Abstract
Major advances in the study of the molecular biology of RNA viruses have resulted from the ability to generate and manipulate full-length genomic cDNAs of the viral genomes with the subsequent synthesis of infectious RNA for the generation of recombinant viruses. Coronaviruses have the largest RNA virus genomes and, together with genetic instability of some cDNA sequences in Escherichia coli, this has hampered the generation of a reverse-genetics system for this group of viruses. In this report, we describe the assembly of a full-length cDNA from the positive-sense genomic RNA of the avian coronavirus, infectious bronchitis virus (IBV), an important poultry pathogen. The IBV genomic cDNA was assembled immediately downstream of a T7 RNA polymerase promoter by in vitro ligation and cloned directly into the vaccinia virus genome. Infectious IBV RNA was generated in situ after the transfection of restricted recombinant vaccinia virus DNA into primary chick kidney cells previously infected with a recombinant fowlpox virus expressing T7 RNA polymerase. Recombinant IBV, containing two marker mutations, was recovered from the transfected cells. These results describe a reverse-genetics system for studying the molecular biology of IBV and establish a paradigm for generating genetically defined vaccines for IBV.
Avian infectious bronchitis virus (IBV) is a highly infectious and contagious pathogen of domestic fowl that replicates primarily in the respiratory tract but also in some epithelial cells of the gut, kidney, and oviduct (8, 10, 16). IBV is an enveloped coronavirus (order Nidovirales, family Coronaviridae, genus Coronavirus) that replicates in the cell cytoplasm and contains an unsegmented, single-stranded, positive-sense RNA genome (12, 15, 34). In the infected cell, in addition to the genomic RNA (27,608 nucleotides [nt]) (6), IBV produces five subgenomic (sg) mRNAs, each possessing a 64-nt leader sequence derived from the 5′ end of the genome. The signals required for the replication and packaging of the virion RNA are located in the terminal regions of the genome (11).
The 5′ two-thirds of a coronavirus genome encompass the replicase gene. The replicase gene products are expressed as two polyproteins, Rep1a and Rep1ab, the latter resulting from a −1 frameshift, that undergo extensive co- and posttranslational proteolytic processing before they are assembled into functional replication-transcription complexes (46). A hallmark of coronavirus replication is the production of a 3′-coterminal nested set of polycistronic sg mRNAs. The sg mRNAs are produced by a discontinuous transcription process in which leader sequences derived from the 5′ end of the genome are fused to the 5′ end of each sg mRNA (4, 32, 33, 35). In general, only the 5′-proximal open reading frame (ORF) of each sg mRNA is translated to produce one of the virus structural proteins, i.e., spike glycoprotein (S), small membrane protein (E), integral membrane protein (M), and nucleocapsid protein (N).
The analysis of recombinant viruses with specific genetic changes has proved to be a powerful method for understanding the molecular biology of RNA viruses and for studying the role of individual genes in pathogenesis. Such reverse genetics systems have been achieved for a number of positive-stranded RNA viruses, with genome sizes ranging from 7 kb (picornaviruses) to 15 kb (arteriviruses) to 20 kb (Citrus tristeza virus of the genus Closterovirus) (26, 30, 31). Despite these successes, the reverse genetics methods used for positive-strand RNA viruses are still not entirely routine procedures; for example, the instability of some virus-derived cDNAs in bacteria has been a problem that has required ingenious strategies for the assembly of full-length cDNAs capable of generating infectious RNAs. Thus, in vitro ligation, without assembly of the full-length cDNA in bacteria, was used to overcome the problem for generating recombinant yellow fever virus (27). Construction of full-length cDNAs in yeast (25) or the introduction of short introns to allow propagation of cDNAs in E. coli (44) have proven successful strategies for the generation of full-length, stable genomic cDNAs of dengue virus and Japanese encephalitis virus.
Coronaviruses have the largest genomes of any known RNA virus and this, together with the observation that some cDNAs derived from regions of the replicase gene are unstable in bacteria, has hampered the generation of full-length coronavirus cDNAs. However, two methods were recently described for the assembly of full-length cDNAs from the genomic RNA of porcine coronavirus transmissible gastroenteritis virus (TGEV). The first method involved the assembly of a TGEV full-length cDNA in a bacterial artificial chromosome (BAC), immediately downstream of a viral RNA polymerase II promoter. Transfection of this construct into susceptible cells resulted in the synthesis of infectious RNA and the recovery of recombinant virus (1). The second system involved the in vitro assembly of the TGEV full-length cDNA by using a series of contiguous cDNAs with engineered unique restriction sites. Bacteriophage T7-RNA polymerase derived RNA transcripts of the cDNA were then used to generate infectious virus (45). In that system TGEV N protein was required for generation of infectious virus. More recently, a third strategy has been developed for the assembly of a full-length cDNA of the human coronavirus (HCoV) 229E (39). This system involved the in vitro assembly of the HCoV cDNA followed by direct cloning into the genome of vaccinia virus. Infectious HCoV RNA synthesised in vitro from the vaccinia virus DNA, again by using bacteriophage T7 RNA polymerase, was transfected into cells and resulted in the recovery of recombinant HCoV. An advantage of this system is the ability to assemble a full-length coronavirus genomic cDNA in a nonbacterial system. As previously observed for TGEV (1, 45), HCoV 229E (39), and murine hepatitis virus (V. Thiel, unpublished results), we have also been unable to assemble a contiguous region of the IBV replicase sequence in bacteria either by using high- or low-copy plasmids or a BAC.
In this study we describe the assembly and successful recovery of the coronavirus IBV, the first group III coronavirus to be successfully recovered by using a reverse genetics system. To date, all of the described coronavirus reverse genetics systems have involved the recovery of a type I mammalian coronavirus. We assembled the IBV full-length cDNA in vitro, followed by direct cloning into the vaccinia virus genome, and have recovered recombinant IBV after the in situ synthesis of infectious IBV RNA by using bacteriophage T7 RNA polymerase expressed from a recombinant fowlpox virus (rFPV-T7 [7]).
MATERIALS AND METHODS
Cells and viruses.
The growth of IBV in 11-day-old embryonated domestic fowl eggs and chick kidney (CK) cells was as described previously (23, 24, 38). IBV Beaudette US (Beau-US [2, 9]) is a Beaudette strain that has been adapted for growth in Vero cells, an African green monkey cell line. IBV Beaudette CK (Beau-CK [9]) is a strain of IBV that was adapted for growth in CK cells but which has not been adapted for growth in Vero cells. Vaccinia virus vNotI/tk (20) and vaccinia virus recombinants were propagated, titrated, and purified on monkey kidney fibroblasts (CV-1) by standard procedures (18). The CV-1 cells were grown in minimum essential medium supplemented with HEPES (25 mM), fetal bovine serum (5 to 10%), and antibiotics. Fowlpox virus (FPV) HP1.441 (19) and rFPV-T7 (fpEFLT7pol [7]), a recombinant expressing the bacteriophage T7 RNA polymerase under the direction of the vaccinia virus P7.5 early-late promoter, were grown in chicken embryo fibroblast cells in medium 199 (M199) supplemented with 2% newborn calf serum.
Recombinant DNA techniques.
Recombinant DNA techniques followed standard procedures (3, 29) or were carried out according to the manufacturers' instructions. All nucleotide and amino acid residue numbers refer to the positions in IBV Beau-CK (6).
Plasmids and bacterial strains.
Three plasmids—pFRAG-1, pFRAG-2 and pFRAG-3 (Fig. 1)—constructed from a variety of cDNAs and representing the complete IBV Beau-CK genome were used to generate a full-length IBV cDNA. The precise details of the various constructs and their uses are available from the authors upon request. The cDNAs for the three plasmids were derived from pKT1a3 (41), pKTF2, pIBV5 (17), pCRScript F4 (41), and pBS/NF5,6 (all a gift from K. Tibbles, University of Cambridge); pIBV179 (6); pIBV136 (6); pIBV322 (6); pCD-91 (23); and pIBV-Vec (13).
FIG. 1.
Schematic diagram for the assembly of the full-length IBV cDNA in the vaccinia virus genome. The positions of the IBV genes within the IBV-derived cDNAs are shown. The strategy used for the assembly of the full-length IBV cDNA and insertion into the vaccinia virus genome is shown with the final orientation of the cDNA in the vaccinia genome. “AP” indicates that the restriction site on the particular cDNA was dephosphorylated. Rep, S, 3, M, 5, and N refer to the IBV genes; T7ψ, HδR, and T7φ refer to the positions of the T7 promoter, hepatitis delta antigenome ribozyme, and T7 terminator sequences, respectively. The three extra G residues at the 5′ end of the IBV sequence forming part of the T7 promoter sequence are indicated. The terminal SalI sites shown represent the 5.7-kb SalI fragment normally containing the 534-bp vaccinia virus tk gene. The right-hand SalI site is ∼1.1 kb from the tk gene, and the left-hand SalI site is ∼4.1 kb from the tk gene. The non-IBV regions are not drawn to scale. Arrowheads indicate the positions of the 14 oligonucleotides used to generate RT-PCR or PCR products from the IBV genome or IBV cDNA in vNotI/IBVFL. The sizes and annotations (A to G) of the fragments are shown.
Plasmid pFRAG-1 was derived from pKT1a3 and a reverse transcription-PCR (RT-PCR) product corresponding to the 5′ end of the IBV genome fused to the T7 RNA polymerase promoter and various restriction endonuclease sites. The IBV cDNA in pFRAG-1 corresponds to genomic nt 1 to 6495, preceded by three additional G nucleotides, the T7 promoter sequence, and restriction sites AscI, Bsp120I, and NotI, inserted into pZSL1190 (23). The T7 promoter sequence and extra G residues have previously been used during the successful rescue of a variety of IBV D-RNAs (11, 13, 23, 24, 37, 38). Plasmid pFRAG-2, derived from pKTF2 and pIBV5, contains IBV cDNA corresponding to genomic nt 5752 to 14474 inserted into pZSL1190. Plasmid pFRAG-3 was essentially generated from two intermediary plasmids that were derived from pIBV5, pCD-91, pCRScriptF4, pIBV179, pBS/NF5,6, IBV136, pIBV322, and pIBV-Vec. The IBV cDNA in pFRAG-3 corresponds to genomic nt 13806 to 27608, followed by a synthetic poly(A) tail of 28 bp, the hepatitis delta virus antigenomic ribozyme (HδR), T7 RNA polymerase termination signal (T7φ), and the restriction sites Bsp120I and NotI. The IBV cDNA was inserted into a modified version of the low-copy plasmid pACNR1180 (28). The cDNA from genomic nt 26857 (an MluI site in the N gene) to the 3′ end of the IBV genome, the synthetic poly(A) tail, HδR, and T7φ termination sequences were derived from pIBV-Vec, which has been successfully used to generate IBV D-RNAs in situ (13).
Plasmid pCi-N (a gift from J. A. Hiscox, University of Reading [14]) contains the Beau-CK N gene inserted into the eukaryotic expression vector pCi-Neo (Promega). Expression of the N gene is under the control of both the cytomegalovirus RNA polymerase II promoter and the T7 RNA polymerase promoter.
Gel electrophoresis.
Smaller DNA fragments were routinely analyzed on 0.6 to 1.0% agarose–TBE (0.1 M Tris, 0.09 M boric acid, 1 mM EDTA) gels. The in vitro ligation products or restricted vaccinia virus DNA were analyzed by pulsed-field gel electrophoresis in 0.8%– or 1.0%–0.5× TBE agarose gels, respectively, at 14°C by using the CHEF-III DR system (Bio-Rad). The gels were run with a 0.1- to 1.0-s switch time for 16 h at 6 V/cm at an angle of 120o for the in vitro ligation products and with a 3.0- to 30.0-s switch time for 16 h at 6 V/cm for vaccinia virus DNA fragments. Exposure to UV irradiation was avoided throughout the preparation of the fragments to reduce DNA damage.
Assembly of a full-length IBV cDNA in vaccinia virus.
The assembly of a full-length cDNA representing the Beau-CK genome was done by using sequential in vitro ligation of cDNA fragments derived from pFRAG-1, pFRAG-2, and pFRAG-3, followed by direct cloning into the genome of vaccinia virus vNotI/tk. The digested fragments were differentially treated with alkaline phosphatase to force ligation of the fragments in the correct order. Preliminary experiments were done to determine the optimal dephosphorylation strategies, and various analytical ligation reactions were done to determine the optimal ratios of the fragments in the ligation reactions.
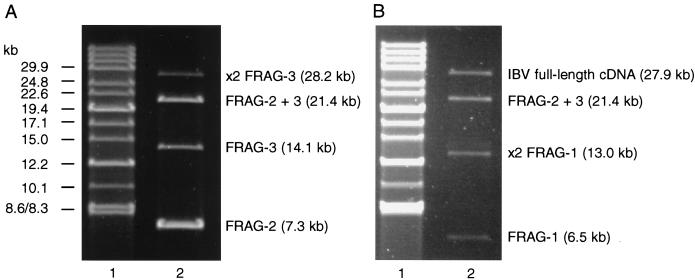
The procedure for the in vitro ligation of the IBV-derived cDNAs and the subsequent cloning of the full-length cDNA are summarized in Fig. 1. Plasmid pFRAG-1 (20 μg) was initially digested with Bsp120I, followed by treatment with 2 U/100 μl of calf intestinal alkaline phosphatase (CIAP; Boehringer Mannheim), and then deproteinized with phenol-chloroform, ethanol precipitated, and digested with SacI. This results in a 6,542-bp Bsp120I-SacI fragment (FRAG-1) with a dephosphorylated Bsp120I end. Plasmid pFRAG-2 (20 μg) was digested with SacI and NheI, followed by CIAP, resulting in a 7,312-bp SacI-NheI fragment (FRAG-2) with dephosphorylated ends. Plasmid pFRAG-3 (40 μg) was initially digested with Bsp120I, treated with CIAP, extracted with phenol-chloroform, ethanol precipitated, and then digested with NheI. This results in a 14,086-bp NheI-Bsp120I fragment (FRAG-3) with a dephosphorylated Bsp120I end. The fragments were purified with QIAEX II resin (Qiagen) after electrophoresis on 0.6% agarose gels. The three fragments were sequentially ligated in vitro, according to the procedure presented in Fig. 1. First, FRAG-2 and FRAG-3, at the molar ratio of 2:1, were ligated with T4 DNA ligase (30 U/μl; MBI Fermentas) overnight at room temperature in ligation buffer (40 mM Tris-HCl, 10 mM MgCl2, 10 mM dithiothreitol, 0.5 mM ATP). The ligation products were separated in a 0.6% agarose gel, and the 21.4-kb SacI-Bsp120I fragment was purified by using Qiaex II resin. Second, FRAG-1 was ligated to the 21.4-kb fragment by using a molar ratio of 1:10 for 3 h at room temperature under the same ligation conditions to generate the 27.9-kb fragment containing the full-length IBV cDNA (Fig. 1). The ligation mixture was analyzed by agarose gel electrophoresis to identify the presence of the 27.9-kb product (Fig. 2) and used without further purification. Vaccinia virus, vNotI/tk, which contains a single NotI site in the thymidine kinase gene, was used as a cloning vector. vNotI/tk DNA was digested with NotI, and the products from the in vitro ligation reaction, containing the 27.9-kb IBV full-length cDNA, were ligated overnight at room temperature to the NotI-digested vNotI/tk genomic arms in the presence of NotI. The T4 DNA ligase was heat inactivated, and the incubation continued for an additional hour at 37°C with supplementary NotI.
FIG. 2.
Analysis of the in vitro assembly of IBV-derived cDNAs for the generation of a full-length cDNA. Samples of the ligation mixtures for assembly of IBV cDNAs FRAG-2 and FRAG-3 (A) and assembly of the full-length IBV cDNA by ligation of FRAG-1 to the intermediary 21.6-kb FRAG-2–FRAG-3 fragment (B) were analyzed by pulsed-field gel electrophoresis in 0.8% agarose gels. The cDNAs are as outlined in Fig. 1. Lanes 1 contained DNA markers (8.3 to 48.5 kb), and lanes 2 contained samples of the ligation mixtures.
Generation of recombinant vaccinia virus.
CV-1 cells (70% confluent) in 30-mm-diameter plates were infected with FPV HP1.441 at a multiplicity of infection (MOI) of 5. After 45 to 60 min, the cells were transfected with the vNotI/tk-IBV cDNA in vitro ligation mixture (1 to 5 μg) by using 10 μg of Lipofectin (Life Technologies/Gibco-BRL). The cells were incubated at 37°C for 1 to 2 h, harvested, placed in 96-well tissue culture dishes with a fourfold excess of fresh CV-1 cells, and incubated at 37°C for 7 to 14 days. Potential recombinant vaccinia viruses were isolated from wells that developed a cytopathic effect (CPE) and then plaque purified and characterized by PCR and Southern blot analyses.
Transfection and recovery of infectious IBV.
CK cells, grown to 50% confluence, in 60-mm-diameter plates were infected with rFPV-T7 (7) at an MOI of 10. After 45 to 60 min, the cells were transfected with 10 μg of DNA isolated from recombinant vaccinia virus (vNotI/IBVFL) containing the 27.9-kb full-length IBV cDNA, previously digested with AscI or SalI, and 5 μg of pCi-Nuc by using 60 μg of LipofecACE (Life Technologies/Gibco-BRL). The transfected cells (P0 cells) were incubated at 37°C for 16 h, after which the transfection medium was replaced with fresh maintenance medium (23). At 3 days posttransfection, the culture medium, potentially containing IBV (V1) was removed, centrifuged for 3 min at 2,500 rpm, filtered through a 0.22-μm (pore-size) filter, to remove any rFPV-T7 (13), and used for serial passage on CK cells. The CK cells (P1 to P5) were incubated until they showed a CPE associated with an IBV infection, and the cell medium, potentially containing IBV (V2 to V6), was removed and treated as described above between passages.
Isolation and analysis of RNAs from infected cells.
Total cellular RNA was extracted from transfected (P0) or infected (P1 to P5) cells by the RNeasy method (Qiagen) and analyzed for the presence of IBV sg mRNAs 3 and 4 by RT-PCR or for all IBV-specific RNAs by Northern blot analysis. In the latter case, RNA was electrophoresed in denaturing 2.2 M formaldehyde–1% agarose gels (as described by Sambrook et al. [29]) and transferred to Hybond XL nylon membranes (Amersham). IBV-derived RNAs were detected nonisotopically by using a 309-bp IBV 3′-untranslated region (UTR) probe corresponding to the proximal 309 nt at the 3′ end of the IBV genome (13). RT-PCR (Ready-To-Go, RT-PCR beads; Amersham Pharmacia Biotech) for detection of IBV sg mRNAs 3 and 4 was done by using the oligonucleotides Leader1 (5′-26CTATTACACTAGCCTTGCGC45-3′), corresponding to part of the IBV leader sequence, and PM5– (5′-24694CAATGTTAAGGGGCCAAAAGCA24673-3′) complementary to sequence within the IBV M gene. The RT-PCRs resulted in two DNA fragments of 900 and 306 bp amplified from IBV mRNAs 3 and 4, respectively.
Confirmation that the recovered IBV was derived from vNotI/IBVFL was investigated by RT-PCR by using oligonucleotides BG-68 (5′-19141GTAAAGACTTAGGCCTAC19158-3′), together with BG-132 (5′-20685ATCTGAAAAATTACAGTGTG20666-3′), and also BG-54 (5′-25941ACCACCTAAAGTCGGTTC25958-3′), together with 93/100 (5′-27607GCTCTAACTCTATACTAGCCT27587-3′). The RT-PCR products, which represented two regions of the IBV genome containing marker mutations, were analyzed by restriction enzyme digestion and sequence determination.
Analysis of vaccinia virus DNA.
CV-1 cells (105), infected with vNotI/tk or vNotI/IBVFL, were incubated until CPEs were evident. The cells were harvested, resuspended in 200 μl of 100 mM Tris-HCl (pH 7.5)–5 mM EDTA–0.2% sodium dodecyl sulfate–200 mM NaCl containing 0.1 mg of proteinase K/ml, incubated at 50°C for 2 h, and extracted with phenol-chloroform, and then any DNA was ethanol precipitated. The resulting DNA was analyzed by restriction enzyme digestion, Southern blot, and PCR analysis. Vaccinia virus DNA was digested overnight at 37°C with HindIII for Southern blot analysis, and the digested DNA was electrophoresed in agarose gels and transferred onto nylon membranes. A mixture of seven PCR fragments (A to G; Fig. 1), which represent the complete IBV genome, were generated from pFRAG-1, pFRAG-2, and pFRAG-3; labeled with [32P]dCTP by the Multiprime DNA-Labeling System (Amersham); and hybridized to the immobilized HindIII-restricted vaccinia virus DNA.
Immunofluorescence assay.
Vero cells (60% confluent) on coverslips in 9.6-cm2 dishes were infected with Beau-CK, Beau-US, and Beau-R at an MOI of 2 to 3 and fixed 18 h postinfection (p.i.) with 50% methanol-acetone. The IBV-infected fixed cells were stained with propidium iodide, to visualize nuclear DNA, and analyzed by indirect immunofluorescence by using rabbit anti-IBV polyclonal sera (21) followed by fluorescein isothiocyanate (FITC)-labeled goat anti-rabbit antibody (Harlan Sera-Lab). Fluorescent images were obtained by using a confocal microscope (Leica).
Sequence analysis.
Sequence analysis of plasmid DNA, PCR products A to G (generated from the IBV cDNA sequence within vNotI/IBVFL), and RT-PCR products from recombinant IBV or from the 3′ end of the replicase to the poly(A) tail of Beau-US was done with an ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems). Oligonucleotide primers used for the sequencing reactions were derived from the Beau-CK sequence (6). Sequences were determined on an Applied Biosystems 377 DNA sequencer. Assembly of the sequences, derived from the plasmid constructs, recombinant IBV and Beau-US, and comparison with the Beau-CK sequence (6) were done by using Gap4 of the Staden Sequence Software Programs (5). The sequences of the recombinant IBV, Beau-R, and from Vero-adapted Beau-US reported in this study have been deposited in the EMBL database under accession numbers AJ311317 (Beau-R) and AJ311362 (Beau-US).
RESULTS
Strategy for the assembly of an IBV full-length cDNA.
At the time that we embarked on producing a full-length IBV cDNA, no successful reverse genetics system for coronaviruses was available. The initial strategy that we adopted involved the production of contiguous plasmids based on the sequence of IBV Beau-CK (6). In addition, we decided not to intentionally introduce mutations into the cDNA, such as those resulting from the introduction of new restriction sites, to aid assembly. This was because the successful generation of an infectious RNA of the arterivirus, equine arteritis virus (EAV) had been affected by a single point mutation within the replicase gene (42). However, during the early cloning stages it became apparent that particular regions from the IBV genome were not compatible for cloning in high-copy-number plasmids in E. coli, as has now been reported for other coronavirus cDNAs (1, 39, 45). Some of these regions were successfully cloned in a low-copy-number plasmid, but the joining of two contiguous IBV cDNAs, corresponding to a region in the replicase gene, was found to be refractory for cloning in E. coli. Attempts to clone this region into the bacterial artificial chromosome pBeloBac11 (43) were also unsuccessful. As a result, we decided to use vaccinia virus as a potential cloning vector by using in vitro ligation for assembly of the full-length IBV cDNA.
Construction of a full-length IBV cDNA in vaccinia virus.
To avoid the introduction of modified sequences, we decided to assemble the full-length cDNA by using natural restriction sites and IBV cDNAs derived from original plasmids used to determine the Beau-CK sequence (6). Several of these cDNAs had also been used for the expression of various domains of the IBV replicase gene (17, 41). Essentially three plasmids, pFRAG-1, pFRAG-2, and pFRAG-3, containing contiguous regions of the IBV genome were constructed for final assembly of the full-length cDNA (Fig. 1).
The IBV cDNAs within plasmids pFRAG-1, pFRAG-2, and pFRAG-3 were sequenced prior to assembly, and two nucleotide substitutions were identified. One substitution, present in pFRAG-3, corresponded to a C→U change at nt 19666 within ORF 1b corresponding to the Rep1ab gene product, resulted in the amino acid substitution Ser6380→Leu and arose during the construction of pCRScriptF4. The second nucleotide substitution, also within pFRAG-3 at nt 27087, corresponded to a A→G change within ORF 6 corresponding to the N gene product, which is translationally silent, and arose during the construction of pIBV-Vec. pIBV-Vec encodes an IBV D-RNA that was successfully replicated (13). Interestingly, the C19666→U substitution resulted in the loss of a BstBI site originally present in the Beau-CK sequence. Sequence analysis of a RT-PCR product from Beau-US, our normal laboratory strain of Beaudette, confirmed the presence of the BstBI site. Therefore, we decided to retain the two nucleotide mutations in FRAG-3 as potential markers for analysis of any recovered virus.
Assembly of a full-length cDNA clone encoding the entire IBV genome downstream of the T7 RNA promoter was achieved by a two-step in vitro ligation method as outlined in Fig. 1. Dephosphorylation of the ends of the various cDNA fragments was used to directionally control the ligation reactions and optimize the yield of the desired fragments. In the first step, FRAG-2 and FRAG-3 were ligated, and analysis of the ligation reaction products identified the resulting intermediary 21.5-kb SacI-Bsp120I cDNA (Fig. 2A). In the second step, FRAG-1 was ligated with the 21.5-kb cDNA to produce a 27.9-kb cDNA (Fig. 2B) that represents a full-length cDNA of the Beau-CK genome under the control of a T7 RNA polymerase promoter and terminated by a HδR-T7 termination sequence distal to the poly(A) tail.
Subsequently, the in vitro ligation products, containing the full-length IBV cDNA with dephosphorylated Bsp102I ends were directly ligated to NotI arms derived from vNotI/tk in the presence of NotI. Although NotI and Bsp120I have compatible cohesive ends, the resultant sequence generated by their ligation is insensitive to cleavage by NotI. This resulted in the irreversible insertion of the IBV cDNA into the vNotI/tk genome, increasing the efficiency of ligation with concomitant reduction in the religation of vNotI/tk genomic DNA. The products of this ligation were used without further purification to recover recombinant vaccinia viruses by using FPV helper virus. We obtained 18 recombinant vaccinia viruses, and Southern blot analysis of DNA isolated from seven of them indicated that one contained an insert of the expected size. Restriction analysis of the DNA from this recombinant vaccinia virus identified that a 27.9-kb DNA had been cloned into the vaccinia virus genome (Fig. 3A). To verify that the 27.9-kb DNA represented a full-length copy of the IBV genome, a series of consecutive overlapping PCR products encompassing the complete IBV genome were generated and shown to be of the expected sizes (Fig. 3B). The PCR products were sequenced, and comparison of the sequences to the Beau-CK sequence confirmed that the 27.9-kb DNA corresponded to a full-length IBV cDNA identical to that of Beau-CK, apart from the two nucleotide substitutions, U19666 and G27087, which are diagnostic for the recombinant IBV genomic cDNA. The orientation of the 27.9-kb DNA present in the recombinant vaccinia virus, termed vNotI/IBVFL, is shown in Fig. 1.
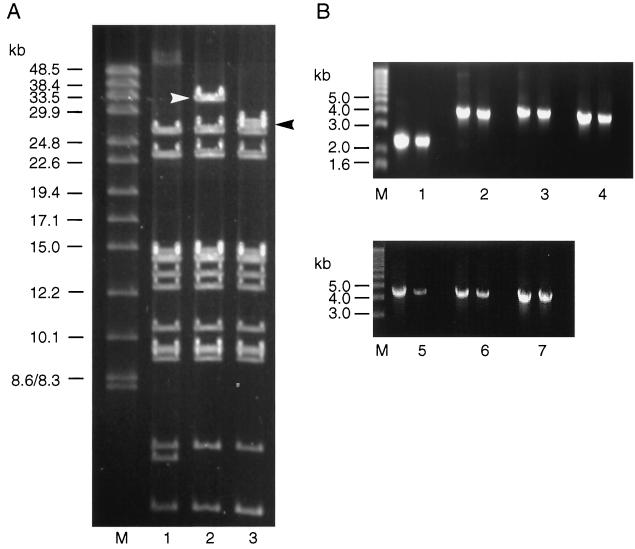
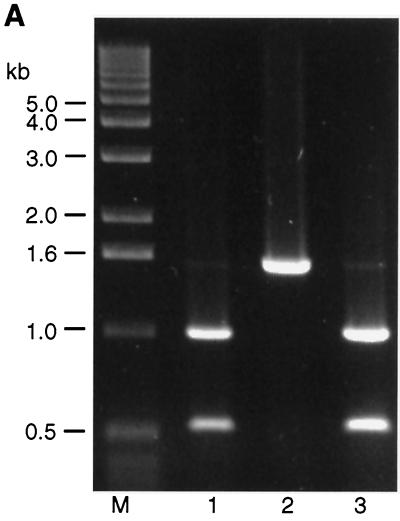
FIG. 3.
Analysis of DNA isolated from the recombinant vaccinia virus vNotI/IBVFL containing the full-length IBV cDNA. (A) Vaccinia virus genomic DNA was analyzed by restriction digestion. Lane 1 contained vNotI/tk DNA digested with SalI, lane 2 contained vNotI/IBVFL DNA digested with SalI, and lane 3 contained vNotI/IBVFL DNA digested with SalI and AscI. The DNA samples were analyzed by pulsed-field gel electrophoresis with 1% agarose gels. The 32-kb vaccinia virus-IBV SalI fragment containing the full-length IBV cDNA (derived from the SalI site following the T7 termination sequence in the IBV cDNA and a SalI site downstream of the vaccinia virus tk gene; Fig. 1) and the 27.9-kb AscI-SalI IBV cDNA fragment are marked by arrows. The AscI site was incorporated upstream of the T7 promoter during the construction of FRAG-1 and is unique in vNotI/IBVFL. The 32-kb SalI fragment contains the 27.9-kb IBV-containing cDNA and the 4.6-kb vaccinia virus-derived DNA containing the tk gene. The 4.6-kb AscI-SalI vaccinia virus-derived DNA comigrates with the smallest vaccinia virus SalI fragment shown at the bottom of the gel. The lane marked M contained DNA markers of 8.3 to 48.5 kb. (B) PCR analysis of DNA extracted from vNotI/IBVFL. Lanes 1 to 7 represent PCR products A to G respectively, as indicated in Fig. 1. Each lane consisted of three tracks in which the first track represented PCR products derived from the appropriate IBV cDNA in pFRAG-1, pFRAG-2, or pFRAG-3; the second track represented PCR products derived from vNotI/IBVFL DNA; and the third track represented PCR products derived from water. The PCR fragments were analyzed by agarose gel electrophoresis with 0.7% agarose. Lane M contained DNA markers with the sizes of the smaller fragments indicated.
Recovery of infectious IBV from full-length cDNA.
Initially, recovery of infectious IBV was attempted by using in vitro T7-derived IBV genomic RNA transcripts generated from vNotI/IBVFL DNA purified from virus particles and restricted with SalI, which is a protocol that had been successfully used for the recovery of recombinant HCoV (39). However, although we were able to produce T7-derived transcripts of the correct length, the amounts and purity of the RNA varied, and attempts to recover recombinant IBV after electroporation of the in vitro RNA into CK cells were not successful. We therefore decided to try an alternative strategy. CK cells were infected with rFPV-T7 to provide cytoplasmic T7 RNA polymerase, and at 1 h p.i. the cells were transfected with SalI- or AscI-restricted vNotI/IBVFL DNA that had been directly isolated from vNotI/IBVFL-infected cells. The vNotI/IBVFL DNA was restricted to prevent the recovery of progeny vaccinia virus by rFPV-T7. Transfection of the restricted vNotI/IBVFL DNA was done with or without pCi-Nuc that expresses the IBV N protein under control of both the T7 promoter and the cytomegalovirus promoter. The transfected cells (P0) were incubated until they showed a CPE, which could result from either rFPV-T7 or from recovered IBV infection. The medium from cells with CPE was filtered to remove any rFPV-T7 and any potential IBV passaged on fresh CK (P1) cells. Previous PCR analyses had demonstrated that filtration of cell medium by using a 0.22-μm (pore-size) filter removed rFPV-T7 (13). PCR analysis with primers within the IBV genomic sequence and the vaccinia virus tk gene to detect the presence of vNotI/IBVFL DNA also demonstrated that vNotI/IBVFL was removed from cell medium following filtration through a 0.22-μm filter (data not shown).
In three independent transfection experiments, in which pCi-Nuc DNA was included in the transfections, 1 of 8, 10 of 10, and 2 of 10 CK (P1) cell monolayers showed a CPE typical of an IBV infection by 36 h p.i. Any recovered IBV present in culture medium from the cells showing CPE was passaged five times (P1 to P5) in CK cells. Analysis of the virus titers recovered in the medium from experiment 1 showed that the titer increased on passage from 2 × 103 at 84 h posttransfection in P0 to 109 PFU/ml at 24 h p.i. in P5 cells. The virus titer was also determined from P1 cells at 72 h p.i. and was found to be 5 × 105 PFU/ml. Therefore, in subsequent experiments, the virus was passaged at 48 h p.i. from P1 and P2 cells and at 24 h p.i. from P3, P4, and P5 cells.
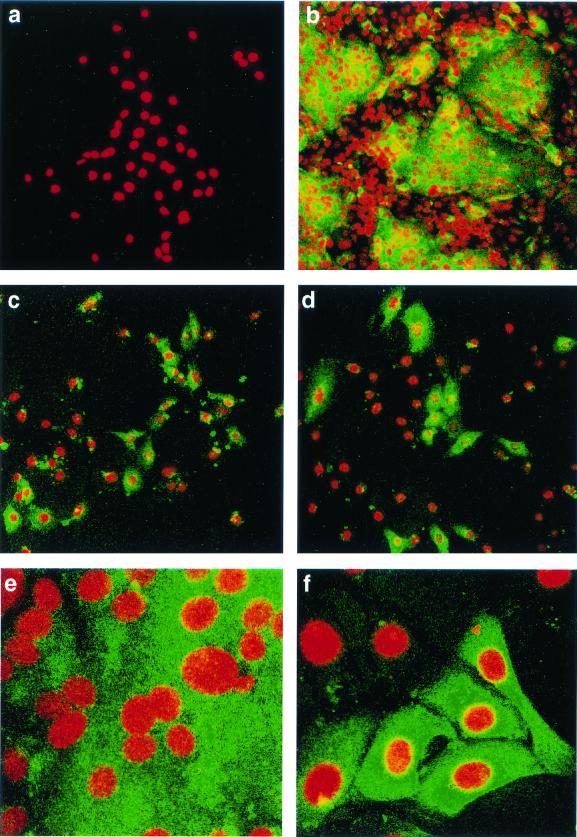
Virus isolated from P5 CK cells was used to infect Vero cells. At 18 h p.i., the cells were analyzed by indirect immunofluorescence by using rabbit anti-IBV polyclonal sera, followed by FITC-labeled goat anti-rabbit antibody. IBV Beau-US is a Vero-adapted isolate and causes extensive syncytia. IBV Beau-CK is able to infect Vero cells but not produce syncytia. As can be seen from Fig. 4, the rabbit antiserum was able to detect cells infected with either Beau-CK, Beau-US, or the virus recovered from P5 CK cells. This indicates that the P5 medium contained recovered IBV (subsequently termed Beau-R). Immunofluorescence analysis of the infected Vero cells showed that, in contrast to Beau-US (Fig. 4b and d), both Beau-CK (Fig. 4c) and the recovered virus (Fig. 4d and f) did not give rise to syncytia and therefore share the same pattern of infection on Vero cells.
FIG. 4.
Detection of IBV in infected Vero cells by indirect immunofluorescence. Vero cells at 60% confluency were infected with IBV, fixed after 18 h with 50% methanol-acetone, analyzed by indirect immunofluorescence with rabbit anti-IBV polyclonal sera, followed by FITC-labeled goat anti-rabbit antibody, and then stained with propidium iodide to visualize the nuclear DNA. (a) Vero cells that had been infected with Beau-R and were analyzed with preimmune rabbit serum. The remaining panels show Vero cells, analyzed with rabbit anti-IBV serum, infected with Beau-US, exhibiting syncytium formation (b and e); Beau-CK (c); or Beau-R (d and f). Magnifications: a to d, ×16; e and f, ×63.
Total RNA isolated from P0 to P5 cell lysates was analyzed by RT-PCR by using the oligonucleotides Leader1 and PM5– to assay for the transcription of IBV sg mRNAs. DNA fragments of 900 and 306 bp, corresponding to RT-PCR products derived from the 5′ end of sg mRNAs 3 and 4, respectively, were identified in the cell lysates from P0 to P5 cells, thus confirming that IBV was replicating in the CK cells (data not shown). IBV was only recovered from cells cotransfected with pCi-Nuc, indicating that the IBV N protein facilitated the recovery of recombinant virus in this system, as was previously observed for the recovery of TGEV (45).
Analysis of recovered IBV.
For further verification that IBV had been recovered, RNA extracted from the cytoplasm of P0 to P5 CK cells after recovery of Beau-R was examined by Northern blot analysis with the 309-bp IBV 3′-UTR probe. The mobilities and relative amounts of the sg mRNAs 1 to 6 were indistinguishable from those of the parental virus (Fig. 5), confirming that the infected cells contained replicating IBV.
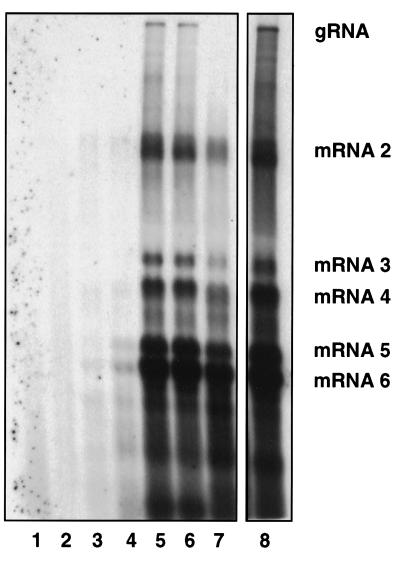
FIG. 5.
Analysis of IBV-specific RNAs after transfection of AscI-restricted vNotI/IBVFL DNA into CK cells infected with rFPV-T7. CK cells infected with rFPV-T7 were transfected with AscI-restricted vNotI/IBVFL DNA (P0 cells), and at 84 h posttransfection the cell medium was filtered to remove any rFPV-T7. Potential IBV (V1) in the filtered medium was used to infect CK cells (P1). This was repeated up to passage 5 (P5) by using any recovered IBV (V2 to V4) in the cell medium. The total cellular RNA was extracted from the transfected (P0) or infected (P1 to P5) CK cells, electrophoresed in denaturing formaldehyde-agarose gels, and Northern blotted, and IBV-derived RNAs were detected nonisotopically with a 309-bp IBV 3′-UTR probe (13). Lane 1, RNA from mock-infected CK cells; lanes 2 to 7, RNA from P0 to P5 CK cells potentially infected with recovered IBV; lane 8, RNA from CK cells infected with Beau-US. The IBV-specific RNAs (indicated by gRNA and mRNAs 2 to 6) represent the IBV genomic RNA and sg mRNAs 2 to 6. The RNAs detected between sg mRNAs 4 and 5 and below sg mRNA 6 are routinely observed from all strains of IBV, as originally identified by (36), and are of unknown origin.
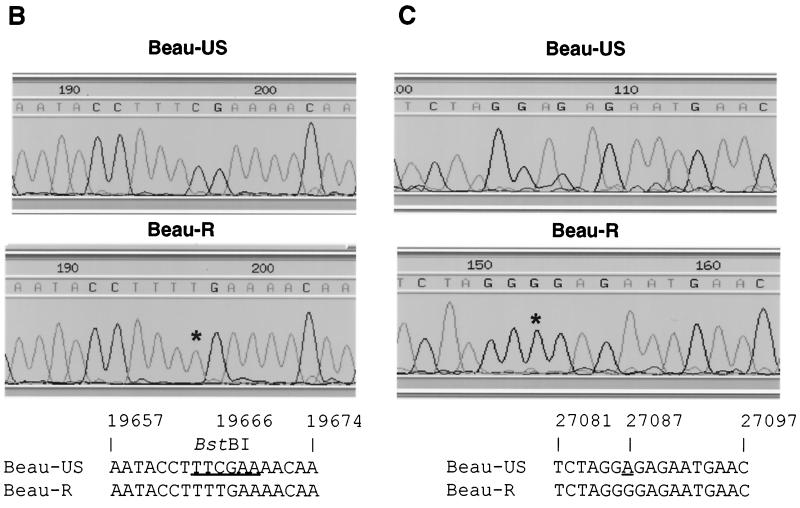
To confirm that IBV Beau-R was derived from the IBV cDNA in vNotI/IBVFL, two RT-PCRs were performed with total cellular RNA derived from CK cells infected with V6 Beau-R, Beau-US, or Beau-CK. The oligonucleotides BG-68 and BG-132 were used to amplify a product of 1,544 bp that encompasses the point mutation at nt 19666, and the oligonucleotides BG-54 and 93/100 were used to amplify a product of 1,666 bp that encompasses the point mutation at nt 27087. Digestion with BstBI of the 1,544-bp RT-PCR product derived from Beau-US and Beau-CK resulted in two fragments of 525 and 1,019 bp, as expected (Fig. 6A). In contrast, the Beau-R-derived 1544 bp RT-PCR product did not digest with BstBI (Fig. 6A), indicating that the C19666→U substitution was indeed present in Beau-R. Sequence analysis of the 1,544- and 1,666-bp products confirmed that Beau-CK contained the published sequences and that the sequence derived from Beau-R contained both of the point mutations (Fig. 6B and C) present in the IBV cDNA in vNotI/IBVFL. Similar RT-PCR analyses on RNA from an additional four independently recovered IBVs (Beau-R2 to -R5) showed that all of the recovered IBVs contained the two point mutations (data not shown). The growth kinetics of Beau-R compared to Beau-CK were very similar, indicating that neither point mutation had a deleterious effect on the growth of the recovered IBV.
FIG. 6.
Analysis of the marker mutations in recombinant IBV Beau-R. (A) BstBI digestion of the 1,544-bp RT-PCR products generated by using oligonucleotides BG-68 and BG-132, corresponding to the region of the BstBI site present in the Beau-CK and Beau-US genomes. Lanes 1, 2, and 3, correspond to the BstBI-restricted RT-PCR products amplified from RNA isolated from CK cells infected with Beau-US, Beau-R, and Beau-CK, respectively. Lane M contained DNA markers. (B) Sequence analysis of IBV genomic RNA, derived from CK cells infected with either Beau-US or Beau-R, representing the BstBI site that contained the C19666→U point mutation in Beau-R. (C) Sequence analysis of IBV genomic RNA, analyzed from CK cells infected with Beau-US and Beau-R, representing the region at the 3′ end of the N gene sequence corresponding to the silent A27087→G point mutation in the Beau-R sequence. The point mutations are marked with an asterisk, and the positions of the mutations within the cDNA sequence are shown. The genomic sequence derived from Beau-CK (6) is the same as that determined for the Beau-US sequence.
The complete genomic sequence analysis of Beau-R has been determined and deposited under accession number AJ311317. This analysis showed that the sequence corresponded to the published Beau-CK sequence (6), apart from the two point mutations described above. However, the Beau-R sequence differed at eight positions within the S gene and seven other positions, including the point mutation at nt 27087, when compared to the corresponding region of Beau-US (Table 1). This further confirmed that Beau-R had been recovered from RNA derived from the IBV cDNA in vNotI/IBVFL.
TABLE 1.
Nucleotide substitutions between CK-adapted Beau-CKa and the Vero-adapted Beau-USb laboratory strain of IBV
| Gene | No. of mutations | Position (nt) | Nucleotide change | Codon change | Amino acid change |
|---|---|---|---|---|---|
| S gene | 8 | 20421 | T→C | GCT→GCC | None |
| 20480 | G→A | AGT→AAT | Ser→Asn | ||
| 20731 | T→A | TTA→ATA | Leu→Ile | ||
| 21403 | T→A | TCA→ACA | Ser→Thr | ||
| 21711 | G→A | GTG→GTA | None | ||
| 22252 | G→A | GGG→AGG | Gly→Arg | ||
| 22415 | A→C | AAT→ACT | Asn→Thr | ||
| 22612 | T→A | TTG→ATG | Leu→Met | ||
| Gene 3 | |||||
| ORF 3a | 1 | 23906 | T→C | TGT→TGC | None |
| ORF 3b | 0 | ||||
| ORF 3c (E) | 1 | 24501 | C→T | CAA→TAA | Gln→Stop |
| M gene | 2 | 24635 | G→C | AGT→ACT | Ser→Thr |
| 24746 | C→T | GCA→GTA | Ala→Val | ||
| Noncoding | 1 | 25227 | C→T | NAd | NA |
| Gene 5 | |||||
| ORF 5a | 1 | 25513 | G→T | AGA→ATA | Arg→Ile |
| ORF 5b | 0 | ||||
| N genec | 0 |
The sequence of Beau-CK was determined previously (6).
The sequence, derived from Vero-adapted Beau-US, has been deposited under accession no. AJ311362.
Beau-R contains the A27087→G substitution not present in Beau-CK or Beau-US.
NA, not applicable.
These results demonstrated that Beau-R differed significantly from our laboratory strain of IBV (Beau-US), differed from the Beau-CK sequence by the two marker mutations, and conclusively confirmed that Beau-R was recovered from the cDNA present in vNotI/IBVFL.
DISCUSSION
In this study we report the first description of a reverse genetic system for the successful recovery of the avian coronavirus, IBV, a group III coronavirus. The IBV cDNA was assembled in vitro and directly cloned in the genome of vaccinia virus in a similar way but by a different assembly strategy, as reported for HCoV (39). However, in contrast to the earlier studies, we have now used a modified method for the generation of infectious IBV RNA. Recombinant vaccinia virus DNA was isolated from infected cells, restricted, and lipofected into cells previously infected with rFPV-T7, an rFPV expressing T7 RNA polymerase (7). Additionally, IBV was only recovered after cotransfection with plasmid DNA encoding the IBV N gene.
In contrast to the assembly of the TGEV and HCoV cDNAs, we chose to assemble the IBV full-length cDNA by utilizing naturally occurring restriction sites rather than by creating new restriction sites in the cDNAs, thereby avoiding the introduction of nucleotide changes into the virus genome associated with such modifications. A potential disadvantage of this method is that the resultant full-length cDNA is identical to the virus genome from which the cDNAs were derived. To overcome this and to eliminate the possibility that any recovered virus resulted from contamination from parental virus, we assembled the IBV cDNA from cDNAs derived from IBV Beau-CK. Beau-CK was never grown in our laboratory prior to the recovery of the recombinant IBV, Beau-R, by using our reverse genetics system. Our normal laboratory strain of IBV, Beau-US, significantly differs from the Beau-CK sequence (Table 1). In addition, we found that one of the cDNAs, FRAG-3, used for the assembly of the full-length cDNA contained two point mutations. One mutation was translationally silent, and the other caused an amino acid substitution and resulted in the loss of a restriction site. We decided to retain the two point mutations as diagnostic markers to prove that any recovered virus was derived from the full-length cDNA and distinguishable from IBV Beau-CK.
In accord with the results reported for the assembly of both TGEV and HCoV full-length cDNAs, we also found that a region of the IBV genome was unstable for propagation in E. coli, irrespective of whether we used high- or low-copy-number plasmids or a BAC vector. We were unable to generate a stable cDNA in E. coli resulting from the ligation of FRAG-1 and FRAG-2. Attempts at joining the 3,623-bp SacI6494-KpnI10117 region from FRAG-2 with FRAG-1 were also unsuccessful in E. coli. However, we were able to isolate a stable cDNA, corresponding to nt 5752 to 12600 of the IBV genome, from the plasmid pKTF2 (K. Tibbles, unpublished results). This indicates that the instability observed for some IBV cDNAs probably results due to the occurrence of a region of sequence preceding nt 5752 (in FRAG-1) and a region of sequence between nt 6494 and 10117 (in FRAG-2) when they are present within the same cDNA. The precise reasons for the instability of certain coronavirus sequences in bacterial vectors have yet to be elucidated.
As described for TGEV (45) and HCoV (39), we decided to assemble the cDNA in vitro to avoid assembly in a bacterial system and to use vaccinia virus as a vector for the full-length IBV cDNA. A significant advantage of using vaccinia virus as a cloning vector for large cDNA clones is that the inserted DNA is very stable and that the cloning procedure, described in this study and for HCoV (39), is very efficient. The approach obviates the need for a selection system or the use of intermediary shuttle-recombination plasmids, and the resulting recombinants provide a base for introducing defined mutations into the IBV-derived cDNA by vaccinia virus-mediated homologous recombination. In this study, we also describe a simplified procedure for generating the infectious recombinant RNA. To do this, we adopted our FPV-T7 RNA polymerase system (7, 13) for the in situ synthesis of T7 RNA polymerase transcripts representing the complete IBV genome. Essentially, restricted recombinant vaccinia virus DNA was transfected into cells previously infected with rFPV-T7. The vaccinia virus DNA was isolated from infected cells without the extensive purification method required for in vitro T7 RNA polymerase transcription (39). This modification has several advantages: (i) highly purified vaccinia DNA is not required, (ii) there is no need to use expensive cap analogues, and (iii) there is no need to electroporate or transfect T7 RNA polymerase transcripts into cells, an inefficient process that is likely to reduce the recovery of recombinant viruses.
In the experiments described here, we only recovered recombinant IBV when we cotransfected the CK P0 cells with restricted recombinant vaccinia virus DNA and plasmid DNA encoding the IBV N protein. The observation that recovery of TGEV by using the BAC system and of HCoV by using the vaccinia vector system did not demonstrate this requirement indicates that N protein is not essential for establishing a productive infection. Also, it has recently been shown that the replication and transcription of the arterivirus EAV (22) and the transcription of sg mRNA from a human coronavirus-based vector (40) occurs in the absence of N protein. However, the data presented here do indicate that exogenous N protein (or N mRNA) has some enhancing effect on the recovery of coronaviruses. The reason for the requirement of either exogenous N protein or the presence of the N mRNA for recovery of either TGEV (45) or IBV (this study) is not known. The binding of N protein to the viral RNA might stabilize the RNA and protect it from nuclease digestion long enough to enable its recruitment by ribosomes.
Our results have confirmed that the vaccinia virus-based reverse genetics system developed for recovery of HCoV (39) is applicable to another coronavirus, IBV. The availability of an IBV reverse genetics system allows us to produce defined, genetically modified viruses. These will be important for the analysis of the molecular biology and pathogenesis of IBV and for the development of defined vaccines to prevent or control infection against new isolates of IBV. The system also has considerable potential for developing IBV as a vector for the expression of heterologous genes, as has been achieved for IBV defective RNAs (37). This would allow IBV to be used not only to protect against IBV, an important world wide veterinary pathogen, but also as an RNA virus vaccine vector against other poultry pathogens.
ACKNOWLEDGMENTS
This work was supported by the Ministry of Agriculture, Fisheries, and Food, United Kingdom, project codes OD1905 and OD0712. R. Casais was the recipient of an EU TMR Marie Curie Research Training Grant and EU Framework Five RTD programme grant QLK2-CT-1999–00002.
We thank the DFG for providing support for R. Casais to work in the laboratory of S. Siddell (DFG SFB 479 Si-B4); Tereza Cardoso, State University of São Paulo, Brazil, for help with the immunofluorescence studies; and J. A. Hiscox, University of Reading, for the confocal microscopy.
REFERENCES
- 1.Almazán F, González J M, Pénzes Z, Izeta A, Calvo E, Plana-Durán J, Enjuanes L. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc Natl Acad Sci USA. 2000;97:5516–5521. doi: 10.1073/pnas.97.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso Caplen F V, Matsuoka Y, Wilcox G E, Compans R W. Replication and morphogenesis of avian coronavirus in Vero cells and their inhibition by monensin. Virus Res. 1984;1:153–167. doi: 10.1016/0168-1702(84)90070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 4.Baric R S, Stohlman S A, Lai M M C. Characterization of replicative intermediate RNA of mouse hepatitis virus: presence of leader RNA sequences on nascent chains. J Virol. 1983;48:633–640. doi: 10.1128/jvi.48.3.633-640.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonfield J K, Smith K F, Staden R. A new DNA sequence assembly program. Nucleic Acids Res. 1995;23:4992–4999. doi: 10.1093/nar/23.24.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boursnell M E G, Brown T D K, Foulds I J, Green P F, Tomley F M, Binns M M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J Gen Virol. 1987;68:57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- 7.Britton P, Green P, Kottier S, Mawditt K L, Pénzes Z, Cavanagh D, Skinner M A. Expression of bacteriophage T7 RNA polymerase in avian and mammalian cells by a recombinant fowlpox virus. J Gen Virol. 1996;77:963–967. doi: 10.1099/0022-1317-77-5-963. [DOI] [PubMed] [Google Scholar]
- 8.Cavanagh D. A nomenclature for avian coronavirus isolates and the question of species status. Avian Pathol. 2001;30:109–115. doi: 10.1080/03079450120044506. [DOI] [PubMed] [Google Scholar]
- 9.Cavanagh D, Davis P J, Pappin D J C, Binns M M, Boursnell M E G, Brown T D K. Coronavirus IBV: partial amino terminal sequencing of spike polypeptide S2 identifies the sequence Arg-Arg-Phe-Arg-Arg at the cleavage site of the spike precursor propolypeptide of IBV strains Beaudette and M41. Virus Res. 1986;4:133–143. doi: 10.1016/0168-1702(86)90037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavanagh D, Naqi S. Infectious bronchitis. In: Calnek B W, Barnes H J, Beard C W, Reid W M, Yoda H W, editors. Diseases of poultry. 10th ed. Ames, Iowa: Iowa State University Press; 1997. pp. 511–526. [Google Scholar]
- 11.Dalton K, Casais R, Shaw K, Stirrups K, Evans S, Britton P, Brown T D, Cavanagh D. cis-Acting sequences required for coronavirus infectious bronchitis virus defective-RNA replication and packaging. J Virol. 2001;75:125–133. doi: 10.1128/JVI.75.1.125-133.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Vries A A F, Horzinek M C, Rottier P J M, de Groot R J. The genome organisation of the Nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin Virol. 1997;8:33–47. doi: 10.1006/smvy.1997.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans S, Cavanagh D, Britton P. Utilizing fowlpox virus recombinants to generate defective RNAs of the coronavirus infectious bronchitis virus. J Gen Virol. 2000;81:2855–2865. doi: 10.1099/0022-1317-81-12-2855. [DOI] [PubMed] [Google Scholar]
- 14.Hiscox J A, Wurm T, Wilson L, Britton P, Cavanagh D, Brooks G. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J Virol. 2001;75:506–512. doi: 10.1128/JVI.75.1.506-512.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai M M, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lambrechts C, Pensaert M, Ducatelle R. Challenge experiments to evaluate cross-protection induced at the trachea and kidney level by vaccine strains and Belgian nephropathogenic isolates of avian infectious bronchitis virus. Avian Pathol. 1993;22:577–590. doi: 10.1080/03079459308418945. [DOI] [PubMed] [Google Scholar]
- 17.Liu D X, Brierley I, Tibbles K W, Brown T D K. A 100-kilodalton polypeptide encoded by open reading frame (ORF) 1b of the coronavirus infectious bronchitis virus is processed by ORF 1a products. J Virol. 1994;68:5772–5780. doi: 10.1128/jvi.68.9.5772-5780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mackett M, Smith G L, Moss B. The construction and characterisation of vaccinia virus recombinants expressing foreign genes. In: Glover D M, editor. DNA cloning: a practical approach. Vol. 2. Oxford, England: IRL Press; 1985. pp. 191–211. [Google Scholar]
- 19.Mayr A, Malicki K. Attenuation of virulent fowlpox virus in tissue culture and characteristics of the attenuated strain. Zentbl Veterinarmedzin. 1966;13:1–13. [PubMed] [Google Scholar]
- 20.Merchlinsky M, Moss B. Introduction of foreign DNA into the vaccinia virus genome by in vitro ligation: recombination-independent selectable cloning vectors. Virology. 1992;190:522–526. doi: 10.1016/0042-6822(92)91246-q. [DOI] [PubMed] [Google Scholar]
- 21.Mockett A P A. Envelope proteins of avian infectious bronchitis virus: purification and biological properties. J Virol Methods. 1985;12:271–278. doi: 10.1016/0166-0934(85)90138-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molenkamp R, van Tol H, Rozier B C, van Der Meer Y, Spaan W J, Snijder E J. The arterivirus replicase is the only viral protein required for genome replication and subgenomic mRNA transcription. J Gen Virol. 2000;81:2491–2496. doi: 10.1099/0022-1317-81-10-2491. [DOI] [PubMed] [Google Scholar]
- 23.Pénzes Z, Tibbles K, Shaw K, Britton P, Brown T D K, Cavanagh D. Characterization of a replicating and packaged defective RNA of avian coronavirus infectious bronchitis virus. Virology. 1994;203:286–293. doi: 10.1006/viro.1994.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pénzes Z, Wroe C, Brown T D, Britton P, Cavanagh D. Replication and packaging of coronavirus infectious bronchitis virus defective RNAs lacking a long open reading frame. J Virol. 1996;70:8660–8668. doi: 10.1128/jvi.70.12.8660-8668.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polo S, Ketner G, Levis R, Falgout B. Infectious RNA transcripts from full-length dengue virus type 2 cDNA clones made in yeast. J Virol. 1997;71:5366–5374. doi: 10.1128/jvi.71.7.5366-5374.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Racaniello V R, Baltimore D. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science. 1981;214:916–919. doi: 10.1126/science.6272391. [DOI] [PubMed] [Google Scholar]
- 27.Rice C M, Grakoui A, Galler R, Chambers T J. Transcription of infectious yellow fever RNA from full-length cDNA templates produced by in vitro ligation. New Biol. 1989;1:285–296. [PubMed] [Google Scholar]
- 28.Ruggli N, Tratschin J D, Mittelholzer C, Hofmann M A. Nucleotide sequence of classical swine fever virus strain Alfort/187 and transcription of infectious RNA from stably cloned full-length cDNA. J Virol. 1996;70:3478–3487. doi: 10.1128/jvi.70.6.3478-3487.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. New York, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 30.Satyanarayana T, Bar-Joseph M, Mawassi M, Albiach-Marti M R, Ayllon M A, Gowda S, Hilf M E, Moreno P, Garnsey S M, Dawson W O. Amplification of Citrus tristeza virus from a cDNA clone and infection of citrus trees. Virology. 2001;280:87–96. doi: 10.1006/viro.2000.0759. [DOI] [PubMed] [Google Scholar]
- 31.Satyanarayana T, Gowda S, Boyko V P, Albiach-Marti M R, Mawassi M, Navas-Castillo J, Karasev A V, Dolja V, Hilf M E, Lewandowski D J, Moreno P, Bar-Joseph M, Garnsey S M, Dawson W O. An engineered closterovirus RNA replicon and analysis of heterologous terminal sequences for replication. Proc Natl Acad Sci USA. 1999;96:7433–7438. doi: 10.1073/pnas.96.13.7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawicki S G, Sawicki D L. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J Virol. 1990;64:1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sawicki S G, Sawicki D L. A new model for coronavirus transcription. Adv Exp Med Biol. 1998;440:215–219. doi: 10.1007/978-1-4615-5331-1_26. [DOI] [PubMed] [Google Scholar]
- 34.Siddell S G. The Coronaviridae. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Publishing, Inc.; 1995. pp. 1–10. [Google Scholar]
- 35.Spaan W J H, Delius H, Skinner M, Armstrong J, Rottier P, Smeekens S, Van der Zeijst B A M, Siddell S G. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 1983;2:1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stern D F, Kennedy S I T. Coronavirus multiplication strategy. I. Identification and characterization of virus-specific RNA. J Virol. 1980;34:665–674. doi: 10.1128/jvi.34.3.665-674.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stirrups K, Shaw K, Evans S, Dalton K, Casais R, Cavanagh D, Britton P. Expression of reporter genes from the defective RNA CD-61 of the coronavirus infectious bronchitis virus. J Gen Virol. 2000;81:1687–1698. doi: 10.1099/0022-1317-81-7-1687. [DOI] [PubMed] [Google Scholar]
- 38.Stirrups K, Shaw K, Evans S, Dalton K, Cavanagh D, Britton P. Leader switching occurs during the rescue of defective RNAs by heterologous strains of the coronavirus infectious bronchitis virus. J Gen Virol. 2000;81:791–801. doi: 10.1099/0022-1317-81-3-791. [DOI] [PubMed] [Google Scholar]
- 39.Thiel V, Herold J, Schelle B, Siddell S G. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J Gen Virol. 2001;82:1273–1281. doi: 10.1099/0022-1317-82-6-1273. [DOI] [PubMed] [Google Scholar]
- 40.Thiel V, Herold J, Schelle B, Siddell S G. Viral replicase gene products suffice for coronavirus discontinuous transcription. J Virol. 2001;75:6676–6681. doi: 10.1128/JVI.75.14.6676-6681.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tibbles K W, Cavanagh D, Brown T D. Activity of a purified His-tagged 3C-like proteinase from the coronavirus infectious bronchitis virus. Virus Res. 1999;60:137–145. doi: 10.1016/S0168-1702(99)00011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Dinten L C, den Boon J A, Wassenaar A L M, Spaan W J M, Snijder E J. An infectious arterivirus cDNA clone: identification of a replicase point mutation that abolishes discontinuous mRNA transcription. Proc Natl Acad Sci USA. 1997;94:991–996. doi: 10.1073/pnas.94.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang K, Boysen C, Shizuya H, Simon M I, Hood L. Complete nucleotide sequence of two generations of a bacterial artificial chromosome cloning vector. BioTechniques. 1997;23:992–994. doi: 10.2144/97236bm04. [DOI] [PubMed] [Google Scholar]
- 44.Yamshchikov V, Mishin V, Cominelli F. A new strategy in design of (+)RNA virus infectious clones enabling their stable propagation in E. coli. Virology. 2001;281:272–280. doi: 10.1006/viro.2000.0793. [DOI] [PubMed] [Google Scholar]
- 45.Yount B, Curtis K M, Baric R S. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J Virol. 2000;74:10600–10611. doi: 10.1128/jvi.74.22.10600-10611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ziebuhr J, Snijder E J, Gorbalenya A E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]