Summary
Antibody–drug conjugates (ADCs) have become an important class of anticancer drugs in solid tumors including drug-resistant gynecologic malignancies. TROP2 is a cell surface antigen that is highly expressed in ovarian carcinoma (OC) but minimally expressed in normal ovarian tissues. In this study, we aimed to identify how TROP2-specific ADC, sacituzumab govitecan (SG), modulates DNA damage response pathways in drug-resistant OC. We found that SG induces G2/M arrest, increases RPA1 foci, and decreases replication fork speed, resulting in replication stress in TROP2-positive cells while these were less evident in TROP2-negative cells. In OC in vitro and in vivo models, SN-38 sensitivity and TROP2 expression play key roles in response to either ATR inhibitor or SG alone, or in combination. Additionally, inhibition of translesion DNA synthesis enhances SG and PARP inhibitor (PARPi) sensitivity in PARPi-resistant OC cells. These findings provide mechanistic insights for clinical development of SG in drug-resistant OC.
Subject areas: Cancer, Molecular biology, Oncology
Graphical abstract
Highlights
-
•
SG shows antitumor activity in TROP2-high and TROP2-low ovarian cancer models
-
•
SG and ATRi have distinct effects on cell cycle and DNA damage response pathways
-
•
SG induces replication stress in TROP2-positive ovarian cancer cells
-
•
SG response is dependent on ssDNA repair, RF progression, and HR responses
Cancer; Molecular biology; Oncology
Introduction
High-grade serous ovarian carcinoma (HGSOC) is the most lethal gynecological malignancy in the United States (U.S.) because of its presentation at late stages and proclivity for drug resistance.1 Approximately 40% of HGSOCs show deficiency in homologous recombination (HR) DNA double-strand break (DSB) repair due to mutations in key tumor suppressor genes (e.g., BRCA1 and BRCA2 [BRCA1/2]),2 which sensitizes them to DNA-damaging agents and PARP inhibitors (PARPis).3 However, despite the clinical benefit of PARPis,3 resistance eventually develops, highlighting the unmet need for innovative treatments in recurrent ovarian cancer (OC).
Antibody-drug conjugates (ADCs) have gained significant momentum in treating patients with drug-resistant tumors.4 ADCs, that use humanized antibodies linked by a cleavable linker to its payload, have helped overcome many of the undesirable immunogenic reactions, adverse effects, and suboptimal target specificity of earlier antibody-based treatments.5 Several ADCs have been approved by the U.S. Food and Drug Administration (FDA) or are under clinical investigation in gynecologic malignancies. Currently, regulatory approvals have been granted to mirvetuximab soravtansine-gynx, a folate receptor alpha (FRα)-specific antibody and microtubule inhibitor conjugate, for the treatment of platinum-resistant OC6,7,8 and tisotumab vedotin, a tissue factor (TF-011)-directed antibody and monomethyl auristatin E (MMAE) conjugate, for metastatic cervical cancer.9,10 Such targeted delivery of cytotoxic drugs can reduce drug doses required to achieve clinical benefit in patients, while potentially reducing off-target toxicities that limit the tolerability of otherwise effective drugs as is the case with DNA topoisomerase 1 inhibitors (TOP1is) e.g., irinotecan or topotecan.11
Trophoblast antigen protein 2 (TROP2) is a 36 kDa transmembrane glycoprotein encoded by the TACSTD2 gene that is involved in intracellular calcium signaling in many cells.12 TROP2 overexpression is associated with tumorigenicity and poor clinical outcome in several cancers, including OC.13,14,15 Bignotti et al. reported that 92% of OC tumors were positive for TROP2 staining, while it was only 15% for samples of normal ovarian surface epithelium.15 TROP2 overexpression also correlated with worse overall survival (OS) and progression free survival (PFS) in OC patients, suggesting TROP2 as a potential therapeutic target.15 Sacituzumab govitecan (SG) is an ADC comprising a TOP1–inhibiting camptothecin, SN-38 (7-ethyl-10-hydroxycamptothecin, the active metabolite of irinotecan), linked to a humanized anti-TROP2 antibody (hRS7).16,17 SG has a high drug-to-antibody (DAR) ratio with ∼7 molecules of moderately toxic SN-38 conjugated to each antibody via the unique hydrolysable and proprietary linker, CL2A.18 The linker allows the intra-tumoral release of therapeutic concentrations of SN-38, as well as extracellular release within the surrounding tumor microenvironment, providing a bystander effect.19 SG monotherapy has been approved by the FDA for multiple indications in breast cancer, as well as in urothelial carcinoma.20,21 Yet, the mechanism by which SG modulates DNA damage response pathways is not well understood. Additionally, the optimal drug combinations involving SG in the treatment of drug-resistant OC and their contributions to the well-characterized target heterogeneous activity of SG have not been established.19,22
Here, we aim to better understand SG’s biology, particularly its effect on DNA damage response pathways. We also investigate the therapeutic potential of SG monotherapy as well as in combination with ATR inhibitor (ATRi) berzosertib in HGSOC in vitro and in vivo models because ATR/CHK1 pathway is also overexpressed in drug-resistant OC.23,24,25 Our results demonstrate that while high TROP2 expression was associated with sensitivity to SG in cell line models, SG induced tumor shrinkage in both high and low TROP2-expressed OC in vivo models. Mechanistically, SG induces G2/M cell-cycle arrest, increases replication protein A1 (RPA1) foci and decelerates replication fork (RF) speed, leading to replication stress in TROP2-positive cells. Also, cell-bound SG and berzosertib were effective as monotherapy in both in vitro and in vivo OC models, unlike previously reported synergism between ATRi and TOP1i.26,27 Collectively, our results provide mechanistic insights to support the clinical use of SG monotherapy in drug-resistant recurrent OC and offer a template for selection of potential inhibitors to improve SG activity in the future.
Results
High-throughput drug combination screening
In the phase I clinical trial,28 a combination of SG with ATRi berzosertib demonstrated safety and preliminary activity in solid tumor patients. We thus conducted high-throughput drug combination screen to validate the combination of TOP1i and ATRi in drug-resistant HGSOC by using PARPi-resistant HGSOC cell lines (PEO1-olaR and PEO1-olaJR29,30). Of note, while both PEO1-olaJR and PEO1-olaR were developed from the parental BRCA2 mutant (BRCA2mut) PEO1, they have distinct mechanisms of PARPi resistance. The former has a gain-of-function heterozygous mutation for BRCA2 and increased drug efflux activity while the latter exhibits a mesenchymal-like morphology, with increased ATR/CHK1 activity, RF stability and restored HR, with a suppressed EZH2/MUS81 axis.29
We performed an initial 6 × 6 matrix screen of 2,450 drugs in combination with an ATRi (ceralasertib) to identify potential drug combinations for synergy. Synergy with ceralasertib was observed with 324 oncology drugs (average ExcessHSA < −20), including inhibitors that target TOP1 and ATR/CHK1 pathways (Table S1). This was expanded to a 10 × 10 matrix screen that also included parental PARPi-sensitive PEO1 and de novo PARPi-resistant PEO4 (BRCA2, silent homozygous mutation, 5193C>T, Y1655Y), which resulted in the identification of 21 “hit” drugs. Notably, two TOP1is, SN-38 (rank 4) and exatecan (rank 3), consistently exhibited synergistic effects with ceralasertib across all four cell lines (Figure 1A; Table S2).
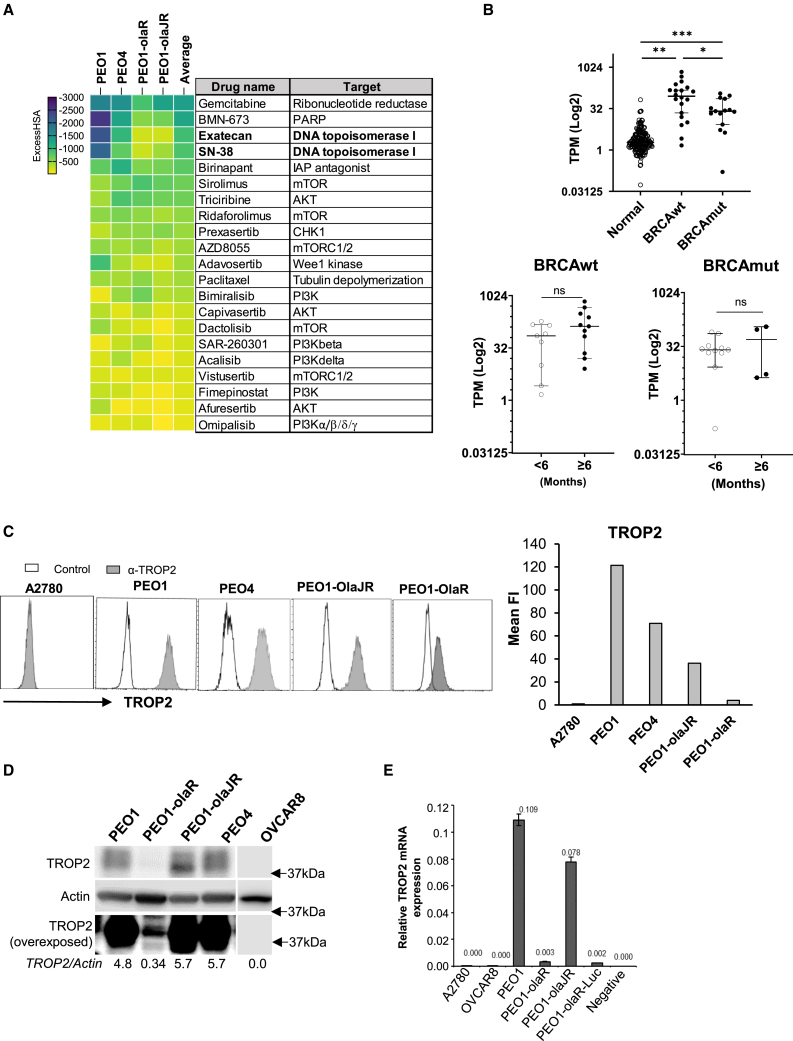
Figure 1.
High-throughput drug combination screens with ATRi and TROP2 expression in patient biopsies and cell lines
(A) The heatmap is a ranked average ExcessHSA from a 10 × 10 combination screen of ATRi (ceralasertib) with various drugs selected from a 6 × 6 screen (Figure S1), in PARPi-sensitive PEO1, PARPi-resistant PEO1-olaJR, and PEO1-olaR and de novo PARPi-resistant PEO4. This experiment was performed once.
(B) Expression values (Log2 transcripts per million, TPM) for TROP2 mRNA (TACSTD2) from both BRCA-mutant (BRCAmut) cohort (n = 15) and BRCA wild-type (BRCAwt) cohort (n = 20) of patients were compared against TPM values of TROP2 from normal ovarian tissues (normal) obtained from the GTEx database (n = 180). TROP2 values for the no-clinical benefit (<6 months PFS) or clinical benefit (≥6 months PFS) groups for cohorts BRCAwt (n = 9 and n = 11 respectively) and BRCAmut (n = 11 and n = 4 respectively) were compared as well. Graphs show TROP2 expression as median TPM (Log2) ± 95% confidence interval (CI) for each cohort or group. Statistical significance was determined using the unpaired Mann-Whitney U-test and is shown as p-values ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001, ns, not significant.
(C) Flow cytometric analysis for TROP2 expression (gray) was performed after incubating cells with recombinant anti-TROP2 hRS7 antibody followed by goat anti-human F(ab)2-AF488 secondary antibody as given in STAR Methods. In control group (white), cells were treated only with secondary antibody. A2780 cell line was used as a TROP2-negative control. Graphs show the value of mean fluorescence intensity (Mean FI) of TROP2 expression peaks divided by that of control peak (n = >8000 events) for each cell line. Figure is representative of 3 biologically independent experiments.
(D) Immunoblotting was performed to screen the baseline levels of TROP2 protein in the same cell lines. Blots were overexposed (bottom panel) to highlight low TROP2 expression in PEO1-olaR. Densitometric quantitation of TROP2 relative to Actin are included below. Closest molecular weight marker positions are shown on the right. Figure is representative of 3 biologically independent experiments.
(E) Quantitative PCR (qPCR) analysis of total RNA from both TROP2 positive and negative cell lines. Graph is representative of 2 biologically independent experiments and shows TROP2 expression values normalized against that of GAPDH for each cell line. A negative control without RNA template is also included as quality control. Data are represented as mean ± SD.
For possible clinical trial development in OC patients, we prioritized berzosertib over other ATRis for the subsequent in vitro and in vivo experiments considering previous safety and activity data of berzosertib in advanced solid tumor patients.24,31,32 We also selected SG that combines the tumor specificity of the antibody with the cytotoxicity of SN-38, for the combination with berzosertib given its less frequent bone marrow toxicities compared to other TOP1-targeting chemotherapeutic drugs. Pharmacokinetic studies in murine models show rapid clearance of intact SG from serum with an alpha-half-life of 11 h.16 Also, the TROP2-specific toxicity of SG is related to cell-bound SG, which is not easily cleared out and subsequently endocytosed by the cells.33
TROP2 expression in HGSOC patient tumors and cell lines
To evaluate the clinical relevance of TROP2 mRNA expression (TACSTD2) in HGSOC, we studied TACSTD2 levels on RNA sequencing (RNA-seq) analysis of tissue samples from HGSOC patients with BRCA mutation (BRCAmut, n = 15) or BRCA wild-type (BRCAwt, n = 20) enrolled in the phase 2 clinical trial of the CHK1 inhibitor (CHK1i) (NCT02203513).34,35 Baseline control TROP2 mRNA expression levels were obtained from normal ovarian tissues in the public Genotype-Tissue Expression Project (GTEx, n = 180). TACSTD2 levels were significantly higher in both BRCAmut and BRCAwt tumors relative to normal tissues (p < 0.01, Figure 1B), suggesting therapeutic potential of TROP2-specific ADC in HGSOC independent of BRCAmut status. No significant association was observed between TROP2 expression and PFS in both BRCAwt and BRCAmut patients with CHK1i treatment (Figure 1B).
TROP2 expression was further evaluated in a panel of PARPi-sensitive (BRCA1-null UWB1.289 and BRCA2mut PEO1), PARPi-resistant (PEO1-olaR,30 PEO-olaJR29 and PEO4) and platinum-resistant cell lines (OVCAR3, OVCAR5, OVCAR8, TOV21G and SKOV3) by flow cytometric and immunoblotting analyses. We also included A2780, a TROP2-negative cell line,36 as a negative control.
Flow cytometric analysis for TROP2 surface expression using the humanized anti-TROP2 hRS7 antibody, revealed differential expression patterns among the cell lines. PEO1, PEO1-olaJR, PEO4, OVCAR3, UWB1.289 and SKOV3 expressed high levels of TROP2. PEO1-olaR, OVCAR5, OVCAR8 and TOV21G showed low or no expression of TROP2 (Figures 1C, S1A, and S1B). Surface expression correlated with total TROP2 protein expression (Figures 1C, 1D, S1A, and S1B) and total mRNA expression by quantitative RT-PCR analysis (qPCR) (Figure 1E).
Immunofluorescence staining was performed to verify TROP2 expression on PEO1-olaR cells but yielded weak signals (Figure S1C). Based on these data, OVCAR5, OVCAR8 and TOV21G were classified as TROP2-negative. TROP2-positive cells were divided into TROP2-high (PEO1, PEO4, PEO1-olaJR, OVCAR3, SKOV3, UWB1.289) and TROP2-low (PEO1-olaR). For the subsequent experiments, we prioritized TROP2-high (PEO1, PEO1-olaJR), TROP2-low (PEO1-olaR) and TROP2-negative (OVCAR8) cells to investigate the mechanisms of action, focusing on SG’s impact on DNA damage response pathways. Using these cell lines, we also explored the therapeutic potential of SG and berzosertib combination.
Distinct contributions of ATRi and SG on growth inhibition in TROP2-positive HGSOC cells
SG’s hydrolysable linker enables partial dissociation into SN-38 and TROP2-specific antibody (RS7) before cellular uptake,16 resulting in tumor cells exhibiting similar apoptotic responses to both SG and free SN-38. Because of this characteristic, we were concerned that longer incubation times might not discern TROP2-specific effects of SG from free SN-38. Hence, we preincubated cells with saturating concentrations of SG (10 μg/mL) for 30 min at 37°C, followed by washes to remove unbound SG, according to previous reports.16,37
Cell growth (XTT) assays were performed on cells pretreated with SG, followed by exposure to a gradient of berzosertib for 48 h. To verify TROP2-specific responses by SG, we included a non-specific control ADC (cADC) in this experiment to account for SG’s TROP2 non-specific bystander effect.16 In TROP2-positive PEO1 and PEO1-olaJR cells, SG significantly inhibited cell growth compared to untreated or cADC (Figures 2A and 2B), attesting to its TROP2 specificity, while berzosertib alone inhibited growth similarly (IC50 ∼0.3 μM) in a concentration-dependent manner. In XTT analyses, addition of berzosertib to SG did not result in additive or synergistic effects with SG (Figure 2A).
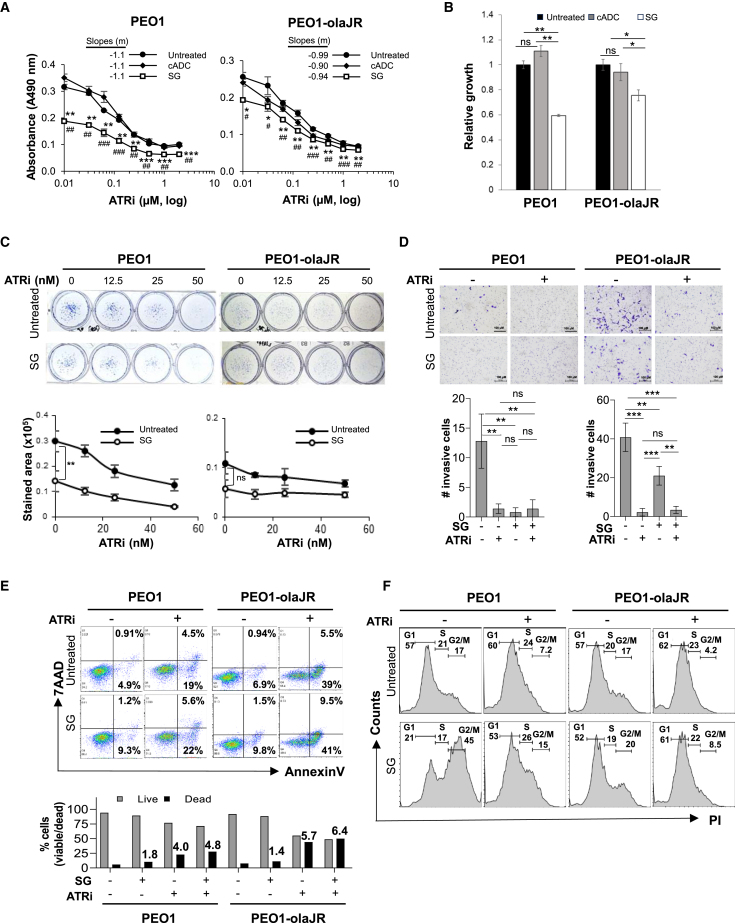
Figure 2.
Effect of antibody drug conjugates (ADCs) on survival and viability in HGSOC cells
(A) Cell growth assays (XTT) were performed on TROP2-positive (PEO1 and PEO1-olaJR) HGSOC cells in triplicate wells, both in normal media or after pretreatment with a nontargeting antibody drug conjugate control (cADC) or SG (10 μg/mL, 30 min at 37°C and washed thrice in PBS), prior to adding ATRi berzosertib (0–2 μM) for 48 h. Graphs show mean absorbance values at 490 nm corrected for background ±SD for each treatment (n = 3 biological replicates) against a concentration gradient (Log2) of ATRi. For all treatments we replaced 0 values for ATRi with 0.01 to enable plotting of absorbance values on a logarithmic X axis. Statistical determinations were done with Student’s t test and shown as p-values. The p-values for SG treatment versus untreated are indicated by asterisks (∗) while SG versus cADC are indicated by hashes (#). Slopes for each curve are derived from linear regression formula “y = mx+b” where y = Y-intercept, x = ATRi (μM) and b = correlation coefficient, calculated from the linear part of the respective curves.
(B) Graphs show mean cell growth values ±SD of cells with cADC or SG alone relative to untreated, derived from panel A (n = 3 biological replicates). Statistical significance (p-values) was determined with Student’s t test.
(C) Clonogenic assays were performed over 7 days with increasing concentrations of berzosertib (ATRi). Graphs show mean ± SD of stained area (n = 3 biological replicates). Statistical significance (p-values) determined using Student’s t test.
(D) Representative images from Matrigel plates after crystal-violet staining are shown. Graphs below image panels show total number of crystal-violet-stained invaded cells from imaged panels ±SD (n = 3 biological replicates). Statistical significance (p-values) was determined using Student’s t test. Experiment was performed twice. Scale bar is 100 μm.
(E) Flow cytometric analysis was conducted to assess cell viability with AnnexinV-FITC and 7AAD reagents on cells pretreated with SG or ATRi berzosertib (1 μM) alone or SG pretreatment followed by ATRi treatment overnight. Percentage of cells positive for AnnexinV alone (early apoptosis) or both AnnexinV and 7AAD (dead) are shown within respective quadrants. Graphs below show percentage of live (gray) and dead cells (black), while numbers above bars indicate fold increase in dead cells for each treatment relative to untreated. Images are representative of 3 biologically independent experiments.
(F) Cells with or without SG pretreatment were treated with ATRi berzosertib (1 μM) for 48 h and then stained with propidium iodide for DNA quantification and cell cycle analysis. Cell populations as fractions of total cells are shown over appropriately marked G1, S, or G2/M phases. Images are representative of 2 experimental replicates. For all images, p-values are ∗, #p < 0.05, ∗∗, ##p < 0.01, and ∗∗∗, ###p < 0.001, ns, not significant.
To evaluate whether this lack of synergy was specific to cell-bound SG, we used cell growth assays (XTT) to compare SN-38 toxicity with equimolar concentrations of free SG (added directly to wells), with or without ATRis (ceralasertib and berzosertib) over 72 h. IC50 values of SN-38 ranged from 2.3 to 5.0 nM across TROP2-negative or positive cell lines (Figure S2A). In combination screening assays, synergy between ATRi and SN-38 was observed in PEO1 and olapR cells (PEO1-olaJR and PEO1-olaR) over a narrow range of concentrations for each drug in combination, while in TROP2 negative OVCAR8, synergy was observed across a wider range of combinations (Figure S2B). Similarly, synergy was also found between berzosertib and equimolar SG similar to that observed for SN-38 in TROP2-positive PEO1, PEO1-olaJR and PEO1-olaR cells (Figure S2C).
To compare the effect of SN-38 alone or equimolar concentrations of free SG on ATRi, XTT assays were performed using predetermined IC50 values (Figure S2A) for SN-38 for OVCAR8 (3 nM), PEO1 (2 nM) and olapR cells (4 nM). SN-38 significantly sensitized all cell lines to ATRi ceralasertib or berzosertib (p < 0.001–0.0001), with free SG demonstrating a lower sensitization potential at equimolar concentrations in all cell lines (Figure S3). Sensitivity to SG was similar to cADC in all cell lines except in TROP2-high PEO1 which exhibited a modestly higher sensitivity to SG.
Effect of cell-bound SG and berzosertib on clonogenicity, viability and invasiveness
We next examined the combination by using other parameters that determine cell growth such as clonogenicity, viability, invasiveness, and proliferation. Since the bystander effect is an integral part of SG’s activity, we excluded the separate treatment arm with cADC alone in subsequent experiments to investigate SG’s impact not driven by bystander effects. In clonogenic assays (>7 days), both cell lines displayed a concentration-dependent response to berzosertib. SG monotherapy significantly reduced colonies in PEO1 compared to control (Figure 2C), while its effect was modest in PEO1-olaJR (Figure 2C). Again, no additive or synergistic effects were observed between cell-bound SG and berzosertib. In cell invasion assay, SG or berzosertib monotherapy caused inhibition of invasiveness in TROP2-positive PEO1 and PEO1-olaJR cells (p < 0.01) with no synergistic or additive effect between the two drugs (Figure 2D).
We next assessed SG’s effect on cell viability through flow cytometric analysis (annexinV/7AAD) over 48 h. In TROP2-positive PEO1 and PEO1-olaJR, SG pretreatment induced modest increase in cell death (1.8- and 1.4-fold, respectively) (Figure 2E). Combination of SG with ATRi berzosertib demonstrated enhanced cell death in both cell lines (4.8- and 6.4-fold, respectively) (Figure 2E). However, given berzosertib alone exhibited a 4.0- and 5.7-fold increase in apoptosis respectively, the combination effect was not sufficient for additivity or synergism.
To compare the effects of free SG and SN-38 in combination with ATRis in cell viability, we treated cells with SN-38 (2 and 4 nM) or concentrations of SG equimolar to SN-38 (0.28 and 0.57 nM) and ATRis (ceralasertib, berzosertib) (Figure S4). Combinations of free SG with either ATRi showed modest increases in cell death compared to ATRi alone. However, these effects were not markedly different from the combination of ATRi with SN-38 in both TROP2-high (PEO1, PEO1-olaJR) and TROP2-low (PEO1-olaR) cells (Figure S4). These results suggest that neither free nor cell-bound SG offers significant advantages over free SN-38 when combined with ATRis.
Berzosertib abrogates SG induced G2/M cell-cycle arrest in HGSOC
Both PARPi-sensitive PEO1 and PARPi-resistant PEO1-olaJR are TP53-mutant cells.29,30,38 In TP53-mutant tumor cells, TOP1is such as irinotecan and topotecan (active metabolite SN-38), induce G2/M checkpoint arrest,32,39 allowing time for DNA damage repair before M-phase transition. We therefore performed cell cycle analysis to study the impact of SG and berzosertib on cell cycle alteration as it might partly account for lack of additive or synergistic effects. We found substantial accumulation of G2/M-populations in SG-pretreated PEO1 and PEO1-olaJR compared to untreated cells (45% and 20% respectively vs. 17% for untreated cell lines, Figure 2F). Consistent with previous reports,24,25,40 ATRi caused cell accumulation in both G1 and S phases while reducing the G2/M accumulation (7.2% and 4.2% respectively vs. 17% for untreated group, Figure 2F). Notably, the addition of ATRi voided SG-induced accumulations in G2/M phase while sustaining G1/S accumulations in both cell lines (Figure 2F).
To further determine the effect of SG on cell cycle regulation in proliferating cells, we conducted bromodeoxyuridine (BrdU) incorporation assays over 48 h. SG alone resulted in cell accumulation in G2/M in both cell lines, but this was mitigated by the addition of ATRi, causing BrdU-positive (BrdU+ve) cells to largely accumulate at G1 phase, similar to the changes made by ATRi alone (Figures 3A and 3B). These findings suggest that ATRi may negate the G2/M accumulation caused by SG by shifting cells to G1/S phases. Given that TOP1is affect RF dynamics,31,32 an important facet of cell cycle progression, we next investigated SG and ATRi’s impact on RF stability and DNA damage response pathways.
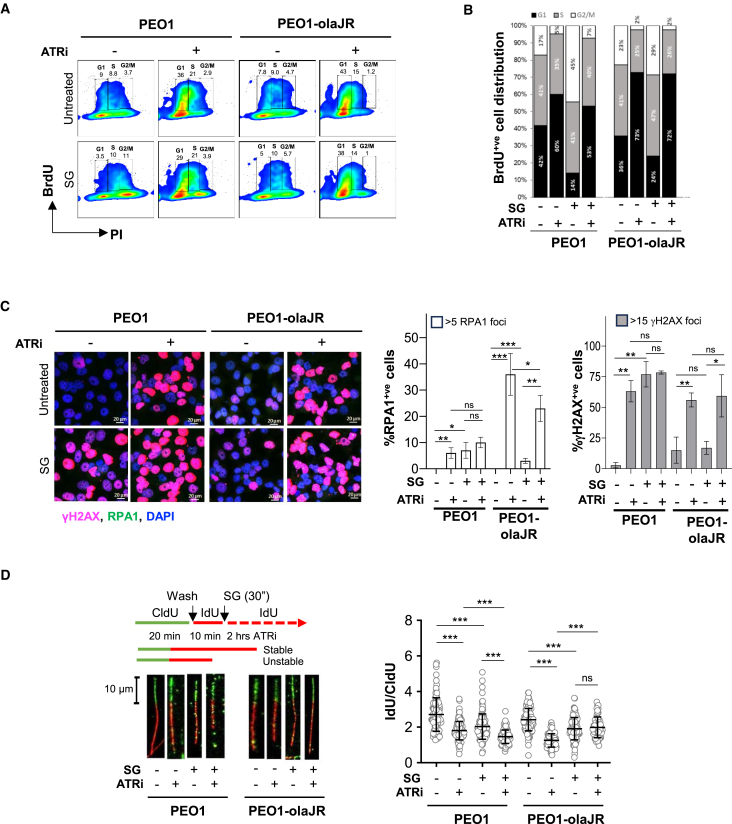
Figure 3.
Effect of SG and berzosertib on tumorigenicity and replication fork stability
(A) PEO1 and PEO1-olaJR were pretreated with SG followed by ATRi (1 μM overnight prior to analyzing them for incorporated BrdU. Images show BrdU-positive (BrdU+ve) cells expressed as percentage of total cells (within boxes) against DNA quantity (PI) as determined by FlowJo software. Images are representative of 3 biologically independent experiments.
(B) Graphs show stacked plots as fractions (as indicated by numbers) of total BrdU+ve cells derived from panel A distributed across G1, S or G2/M phases.
(C) Immunofluorescent confocal microscopic analysis for RPA1 (green) and γH2AX-S139 (pink) of cells pretreated with SG (10μg/ml for 30 min at 37°C, washed 3 times with PBS) and then exposed to berzosertib (ATRi) (1 μM) overnight. Approximately 100–300 cells were analyzed over 3–5 sampled microscopic fields. Graphs show mean ± SD of % cells positive for RPA1 (>5 foci) or γH2AX foci (>15 foci) relative to total cells from each sampled field (n = 3–5 biological replicates). Statistical significance was determined with Student’s t test (p-values). Results are representative of 2 biologically independent experiments. Scale bar is 20 μm.
(D) DNA fiber assays was performed as described in STAR Methods. About 200 DNA fibers were selected and the length of both red (IdU) and green (CldU) segments of each strand were measured using Fiji software. Representative strands are shown to the left. Graphs on the right show mean ± SD of the ratios of IdU/CldU from all strands (n = ∼200) per treatment using GraphPad prism. Statistical significance was determined with Student’s t test and represented as p-values, ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001, ns, not significant. Results are representative of 2 biologically independent experiments. Scale bar is 10 μm.
SG and berzosertib induce RF instability and DNA damage
Unstable RFs are the consequence of unrepaired DNA damage and marked by the formation of single-stranded DNA (ssDNA) lesions that recruit replication protein A (RPA1, ssDNA marker)41 followed by other DNA damage response proteins such as γH2AX (DNA DSB marker).42 We used immunofluorescent microscopy to study SG-induced DNA damage and RF progression with or without ATRi, given that TOP1i payload has been shown to cause RF stalling and consequent replication stress.11
Confocal image analysis revealed that ATRi caused an increased proportion of cells with >5 RPA1 foci (RPA1-positive [RPA1+ve] cells) (Figure 3C, middle). SG pretreatment did not further increase ATRi-induced RPA1+ve cells or cells with >15 γH2AX foci (γH2AX-positive [γH2AX+ve] cells) in TROP2-high PEO1 and PEO1-olaJR (Figure 3C). PEO1 and PEO1-olaJR showed differential responses. In PEO1 cells, berzosertib monotherapy (1 μM) or SG pretreatment alone increased γH2AX+ve cells (63% and 77%, respectively) but the effect of combination treatment (78% γH2AX+ve) was not significantly different compared to either monotherapy (Figure 3C, right). In PEO1-olaJR, only berzosertib monotherapy increased γH2AX+ve cells and the combination effect was not different compared to berzosertib alone (Figure 3C, right).
We also evaluated γH2AX+ve cells at various berzosertib concentrations (0.25, 0.5, and 1μM). In PEO1 cells, the robust response to SG (60% γH2AX+ve) was not further enhanced by increasing concentrations of berzosertib (Figure S5C, left), aligning with earlier observations (Figure 3C, right). In PEO1-olaJR cells, SG demonstrated moderate sensitivity (8.7% vs. 4.7% γH2AX+ve cells in untreated) but did not significantly enhance the linear response to ATRi (Figure S5C, right).
We then examined the effects of SG on RF dynamics. DNA fiber analysis showed significantly lower IdU/CldU ratios with SG pretreatment or berzosertib alone than in control in both cell lines, indicating stalled RFs (Figure 3D). In PEO1 cells, adding SG to berzosertib further lowered this ratio compared to either SG or ATRi alone (Figure 3D). However, in PEO1-olaJR cells, the IdU/CldU ratio for the combination was not lower than SG or ATRi alone, suggesting no advantages over individual treatments with RF stalling. These differential effects led us to speculate that in the two TROP2-positive cell lines, underlying PARPi resistance mechanisms and key DNA repair molecules may have contributed to different outcomes on RF dynamics. To better understand the varying impacts of SG and berzosertib in PARPi-sensitive and PARPi-resistant settings, we studied the expression levels of proteins governing DNA damage repair and HR known to be modulated by SG treatment.43
Effect of SG and ATRi on regulators of DNA damage response pathways
In response to DNA DSBs, ATM, a sensor of DSB, is activated and induces phosphorylation and activation of DNA damage response proteins including γH2AX.44 As such, SG pretreatment and/or berzosertib increased levels of activated ATM (pATM-S1981, 3.5- to 6.6-fold) as well as phosphorylated KAP1 (pKAP1-S824, 1.3- to 9.0-fold), a specific substrate of ATM, in both cell lines (Figure 4A). We also found that SG increased levels of γH2AX in PEO1 (6.1-fold), and less evidently in PEO1-olaJR (1.3-fold) (Figure 4A) while decreasing levels of RAD51 in PEO1. In both cell lines, ATRi with or without SG increased γH2AX levels and reduced RAD51. Of note, we observed a moderate increase in the levels of ATR substrate pCHK1-S345 and ATM substrate pCHK2-T68 (Figure 4B) upon ATRi treatment with or without SG, possibly due to compensatory upregulation by ATM.
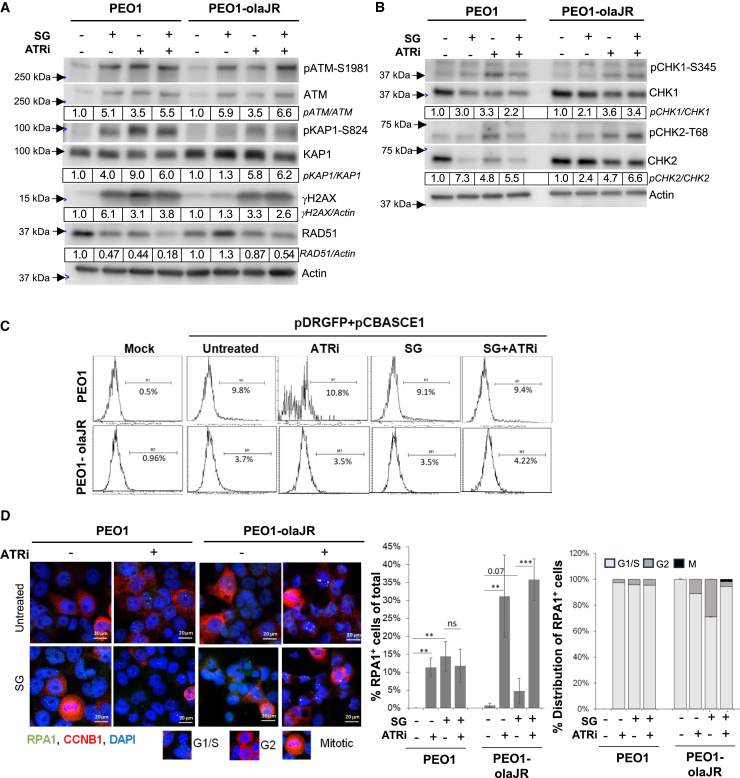
Figure 4.
Impact of SG in DNA damage repair pathways
Cells were left untreated or pretreated with SG (10 μg/mL for 30 min/37°C, washed thrice with PBS) prior to treatment with ATRi berzosertib for all experiments.
(A and B) Western blot analysis for DNA damage markers of lysates from cells pretreated with SG and treated with ATRi (1 μM) overnight. Densitometric quantitation was performed as detailed in the STAR Methods. Densitometric values of phosphorylated proteins relative to total proteins are further normalized to untreated (value of 1) for each cell line and shown below respective images. Values were rounded to two significant numbers. Representative of 3 biologically independent experiments.
(C) Graphs show results from homologous recombination (HR) repair activity assay. After cells were pretreated with SG, they were cultured overnight in plain media and then transfected with HR reporter plasmids overnight, followed by treatment with ATRi or in plain media for 48 h prior to flow cytometric analysis for GFP expression. Mock transfection without plasmids were also performed as a control for transfection efficiency. Flow data was analyzed on Flowjo and plotted as histograms. Percent GFP-positive (GFP+ve) cells for histograms are shown under each range gate. This is representative of two experimental replicates.
(D) Representative images of cells pretreated with SG, washed and incubated overnight with ATRi (1 μM) prior to co-staining for targets RPA1 (green foci) and Cyclin B1 (CCNB1, red). For reference, images of cell cycle phases as inferred from CCNB1 staining is given below image panels. The graph (middle panel) shows the mean ratio ±SD of RPA1+ve cells (>5 foci) to total cells (n = 100–300) from all sampled fields (n = 3–5 biological replicates) for each treatment. The stacked plot on the right show percent distribution of RPA1+ve cells across cell cycle phases as defined by staining pattern of nuclear CCNB1 from the sampled fields (n = 3–5 biological replicates per treatment). Representative of n = 2 biologically independent experiments. ∗∗p < 0.01, and ∗∗∗p < 0.001, ns, not significant. Scale bar is 20 μm.
Because of low RAD51 levels, we conducted HR activity assay to determine HR repair ability and no difference in HR activity was found across all treatments when compared to untreated cells (Figure 4C). We therefore hypothesized that activity of SG or ATRi was unlikely to be mediated via downregulation of HR repair but more likely through RF destabilization. Also, we reasoned that DNA damage induced by SG or ATRi during the G1/S phase, would likely be repaired before entering G2, making cells less dependent upon HR repair.
To test this idea, we examined the dynamics of RPA145 within cell cycle progression because formation of RPA1 foci (marker of ssDNA gaps) predominantly occurs during S phase and resolves prior to G2 entry.45,46 Confocal microscopic analysis of cells co-stained with fluorescent-tagged antibodies against RPA1 and cell cycle marker cyclin B1 (CCNB1)47 demonstrated an accumulation of RPA1 foci predominantly in G1/S phases in PEO1 cells when treated with SG or ATRi or the combination (Figure 4D). Conversely, in PEO1-olaJR cells, SG alone caused a marked increase in number of RPA1+ve cells in G2, which was abrogated in the presence of ATRi (Figure 4D). We hypothesized that this difference between PEO1 and PEO1-olaJR cells might be associated with increased transitioning of the later cells with RPA1 foci into G2, while resolution of ssDNA gaps takes place at a later stage prior to mitotic entry. Thus, we investigated the error-prone translesion DNA synthesis (TLS) pathway as TLS repair might be sufficient to handle SG-induced ssDNA gaps without requiring further increases in its activity in PEO1-olaJR cells.
SG may overwhelm functional TLS repair to resensitize HGSOC to PARPi
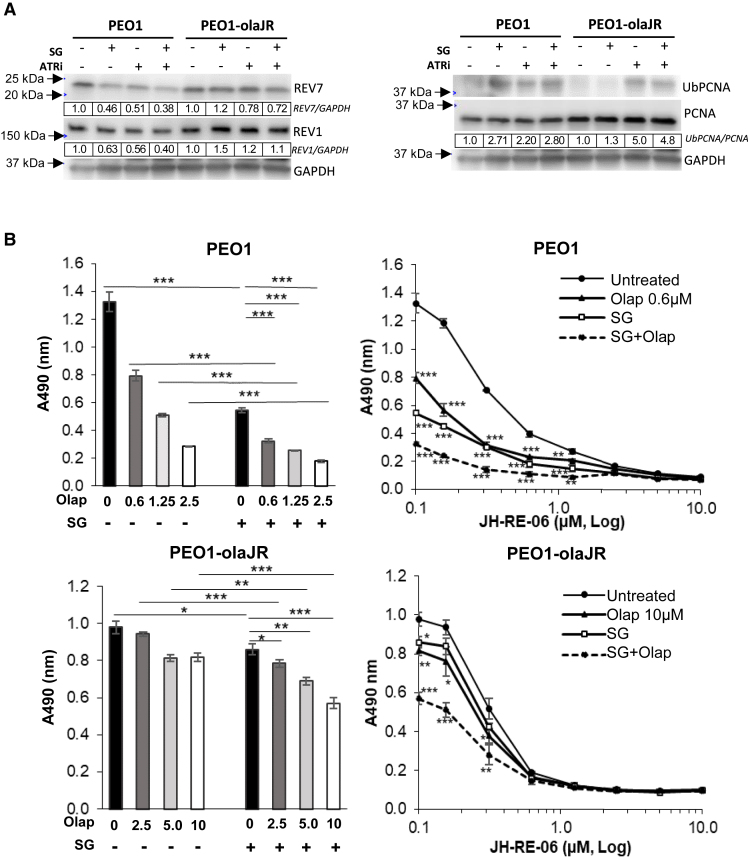
TLS polymerases help cells bypass ssDNA gaps in concert with mono-ubiquitinated proliferating cell nuclear antigen (UbPCNA) to sustain continued DNA replication while these lesions are repaired by other TLS polymerases prior to mitotic entry.48,49 UbPCNA levels were measured to determine TLS activity.49,50 We noted that SG induced elevated levels of UbPCNA in PEO1 but not in PEO1-olaJR cells (Figure 5A). However, in both cell lines, ATRi increased UbPCNA levels independent of SG (Figure 5A). Densitometric analysis of expression levels of REV7 and REV1, components of TLS polymeraseζ,49 demonstrate a 40–60% decrease in their levels relative to untreated cells in PEO1 cells when treated with SG or ATRi alone or in combination (Figure 5A).
Figure 5.
Translesion DNA synthesis (TLS) pathway inhibition improves SG and olaparib activity in HGSOC
(A) Western analysis of protein lysates from cells with or without SG pretreatment (10 μg/mL for 30 min/37°C, washed thrice with PBS) followed by overnight incubation with ATRi (1 μM) were examined for markers of TLS activity, REV1, REV7 (components of TLS polymeraseζ) and its marker of activity ubiquitinylated PCNA (UbPCNA). As described in STAR Methods, ratio of total chemiluminescence signal (ImageStudio) from each UbPCNA band to that of total PCNA was then further normalized to untreated (value of 1) for each cell line, and values shown below rounded to two significant numbers. Images are representative of 3 biologically independent experiments.
(B) The graphs on the left show mean cell growth ±SD (n = 3 biological replicates per treatment) from XTT assays on the effect of SG on sensitivity to a gradient of olaparib. On the right, the graphs show mean ± SD (n = 3 biological replicates) cell growth with or without SG pretreatment followed by olaparib in the presence of a gradient of specific TLS inhibitor JH-RE-06 over 5 days. During the assay, cells were retreated with SG in fresh media on day 3, washed twice with PBS, prior to reincubation with olaparib and JH-RE-06 for another 2 days. The Y axis shows actual background-corrected absorbance at 490nm. Untreated cell values (0 μM of JH-RE-06) are plotted at 0.1 μM to enable representation on the log X axis. Statistical significance was determined using Student’s t-test. The p-values for olaparib or SG were determined against untreated while those for the combination (SG + olaparib) (dotted line) were determined against olaparib monotherapy. ∗∗p < 0.01, and ∗∗∗p < 0.001, ns, not significant.
To determine whether TLS is a dominant mechanism for repairing DNA damage induced by SG or PARPi, we performed XTT assays over 5 days using JH-RE-06,51 a TLS inhibitor with or without SG and PARPi olaparib. We observed that both PEO1 and PEO1-olaJR showed similar sensitivity to JH-RE-06 (IC50 = 0.5–0.6 μM) in the absence of SG or PARPi (Figure S6A). SG conferred increased sensitivity to olaparib in PEO1 and partly reversed resistance to olaparib in PEO1-olaJR (Figure 5B, left). TLS inhibition further increased growth inhibition by either SG and/or olaparib in a concentration-dependent manner in both cell lines (Figure 5B, right). This suggests that TLS pathway could be a potential therapeutic target to enhance SG’s ability in sensitizing PARPi-resistant cells to olaparib.
Effect of SG and ATRi on DNA damage and survival of TROP2-low or negative HGSOC cells
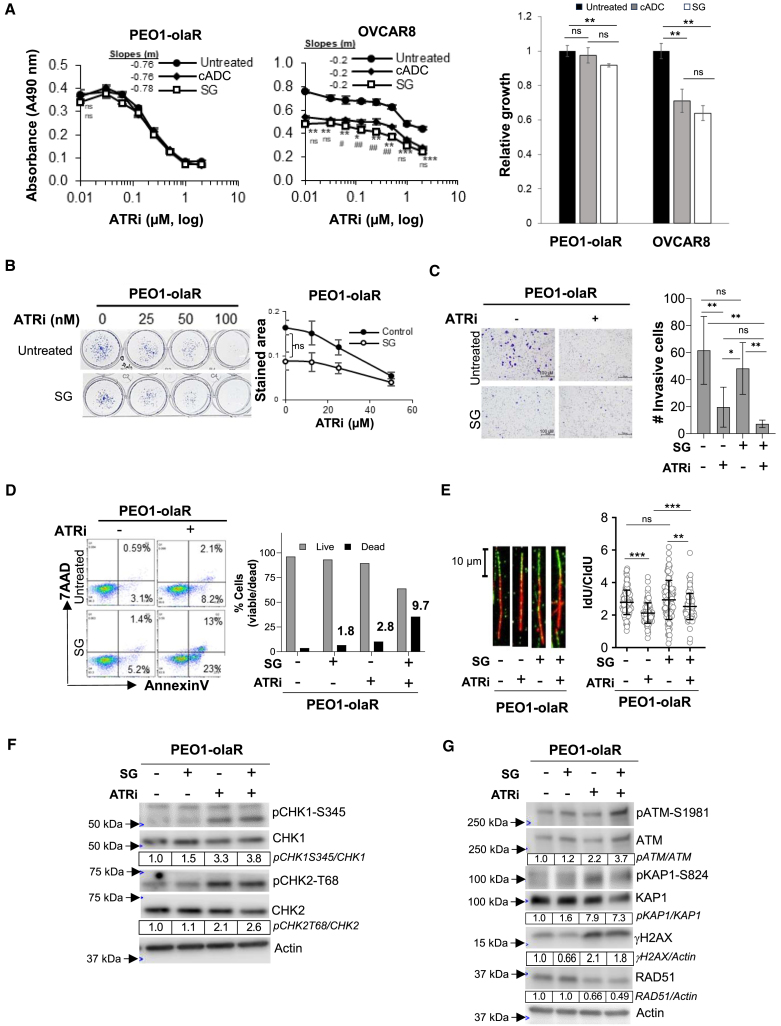
Lastly, to determine whether the changes on cell cycle and DNA damage response pathways induced by SG and berzosertib are limited to TROP2-high cell lines, we tested the TROP2-low HR proficient, BRCA2mut PEO1-olaR,30 and the TROP2-negative BRCA1 hypermethylated but HR proficient OVCAR8.35 XTT assays revealed that pretreatment with SG or cADC alone did not induce cytotoxicity in PEO1-olaR cells, but both SG and cADC showed similar cytotoxicity in OVCAR8 suggesting bystander effects (Figure 6A). In PEO1-olaR cells, while those treated with SG showed no significant difference in clonogenic growth or invasiveness (Figures 6B and 6C), there was a modest increase (1.8-fold) in cell death, which was further enhanced by 10-fold upon addition of ATRi compared to untreated group (Figure 6D).
Figure 6.
SG activity in TROP2-negative OVCAR8 and TROP2-low PEO1-olaR
Cells were left untreated or pretreated with SG or cADC (10 μg/mL, 30 min at 37°C and washed thrice with PBS) prior to use in all experiments below. Student’s t test was used to measure significance in all experiments below.
(A) Graphs show mean ± SD (n = 3 biological replicates per treatment) cell growth inhibition from XTT assays for TROP2-negative OVCAR8 and TROP2-low PEO1-olaR cells that were untreated or pretreated with either SG or cADC followed by treatment with a gradient of ATRi (0–10 μM) over 48 h. Untreated cell values (0 μM ATRi) are plotted at 0.01 μM to enable representation on the log X axis. Statistical significance (p-value) was determined by Student’s t test. p-values between untreated and SG plots are indicated by asterisks (∗) while between SG and cADC plots are indicated by hashes (#). Slopes for each curve are derived from linear regression formula “y = mx+b” where y = Y-intercept, x = ATRi (μM) and b = correlation coefficient, calculated from the linear part of the respective curves.
(B) Clonogenic assays were performed over 7 days with untreated or SG pretreated cells against increasing concentrations of berzosertib (ATRi). Representative images are shown on the left while graphs on the right show mean ± SD from 3 biological replicates.
(C) Representative images from one set of Matrigel plates after crystal-violet staining are shown. Following appropriate treatments, invaded cells on underside of inserts were quantified after crystal-violet staining using ImageJ (Fiji) software as detailed in STAR Methods. The plots show mean ± SD for 3 biological replicates. Statistical significance was determined by Student’s t test. Scale bar is 100 μm.
(D) Flow cytometric analysis of viability (AnnexinV/7-AAD) for PEO1-olaR treated with SG or ATRi monotherapy or a combination of both. Dead and live cell proportions are plotted on the right as bar charts from >8000 events. Numbers above bars indicate fold increase in dead cells for each treatment relative to untreated. Results are representative of 3 biologically independent experiments.
(E) DNA fiber assays was performed as explained in STAR Methods. Lengths of both red (IdU) and green (CldU) segments of each strand (∼200 DNA strands per treatment) were measured using Fiji software, and the mean ± SD of the ratio of IdU/CldU from all strands (n = ∼200) were plotted as column charts using GraphPad prism. Representative strands are shown to the left from 2 biologically independent experiments. Statistical significance was determined by Student’s t test. Scale bar is 10 μm.
(F and G) Western blot analysis of lysates from cells pretreated with SG alone or followed by overnight treatment with ATRi berzosertib (1 μM) and analyzed for DNA damage and cell cycle markers. Densitometric quantitation was performed as detailed in the STAR Methods. Densitometric values of phosphorylated proteins relative to total proteins are further normalized to untreated (value of 1) for each cell line and shown below respective images. Values were rounded to two significant numbers. Representative of 3 biologically independent experiments. For panels A, B, C and E, p-values are ∗, #p < 0.05, ∗∗, ##p < 0.01, and ∗∗∗, ###p < 0.001, ns, not significant.
With respect to RPA1 foci and γH2AX formation, SG pretreatment alone or in combination with berzosertib slightly increased the proportion of cells with RPA1 foci in PEO1-olaR (5% and 15%) though it was substantially lower compared to OVCAR8 (17% and 41%) (Figures S5A and S5B). Also, the proportion of γH2AX+ve cells induced by the combination (15%) in PEO1-olaR was not significantly different from SG alone (11%) while in OVCAR8 the combination was higher (12%) than SG alone (1.2%) (p < 0.001) (Figures S5A and S5B). Also, we tested the DNA damage response to varying concentrations of ATRi. In PEO1-olaR cells, the increase of γH2AX+ve cells was berzosertib dose-dependent while SG did not significantly enhance this effect in both PEO1-olaR and PEO1-olaJR cells (Figure S5C). In TROP2-low PEO1-olaR cells, confocal analysis for RPA1 and cell cycle marker CCNB1 showed increased accumulation of RPA1+ve cells in G2 (Figure S5D) similar to TROP2-high PEO1-olaJR cells (Figure 4D) suggesting the role of TROP2 specificity in this effect.
For RF dynamics and DNA damage response endpoints, SG pretreatment did not affect RF progression and its combination with ATRi did not further enhance stalled RFs induced by ATRi in PEO1-olaR (Figure 6E). Immunoblotting results also exhibited similar levels of pCHK1-S345 and pCHK2-T68 in PEO1-olaR treated with SG and ATRi (Figure 6F). Although activation of γH2AX and ATM activity was found in PEO1-olaR, it was at lower intensity than that seen in PEO1 and PEO1-olaJR, suggesting a possible bystander effect of SG attributed to free SN-38 (Figure 6G). Additionally, PEO1-olaR exhibited modest sensitivity to the TLS inhibitor JH-RE-06 (IC50 = 2.2 μM) (Figure S6A) and limited changes in UbPCNA levels relative to total PCNA, when compared with PEO1 or PEO1-olaJR although UbPCNA levels increased upon ATRi treatment in PEO1-olaR (Figure S6B), like PEO1-olaJR (Figure 5A). We noted sensitization to olaparib by JH-RE-06 alone was moderate and only significant at higher concentrations of olaparib (>5 μM) (Figure S6C). In combination with SG, this toxicity was enhanced (p < 0.01) in a JH-RE-06 concentration-dependent manner. Over the 5-day assay, SG alone showed a modest inhibition of cell growth in PEO1-olaR (p < 0.001) (Figures S6C and S6D).
SG decreases tumor growth in both TROP2-high and TROP2-low HGSOC animal models
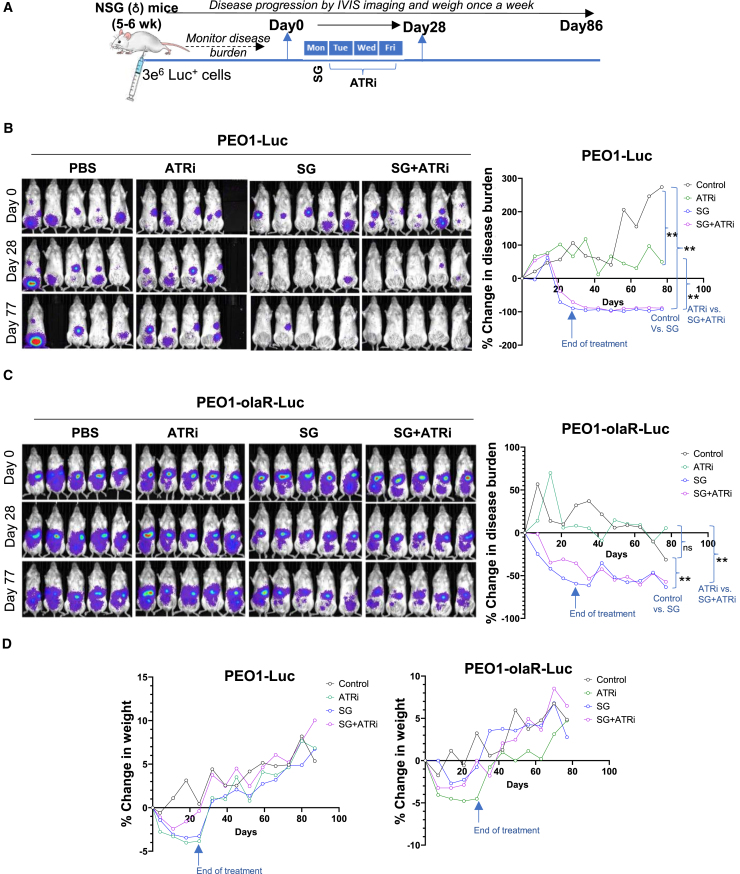
To evaluate in vivo activity of the combination treatment, we used metastatic murine models of PEO1 and its olapR variants that stably express luciferase. These luciferase clones were confirmed to have similar TROP2 protein and mRNA expression profiles as their respective parental cell lines (Figures 1C, 1E, and S7A). In mice injected with TROP2-high PEO1 cells, treatment with SG alone led to a sustained complete resolution of disease as early as 4 weeks into drug treatment, with no visible reversal after 2 months following drug treatments (p < 0.01), while no advantage was observed with the addition of berzosertib over SG alone (Figures 7A and 7B).
Figure 7.
Evaluation of berzosertib efficacy in combination with SG in murine models of PARPi-resistant HGSOC
(A) Schematic of treatment strategy for mice treated with SG or berzosertib (ATRi) or a combination of both over a period of 4 weeks as described in STAR Methods. Each treatment group represents 10 mice (biological replicates) on day 0. Groups were eliminated if mice number dropped below three due to death or when euthanasia criteria was reached.
(B and C) Representative images of mice injected with PEO1-Luc (B) or PEO1-olaR-luc (C), imaged for luciferase activity are shown from day 0 to day 77. Line plots to the right represent % change in disease burden against time on X axis (days 0–77) measured at weekly intervals (n = 8–10 mice per group). Statistical significance was determined using Mann-Whitney U-test (unpaired) to generate p-values between relevant groups and shown on the right of the plots. Error bars are not shown for better visualization of curves. ∗∗p < 0.01, ns, not significant.
(D) Line plots showing % change in mice weight from day 0 recorded weekly for PEO1-Luc and for PEO1-olaR-Luc (n = 8–10 mice per group).
Significantly decreased tumor burden was also observed in the TROP2-low PEO1-olaR xenograft model, though no difference was noted between SG monotherapy and its combination with berzosertib (Figure 7C). In TROP2-high PEO1-olaJR models, SG showed a trend toward lower disease burden, but it did not differ from the combination (Figure S7C). Nevertheless, the line charts of individual mice showed that SG tended to reduce disease burden as determined based on the RECIST 1.1 criteria52 (<30% tumor burden relative to Day 0) without showing an advantage in combination with berzosertib (Figure S7C). No significant adverse effects were observed, and mouse weights returned to baseline within two weeks of treatments (Figures 7D and S7C). Mice survival remained largely unaffected throughout the experiments, with only a few mice being euthanized because of low weight not related to the treatments or disease.
Discussion
The recent success of ADCs in the treatment of hematological malignancies and some solid tumors, i.e., breast cancer, stems from their specific tumor-targeted drug delivery and reduced toxicity compared to conventional cytotoxic drugs.4 Here, we report SG’s monotherapy activity in HGSOC preclinical models. While SG induced DNA damage and replication stress primarily in TROP2-positive cell lines, SG alone caused substantial tumor shrinkage in both TROP2-high and TROP2-low xenograft models, suggesting significant contributions from both TROP2-specific and bystander effects of SG. Our findings align with previous reports.19,24,53 Perrone et al. demonstrated SG’s bystander effects in both TROP2-positive KRCH31 OC cells and TROP2-negative ARK4 cells in co-cultures,19 and Cardillo et al. reported significant cytotoxic activity in breast cancer cell lines where the levels of TROP2 expression were heterogeneous.22 Hence, our data represent an important therapeutic opportunity of SG in HGSOC independent of TROP2 expression.
While ATRi and TOP1i combination has shown synergistic or additive effects in many preclinical cancer models,27,31 combination of SG and berzosertib showed limited effects in our HGSOC in vitro and mouse models. Indeed, we identified mutually exclusive actions of SG and ATRi affecting different parameters of cell survival, particularly cell cycle regulation. Berzosertib’s counter-regulatory effect of transitioning SG-induced G2/M accumulated cells back to G1/S phases is consistent with a previous report in non-small cell lung cancer where berzosertib shifts cells in irradiation-induced G2 arrest into G1 phase.40 However, it is possible that the lack of synergism between SG and ATRi in vitro, may be in part due to a brief drug exposure and washout strategies for SG, allowing cells more time to recover from DNA damages. Nonetheless, our data require further investigation as it may have direct clinical implications. For instance, although the recent phase 1 study of SG and berzosertib demonstrated the safety and limited activity in patients with solid tumors,28 the individual contributions or the mechanistic basis of this collective activity of two drugs need further studies in PARPi-resistant HGSOC. In another clinical study, concurrent dosing of SG and talazoparib (TLZ) was not tolerated clinically due to a narrow therapeutic window while sequential dosing of SG and TLZ was successful.54 This might be the way forward for SG plus ATRi combination therapies.
In this study, we identified mechanisms of action of SG, particularly on RF dynamics and DNA damage response pathways. While ATRi is expected (or known) to stall RFs across all cells line,31 SG-induced RF stall was specific to SN-38 sensitive TROP2-high cells. Also, besides TROP2 expression, our data suggest that other factors like HR deficiency status, may influence SG specificity. In our PARPi-resistant HGSOC cells, significant delays in the transition of ssDNA gaps to DSBs were found when treated with SG relative to PARPi-sensitive PEO1 cells. This finding may indicate that either ssDNA gaps are repaired in a timely manner before progressing into DSBs, or they are bypassed for repair at a later stage, requiring further investigation.
SG’s distinct mechanisms of action in PARPi-resistant cells involve complex interactions between ssDNA repair, RF progression and HR response. It has been shown that error-prone repair of DNA damage induces chemoresistance in B-cell lymphoma and PARPi resistance in small cell lung cancer.55,56 As such, in PARPi-resistant HSGOC cells, TLS may be a responding pathway as shown in our confocal data on the G2 resident RPA1+ve cells.50 TLS pathway helps sustain DNA replication while resolving ssDNA gaps, contributing to PARPi resistance.56 We therefore reasoned that the ubiquitous levels of TLS machinery might be sufficient to resolve SG-induced ssDNA gaps since TLS activity measured by UbPCNA levels was not altered upon SG treatment in PARPi-resistant cells. Notably, ATRi induced a substantial increase in ssDNA gaps in both PARPi-sensitive and -resistant cells independent of SG, with concomitant increases in UbPCNA levels. Hence, it is possible that inhibiting TLS activity may sensitize HGSOC cells to SG and/or PARPi. Further investigation into the TLS pathway as a potential therapeutic target or biomarker for SG-based combination therapy in PARPi-resistant HGSOC is warranted.
In conclusion, our studies demonstrate mechanisms of action of SG in HGSOC preclinical models and helps delineate parameters that influence the TROP2 heterogeneous activity of SG. TROP2 specific effect of SG has a dominant negative effect on invasiveness, RF stalling and proliferation in both PARPi-sensitive and PARPi-resistant HGSOC cells. The TROP2 non-specific activities of SG, like G2/M delays or TLS mediated ssDNA resolution might have more to do with its bystander effect, payload sensitivity and/or prevalent PARPi resistance pathways than any TROP2-specific activity. Moreover, although the lack of synergistic effect between SG and berzosertib was unanticipated, it is important to note that SG alone can induce antitumor activity in HGSOC mouse models even with very low TROP2 expression. Our findings thus offer an important clinical implication of SG monotherapy in HGSOC by avoiding unnecessary side effects from combination therapies.
Limitations of the study
The number of cell lines and mouse models are limited to three. Mechanistic studies were done primarily in BRCA2mut HGSOC which may not fully reflect the complexity of PARPi-resistant HGSOC. In murine models, we limited our studies to one dosage of SG that was optimized for its moderate effect on tumor regression. Also, the spontaneous hydrolysis of SG into free SN-38 may have introduced unknown toxicities. Mechanistic events occurring in murine tumors in response to SG were not explored in this study and exploration of other error-prone DNA repair pathways besides TLS is needed. These limitations are currently under investigation and will be addressed in future studies.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead contact, Jayakumar R. Nair (nairjr@nih.gov).
Materials availability
SG (hRS7-CL2A-SN-38) and the nontargeting control ADC (cADC, h679-CL2A-SN-38) were obtained from Gilead Sciences, Inc through a Materials Transfer Agreement between NCI and Gilead Sciences.
Data and code availability
-
•
Processed and raw RNA-seq data have been deposited at Gene Expression Omnibus (GEO) collection (https://www.ncbi.nlm.nih.gov/geo/) as GSE278664 and are publicly available as of the date of the publication.
-
•
This paper does not report original code.
-
•
Original uncropped western blots are provided as Data S1. Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from lead contact upon request.
Acknowledgments
This research was supported by the Intramural Research Program of NCI, CCR (JML, #ZIA BC011525). We thank Gilead for providing S.G. and its control nontargeting ADC for our experiments through the material transfer agreement between Gilead and NCI, and for valuable research input. The authors thank Yolanda L. Jones, NIH Library Editing Services, for editing assistance. Graphical abstract was created in BioRender. Nair, J. (2024) BioRender.com/n12r489.
Author contributions
Conceptualization, J.M.L., T.M.C., S.H., and J.R.N.; Methodology, J.R.N., J.M.L., and A.S.; Investigation, J.R.N.; Formal analysis, J.R.N., T.T.H., C.Y.C., K.C.C., and K.W.; Writing–original draft, J.R.N.; Writing–Review and Editing, J.R.N., T.T.H., J.M.L., T.M.C., M.R.P., K.R.I., and S.H.; Funding acquisition, J.M.L.; Resources, J.M.L., T.M.C., and S.H. Supervision, J.M.L.
Declaration of interests
T.M.C. is a paid contract employee of Gilead Sciences, Inc. J.M.L. has research grant funding from AstraZeneca and Acrivon Therapeutics (paid to institution) and is on the Scientific Advisory Board of Acrivon Therapeutics and Genentech (unpaid). All other authors declare no conflict of interest.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Recombinant anti-TROP2 humanized RS7 antibody | Gilead pharma | Cat #GS-1170562 |
| Goat anti-Human IgG F(ab')2 Secondary Antibody | ThermoFisher | Cat # 31628; RRID: AB_429704 |
| Annexin V-FITC | BioLegend | Cat #640906 |
| Mouse anti-BrdU-FITC conjugate | BioLegend | Cat #364104; RRID: AB_2564481 |
| Mouse anti-H2AX (S139), AF647 conjugate | BioLegend | Cat #613408; RRID: AB_2295046 |
| Goat anti-mouse IgG, AF488 conjugate. | ThermoFisher | Cat # A-11001; RRID: AB_2534069 |
| Donkey anti-Rabbit IgG, AF555 conjugate. | ThermoFisher | Cat #A-21428; RRID: AB_2535849 |
| Goat anti-rat IgG(H+L), AF488 conjugate | ThermoFisher | Cat # A-11006; RRID: AB_2534074 |
| Goat anti-mouse IgG (H+L), AF594 conjugate | ThermoFisher | Cat # A-11005; RRID: AB_2534073 |
| Rabbit anti-TROP2 | Cell Signaling Tech. | Cat #47866; RRID: AB_2938529 |
| Rabbit anti- pCHK1-S296 | Cell Signaling Tech. | Cat #2349; RRID: AB_2080323 |
| Rabbit anti-pCHK1-S345 | Cell Signaling Tech. | Cat #2348; RRID: AB_331212 |
| Mouse anti-CHK1 | Cell Signaling Tech. | Cat #2360; RRID: AB_2080320 |
| Mouse anti-GAPDH | Cell Signaling Tech. | Cat #97166; RRID: AB_2756824 |
| Rabbit anti-βActin | Cell Signaling Tech. | Cat #8457; RRID: AB_10950489 |
| Mouse anti-βActin | Cell Signaling Tech. | Cat #3700; RRID: AB_2242334 |
| Rabbit anti-pCHK2-T68 | Cell Signaling Tech. | Cat #2661; RRID: AB_331479 |
| Rabbit anti-CHK2 | Cell Signaling Tech. | Cat #6334; RRID: AB_11178526 |
| Rabbit anti-γH2AX (S139) | Cell Signaling Tech. | Cat #9718; RRID: AB_2118009 |
| Rabbit anti-pKAP1-S824 | Cell Signaling Tech. | Cat #4127; RRID: AB_2209906 |
| Mouse anti-KAP1 | Cell Signaling Tech. | Cat # 5868; RRID: AB_10707324 |
| Rabbit anti-CyclinB1 | Cell Signaling Tech. | Cat #4138; RRID: AB_2072132 |
| Rabbit anti-ATM | Cell Signaling Tech. | Cat #2873; RRID: AB_2062659 |
| Rabbit anti-Ubiquityl PCNA (L164) | Cell Signaling Tech. | Cat #13439; RRID: AB_2798219 |
| Mouse anti-PCNA | Cell Signaling Tech. | Cat #2586; RRID: AB_2160343 |
| Rabbit anti-RAD51 | Cell Signaling Tech. | Cat #8875; RRID: AB_2721109 |
| Anti-rabbit IgG, HRP-linked | Cell Signaling Tech. | Cat #7074P2; RRID: AB_2099233 |
| Anti-mouse IgG, HRP-linked | Cell signaling Tech. | Cat #7076P2; RRID: AB_330924 |
| Rabbit anti-RPA70, AF488 conjugate | Abcam | Cat #Ab199097 |
| Rat anti-CldU | Abcam | Cat #ab6326; RRID: AB_305426 |
| Rabbit anti-REV1 | ProteinTech. | Cat #17703-1-AP; RRID: AB_3085535 |
| Rabbit anti- MAD2L2/REV7 | ProteinTech | Cat #12683-1-AP; RRID: AB_2139530 |
| Mouse anti-IdU | Novus biological | Cat #NBP2-44056; RRID: AB_3307851 |
| Biological samples | ||
| Patient biopsies | Clinical trial (NCI). | NCT02203513 |
| Chemicals, peptides, and recombinant proteins | ||
| 7-AAD Viability Staining Solution | BioLegend | Cat #420404 |
| Crystal-violet | Sigma-Aldrich | Cat #C0775 |
| 5-Iodo-2'-deoxyuridine (IdU) | Sigma-Aldrich | Cat #I7125 |
| 5-Chloro-2'-deoxyuridine (CldU) | Sigma-Aldrich | Cat #C6891 |
| DAPI nuclear stain | Sigma-Aldrich | Cat #D9542 |
| Propidium iodide | ThermoFisher | Cat #P3566 |
| Prolong glass antifade mountant | ThermoFisher | Cat #P36980 |
| XTT (2,3-Bis-(2-Methoxy-4-Nitro-5-Sulfophenyl)-2H-Tetrazolium-5-Carboxanilide) | ThermoFisher | Cat #X6493 |
| RPMI 1640 medium with glutamine | ThermoFisher | Cat #11835055 |
| Restore™ PLUS Western Blot Stripping Buffer | ThermoFisher | Cat #46430 |
| Bromodeoxyuridine (BrdU) | Fisher Scientific | Cat #BDB550891 |
| Berzosertib | Medkoo biosciences | Cat #406258 |
| Sacituzumab govitecan (SG) | Gilead | N/A |
| Control ADC (h679-CL2A-SN-38) | Gilead | N/A |
| D-α-Tocopherol polyethylene glycol 1000 succinate (TPGS) | Sigma-Aldrich | Cat #57668 |
| JH-RE-06, TLS inhibitor | Selleckchem | Cat # S8850 |
| Olaparib | Medchem express | Cat # HY-10162 |
| Fetal Bovine serum | BioWest LLC | Cat # S1480 |
| Critical commercial assays | ||
| Ssoadvanced SYBR® Green supermix | BioRad | Cat #1725270 |
| PrimePCR™ SYBR® Green Assay: GAPDH | BioRad | Cat #10025636 |
| iScript™ Advanced cDNA Synthesis Kit | BioRad | Cat #1725037 |
| Deposited data | ||
| RNAseq datasets | Gene expression omnibus (GEO) | GSE278664 |
| Experimental models: Cell lines | ||
| PEO1 | Sigma-Aldrich | #10032308 |
| PEO1-olaJR | Lab stored | Huang et al.29 |
| PEO1-olaR | Lab stored | Huang et al.29 |
| PEO4 | Sigma-Aldrich | #10032309 |
| OVCAR8 | NCI-60 collection | N/A |
| PEO1-Luc | Lab stored | This study |
| PEO1-olaR-Luc | Benjamin Bitler lab | Yamamoto et al.30 |
| PEO1-olaJR-Luc | Lab stored | This study |
| OVCAR3 | American Type Culture Collection (ATCC) | #HTB-161 |
| OVCAR5 | NCI-60 collection | N/A |
| UWB1.289 | American Type Culture Collection (ATCC) | #CRL-2945 |
| TOV21G | American Type Culture Collection (ATCC) | #CRL-3577 |
| SKOV3 | NCI-60 collection | N/A |
| A2780 | Christina Annunziata Lab | Hernandez et al.57 |
| Experimental models: Mice | ||
| NSG mouse: NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ | NCI inhouse | N/A |
| Recombinant DNA | ||
| pDRGFP, HR assay | Addgene | Cat #26475 |
| pCBASce1, HR assay | Addgene | Cat #26477 |
| Software and algorithms | ||
| GraphPad Prism V. 10.1.1 | Graphpad software LLC | https://www.graphpad.com/ |
| Image Studio | LI-COR instruments | https://www.licor.com |
| Zen Blue | Carl-Zeiss microscopy | https://www.zeiss.com/ |
| Image-J | National Cancer Institute. | https://imagej.net/ij/ |
| Aura image analysis software | Spectral Instruments Imaging LLC. | https://spectralinvivo.com/aura-imaging-software/ |
| FlowJo V. 10.7.2 | BD biosciences | https://www.flowjo.com/ |
Experimental model and study participant details
Cell lines
PEO1 (5193C>G [Y1655X] BRCA2mut HGSOC) was purchased from Sigma-Aldrich. Acquired PARPi olaparib-resistant cell line PEO1-olaJR was developed from PEO1 in-house as explained earlier,29 while PEO1-olaR, another PARPi olaparib-resistant HGSOC cell line was obtained from Dr. Bitler’s group.29,30 The OVCAR8 (BRCA1 promoter methylation), SKOV3 and OVCAR5 were obtained from the NCI-60 collection at the National Cancer Institute (NCI), National Institutes of Health (NIH), Frederick, MD. The cell lines OVCAR3, UWB1.289 (BRCA1-null) and TOV21G were obtained from American Type Culture Collection (ATCC). All cell lines were cultured in RPMI1640 media with L-glutamine (Life Technologies) containing 10% FBS, 1000 U/ml penicillin/streptomycin, 1 mM sodium pyruvate, and 5 μg/ml of insulin from bovine pancreas (Sigma-Aldrich).
Murine models
PEO1, PEO1-olaJR and PEO1-olaR cells were transfected with pGL4.51-Luc2/CMV/Ne plasmid (Promega) using Dharmafect Kb® reagent (#T-2006-01, Horizon discovery, UK) and clonal populations that stably express luciferase for over 3 months were expanded for use in murine studies. Heavily immunocompromised mice (5–6 weeks, female) (NSG) were injected intraperitoneally (IP) with 3 × 106 cells of luciferase expressing PEO1, PEO1-olaR and PEO1-olaJR. Disease burden was monitored weekly with an IVIS imager (Perkin Elmer) and analyzed using Aura® software (Spectral Instruments imaging LLC). Once an appreciable disease burden that shows a linear increase over 2 weeks was observed, mice were grouped randomly and subjected to appropriate treatments. The control group was treated with sterile 0.9% saline. SG (DAR values of ∼7) was prepared in sterile 0.9% saline as 2.5 mg/ml stocks and injected at 250 μg per mouse IP on Mondays. Berzosertib for murine work was obtained from the NCI drug repository and dissolved in 10% D-α-Tocopherol polyethylene glycol 1000 succinate (TPGS, Sigma-Aldrich), sterile filtered and administered via oral gavage (25 mg/kg) in mice on Tuesdays, Wednesdays, and Fridays for 4 weeks. Drug dosages were optimized in murine models of PEO1-luc over a range of berzosertib and SG concentrations prior to selecting sublethal dosages of both for combination treatments (Data not shown). Disease burden and mice weights were measured weekly. After 4 weeks of drug treatment, both disease burden and mice weights were recorded for a further 6–8 weeks. Mice that did not meet health guidelines outlined in the animal protocols were euthanized during the experiment. All cell lines were tested for microbial contamination at the Frederick national laboratory for cancer research (Leidos Inc.) prior to mice injections.
Study approval
All animal procedures reported in this study that were performed by NCI-CCR affiliated staff were approved by the NCI Animal Care and Use (ACU) committee, and in accordance with federal regulatory requirements and standards (protocol #WMB-002). All components of the intramural NIH ACU program are accredited by Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC).
Patient tumor samples
We collected fresh frozen core biopsy samples prior to treatment from adult female HGSOC patients with BRCA mutation (BRCAmut, n = 15) or BRCA wild-type (BRCAwt, n = 20) enrolled in the phase 2 clinical trial of the CHK1 inhibitor (CHK1i) (NCT02203513). The characteristics, relevant to the study, of the patients whose specimens were used in this study is listed in the table below. Tumor samples were verified by board certified pathologists at the institution.
| Characteristics of HGSOC patients and specimens | ||
|---|---|---|
| Characteristics | Number (n = 35) | (%) |
| Age, y | ||
| 40 – 60 | 15 | 43 |
| 60 – 80 | 19 | 54 |
| >80 | 1 | 3 |
| BRCA mutation status | ||
| Mutant | 15 | 43 |
| Wild-Type | 20 | 57 |
| Prior PARPi therapy | ||
| Yes | 21 | 60 |
| No | 14 | 40 |
Study approval
The Phase II clinical trial of CHK1 inhibitor prexasertib monotherapy in patients with recurrent HGSOC (NCT02203513) was reviewed and approved by the Institutional Review Board of the Center for Cancer Research, NCI (IRB study number is 14C0156). Written informed consent was obtained from all patients.
Method details
High-throughput drug screening
High-throughput drug combination screening was performed as previously reported.58,59 Briefly, we first screened two PARPi-resistant HGSOC cells (PEO1-olaR and PEO1-olaJR) treated with ATRi against 2,450 combinations using a 96-hour cell proliferation assay with an ATP-based readout CellTiter-Glo (#G7570, Promega) to determine activity and synergy of compounds in a dose-response manner. The concentration range of each drug was 0 to 5 μM for initial 6 × 6 matrix screen. For the subsequent 10 × 10 validation screen, the concentration range was 0 to 20 μM for ceralasertib, 0 to 50 nM for SN-38, gemcitabine, paclitaxel, prexasertib, exatecan, 0 to 5 μM for adavosertib, and 0-10 μM for the remaining drugs. Each matrix was scored by the sum of ExcessHSA for evidence of synergistic (ExcessHSA score < − 20), additive (− 20 ≤ ExcessHSA score ≤ 20), or antagonistic effects (ExcessHSA score > 20), and the average ExcessHSA score of each compound was ranked accordingly. CellTiter-Glo was added after 96 hours of compound incubation were utilized to inform on cell viability as described in manufacturers. Luminescence signal was measured for 6 × 6 and 10 × 10 matrix screening using a Pherastar (BMG Labtech) and a ViewLux (Perkin-Elmer) reader, respectively.
RNA-seq analysis
Total RNA was prepared from pretreatment frozen core biopsies from recurrent HGSOC patients with and without BRCA mutation and RNA-seq was performed as described.34 Briefly, each sample (20-100 ng) was preprocessed with NEBnext rDNA depletion kit (New England Biolabs, Ipswich, MA) to remove ribosomal RNA, barcoded and pooled to ensure at least 100 million reads per sample on a HiSeq3000 sequencing system (Illumina, San Diego, CA). Reads were aligned against the Human reference genome Hg38 and gene expression data was generated as counts per million mapped reads (CPM) values. Quality check of sample and sequencing outputs was performed by the Center for Cancer Research (CCR) sequencing facility and CCR collaborative bioinformatics resource at National Cancer Institute, Bethesda, MD and ensured to be within recommended parameters.For comparisons that include normal ovarian tissues, we used transcripts per million (TPM) values of the patient RNA-seq dataset against TPM values for normal ovarian tissues downloaded from the GTEx database (GTEx project, NIH; https://www.gtexportal.org/home/).60
Quantitative reverse-transcriptase PCR (qPCR)
The qPCR was performed from total RNA isolated from untreated logarithmically growing cells. Using primer sequences and PCR conditions derived from literature,61 TROP2 mRNA levels were estimated against GAPDH control primer set (BioRad) and QPCR reagents from BioRad. Ct values were normalized against GAPDH for each cell line, and the resultant relative mRNA levels for TROP2 were plotted as bar charts.
Flow cytometry
Flow cytometry for TROP2 was performed with recombinant anti-human TROP2 antibody hRS7 (gift from Gilead) (2.5 μg/ml) in cell staining buffer for 2 hours, followed by treatment with goat anti-Human IgG F(ab')2 secondary antibody for 1 hour, washed and then fixed in 2% paraformaldehyde in PBS. Control cells were treated with secondary antibody alone. DNA content measurement for cell cycle analysis was performed as described earlier.62 Briefly, cells were fixed in 70% ethanol for at least 1 h at 4°C, washed twice with PBS and resuspended in 200 μl of propidium iodide (PI) solution (25 μg/ml), and 50 μl of RNAse A solution (0.5 μg/ml). After incubation for 1 hour at room temperature, acquisition was performed on a BD FACScanto™II (BD Biosciences) and analyzed using FlowJo® software (FlowJo LLC). Flow cytometric analysis for viability was performed using Annexin V-FITC and 7AAD after suspending cells in Annexin V binding buffer as per manufacturer’s instructions (Biolegend).
Clonogenic assays
Assays to measure cellular ability to form colonies in the absence or presence of inhibitors were performed in 12-well plates in triplicate against predetermined sublethal concentrations of ATRi berzosertib optimized for each cell line. After overnight incubation in the plates (5,000 cells per well), cells were incubated with SG (10 μg/ml in normal media) for 30 minutes at 37°C. Wells were washed twice with sterile PBS prior to incubation with ATRi. On Day 3 after ATRi addition, wells were again pretreated with ADC followed by ATRi incubation for another 4 days, after which wells were washed in PBS, fixed with methanol (10 minutes), and then stained with 0.5% crystal-violet in 20% methanol (20 minutes), followed by washing in distilled water and imaged. Staining was quantified using ImageJ™ and the mean of triplicate wells plotted as a line graph.
HR reporter assay
This assay was used to measure HR repair activity as reporter earlier.35 Briefly, cells were transfected with plasmid-carrying pDRGFP (#26475, Addgene, Cambridge) and pCBASceI (#26477, Addgene) plasmid with Lipofectamine 2000 transfection reagent (#11668019, ThermoFisher Scientific). After overnight transfection, cells were subjected to drug treatment for 48 hours, cells were collected, fixed in 4% paraformaldehyde (in cell staining buffer), and examined by BD LSRFortessa (BD biosciences) to quantify the GFP-positive cells.
Cell growth assays
XTT assay was done as detailed.35 Plates were read on a BioTek SynergyHT™ plate reader (BioTek Instruments) and analyzed on Gen5™ software. Absorbance measured at 490 nm was plotted as absolute values (corrected for background) or relative to untreated control. Zero values for drug concentrations were made non-zero to enable plotting them on a logarithmic scale, as mentioned in the respective figure legends.
BrdU proliferation assays
Proliferation assays for the analysis of DNA synthesis in proliferating cells were performed in 6-well plates on cells that were plated to 50% confluency 24 hours earlier. Cells were pretreated with SG as detailed earlier and then with ATRi berzosertib (1 μM) overnight. Next day, fresh BrdU was added to a final concentration of 10 μM, incubated for 3 hours and cells harvested by trypsinization into tubes for flow cytometry. After washing with PBS, cells were treated with BD cytofix/cytoperm buffer (BD fixation/permeabilization kit, #554714, BD biosciences) for 20 minutes on ice, washed with perm/wash buffer, permeabilized with BD cytoperm permeabilization buffer plus for 10 minutes on ice, washed once more, refixed with cytofix/cytoperm buffer for 5 minutes, washed and then treated with DNase1 (300 μg/ml) (#D4513, Sigma-Aldrich) to expose incorporated BrdU. Cells were washed once with perm/wash buffer and then treated with anti-BrdU-FITC antibody (1:10) in cell staining buffer (CSB) (Biolegend) for 30 minutes at room temperature. Cells were washed in CSB and then suspended briefly in 50 μl of RNAseA (100μg/ml) (#10109169001, Roche Diagnostics) and then stained with 200 μl of DNA dye propidium iodine (PI) (25 μg/ml in CSB) for 1 hour before flow cytometric analysis on BD LSRFortessa machine (BD biosciences).
Matrigel invasion assays
Invasion assays were performed in 24 well Biocoat® Matrigel™ invasion chamber assay plates from Corning™ (Thermofisher scientific) prepared as per manufacturer’s instructions. Briefly, 1 × 105 cells with or without pretreatment with SG, were plated in RPMI media without serum with or without ATRi berzosertib (1 μM). The lower wells contained media with serum with or without ATRi and plates were incubated overnight, and subsequently processed and stained with crystal-violet stain as per manufacturer’s instructions. Invaded cells that stain violet observed on the bottom surface of the insert membrane were quantified and plotted.
Western immunoassays
All western blots were performed with appropriate antibodies mentioned above as described.63 Briefly, cell pellets were washed with PBS and lysed on ice for 20 minutes in RIPA buffer (#89901, Life Technologies) that was supplemented with protease and phosphatase inhibitors (Roche Diagnostics). Proteins were separated on a 4–15% Mini-PROTEAN® TGX™ Precast Protein Gels (Bio-Rad) using Mini PROTEAN® tetra cells as per manufacturers’ instructions. Western transfers were performed using Mini PROTEAN® Tetra cell respectively onto 0.2μ PVDF membranes (Immobilon-PSQ®, Sigma-aldrich), using Towbin’s buffer (25 mM Tris-HCL, 192 mM Glycine and 20% methanol) at 90V for 30 min, followed by blocking with TBS-T (Tris buffered saline+0.05% Tween-20) with 5% BSA. After antibody hybridization and washes, blots were incubated in SuperSignal™ West Dura extended duration substrate (Thermoscientific) prior to image acquisition on a LI-COR imaging system (Odyssey Fc. LI-COR biosciences) using the ImageStudio™ software. We used ImageStudio™ software (LI-COR) to quantify western blot bands, measuring signal intensity with background subtraction. Signal intensities were normalized against loading controls for total proteins or corresponding total protein bands for phosphorylated proteins. Relative changes were calculated by comparing the normalized values to the untreated baseline, ensuring consistent and reliable quantification across all samples. The raw uncropped western blots are provided as supplemental information.
DNA fiber assays
DNA fiber assays were performed as reported.35 Briefly, cells were labeled with 100 μM 5-chloro-2'-deoxyuridine (CldU) for 20 minutes, washed, treated with 200 μM 5-Iodo-2′-deoxyuridine (IdU) for 10 minutes, washed and then treated with SG at 10 μg/ml for 30 minutes in the incubator, washed twice with PBS and then treated for 2 hours with berzosertib in the presence of IdU. Cells were harvested and processed as described.29 Briefly, cells were lysed with lysis buffer (1% sodium dodecyl sulfate, 100 mM Tris-HCl [pH 7.4], 50 mM EDTA). Labeled DNAs with CldU and IdU were stained with mouse anti-IdU primary antibody (1:250) and rat anti-CldU primary (1:200) respectively. Anti-rat Alexa Fluor 488 (1:250) and anti-mouse Alexa 594 (1:250) were used for secondary antibodies. Images were captured with a Zeiss LSM 780 confocal microscope. Fiber length was measured using ImageJ software.
Immunofluorescent microscopy
Cells were prepared as detailed before3 using appropriate antibodies mentioned in the resource table. For RPA1, γH2AX or CCNB1 immunofluorescence, cells were cultured in chamber slides (Ibidi GmbH) overnight before treating it with inhibitors. Slides were then fixed in 4% paraformaldehyde, for 10 minutes at room temperature, permeabilized in 0.5% Triton X-100 in PBS, washed and blocked in blocking buffer (1% BSA, 1% goat serum in PBS). Slides were incubated in primary antibody overnight at 4°C, washed briefly in PBS containing 0.1% Triton X-100, followed by PBS washes twice followed by conjugated secondary antibody (1:100) incubations if required, before staining with DAPI (300 nM) for 4 minutes in the dark. Slides were washed three times prior to mounting with Prolong anti-Fade® mountant (Invitrogen) and visualized on a Zeiss 780 laser confocal microscope. Images were analyzed with Zen® software (Carl Zeiss microscopy, GmbH) or with Fiji™ (a.k.a. ImageJ), NIH. Cells with > 5 RPA1 or >15 γH2AX foci were counted as positive and an average of at least three to five images (100 - 300 cells) were plotted with standard deviation of the population shown as error bars. Distribution of RPA1+ve cells between different cell cycle phases were identified by the pattern of nuclear staining of CCNB1 as described by Lindqvist et al.47 For microscopic analysis of TROP2 surface expression, cells plated in 8-well chamber slides were fixed with 4% paraformaldehyde in PBS for 15 minutes at room temperature, washed three times, blocked with blocking solution (1% goat serum in PBS) for 30 minutes, followed by incubation for 2 hours at 7°C with rabbit anti-TROP2 antibody (Cell Signal) and then stained with secondary antibody conjugated to AF488 for 1 hour, washed, stained with DAPI, and mounted as explained previously.
Quantification and statistical analysis
Student’s t-test was used to determine statistical significance for paired groups of data whereas an unpaired Mann-Whitney t-test was performed for DNA fiber, RNA-seq data analyses and murine work (GraphPad Prism). Minimum sample sizes for statistical significance were determined in consultation with biostatisticians. For all plots shown in the study, p values < 0.05 are considered significant and are shown as asterisks ∗ p < 0.05, ∗∗ p < 0.01 and ∗∗∗ p < 0.001 unless otherwise mentioned in the figure legends.
Additional resources
-
•
The Phase II clinical trial of CHK1 inhibitor prexasertib monotherapy in patients with recurrent HGSOC can be found using identifier NCT02203513 at https://clinicaltrials.gov/.
Published: October 29, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.111283.
Supplemental information
References
- 1.Kurman R.J., Shih I.M. Pathogenesis of ovarian cancer: lessons from morphology and molecular biology and their clinical implications. Int. J. Gynecol. Pathol. 2008;27:151–160. doi: 10.1097/PGP.0b013e318161e4f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Konstantinopoulos P.A., Spentzos D., Karlan B.Y., Taniguchi T., Fountzilas E., Francoeur N., Levine D.A., Cannistra S.A. Gene expression profile of BRCAness that correlates with responsiveness to chemotherapy and with outcome in patients with epithelial ovarian cancer. J. Clin. Oncol. 2010;28:3555–3561. doi: 10.1200/JCO.2009.27.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brill E., Yokoyama T., Nair J., Yu M., Ahn Y.R., Lee J.M. Prexasertib, a cell cycle checkpoint kinases 1 and 2 inhibitor, increases in vitro toxicity of PARP inhibition by preventing Rad51 foci formation in BRCA wild type high-grade serous ovarian cancer. Oncotarget. 2017;8:111026–111040. doi: 10.18632/oncotarget.22195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chau C.H., Steeg P.S., Figg W.D. Antibody-drug conjugates for cancer. Lancet. 2019;394:793–804. doi: 10.1016/S0140-6736(19)31774-X. [DOI] [PubMed] [Google Scholar]
- 5.Cheng X., Li J., Tanaka K., Majumder U., Milinichik A.Z., Verdi A.C., Maddage C.J., Rybinski K.A., Fernando S., Fernando D., et al. MORAb-202, an antibody--drug conjugate utilizing humanized anti-human FR$∖alpha$ farletuzumab and the microtubule-targeting agent eribulin, has potent antitumor activity. Mol. Cancer Therapeut. 2018;17:2665–2675. doi: 10.1158/1535-7163.MCT-17-1215. [DOI] [PubMed] [Google Scholar]
- 6.Dilawari A., Shah M., Ison G., Gittleman H., Fiero M.H., Shah A., Hamed S.S., Qiu J., Yu J., Manheng W., et al. FDA approval summary: mirvetuximab soravtansine-gynx for FR$∖alpha$-positive, platinum-resistant ovarian cancer. Clin. Cancer Res. 2023;29:3835–3840. doi: 10.1158/1078-0432.CCR-23-0991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fowler M. Mirvetuximab Soravtansine Combination Yields Encouraging Response Rates in Ovarian Cancer. Oncology. 2020;34:250. [PubMed] [Google Scholar]
- 8.Matulonis U.A., Lorusso D., Oaknin A., Pignata S., Dean A., Denys H., Colombo N., Van Gorp T., Konner J.A., Marin M.R., et al. Efficacy and Safety of Mirvetuximab Soravtansine in Patients With Platinum-Resistant Ovarian Cancer With High Folate Receptor Alpha Expression: Results From the SORAYA Study. J. Clin. Oncol. 2023;41:2436–2445. doi: 10.1200/JCO.22.01900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De S.K. Tisotumab Vedotin: The First FDA-Approved Antibody-Drug Conjugate for Cervical Cancer. Anti Cancer Agents Med. Chem. 2022;22:2808–2810. doi: 10.2174/1871520622666220421095240. [DOI] [PubMed] [Google Scholar]
- 10.Markham A. Tisotumab Vedotin: First Approval. Drugs. 2021;81:2141–2147. doi: 10.1007/s40265-021-01633-8. [DOI] [PubMed] [Google Scholar]
- 11.Ray Chaudhuri A., Hashimoto Y., Herrador R., Neelsen K.J., Fachinetti D., Bermejo R., Cocito A., Costanzo V., Lopes M. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat. Struct. Mol. Biol. 2012;19:417–423. doi: 10.1038/nsmb.2258. [DOI] [PubMed] [Google Scholar]
- 12.Fornaro M., Dell'Arciprete R., Stella M., Bucci C., Nutini M., Capri M.G., Alberti S. Cloning of the gene encoding Trop-2, a cell-surface glycoprotein expressed by human carcinomas. Int. J. Cancer. 1995;62:610–618. doi: 10.1002/ijc.2910620520. [DOI] [PubMed] [Google Scholar]
- 13.Ambrogi F., Fornili M., Boracchi P., Trerotola M., Relli V., Simeone P., La Sorda R., Lattanzio R., Querzoli P., Pedriali M., et al. Trop-2 is a determinant of breast cancer survival. PLoS One. 2014;9 doi: 10.1371/journal.pone.0096993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sin S.T.K., Li Y., Liu M., Yuan Y.F., Ma S., Guan X.Y. Down-regulation of TROP-2 Predicts Poor Prognosis of Hepatocellular Carcinoma Patients. Hepatol. Commun. 2018;2:1408–1414. doi: 10.1002/hep4.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bignotti E., Todeschini P., Calza S., Falchetti M., Ravanini M., Tassi R.A., Ravaggi A., Bandiera E., Romani C., Zanotti L., et al. Trop-2 overexpression as an independent marker for poor overall survival in ovarian carcinoma patients. Eur. J. Cancer. 2010;46:944–953. doi: 10.1016/j.ejca.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 16.Cardillo T.M., Govindan S.V., Sharkey R.M., Trisal P., Arrojo R., Liu D., Rossi E.A., Chang C.H., Goldenberg D.M. Sacituzumab Govitecan (IMMU-132), an Anti-Trop-2/SN-38 Antibody-Drug Conjugate: Characterization and Efficacy in Pancreatic, Gastric, and Other Cancers. Bioconjugate Chem. 2015;26:919–931. doi: 10.1021/acs.bioconjchem.5b00223. [DOI] [PubMed] [Google Scholar]
- 17.Perrone E., Manara P., Lopez S., Bellone S., Bonazzoli E., Manzano A., Zammataro L., Bianchi A., Zeybek B., Buza N., et al. Sacituzumab govitecan, an antibody-drug conjugate targeting trophoblast cell-surface antigen 2, shows cytotoxic activity against poorly differentiated endometrial adenocarcinomas in vitro and in vivo. Mol. Oncol. 2020;14:645–656. doi: 10.1002/1878-0261.12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cardillo T.M., Govindan S.V., Sharkey R.M., Trisal P., Goldenberg D.M. Humanized anti-Trop-2 IgG-SN-38 conjugate for effective treatment of diverse epithelial cancers: preclinical studies in human cancer xenograft models and monkeys. Clin. Cancer Res. 2011;17:3157–3169. doi: 10.1158/1078-0432.CCR-10-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perrone E., Lopez S., Zeybek B., Bellone S., Bonazzoli E., Pelligra S., Zammataro L., Manzano A., Manara P., Bianchi A., et al. Sacituzumab govitecan, an antibody-drug conjugate targeting trophoblast cell-surface antigen 2, shows cytotoxic activity against poorly differentiated endometrial adenocarcinomas in vitro and in vivo. Front. Oncol. 2020;10:118. doi: 10.3389/fonc.2020.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tagawa S.T., Balar A.V., Petrylak D.P., Kalebasty A.R., Loriot Y., Fléchon A., Jain R.K., Agarwal N., Bupathi M., Barthelemy P., et al. TROPHY-U-01: A Phase II Open-Label Study of Sacituzumab Govitecan in Patients With Metastatic Urothelial Carcinoma Progressing After Platinum-Based Chemotherapy and Checkpoint Inhibitors. J. Clin. Oncol. 2021;39:2474–2485. doi: 10.1200/JCO.20.03489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Shaughnessy J., Brufsky A., Rugo H.S., Tolaney S.M., Punie K., Sardesai S., Hamilton E., Loirat D., Traina T., Leon-Ferre R., et al. Analysis of patients without and with an initial triple-negative breast cancer diagnosis in the phase 3 randomized ASCENT study of sacituzumab govitecan in metastatic triple-negative breast cancer. Breast Cancer Res. Treat. 2022;195:127–139. doi: 10.1007/s10549-022-06602-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cardillo T.M., Rossi D.L., Zalath M.B., Liu D., Arrojo R., Sharkey R.M., Chang C.H., Goldenberg D.M. Predictive biomarkers for sacituzumab govitecan efficacy in Trop-2-expressing triple-negative breast cancer. Oncotarget. 2020;11:3849–3862. doi: 10.18632/oncotarget.27766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huntoon C.J., Flatten K.S., Wahner Hendrickson A.E., Huehls A.M., Sutor S.L., Kaufmann S.H., Karnitz L.M. ATR inhibition broadly sensitizes ovarian cancer cells to chemotherapy independent of BRCA status. Cancer Res. 2013;73:3683–3691. doi: 10.1158/0008-5472.CAN-13-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim H., George E., Ragland R., Rafail S., Zhang R., Krepler C., Morgan M., Herlyn M., Brown E., Simpkins F. Targeting the ATR/CHK1 axis with PARP inhibition results in tumor regression in BRCA-mutant ovarian cancer models. Clin. Cancer Res. 2017;23:3097–3108. doi: 10.1158/1078-0432.CCR-16-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim H., Xu H., George E., Hallberg D., Kumar S., Jagannathan V., Medvedev S., Kinose Y., Devins K., Verma P., et al. Combining PARP with ATR inhibition overcomes PARP inhibitor and platinum resistance in ovarian cancer models. Nat. Commun. 2020;11:3726. doi: 10.1038/s41467-020-17127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas A., Takahashi N., Rajapakse V.N., Zhang X., Sun Y., Ceribelli M., Wilson K.M., Zhang Y., Beck E., Sciuto L., et al. Therapeutic targeting of ATR yields durable regressions in small cell lung cancers with high replication stress. Cancer Cell. 2021;39:566–579.e7. doi: 10.1016/j.ccell.2021.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hur J., Ghosh M., Kim T.H., Park N., Pandey K., Cho Y.B., Hong S.D., Katuwal N.B., Kang M., An H.J., Moon Y.W. Synergism of AZD6738, an ATR Inhibitor, in Combination with Belotecan, a Camptothecin Analogue, in Chemotherapy-Resistant Ovarian Cancer. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22031223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abel M.L., Takahashi N., Peer C., Redon C.E., Nichols S., Vilimas R., Lee M.J., Lee S., Shelat M., Kattappuram R., et al. Targeting Replication Stress and Chemotherapy Resistance with a Combination of Sacituzumab Govitecan and Berzosertib: A Phase I Clinical Trial. Clin. Cancer Res. 2023;29:3603–3611. doi: 10.1158/1078-0432.CCR-23-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang T.T., Burkett S.S., Tandon M., Yamamoto T.M., Gupta N., Bitler B.G., Lee J.M., Nair J.R. Distinct roles of treatment schemes and BRCA2 on the restoration of homologous recombination DNA repair and PARP inhibitor resistance in ovarian cancer. Oncogene. 2022;41:5020–5031. doi: 10.1038/s41388-022-02491-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamamoto T.M., McMellen A., Watson Z.L., Aguilera J., Ferguson R., Nurmemmedov E., Thakar T., Moldovan G.L., Kim H., Cittelly D.M., et al. Activation of Wnt signaling promotes olaparib resistant ovarian cancer. Mol. Carcinog. 2019;58:1770–1782. doi: 10.1002/mc.23064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jossé R., Martin S.E., Guha R., Ormanoglu P., Pfister T.D., Reaper P.M., Barnes C.S., Jones J., Charlton P., Pollard J.R., et al. ATR inhibitors VE-821 and VX-970 sensitize cancer cells to topoisomerase i inhibitors by disabling DNA replication initiation and fork elongation responses. Cancer Res. 2014;74:6968–6979. doi: 10.1158/0008-5472.CAN-13-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas A., Redon C.E., Sciuto L., Padiernos E., Ji J., Lee M.J., Yuno A., Lee S., Zhang Y., Tran L., et al. Phase I Study of ATR Inhibitor M6620 in Combination With Topotecan in Patients With Advanced Solid Tumors. J. Clin. Oncol. 2018;36:1594–1602. doi: 10.1200/JCO.2017.76.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kopp A., Hofsess S., Cardillo T.M., Govindan S.V., Donnell J., Thurber G.M. Antibody-Drug Conjugate Sacituzumab Govitecan Drives Efficient Tissue Penetration and Rapid Intracellular Drug Release. Mol. Cancer Therapeut. 2023;22:102–111. doi: 10.1158/1535-7163.MCT-22-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J.M., Nair J., Zimmer A., Lipkowitz S., Annunziata C.M., Merino M.J., Swisher E.M., Harrell M.I., Trepel J.B., Lee M.J., et al. Prexasertib, a cell cycle checkpoint kinase 1 and 2 inhibitor, in BRCA wild-type recurrent high-grade serous ovarian cancer: a first-in-class proof-of-concept phase 2 study. Lancet Oncol. 2018;19:207–215. doi: 10.1016/S1470-2045(18)30009-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nair J., Huang T.T., Murai J., Haynes B., Steeg P.S., Pommier Y., Lee J.M. Resistance to the CHK1 inhibitor prexasertib involves functionally distinct CHK1 activities in BRCA wild-type ovarian cancer. Oncogene. 2020;39:5520–5535. doi: 10.1038/s41388-020-1383-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu N., Zhang Z., Zhu J., Xu L., Li Y., Duan L., Mao Y., Li H. Overexpression of trophoblast cell surface antigen 2 as an independent marker for a poor prognosis and as a potential therapeutic target in epithelial ovarian carcinoma. Int. J. Exp. Pathol. 2016;97:150–158. doi: 10.1111/iep.12174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldenberg D.M., Cardillo T.M., Govindan S.V., Rossi E.A., Sharkey R.M. Trop-2 is a novel target for solid cancer therapy with sacituzumab govitecan (IMMU-132), an antibody-drug conjugate (ADC) Oncotarget. 2015;6:22496–22512. doi: 10.18632/oncotarget.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beaufort C.M., Helmijr J.C.A., Piskorz A.M., Hoogstraat M., Ruigrok-Ritstier K., Besselink N., Murtaza M., van I.W.F., Heine A.A.J., Smid M., et al. Ovarian cancer cell line panel (OCCP): clinical importance of in vitro morphological subtypes. PLoS One. 2014;9 doi: 10.1371/journal.pone.0103988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y., Yang L., Zhang J., Zhou M., Shen L., Deng W., Liang L., Hu R., Yang W., Yao Y., et al. Radiosensitization by irinotecan is attributed to G2/M phase arrest, followed by enhanced apoptosis, probably through the ATM/Chk/Cdc25C/Cdc2 pathway in p53-mutant colorectal cancer cells. Int. J. Oncol. 2018;53:1667–1680. doi: 10.3892/ijo.2018.4514. [DOI] [PubMed] [Google Scholar]
- 40.Baschnagel A.M., Elnaggar J.H., VanBeek H.J., Kromke A.C., Skiba J.H., Kaushik S., Abel L., Clark P.A., Longhurst C.A., Nickel K.P., et al. ATR Inhibitor M6620 (VX-970) Enhances the Effect of Radiation in Non-Small Cell Lung Cancer Brain Metastasis Patient-Derived Xenografts. Mol. Cancer Therapeut. 2021;20:2129–2139. doi: 10.1158/1535-7163.MCT-21-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zou L., Elledge S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 42.Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 43.Cardillo T.M., Sharkey R.M., Rossi D.L., Arrojo R., Mostafa A.A., Goldenberg D.M. Synthetic lethality exploitation by an anti--Trop-2-SN-38 antibody--drug conjugate, IMMU-132, plus PARP inhibitors in BRCA1/2--wild-type triple-negative breast cancer. Clin. Cancer Res. 2017;23:3405–3415. doi: 10.1158/1078-0432.CCR-16-2401. [DOI] [PubMed] [Google Scholar]
- 44.Burma S., Chen B.P., Murphy M., Kurimasa A., Chen D.J. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 45.Ruff P., Donnianni R.A., Glancy E., Oh J., Symington L.S. RPA Stabilization of Single-Stranded DNA Is Critical for Break-Induced Replication. Cell Rep. 2016;17:3359–3368. doi: 10.1016/j.celrep.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Porter L.A., Cukier I.H., Lee J.M. Nuclear localization of cyclin B1 regulates DNA damage-induced apoptosis. Blood. 2003;101:1928–1933. doi: 10.1182/blood-2002-04-1103. [DOI] [PubMed] [Google Scholar]
- 47.Lindqvist A., van Zon W., Karlsson Rosenthal C., Wolthuis R.M.F. Cyclin B1-Cdk1 activation continues after centrosome separation to control mitotic progression. PLoS Biol. 2007;5 doi: 10.1371/journal.pbio.0050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie K., Doles J., Hemann M.T., Walker G.C. Error-prone translesion synthesis mediates acquired chemoresistance. Proc. Natl. Acad. Sci. USA. 2010;107:20792–20797. doi: 10.1073/pnas.1011412107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quinet A., Lerner L.K., Martins D.J., Menck C.F.M. Filling gaps in translesion DNA synthesis in human cells. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2018;836:127–142. doi: 10.1016/j.mrgentox.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 50.Kannouche P.L., Lehmann A.R. Ubiquitination of PCNA and the polymerase switch in human cells. Cell Cycle. 2004;3:1011–1013. [PubMed] [Google Scholar]
- 51.Chatterjee N., Whitman M.A., Harris C.A., Min S.M., Jonas O., Lien E.C., Luengo A., Vander Heiden M.G., Hong J., Zhou P., et al. REV1 inhibitor JH-RE-06 enhances tumor cell response to chemotherapy by triggering senescence hallmarks. Proc. Natl. Acad. Sci. USA. 2020;117:28918–28921. doi: 10.1073/pnas.2016064117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Therasse P., Arbuck S.G., Eisenhauer E.A., Wanders J., Kaplan R.S., Rubinstein L., Verweij J., Van Glabbeke M., van Oosterom A.T., Christian M.C., Gwyther S.G. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J. Natl. Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 53.Bardia A., Mayer I.A., Diamond J.R., Moroose R.L., Isakoff S.J., Starodub A.N., Shah N.C., O'Shaughnessy J., Kalinsky K., Guarino M., et al. Efficacy and Safety of Anti-Trop-2 Antibody Drug Conjugate Sacituzumab Govitecan (IMMU-132) in Heavily Pretreated Patients With Metastatic Triple-Negative Breast Cancer. J. Clin. Oncol. 2017;35:2141–2148. doi: 10.1200/JCO.2016.70.8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bardia A., Sun S., Thimmiah N., Coates J.T., Wu B., Abelman R.O., Spring L., Moy B., Ryan P., Melkonyan M.N., et al. Antibody-Drug Conjugate Sacituzumab Govitecan Enables a Sequential TOP1/PARP Inhibitor Therapy Strategy in Patients with Breast Cancer. Clin. Cancer Res. 2024;30:2917–2924. doi: 10.1158/1078-0432.CCR-24-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stanzione M., Zhong J., Wong E., LaSalle T.J., Wise J.F., Simoneau A., Myers D.T., Phat S., Sade-Feldman M., Lawrence M.S., et al. Translesion DNA synthesis mediates acquired resistance to olaparib plus temozolomide in small cell lung cancer. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abn1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jackson L.M., Moldovan G.L. Mechanisms of PARP1 inhibitor resistance and their implications for cancer treatment. NAR Cancer. 2022;4 doi: 10.1093/narcan/zcac042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hernandez L., Kim M.K., Lyle L.T., Bunch K.P., House C.D., Ning F., Noonan A.M., Annunziata C.M. Characterization of ovarian cancer cell lines as in vivo models for preclinical studies. Gynecol. Oncol. 2016;142:332–340. doi: 10.1016/j.ygyno.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin G.L., Wilson K.M., Ceribelli M., Stanton B.Z., Woo P.J., Kreimer S., Qin E.Y., Zhang X., Lennon J., Nagaraja S., et al. Therapeutic strategies for diffuse midline glioma from high-throughput combination drug screening. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aaw0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang T.T., Brill E., Nair J.R., Zhang X., Wilson K.M., Chen L., Thomas C.J., Lee J.M. Targeting the PI3K/mTOR Pathway Augments CHK1 Inhibitor-Induced Replication Stress and Antitumor Activity in High-Grade Serous Ovarian Cancer. Cancer Res. 2020;80:5380–5392. doi: 10.1158/0008-5472.CAN-20-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ohmachi T., Tanaka F., Mimori K., Inoue H., Yanaga K., Mori M. Clinical significance of TROP2 expression in colorectal cancer. Clin. Cancer Res. 2006;12:3057–3063. doi: 10.1158/1078-0432.CCR-05-1961. [DOI] [PubMed] [Google Scholar]
- 62.Darzynkiewicz Z., Juan G. DNA content measurement for DNA ploidy and cell cycle analysis. Curr. Protoc. Cytom. 2001;7:7.5.1–7.5.24. doi: 10.1002/0471142956.cy0705s00. [DOI] [PubMed] [Google Scholar]
- 63.Matsumoto K., Katsumata N., Shibata T., Satoh T., Saitou M., Yunokawa M., Takano T., Nakamura K., Kamura T., Konishi I. Phase II trial of oral etoposide plus intravenous irinotecan in patients with platinum-resistant and taxane-pretreated ovarian cancer (JCOG0503) Gynecol. Oncol. 2015;136:218–223. doi: 10.1016/j.ygyno.2014.10.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Processed and raw RNA-seq data have been deposited at Gene Expression Omnibus (GEO) collection (https://www.ncbi.nlm.nih.gov/geo/) as GSE278664 and are publicly available as of the date of the publication.
-
•
This paper does not report original code.
-
•
Original uncropped western blots are provided as Data S1. Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from lead contact upon request.