Abstract
Background & Aims
Small noncoding vault RNAs (vtRNAs) are involved in many cell processes important for health and disease, but their pathobiological functions in the intestinal epithelium are underexplored. Here, we investigated the role of human vtRNA1-1 in regulating intestinal epithelial renewal and barrier function.
Methods
Studies were conducted in vtRNA1-1 transgenic (vtRNA1-1Tg) mice, primary enterocytes, and Caco-2 cells. Extracellular vesicles (EVs) were isolated from the serum of shock patients and septic mice. Intestinal organoids (enteroids) were prepared from vtRNA1-1Tg and littermate mice. Mucosal growth was measured by Ki67 immunostaining or BrdU incorporation, and gut permeability was assessed using the FITC-dextran assay.
Results
Intestinal tissues recovered from shock patients and septic mice evidenced mucosal injury and gut barrier dysfunction; vtRNA levels were elevated in EVs isolated from their sera. In mice, intestinal epithelial-specific transgenic expression of vtRNA1-1 inhibited mucosal growth, reduced Paneth cell numbers and intercellular junction (IJ) protein expression, and increased gut barrier vulnerability to lipopolysaccharide exposure. Conversely, in vitro silencing of vtRNA1-1 increased IJ protein levels and enhanced epithelial barrier function. Exposing enteroids to vtRNA1-1-rich EVs augmented paracellular permeability. Mechanistically, vtRNA1-1 interacted with CUG-binding protein 1 (CUGBP1) and increased CUGBP1 association with claudin-1 and occludin mRNAs, thereby inhibiting their expression.
Conclusions
These findings indicate that elevated levels of vtRNA1-1 in EVs and mucosal tissues repress intestinal epithelial renewal and barrier function. Notably, this work reveals a novel role for dysregulation of the vtRNA1-1/CUGBP1 axis in the pathogenesis of gut mucosal disruption in critical illness.
Keywords: Gut Permeability, Mucosal Growth, Paneth Cells, RNA-binding proteins, Small Noncoding RNAs
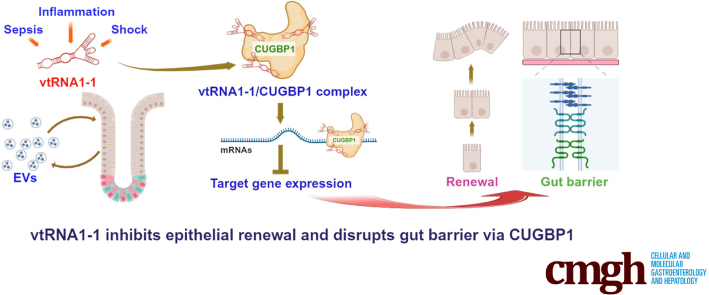
Graphical abstract
Summary.
We investigated the role of noncoding vault RNAs in intestinal epithelium homeostasis. Our results demonstrate that increased levels of vtRNA1-1, either in extracellular vesicles of serum or mucosal tissues, disrupts epithelial renewal and barrier function via interaction with CUG-binding protein 1.
The mammalian intestinal epithelium is a specialized domain that interacts with and protects subepithelial tissue against luminal noxious substances and the gut microbiome.1,2 The maintenance of intestinal epithelium homeostasis is a dynamic process that requires epithelial cells to alter gene expression patterns quickly to modulate apoptosis, proliferation, migration, differentiation, and cell-to-cell interactions.3,4 Disrupted intestinal epithelial integrity is common in critical disorders (eg, shock, sepsis, major trauma, thermal injury, and extensive surgery), resulting in epithelial barrier dysfunction.5 The resulting ‘leaky’ gut permits the translocation of luminal toxic substances and bacteria and their metabolites into the bloodstream and, in some instances, leads to multiple organ dysfunction syndrome and death.6,7 Effective therapies to preserve intestinal epithelial integrity and protect gut barrier function are limited, primarily because of incomplete understanding of the mechanisms underlying mucosal disruption in critical illness.
Emerging evidence indicates that control of noncoding RNAs (ncRNAs) and RNA-binding proteins (RBPs) plays an important role in maintaining intestinal epithelial homeostasis in stressful environments, whereas dysregulation of specific ncRNAs and RBPs impairs mucosal renewal, compromises epithelial host defenses, and thereby disrupts intestinal barrier function.4,8, 9, 10 However, little is known regarding how dysregulated ncRNAs and RBPs in intestinal epithelial cells (IECs) in the critically ill impact the function of neighboring and remote tissues. Intercellular communication directed by the secretion of extracellular vesicles (EVs) is gaining increasing attention.11,12 EVs released from many cell types, including IECs, can transfer a variety of bioactive molecules to neighboring or distant tissues, thereby playing previously unrecognized functional roles.13,14 The involvement of IEC-derived EVs and their cargo ncRNAs in regulating intestinal epithelial integrity is particularly important in critically ill surgical patients because they often exhibit systemic gut barrier dysfunction rather than only localized changes in paracellular permeability.6,8
Vault RNAs (vtRNAs), small (∼100 nucleotide) ncRNAs transcribed by RNA polymerase III, are highly conserved across mammalian genomes. Humans produce 4 vtRNA paralogs, vtRNA1-1, vtRNA1-2, vtRNA1-3, and vtRNA2-1, whereas mice express only 1 vtRNA.15 vtRNAs are associated with giant cytoplasmic ribonucleoprotein (RNP) particles termed vaults and commonly identified in EVs.12,16 Moreover, vtRNAs play important roles outside of vault RNPs and can function independently of vault particles.15,17 vtRNAs are implicated in many cellular processes such as mRNA splicing, nuclear transport, drug resistance, synaptogenesis, lysosome function, apoptosis, influenza virus replication, and tumorigenesis,15,18, 19, 20, 21, 22, 23 although only about 5% of total cellular vtRNA is incorporated into vaults.18 The 4 human vtRNAs differ only slightly in their primary and second structures, but they have distinct biological functions via different targets and mechanisms.15,17 For example, vtRNA1-1 selectively regulates autophagy by interacting directly with RBP p62,17 whereas vtRNA2-1 acts as a tumor suppressor by associating with protein kinase R in a wide range of cancer cells.23,24 Expression of vtRNA1-1 is often upregulated in cancer cells due to dysregulation of oncogenic signals including PI3K/Akt and MAPK pathways and plays an important role in cancer progression by altering apoptosis, autophagy, and sensitivity to chemotherapy.20 The levels of vtRNA1-1 also increase after infection with the influenza A virus, which enhances viral replication by suppressing PKR-mediated innate immunity.25 Although vtRNA2-1 is involved in regulating gut barrier function via interaction with HuR,26 the exact role of vtRNA1-1 and specifically its presence in EVs remains unknown in the context of intestinal epithelial homeostasis and dysfunction.
In this study, we provide evidence that vtRNA1-1 modulates intestinal epithelial homeostasis by interacting with CUG-binding protein 1 (CUGBP1). We found that EVs isolated from the serum of hemorrhagic shock patients exhibited increased vtRNAs, including vtRNA1-1, that were associated with disrupted gut epithelial integrity. Transgenic expression of human vtRNA1-1 in mice inhibited small intestinal mucosal growth and increased gut barrier vulnerability to stress. Our results further show that exposing intestinal organoids (enteroids) to vtRNA1-1-rich EVs increased paracellular permeability ex vivo. Moreover, vtRNA1-1 interacted with and enhanced CUGBP1 binding activity, thus inhibiting the expression of tight junction (TJ) proteins such as claudin-1 and occludin. These findings reveal that vtRNA1-1 functions as a repressor of intestinal epithelial renewal and barrier function and identify the vtRNA1-1/CUGBP1 axis as a novel therapeutic target for interventions to promote intestinal mucosal regeneration and enhance gut barrier function in the critically ill.
Results
vtRNAs Are Enriched in EVs From Shock Patients and Septic Mice
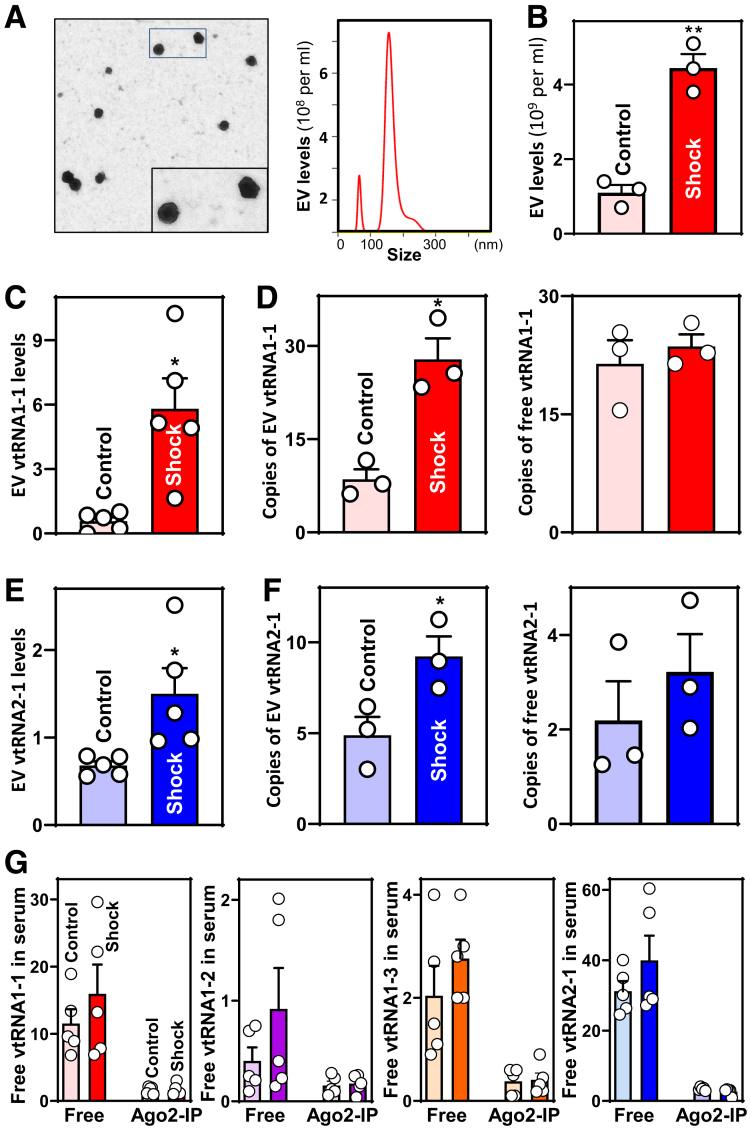
To explore the involvement of EV vtRNAs in mediating the disrupted integrity of the intestinal epithelium in the critically ill, blood samples were collected from healthy subjects (controls) and patients with hemorrhagic shock (systolic blood pressure < 90 mmHg), a condition associated with intestinal mucosal injury and gut barrier dysfunction. EVs were isolated using protocols described previously27,28 and collected as pellets resuspended in phosphate-buffered saline (PBS). The presence of EVs from human serum was confirmed by electron microscopy and NanoSight particle (NSP) analysis that revealed 50- to 200-nm diameter particles (Figure 1A). Interestingly, compared with control EVs, those isolated from the serum of shock patients were both more concentrated (Figure 1B) and contained more vtRNA1-1 and vtRNA2-1 (Figure 1C, E). In contrast, the levels of EV vtRNA1-2 and vtRNA1-3 were low in EVs and exhibited no significant differences between the 2 groups. Droplet digital real-time PCR (ddPCR) analysis showed that copy numbers of EV vtRNA1-1 and vtRNA2-1 increased dramatically in subjects with hemorrhagic shock relative to controls (Figure 1D and F, left panels). In this experiment, for quantification, vtRNA1-1 and vtRNA2-1 levels were measured in at least 10,000 droplets/well of EV sample. We also examined changes in the levels of several microRNAs including (miR)-29b, miR-195, and miR-222 and long ncRNAs (lncRNAs) such as uc.173, uc.230, H19, and SPRY4-IT1 in EVs isolated from serum of shock patients and controls, because these ncRNAs play an important role in the intestinal epithelium homeostasis and gut barrier function.4,29, 30, 31, 32, 33, 34, 35 Our results revealed no meaningful differences in the levels of EV miR-29b, miR-195, and miR-222 between shock and control subjects, and in EVs from both groups the levels of lncRNAs uc.173, uc.230, H19, and SPRY4-IT1 were below the limits of detection.
Figure 1.
Increased levels of EV vtRNAs in the serum of patients with hemorrhagic shock. (A) Characterization of EVs isolated from human serum. Left panel, EVs under electron microscope; right panel, EVs as examined by NSP analysis. Serum samples from 3 patients were examined and showed similar results. (B) EV concentrations in the serum from shock patients and control subjects. Values are the means ± SED (n = 3). ∗∗P < .01 compared with controls. (C, D) Levels of EV vtRNA1-1 and its copy numbers in serum described in B, as measured by Q-PCR and ddPCR analyses. ∗P < .05 compared with controls (n = 3 or 5). (E, F) Levels of EV vtRNA2-1 and its copy numbers in human serum described in B. ∗P < .05 compared with controls (n = 3 or 5). (G) Levels of free ‘floating’ vtRNAs in the serum from shock patients and control individuals. The levels of non-EV-containing vtRNAs bound to Ago2 were measured by IP/Q-PCR analysis using anti-Ago2 antibody. Values were means ± SED (n = 5).
We examined and compared ‘free floating’ (non-EV-containing) vtRNAs in serum from shock patients and control subjects and found no significant differences in the levels of any of the 4 human vtRNAs (Figure 1D and F, right panels; G). Non-EV-containing vtRNAs bound to Ago2 were also detectable in the serum, but the levels of these Ago2-bound vtRNAs in shock patients were indistinguishable from levels in control samples.
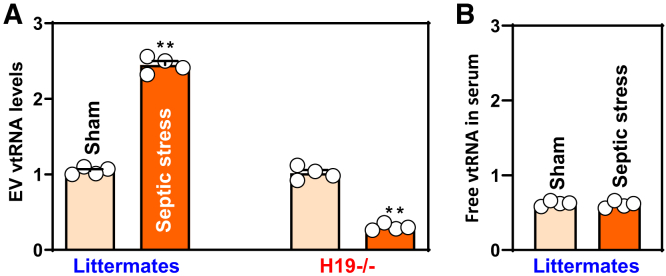
Consistent with observations in humans, mice exposed to septic stress induced by cecal ligation and puncture (CLP) for 24 h also exhibited increased serum levels of EV vtRNA (Figure 2A, left), associated with increased gut permeability; the latter had been reported previously.36 CLP did not alter the abundance of free vtRNA in serum (Figure 2B) but did increase the levels of small intestinal mucosal vtRNA, as in our previous report.26 Because targeted deletion of the H19 gene in mice protects the gut barrier against CLP-induced injury,29 we examined changes in EV vtRNA in H19-deficient mice and found significantly reduced levels of EV vtRNA after CLP compared with littermate controls (Figure 2A, right). These findings strongly implicate vtRNA-containing EVs, but not ‘free floating’ serum vtRNAs, in gut mucosal pathology in critical surgical disorders.
Figure 2.
Effect of septic stress on the levels of EV vtRNA in mice. (A) Levels of EV vtRNA in littermate and H19-deficient (H19-/-) mice after exposure to septic stress induced by CLP. Serum was harvested 24 hours after CLP, and EVs were isolated. Values are the means ± SED (n = 4). ∗∗P < .01 compared with sham. (B) Levels of free ‘floating’ vtRNA in the serum of littermate mice described in A. In all experiments, statistical significance was analyzed using unpaired, 2-tailed Student’s t-test.
vtRNA1-1 Inhibits Epithelial Renewal in Small Intestine and Compromises Gut Barrier Function
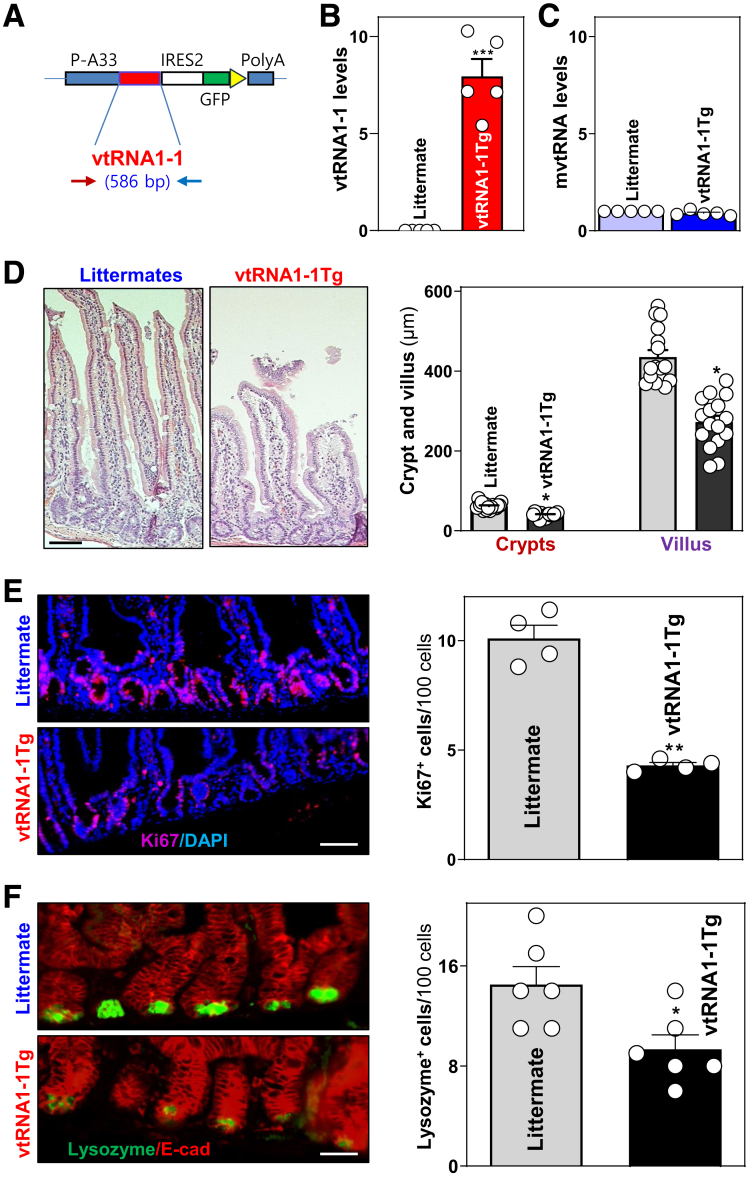
Although vtRNA2-1 down-regulates gut epithelial barrier function,26 the exact role of vtRNA1-1 in intestinal epithelial homeostasis remains unknown. To examine the in vivo function of vtRNA1-1 in the intestinal epithelium, we utilized a gain-of-function transgenic approach and generated mice that highly express human vtRNA1-1 (vtRNA1-1Tg). The regulatory elements selected to drive vtRNA1-1 expression were derived from the A33 antigen gene because of its temporally and spatially restricted expression pattern.34 As described previously,37,38 we used the A33 promoter to drive intestinal epithelial tissue-specific expression of vtRNA1-1. The A33-vtRNA1-1 transgene construct (Figure 3A) was microinjected into the male pronucleus of fertilized oocytes from mice on a pure C57BL/6 genetic background. A total of 6 transgenic founders was generated; however, only 2 (mouse #3 and #5) transmitted the transgene to their progeny. Male and female, 3- to 4-month-old vtRNA1-1Tg and littermate control mice were used for phenotype comparisons. As measured by quantitative real-time polymerase chain reaction (Q-PCR) analysis, vtRNA1-1Tg mice exhibited specific vtRNA1-1 overexpression in intestinal mucosal tissues (Figure 3B), but not in tissues from heart, lung, liver, kidney, and spleen. Transgenic expression of human vtRNA1-1 in mice did not alter the levels of endogenous mouse vtRNA (mvtRNA) in the small intestinal mucosa (Figure 3C). vtRNA1-1Tg mice appeared grossly normal, and there were no significant differences in gastrointestinal gross morphology between vtRNA1-1Tg mice and littermates, although vtRNA1-1Tg mouse body weight was slightly less than that of control mice at 8 weeks after birth.
Figure 3.
Transgenic expression of the vtRNA1-1 inhibits mucosal renewal and impairs Paneth cell function in the small intestine of mice. (A) The A33/vtRNA1-1/GFP construct used to be injected into fertilized oocytes for generating vtRNA1-1Tg mice. (B) Levels of mucosal vtRNA1-1 in littermate control and vtRNA1-1Tg mice as measured Q-PCR analysis. Values are the means ± SEM (n = 5). ∗∗∗P < .001 compared with littermate controls. (C) Levels of mouse vtRNA (mvtRNA) in the small intestinal mucosa described in B. (D) Photomicrographs of hematoxylin and eosin (left) and changes in the length of villi and crypts (right) of the mucosa described in B. Scale bars: 25 μm. ∗P < .05 compared with littermates. (E) Proliferating cells in small intestinal crypts as measured by Ki67 immunostaining (left). Red, Ki67; blue, DAPI. Right panel, quantitative data of Ki67-positive cells. ∗∗P < .01 compared with littermates. (F) Changes in Paneth cells (lysozyme-positive cells) in the mucosa described in B. ∗P < .05 compared with littermates. Statistical significance was analyzed using unpaired, 2-tailed Student’s t-test.
Importantly, vtRNA1-1Tg mice (the line generated from founder #3) displayed substantial inhibition of small intestinal mucosal growth, marked by histological features and the proliferating crypt cell population. As shown in Figure 3D, crypts and villi in the small intestinal mucosa of vtRNA1-1Tg mice were substantially diminished relative to those from control littermates. Quantification of these reductions indicated that the lengths of crypts and villi in the mucosa of vtRNA1-1Tg mice were decreased by ∼25% and ∼35%, respectively. Consistently, the numbers of Ki67-positive cells decreased by ∼58% in vtRNA1-1Tg mice compared with those observed in control littermates (Figure 3E). Increasing vtRNA1-1 levels in the intestinal epithelium also impacted Paneth cells (Figure 3D). Staining of whole mounts of the small intestine demonstrated that in control littermate mice, lysozyme-positive Paneth cells were located at crypt bases; compared with littermate controls, Paneth cell numbers were reduced by ∼36% in the small intestinal mucosa of vtRNA1-1Tg mice. However, increased vtRNA1-1 levels failed to alter small intestinal enterocyte differentiation as determined by villin immunostaining analysis and did not alter growth of colonic mucosa.
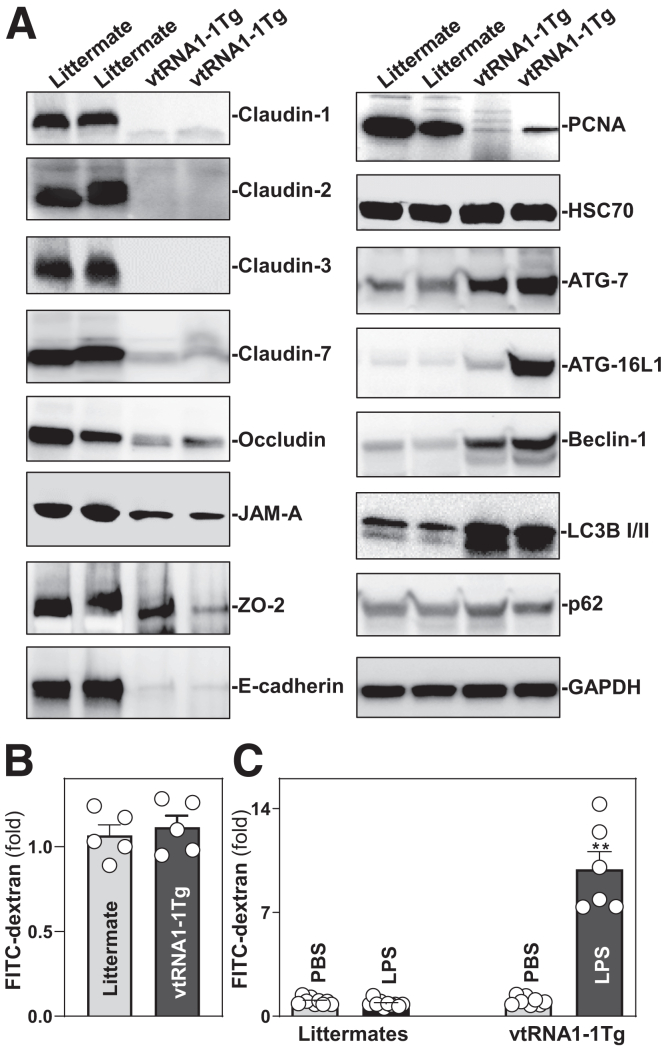
Intestinal epithelium-specific transgenic expression of vtRNA1-1 in mice reduced the expression of small intestinal mucosal TJs and adherens junction (AJ) proteins. As shown in Figure 4A (left), the small intestinal mucosa of vtRNA1-1Tg mice exhibited decreased levels of several TJ proteins, including claudin-1, claudin-2, claudin-3, claudin-7, occludin, JAM-A, and ZO-2, and AJ protein E-cadherin. Locally increasing the levels of vtRNA1-1 in the intestinal epithelium also decreased the levels of the proliferation-associated protein PCNA (Figure 4A, right-top). Because autophagy is crucial for host intestinal epithelial defense against invasive pathogens and to maintain gut barrier function,39,40 we examined changes in the levels of autophagy-related proteins in the intestinal epithelium of vtRNA1-1Tg mice. Unexpectedly, transgenic expression of vtRNA1-1 in the intestinal epithelium increased the abundances of mucosal ATG-7, ATG-16L1, LC3B I/II, and beclin-1 proteins (Figure 4A, right). Although the exact roles of altered levels of mucosal autophagy-associated proteins in vtRNA1-1Tg mice remain unknown, this upregulation of autophagy likely represents a compensatory mechanism to maintain the intestinal epithelial barrier function following reduction in the levels of TJ/AJ proteins. In addition, transgenic expression of vtRNA1-1 failed to alter the content of HSC70 and p62 proteins in the intestinal mucosa. We examined another line of vtRNA1-1Tg mice generated from an independent founder (founder #5) and showed similar decreases in intestinal epithelial renewal and the levels of TJ and AJ proteins.
Figure 4.
Transgenic expression of vtRNA1-1 decreases the levels of intercellular junction proteins and increases vulnerability of the gut barrier to LPS. (A) Changes in the levels of tight junctions claudin 1, occludin, ZO-2, and JAM-A, adherens junction E-cadherin, and autophagy-associated proteins in the small intestinal mucosa of littermate and vtRNA1-1Tg mice. GAPDH immunoblotting was performed as an internal control for equal loading. (B) Gut permeability in the mice described in A. FITC dextran was given orally, and blood samples were collected 4 hours thereafter for measurement. Values are the means ± SEM (n = 5). (C) Increased vulnerability of the gut barrier in response to LPS. Mice were intraperitoneally injected with LPS (0.1 mg/kg) in 200 μl PBS once daily for 5 days. Gut permeability was measured 24 hours after treatment with the last dose of LPS. Values are the means ± SEM (n = 6 or 8). ∗∗P < .01 compared with PBS or littermate controls exposed to LPS. In B and C, statistical significance was analyzed using unpaired, 2-tailed Student’s t-test except for results in A. All experiments in A were repeated 3 times with similar results.
Transgenic expression of vtRNA1-1 in mice did not alter baseline gut permeability as measured by fluorescein isothiocyanate (FITC)-conjugated dextran assays (Figure 4B), but it increased vulnerability of the gut barrier in response to the bacterial product lipopolysaccharide (LPS). There was a significant increase in gut permeability in vtRNA1-1Tg mice compared with littermates after exposure to the same doses of LPS (Figure 4C). Treatment of littermate mice with a low dose of LPS (0.1 mg/kg) for 5 days failed to alter gut permeability but induced remarkable gut barrier dysfunction in vtRNA1-1Tg mice. In addition, transgenic vtRNA1-1 expression did not induce apoptosis in the intestinal epithelium, as no significant increase in cell death was observed in vtRNA1-1Tg mice. As measured by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining, rates of apoptotic cell death were <2% in both littermate control and vtRNA1-1Tg mice.
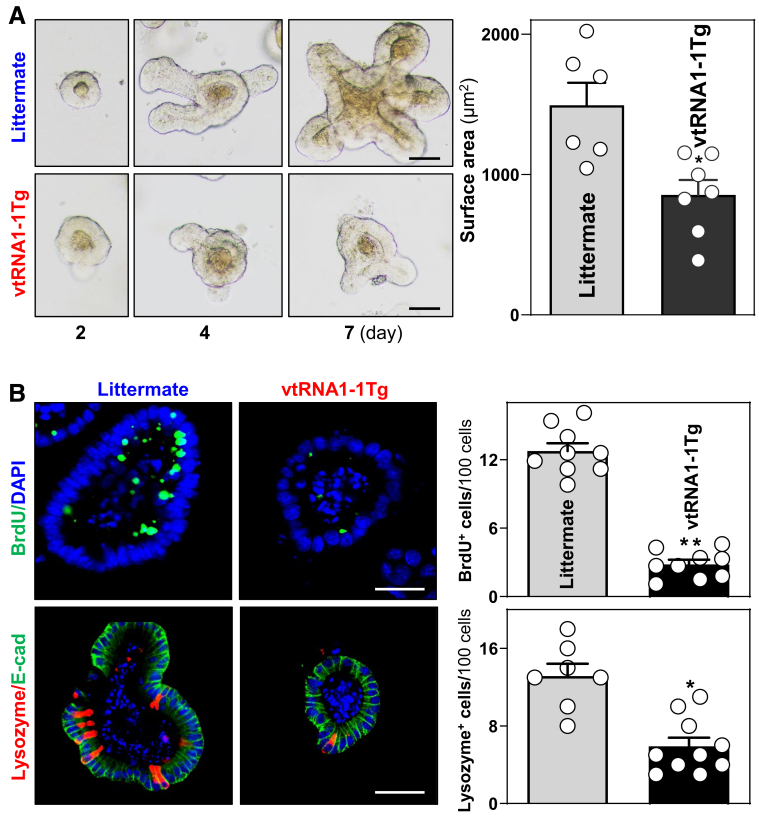
As another approach to gauge the impact of transgenic vtRNA1-1 expression on gut epithelial homeostasis, we examined the growth of enteroids generated from vtRNA1-1Tg and littermate control mice. Using methods described previously,41 enteroids were derived from proliferating crypts of the mucosa of the mouse small intestine. Enteroids derived from both littermate control and vtRNA1-1Tg mice consisted of multiple buds and cells on day 4 after initial culture. However, enteroids derived from vtRNA1-1Tg mice grew much slower than those from littermate mice, as evidenced by a significant decrease in enteroid surface area (Figure 5A) and BrdU incorporation (Figure 5B, top) in enteroids derived from vtRNA1-1Tg mice compared with those generated from control littermates. In agreement with this observation, enteroids derived from vtRNA1-1Tg mice also exhibited reduced numbers of Paneth cells (Figure 5B, bottom). Because enteroids lack stromal, immune, and neural cells, these results indicate that the disrupted renewal of intestinal epithelial cells in vtRNA1-1Tg mice results primarily from an epithelial cell-autonomous regulatory effect of this vtRNA and not from the effects of secreted factors from other cell types in the gut. Together, the results from these in vivo and ex vivo experiments strongly support the concept that vtRNA1-1 is a negative regulator of intestinal epithelial renewal and gut barrier function.
Figure 5.
Increased vtRNA1-1 inhibits the growth of enteroids and disrupts Paneth cell function ex vivo. (A) Growth of enteroids derived from the proximal small intestine of littermate control and vtRNA1-1Tg mice. Images were taken at different times after culture. Scale bars: 50 μM. Values are the means ± SEM (n = 6 or 7). ∗P < .05 compared with littermate controls. (B) Proliferating cells (top) and Paneth cells (bottom) in enteroids on day 3 after culture, as measured by BrdU labeling and immunostaining of lysozyme as marker of Paneth cells. ∗P <0.05; ∗∗P < .01 compared with littermate controls. In all experiments, statistical significance was analyzed using unpaired, 2-tailed Student’s t-test.
vtRNA1-1 Decreases Intercellular Junctions and Impairs Epithelial Barrier Function In Vitro
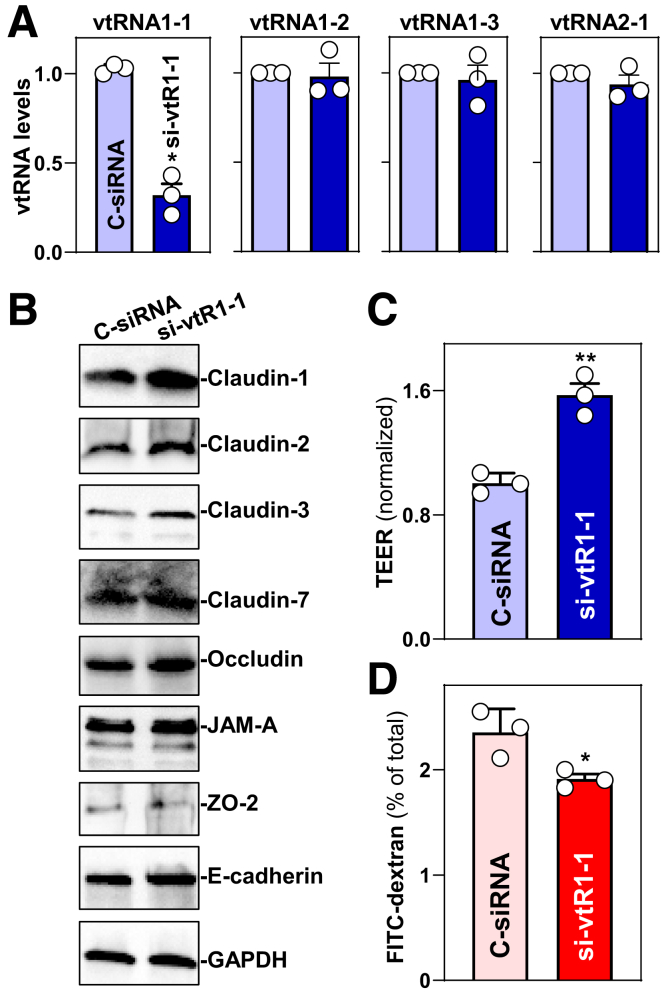
To further define the role of vtRNA1-1 in the regulation of gut barrier function, 2 sets of experiments were performed in Caco-2 cells. First, we examined the effect of vtRNA1-1 silencing on TJ/AJ protein expression and epithelial barrier function. The levels of cellular vtRNA1-1 decreased specifically 48 hours after transfection with small interfering RNA (siRNA) targeting vtRNA1-1 (si-vtR1-1); there were no significant differences in the levels of vtRNA1-2, vtRNA1-3, and vtRNA2-1 between cells transfected with si-vt1-1 or control siRNA (Figure 6A). This decrease in vtRNA1-1 levels by si-vtR1-1 transfection increased the abundances of claudin-1, claudin-2, claudin-3, claudin-7, and occludin, although it failed to alter the levels of JAM-A, ZO-2, and E-cadherin (Figure 6B). Importantly, vtRNA1-1 silencing enhanced epithelial barrier function, as revealed by an increase in transepithelial electrical resistance (TEER) (Figure 6C) and a decrease in the levels of paracellular flux of FITC-dextran (Figure 6D). Transfection with sivtR1-1 did not alter cell viability.
Figure 6.
vtRNA1-1 silencing improves intestinal epithelial barrier function in vitro. (A) Levels of vtRNAs in Caco-2 cells 48 hours after transfection with si-vtR1-1. Values are the means ± SEM (n = 3). ∗P < .05 compared with C-siRNA. (B) Immunoblots of claudins, occludin, ZO-2, JAM-A, and E-cadherin in cells described in A. GAPDH immunoblotting was performed as an internal control for equal loading. Three separate experiments were performed and showed similar results. (C, D) Changes in TEER and FITC-dextran paracellular permeability in cells described in A. TEER assays were performed on 12-mm Transwell filters; paracellular permeability was assayed by adding the membrane-impermeable trace molecule FITC-dextran to the insert medium. ∗P < .05; ∗∗P < .01 compared with C-siRNA. In A, C, and D, statistical significance was analyzed using unpaired, 2-tailed Student’s t-tests.
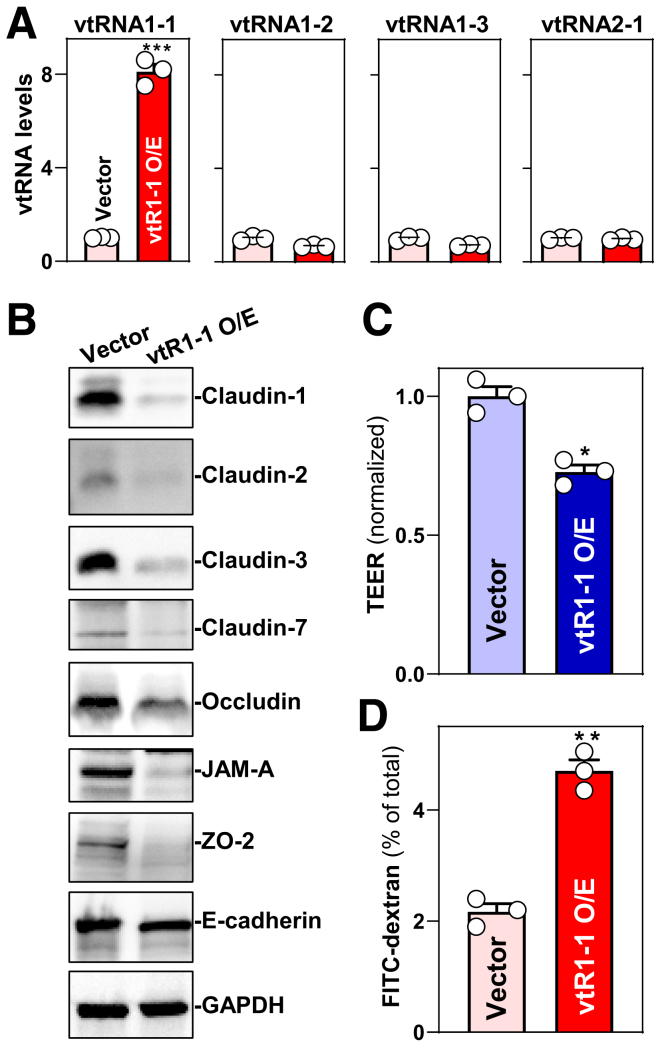
Second, we examined the influence of increased vtRNA1-1 levels on the expression of TJ and AJ proteins. Transfection of a plasmid expressing vtRNA1-1 under control of the pCMV promoter (pcDNA3.1 backbone) increased the levels of cellular vtRNA1-1 but did not alter the abundances of vtRNA1-2, vtRNA1-3, and vtRNA2-1 (Figure 7A). Ectopically expressed vtRNA1-1 decreased the levels of TJs claudin-1, claudin-2, claudin-3, claudin-7, occludin, JAM-A, and ZO-2 (Figure 7B) but did not alter E-cadherin expression levels. Moreover, elevating vtRNA1-1 levels disrupted epithelial barrier function because it decreased TEER (Figure 7C) and increased paracellular flux of FITC-dextran (Figure 7D). These results indicate that vtRNA1-1 impairs intestinal epithelial barrier function by lowering the levels of several TJ proteins.
Figure 7.
Ectopically expressed vtRNA1-1 disrupts intestinal epithelial barrier function in vitro. (A) Levels of vtRNAs in Caco-2 cells 48 h after transfection with the vtRNA1-1 expression vector (vtR1-1). Values are the means ± SEM (n = 3). ∗∗∗P < .001 compared with control vector. (B) Immunoblots of claudins, occludin, ZO-2, JAM-A, and E-cadherin in cells described in A. GAPDH immunoblotting was performed as an internal control for equal loading. Three separate experiments were performed and showed similar results. (C, D) Changes in TEER and FITC-dextran paracellular permeability in cells described in A. ∗P < .05; ∗∗P < .01 compared with control vector. In A, C, and D, statistical significance was analyzed using unpaired, 2-tailed Student’s t-tests.
vtRNA-rich EVs Increase Enteroid Paracellular Permeability
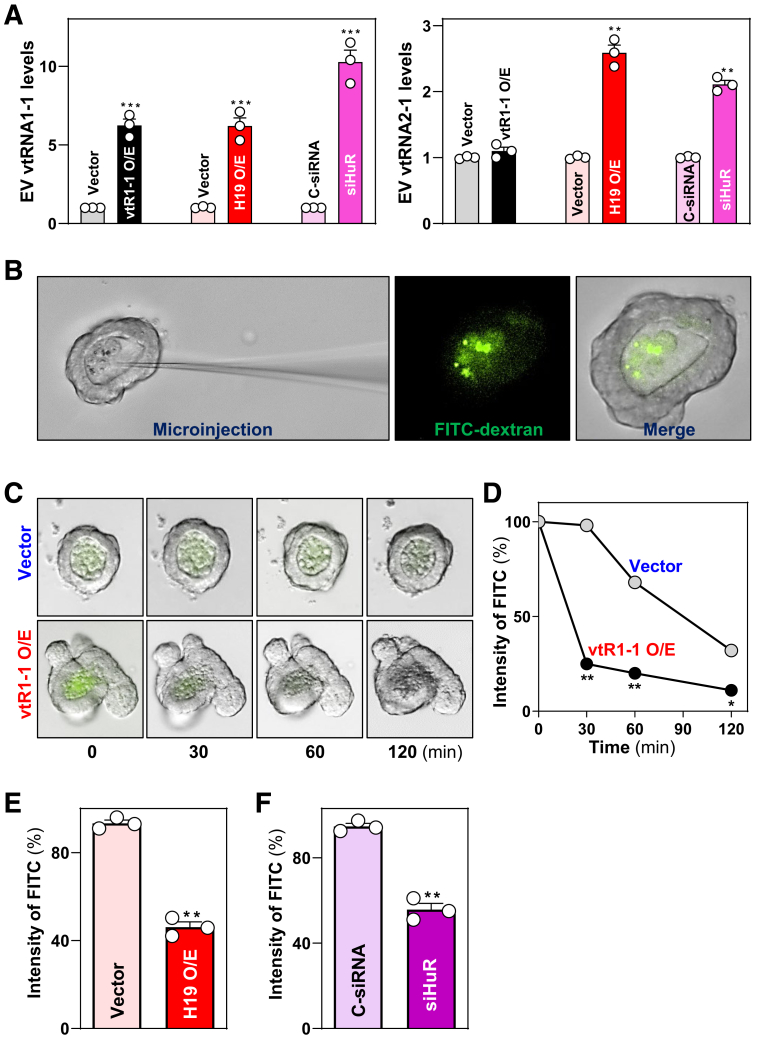
To elucidate the role of EV vtRNA1-1 in regulating intestinal epithelial barrier function, we microinjected FITC-dextran into 3-dimensional enteroids as described previously.42,43 To harvest vtRNA-rich EVs from IECs, Caco-2 cells were transfected with the vector expressing vtRNA1-1 or H19 or with siRNA specifically targeting HuR (siHuR). Ectopically overexpressed H19 and HuR silencing were also chosen to generate vtRNA-rich EVs from Caco-2 cells, because H19 knockout decreased EV vtRNA in mice exposed to CLP (Figure 2A), and because targeted deletion of HuR from the intestinal epithelium inhibits autophagy that modulates EV release.2,10 The levels of vtRNA1-1 increased dramatically in EVs isolated from conditional media incubated with cells overexpressing vtRNA1-1 or H19 and with HuR-silenced cells for 48 h (Figure 8A, left). The levels of vtRNA2-1 increased only in EVs from the conditional media incubated with cells transfected with H19 expression vector or siHuR but not from cells overexpressing vtRNA1-1 (Figure 8A, right). The levels of vtRNA1-2 and vtRNA1-3 were low or undetectable in EVs isolated from all 3 conditional media incubated with cells overexpressing vtRNA1-1 or H19 and with HuR-silenced cells. On the other hand, EVs isolated from the medium incubated with cells transfected with control vector or control siRNA (C-siRNA) did not alter the levels of EV vtRNA1-1 and served as controls.
Figure 8.
vtRAN1-1-rich EVs increase epithelial paracellular permeability in primarily cultured enteroids. (A) Levels of vtRNA1-1 (left) and vtRNA2-1 (right) in EVs isolated from the conditional media. Caco-2 cells were transfected with the vectors expressing vtRNA1-1 or lncRNA H19, or with siHuR. Conditional media were collected 48 hours after transfection, and EVs were isolated. Values are the means ± SEM (n = 3). ∗∗P < .01; ∗∗∗P < .001 compared with control vectors or C-siRNA. (B) Pictures of microinjection (left) and luminal FITC-dextran (right) in enteroids derived from the proximal small intestine of wild-type mice. (C) Images of enteroids 24 hours after incubation with EVs generated by cells overexpressing vtRNA1-1 (vtR1-1 O/E) or EVs from control group (Vector). Pictures were taken at different times after microinjection with FITC-dextran. (D) Changes in paracellular permeability in enteroids described in C. Values are the means ± SEM (n = 3). ∗P < .05; ∗∗P < .01 compared with controls. (E, F) Changes in epithelial permeability in enteroids incubated with EVs generated from cells overexpressing H19 (H19 O/E) or siHuR-transfected cells. Paracellular permeability was examined 30 minutes after microinjection with FITC-dextran as described in B. ∗∗P < .01 compared with vector or C-siRNA. In A, E, and F, statistical significance was analyzed using unpaired, 2-tailed Student’s t-test. In D, statistical comparison between time-course curves was by 1-way ANOVA with Bonferroni’s hoc test.
To test the function of EV vtRNAs ex vivo, enteroids were seeded in a basement matrix, embedded in standard growth medium for 2 days, and then cultured in medium containing EVs at concentrations of 50 ng/μl protein, equivalent to the constant EV number 1 × 107 as measured by NSP analysis. Twenty-four hours after administering EVs, FITC-dextran in 2 μl PBS was microinjected into the enteroid lumen (Figure 8B). Paracellular permeability was examined at different times after microinjection. As shown, treatment of enteroids with EVs from vtRNA1-1 overexpressed cells increased paracellular permeability significantly, as evidenced by rapidly decreasing luminal FITC intensity in enteroids exposed to vtRNA1-1-rich EVs, compared with EVs from control cells (transfected with control vector) (Figure 8C, D). Exposure to EVs from cells overexpressing H19 or HuR-silenced cells also increased paracellular permeability in enteroids, which exhibited a similar pattern in the decrease of luminal FITC intensity after microinjection of FITC-dextran (Figure 8E, F). These results suggest that EVs transfer vtRNAs generated from IECs, thus contributing to the increased levels of intracellular vtRNA1-1 in neighboring and distant intestinal epithelium and subsequent systemic gut barrier dysfunction in shock patients.
vtRNA1-1 Regulates TJ Expression and Barrier Function Via Interaction With CUGBP1
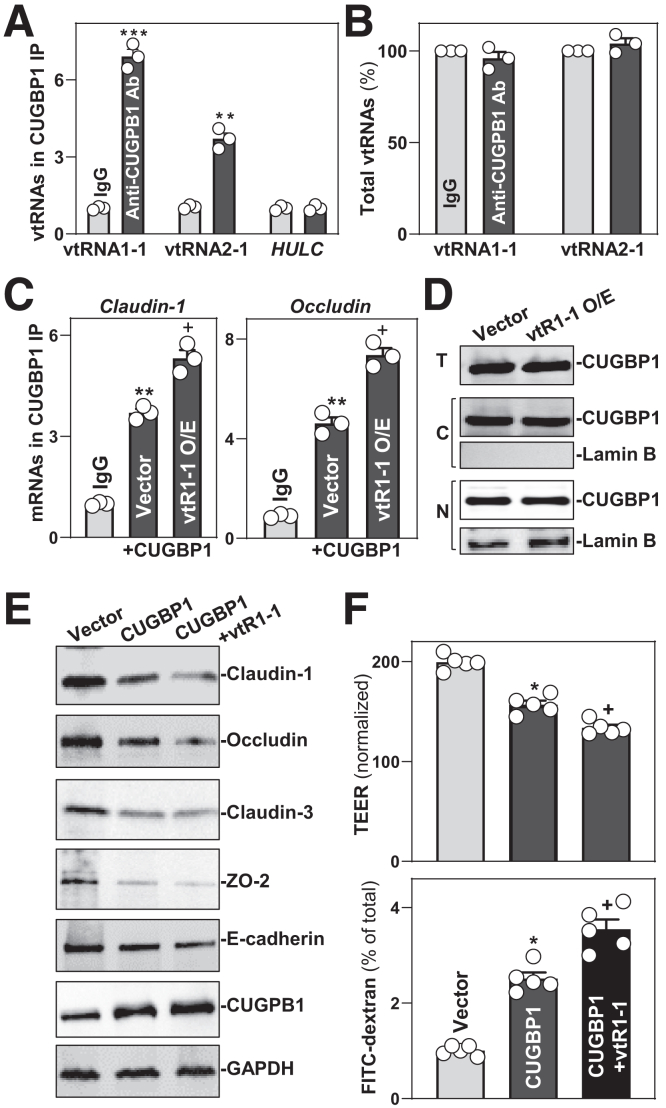
To define the mechanism underlying the actions of vtRNA1-1 in regulating intestinal epithelial barrier function, we tested the hypothesis that vtRNA1-1 represses TJ expression by interacting with the RBP CUGBP1. It has been reported that CUGBP1 regulates expression of TJ proteins such as claudins and occludin by binding directly to the 3′-untranslated regions of their mRNAs.4,31,36 First, we examined the association of endogenous vtRNA1-1 with endogenous CUGBP1 by RNP immunoprecipitation (RIP) assays using anti-CUGBP1 and control IgG antibodies, followed by isolation of bound RNA in both RIP reactions. After reverse transcription, Q-PCR analysis was used to measure the levels of vtRNA enrichment in the CUGBP1 IP relative to IgG IP, as described.32 As shown, vtRNA1-1 and vtRNA2-1 were highly enriched in CUGBP1 IP compared with control IgG IP (Figure 9A), although there were no significant changes in the levels of total input vtRNAs (Figure 9B). These results demonstrate the interaction between vtRNA1-1 and CURGP1 in IECs.
Figure 9.
vtRNA1-1 regulates TJ protein expression and epithelial barrier function via interaction with CUGBP1. (A) vtRNA1-1 binds to CUGBP1. Association of endogenous CUGBP1 with endogenous vtRNA1-1 and vtRNA2-1 in Caco-2 cells was measured by RIP/Q-PCR analysis using anti-CUGBP1 antibody (Ab). Values are means ± SEM (n = 3). ∗∗P < .01; ∗∗∗P < .001 compared with IgG. (B) Total input vtRNAs in cells described in A. (C) Levels of TJ protein mRNAs in the materials pulled down by anti-CUGBP1 Ab in cells transfected with a vtRNA1-1 expression vector. Interactions of the mRNAs with CUGBP1 were examined 48 hours after vtRNA1-1 overexpression. ∗∗P < .01 compared with IgG. +P < .05 compared with Vector. (D) Levels and cellular distribution of CUGBP1 48 hours after cells were transfected with vtRNA1-1 expression vector (vtR1-1). Total (T), cytoplasmic (C), and nuclear (N) proteins were isolated and used for Western blotting analysis. Lamin B (a nuclear protein) was examined to monitor the quality of the nuclear and cytoplasmic fractions. (E) Immunoblots of TJ and CUGBP1 proteins in cells 48 hours after transfection with CUGBP1 alone or co-transfected with CUGBP1 expression vector and vtR1-1. GAPDH immunoblotting was performed as an internal control for equal loading. (F) Changes in the levels of TEER and FITC-dextran paracellular permeability in cells treated as described in C. ∗,+P < .05 compared with control vector or CUGBP1 alone, respectively. In A, B, and C, statistical significance was analyzed using unpaired, 2-tailed Student’s t-tests, whereas comparison of means between more than 2 groups were performed by 1-way ANOVA with Bonferroni’s hoc test for F. All experiments in D and E were repeated 3 times with similar results.
Second, we examined if association of vtRNA1-1 with CUGBP1 altered the abundance of TJ proteins. As reported previously,36,44 CUGBP1 bound to the mRNAs encoding claudin-1 and occludin, but this association was increased by vtRNA1-1 overexpression (Figure 9C). The levels of CUGBP1/claudin-1 or CUGBP1/occludin mRNA complexes were greatly augmented in vtRNA1-1 transfected cells relative to cells transfected with control vector. On the other hand, vtRNA1-1 overexpression did not alter total CUGBP1 abundance or its subcellular distribution (Figure 9D). As expected, ectopically overexpressed CUGBP1 decreased the levels of claudin-1 and occludin proteins, but vtRNA1-1 and CUGBP1 synergistically inhibited TJ protein expression, as evidenced by an additional decrease in the levels of claudin-1 and occludin in cells co-transfected with CUGBP1 and vtRNA1-1 expression vectors when compared with cells transfected with CUGBP1 alone (Figure 9E).
Finally, we examined the role of vtRNA1-1/CUGBP1 interaction in the regulation of epithelial barrier function in vitro. Consistent with our previous study,45 ectopically expressed CUGBP1 impaired epithelial barrier function, as shown by decreased TEER and increased paracellular flux of FITC-dextran in CUGBP1-transfected cells relative to cells transfected with control vector (Figure 9F). However, this disruption of epithelial barrier function by CUGBP1 was enhanced by vtRNA1-1 overexpression, because decreased TEER and increased paracellular flux of FITC-dextran were enhanced in cells co-transfected with CUGBP1 and vtRNA1-1 expression vectors compared with cells transfected with CUGBP1 alone. These findings indicate that vtRNA1-1 disrupts intestinal barrier function at least partially by interaction with CUGBP1.
Discussion
Constant epithelial renewal is essential to maintaining intestinal epithelial integrity and effective gut barrier function particularly in response to stress.46,47 Nonetheless, the precise mechanisms underlying this process remain largely unknown. In the present study, we provide strong evidence that vtRNA1-1 plays an important role in regulating intestinal mucosal homeostasis and gut permeability. Tissue-specific transgenic expression of human vtRNA1-1 in the intestinal epithelium inhibited mucosal renewal in the small intestine, resulting in the dysregulation of IJ protein expression and predisposing the gut barrier to pathological stress in mice. Experiments aimed at identifying the mechanisms underlying the actions of vtRNA1-1 in this process revealed that vtRNA1-1 interacts physically with CUGBP1; this association reduced expression of the TJ proteins claudin-1 and occludin by enhancing CUGBP1 binding to their mRNAs. The findings that vtRNA1-1 is highly enriched in EVs from hemorrhagic shock patients in vivo, that silencing vtRNA1-1 expression in vitro enhances epithelial barrier function, and that exposing murine enteroids to vtRNA1-1-rich EVs ex vivo increases paracellular permeability suggest that vtRNAs play an important role in maintaining gut epithelial homeostasis. These findings also position vtRNA1-1, expressed either in EVs or mucosal tissues, as a key player in intestinal epithelial pathology in patients with critical illness.
We identified a substantial increase in the levels of EV vtRNA1-1 and vtRNA2-1, but not vtRNA1-2 and vtRNA1-3, in serum obtained from hemorrhagic shock patients relative to that obtained from healthy individuals. Compared with those from control mice, EVs isolated from mice with CLP-induced sepsis also displayed increased levels of EV vtRNA. Consistently, levels of intestinal mucosal vtRNAs were markedly increased in tissues from critically ill patients and septic mice, conditions associated with mucosal injury and gut barrier dysfunction, as reported previously.26 A murine genetic model with tissue-specific transgenic gene expression revealed that increasing the levels of human vtRNA1-1 locally in the intestinal epithelium inhibited mucosal renewal of the small intestine and impaired epithelial defenses, highlighted by reduced Paneth cell numbers. Moreover, enteroids derived from vtRNA1-1Tg mice grew much more slowly than those from littermate control mice and this inhibition of mucosal growth ex vivo was associated with defective Paneth cell function. In contrast, there were no significant differences in the rates of colonic mucosal growth between vtRNA1-1Tg and littermate control mice, even though vtRNA1-1 levels were also increased in the colonic epithelium of vtRNA1-1Tg mice. Although the exact mechanism underlying the failure of vtRNA1-1 overexpression to alter colonic mucosal growth remains unknown, it could be related to the fact that turnover of the colon mucosa is slower than that observed in the small intestine.33,46 Consistent with the current findings, we have reported that targeted deletion of the RBP HuR, ablation of the circular RNA Cdrlas locus, H19 knockout, or decreasing the levels of uc.173 in the intestinal epithelium only affects the growth of small intestinal mucosa without affecting renewal of colonic mucosa.29,41,47,48
Ectopically expressed vtRNA1-1 in the intestinal epithelium not only decreased levels of small intestinal intercellular TJ and AJ proteins but also increased gut barrier vulnerability to pathological stress such as low-dose LPS. Further studies in cultured cells and enteroids strongly support the view that vtRNA1-1 functions as a repressor of intestinal epithelial barrier function. vtRNA1-1 silencing by transfecting Caco-2 cells with si-vtR1-1 enhanced epithelial barrier function, as demonstrated by an increase in the levels of several TJ proteins and TEER values and a decrease in paracellular flux of FITC-dextran. Conversely, vtRNA1-1 overexpression disrupted epithelial barrier function by reducing TJ protein levels in vitro. Microinjecting FITC-dextran in 3-dimensional enteroids showed that EV vtRNAs negatively affect gut barrier function, because treating enteroids with vtRNA-enriched EVs increased paracellular permeability ex vivo. On the other hand, transgenic expression of vtRNA1-1 in the intestinal epithelium did not directly cause gut barrier dysfunction in the absence of pathological stress, although it decreased basal TJ protein expression. This inconsistency might result from altered autophagy in the epithelium of vtRNA1-1Tg mice, because increasing the levels of mucosal vtRNA1-1 induced the abundances of several autophagy-associated proteins. In support of this possibility, inducing autophagy improves epithelium-host defense and enhances gut barrier integrity.40,49
Our results also suggest that vtRNA1-1 regulates gut barrier function by interacting with CUGBP1, an evolutionally conserved molecule that plays an essential role in post-transcriptional gene regulation.50 CUGBP1 contains 3 RNA recognition motifs through which it associates with specific mRNAs that often contain GU-rich elements in their 3'-untranslated regions or coding regions.44,50 Our previous studies demonstrated that CUGBP1 acts as a master regulator of intestinal epithelium homeostasis and that increasing CUGBP1 levels causes gut barrier dysfunction and inhibits mucosal growth in vitro and in vivo by inhibiting the translation of TJ proteins such as occludin and claudin-1 and proliferation-associated proteins including c-Myc.36,45,51 CUGBP1 also interacts with miR-22235 and miR-195,52 long ncRNA uc230,4 and HuR45 to jointly regulate target transcripts in the intestinal epithelium antagonistically or synergistically. vtRNA1-1 bound to CUGBP1 and its overexpression enhanced CUGBP1 association with claudin-1 and occludin mRNAs, thus enhancing CUGBP1-mediated repression of TJ protein expression. In addition, CUGBP1 inhibits intestinal epithelial renewal by post-transcriptionally suppressing expression of c-Myc, cyclin-dependent kinase 4, and insulin-like growth factor receptor.31,51,52 Nonetheless, the role of the interaction of vtRNA1-1 with CUGBP1 in vtRNA1-1-induced inhibition of small intestinal mucosal growth remains to be fully elucidated. Because CUGBP1 is highly expressed in the intestinal epithelium and its cellular levels and binding affinity for target mRNAs are tightly regulated by RBPs and ncRNAs including vtRNA1-1,4,36,31,52 it is likely that vtRNA1-1 affects gene regulatory programs governing intestinal epithelium homeostasis and barrier function at least partially by interacting with CUGBP1 or other RBPs.
Our results may have important clinical implications because the levels of vtRNA1-1 and vtRNA2-1 are markedly increased in EVs from patients with hemorrhagic shock. Abnormalities in the abundances of tissue vtRNAs also occur in patients with inflammatory bowel disease, viral infection, cancer, drug resistance, and preterm birth.24, 25, 26,53 This dysregulation of vtRNA expression contributes to the pathogenesis of different human disorders by altering autophagy, proliferation, apoptosis, cell-to-cell interaction, and host defense. In the present study, we further found that increasing the levels of vtRNA1-1 inhibited intestinal mucosal growth and compromised gut barrier function, suggesting the potential of controlling vtRNA1-1 expression to maintain intestinal epithelial homeostasis in pathological conditions. vtRNA1-1 interacted with and enhanced CUGBP1-mediated inhibitory effect on TJ protein expression. In this regard, vtRNA2-1 associates with HuR but prevents HuR binding to its target mRNAs in IECs.26 These exciting findings showing the interaction between vtRNAs and RBPs advance our understanding of vtRNAs and their function in regulating intestinal epithelium homeostasis. It is possible that the regulation of RBP activity by vtRNAs could represent a general mechanism underlying their biological functions in the gut epithelium.
In summary, the results of the present study indicate that by regulating intestinal epithelial renewal and barrier function, vtRNA1-1 plays an essential role in altering intestinal epithelial homeostasis. Because EVs isolated from the serum of shock patients contained more vtRNA1-1 and treatment of enteroids with vtRNA1-1-rich EVs increased paracellular permeability ex vivo, these findings suggest that EVs can transfer vtRNA1-1, thus contributing to the increased intracellular vtRNA1-1 in neighboring and distant IECs of the epithelium and subsequent systemic gut barrier dysfunction in shock patients. Our results also suggest that the vtRNA1-1/CUGBP1 axis is a potential therapeutic target to preserve or reestablish intestinal epithelial integrity in patients with critical illness.
Materials and Methods
Chemicals and Cell Cultures
Biochemicals were purchased from Sigma, and culture medium and fetal bovine serum were from Invitrogen. Antibodies recognizing claudin-1 (CST-13255), claudin-2 (CST-48120), claudin-3 (CST-83607), claudin-7 (Inv-34-9100), occludin (sc-133256), JAM-A (sc-56323), ZO-2 (sc-33725), E-cadherin (sc-8426), lysozyme (sc-518012), CUGBP1 (sc-5261), p62 (sc-28359), HSC70 (MABE1120), and GAPDH (G9545) were obtained from Cell Signaling Technology, Invitrogen, Santa Cruz Biotechnology, and BD Biosciences. The secondary antibody conjugated to horseradish peroxidase was purchased from Sigma. All antibodies utilized in this study were validated for species specificity. Antibody dilutions used for Western blots of claudins, occludin, JAM-A, ZO-2, E-cadherin, CUGBP1, p62, HSP70, and GAPDH were 1:800 or 1000 (first Ab) and 1:2000 (second Ab), respectively, whereas antibody dilutions for immunostaining were 1:200 (first) and 1:2000 (second). Relative protein levels were analyzed by using Biorad Chemidoc and XRS system equipped with Image lab software (version 4.1). We also utilized “Quantity tool” to determine the band intensity volume; the values were normalized with internal loading control GAPDH. Caco-2 cells were purchased from the American Type Culture Collection and were maintained under standard culture conditions.41
Generation of vtRNA1-1 Transgenic Mice and Animal Studies
To create a gain-of-function mouse model for vtRNA1-1, transgenic mice that specifically overexpressed the human vtRNA1-1 in the intestinal epithelium were generated using A33-promoter as described in our previous studies.30,34 Briefly, the 586-bp fragment including the human vtRNA1-1 locus and β-globin intron (222-bp 5' upstream sequence and 259-bp 3' sequence) were cloned into the pIRES-AcGFP1-Nuc vector, and the final vtRNA1-1 expression vector, A33-vtRNA1-1/GFP, was used for microinjection into fertilized eggs.30,34,37 Transgenic founders on a pure C57BL/6J genetic background were established by pronuclear injection through a commercial service provided by the Genome Modification Facility at Harvard University. Genotyping was performed by PCR in DNA extracted from tail clippings to identify the first generation of recombinant mice with human vtRNA1-1/GFP bicistronic RNA. Two founders were further characterized for the transmission to subsequently establish transgenic colonies. Both male and female and age-matched (6 or 8-week-old) vtRNA1-1Tg and control littermate mice were housed in a specific pathogen-free breeding barrier and cared for by trained technicians and veterinarians.
All animal experiments were performed in accordance with National Institutes of Health guidelines and were approved by the Institutional Animal Care and Use Committee of University Maryland School of Medicine and Baltimore VA hospital. Animals were deprived of food but allowed free access to tap water for 24 hours. Two portions of the small intestine taken from 0.5 cm distal to the ligament of Trietz were removed, one for histological examination and the other for extraction of protein and RNA. Intestinal mucosal growth was examined by immunostaining of Ki67, whereas the mucosa was scraped with a glass slide for various measurements of RNAs and proteins as described previously.33 Representative results from 2 independent founders were reported here and compared with those obtained from control littermate mice. To generate septic stress model induced by CLP, age-matched male and female mice were anesthetized by Nembutal, and CLP was performed as described.54
Human Samples and EV Isolation
Blood was collected from shock patients and healthy control subjects in Shock Trauma Center, University of Maryland Health Science Center. The study was approved by the University Maryland Institutional Review Board. Blood samples were stored in pyrogen-free tubes and centrifuged at 4 °C and 2000 rpm for 10 minutes, then divided into sterile cryotubes and stored at −80 °C until analysis. EVs were isolated from the serum by the procedures with multiple steps of centrifugation as described.27,28 EV quality control was performed following Methodological Guidelines to Study EVs recommended by the American Heart Association.28 The final pellets containing EVs were resuspended in PBS and characterized by NSP analysis and immunoblotting analysis of EV markers Alix and CD63. In addition, vtRNA-rich EVs were also isolated from serum-free conditional media collected 48 hours after cells were transfected with the vtRNA1-1 expression vector or others. To isolate EVs from mice, C57BL/6J mice were exposed to CLP, and blood samples were collected 24 hours afterwards.
Enteroid Isolation and Culture
Isolation and culture of primary enterocytes were conducted following the method provided by Stem Cell Technologies, with minor modifications as described previously.41,55
Briefly, primary crypts were released from the small intestinal mucosa in mice; then, the isolated crypts were mixed with Matrigel and cultured in Advanced Dulbecco’s modified Eagle medium/F12 medium. The growth of enteroids was examined under phase-contrast microscopy as described.48
Plasmid Construction and RNA Interference
An expression vector containing vtRNA1-1 cDNA under control of pCMV-promoter was constructed and used to increase vtRNA1-1 in Caco-2 cells, whereas a vector containing a scrambled sequence of vtRNA1-1 was used as control. Transient transfections were performed using the Lipofectamine reagent following the manufacturer’s recommendations (Invitrogen). Forty-eight hours after transfection using LipofectAMINE, cells were harvested for analysis. Expression of vtRNA1-1 was silenced by transfection with specific si-vtR1-1 as described.26,56 The si-vtR1-1 and C-siRNA (a scrambled version of si-vt1-1) were purchased from Santa Cruz Biotechnologies. For each 60-mm cell culture dish, 15 μl of the 20 μM stock duplex si-vtR1-1 or C-siRNA was used. Forty-eight hours after transfection using LipofectAMINE (Invitrogen 116668019), cells were harvested for analysis. An expression vector containing the human CUGBP1 cDNA under the control of the pCMV promoter was purchased from Origene and used to increase cellular CUGBP1 levels as described previously.52
Histology and Immunofluorescence Staining
Dissected and opened intestines were mounted onto a solid surface and fixed in formalin and paraffin. Sections of 5-μm thickness were stained with hematoxylin and eosin (H&E) for general histology. Using a grade micrometer eyepiece, the overall length of villus and crypts of each section was measured, as reported previously.4 The immunofluorescence staining procedure was carried out according to the method described previously.41,57 For experiments using mucosal tissue samples from mice, more than 5 slides (5-μm thickness section) in each tissue sample were prepared for immunofluorescence staining. For studies in cultured enteroids, slides were fixed in 3.7% formaldehyde in PBS and rehydrated. All slides were incubated with a primary antibody recognizing lysozyme, or E-cadherin in blocking buffer overnight and then incubated with secondary antibody conjugated with Alexa Fluor-594 (Cat#: A32754, Molecular Probes) for 2 hours at room temperature. After rinsing 3 times, the slides were incubated with DAPI (Cat#: D1306, Molecular Probes) at a concentration of 1 μM for 10 minutes to stain cell nuclei. Finally, the slides were washed, mounted, and viewed through a Zeiss confocal microscope (model LSM700). Slides were examined in a blinded, coded fashion, and they were decoded only after examination was completed. Images were processed using Photoshop software (Adobe).
Measurement of Gut Permeability
Epithelial barrier function in vitro was examined by using the 12-mm Transwell plate as described.45,35 FITC-dextran (70 kDa; Sigma-Aldrich), a membrane-impermeable molecule, served as the paracellular tracer and was added at a final concentration of 0.25 mM to the apical bath wells that contained 0.5 mL of medium. The basal bath well had no added tracers and contained 1.5 mL of the same flux assay medium as the apical compartment. All flux assays were performed at 37 °C, and the basal medium was collected at different times after addition of FITC-dextran. The concentration of FITC-dextran in the basal medium was determined using a fluorescence plate reader with an excitation wavelength at 490 nm and an emission wavelength of 530 nm. TEER was measured with an epithelial voltmeter under open-circuit conditions (WPI) as described,35 and the TEER of all monolayers was normalized to that of control monolayers in the same experiment.
Gut permeability in vivo was determined by examining the appearance in blood of FITC-dextran administered by gavage as described.36 Briefly, mice were gavaged with FITC-dextran at a dose of 60 mg/100-g weight 4 hours before harvest. Blood sample was collected by cardiac puncture. The serum concentration of FITC-dextran was determined using a fluorescence plate reader as described above. In experiments in 3-dimensional enteroids, epithelial permeability ex vivo was assessed by microinjecting FITC-dextran. On day 3 after primary culture, FITC-dextran was injected into the lumen of enteroids using a micromanipulator and microinjector (Eppendorf FemtoJet 4X) under a stereomicroscope (Nikon Ti TE2000) in a biosafety cabinet, as described.42,43 Rates of epithelial permeability were monitored by examining the diffusion speed of luminal FITC-dextran as analyzed with software NIS-Elements AR5.30.05.
Q-PCR and ddPCR Analyses
Total RNA was isolated by using the RNeasy mini kit (Qiagen) and used in reverse transcription (RT) and PCR amplification reactions as described.48 Q-PCR analysis was performed using Step-one-plus Systems with specific primers, probes, and software (Applied Biosystems). To measure copy numbers of vtRNAs, ddPCR analysis was performed by using QX200 Droplet Digital PCR System (Bio-Rad) as described.56,58 Briefly, PCR reaction mixture containing cDNA was partitioned into aqueous droplets in oil via the QX100 Droplet Generator, and then transferred to a 96-well PCR plate. A 2-step thermocycling protocol (95 °C × 10 minutes; 40 cycles of [94 °C × 30 seconds, 60 °C × 60 seconds], 98 °C ×10 minutes) was undertaken in a Bio-Rad C1000 (Bio-Rad). The PCR plate was then transferred to the QX100 Droplet Reader for automatic reading of samples in all wells. Copy number of each vtRNA per μl PCR reaction was directly determined. QuantaSoft 1.7.4 analysis software (Bio-Rad) and Poisson statistics were used to compute droplet concentrations (copies/ng RNA).
Western Immunoblotting and RIP Pull-down Assays
Whole-cell lysates were prepared by using the RIPA buffer (50 mM Tris pH 7.4, 150 mM NaCl) containing 1% sodium dodecyl sulfate (SDS), sonicated, and centrifuged at 4°C for 15 min. The supernatants were boiled and size-fractionated by SDS-PAGE. After the blots were incubated with primary antibody and then secondary antibodies, immunocomplexes were developed by using chemiluminescence.
IP of RNP complexes was carried out to assess the association of endogenous CUGBP1 with endogenous vtRNAs or mRNAs encoding intercellular junction proteins as described.51 Twenty million cells were collected per sample, and lysates were used for IP for 4 hours at room temperature in the presence of excess (30 μg) IP antibody (IgG, or anti-CUGBP1). RNA in IP materials was used in RT reactions followed by Q-PCR analysis. The amplification of Gapdh mRNA, found in all samples as low-level contaminating housekeeping transcripts (not CUGBP1 target), served to monitor the evenness of sample input, as reported previously.45,51
Statistical Analysis
All values were expressed as the means ± standard error of the mean (SEM) or standard error of difference (SED). Unpaired, 2-tailed Student’s t-test was used when indicated, with P < .05 considered significant. When assessing multiple groups, 1-way analysis of variance (ANOVA) was utilized with Tukey’s post hoc test.59 The statistical software used was GraphPad Instat Prism 9.0. For non-parametric analysis rank comparison, we used the Kruskal-Wallis test.
Acknowledgments
CRediT Authorship Contributions
Shweta Sharma, PhD (Conceptualization: Equal; Data curation: Equal; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Software: Supporting; Validation: Equal)
Lan Xiao, MD (Conceptualization: Equal; Data curation: Equal; Formal analysis: Equal; Investigation: Lead; Methodology: Lead; Software: Supporting; Supervision: Supporting; Validation: Equal)
Hee K. Chung (Data curation: Supporting; Formal analysis: Supporting; Investigation: Equal; Methodology: Equal; Supervision: Supporting)
Ting Chen, PhD (Conceptualization: Supporting; Data curation: Equal; Investigation: Equal; Methodology: Supporting)
Caroline G Mallard, BS (Data curation: Supporting; Investigation: Equal; Methodology: Equal; Project administration: Supporting)
Bridgette Warner, BS (Formal analysis: Supporting; Investigation: Equal; Methodology: Supporting)
Ting-Xi Yu, PhD (Formal analysis: Supporting; Investigation: Equal; Methodology: Supporting)
Min S. Kwon, PhD (Data curation: Supporting; Formal analysis: Supporting; Investigation: Equal; Methodology: Supporting)
Songah Chae, PhD (Data curation: Supporting; Formal analysis: Supporting; Investigation: Equal; Methodology: Supporting)
Jean-Pierre Raufman, MD (Conceptualization: Supporting; Formal analysis: Supporting; Investigation: Supporting; Writing – original draft: Supporting)
Rosemary Kozar, MD (Conceptualization: Supporting; Formal analysis: Equal; Investigation: Supporting; Resources: Equal)
Jian-Ying Wang, MD, PhD (Conceptualization: Lead; Data curation: Equal; Formal analysis: Lead; Funding acquisition: Lead; Investigation: Equal; Methodology: Equal; Project administration: Lead; Resources: Lead; Software: Supporting; Supervision: Lead; Validation: Equal; Visualization: Equal; Writing – original draft: Lead)
Footnotes
Conflicts of interest This author discloses the following: Jian-Ying Wang is a Senior Research Career Scientist at the Biomedical Laboratory Research and Development Service (US Department of Veterans Affairs). The remaining authors disclose no conflicts.
Funding This work was supported by Merit Review Awards (BX000332 to Jian-Ying Wang; BX004890 to Jean-Pierre Raufman) from the United States Department of Veterans Affairs; and grants from the National Institutes of Health (DK57819, DK61972, DK68491 to Jian-Ying Wang; GM140983 to Rosemary Kozar).
References
- 1.Villablanca E.J., Selin K., Hedin C.R.H. Mechanisms of mucosal healing: treating inflammatory bowel disease without immunosuppression? Nat Rev Gastroenterol Hepatol. 2022;19:493–507. doi: 10.1038/s41575-022-00604-y. [DOI] [PubMed] [Google Scholar]
- 2.Sharma S., Xiao L., Wang J.Y. HuR and its interactions with noncoding RNAs in gut epithelium homeostasis and diseases. Front Biosci (Landmark Ed) 2023;28:262. doi: 10.31083/j.fbl2810262. [DOI] [PubMed] [Google Scholar]
- 3.Goldenring J.R., Mills J.C. Cellular plasticity, reprogramming, and regeneration: metaplasia in the stomach and beyond. Gastroenterology. 2022;162:415–430. doi: 10.1053/j.gastro.2021.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu T.X., Kalakonda S., Liu X., et al. Long noncoding RNA uc.230/CUG-binding protein 1 axis sustains intestinal epithelial homeostasis and response to tissue injury. JCI Insight. 2022;7 doi: 10.1172/jci.insight.156612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Payen D. The gut as a hidden source of sepsis. Minerva Anestesiol. 2020;86:662–669. doi: 10.23736/S0375-9393.20.14302-5. [DOI] [PubMed] [Google Scholar]
- 6.Carter S.R., Zahs A., Palmer J.L., et al. Intestinal barrier disruption as a cause of mortality in combined radiation and burn injury. Shock. 2013;40:281–289. doi: 10.1097/SHK.0b013e3182a2c5b5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camilleri M. Leaky gut: mechanisms, measurement and clinical implications in humans. Gut. 2019;68:1516–1526. doi: 10.1136/gutjnl-2019-318427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cairns C.A., Xiao L., Wang J.Y. Posttranscriptional regulation of intestinal mucosal growth and adaptation by noncoding RNAs in critical surgical disorders. J Invest Surg. 2024;37 doi: 10.1080/08941939.2024.2308809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu L., Zhuang R., Xiao L., et al. HuR enhances early restitution of the intestinal epithelium by increasing Cdc42 translation. Mol Cell Biol. 2017;37 doi: 10.1128/MCB.00574-16. e00574–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X.X., Xiao L., Chung H.K., et al. Interaction between HuR and circPABPN1 modulates autophagy in the intestinal epithelium by altering ATG16L1 translation. Mol Cell Biol. 2022;40 doi: 10.1128/MCB.00492-19. e00492–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das S., Extracellular RNA Communication Consortium, Ansel K.M., Bitzer M., et al. The extracellular RNA communication consortium: establishing foundational knowledge and technologies for extracellular RNA research. Cell. 2019;177:231–242. doi: 10.1016/j.cell.2019.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeppesen D.K., Fenix A.M., Franklin J.L., et al. Reassessment of exosome composition. Cell. 2019;177:428–445.e18. doi: 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hessvik N.P., Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci. 2018;75:193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caparrós E., García-Martinez I., Zapater Pedro, et al. An altered expression of miR-376a-3p and miR-20a-5p in peripheral blood exosomes regulates the autophagy and inflammatory systemic substrates, and relates to the smoking habit and age in Crohn’s disease. FASEB J. 2024;38 doi: 10.1096/fj.202301761R. [DOI] [PubMed] [Google Scholar]
- 15.Berger W., Steiner E., Grusch M., Elbling L., Micksche M. Vaults and the major vault protein: novel roles in signal pathway regulation and immunity. Cell Mol Life Sci. 2009;66:43–61. doi: 10.1007/s00018-008-8364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lässer C. Mapping extracellular RNA sheds lights on distinct carriers. Cell. 2019;177:228–230. doi: 10.1016/j.cell.2019.03.027. [DOI] [PubMed] [Google Scholar]
- 17.Horos R., Büscher M., Kleinendorst R., et al. The small non-coding vault RNA1-1 acts as a riboregulator of autophagy. Cell. 2019;176:1054–1067.e12. doi: 10.1016/j.cell.2019.01.030. [DOI] [PubMed] [Google Scholar]
- 18.Kickhoefer V.A., Rajavel K.S., Scheffer G.L., Dalton W.S., Scheper R.J., Rome L.H. Vaults are up-regulated in multidrug-resistant cancer cell lines. J Biol Chem. 1998;273:8971–8974. doi: 10.1074/jbc.273.15.8971. [DOI] [PubMed] [Google Scholar]
- 19.Kolev N.G., Rajan K.S., Tycowski K.T., et al. The vault RNA of Trypanosoma brucei plays a role in the production of trans-spliced mRNA. J Biol Chem. 2019;294:15559–15574. doi: 10.1074/jbc.RA119.008580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bracher L., Ferro I., Pulido-Quetglas C., Ruepp M.D., Johnson R., Polacek N. Human vtRNA1-1 levels modulate signaling pathways and regulate apoptosis in human cancer cells. Biomolecules. 2020;10:614. doi: 10.3390/biom10040614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wakatsuki S., Ohno M., Araki T. Human vault RNA1-1, but not vault RNA2-1, modulates synaptogenesis. Commun Integr Biol. 2021;14:61–65. doi: 10.1080/19420889.2021.1909229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wakatsuki S., Araki T. Novel molecular basis for synapse formation: small non-coding vault RNA functions as a riboregulator of MEK1 to modulate synaptogenesis. Front Mol Neurosci. 2021;14 doi: 10.3389/fnmol.2021.748721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferro I., Gavini J., Gallo S., et al. The human vault RNA enhances tumorigenesis and chemoresistance through the lysosome in hepatocellular carcinoma. Autophagy. 2022;18:191–203. doi: 10.1080/15548627.2021.1922983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee H.S., Lee K., Jang H.J., et al. Epigenetic silencing of the non-coding RNA nc886 provokes oncogenes during human esophageal tumorigenesis. Oncotarget. 2014;5:3472–3481. doi: 10.18632/oncotarget.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li F., Chen Y., Zhang Z., et al. Robust expression of vault RNAs induced by influenza A virus plays a critical role in suppression of PKR-mediated innate immunity. Nucleic Acids Res. 2015;43:10321–10337. doi: 10.1093/nar/gkv1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma X.X., Xiao L., Wen S.J., et al. Small noncoding vault RNA2-1 disrupts gut epithelial barrier function via interaction with HuR. EMBO Rep. 2023;24 doi: 10.15252/embr.202254925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeppesen D.K., Hvam M.L., Primdahl-Bengtson B., et al. Comparative analysis of discrete exosome fractions obtained by differential centrifugation. J Extracell Vesicles. 2014;3 doi: 10.3402/jev.v3.25011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coumans F.A.W., Brisson A.R., Buzas E.I., et al. Methodological guidelines to study extracellular vesicles. Circ Res. 2017;120:1632–1648. doi: 10.1161/CIRCRESAHA.117.309417. [DOI] [PubMed] [Google Scholar]
- 29.Yu T.X., Chung H.K., Xiao L., et al. Long noncoding RNA H19 impairs the intestinal barrier by suppressing autophagy and lowering Paneth and goblet cell function. Cell Mol Gastroenterol Hepatol. 2020;9:611–625. doi: 10.1016/j.jcmgh.2019.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S.R., Rathor N., Kwon M.S., et al. miR-195 regulates intestinal epithelial restitution after wounding by altering actin-related protein-2 translation. Am J Physiol Cell Physiol. 2022;322:C712–C722. doi: 10.1152/ajpcell.00001.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao L., Cui Y.H., Rao J.N., et al. Regulation of cyclin-dependent kinase 4 translation through CUG-binding protein 1 and microRNA-222 by polyamines. Mol Biol Cell. 2011;22:3055–3069. doi: 10.1091/mbc.E11-01-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao L., Rao J.N., Cao S., et al. Long noncoding RNA SPRY4-IT1 regulates intestinal epithelial barrier function by modulating the expression levels of tight junction proteins. Mol Biol Cell. 2016;27:617–626. doi: 10.1091/mbc.E15-10-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao L., Wu J., Wang J.Y., et al. Long noncoding RNA uc.173 promotes renewal of the intestinal mucosa by inducing degradation of microRNA 195. Gastroenterology. 2018;154:599–611. doi: 10.1053/j.gastro.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung H.K., Chen Y., Rao J.N., et al. Transgenic expression of miR-222 disrupts intestinal epithelial regeneration by targeting multiple genes including Frizzled-7. Mol Med. 2015;21:676–687. doi: 10.2119/molmed.2015.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou T., Jaladanki S.K., Liu L., et al. H19 long noncoding RNA regulates intestinal epithelial barrier function via microRNA 675 by interacting with RNA-binding protein HuR. Mol Cell Biol. 2016;36:1332–1341. doi: 10.1128/MCB.01030-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu T.X., Wang P.Y., Rao J.N., et al. Chk2-dependent HuR phosphorylation regulates occludin mRNA translation and epithelial barrier function. Nucleic Acids Res. 2011;39:8472–8487. doi: 10.1093/nar/gkr567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flentjar N., Chu P.Y., Ng A.Y., et al. TGF-βRII rescues development of small intestinal epithelial cells in Elf3-deficient mice. Gastroenterology. 2007;132:1410–1419. doi: 10.1053/j.gastro.2007.02.054. [DOI] [PubMed] [Google Scholar]
- 38.Kwon M.S., Chung H.K., Xiao L., et al. MicroRNA-195 regulates Tuft cell function in the intestinal epithelium by altering translation of DCLK1. Am J Physiol Cell Physiol. 2021;320:C1042–C1054. doi: 10.1152/ajpcell.00597.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bel S., Pendse M., Wang Y., et al. Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science. 2017;357:1047–1052. doi: 10.1126/science.aal4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haq S., Grondin J., Banskota S., Khan W.I. Autophagy: roles in intestinal mucosal homeostasis and inflammation. J Biomed Sci. 2019;26:19. doi: 10.1186/s12929-019-0512-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao L., Li X.X., Chung H.K., et al. RNA-binding protein HuR regulates Paneth cell function by altering membrane localization of TLR2 via posttranscriptional control of CNPY3. Gastroenterology. 2019;157:731–743. doi: 10.1053/j.gastro.2019.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartfeld S., Clevers H. Organoids as model for infectious diseases: culture of human and murine stomach organoids and microinjection of Helicobacter pylori. J Vis Exp. 2015;105 doi: 10.3791/53359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bardenbacher M., Ruder B., Britzen-Laurent N., et al. Permeability analyses and three dimensional imaging of interferon gamma-induced barrier disintegration in intestinal organoids. Stem Cell Res. 2019;35 doi: 10.1016/j.scr.2019.101383. [DOI] [PubMed] [Google Scholar]
- 44.Chung H.K., Rao J.N., Wang J.Y. In: Kenakin T., editor. Vol. 5. Elsevier; 2022. Regulation of gut barrier function by RNA-binding proteins and noncoding RNAs; pp. 194–213. (Comprehensive Pharmacology). [Google Scholar]
- 45.Yu T.X., Rao J.N., Zou T., et al. Competitive binding of CUGBP1 and HuR to occludin mRNA controls its translation and modulates epithelial barrier function. Mol Biol Cell. 2013;24:85–99. doi: 10.1091/mbc.E12-07-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hagen S.J. Gastroduodenal injury and repair: novel targets for therapeutic intervention. Curr Opin Gastroenterol. 2022;38:607–612. doi: 10.1097/MOG.0000000000000883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu L., Christodoulou-Vafeiadou E., Rao J.N., et al. RNA-binding protein HuR promotes growth of small intestinal mucosa by activating the Wnt signaling pathway. Mol Biol Cell. 2014;25:3308–3318. doi: 10.1091/mbc.E14-03-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chung H.K., Xiao L., Han N., et al. Circular RNA Cdr1as inhibits proliferation and delays injury-induced regeneration of the intestinal epithelium. JCI Insight. 2024;9 doi: 10.1172/jci.insight.169716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiao L., Rao J.N., Wang J.Y. RNA-binding proteins and long noncoding RNAs in intestinal epithelial autophagy and barrier function. Tissue Barriers. 2021;9 doi: 10.1080/21688370.2021.1895648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qin W.J., Shi J.J., Chen R.Y., et al. Curriculum vitae of CUG binding protein 1 (CELF1) in homeostasis and diseases: a systematic review. Cell Mol Biol Lett. 2024;29:32. doi: 10.1186/s11658-024-00556-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu L., Ouyang M., Rao J.N., et al. Competition between RNA-binding proteins CELF1 and HuR modulates MYC translation and intestinal epithelium renewal. Mol Biol Cell. 2015;26:1797–1810. doi: 10.1091/mbc.E14-11-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y., Zhang Y., Xiao L., et al. Cooperative repression of insulin-like growth factor type 2 receptor translation by microRNA 195 and RNA-binding protein CUGBP1. Mol Cell Biol. 2017;37 doi: 10.1128/MCB.00225-17. e00225–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.You Y.A., Kwon E.J., Hwang H.S., Choi S.J., Choi S.K., Kim Y.J. Elevated methylation of the vault RNA2-1 promoter in maternal blood is associated with preterm birth. BMC Genomics. 2021;22:528. doi: 10.1186/s12864-021-07865-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hubbard W.J., Choudhry M., Schwacha M.G., et al. Cecal ligation and puncture. Shock. 2005;24(Suppl 1):52–57. doi: 10.1097/01.shk.0000191414.94461.7e. [DOI] [PubMed] [Google Scholar]
- 55.Lindemans C.A., Calafiore M., Mertelsmann A.M., et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 2015;528:560–564. doi: 10.1038/nature16460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiao L., Ma X.X., Luo J., et al. Circular RNA circHIPK3 promotes homeostasis of the intestinal epithelium by reducing microRNA 29b function. Gastroenterology. 2021;161:1303–1317.e3. doi: 10.1053/j.gastro.2021.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao L., Warner B., Mallard C.G., et al. Control of Paneth cell function by HuR regulates gut mucosal growth by altering stem cell activity. Life Sci Alliance. 2023;6 doi: 10.26508/lsa.202302152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vasseur A., Cabel L., Hego C., et al. Fulvestrant and everolimus efficacy after CDK4/6 inhibitor: a prospective study with circulating tumor DNA analysis. Oncogene. 2024;43:1214–1222. doi: 10.1038/s41388-024-02986-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harter H.L. Critical values for Duncan’s new multiple range test. Biometrics. 1960;16:671–685. [Google Scholar]