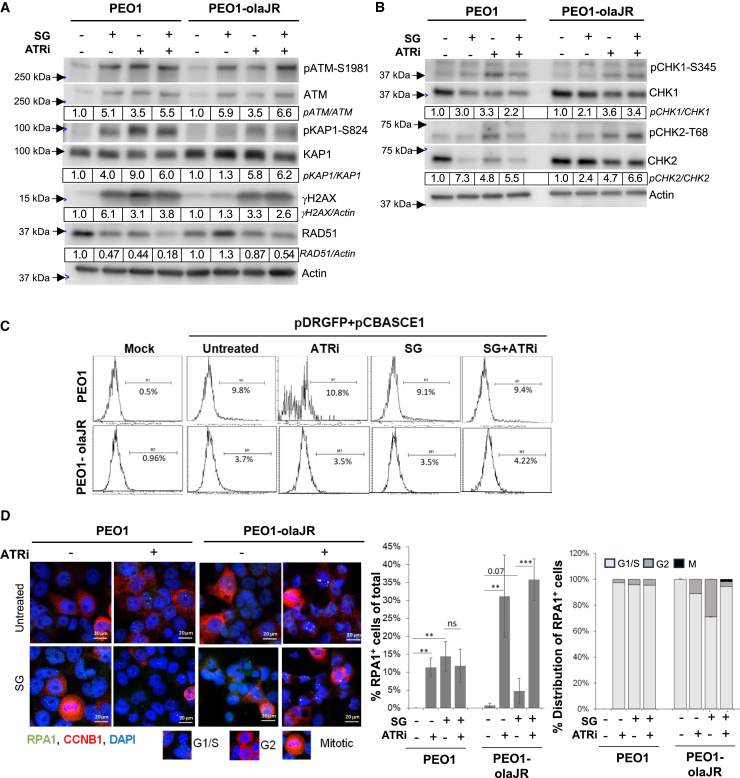
Figure 4.
Impact of SG in DNA damage repair pathways
Cells were left untreated or pretreated with SG (10 μg/mL for 30 min/37°C, washed thrice with PBS) prior to treatment with ATRi berzosertib for all experiments.
(A and B) Western blot analysis for DNA damage markers of lysates from cells pretreated with SG and treated with ATRi (1 μM) overnight. Densitometric quantitation was performed as detailed in the STAR Methods. Densitometric values of phosphorylated proteins relative to total proteins are further normalized to untreated (value of 1) for each cell line and shown below respective images. Values were rounded to two significant numbers. Representative of 3 biologically independent experiments.
(C) Graphs show results from homologous recombination (HR) repair activity assay. After cells were pretreated with SG, they were cultured overnight in plain media and then transfected with HR reporter plasmids overnight, followed by treatment with ATRi or in plain media for 48 h prior to flow cytometric analysis for GFP expression. Mock transfection without plasmids were also performed as a control for transfection efficiency. Flow data was analyzed on Flowjo and plotted as histograms. Percent GFP-positive (GFP+ve) cells for histograms are shown under each range gate. This is representative of two experimental replicates.
(D) Representative images of cells pretreated with SG, washed and incubated overnight with ATRi (1 μM) prior to co-staining for targets RPA1 (green foci) and Cyclin B1 (CCNB1, red). For reference, images of cell cycle phases as inferred from CCNB1 staining is given below image panels. The graph (middle panel) shows the mean ratio ±SD of RPA1+ve cells (>5 foci) to total cells (n = 100–300) from all sampled fields (n = 3–5 biological replicates) for each treatment. The stacked plot on the right show percent distribution of RPA1+ve cells across cell cycle phases as defined by staining pattern of nuclear CCNB1 from the sampled fields (n = 3–5 biological replicates per treatment). Representative of n = 2 biologically independent experiments. ∗∗p < 0.01, and ∗∗∗p < 0.001, ns, not significant. Scale bar is 20 μm.