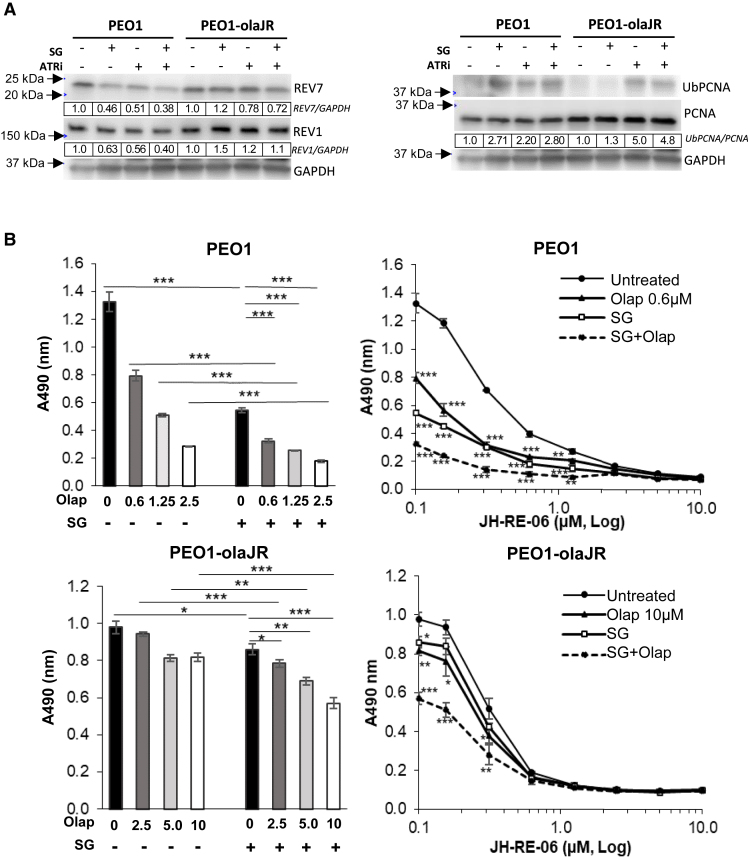
Figure 5.
Translesion DNA synthesis (TLS) pathway inhibition improves SG and olaparib activity in HGSOC
(A) Western analysis of protein lysates from cells with or without SG pretreatment (10 μg/mL for 30 min/37°C, washed thrice with PBS) followed by overnight incubation with ATRi (1 μM) were examined for markers of TLS activity, REV1, REV7 (components of TLS polymeraseζ) and its marker of activity ubiquitinylated PCNA (UbPCNA). As described in STAR Methods, ratio of total chemiluminescence signal (ImageStudio) from each UbPCNA band to that of total PCNA was then further normalized to untreated (value of 1) for each cell line, and values shown below rounded to two significant numbers. Images are representative of 3 biologically independent experiments.
(B) The graphs on the left show mean cell growth ±SD (n = 3 biological replicates per treatment) from XTT assays on the effect of SG on sensitivity to a gradient of olaparib. On the right, the graphs show mean ± SD (n = 3 biological replicates) cell growth with or without SG pretreatment followed by olaparib in the presence of a gradient of specific TLS inhibitor JH-RE-06 over 5 days. During the assay, cells were retreated with SG in fresh media on day 3, washed twice with PBS, prior to reincubation with olaparib and JH-RE-06 for another 2 days. The Y axis shows actual background-corrected absorbance at 490nm. Untreated cell values (0 μM of JH-RE-06) are plotted at 0.1 μM to enable representation on the log X axis. Statistical significance was determined using Student’s t-test. The p-values for olaparib or SG were determined against untreated while those for the combination (SG + olaparib) (dotted line) were determined against olaparib monotherapy. ∗∗p < 0.01, and ∗∗∗p < 0.001, ns, not significant.