Abstract
BACKGROUND:
Osteoarthritis (OA) and osteoporosis (OS) are the most common orthopedic diseases.
OBJECTIVE:
To identify important genes as biomarkers for the pathogenesis of OA and OS.
METHODS:
Microarray data for OA and OS were downloaded from the Gene Expression Omnibus database. Differentially expressed genes (DEGs) between the OA and healthy control groups and between the OS and healthy control groups were identified using the Limma software package. Overlapping hub DEGs were selected using MCC, MNC, DEGREE, and EPC. Weighted gene co-expression network analysis (WGCNA) was used to mine OA- and OS-related modules. Shared hub DEGs were identified, human microRNA disease database was used to screen microRNAs associated with OA and OS, and an miRNA-target gene network was constructed. Finally, the expression of shared hub DEGs was evaluated.
RESULTS:
A total of 104 overlapping DEGs were identified in both the OA and OS groups, which were mainly related to inflammatory biological processes, such as the Akt and TNF signaling pathways Forty-six hub DEGs were identified using MCC, MNC, DEGREE, and EPC modules using different algorithms. Seven modules with 392 genes that highly correlated with disease were identified in the WGCNA. Furthermore, 10 shared hub DEGs were identified between the OA and OS groups, including OGN, FAP, COL6A3, THBS4, IGFBP2, LRRC15, DDR2, RND3, EFNB2, and CD48. A network consisting of 8 shared hub DEGs and 55 miRNAs was constructed. Furthermore, CD48 was significantly upregulated in the OA and OS groups, whereas EFNB2, DR2, COL6A3, and RND3 were significantly downregulated in OA and OS. Other hub DEGs were significantly upregulated in OA and downregulated in OS.
CONCLUSIONS:
The ten genes may be promising biomarkers for modulating the development of both OA and OS.
Keywords: Osteoarthritis, osteoporosis, hub DEGs
1. Introduction
Osteoarthritis (OA) and osteoporosis (OS) are the most common skeletal disorders associated with aging. OA, a prevalent degenerative joint disorder, manifests as joint pain, stiffness, and restricted mobility [1]. It stands as a leading contributor to global disability, impacting over half a billion individuals [2]. Recent years have witnessed some progress in the treatment of OA, yet these treatments invariably lead to complications such as fractures during and after surgery, loss of correction, and nonunion [3]. OS, a systemic skeletal disease frequently occurring in the population, is associated with aging and is typified by reduced bone strength, degradation of bone tissue microstructure, and an increased likelihood of fractures [4]. More than 8. 9 million bone fractures occur annually, and the morbidity continues to rise [5]. The primary strategies for OS prevention and treatment involve essential bone health supplementation and pharmacological interventions, both of which incur substantial costs, thereby exacerbating the economic strain on families and society at large due to OS and related fractures [6, 7]. Although OA and OS are the two most common skeletal disorders, the relationship between them remains controversial.
The genetic backgrounds of OA and OS are complex, and genetic variants have been demonstrated to be responsible for a small proportion of bone diseases Recently, the role of epigenetics in OA and OS has attracted considerable attention [8]. Visconti et al. showed that DNA methylation, one of the most important epigenetic mechanisms for modulating gene expression, is strongly associated with the pathogenesis of aging-related bone diseases, including OA and OS [9]. Pertusa et al. reported that miR-122-5p, miR-125b-5p, and miR-21-5p are valuable biomarkers for modulating bone pathologies [10]. Osteopontin, a vital molecule in bone homeostasis, is associated with OA and OS progression [11]. Although some biomarkers associated with OA and OS development have been identified, more molecules or pathological factors need to be explored to improve the treatment strategies for OA and OS.
Thus, four profiles, each of OA and OS, were used to explore the molecules involved in both OA and OS. Differentially expressed genes (DEGs) in OA vs. healthy controls and OS vs. healthy controls were screened using the limma package. The overlapping genes of hub DEGs were selected using MCC, MNC, DEGREE, and EPC, and genes involved in the modules were selected as important genes in OA and OS, and their expression levels were also displayed. Finally, ten genes were identified as promising candidates for OA and OS development.
2. Materials and methods
2.1. Gene expression omnibus (GEO) datasets
OA and OS expression profiles of were downloaded from the NCBI GEO database [12] (http://www. ncbi.nlm.nih.gov/geo/). Four OA profiles were downloaded: GSE55457 [13], GSE55235 [13], GSE12021 [14], and GSE82107 [15]. GSE55457 and GSE55235 included 20 synovial tissue samples from 10 OA patients and 10 healthy controls, respectively Ten OA synovial tissue samples and nine synovial tissue samples from healthy controls were included in GSE12021. GSE82107 included 10 OA synovial tissue samples and seven synovial tissue samples from healthy controls. The detection platform for GSE55457, GSE55235, and GSE12021 was the [HG-U133A] Affymetrix Human Genome U133A Array, and the detection platform for GSE82107 was the [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2. 0 Array.
Four OS profiles were downloaded: GSE56814 [16, 17, 18], GSE62402, GSE7158, and GSE13850. A total of 73 blood samples from 31 OS patients and 42 healthy controls were included in GSE56814, and 10 blood samples from five OS patients and five healthy controls were included in GSE62402. Both profiles were sequenced using the [HuEx-1_0-st] Affymetrix Human Exon 1. 0 ST Array. A total of 26 blood samples from 12 OS patients and 14 healthy controls were included in GSE7158, based on the [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2. 0 Array Forty blood samples from 20 OS patients and 20 healthy controls were included in GSE13850, based on the [HG-U133A] Affymetrix Human Genome U133A Array.
GSE55457, GSE55235, and GSE12021 were used as training datasets for OA, and GSE56814, GSE62402, and GSE7158 were used as training datasets for OS. SVA (http://www.bioconductor.org/ packages/release/bioc/html/sva.html, version 3.38.0) in the R4.3.1 language package [19] was used to remove batch effects from the OA and OS training datasets. GSE82107 and GSE13850 were used as validation datasets.
2.2. Screening of DEGs
DEGs between the OA and healthy control groups and the OS and healthy control groups were screened using the limma package (version 3.34.7) of R4.3.1 in the OA and OS training datasets, respectively [20]. FDR values less than 0. 05 and Log2FC0. 263 were defined as the screening threshold. The pheatmap package of R4.3.1 [21] was used to perform bidirectional hierarchical clustering based on the expression values of DEGs in the OA and OS groups and it was visualized using heatmaps.
Overlapping DEGs between the OS and OA groups were retained as significant DEGs related to both diseases. DAVID (version 6.8) [22, 23, 24] was used to annotate the GO function and Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathway enrichment analysis, and a p-value less than 0.05 was selected as the threshold for enrichment significance.
2.3. Construction of PPI network and selection of hub DEGs
The interaction relationships between the protein products of overlapping DEGs were constructed using STRING (version 11.0) [25] and visualized using Cytoscape (version 3.9.0) [26]. Subsequently, hub DEGs were selected based on four topology analysis algorithms, MCC, MNC, DEGREE, and EPC, using the cytoHubba module recognition plugin (version 0.1) [27] in Cytoscape 3.9.0. The important genes screened by each of the four algorithms were compared and the overlapping parts were retained as hub DEGs.
2.4. Selection of modules associated with disease status using weighted gene co-expression network analysis (WGCNA) algorithm
WGCNA [28] is a bioinformatics algorithm used for constructing co-expression networks that are used to identify modules associated with diseases and screen important pathogenic mechanisms or potential therapeutic targets. Based on the expression levels of all genes in the merged OA- and OS-related dataset samples, we used the WGCNA package (version 1.61) [29] in R4.3.1 to screen modules significantly correlated with the disease status of the sample.
The threshold for module-partitioning screening was set as follows: the module set should contain at least 200 genes and cutHeight was set to 0.995. Subsequently, disease-related modules were selected from the OA- and OS-related datasets and the gene sets of each module were compared. The overlapping parts were selected as genes significantly associated with both the diseases, and hub DEGs were identified in the interaction network to obtain the shared hub DEGs related to both diseases.
2.5. Construction of miRNA regulation network
miRNAs related to all diseases were downloaded from the human microRNA disease database (HMDD) (version 3.2) [30], and miRNAs related to both OA and OS were selected for further analysis. Genes targeted by the selected miRNAs were screened using miRWalk 3.0 [31]. The intersection of targeted genes and shared hub DEGs was retained, and an miRNA-targeted gene network was constructed and visualized using Cytoscape (version 3.9.0) [26].
2.6. Analysis of expression levels of hub DEGs in training and validation datasets
The expression levels of hub DEGs in the combined OA and OS training datasets were selected. The expression levels of hub DEGs in the validation datasets (GSE82107 and GSE13850) were also extracted.
3. Results
3.1. A total of 104 overlapping DEGs in OS and OA were identified
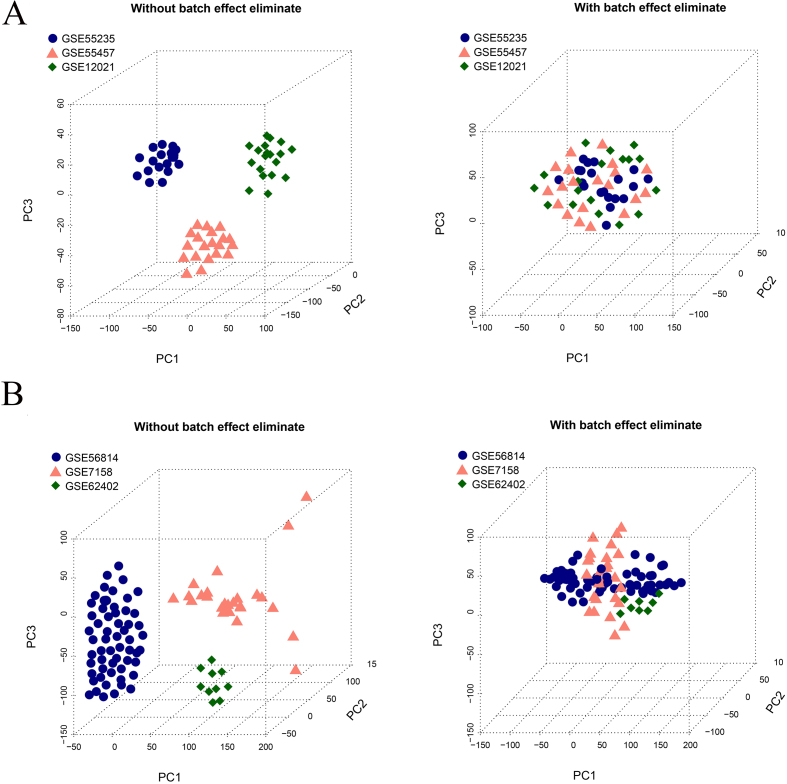
Three profiles of OA, GSE55457, GSE55235, and GSE12021 were combined as a training dataset, and the batch effect was removed using SVA. Similarly, the three OS profiles (GSE56814, GSE62402, and GSE7158) are used as a training dataset and SVA was used to remove the batch effect. After SVA, the samples in the OA and OS combined training datasets showed similar levels (Fig. 1A and B).
Figure 1.
Sample distribution map. A. Sample distribution map of OA before and after removing batch effects using SVA. B. Sample distribution map of OS before and after removing batch effects using SVA.
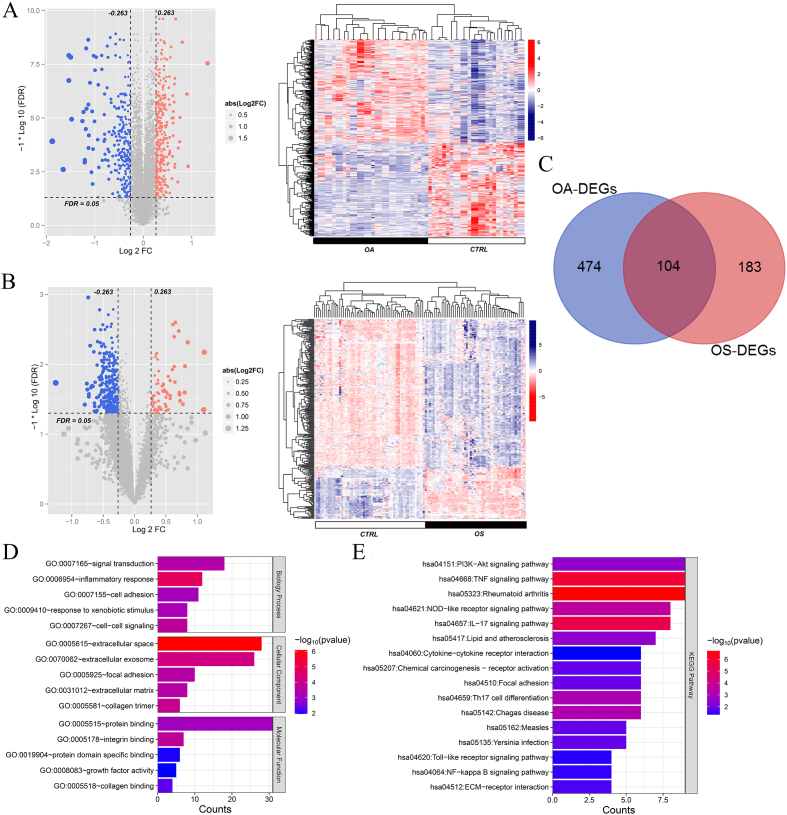
A total of 578 and 287 DEGs were identified between the OA and healthy control groups and the OS and healthy control groups, respectively. The volcano map and bidirectional hierarchical clustering of DEGs in the OA and OS groups are shown in Fig. 2A and B, respectively. The results showed that the levels of upregulated DEGs differed from those of downregulated DEGs. A total of 104 overlapping DEGs were identified between the OA and OS groups (Fig. 2C).
Figure 2.
A total of 104 overlapping differentially expressed genes (DEGs) in osteoporosis (OS) and osteoarthritis (OA) were identified. A: Volcano plot and heatmap of DEGs in OA; B: Volcano plot and heatmap of DEGs in OA. Left: Effect size (log2FC)-log10 (FDR) volcano plot. Red and blue dots represent significantly upregulated and downregulated DEGs, respectively. The horizontal dashed line indicates a false discovery rate (FDR) 0. 05. Right: Gene clustering heatmap based on the expression levels of DEGs in each group. C: Venn map of DEGs in OA and OS; D: Gene ontology (GO) item enriched by overlapping DEGs in OA and OS; E: Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched by overlapping DEGs in OA and OS. The horizontal axis represents the number of DEGs, the vertical axis represents the item name, and the color represents the correlation level.
Functional analysis of these 104 overlapping genes revealed that they were significantly enriched in 84 GO items, including 58 biological processes such as signal transduction and inflammation, 13 cellular components such as extracellular space and extracellular exosomes, and 13 molecular functions such as protein binding and integrin binding (Fig. 2D). Moreover, 16 KEGG pathways were enriched in the overlapping genes, including the Akt signaling pathway, TNF signaling pathway, and rheumatoid arthritis (Fig. 2E).
3.2. A total of 46 hub DEGs were screened in the OA and OS groups
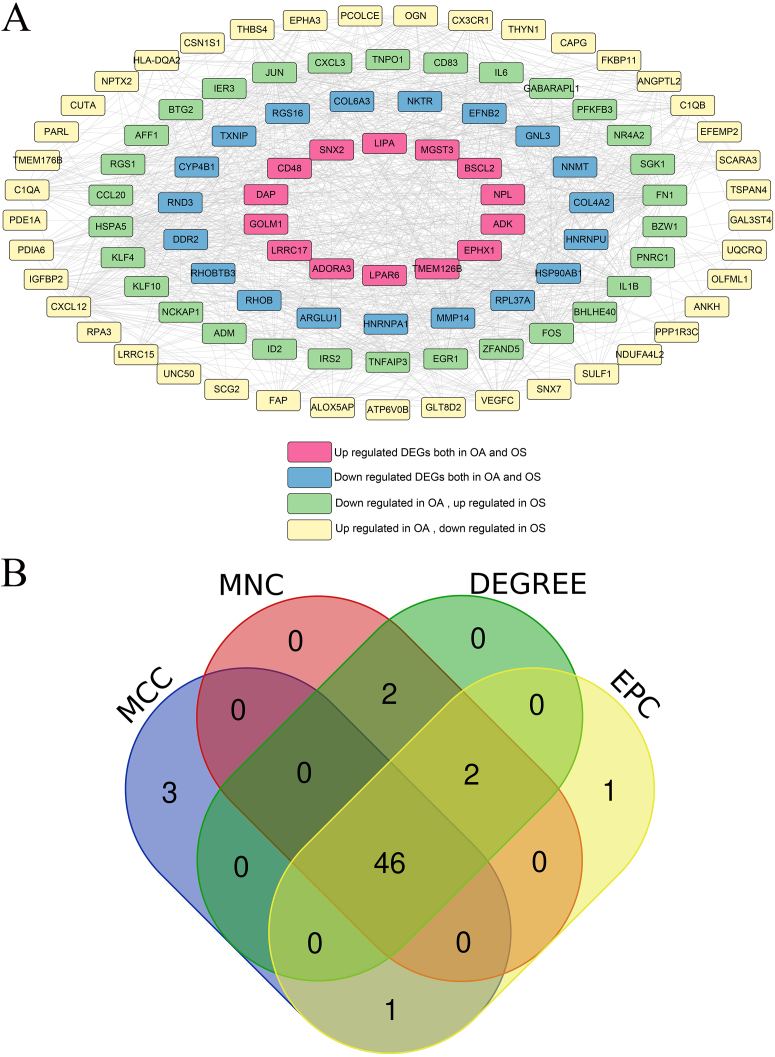
Using the STRING database, interactions among the proteins encoded by 104 overlapping DEGs were identified and interactions with scores higher than 0.4 were selected A total of 984 pairs of interactions were obtained and a PPI network was constructed based on these pairs (Fig. 3A). Hub DEGs in both the OA and OS groups were screened using four topology analysis algorithms: MCC, MNC, DEGREE, and EPC. The top 50 genes in the four topology analysis algorithms were selected, and 46 overlapping genes were identified as hub DEGs in the OA and OS groups, including PFKFB3, MMP14, HSP90AB1, LRRC15, DDR2, EGR1, FOS, BTG2, CD48, and HSPA5 (Fig. 3B).
Figure 3.
Protein-protein interaction (PPI) network and Venn map for selecting hub differentially expressed genes (DEGs). A: PPI network, and nodes of different colors represent genes with different differences; B: Venn map for hub DEGs, selected using MCC, MNC, DEGREE, and EPC.
3.3. Ten genes were selected as valuable genes in the OA and OS groups
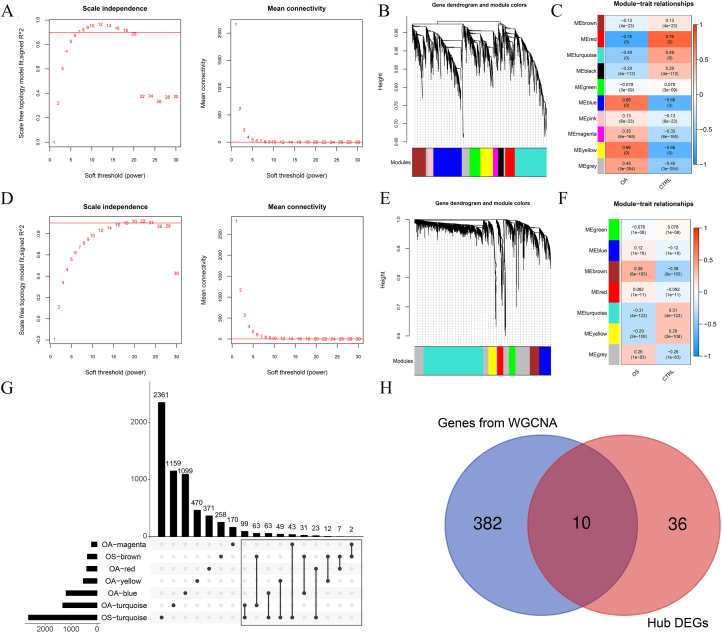
When the power was 7 or 18, the signed R2 was 0.9. The mean connectivity was 1, which is consistent with the nature of small-world networks (Fig. 4A and B). The coefficient of dissimilarity between gene points was then calculated, and a systematic clustering tree was obtained When we set the minimum number of genes in each module to 200 and cutHeight 0.995, ten modules in the OA group (Fig. 4C) and seven modules in the OS group were obtained (Fig. 4D). Figure 4E shows the module-trait relationships for the OA group. Five modules had absolute correlation values higher than 0.3: OA-blue, OA-magenta, OA-red, OA-turquoise, and OA-yellow. The module-trait relationships in the OS group are shown in Fig. 4F, with absolute correlation values higher than 0.3 for the OS-brown and OS-turquoise modules.
Figure 4.
Ten genes were selected as valuable genes in the osteoporosis (OS) and osteoarthritis (OA) groups. A: Left: Adjacency matrix weight parameter power selection chart for OA. The horizontal axis represents the weight parameter power and the vertical axis represents the square of the correlation coefficients between log (k) and log (p (k)) in the corresponding network. The red line represents the standard line, where the square value of the correlation coefficient reaches 0.9. Right figure: Schematic diagram of average gene connectivity under different power parameters of OA. The red line represents the value of average network node connectivity under the weight parameter power of the adjacency matrix in the left figure (1); B: Module partition tree diagram with each color representing different modules in OA; C: Module-trait correlation heatmap in OA; D: Left: Adjacency matrix weight parameter power selection chart for OS. The horizontal axis represents the weight parameter power and the vertical axis represents the square of the correlation coefficients between log (k) and log (p (k)) in the corresponding network. The red line represents the standard line, where the square value of the correlation coefficient reaches 0.9. Right figure: Schematic diagram of average gene connectivity under different power parameters of OS. The red line represents the value of average network node connectivity under the weight parameter power of the adjacency matrix in the left figure (1); E: Module partition tree diagram with each color representing different modules in OS; F: Module-trait correlation heatmap in OS; G: Comparison chart of gene sets involved in five OA-related modules and two OS-related modules; H: Venn map of hub differentially expressed genes (DEGs) and genes selected by WGCNA.
Among these modules, 392 genes were associated with both OA and OS (Fig. 4G). These 392 genes and 46 hub DEGs were compared, and 10 shared hub DEGs were identified between the OA and OS groups, including OGN, FAP, COL6A3, THBS4, IGFBP2, LRRC15, DDR2, RND3, EFNB2, and CD48 (Fig. 4H).
3.4. Eight shared hub DEGs and 114 miRNAs were involved in the miRNA-target gene network
A total of 211 miRNAs associated with OA and 230 miRNAs associated with OS were screened using the HMDD (v3.2). Among them, 114 miRNAs were associated swith both OA and OS.
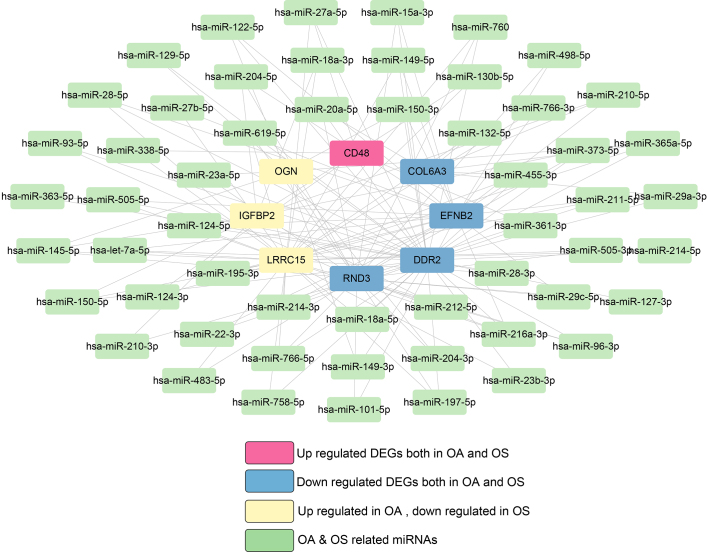
The genes targeting 114 miRNAs associated with both OA and OS were analyzed using miRWalk. Subsequently, ten shared hub DEGs in the OA and OS groups were mapped to these target genes, resulting in 140 connection pairs (Fig. 5), including eight shared hub DEGs: OGN, COL6A3, IGFBP2, LRRC15, DDR2, RND3, EFNB2, and CD48.
Figure 5.
miRNA-target gene network in osteoporosis (OS) and osteoarthritis (OA).
Two KEGG pathways were significantly enriched in these eight shared hub DEGs, including ECM-receptor interaction and focal adhesion.
3.5. Similarly dysregulated expression trends were observed in the training and validation datasets in the OA and OS groups
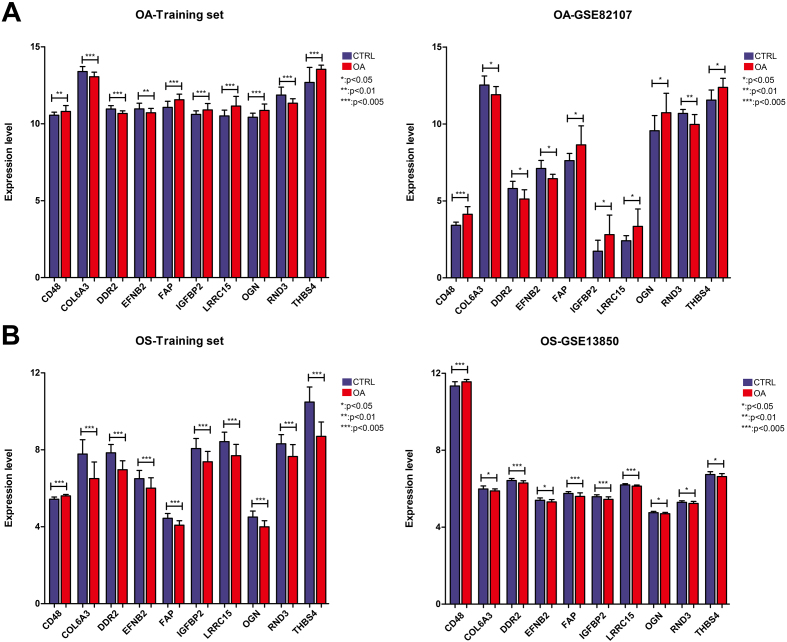
The expression levels of the 10 shared hub DEGs in the OA and OS groups are shown in Fig. 6. Figure 6A shows that the levels of 10 shared hub DEGs in the OA training dataset were consistent with those in GSE82107. Similarly, the levels of 10 shared hub DEGs in the OS training dataset align with those in GSE13850 (Fig. 6B). Furthermore, we observed that CD48 was significantly upregulated in the OA and OS groups and that four genes, EFNB2, DR2, COL6A3, and RND3, were significantly downregulated. The other five genes, THBS4, LRRC15, OGN, FAP, and IGFBP2, were significantly upregulated in the OA group and downregulated in the OS group.
Figure 6.
Expression levels of ten important genes in the osteoporosis (OS) and osteoarthritis (OA) groups. A: Expression levels of ten important genes in the OA training and validation datasets of GSE82107; B: Expression levels of ten important genes in the OS training and validation datasets of GSE13850.
4. Discussion
OA and OS are chronic diseases with prevalence rates that escalate with advancing age, both capable of inflicting pain and functional impairment, thereby diminishing patient quality of life [32, 33]. The relationship between OA and OS – whether they are positively correlated [34] or inversely correlated [35] – has been a point of contention. Emerging research has provided new insights into convergent and divergent risk factors for OA and OS, leading to revised understandings of the roles of bone mineral density, body mass index, falls, genetics, and epigenetics in the pathophysiology of these diseases and their associated increased fracture risks [36]. In recent years, a plethora of bioinformatics approaches has been extensively applied to the analysis of potential biomarkers related to OA and OS, leading to the discovery of numerous genes associated with the incidence and prognosis of these conditions. For instance, Wang et al. discovered an upregulation of CRIP1 in synovial, blood, and cartilage samples through the analysis of the GEO dataset, suggesting its potential as a diagnostic biomarker for OA [37]. Zhou et al. recognized ten pivotal genes associated with OA via gene expression microarray data, potentially aiding in the elucidation of molecular mechanisms of OA [38]. Yu et al. ascertained eight key genes through bioinformatics analysis conducive to the construction of an early diagnostic model for OS [39]. Chen et al. developed a diagnostic model based on nine key genes that reliably distinguished OS patients from healthy subjects, providing new insights into the diagnosis of OS [40]. Although the application of bioinformatics in OA and OS has been extensively documented, there is a paucity of research exploring the common mechanisms between OA and OS at the gene level. Hence, we conducted a bioinformatics analysis aimed at investigating the shared molecular mechanisms between OS and OA. In the present analysis, 104 overlapping DEGs were identified in the OA and OS groups, which were mainly related to inflammatory biological processes, such as the Akt and TNF signaling pathways Furthermore, 10 important genes were identified in the OA and OS groups. Among them, CD48 was significantly upregulated in the OA and OS groups, whereas four genes, EFNB2, DR2, COL6A3, and RND3, were significantly downregulated. The other five genes, THBS4, LRRC15, OGN, FAP, and IGFBP2, were significantly upregulated in the OA group and downregulated in the OS group Overall, ten shared hub DEGs were closely associated with the pathogenic mechanism of both OA and OS.
Multiple convergent and divergent factors, including genetics, mechanical load, and inflammation, have been reported in OS and OA patients. Both these diseases are polygenic. In the present analysis, 104 overlapping genes of 578 DEGs in the OA group and 287 DEGs in the OS group were mainly related to inflammatory biological processes, such as the Akt and TNF signaling pathways The PI3K/AKT and TNF signaling pathways are essential for normal metabolism of bone tissues, such as bone metabolism and immune responses. TNF- is an important pathogenic factor associated with immune-mediated bone diseases [41]. Bone marrow mesenchymal stem cells inhibit neuroinflammation by promoting the PI3K/AKT signaling pathway and suppressing the NF-B signaling pathway [42]. Sun et al. showed that the PI3K/AKT signaling pathway is critical for modulating synovial inflammation, cartilage degradation, and subchondral bone dysfunction [43]. Thus, genes related to inflammation may be involved in important mechanisms that lead to OA and OS.
Among the ten shared hub DEGs, OGN, FAP, COL6A3, THBS4, IGFBP2, LRRC15, DDR2, RND3, EFNB2, and CD48, the roles of some genes in the bone mechanism have been well demonstrated Previous evidence suggests that the progression of OA and OS is related to multiple factors such as nutritional factors, physical activity, and genetic factors. Increased subchondral bone loss is a characteristic feature of early stages of OA and OS [44]. CD48 is an important component of T cell activation pathways, and the activation of T cells is related to bone fragility and destruction [45]. Enhancement of the EFNB2-EphB4 signaling cascade inhibits osteoclastogenesis, thereby preventing bone loss in ovariectomized rats, whereas ablation of EFNB2 expression reverses the inhibitory effects on osteoclasts [46]. Furthermore, modulating the balance of the EphB4-EFNB2 axis may ameliorate the pathological profile of OS [46, 47]. Some genes are involved in the differentiation of bone marrow-derived stem cells, which may provide novel therapeutic strategies for OA and OS. For example, COL6A3 enhances osteogenic and adipogenic lineage differentiation by modulating mitochondrial health maintenance and promoting mitophagy [48]. Rnd3/RhoE are well known key regulators of the actin cytoskeleton in various cell types [49]. LRRC15, an evolutionarily conserved leucine-rich transmembrane protein, is highly expressed in human OA cartilage and osteoclasts derived from patients with rheumatoid arthritis [50, 51]. Moreover, differential methylation and LRRC15 expression in OA cartilage are associated with cellular stress responses and dysregulated extracellular matrix remodeling [52]. OGN is associated with bone formation, with elevated OGN levels found in both synovial fluid and damaged cartilage in OA patients [53, 54]. A series of studies have revealed the role of OGN in OA, suggesting that high expression of OGN may promote ossification and repair of the articular cartilage [55, 56]. In bone marrow stromal cells, FAP is significantly upregulated, and functions of the gene play as an osteogenic suppressor. Moreover, THBS4 is involved in primary keratinocyte migration and is upregulated in bone marrow lesions [57]. Although the roles of other genes, including IGFBP2 and DDR2, have not been widely studied in OS and OA, it is recommended to verify their functions in both OS and OA.
This study had some limitations. First, the conclusions were based on an open-access online dataset. Meanwhile, the background information of the enrolled subjects, such as the clinical backgrounds of patients and disease subtypes, was lacking. To filter out the noise signal induced by the background information, this conclusion should be verified using a large number of samples and clinical research. Second, the potential role of the shared hub DEGs in OA and OS was confirmed by western blotting and polymerase chain reaction. Furthermore, the functions of these genes should be explored in animal models with gene silencing or overexpression Despite these limitations, four OA and OS datasets were used in this study, and the expression levels of the selected hub DEGs were similar in both the training and validation datasets.
In conclusion, we showed that ten shared hub DEGs, including OGN, FAP, COL6A3, THBS4, IGFBP2, LRRC15, DDR2, RND3, EFNB2, and CD48, may be valuable biomarkers for both OA and OS, which may improve our understanding of the molecular mechanisms underlying OA and OS.
Conflict of interest
The authors declare that they have no conflict of interest.
Funding
None.
References
- [1]. Yao Q, Wu X, Tao C, Gong W, Chen M, Qu M, et al. Osteoarthritis: pathogenic signaling pathways and therapeutic targets. Signal Transduct Target Ther. 2023; 8(1): 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2]. Yao Q, Wu X, Tao C, Gong W, Chen M, Qu M, et al. Osteoarthritis: pathogenic signaling pathways and therapeutic targets. Signal Transduction and Targeted Therapy. 2023; 8(1): 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3]. Miltenberg B, Puzzitiello RN, Ruelos VCB, Masood R, Pagani NR, Moverman MA, et al. Incidence of Complications and Revision Surgery After High Tibial Osteotomy: A Systematic Review. Am J Sports Med. 2024; 52(1): 258-68. [DOI] [PubMed] [Google Scholar]
- [4]. Kim D, Pirshahid AA, Li Y, Varghese T, Pope JE. Prevalence of osteoporosis in osteoarthritis: a systematic review and meta-analysis. Osteoporos Int. 2022; 33(8): 1687-93. [DOI] [PubMed] [Google Scholar]
- [5]. Wong RMY, Wong PY, Liu C, Wong HW, Chung YL, Chow SKH, et al. The imminent risk of a fracture – existing worldwide data: a systematic review and meta-analysis. Osteoporosis International. 2022; 33(12): 2453-66. [DOI] [PubMed] [Google Scholar]
- [6]. Li N, Zheng B, Liu M, Zhou H, Zhao L, Cai H, et al. Cost-effectiveness of antiosteoporosis strategies for postmenopausal women with osteoporosis in China. Menopause. 2019; 26(8): 906-14. [DOI] [PubMed] [Google Scholar]
- [7]. Clynes MA, Harvey NC, Curtis EM, Fuggle NR, Dennison EM, Cooper C. The epidemiology of osteoporosis. Br Med Bull. 2020; 133(1): 105-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. Kim K-T, Lee Y-S, Han I. The role of epigenomics in osteoporosis and osteoporotic vertebral fracture. International journal of molecular sciences. 2020; 21(24): 9455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9]. Visconti VV, Cariati I, Fittipaldi S, Iundusi R, Gasbarra E, Tarantino U, et al. DNA Methylation Signatures of Bone Metabolism in Osteoporosis and Osteoarthritis Aging-Related Diseases: An Updated Review. Int J Mol Sci. 2021; 22(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10]. Pertusa C, Tarin JJ, Cano A, Garcia-Perez MA, Mifsut D. Serum microRNAs in osteoporotic fracture and osteoarthritis: a genetic and functional study. Sci Rep. 2021; 11(1): 19372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11]. Bai R-J, Li Y-S, Zhang F-J. Osteopontin, a bridge links osteoarthritis and osteoporosis. Frontiers in Endocrinology. 2022; 13(1012508). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]. Clough E, Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, et al. NCBI GEO: archive for gene expression and epigenomics data sets: 23-year update. Nucleic Acids Research. 2024; 52(D1): D138-D44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13]. Woetzel D, Huber R, Kupfer P, Pohlers D, Pfaff M, Driesch D, et al. Identification of rheumatoid arthritis and osteoarthritis patients by transcriptome-based rule set generation. Arthritis Res Ther. 2014; 16(2): R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14]. Huber R, Hummert C, Gausmann U, Pohlers D, Koczan D, Guthke R, et al. Identification of intra-group, inter-individual, xand gene-specific variances in mRNA expression profiles in the rheumatoid arthritis synovial membrane. Arthritis Res Ther. 2008; 10(4): R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15]. Broeren MG, de Vries M, Bennink MB, van Lent PL, van der Kraan PM, Koenders MI, et al. Functional Tissue Analysis Reveals Successful Cryopreservation of Human Osteoarthritic Synovium. PLoS One. 2016; 11(11): e0167076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16]. Zhou Y, Zhu W, Zhang L, Zeng Y, Xu C, Tian Q, et al. Transcriptomic Data Identified Key Transcription Factors for Osteoporosis in Caucasian Women. Calcif Tissue Int. 2018; 103(6): 581-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17]. Zhou Y, Xu C, Zhu W, He H, Zhang L, Tang B, et al. Long Noncoding RNA Analyses for Osteoporosis Risk in Caucasian Women. Calcif Tissue Int. 2019; 105(2): 183-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18]. Zhou Y, Gao Y, Xu C, Shen H, Tian Q, Deng HW. A novel approach for correction of crosstalk effects in pathway analysis and its application in osteoporosis research. Sci Rep. 2018; 8(1): 668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19]. Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012; 28(6): 882-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20]. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43(7): e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. Wang L, Cao C, Ma Q, Zeng Q, Wang H, Cheng Z, et al. RNA-seq analyses of multiple meristems of soybean: novel and alternative transcripts, evolutionary and functional implications. BMC Plant Biol. 2014; 14(169). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22]. Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic acids research. 2022; 50(W1): W216-W21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23]. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009; 4(1): 44-57. [DOI] [PubMed] [Google Scholar]
- [24].Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009; 37(1): 1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25]. Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, et al. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic acids research. 2021; 49(D1): D605-D12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26]. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13(11): 2498-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27]. Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014; 8(Suppl 4): S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28]. Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HY, Chen R, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012; 148(6): 1293-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29]. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008; 9(559). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30]. Huang Z, Shi J, Gao Y, Cui C, Zhang S, Li J, et al. HMDD v3.0: a database for experimentally supported human microRNA-disease associations. Nucleic Acids Res. 2019; 47(D1): D1013-D7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31]. Li XJ, Wen R, Wen DY, Lin P, Pan DH, Zhang LJ, et al. Downregulation of miR193a3p via targeting cyclin D1 in thyroid cancer. Mol Med Rep. 2020; 22(3): 2199-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32]. Bi J, Zhang C, Lu C, Mo C, Zeng J, Yao M, et al. Age-related bone diseases: Role of inflammaging. Journal of Autoimmunity. 2024; 143(103169). [DOI] [PubMed] [Google Scholar]
- [33]. Fukase N, Stake IK, Murata Y, Hambright WS, Ravuri S, Philippon MJ, et al. Interventional Strategies to Delay Aging-Related Dysfunctions of the Musculoskeletal System. Muscle Cell and Tissue-Novel Molecular Targets and Current Advances: IntechOpen; 2021. [Google Scholar]
- [34]. Li D, Wan Y, Sun Y, Wu X. Clinical Study of Correlation Between Osteoporosis and Osteoarthritis of Knee Joint Using Gold Nanomaterial Contrast Agent. Journal of Nanoscience and Nanotechnology. 2020; 20(12): 7761-8. [DOI] [PubMed] [Google Scholar]
- [35]. Lin L, Luo P, Yang M, Wang J, Hou W, Xu P. Causal relationship between osteoporosis and osteoarthritis: A two-sample Mendelian randomized study. Frontiers in Endocrinology. 2022; 13(1011246). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36]. Li Y, Xie B, Jiang Z, Yuan B. Relationship between osteoporosis and osteoarthritis based on DNA methylation. Int J Clin Exp Pathol. 2019; 12(9): 3399-407. [PMC free article] [PubMed] [Google Scholar]
- [37]. Wang X, Yu Y, Huang Y, Zhu M, Chen R, Liao Z, et al. Identification of potential diagnostic gene biomarkers in patients with osteoarthritis. Scientific Reports. 2020; 10(1): 13591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38]. Zhou J, Zou D, Wan R, Liu J, Zhou Q, Zhou Z, et al. Gene Expression Microarray Data Identify Hub Genes Involved in Osteoarthritis. Front Genet. 2022; 13(870590). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39]. Deng Y, Wang Y, Shi Q, Jiang Y. Identification of hub genes associated with osteoporosis development by comprehensive bioinformatics analysis. Front Genet. 2023; 14(1028681). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40]. Chen X, Liu G, Wang S, Zhang H, Xue P. Machine learning analysis of gene expression profile reveals a novel diagnostic signature for osteoporosis. J Orthop Surg Res. 2021; 16(1): 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41]. Wang T, He C. TNF-alpha and IL-6: The Link between Immune and Bone System. Curr Drug Targets. 2020; 21(3): 213-27. [DOI] [PubMed] [Google Scholar]
- [42]. Zhong Z, Chen A, Fa Z, Ding Z, Xiao L, Wu G, et al. Bone marrow mesenchymal stem cells upregulate PI3K/AKT pathway and down-regulate NF-kappaB pathway by secreting glial cell-derived neurotrophic factors to regulate microglial polarization and alleviate deafferentation pain in rats. Neurobiol Dis. 2020; 143(104945). [DOI] [PubMed] [Google Scholar]
- [43]. Sun K, Luo J, Guo J, Yao X, Jing X, Guo F. The PI3K/AKT/mTOR signaling pathway in osteoarthritis: a narrative review. Osteoarthritis Cartilage. 2020; 28(4): 400-9. [DOI] [PubMed] [Google Scholar]
- [44]. Chu L, Liu X, He Z, Han X, Yan M, Qu X, et al. Articular cartilage degradation and aberrant subchondral bone remodeling in patients with osteoarthritis and osteoporosis. Journal of Bone and Mineral Research. 2020; 35(3): 505-15. [DOI] [PubMed] [Google Scholar]
- [45]. Cifuentes-Mendiola SE, Solis-Suarez DL, Martinez-Davalos A, Godinez-Victoria M, Garcia-Hernandez AL. CD4(+) T-cell activation of bone marrow causes bone fragility and insulin resistance in type 2 diabetes. Bone. 2022; 155(116292). [DOI] [PubMed] [Google Scholar]
- [46]. Zhang Y, Kou Y, Yang P, Rong X, Tang R, Liu H, et al. ED-71 inhibited osteoclastogenesis by enhancing EphrinB2–EphB4 signaling between osteoclasts and osteoblasts in osteoporosis. Cellular Signalling. 2022; 96(110376). [DOI] [PubMed] [Google Scholar]
- [47]. Huang M, Wang Y, Peng R. Icariin Alleviates Glucocorticoid-Induced Osteoporosis through EphB4/Ephrin-B2 Axis. Evid Based Complement Alternat Med. 2020; 2020(2982480). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48]. Wang K, Peng X, Zhang R, Wu X, Mao L. COL6A3 enhances the osteogenic differentiation potential of BMSCs by promoting mitophagy in the osteoporotic microenvironment. Mol Biol Rep. 2024; 51(1): 206. [DOI] [PubMed] [Google Scholar]
- [49]. Abbey CA, Duran CL, Chen Z, Chen Y, Roy S, Coffell A, et al. Identification of new markers of angiogenic sprouting using transcriptomics: New role for RND3. Arteriosclerosis, Thrombosis, and Vascular Biology. 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50]. Ji Q, Zheng Y, Zhang G, Hu Y, Fan X, Hou Y, et al. Single-cell RNA-seq analysis reveals the progression of human osteoarthritis. Ann Rheum Dis. 2019; 78(1): 100-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51]. He X, Yin J, Yu M, Wang H, Qiu J, Wang A, et al. Identification and validation of hub genes for predicting treatment targets and immune landscape in rheumatoid arthritis. BioMed Research International. 2022; 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52]. Singh P, Wang M, Mukherjee P, Lessard SG, Pannellini T, Carballo CB, et al. Transcriptomic and epigenomic analyses uncovered Lrrc15 as a contributing factor to cartilage damage in osteoarthritis. Sci Rep. 2021; 11(1): 21107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53]. Zheng H, Aihaiti Y, Cai Y, Yuan Q, Yang M, Li Z, et al. The m6A/m1A/m5C-Related Methylation Modification Patterns and Immune Landscapes in Rheumatoid Arthritis and Osteoarthritis Revealed by Microarray and Single-Cell Transcriptome. Journal of Inflammation Research. 2023; 5001-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54]. Delgado-Enciso I, Paz-Garcia J, Valtierra-Alvarez J, Preciado-Ramirez J, Olmedo-Buenrostro BA, Delgado-Enciso J, et al. A novel cell-free formulation for the treatment of knee osteoarthritis generates better patient-reported health outcomes in more severe cases. Journal of Orthopaedic Surgery. 2020; 28(2): 2309499020938121. [DOI] [PubMed] [Google Scholar]
- [55]. Yan CL, An FY, Wang YF, Jin H, Zhao CB, Wang TX, et al. [Intervention effect of Youguiwan on rats with knee osteoarthritis and its mechanism]. Zhongguo Ying Yong Sheng Li Xue Za Zhi. 2020; 36(5): 511-6. [DOI] [PubMed] [Google Scholar]
- [56]. Cao J, Ding H, Shang J, Ma L, Wang Q, Feng S. Weighted gene co-expression network analysis reveals specific modules and hub genes related to immune infiltration of osteoarthritis. Ann Transl Med. 2021; 9(20): 1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57]. Maemets-Allas K, Klaas M, Cardenas-Leon CG, Arak T, Kankuri E, Jaks V. Stimulation with THBS4 activates pathways that regulate proliferation, migration and inflammation in primary human keratinocytes. Biochem Biophys Res Commun. 2023; 642(97-106). [DOI] [PubMed] [Google Scholar]