Abstract
Recently, small nucleolar RNAs (snoRNAs) have transcended the genomic “noise” to emerge as pivotal molecular markers due to their essential roles in tumor progression. Substantial evidence indicates a strong association between snoRNAs and critical clinical features such as tumor pathology and drug resistance. Historically, snoRNA research has concentrated on two classical mechanisms: 2'-O-ribose methylation and pseudouridylation. This review specifically summarizes the novel regulatory mechanisms and functional patterns of snoRNAs in tumors, encompassing transcriptional, post-transcriptional, and post-translational regulation. We further discuss the synergistic effect between snoRNA host genes (SNHGs) and snoRNAs in tumor progression. More importantly, snoRNAs extensively contribute to the development of tumor cell resistance as oncogenes or tumor suppressor genes. Accordingly, we provide a comprehensive review of the clinical diagnosis and treatment associated with snoRNAs and explore their significant potential as novel drug targets.
Keywords: snoRNA, Cancer, snoRNA host gene, Multidrug resistance, Targeted therapy, Tumor immune microenvironment
Graphical abstract
Highlights
-
•
SnoRNAs have two classical mechanisms: 2'-O-ribose methylation and pseudouridylation.
-
•
SnoRNAs have novel regulatory mechanisms and functional patterns of snoRNAs in tumors.
-
•
SnoRNAs contribute to the development of tumor cell resistance as oncogenes or tumor suppressor genes.
-
•
SnoRNAs play a critical role in cancer biology and have significant clinical implications.
1. Introduction
Small nucleolar RNAs (snoRNAs) were previously regarded as housekeeping genes and often neglected. Nonetheless, the human genome is increasingly being annotated with snoRNAs [[1], [2], [3]]. Eukaryotic cells mainly harbor snoRNAs in the nucleolus, typically ranging in length from 60 to 250 nucleotides [4]. These can be divided into box C/D snoRNAs, which govern 2'-O-ribose methylation; and box H/ACA snoRNAs, which govern pseudouridylation [[4], [5], [6]]. Moreover, another particular subclass of snoRNAs is known as small Cajal body-specific RNAs (scaRNAs). ScaRNAs govern the alteration of small nuclear RNA (snRNA) 2'-O-ribose methylation and pseudouridylation in the subnuclear Cajal body [7,8]. SnoRNAs are mainly derived from introns of protein-coding genes or non-protein coding genes, thus these genes are also known as snoRNA host genes (SNHGs) [[9], [10], [11]]. Afterwards, mature snoRNAs are cleaved and processed, giving rise to metabolically durable snoRNA fragments recognized as snoRNA-derived RNAs (sdRNAs), a subset of which share features with microRNAs (miRNAs) [[12], [13], [14], [15], [16]]. Furthermore, many snoRNAs without ribosomal RNA (rRNA) or snRNA recognition targets are labeled as “orphan snoRNAs".
The participation of snoRNAs in cancer development has been associated with multiple biological processes, including cell proliferation, invasion, apoptosis, and metastasis [[17], [18], [19], [20]]. There is a growing interest regarding the involvement of snoRNAs in different cancer types. Previous studies have primarily focused on the two traditional mechanisms of snoRNAs: 2'-O-ribose methylation and pseudouridylation of rRNAs. Recent research has suggested that snoRNAs have additional non-classical functions, such as the regulations of transcriptional, post-transcriptional, and post-translational processes. These findings expand the downstream of snoRNAs and indicate the additional implications of snoRNAs in the onset and progression of cancer. Consequently, this review aimed to summarize the classical and novel non-classical functional pathways of snoRNAs in tumor formation, as well as the significant implications of SNHGs on cancer progression. This review also summarized recent studies on snoRNAs involved in mediating resistance to anti-cancer drugs, highlighting the potential clinical applications of snoRNAs, as biomarkers, and therapeutic targets for anti-cancer therapy. Finally, we proposed diagnosis and treatment strategies to effectively combat tumors with snoRNAs involved. With these scientific advances, our review provided directions and insights for future investigations, facilitating a deeper comprehension of snoRNA function and advancements in the application in tumor treatment.
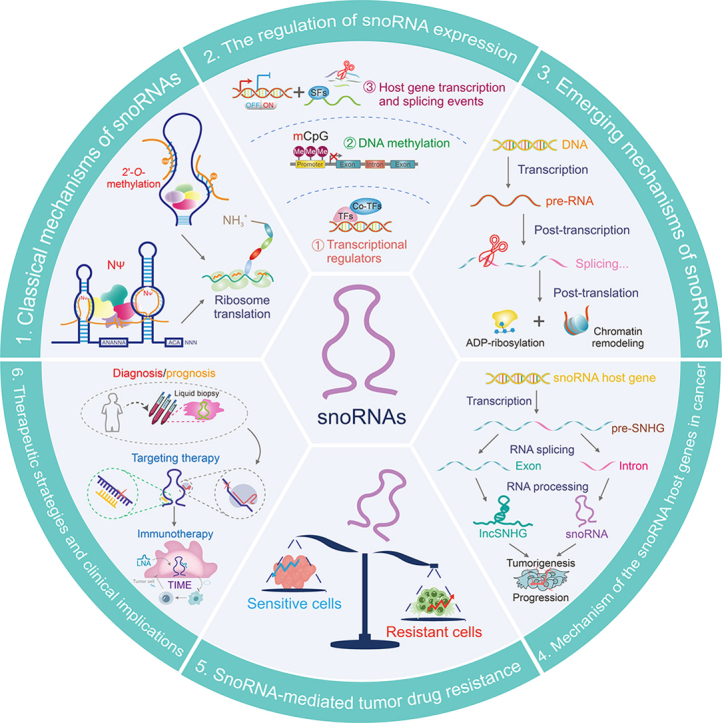
2. Classical mechanisms of snoRNAs in cancer
SnoRNAs can be categorized into two families, namely C/D box snoRNAs (SNORDs) and H/ACA box snoRNAs (SNORAs), based on their secondary structures and conserved sequence motifs. SNORDs perform site-specific 2'-O-methylation of ribose, whereas SNORAs facilitate uridine isomerization [[21], [22], [23]]. These classical modification mechanisms influence the folding and stability of rRNA, consequently affecting the onset and progression of cancer [[24], [25], [26]]. SnoRNAs dynamically interact with proteins to generate catalytically active small nucleolar ribonucleoprotein (snoRNP) complexes, which are essential for RNA synthesis, transport, and function [6,[27], [28], [29]]. Each snoRNA family contains a distinct set of binding proteins. The SNORDs family comprises four highly conserved core proteins: fibrillarin (FBL), nucleolar protein 56 (NOP56), NOP58, and 15.5KD [[30], [31], [32]]. The binding proteins of SNORAs, including dyskerin pseudouridine synthase 1 (DKC1), GAR1 ribonucleoprotein (GAR1), NHP2 ribonucleoprotein (NHP2), and nucleolar protein 10 (NOP10), are conserved and crucial for pseudouridylation processes [16,33,34]. The classical mechanisms of snoRNAs are shown in Fig. 1.
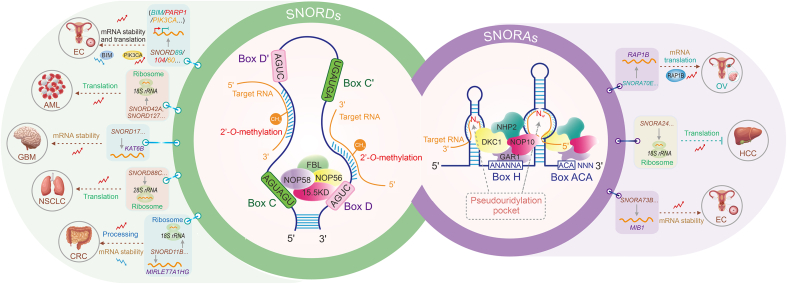
Fig. 1.
The classical mechanisms of small nucleolar RNAs (snoRNAs) involved in tumor initiation and development. Based on conserved sequence elements, snoRNAs are classified as C/D box snoRNAs (SNORDs) and H/ACA box snoRNAs (SNORAs). SNORDs contain sequence elements termed C (UGAUGA) and D (CUGA) boxes, usually present in duplicates (C' and D' boxes). SNORAs contain sequence elements termed H (ANANNA) and ACA boxes. SNORDs are responsible for the 2'-O-methylation modification of target RNAs by binding with proteins fibrillarin (FBL), nucleolar protein 56 (NOP56), NOP58, and 15.5KD. SNORAs are responsible for the pseudouridine modification of target RNAs by binding with proteins dyskerin pseudouridine synthase 1 (DKC1), GAR1 ribonucleoprotein (GAR1), NHP2 ribonucleoprotein (NHP2), and nucleolar protein 10 (NOP10). BIM: B cell lymphoma-2 (BCL-2)-like protein 11; PARP1: poly (adenosine diphosphate (ADP)-ribose) polymerase 1; PIK3CA: phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; mRNA: messenger RNA; EC: endometrial cancer; 18S rRNA: 18S ribosomal RNA; AML: acute myeloid leukemia; KAT6B: lysine acetyltransferase 6B; GBM: glioblastoma; NSCLC: non-small cell lung cancer; CRC: colorectal cancer; RAP1B: RAS-related protein 1B; OV: ovarian cancer; HCC: hepatocellular carcinoma; MIB1: mindbomb E3 ubiquitin protein ligase 1.
More specifically, snoRNAs are primarily recognized as mediators of 2'-O-methylation and pseudouridylation in rRNA and snRNAs. However, recent discoveries of their roles in modifying messenger RNAs (mRNAs) are highly significant. This highlights the evolving landscape of snoRNAs in post-transcriptional regulation and offers a new perspective for understanding the molecular mechanisms driving cancer progression.
2.1. SNORDs mediate 2'-O-methylation modification of target RNA
Most SNORDs use antisense complementarity to direct 2'-O-methylation modifications at specific sites on rRNA [35]. Antisense complementarity refers to the ability of a 10−21 nt segment upstream of the C/D box to form base pairs with the target RNA, resulting in site-specific modifications [36,37]. Elevated expression of SNORD89 in endometrial cancer (EC) cells interacts with FBL to hinder the translation of B cell lymphoma-2 (BCL-2)-like protein 11 (BIM) through 2'-O-methylation. This leads to decreased BIM protein levels, which promotes the proliferation and migration of EC cells [38]. SNORD104 binds to FBL to increase the stability and nuclear localization of poly(adenosine diphosphate (ADP)-ribose) polymerase 1 (PARP1) mRNA via 2'-O-methylation, promoting EC proliferation [39]. SNORD60 interacts with FBL to enhance the stability and expression of PIK3CA mRNA through 2'-O-methylation, thereby regulating phosphatidylinositol 3-kinase/protein kinase B/mammalian target of rapamycin (PI3K/AKT/mTOR) pathway and promoting the progression of EC [40]. Additionally, SNORD127 specifically recognizes the Gm1447 site on 18S rRNA, upregulating the 2'-O-methylation levels of Gm1447 and subsequently promoting the activity of leukemia stem cells [41]. In glioblastoma (GBM), SNORD17 methylates KAT6B via 2'-O-methylation, promoting angiogenesis and providing potential avenues for targeted therapy in clinical settings [42]. SNORD11B mediates 2'-O-methylation at the G509 site on 18S rRNA in colorectal cancer (CRC), promoting the processing and maturation of 18S rRNA. Furthermore, SNORD11B induces 2'-O-methylation modification at the G225 site on MIRLET7A1HG (pri-let-7a). This modification leads to pri-let-7a degradation, reduced expression of mature tumor suppressor let-7a-5p, and the upregulation of downstream oncogene cellular MYC (c-MYC) and rat sarcoma (RAS) translation, thereby promoting colorectal tumorigenesis [43].
Collectively, SNORDs can influence tumor onset and development through 2'-O-methylation modifications, providing novel insights into cancer regulation and treatment.
2.2. SNORAs mediate pseudouridine modification of target RNA
SNORAs are crucial for pseudouridylation, a process that modifies specific sites in eukaryotic rRNA and snRNA. Pseudouridylation involves the conversion of uridine to pseudouridine in the targeted RNA molecules [22,44]. SNORA molecules possess a conserved secondary structure known as the “hairpin-hinge hairpin tail.” The guiding sequence of SNORA is located in one or two hairpins within this structure, forming the “pseudouridylation pocket” [44]. This pocket facilitates non-canonical base pairing with the target uridine through interactions with pre-rRNA. In ovarian cancer (OV), SNORA70E interacts with dyskerin and induces pseudouridylation of RAP1B mRNA, subsequently leading to increased RAP1B protein levels and promoting the proliferation, invasion, and migration of OV cells [45]. SNORA73B binds to DKC1, enhancing the stability and expression of MIB1 mRNA through pseudouridylation. This process leads to increased Jagged 1 (JAG1) ubiquitin levels and activation of the neurogenic locus notch homolog (NOTCH) pathway, ultimately promoting the development of EC [46]. Therefore, SNORAs that mediate pseudouridine modification of target RNA offer a novel perspective for cancer diagnosis and treatment.
2.3. Increasingly important role in ribosome translation of snoRNA-mediated classical modifications
The ribosome, often referred to as the “protein-producing machine”, facilitates the dehydration condensation of amino acids to form peptide chains during the translation process [47,48]. Thus, modifications in rRNA, particularly 2'-O-methylation and pseudouridylation, contribute to the plasticity of ribosomal translational functions, a critical factor given the evidence that these modifications regulate cancer cell translational programs [21]. For example, rRNA with 2'-O-methylation has been shown to affect the ribosome's ability to translate mRNA in HeLa cells [21]. Reduced pseudouridylation levels alter ribosomal activity, particularly impacting translation mediated by internal ribosome entry site (IRES) and overall translation fidelity in breast cancer (BRCA) cells [49]. Aberrant rRNA methylation, regulated by the P53/FBL axis, compromises ribosomal translation fidelity, activating the translation of oncogenes such as IGF1R dependent on the IRES, thereby promoting BRCA progression [49]. In recent years, mounting evidence suggests that snoRNA-mediated classical modifications play a significant role in ensuring the fidelity of ribosome translation, potentially influencing cancer onset and progression [[24], [25], [26]]. SNORD42A guides 2'-O-methylation at the U116 site on 18S rRNA, promoting the translation of ribosomal proteins in acute myeloid leukemia (AML) and enhancing leukemia cell proliferation and survival [50]. SNORD45C induces 2'-O-methylation modification at the C174 site of 18S rRNA, remodeling mRNA translation in cervical cancer (CC) cells [51]. SNORD127 mediates 2'-O-methylation at the G1447 site on 18S rRNA to promote preferential translation of mRNAs, driving AML stem cell phenotypes [41]. SNORA24 guides the modification of pseudouridines in 18S rRNA to decrease translational miscoding and stop codon readthrough frequencies, resulting in the suppression of hepatocellular carcinoma (HCC) development during RAS-induced aging [52]. Furthermore, SNORD88C promotes 2'-O-methylation at the C3680 site on 28S rRNA in non-small cell lung cancer (NSCLC), which increases the translation of the downstream lipogenic enzyme stearoyl-CoA desaturase 1 (SCD1), thereby promoting NSCLC proliferation and metastasis [53]. In conclusion, the significance of snoRNA-mediated classical modifications for ribosome translation is steadily growing, providing valuable insights into tumor regulation patterns. The classical mechanisms of those crucial snoRNAs in tumor initiation and development are summarized as shown in Table 1 [[38], [39], [40], [41], [42], [43], [45], [46], [50], [51], [52], [53]] and Fig. 1.
Table 1.
The classical mechanisms of small nucleolar RNAs (snoRNAs) involved in tumorigenesis and progression.
| SnoRNA | Mechanism | Target RNA | Biological function | Related tumor type | Refs. |
|---|---|---|---|---|---|
| SNORD89 | Regulating 2’-O-methylation modification | BIM mRNA | Promoting tumor occurrence and development |
EC | [38] |
| SNORD104 | Regulating 2’-O-methylation modification | PARP1 mRNA | Promoting tumor growth | EC | [39] |
| SNORD60 | Regulating 2’-O-methylation modification | PIK3CA mRNA | Promoting tumorigenesis and progression | EC | [40] |
| SNORD127 | Regulating 2’-O-methylation modification | 18S rRNA | Promoting the activity of cancer stem cells | AML | [41] |
| SNORD17 | Regulating 2’-O-methylation modification | KAT6B mRNA | Promoting vasculogenic mimicry | Glioblastoma | [42] |
| SNORD11B | Regulating 2’-O-methylation modification |
18S rRNA and MIRLET7A1HG(pri-let-7a) mRNA |
Promoting tumor cell proliferation and invasion and inhibiting apoptosis | CRC | [43] |
| SNORA70E | Guiding pseudouridine modification | RAP1B mRNA | Promoting tumor occurrence and development | OV | [45] |
| SNORA73B | Guiding pseudouridine modification | MIB1 mRNA | Promoting tumor cell proliferation, migration, and invasion and inhibiting apoptosis | EC | [46] |
| SNORD42A | Regulating 2’-O-methylation modification | 18S rRNA | Promoting tumor cell proliferation | AML | [50] |
| SNORD45C | Regulating 2’-O-methylation modification | 18S rRNA | / | CC | [51] |
| SNORA24 | Guiding pseudouridine modification | 18S rRNA | Inhibiting tumor development | HCC | [52] |
| SNORD88C | Regulating 2’-O-methylation modification | 28S rRNA | Promoting tumor growth and metastasis and inhibiting autophagy | NSCLC | [53] |
/: no data. SNORDs: C/D box snoRNAs; BIM: B cell lymphoma-2 (BCL-2)-like protein 11; mRNA: messenger RNA; EC: endometrial cancer; PARP1: poly(adenosine diphosphate (ADP)-ribose) polymerase 1; PIK3CA: phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; rRNA: ribosomal RNA; AML: acute myeloid leukemia; KAT6B: lysine acetyltransferase 6B; CRC: colorectal cancer; RAP1B: RAS-related protein 1B; OV: ovarian cancer; MIB1: mindbomb E3 ubiquitin protein ligase 1; CC: cervical cancer; HCC: hepatocellular carcinoma; NSCLC: non-small cell lung cancer.
3. The regulation of snoRNA expression
SnoRNAs are metabolically stable products due to their conservative secondary structure and ability to form complexes with proteins and RNA [31]. SnoRNAs exhibit relatively longer half-lives (24–48 h) compared to mRNAs (less than 6 h), with their levels regulated by both synthesis and degradation mechanisms in the cell [2,54,55]. It is now widely recognized that abundant functional snoRNAs are dysregulated in cancer, showing differential expression across cancer types, stages, and metastasis, and actively influencing tumor progression [[56], [57], [58]]. Recent studies have indicated that snoRNA expression can be transcriptionally regulated through interactions with transcription regulators and DNA methylation. Additionally, most snoRNAs are coded with introns of coding and non-coding genes, making their expression dependent on host gene transcription and splicing events [59,60]. Thus, elucidating the regulatory mechanisms of snoRNA expression in tumor development is both important and intriguing.
3.1. SnoRNA expression regulated by transcription regulators
There is increasing evidence that transcription factors play a pivotal role in modulating snoRNA expression, which is crucial for maintaining cellular homeostasis and ultimately influencing cancer progression [57,58,61,62] (Fig. 2). In mice with P53 mutation and osteosarcoma (OS), snoRNAs were upregulated through E26 transformation-specific sequence 2 (ETS2)-dependent transcriptional regulation, facilitating tumor metastasis and suggesting ETS2 as a potential therapeutic target for P53-mutant osteosarcoma [57]. P53, acting as a tumor suppressor, can inhibit SNORD17 transcription, thereby suppressing the progression of HCC [58]. Additionally, amino-terminal enhancer of split (AES) has been identified as a co-transcriptional factor that affects multiple RNA processing pathways at the transcriptional level in acute myeloid leukemia 1-eight-twenty-one (AML1-ETO)-induced leukemogenesis [61,62].
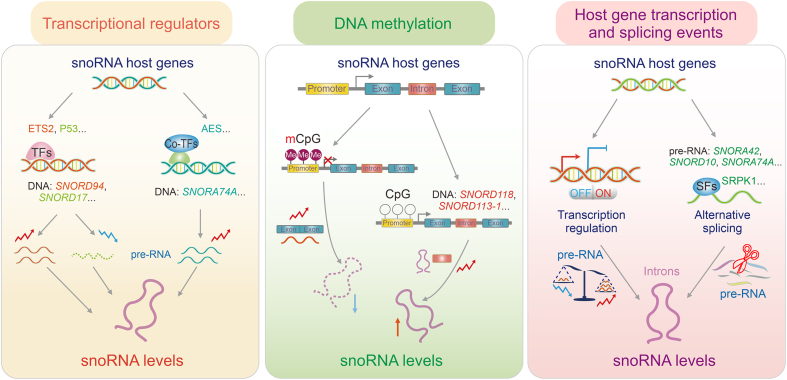
Fig. 2.
Small nucleolar RNAs (SnoRNA) expression regulated by transcription regulators. SnoRNA expression is regulated mainly through three patterns: dynamic regulation by transcription regulators, transcriptional inhibition by DNA methylation, and dependence on host gene transcription and splicing events. ETS2: E26 transformation-specific sequence 2; TFs: transcription factors; SNORDs: C/D box snoRNAs; AES: amino-terminal enhancer of split; mCpG: methylated cytosine-phosphate-guanine (CpG); SRPK1: serine-arginine protein kinase 1; SFs: splicing factors.
3.2. Inhibition of snoRNA transcription by DNA methylation
DNA methylation is a common epigenetic mechanism that regulates gene expression. DNA methylation primarily occurs on cytosine-phosphate-guanine (CpG) islands in the gene promoter regions. High levels of CpG island methylation play an important role in cancer induction [63] (Fig. 2). High-throughput sequencing has revealed that DNA methylation inhibits snoRNA transcription, affecting cancer progression [56,64,65]. Gong et al. [64] analyzed snoRNA expression in 31 cancer types and observed a negative association between the methylation level of the cg06726167 site upstream of SNORD118 host gene and its expression in diverse cancers. In HCC patients, CpG island methylation in the gene promoter region of SNORD113-1 was significantly higher compared to adjacent non-tumor tissue, leading to a marked downregulation of its expression. SNORD113-1 exhibits tumor suppressor properties by inhibiting the phosphorylation of extracellular signal-regulated kinase 1/2 (ERK1/2) and mothers against decapentaplegic homolog 2/3 (SMAD2/3) in the mitogen-activated protein kinase (MAPK/ERK) and transforming growth factor beta (TGF-β) pathways, thus suppressing the proliferation of HepG2 and Huh7 cancer cells [56]. Furthermore, extensive evidence shows the widespread occurrence of highly methylated 5'-CpG islands in host genes, including SNORD123, U70C, and ACA59B in colorectal and lung cancer cell lines, leading to epigenetic silencing of the associated snoRNAs [65].
3.3. Dependence of snoRNA expression on their host gene transcription and splicing events
Recent studies have revealed that most snoRNAs are derived from introns of coding or non-coding genes, with their expression relying on the transcription levels and splicing events of the host genes [59,60] (Fig. 2). For instance, Fafard-Couture et al. [2,2] recently demonstrated that the transcription and splicing activities of host genes determine the expression and conservation of encoded snoRNAs through comprehensive analysis. Similarly, Boivin et al. [66] found the snoRNAs embedded within introns of protein-coding genes are generated via the transcription and splicing activities of their host genes.
The production of full-length transcripts (also known as pre-RNA), including exons and introns, is encoded by host genes. Among these transcripts, introns with nuclear localization are further processed into smaller molecules ranging from 65 to 300 nucleotides, thus defined as snoRNA [60]. Consequently, the abundance of the encoded snoRNAs directly depends on the transcriptional regulation of their host genes. For instance, snoRNA U14–U24 and E3 are derived from SNHG1, which is actively transcribed into pre-RNA and released through the processing of excised introns [9]. Additionally, existing evidence suggests co-expression between intronic snoRNAs and their host genes, further confirming their transcriptional regulatory pattern [11,67].
Furthermore, the splicing patterns of host genes significantly influence the expression of their encoded snoRNAs [59]. Specifically, transcripts from genes encoding multiple snoRNAs typically generate alternative transcript isoforms, allowing for the differential expression of individual co-encoded snoRNAs [2]. A representative study confirms that the splicing factor serine-arginine protein kinase 1 (SRPK1) upregulates the expression of SNORA42, SNORD10, and SNORA74A, which is associated with increased gastric cancer cell proliferation [68]. Additionally, splicing events in host genes produce considerable amounts of nonsense-mediated mRNA decay-sensitive splice variants with premature termination codons, impacting the levels of encoded snoRNAs [59,69]. Interestingly, multi-SNHGs can fine-tune the levels of their encoded individual snoRNAs through post-transcriptional alternative splicing events [69]. For instance, the gene C17orf76-AS1 encodes and regulates multiple individual snoRNAs, such as SNORD49B, SNORD49A, and SNORD65 by yielding alternative transcript isoforms, enabling their differential expression [69].
In summary, these studies have elucidated the complex regulatory relationship between host genes and their intron-encoded snoRNAs, offering intriguing insights into the mechanisms governing snoRNA expression (Table S1 [[56], [57], [58], [62], [65], [68]] and Fig. 2).
4. Emerging mechanisms of snoRNAs in cancer
SnoRNAs significantly influence tumor development and progression. In recent years, novel regulatory mechanisms have been discovered that are crucial in the development and occurrence of tumors, in addition to the two traditional mechanisms. The primary regulatory mechanisms include the transcriptional and post-transcriptional as well as post-translational regulation of snoRNAs (Fig. 3). Multiple studies have indicated that snoRNAs could indirectly regulate tumor development and progression by selectively modifying target RNAs, whereas direct binding of snoRNAs to target RNAs regulates their post-transcriptional modifications and contributes to post-translational regulation.
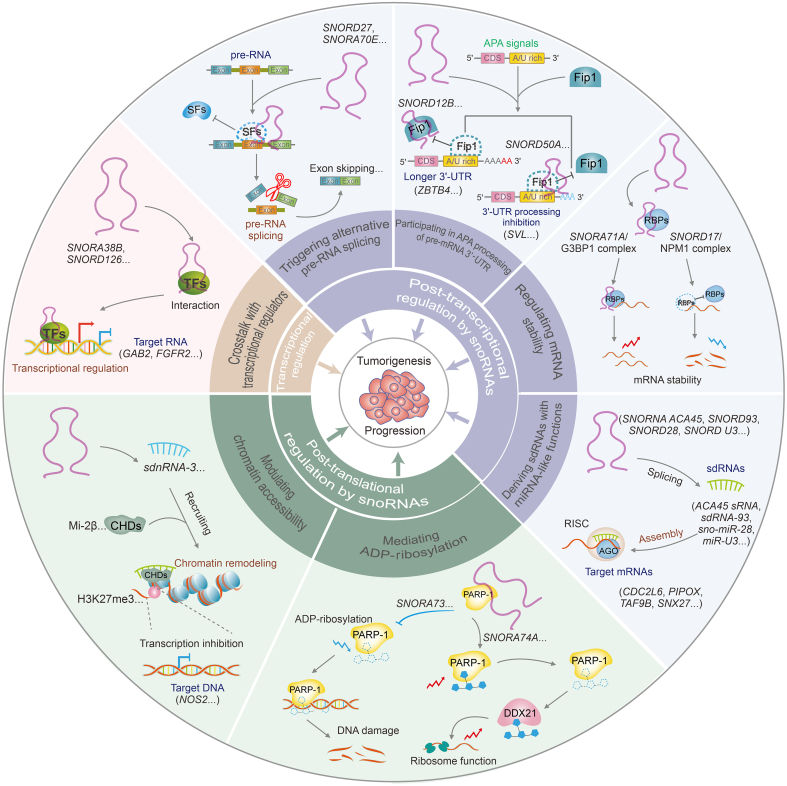
Fig. 3.
The emerging mechanisms of small nucleolar RNAs (snoRNAs) involved in tumorigenesis and progression. SnoRNAs affect the biological functions of multiple target genes, and thus tumor progression, through three main regulatory mechanisms. The first is the transcriptional regulation of snoRNAs, including the crosstalk between snoRNAs and transcription regulators. The second is the post-transcriptional regulation of snoRNAs, including snoRNAs triggering alternative pre-RNA splicing, participating in alternative polyadenylatio (APA) processing of the pre-messenger RNA (mRNA) 3'-untranslated region (UTR), regulating mRNA stability, and deriving snoRNA-derived RNAs (sdRNAs) with microRNAs (miRNA)-like functions. The last is the post-translational regulation of snoRNAs, including snoRNAs mediating adenosine diphosphate (ADP)-ribosylation and modulating chromatin remodeling. FGFR2: fibroblast growth factor receptor 2; TFs: transcription factors; SFs: splicing factors; CDS: coding sequence; SNORDs: C/D box snoRNAs; RBPs: RNA binding proteins; G3BP1: G3BP stress granule assembly factor 1; NPM1: nucleophosmin 1; RISC: RNA-induced silencing complex; AGO: argonaute; PARP1: poly(ADP-ribose) polymerase 1; DDX21: DEAD-box helicase 21; sdnRNA-3: small nucleolar-derived RNA 3; CHDs: chromodomain helicase DNA binding proteins; H3K27me3: tri-methylation at lysine 27 of histone H3.
4.1. Transcriptional and post-transcriptional regulation by snoRNAs
4.1.1. SnoRNAs regulate gene transcription by crosstalk with transcription factors
The interaction between snoRNAs and transcription regulators could modulate the expression levels of crucial cancer-related genes, thereby initiating malignant tumor development [70,71] (Fig. 3). For instance, in NSCLC, SNORA38B facilitates the transcription of the downstream GAB2 gene by directly interacting with the transcription factor E2F transcription factor 1 (E2F1), leading to activation of the AKT/mTOR pathway and driving the progression of NSCLC [70]. In addition, SNORD126 can directly bind to heterogeneous nuclear ribonucleoprotein K (hnRNPK), resulting in the upregulation of FGFR2 transcription and activation of the PI3K-AKT pathway, thereby facilitating the progression of HCC [71]. Thus, understanding the interactions between snoRNAs and transcription regulators is crucial for understanding the molecular basis of cancer.
4.1.2. SnoRNAs trigger alternative pre-RNA splicing
SnoRNAs are a type of snRNA molecules that play a crucial role in regulating gene expression. Recent studies have demonstrated that snoRNAs can induce diverse biological alterations in tumors by affecting target splicing and RNA levels [12,45,[72], [73], [74]] (Fig. 3). SNORA70E is a splicing regulatory factor that influences the selective splicing of PARPBP in OV, resulting in the exclusion of exon 4 of the PARPBP-88 transcript and generation of a novel transcript, PARPBP-15. This action subsequently facilitates the invasion, migration, and proliferation of OV cells [45]. Similarly, SNORD27 competitively binds to splicing factors in HeLa cells, resulting in the exclusion of exon 12 of E2F7 pre-mRNA, while inhibiting the inclusion of silencing exons in FER, ZBTB37, MAP4K3, and ABCA8 pre-mRNA. This ultimately contributes to cell proliferation [72]. SNORD88C (also known as HBII-180C) regulates the splicing of fibroblast growth factor receptor 3 (FGFR3) mRNA, contributing to the development of various cancers [12,75,76]. Moreover, a recent study showed that SNORD75 can regulate the splicing of GAS pre-long non-coding RNA (lncRNA) based on N6-methyladenosine (m6A) modification factors by recruiting and binding to the methyltransferase-like 3 (METTL3)/METTL14 complex [73]. These findings suggest potential avenues for cancer treatment and highlight the crucial role of snoRNAs in regulating gene expression and cancer progression.
4.1.3. SnoRNAs participate in alternative polyadenylation (APA) processing of the pre-mRNA 3'-untranslated region (UTR)
APA is a crucial post-transcriptional regulatory mechanism in the mRNA maturation process of eukaryotic cells. It is used to modify and process the 3'-end of precursor mRNA [77]. SnoRNAs regulate APA modification, which in turn regulates mRNA 3'-end processing and affects the expression of cancer-related genes [78,79] (Fig. 3). SNORD12B is upregulated in GBM cells. FIP1L1 (also known as FIP1) binding facilitates the use of the distal poly(A) site of the transcriptional repressor ZBTB4 during APA, resulting in an increased proportion of the long 3'-UTR transcript of ZBTB4, which downregulates ZBTB4 expression, ultimately inhibiting glucose and lipid metabolism and the proliferation of GBM cells [78]. Additionally, SNORD50A functions as a direct APA regulator that inhibits SVL mRNA 3'-end processing in HeLa cells by disrupting the FIP1-PAS interaction [79,80].
4.1.4. SnoRNAs regulate mRNA stability
Several studies have shown that snoRNAs can function as post-transcriptional regulators that influence mRNA stability, thereby contributing to cancer development [58,81] (Fig. 3). For example, SNORA71A binds to G3BP1, a protein that regulates mRNA stability, increases its stability, and upregulates ROCK2, a negative regulatory factor of TGF-β. This promotes the growth and metastasis of BRCA [81]. Upregulated SNORD17 binds to NPM1, reducing the stability of the tumor suppressor protein, P53, and promoting the onset and metastasis of HCC [58]. Therefore, snoRNAs play a significant role in cancer progression by regulating mRNA stability and influencing specific signaling pathways. Further in-depth studies will provide a clearer understanding of the specific mechanisms by which these molecules affect various diseases.
4.1.5. sdRNAs with miRNA-like functions
SnoRNAs can generate smaller molecules that bind to the argonaute (AGO) protein, exhibiting a function similar to that of miRNA. These small molecules are referred to as sdRNAs or sno-miRNAs. Preliminary research has shown that the snoRNA ACA45 can produce stable sdRNA, specifically ACA45 sRNA, in HEK293 cells [14]. Similar to miRNAs, this molecule can bind complementarily to the 3'-UTR of CDC2L6 and inhibit its expression [14]. SdRNAs can mimic the functions of miRNAs by completely or partially binding to target mRNA, promoting gene silencing, and affecting the onset and development of tumors [[82], [83], [84]] (Fig. 3). For example, snoRNA93-derived sdRNA-93 (also known as HBII-336) exhibits miRNA-like characteristics. In BRCA cells, this highly expressed sdRNA-93 can bind to the PIPOX 3'-UTR and suppress its expression, thereby inducing invasive characteristics in specific BRCA cell types (e.g., MCF-7 and MDA-MB-231) [82]. In addition, sno-miR-28 contributes to the proliferation of BRCA cells by directly targeting the upregulated TAF9B 3'-UTR and indirectly reducing P53 stability [83]. Box C/D snoRNA U3 is overexpressed in multiple types of cancer [85]. Lemus-Diaz et al. [84] observed the transformation of U3 snoRNA into miR-U3 in HCT116 cells, sharing characteristics with miRNA. This enables miR-U3 to bind to the 3'-UTR of SNX27 mRNA, potentially influencing various cellular functions. Similarly, miR-U3 also binds to the 3'-UTR of ZBTB4, reducing the stability of ZBTB7A mRNA. This results in downregulation of ZBTB7A protein expression, promoting glioblastoma cell glycolytic capacity and proliferation [86]. Targeting these functional snoRNAs and their smaller molecular derivatives, such as with antagonistic antisense oligonucleotides (ASOs) and small molecules, could be an effective strategy, similar to the inhibition of oncogenic miRNAs [87,88]. Thus, these findings provide insights into the development of novel cancer treatments.
4.2. Post-translational regulation by snoRNAs
4.2.1. SnoRNAs mediate ADP-ribosylation
SnoRNAs affect the post-translational modification of nuclear proteins by influencing the catalytic activity of PARP1, which has implications for cancer occurrence and development [89,90] (Fig. 3). Recent research has revealed that SNORA73, a chromatin-related orphan snoRNA, can bind to PARP1 to form snoRNP, which inhibits the automatic poly(ADP-ribosyl)ation of PARP1, thereby affecting the levels of DNA damage. Consequently, this process contributes to increased genomic instability and AML progression [89]. Kim et al. [90] found that SNORA74A could activate PARP-1 self-modification in BRCA1/2 wild-type BRCA cells. This activation leads to site-specific ADP-ribosylation of DEAD-box helicase 21 (DDX21) via its interaction with PARP-1 and the RNA helicase DDX21. Consequently, this process promotes the growth of BRCA cells by enhancing ribosome biosynthesis and protein translation. Further in-depth research is warranted to investigate the regulatory mechanisms of these snoRNAs on PARP1 and DNA damage repair (DDR) pathways to improve our understanding of the significant roles in cancer progression.
4.2.2. SnoRNAs and sdRNAs modulate chromatin remodeling
SnoRNAs facilitate the selective recruitment and binding of chromatin modifiers to regulate chromatin accessibility. Schubert et al. [91] found that snoRNAs can interact with drosophila decondensation factor 31 (DF31) to form ribonucleoprotein (RNP) complexes, resulting in the decondensation of higher-order chromatin structures and maintenance of high-level chromatin structures. Thus, snoRNAs and their derivatives may influence cancer development through chromatin structural modifications [65,92] (Fig. 3). Bisphenol A promotes the development of prostate cancer by inhibiting the activity of SNORD59A, SNORD82, SNORD116, and SNORD117 through the recruitment of histone H3 lysine 9 trimethylation (H3K9me3), H3K4me3, and H3K27me3 [92]. Recent studies have identified dicer-independent small non-coding RNA (sncRNA) derived from snoRNAs (sdnRNA)-sdnRNA-3 and snRNAs/snoRNAs that inhibit NOS2 transcription in macrophages. This is accomplished by recruiting the Mi-2β protein, thereby blocking the accessibility of the NOS2 gene promoter and promoting tumor growth [93]. These findings provide novel insights into the regulation of macrophage function in cancer immunity.
In summary, the regulatory mechanisms of snoRNAs in tumor development are complex and comprehensive, providing novel insights into these conditions (Table S1 [[12], [14], [45], [70], [71], [72], [73], [78], [79], [80], [81], [82], [83], [84], [86], [89], [90], [92], [93]] and Fig. 3).
5. Mechanisms of SNHGs in cancer
As mentioned above, unlike independent transcription pattern, a significant portion of snoRNAs is transcribed from sequences within introns of protein-coding or non-coding host genes [2,59]. SnoRNAs show a strong correlation in abundance with SNHGs; however, the expression relationship between snoRNAs and their coding host genes is largely uncorrelated [2]. Fafard-Couture et al. [2] explain that this discrepancy is due to the presence of more dual-initiation promoters in protein-coding host gene/snoRNA pairs than in non-coding host gene/snoRNA pairs. Interestingly, the functional roles also differ significantly between coding genes, many of which are related to the translational apparatus, and non-coding SNHGs. The architecture of SNHGs varies to uncouple the expressions of the host genes and snoRNAs, thus meeting diverse snoRNA abundance levels and functional requirements. This suggests that the functions of SNHGs may actually reflect those of snoRNAs [2]. Therefore, clarifying the pivotal roles of SNHGs is essential to uncovering the intricate roles of snoRNAs in tumors.
SNHGs have been implicated in the regulation of cellular homeostasis and cancer biology, in addition to contributing to the etiology of cancer, as shown in Table 2 [[94], [95], [96], [97]] and Fig. 4A. SNHG5, the host gene of snoRNA U50, serves as a tumor suppressor in gastric cancer by sequestering metastasis-associated protein 2 (MTA2) in the cytoplasm, thereby impeding cancer progression. This finding indicates that SNHG5 may be a promising therapeutic target for gastric cancer [94]. The lncRNA SNHG6 promotes CRC cell migration, invasion, and epithelial-mesenchymal transition by regulating the miR-26a/enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) axis [95]. SNHG15 has been implicated in prostate cancer development via the miR-338-3p/FK506-binding protein (FKBP) prolyl isomerase 1A (FKBP1A) axis [96]. SNHG12 promotes the proliferation and invasiveness of papillary thyroid cancer cells by targeting miR-16-5p [97]. SNHG1, SNHG6, SNHG16, and SNHG20 play important roles in HCC progression via various regulatory mechanisms [98]. Additionally, the majority of SNHGs (SNHG1, SNHG12, SNHG20, SNHG15, and SNHG16) function by sequestering tumor-suppressing microRNAs, thereby enabling the production of oncogene transcripts in malignancies [99].
Table 2.
Mechanisms of small nucleolar RNAs (snoRNA) host genes (SNHGs) in cancer.
| SNHG | snoRNA | Mechanism | Biological function | Tumor type | Refs. |
|---|---|---|---|---|---|
| SNHG5 | U50 | Trapping MTA2 in the cytosol | Inhibiting tumor development | Gastric cancer | [94] |
| SNHG6 | / | Sponging miR-26a | Promoting tumorigenesis and metastasis | CRC | [95] |
| SNHG15 | / | Sponging miR-338-3p | Promoting proliferation | Prostate cancer | [96] |
| SNHG12 | / | Sponging miR-16-5p | Promoting growth and invasion | Papillary thyroid cancer | [97] |
| SNHG17 | SNORA71B | Sponging miR-339-5p | Promoting tumorigenesis and metastasis | Prostate cancer | [100] |
| SNHG10 | SCARNA13 | Sponging miR-150-5p | Promoting tumorigenesis and metastasis | Hepatocellular cancer | [101] |
| SNHG5 | SNORA50C | Recruiting DKC1 | Promoting tumorigenesis and metastasis | Hepatocellular cancer | [102] |
/: no data. MTA2: metastasis-associated protein 2; CRC: colorectral cancer; DKC1: dyskerin pseudouridine synthase 1.
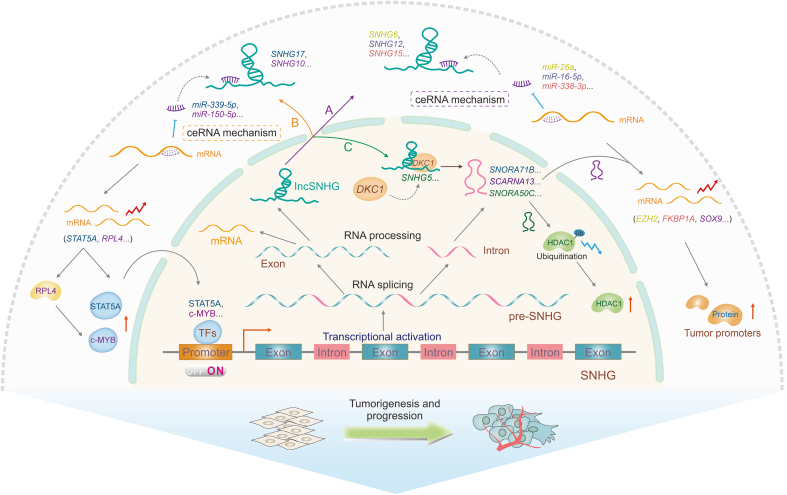
Fig. 4.
Regulatory mechanisms of small nucleolar RNAs (snoRNAs) derived from long non-coding snoRNA host gene (lncSNHGs) induce consistent biological functions in cancer. Long non-coding RNA snoRNA host genes (lncRNA SNHGs) and their homologous snoRNAs are derived from the same primary transcript, pre-SNHGs. SnoRNAs are usually derived from introns and lncRNA SNHGs are usually derived from exons. LncRNA SNHGs and snoRNAs participate in tumorigenesis and development through three regulatory pathways. (A) LncRNA SNHGs sponge microRNAs (miRNAs) through the ceRNA mechanism and inhibit the degradation of downstream messenger RNAs (mRNAs), which in turn promotes tumor progression. (B) LncRNA SNHGs, through the competing endogenous RNA (ceRNA) mechanism, upregulate transcription factors, forming a positive feedback loop to upregulate their homologous snoRNAs. (C) LncRNA SNHG5 recruits dyskerin pseudouridine synthase 1 (DKC1) to promote SNORA50C accumulation and inhibits histone deacetylase 1 (HDAC1) ubiquitination, promoting hepatocellular cancer progression. The yellow color denotes SNHG6/miR-26a/enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) axis. The grey color denotes SNHG12/miR-16-5p axis. The peach color denotes SNHG15/miR-338-3p/FK506-binding protein (FKBP) prolyl isomerase 1A (FKBP1A) axis. The blue color denotes SNHG17/miR-339-5p/signal transducer and activator of transcription 5A (STAT5A)/SNORA71B axis. The purple denotes SNHG10/miR-150-5p/ribosomal protein L4 (RPL4)/MYB proto-oncogene, transcription factor (c-Myb)/SCARNA13/SRY-box transcription factor 9 (SOX9) axis. The green denotes SNHG5/DKC1/SNORA50C/HDAC1 axis. TFs: transcription factors.
Moreover, snoRNAs and their associated SNHGs are significantly involved in human cancer progression. SNHGs regulate the expression of homologous snoRNAs through positive feedback loops, thus influencing cancer onset and progression, as shown in Table 2 [100,101] and Fig. 4B. The lncRNA SNHG17 positively regulates its homologous gene SNORA71B in prostate cancer cells [100]. SNHG17 functions as a sponge for miR-339-5p, resulting in the upregulation of signal transducer and activator of transcription 5A (STAT5A) and subsequent activation of SNORA71B, thereby exacerbating the progression of prostate cancer [100]. In HCC, SNHG10 expression is positively correlated with its corresponding snoRNA, SCARNA13. SNHG10 regulates SCARNA13 expression through a positive feedback loop involving miR-150-5p, ribosomal protein L4 (RPL4), and MYB proto-oncogene, transcription factor (c-MYB). Conversely, SCARNA13 regulates the oncogenic activity of SRY-box transcription factor 9 (SOX9) in HCC by promoting cellular proliferation, invasion, and migration [101]. Besides, SNHGs can interact directly with snoRNAs and affect their expression, as shown in Table 2 [102] and Fig. 4C. In neuroblastoma (NB), SNHG5 promotes the accumulation of its homologous gene SNORA50C and assembly of the associated snoRNP by recruiting DKC1. SNORA50C suppresses the ubiquitination of histone deacetylase 1 (HDAC1) and upregulates the expression of the oncogenic factor HDAC1 in NB cells. This process facilitated the proliferation and migration of NB cells [102]. Taken together, research on SNHGs and snoRNAs offers preliminary insights into cancer occurrence and progression, potentially advancing diagnosis and treatment strategies for cancer patients. However, further studies are needed to elucidate the distinct roles of snoRNAs and their host genes. Specifically, research distinguishing the contributions of snoRNAs from those of their host genes would be particularly valuable for understanding their individual impacts on cancer.
6. SnoRNA-mediated tumor drug resistance
6.1. SnoRNAs mediate multidrug resistance by regulating tumor cell stemness
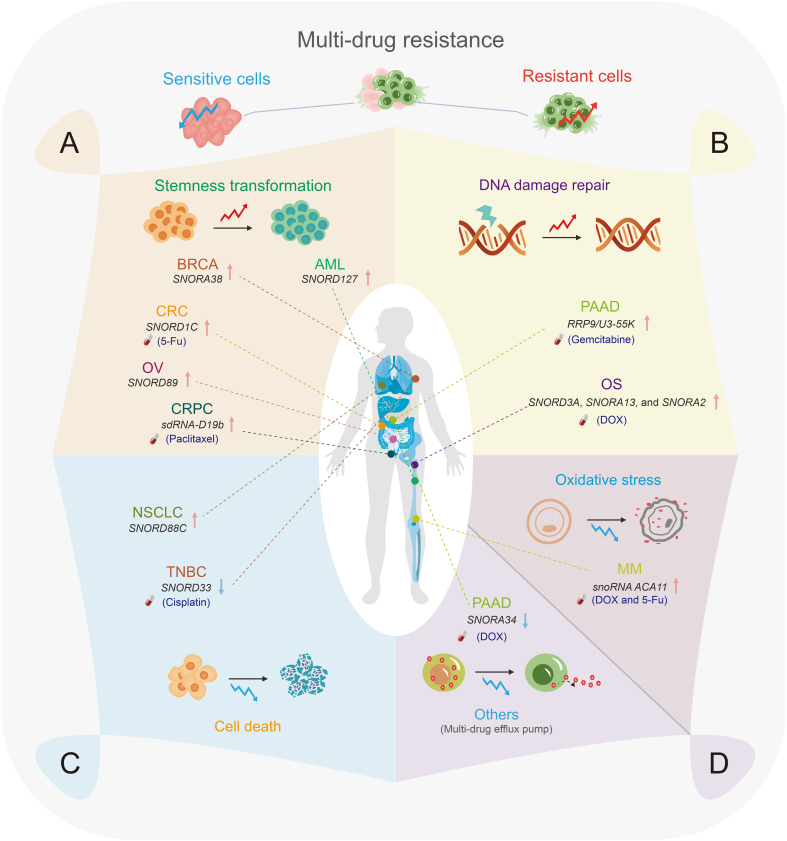
Multiple studies have demonstrated the regulatory role of snoRNAs in the maintenance of tumor cell stemness, which has significant implications for tumor onset and progression [41,103,104] (Fig. 5). Remarkably, SNORD127 triggers preferential translation of amino acid transporter mRNAs in a codon-dependent manner to drive AML stem cell phenotypes, such as self-renewal [41]. Similarly, SNORD89 activates the NOTCH1/cellular MYC (c-MYC) pathway and induces stemness in OV cells, as shown in Table 3 [103]. SNORA38 positively correlates with OCT-4 stem cell marker and regulates the stemness of BRCA cells [104]. Moreover, snoRNAs can transform tumor stem cells by regulating the expression of stem cell markers, resulting in uncontrolled tumor proliferation and drug resistance, as shown in Table 3 [105,106]. SNORD1C overexpression in CRC activates the Wnt/β-catenin pathway, resulting in the upregulation of CD44, SOX2, and other stem cell genes. This activation enhances colon cancer cell proliferation and cell cycle progression and contributes to 5-fluorouracil (5-FU) resistance in these cells [105]. In addition, sdRNA-D19b increased the proliferation and metastasis of castration-resistant prostate cancer (CRPC) cells by selectively targeting the 3'-UTR of the CD44 mRNA. This mechanism confers resistance to paclitaxel drugs [106]. This study proposed sdRNAs as potential biomarkers and therapeutic targets for future clinical interventions in CRPC [106].
Fig. 5.
Small nucleolar RNAs (snoRNAs) mediating tumor multi-drug resistance. (A) Some snoRNAs promote tumor drug resistance by promoting cell stemness transformation. (B) Some snoRNAs promote tumor drug resistance by promoting DNA damage repair (DDR). (C) Some snoRNAs mediate tumor drug resistance by regulating cell death. (D) Some snoRNAs promote tumor multi-drug resistance by inhibiting oxidative stress. Others, such as SNORA34, can downregulate the membrane transporter protein adenosine triphosphate (ATP) binding cassette subfamily C member 1 (ABCC1) to inhibit tumor drug resistance. Red arrows denote promoting drug resistance and blue arrows denote reversing drug resistance. BRCA: breast cancer; AML: acute myeloid leukemia; CRC: colorectal cancer; 5-Fu: 5-fluorouracil; OV: ovarian cancer; SNORDs: C/D box snoRNAs; CRPC: castration-resistant prostate cancer; sdRNA: snoRNA-derived RNAs; PAAD: pancreatic adenocarcinoma; RRP9: ribosomal RNA (rRNA) processing 9; OS: osteosarcoma; NSCLC: non-small cell lung cancer; TNBC: triple-negative BRCA; MM: multiple myeloma; 5-FU: 5-fluorouracil; DOX: doxorubicin.
Table 3.
Small nucleolar RNAs (snoRNAs) involved in tumor drug resistance.
| SnoRNA | Mechanism | Biological function | Drug sensitivity | Related tumor type | Refs. |
|---|---|---|---|---|---|
| SNORD88C | Guiding 2'-O-methylation of 28S rRNA and upregulating SCD1 | Inhibiting autophagy and promoting growth and metastasis | / | NSCLC | [53] |
| SNORD89 | Activating the NOTCH1/c-MYC pathway | Promoting tumor cell proliferation and self-renewal ability | / | OV | [103] |
| SNORD1C | Activating the Wnt/β-catenin pathway | Promoting tumor cell proliferation, migration, and invasion | / | CRC | [105] |
| sdRNA-D19b | Targeting the 3'-UTR of CD44 genes | Promoting tumor cell proliferation and migration | Paclitaxel (−) | CRPC | [106] |
| SNORD3A, SNORA13, and SNORA2 | Downregulating TOP2A and upregulating GADD45A | Promoting tumor development | DOX (−) | Human osteosarcoma | [111] |
| U3 snoRNA-related protein, RRP9/U3-55K | Activating the AKT signaling pathway | Promoting tumor growth | Gemcitabine (−) | Pancreatic cancer | [112] |
| SNORD33 | Binding to MeCP2 | Promoting tumor cell apoptosis | Cisplatin (+) | Triple-negative BRCA | [116] |
| SNORNA ACA11 | Inhibiting NRF2 transcriptional activity | Promoting tumor development | DOX (−) and 5-FU (−) | MM | [122] |
| SNORA34 | Deriving hsa-miR-1291 to target 3'-UTR of ABCC1 | Promoting tumor cell sensitivity | DOX (+) | Pancreatic cancer | [125] |
/: no data. (−) refers to downregulation and (+) refers to upregulation. SNORDs: C/D box snoRNAs; rRNA: ribosomal RNA; SCD1: stearoyl-CoA desaturase 1; NSCLC: non-small cell lung cancer; NOTCH1: neurogenic locus notch homolog protein 1; c-MYC: cellular MYC; OV: ovarian cancer; CRC: colorectal cancer; CD44: CD44 molecule; CRPC: castration-resistant prostate cancer; TOP2A: DNA topoisomerase II alpha; GADD45A: growth arrest and DNA damage inducible 45A; DOX: doxorubicin; RRP9: ribosomal RNA processing 9; AKT: protein kinase B; MeCP2: methyl cytosine-phosphate-guanine-binding protein 2; BRCA: breast cancer; NRF2: NF-E2-related factor 2; 5-FU: 5-Fluorouracil; MM: multiple myeloma; ABCC1: ATP binding cassette subfamily C member 1.
6.2. SnoRNAs or their associated regulatory proteins mediate tumor chemoresistance via the DDR pathway
Tumor cells possess a robust DNA repair mechanism that effectively repairs chemotherapy-induced DNA damage, thereby enhancing drug resistance [[107], [108], [109]]. Doxorubicin (Dox)-induced DNA damage in colon cancer cells results in the expression of growth arrest-specific transcript 5 (GAS5)-derived snoRNA in a P53-dependent pathway, suggesting that snoRNA may influence DNA damage [110]. SnoRNAs and their associated proteins are involved in DDR, which has implications for drug resistance, as shown in Table 3 [111,112] and Fig. 5. The upregulation of SNORD3A, SNORA13, and SNORA2 expression in Dox-resistant osteosarcoma cells is crucial for the synthesis of Dox-resistant proteins by downregulating the Dox target DNA topoisomerase II alpha (TOP2A), upregulating the DNA damage sensor growth arrest and DNA damage inducible 45A (GADD45A), and regulating the mechanisms involved in DNA repair and ribosome formation. Consequently, these molecular mechanisms contribute to the development of Dox resistance in osteosarcoma [111]. Therefore, the identification and targeting of specific snoRNAs or their downstream genes present novel therapeutic prospects for chemoresistant osteosarcoma. In addition, gemcitabine resistance in pancreatic adenocarcinoma (PAAD) is induced by the U3 snoRNA-related protein, rRNA processing 9 (RRP9)/U3-55K. This protein activates the AKT signaling pathway, reducing DNA damage and impeding cell apoptosis [112]. Therefore, the combination of the AKT inhibitor MK-2206 and gemcitabine shows potential as a therapeutic strategy for patients with gemcitabine-resistant pancreatic cancer.
6.3. SnoRNAs mediate chemoresistance by regulating cell death
Cell death pathways such as apoptosis, autophagy, and necrosis have distinct morphological and biochemical characteristics. Apoptosis is typically associated with cell contraction, whereas autophagy is characterized by cytoplasmic vacuolization and autophagosome formation. These autophagosomes facilitate the elimination of substances via lysosomes [[113], [114], [115]]. SnoRNAs have the potential to influence tumor progression by regulating cell death, as shown in Table 3 [53,116] and Fig. 5. SNORD88C promotes the growth and metastasis of NSCLC by inducing 2'-O-methylation of 28S rRNA. This process enhances the translation activity of the downstream target gene SCD1, reduces the number of autophagosomes, and inhibits autophagy [53]. And the resistance of tumor cells to anti-cancer drugs is influenced by disruption of the snoRNA-mediated apoptotic mechanism [116]. SNORD33 binds to methyl CpG-binding protein 2 (MeCP2) and disrupts its interaction with target genes. Therefore, the expression of anti-apoptotic proteins (myeloid leukemia 1 (MCL-1) and BCL-2) decreases, whereas that of pro-apoptotic proteins (caspase-3 (Cas3) and Cas9) increases. Therefore, the promotion of tumor cell apoptosis leads to the reversal of cisplatin resistance in triple-negative BRCA cells [116].
6.4. SnoRNAs mediate chemoresistance by regulating reactive oxygen species (ROS)
ROS are generated during oxidative stress. Increased cellular ROS levels activate multiple antioxidant mechanisms in tumor cells [117,118]. This suppresses ROS generation, resulting in increased resistance to cancer therapies [[119], [120], [121]]. Previous studies have demonstrated the involvement of snoRNAs in regulating cellular oxidative stress and promoting tumor resistance [[122], [123], [124]] (Fig. 5). ACA11, an orphan snoRNA, inhibits oxidative stress, promotes MM cell proliferation, and enhances resistance to Dox and 5-FU, as shown in Table 3 [122]. ACA11 inhibits nuclear factor erythroid 2 (NF-E2)-related factor 2 (NRF2) transcriptional activity in multiple myeloma (MM). This inhibition increases ROS-dependent biological processes, including pre-47S rRNA synthesis, protein synthesis, and ribosome biogenesis [123,124]. Moreover, ACA11 overexpression enhances MM cell responsiveness to the proteasome inhibitor bortezomib [124].
6.5. Others
SnoRNAs regulate multidrug resistance-associated protein (MRP1)/adenosine triphosphate (ATP) binding cassette subfamily C member 1 (ABCC1) expression, affecting intracellular drug transport and promoting multidrug resistance in cancer cells. In pancreatic cancer, hsa-miR-1291 from SNORA34 specifically targets the 3'-UTR of the membrane transport protein ABCC1, leading to the downregulation of its expression. Consequently, drug accumulation increases within cells, causing a reversal of mitoxantrone resistance in cancer cells, as shown in Table 3 [125].
7. Therapeutic strategies and clinical implications of cancer-associated snoRNAs
7.1. SnoRNAs as diagnostic and prognostic biomarkers in cancer
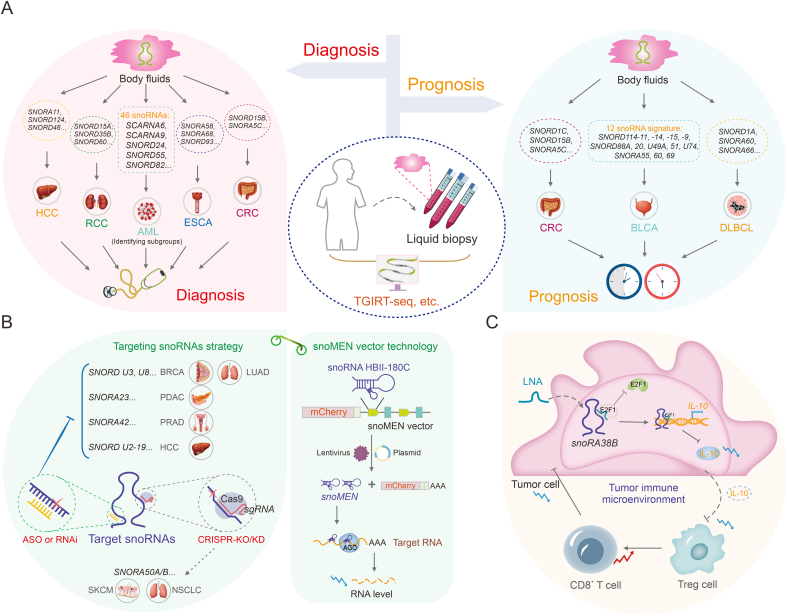
SnoRNAs play a significant role in cancer and can be readily detected in different body fluids, including plasma and serum [[126], [127], [128]] (Fig. 6). SnoRNAs have emerged as promising diagnostic and prognostic biomarkers owing to advancements in high-throughput sequencing and bioinformatics technology, which have radically changed in recent years [129]. Notably, thermostable group II intron reverse transcriptases RNA sequencing (TGIRT-seq) exhibits high fidelity, processivity, and strand-displacement activity, along with proficient template-switching capabilities. Evidence indicates that TGIRT-seq allows comprehensive profiling of structured sncRNAs, including snoRNAs, revolutionizing our understanding of snoRNA expression. This method offers a novel strategy for evaluating tumorigenesis and outcomes [2,130]. Further, Xie et al. [131] identified SNORA11, SNORD124, and SNORD46 as key snoRNAs with potential implications in HCC diagnosis and treatment. This discovery was made by analyzing transcriptome data from The Cancer Genome Atlas (TCGA) and The International Cancer Genome Consortium (ICGC) datasets. Similarly, SNORD15A, SNORD35B, and SNORD60 are significantly overexpressed in cancerous tissues and urine, making them useful biomarkers for the early diagnosis and potential therapeutic targets for renal cell carcinoma (RCC) [132]. In esophageal cancer (ESCA), the upregulation of SNORA58, SNORA68, and SNORD93 is observed in both ESCA patients and early-stage patients. Additionally, combining snoRNAs with the existing tumor marker carcinoembryonic antigen is shown to enhance the diagnostic ability in ESCA patients [133]. In diffuse large B-cell lymphoma (DLBCL), a triple snoRNA prognostic risk model consisting of SNORD1A, SNORA60, and SNORA66 was constructed. The study revealed that the upregulation of SNORD1A and SNORA60 was associated with poor prognosis in DLBCL patients, while SNORA66 was upregulated in the low-risk group, serving as a protective factor. This study provides novel insights into the prognostic significance of snoRNAs in DLBCL [134].
Fig. 6.
Therapeutic strategies and clinical implications of cancer-associated small nucleolar RNAs (snoRNAs). (A) SnoRNAs act as liquid biopsy biomarkers for tumor early diagnosis and prognosis evaluation. (B) Current cancer therapeutic strategies for targeting oncogenic snoRNAs include antisense oligonucleotide (ASO) technology, small interfering RNA (siRNA), short hairpin RNA (shRNA), clustered regularly interspaced short palindromic repeats (CRISPR)/caspase-9 (Cas9) technology, and snoRNA modulator of gene expression (snoMEN) vector technology. (C) Targeting tumor immune microenvironment (TIME)-related SNORA38B by locked nucleic acid (LNA) increases the infiltration of CD8+ T cells, and inhibits proliferation of non-small cell lung cancer (NSCLC) cells. SNORAs: H/ACA box snoRNAs; SNORDs: C/D box snoRNAs; HCC: hepatocellular carcinoma; RCC: renal cell carcinoma; AML: acute myeloid leukemia; ESCA: esophageal cancer; CRC: colorectal cancer; TGIRT-seq: thermostable group II intron reverse transcriptases RNA-seq; BLCA: bladder cancer; DLBCL: diffuse large B-cell lymphoma; BRCA: breast cancer; LUAD: lung adenocarcinoma; PDAC: pancreatic ductal adenocarcinoma; PRAD: prostate adenocarcinoma; RNAi: RNA interference; sgRNA: single guide RNA; KO/KD: knockout/knockdown; SKCM: skin cutaneous melanoma; NSCLC: non-small cell lung cancer; AGO: argonaute; LNA: locked nucleic acid; E2F1: E2F transcription factor 1; IL-10: interleukin-10; Treg cell: regulatory T cell.
The oncogenic properties of SNORD15B and SNORA5C in CRC suggest that these two snoRNAs could serve as promising biomarkers for the diagnosis and prognostic prediction of CRC [135]. Moreover, distinct B-cell malignancies exhibit the abnormal expression of specific snoRNAs [136,137]. Teittinen et al. [137] identified 46 snoRNAs with differential expression between B-cell precursor acute lymphoblastic leukemia (BCP-ALL) and T-cell acute lymphoblastic leukemia (T-ALL) using large-scale parallel sequencing (sequencing by oligonucleotide ligation and detection (SOLiD)). These snoRNAs have potential applications as diagnostic biomarkers or therapeutic targets.
SnoRNAs exhibit diverse characteristics in cancer and can serve as reliable prognostic markers [138,139] (Fig. 6). A risk assessment classifier was developed in bladder cancer (BLCA), using the expression profiles of 12 survival-related snoRNAs. Clinical outcomes vary between high- and low-risk patient groups. This characteristic makes it a reliable prognostic predictor for patients with BLCA [138]. Cai et al. [139] identified TIIsno-snoRNA characteristics associated with immune infiltration by analyzing integrated expression profiles of snoRNAs in 21 purified immune cell lines, 43 colon cancer cell lines, and 3 data sets. Patients with lower TIIsno scores exhibited better responses to immune therapy. This feature may serve as an indicator for predicting prognosis and response to immune treatment in patients with colon cancer.
7.2. Application of snoRNAs in targeting cancer therapy
SnoRNAs are implicated in carcinogenesis through diverse regulatory mechanisms. These molecules can be targeted for cancer therapy owing to their oncogenic potential [140]. Several cancer therapeutic strategies have been developed to target snoRNAs, including clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9, small interfering RNA (siRNA), short hairpin RNA (shRNA), and ASO technologies [87]. ASOs can effectively silence non-coding RNA. ASOs may serve as effective and targeted therapeutic interventions that target specific cancer-related snoRNAs. Additionally, ASOs can inhibit cancer cell tumorigenicity, both in vitro and in vivo. In vitro experiments have demonstrated that targeting box C/D snoRNAs U3 and U8 can eliminate tumorigenicity in lung (H1944) and BRCA (MCF-7) cells [85,141]. SNORA23 upregulates SYNE2 expression, promoting increased survival and invasiveness in pancreatic ductal adenocarcinoma (PDAC) cells. Targeting SNORA23 with ASOs reduces the growth and metastasis of xenograft tumors [142]. Therefore, ASO technology shows promise as a novel cancer therapy that targets tumor-associated snoRNAs. Both siRNA and shRNA induce degradation of target genes and inhibit tumor progression by degrading oncogenes [88,143,144]. In fact, siRNA and shRNA have been shown to be quite inefficient on the highly structured mRNA but can precisely silence snoRNA due to its conserved secondary structure and crucial RNA-binding sequences. For example, SNORA42 expression is significantly higher in prostate cancer tissues than in adjacent tissues. Transfection with anti-SNORA42 siRNA significantly reduced the growth rate of DU145 and PC3 prostate cancer cells and induced apoptosis [143]. Wang et al. [144] observed a correlation between increased snoU2_19 expression and invasive phenotypes in HCC patients. Additionally, the use of snoU2_19-shRNAs to knock out snoU2_19 effectively inhibited cell cycle proliferation and progression in SK-HEP-1 and HCCLM3 HCC cells. Furthermore, CRISPR/Cas9 gene editing technology has been used to remove snoRNAs, specifically SNORA50 A/B, resulting in increased tumorigenicity in CHL-1 melanoma, A549 lung cancer, and NCI-H23 cells [145].
Additionally, the snoRNA gene expression regulator (snoRNA modulator of gene expression (snoMEN)), a vector technology derived from snoRNA, may also benefit targeted cancer therapy [146]. The snoMEN technology functions as an antisense gene suppression technique that targets intron sequences of nuclear pre-mRNAs, resulting in the downregulation of genes and the regulation of RNA targets [147]. The snoMEN vector selectively induced apoptotic death in miR21-overexpressed human lung adenocarcinoma cells by targeting specific sequences in pri-miR21 [148]. Therefore, snoMEN vectors have potential for future anti-cancer therapies.
7.3. Tumor immune microenvironment (TIME)-related snoRNAs in immunotherapy
SnoRNAs have been linked to several components of the TIME, including CD8+ T cell infiltration, cytotoxic T cell function, and tumor vasculature [149,150]. SnoRNA targeting can improve the efficacy of cancer immunotherapy [70]. SNORA38B promotes the recruitment of CD3+FOXP3+ regulatory T cells (Tregs) to the NSCLC tumor microenvironment by stimulating interleukin-10 (IL-10) secretion. This results in decreased infiltration of CD3+CD8+ T cells, promoting tumor development and immune suppression. The application of locked nucleic acids (LNAs) targeting SNORA38B can significantly improve the TIME and increase the responsiveness of NSCLC cells to immune checkpoint blockade therapy [70]. Therefore, targeting TIME-associated snoRNAs has the potential to modulate cancer sensitivity to immunotherapy, providing novel modalities and insights into combination therapies for cancer.
8. Conclusion
SnoRNAs were previously disregarded as a subtype of non-coding RNAs (ncRNAs) in cancer cells. In recent years, there has been an increasing interest in the study of snoRNAs and their function in tumor development, specifically in 2'-O-ribose methylation and pseudouridylation. However, a comprehensive understanding of the intricate link between snoRNAs and cancer remains limited. Several studies have revealed that snoRNAs display atypical characteristics and expression patterns. This review introduces the regulation of snoRNA expression from three perspectives: dynamic regulation by transcription regulators, transcriptional inhibition by DNA methylation, and dependence on host gene transcription and splicing events. Importantly, we provide a concise overview and explanation of the three novel molecular mechanisms regulated by cancer-associated snoRNAs: 1) the transcriptional regulation of targets by snoRNAs including the crosstalk between snoRNAs and transcription regulators (TRs); 2) the regulation by snoRNAs in post-transcriptional processes, including pre-RNA splicing, mRNA 3'-end APA, mRNA stability, and formation of sdRNAs with miRNA-like functions, indirectly regulating tumor occurrence and development; and 3) the effects of snoRNAs on ADP-ribosylation and chromatin remodeling following target RNA translation. The non-classical cancer regulatory mechanisms of snoRNAs undoubtedly enhance our understanding of cancer and open novel avenues for tumor treatment. SnoRNAs may influence drug resistance in tumor cells by regulating cellular stemness, DDR, cell death pathways, and oxidative stress. Consequently, they could be considered as promising therapeutic targets for cancer treatment, particularly in relation to drug resistance. Furthermore, the cross-talk and feedback mechanisms between SNHGs and snoRNAs have significant implications for cancer progression, warranting further research.
SnoRNAs play a critical role in cancer biology and have significant clinical implications. Dysregulated snoRNAs are potential therapeutic targets for cancer treatment and serve as diagnostic and prognostic markers. Silencing of snoRNAs, which are recognized for their oncogenic characteristics, can potentially lead to the reversal of cancer clinical outcomes. Additionally, targeting tumor immunity-associated snoRNAs has the potential to improve the efficacy of cancer immunotherapy. Therefore, comprehending the mechanisms driving snoRNAs is imperative to enhance our understanding of cancer and holds immense potential for diverse applications in the field. The study of snoRNAs in cancer is an emerging research area. Advancements in high-throughput sequencing technology have facilitated the identification of numerous abnormally expressed snoRNAs in cancers. This technology has also facilitated the development of tools for experimental research. At the current stage, researchers are diligently working to compile a comprehensive overview of snoRNA genetics and pharmacogenomics, with the aim of serving as a clinical reference guide and establishing a foundation for future advancements in snoRNA-targeted therapy. However, the implementation of snoRNA-based treatments in clinical practice may require considerable time. Further research will contribute to a better understanding of the clinical significance of these snoRNAs.
CRediT authorship contribution statement
Xiaoyun Hu: Writing – original draft, Conceptualization. Wanlin Cui: Writing – original draft. Min Liu: Writing – original draft. Fangxiao Zhang: Data curation, Writing – original draft. Yingqi Zhao: Writing – review & editing. Mingrong Zhang: Writing – review & editing. Yuhang Yin: Writing – review & editing. Yalun Li: Writing – review & editing. Ying Che: Writing – review & editing. Xianglong Zhu: Writing – review & editing. Yuxuan Fan: Writing – review & editing. Xiaolan Deng: Writing – review & editing. Minjie Wei: Writing – review & editing. Huizhe Wu: Writing – review & editing.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81872905, 82272915, and 82073884), the Project of the Fourth Batch of Science and Technology Plan of Liaoning Province, China (Grant No.: 2021JH210300133), Science and Technology Innovation Team Project of China Medical University, China (Grant No.: CXTD2022007), the Supporting the High-quality Development of Science and Technology Funding Projects at China Medical University (Grant No.: 2022011963-JH2/202), China Postdoctoral Science Foundation (Grant No.: 2023MD734246), and Liaoning Province Natural Science Foundation Joint Fund (Grant No.: 2023-MSLH-396). We thank Bullet Edits Limited for the linguistic editing and proofreading of the manuscript.
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2024.101064.
Contributor Information
Xiaolan Deng, Email: xideng@coh.org.
Minjie Wei, Email: mjwei@cmu.edu.cn.
Huizhe Wu, Email: wuhz@cmu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bouchard-Bourelle P., Desjardins-Henri C., Mathurin-St-Pierre D., et al. snoDB: An interactive database of human snoRNA sequences, abundance and interactions. Nucleic Acids Res. 2020;48:D220–D225. doi: 10.1093/nar/gkz884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fafard-Couture É., Bergeron D., Couture S., et al. Annotation of snoRNA abundance across human tissues reveals complex snoRNA-host gene relationships. Genome Biol. 2021;22:172. doi: 10.1186/s13059-021-02391-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergeron D., Paraqindes H., Fafard-Couture É., et al. snoDB 2.0: An enhanced interactive database, specializing in human snoRNAs. Nucleic Acids Res. 2023;51:D291–D296. doi: 10.1093/nar/gkac835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dieci G., Preti M., Montanini B. Eukaryotic snoRNAs: A paradigm for gene expression flexibility. Genomics. 2009;94:83–88. doi: 10.1016/j.ygeno.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Bachellerie J.P., Cavaillé J., Hüttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- 6.Reichow S.L., Hamma T., Ferré-D'Amaré A.R., et al. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–1464. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darzacq X., Jády B.E., Verheggen C., et al. Cajal body-specific small nuclear RNAs: A novel class of 2'-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21:2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deryusheva S., Gall J.G. scaRNAs and snoRNAs: Are they limited to specific classes of substrate RNAs? RNA. 2019;25:17–22. doi: 10.1261/rna.068593.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tycowski K.T., Shu M.D., Steitz J.A. A mammalian gene with introns instead of exons generating stable RNA products. Nature. 1996;379:464–466. doi: 10.1038/379464a0. [DOI] [PubMed] [Google Scholar]
- 10.Williams G.T., Farzaneh F. Are snoRNAs and snoRNA host genes new players in cancer? Nat. Rev. Cancer. 2012;12:84–88. doi: 10.1038/nrc3195. [DOI] [PubMed] [Google Scholar]
- 11.Zimta A.A., Tigu A.B., Braicu C., et al. An emerging class of long non-coding RNA with oncogenic role arises from the snoRNA host genes. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott M.S., Ono M., Yamada K., et al. Human box C/D snoRNA processing conservation across multiple cell types. Nucleic Acids Res. 2012;40:3676–3688. doi: 10.1093/nar/gkr1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taft R.J., Glazov E.A., Lassmann T., et al. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ender C., Krek A., Friedländer M.R., et al. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 15.Brameier M., Herwig A., Reinhardt R., et al. Human box C/D snoRNAs with miRNA like functions: Expanding the range of regulatory RNAs. Nucleic Acids Res. 2011;39:675–686. doi: 10.1093/nar/gkq776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wajahat M., Bracken C.P., Orang A. Emerging Functions for snoRNAs and snoRNA-Derived Fragments. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms221910193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rahimy E., Kuo S.Z., Ongkeko W.M. Evaluation of non-coding RNAs as potential targets in head and neck squamous cell carcinoma cancer stem cells. Curr. Drug Targets. 2014;15:1247–1260. doi: 10.2174/1389450115666141024113446. [DOI] [PubMed] [Google Scholar]
- 18.Lin L., Pan Q., Sun Y., et al. Small nucleolar RNA is potential as a novel player in leukemogenesis and clinical application. Blood Sci. 2021;3:122–131. doi: 10.1097/BS9.0000000000000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dsouza V.L., Adiga D., Sriharikrishnaa S., et al. Small nucleolar RNA and its potential role in breast cancer – A comprehensive review. Biochim. Biophys. Acta BBA Rev. Cancer. 2021;1875 doi: 10.1016/j.bbcan.2020.188501. [DOI] [PubMed] [Google Scholar]
- 20.van der Werf J., Chin C.V., Fleming N.I. SnoRNA in cancer progression, metastasis and immunotherapy response. Biology. 2021;10 doi: 10.3390/biology10080809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erales J., Marchand V., Panthu B., et al. Evidence for rRNA 2'-O-methylation plasticity: Control of intrinsic translational capabilities of human ribosomes. Proc. Natl. Acad. Sci. U S A. 2017;114:12934–12939. doi: 10.1073/pnas.1707674114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penzo M., Guerrieri A.N., Zacchini F., et al. RNA pseudouridylation in physiology and medicine: For better and for worse. Genes. 2017;8 doi: 10.3390/genes8110301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monaco P.L., Marcel V., Diaz J.J., et al. 2'-O-methylation of ribosomal RNA: Towards an epitranscriptomic control of translation? Biomolecules. 2018;8 doi: 10.3390/biom8040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang J., Wen J., Huang Z., et al. Small nucleolar RNAs: Insight into their function in cancer. Front. Oncol. 2019;9 doi: 10.3389/fonc.2019.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao L., Wang J., Ju S., et al. Disorders and roles of tsRNA, snoRNA, snRNA and PiRNA in cancer. J. Med. Genet. 2022;59:623–631. doi: 10.1136/jmedgenet-2021-108327. [DOI] [PubMed] [Google Scholar]
- 26.Barros-Silva D., Klavert J., Jenster G., et al. The role of OncoSnoRNAs and ribosomal RNA 2'-O-methylation in cancer. RNA Biol. 2021;18:61–74. doi: 10.1080/15476286.2021.1991167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mleczko A.M., Bąkowska-Żywicka K. When small RNAs become smaller: Emerging functions of snoRNAs and their derivatives. Acta Biochim. Pol. 2016;63:601–607. doi: 10.18388/abp.2016_1330. [DOI] [PubMed] [Google Scholar]
- 28.Abel Y., Rederstorff M. SnoRNAs and the emerging class of sdRNAs: Multifaceted players in oncogenesis. Biochimie. 2019;164:17–21. doi: 10.1016/j.biochi.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 29.Khoshnevis S., Dreggors R.E., Hoffmann T.F.R., et al. A conserved Bcd1 interaction essential for box C/D snoRNP biogenesis. J. Biol. Chem. 2019;294:18360–18371. doi: 10.1074/jbc.RA119.010222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Omer A.D., Lowe T.M., Russell A.G., et al. Homologs of small nucleolar RNAs in Archaea. Science. 2000;288:517–522. doi: 10.1126/science.288.5465.517. [DOI] [PubMed] [Google Scholar]
- 31.Falaleeva M., Stamm S. Processing of snoRNAs as a new source of regulatory non-coding RNAs: SnoRNA fragments form a new class of functional RNAs. Bioessays. 2013;35:46–54. doi: 10.1002/bies.201200117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ojha S., Malla S., Lyons S.M. snoRNPs: Functions in ribosome biogenesis. Biomolecules. 2020;10 doi: 10.3390/biom10050783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lafontaine D.L., Bousquet-Antonelli C., Henry Y., et al. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu X., Xie W., Meng S., et al. Small nucleolar RNAs and their comprehensive biological functions in hepatocellular carcinoma. Cells. 2022;11 doi: 10.3390/cells11172654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cavaillé J., Nicoloso M., Bachellerie J.P. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- 36.Bachellerie J.P., Michot B., Nicoloso M., et al. Antisense snoRNAs: A family of nucleolar RNAs with long complementarities to rRNA. Trends Biochem. Sci. 1995;20:261–264. doi: 10.1016/s0968-0004(00)89039-8. [DOI] [PubMed] [Google Scholar]
- 37.Elliott B.A., Ho H.T., Ranganathan S.V., et al. Modification of messenger RNA by 2'-O-methylation regulates gene expression in vivo. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-11375-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bao H., Chen X., Liu X., et al. Box C/D snoRNA SNORD89 influences the occurrence and development of endometrial cancer through 2'-O-methylation modification of Bim. Cell Death Discov. 2022;8 doi: 10.1038/s41420-022-01102-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu B., Chen X., Liu X., et al. C/D box small nucleolar RNA SNORD104 promotes endometrial cancer by regulating the 2'-O-methylation of PARP1. J. Transl. Med. 2022;20 doi: 10.1186/s12967-022-03802-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu W., Chen X., Liu X., et al. SNORD60 promotes the tumorigenesis and progression of endometrial cancer through binding PIK3CA and regulating PI3K/AKT/mTOR signaling pathway. Mol. Carcinog. 2023;62:413–426. doi: 10.1002/mc.23495. [DOI] [PubMed] [Google Scholar]
- 41.Zhou F., Aroua N., Liu Y., et al. A dynamic rRNA ribomethylome drives stemness in acute myeloid leukemia. Cancer Discov. 2023;13:332–347. doi: 10.1158/2159-8290.CD-22-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cui J., Liu X., Dong W., et al. SNORD17-mediated KAT6B mRNA 2'-O-methylation regulates vasculogenic mimicry in glioblastoma cells. Cell Biol. Toxicol. 2023;39:2841–2860. doi: 10.1007/s10565-023-09805-w. [DOI] [PubMed] [Google Scholar]
- 43.Bian Z., Xu C., Xie Y., et al. SNORD11B-mediated 2'-O-methylation of primary let-7a in colorectal carcinogenesis. Oncogene. 2023;42:3035–3046. doi: 10.1038/s41388-023-02808-1. [DOI] [PubMed] [Google Scholar]
- 44.Duan J., Li L., Lu J., et al. Structural mechanism of substrate RNA recruitment in H/ACA RNA-guided pseudouridine synthase. Mol. Cell. 2009;34:427–439. doi: 10.1016/j.molcel.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 45.Chen S., Li Q., Chen X., et al. SNORA70E promotes the occurrence and development of ovarian cancer through pseudouridylation modification of RAP1B and alternative splicing of PARPBP. J. Cell. Mol. Med. 2022;26:5150–5164. doi: 10.1111/jcmm.17540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen X., Li Q., Xie B., et al. SNORA73B promotes endometrial cancer progression through targeting MIB1 and regulating host gene RCC1 alternative splicing. J. Cell. Mol. Med. 2023;27:2890–2905. doi: 10.1111/jcmm.17850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Penzo M., Rocchi L., Brugiere S., et al. Human ribosomes from cells with reduced dyskerin levels are intrinsically altered in translation. FASEB J. 2015;29:3472–3482. doi: 10.1096/fj.15-270991. [DOI] [PubMed] [Google Scholar]
- 48.Pelletier J., Thomas G., Volarević S. Ribosome biogenesis in cancer: New players and therapeutic avenues. Nat. Rev. Cancer. 2018;18:51–63. doi: 10.1038/nrc.2017.104. [DOI] [PubMed] [Google Scholar]
- 49.Marcel V., Ghayad S.E., Belin S., et al. p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell. 2013;24:318–330. doi: 10.1016/j.ccr.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pauli C., Liu Y., Rohde C., et al. Site-specific methylation of 18S ribosomal RNA by SNORD42A is required for acute myeloid leukemia cell proliferation. Blood. 2020;135:2059–2070. doi: 10.1182/blood.2019004121. [DOI] [PubMed] [Google Scholar]
- 51.Jansson M.D., Häfner S.J., Altinel K., et al. Regulation of translation by site-specific ribosomal RNA methylation. Nat. Struct. Mol. Biol. 2021;28:889–899. doi: 10.1038/s41594-021-00669-4. [DOI] [PubMed] [Google Scholar]
- 52.McMahon M., Contreras A., Holm M., et al. A single H/ACA small nucleolar RNA mediates tumor suppression downstream of oncogenic RAS. eLife. 2019;8 doi: 10.7554/eLife.48847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang K., Wang S., Zhang Y., et al. SNORD88C guided 2'-O-methylation of 28S rRNA regulates SCD1 translation to inhibit autophagy and promote growth and metastasis in non-small cell lung cancer. Cell Death Differ. 2023;30:341–355. doi: 10.1038/s41418-022-01087-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang E., van Nimwegen E., Zavolan M., et al. Decay rates of human mRNAs: Correlation with functional characteristics and sequence attributes. Genome Res. 2003;13:1863–1872. doi: 10.1101/gr.1272403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sklias A., Cruciani S., Marchand V., et al. Comprehensive map of ribosomal 2'-O-methylation and C/D box snoRNAs in Drosophila melanogaster. Nucleic Acids Res. 2024;52:2848–2864. doi: 10.1093/nar/gkae139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu G., Yang F., Ding C.-L., et al. Small nucleolar RNA 113-1 suppresses tumorigenesis in hepatocellular carcinoma. Mol. Cancer. 2014;13 doi: 10.1186/1476-4598-13-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pourebrahim R., Zhang Y., Liu B., et al. Integrative genome analysis of somatic p53 mutant osteosarcomas identifies Ets2-dependent regulation of small nucleolar RNAs by mutant p53 protein. Genes Dev. 2017;31:1847–1857. doi: 10.1101/gad.304972.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liang J., Li G., Liao J., et al. Non-coding small nucleolar RNA SNORD17 promotes the progression of hepatocellular carcinoma through a positive feedback loop upon p53 inactivation. Cell Death Differ. 2022;29:988–1003. doi: 10.1038/s41418-022-00929-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bergeron D., Faucher-Giguère L., Emmerichs A.K., et al. Intronic small nucleolar RNAs regulate host gene splicing through base pairing with their adjacent intronic sequences. Genome Biol. 2023;24 doi: 10.1186/s13059-023-03002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiao H., Feng X., Liu M., et al. SnoRNA and lncSNHG: Advances of nucleolar small RNA host gene transcripts in anti-tumor immunity. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1143980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khalaj M., Park C.Y. snoRNAs contribute to myeloid leukaemogenesis. Nat. Cell Biol. 2017;19:758–760. doi: 10.1038/ncb3566. [DOI] [PubMed] [Google Scholar]
- 62.Zhou F., Liu Y., Rohde C., et al. AML1-ETO requires enhanced C/D box snoRNA/RNP formation to induce self-renewal and leukaemia. Nat. Cell Biol. 2017;19:844–855. doi: 10.1038/ncb3563. [DOI] [PubMed] [Google Scholar]
- 63.Yang X., Han H., De Carvalho D.D., et al. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gong J., Li Y., Liu C.-J., et al. A pan-cancer analysis of the expression and clinical relevance of small nucleolar RNAs in human cancer. Cell Rep. 2017;21:1968–1981. doi: 10.1016/j.celrep.2017.10.070. [DOI] [PubMed] [Google Scholar]
- 65.Ferreira H.J., Heyn H., Moutinho C., et al. CpG island hypermethylation-associated silencing of small nucleolar RNAs in human cancer. RNA Biol. 2012;9:881–890. doi: 10.4161/rna.19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boivin V., Deschamps-Francoeur G., Scott M.S. Protein coding genes as hosts for noncoding RNA expression. Semin. Cell Dev. Biol. 2018;75:3–12. doi: 10.1016/j.semcdb.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 67.Li T., Zhou X., Wang X., et al. Identification and characterization of human snoRNA core promoters. Genomics. 2010;96:50–56. doi: 10.1016/j.ygeno.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 68.Li Y., Yu S., Wang X., et al. SRPK1 facilitates tumor cell growth via modulating the small nucleolar RNA expression in gastric cancer. J. Cell. Physiol. 2019;234:13582–13591. doi: 10.1002/jcp.28036. [DOI] [PubMed] [Google Scholar]
- 69.Lykke-Andersen S., Chen Y., Ardal B.R., et al. Human nonsense-mediated RNA decay initiates widely by endonucleolysis and targets snoRNA host genes. Genes Dev. 2014;28:2498–2517. doi: 10.1101/gad.246538.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhuo Y., Li S., Hu W., et al. Targeting SNORA38B attenuates tumorigenesis and sensitizes immune checkpoint blockade in non-small cell lung cancer by remodeling the tumor microenvironment via regulation of GAB2/AKT/mTOR signaling pathway. J. Immunother. Cancer. 2022;10 doi: 10.1136/jitc-2021-004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu W., Wu Y., Fang X., et al. SnoRD126 promotes the proliferation of hepatocellular carcinoma cells through transcriptional regulation of FGFR2 activation in combination with hnRNPK. Aging. 2021;13:13300–13317. doi: 10.18632/aging.203014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Falaleeva M., Pages A., Matuszek Z., et al. Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proc. Natl. Acad. Sci. U S A. 2016;113:E1625–E1634. doi: 10.1073/pnas.1519292113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Filippova J.A., Matveeva A.M., Zhuravlev E.S., et al. Are small nucleolar RNAs "CRISPRable"? A report on box C/D small nucleolar RNA editing in human cells. Pharmacol. 2019;10 doi: 10.3389/fphar.2019.01246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Deschamps-Francoeur G., Couture S., Abou-Elela S., et al. The snoGloBe interaction predictor reveals a broad spectrum of C/D snoRNA RNA targets. Nucleic Acids Res. 2022;50:6067–6083. doi: 10.1093/nar/gkac475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chew N.J., Nguyen E.V., Su S.P., et al. FGFR3 signaling and function in triple negative breast cancer. Cell Commun. Signal. 2020;18:13. doi: 10.1186/s12964-019-0486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kacew A., Sweis R.F. FGFR3 alterations in the era of immunotherapy for urothelial bladder cancer. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.575258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tian B., Manley J.L. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 2017;18:18–30. doi: 10.1038/nrm.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dong W., Liu X., Yang C., et al. Glioma glycolipid metabolism: MSI2-SNORD12B-FIP1L1-ZBTB4 feedback loop as a potential treatment target. Clin. Transl. Med. 2021;11 doi: 10.1002/ctm2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang C., Shi J., Guo Y., et al. A snoRNA modulates mRNA 3' end processing and regulates the expression of a subset of mRNAs. Nucleic Acids Res. 2017;45:8647–8660. doi: 10.1093/nar/gkx651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shi J., Huang C., Huang S., et al. snoRNAs associate with mRNA 3' processing complex: New wine in old bottles. RNA Biol. 2018;15:194–197. doi: 10.1080/15476286.2017.1416278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hu T., Lu C., Xia Y., et al. Small nucleolar RNA SNORA71A promotes epithelial-mesenchymal transition by maintaining ROCK2 mRNA stability in breast cancer. Mol. Oncol. 2022;16:1947–1965. doi: 10.1002/1878-0261.13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Patterson D.G., Roberts J.T., King V.M., et al. Human snoRNA-93 is processed into a microRNA-like RNA that promotes breast cancer cell invasion. NPJ Breast Cancer. 2017;3:25. doi: 10.1038/s41523-017-0032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu F., Bracken C.P., Pillman K.A., et al. p53 represses the oncogenic sno-MiR-28 derived from a SnoRNA. PLoS One. 2015;10 doi: 10.1371/journal.pone.0129190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lemus-Diaz N., Ferreira R.R., Bohnsack K.E., et al. The human box C/D snoRNA U3 is a miRNA source and miR-U3 regulates expression of sortin nexin 27. Nucleic Acids Res. 2020;48:8074–8089. doi: 10.1093/nar/gkaa549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Langhendries J.L., Nicolas E., Doumont G., et al. The human box C/D snoRNAs U3 and U8 are required for pre-rRNA processing and tumorigenesis. Oncotarget. 2016;7:59519–59534. doi: 10.18632/oncotarget.11148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dong W., Liu Y., Wang P., et al. U3 snoRNA-mediated degradation of ZBTB7A regulates aerobic glycolysis in isocitrate dehydrogenase 1 wild-type glioblastoma cells, CNS Neurosci. Ther. 2023;29:2811–2825. doi: 10.1111/cns.14218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ideue T., Hino K., Kitao S., et al. Efficient oligonucleotide-mediated degradation of nuclear noncoding RNAs in mammalian cultured cells. RNA. 2009;15:1578–1587. doi: 10.1261/rna.1657609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang W.-Q., Zhang Y. RNAi-mediated gene silencing in cancer therapy. Expert Opin. Biol. Ther. 2012;12:1495–1504. doi: 10.1517/14712598.2012.712107. [DOI] [PubMed] [Google Scholar]
- 89.Han C., Sun L., Luo X., et al. Chromatin-associated orphan snoRNA regulates DNA damage-mediated differentiation via a non-canonical complex. Cell Rep. 2022;38 doi: 10.1016/j.celrep.2022.110421. [DOI] [PubMed] [Google Scholar]
- 90.Kim D.S., Camacho C.V., Nagari A., et al. Activation of PARP-1 by snoRNAs controls ribosome biogenesis and cell growth via the RNA helicase DDX21. Mol. Cell. 2019;75:1270–1285.e14. doi: 10.1016/j.molcel.2019.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schubert T., Pusch M.C., Diermeier S., et al. Df31 protein and snoRNAs maintain accessible higher-order structures of chromatin. Mol. Cell. 2012;48:434–444. doi: 10.1016/j.molcel.2012.08.021. [DOI] [PubMed] [Google Scholar]
- 92.Ho S.M., Cheong A., Lam H.M., et al. Exposure of human prostaspheres to bisphenol A epigenetically regulates SNORD family noncoding RNAs via histone modification. Endocrinology. 2015;156:3984–3995. doi: 10.1210/en.2015-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shi Y., Shi Q., Shen Q., et al. Dicer-independent snRNA/snoRNA-derived nuclear RNA 3 regulates tumor-associated macrophage function by epigenetically repressing inducible nitric oxide synthase transcription. Cancer Commun. 2021;41:140–153. doi: 10.1002/cac2.12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao L., Guo H., Zhou B., et al. Long non-coding RNA SNHG5 suppresses gastric cancer progression by trapping MTA2 in the cytosol. Oncogene. 2016;35:5770–5780. doi: 10.1038/onc.2016.110. [DOI] [PubMed] [Google Scholar]
- 95.Zhang M., Duan W., Sun W. LncRNA SNHG6 promotes the migration, invasion, and epithelial-mesenchymal transition of colorectal cancer cells by miR-26a/EZH2 axis, Onco. Targets Ther. 2019;12:3349–3360. doi: 10.2147/OTT.S197433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang Y., Zhang D., Lv J., et al. LncRNA SNHG15 acts as an oncogene in prostate cancer by regulating miR-338-3p/FKBP1A axis. Gene. 2019;705:44–50. doi: 10.1016/j.gene.2019.04.033. [DOI] [PubMed] [Google Scholar]
- 97.Feng X., Dong X., Wu D., et al. Long noncoding RNA small nucleolar RNA host gene 12 promotes papillary thyroid carcinoma cell growth and invasion by targeting miR-16-5p. Histol. Histopathol. 2020;35:217–224. doi: 10.14670/HH-18-155. [DOI] [PubMed] [Google Scholar]
- 98.Han S., Yang X., Qi Q., et al. Can small nucleolar RNA be a novel molecular target for hepatocellular carcinoma? Gene. 2020;733 doi: 10.1016/j.gene.2020.144384. [DOI] [PubMed] [Google Scholar]
- 99.Huldani H., Gandla K., Asiri M., et al. A comprehensive insight into the role of small nucleolar RNAs (snoRNAs) and SNHGs in human cancers. Pathol. Res. Pract. 2023;249 doi: 10.1016/j.prp.2023.154679. [DOI] [PubMed] [Google Scholar]
- 100.Wu G., Hao C., Qi X., et al. LncRNA SNHG17 aggravated prostate cancer progression through regulating its homolog SNORA71B via a positive feedback loop. Cell Death Dis. 2020;11 doi: 10.1038/s41419-020-2569-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lan T., Yuan K., Yan X., et al. LncRNA SNHG10 facilitates hepatocarcinogenesis and metastasis by modulating its homolog SCARNA13 via a positive feedback loop. Cancer Res. 2019;79:3220–3234. doi: 10.1158/0008-5472.CAN-18-4044. [DOI] [PubMed] [Google Scholar]
- 102.Zeng H., Pan J., Hu C., et al. SNHG25 facilitates SNORA50C accumulation to stabilize HDAC1 in neuroblastoma cells. Cell Death Dis. 2022;13 doi: 10.1038/s41419-022-05040-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhu W., Niu J., He M., et al. SNORD89 promotes stemness phenotype of ovarian cancer cells by regulating Notch1-c-Myc pathway. J. Transl. Med. 2019;17 doi: 10.1186/s12967-019-2005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Song J., Zheng A., Li S., et al. Clinical significance and prognostic value of small nucleolar RNA SNORA38 in breast cancer. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.930024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu Y., Zhao C., Wang G., et al. SNORD1C maintains stemness and 5-FU resistance by activation of Wnt signaling pathway in colorectal cancer. Cell Death Discov. 2022;8 doi: 10.1038/s41420-022-00996-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Coley A.B., Stahly A.N., Kasukurthi M.V., et al. MicroRNA-like snoRNA-derived RNAs (sdRNAs) promote castration-resistant prostate cancer. Cells. 2022;11 doi: 10.3390/cells11081302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Goldstein M., Kastan M.B. The DNA damage response: Implications for tumor responses to radiation and chemotherapy. Annu. Rev. Med. 2015;66:129–143. doi: 10.1146/annurev-med-081313-121208. [DOI] [PubMed] [Google Scholar]
- 108.Khanna A. DNA damage in cancer therapeutics: A boon or a curse? Cancer Res. 2015;75:2133–2138. doi: 10.1158/0008-5472.CAN-14-3247. [DOI] [PubMed] [Google Scholar]
- 109.Pilié P.G., Tang C., Mills G.B., et al. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat. Rev. Clin. Oncol. 2019;16:81–104. doi: 10.1038/s41571-018-0114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Krell J., Frampton A.E., Mirnezami R., et al. Growth arrest-specific transcript 5 associated snoRNA levels are related to p53 expression and DNA damage in colorectal cancer. PLoS One. 2014;9 doi: 10.1371/journal.pone.0098561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Godel M., Morena D., Ananthanarayanan P., et al. Small nucleolar RNAs determine resistance to doxorubicin in human osteosarcoma. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21124500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang Z., Yu H., Yao W., et al. RRP9 promotes gemcitabine resistance in pancreatic cancer via activating AKT signaling pathway, Cell Commun. Signal. 2022;20 doi: 10.1186/s12964-022-00974-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Green D.R. The coming decade of cell death research: Five riddles. Cell. 2019;177:1094–1107. doi: 10.1016/j.cell.2019.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Strasser A., Vaux D.L. Cell death in the origin and treatment of cancer. Mol. Cell. 2020;78:1045–1054. doi: 10.1016/j.molcel.2020.05.014. [DOI] [PubMed] [Google Scholar]
- 115.Snyder A.G., Oberst A. The antisocial network: Cross talk between cell death programs in host defense. Annu. Rev. Immunol. 2021;39:77–101. doi: 10.1146/annurev-immunol-112019-072301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang B., Zhao Y., Li Y., et al. A plasma SNORD33 signature predicts platinum benefit in metastatic triple-negative breast cancer patients. Mol. Cancer. 2022;21 doi: 10.1186/s12943-022-01504-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Moloney J.N., Cotter T.G. ROS signalling in the biology of cancer. Semin. Cell Dev. Biol. 2018;80:50–64. doi: 10.1016/j.semcdb.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 118.Saleh E.A.M., Al-Dolaimy F., Qasim Almajidi Y., et al. Oxidative stress affects the beginning of the growth of cancer cells through a variety of routes. Pathol. Res. Pract. 2023;249 doi: 10.1016/j.prp.2023.154664. [DOI] [PubMed] [Google Scholar]
- 119.Yang H., Villani R.M., Wang H., et al. The role of cellular reactive oxygen species in cancer chemotherapy. J. Exp. Clin. Cancer Res. 2018;37 doi: 10.1186/s13046-018-0909-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Arfin S., Jha N.K., Jha S.K., et al. Oxidative stress in cancer cell metabolism. Antioxidants (Basel) 2021;10 doi: 10.3390/antiox10050642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chun K.S., Kim D.H., Surh Y.J. Role of reductive versus oxidative stress in tumor progression and anticancer drug resistance. Cells. 2021;10 doi: 10.3390/cells10040758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chu L., Su M.Y., Maggi L.B., Jr., et al. Multiple myeloma-associated chromosomal translocation activates orphan snoRNA ACA11 to suppress oxidative stress. J. Clin. Invest. 2012;122:2793–2806. doi: 10.1172/JCI63051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mahajan N., Wu H.-J., Bennett R.L., et al. Sabotaging of the oxidative stress response by an oncogenic noncoding RNA. FASEB J. 2017;31:482–490. doi: 10.1096/fj.201600654R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Oliveira V., Mahajan N., Bates M.L., et al. The snoRNA target of t(4;14) in multiple myeloma regulates ribosome biogenesis. FASEB bioadv. 2019;1:404–414. doi: 10.1096/fba.2018-00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pan Y., Zhou A., Hu Z., et al. Small nucleolar RNA-derived microRNA hsa-miR-1291 modulates cellular drug disposition through direct targeting of ABC transporter ABCC1. Drug Metab. Dispos. 2013;41:1744–1751. doi: 10.1124/dmd.113.052092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Okugawa Y., Toiyama Y., Toden S., et al. Clinical significance of SNORA42 as an oncogene and a prognostic biomarker in colorectal cancer. Gut. 2017;66:107–117. doi: 10.1136/gutjnl-2015-309359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yoshida K., Toden S., Weng W., et al. SNORA21 − An oncogenic small nucleolar RNA, with a prognostic biomarker potential in human colorectal cancer. EBioMedicine. 2017;22:68–77. doi: 10.1016/j.ebiom.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wu L., Chang L., Wang H., et al. Clinical significance of C/D box small nucleolar RNA U76 as an oncogene and a prognostic biomarker in hepatocellular carcinoma. Clin. Res. Hepatol. Gastroenterol. 2018;42:82–91. doi: 10.1016/j.clinre.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 129.Nottingham R.M., Wu D.C., Qin Y., et al. RNA-seq of human reference RNA samples using a thermostable group II intron reverse transcriptase. RNA. 2016;22:597–613. doi: 10.1261/rna.055558.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Xu H., Nottingham R.M., Lambowitz A.M. TGIRT-seq protocol for the comprehensive profiling of coding and non-coding RNA biotypes in cellular, extracellular vesicle, and plasma RNAs. Bio Protoc. 2021;11 doi: 10.21769/BioProtoc.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xie Q., Zhang D., Ye H., et al. Identification of key snoRNAs serves as biomarkers for hepatocellular carcinoma by bioinformatics methods. Medicine. 2022;101 doi: 10.1097/MD.0000000000030813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang Y., Shang X., Yu M., et al. A three-snoRNA signature: SNORD15A, SNORD35B and SNORD60 as novel biomarker for renal cell carcinoma. Cancer Cell Int. 2023;23 doi: 10.1186/s12935-023-02978-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang Q., Bi Z., Song X., et al. Tumor-educated platelet SNORA58, SNORA68 and SNORD93 as novel diagnostic biomarkers for esophageal cancer. Future Oncol. 2023;19:651–661. doi: 10.2217/fon-2023-0129. [DOI] [PubMed] [Google Scholar]
- 134.Li M., Huang F., Xie Z., et al. Identification of three small nucleolar RNAs (snoRNAs) as potential prognostic markers in diffuse large B-cell lymphoma. Cancer Med. 2023;12:3812–3829. doi: 10.1002/cam4.5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shen L., Lu W., Huang Y., et al. SNORD15B and SNORA5C: Novel diagnostic and prognostic biomarkers for colorectal cancer. BioMed Res. Int. 2022;2022 doi: 10.1155/2022/8260800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Verbeek M.W.C., Erkeland S.J., van der Velden V.H.J. Dysregulation of small nucleolar RNAs in B-cell malignancies. Biomedicines. 2022;10 doi: 10.3390/biomedicines10061229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Teittinen K.J., Laiho A., Uusimäki A., et al. Expression of small nucleolar RNAs in leukemic cells. Cell. Oncol. (Dordr.) 2013;36:55–63. doi: 10.1007/s13402-012-0113-5. [DOI] [PubMed] [Google Scholar]
- 138.He R., Huang Z., Zhai G., et al. Small nucleolar RNAs (snoRNAs)-based risk score classifier predicts overall survival in bladder carcinoma. Med. Sci. Monit. 2020;26 doi: 10.12659/MSM.926273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cai C., Peng Y., Shen E., et al. Identification of tumour immune infiltration-associated snoRNAs (TIIsno) for predicting prognosis and immune landscape in patients with colon cancer via a TIIsno score model. EBioMedicine. 2022;76 doi: 10.1016/j.ebiom.2022.103866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rabany O., Nachmani D. Small nucleolar (sno)RNA: Therapy lays in translation. Noncoding RNA. 2023;9 doi: 10.3390/ncrna9030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Su H., Xu T., Ganapathy S., et al. Elevated snoRNA biogenesis is essential in breast cancer. Oncogene. 2014;33:1348–1358. doi: 10.1038/onc.2013.89. [DOI] [PubMed] [Google Scholar]
- 142.Cui L., Nakano K., Obchoei S., et al. Small nucleolar noncoding RNA SNORA23, up-regulated in human pancreatic ductal adenocarcinoma, regulates expression of spectrin repeat-containing nuclear envelope 2 to promote growth and metastasis of xenograft tumors in mice. Gastroenterology. 2017;153:292–306.e2. doi: 10.1053/j.gastro.2017.03.050. [DOI] [PubMed] [Google Scholar]
- 143.Yi C., Wan X., Zhang Y., et al. SNORA42 enhances prostate cancer cell viability, migration and EMT and is correlated with prostate cancer poor prognosis. Int. J. Biochem. Cell Biol. 2018;102:138–150. doi: 10.1016/j.biocel.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 144.Wang H., Ma P., Liu P., et al. Small nucleolar RNA U2_19 promotes hepatocellular carcinoma progression by regulating Wnt/β-catenin signaling. Biochem. Biophys. Res. Commun. 2018;500:351–356. doi: 10.1016/j.bbrc.2018.04.074. [DOI] [PubMed] [Google Scholar]
- 145.Siprashvili Z., Webster D.E., Johnston D., et al. The noncoding RNAs SNORD50A and SNORD50B bind K-Ras and are recurrently deleted in human cancer. Nat. Genet. 2016;48:53–58. doi: 10.1038/ng.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ono M., Yamada K., Avolio F., et al. Analysis of human small nucleolar RNAs (snoRNA) and the development of snoRNA modulator of gene expression vectors. Mol. Biol. Cell. 2010;21:1569–1584. doi: 10.1091/mbc.E10-01-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ono M., Yamada K., Endo A., et al. Analysis of human protein replacement stable cell lines established using snoMEN-PR vector. PLoS One. 2013;8 doi: 10.1371/journal.pone.0062305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ono M., Yamada K., Avolio F., et al. Targeted knock-down of miR21 primary transcripts using snoMEN vectors induces apoptosis in human cancer cell lines. PLoS One. 2015;10 doi: 10.1371/journal.pone.0138668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chow R.D., Chen S. Sno-derived RNAs are prevalent molecular markers of cancer immunity. Oncogene. 2018;37:6442–6462. doi: 10.1038/s41388-018-0420-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Roy R.K., Yadav R., Sharma U., et al. Impact of noncoding RNAs on cancer directed immune therapies: Now then and forever. Int. J. Cancer. 2022;151:981–992. doi: 10.1002/ijc.34060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.