fl; Atoh1 Cre+/−). Scale bar, 200μm. (h’, h’’, i’, and i’’) Enlarged view of the boxed region from figures H and I respectively.
(J) Quantification of the number of BrdU+ive and Pax6+ive granule cells in the EGL layer of cerebellar cortex at E16 (n = 9 for both genotypes).
(K) Quantification of the number of BrdU+ive and Pax6+ive granule cells in the EGL layer of cerebellar cortex at PN7 (n = 9 for both genotypes). Data are represented as mean ± SD. ∗p < 0.05, ∗∗∗p < 0.001 (Unpaired-t test). See also Figures S1–S6.
Next, we checked if TLX3 has any role in regulating the proliferation of cGNPs at E16 and PN7 stages using a 12-h BrdU pulse-chase. We chose E16 and PN7 stages since E16 represents the beginning of the proliferation or genesis of GNPs, whereas PN7 represents the proliferation peak of cGNPs and harbors a maximum number of GNPs in the EGL. Immunohistochemical analysis showed that the number of BrdU-incorporated Pax6+ive cGNPs was significantly downregulated at both E16 and PN7 stages (Figures 1F–1I and 1J–1K). We also analyzed the proliferation marker Ki67 at both the stages to confirm these results. Here, too, we found a consistently reduced proliferation of the GNPs in the Tlx3 cKO cerebellum compared to the control (Figures S4A–S4F). We then carried out a short duration (2-h) BrdU pulse-chase experiment at the PN7 stage and found a similar reduction of cGNPs (Figures S5A–S5C). Surprisingly, we observed an overall decrease in proliferation of cGNPs in the anterior lobes at the PN7 stage, where TLX3 was not expressed which needs further exploration (Figures S6A and S6B). Together, these results confirmed that TLX3 depletion affects the proliferation of GNPs at both early and later stages of GNP development.
Having found that the deletion of Tlx3 from GNPs affected its proliferation rate, we next sought to evaluate other biological consequences of reduced cGNP proliferation during cerebellum development. We evaluated the cerebellar patterning with DAPI nuclear staining at multiple cerebellum developmental time points (Figures 2A–2H). Here, we observed a shift in the primary fissure in E18 Tlx3 cKO compared to the control (Figures 2c’and d’). A prominent cerebellar patterning defect along the anterior-posterior axis, starting mainly in the 4/5th lobe and continuing to the central and posterior lobes, was observed between PN7 and PN21 stages (Figures 2E–2H). Further, analyzing the whole brain, we observed an apparent difference in lobule arrangement toward the cerebellum’s vermis region of Tlx3 cKO mice at PN21 stage (Figure 2I). However, weight of the whole brain and cerebellum did not differ among the genotypes (Figure 2J). Nevertheless, at PN21, a stage at which cerebellar neurogenesis is almost complete, we observed a significant reduction in the total cerebellum area in Tlx3 cKO mice compared to controls, indicating the onset of cerebellar hypoplasia (Figure 2K). Our results thus imply that Tlx3 has a significant role in maintaining the proliferation of GNPs at the early stages of cerebellum development.
Figure 2.
TLX3 is a crucial determinant for patterning along the anterior-posterior axis of developing cerebellum
(A–H) DAPI staining of sagittal sections of the cerebellum including the vermis of control (Tlx3fl/fl; Atoh1 Cre−/−) and Tlx3 conditional knockout (Tlx3fl/fl; Atoh1 Cre+/−) from various developmental time points such as E16, E18, PN7, and PN21. Scale bar, 100μm. (c’and d’) Enlarged images of E18.
(C and D) DAPI staining from the primary fissure. Arrow and arrowheads indicate changes in the cerebellar lobe pattern.
(I) Appearance of the dorsal view of brain isolated from control (Tlx3fl/fl; Atoh1 Cre−/−) and Tlx3 conditional knockout mice (Tlx3fl/fl; Atoh1 Cre+/−) at PN21 stage. Arrows indicate the changes in the cerebellar lobule arrangement.
(J) Weight of the whole brain without olfactory bulb and cerebellum from control (Tlx3fl/fl;Atoh1 Cre−/−) and Tlx3 conditional knockout mice (Tlx3fl/fl; Atoh1 Cre+/−) at PN21 stage (n = 3 for both genotypes).
(K) Quantitative measurement of cerebellar cross-sectional area of control (Tlx3fl/fl; Atoh1 Cre−/−) and Tlx3 conditional knockout mice (Tlx3fl/fl; Atoh1 Cre+/−) at E16, E18, PN7 and PN21 stages (n = 9 for both genotypes). Images (A–H) were generated through stitching multiple images using Photoshop software. Nuclei were stained with DAPI. Data are represented as mean ± SD. ∗∗∗p < 0.001 (Unpaired-t test).
The absence of TLX3 during the window of GNP proliferation/differentiation alters the balance between granule neurons and Purkinje neurons
Next, we were curious about the underlying molecular mechanism by which Tlx3 maintains the proliferation of GNPs. To understand this, we performed RNA sequencing on the posterior and anterior cerebellum of PN7 control and Tlx3 cKO mice. Our data showed a total of 292 genes that are differentially expressed in the posterior cerebellum and 295 in the anterior cerebellum (false discovery rate [FDR]-adjusted p < 0.05) of the Tlx3 cKO mice compared to controls (Figures S7A and S7C; Tables S1, S2, S3, and S4). Among the 292 differentially expressed genes (DEGs) in the posterior cerebellum, we found a down regulation of 206 genes and upregulation of 86 genes. Gene set enrichment analysis (GSEA) signature analysis of cell types in the posterior cerebellum shows a significant down regulation for “cerebellar granule neuron” genes including Grin2a, Kcna1, Grik2, Bmp5, and Ntf3. The gene ontology (GO) enrichment results revealed a significant reduction of “DNA replication checkpoint signaling” including Cdc6 and Orc1,”negative regulation of WNT signaling pathway” and “collagen containing extracellular matrix” (Figures S7B and S8A–S8D).
Similarly, we performed GO and GSEA analysis for cell type and GO in the anterior cerebellum which showed that consistent with the posterior cerebellum, there was a significant reduction in the “cGN” in the anterior cerebellum too that included genes Grin2a, Grik2, Bmp5, Pcsk9, and Kcna1 (Figure S7D). Hence, our data indicated that absence of Tlx3 caused an overall reduction of the GN in both anterior and posterior cerebellum, and genes such as Grin2a, Grik2, Bmp5, and Kcna1 are differentially expressed in both regions. Interestingly, GSEA analysis on the anterior cerebellum did not show enriched downregulation of DNA replication checkpoint signaling but showed an increase in the enrichment of genes in “cytoplasmic translation” and downregulation of genes in “voltage gated channel activity” including Cacna2d1, and Lrrc26 (Figures S8E–S8F). These findings indicated that Tlx3 can regulate DNA replication checkpoint signaling genes in the posterior lobes where it is expressed. In conclusion, the findings from RNA sequencing shows that Tlx3 deletion from early cerebellar GNPs specifically resulted in the significant downregulation of DNA-replication checkpoint genes, which resulted in the reduced proliferation of GN in the posterior cerebellum.
Next, to understand the molecular mechanism of GNP proliferation mediated by Tlx3 explicitly in the cGN, we carried out single-cell RNA sequencing of the PN4 posterior cerebellum using the 10× Genomics platform. We chose the PN4 stage to get the representation of two populations of GN in the cerebellum, including proliferating GNPs and differentiated GN. Single-cell sequencing was carried out with genotype confirmed control (Tlx3 fl/fl; Atoh1 Cre−/−) and Tlx3 cKO (Tlx3 fl/fl; Atoh1 Cre+/−) cerebellum. After clustering using distinct transcriptional profiles, we identified a total of ten clusters among the genotypes, including GNP (GNP), GN, GABAergic interneuron, PC, Bergmann Glia, oligodendrocytes, endothelial precursor cells, erythrocytes, and endothelial cells (Figure 3A). As expected, we identified two clusters for GN, designated as GNP and GN, due to modest variability in the marker genes, with early progenitor markers in GNP and mature markers in GN (Figures 3B and S9–S12). Upon identifying the clusters, we first confirmed the synchronous expression of Atoh1 and Cre recombinase expression among the clusters. Cre expression was only present in the GNP and GN clusters of Tlx3 cKO samples and was absent in these specific clusters of control. The Tlx3 expression remained unaffected in the GNP and GN clusters of the control and Tlx3 cKO samples since we only deleted exon 2 of the Tlx3 gene, which contains the homeobox binding domain (Figure 3B; Figures S9A–S9D).
Figure 3.
scRNA Sequencing profiling of PN4 cerebellum reveals cell type proportion changes in Tlx3 cKOs
(A) UMAP representation of the scRNA-seq of PN4 posterior cerebellum from control (Tlx3fl/fl; Atoh1 Cre−/−) and Tlx3 conditional knockouts (Tlx3fl/fl; Atoh1 Cre+/−). Dots represents the individual cells and colors indicate the cell clusters.
(B) Expression of Tlx3, Atoh1, Cre and specific marker genes for each cluster identification from control (Tlx3fl/fl; Atoh1 Cre−/−) and Tlx3 conditional knockout (Tlx3fl/fl; Atoh1 Cre+/−). Dot sizes represent the percentage of cells expressing a gene of interest. Colors show cell clusters, cell cycle phase and genotype of control and cKO.
(C) Cell type proportion analysis of each clusters of control (Tlx3fl/fl; Atoh1 Cre−/−) and Tlx3 conditional knockout (Tlx3fl/fl; Atoh1 Cre+/−). ∗p < 0.05, ∗∗∗p < 0.001(FisherTest).
(D and E) Immunofluorescence staining of Pax6 (marker of cerebellar granule neurons) in the sagittal section of cerebellum at PN7 stage. Scale bar, 100μm.
(F and G) Immunofluorescence staining of calbindin (marker of Purkinje neurons) in the sagittal section of cerebellum at PN7. Scale bar,100μm.
(H) Quantification of the number of Pax6+ive granule cells in the EGL layer of cerebellar (n = 9 for both genotypes, Unpaired T-Test).
(I) Quantification of the number of calbindin+ive Purkinje cells in the cerebellar cortex (n = 9 for both genotypes, Unpaired T-Test).
(J and K) Volcano plots showing the differential gene expression (DGE) between control (Tlx3fl/fl; Atoh1 Cre−/−) and cKO (Tlx3fl/fl; Atoh1 Cre+/−) in clusters of granule neuron progenitors (GNP) and granule neurons (GN). Dots represent the genes. Dashed lines depicts the adjusted p-value down to ∗∗∗p < 0.0001.
(L) Gene Ontology overrepresentation analysis of up regulated (Control (Tlx3fl/fl; Atoh1 Cre−/−) vs. cKO (Tlx3fl/fl; Atoh1 Cre+/−) genes in GNP and GN clusters. Dot size represents the fold enrichment of genes referring to respective GO-term.
(H and I) Data are represented as mean ± SD. ∗∗p < 0.01, ∗∗∗p < 0.001(Un-paired t test). The number of animals is represented by “n”. See also Figures S7–S13.
Next, we examined whether the deletion of Tlx3 from the early cerebellum induced any changes in the cell type proportion in these clusters. The GN proportion in the Tlx3 cKO were significantly decreased compared to the control (Figure 3C). This result is consistent with RNA-seq analysis of the posterior cerebellum, as cell type enrichment showed a significant reduction of GN. Apart from that, we also found a drastic increase in Purkinje cell clusters in the Tlx3 cKO (Figure 3C). To further evaluate this change in the cell type ratio, we quantified the granule neuron number at PN7 (Pax6+ive) and at PN21 (NeuN+ive) using immunohistochemical analysis. Consistent with scRNA-seq findings, our immunofluorescence analysis showed a significant reduction in the number of GN in the Tlx3 cKO at both the stages compared to their respective controls (Figures 3D, 3E, and 3H; Figures S13A–S13E).
Similarly, we also examined the Purkinje neurons of the cerebellum using the specific marker calbindin at PN7 and PN21 stages. Consistently, we found a drastic increase in the number of Purkinje neurons at both the stages in the Tlx3 cKO compared to controls (Figures 3F, 3G, and 3I; Figures S13F–S13J). Our data indicated that loss of proliferation of cGNPs due to the absence of Tlx3 indirectly contributes to the increased generation of Purkinje neurons, which needs to be addressed further.
Following cell type proportion analysis, we next performed pseudo-bulk differential gene expression in GNP and GN clusters between control and Tlx3 cKO to identify the DEGs in posterior cerebellum. A total of 38 genes were differentially expressed in the GNP cluster, where 28 were upregulated and 10 were down regulated (false discovery rate [FDR]-adjusted p < 0.05) (Tables S5 and S6). In GN cluster, 123 genes were differentially expressed, including 120 upregulated and 3 downregulated genes (false discovery rate [FDR]-adjusted p < 0.05) (Figures 3J and 3K; Tables S7 and S8). We chose up regulated genes from the GNP and GN cluster and conducted GO analysis to identify the processes affected by Tlx3 deletion (Figure 3L). Results demonstrated that GO terms significantly enriched for “cell proliferation”, “axon”, “synapse”, and “negative regulation of Wnt signaling pathway” for GNP clusters, including genes Tgfb2 (TGF β2), Ccnd2 (Cyclin D2), Mab21L2, Ctnnb1(beta catenin1), and Insm1. Together, findings from RNA and scRNA-seq analysis indicated that Tlx3 deletion from early cGNPs leads to the alteration in GN to Purkinje cell ratio by reducing the number of GN.
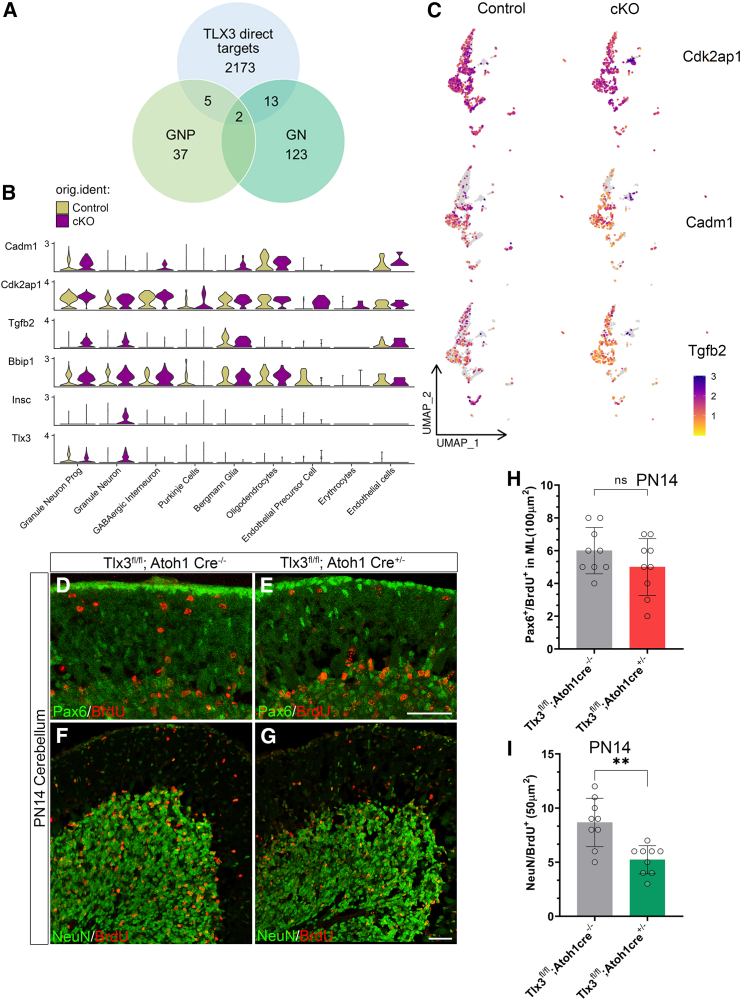
TLX3 directly regulates the expression of anti-proliferative genes to maintain the homeostasis of granule neuron progenitors
Next, to gain insights into the direct binding targets and molecular mechanism through which TLX3 regulates the proliferation of the cGNPs, we analyzed the DEGs of GNP and GN with binding targets of TLX3. The data regarding the binding sites of TLX3 generated using a structure-based computational algorithm (TF2DNA) was already available.25 Using this information, we obtained 2173 direct target genes of TLX3 and overlapped these genes with DEGs of GNP (37) and GN (123). Among the DEGs, we identified 5 upregulated genes in GNP and 13 upregulated genes in GNs identified as direct targets of TLX3 (Figure 4A). Here, we found an interesting observation that anti-proliferative genes Tgfb2 (TGF β2) and Cdk2ap1 were significantly upregulated in the GNP and GN clusters (Figures 4B and 4C). In addition, we also found a significant upregulation of a few genes that are anti-proliferative and also involved in microtubule stability, including Cadm1, Insc, and Bbip1 as direct targets of TLX3 in the cKO (Figures 4B and 4C; Figures S14A and S14B). We further assessed the expression of these genes including Cadm1, Cdk2ap1, Tgfb2, Bbip1, and Insc using real-time PCR analysis in PN4 control and Tlx3 cKO posterior cerebellum. The results recapitulated the scRNA-seq analysis except for the gene Cdk2ap1 where the fold change was negligible (Figure S13C). This could be due to the ubiquitous expression of Cdk2ap1 in all clusters of cerebellar cell types (Figure 4B). For qPCR we have isolated RNA from the whole cerebellum and therefore the differential expression of Cdk2ap1 in the GN clusters will be compensated due to its ubiquitous expression in other cell types (Figure S14C). Together, these results imply that TLX3 promoted the proliferation of GNPs by negatively regulating anti-proliferative genes at the transcriptional level during the early development window.
Figure 4.
TLX3 directly regulates the expression of anti-proliferative genes and its dysfunction leads to the decreased differentiation of granule neurons
(A) Venn diagram showing the overlap of genes between TLX3 direct targets (2173 genes) and differentially expressed genes from control (Tlx3fl/fl; Atoh1 Cre−/−) and cKO (Tlx3fl/fl; Atoh1 Cre+/−) in clusters GNP (37) and GN (123).
(B) Violin plot representing the expression of anti-proliferative (Cadm1, Cdk2ap1, Tgfb2) and microtubule stabilizing genes (Bbip1 and Insc) in each clusters identified from scRNA seq of control (Tlx3fl/fl; Atoh1 Cre−/−) and cKO (Tlx3fl/fl; Atoh1 Cre+/−).
(C) UMAP plot representing the anti-proliferative genes (Cadm1, Cdk2ap1, Tgfb2) obtained from overlap analyses.
(D and E) Immunofluorescence staining of BrdU+ive and Pax6+ive granule cells in the migrating EGL layer of cerebellar cortex at PN14 after 7 days of BrdU pulsing. Scale bar, 50μm.
(F and G) Immunofluorescence staining of BrdU+ive and NeuN+ive granule cells in the differentiated IGL layer of cerebellar cortex at PN14 after 7 days of BrdU pulsing. Scale bar, 50μm.
(H) Quantification of the number of BrdU+ive and Pax6+ive granule cells in the migrating EGL layer of cerebellar cortex at PN14 corresponding to D and E.
(I) Quantification of the number of BrdU+ive and NeuN+ive granule cells in the differentiated IGL layer of cerebellar cortex at PN14 corresponding to F and G. (n = 9 for both genotypes). (H and I) Data are represented as mean ± SD.∗∗p < 0.01, ns = 0.198(Un-paired t test). The number of animals is represented by “n”. See also Figures S14 and 15.
Given that TLX3 regulated the genes involved in anti-proliferation thereby promoting the proliferation of cGNPs, we next evaluated the role of TLX3 on the migration and differentiation potential of cGNPs using BrdU pulse-chase experiment. We labeled highly proliferating GN at PN7 with BrdU and analyzed the migrating GN in the molecular layer of control and Tlx3 cKO at the PN14 stage. We did not find any difference in the number of migrating GN in the molecular layer among the genotypes in any of the lobes (Figures 4D, 4E, and 4H). Further, we assessed the differentiation of GN labeled from the PN7 stage, using a granule neuron differentiation marker, NeuN, and analyzed the number of BrdU-incorporated NeuN+ive cells in the IGL layer at the PN14 stage. Here, we observed fragmented and unfragmented BrdU+ive cells in the IGL layer, which indicated the multiple rounds of divisions undergone by the granule neuron after incorporating BrdU from the PN7 stage. We included fragmented and unfragmented cells to calculate the differentiation rate and assessed the number of BrdU+ive/NeuN+ive cells in the IGL. We noticed that BrdU+ive/NeuN+ive cells in the Tlx3 cKO were significantly reduced in all the lobes at PN14 stage as compared to the control (Figures 4F, 4G, and 4I). Consistent with the rate of proliferation, deletion of Tlx3 affected the differentiation of GN in all the lobes of the cerebellum (Figure S15). Overall, our results suggested that deletion of Tlx3 from the early window of cerebellar granule progenitor development affected the proliferation of cGNP by directly regulating anti-proliferation genes. This subsequently led to reduced differentiation of GN in the whole cerebellum. Thus the loss of Tlx3 from the early window of cerebellar development would disrupt the homeostasis of granule neuron development, and can further lead to other consequent dysfunctions in the Tlx3 cKO. Although the direct link of TLX3 in regulating the proliferation and differentiation of cGNP is evident in the posterior lobes, its influence on the anterior lobes which lack TLX3 expression, is still unclear.
Dynamic homeostasis of GNPs maintained through TLX3 is crucial for the formation of proper synaptic connections between granule and Purkinje neurons
Since we now know that absence of TLX3 can alter the ratio of GN to PC in the cerebellum, we next assessed the primary synaptic connections in the cerebellum. PC are innervated and synaptically connected by climbing fibers (CF) and granule neuron extension parallel fibers (PF). Compared to the control, PN7 Tlx3 cKO had significantly more parallel fiber puncti marked by vesicular glutamate transporter 1(VGLUT1) onto the Purkinje cell body, as evidenced by the parvalbumin expression (Figures 5A, 5B, and 5G). We also evaluated the synaptic connections during PN21; the results corroborated our prior findings that VGLUT1 puncti on the Purkinje cell body were significantly increased in the Tlx3 cKO (Figures 5C–5F, e’, f’ and 5G). Similarly, we evaluated climbing fiber innervations marked by vesicular glutamate transporter 2 (VGLUT2) connecting to Purkinje and observed that VGLUT2 puncta on to Purkinje cell was elevated in Tlx3 cKO mice at PN7 and PN21 stages (Figures 5H–5M, h’, i’, 5N, and 5O). These findings indicated that synaptic connections PC-PF and PC-CF were increased and that each Purkinje cell was highly innervated and connected with enhanced parallel fiber and climbing fiber puncta following Tlx3 deletion. However, we did not find any synapse-related genes which are direct target for TLX3 in single-cell sequencing analysis except Snca, and cadm1 which was found to be upregulated. Thus, the altered synaptic connections could be an indirect, compensatory mechanism due to the fewer GN in the Tlx3 cKOs. There are enough reports to prove the interactions between GN and Purkinje, and these interactions are critical for the synaptic connections.26,27 In addition, reports have also shown that the final phase of supernumerary elimination of CF from PC was influenced by GC and their parallel fiber projections. Partial elimination of parallel fiber inputs into Purkinje by disrupting postsynaptic glutamate receptors also results in the multi-innervation of climbing fiber extensions into Purkinje.26,28,29 Hence, the excess-connectivity of synaptic connections between PC and CF could be due to impaired signaling from Tlx3-deficient GN. To eliminate the possibility of cell death within the Tlx3 cKO cerebellum at different stages of development, we carried out a TUNEL assay. We did not find any significant difference in the number of TUNEL+ive cells in the cerebellum at any developmental stages between the genotypes, indicating negligible cell death in the Tlx3 cKO cerebellum (Figures S16A–S16Q).
Figure 5.
TLX3 is crucial for forming the correct synaptic connection architecture of the cerebellum
(A–F) Immunofluorescence staining of parvalbumin (marker for Purkinje neurons) and VGLUT1 (marker of parallel fiber extension of granule neurons) in the sagittal section of cerebellum at PN7 and PN21 in control (Tlx3fl/fl; Atoh1 Cre−/−) and Tlx3 conditional knockouts (Tlx3fl/fl; Atoh1 Cre+/−). Scale bar, 20μm (A and B), 50 μm (C–F).
(G) Quantification of the number of VGLUT1 puncta on to each Purkinje cell body of images A–F. (n = 9 for both genotypes).
(H–M) Immunofluorescence staining of parvalbumin (marker for Purkinje neurons) and VGLUT2 (marker of climbing fiber) in the sagittal section of cerebellum at PN7 and PN21 in control (Tlx3fl/fl; Atoh1 Cre−/−) and Tlx3 conditional knockouts (Tlx3fl/fl; Atoh1 Cre+/−). Scale bar, 20μm (H and I), 50 μm (J-M). (a’,b’, e’ f’, h’, and i’) Enlarged images of A, B, E, F, H, and I, respectively.
(N) Quantification of the number of VGLUT2 puncti on to each Purkinje cell body of images H–M. (n = 9 for both genotypes).
(O) Violin plot showing the percentage of climbing fiber extensions into each Purkinje neurons at PN21 in control (Tlx3fl/fl; Atoh1 Cre−/−) and Tlx3 conditional knockout (Tlx3fl/fl; Atoh1 Cre+/−). (n = 9 for both genotypes). (G, N, and O) Data are represented as mean ± SD. ∗∗p < 0.01, ∗∗∗p < 0.001(Un-paired t test). The number of animals is represented by “n”. Arrow indicate puncti. See also Figure S16.
Tlx3 deficient mice exhibit behavioral deficits in motor, and social interaction similar to ASD
Having established that the deletion of Tlx3 from the GNPs caused significant structural and molecular changes in the cerebellum, we next sought to evaluate the various behavioral deficits in Tlx3 cKO. The cerebellum is associated with discrete behavioral activities, such as fine motor coordination, cognitive abilities, and social functions. Furthermore, the posterior cerebellum is implicated in cognition and social development, whereas the anterior cerebellum contributed to fine motor development.30,31,32,33,34 Hence, we assessed motor, cognitive, and social behavioral functions in Tlx3 cKO. The results showing the latency to fall from wire hang and rotarod tests revealed a significant reduction in the time, where Tlx3 cKO mice were unable to hang in the wire or on the accelerating rotarod, which signifies a substantial loss of motor coordination in Tlx3 cKO compared to control (Figures 6A–6C). However, the total distance traveled in the open-field test did not differ between control and Tlx3 cKO, indicating that normal movement is not impaired (Figure 6D).
Figure 6.
TLX3 deficient mice exhibit defects in motor and social function
(A) Latency to fall in the two limb wire hang test for control (Tlx3fl/fl; Atoh1 Cre−/−, n = 12) and Tlx3 conditional knockouts (Tlx3fl/fl; Atoh1 Cre+/−, n = 9).
(B) Latency to fall in the four limb wire hang test for control (Tlx3fl/fl; Atoh1 Cre−/−, n = 12) and Tlx3 conditional knockouts (Tlx3fl/fl; Atoh1 Cre+/−, n = 14).
(C) Latency to fall in the rotarod test for control (Tlx3fl/fl; Atoh1 Cre−/−, n = 8) and Tlx3 conditional knockout (Tlx3fl/fl; Atoh1 Cre+/−, n = 8).
(D) Total distance traveled by control (Tlx3fl/fl; Atoh1 Cre−/−, n = 11) and Tlx3 conditional knockouts (Tlx3fl/fl; Atoh1 Cre+/−, n = 9) in the open maze chamber.
(E) Time spent in the open maze central zone by control (Tlx3fl/fl; Atoh1 Cre−/−, n = 12) and Tlx3 conditional knockouts (Tlx3fl/fl; Atoh1 Cre+/−, n = 10).
(F) Time spent in the open arm of elevated plus maze by control (Tlx3fl/fl; Atoh1 Cre−/−, n = 13) and Tlx3 conditional knockouts (Tlx3fl/fl; Atoh1 Cre+/−, n = 13).
(G) Interaction time of control (Tlx3fl/fl; Atoh1 Cre−/−, n = 6) and Tlx3 conditional knockouts (Tlx3fl/fl; Atoh1 Cre+/−, n = 6) with familiar and novel object in the Novel object recognition test.
(H) Number of marbles buried by control (Tlx3fl/fl; Atoh1 Cre−/−, n = 13) and Tlx3 conditional knockouts (Tlx3fl/fl; Atoh1 Cre+/−, n = 15) in the marble burying test.
(I) Social preference test results of control (Tlx3fl/fl; Atoh1 Cre−/−, n = 8) and Tlx3 conditional knockouts (Tlx3fl/fl; Atoh1 Cre+/−, n = 7) which shows the interaction time of each mouse with social stimuli and empty cup.
(J) Social novelty test results of control (Tlx3fl/fl; Atoh1 Cre−/−, n = 8) and Tlx3 conditional knockouts (Tlx3fl/fl; Atoh1 Cre+/−, n = 7) that shows the interaction time taken by each mouse with familiar social stimuli and novel social stimuli.
(K) Rate of calling of control (Tlx3fl/fl; Atoh1 Cre−/−, n = 6) and Tlx3 conditional knockouts (Tlx3fl/fl; Atoh1 Cre+/−, n = 5) upon introduction of intruder mice in the resident –intruder test and ultra-sonic vocalization recording.
(L) Peak frequency of the calls produced by control (Tlx3fl/fl; Atoh1 Cre−/−, n = 6) and Tlx3 conditional knockouts (Tlx3fl/fl; Atoh1 Cre+/−, n = 5) during resident –intruder test and ultra-sonic vocalization recording.
(M) Latency for initiating the first ultrasonic call when intruder is introduced with the control (Tlx3fl/fl; Atoh1 Cre−/−, n = 6) and Tlx3 conditional knockouts (Tlx3fl/fl; Atoh1 Cre+/−, n = 5). (A–M) Data are represented as mean ± SD. (A–F, H, and M) Data are represented as box and whiskers plot (Mann-Whitney test Unpaired(U)-Tests in A–F and H, Mann-Whitney test U-Tests and Ordinary one-way ANOVA for group analysis in H–J and Unpaired-t test in K–M). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). The number of animals is represented by “n”. See also Figures S17–S19.
Further, to test anxiety-like behavior, open maze, and elevated plus maze tests were performed. We observed that the Tlx3 cKO mice spent comparatively more time in the central zone and open arms when compared to the control. These observations indicated that the animals were less anxious than the control (Figures 6E and 6F). Next, we evaluated the cognitive skill of Tlx3 cKO using a novel object recognition test. We found that, unlike controls, Tlx3 cKO mice could not distinguish between familiar and novel objects, suggesting a cognitive impairment (Figure 6G). Tlx3 cKO mice have spent nearly equal time with familiar and novel objects and did not prefer novel object-like controls.
Next, we conducted three other behavioral tests, such as marble burying, three-chamber, and resident intruder tests, followed by the recording of ultrasonic vocalizations, to investigate the social function in Tlx3 cKO mice. Tlx3 cKO mice buried substantially more marbles than control mice in the marble burying test, demonstrating their repetitive digging activity (Figures 6H and S17). In the three-chamber social preference test, control mice tend to prefer social stimuli over empty apparatus, whereas Tlx3 cKO mice failed to exhibit a preference for social stimuli or instead spent nearly equal amounts of time with both stimuli (Figure 6I). However, both genotypes significantly preferred novel social stimuli in the social novelty test (Figure 6J). These observations indicated that the deletion of Tlx3 from the window of early cerebellar development influenced social preference function and has no significant effect on social memory. Even though, we observed a significant cognitive impairment in the novel object recognition test of Tlx3 cKO mice, social memory was found to be intact. Thus, it indicates a mild cognitive impairment in the Tlx3 cKOs.
Further, ultrasonic vocalizations were recorded and analyzed with a resident-intruder interaction for quantitative and qualitative alterations in vocal call types in the control and Tlx3 cKO. Quantitative analyses revealed that the call rate and peak frequency of calls emitted did not differ significantly between control and Tlx3 cKO mice (Figures 6K and 6L). Nevertheless, we found that the Tlx3 cKO mice have a longer latency to emit their first call or vocalization. In contrast, the control mice began emitting calls when an intruder was detected (Figure 6M). Following this, we examined the qualitative profile of call types emitted and observed that flat calls are significantly increased in the Tlx3 cKO (Figures S18A and S18B). Flat calls are often referred to as aversive calls,35 indicating that Tlx3 cKO mice experience irritability in the presence of a stranger mouse. In conclusion, the resident intruder test demonstrated that Tlx3 cKO mice display a delay in initiating the first call and variations in the call types emitted, confirming the impaired social function of these mice. Finally, we carried out a clasping test to assess for any severe neurodegenerative symptoms. We found no significant difference in clasping scores between the genotypes. We also conducted a clasping test on the 7-9 month-old mice to identify any neurological signs at the later stages; however, we did not observe a significant difference in the clasping score between genotypes (Figures S19A and S19B). In conclusion, results from the behavior tests demonstrates that Tlx3 cKO mice exhibits a severe loss in both motor and social function, which in turn resembles a typical behavioral dysfunction associated with ASD. This indicated that loss of Tlx3 in the window of early development reduces proliferation through direct regulation of anti-proliferative genes that leads to reduced cGN generation and defects in synaptic connections, leading to developmental disorders including ASD-like behavior.
Mutation and structural variations of the TLX3 in human population indicate a possible link to ASD and other neurodevelopmental disorders
The findings of our comprehensive behavioral assessment unveiled the compromised motor, and social functions of Tlx3 cKO mice resembling ASD. Therefore, we next evaluated the expression profile of ASD candidate genes in the Tlx3 cKO cerebellum. To assess the expression of ASD candidate genes in Tlx3 cKO, we overlapped the DEGs from granule neuron clusters (GNP and GN) of scRNA-seq with genes from the ASD Simons Foundation Autism Research Initiative (SFARI) database. The syndromic and non-syndromic genes from the database overlapped with the DEGs of GN clusters, and we found ten genes that overlapped with high confidence along with a strong candidate gene score (Figure 7A). These genes include Ctnnb1, Rora, Atp1a3, Dip2a, Cadm1, Snap25, Ptbp2, Zswim6, Dvl3, and Rheb. We also assessed the expression of these ten genes in the remaining clusters and found a differential expression among the clusters. All of these genes were differentially expressed in many of the clusters of Tlx3 cKO (Figure 7B). Interestingly, we found that Cadm1 was a direct target of TLX3, as shown in (Figures 4B and 4C), and the remaining genes were indirect targets of TLX3.
Figure 7.
Deletion of Tlx3 shows altered expression of the ASD Candidate genes in GNPs of Cerebellum
(A) Dot plot visualizing the genes obtained from overlap analysis of differentially expressed genes from scRNA Seq of GNP and GN clusters with SFARI ASD database. Size of the dot is determined by the gene score.
(B) Violin plot representing the expression of overlapping genes obtained from overlap analysis of differentially expressed genes from scRNA Seq of GNP and GN clusters of control (Tlx3fl/fl; Atoh1 Cre−/−) and Tlx3 conditional knockouts (Tlx3fl/fl; Atoh1 Cre+/−) with SFARI ASD database. Colors show genotype of control and KO.
(C) Dot plot depicting the distribution of missense and structural variations of the TLX3 gene in the global population, determined by the analysis of minor allele frequencies.
(D) Pie diagram showing the percentage of TLX3 variations in association with various pathological conditions curated from published literature and ClinVar database.
Having established the potential link of TLX3 in association with ASD, we next evaluated for any mutation/structural variation in TLX3 allele in the human population. For this, we initially examined the possibility for mutation and pathogenic variations in the TLX3 allele of the global population. Here, we found that the minor allele frequency of TLX3 in distinct populations has 29 genetic variations associated with the TLX3 gene, consisting of 10 missense variants and 19 structural variants (SVs, Figure 7C). Additionally, we identified 18 potential pathogenic variations, and based on the ACMG (American College of Medical Genetics and Genomics) and AMP (Association for Molecular Pathology) criteria, 4 were confirmed as pathogenic, while 26 others were classified as variants of uncertain significance (VUS). Thus, after confirming the existence of pathogenic TLX3 allele in the global population, we next assessed the TLX3 variations in the ClinVar database and published literature. The results demonstrate that TLX3 variants were associated with 9 cases of ASD, and the others were associated with developmental disability and other NDDs (Figure 7D).
Discussion
Our study utilizing cKO mouse model has demonstrated that TLX3 is crucial for the proliferation of GNPs of the outer EGL and its dysfunction can cause Autism. Here, TLX3 is essential for maintaining the homeostasis of GNs in the posterior lobes of the cerebellum. Ablation of Tlx3 in the window of early cerebellum development (E15-PN20) severely affects the proliferation of the GNPs and affects the number of GNs generated within this window. Genesis of a definite number of GNs during this window is essential to form proper downstream connections with the Purkinje neurons.26 TLX3 controls the genesis of a particular number of GNs during this window by regulating anti-proliferative genes such as Cadm1, Cdk2ap1, and Tgfb2. We have observed the downstream consequence of the reduced proliferation of GNPs having a cascading effect on the structural development of the cerebellum and also on its downstream cellular connections. The Purkinje neurons collect all the complex inputs from the cerebellar cortex majorly through PF- extensions of GN as well as CF from the inferior olive, integrate the signal and serve as the sole output of the cerebellum. Therefore, any altered connection to the Purkinje neurons will affect the complete functionality of the cerebellum and, in turn, affect the cortico-cerebellar connectivity. The deletion of Tlx3 from the GN has led to the increased Purkinje cell number as well as excess-connectivity of parallel fiber and climbing fiber extensions into the PC. This demonstrates the altered synaptic connections originating from the PC of the Tlx3 knockout cerebellum.
Meanwhile, interestingly, we observed that GABAergic Purkinje marker gene Calb1 (calbindin) is misexpressed and are upregulated in the granule neuron clusters in Tlx3 cKOs (Figures 3J and 3K). Calbindin is exclusively expressed in the PC of the cerebellum, and it is a calcium-binding protein that helps in the buffering of intracellular free calcium.36,37,38,39 Tlx3 is implicated and well-reported for its function as a glutamatergic selector gene, and its downregulation can lead to the upregulation of GABAergic cell fate.11,14,40,41 The upregulation of the GABAergic Calb1 gene in the clusters of GN after Tlx3 deletion demonstrates a possible analogous function of Tlx3 in regulating the glutamatergic fate of GN in the cerebellum. The intrinsic molecular mechanism or downstream genes that govern glutamatergic cell type specification would have switched upon Tlx3 deletion, and this could have resulted in a shift from glutamatergic to GABAergic marker gene expression in the GN. Molecular analysis from RNA sequencing of the posterior cerebellum has also revealed that glutamate transporter genes such as Grik2 and Kcnd2 were downregulated in the Tlx3 cKO. However, we have not evaluated the functional output or connectivity of Tlx3-depleted GN to determine the possibility of trans-differentiation into Purkinje neurons or a shift in the fate of GN. Several studies have reported a change in the expression of marker genes related to glutamatergic and GABAergic signaling after the deletion or overexpression of various fate-specifying genes, subsequently leading to a shift in the specification of cell types.42,43,44 Therefore, all our observation leads to the conclusion that GN starts expressing a Purkinje marker and may not function as an excitatory neuron. Further investigation is required to evaluate the possibility of trans-differentiation or fate change of GN in Tlx3 cKOs.
Another intriguing observation that remains unanswered is the reduction of GNs in the anterior lobes of the cerebellum, similar to that of the posterior lobes where Tlx3 is knocked out. This could be due to the early deletion of Tlx3 from cGNPs and this synergetic effect is quite exciting and needs to be explored further. Alike Tlx3, other genes, such as Otx2 and Lmx1a are exclusively expressed in the posterior lobes of the cerebellum.45,46 Nevertheless, deleting these genes, particularly from the posterior lobe, had distinct consequences on both the anterior and posterior cerebellum. Meanwhile, we also found a 20% mortality of Tlx3 cKO at the age of 6–9 months and observed severe skin lesions in the neck and near the tail in the deceased. However, the precise cause of death and the persistent possibility for the prevalence of mortality among the Tlx3 cKO needs to be explored further.
Finally, we found that all these structural abnormalities, cell type imbalance and altered synaptic connectivity have culminated into autistic-like phenotype in Tlx3 cKO mouse. It is known that altered development of the cerebellum is associated with neurodevelopmental disorders (NDDs), such as ASD, ADHD, developmental dyslexia(DD), intellectual disability (ID), communication disorders, and childhood motor disorders.30,47,48 Apart from other NDDs, ASD is characterized by structural and functional abnormalities in the cerebellum, eventually resulting in altered cerebello-cortical connectivity.48,49 Furthermore, ASD are intricate neurodevelopmental disorders characterized by a range of neuropsychiatric symptoms, such as repetitive behavior, linguistic impairments, social dysfunction, and cognitive challenges, including difficulty with motor function in severe cases.50,51,52 Many reports have shown that the genes involved in the early development of the cerebellum, such as Engrailed homeobox 2, Rorα, Foxp2, Reln, Pten, Cadps2, and Gabrb3 if misexpressed at the early developmental stages, can lead to ASD.34,53,54,55,56,57,58,59,60,61,62,63 Therefore, there exists a clear link between cerebellar development and ASD. We have observed a similar link in the case of the homeobox gene Tlx3. This information is relatively new since, to date, pathological involvement of Tlx3 has only been limited to T cell leukemia due to a recurrent interchromosomal translocation in T cell acute lymphoblastic leukemia cells.64,65 Previous studies on Tlx3 in the spinal cord and brainstem did not show association with any pathological condition.
The cerebellar structural and connectivity abnormalities found in ASD majorly include hypoplasia in the posterior vermis (lobules VI–VII) and excitatory/inhibitory imbalance due to the alterations in Purkinje outputs concerning cortico-cerebellar connectivity.66 Structural imaging analysis of ASD patients, particularly in early age, i.e., under 10, showed a decreased volume of the posterior vermis region of the cerebellum.67,68,69 These abnormalities are similar to that we observed in Tlx3 cKO mouse. Together with these, many studies have also showed that the number and morphology of PC are reduced in the cerebellum of individuals with ASD.66,70 However, here, we found that Tlx3 dysfunction has led to an increased number of Purkinje neurons. The subsequent increase of Purkinje neurons could have occurred as a compensatory response to the depletion of GN in the Tlx3 cKO cerebellum. This needs further investigation. In addition, global human genome data analysis has shown potential pathogenic variation in the TLX3 allele with missense and structural mutations. We could identify nine ASD cases involving TLX3 mutations, however, the mutated DNA region of TLX3 remained uncharacterized and was merely classified as a structural mutation. There are also no reports regarding how these variations could lead to ASD. Hence, it warrants further investigation.
In conclusion, we emphasize that all the defects in proper differentiation of the cerebellum during embryonic development due to the loss of Tlx3 manifest into ASD-like behaviors in adults. The most prominent behavioral deficits were losing motor functions, social interactions, and repetitive behavior. Cerebellar dysfunction or any abnormality during sensitive periods of development, specifically in postnatal weeks, results in ASD-like outcomes.67 Recent finding has also shown that pre-term birth of babies will lead to atypical connectivity in the brain (Cortex) and is associated with impaired social, sensory, and repetitive behaviors in toddlers at 18 months of age.66 Therefore, it is evident that premature birth disrupts the pattern of functional brain dynamics, and an early insult during this critical time of development might result in the manifestation of symptoms like autism in the early stages of toddlerhood. These observations point toward the fact that most of the developmental disorders including ASDs have their origin during early embryonic development involving an array of genes required for proper development of cerebral cortex and cerebellum. Our findings establish the role of Tlx3 in the development of the cerebellum and its absence could be considered as one of the causative factors for ASD pathogenesis and manifestation.
Limitations of the study
Our study demonstrates that TLX3 is a crucial component involved in the proliferation of cGNPs, and the altered homeostasis of GN due to dysfunction of TLX3 results into typical autism phenotype in mice. We have shown the reason behind the reduced proliferation of cGNPs and it occurs through anti-proliferative genes. Following that, we assessed the consequences of altered granule neuron generation in terms of cerebellar structure, cell type proportion, primary synaptic connections, and finally, the behavioral function of the mice. However, we have not unveiled other pathways by which TLX3 affects the generation of other cell types and synaptic connections in the cerebellum, as the primary focus of this current study aimed to evaluate how TLX3 is involved in cGNP proliferation. Setting that aside, Tlx3 expression is absent in other cell types, and our scRNA sequencing analysis showed that DEGs are present majorly in the granule neuron clusters. Therefore, the alteration in the primary synaptic connections and cell type proportion change could be due to the disturbed generation of cGNPs. However, more data is required to delineate the same. In addition, we could not find the exact mechanism by which TLX3 affects the granule neuron reduction in the anterior lobe of the cerebellum, where TLX3 is absent during cerebellar neurogenesis and warrants further investigation. Finally, we assessed the potential possibility of TLX3 mutation in ASD human subjects and found the presence of TLX3 mutation associated with ASD patients. Here, we only evaluated the potential possibility of TLX3 mutation in ASD human subjects and identified that it is not restricted exclusively to mice but also appears in humans. However, a genome-wide global cohort study is required to evaluate the frequency of TLX3 mutation and how the variation is associated with distinct populations, as ASD is a typical NDD known to have significant variation within and among different socio-demographic groups.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jackson James (jjames@rgcb.res.in).
Materials availability
Tlx3 floxed mouse used for this study was originally obtained from Dr Zijing Liu, Beijing Institute of Biotechnology, Beijing, China. Commercially available mouse lines and reagents were listed in the key resources table.
Data and code availability
-
•
Data: RNA-seq and Single-cell RNA-seq data have been deposited at NCBI Sequence Read Archive (SRA) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
Code: All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOI is listed in the key resources table.
-
•
Additional information: Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. All additional softwares used for this study are listed in the key resources table.
Acknowledgments
We thank Dr Zijing Liu, Beijing Institute of Biotechnology, Beijing, China for generously providing us the Tlx3fl/fl parent mouse line. We thank 10X genomics, Premas Life science, Bangalore, India for support with scRNA sequencing. We thank Bioinformatics facility, Rajiv Gandhi Center for Biotechnology for providing access to their high-performance computing facility. We appreciate the help extended by IGE3 Genomics platform, University of Geneva by carrying out the RNA sequencing. We also thank Ms. Sreedevi L.R for her help in animal genotyping and the Animal Research Facility (ARF), Rajiv Gandhi Center for Biotechnology. We appreciate the help extended by Ms. Reena Sarah Jacob for animal behavior experiments. We thank Dr. Ani V Das and Ms. Aryasree R for critically evaluating the manuscript. This work was supported by Intramural grants to J.J. from Rajiv Gandhi Centre for Biotechnology (BRIC-RGCB). S.P, (CSIR-09/716(0161)/2015-EMR-I), B.B., (UGC-332486), S.S., (UGC-316695), R.J., (CSIR-09/0716(13765)/2022-EMR-1), V.M(CSIR-09/716(0168)/2016-EMR-I), P.A.R, (CSIR-09/716(0156)/2015-EMR-I) and N.P.J. (UGC-366288) were supported by research fellowships from Council for Scientific and Industrial Research (CSIR), Government of India and University Grants Commission, Government of India.
Author contributions
Conceptualization: S.P. and J.J.; methodology: S.P., B.B., S.S., V.M., P.A.R., N.P.J., R.S., V.P., N.R., B.S.N., K.K.G., M.K., B.K.B., and J.J.; software: B.B., R.J.,V.P., M.K., and B.K.B.; validation: S.P. and J.J.; formal analysis: S.P., B.B., and J.J.; investigation: S.P. and J.J.; resources: J.J.; writing—original draft: S.P. and J.J.; writing—review and editing: S.P., B.B., S.S., P.A.R., N.P.J., B.K.B., and J.J.; visualization: S.P., B.B., R.J., M.K., B.K.B., and J.J.; supervision: J.J.; project administration: J.J.; funding acquisition: J.J.
Declaration of interests
N.R. and R.S. have been employees of Genentech since 2022.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Guinea pig anti –Tlx3 | Gift from Dr. Carmen Birchmeier, Germany | N/A |
| Rabbit polyclonal anti-Pax6 | Millipore | Cat# AB2237; RRID:AB_1587367 |
| Rat monoclonal anti-BrdU | Abcam | Cat# ab6326; RRID:AB_305426 |
| Mouse monoclonal anti-Ki67 | BD Biosciences | Cat# 550609; RRID:AB_393778 |
| Rabbit polyclonal anti-NeuN | Abcam | Cat# ab104225; RRID:AB_10711153 |
| Rabbit polyclonal anti-Parvalbumin | Abcam | Cat# ab11427; RRID:AB_298032 |
| Mouse monoclonal anti-Calbindin | Sigma | Cat# C9848; RRID:AB_476894 |
| Mouse monoclonal anti-vGlut1 | Millipore | Cat# MAB5502; RRID:AB_262185 |
| Mouse monoclonal anti-vGlut2 | Millipore | Cat# MAB5504; RRID:AB_2187552 |
| Goat Anti-Guinea Pig IgG Cy3 | Jackson Immuno Research | RRID: AB_2337423 |
| Goat Anti-Mouse IgG Cy3 | Jackson Immuno Research | RRID: AB_2338680 |
| Goat Anti-Rabbit IgG Cy3 | Jackson Immuno Research | RRID: AB_2338007 |
| Goat Anti-Rat IgG Cy3 | Jackson Immuno Research | RRID: AB_2338254 |
| Goat anti-mouse Alexa Flour 488 | Molecular probes | Cat# A11001; RRID:AB_2534069 |
| Goat anti-rabbit Alexa Fluor 488 | Molecular probes | Cat# A11008; RRID:AB_143165 |
| Chemicals, peptides, and recombinant proteins | ||
| HCL | Thermofisher Scientific | Cat #Q29145 |
| NaCl | Merck Life science Private.Ltd | Cat #MG2M720652 |
| KCl | SRL | Cat #1644133 |
| Na2HPO4 | SRL | Cat #1949146 |
| KH2PO4 | Merck Life science Private.Ltd | Cat #DH8D682240 |
| Tris | Merck Life science Private.Ltd | Cat #DJ8D682223 |
| EDTA | Merck Life science Private.Ltd | Cat #DG8D681653 |
| SDS | SRL | Cat #1948101 |
| Paraformaldehyde | Sigma-Aldrich | Cat #P6148 |
| Sucrose | Merck Life science Private.Ltd | Cat #DF8D681300 |
| OCT | TissueTek | Cat # 4583 |
| Normal goat serum | Jackson Immuno Research | Cat # 005000121 |
| Triton X-100 | Sigma-Aldrich | Cat # 8787 |
| DAPI | Sigma-Aldrich | Cat #D9564 |
| Fluoromount G | Electron Microscopy Sciences | Cat #17984-25 |
| BrdU | Sigma-Aldrich | Cat #B5002 |
| Boric acid | Sigma-Aldrich | Cat #B6768 |
| Ovomucoid | Sigma-Aldrich | Cat #T9253 |
| Papain | Sigma-Aldrich | Cat #P4762 |
| L-Cysteine | Millipore | Cat #C-7352 |
| DNaseI | Sigma-Aldrich | Cat #D4513 |
| Neurobasal medium | Thermofisher Scientific | Cat # 21103049 |
| Phenol | Sigma-Aldrich | Cat #P4557 |
| Chloroform | Merck Life science Private.Ltd | Cat #DI1P711147 |
| Isoamyl alcohol | Merck Life science Private.Ltd | Cat #SH8S680463 |
| Diluent for DNA Extraction | Himedia | Cat #MB228 |
| Proteinase K | Sigma-Aldrich | Cat #49936 |
| RNaseA | Merck Life science Private.Ltd | Cat # 10109142001 |
| Hank’s Balanced Salt Solution | Thermofisher Scientific | Cat #14170112 |
| Tri sodium citrate dihydrate | Merck Life science Private.Ltd | Cat #DEOD701038 |
| Tween 20 | Sigma-Aldrich | Cat #P9416 |
| Taq DNA Polymerase, Buffer,Mgcl2 | Sigma-Aldrich | Cat #D4545 |
| dNTPs | Sigma-Aldrich | Cat #A4036,T1875,G6264,C4654 |
| TRI reagent | Sigma-Aldrich | Cat# T9424 |
| DNaseI | Ambion | Cat# 8169G2 |
| DNaseI Inactivation reagent | Ambion | Cat# 8174G |
| Random Hexamer | Promega | Cat#C118A |
| Reverse Transcriptase Superscript III | Invitrogen | Cat#P/N100004295 |
| DTT | Invitrogen | Cat#P/NY00147 |
| Rnasin | Promega | Cat#N2611 |
| Critical commercial assays | ||
| RNA Extraction, RNeasy kit | Qiagen | Cat # 11684795910 |
| Chromium Next GEM Single Cell 3′ LT Reagent Kits v3.1(Dual index) | 10× Genomics | Cat # PN-1000325 |
| In Situ Cell Death Detection Kit, Fluorescein | Roche | Cat # 11684795910 |
| Experimental models: Organisms/strains | ||
| Mouse: Tlx3fl/fl | Gift from Dr Zijing Liu, Beijing Institute of Biotechnology, Beijing, China Lopes et al.71 |
N/A |
| Mouse: Atoh1 Cre | The Jackson Laboratory, USA | RRID:IMSR_JAX:011104 (B6.Cg-Tg(Atoh1-Cre)1Bfri/J) |
| Mouse: C57BL/6J | Animal facility, RGCB | NA |
| Oligonucleotides | ||
|
Tlx3 floxed genotyping primer F-TGTTTCGCCTCCTTTGCTCG R-GTTGGATGGAAGCAAAGATAG |
Lopes et al.71 | N/A |
|
Atoh1 Cre genotyping primer (Transgene) F-CCGGCAGAGTTTACAGAAGC R-ATGTTTAGCTGGCCCAAATG |
The Jackson Laboratory, USA | N/A |
|
Atoh1 Cre genotyping primer (Internal positive control) F-CTAGGCCACAGAATTGAAAGATCT R-GTAGGTGGAAATTCTAGCATCATCC |
The Jackson Laboratory, USA | N/A |
| Mouse Tlx3 qPCR primer F-GCGCATCGGCCACCCCTACCAGA R- CCGCTCCGCCTCCCGCTCCTC |
Divya et al.16 | N/A |
| Mouse Cdk2ap1 qPCR primer F- ACCCAGGGAACTGGAAATAG R- AATGATGCCTCGTTTTAGCC |
This paper | N/A |
| Mouse Cadm1 qPCR primer F- CACCATCCTTACCATCATCAC R- ATAGCATGGCAAACACCAC |
This paper | N/A |
| Mouse Tgfb2 qPCR primer F- TCTACAACAGTACCAGGGAC R- TGTTGAGACATCAAAGCGG |
This paper | N/A |
| Mouse Bbip1 qPCR primer F- TGAAGTCGATGTTCCGAGAAG R- TGTATCCTGTGCTGCTTGC |
This paper | N/A |
| Mouse Insc qPCR primer F- GACATCATTCAGGAGAACGG R- CAATCAGGCGAGACATACAG |
This paper | N/A |
| Software and algorithms | ||
| R | 4.2.0 | https://www.r-project.org/ |
| Cell Sens | Olympus FV3000 | https://www.olympus-lifescience.com/en/software/cellsens/ |
| ImageJ | ImageJ 1.53t NIH,USA |
ImageJ.net |
| EthoVision XT 8.5.614 | Noldus Information Technology by Netherlands | https://www.noldus.com/ethovision-xt |
| GraphPad Prism Version 8 | GraphPad | https://www.graphpad.com/scientificsoftware/prism/ |
| Deposited data | ||
| RNA-Seq raw data: PN7 Anterior cerebellum control | NCBI Sequence Read Archive (SRA) | NCBI SRA: SRR30996787; NCBI SRA: SRR30996793 (Biological replicates) |
| RNA-Seq raw data: PN7 Anterior cerebellum Tlx3 cKO | NCBI Sequence Read Archive (SRA) | NCBI SRA: SRR30996785; NCBI SRA: SRR30996786 (Biological replicates) |
| RNA-Seq raw data: PN7 Posterior cerebellum control | NCBI Sequence Read Archive (SRA) | NCBI SRA: SRR30996799; NCBI SRA: SRR30996797 (Biological replicates) |
| RNA-Seq raw data: PN7 Posterior cerebellum Tlx3 cKO | NCBI Sequence Read Archive (SRA) | NCBI SRA: SRR30996788; NCBI SRA: SRR30996789 (Biological replicates) |
| Single-cell RNA-Seq raw data: PN4 Posterior cerebellum control | NCBI Sequence Read Archive (SRA) | NCBI SRA: SRR30996796; NCBI SRA: SRR30996798; NCBI SRA: SRR30996794; NCBI SRA: SRR30996784 (Technical replicates) |
| Single-cell RNA-Seq raw data: PN4 Posterior cerebellum Tlx3 cKO | NCBI Sequence Read Archive (SRA) | NCBI SRA: SRR30996790; NCBI SRA: SRR30996791; NCBI SRA: SRR30996792; NCBI SRA: SRR30996795 (Technical replicates) |
| Complete dataset | NCBI BioProject database | NCBI BioProject: PRJNA1173099 |
| Code: | Zenodo | https://doi.org/10.5281/zenodo.13934594 |
Experimental model and study participant details
Generation of Tlx3fl/fl; Atoh1 Cre conditional knockout mice
The generation of Tlx3fl/fl mice was described previously.71 The mouse was a generous gift from Dr Zijing Liu, Beijing Institute of Biotechnology, Beijing, China. The loxP sites were inserted within Intron 1 and Intron 2 of the Tlx3 gene to target the conditional deletion of Exon 2 (contains the homeobox DNA binding domain) using Cre recombinase. All experiments were conducted as per the ethical guidelines approved by the Institutional Animal Ethics Committee (IAEC) of Rajiv Gandhi Center for Biotechnology (IAEC/792/JAC/2020). All the animals were maintained in pathogen free condition in individually ventilated cages (IVC), with 12h light/dark cycle, standard temperature, humidity and were fed with standard feed and water ad libitum. Tlx3fl/fl mice were bred with Atoh1-Cre (Jackson Laboratory, USA)72 heterozygous mice to produce heterozygous mutants. Homozygous male mutants generated after the second round of breeding were bred with Tlx3fl/fl female to produce mixed genotypes of homozygous Tlx3 cKO (Tlx3fl/fl; Atoh1 Cre+/−) mice and Cre negative control (Tlx3fl/fl; Atoh1 Cre−/−) mice from the same litter (Figure 1D). For further experiments, pregnant Tlx3fl/fl mouse were sacrificed at embryonic days, E15-16 and E17-18, and embryos were collected for qRT PCR and immunohistochemistry analysis. Male and female mice at PN7, PN14, and PN21 were also collected for BrdU pulse-chase experiments and immunohistochemical analysis. The comprehensive list of the total number of mice used in each immunohistochemistry image and behavior study is included in Tables S10 and S11. Cerebellum from PN4 and PN7 pups were used for single-cell transcriptomics and for RNA-Seq respectively. Male/female control and cKO mice were allowed to grow till 3 months of age and were used for all behavior analysis. Genotyping PCR was carried out with tail tips to confirm the genotypes of the mice and also knocking out of Tlx3.
Method details
Tail DNA extraction and genotyping PCR
We used a modified version of Jackson Laboratory’s protocol to isolate DNA from the tail tips and perform genotyping PCR. Briefly, a small 0.5 mm tail tip was cut and treated with 500 μL lysis buffer containing proteinase K and incubated overnight at 50°C. Further, an equal volume (500 μL) of phenol/chloroform/iso-amyl alcohol in a ratio of 25:24:1 was added to the samples. The upper aqueous phase containing DNA was collected after centrifugation, and the resultant DNA was precipitated using 100% ethanol and subsequent centrifugation. The pellet was purified using 70% ethanol wash, air dried at room temperature, and dissolved in 20 μL sterile water. The resultant purified DNA was then used as the template for genotyping PCR. Specific strains, control and Tlx3 cKO were identified using respective genotyping primers as listed in the key resources table.
Survivorship curves
Mice were monitored starting from birth, allowed to grow until 9 months old, and euthanized after this time point. Survivorship curves were generated from this period based on survival, plotted using GraphPad Prism 8.0.1, and plotted as percentage survival.
Histology and immunohistochemical analysis
Mouse brains from respective stages were collected and fixed in 4% paraformaldehyde made in phosphate-buffered saline overnight and then dehydrated using 30% sucrose as described previously.16 Fixed, dehydrated whole brains were embedded in OCT, and 14 μm cryosections were collected for immunohistochemistry. Frozen sections were washed in 1X PBS, and then blocked with 5% Normal Goat Serum (NGS) containing 0.1%/0.2%/0.4% Triton X-100 (based on nuclear/cytoplasmic localization of the target protein) for 45 min at room temperature before incubating with primary antibodies (Listed in key resources table) overnight at 4°C. Immune complexes were detected using secondary antibodies conjugated to Cy3/Alexa Fluor 488. Finally, sections were counterstained with DAPI and mounted using Fluromount G. Images were captured and analyzed using Confocal laser scanning microscope with a high-sensitivity spectral detector (Olympus FV3000) and CellSens software or using Olympus BX61 upright fluorescence microscope with a cooled CCD camera (Andor 885). The total number of mice used per genotypes of control and Tlx3 cKO for Immunohistochemical analysis was listed in Table S10.
BrdU labeling
BrdU pulse labeling were performed by injecting BrdU (100 mg/kg) into mice at the E15-E16 and PN7 stage for 2 h (short-term) and 12 h. Long-term tracking of BrdU was carried out by injecting at the PN7 stage and subsequent collection at PN14. Tissues were processed as mentioned earlier. Briefly, after washing with 1× PBS, sections were treated with IN HCL for 45 min at 37°C. Tissue sections were then neutralized using boric acid in IX PBS, pH 8.5, for 10 min. BrdU was detected using an anti-rat BrdU antibody. Fluorescence was analyzed and captured using Confocal laser scanning microscope with a high-sensitivity spectral detector (Olympus FV3000) and CellSens software. The total number of mice used per genotypes of control and Tlx3 cKO for BrdU-pulse chase analysis was listed in Table S10.
TUNEL assay
Cell death was assessed using the manufacturer’s protocol as described (In Situ Cell Death Detection Kit, Fluorescein). Frozen tissue samples were permeabilized using 10 mM sodium citrate buffer at pH 6 for 5–15 min based on the developmental stages. Briefly, permeabilized sections were washed in 1× PBS, and incubated with 50 μL TUNEL analysis solution for 60 min at 37°C in a humidified chamber in the dark. Positive (DNase digested) and negative control (only label solution) for the experiment were also processed with all the samples. Fluorescence was analyzed and captured using a Confocal laser scanning microscope with a high-sensitivity spectral detector (Olympus FV3000) and CellSens software.
Measurement of cerebellar area and cell count
The total area of the cerebellum for each developmental stage (E16, E18, PN7, PN21) were measured from sagittal cerebellar sections after DAPI staining using ImageJ software (version 1.53t). Each section was subdivided into equal bins (constituting the same area) using Photoshop, and the cell count was obtained using the counting tool. Genotype-blind quantifications were performed for all the analyses. The cell count obtained from respective replicates of each stage and each marker was then evaluated for statistical significance, and the results were plotted using GraphPad Prism 8.0.1.
Meta-analysis of cerebellar single-cell transcriptome data across developmental points
The data was obtained from R. A. Carter et al., 201821 (European Nucleotide Archive: PRJEB23051). The output files from 10× Genomics CellRanger pipeline (obtained from GSE224466) were analyzed using Seurat73 and R. The cells were filtered based on quality, normalized, and clustered using principal components (PCs). The number of PCs to be used was determined using Elbow plot. The cluster identities were determined using the DEGs identified in Carter et al.,21 except Tlx3. The non-cerebellar cell types were eliminated in further analysis. The aggregate expression of genes was calculated for different cell types across developmental points and was plotted as a heatmap using R.
RNA sequencing
RNA Sequencing was carried out between control and Tlx3 cKO posterior and anterior cerebellum at PN7 stage and total RNA was isolated using RNeasy kit (Qiagen) following manufacturer’s protocol. A minimum of 100 ng of isolated mRNA was used to prepare sequencing libraries with the TruSeq stranded mRNA reagent (Illumina), and sequencing was performed on a HiSeq Illumina Genome Analyzer. The RNA-Seq paired-end reads were trimmed using Trim-galore version 0.6.4 (https://github.com/FelixKrueger/TrimGalore). Trimmed reads were then aligned on both the mouse genome and transcriptome assembly GRCm39/mm39) with STAR version 2.5.0.74 Transcript quantification was done using rsem version 1.3.0.75 The resulting counts matrix was used for further analysis. Analysis was conducted in R. Genes were filtered based on the rule of 1 count per million (cpm) in at least 1 sample. Library sizes were scaled using TMM normalization. edgeR, a Bioconductor package for differential expression analysis of digital gene expression data, were utilized and log-transformed with limma voom function.76,77 Normalized data was corrected for batch effect using remove Batch Effect function (limma). Also, unwanted variation was removed using RUVr78 (RUVseq package version 1.24.0). Differential expression was computed directly on the lRUVr batch-corrected values using limma by fitting a linear model on the selected samples. Statistically significant DEGs were identified from log-transformed, TMM-scaled values using lmFit of the Limma package through fitting a linear model on the selected samples. Further, gene set enrichment analysis (GSEA) was performed on the gene list ranked based on a custom metric score, where score = {-log10(P-value) ∗ sign(log2(FoldChange)} using fgsea package. The ontology gene set and cell type signature gene set were downloaded from the Molecular Signature Database (MSigDB) using msigdbr package. The plots were made using ‘ggplot2’ and ‘enrichplot’ packages.
Single-cell RNA sequencing
Single-cell RNA-Sequencing was carried out between PN4 control and Tlx3 cKO posterior cerebellum. Posterior cerebellar tissue was freshly dissociated using Papain solution in HBSS containing L-Cysteine, and papain was inactivated using ovomucoid solution. Following this, samples were centrifuged at 1800 rpm for 5 min. Supernatant were carefully removed and cells was resuspended in 1mL of 10 mg/mL BSA. After another round of centrifugation at 1800 rpm for 5 min, cells were resuspended in 1mL 1× PBS. Cell death was analyzed using trypan blue and cells were counted using a hemocytometer. Approximately 1000–2000 cells of control and Tlx3 cKO single-cell suspensions were loaded onto a 10× chromium platform for generating single-cell Gel Bead-In-EMulsion (GEMs). Libraries were generated with Chromium Next GEM Single Cell 3′ LT Reagent Kits v3.1 (Dual index) as per 10× genomics protocol and were sequenced.
The sequencing of libraries was performed on an Illumina Novaseq platform. The software 10× Genomics Cell Ranger (version 3.1.0) was used to align to a custom reference mouse genome mm39 with Cre gene added. Post alignment a count matrix was generated. Next, the percentage of mitochondrial RNA per cell was calculated, filtered and the cells with higher quality (mitochondrial reads <7.5%, 200 < genes <7000) were used. We found the female sex specific Xist expression in the KO sample. After removing low-quality cells, we performed the global-scaling normalization “LogNormalize”, which normalizes gene expression measurements for each cell concerning the total expression, multiplied by a scale factor (10,000 by default), and log-transforms the outcomes. We next calculated 2000 most variable genes using Seurat function ‘FindVariableFeature’ with the default variance stabilizing process. Further we performed integration of control and KO samples based on integration anchors using Seurat function ‘IntegrateData’. Next, we identified clusters of cells in each sample using Seurat, which aggregated all samples using canonical correlation analysis and mutual adjacent neighbor analysis.79 The optimal number of principal components was then determined by utilizing the ElbowPlot and DimHeatmap functions. Dimensionality reduction was performed using the RunUMAP function and cluster resolution of 0.4 was visualized. Cell clusters were annotated utilizing canonical markers of known cell types along with the identified distinct marker signatures. We identified DEGs between two groups of all the clusters using the FindMarkers function in Seurat (Wilcoxon rank-sum test). Genes with an adjusted p value less than 0.05 and a log2(fold change) greater than 0.25 were considered differentially expressed. Following this, enrichment analysis was carried out in identified clusters. The cell cycle scoring was measured based on expression of G2/M and S phase markers. The gene ontology (GO) enrichment analysis, and Kyoto encyclopaedia of genes and genomes (KEGG) enrichment analyses were performed to annotate the clusters. The identified clusters were also confirmed with the gene set enrichment analysis (GSEA) using fgsea package. The gene sets used for fgsea were downloaded from MSigDB using msigdbr package (v7.5.1). GO analysis were conducted specifically on up regulated genes of granule neuron and granule neuron progenitor clusters using DAVID and the results were plotted using R.
Data visualization
Basic data analysis was performed using ‘dplyr’ and ‘tidyr’ packages. Base plots were made using R 4.1.0. Venn diagram was made using ‘VennDiagram’ package. Data visualization of single-cell transcriptomic analysis was performed using Seurat: v4.3.0. (https://github.com/satijalab/seurat), SCP: v0.4.7 (https://github.com/zhanghao-njmu/SCP), scCustomize: v1.1.3 (https://github.com/samuel-marsh/scCustomize), fgsea: v1.20.0 (https://github.com/ctlab/fgsea), ggplot2: v3.4.4 (https://github.com/tidyverse/ggplot2), and ComplexHeatmap: v2.15.4 (https://github.com/jokergoo/ComplexHeatmap).
RNA isolation and real-time PCR analysis
The deletion of Tlx3 and anti-proliferative genes obtained from scRNA-seq was confirmed using real-time PCR analysis as described previously.16 Briefly, RNA was isolated from the posterior cerebellar tissue of control and Tlx3 cKO at the PN4 stage using Tri reagent (Sigma-Aldrich). The isolated RNA then underwent DNAse treatment, and the quality of isolated RNA was quantified and assessed using a Nanodrop 2000 UV Visible Spectrophotometer (Thermo Scientific). After quantification, an equal amount of RNA from control and Tlx3 cKO was taken and reverse transcribed into cDNA using superscript RT-II on ProFlex PCR System (Applied Biosystems by Life technologies). Real-time PCR analysis of selected genes was conducted using SYBR green mix (Bio-Rad) on QuantStudio 3 (Applied Biosystems by Thermo Fisher Scientific). The expression levels of selected genes were analyzed using the 2−ΔΔCt method and used β -actin as an internal control. The list of reagents, enzymes, and oligonucleotides used for isolating RNA, cDNA synthesis, and q-PCR analysis were listed in the key resources table.
General protocol for behavior tests
Tlx3 cKO mice and littermate control mice were housed in the same cages after weaning at the PN21 stage, except for the ultrasonic vocalization, where each mouse was individually housed for 21 days prior to the test. As described previously, each group followed a 12-h dark and 12-h light cycle. All behavior tests were conducted using mature adult male and female mice of 3 months age between 9a.m. and 6p.m. Before each test and between each trial, all apparatus used for each experiment was cleaned with hypochlorite solution and 90% alcohol to avoid any olfactory cues to remove any bias caused by them. In addition, mice were given a 1-h acclimatization period before each test to adjust to the experimental area. Between each experiment and each trial, video recordings of each mouse was carried out. Detailed information of the total number of mice that have undergone each behavior test is listed in Table S11.
Motor function tests
As described previously, 3 month old male/female control and Tlx3 cKO mouse were used for the experiments.80 The two-limb wire hang test was conducted to assess the grip strength, coordination, and individual balance for the forelimb. Furthermore, forelimb and hindlimb strength and coordination were evaluated using the four-limb wire hang test.
Two-limb hanging test
The two-limb wire hang test was performed using a 55 cm wide, 2 mm thick metallic cloth hanger fixed on a wooden rod attached to the wall and kept 37 cm above a layer of bedding. Each mouse was individually placed on the wire and allowed to hang with its forepaws for a maximum of 600 s, which was manually monitored and recorded using a timer. Mice that fall off before this time were given two more attempts, and the third fall was considered the maximum hanging time. Mice that do not fall within this time were returned to the cage after 600 s.
Four-limb hanging test
A rat cage lid was used for the four-limb hanging test. The grid was positioned 25 cm above the bedding layer and tightly secured to a supporting wall. As previously described, each mouse was tested separately by placing it on the grid, allowing it to grab with all four paws, in an upside position. Similar to the two-limb test, each mouse was given a maximum of 600 s and mice that had fallen before this time were given two more chances and were scored for the maximum hanging time. In both the two- and four-limb tests, each mouse is given three consecutive sessions separated by one week gap, and the average hanging time from these three sessions was used for further analysis.
Rotarod test
The rotarod test was conducted as described previously.80,81 Rotarod tests determine the mouse’s muscular strength, coordination, balance, and motor condition. The apparatus consists of three compartments with 3 cm-diameter rotating rods positioned 50 cm above the ground (Rotarod 3 Compartment; Diwakar Instruments Company, India). Each mouse was placed on a rotating rod with a minimum speed of 3–6 rpm and the rpm was increased slowly to a maximum of 24 rpm within 100 s. The maximum hanging time for one trial was 300 s, and mice that fall off before this time were scored. They underwent three consecutive trials in a single day. Similarly, two sessions were conducted with a gap of one week, and the average of six trials were scored and used for further analysis.
Anxiety assessment tests
Open-maze test
The apparatus consists of one chamber measuring 50 cm (length) x 50 cm (width) x 38 cm (height) and was painted white as described earlier.82 After a 1-h acclimatization period in the experimental area, the mouse was transferred into the chamber and recorded for 10 min. Total distance traveled and time spent in the central zone was measured using Noldus EthoVision XT software (Noldus Information Technology, Wageningen, The Netherlands) using the recorded video files.
Elevated plus-maze test
As previously described,83 the device has a Plus layout and consists of two open arms (25 × 5 × 0.5 cm), two closed arms (25 × 16 cm), and a central platform (5 × 5 × 0.5 cm). The closed-arm walls were transparent and made from transparent plexiglass. The whole apparatus was supported by a wooden log and elevated 50 cm from the floor. Similar to the open maze test, individual mice were transferred to the central platform of the apparatus and recorded for 10 min. Video files were analyzed using the Noldus EthoVision XT software. Time spent in open arms was measured to assess open-space-induced anxiety.
Cognition test
Novel object recognition test
The novel object recognition test was done as described previously.84,85 Here, the mouse was exposed to two similar objects in the first session, and in the second session, one of the two objects was replaced with a new one. The total time spent exploring the new object is a measure of recognition memory. We used the open maze chamber, which was used in the open maze test, and each mouse had a short habituation period in the empty open field prior to the test. A falcon tissue culture flask filled with sand and a tower of Lego bricks were taken as two typical objects. During the first familiarization session, the mouse was presented with two identical objects and allowed it to explore the objects for 10 min. After a short 1-h gap period, the mouse was tested for recognition memory in the test session, where one of the familiar objects was replaced by a novel object. The session was recorded for 10 min. The video recordings were analyzed using The Noldus EthoVision XT software, and the total time spent exploring the novel object was calculated and used for further analysis.
Repetitive behavior test
Marble burying test
The marble burying test was carried out to assess the repetitive behaviors in mice as described previously.86 Standard polycarbonate rat cages (26 cm × 48 cm × 20 cm) with fitted filter-top covers were used for the test. Fresh, odorless bedding was added to the cage to a depth of 5 cm, and the surface was leveled to create parallel lines for keeping the marbles. Five rows of four standard glass toy marbles of 15 mm diameter, and 5.2 g weight, in varied styles and colors, were gently placed on the surface of the bedding. Each mouse was given 20 marbles and permitted to stay in the cage undisturbed for 30 min. Following the test period, the mouse was returned to its home cage without disturbing or dislodging the marbles. The number of buried marbles was then counted (Figure S17). A marble is considered buried if two-thirds of its surface is covered by the bedding. Total scores for each mouse were taken and used for further analysis.
Social interaction tests
Three chamber test (Social preference and social novelty test)
The three-chamber apparatus consists of three chambers each chamber had a dimension of 19 × 45 cm, and the dividing walls were made from clear Plexiglas, with an open middle section. This allows free access to each chamber as described previously.87 The three-chamber test evaluates the social affiliation and social memory in mice with two consecutive sessions. The mice will get free choice to spend time with a subject mouse/social stimuli/novel social stimuli and empty space in any of the three chambers in two consecutive sessions. In the first session, the mouse was tested for social preference using the S-E social preference protocol as described previously.88 To score this, the mouse was given social stimuli in one chamber and an empty cup in the other chamber, and after an acclimatization period of 5 min, the mouse was free to interact with any of these subjects. The session was recorded for 10 min. The unfamiliar mouse used for this test was age, sex, and background matched BL6J strain, it was placed in a small wire containment cup that allowed nose contact between the mouse and prevented them from fighting and freely moving around. The test mouse was placed in the center chamber and given a habituation period of 5 min. Subsequently, after the habituation, dividing walls were removed and mouse were allowed to interact with the social stimuli in one chamber and empty cup in another chamber. This session was recorded for 10 min. Immediately after this, the empty cup was replaced by a novel social stimulus/novel mouse. Similar to the first session, the mouse was allowed to interact with both stimuli for 10 min. The two recorded sessions of each mouse were then analyzed for total time spent between social stimuli vs. empty cup and familiar social stimuli vs. novel social stimuli using The Noldus EthoVision XT software.
Resident-Intruder test followed by Ultrasonic vocalization recording
Emission of USVs is a consistent and a robust behavior during social interactions between adult mice, and it is regarded as a sign of social interest and motivation.89 As mentioned previously, each mouse was individually housed for 21 days prior to the test to increase social motivation. Similar to the three chamber test, we have used age, sex, and background-matched BL6J mouse as an intruder.90,91 The test mouse was placed in a test cage made of Plexiglass with a dimension of 36 cm × 25 cm × 30 cm, in a soundproof chamber and the microphone was placed 20 cm above the cage. The test mouse had a habituation period of 20 min in the test cage, and after that, the intruder mouse was introduced and allowed to interact for 4 min. During this whole session, ultrasonic vocalizations were recorded using ultrasound microphone CM16/CMPA, the interface Ultrasound Gate 416H, and the software Avisoft SASLab Pro Recorder from Avisoft Bioacoustics (sampling frequency: 300 kHz; fast Fourier transform length: 1024 points; 16-bit format). The adult mouse call types were identified and analyzed qualitatively and quantitatively.92
Neurodegeneration test
Clasping test (hindlimb)
This test is a marker for neurodegeneration, and was carried out as described previously.93,94 Hindlimb clasping is tested by lifting the animal with the tail in an inverted position for 10 s. Hindlimb positions were observed, and the sessions were video recorded and scored. If the hind limbs are consistently splayed outward, away from the abdomen, it is assigned a score of 0. If one hindlimb is retracted toward the abdomen for more than 50% of the time suspended, it receives a score of 1. If both hind limbs are partially retracted toward the abdomen for more than 50% of the time suspended, it receives a score of 2. If its hind limbs are entirely retracted and touching the abdomen for more than 50% of the time suspended, it receives a score of 3.
Enrichment of SFARI ASD associated genes
The combined DEGs in cKO among the granule neuron, granule neuron progenitor and Purkinje clusters were compared with the SFARI ASD associated genes using package ‘geneLists’. Hypergeometric test was performed for enrichment of ASD associated genes using ‘hyperTest’ function of geneLists package. p value <0.05 was considered significant enrichment in the differentially expressed gene list. Results were analyzed and plotted using R.
TLX3 variant curation and ACMG classification
TLX3 variants were curated from public domain, ClinVar and scientific literature published in English language. The variants have been curated and validated using variants validator tools. Thereafter annotation of TLX3 gene variant were performed according to standard guidelines of ACMG. We conducted a thorough assessment of minor allele frequencies (MAF) of variants across the major global populations, including Africans, Americans, Europeans (of Finnish and non-Finnish origin), Middle Eastern individuals, the Amish community, East Asians and South Asians, utilizing data from the gnomAD resource. The plot was prepared on ggplot package using R program.
Quantification and statistical analysis
Statistical significance between the two groups was calculated using an unpaired Student’s t test and a Mann-Whitney test based on the data distribution. Fisher test was conducted for analyzing the statistical significance for scRNA Seq. An outlier test was also conducted for all behavior experiment data and after the outliers were removed, a p-value<0.05 was considered statistically significant. The multiple-group comparison was conducted using an ordinary one-way ANOVA. The results obtained were presented as mean ± SD. All the statistical tests except that for RNA and scRNA sequencing were conducted, and the graphs were plotted using GraphPad Prism 8.0.1. In the Figures,∗ indicates p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Published: November 5, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.111260.
Supplemental information
Italic font is used to indicate the gene names (Excel format).
Italic font is used to indicate the gene names (Excel format).
Italic font is used to indicate the gene names (Excel format).
Italic font is used to indicate the gene names (Excel format).
References
- 1.Holland P.W.H., Takahashi T. The evolution of homeobox genes: Implications for the study of brain development. Brain Res. Bull. 2005;66:484–490. doi: 10.1016/j.brainresbull.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Cheng S.H., Mak T.W. Molecular Characterisation of Three Murine HOX11-Related Homeobox Genes, Tlx-1, −2, and −3, and Restricted Expression of Tlx-1 during Embryogenesis. Dev. Growth Differ. 1993;35:655–663. doi: 10.1111/j.1440-169X.1993.00655.x. [DOI] [PubMed] [Google Scholar]
- 3.Dear T.N., Sanchez-Garcia I., Rabbitts T.H. The HOX11 gene encodes a DNA-binding nuclear transcription factor belonging to a distinct family of homeobox genes. Proc. Natl. Acad. Sci. USA. 1993;90:4431–4435. doi: 10.1073/pnas.90.10.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dear T.N., Colledge W.H., Carlton M.B., Lavenir I., Larson T., Smith A.J., Warren A.J., Evans M.J., Sofroniew M.V., Rabbitts T.H. The Hox11 gene is essential for cell survival during spleen development. Development. 1995;121:2909–2915. doi: 10.1242/dev.121.9.2909. [DOI] [PubMed] [Google Scholar]
- 5.Raju K., Tang S., Dubé I.D., Kamel-Reid S., Bryce D.M., Breitman M.L. Characterization and developmental expression of Tlx-1, the murine homolog of HOX11. Mech. Dev. 1993;44:51–64. doi: 10.1016/0925-4773(93)90016-Q. [DOI] [PubMed] [Google Scholar]
- 6.Roberts C.W., Sonder A.M., Lumsden A., Korsmeyer S.J. Development expression of Hox11 and specification of splenic cell fate. Am. J. Pathol. 1995;146:1089–1101. [PMC free article] [PubMed] [Google Scholar]
- 7.Logan C., Wingate R.J., McKay I.J., Lumsden A. Tlx-1 and Tlx-3 Homeobox Gene Expression in Cranial Sensory Ganglia and Hindbrain of the Chick Embryo: Markers of Patterned Connectivity. J. Neurosci. 1998;18:5389–5402. doi: 10.1523/JNEUROSCI.18-14-05389.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts C.W., Shutter J.R., Korsmeyer S.J. Hox11 controls the genesis of the spleen. Nature. 1994;368:747–749. doi: 10.1038/368747a0. [DOI] [PubMed] [Google Scholar]
- 9.Shirasawa S., Yunker A.M., Roth K.A., Brown G.A., Horning S., Korsmeyer S.J. Enx (Hox11L1)-deficient mice develop myenteric neuronal hyperplasia and megacolon. Nat. Med. 1997;3:646–650. doi: 10.1038/nm0697-646. [DOI] [PubMed] [Google Scholar]
- 10.Shirasawa S., Arata A., Onimaru H., Roth K.A., Brown G.A., Horning S., Arata S., Okumura K., Sasazuki T., Korsmeyer S.J. Rnx deficiency results in congenital central hypoventilation. Nat. Genet. 2000;24:287–290. doi: 10.1038/73516. [DOI] [PubMed] [Google Scholar]
- 11.Cheng L., Arata A., Mizuguchi R., Qian Y., Karunaratne A., Gray P.A., Arata S., Shirasawa S., Bouchard M., Luo P., et al. Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates. Nat. Neurosci. 2004;7:510–517. doi: 10.1038/nn1221. [DOI] [PubMed] [Google Scholar]
- 12.Qian Y., Fritzsch B., Shirasawa S., Chen C.-L., Choi Y., Ma Q. Formation of brainstem (nor)adrenergic centers and first-order relay visceral sensory neurons is dependent on homeodomain protein Rnx/Tlx3. Genes Dev. 2001;15:2533–2545. doi: 10.1101/gad.921501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Logan C., Millar C., Bharadia V., Rouleau K. Onset of Tlx-3 expression in the chick cerebellar cortex correlates with the morphological development of fissures and delineates a posterior transverse boundary. J. Comp. Neurol. 2002;448:138–149. doi: 10.1002/cne.10234. [DOI] [PubMed] [Google Scholar]
- 14.Cheng L., Samad O.A., Xu Y., Mizuguchi R., Luo P., Shirasawa S., Goulding M., Ma Q. Lbx1 and Tlx3 are opposing switches in determining GABAergic versus glutamatergic transmitter phenotypes. Nat. Neurosci. 2005;8:1510–1515. doi: 10.1038/nn1569. [DOI] [PubMed] [Google Scholar]
- 15.Xu Y., Lopes C., Qian Y., Liu Y., Cheng L., Goulding M., Turner E.E., Lima D., Ma Q. Tlx1 and Tlx3 Coordinate Specification of Dorsal Horn Pain-Modulatory Peptidergic Neurons. J. Neurosci. 2008;28:4037–4046. doi: 10.1523/JNEUROSCI.4126-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Divya T.S., Lalitha S., Parvathy S., Subashini C., Sanalkumar R., Dhanesh S.B., Rasheed V.A., Divya M.S., Tole S., James J. Regulation of Tlx3 by Pax6 is required for the restricted expression of Chrnα3 in Cerebellar Granule Neuron progenitors during development. Sci. Rep. 2016;6 doi: 10.1038/srep30337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sillitoe R.V., Joyner A.L. Morphology, molecular codes, and circuitry produce the three-dimensional complexity of the cerebellum. Annu. Rev. Cell Dev. Biol. 2007;23:549–577. doi: 10.1146/annurev.cellbio.23.090506.123237. [DOI] [PubMed] [Google Scholar]
- 18.Buffo A., Rossi F. Origin, lineage and function of cerebellar glia. Prog. Neurobiol. 2013;109:42–63. doi: 10.1016/j.pneurobio.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Hoshino M. Neuronal subtype specification in the cerebellum and dorsal hindbrain. Dev. Growth Differ. 2012;54:317–326. doi: 10.1111/j.1440-169X.2012.01330.x. [DOI] [PubMed] [Google Scholar]
- 20.Stoodley C.J. The Cerebellum and Neurodevelopmental Disorders. Cerebellum. 2016;15:34–37. doi: 10.1007/s12311-015-0715-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carter R.A., Bihannic L., Rosencrance C., Hadley J.L., Tong Y., Phoenix T.N., Natarajan S., Easton J., Northcott P.A., Gawad C. A Single-Cell Transcriptional Atlas of the Developing Murine Cerebellum. Curr. Biol. 2018;28:2910–2920.e2. doi: 10.1016/j.cub.2018.07.062. [DOI] [PubMed] [Google Scholar]
- 22.Akazawa C., Ishibashi M., Shimizu C., Nakanishi S., Kageyama R. A Mammalian Helix-Loop-Helix Factor Structurally Related to the Product of Drosophila Proneural Gene atonal Is a Positive Transcriptional Regulator Expressed in the Developing Nervous System. J. Biol. Chem. 1995;270:8730–8738. doi: 10.1074/jbc.270.15.8730. [DOI] [PubMed] [Google Scholar]
- 23.Ben-Arie N., Bellen H.J., Armstrong D.L., McCall A.E., Gordadze P.R., Guo Q., Matzuk M.M., Zoghbi H.Y. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- 24.Consalez G.G., Goldowitz D., Casoni F., Hawkes R. Origins, Development, and Compartmentation of the Granule Cells of the Cerebellum. Front. Neural Circuits. 2020;14 doi: 10.3389/fncir.2020.611841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pujato M., Kieken F., Skiles A.A., Tapinos N., Fiser A. Prediction of DNA binding motifs from 3D models of transcription factors; identifying TLX3 regulated genes. Nucleic Acids Res. 2014;42:13500–13512. doi: 10.1093/nar/gku1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Der Heijden M.E., Sillitoe R.V. Interactions Between Purkinje Cells and Granule Cells Coordinate the Development of Functional Cerebellar Circuits. Neuroscience. 2021;462:4–21. doi: 10.1016/j.neuroscience.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L., Liu Y. Signaling pathways in cerebellar granule cells development. Am. J. Stem Cells. 2019;8:1–6. [PMC free article] [PubMed] [Google Scholar]
- 28.Hashimoto K., Kano M. Functional Differentiation of Multiple Climbing Fiber Inputs during Synapse Elimination in the Developing Cerebellum. Neuron. 2003;38:785–796. doi: 10.1016/S0896-6273(03)00298-8. [DOI] [PubMed] [Google Scholar]
- 29.Kano M., Hashimoto K. Activity-Dependent Maturation of Climbing Fiber to Purkinje Cell Synapses during Postnatal Cerebellar Development. Cerebellum. 2012;11:449–450. doi: 10.1007/s12311-011-0337-3. [DOI] [PubMed] [Google Scholar]
- 30.Stoodley C.J., Schmahmann J.D. Evidence for topographic organization in the cerebellum of motor control versus cognitive and affective processing. Cortex. 2010;46:831–844. doi: 10.1016/j.cortex.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Badura A., Verpeut J.L., Metzger J.W., Pereira T.D., Pisano T.J., Deverett B., Bakshinskaya D.E., Wang S.S.-H. Normal cognitive and social development require posterior cerebellar activity. Elife. 2018;7 doi: 10.7554/eLife.36401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D’Mello A.M., Stoodley C.J. Cerebro-cerebellar circuits in autism spectrum disorder. Front. Neurosci. 2015;9 doi: 10.3389/fnins.2015.00408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmahmann J.D. The cerebellum and cognition. Neurosci. Lett. 2019;688:62–75. doi: 10.1016/j.neulet.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 34.Mapelli L., Soda T., D’Angelo E., Prestori F. The Cerebellar Involvement in Autism Spectrum Disorders: From the Social Brain to Mouse Models. Int. J. Mol. Sci. 2022;23:3894. doi: 10.3390/ijms23073894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Portfors C.V. Types and Functions of Ultrasonic Vocalizations in Laboratory Rats and Mice. J. Am. Assoc. Lab. Anim. Sci. 2007;46:28–34. [PubMed] [Google Scholar]
- 36.Barski J.J., Hartmann J., Rose C.R., Hoebeek F., Mörl K., Noll-Hussong M., De Zeeuw C.I., Konnerth A., Meyer M. Calbindin in Cerebellar Purkinje Cells Is a Critical Determinant of the Precision of Motor Coordination. J. Neurosci. 2003;23:3469–3477. doi: 10.1523/JNEUROSCI.23-08-03469.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scotti A.L. Calbindin D28k in the olivocerebellar projection. A light and electron microscope study. J. Anat. 1995;187:649–659. [PMC free article] [PubMed] [Google Scholar]
- 38.Arnold D.B., Heintz N. A calcium responsive element that regulates expression of two calcium binding proteins in Purkinje cells. Proc. Natl. Acad. Sci. 1997;94:8842–8847. doi: 10.1073/pnas.94.16.8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Celio M.R. Calbindin D-28k and parvalbumin in the rat nervous system. Neuroscience. 1990;35:375–475. doi: 10.1016/0306-4522(90)90091-h. [DOI] [PubMed] [Google Scholar]
- 40.Kondo T., Sheets P.L., Zopf D.A., Aloor H.L., Cummins T.R., Chan R.J., Hashino E. Tlx3 exerts context-dependent transcriptional regulation and promotes neuronal differentiation from embryonic stem cells. Proc. Natl. Acad. Sci. 2008;105:5780–5785. doi: 10.1073/pnas.0708704105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Indulekha C.L., Divya T.S., Divya M.S., Sanalkumar R., Rasheed V.A., Dhanesh S.B., Sebin A., George A., James J. Hes-1 regulates the excitatory fate of neural progenitors through modulation of Tlx3 (HOX11L2) expression. Cell. Mol. Life Sci. 2012;69:611–627. doi: 10.1007/s00018-011-0765-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leto K., Arancillo M., Becker E.B.E., Buffo A., Chiang C., Ding B., Dobyns W.B., Dusart I., Haldipur P., Hatten M.E., et al. Consensus Paper: Cerebellar Development. Cerebellum. 2016;15:789–828. doi: 10.1007/s12311-015-0724-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamada M., Seto Y., Taya S., Owa T., Inoue Y.U., Inoue T., Kawaguchi Y., Nabeshima Y.I., Hoshino M. Specification of Spatial Identities of Cerebellar Neuron Progenitors by Ptf1a and Atoh1 for Proper Production of GABAergic and Glutamatergic Neurons. J. Neurosci. 2014;34:4786–4800. doi: 10.1523/JNEUROSCI.2722-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Consalez G.G., Hawkes R. The compartmental restriction of cerebellar interneurons. Front. Neural Circuits. 2012;6 doi: 10.3389/fncir.2012.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El Nagar S., Chakroun A., Le Greneur C., Figarella-Branger D., Di Meglio T., Lamonerie T., Billon N. Otx2 promotes granule cell precursor proliferation and Shh-dependent medulloblastoma maintenance in vivo. Oncogenesis. 2018;7:60. doi: 10.1038/s41389-018-0070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chizhikov V.V., Lindgren A.G., Mishima Y., Roberts R.W., Aldinger K.A., Miesegaes G.R., Currle D.S., Monuki E.S., Millen K.J. Lmx1a regulates fates and location of cells originating from the cerebellar rhombic lip and telencephalic cortical hem. Proc. Natl. Acad. Sci. USA. 2010;107:10725–10730. doi: 10.1073/pnas.0910786107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mackie S., Shaw P., Lenroot R., Pierson R., Greenstein D.K. Cerebellar Development and Clinical Outcome in Attention Deficit Hyperactivity Disorder. Am. J. Psychiatry. 2007;164:647–655. doi: 10.1176/ajp.2007.164.4.647. [DOI] [PubMed] [Google Scholar]
- 48.Sathyanesan A., Zhou J., Scafidi J., Heck D.H., Sillitoe R.V., Gallo V. Emerging connections between cerebellar development, behaviour and complex brain disorders. Nat. Rev. Neurosci. 2019;20:298–313. doi: 10.1038/s41583-019-0152-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fatemi S.H., Aldinger K.A., Ashwood P., Bauman M.L., Blaha C.D., Blatt G.J., Chauhan A., Chauhan V., Dager S.R., Dickson P.E., et al. Consensus Paper: Pathological Role of the Cerebellum in Autism. Cerebellum. 2012;11:777–807. doi: 10.1007/s12311-012-0355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abrahams B.S., Geschwind D.H. Connecting Genes to Brain in the Autism Spectrum Disorders. Arch. Neurol. 2010;67:395–399. doi: 10.1001/archneurol.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lord C., Elsabbagh M., Baird G., Veenstra-Vanderweele J. Autism spectrum disorder. Lancet Lond. Engl. 2018;392:508–520. doi: 10.1016/S0140-6736(18)31129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khan N.Z., Gallo L.A., Arghir A., Budisteanu B., Budisteanu M., Dobrescu I., Donald K., El-Tabari S., Hoogenhout M., Kalambayi F., et al. Autism and the grand challenges in global mental health. Autism Res. 2012;5:156–159. doi: 10.1002/aur.1239. [DOI] [PubMed] [Google Scholar]
- 53.Gharani N., Benayed R., Mancuso V., Brzustowicz L.M., Millonig J.H. Association of the homeobox transcription factor, ENGRAILED 2, 3, with autism spectrum disorder. Mol. Psychiatry. 2004;9:474–484. doi: 10.1038/sj.mp.4001498. [DOI] [PubMed] [Google Scholar]
- 54.Gold D.A., Gent P.M., Hamilton B.A. RORα in genetic control of cerebellum development: 50 staggering years. Brain Res. 2007;1140:19–25. doi: 10.1016/j.brainres.2005.11.080. [DOI] [PubMed] [Google Scholar]
- 55.Sarachana T., Hu V.W. Genome-wide identification of transcriptional targets of RORA reveals direct regulation of multiple genes associated with autism spectrum disorder. Mol. Autism. 2013;4:14. doi: 10.1186/2040-2392-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sayad A., Noroozi R., Omrani M.D., Taheri M., Ghafouri-Fard S. Retinoic acid-related orphan receptor alpha (RORA) variants are associated with autism spectrum disorder. Metab. Brain Dis. 2017;32:1595–1601. doi: 10.1007/s11011-017-0049-6. [DOI] [PubMed] [Google Scholar]
- 57.Shu W., Cho J.Y., Jiang Y., Zhang M., Weisz D., Elder G.A., Schmeidler J., De Gasperi R., Sosa M.A.G., Rabidou D., et al. Altered ultrasonic vocalization in mice with a disruption in the Foxp2 gene. Proc. Natl. Acad. Sci. USA. 2005;102:9643–9648. doi: 10.1073/pnas.0503739102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fujita E., Tanabe Y., Shiota A., Ueda M., Suwa K., Momoi M.Y., Momoi T. Ultrasonic vocalization impairment of Foxp2 (R552H) knockin mice related to speech-language disorder and abnormality of Purkinje cells. Proc. Natl. Acad. Sci. USA. 2008;105:3117–3122. doi: 10.1073/pnas.0712298105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bowers J.M., Konopka G. The Role of the FOXP Family of Transcription Factors in ASD. Dis. Markers. 2012;33:251–260. doi: 10.1155/2012/456787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fatemi S.H., Snow A.V., Stary J.M., Araghi-Niknam M., Reutiman T.J., Lee S., Brooks A.I., Pearce D.A. Reelin signaling is impaired in autism. Biol. Psychiatry. 2005;57:777–787. doi: 10.1016/j.biopsych.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 61.Cupolillo D., Hoxha E., Faralli A., De Luca A., Rossi F., Tempia F., Carulli D. Autistic-Like Traits and Cerebellar Dysfunction in Purkinje Cell PTEN Knock-Out Mice. Neuropsychopharmacology. 2016;41:1457–1466. doi: 10.1038/npp.2015.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sadakata T., Kakegawa W., Mizoguchi A., Washida M., Katoh-Semba R., Shutoh F., Okamoto T., Nakashima H., Kimura K., Tanaka M., et al. Impaired Cerebellar Development and Function in Mice Lacking CAPS2, a Protein Involved in Neurotrophin Release. J. Neurosci. 2007;27:2472–2482. doi: 10.1523/JNEUROSCI.2279-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Homanics G.E., DeLorey T.M., Firestone L.L., Quinlan J.J., Handforth A., Harrison N.L., Krasowski M.D., Rick C.E., Korpi E.R., Mäkelä R., et al. Mice devoid of γ-aminobutyrate type A receptor β3 subunit have epilepsy, cleft palate, and hypersensitive behavior. Proc. Natl. Acad. Sci. USA. 1997;94:4143–4148. doi: 10.1073/pnas.94.8.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bernard O.A., Busson-LeConiat M., Ballerini P., Mauchauffé M., Della Valle V., Monni R., Nguyen Khac F., Mercher T., Penard-Lacronique V., Pasturaud P., et al. A new recurrent and specific cryptic translocation, t(5;14)(q35;q32), is associated with expression of the Hox11L2 gene in T acute lymphoblastic leukemia. Leukemia. 2001;15:1495–1504. doi: 10.1038/sj.leu.2402249. [DOI] [PubMed] [Google Scholar]
- 65.Van Vlierberghe P., Homminga I., Zuurbier L., Gladdines-Buijs J., Van Wering E.R., Horstmann M., Beverloo H.B., Pieters R., Meijerink J.P.P. Cooperative genetic defects in TLX3 rearranged pediatric T-ALL. Leukemia. 2008;22:762–770. doi: 10.1038/sj.leu.2405082. [DOI] [PubMed] [Google Scholar]
- 66.Sydnor L.M., Aldinger K.A. Structure, Function, and Genetics of the Cerebellum in Autism. J. Psychiatr. Brain Sci. 2022;7 doi: 10.20900/jpbs.20220008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang S.S.-H., Kloth A.D., Badura A. The Cerebellum, Sensitive Periods, and Autism. Neuron. 2014;83:518–532. doi: 10.1016/j.neuron.2014.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Courchesne E., Mouton P.R., Calhoun M.E., Semendeferi K., Ahrens-Barbeau C., Hallet M.J., Barnes C.C., Pierce K. Neuron Number and Size in Prefrontal Cortex of Children With Autism. JAMA. 2011;306:2001–2010. doi: 10.1001/jama.2011.1638. [DOI] [PubMed] [Google Scholar]
- 69.Stanfield A.C., McIntosh A.M., Spencer M.D., Philip R., Gaur S., Lawrie S.M. Towards a neuroanatomy of autism: A systematic review and meta-analysis of structural magnetic resonance imaging studies. Eur. Psychiatry. 2008;23:289–299. doi: 10.1016/j.eurpsy.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 70.Whitney E.R., Kemper T.L., Rosene D.L., Bauman M.L., Blatt G.J. Density of cerebellar basket and stellate cells in autism: Evidence for a late developmental loss of Purkinje cells. J. Neurosci. Res. 2009;87:2245–2254. doi: 10.1002/jnr.22056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lopes C., Liu Z., Xu Y., Ma Q. Tlx3 and Runx1 Act in Combination to Coordinate the Development of a Cohort of Nociceptors, Thermoceptors, and Pruriceptors. J. Neurosci. 2012;32:9706–9715. doi: 10.1523/JNEUROSCI.1109-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Matei V., Pauley S., Kaing S., Rowitch D., Beisel K.W., Morris K., Feng F., Jones K., Lee J., Fritzsch B. Smaller inner ear sensory epithelia in Neurog1 null mice are related to earlier hair cell terminal mitosis. Dev. Dyn. 2005;234:633–650. doi: 10.1002/dvdy.20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hao Y., Stuart T., Kowalski M.H., Choudhary S., Hoffman P., Hartman A., Srivastava A., Molla G., Madad S., Fernandez-Granda C., Satija R. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 2024;42:293–304. doi: 10.1038/s41587-023-01767-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Van De Geijn B., McVicker G., Gilad Y., Pritchard J.K. WASP: allele-specific software for robust molecular quantitative trait locus discovery. Nat. Methods. 2015;12:1061–1063. doi: 10.1038/nmeth.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:1–16. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR : a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Risso D., Ngai J., Speed T.P., Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 2014;32:896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kozareva V., Martin C., Osorno T., Rudolph S., Guo C., Vanderburg C., Nadaf N., Regev A., Regehr W.G., Macosko E. A transcriptomic atlas of mouse cerebellar cortex comprehensively defines cell types. Nature. 2021;598:214–219. doi: 10.1038/s41586-021-03220-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aartsma-Rus A., Van Putten M. Assessing Functional Performance in the Mdx Mouse Model. J. Vis. Exp. 2014;51303 doi: 10.3791/51303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Deacon R.M.J. Measuring Motor Coordination in Mice. J. Vis. Exp. 2013;2609 doi: 10.3791/2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Seibenhener M.L., Wooten M.C. Use of the Open Field Maze to Measure Locomotor and Anxiety-like Behavior in Mice. J. Vis. Exp. 2015;52434 doi: 10.3791/52434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Komada M., Takao K., Miyakawa T. Elevated Plus Maze for Mice. J. Vis. Exp. 2008;1088 doi: 10.3791/1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Leger M., Quiedeville A., Bouet V., Haelewyn B., Boulouard M., Schumann-Bard P., Freret T. Object recognition test in mice. Nat. Protoc. 2013;8:2531–2537. doi: 10.1038/nprot.2013.155. [DOI] [PubMed] [Google Scholar]
- 85.Vogel-Ciernia A., Wood M.A. Examining Object Location and Object Recognition Memory in Mice. Curr. Protoc. Neurosci. 2014;69:8–31. doi: 10.1002/0471142301.ns0831s69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Angoa-Pérez M., Kane M.J., Briggs D.I., Francescutti D.M., Kuhn D.M. Marble Burying and Nestlet Shredding as Tests of Repetitive, Compulsive-like Behaviors in Mice. J. Vis. Exp. 2013;50978 doi: 10.3791/50978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kaidanovich-Beilin O., Lipina T., Vukobradovic I., Roder J., Woodgett J.R. Assessment of Social Interaction Behaviors. J. Vis. Exp. 2011;2473 doi: 10.3791/2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rein B., Ma K., Yan Z. A standardized social preference protocol for measuring social deficits in mouse models of autism. Nat. Protoc. 2020;15:3464–3477. doi: 10.1038/s41596-020-0382-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Caruso A., Ricceri L., Scattoni M.L. Ultrasonic vocalizations as a fundamental tool for early and adult behavioral phenotyping of Autism Spectrum Disorder rodent models. Neurosci. Biobehav. Rev. 2020;116:31–43. doi: 10.1016/j.neubiorev.2020.06.011. [DOI] [PubMed] [Google Scholar]
- 90.Ferhat A.-T., Torquet N., Le Sourd A.-M., De Chaumont F., Olivo-Marin J.-C., Faure P., Bourgeron T., Ey E. Recording Mouse Ultrasonic Vocalizations to Evaluate Social Communication. J. Vis. Exp. 2016;53871 doi: 10.3791/53871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schmeisser M.J., Ey E., Wegener S., Bockmann J., Stempel A.V., Kuebler A., Janssen A.-L., Udvardi P.T., Shiban E., Spilker C., et al. Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature. 2012;486:256–260. doi: 10.1038/nature11015. [DOI] [PubMed] [Google Scholar]
- 92.Scattoni M.L., Ricceri L., Crawley J.N. Unusual repertoire of vocalizations in adult BTBR T+tf/J mice during three types of social encounters. Genes Brain Behav. 2011;10:44–56. doi: 10.1111/j.1601-183X.2010.00623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guyenet S.J., Furrer S.A., Damian V.M., Baughan T.D., La Spada A.R., Garden G.A. A Simple Composite Phenotype Scoring System for Evaluating Mouse Models of Cerebellar Ataxia. J. Vis. Exp. 2010;1787 doi: 10.3791/1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Miedel C.J., Patton J.M., Miedel A.N., Miedel E.S., Levenson J.M. Assessment of Spontaneous Alternation, Novel Object Recognition and Limb Clasping in Transgenic Mouse Models of Amyloid-β and Tau Neuropathology. J. Vis. Exp. 2017;123 doi: 10.3791/55523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Italic font is used to indicate the gene names (Excel format).
Italic font is used to indicate the gene names (Excel format).
Italic font is used to indicate the gene names (Excel format).
Italic font is used to indicate the gene names (Excel format).
Data Availability Statement
-
•
Data: RNA-seq and Single-cell RNA-seq data have been deposited at NCBI Sequence Read Archive (SRA) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
Code: All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOI is listed in the key resources table.
-
•
Additional information: Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. All additional softwares used for this study are listed in the key resources table.