Abstract
orf101 is a late gene of Autographa californica nucleopolyhedrovirus (AcMNPV). It encodes a protein of 42 kDa which is a component of the nucleocapsid of budded virus (BV) and occlusion-derived virus (ODV). To reflect this viral localization, the product of orf101 was named BV/ODV-C42 (C42). C42 is predominantly detected within the infected-cell nucleus: at 24 h postinfection (p.i.), it is coincident with the virogenic stroma, but by 72 h p.i., the stroma is minimally labeled while C42 is more uniformly located throughout the nucleus. Yeast two-hybrid screens indicate that C42 is capable of directly interacting with the viral proteins p78/83 (1629K) and ODV-EC27 (orf144). These interactions were confirmed using blue native gels and Western blot analyses. At 28 h p.i., C42 and p78/83 are detected in two complexes: one at approximately 180 kDa and a high-molecular-mass complex (500 to 600 kDa) which also contains EC27.
Occlusion-derived virus (ODV) of Autographa californica nucleopolyhedrovirus (AcMNPV) initiates primary infection through gut cells of the susceptible insect, where the columnar cells are the primary sites of infection (13, 31). Columnar cells, however, are in a differentiated state (7). Thus, AcMNPV must have the ability to progress the infected columnar cell from G0 to G1 and further progress the cell to a phase conducive to viral DNA replication (S phase). These events occur rapidly: in virus-challenged Trichoplusia ni larvae, midgut infection is detectable by 4 h postinoculation and systemic infection is detected by 12 h (31).
Viral regulation of the host cell cycle could be accomplished through several mechanisms. One mechanism is the early transcription-translation of viral genes which interact with cellular proteins or protein complexes. Some evidence suggests that AcMNPV may be utilizing this mechanism. The ie-2 gene is sufficient to arrest transfected Sf9 cells in S phase: the arrested cells do not undergo mitosis, have abnormally large nuclei, and contain greater than 4N DNA content (19). Infected Sf9 cells, however, show only a transient increase of cells in S phase, and the cells quickly progress to G2/M phase, where they remain arrested throughout infection (4). A second viral strategy to regulate the host cell cycle is to present, as structural components of the virus, a protein(s) or protein complexes that interact with cellular proteins immediately upon infection. For AcMNPV, the structural protein ODV-EC27 (EC27) is a candidate for such a strategy: it has amino acid similarity with cellular cyclins, and it coprecipitates with cellular cyclin kinases (1, 4). Its introduction into the cell at the time of infection may allow it to interact immediately with cellular cyclin kinases or function as a cyclin homologue in a manner analogous to that of other viral cyclin homologues. In our continuing study of the function of AcMNPV structural proteins and their interactions with host cell proteins, we have identified a new structural protein of budded virus (BV) and ODV, BV/ODV-C42 (C42; orf101), and show that this protein is present in complexes that also contain the viral proteins EC27 and p78/83.
MATERIALS AND METHODS
Insect cell line and virus.
Spodoptera frugiperda IPLB-Sf21-AE clonal isolate 9 (Sf9) cells were cultured in suspension at 27°C in TNMFH medium (26) supplemented with 10% fetal bovine serum and 1% pluronic F68 (complete medium). AcMNPV (strain E2) was used to infect cells at a defined multiplicity of infection (MOI), with time zero set at the time of virus addition. After 1 h of adsorption, cells were washed and resuspended in fresh, complete medium.
Primer extension.
Sf9 cells were infected (MOI, 10), and mRNA was purified using the Poly(A)Pure mRNA isolation kit (Ambion, Inc., Austin, Tex.). Two oligonucleotides (PE1, 5′-GCATAGGCTCGTCTATTTTTAACCGC-3′; PE2, GTCTAACACGTTGACCGTTTCG) were 5′ end labeled with T4 polynucleotide kinase and probed against 3 μg of cellular mRNA. Extension products were generated using Superscript II reverse transcriptase (Life Technologies, Gaithersburg, Md.) in the presence of 2.3 μg of actinomycin D, 28 U of RNasin, and 0.5 mM deoxynucleoside triphosphates for 60 min at 45°C and then precipitated and digested with 0.1 N NaOH for 30 min at room temperature, resuspended in 80% formamide loading buffer, and boiled for 3 min before being loaded onto a denaturing gel (7.0 M urea, 6% polyacrylamide, 100 mM Tris-borate, 20 mM EDTA, pH 8.3). Transcript initiation sites were identified by comparison with a DNA sequence by using the same oligonucleotide primers as used for generating the extension products and run concurrently on the denaturing gel with the extension products.
Western blot analysis of infected cells, virus, and virus fractionation.
Sf9 cells were infected with AcMNPV (MOI, 10), and at the indicated time postinfection (p.i.), cells were collected and washed once with phosphate-buffered saline (PBS). Cell pellets were resuspended in PBS containing protease inhibitors (20 μg of leupeptin/ml, 20 μg of aprotinin/ml, 20 μg of pepstatin A/ml, 1 mM E64). Cells were broken by sonication, and protein concentration was determined by the method of Bradford (3).
BV was purified from the cell culture supernatant of infected cells (36 h p.i.) by the technique described in the work of Braunagel and Summers (5). Occlusions were purified from infected Sf9 cells by a modified technique of Summers and Egawa (25) and Braunagel and Summers (5). The occlusions were then purified from the pellet by using sucrose gradients as described in the work of Braunagel and Summers (5). ODV was purified from occlusions, and purified BV and ODV were fractionated into envelope and nucleocapsid preparations according to the method described in the work of Braunagel and Summers (5). The purified virus and viral fractions were analyzed using known viral markers (p39, E66, and gp67) to verify purity.
Vertical slab gel electrophoresis was performed according to the method of Laemmli (14). A 4% stacking gel was used above a 12.5% separating gel. Samples were incubated in 1.5% sodium dodecyl sulfate (SDS)–0.5% β-mercaptoethanol–25 mM Tris-HCl (pH 6.8)–7% glycerol for 15 min at 65°C before loading and separated by electrophoresis. Western analysis was performed using protein blotted onto polyvinylidene difluoride (PVDF) membranes (Immobilon-P; Millipore, Bedford, Mass.). The membranes were blocked, and antibody binding was performed in TTBS-BLOTTO (150 mM NaCl, 10 mM Tris, 1 to 3% nonfat dry milk, 0.1% Tween 20, pH 8.0). Primary antibody was bound overnight, blots were washed three times (TTBS), and secondary antibody (anti-rabbit, horseradish peroxidase-linked immunoglobulin G [IgG]) was reacted for 1 h (1:10,000; room temperature). Blots were washed three times with Tris-buffered saline (15 min) and treated for 1 min with Renaissance chemiluminescence reagent (NEN Life Science Products, Boston, Mass.), and a positive reaction was detected by exposure to X-ray film.
Yeast two-hybrid library construction and screen.
Sf9 cells were infected (MOI, 20), and at 18 or 24 h p.i., cells were collected and mRNA was isolated by using either the Poly A Tract System 1000 (Promega, Madison, Wis.) or the Poly(A)Pure mRNA isolation kit (Ambion). The cDNA library was generated using the Two Hybrid cDNA Construction kit (Clontech, Palo Alto, Calif.), and thus, the genomic fragments were cloned into the vector pGAD10. The libraries were amplified, and the resulting titers of the amplified libraries were as follows: at 18 h p.i., 1.31 × 1012, and at 24 h p.i., 3.25 × 1013. To harvest large quantities of DNA from each library, a 1-ml aliquot of amplified library was diluted and grown on 200 Luria broth-ampicillin-supplemented plates (150-mm diameter), bacteria were harvested, and plasmid DNA was purified using the Plasmid Giga kit (Qiagen, Valencia, Calif.). Transfection and chromogenic reactions were performed according to the manufacturer's protocol (Clontech).
Clones for yeast two-hybrid screens.
orf144 (EC27), 1629K (p78/83), and orf101 (C42) were cloned into the yeast binding domain vector pAS2-1. Both orf144 and orf101 were amplified from genomic fragments using the appropriate PCR primers and cloned into pAS2-1 such that the inserted gene was placed in frame using the EcoRI site provided by the multicloning region. The fusion region for the EC27 clone was EFELGTRGS-Met-144, while the fusion for C42 was EFR-Met-101 (fusion amino acids E and F were provided by the EcoRI cloning site, and the first amino acid encoded by the cloned gene is underlined). The resultant insert was fully sequenced to verify frame and to assure the fidelity of the polymerase reaction. A copy of the AcMNPV E2 strain 1629K gene was provided by C. Richardson (McGill University, Montreal, Quebec, Canada [28]), and it was directly subcloned into pAS2-1. The resultant clone had the Met start provided by an NdeI site with an N-terminal His tag sequence; thus, the fusion region was MHHHHHHLVPRGSGI-Thr-1629K.
Production of antibodies.
For C42, orf101 was digested from pAS2-1 (described above) and cloned into pGEX 5X-1 using a 5′ EcoRI site and a 3′ SalI site. The frame was established by the EcoRI site; thus, the fusion site was EFR-Met-101. The clone was transformed into BL21 cells, and protein was induced using 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and either purified by excision from an SDS-polyacrylamide gel (denatured) or purified using glutathione-agarose beads (native) according to the manufacturer's instructions (Pharmacia Biotech, Arlington Heights, Ill.). After antigen purification, three rabbits were injected for each antigen (native or denatured). The first injection (intramuscular) was approximately 200 μg of protein diluted with RIBI adjuvant (MPL+TDM+CWS emulsion; RIBI Immunochem Research Inc., Hamilton, Mont.). Two additional injections followed at 28-day intervals, and each injection contained one-half the amount of the previous injection. Twelve days after the third injection, the sera were tested, and 2 days later, animals were exsanguinated and plasma was collected.
Microscopy. (i) Confocal analysis.
Sf9 cells were infected (MOI, 10) and collected at the appropriate time. The harvested cells were washed once with Grace's medium, and 2.1 × 105 cells were transferred and allowed to attach to the slide using a one-well Cytofuge concentrator (StatSpin Technologies, Norwood, Mass.). The cells were fixed with 3.7% paraformaldehyde in PBS (20 mM phosphate, 140 mM NaCl, pH 7.2) for 10 min at room temperature. The cells were washed three times with PBS, permeabilized by sequential treatments with methanol (10 min) and Triton X-100 (0.5%, 10 min), and washed three times with PBS. The cells were then blocked for 1 h with blocking solution (1% chicken serum and 3% bovine serum albumin in PBS) and incubated overnight at 4°C with primary antibody (C42; pAb 10910; 1:2,000). Primary antibody was removed by washing three times with PBS, the cells were then incubated with Alexa-488-linked anti-rabbit IgG for 2 h (1:2,000; Molecular Probes, Eugene, Oreg.) and washed three times with PBS, and DNA was stained with DAPI (4′,6-diamidino-2-phenylindole; 0.1 μg/ml in PBS; 5 s). Cells were washed three times with PBS and viewed with a Zeiss Axiovert CARV 135 confocal microscope. Hundreds of cells were viewed, and approximately 15 fields of view containing 30 to 50 cells were collected, along with representative single cells. Representative cells are shown. Z-stack sections were collected at 0.75-μm intervals. Deconvolution was performed using Zeiss KS 4.0 software.
(ii) Immunoelectron microscopy (IEM).
Sf9 cells were infected (MOI, 20), and virus was adsorbed for 1 h, removed, and replaced with complete medium. At the designated time p.i., cells were pelleted and fixed and ultrathin sections were prepared for antibody reactions as described by Hong et al. (11). Sections were blocked with TTBS-bovine serum albumin (1%) for 1 h, reacted with antibody (C42; pAb 10910; 1:1,000; overnight; 4°C), washed with Tris-buffered saline, and reacted with secondary anti-rabbit, gold-conjugated IgG (30 nm, 1:15 dilution; Electron Microscopy Sciences, Washington, Pa.) for 1 h at room temperature. The sections were stained with uranyl acetate (2) and lead citrate (27) and visualized with a Zeiss 10C transmission electron microscope.
Blue native gel electrophoresis.
Blue native gel electrophoresis was adapted from the work of Schagger and von Jagow (24). Sf9 cells were infected (MOI, 20), collected at 28 h p.i. (2 × 107 cells per sample), and washed with PBS, and the pellet was frozen until use. The pellet was resuspended in water containing 200 U of DNase I, passed through a 27-gauge needle 10 times, sonicated for 45 s, and incubated on ice for 2 h. The cells were then microcentrifuged (16,000 × g) for 10 min (4°C), and the soluble fraction (supernatant) was transferred to a fresh tube and mixed with an equal volume of 3× gel buffer (1.5 M aminocaproic acid, 150 mM Bis-Tris, pH 7.0). To remove intact viral nucleocapsids, the soluble fraction was centrifuged (100,000 × g, 30 min; Beckman TLA100, 50,000 rpm), and to further remove large protein complexes (i.e., ribosomes [30]), the soluble fraction was collected and further centrifuged (400,000 × g, 15 min; Beckman TLA100, 95,000 rpm). The supernatant was collected, and for every 200 μl of sample, 10 μl of dye was added (5% Serva Blue G, 500 mM aminocaproic acid). A linear 6 to 13% acrylamide gradient gel was generated (6%, 1.43 ml of 48:1.5 acrylamide:bisacrylamide, 4 ml of 3× gel buffer, 6.57 ml of H2O, 38 μl of 10% ammonium persulfate, 1.2 μl of TEMED [N,N,N′,N′-tetramethylethylenediamine]; 13%, 3.14 ml of acrylamide:bisacrylamide, 4 ml of 3× gel buffer, 2.4 ml of glycerol, 2.46 ml of H2O, 38 μl of 10% ammonium persulfate, 12 μl of TEMED). After polymerization, the gel was overlaid with a 4% stack (0.6 ml of acrylamide:bisacrylamide, 2.5 ml of 3× gel buffer, 4.33 ml of H2O, 60 μl of 10% ammonium persulfate, 6 μl of TEMED). The gel was run with the cathode buffer containing 50 mM Tricine, 15 mM Bis-Tris, and 0.02% Serva Blue G (pH 7.0) and the anode buffer containing 50 mM Bis-Tris, pH 7.0. Samples were loaded (30 μl per well) and run at a constant voltage (200 V) for 2 h, at which time the cathode buffer was discarded and replaced with the same buffer minus Serva Blue G. The gel was then run overnight at 4°C.
After completion, the gel was transferred to Immobilon P using Western blotting procedures and probed with antibody (EC27, pAb 7351, 1:1,000; C42, pAb 10910, 1:10,000; and p78/83 [provided by C. Richardson], 1:2,000). All primary antibody reactions were performed overnight (4°C), secondary reactions were performed using the appropriate secondary horseradish peroxidase-linked IgG antibody (1:10,000) for 2 h at room temperature, reaction mixtures were treated for 1 min with Renaissance chemiluminescence reagent (NEN Life Science Products), and positive reactions were detected by exposure to X-ray film.
RESULTS
orf101 is a late gene that codes for a highly conserved 42-kDa structural protein of the baculovirus nucleocapsid.
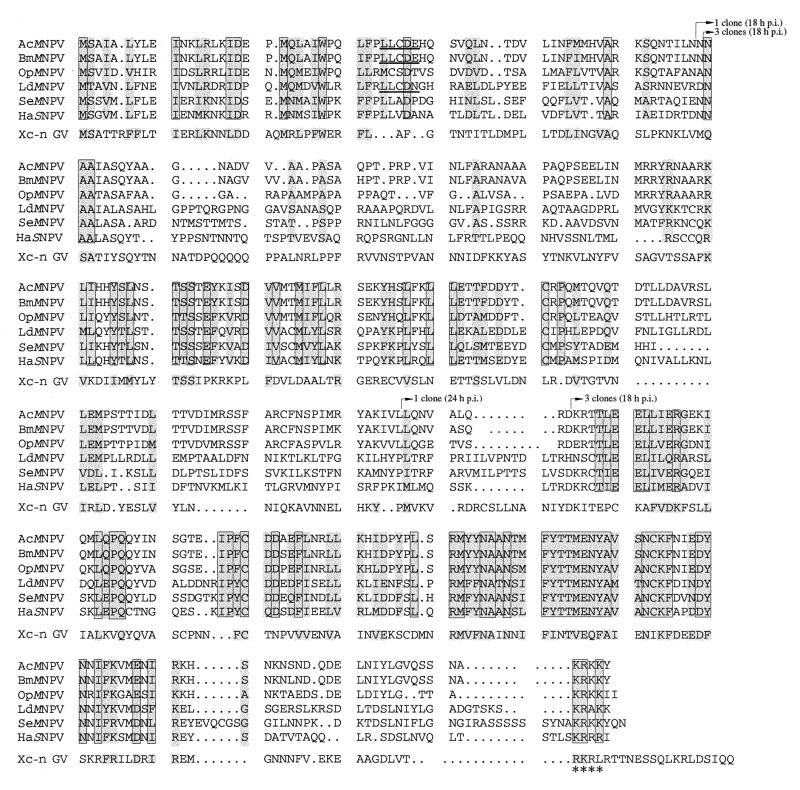
A homologue of orf101 is found in all the baculovirus genome databases, including the most divergent genome, Xestia c-nigrum granulovirus (Xc-n GV). A comparison of the predicted amino acid sequences shows that several regions of the orf101 gene product are highly conserved among the nucleopolyhedroviruses (Fig. 1). Three conserved regions are significant, the N-terminal region (amino acids 1 to 45) and amino acids 130 to 200 and 280 to 360, and these regions share the highest degree of conservation with Xc-n GV. A conserved classical putative nuclear localization signal (KRKK) is located at the C terminus of all the proteins (Fig. 1, asterisk) and SOSUI analysis (10) predicts that the protein product of orf101 would be soluble. The canonical binding motif for the family of pocket proteins (pRB, p130, and p107 [reviewed in reference 9]) is found in the N-terminal conserved region of AcMNPV and Bombyx mori nucleopolyhedrovirus (LxCxE [Fig. 1, underlined]). This motif is not found in Lymantria dispar nucleopolyhedrovirus, Orgyia pseudotsugata nucleopolyhedrovirus (OpMNPV), Spodoptera exigua nucleopolyhedrovirus, Helicoverpa armigera nucleopolyhedrovirus, or Xc-n GV.
FIG. 1.
Amino acid sequence comparison of orf101. Identical amino acids of the nucleopolyhedroviruses are shaded and outlined, while conservative changes are shown in shaded regions. Because it is the most divergent, Xc-n GV C42 was treated separately, and both identical and conserved amino acids are shaded. The clones that were identified as interacting with orf144 using yeast two-hybrid library screening are noted with arrows above the sequence. The location of the LxCxE motif (canonical binding sequence for pocket proteins) is underlined, while the nuclear localization signal (KRKK) is noted with asterisks. Rules used to assign conservation are as follows: A = G = S = T, V = L = I = M = F = Y = W, N = Q = D = E, and R = K = H. Accession numbers: AcMNPV, L22858 (nucleotides 88004 to 86921); BmMNPV (B. mori nucleopolyhedrovirus), L33180 (nucleotides 81679 to 80591); OpMNPV, U79530 (nucleotides 85649 to 85485); LdMNPV (L. dispar nucleopolyhedrovirus), U58676 (nucleotides 101348 to 100203); SeMNPV (S. exigua nucleopolyhedrovirus), AF169823 (nucleotides 61806 to 62972); HaSNPV (H. armigera single nucleopolyhedrovirus), AF271059 (nucleotides 82544 to 83653); and Xc-n GV, AF162221 (nucleotides 86182 to 87300).
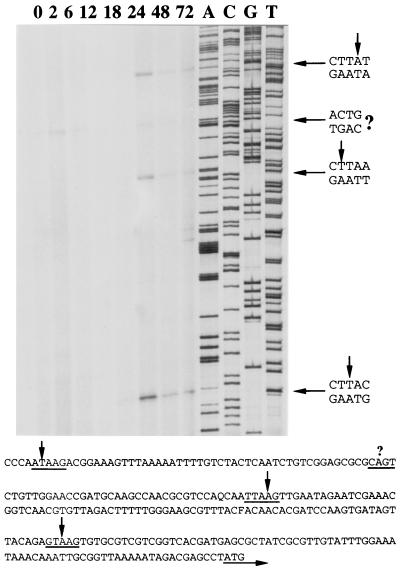
5′-primer extension analysis was performed to determine the temporal pattern of orf101 transcription. Because three late transcription initiation motifs (TAAG) are present from −80 to −254, two primers were designed and tested using the appropriate sequencing ladder to define nucleotide initiation sites; all primers gave the same results. In Fig. 2, use of a representative analysis and the first ATG as the putative sequence for translation initiation shows that all of the TAAG sequences serve as initiation sites (−80, −164, and −254). Transcripts were detected at 18 h p.i., with levels increasing by 24 h p.i., and were still detectable at 72 h p.i. There was an early consensus initiation sequence (CAGT) located at −205, and the primer extension results suggested that this site was recognized by 2 h p.i.; thereafter, transcript levels decreased rapidly but were still detectable at 24 h p.i. We report these data in Fig. 2 with a question mark because trace amounts of transcripts of a similar size were also detected in the uninfected control. The use of other primers, various gel exposures, or increased amounts of RNA in the analysis did not give more conclusive data. We conclude from this experiment that orf101 is a late gene with transcripts being produced from all three upstream TAAG motifs, and that transcripts may be produced at low levels early in infection.
FIG. 2.
Primer extension analysis of orf101. Transcription initiation was mapped using 3 μg of mRNA isolated from infected-cell extracts. Two oligonucleotides were used to accurately assign nucleotide initiation sites, although only the results from oligonucleotide 1 are shown here. The initiation sites of the extension products are indicated to the right and in the sequence shown below. Because of the high degree of amino acid sequence conservation (Fig. 1), the first ATG is tentatively assigned as the start codon.
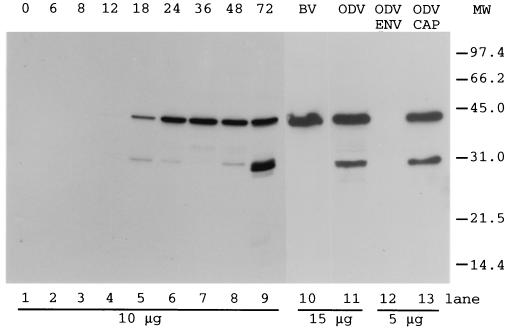
orf101 was cloned into pGEX 5X-1, and rabbit polyclonal antibodies to two fusion antigens (purified from SDS-polyacrylamide gels and purified in a nondenatured state using glutathione-agarose) were produced. Antibodies reacted with whole-cell lysates that were collected throughout the time course of infection detected a predominant band of 42 kDa (relative molecular mass) (Fig. 3, lanes 1 to 9) that correlated with the predicted molecular mass for orf101 (41.5 kDa). This band was detected at low levels by 12 h p.i., increased by 18 h p.i., and continued to accumulate through 72 h p.i. A second band of a lower molecular mass (relative molecular mass, 30 kDa) was also detected during the time course, with the greatest accumulation occurring late in infection (Fig. 3, lane 9). The quantity of the lower-molecular-mass product varied when different preparations of infected-cell lysates were analyzed, always showing the greatest amount in cell lysates collected after 48 h p.i. We do not know if this band represents alternate initiation from an internal methionine, a degradation product, or a modified but functional form.
FIG. 3.
Temporal analysis of appearance and accumulation of C42. Preparations of uninfected- and infected-cell extracts collected at various times p.i. (noted above lanes 1 to 9) and purified BV and ODV and respective envelope (ENV) and nucleocapsid (CAP) preparations were separated by SDS-polyacrylamide gel electrophoresis, transferred to a PVDF membrane, and tested with antisera to C42 (pAb 10910, 1:5,000). Sample concentrations are noted on the bottom (micrograms), and migration of molecular weight (MW; in thousands) markers is noted on the right. The purity of the envelope and nucleocapsid preparations was verified using antibodies to the marker proteins ODV-E66, gp67, and p39 (data not shown).
The 42-kDa protein was present in both purified BV and ODV (Fig. 3, lanes 10 and 11). The amount of the 30-kDa form varied with the ODV preparation, while BV preparations contained very small amounts, nearly undetectable in most preparations (Fig. 3, lane 10). When ODV was fractionated into envelope and nucleocapsid preparations, the 42-kDa protein was detected only in the nucleocapsid fraction (Fig. 3, lanes 12 and 13), which was also true for BV (data not shown). The purity of the viral fractionation was verified by using antibodies to the marker proteins p39 (capsid), gp67 (BV-env), and ODV-E66 (ODV-env) (data not shown). The product of orf101 is named BV/ODV-C42 (C42), to reflect its relative molecular mass (42 kDa) and localization in the nucleocapsids of BV and ODV.
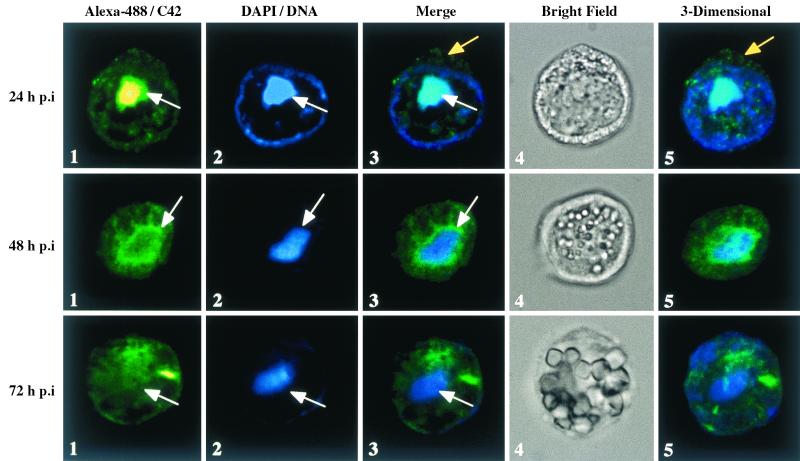
Western blot analysis demonstrated that C42 was abundantly detected in infected cells at 24 h p.i. (Fig. 3). At 24 h p.i., confocal microscopy showed C42 to be predominantly located within the DNA-rich virogenic stroma (Fig. 4, 24 h p.i., columns 1 to 5, white arrow). Low levels of C42 were also detected within the cytoplasm, with this labeling most visible in the merged view and the three-dimensional reconstruction of the Z sections (24 h p.i., columns 3 and 5, yellow arrow). By 48 h p.i., the general character of C42 localization was at the edge of the virogenic stroma; however, C42 was also detected in a more diffuse pattern throughout the nucleus (Fig. 4, 48 h p.i., columns 1 to 5, arrow). By 72 h p.i., C42 was detected throughout the nucleus with very little associated with the virogenic stroma (Fig. 4, 72 h p.i., columns 1 to 5). The three-dimensional reconstruction shows the overall representation of the cellular localization of C42 at each time point (Fig. 4, column 5).
FIG. 4.
Confocal analysis of C42 localization in infected Sf9 cells. Infected Sf9 cells were collected at 24, 48, and 72 h p.i. and reacted with primary antibody to C42 (pAb 10910) and secondary antibody labeled with Alexa-488 IgG (Alexa-488; column 1). DNA was stained using DAPI (column 2). The merged image includes Alexa-488 and DAPI (column 3), while the bright-field image shows the gross structure of the cell (column 4). Z-stack sections from representative cells are shown, and column 5 shows a single view of the three-dimensional reconstruction. The white arrow indicates the location of the virogenic stroma, while the yellow arrow (24 h p.i., columns 3 and 4) shows the cytoplasmic label of C42.
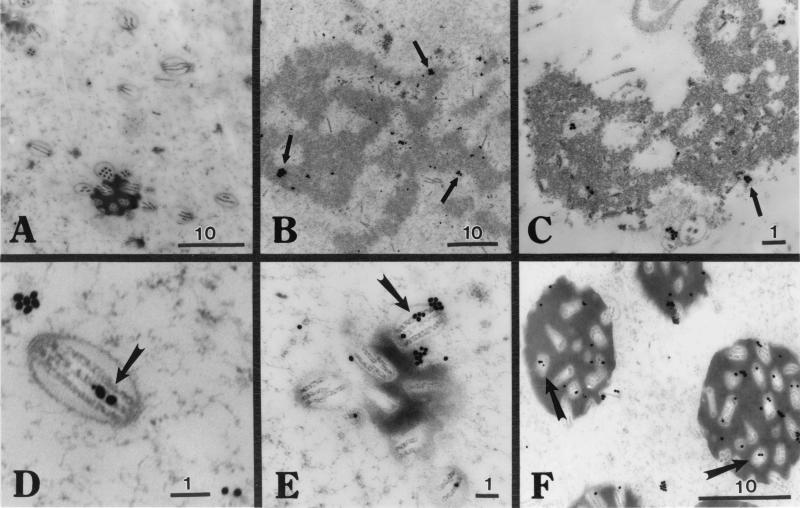
Fine-structure localization of C42 (IEM) at 48 h p.i. detected gold-labeled clusters of C42 at the periphery and clear areas within the virogenic stroma (Fig. 5B, arrow), while less label was detected in association with the virogenic stroma at 72 h p.i. (Fig. 5C, arrow). These data are consistent with observations generated using confocal microscopy. Gold-labeled C42 was detected with nucleocapsids of ODV (Fig. 5D, arrow); this label was easier to see when the ODV nucleocapsids were grouped in the process of being occluded (Fig. 5E, arrow) or in mature occlusions (Fig. 5F, arrows). The preimmune serum control showed limited background cross-reactivity when tested against infected cells (Fig. 5A).
FIG. 5.
IEM analyses of C42 localization in infected Sf9 cells. Infected Sf9 cells were collected at 24, 48, and 72 h p.i. and processed for IEM. Primary antibody was C42 (pAb 10910), and secondary antibody was anti-rabbit 30-nm-gold-labeled IgG. (A) Preimmune control serum (48 h p.i.). (B and C) Labeling of virogenic stroma (48 h p.i. [B] and 72 h p.i. [C]). (D to F) Labeling of C42 at the nucleocapsid of ODV (48 h p.i.). Size designations are in micrometers. Arrows show immunogold labeling of C42.
C42 is present in a complex with p78/83 and EC27.
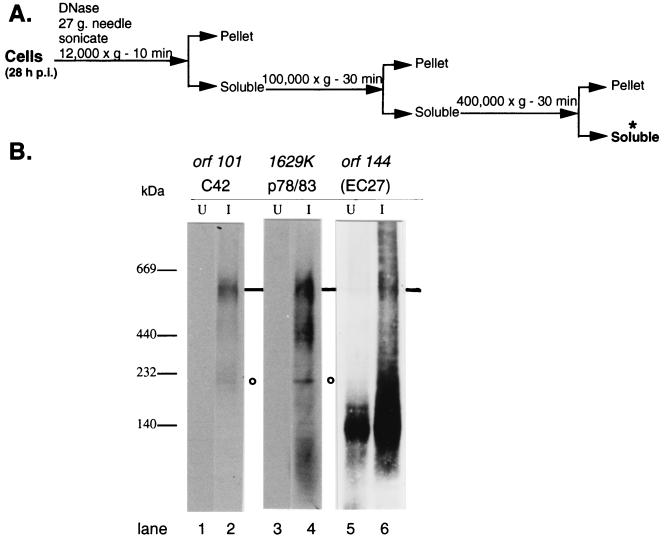
orf101 (C42) was first drawn to our attention when a yeast two-hybrid screen of infected-cell cDNA libraries (18 and 24 h p.i.), using orf144 (EC27) as bait, identified eight interacting clones encoded by orf101 (noted in Fig. 1, arrows). In a similar screen, when 1629K (p78/83) was used as bait to probe a 24 h-p.i. Sf9 cDNA library, this screen identified orf101 (C42) seven times. Sequence analysis showed that all seven orf101 library clones started at the first methionine. To confirm the yeast two-hybrid predictions that C42:EC27 and C42:p78/83 interact directly to form complexes, we needed a procedure that would (i) identify protein interactions which occur prior to virus assembly and/or (ii) remove assembled nucleocapsids and virus, while retaining conditions that allow protein complexes to remain intact. For this, we used blue native electrophoresis. This technique separates both acidic and basic protein complexes and provides a tentative assignment of the complex's molecular mass (17, 23, 24). We chose to use a 28-h-p.i. infected-cell sample because EC27, C42, and p78/83 are present, and studies using other viral structural proteins (ODV-E66, ODV-E25, and FP25K) show that these proteins are being translated at their maximal rate (20). Figure 6A gives a summary of sample preparation. After infected cells were harvested, DNA was degraded (DNase, sonication, and shear forces), and virus, nucleocapsids (100,000 × g), and very large complexes (e.g., ribosomes, 400,000 × g [30]) were removed by sequential centrifugation. All fractions were analyzed, and C42 was detected primarily in the 400,000 × g soluble fraction (Fig. 6A, asterisk). The blue native gel analysis of this fraction is shown in Fig. 6B. A complex with an approximate molecular mass of 500 to 600 kDa which was immunoreactive with antibodies to C42 (lane 2), p78/83 (lane 4), and EC27 (lane 6) was detected. Additionally, complexes showing positive interactions between C42 and p78/83 were also detected at lower masses, with the most prominent of these being at ∼180 kDa (lanes 2 and 4, circle). This experiment was repeated three times with cell extracts isolated from different infections, and the immunoreactive profiles were the same. To confirm the specificity of the interactions detected by native gel electrophoresis, we performed control antibody reactions with E66, FP25K, and E25, and none of these antibodies were detected in complexes with molecular masses corresponding to those shown in Fig. 6 (data not shown). We note that the antibody to EC27 strongly reacts to a cellular complex at approximately 140 kDa; however, we do not know the significance of this observation.
FIG. 6.
Native gel electrophoresis. (A) Summary overview of purification protocol of 28-h-p.i. infected-cell extracts. (B) The samples of the soluble fraction were separated using blue native electrophoresis, blotted onto a PVDF membrane, and probed with the indicated antibody. Arrows point to a complex containing EC27, C42, and p78/83, while the circles show a complex containing only C42 and p78/83. U, uninfected-cell lysate; I, infected-cell lysate. Approximate molecular masses are indicated on the left (kilodaltons).
DISCUSSION
Transcriptional analyses show that orf101 is a late gene with maximal levels of transcripts accumulating at 24 h p.i. (Fig. 2). This agrees with the data of Russell and Rohrmann (22) showing that, during infection by OpMNPV, p40 (orf3, the gene homologue to AcMNPV orf101) transcripts are detected at 24 h p.i., steady-state transcript levels decrease by 36 h p.i., and transcription initiates from a conserved TAAG sequence. Our study suggests that, if transcripts are made early during infection (CAGT), their steady-state levels are low and protein accumulation is not detectable (Fig. 2 and 3). orf101 encodes a protein with a relative molecular mass of 42 kDa that is a component of both BV and ODV nucleocapsid (Fig. 2 and 3). We have designated this gene product BV/ODV-C42 (C42).
C42 is conserved among baculoviruses and contains a classical nuclear localization sequence located at the C terminus (Fig. 1). Consistent with the presence of this motif, C42 is predominantly detected within the infected-cell nucleus. At 24 h p.i., C42 is associated with the virogenic stroma; by 48 h p.i., it locates at the edge of the stroma; and at later time points, it is located in a dispersed pattern throughout the nucleus which is consistent with its incorporation into ODV (Fig. 4). These observations were confirmed by IEM, which shows the localization of C42 within the virogenic stroma and the nucleocapsid of the virion (Fig. 5).
Yeast two-hybrid analysis, using orf144 as bait, identified eight interacting clones containing truncated orf101 (C42). Sequence analysis showed that these corresponded to four independent clones of orf101, and the details of these clones are shown in Fig. 1. Yeast two-hybrid screens using 1629K (p78/83) as bait also identified orf101 seven times, and sequence analysis revealed that these represented the same clone, all containing the methionine start codon for C42. To confirm that the interactions predicted by yeast two-hybrid analysis reflected interactions which occurred in vivo, blue native gel and Western blot analysis was performed using fractions prepared from infected cells (28 h p.i.). This analysis showed that C42 and p78/83 (p78/83 is also a structural protein of baculovirus [18, 21]) comigrate, consistent with complex formation, and that they are present in two complexes, a complex at 180 kDa and a larger complex (approximately 500 to 600 kDa) that also contains EC27.
The primary sequence of AcMNPV C42 contains the canonical binding motif of the pocket proteins (LxCxE). Interaction of viral proteins with pocket proteins (pRB, p130, and p107) plays an essential role in viral infectivity of other viruses. Simian virus 40 T, human papillomavirus E7, and adenovirus E1A (reviewed in reference 9) interact with pocket proteins to stimulate activity of the transcription factor E2F. Indeed, mutations within the pRB binding motif of the rubella virus gene NSP90 decreased viral DNA replication to <0.5%, while deletion of the motif was lethal (8). If such a mechanism is utilized by baculoviruses, one might predict that the pocket protein binding motif of C42 would be conserved among all baculoviruses; however, genome sequences show that this is not the case (Fig. 1). Thus, if C42 functionally interacts with pocket proteins, it is possible that it also performs other functions, that this function is host specific, or that other viruses and/or viral genes encode proteins with analogous function. Other examples of control of cell cycle and viral DNA replication by viruses include the coding of cellular cyclin homologues. Human herpesvirus (k-cyclin [6]), herpesvirus saimiri (v-cyclin [12]), murine gammaherpesvirus 68 (m-cyclin [29]), and walleye fish dermal sarcoma retrovirus (16) all encode cyclin-like proteins. The viral k- and v-cyclins complex with cdk6, increase the resistance of cyclin-dependent kinases to inhibitors, and enhance substrate range, and the viral cyclin-cdk6 complex directly triggers the initiation of DNA synthesis in isolated late-G1 nuclei (15).
It is possible that C42, EC27, and p78/83 together, or individually, interact with cellular components to generate an S-phase-like environment and stimulate in vivo DNA replication. Like the putative interaction of EC27 with cellular cyclin kinases, interaction of C42 with the pocket family of proteins would be a good candidate for regulation of the cell cycle and may play a direct role in viral DNA replication in vivo. Current studies are focused on more fully understanding the nature and function of the viral protein complexes containing EC27, C42, and p78/83 and determining if C42 functionally interacts with pocket proteins. Such interactions would strongly suggest that AcMNPV presents proteins (as part of its structural composition) that interact with cellular regulatory proteins immediately upon infection. These interactions likely play a critical role in regulating the cell cycle and enhancing viral infectivity.
ACKNOWLEDGMENTS
We thank Gabriela Marcano and Shawn Williamson for the development of the yeast two-hybrid cDNA libraries and Bridget Sweeney and Erin Pickerton for the library screens. We thank C. Richardson (Amgen Institute, Toronto, Ontario, Canada) for providing antibodies to p78/83 and a plasmid containing the 1629K gene and Tina Heyman and Jared Burks for their expert assistance with confocal microscopy. Electron microscopy was performed using the facilities of the Electron Microscopy Center at Texas A&M University.
This work was supported in part by National Institutes of Health grant 2RO1GM47552 (M.D.S. and S.C.B.) and Texas Agricultural Experiment Station project TEXO8078 (M.D.S.).
REFERENCES
- 1.Belyavskyi M, Braunagel S C, Summers M D. The structural protein ODV-EC27 of Autographa californica nucleopolyhedrovirus is a multifunctional viral cyclin. Proc Natl Acad Sci USA. 1998;95:11205–11210. doi: 10.1073/pnas.95.19.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bozzola J J, Russell L D. Electron microscopy. Boston, Mass: Jones and Bartlett; 1992. [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Braunagel S C, Parr R, Belyavskyi M, Summers M D. Autographa californica nucleopolyhedrovirus infection results in Sf9 cell cycle arrest at G2/M phase. Virology. 1998;244:195–211. doi: 10.1006/viro.1998.9097. [DOI] [PubMed] [Google Scholar]
- 5.Braunagel S C, Summers M D. Autographa californica nuclear polyhedrosis virus, PDV, and ECV viral envelopes and nucleocapsids: structural proteins, antigens, lipid and fatty acid profiles. Virology. 1994;202:315–328. doi: 10.1006/viro.1994.1348. [DOI] [PubMed] [Google Scholar]
- 6.Chang Y, Moore P S, Talbot S J, Boshoff C H, Zarkowska T, Godden-Kent D, Paterson H, Weiss R A, Mittnacht S. Cyclin encoded by KS herpesvirus. Nature (London) 1996;382:410. doi: 10.1038/382410a0. [DOI] [PubMed] [Google Scholar]
- 7.Dow J A T. Insect midgut function. Adv Insect Physiol. 1986;19:197–328. [Google Scholar]
- 8.Forng R-Y, Atreya C D. Mutations in the retinoblastoma protein-binding LXCXE motif of rubella virus putative replicase affect virus replication. J Gen Virol. 1999;80:327–332. doi: 10.1099/0022-1317-80-2-327. [DOI] [PubMed] [Google Scholar]
- 9.Funk J O, Galloway D A. Inhibiting CDK inhibitors: new lessons from DNA tumor viruses. Trends Biochem Sci. 1998;23:337–341. doi: 10.1016/s0968-0004(98)01242-0. [DOI] [PubMed] [Google Scholar]
- 10.Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 11.Hong T, Braunagel S C, Summers M D. Transcription, translation, and cellular localization of PDV-E66: a structural protein of the PDV envelope of Autographa californica nuclear polyhedrosis virus. Virology. 1994;204:210–222. doi: 10.1006/viro.1994.1525. [DOI] [PubMed] [Google Scholar]
- 12.Jung J U, Stager M, Desrosiers R C. Virus-encoded cyclin. Mol Cell Biol. 1994;14:7235–7244. doi: 10.1128/mcb.14.11.7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keddie B A, Aponte G W, Volkman L E. The pathway of infection of Autographa californica nuclear polyhedrosis virus in an insect host. Science. 1989;243:1728–1730. doi: 10.1126/science.2648574. [DOI] [PubMed] [Google Scholar]
- 14.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 15.Laman H, Coverly D, Krude T, Laskey R, Jones N. Viral cyclin–cyclin-dependent kinase 6 complexes initiate nuclear DNA replication. Mol Cell Biol. 2001;21:624–635. doi: 10.1128/MCB.21.2.624-635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LaPierre L A, Casey J W, Holzschu D L. Walleye retroviruses associated with skin tumors and hyperplasias encode cyclin D homologs. J Virol. 1998;72:8765–8771. doi: 10.1128/jvi.72.11.8765-8771.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neff D, Dencher N A. Purification of multisubunit membrane protein complexes: isolation of chloroplast FoF1-ATP synthase, Cfo and CF1 by blue native electrophoresis. Biochem Biophys Res Commun. 1999;259:569–575. doi: 10.1006/bbrc.1999.0820. [DOI] [PubMed] [Google Scholar]
- 18.Pham D Q-D, Hice R H, Sivasubramanian N, Federici B A. The 1629-bp open reading frame of the Autographa californica multinucleocapsid polyhedrosis virus encodes a virion structural protein. Gene. 1993;137:275–280. doi: 10.1016/0378-1119(93)90020-4. [DOI] [PubMed] [Google Scholar]
- 19.Prikhod'ko E A, Miller L K. Role of baculovirus IE2 and its RING finger in cell cycle arrest. J Virol. 1998;72:684–692. doi: 10.1128/jvi.72.1.684-692.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosas-Acosta G, Braunagel S C, Summers M D. Effects of deletion and overexpression of the Autographa californica nuclear polyhedrosis virus FP25K gene on synthesis of two occlusion-derived virus envelope proteins and their transport into virus-induced intranuclear membranes. J Virol. 2001;75:10829–10842. doi: 10.1128/JVI.75.22.10829-10842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russell R L Q, Funk C J, Rohrmann G F. Association of a baculovirus-encoded protein with the capsid basal region. Virology. 1997;227:142–152. doi: 10.1006/viro.1996.8304. [DOI] [PubMed] [Google Scholar]
- 22.Russell R L Q, Rohrmann G F. The p6.5 gene region of a nuclear polyhedrosis virus of Orgyia pseudotsugata: DNA sequence and transcriptional analysis of four late genes. J Gen Virol. 1990;71:551–560. doi: 10.1099/0022-1317-71-3-551. [DOI] [PubMed] [Google Scholar]
- 23.Schagger H, Cramer W A, von Jagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- 24.Schagger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 25.Summers M D, Egawa K. Physical and chemical properties of Trichoplusia ni granulosis virus granulin. J Virol. 1973;12:1092–1103. doi: 10.1128/jvi.12.5.1092-1103.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Summers M D, Smith G E. A manual of methods for baculovirus vectors and insect cell culture procedures. Texas Agricultural Experiment Station Bulletin 1555. College Station: Texas A&M University; 1987. [Google Scholar]
- 27.Venable J H, Coggeshall R. A simplified lead citrate stain for use in electron microscopy. J Cell Biol. 1965;25:407–408. doi: 10.1083/jcb.25.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vialard J E, Richardson C D. The 1,629-nucleotide open reading frame located downstream of the Autographa californica nuclear polyhedrosis virus polyhedrin gene encodes a nucleocapsid-associated phosphoprotein. J Virol. 1993;67:5859–5866. doi: 10.1128/jvi.67.10.5859-5866.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Virgin H W, Latreille P, Wamsley P, Hallsworth K, Weck K E, Dal Canto A J, Speck S H. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Dobberstein B. Oligomeric complexes involved in translocation of proteins across the membrane of the endoplasmic reticulum. FEBS Lett. 1999;457:316–322. doi: 10.1016/s0014-5793(99)01075-3. [DOI] [PubMed] [Google Scholar]
- 31.Washburn J O, Kirkpatrick B A, Volkman L E. Comparative pathogenesis of Autographa californica M nuclear polyhedrosis virus in larvae of Trichoplusia ni and Heliothis virescens. Virology. 1995;209:561–568. doi: 10.1006/viro.1995.1288. [DOI] [PubMed] [Google Scholar]