Abstract
The suprachiasmatic nucleus (SCN) is the master clock that directly dictates behavioural rhythms to anticipate the earth's light/dark cycles. Although post‐transcriptional regulators called microRNAs have been implicated in physiological SCN function, how the absence of the entire mature miRNome impacts SCN output has not yet been explored. To study the behavioural consequences of miRNA depletion in the SCN, we first generated a mouse model in which Dicer is inactivated in the SCN by crossing Syt10 Cre mice with Dicer flox mice to study behavioural consequences of miRNA depletion in the SCN. Loss of all mature miRNAs in the SCN shortened the circadian period length by ~37 minutes at the tissue level and by ~45 minutes at the locomotor activity level. Moreover, knockout animals exhibited a reduction in the precision of the circadian rhythm with more variable activity onsets under both LD 12:12 and DD conditions. We also observed that knockouts with higher onset variations were inclined to develop ultradian rhythms under constant light. In a second mouse model, recombination of Dicer flox via Cre delivery specifically in the SCN resulted in loss of behavioural rhythms in some animals depending on the injection efficiency. Together, our observations highlight the importance of microRNAs for a physiological SCN function and their pivotal role in robust circadian oscillations.
Keywords: circadian rhythms, Dicer knockout, locomotor activity, miRNAs, period length, SCN
We report here the behavioural phenotypes of conditional Dicer knockout in the suprachiasmatic nucleus. Both at tissue and organismal levels, clocks run faster when miRNAs are depleted in the master clock. Knockout mice also have less precise clocks as assessed by onset variations under both DD and LD. Finally, higher onset variations correlate with ultradian rhythms under LL in knockouts.
Abbreviations
- AAV
adenovirus‐associated virus
- AVP
Arginine vasopressin
- BMAL
brain and muscle Arnt‐like protein
- Chi
chi‐square periodogram
- CT
circadian time
- DD
constant darkness
- Fourier
Fourier analysis
- hSyn
human synapsin
- IR
infrared sensor
- KI
knockin
- KO
knockout
- LD
light dark
- LS
Lomb‐Scargle periodogram
- LL
constant light
- miRNAs
microRNAs
- PCR
polymerase chain reaction
- Per2
Period2
- Luc
luciferase
- POMC
pro‐opiomelanocortin
- RW
running wheel
- RT‐qPCR
reverse‐transcription quantitative polymerase chain reaction
- SCN
suprachiasmatic nucleus
- Syt10
synaptotagmin X
- VIP
vasoactive intestinal peptide
- ZT
zeitgeber time
1. INTRODUCTION
Circadian rhythmicity relies on a hierarchical system of clocks coordinated by a master clock residing in the brain region called the suprachiasmatic nucleus (SCN) (Moore, 1995). This small nucleus lies just above the optic chiasm and receives direct photic information from the ganglion cells of the retina (Abrahamson & Moore, 2001). The utmost outputs of the SCN are vital behaviours such as sleep/wake cycles, and feeding/fasting rhythms (Kalsbeek et al., 2006; Patton & Hastings, 2023; Weaver, 1998). At the molecular level, the output of all autonomous clocks present throughout the body is rhythmic gene expression with a period of about 24 hours (Partch et al., 2014). Interestingly, a large proportion of the transcriptome, ~43% in mouse (Zhang et al., 2014), 44% in humans (Ruben et al., 2018) and 82% in primates (Mure et al., 2018), is rhythmically expressed somewhere in the body.
Circadian gene expression originates not only from circadian transcription but also from circadian post‐transcriptional and post‐translational mechanisms (Anna & Kannan, 2021; Crosby & Partch, 2020). A recent meta‐analysis and modelling approach estimated that 30% of circadian transcripts are regulated post‐transcriptionally in mouse liver (Lück et al., 2014). Among the known post‐transcriptional mechanisms, the short (19–25 nucleotides) non‐coding RNA molecules coined microRNAs (miRNAs) play a crucial role in shaping the dynamics of gene expression, by regulating both mRNA degradation and translation of a multitude of target genes. There are more than 1000 miRNA genes in the human genome that target up to 60% or protein‐coding genes (Helwak et al., 2013; Krol et al., 2010), with one miRNA targeting from a dozen to hundreds of mRNA targets (Liu & Wang, 2019). In the last two decades, miRNAs have emerged as important players in regulating all kinds of biological processes, including circadian rhythms, from the molecular to the behavioural level.
The involvement of miRNAs in circadian oscillations has been reported in cell lines (Park et al., 2020), in peripheral tissues (Xu et al., 2007), as well as in the SCN (Alvarez‐Saavedra et al., 2011; Mehta & Cheng, 2013). However, most studies focus on the role of an individual miRNA in a specific context. Informative as it may be, this approach often cannot showcase the magnitude of miRNA influence on a biological system. We previously demonstrated a comprehensive view of miRNA regulation of the hepatic transcriptome employing a genetic mouse model in which miRNA biosynthesis was inactivated (liver‐specific Dicer knockout) (Du et al., 2014). In our hands, miRNAs played an essential role in adjusting the phase and amplitude of 30% of the circadian transcriptome in mouse liver.
To study the roles of miRNA ensemble in shaping the function of the master clock, we have now extended the use of the tissue‐specific Dicer knockout model from the liver to the SCN. We report here circadian behavioural phenotypes of Dicer knockout in the SCN in two different mouse models, conditional and inducible knockout models. We also provide suggestive evidence for sex differences in the conditional knockout model.
2. MATERIALS AND METHODS
2.1. Animals
All animal experiments were performed according to the cantonal guidelines of either the Canton of Vaud, Switzerland, license 2376.1, or the Canton of Zurich, Switzerland, license 060/2017. Animals were allowed to access food and water ad libitum under a 12:12‐hr light‐dark (LD) cycle, unless otherwise stated. Dicer flox mice (IMSR_JAX:006366) were gift from Professor David Gatfield (Du et al., 2014), Syt10 Cre knock‐in mice (MGI:5286607) were gift from Dr. Henrik Oster (Husse et al., 2011), Period2::Luciferase (Per2::Luc) knock‐in mice (MGI:3040876) were gift from Dr. Joseph Takahashi (Yoo et al., 2004). Genotype of the animals was examined by PCR as described in the original publication of each mouse strain. Due to the expression of Syt10 in the testis, males homozygous for Syt10 Cre should not be used for mating with Dicer flox mice because this will create whole‐body knockout. Therefore, the male of genotype Dicer flox/flox ; Syt10 +/+ were crossed with female Dicer flox/flox ; Syt10 Cre/+ to create knockout of genotype Dicer flox/flox ; Syt10 Cre/+, dubbed conditional knockouts and control of genotype Dicer flox/flox ; Syt10 +/+. Due to the low pregnancy rate that prevented the use of same age animals, males aged between 2 and 6 months were used for various experiments. Litter mates or animals of similar ages were used for the same experimental conditions.
2.2. Genotyping
Polymerase chain reaction (PCR) was used for genomic DNA extracted from either the tail, ear or olfactory bulb to genotype each tissue or animal. To genotype alleles of Dicer, the following two primers were used: DicerR1, AAACATGACTCTTCAACTCAAACTCAAACG, and DicerF1, AATATTAATCCTGACAGTGACGGTCCAAAG. To confirm deletion of exon 23 using PCR, primer DicerF1 and DicerDel, GGGCAGCCCCATCTCAAAGGCCTACCTGAG were used. To genotype alleles of Syt10, the following three primers were used: Syt10 F, AGACCTGGCAGCAGCGTCCGTTGG; Syt10 R, AAGATAAGCTCCAGCCAGGAAGTC; Syt10 KI, GGCGAGGCAGGCCAGATCTCCTGTG. To genotype alleles of Per2::Luc, the following three primers were used: P1, CTGTGTTTACTGCGAGAGT; P2, GGGTCCATGTGATTAGAAAC; P3, TAAAACCGGGAGGTAGATGAGA.
2.3. RT‐qPCR confirmation of recombination
cDNA was synthesized from 60 ng of total RNA from the SCN of KO and control mice (for both conditional and inducible models), using random hexamers and SuperScript IV reverse transcriptase (Cat #18090010, Invitrogen) according to manufacturer's instructions. cDNA was PCR‐amplified using PowerUp™ SYBR™ Green Master Mix (Cat #A25741, Thermo Fischer Scientific). To quantify the appearance of exon 23–25 spliced transcript upon knockout of exon 24, we used primers targeting the junction between exon 23 and 25: E23/25_3 (Forward:
AACACTATCACTGCTTAGGAGAT and Reverse: TGGGGACTTCGATATCCTCTTC). Expression of the skipped exon 23–25 was normalized to that of exon 21, taken as an internal reference (Forward: GAGTCACCTGCTAAGCTCCA and Reverse: ACATTCATTGCTGGGCTTGG). Exon 21 expression was normalized to three reference genes: Eef1a1 (Forward:
TCCACTTGGTCGCTTTGCT and Reverse: CTTCTTGTCCACAGCTTTGATGA),
Nudt4 (Forward: AAGTTCAAGCCCAACCAGACG and Reverse: TCCTGGGACAATCCATTGGTC) and Trip12 (Forward: TCCCTGGGATCTACAACACCA and Reverse:
ATCATCTCTGCTATGCTGCAAG). The mean expression level for each exon in KO samples was expressed relative to that of the control samples (n = 2/genotype).
2.4. Tissue explants and bioluminescence recording and analysis
Dicer flox ; Syt10 Cre mice were crossed to Per2::Luc mice for bioluminescence recording of tissue explants. Except for the preliminary results of female explants, all other explants were from males. To prepare tissue explants, animals were sacrificed and each tissue was collected into ice‐cold HBSS (Cat# 14025, Life Technologies, CA) with 10 mM Hepes (Cat# H0887, Sigma, MO). For SCN and pituitary, brains were sliced at 300 μm intervals with a McILWAIN Tissue Chopper, and each area was isolated separately in ice‐cold HBSS buffer with Hepes. For kidney, liver, tail and lung, pieces of tissues were sliced at 300 μm interval. Each tissue explant was cultured on either a Millicell (#PICMORG, Merckmillipore, MA) or a piece of hydrophilic PTFE‐membrane (#BGCM00010, Merckmillipore, MA) submerged with DMEM (Cat# D2902, Sigma, MO) with 0.035% Sodium bicarbonate (Cat# S8761, Sigma, MO), 10 mM Hepes (Cat# H0887, Sigma, MO), 4.5 g/l D‐glucose (Cat# G8769, Sigma, MO), 1.0% Penicillin–Streptomycin (Cat# 15070–063, Life Technologies, CA), 2% B27 (Cat# 17504044, Gibco) and 0.1 mM D‐Luciferin (Promega, WI). Circadian bioluminescence was recorded with photomultiplier tubes (PMTs) every 48 mins at 34.5°C with 5% CO2. To assess the period length of the bioluminescence ex vivo, the original data were subtracted with a 24‐hr running average and sinusoidal curve fitting was applied using Lumicycle analysis software (Actimetrics).
2.5. Locomotor activity recordings and analysis
Male mice were individually housed in cages containing running wheels, or with infrared detectors as indicated, with ad libitum access to food and water. Light intensity at the cage level was 60 lx. Locomotor activity was recorded as counts per minute using ClockLab software (Actimetrics). To analyse the locomotor activity, the activity counts were binned to every five minutes and exported as a csv file. The text files were used for further analysis in R. Actograms were plotted using geom_density_ridges function from ggridges package in R. To analyse activity onsets, actograms were plotted using the shiny package in addition to the above‐mentioned packages. To obtain consistent onset time, the first three or four days after being transferred to each light‐dark condition were removed, and then the onset time for seven to ten days afterwards was used for analyses. In case the animals were under light/dark conditions, activity onset time was compared to the light‐off time as reference time. In case the animals were in constant darkness, activity onset time was compared to CT12 as reference time. Note that CT12 in constant darkness is defined with the free‐running period of each animal obtained from chi‐square in ActogramJ and the last light‐off time (ZT12) in LD directly before DD. For each animal, the absolute difference between onset time and reference time was calculated, dubbed onset deviation. For each animal, the standard deviation of the absolute difference between onset time and reference time was calculated, dubbed daily onset variation. To obtain a periodogram, the first three or four days after being transferred to each light‐dark condition were removed, then the activity data of the ten days afterwards were analysed in ActogramJ v1.0 (Schmid et al., 2011). The activity counts were smoothed with a Gaussian kernel set to 15 to avoid many small sharp peaks. The Gaussian value was applied to all three methods: chi‐square, Lomb‐Scargle and Fourier. The phase shift half‐time was defined as the time at which half the phase shift was completed and was extracted using drda R package (Malyutina et al., 2023).
2.6. Histological assessment
Animals were deeply anaesthetized with Pentobarbital and intracardially perfused with 10 ml of ice‐cold saline, followed by 20 ml of ice‐cold 4% paraformaldehyde/0.1 M phosphate buffer (PB, pH 7.4). Brains were collected and post‐fixed in 4% paraformaldehyde with 0.1 M PB for overnight at 4°C followed by cryoprotection in 20% sucrose/0.1 M PB for 48 hrs at 4°C. The brains were sectioned by 30 μm with cryostat at −17°C and washed in 0.1 M PB at room temperature. The sections were incubated in DAPI/0.1 M PB, washed in 0.1 M PB, mounted onto gelatin‐coated microscope slides, air‐dried and then dehydrated with Fluoromount™ Aqueous Mounting Medium (Cat# S3023, Dako). Fluorescent images were obtained with a widefield microscope Apotome (Zeiss, Germany). Sections were acquired from the same coronal plane, as assessed by the presence of other topological landmarks, such as the paraventricular nucleus (PVH) and the optic chiasm.
2.7. Injection of AAV constructs expressing hSyn‐Cre in the SCN
Male Dicer flox mice (12–16 weeks old) were stereotactically (Kopf Instruments, CA, USA) injected under isofluorane anaesthesia, bilaterally at the SCN (ML = ± 0.18 mm; AP = 0.46 mm; DV = 5.8 mm; relative to bregma). The following viruses were injected at a volume of 300 nl with a rate of 150 nl/min: for knockouts (n = 6), ssAAV9/2‐hSyn1‐chl‐mCherry_2A_iCre‐WPRE‐SV40p(A) (produced by Viral Vector Facility at the University of Zurich, 7.9 × 1012 viral particles/ml; iCre: Addgene #24593) and for controls (n = 7), ssAAV9/2‐hSyn1‐chl‐mCherry‐WPRE‐SV40p(A) (produced by Viral Vector Facility at the University of Zurich, 4.8 × 1012 viral particles/ml). Post‐surgery, mice were returned to the housing cage, and allowed to recover. Following the completion of the activity recording using a running wheel, mice were perfused with 4% paraformaldehyde, their brains were extracted and sites of injection were confirmed through mCherry expression with confocal microscopy.
3. RESULTS
3.1. Generation of a mouse model with inactivated Dicer in the SCN
To study the function of miRNAs in the SCN, we generated a mouse model in which miRNA biogenesis is inactivated in the majority of the SCN cells. We bred mice carrying conditional knockout alleles for the Dicer1 gene (referred to as Dicer flox in the following), and mice carrying Cre recombinase cDNA inserted into the Synaptotagmin 10 locus (referred to as Syt10 Cre ), a gene strongly expressed in the SCN (Husse et al., 2011), and obtained Dicer flox ; Syt10 Cre mice, called here conditional Dicer knockout (KO) mice. It is worth noting that although Syt10 is abundantly expressed in the SCN, it is also found in neuronal cells in other brain regions such as the olfactory bulb, the pituitary, the cortex, the hypothalamus, the cerebellum and the spinal cord (Husse et al., 2011; Lein et al., 2007; Roper et al., 2015). In addition to neuronal cells, Syt10 is expressed in the seminiferous tubules of the testis but not in any other peripheral tissues (Husse et al., 2011). The insertion of the Cre recombinase gene in the Syt10 locus results in a knockout of endogenous Syt10. The Dicer flox mouse carries loxP sites flanking exon 24, which encodes most of the second RNaseIII domain of the Dicer1 gene (Harfe et al., 2005). We evaluated recombination using two methods. For the first method, due to the small size of the SCN and potential contamination from surrounding tissues that limits its use in PCR analysis, the olfactory bulb, which also expresses Syt10, was used for PCR analysis to confirm successful recombination at the Dicer flox locus (Figure S1A, B). PCR analysis of the ear showed no detection of the recombined allele as expected. For the second method, we quantified the splicing of exon 23 onto exon 25 using RT‐qPCR as previously described (Mang et al., 2015). While the expression of exon 21 in the SCN (Figure S1C, left panel), served as positive control, and was detected similarly in controls (n = 3) and knockouts (n = 2), exon 23–25 splicing was higher in knockouts compared to controls (Figure S1C, middle and right panel). Note that the CT values for controls were close to the detection limit (34.84 ± 0.97) while the CT values for knockouts were much lower (28.15 and 28.61).
3.2. Conditional Dicer knockout mice showed shorter free‐running period with variable onsets
We found that conditional knockout animals had shorter free‐running periods than control animals (Figure 1). Three different methods were used to calculate the free‐running period from the actogram: chi‐square periodogram, Fourier analysis and Lomb‐Scargle periodogram, which all gave similar results. The period lengths of knockouts were 23.00 ± 0.50 h, 23.00 ± 0.51 h and 23.02 ± 0.49 h, by chi‐square periodogram, Fourier analysis and Lomb‐Scargle periodogram, respectively. The period lengths of controls were 23.77 h ± 0.24 h, 23.74 ± 0.26 h and 23.77 ± 0.26 h, by chi‐square periodogram, Fourier analysis and Lomb‐Scargle periodogram, respectively. Welch two‐sample t‐tests revealed a statistically significant difference between the mean period length of knockouts and controls calculated by all three methods (p < 0.0005). Because the period length calculated by the three methods was similar to each other, for the rest of the activity recordings, only results for the chi‐square periodogram were used for further analyses.
FIGURE 1.
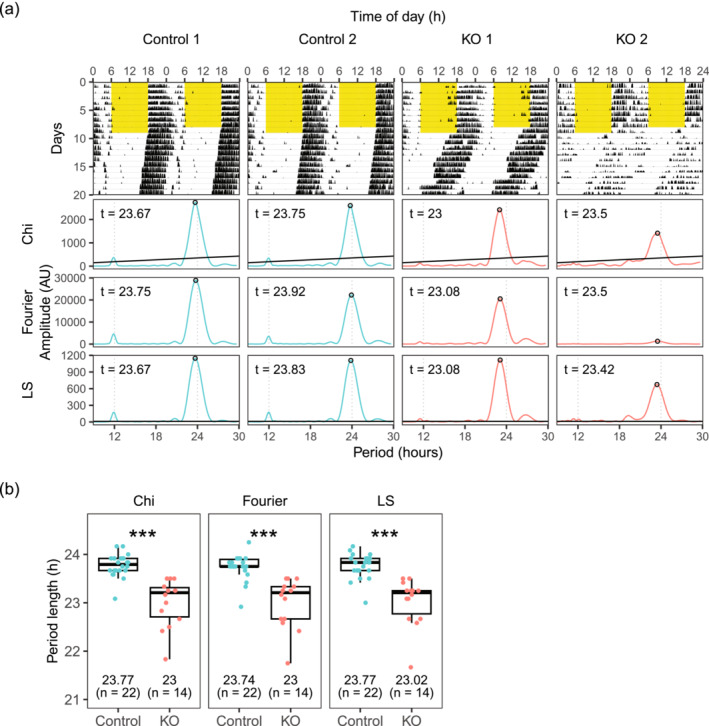
Conditional Dicer knockouts have shorter period length. (a) Representative actograms and periodograms under constant darkness (DD). Yellow part in the actograms denotes when light was on. (b) Free running period length in constant darkness calculated by three different methods: chi‐square periodogram (Chi), Fourier analysis (Fourier) and Lomb‐Scargle periodogram (LS).
We next evaluated the re‐entrainment of conditional knockouts to jetlag. A subset of animals used for Figure 1 was used for this purpose. We found that knockout animals re‐entrained almost immediately to a new light–dark cycle after 6 h phase advance (Figure 2a–c). Phase shift half time, i.e., time required to reach half of the phase shift, was analysed in two different ways to quantify jetlag re‐entrainment. When all animals were analysed as a group, the phase shifts half‐time for the knockout group (n = 5) and the control group (n = 9) were 1.15 days and 2.48 days, respectively (Figure 2b). When the phase shift half‐time was calculated for each animal, the values for the controls and the knockouts were 1.27 ± 0.59 days and 2.75 ± 1.06 days, respectively (Figure 2c). A Welch two‐sample t‐test revealed a statistically significant difference between the two groups, T(11.98) = 3.35, p = 0.006. When animals were placed under 6 h phase delay schedule (Figure 2d), there was no statistically significant difference between knockouts and controls. The phase shift half times as a group for knockout (n = 3) and control (n = 5) were 0.61 days and 0.51 days, respectively (Figure 2e). The average phase shift half times for each animal for knockouts and controls were 1.74 ± 1.18 days and 0.70 ± 0.31 days, respectively (Figure 2f). A Welch two‐sample t‐test revealed that there was no statistically significant difference between the two groups, T(2.10) = 1.52, p = 0.26.
FIGURE 2.
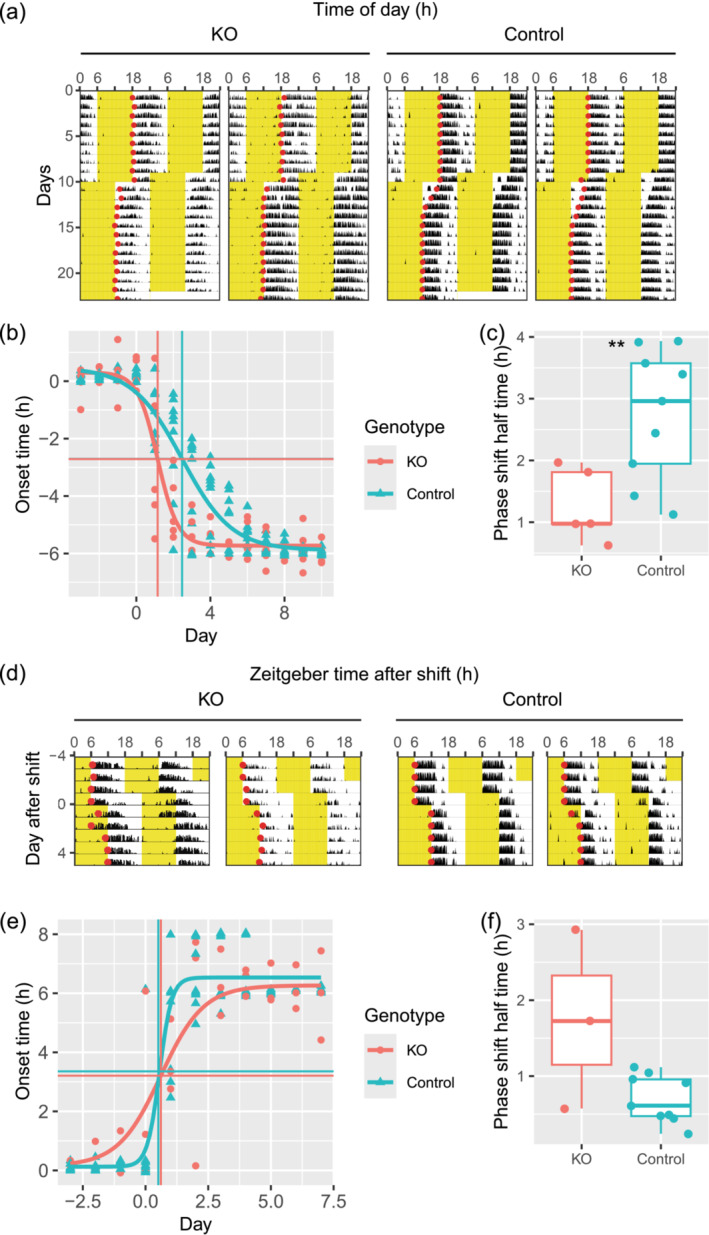
Conditional Dicer knockouts re‐entrain faster to jetlag. (a) Representative actograms with detected onsets (red circles) under 6 h‐advance jetlag. Yellow part in the actograms denotes when light was on. (b) Activity onsets under 6 h‐advance jetlag was plotted against day since jetlag. Y‐axis shows onset time with ZT12 before jetlag as time 0. X‐axis shows the day since jetlag with day 0 as the day when the light was shifted 6 h in advance. The fitted curve shows the dynamic of onset time for the whole animal population. The vertical lines show the phase‐shift half‐time of the population calculated from the fit. The horizontal lines show the corresponding onset times calculated from the fit. (c) Phase shift half time of 6 h‐advance jetlag calculated for each animal separately. (d) Representative actograms with detected onsets (red circles) under 6 h‐delay jetlag on day 0. Yellow part in the actograms denotes when light was on. (e) Activity onsets under 6 h‐delay jetlag was plotted against day since jetlag. Y‐axis shows onset time with ZT12 before jetlag as time 0. X‐axis shows day since jetlag with day 0 as the day when the light was shifted 6 h delay. The fitted curve shows the dynamic of onset time for the whole animal population. The vertical lines show the phase‐shift half‐time of the population calculated from the fit. The horizontal lines show the corresponding onset times calculated from the fit. (f) Phase shift half time of 6 h‐ delay jetlag calculated for each animal separately. **: p < 0.01.
We noticed that knockouts had more variable onsets than controls. Due to the limited availability of running wheel recording apparatus, although all animals were kept in LD 12:12 for 2 weeks before being released under DD, not all animals in Figure 1 had recordings under LD 12:12. We thus analysed onsets for a subset of animals presented in Figure 1 whose activities under LD 12:12 were available. Under LD 12:12, daily activity‐onsets could precede or follow dark‐onset for both control and knockout mice (Figure S2A). Knockout mice, however, had a broader distribution of onset time than controls which we further quantified. To remove the direction of the relationship between activity‐onsets and dark‐onset, we calculated the absolute difference between activity‐onsets and dark‐onset for every day, dubbed onset deviation and took the average of these values for each animal. We found that knockouts showed an onset deviation of 62.42 ± 23.65 minutes, while controls showed only 8.44 ± 4.04 minutes activity‐onset deviation (Figure 3a, T[4.10] = 5.07, p = 0.007, Welch two‐sample t‐test). In addition to the larger deviation of activity onsets to the dark onset, knockout animals showed larger variation in their daily activity onsets compared to control animals. For each animal, from the daily onset deviations, we calculated the standard deviation of these values. Under LD = 12:12, the standard deviation of onset for each animal was 50.29 ± 25.78 minutes for knockouts and 6.96 ± 6.72 minutes for controls (Figure 3b, T[12.14] = 3.93 p = 0.002, Welch two‐sample t‐test). Since the running wheel might change the animals' behaviours, we measured the onset in another cohort of animals using an infrared detector under LD 12:12 (Figure 3c, Figure S2B). In spite of the different devices, we found again that knockout animals had a larger deviation of onset time to the light‐off time (41.87 ± 18.86 minutes), compared to controls (5.79 ± 4.34 minutes, Figure 3c, T[3.18] = 3.77, p = 0.03, Welch two‐sample t‐test). Though no significant differences were observed (Figure 3d; T[3.14] = 2.07, p = 0.13, Welch two‐sample t‐test), there was a tendency toward larger standard deviation of activity onsets in knockouts (30.32 ± 22.94 minutes) vs controls (6.25 ± 4.68 minutes). In conclusion, the precision of onset time under LD 12:12, either quantified by deviation to the light‐off time, or by daily variation for each animal, was lower for knockouts compared to controls.
FIGURE 3.
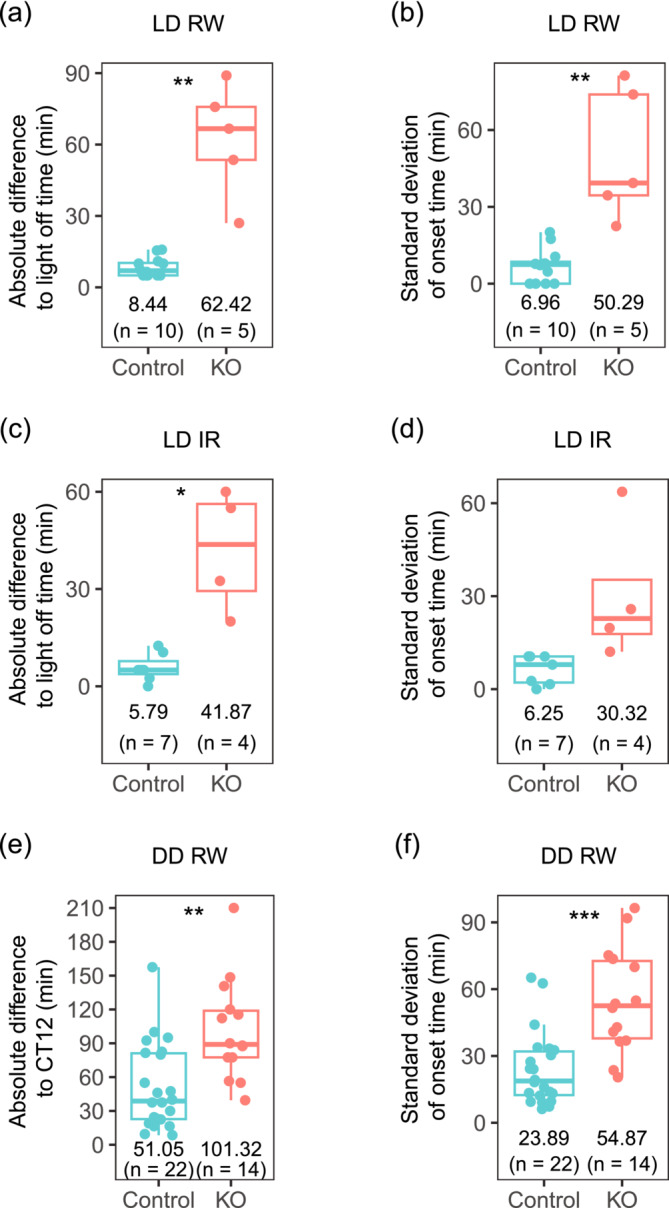
Conditional KO mice have larger onset variation than controls. (a) Absolute difference to light‐off time under LD 12:12 condition measured with a running wheel (RW). (b) Standard deviation of difference to light‐off time under LD 12:12 condition measured with RW. (c) Absolute difference to light‐off time under LD 12:12 condition measured with infrared sensor (IR). (d) Standard deviation of difference to light‐off time under LD 12:12 condition measured with IR. (e) Absolute difference to CT12 under constant darkness condition (DD) measured with RW. (f) Standard deviation of difference to CT12 under constant darkness condition (DD) measured with RW.
Interestingly, the larger variation of activity onsets in knockouts vs controls was also observed under the DD condition. For each animal, circadian time under DD was calculated based on its free‐running period length and the last light‐off time in LD directly before DD. Similar to when under LD, daily activity onsets could precede or be after CT12 for both controls and knockouts (Figure S2C). We calculated the absolute difference between activity‐onsets and CT12 for each animal, i.e. onset deviation. Knockouts showed activity onsets that were different absolutely to CT12 by 101.32 ± 44.60 minutes, while controls showed only 51.05 ± 38.01 minutes differences (Figure 3e, T[24.55] = 3.49, p = 0.002, Welch two‐sample t‐test). Under DD, the standard deviation of onset deviation was 54.87 ± 23.63 minutes for knockouts and 23.89 ± 16.51 minutes for controls (Figure 3f, T[21.07] = 4.29, p = 0.0003, Welch two‐sample t‐test). The more variable onsets in knockouts under both LD 12:12 and DD conditions suggested that the circadian clocks in knockouts are less precise than those in controls.
3.3. Lack of mature miRNAs in the SCN caused heterogeneous phenotypes under constant light condition that correlated with their onset variations
Reasoning that less precise clocks would be more easily disrupted under challenging conditions, we placed animals under different light schedules. Since constant light condition (LL) is known to disrupt circadian rhythms (Ohta et al., 2005), activities of mice lacking Dicer in their SCN (n = 10) and controls (n = 11) were recorded under LL condition (Figure 4). We observed that control animals under LL exhibited longer period lengths that changed with time (Figure 4, Figure S3). The period lengths of circadian rhythm from day 4 to day 13 in LL were 24.79 ± 0.35 h, and became shorter to 24.31 ± 0.60 h during day 14 to day 23 in LL (T[16.15] = 2.25, p = 0.04, Welch two sample t‐test). In contrast, knockout animals showed three different phenotypes based on the activities from day 14 to day 23 in LL (Figure 4, Figure S3). Three knockouts showed ultradian rhythms of very low chi‐squared statistical amplitudes (Figure 4, KO1 as representative, and Figure S3). These three knockouts had a tendency towards higher activities over 24 h (18,993 ± 10,081 counts) than controls (7160 ± 5591 counts) but did not reach statistical significance, T(2.35) = 1.95, p = 0.17, Welch two‐sample t‐test. Four animals showed ultradian rhythms of high chi‐squared statistical amplitudes that fell into three main period lengths, one between 7 and 10 h, one between 15 and 20 h and one between 24 and 29 h (Figure 4, KO2 as representative, and Figure S3). Because the ultradian rhythms changed over time, we also analysed the data in three consecutive windows, day 3–9, day 10–16 and day 17–23 (Figure S3B, KO2–1 to KO2–4). The ultradian rhythms in the last window, day 17–23 remained similar to those during day 14–23. We could also see that the ultradian rhythms started to form on day 10–16. In terms of total activity, these four knockouts had higher activity over 24 h (18,235 ± 6010 counts) than controls (7160 ± 5591 counts), T(5.04) = 3.21, p = 0.02, Welch two‐sample t‐test. Lastly, three animals showed one main period length (24.28 ± 0.33 h, Figure 4, KO3 as representative, and Figure S3) that was similar to controls (24.31 ± 0.60 h, Welch two‐sample t‐test, p = 0.9), dubbed control‐like. Although the period length phenotypes were similar to controls, these three knockouts also had higher activity over 24 h (19,394 ± 10,158 counts) than controls (7160 ± 5591 counts), T(3.26) = 3.43, p = 0.04, Welch two‐sample t‐test. Overall, the different behavioural phenotypes of knockouts in LL suggested that they have larger inter‐individual differences than controls.
FIGURE 4.
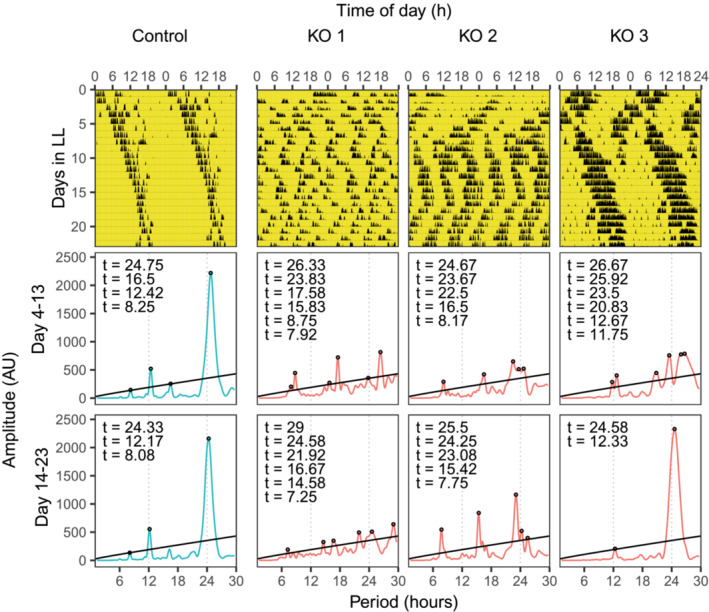
Conditional KO mice show heterogeneous phenotypes under constant light (LL). Representative actograms (upper panel) and periodograms (middle and lower panel) are shown. Yellow part denotes when light was on. Periodograms were plotted for the two durations, from day 4 to day 13, and from day 14 to day 23.
The three behavioural phenotypes of knockouts in LL ranged from less robust ultradian period length, to more robust ultradian with three main period lengths, to robust one main period length as similarly observed in controls. We, therefore, hypothesized that their phenotypes in LL correlate with the precision of their clock evaluated by onset variations. Among the 10 knockouts kept in LL, 9 of them had been recorded under DD condition. Due to the low animal number, the two phenotypes that were different to controls, i.e., those that had ultradian under LL, were grouped together, dubbed “other” phenotype. We found that compared to a control‐like group, animals in group “other” had a higher deviation of onset time to the light‐off time (Table S1, Figure S4A, 127.71 ± 46.26 minutes and 57.34 ± 19.10 minutes for “other” and control‐like, respectively), Welch two‐sample t‐test, T(6.97) = 3.22, p = 0.015. Similarly, animals in group “other” had a higher standard deviation of daily onset time (Table S1, Figure S4B, 64.42 ± 20.79 minutes) vs control‐like (26.98 ± 8.73 minutes), T(6.98) = 3.79, p = 0.007, Welch two‐sample t‐test. Thus, animals with less precise clocks exhibited less robust LL phenotypes.
3.4. Conditional Dicer knockout reduced their activities during light phase less than controls under challenging light conditions
It is known that nocturnal animals reduce activity under constant light conditions. We observed that controls reduced their daily total activities by 78.2 ± 24.3% under LL compared to LD 12:12. In contrast, knockouts reduced their activities much less than controls, by only 35.0 ± 20%, Welch two‐sample t‐test, T(9.96) = 3.48, p = 0.006. In addition, we also observed that already under LD 12:12, knockouts had higher activities during the light phase compared to the controls. To remove the difference in total activities, we calculated the ratio of activities during the light phase to those in the dark phase for each animal. Knockouts had higher ratio (0.083 ± 0.051) than controls (0.029 ± 0.033), T(12.71) = 3.06, p = 0.009, Welch two‐sample t‐test.
Reduced activity can be observed during the light phase under other light conditions such as in LD 3:3 or LD 2:2 as well. A new cohort of animals was placed under the LD 3:3 or LD 2:2 condition. When control animals were placed under LD 3:3, their activities were reduced during light phases. In addition, they chose one of the light phases as the starting time for their “night time”, and reduced their activities in the third or/and fourth light phase after this starting time. They also exhibited a period length slightly longer than 24 h (Figure 5, Figure S5, n = 5, 24.17 ± 0.33 h), although to a lesser extent than was reported previously (Oishi et al., 2015). In contrast, knockout animals showed two different phenotypes (Figure 5, Figure S5). Seven knockouts reduced their activities during light phases similarly to control animals (Figure 5, KO1 as representative, and Figure S5). In order to remove the differences in total activity levels, we calculated the ratio of activity levels during the light phases to those during the dark phases. We did not find a statistically significant difference between these seven knockouts and controls. However, in contrast to controls, these knockouts were active during all dark phases, which resulted in ultradian rhythms with period lengths close to multiples of six, i.e., period lengths of around 6, 12, 18 and 24 hours. In addition to these animals, we found five knockouts free‐ran with a period length shorter than 24 h, and more similar to their shorter free‐running period measured under DD (Figure 5, KO2 as representative, and Figure S5, n = 5, 22.25 ± 0.47 h). We again calculated the ratio of activity during the light phases to those during the dark phases for these five knockouts. We found that these knockouts had higher ratios (0.48 ± 0.20) than controls (0.22 ± 0.12), T(6.43) = 2.51, p = 0.043, Welch two‐sample t‐test.
FIGURE 5.
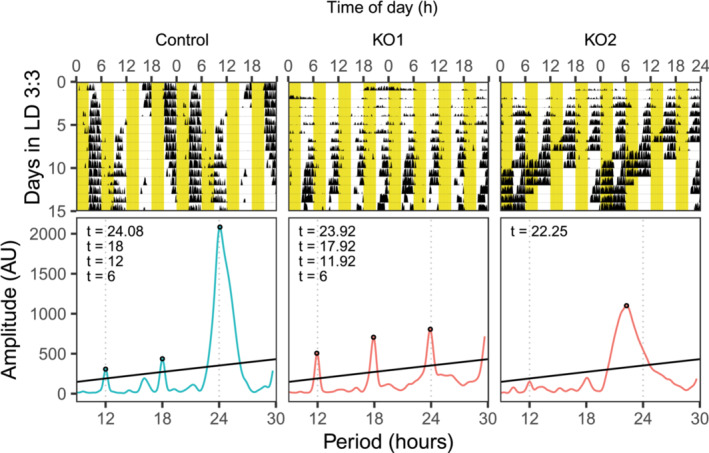
Conditional KO mice show heterogeneous phenotypes with reduced activities during light phases under LD 3:3. Representative actograms and periodograms are shown. Yellow part denotes when light was on.
When control animals were placed under LD 2:2, they seemed to switch their night starting time gradually and as a result exhibited a longer period length (n = 3, Figure 6, 24.92 ± 0.25 h), similar to what was reported under LD 4:4 (Park et al., 2012). However, knockout animals again showed heterogeneous phenotypes. Four knockouts showed ultradian rhythms with random period lengths that are not related to multiples of four (Figure 6, KO 3 to KO 6). These four animals were the ones that had ultradian rhythms that are multiples of six under LD 3:3. One knockout showed arrhythmicity (Figure 6, KO 2) while it was rhythmic with a period length of 22.42 h under LD 3:3 (Figure S5B, KO 2–5). One knockout (Figure 6, KO1) showed a shorter period length that is close to the shorter period length when the animals were placed under LD 3:3 (Figure S5B, KO 2–4). Interestingly, while controls still reduced their activities during the light‐on times, knockouts seemed to mostly ignore when the light was on. For all knockouts, the ratio of activity during light phases to those during dark phases was higher (0.66 ± 0.29) when compared to controls (0.27 ± 0.16), T(6.60) = 2.44, p = 0.046, Welch two‐sample t‐test. Overall, these results suggested that knockout animals could perceive light and respond to it by reducing their activities under some light conditions, but their responses were different and more heterogeneous than controls.
FIGURE 6.
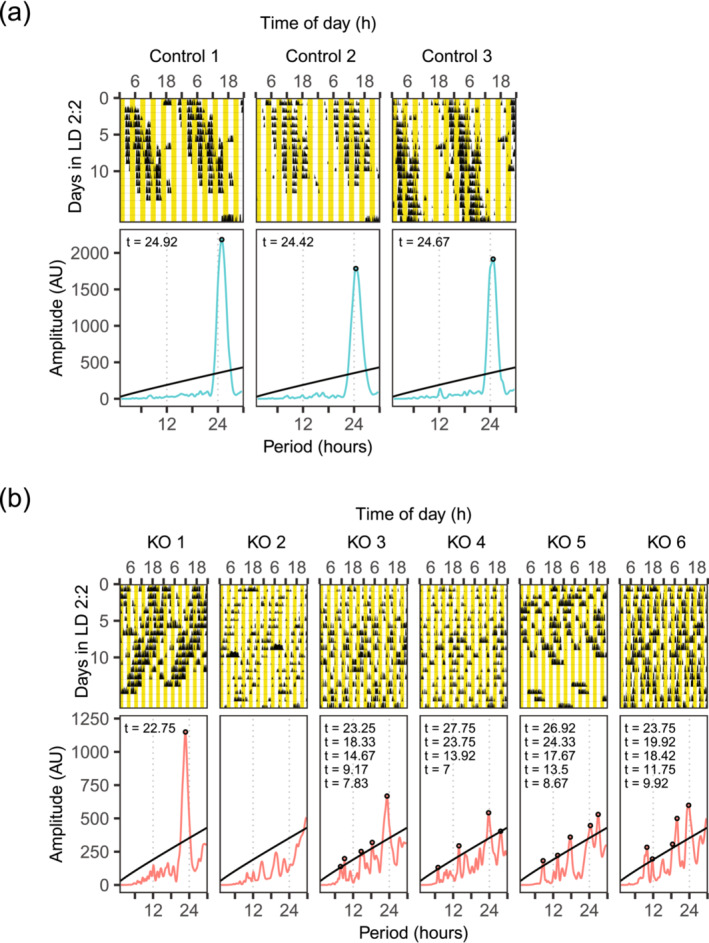
Conditional KO mice showed heterogeneous phenotypes but more towards ultradian rhythms/arrhythmic under LD 2:2. Actograms and periodograms of all animals are shown. Yellow part denotes when light was on.
3.5. SCN tissue explants from conditional Dicer knockout also showed shorter period length
To confirm if the short circadian period in the behaviour of knockout animals was due to the disrupted SCN, we bred Dicer flox ; Syt10 Cre mice with Period2::Luciferase (Per2::Luc) knock‐in mice and cultured their tissue explants from different tissues (Figure 7). SCN from knockout animals indeed showed shorter period length compared to controls (Figure 7a left panel, knockout = 24.21 ± 0.58 h, control = 24.83 ± 0.45 h, T[25.4] = 3.46, p = 0.002, Welch two sample t‐test). We next evaluated the period lengths measured in other tissues. For this analysis, we chose animals whose explants from all three tissues, SCN, lung and pituitary were available from the same animals (Figure 7a right panel). A two‐way mixed analysis of variance was performed to analyse the effect of genotype and tissue on period length. We found that the interaction effect between genotype and tissue on period length was not significant, F (2,16) = 3.43, p = 0.058. We then compared the period lengths of different tissues within each genotype. Within controls' tissue explants, there was no statistically significant difference in period lengths. Within knockouts' tissue explants, SCN ones had a shorter period length (24.59 ± 0.52 h) than lung ones, where Syt10 is not expressed (Husse et al., 2011), (25.67 ± 0.50 h, T (5) = 7.45, p < 0.01, paired t‐test). Interestingly in knockout animals, pituitary explants, where Syt10 is expressed (Roper et al., 2015), also had shorter period length (23.78 ± 0.49 h) than lung ones (25.67 ± 0.50 h, T (5) = 47.75, p < 0.001, paired t‐test). Finally, knockout pituitary explants had shorter period length (23.78 ± 0.49 h) than SCN ones (24.59 ± 0.52 h, T (5) = 5.00, p < 0.05, paired t‐test). It is worth noting that during SCN explant preparation, SCN from knockout animals detached more easily from the optic chiasm. We also evaluated the damping rate and amplitude of the SCN explants' oscillations (Figure 7b,c). Welch two‐sample t‐test revealed that there was no statistically significant difference in the damping rate between knockouts and controls (p = 0.17). However, knockout explants showed lower amplitude (30.86 ± 39.98 AU) than controls' ones (111.27 ± 85.53 AU, T[31.73] = 3.84, p = 0.0006, Welch two sample t‐test).
FIGURE 7.
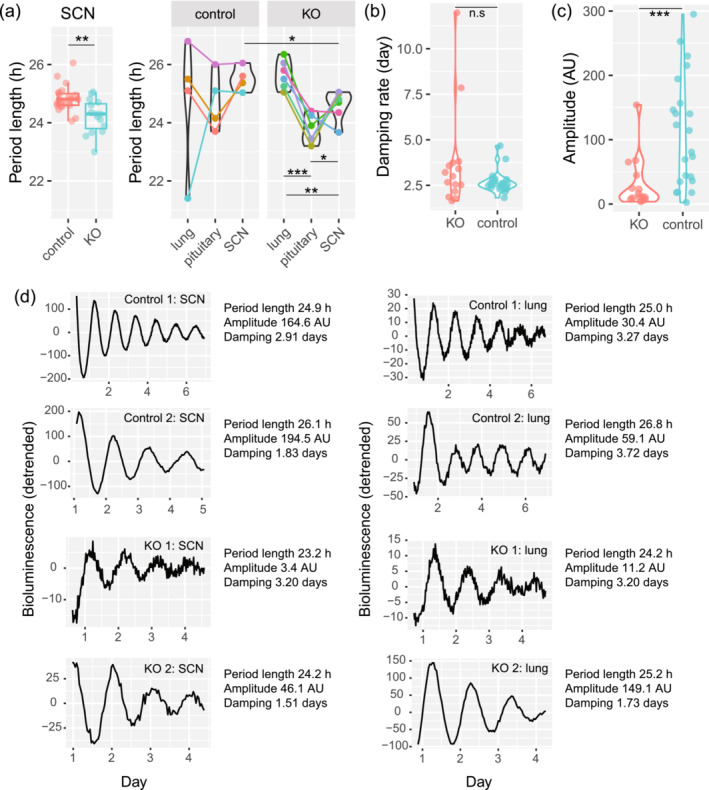
SCN explants in culture from conditional KO mice have shorter period length. (a) Period lengths of Per2::Luc are shown. Tissue explants from the same animals share the same colours and are connected by lines. (b) Damping rate of circadian oscillation. (c) Amplitude of circadian oscillation. (d) Representative rhythms from SCN and lung. SCN rhythms from KO mice were more difficult to obtain than those from controls. **: p < 0.01, ***: p < 0.001.
3.6. Inducible SCN‐specific Dicer knockout showed heterogeneous behavioural phenotypes that correlated with Cre injection efficiency
To rule out the effect of extra‐SCN expression of Syt10, as well as the effect of possible developmental process, on the period length phenotype observed in our knockout model, we induced Dicer knockout in the SCN by injecting AAV expressing hSyn‐Cre to the SCN. Induction of Cre recombination was tested 50 days after injection (two knockouts and two controls) by RT‐qPCR as has been done for conditional knockout. Expression of exon 21 (Figure S7A) was similar between knockouts (1.57 and 0.66 [AU]) and controls (0.48 and 2.08 [AU]). Splicing of exon 23 onto exon 25 normalized to reference genes (Figure S7B) was higher in knockouts (3.29 and 11.34 [AU]) vs controls (0.40 and 2.48 [AU]). When exon 23–25 splicing was normalized to the expression of exon 21, knockouts (2.10 and 17.45 [AU]) still showed higher levels than controls (0.84 and 1.20 [AU]). Similar to conditional knockouts, one should be careful when interpreting the fold change of exon 23–25 splicing products between knockouts (CT values were 33.90 and 31.09) and controls due to low levels in controls (CT values were 35.54 and 34.04). In addition, although the fold changes of exon 23–25 splicing between knockouts and controls could not be reliably compared, the fold changes for the tested inducible knockouts were lower than the tested conditional knockouts.
Another cohort of animals was injected to investigate behaviours of induced SCN‐specific Dicer knockout. Two weeks after injection, the free‐running period length under DD was assessed by recording wheel‐running activities (Figure 8, Figure S6). In contrast to animals with control injection (n = 6, free‐running period length = 23.88 ± 0.07 h), we found that hSyn‐Cre injected animals exhibited heterogeneous phenotypes. Two knockout animals showed rhythms that could be categorized as arrhythmic (one did not have any period length above the threshold, the other had one period length just above the threshold [12% above a threshold, while the values for controls were 600 ± 90%]). Three knockout animals showed period length of slightly longer than 24 hours (24.08 ± 0.14 h). Four knockout animals showed a period length shorter than 24 hours (23.44 ± 0.57 h), with one animal having 22.58 h period length.
FIGURE 8.
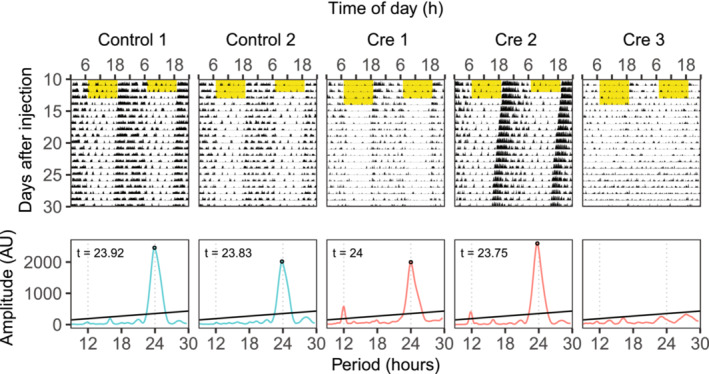
KOs induced by injection of Dicer flox mice with AVV expressing hSyn‐Cre have variable phenotypes including arrhythmic. Representative actograms and periodograms are shown. Yellow part denotes when light was on.
At the end of the recording, we confirmed injection efficiency in a subset of animals that represented each phenotype by confirming the area of expression of the reporter gene mCherry at the injection site (Figure S8). In the control group (Figure S8A), three out of six animals were imaged and they showed successful targeting in at least one lateral side of the SCN. SCN of all three imaged animals were well preserved. Thus, we did not detect any damage due to the injection procedure. For the Cre injection, we found that two animals with period lengths shorter than 24 h did not express mCherry in the SCN, and the SCNs were well preserved, suggesting that the injection missed the SCN (Figure S8B). This explained the normal period length of these animals (23.67 h and 23.75 h). For the animal with a very short period length (22.58 h) that is close to the period length of conditional knockout animals, Cre injection was confirmed in at least one side of the SCN (Figure S8C). In addition, the SCN was much less well preserved than any of the above‐mentioned animals. For all three knockouts with period lengths slightly longer than 24 h, mCherry expression was confirmed in part of the two SCNs (Figure S8D). The SCNs of these animals were less well preserved than the animals with missed or control injection, suggesting that successful Cre injection affected the SCN, potentially underlying the slightly longer period length observed. Finally, for the two animals with arrhythmic phenotype, Cre injection was confirmed on both sides of the position where the SCN should be (Figure S8E). Considering the surrounding structures, such as the optic chiasm and paraventricular nucleus, a typical SCN structure in the brain sections could not be detected. This suggests that a completely successful Cre injection resulted in the destruction of the SCN structure. Therefore, it is likely that the heterogeneous phenotypes of the Cre‐injected animals were likely caused by different injection efficiencies, with successful Dicer inactivation in both SCNs of adult mice resulting in arrhythmicity, a typical SCN lesion phenotype.
3.7. Altered SCN structure in conditional knockouts
The different SCN structures observed in inducible knockout animals prompted us to investigate the structure in conditional ones. DAPI staining of brain slices from control confirmed the presence of SCN structure (Figure S9A). In contrast, we found a much smaller SCN structure in the slices from knockout animals (Figure S9B). Therefore, together with the morphology phenotype in inducible knockouts, these results suggest that a large portion of the SCN cells died upon conditional miRNA depletion in the SCN during early development.
3.8. Preliminary observations of conditional knockout females
We observed that conditional knockout females reached extreme weight (Figure S10A at 13‐month‐old, knockout females weighed 59.9 g, while control females weighed 32.7 g, both n = 2). Knockout females were also less fertile. Over a period of one year, from 10 breeding pairs between control males and knockout females, one pair produced 4 litters, four pairs produced 2 litters and five pairs produced only one litter before pausing pregnancy. Regarding the circadian period, SCN tissue explants from KO females showed a tendency towards shorter periods compared to controls, similar direction as in males (Figure S10B, knockout SCN = 24.50 ± 0.90 h, control SCN = 25.36 ± 0.41 h, T[2.2] = 1.60, p = 0.12, Welch two sample t‐test). Surprisingly, we found that tissue explants from pituitary gland of control females (23.07 ± 0.55 h) showed shorter period compared to their SCN counterparts (25.68 ± 0.82 h, T (2) = 4.15, p = 0.03, paired t‐test), while the two tissues from control males showed similar period (male pituitary = 24.74 ± 1.02 h, male SCN = 24.83 ± 0.45 h, p > 0.05, paired t‐test). In contrast, in knockout females, tissue explants from the pituitary gland exhibited similar period length (knockout female pituitary = 24.33 ± 1.40 h) compared to their SCN counterparts (24.50 ± 0.90 h, p > 0.05, paired t‐test).
4. DISCUSSION
In the current study, we explored the circadian consequences of depleting mature miRNAs in the master clock of the mouse brain. We report here a shorter period length at the locomotor activity level which was corroborated at the tissue level in the SCN explants in conditional knockout animals. Interestingly, the shorter period length was also observed in pituitary explants, where Syt10 is also expressed. We reported previously that conditional Dicer knockout in the liver leads to a tendency towards longer period length in hepatocytes (Du et al., 2014). Thus, the shorter period length observed in explants from two brain regions suggested that the impact of Dicer knockout on period length phenotype is tissue‐specific. In addition, conditional knockouts also re‐entrained faster to a 6‐hour phase advance of light onset compared to controls, which was probably due to their faster clocks. Although not statistically significant, knockouts also appeared slower in adjusting to the phase delay compared to controls, which would be consistent with their shorter period length. This empirical observation should be confirmed in future studies with a larger number of animals.
To complement the disadvantage of the conditional knockout model (since Dicer was knocked out in the olfactory bulb, pituitary gland and testis in addition to the SCN, and potential compensation during the development process), we attempted to confirm the period length phenotype in an inducible model. Molecular validation of the recombination suggested that it is less efficient in the inducible knockout animals used for this experiment than the conditional ones. Moreover, inducible knockout animals showed heterogeneous phenotypes that seemed to correlate with injection efficiency evaluated by expression of the reporter gene mCherry.
We observed smaller SCN structures for some of the conditional knockout animals for which histological assessments were performed. The inducible knockout model also showed SCN structure changes that correlate with injection efficiency and thus probably with miRNA depletion efficiency. In the cases when injection was successful in both SCNs for two inducible knockouts, the SCN structure was hardly detected, and the animals showed arrhythmicity that resembles an SCN lesion. Overall, these results suggest that Dicer inactivation in the SCN leads to loss of SCN cells: for the conditional knockout a partial loss, and for the inducible knockout a Cre dose‐dependent cell loss.
Relatively mild behavioural phenotype and less cell loss in the SCN in conditional knockout animals suggest incomplete deletion of Dicer in the SCN of the conditional knockouts compared to the inducible ones. Indeed, it was previously shown that when using a Syt10 Cre mouse model to obtain SCN‐specific Bmal1 knockouts, BMAL1 expression was deleted in the SCN in a Cre dose‐dependent manner (Husse et al., 2011). However, due to the expression of Syt10 in the testis and that whole‐body Dicer knockout is embryonic lethal (Bernstein et al., 2003; Chen et al., 2013; Husse et al., 2011), it is not possible to obtain tissue‐specific Dicer knockout mice that are homozygous for Syt10 Cre . Complete Dicer deletion might then lead to cell apoptosis in the SCN as has been seen in the excitatory forebrain neuron‐specific Dicer knockout model (Davis et al., 2008). We cannot exclude the possibility that conditional knockouts did not show arrhythmicity under DD due to compensation by neuronal plasticity during development. This can be confirmed, for example, by knocking out Dicer in neonatal SCN slices by infection with AAV expressing hSyn‐Cre. It would be interesting then to compare different doses of AAV infection with the resulting period lengths.
Despite a large portion of the SCN structure was missing, the conditional knockout animals could still maintain rhythmic behaviours. It would be important to investigate if the whole structure, or only a specific SCN region, was reduced in size. It was shown that AVP deficiency does not affect behavioural period length in males, but increases period in females (Rohr et al., 2021). At the tissue level, SCN explants from both AVP deficient males and females had longer period length compared to control ones. However, the deletion of VIP neurons renders mice shorter period length (Mazuski et al., 2020), which is similar to the shorter period length expressed by our conditional Dicer knockout animals. For future studies, one can assess Dicer inactivation in a VIP‐Cre mouse at the level of SCN to confirm whether Dicer loss in VIP neurons is mainly responsible for our observed phenotypes.
Surprisingly, the genetic Dicer knockout also exhibited a female‐specific phenotype, namely obesity and compromised fertility. As reported in the original paper (Husse et al., 2011) and elsewhere, Syt10 is highly expressed in the SCN, although it can be found also in the olfactory bulb and in the pituitary (Husse et al., 2011; Roper et al., 2015). Therefore, we cannot rule out that the expression of Syt10 outside of the SCN contributed to the above female Dicer knockout phenotypes. Furthermore, we found that wild‐type female mice's tissue explants from the pituitary showed shorter period length compared to the SCN tissue explants from the same animals, a phenotype that was not observed in wild‐type males. In fact, sex differences in circadian phenotypes have been described previously. For example, females re‐entrain to a new light–dark cycle rapidly at proestrus than at metestrus (Pilorz et al., 2020). The fast re‐entrainment of locomotor activity is accompanied by fast clock phase shifts in peripheral tissues but not the SCN. The observed phenotypes could be conveyed also via pro‐opiomelanocortin (POMC) neurons, which receive direct input from the SCN, and have been previously implicated both in hyperphagia and obesity (Jais et al., 2020), as well as sexually dimorphic functions in the context of energy homeostasis (Wang et al., 2018). Future studies should explore the relationship between SCN and extra‐SCN regions when using Syt10 Cre mouse model, in both males and females.
Determining the responsible miRNA(s) is direct evidence to understand miRNA‐dependent regulation of the SCN functions. Recently, whole‐body deficiency in miR‐183/96/182 cluster was shown to affect locomotor activity as well as circadian oscillations at tissue levels (Zhou et al., 2021). However, in this mouse model, even though the mice are behaviourally arrhythmic under constant darkness, SCN tissue explants are rhythmic with the same period length as controls. Taking into consideration that miR183/96/182 cluster was inactivated throughout the whole body, it is therefore highly unlikely that they are driving the behavioural alterations and SCN cell loss observed in our Dicer deficient models. Nevertheless, several miRNAs, such as miR‐219 (Cheng et al., 2007), miR‐132 (Cheng et al., 2007) and miR‐17 (Gao et al., 2016), have recently been reported to be expressed rhythmically in the SCN. miR‐7a, whose predicted targets include GABA B receptor 1, and Cry2, is reported to be enriched in the SCN (Herzer et al., 2012). It might be also interesting to explore the role of miRNAs whose functions are specific to different regions in the SCN. Exploring a broad set of responsible miRNAs working synergistically and profiling transcriptomic changes upon miRNA depletion in different SCN neuronal populations will help to answer the open questions that could not be addressed in our previous studies.
Although the usage of conditional or inducible Dicer knockouts in the exploration of miRNA function allows for a topologically more accurate approach than whole‐body genetic knockouts, it does not come without limitations. Inactivation of Dicer during early development may prevent the physiological formation of neuronal networks, and even cause cell death. Moreover, Syt10 is expressed as early as E13.5 during mouse development (Thompson et al., 2014), and knocking out Syt10 has previously been reported to result in reduced neuron and synapse numbers (Cao et al., 2011; Greenman et al., 2013). We thus have to be explicit that some of our observed phenotypes in the two Dicer knockout mouse models may be potentially not specific to the absence of mature miRNAs, but rather due to a postnatally underdeveloped SCN network or loss of Syt10‐expressing neurons. In the same context, Dicer function is involved not only in miRNA biogenesis but also in the production of some non‐coding RNA such as tRNAs or tRNA‐like precursors (Babiarz et al., 2008), small nucleolar RNAs (Ender et al., 2008) or small nuclear RNA‐like viral RNAs (Cazalla et al., 2011). Therefore, we cannot exclude the possibility that the phenotypes observed here were not exclusively related to miRNAs. Our observations should thus be validated in future studies by knocking out other miRNA biosynthesis regulators such as Drosha or DGCR8, or via the use of targeted miRNA inhibitors, which could provide evidence that are more strictly related to miRNA‐based regulation.
AUTHOR CONTRIBUTIONS
Ngoc‐Hien Du: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; software; supervision; validation; visualization; writing—original draft. Konstantinos Kompotis: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; software; supervision; validation; visualization; writing—original draft. Miho Sato: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; software; supervision; validation; visualization; writing—original draft. Erica Pedron: Validation. Sabrina Androvic: Validation. Steven Brown: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; software; supervision; validation; visualization.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
PEER REVIEW
The peer review history for this article is available at https://www.webofscience.com/api/gateway/wos/peer‐review/10.1111/ejn.16605.
Supporting information
Figure S1: Confirmation of recombination by PCR analysis. (A) Schematic of the genomic locus. The Dicerflox mouse carries loxP sites flanking exon 24, which encodes most of the second RNaseIII domain of the Dicer1 gene. Primers DicerF1 and DicerDel produce a fragment of 601 nt length. (B) Gel picture of the recombined Dicer fragment detected in the olfactory bulb of the knockout (KO) animal but not in the ear. * unspecific bands that were amplified in all samples. (C) RT‐qPCR confirmation of recombination in the conditional knockout model. Left panel shows expression of exon 21 normalized to reference genes. Middle panel shows splicing level of exon 23 onto 25. Right panel shows exon 23–25 splicing level normalized to the expression of exon 21. The average expression in controls was set to 1.
Figure S2: (A) Histogram of daily onset time under LD 12:12 in comparison to ZT12 (set at 0). Daily onset time for all animals were shown (n = 10 for control, n = 5 for KO). Each animal had at least 10 days of recording. (B) Examples of onset detection (red circles) in KO and control mice. Activities were recorded using infrared detectors. Onsets of the first three days were omitted to avoid artefacts caused by the previous light schedule. (C) Histogram of daily onset time under DD in comparison to CT12 (set at 0). Daily onset time for all animals were shown (n = 22 for control, n = 14 for KO). Each animal had at least 10 days of recording.
Figure S3: Actograms and periodograms of all control (A) and knockout (B) animals under constant light condition (LL).
Figure S4: Knockout phenotype under LL correlates with their clock precision evaluated by onset variations. (A) Absolute difference to CT12 under DD. (B) Standard deviation of difference to CT12 under DD.
Figure S5: Actograms and periodograms of all control (A) and knockout (B) animals under LD 3:3. Yellow part denotes when light was on.
Figure S6: Actograms and periodograms of all control (A) and knockout (B) animals for the inducible knockout model. Yellow part denotes when light was on.
Figure S7: RT‐qPCR confirmation of recombination in the inducible knockout model. (A) Expression of exon 21 normalized to reference genes. (B) Splicing level of exon 23 onto 25. (C) Exon 23–25 splicing level normalized to the expression of exon 21.
Figure S8: Confirmation of Cre injection. (A) Representative of mCherry expression in a control brain slice. (B) Representative of mCherry expression in a missed Cre injection. (C) mCherry expression in a Cre‐injected animal with period length of 22.58 h under DD. (D) Representative of mCherry expression in a Cre‐injected animal with period length of 24.25 h. (E) Representative of mCherry expression in a Cre‐injected animal with arrhythmicity.
Figure S9: Altered SCN structure in conditional knockout animals. (A) DAPI staining of control brain slices. (B) DAPI staining of conditional knockout brain slices.
Figure S10: Gender differences in phenotypes of knockout animals. (A) Female KO were extremely overweight. Left: control, right: KO female. (B) Period length of tissue explants from control and KO females. Tissue explants from the same animals share the same colours and are connected by lines.
Table S1. Supporting Information related to Figure S4.
ACKNOWLEDGEMENTS
NH. D. designed and performed the study. K. K. performed the injections for the inducible knockout model and molecular validation of Dicer inactivation for both models. E. P. and S. A. performed the histological assessments. NH. D., K. K., and M. S. carried out the analyses and wrote the manuscript. We thank Professor David Gatfield for providing the Dicer flox mice as well as the infrastructure to carry out part of the wheel‐running experiments. We thank Professor Steven Brown for the supervision of the project and mentorship. Open access funding provided by Universitat Zurich.
Du, N.‐H. , Kompotis, K. , Sato, M. , Pedron, E. , Androvic, S. , & Brown, S. (2024). Behavioural phenotypes of Dicer knockout in the mouse SCN. European Journal of Neuroscience, 60(11), 6634–6651. 10.1111/ejn.16605
Ngoc‐Hien Du, Konstantinos Kompotis and Miho Sato contributed equally.
Edited by: Hanspeter Landolt
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Abrahamson, E. E. , & Moore, R. Y. (2001). Suprachiasmatic nucleus in the mouse: Retinal innervation, intrinsic organization and efferent projections. Brain Research, 916, 172–191. 10.1016/S0006-8993(01)02890-6 [DOI] [PubMed] [Google Scholar]
- Alvarez‐Saavedra, M. , Antoun, G. , Yanagiya, A. , Oliva‐Hernandez, R. , Cornejo‐Palma, D. , Perez‐Iratxeta, C. , Sonenberg, N. , & Cheng, H. Y. M. (2011). miRNA‐132 orchestrates chromatin remodeling and translational control of the circadian clock. Human Molecular Genetics, 20, 731–751. 10.1093/hmg/ddq519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anna, G. , & Kannan, N. N. (2021). Post‐transcriptional modulators and mediators of the circadian clock. Chronobiology International, 38, 1244–1261. 10.1080/07420528.2021.1928159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz, J. E. , Ruby, J. G. , Wang, Y. , Bartel, D. P. , & Blelloch, R. (2008). Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor‐independent, dicer‐dependent small RNAs. Genes & Development, 22, 2773–2785. 10.1101/gad.1705308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, E. , Kim, S. Y. , Carmell, M. A. , Murchison, E. P. , Alcorn, H. , Li, M. Z. , Mills, A. A. , Elledge, S. J. , Anderson, K. V. , & Hannon, G. J. (2003). Dicer is essential for mouse development. Nature Genetics, 35, 215–217. 10.1038/ng1253 [DOI] [PubMed] [Google Scholar]
- Cao, P. , Maximov, A. , & Südhof, T. C. (2011). Activity‐dependent IGF‐1 exocytosis is controlled by the Ca(2+)‐sensor synaptotagmin‐10. Cell, 145, 300–311. 10.1016/j.cell.2011.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla, D. , Xie, M. , & Steitz, J. A. (2011). A primate herpesvirus uses the integrator complex to generate viral microRNAs. Molecular Cell, 43, 982–992. 10.1016/j.molcel.2011.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R. , D'Alessandro, M. , & Lee, C. (2013). miRNAs are required for generating a time delay critical for the circadian oscillator. Current Biology, 1–10, 1959–1968. 10.1016/j.cub.2013.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, H.‐Y. M. , Papp, J. W. , Varlamova, O. , Dziema, H. , Russell, B. , Curfman, J. P. , Nakazawa, T. , Shimizu, K. , Okamura, H. , Impey, S. , & Obrietan, K. (2007). microRNA modulation of circadian‐clock period and entrainment. Neuron, 54, 813–829. 10.1016/j.neuron.2007.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby, P. , & Partch, C. L. (2020). New insights into non‐transcriptional regulation of mammalian core clock proteins. Journal of Cell Science, 133(18), jcs241174. 10.1242/jcs.241174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, T. H. , Cuellar, T. L. , Koch, S. M. , Barker, A. J. , Harfe, B. D. , McManus, M. T. , & Ullian, E. M. (2008). Conditional loss of dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. The Journal of Neuroscience, 28, 4322–4330. 10.1523/JNEUROSCI.4815-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, N.‐H. , Arpat, A. B. , De Matos, M. , & Gatfield, D. (2014). MicroRNAs shape circadian hepatic gene expression on a transcriptome‐wide scale. eLife, 3, e02510. 10.7554/eLife.02510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender, C. , Krek, A. , Friedländer, M. R. , Beitzinger, M. , Weinmann, L. , Chen, W. , Pfeffer, S. , Rajewsky, N. , & Meister, G. (2008). A human snoRNA with microRNA‐like functions. Molecular Cell, 32, 519–528. 10.1016/j.molcel.2008.10.017 [DOI] [PubMed] [Google Scholar]
- Gao, Q. , Zhou, L. , Yang, S.‐Y. , & Cao, J.‐M. (2016). A novel role of microRNA 17‐5p in the modulation of circadian rhythm. Scientific Reports, 6, 30070. 10.1038/srep30070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman, Y. , Kuperman, Y. , Drori, Y. , Asa, S. L. , Navon, I. , Forkosh, O. , Gil, S. , Stern, N. , & Chen, A. (2013). Postnatal ablation of POMC neurons induces an obese phenotype characterized by decreased food intake and enhanced anxiety‐like behavior. Molecular Endocrinology, 27, 1091–1102. 10.1210/me.2012-1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe, B. D. , McManus, M. T. , Mansfield, J. H. , Hornstein, E. , & Tabin, C. J. (2005). The RNaseIII enzyme dicer is required for morphogenesis but not patterning of the vertebrate limb. Proceedings of the National Academy of Sciences of the United States of America, 102, 10898–10903. 10.1073/pnas.0504834102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helwak, A. , Kudla, G. , Dudnakova, T. , & Tollervey, D. (2013). Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell, 153, 654–665. 10.1016/j.cell.2013.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzer, S. , Silahtaroglu, A. , & Meister, B. (2012). Locked nucleic acid‐based in situ hybridisation reveals miR‐7a as a hypothalamus‐enriched MicroRNA with a distinct expression pattern. Journal of Neuroendocrinology, 24, 1492–1504. 10.1111/j.1365-2826.2012.02358.x [DOI] [PubMed] [Google Scholar]
- Husse, J. , Zhou, X. , Shostak, A. , Oster, H. , & Eichele, G. (2011). Synaptotagmin10‐Cre, a driver to disrupt clock genes in the SCN. Journal of Biological Rhythms, 26, 379–389. 10.1177/0748730411415363 [DOI] [PubMed] [Google Scholar]
- Jais, A. , Paeger, L. , Sotelo‐Hitschfeld, T. , Bremser, S. , Prinzensteiner, M. , Klemm, P. , Mykytiuk, V. , Widdershooven, P. J. M. , Vesting, A. J. , Grzelka, K. , Minère, M. , Cremer, A. L. , Xu, J. , Korotkova, T. , Lowell, B. B. , Zeilhofer, H. U. , Backes, H. , Fenselau, H. , Wunderlich, F. T. , … Brüning, J. C. (2020). PNOCARC neurons promote hyperphagia and obesity upon high‐fat‐diet feeding. Neuron, 106, 1009–1025.e10. 10.1016/j.neuron.2020.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsbeek, A. , Palm, I. F. , la Fleur, S. E. , Scheer, F. A. J. L. , Perreau‐Lenz, S. , Ruiter, M. , Kreier, F. , Cailotto, C. , & Buijs, R. M. (2006). SCN outputs and the hypothalamic balance of life. Journal of Biological Rhythms, 21, 458–469. 10.1177/0748730406293854 [DOI] [PubMed] [Google Scholar]
- Krol, J. , Loedige, I. , & Filipowicz, W. (2010). The widespread regulation of microRNA biogenesis, function and decay. Nature Reviews. Genetics, 11, 597–610. 10.1038/nrg2843 [DOI] [PubMed] [Google Scholar]
- Lein, E. S. , Hawrylycz, M. J. , Ao, N. , Ayres, M. , Bensinger, A. , Bernard, A. , Boe, A. F. , Boguski, M. S. , Brockway, K. S. , Byrnes, E. J. , Chen, L. , Chen, L. , Chen, T. M. , Chi Chin, M. , Chong, J. , Crook, B. E. , Czaplinska, A. , Dang, C. N. , Datta, S. , … Jones, A. R. (2007). Genome‐wide atlas of gene expression in the adult mouse brain. Nature, 445, 168–176. 10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- Liu, W. , & Wang, X. (2019). Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biology, 20, 18. 10.1186/s13059-019-1629-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lück, S. , Thurley, K. , Thaben, P. F. , & Westermark, P. O. (2014). Rhythmic degradation explains and unifies circadian transcriptome and proteome data. Cell Reports, 741–751, 741–751. 10.1016/j.celrep.2014.09.021 [DOI] [PubMed] [Google Scholar]
- Malyutina, A. , Tang, J. , & Pessia, A. (2023). Drda: An R package for dose‐response data analysis using logistic functions. Journal of Statistical Software, 106, 1–26. 10.18637/jss.v106.i04 37138589 [DOI] [Google Scholar]
- Mang, G. M. , Pradervand, S. , du, N. H. , Arpat, A. B. , Preitner, F. , Wigger, L. , Gatfield, D. , & Franken, P. (2015). A neuron‐specific deletion of the microRNA‐processing enzyme DICER induces severe but transient obesity in mice. PLoS ONE, 10, e0116760. 10.1371/journal.pone.0116760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazuski, C. , Chen, S. P. , & Herzog, E. D. (2020). Different roles for VIP neurons in the neonatal and adult suprachiasmatic nucleus. Journal of Biological Rhythms, 35, 465–475. 10.1177/0748730420932073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, N. , & Cheng, H. Y. M. (2013). Micro‐managing the circadian clock: The role of microRNAs in biological timekeeping. Journal of Molecular Biology, 425, 3609–3624. 10.1016/j.jmb.2012.10.022 [DOI] [PubMed] [Google Scholar]
- Moore, R. Y. (1995). Organization of the mammalian circadian system. Ciba Foundation Symposium 183, 86–88. [PubMed] [Google Scholar]
- Mure, L. S. , Le, H. D. , Benegiamo, G. , Chang, M. W. , Rios, L. , Jillani, N. , Ngotho, M. , Kariuki, T. , Dkhissi‐Benyahya, O. , Cooper, H. M. , & Panda, S. (2018). Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science, 359(6381), eaao0318. 10.1126/science.aao0318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, H. , Yamazaki, S. , & McMahon, D. G. (2005). Constant light desynchronizes mammalian clock neurons. Nature Neuroscience, 8, 267–269. 10.1038/nn1395 [DOI] [PubMed] [Google Scholar]
- Oishi, K. , Higo‐Yamamoto, S. , Yamamoto, S. , & Yasumoto, Y. (2015). Disrupted light–dark cycle abolishes circadian expression of peripheral clock genes without inducing behavioral arrhythmicity in mice. Biochemical and Biophysical Research Communications, 458, 256–261. 10.1016/j.bbrc.2015.01.095 [DOI] [PubMed] [Google Scholar]
- Park, I. , Kim, D. , Kim, J. , Jang, S. , Choi, M. , Choe, H. K. , Choe, Y. , & Kim, K. (2020). microRNA‐25 as a novel modulator of circadian Period2 gene oscillation. Experimental & Molecular Medicine, 52, 1614–1626. 10.1038/s12276-020-00496-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, N. , Cheon, S. , Son, G. H. , Cho, S. , & Kim, K. (2012). Chronic circadian disturbance by a shortened light‐dark cycle increases mortality. Neurobiology of Aging, 33(1122), e11–1122.e22. 10.1016/j.neurobiolaging.2011.11.005 [DOI] [PubMed] [Google Scholar]
- Partch, C. L. , Green, C. B. , & Takahashi, J. S. (2014). Molecular architecture of the mammalian circadian clock. Trends in Cell Biology, 24, 90–99. 10.1016/j.tcb.2013.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, A. P. , & Hastings, M. H. (2023). The mammalian circadian time‐keeping system. Journal of Huntington's Disease, 12, 91–104. 10.3233/JHD-230571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilorz, V. , Kolms, B. , & Oster, H. (2020). Rapid jetlag resetting of behavioral, physiological, and molecular rhythms in proestrous female mice. Journal of Biological Rhythms, 35, 612–627. 10.1177/0748730420965291 [DOI] [PubMed] [Google Scholar]
- Rohr, K. E. , Telega, A. , Savaglio, A. , & Evans, J. A. (2021). Vasopressin regulates daily rhythms and circadian clock circuits in a manner influenced by sex. Hormones and Behavior, 127, 104888. 10.1016/j.yhbeh.2020.104888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper, L. K. , Briguglio, J. S. , Evans, C. S. , Jackson, M. B. , & Chapman, E. R. (2015). Sex‐specific regulation of follicle‐stimulating hormone secretion by synaptotagmin 9. Nature Communications, 6, 8645. 10.1038/ncomms9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruben, M. D. , Wu, G. , Smith, D. F. , Schmidt, R. E. , Francey, L. J. , Lee, Y. Y. , Anafi, R. C. , & Hogenesch, J. B. (2018). A database of tissue‐specific rhythmically expressed human genes has potential applications in circadian medicine. Science Translational Medicine, 10, 1–8. 10.1126/scitranslmed.aat8806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, B. , Helfrich‐Förster, C. , & Yoshii, T. (2011). A new ImageJ plug‐in ‘ActogramJ’ for chronobiological analyses. Journal of Biological Rhythms, 26, 464–467. 10.1177/0748730411414264 [DOI] [PubMed] [Google Scholar]
- Thompson, C. L. , Ng, L. , Menon, V. , Martinez, S. , Lee, C. K. , Glattfelder, K. , Sunkin, S. M. , Henry, A. , Lau, C. , Dang, C. , Garcia‐Lopez, R. , Martinez‐Ferre, A. , Pombero, A. , Rubenstein, J. L. R. , Wakeman, W. B. , Hohmann, J. , Dee, N. , Sodt, A. J. , Young, R. , … Jones, A. R. (2014). A high‐resolution spatiotemporal atlas of gene expression of the developing mouse brain. Neuron, 83, 309–323. 10.1016/j.neuron.2014.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , He, Y. , Xu, P. , Yang, Y. , Saito, K. , Xia, Y. , Yan, X. , Hinton Jr, A. , Yan, C. , Ding, H. , Yu, L. , Shu, G. , Gupta, R. , Wu, Q. , Tong, Q. , Lagor, W. R. , Flores, E. R. , & Xu, Y. (2018). TAp63 contributes to sexual dimorphism in POMC neuron functions and energy homeostasis. Nature Communications, 9, 1544. 10.1038/s41467-018-03796-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver, D. R. (1998). The suprachiasmatic nucleus: A 25‐year retrospective. Journal of Biological Rhythms, 13, 100–112. 10.1177/074873098128999952 [DOI] [PubMed] [Google Scholar]
- Xu, S. , Witmer, P. D. , Lumayag, S. , Kovacs, B. , & Valle, D. (2007). MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ‐specific miRNA cluster. The Journal of Biological Chemistry, 282, 25053–25066. 10.1074/jbc.M700501200 [DOI] [PubMed] [Google Scholar]
- Yoo, S.‐H. , Yamazaki, S. , Lowrey, P. L. , Shimomura, K. , Ko, C. H. , Buhr, E. D. , Siepka, S. M. , Hong, H. K. , Oh, W. J. , Yoo, O. J. , Menaker, M. , & Takahashi, J. S. (2004). PERIOD2::LUCIFERASE real‐time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proceedings of the National Academy of Sciences of the United States of America, 101, 5339–5346. 10.1073/pnas.0308709101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, R. , Lahens, N. F. , Ballance, H. I. , Hughes, M. E. , & Hogenesch, J. B. (2014). A circadian gene expression atlas in mammals: Implications for biology and medicine. Proceedings of the National Academy of Sciences, 2–7, 16219–16224. 10.1073/pnas.1408886111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, L. , Miller, C. , Miraglia, L. J. , Romero, A. , Mure, L. S. , Panda, S. , & Kay, S. A. (2021). A genome‐wide microRNA screen identifies the microRNA‐183/96/182 cluster as a modulator of circadian rhythms. Proceedings of the National Academy of Sciences of the United States of America, 118, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Confirmation of recombination by PCR analysis. (A) Schematic of the genomic locus. The Dicerflox mouse carries loxP sites flanking exon 24, which encodes most of the second RNaseIII domain of the Dicer1 gene. Primers DicerF1 and DicerDel produce a fragment of 601 nt length. (B) Gel picture of the recombined Dicer fragment detected in the olfactory bulb of the knockout (KO) animal but not in the ear. * unspecific bands that were amplified in all samples. (C) RT‐qPCR confirmation of recombination in the conditional knockout model. Left panel shows expression of exon 21 normalized to reference genes. Middle panel shows splicing level of exon 23 onto 25. Right panel shows exon 23–25 splicing level normalized to the expression of exon 21. The average expression in controls was set to 1.
Figure S2: (A) Histogram of daily onset time under LD 12:12 in comparison to ZT12 (set at 0). Daily onset time for all animals were shown (n = 10 for control, n = 5 for KO). Each animal had at least 10 days of recording. (B) Examples of onset detection (red circles) in KO and control mice. Activities were recorded using infrared detectors. Onsets of the first three days were omitted to avoid artefacts caused by the previous light schedule. (C) Histogram of daily onset time under DD in comparison to CT12 (set at 0). Daily onset time for all animals were shown (n = 22 for control, n = 14 for KO). Each animal had at least 10 days of recording.
Figure S3: Actograms and periodograms of all control (A) and knockout (B) animals under constant light condition (LL).
Figure S4: Knockout phenotype under LL correlates with their clock precision evaluated by onset variations. (A) Absolute difference to CT12 under DD. (B) Standard deviation of difference to CT12 under DD.
Figure S5: Actograms and periodograms of all control (A) and knockout (B) animals under LD 3:3. Yellow part denotes when light was on.
Figure S6: Actograms and periodograms of all control (A) and knockout (B) animals for the inducible knockout model. Yellow part denotes when light was on.
Figure S7: RT‐qPCR confirmation of recombination in the inducible knockout model. (A) Expression of exon 21 normalized to reference genes. (B) Splicing level of exon 23 onto 25. (C) Exon 23–25 splicing level normalized to the expression of exon 21.
Figure S8: Confirmation of Cre injection. (A) Representative of mCherry expression in a control brain slice. (B) Representative of mCherry expression in a missed Cre injection. (C) mCherry expression in a Cre‐injected animal with period length of 22.58 h under DD. (D) Representative of mCherry expression in a Cre‐injected animal with period length of 24.25 h. (E) Representative of mCherry expression in a Cre‐injected animal with arrhythmicity.
Figure S9: Altered SCN structure in conditional knockout animals. (A) DAPI staining of control brain slices. (B) DAPI staining of conditional knockout brain slices.
Figure S10: Gender differences in phenotypes of knockout animals. (A) Female KO were extremely overweight. Left: control, right: KO female. (B) Period length of tissue explants from control and KO females. Tissue explants from the same animals share the same colours and are connected by lines.
Table S1. Supporting Information related to Figure S4.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


