Summary
As advantageous as sexual reproduction is during progeny generation, it is also an expensive and treacherous reproductive strategy. The viviparous eukaryote has evolved to survive stress before, during, and after pregnancy. An important and conserved intracellular pathway for the control of metabolic stress is autophagy. The autophagy process occurs in multiple stages through the coordinated action of autophagy-related genes. This review summarizes the evidence that autophagy is an integral component of reproduction. Additionally, we discuss emerging in vitro techniques that will enable cellular and molecular studies of autophagy and its associated pathways in reproduction. Finally, we discuss the role of autophagy in the pathogenesis and progression of several pregnancy-related disorders such as preterm birth, preeclampsia, and intra-uterine growth restriction, and its potential as a therapeutic target.
Subject areas: physiology, pathophysiology, molecular biology, cell biology
Graphical abstract
Physiology; Pathophysiology; Molecular biology; Cell biology.
Introduction
Sexual reproduction leads to genetic material exchange during progeny generation.1 Shuffling and rearranging parental genomes provide, in most effective cases, enough genetic diversity in the offspring to adapt to and thrive in new environments,2 even though theoretical models cannot fully support some of these benefits.3,4 This apparent evolutionary advantage over asexual reproduction imposes many costly challenges. It requires mating, increased time and energy, and is intrinsically and molecularly more complex. The development of gonads that produce competent gametes, the physical need of such gametes to encounter the correct spatiotemporal conditions, and the proper generation of a new organism are all extremely critical steps of sexual reproduction.5 In particular, viviparous eukaryotes face extra challenges in comparison to those with external organismal development; the former requires the evolution of pregnancy-specific mechanisms of protection, nurturing, survival, and delivery.6,7 Thus, it is not surprising that the rate of successful pregnancies is low in some species, and that survival mechanisms in response to stress (metabolic, infection, and so forth) have evolved with pregnancy to allow eukaryotic cells to cope with different assaults before, during, and after pregnancy.8,9
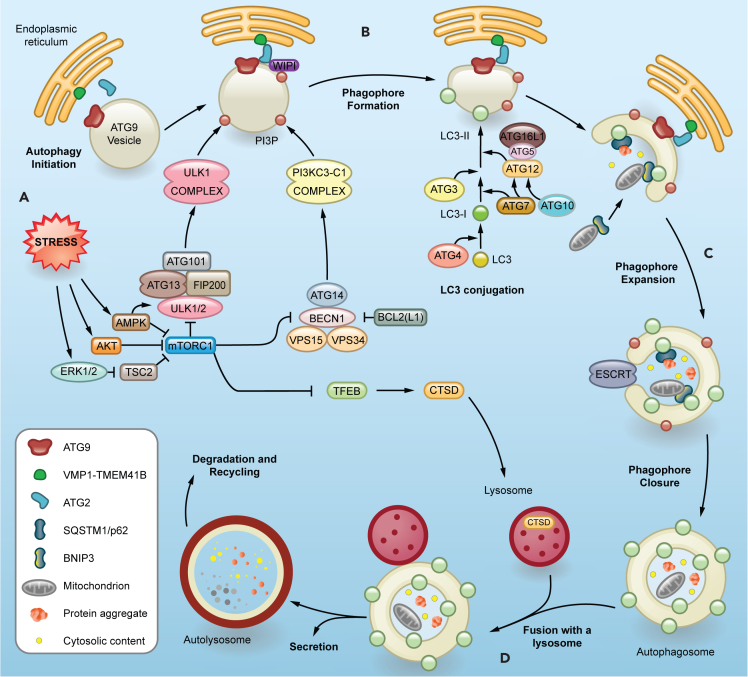
One highly conserved and ubiquitous mechanism of cellular and organismal survival is autophagy. The autophagic process involves the engulfment of intracellular materials in double-membraned autophagosomes that fuse with lysosomes for degradation (previously reviewed10,11,12). Autophagy proceeds over several stages with the coordination of core autophagy-related (ATG) genes and key accessory genes to perform essential functions (Figure 1). The initiation of autophagy requires the activation (AMPK, ULK1) and inhibition (AKT, mTOR) of several kinases involved in transducing intra- and extracellular signals of stress13,14 (Figure 1A). Some examples of these stressors are low levels of free amino acids, glucose, and oxygen, the presence of exogenous pathogens, misfolded proteins, and damaged organelles. In response to these stressors, the ULK1 complex (FIP200, ULK1 or ULK2, ATG13, and ATG101) is recruited to a pre-autophagosomal structure (PAS) rich in phosphatidylinositol 3-phosphate (PI3P) that forms in close vicinity to an "omegasome," a subdomain of the endoplasmic reticulum (ER).15,16,17 The main autophagy proteins involved in PI3P generation on the PAS are part of the class III phosphatidylinositol 3-kinase (PI3K) complex I (PI3KC3-C1) composed of BECN1, PIK3R4/VPS15, PIK3C3/VPS34, and ATG14.18,19 Next, WD-repeat protein interacting with phosphoinositides (WIPI) 1–4 proteins bind to the PI3P in the PAS and recruit the next two important components of phagophore expansion: ATG2 proteins and the ATG16L1 complex.20,21 ATG2A and ATG2B are cytosolic lipid transfer proteins that transport phospholipids to the PAS and growing autophagosome.22 They work together with a set of scramblases to keep membrane integrity at the transfer sites23 (Figure 1A). VMP1 and TMEM41B function as scramblases in the ER,24 while ATG9 proteins fulfill the same function in small Golgi-derived vesicles.25,26,27 Recent discoveries suggest that ATG9-containing vesicles are the actual PAS where PI3P and its effectors are recruited28,29,30 (Figure 1A). In addition, other ER subdomains and membranous organelles often surround nascent phagophores, likely contributing to their growth and maturation, though how the cell coordinates the different roles of these components is still an area of intense research.31,32,33,34,35,36,37 While the phagophore membrane expands, a ubiquitin-conjugation-like machinery (ATG16L1 complex) decorates it with the ATG8/LC3-family proteins and cargo receptors38 (Figure 1B). ATG7 (E1), ATG3 and ATG10 (E2), and ATG5-ATG12-ATG16L1 (E3) coordinate the covalent conjugation between the C-terminus of ATG8/LC3 (ubiquitin-like) and the amino group of phophatidylethanolamine enriched in the phagopore by the assistance of ATG4. The lipidation of ATG8 is required for an efficient phagophore formation, cargo recruitment, and downstream fusion with lysosomes.39 The rapid expansion of the phagophore can engulf mostly cytosolic content nonspecifically.40 Perhaps more importantly, phagophore generation and growth can also occur around and specifically recruit cytoplasmic content tagged for degradation during selective autophagy41 (Figure 1C). This is mediated by cargo receptors, such as sequestosome 1 (SQSTM1/p62), that recognize both cargo and autophagosome components.42 Each one of these selective autophagy pathways requires specific cargo receptors that contain an LC3-interaction region (LIR) motif. The canonical LIR sequence follows the [W/F/Y]-X-X-[L/I/V] pattern (X represents any amino acid preferably small and flexible). Most of the autophagy cargo receptors can oligomerize43 in order to recruit cargo to the phagophore through direct interaction with ubiquitin and lipidated LC3 proteins. Examples of selective autophagy are mitophagy, ER-phagy, lysophagy, and lipophagy, which represent the specific degradation of mitochondria, ER, lysosomes, and lipid droplets, respectively, among others.44 In case of selective elimination of damaged mitochondria, for example, the ubiquitin kinase PTEN induced kinase 1 (PINK1) recruits, stabilizes, and phosphorylates Parkin (PRKN), an E3 ubiquitin protein ligase, which in turn ubiquitinates several outer membrane mitochondrial proteins. These ubiquitin tags are recognized by LIR domain-containing autophagy receptors, which direct the selective mitochondria targeting to autophagosomes.45 BNIP3 and BNIP3L/NIX are well-known selective mitophagy receptors for the delivery of damaged mitochondria to autophagosomes.46,47 Less studied are the processes that lead to phagophore closure, which generates a mature autophagosome,48,49 but a clear role of the endosomal sorting complexes required for transport (ESCRT) machinery has been established.50 Once fully formed, the autophagosomes are capable of fusing to late endosomes and lysosomes to assist with the degradation of the encapsulated material,51 their secretion,52 and recycling53 (Figure 1D). This allows the cell to control the stressful insult that prompts the autophagic response.
Figure 1.
Molecular players in the autophagy pathway
Schematic overview of the mechanism of autophagy.
(A) Cellular stress results in the formation of pre-autophagosomal structures. Autophagy initiation involves the mTOR inhibition and activation of the ULK complex, which recruits class III phosphatidylinositol 3-kinase complex C1 (PI3KC3-C1) to the phagophore formation site. This lipid kinase complex is responsible for phosphatidylinositol 3-phosphate (PI3P) generation on ATG9 vesicles, the now speculated membranous seed for phagophore formation. WIPI proteins bind to PI3P and subsequently recruit ATG2, which controls phospholipid trafficking into the growing phagophore from other membrane sources, mainly the endoplasmic reticulum.
(B) The growing phagophore membrane is decorated with the ATG8/LC3 family of proteins and cargo receptors by a ubiquitin-conjugation-like machinery. LC3 is first processed from its precursor form to LC3-I through cleavage by ATG4, exposing a C-terminal glycine. LC3-I is then lipidated with phosphatidylethanolamine to form LC3-II, which integrates into autophagosomal membranes.
(C) Sequestration of selectively ubiquitin-tagged or non-selective cargo recruitment into the expanding phagophore. This is followed by the sealing of the phagophore membrane by ESCRT machinery.
(D) A fully formed double membrane autophagosome fuses with a lysosome to assist in the degradation, recycling, and secretion of all the encapsulated material.
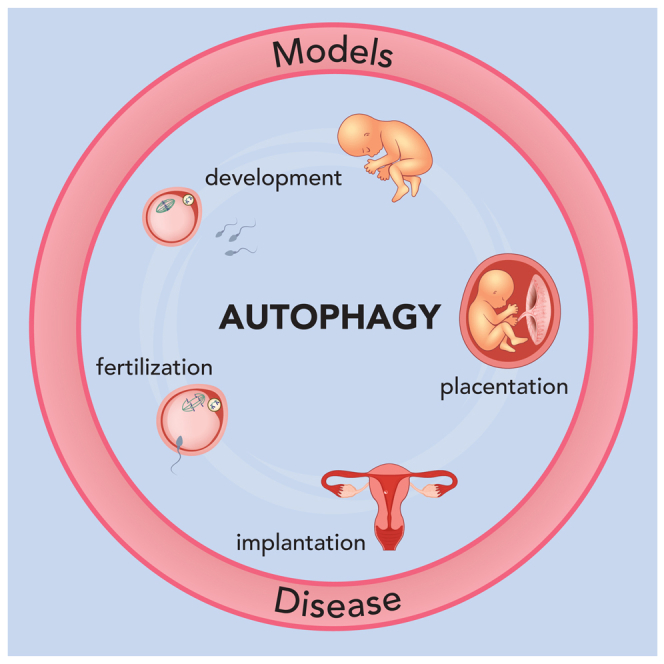
In this review, we summarize the essential role of the autophagy machinery at different stages of a healthy pregnancy (Table 1) while exploring the central concept that autophagy serves as a surveillance mechanism that secures positive reproductive outcomes by monitoring resource availability during cellular stress. We explore the evidence and gaps in knowledge for the role of autophagy in the formation of competent gametes, fertilization and implantation, the development of the placenta, and parturition. The role of autophagy in mammalian embryonic development and organogenesis has been extensively reviewed and readers are directed to these articles for more information.54,55 Additionally, we provide an overview of the latest tools that can be used to interrogate the function of autophagy during early stages of human development in vitro. Finally, we review the role of autophagy in pathogenesis and disease progression for a myriad of pregnancy-related disorders. With a focus on murine and human models, this review highlights the most fundamental and recent findings in the field.
Table 1.
A summary of important studies on autophagy in pregnancy
| Stage | Study with the main phenotype | Citation |
|---|---|---|
| Sperm development |
|
Ref.56,57 |
|
Ref.58 | |
|
Ref.59 | |
| Oocyte development |
|
Ref.60 |
|
Ref.61 | |
|
Ref.62 | |
|
Ref.63 | |
| Embryo implantation |
|
Ref.64 |
|
Ref.65,66 | |
| Fetus and Placenta development |
|
Ref.67,68 |
|
Ref.69 | |
|
Ref.70 | |
|
Ref.71,72,73 | |
|
Ref.74 | |
|
Ref.75 | |
|
Ref.76 | |
|
Ref.77 | |
|
Ref.78 | |
|
Ref.79 | |
|
Ref.80 | |
|
Ref.81 | |
|
Ref.63 | |
|
Ref.82 | |
|
Ref.83 | |
|
Ref.84 | |
|
Ref.85 | |
|
Ref.78,82 | |
|
Ref.86 | |
| Birth |
|
Ref.87,88 |
|
Ref.87,89,90,91,92,93,94 | |
|
Ref.79,87,90,91,92,95 | |
|
Ref.96,97,98 | |
|
Ref.99,100 |
Gamete formation and hormone regulation
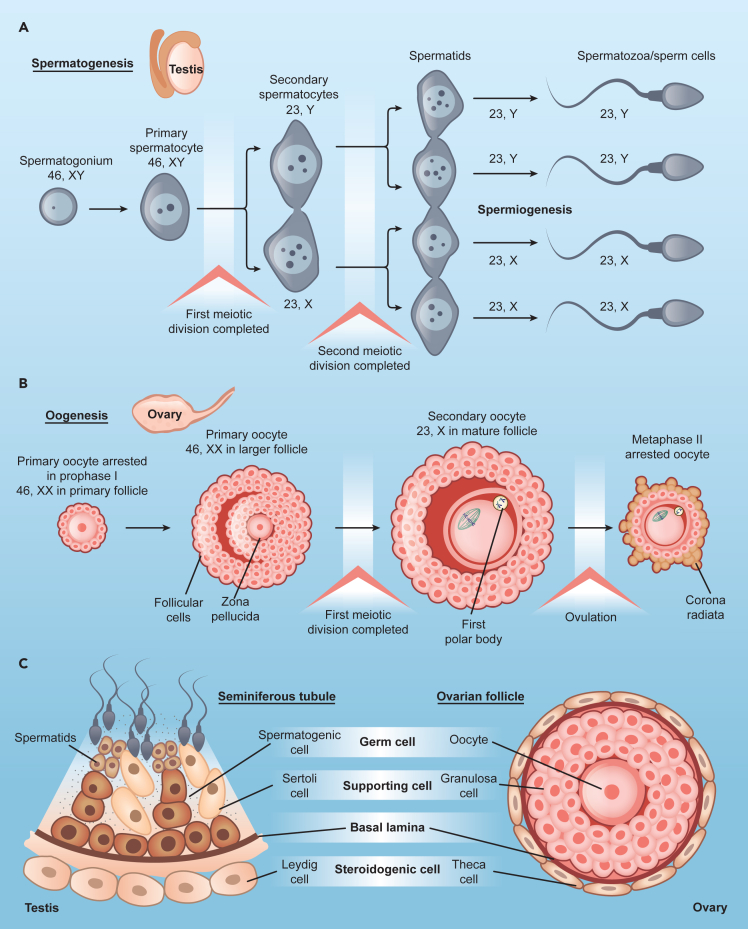
Central to sexual reproduction, gametogenesis (Figure 2) involves maintaining a balance between degradation and metabolic supply until gametes encounter fertilization. Deficient progenitor cells or with developmental disruptions do not produce viable or fertile gametes. In addition to potential problems during meiosis, each germ cell undergoes an enormous number of structural and functional changes to become a competent gamete. Not surprisingly, autophagy contributes to various aspects of gamete development and maturation.
Figure 2.
Schematic of gametogenesis
The formation of an embryo after fertilization requires the proper development of a healthy egg and sperm in the process of gametogenesis.
(A) The generation of sperm in males is known as spermatogenesis. The diploid primary spermatocytes arise from diploid spermatogonia which undergo two meiotic divisions to form haploid spermatozoa. First meiotic division produces two haploid secondary spermatocytes, each of which undergo second meiotic division to form two haploid spermatids. These spermatids acquire flagella to mature into spermatozoa.
(B) In females, the formation of oocytes is known as oogenesis. The diploid primary oocyte undergoes two rounds of meiotic divisions to form a haploid secondary oocyte and a polar body. The primary oocyte is surrounded by follicular cells within the ovarian follicle. The follicle matures during the process of oogenesis to form a mature follicle which releases the egg during ovulation.
(C) A comparison of seminiferous tubules in the testis and ovarian follicles highlights the different architecture and cell types surrounding the germ cells. The steroidogenic and supporting cells in each gonad are critical for the whole gametogenesis process and autophagy deficiency in these cells severely impacts the development of the gametes.
In male germ cells, autophagy is essential for normal sperm development and function (Figure 2A). Murine male germ cells lacking Atg5 showed reduced levels of conjugated LC3A/B and accumulated cargo receptor SQSTM1/p62.56 At a cellular level, these gametes had numerous, large residual bodies in different stages of spermatogenesis after their normal resorption within the seminiferous epithelium.56 Sperm had abnormal morphology, including disrupted head formation, abnormal acrosome and mid-piece formation, and abnormal mitochondria.56 This suggests a clear involvement of autophagy in spermiogenesis, the last and longest step of spermatogenesis (Figure 2A). Additionally, male germ cells lacking Atg7 presented serious defects in acrosome biogenesis, resulting in round-headed sperm, which made the sperm appear globozoospermic.57 Interestingly, this conditional knockout (KO) only affected later stages of spermatogenesis, suggesting specific temporal roles of autophagy during sperm differentiation. Examination of Atg7 null mouse showed the disruption of flagella biogenesis, with a reduction in sperm motility, retention of extra cytoplasm on the mature sperm head, and a disorganized cytoskeleton, probably due to failure of selective autophagy.58 This study demonstrated that autophagy in nurse and supporting cells ensures the correct development of the sperm (Figure 2C). In fact, Atg5 and Atg7 loss in Leydig cells affects sexual behavior in mice due to a decline in cholesterol trafficking and testosterone production, suppressing spermatogenesis.59 Additionally, using single-cell RNA sequencing analysis, transcriptional signatures of ATG genes were found to be dysregulated in sperm samples of infertile men presented with dysfunctional spermatogenesis.101 Therefore, autophagy is important for maintaining male fertility.
In the ovary, autophagy plays a role in the development of the primordial follicle pool, folliculogenesis, and progesterone synthesis (Figure 2B). At the time of birth, during the normal development of the primordial follicle pool in the mouse ovaries, a significant loss of the germ cell pool occurs.102,103 Autophagy protects against the excessive loss of oocytes during neonatal severe nutrient starvation which occurs just after birth and before feeding. Treatment with rapamycin (an mTOR inhibitor and thus, an autophagy enhancer) increases the number of surviving oocytes by 25–30% during mouse primordial follicle assembly.104 A high level of autophagy is thus associated with the suppression of apoptosis to prevent the loss of oocytes.104 Further, Becn1+/− murine ovaries showed a 56% reduction of germ cells on postnatal day 1, and Atg7-deficient ovaries resulted in total loss of germ cells, demonstrating a role of autophagy in oogenesis.60 Germ cell-specific Atg7 KO also showed more than 50% oocyte loss during the neonatal transition period.61 During folliculogenesis, only 1% of all follicles develop into mature oocytes, whereas other follicles undergo degeneration (atresia) at different developmental stages. Follicle atresia is thought to be triggered by massive granulosa cell (GC) apoptosis. Interestingly, autophagy is involved in the regulation of atresia and the development of the dominant follicle in different species, such as rats105,106 and humans.107 This was shown to be controlled in ovarian GCs by the mitochondrial voltage dependent anion channel 2 (VDAC2) protein which stabilizes the interaction between BECN-1 and BCL2L1 (an anti-apoptotic factor), leading to the inhibition of autophagy. This balance is a key switch for proper follicular development, offering potential strategies for enhancing female fertility by autophagy suppression.108 Interestingly, endolysosomal vesicular assemblies (ELVAs) in mouse oocytes were recently discovered.109 This unique structure accumulates autophagosomes, proteasomes, and protein aggregates that are only degraded at the final stages of oocyte growth and maturation. The dynamic of degradation and disassembly of these structures offers a potential explanation for how growing oocytes rely on autophagy only after maturation and fertilization (see next section).
The process of autophagy plays a critical role in regulating the synthesis of steroid hormones during gamete formation. Knockdown (KD) of ATG5 and BECN1 results in decreased expression of genes associated with the differentiation of ovarian GCs and those encoding steroidogenic enzymes hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 (HSD3B1) and steroidogenic acute regulatory protein (STAR).62 This decrease is due to the accumulation of Wilms’ tumor 1 (WT1) transcription factor which contains an LIR motif and is degraded through autophagy. Furthermore, GCs from patients with biochemical premature ovarian insufficiency (bPOI) also show a reduction in the expression of key autophagy genes.62 Women with ovarian endometriosis exhibit an elevated ratio of serum progesterone/estradiol (P4/E2) and the P4-to-follicle index along with higher expression of P4 biosynthesis proteins.110 This increase is accompanied by a rise in BECN1 levels in GCs. The authors of this work suggested that in the case of ovarian endometriosis, increased activity of BECN1 results in increased hydrolyzation of low-density lipoproteins (LDL), cholesterol utilization, and lysosomal activity, resulting in elevated progesterone secretion.110 A similar study using a Becn1-deficient mouse also indicates Becn1 is essential in lipid regulation and progesterone synthesis in luteal cells.63 Additionally, autophagy also degrades the negative regulators of cholesterol uptake pathways in Leydig cells, increasing cholesterol uptake for testosterone production in males.59 Finally, lipophagy is involved in gonadotropin-stimulated sex steroid synthesis in the human ovary (and testis).111 Hence, autophagy is essential to hormone-regulated successful gametogenesis by maintaining lipid supply for steroid synthesis, although the precise mechanisms remain unclear.
Fertilization and preimplantation
The onset of fertilization triggers several critical processes, and autophagy is essential to their successful completion. Because autophagy has not been observed in unfertilized oocytes, the induction of autophagy is thought to be fertilization-dependent.112 Autophagy plays a role in providing nutrients and energy to the developing zygote and degrading paternal mitochondria to allow for maternal mitochondria transmission. The elimination of sperm-borne mitochondria during fertilization ensures normal preimplantation development.113 In C. elegans, paternal mitochondria are present in the embryo post-fertilization, which are marked by LGG-1/LGG-2 (ATG8-like proteins), signaling their autophagy degradation. Although these mitochondria are sequestered in double membrane vesicles indicating mitophagy, they are not ubiquitinated.114 In mammals, sperm mitochondria are ubiquitinated and tagged with several autophagosomal and lysosomal markers post-fertilization,114,115 which suggests the evolution of autophagy pathways to accommodate more complex fertilization events in mammals. Mice embryos expressing shRNA against Sqstm1/p62, Tbc1d15, Pink1, and mitochondrial E3 ligase Mul1 exhibit suppression of paternal mitochondrial colocalization with LC3114,116,117 suggesting active mitophagy. Studies in porcine fertilization demonstrated that SQSTM1 participates in sperm mitophagy but not LC3, indicating the involvement of noncanonical ubiquitin-recognizing autophagy.115 Interestingly, a recent study using RFP-labelled mitochondria transgenic mice has found that the sperm mitochondria persisted until the morula stage and that autophagy did not participate in paternal mitochondria degradation post-fertilization,118 suggesting the involvement of other mechanisms besides autophagy.
Once the oocyte is fertilized, maternal mRNA and protein content are degraded in preparation for the new embryonic proteome. This process is part of what is known as the maternal-to-zygotic transition (MZT).119 Autophagy increases in the 1-cell stage embryo before a transient decrease and is then reactivated in the 2-cell stage embryo and maintained throughout the blastocyst stage.64,65,112 In mice, wild-type (WT) sperm fertilization of Atg5 null oocytes was not affected, but Atg5 null sperm fertilization caused failure during development beyond the 8-cell stage.64 This study indicates a critical role for autophagy in preimplantation embryogenesis and assistance in cytosolic remodeling during MZT stages. At the blastocyst stage, autophagy becomes particularly important for the development of the trophectoderm (the most external embryo layer). These cells produce 80% of the energy and amino acid turnover required for proper development and successful implantation of the blastocyst, while the inner cell mass remains quiescent.120
Temporary suspension of embryonic development, known as embryonic diapause, occurs prior to implantation and is conserved across mammals. As an evolutionary strategy, diapause ensures the survival of the embryo so optimal conditions of implantation and embryogenesis can be met.121 Studies have shown that autophagy is activated during diapause and that rapamycin reversibly delays blastocyst development.65,66 Despite this, researchers found that suppressing mTORC1 is not necessary nor sufficient to initiate this surge of autophagy.122 These results suggest that diapause-induced autophagy contributes to the maintenance of the blastocyst pluripotent state before implantation, but the mechanistic details of autophagy regulation at this developmental stage require future investigations.
Overall, autophagy facilitates the elimination of paternal mitochondria after fertilization, cytosolic remodeling during MTZ, and embryonic development, particularly at the blastocyst stage and embryonic diapause. As these dramatic cellular processes unfold, autophagy plays a crucial role in properly allocating resources and handling external challenges intracellularly until implantation and placentation occur.
Implantation
Embryo attachment to the uterine lining is a critical factor for successful mammalian pregnancy. Autophagy plays a pivotal role in maintaining endometrial homeostasis. In the mouse, autophagy markers are increased in the endometrium at the time of implantation and rapidly decrease afterward.123,124 If autophagy is inhibited during pregnancy, the number of implantation sites are reduced.124 Essential to the implantation of the blastocyst is the process of decidualization where the endometrial stromal cells differentiate into secretory decidual cells to support embryo attachment and placenta formation.125,126 Unsuccessful decidual remodeling results in recurrent implantation failure and pregnancy loss.127 Using Rb1cc1/FIP200 conditional KO in the uterus of mice and human endometrial stromal cells (hESC) in culture67 and a conditional Atg16l1 KO female mice,68 fertility was reduced through a decrease in implantation rate, due to improper decidualization. Further, ATG9A regulates mitophagy to increase prolactin (PRL) secretion and induce decidualization in hESC,128 while KD of ATG7 and ATG5 hinders this process.69 A recent study in mice showed that Becn1-deficient uteri resulted in loss of endometrial progenitor stem cells, impacting proper uterine development.70 Altogether, autophagy is required for decidualization, a fundamental uterine cell differentiation process that assures implantation and placentation. There is still a need to understand how autophagy is precisely regulated in the various cells of the endometrium, how it supports stromal cell differentiation, how it participates in the menstrual cycle, why it is downregulated following implantation, and what role it has in many uterine diseases.129
Placenta development and function
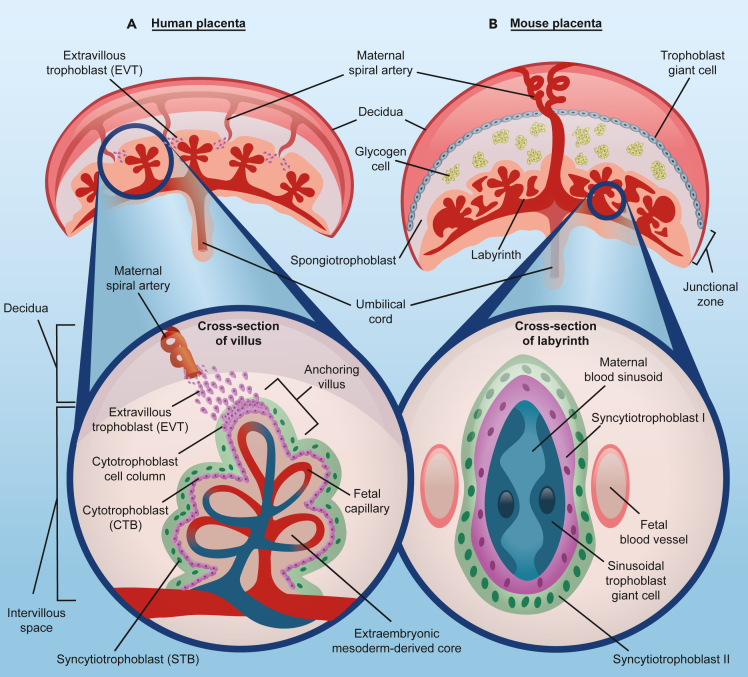
After blastocyst implantation in the uterine endometrium, trophectoderm cells aggressively proliferate, invade, and nest in the decidual bed. Once anchored, the placenta is formed as the physical and biochemical interface between the host and the developing embryo (Figure 3). Placental mammals have evolved species-specific structures and strategies to form a functional placenta depending on the developing environment and the needs of the embryo.130,131 In humans, the proliferative epithelial stem trophoblasts, the cytotrophoblasts (CTBs), fuse to form the syncytiotrophoblast (STB), and later the characteristic villous structure for gas and nutrient exchange with the maternal interface. Additionally, CTBs differentiate into extravillous trophoblasts (EVTs), which migrate and invade the maternal uterine spiral arteries to control maternal blood flow during pregnancy132 (Figure 3A). In mice, embryonic mesoderm differentiates to vascularize the yolk sac that is eventually replaced by the chorioallantoic placenta (Figure 3B).133
Figure 3.
Structure of the fully developed human and mouse placentas
(A) Structure of the human placenta - It is composed of three main cell types-extravillous trophoblast (EVT), cytotrophoblast (CTB), and syncytiotrophoblast (STB). The maternal decidua is the site where maternal circulation is established and where the placenta anchors to the uterine lining. The invasive EVTs penetrate the uterine wall to remodel the spiral arteries to ensure adequate blood supply to the growing fetus. Cross section of the human placental chorionic villi illustrates the cells in the fetal-maternal interface. The fetal blood vessels are surrounded by the layer of CTBs. Some CTBs detach from the basement membrane to form CTB cell columns, which contribute to the formation of the anchoring villi. The mononucleated CTBs in the villi fuse together to form the STB, a syncytium layer of nuclei and cytoplasmic content. This layer is in direct contact with maternal blood, secretes protein and hormones, and is involved in nutrient exchange with the underlying fetal circulation.
(B) Structure of the mouse placenta - It is composed of three layers. 1) Maternal decidua which consists of invaded trophoblast giant cells that anchors the placenta to the uterus; 2) Junctional zone which is formed by glycogen cells and spongiotrophoblast. These cells control the metabolism and secretory functions of the placenta in the mouse; 3) Labyrinth zone is the region where fetal-maternal exchange occurs. The cross-section of the labyrinth illustrates the multiple cell layers between the maternal sinusoids and fetal blood vessels. Sinusoidal trophoblast giant cells line the maternal sinusoids, in direct contact with syncytiotrophoblast-I and -II cells. These cells form the barrier between maternal and fetal circulation, regulating nutrient exchange.
Initial human placenta formation triggers a persistent and severe hypoxic environment driven by trophoblasts plugging the uterine spiral arteries that last up to 10 weeks.134 After the first trimester, the plug dissolves, and EVTs replace endothelial cells remodeling maternal spiral arteries to gain access to maternal blood causing oxygen levels to return to normoxia.132,135,136 The low oxygen tension limits metabolism and reactive oxygen species (ROS) production in the embryonic compartment137 but it also triggers trophoblast differentiation and proliferation. Autophagy activation is observed in EVTs as an adaptation to hypoxia to maintain cellular homeostasis via a reduction in mTOR activity.71,138 Hypoxia inducible factor 1 subunit alpha (HIF1α), a master transcriptional regulator of the adaptive response to hypoxia, also regulates the invasion of trophoblast cells by increasing autophagy.139 Autophagy-deficient EVT-derived placental cells fail to show invasion, with decreased adenosine triphosphate (ATP) content following hypoxic conditions.71,72,73 Once EVTs adopt endothelial characteristics and remodel the spiral arteries, metabolic adaptations occur under these aerobic conditions in the placenta. Efficient remodeling is necessary to prevent trophoblast damage due to ischemia-reoxygenation injury and placental oxidative stress, including ER stress and inflammatory stress.140 Excessive oxidative stress results in trophoblast senescence by autophagy inhibition and decreased autophagic flux in a p53/mTOR-dependent fashion.141 Yet, oxidative stress was also shown to induce autophagy within trophoblasts and endothelial cells, reducing trophoblast invasion and placental vasculature.142 These contradictory observations require further dissection of autophagy responses to oxidative stress during placenta development.
In both mice and humans, STBs are responsible for exchanging essential molecules between fetal and maternal blood, among other secretory functions.143 This multinucleated STB (two contiguous layers in the mouse) is formed by cell-cell fusion of precursor cells by a process known as syncytialization.144 Failure of cell-cell fusion is implicated in placental pathology. The exact mechanism is still not clear; however, several players have been identified including fusogenic proteins (syncytins), various cytoskeleton elements, galectins, and phosphatidylserine exposure.145,146,147 Autophagy is considered a contributing mechanism to trophoblast syncytialization.148 Placental CTBs utilize autophagy for energy replenishment and cytoplasm remodeling when experiencing strong stimuli for syncytialization.73,75 Blocking autophagy flux by using bafilomycin A1 suppresses CTB fusion as shown by a decrease in human chorionic gonadotropin (hCG) secretion from STBs.74 On the other hand, activating autophagy using rapamycin results in excessive syncytialization in a pregnant mouse model.75 A recent study examined the expression of autophagy markers during a 96-hour time course of syncytialization in villous CTB cells derived from the human term placenta.76 They observed a gradual increase in the formation of acidic compartments and the autophagy marker LC3B. By manipulating the levels of BECN1, authors found that the inhibition or overactivation of autophagy led to a decrease in the cell fusion index, indicating the importance of tightly regulated autophagy during syncytialization.
Lysosomes are key organelles that respond to nutrients and cellular stress by activating multiple signaling events to recover from cellular damage and have an essential role in the late stages of autophagy149 (Figure 1). Although very low levels of the transcription factor EB (TFEB), a master transcriptional regulator of lysosomal biogenesis,150 are found in embryos, its expression is very evident in the trophectoderm and placenta,151 specifically in the labyrinthine trophoblast cells of mice.77 Homozygous mice carrying a germline mutation in Tfeb die between 9.5 and 10.5 days of embryonic development as they fail to express vascular endothelial growth factor (VEGF). Therefore, these mice lose normal placental vascularization.77 In vitro trophoblasts isolated from third trimester placenta and cultured under hypoxia showed a decrease in the levels of TFEB and several lysosomal proteins such as lysosomal associated membrane protein 1 and 2 (LAMP1, LAMP2), and cathepsin D (CTSD), with the concomitant impairment of autophagic flux, as observed by a decrease in the number of autophagic vacuoles.78 This was further verified in TCL-1 cells (third trimester EVT cell line) and an autophagy-deficient EVT trophoblast cell line (HchEpClb-ATG4BC74A), pointing to the role of autophagy in TFEB localization and lysosomal function in the placenta.78 Remarkably, three recent studies have found TFEB as a direct transcriptional regulator of trophoblast fusion proteins such as syncytins, independently from its canonical lysosomal targets,152,153,154 highlighting a novel placenta-specific dual gene program of TFEB and a better explanation for murine Tfeb KO vascular phenotypes.
In addition to its role in maintaining cellular homeostasis and trophoblast differentiation, autophagy plays a crucial role in safeguarding the feto-placenta unit against pathogens. Selective autophagy of pathogens is involved in recognizing and removal of invading intracellular bacteria and viruses.155 Placental explants deficient in ATG16L1 exhibit heightened susceptibility to infections caused by pathogenic E. coli.79 On the other hand, the infection of human trophoblast cells and mouse placentas by Zika virus (ZIKV) induced the activation of autophagy.80 In mouse models with Atg16l1 gene deficiency, ZIKV infection is reduced in both placentas and fetuses, resulting in improved fetal outcomes.80 However, in another study using human and non-human primate placenta-derived STBs, authors observed a decrease in expression of ATG5 and SQSTM1 and a decrease in protein levels of LC3B and SQSTM1 in cells with active ZIKV replication.81 This highlights the differential ability of various pathogens to exploit the autophagy mechanism for their intracellular survival in different species, underscoring the importance of autophagy in favoring or preventing the vertical transmission of infections from the placenta to the fetus.
Parturition
At the end of every pregnancy, the process of labor begins and includes several stages: early or latent (body preparation), active (delivery of the fetus and expulsion of the placenta), and fetal-to-neonate transition. These processes are propelled by hormones and require very intense physiological responses for both the host and the fetus. Autophagy, despite being studied for its role in earlier stages of pregnancy, has not been extensively studied during delivery given practical restrictions. One study in humans points to more LC3-II in the placentas of non-labor cesarean birth compared to labor deliveries.156 Similarly, fetal membranes obtained after spontaneous term labor showed lower expression of the autophagy proteins BECN1, ATG3, ATG5, ATG7, ATG12, and ATG16L1 compared with term non-labored deliveries.157 However, these phenotypes could be due to patients fasting prior to the surgery and/or the use of anesthetics during cesarean section.158 No changes in the expression of several autophagy markers were observed in the placentas from the spontaneous onset of labor or pharmacologically induced parturition.159 ATG16L1 affects the timing of delivery during labor induction.160 Single nucleotide polymorphisms in ATG16L1 in the maternal genetic makeup were also found to be associated with pre-term delivery when hCG levels during the first 12 weeks were elevated.161
Autophagy is activated in the human uterine myometrium during labor and might play an important role in maintaining uterine contraction function.162 RNA-seq data analysis in the myometrium cells during labor showed significant enrichment in protein ubiquitination pathways, which is correlated with labor duration. Expression of genes involved in autophagy and vesicle transport proteins were also upregulated during the active phase of labor onset, normally lower in the latent phase of labor.163
Early neonatal survival depends on autophagy. Two fundamental studies in Atg5 KO87 and Ras related GTP binding A (small GTPase that regulates mTORC1 activity) RragaQ66L mutant mice88 demonstrated that the interruption of sudden nutrient supply through the placenta during delivery triggers AMPK- and mTORC1-dependent autophagy in the neonate to maintain an adequate pool of nutrients until lactation takes over. Autophagy is actively induced right after birth in almost all the tissues87 and in line with this observation, conventional Atg3, Atg5, Atg7, Atg9a, Atg12, and Atg16l1 null mice present fetal growth restriction, decreased plasma amino acid levels, and lead to pre-natal or neonatal lethality within one day of birth.87,89,90,91,92,93,94
In conclusion, the process of labor involves multiple stages and intense physiological responses in both the host and fetus. Autophagy, although less studied during delivery, has been found to have potential involvement and novel experimental approaches are needed to fully understand the mechanisms and significance of autophagy in the context of labor and delivery.
Models to study human pregnancy in a dish
This section summarizes the development of different in vitro tools focusing primarily on models to mimic the human maternal-fetal interface. The development of models for gonads, germ cells, and embryos (or embryoids) is covered in several other specialized articles.164,165,166,167,168,169 Similarly, trophoblast monolayer cultures have been employed for over 50 years, providing valuable insights into placental barrier function, trophoblast invasion, toxicology, and disease modeling in vitro, are extensively reviewed elsewhere170 and therefore we will focus on recent innovations in cell lines and organoid models for studying human placenta physiology and pathology.
In vitro models to study autophagy in the placenta
Recently, a breakthrough in the derivation of human trophoblast stem cells (TSCs) has been reported.171 Okae and colleagues demonstrated the derivation of TSCs from CTBs isolated from first-trimester human placental villi by manipulating signaling pathways (activation of Wnt and EGF pathways, inhibition of TGFB, and histone deacetylase-mediated inhibition of ROCK). These TSCs can be maintained in culture long-term (approximately 80 passages) and undergo directed differentiation into EVT- and STB-like cells. This study provides a detailed description of culture conditions and shows that the differentiated cell types closely resemble transcriptomes and epigenomes of in vivo human EVTs and STBs. The authors were unable to derive human TSCs from the term placenta, speculating that the CTBs that served as the origin for first trimester-derived lines are lost during the second trimester of pregnancy. This suggests the possibility of multiple CTB subpopulations throughout human pregnancy, although this has not been investigated yet. However, several years later, another group reported isolation and prolonged culture of trophoblasts from term placenta – a tissue that is easier to obtain compared to first-trimester placenta.172 Both first trimester- and term-derived trophoblast cultures as well as stem-cells-derived methods173,174,175 are great tools to investigate autophagy mechanisms as they have laid the groundwork for the establishment of 3D models to study placenta development.
Shortly after the emergence of methods to isolate and culture human trophoblasts, two groups generated trophoblast organoid models. In the first model, Haider et al. used purified first-trimester CTBs to establish a human placental organoid culture system.176 Similar to the Okae 2D culture, these organoids spontaneously differentiate into STBs. However, when self-renewal factors are removed from the culture, CTBs begin to express NOTCH1 (known EVT progenitor marker) and eventually form adjacent, distally located EVTs. In a parallel experiment, Turco et al. also successfully generated placental organoid cultures that follow similar developmental characteristics under specific conditions.177 Both methods rely on access to first-trimester tissue. However, more recent studies offered new alternative sources of these cells to generate placenta organoids: term placentas178 and stem cells.179,180,181
The current limitation of original 3D models is the inverted architecture of organoids – the STB lacks an outer apical membrane exposed to the culture medium and therefore does not recapitulate the in vivo shedding, making it difficult to study the maternal-fetal exchange of nutrients and gases. Interestingly, placental organoids are among many other in vitro tissue systems that face this challenge. Scientists are currently pursuing a workaround for this problem and a recent work shows a promising solution by culturing the organoid first in extracellular matrix domes and then resuspending it for proper cellular orientation.182 Other approaches implement several “organ-on-a-chip” systems, commonly referred to as “placenta-on-a-chip.”183 Although these systems have not yet solved the impediment to study syncytialization in 3D, they do provide new tools to investigate EVT invasion. Multiple labs have recently demonstrated new capabilities of reconstructing the 3D structure of the maternal-fetal interface to study in vivo-like invasion of EVTs into the host uterus. In one study,184 through testing multiple variations of cellular environments, authors employ such a system to show the importance of decidualized stromal cells in regulating EVT invasion and provide new evidence for the role that pre-implantation maternal immune cells play in this process. Another study focused on examining the engineering aspects of the model and its potential applications.185 When combined with fluorescent cell tagging and flow cytometry, the platform allows the collection of the invasive cells,185 opening doors for researchers to manipulate the autophagic network in ways that have previously been impossible in live animal models. The effects of these manipulations can be further investigated by biochemical profiling control and experimental trophoblast cells.
In vitro models to study endometrium-placenta interactions
Perhaps a more detailed investigation of the autophagy network can be achieved by modeling trophoblast-endometrium interactions in 3D. To date, several in vitro systems have been designed to model such interactions.186 The first 3D models began to emerge in the mid 1980s and have become an invaluable tool to study mechanisms of embryo implantation, drug development, and disease modeling. These models often involve co-culturing primary cells and establishing cell lines in the presence of collagen, agarose gels, or Matrigel to recreate physiologic architecture. Supplementation of culture media with estrogen and progesterone is used to mimic physiological conditions.187,188,189,190 Models of endometrium have been used to investigate the impact of pharmaceutical compounds on endometrial receptivity191,192 and can be similarly employed to study the mechanism of autophagy. An alternative to cell-based 3D cultures to model endometrium is an ex vivo tissue approach.193 This model uses human endometrial tissue explants and incorporates an air-liquid interface into a 3D matrix scaffold using a type I collagen gel. Furthermore, a combined 2D/3D model to study implantation has been reported.194 In this model, three cell lines (HEC-1, Ishikawa, and RL95-2) were seeded in Matrigel until solidified (3D) and then overlaid with the same cell type (2D). Finally, a non-invasive method for the isolation of endometrial epithelial organoids and stromal cells from menstrual fluid has also been generated.195 Similar approaches can be combined in the future to fully understand human placental and endometrium interactions at the different stages of pregnancy.
The in vitro models described above provide a valuable platform for monitoring autophagy regulation, where it is difficult to obtain primary human tissue. Additionally, these novel cellular systems are more amenable to interrogation using classical tools and assessments196,197,198,199 such as autophagy flux assays, electron microscopy, and live-cell imaging of fluorescent reporters. Moreover, unbiased functional screens such as CRISPR or siRNA can be employed to identify novel cell-specific regulators of autophagy, broadening our understanding of its regulation, such as recently proven useful using human trophoblast stem cells.200,201 The novel hits from those screens can be further validated through orthogonal assays both in vitro and in vivo. Taking the advantage of these modern systems can help us better understand the role of autophagy in various aspects of reproductive biology.
Autophagy in human pregnancy disorders
Autophagy has been implicated in the most prevalent human pregnancy disorders. Here we summarize such evidence from both mouse and human studies.
Placenta accreta spectrum
The placenta accreta spectrum pathology is characterized by abnormal trophoblast invasion, permitting the placenta to deeply invade into the myometrium.202 This process is often compared to the behavior of invasive cancerous cells which involves epithelial-to-mesenchymal transition (EMT). Dysfunction of trophoblast cells or defect in endometrial-myometrial interface from failure in proper decidualization, results in placenta accreta pathology. Limited research points toward potential disease association with autophagy. Microarray analysis on normal and placenta accreta placentas revealed transcriptomic differences in genes related to autophagy.203 Chen et al. observed high expression levels of HIF1α, BECN1, SQSTM1, and LC3B in trophoblasts from pathologic placentas.204 Whether autophagy upregulation is hypoxia-driven or the cause for the invasive trophoblast phenotype in the placenta was not addressed. Evidently, more studies on these placental invasive phenotypes are required to understand the role, if any, of autophagy in both the pathogenesis and progression of placenta accreta pathology.
Preterm birth
Preterm birth (PTB) refers to births occurring before 37 weeks of pregnancy, characterized by the weakening of fetal membranes. The majority of PTBs do not have clearly defined risk factors.205 Altered steroid levels during pregnancy, oxidative stress, and infection-driven inflammation mark major risks for PTB.206,207 These stressors are associated with aberrant EMT in cells of the fetal membranes leading to pregnancy complications associated with PTB.208 In a recent study using transcriptome analysis of amniochorion membranes from preterm pregnancies revealed the dysregulation of autophagy and EMT in fetal membranes.209 Brickle et al. utilized human placenta tissues obtained from spontaneous term and preterm labor and observed a decrease in protein expression of BECN1 and ATG7 in spontaneous preterm labor fetal membranes.157 A decrease in autophagy was further associated with high IL-1β levels and increased inflammasome activation, which contributed to spontaneous PTB.157 Infection-driven inflammatory conditions also affect autophagy flux, with a decrease in levels of ATG7, ATG16L1, and ATG4, and increased accumulation of LC3B.79,207 Studies with Becn1 KO in precursor follicular granulosa murine cells show defects in progesterone production in the corpus luteum resulting in failure of decidualization and predisposition to preterm labor.63 Despite the largely unknown risk factors for PTB, autophagy appears to play a significant role, but its extent remains unclear.
Intra-uterine growth restriction
In intra-uterine growth restriction (IUGR), the fetus fails to reach its full genetic growth potential resulting in pre- and postnatal complications. It is also one of the risk factors for preterm labor. Among various factors that lead to IUGR pathology, abnormal placenta development contributes as a major determinant.210 These placentas are smaller in size and have defective patterns of CTB invasion and proliferation, with the impaired remodeling of the maternal spiral arteries resulting in low levels of nutrient supply to the growing fetus. Marked changes in placenta autophagy have been observed in pregnancies resulting in IUGR. Placenta-specific Atg7 KO mice showed a reduction in fetal growth, which was due to reduced placental weight.82 Interestingly, STB-specific Atg7 KO inhibited fetal growth without affecting placental weight.83 Fetuses from KO mouse models of autophagy essential genes such as Becn1, Atg3, Atg5, Atg7, Atg9a, and Atg16l1 showed an IUGR phenotype and died on day one after birth, thus implying the necessity of the autophagy process soon after birth.79,87,90,91,92,95 Remarkably, IUGR placentas showed an increase in the total number of autophagosomes in STBs.211 Further, primary CTBs from these placentas showed higher levels of ER stress, lower levels of mTOR phosphorylation, and increased expression of mTOR inhibitor TSC2 when subjected to oxygen-glucose deprivation (OGD) treatment.96 mTOR activity in fetal growth-restricted pregnancies is regulated by placental AKT and AMPK.96 However, these effects were not observed in in vitro-cultured CTB cells exposed to OGD. Loss of placental-specific mTOR function (mTOR-KOplacenta) in a mouse model leads to reduced placental function and impaired metabolic health of the offspring.97 In conclusion, these phenotypes suggest that autophagy deficiency can cause the IUGR by impairing nutrient supply and placental function.
Gestational diabetes mellitus
Gestational diabetes mellitus (GDM) is a type of diabetes that occurs during pregnancy, affecting both the host and the fetus. It alters fetal metabolism and placental uptake of nutrients, increasing the chances of hypertensive pregnancy disorders and risks to the fetus, including macrosomia and the development of diabetes and cardiovascular disease as an adult.212 Changes in nutrient availability are often sensed by cellular autophagy, which is associated with the development of insulin resistance in hosts during gestational diabetes.213 In GDM, fetal pancreatic beta-cells and placental STBs exhibit heightened LC3 and SQSTM1 expression, while placental BECN1 decreases, suggesting autophagy inhibition.213 Lower levels of ATG7 in the placenta are associated with risk factors of GDM in a population study of Chinese women.214 KD of ATG5 in HTR8/SVneo cells exposed to high glucose levels showed decreased apoptosis and increased invasion. This suggests that a deficiency in autophagy may contribute to improved invasion capabilities.84 Increased phosphorylated mTOR and associated regulator molecules such as pAMPK were observed in the trophoblast villi of the placenta in GDM.215 Placentas from obese and GDM-affected pregnancies were shown to weigh heavier, which can lead to placental insufficiency and hypoxia resulting in poor feto-maternal exchange.216
Preeclampsia
Preeclampsia (PE) is a hypertensive disorder of pregnancy, characterized by hypertension and proteinuria around 20 weeks of gestation. The exact mechanism is not understood; however, placental dysfunction is often associated with PE217 and autophagy may contribute to the development of this syndrome. Several studies have investigated the association between autophagy-related markers and hypertensive disorders in the placenta. Studies on human placentas showed that hypertensive disorders were associated with increased levels of LC3-II and decreased levels of SQSTM1.218,219,220,221 Enhanced expression of LC3 and BECN1 was found in placental STBs and vascular endothelial cells in patients with early-onset PE.142 In preeclamptic placentas, oxidative stress initiates an overproduction of ceramides.85 This, in turn, activates the expression of TFEB, leading to an increase in lysosome biogenesis and exocytosis. Accordingly, JEG-3 trophoblast cells exposed to external ceramides display heightened autophagy activation.85 These findings suggest that elevated autophagy may be a distinguishing feature of preeclamptic placentas. However, contrasting reports of autophagy impairment with higher SQSTM1 expression in PE-placentas have been described.222,223 In a placenta-specific Atg7 KO mouse, which shows pregnancy-specific hypertension symptoms, an increase in SQSTM1 was observed in parietal trophoblast giant cells and the spongiotrophoblast layer of the placenta.82 These authors also observed lower levels of TFEB expression in both the labyrinth and junctional zone and high protein aggregates in the junctional zone.78 These findings suggest impaired autophagy and lysosomal function in PE placentas, contributing to protein aggregate accumulation. Although maternal blood pressure increased in the KO model, no proteinuria or IUGR was observed in the dams.82 Another autophagy regulator, PKCβ, a serine/threonine protein kinase, is downregulated in human PE placentas.86 Inhibition of PKCβ resulted in enhanced autophagic flux and an angiogenic imbalance in mouse placentas, which could be reversed by autophagic inhibition with 3-methyladenine (3-MA) treatment.86
Excessive protein aggregates were observed in trophoblast layers of villi and EVTs in placenta tissues from patients with PE.78 Lysosomes are key regulators for the clearance of protein aggregates. In the comparison of trophoblast cells from PE placentas to those of placentas from normal pregnancy and gestational age-matched deliveries, lysosomal markers LAMP1 and LAMP2 were notably absent and TFEB protein levels and nuclear translocation were reduced in trophoblast and anchoring EVTs.78 Studies from preeclamptic patients revealed the presence of aggregated transthyretin (TTR) in the placental tissue,224 while serum levels were reduced.225 In addition, aggregated TTR was also found in extracellular vesicles secreted from PE placenta tissue.224 TTR is a transporter of thyroxine and retinol that supplies thyroxine for the development of the fetal central nervous system.226 This protein was previously implicated in amyloid disease and associated aggregation in Alzheimer’s disease. It is also present in human trophoblastic cells and at the implantation site in mice.227 Moreover, studies have reported the presence of TTR protein aggregates in hypoxia/reoxygenation-exposed trophoblasts and preeclamptic placentas.222 Overexpression of human TTR in transgenic mice results in aggregated TTR in the placenta and PE-like features with an excessive unfolded protein response.222 Related to the presence of protein aggregates in pathological human placentas, fragments of amyloid precursor protein (APP) along with prototype APP processing enzymes α-secretase ADAM10, β-secretases BACE1 and BACE2, and the γ-secretase presenilin-1 were all upregulated in PE, which could be detected in urine samples.228 Accumulation of unfolded and misfolded proteins can activate ER stress-response pathways, leading to placental dysfunction. This stress contributes to early-onset but not late-onset PE.229 Further investigation is necessary to better understand the relationship between ER stress, autophagy, and placental dysfunction in PE.
Spontaneous miscarriage
Spontaneous miscarriage (SM) is the loss of pregnancy before the 20th week of gestation. A number of factors such as poor decidualization, uterine abnormalities, endocrine dysfunctions, genetic defects, immunological factors, and environmental stress can be responsible for the etiology of SM.230 Autophagy can compensate for a variety of stressors during placenta development. Various studies have shown an increased expression of autophagy markers LC3, ATG5, and BECN1, along with decreased mitofusin-2 (MFN2), a protein involved in mitochondrial dynamics, in trophoblastic villi of the miscarriage placentas, indicating dysregulated autophagy in SM.231,232 Avagliano et al. demonstrated that STB from SM placentas exhibit increased LC3 levels as compared to normal placentas and a higher prevalence of autophagic structures by electron microscopy. However, additional experiments are warranted to understand the underlying cause of the high autophagy in SM.159 SM placentas also showed high levels of HIF-1α in villi and decidua, pointing to autophagy as a protector from hypoxia-induced apoptosis.159 Macrophage metabolite lysophosphatidic acid (LPA) can enhance the formation of autolysosomes, disruption of which was shown to induce SM by impairing trophoblast invasion and placenta development.99 Rapamycin may reduce the risk of miscarriage by enhancing endometrial and decidual macrophage autophagy which improved embryo absorption in SM mice models.99,100 As a result, autophagy activation might be a compensatory mechanism for mitigating SM-induced cellular stress.
Conclusions
The environment has a significant effect on reproductive processes and plays a vital role in producing healthy offspring. Metabolic decisions are intricately linked to resource availability and environmental cues requiring a careful balance. When organisms face adverse conditions, autophagy becomes a key mechanism of cellular survival. Serving as a molecular gatekeeper, autophagy facilitates cellular function and development under stress by degrading and recycling cellular components, particularly during nutrient deprivation. Its tight regulation ensures the continuity of essential cellular functions, even amidst unfavorable external conditions. Our review shows that autophagy not only functions as a homeostatic process but may act as a dynamic regulator that senses and adapts to the shifting metabolic and environmental stressors at each step in reproduction. Mechanistically, autophagy can control the timing of cell differentiation, the amount of competent germ cells and fertilized embryos, the window and efficiency of implantation, the correct development of the placenta and its function while the fetus grows, and the expulsion of a healthy offspring, ready to survive outside the host. It is even required for cellular defense during pregnancy and immediately after delivery, controlling the maternal immune system from fetal rejection and excessive inflammation and protecting the new organism from infection. Autophagy appears to play a dual role: one that balances cellular demands with resource availability while also guiding complex signaling pathways that determine cellular fate. This layered control mechanism may represent an evolved strategy where autophagy ensures reproductive success by continuously monitoring and responding to the delicate interplay between metabolic shifts and cellular stress during early pregnancy.
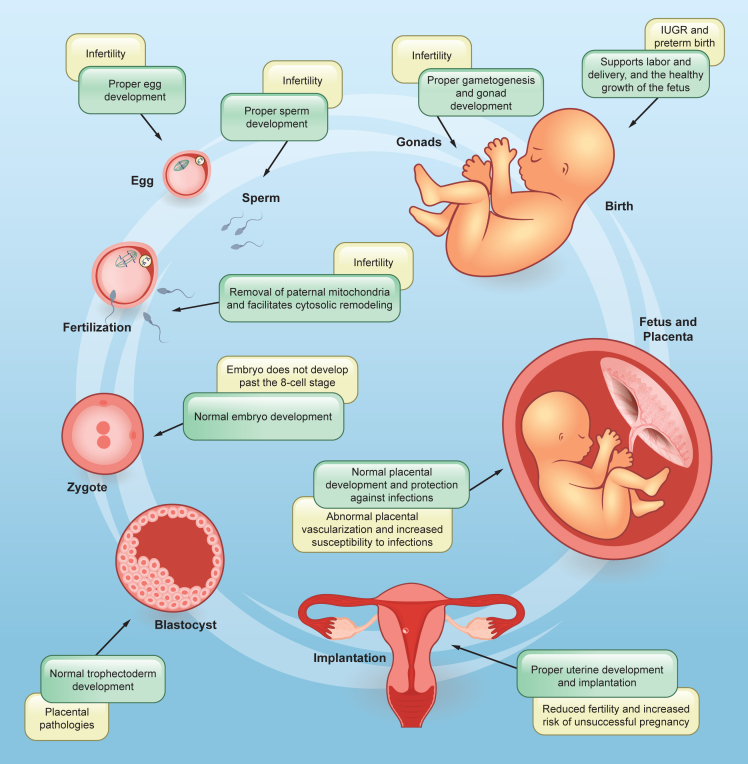
Although such a role in cell survival seems to be so essential, our review shows that autophagy is largely unexplored and understudied in the field of reproduction (Figure 4). Here are some examples of research gaps that, when addressed, could have a significant impact on the field: 1) Whether research has been conducted using only a few classical ATG genes (ATG5, ATG7, and BECN1) or in a handful of specific cell types, the picture is far from complete. A combination of tools and models must be used to address autophagy systematically, taking into account the developmental timing and cell type of each phenotype, the role of early events (autophagosome formation and LC3 conjugation) as well as late players (lysosomes); 2) The lack of good models to study the different aspects of reproduction in vitro or the challenges to properly study early stages of implantation and placental and fetal development in humans make this particular field slower than others where autophagy has been explored in much more detail (e.g., neurodegeneration). Developing new in vitro models that ethically and practically allow researchers to study this phenomenon will certainly solve this dilemma. In the meantime, animal models remain an invaluable tool; 3) A significant role for reproductive hormones in regulating metabolism and autophagy in the gonads, as well as proper steroid formation, needs further exploration, given mouse models with clear infertility phenotypes; 4) Similarly, selective autophagy mechanisms and the different cargo receptors largely identified in the recent years have not been explored in the context of reproductive cells. Studies involving the silencing of such specific pathways may provide mechanistic insights into how autophagy of cytosolic content can cause some of the phenotypes described in this review; 5) In vitro studies using placental cell lines and in vivo studies using mouse models have demonstrated the involvement of autophagy in regulating placental development and function. However, most studies focus on specific time points or pathological conditions, providing limited insight into the dynamic changes in autophagy during the entire course of placental development. Research involving human placental samples from different gestational stages and other animal models, including non-human primates, is necessary to validate the findings observed in experimental models; and 6) There has been limited progress in understanding how autophagy (and its associated pathways) function during delivery, the most physiologically stressful event of a pregnancy. The use of animal models to enhance labor and delivery interrogation, combined with cellular approaches to study placenta-uterine communication, cervical remodeling, placental detachment, and uterine involution, presents promising research directions. Additionally, clinical and epidemiological studies that identify autophagy-specific disease patterns, risk factors, and causal relationships to be tested in the laboratory could further help bridge existing knowledge gaps.
Figure 4.
Key roles of autophagy in the different stages of pregnancy and associated pathologies
The process of pregnancy requires strict mechanisms in order to ensure quality gametes for fertilization, successful implantation, formation of a healthy placenta and fetus, and delivery. Alterations in autophagy pathway genes are associated with various abnormalities observed at different stages of pregnancy.
It is still unclear whether autophagy plays a role in pathogenesis or is a potential target for several major pregnancy disorders. The reason for this is that autophagy appears to play a dual role, a characteristic that has long been observed in cancer.233,234,235 In conditions such as placenta accreta spectrum and preterm birth, it may contribute to the pathology, while in others, it can be a response to cellular stress and damage caused by other mechanisms failing. The complexity of interpretation is a result of the intricate nature of each disorder: its manifestation and degree depend on a number of variables, including timing, specific cellular failure or environmental triggers, and the inter-organ or systemic response to the failure. It may be possible to begin studying autophagy’s role in specific diseases by mechanistically examining the pathogenesis of the disease, using animal and human cell models that use conditional KO lines, autophagy reporters, and activators and inhibitors to test different stressors.197,198,199 Different cells control autophagy levels under different stress conditions. The placenta, for example, has evolved to proliferate and differentiate under hypoxic conditions where other cells may struggle. Recent studies have demonstrated the complexity of autophagy regulation, and its connection to cell differentiation, intracellular pathways, and apoptosis, highlighting the need to study this pathway not in isolation, but as a whole cell biology phenomenon that spans a cell’s entire life cycle. The methods for evaluating autophagy, or the way samples are handled, can significantly impact the consistency of data. Future studies should focus on large scale studies and well-designed protocols to address the challenging questions in the field.
While many questions remain unaddressed, there are promising efforts being made in this field to demystify the role of autophagy in pregnancy-related diseases. The emergence of several in vitro organoid techniques, the improvement of cell lines and models to mimic human reproductive cells, the advancement of non-invasive techniques for monitoring pregnancy, and the invaluable resource that several animal models continue to provide for scientists make this field fertile soil for more discoveries and innovative approaches to therapies and diagnostic tools that could revolutionize the future of the reproductive biology field. Although numerous clinical trials are actively investigating various drug targets within the autophagy pathway for various diseases,236 the potential to apply this approach within the context of pregnancy, reproductive development, and related disorders remains largely unexplored, with a few exceptions.129 Thus, understanding the importance of these crucial phases in human life provides an opportunity to dive deeper into autophagy’s involvement in regulating these important developmental stages which may unlock new insights for future clinical trials and prevention strategies. The world will keep changing and understanding how the cell adapts to each new environment, no matter the stage in the reproductive cycle, could be the solution to the next generation’s problems.
Acknowledgments
We thank members of the Guardia’s lab and Drs. Carmen Williams and Martin Estermann for helpful discussions, and Paul Windsor from the NIEHS Office of Communications and Public Liaison for assistance with figures. As space limitations and our focus on the most recent research in the field prevented us from including all important studies, we apologize for any omissions. We thank NIH Fellows Editorial Board for reviewing the article. This work was supported by the Intramural Program of NIEHS (ZIA ES103370-01).
Author contributions
Conceptualization: A.S. and C.M.G.; writing – original draft: A.S., M.L.P, O.K, E.P.-B., and C.M.G.; writing – review and editing: A.S., M.L.P, and C.M.G.; visualization: M.L.P. and C.M.G.; project administration and supervision: C.M.G.; funding acquisition: C.M.G. All authors have read and agreed to the published version of the article.
Declaration of interests
All authors declare no competing interests.
References
- 1.Michod R.E., Levin B.R. The evolution of sex: an examination of current ideas. Sinauer Associates, Incorporated; 1988. [Google Scholar]
- 2.de Visser J.A.G.M., Elena S.F. The evolution of sex: empirical insights into the roles of epistasis and drift. Nat. Rev. Genet. 2007;8:139–149. doi: 10.1038/nrg1985. [DOI] [PubMed] [Google Scholar]
- 3.Otto S.P., Lenormand T. Resolving the paradox of sex and recombination. Nat. Rev. Genet. 2002;3:252–261. doi: 10.1038/nrg761. [DOI] [PubMed] [Google Scholar]
- 4.Otto S.P. The advantages of segregation and the evolution of sex. Genetics. 2003;164:1099–1118. doi: 10.1093/genetics/164.3.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harshman L.G., Zera A.J. The cost of reproduction: the devil in the details. Trends Ecol. Evol. 2007;22:80–86. doi: 10.1016/j.tree.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Saldívar-Lemus Y., Macías Garcia C. Conflict and the evolution of viviparity in vertebrates. Behav. Ecol. Sociobiol. 2022;76:68. doi: 10.1007/s00265-022-03171-z. [DOI] [Google Scholar]
- 7.Blackburn D.G. Evolution of vertebrate viviparity and specializations for fetal nutrition: a quantitative and qualitative analysis. J. Morphol. 2015;276:961–990. doi: 10.1002/jmor.20272. [DOI] [PubMed] [Google Scholar]
- 8.Spencer T.E. Early pregnancy: concepts, challenges, and potential solutions. Anim. Front. 2013;3:48–55. doi: 10.2527/af.2013-0033. [DOI] [Google Scholar]
- 9.Mader S.L., Libal N.L., Pritchett-Corning K., Yang R., Murphy S.J. Refining timed pregnancies in two strains of genetically engineered mice. Lab. Anim. 2009;38:305–310. doi: 10.1038/laban0909-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu L., Chen Y., Tooze S.A. Autophagy pathway: Cellular and molecular mechanisms. Autophagy. 2018;14:207–215. doi: 10.1080/15548627.2017.1378838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bento C.F., Renna M., Ghislat G., Puri C., Ashkenazi A., Vicinanza M., Menzies F.M., Rubinsztein D.C. Mammalian autophagy: how does it work? Annu. Rev. Biochem. 2016;85:685–713. doi: 10.1146/annurev-biochem-060815-014556. [DOI] [PubMed] [Google Scholar]
- 12.Melia T.J., Lystad A.H., Simonsen A. Autophagosome biogenesis: From membrane growth to closure. J. Cell Biol. 2020;219 doi: 10.1083/jcb.202002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J., Kundu M., Viollet B., Guan K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russell R.C., Tian Y., Yuan H., Park H.W., Chang Y.Y., Kim J., Kim H., Neufeld T.P., Dillin A., Guan K.L. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ylä-Anttila P., Vihinen H., Jokitalo E., Eskelinen E.-L. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 16.Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., Ktistakis N.T. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 18.Itakura E., Kishi C., Inoue K., Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell. 2008;19:5360–5372. doi: 10.1091/mbc.e08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Obara K., Sekito T., Ohsumi Y. Assortment of phosphatidylinositol 3-kinase complexes--Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol. Biol. Cell. 2006;17:1527–1539. doi: 10.1091/mbc.e05-09-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimizu T., Tamura N., Nishimura T., Saito C., Yamamoto H., Mizushima N. Comprehensive analysis of autophagic functions of WIPI family proteins and their implications for the pathogenesis of β-propeller associated neurodegeneration. Hum. Mol. Genet. 2023;32:2623–2637. doi: 10.1093/hmg/ddad096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Almannai M., Marafi D., El-Hattab A.W. WIPI proteins: Biological functions and related syndromes. Front. Mol. Neurosci. 2022;15 doi: 10.3389/fnmol.2022.1011918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valverde D.P., Yu S., Boggavarapu V., Kumar N., Lees J.A., Walz T., Reinisch K.M., Melia T.J. ATG2 transports lipids to promote autophagosome biogenesis. J. Cell Biol. 2019;218:1787–1798. doi: 10.1083/jcb.201811139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghanbarpour A., Valverde D.P., Melia T.J., Reinisch K.M. A model for a partnership of lipid transfer proteins and scramblases in membrane expansion and organelle biogenesis. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2101562118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y.E., Wang Y., Du X., Zhang T., Mak H.Y., Hancock S.E., McEwen H., Pandzic E., Whan R.M., Aw Y.C., et al. TMEM41B and VMP1 are scramblases and regulate the distribution of cholesterol and phosphatidylserine. J. Cell Biol. 2021;220 doi: 10.1083/jcb.202103105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guardia C.M., Tan X.-F., Lian T., Rana M.S., Zhou W., Christenson E.T., Lowry A.J., Faraldo-Gómez J.D., Bonifacino J.S., Jiang J., Banerjee A. Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maeda S., Yamamoto H., Kinch L.N., Garza C.M., Takahashi S., Otomo C., Grishin N.V., Forli S., Mizushima N., Otomo T. Structure, lipid scrambling activity and role in autophagosome formation of ATG9A. Nat. Struct. Mol. Biol. 2020;27:1194–1201. doi: 10.1038/s41594-020-00520-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matoba K., Kotani T., Tsutsumi A., Tsuji T., Mori T., Noshiro D., Sugita Y., Nomura N., Iwata S., Ohsumi Y., et al. Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat. Struct. Mol. Biol. 2020;27:1185–1193. doi: 10.1038/s41594-020-00518-w. [DOI] [PubMed] [Google Scholar]
- 28.Feng Y., Backues S.K., Baba M., Heo J.-M., Harper J.W., Klionsky D.J. Phosphorylation of Atg9 regulates movement to the phagophore assembly site and the rate of autophagosome formation. Autophagy. 2016;12:648–658. doi: 10.1080/15548627.2016.1157237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawa-Makarska J., Baumann V., Coudevylle N., von Bülow S., Nogellova V., Abert C., Schuschnig M., Graef M., Hummer G., Martens S. Reconstitution of autophagosome nucleation defines Atg9 vesicles as seeds for membrane formation. Science. 2020;369 doi: 10.1126/science.aaz7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olivas T.J., Wu Y., Yu S., Luan L., Choi P., Guinn E.D., Nag S., De Camilli P.V., Gupta K., Melia T.J. ATG9 vesicles comprise the seed membrane of mammalian autophagosomes. J. Cell Biol. 2023;222 doi: 10.1083/jcb.202208088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graef M., Friedman J.R., Graham C., Babu M., Nunnari J. ER exit sites are physical and functional core autophagosome biogenesis components. Mol. Biol. Cell. 2013;24:2918–2931. doi: 10.1091/mbc.E13-07-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., Hiraoka Y., et al. Autophagosomes form at ER–mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 33.Biazik J., Ylä-Anttila P., Vihinen H., Jokitalo E., Eskelinen E.-L. Ultrastructural relationship of the phagophore with surrounding organelles. Autophagy. 2015;11:439–451. doi: 10.1080/15548627.2015.1017178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nascimbeni A.C., Giordano F., Dupont N., Grasso D., Vaccaro M.I., Codogno P., Morel E. ER–plasma membrane contact sites contribute to autophagosome biogenesis by regulation of local PI 3P synthesis. EMBO J. 2017;36:2018–2033. doi: 10.15252/embj.201797006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puri C., Vicinanza M., Ashkenazi A., Gratian M.J., Zhang Q., Bento C.F., Renna M., Menzies F.M., Rubinsztein D.C. The RAB11A-positive compartment is a primary platform for autophagosome assembly mediated by WIPI2 recognition of PI3P-RAB11A. Dev. Cell. 2018;45:114–131.e8. doi: 10.1016/j.devcel.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ravikumar B., Moreau K., Jahreiss L., Puri C., Rubinsztein D.C. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puri C., Gratian M.J., Rubinsztein D.C. Mammalian autophagosomes form from finger-like phagophores. Dev. Cell. 2023;58:2746–2760.e5. doi: 10.1016/j.devcel.2023.08.016. [DOI] [PubMed] [Google Scholar]
- 38.Johansen T., Lamark T. Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors. J. Mol. Biol. 2020;432:80–103. doi: 10.1016/j.jmb.2019.07.016. [DOI] [PubMed] [Google Scholar]
- 39.Kriegenburg F., Ungermann C., Reggiori F. Coordination of autophagosome–lysosome fusion by ATG8 family members. Curr. Biol. 2018;28:R512–R518. doi: 10.1016/j.cub.2018.02.034. [DOI] [PubMed] [Google Scholar]
- 40.Bieber A., Capitanio C., Erdmann P.S., Fiedler F., Beck F., Lee C.-W., Li D., Hummer G., Schulman B.A., Baumeister W., Wilfling F. In situ structural analysis reveals membrane shape transitions during autophagosome formation. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2209823119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gatica D., Lahiri V., Klionsky D.J. Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 2018;20:233–242. doi: 10.1038/s41556-018-0037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adriaenssens E., Ferrari L., Martens S. Orchestration of selective autophagy by cargo receptors. Curr. Biol. 2022;32:R1357–R1371. doi: 10.1016/j.cub.2022.11.002. [DOI] [PubMed] [Google Scholar]
- 43.Kirkin V., Rogov V.V. A diversity of selective autophagy receptors determines the specificity of the autophagy pathway. Mol. Cell. 2019;76:268–285. doi: 10.1016/j.molcel.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 44.Kirkin V. History of the selective autophagy research: how did it begin and where does it stand today? J. Mol. Biol. 2020;432:3–27. doi: 10.1016/j.jmb.2019.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamano K., Matsuda N., Tanaka K. The ubiquitin signal and autophagy: an orchestrated dance leading to mitochondrial degradation. EMBO Rep. 2016;17:300–316. doi: 10.15252/embr.201541486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Novak I., Kirkin V., McEwan D.G., Zhang J., Wild P., Rozenknop A., Rogov V., Löhr F., Popovic D., Occhipinti A., et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bunker E.N., Le Guerroué F., Wang C., Strub M.P., Werner A., Tjandra N., Youle R.J. Nix interacts with WIPI2 to induce mitophagy. EMBO J. 2023;42 doi: 10.15252/embj.2023113491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knorr R.L., Lipowsky R., Dimova R. Autophagosome closure requires membrane scission. Autophagy. 2015;11:2134–2137. doi: 10.1080/15548627.2015.1091552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takahashi Y., He H., Tang Z., Hattori T., Liu Y., Young M.M., Serfass J.M., Chen L., Gebru M., Chen C., et al. An autophagy assay reveals the ESCRT-III component CHMP2A as a regulator of phagophore closure. Nat. Commun. 2018;9:2855. doi: 10.1038/s41467-018-05254-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang W., Chen X., Ji C., Zhang W., Song J., Li J., Wang J. Key regulators of autophagosome closure. Cells. 2021;10:2814. doi: 10.3390/cells10112814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fader C.M., Sánchez D., Furlán M., Colombo M.I. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 2008;9:230–250. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- 52.Ponpuak M., Mandell M.A., Kimura T., Chauhan S., Cleyrat C., Deretic V. Secretory autophagy. Curr. Opin. Cell Biol. 2015;35:106–116. doi: 10.1016/j.ceb.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu L., McPhee C.K., Zheng L., Mardones G.A., Rong Y., Peng J., Mi N., Zhao Y., Liu Z., Wan F., et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wada Y., Sun-Wada G.H., Kawamura N., Aoyama M. Role of autophagy in embryogenesis. Curr. Opin. Genet. Dev. 2014;27:60–66. doi: 10.1016/j.gde.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 55.Tatsumi T., Tsukamoto S. Autophagy in Health and Disease. Elsevier; 2022. Role of autophagy in embryogenesis; pp. 113–123. [DOI] [Google Scholar]
- 56.Huang Q., Liu Y., Zhang S., Yap Y.T., Li W., Zhang D., Gardner A., Zhang L., Song S., Hess R.A., Zhang Z. Autophagy core protein ATG5 is required for elongating spermatid development, sperm individualization and normal fertility in male mice. Autophagy. 2021;17:1753–1767. doi: 10.1080/15548627.2020.1783822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang H., Wan H., Li X., Liu W., Chen Q., Wang Y., Yang L., Tang H., Zhang X., Duan E., et al. Atg7 is required for acrosome biogenesis during spermatogenesis in mice. Cell Res. 2014;24:852–869. doi: 10.1038/cr.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shang Y., Wang H., Jia P., Zhao H., Liu C., Liu W., Song Z., Xu Z., Yang L., Wang Y., Li W. Autophagy regulates spermatid differentiation via degradation of PDLIM1. Autophagy. 2016;12:1575–1592. doi: 10.1080/15548627.2016.1192750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao F., Li G., Liu C., Gao H., Wang H., Liu W., Chen M., Shang Y., Wang L., Shi J., et al. Autophagy regulates testosterone synthesis by facilitating cholesterol uptake in Leydig cells. J. Cell Biol. 2018;217:2103–2119. doi: 10.1083/jcb.201710078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gawriluk T.R., Hale A.N., Flaws J.A., Dillon C.P., Green D.R., Rucker E.B. Autophagy is a cell survival program for female germ cells in the murine ovary. Reproduction. 2011;141:759–765. doi: 10.1530/REP-10-0489. [DOI] [PubMed] [Google Scholar]
- 61.Song Z.H., Yu H.Y., Wang P., Mao G.K., Liu W.X., Li M.N., Wang H.N., Shang Y.L., Liu C., Xu Z.L., et al. Germ cell-specific Atg7 knockout results in primary ovarian insufficiency in female mice. Cell Death Dis. 2015;6:e1589. doi: 10.1038/cddis.2014.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shao T., Ke H., Liu R., Xu L., Han S., Zhang X., Dang Y., Jiao X., Li W., Chen Z.-J., et al. Autophagy regulates differentiation of ovarian granulosa cells through degradation of WT1. Autophagy. 2022;18:1864–1878. doi: 10.1080/15548627.2021.2005415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gawriluk T.R., Ko C., Hong X., Christenson L.K., Rucker E.B., 3rd Beclin-1 deficiency in the murine ovary results in the reduction of progesterone production to promote preterm labor. Proc. Natl. Acad. Sci. USA. 2014;111:E4194–E4203. doi: 10.1073/pnas.1409323111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsukamoto S., Kuma A., Mizushima N. The role of autophagy during the oocyte-to-embryo transition. Autophagy. 2008;4:1076–1078. doi: 10.4161/auto.7065. [DOI] [PubMed] [Google Scholar]
- 65.Lee J.-E., Oh H.-A., Song H., Jun J.H., Roh C.-R., Xie H., Dey S.K., Lim H.J. Autophagy regulates embryonic survival during delayed implantation. Endocrinology. 2011;152:2067–2075. doi: 10.1210/en.2010-1456. [DOI] [PubMed] [Google Scholar]
- 66.Bulut-Karslioglu A., Biechele S., Jin H., Macrae T.A., Hejna M., Gertsenstein M., Song J.S., Ramalho-Santos M. Inhibition of mTOR induces a paused pluripotent state. Nature. 2016;540:119–123. doi: 10.1038/nature20578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oestreich A.K., Chadchan S.B., Medvedeva A., Lydon J.P., Jungheim E.S., Moley K.H., Kommagani R. The autophagy protein, FIP200 (RB1CC1) mediates progesterone responses governing uterine receptivity and decidualization. Biol. Reprod. 2020;102:843–851. doi: 10.1093/biolre/ioz234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oestreich A.K., Chadchan S.B., Popli P., Medvedeva A., Rowen M.N., Stephens C.S., Xu R., Lydon J.P., Demayo F.J., Jungheim E.S., et al. The autophagy gene Atg16L1 is necessary for endometrial decidualization. Endocrinology. 2020;161 doi: 10.1210/endocr/bqz039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Citrinovitz A.C.M., Strowitzki T., Germeyer A. Decreased autophagy impairs decidualization of human endometrial stromal cells: a role for ATG proteins in endomtrial physiology. Int. J. Mol. Sci. 2019;20:3066. doi: 10.3390/ijms20123066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Popli P., Tang S., Chadchan S.B., Talwar C., Rucker E.B., 3rd, Guan X., Monsivais D., Lydon J.P., Stallings C.L., Moley K.H., et al. Beclin-1-dependent autophagy, but not apoptosis, is critical for stem-cell-mediated endometrial programming and the establishment of pregnancy. Dev. Cell. 2023;58:885–897.e4. doi: 10.1016/j.devcel.2023.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nakashima A., Yamanaka-Tatematsu M., Fujita N., Koizumi K., Shima T., Yoshida T., Nikaido T., Okamoto A., Yoshimori T., Saito S. Impaired autophagy by soluble endoglin, under physiological hypoxia in early pregnant period, is involved in poor placentation in preeclampsia. Autophagy. 2013;9:303–316. doi: 10.4161/auto.22927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fujita N., Hayashi-Nishino M., Fukumoto H., Omori H., Yamamoto A., Noda T., Yoshimori T. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol. Biol. Cell. 2008;19:4651–4659. doi: 10.1091/mbc.e08-03-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamanaka-Tatematsu M., Nakashima A., Fujita N., Shima T., Yoshimori T., Saito S. Autophagy induced by HIF1α overexpression supports trophoblast invasion by supplying cellular energy. PLoS One. 2013;8 doi: 10.1371/journal.pone.0076605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Furuta A., Shima T., Kawaguchi M., Yamaki-Ushijima A., Yasuda I., Tsuda S., Yoneda S., Higashisaka K., Cheng S.-B., Matsumoto K., et al. The Autophagy-Lysosomal Machinery Enhances Cytotrophoblast–Syncytiotrophoblast Fusion Process. Reprod. Med. (Basel) 2022;3:112–126. doi: 10.3390/reprodmed3020010. [DOI] [Google Scholar]
- 75.Shao X., Cao G., Chen D., Liu J., Yu B., Liu M., Li Y.-X., Cao B., Sadovsky Y., Wang Y.-L. Placental trophoblast syncytialization potentiates macropinocytosis via mTOR signaling to adapt to reduced amino acid supply. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2017092118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bastida-Ruiz D., Yart L., Wuillemin C., Ribaux P., Morris N., Epiney M., Martinez de Tejada B., Cohen M. The fine-tuning of endoplasmic reticulum stress response and autophagy activation during trophoblast syncytialization. Cell Death Dis. 2019;10:651. doi: 10.1038/s41419-019-1905-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Steingrímsson E., Tessarollo L., Reid S.W., Jenkins N.A., Copeland N.G. The bHLH-Zip transcription factor Tfeb is essential for placental vascularization. Development. 1998;125:4607–4616. doi: 10.1242/dev.125.23.4607. [DOI] [PubMed] [Google Scholar]
- 78.Nakashima A., Cheng S.-B., Ikawa M., Yoshimori T., Huber W.J., Menon R., Huang Z., Fierce J., Padbury J.F., Sadovsky Y., et al. Evidence for lysosomal biogenesis proteome defect and impaired autophagy in preeclampsia. Autophagy. 2020;16:1771–1785. doi: 10.1080/15548627.2019.1707494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cao B., Macones C., Mysorekar I.U. ATG16L1 governs placental infection risk and preterm birth in mice and women. JCI Insight. 2016;1 doi: 10.1172/jci.insight.86654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cao B., Parnell L.A., Diamond M.S., Mysorekar I.U. Inhibition of autophagy limits vertical transmission of Zika virus in pregnant mice. J. Exp. Med. 2017;214:2303–2313. doi: 10.1084/jem.20170957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McKinney J.R., Seferovic M.D., Major A.M., Suter M.A., Tardif S.D., Patterson J.L., Castro E.C.C., Aagaard K.M. Placental Autophagy and Viral Replication Co-localize in Human and Non-human Primate Placentae Following Zika Virus Infection: Implications for Therapeutic Interventions. Front. Virol. 2021;1 doi: 10.3389/fviro.2021.720760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aoki A., Nakashima A., Kusabiraki T., Ono Y., Yoshino O., Muto M., Kumasawa K., Yoshimori T., Ikawa M., Saito S. Trophoblast-specific conditional Atg7 knockout mice develop gestational hypertension. Am. J. Pathol. 2018;188:2474–2486. doi: 10.1016/j.ajpath.2018.07.021. [DOI] [PubMed] [Google Scholar]
- 83.Muralimanoharan S., Gao X., Weintraub S., Myatt L., Maloyan A. Sexual dimorphism in activation of placental autophagy in obese women with evidence for fetal programming from a placenta-specific mouse model. Autophagy. 2016;12:752–769. doi: 10.1080/15548627.2016.1156822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ji L., Chen Z., Xu Y., Xiong G., Liu R., Wu C., Hu H., Wang L. Systematic characterization of autophagy in gestational diabetes mellitus. Endocrinology. 2017;158:2522–2532. doi: 10.1210/en.2016-1922. [DOI] [PubMed] [Google Scholar]
- 85.Melland-Smith M., Ermini L., Chauvin S., Craig-Barnes H., Tagliaferro A., Todros T., Post M., Caniggia I. Disruption of sphingolipid metabolism augments ceramide-induced autophagy in preeclampsia. Autophagy. 2015;11:653–669. doi: 10.1080/15548627.2015.1034414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhao H., Gong L., Wu S., Jing T., Xiao X., Cui Y., Xu H., Lu H., Tang Y., Zhang J., et al. The inhibition of protein kinase c β contributes to the pathogenesis of preeclampsia by activating autophagy. EBioMedicine. 2020;56 doi: 10.1016/j.ebiom.2020.102813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 88.Efeyan A., Zoncu R., Chang S., Gumper I., Snitkin H., Wolfson R.L., Kirak O., Sabatini D.D., Sabatini D.M. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 2013;493:679–683. doi: 10.1038/nature11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sou Y.-s., Waguri S., Iwata J.-i., Ueno T., Fujimura T., Hara T., Sawada N., Yamada A., Mizushima N., Uchiyama Y., et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol. Biol. Cell. 2008;19:4762–4775. doi: 10.1091/mbc.e08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Saitoh T., Fujita N., Hayashi T., Takahara K., Satoh T., Lee H., Matsunaga K., Kageyama S., Omori H., Noda T., et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl. Acad. Sci. USA. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kojima T., Yamada T., Akaishi R., Furuta I., Saitoh T., Nakabayashi K., Nakayama K.I., Nakayama K., Akira S., Minakami H. Role of the Atg9a gene in intrauterine growth and survival of fetal mice. Reprod. Biol. 2015;15:131–138. doi: 10.1016/j.repbio.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 93.Malhotra R., Warne J.P., Salas E., Xu A.W., Debnath J. Loss of Atg12, but not Atg5, in pro-opiomelanocortin neurons exacerbates diet-induced obesity. Autophagy. 2015;11:145–154. doi: 10.1080/15548627.2014.998917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Saitoh T., Fujita N., Jang M.H., Uematsu S., Yang B.-G., Satoh T., Omori H., Noda T., Yamamoto N., Komatsu M., et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 95.Yue Z., Jin S., Yang C., Levine A.J., Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hung T.H., Wu C.P., Chen S.F. Differential Changes in Akt and AMPK Phosphorylation Regulating mTOR Activity in the Placentas of Pregnancies Complicated by Fetal Growth Restriction and Gestational Diabetes Mellitus With Large-For-Gestational Age Infants. Front. Med. 2021;8 doi: 10.3389/fmed.2021.788969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Akhaphong B., Baumann D.C., Beetch M., Lockridge A.D., Jo S., Wong A., Zemanovic T., Mohan R., Fondevilla D.L., Sia M., et al. Placental mTOR complex 1 regulates fetal programming of obesity and insulin resistance in mice. JCI Insight. 2021;6 doi: 10.1172/jci.insight.149271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hung T.-H., Hsieh T.T., Wu C.P., Li M.J., Yeh Y.L., Chen S.F. Mammalian target of rapamycin signaling is a mechanistic link between increased endoplasmic reticulum stress and autophagy in the placentas of pregnancies complicated by growth restriction. Placenta. 2017;60:9–20. doi: 10.1016/j.placenta.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 99.Yang H.-L., Lai Z.-Z., Shi J.-W., Zhou W.-J., Mei J., Ye J.-F., Zhang T., Wang J., Zhao J.-Y., Li D.-J., Li M.Q. A defective lysophosphatidic acid-autophagy axis increases miscarriage risk by restricting decidual macrophage residence. Autophagy. 2022;18:2459–2480. doi: 10.1080/15548627.2022.2039000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lu H., Yang H.-L., Zhou W.-J., Lai Z.-Z., Qiu X.-M., Fu Q., Zhao J.-Y., Wang J., Li D.-J., Li M.-Q. Rapamycin prevents spontaneous abortion by triggering decidual stromal cell autophagy-mediated NK cell residence. Autophagy. 2021;17:2511–2527. doi: 10.1080/15548627.2020.1833515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang M., Xu Y., Zhang Y., Chen Y., Chang G., An G., Yang X., Zheng C., Zhao J., Liu Z., et al. Deciphering the autophagy regulatory network via single-cell transcriptome analysis reveals a requirement for autophagy homeostasis in spermatogenesis. Theranostics. 2021;11:5010–5027. doi: 10.7150/thno.55645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McClellan K.A., Gosden R., Taketo T. Continuous loss of oocytes throughout meiotic prophase in the normal mouse ovary. Dev. Biol. 2003;258:334–348. doi: 10.1016/s0012-1606(03)00132-5. [DOI] [PubMed] [Google Scholar]
- 103.Pepling M.E., Spradling A.C. Mouse ovarian germ cell cysts undergo programmed breakdown to form primordial follicles. Dev. Biol. 2001;234:339–351. doi: 10.1006/dbio.2001.0269. [DOI] [PubMed] [Google Scholar]
- 104.Sun Y.-C., Wang Y.-Y., Sun X.-F., Cheng S.-F., Li L., Zhao Y., Shen W., Chen H. The role of autophagy during murine primordial follicle assembly. Aging (Albany NY) 2018;10:197–211. doi: 10.18632/aging.101376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Choi J., Jo M., Lee E., Choi D. Induction of apoptotic cell death via accumulation of autophagosomes in rat granulosa cells. Fertil. Steril. 2011;95:1482–1486. doi: 10.1016/j.fertnstert.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 106.Choi J., Jo M., Lee E., Choi D. AKT is involved in granulosa cell autophagy regulation via mTOR signaling during rat follicular development and atresia. Reproduction. 2014;147:73–80. doi: 10.1530/REP-13-0386. [DOI] [PubMed] [Google Scholar]
- 107.Serke H., Vilser C., Nowicki M., Hmeidan F.A., Blumenauer V., Hummitzsch K., Lösche A., Spanel-Borowski K. Granulosa cell subtypes respond by autophagy or cell death to oxLDL-dependent activation of the oxidized lipoprotein receptor 1 and toll-like 4 receptor. Autophagy. 2009;5:991–1003. doi: 10.4161/auto.5.7.9507. [DOI] [PubMed] [Google Scholar]
- 108.Yuan J., Zhang Y., Sheng Y., Fu X., Cheng H., Zhou R. MYBL2 guides autophagy suppressor VDAC2 in the developing ovary to inhibit autophagy through a complex of VDAC2-BECN1-BCL2L1 in mammals. Autophagy. 2015;11:1081–1098. doi: 10.1080/15548627.2015.1040970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zaffagnini G., Cheng S., Salzer M.C., Pernaute B., Duran J.M., Irimia M., Schuh M., Böke E. Mouse oocytes sequester aggregated proteins in degradative super-organelles. Cell. 2024;187:1109–1126.e21. doi: 10.1016/j.cell.2024.01.031. [DOI] [PubMed] [Google Scholar]
- 110.Ding Y., Zhu Q., He Y., Lu Y., Wang Y., Qi J., Wu H., Xu R., Li J., Li X., Sun Y. Induction of autophagy by Beclin-1 in granulosa cells contributes to follicular progesterone elevation in ovarian endometriosis. Transl. Res. 2021;227:15–29. doi: 10.1016/j.trsl.2020.06.013. [DOI] [PubMed] [Google Scholar]
- 111.Esmaeilian Y., Hela F., Bildik G., İltumur E., Yusufoglu S., Yildiz C.S., Yakin K., Kordan Y., Oktem O. Autophagy regulates sex steroid hormone synthesis through lysosomal degradation of lipid droplets in human ovary and testis. Cell Death Dis. 2023;14:342. doi: 10.1038/s41419-023-05864-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tsukamoto S., Kuma A., Murakami M., Kishi C., Yamamoto A., Mizushima N. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–120. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- 113.Sasaki T., Sato M. Degradation of paternal mitochondria via mitophagy. Biochim. Biophys. Acta Gen. Subj. 2021;1865 doi: 10.1016/j.bbagen.2021.129886. [DOI] [PubMed] [Google Scholar]
- 114.Sato M., Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011;334:1141–1144. doi: 10.1126/science.1210333. [DOI] [PubMed] [Google Scholar]
- 115.Song W.-H., Yi Y.-J., Sutovsky M., Meyers S., Sutovsky P. Autophagy and ubiquitin–proteasome system contribute to sperm mitophagy after mammalian fertilization. Proc. Natl. Acad. Sci. USA. 2016;113:E5261–E5270. doi: 10.1073/pnas.1605844113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Al Rawi S., Louvet-Vallée S., Djeddi A., Sachse M., Culetto E., Hajjar C., Boyd L., Legouis R., Galy V. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334:1144–1147. doi: 10.1126/science.1211878. [DOI] [PubMed] [Google Scholar]
- 117.Rojansky R., Cha M.-Y., Chan D.C. Elimination of paternal mitochondria in mouse embryos occurs through autophagic degradation dependent on PARKIN and MUL1. Elife. 2016;5 doi: 10.7554/eLife.17896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Luo S.-M., Ge Z.-J., Wang Z.-W., Jiang Z.-Z., Wang Z.-B., Ouyang Y.-C., Hou Y., Schatten H., Sun Q.-Y. Unique insights into maternal mitochondrial inheritance in mice. Proc. Natl. Acad. Sci. USA. 2013;110:13038–13043. doi: 10.1073/pnas.1303231110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li L., Lu X., Dean J. The maternal to zygotic transition in mammals. Mol. Aspect. Med. 2013;34:919–938. doi: 10.1016/j.mam.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Houghton F.D. Energy metabolism of the inner cell mass and trophectoderm of the mouse blastocyst. Differentiation. 2006;74:11–18. doi: 10.1111/j.1432-0436.2006.00052.x. [DOI] [PubMed] [Google Scholar]
- 121.Renfree M.B., Fenelon J.C. The enigma of embryonic diapause. Development. 2017;144:3199–3210. doi: 10.1242/dev.148213. [DOI] [PubMed] [Google Scholar]
- 122.Yamamoto A., Mizushima N., Tsukamoto S. Fertilization-induced autophagy in mouse embryos is independent of mTORC1. Biol. Reprod. 2014;91:7. doi: 10.1095/biolreprod.113.115816. [DOI] [PubMed] [Google Scholar]
- 123.Choi S., Shin H., Song H., Lim H.J. Suppression of autophagic activation in the mouse uterus by estrogen and progesterone. J. Endocrinol. 2014;221:39–50. doi: 10.1530/JOE-13-0449. [DOI] [PubMed] [Google Scholar]
- 124.Su Y., Zhang J.-J., He J.-L., Liu X.-Q., Chen X.-M., Ding Y.-B., Tong C., Peng C., Geng Y.-Q., Wang Y.-X., Gao R.F. Endometrial autophagy is essential for embryo implantation during early pregnancy. J. Mol. Med. 2020;98:555–567. doi: 10.1007/s00109-019-01849-y. [DOI] [PubMed] [Google Scholar]
- 125.Muter J., Lynch V.J., McCoy R.C., Brosens J.J. Human embryo implantation. Development. 2023;150 doi: 10.1242/dev.201507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ng S.-W., Norwitz G.A., Pavlicev M., Tilburgs T., Simón C., Norwitz E.R. Endometrial decidualization: the primary driver of pregnancy health. Int. J. Mol. Sci. 2020;21:4092. doi: 10.3390/ijms21114092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rawlings T.M., Makwana K., Taylor D.M., Molè M.A., Fishwick K.J., Tryfonos M., Odendaal J., Hawkes A., Zernicka-Goetz M., Hartshorne G.M., et al. Modelling the impact of decidual senescence on embryo implantation in human endometrial assembleds. Elife. 2021;10 doi: 10.7554/eLife.69603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li J., Zhang S., Zhang Y., Dai Y., Zhang Y., Yang A., Hong F., Pan Y. Atg9A-mediated mitophagy is required for decidual differentiation of endometrial stromal cells. Reprod. Biol. 2022;22 doi: 10.1016/j.repbio.2022.100707. [DOI] [PubMed] [Google Scholar]
- 129.Popli P., Sun A.J., Kommagani R. The multifaceted role of autophagy in endometrium homeostasis and disease. Reprod. Sci. 2022;29:1054–1067. doi: 10.1007/s43032-021-00587-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Roberts R.M., Green J.A., Schulz L.C. The evolution of the placenta. Reproduction. 2016;152:R179–R189. doi: 10.1530/REP-16-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wildman D.E., Chen C., Erez O., Grossman L.I., Goodman M., Romero R. Evolution of the mammalian placenta revealed by phylogenetic analysis. Proc. Natl. Acad. Sci. USA. 2006;103:3203–3208. doi: 10.1073/pnas.0511344103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pijnenborg R., Vercruysse L., Hanssens M. The uterine spiral arteries in human pregnancy: facts and controversies. Placenta. 2006;27:939–958. doi: 10.1016/j.placenta.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 133.Elmore S.A., Cochran R.Z., Bolon B., Lubeck B., Mahler B., Sabio D., Ward J.M. Histology atlas of the developing mouse placenta. Toxicol. Pathol. 2022;50:60–117. doi: 10.1177/01926233211042270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Burton G.J., Cindrova-Davies T., Yung H.W., Jauniaux E. Hypoxia and reproductive health: Oxygen and development of the human placenta. Reproduction. 2021;161:F53–F65. doi: 10.1530/REP-20-0153. [DOI] [PubMed] [Google Scholar]
- 135.Burton G.J., Jauniaux E., Watson A.L. Maternal arterial connections to the placental intervillous space during the first trimester of human pregnancy: the Boyd collection revisited. Am. J. Obstet. Gynecol. 1999;181:718–724. doi: 10.1016/s0002-9378(99)70518-1. [DOI] [PubMed] [Google Scholar]
- 136.Genbacev O., Zhou Y., Ludlow J.W., Fisher S.J. Regulation of human placental development by oxygen tension. Science. 1997;277:1669–1672. doi: 10.1126/science.277.5332.1669. [DOI] [PubMed] [Google Scholar]
- 137.Burton G.J. Oxygen, the Janus gas; its effects on human placental development and function. J. Anat. 2009;215:27–35. doi: 10.1111/j.1469-7580.2008.00978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chen B., Longtine M.S., Nelson D.M. Hypoxia induces autophagy in primary human trophoblasts. Endocrinology. 2012;153:4946–4954. doi: 10.1210/en.2012-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Choi J.-H., Lee H.-J., Yang T.-H., Kim G.J. Effects of hypoxia inducible factors-1α on autophagy and invasion of trophoblasts. Clin. Exp. Reprod. Med. 2012;39:73–80. doi: 10.5653/cerm.2012.39.2.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Aye I.L.M.H., Aiken C.E., Charnock-Jones D.S., Smith G.C.S. Placental energy metabolism in health and disease—significance of development and implications for preeclampsia. Am. J. Obstet. Gynecol. 2022;226:S928–S944. doi: 10.1016/j.ajog.2020.11.005. [DOI] [PubMed] [Google Scholar]
- 141.Li Z., Wang S., Li L. Advanced Oxidative Protein Products Drive Trophoblast Cells Into Senescence by Inhibiting the Autophagy: The Potential Implication of Preeclampsia. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.810282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gao L., Qi H.-B., Kamana K.C., Zhang X.-M., Zhang H., Baker P.N. Excessive autophagy induces the failure of trophoblast invasion and vasculature: possible relevance to the pathogenesis of preeclampsia. J. Hypertens. 2015;33:106–117. doi: 10.1097/HJH.0000000000000366. [DOI] [PubMed] [Google Scholar]
- 143.Chang C.-W., Wakeland A.K., Parast M.M. Trophoblast lineage specification, differentiation and their regulation by oxygen tension. J. Endocrinol. 2018;236:R43–R56. doi: 10.1530/JOE-17-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cross J.C., Werb Z., Fisher S.J. Implantation and the placenta: key pieces of the development puzzle. Science. 1994;266:1508–1518. doi: 10.1126/science.7985020. [DOI] [PubMed] [Google Scholar]
- 145.Lyden T.W., Ng A.K., Rote N.S. Modulation of phosphatidylserine epitope expression by BeWo cells during forskolin treatment. Placenta. 1993;14:177–186. doi: 10.1016/s0143-4004(05)80259-0. [DOI] [PubMed] [Google Scholar]
- 146.Vargas A., Moreau J., Landry S., LeBellego F., Toufaily C., Rassart É., Lafond J., Barbeau B. Syncytin-2 plays an important role in the fusion of human trophoblast cells. J. Mol. Biol. 2009;392:301–318. doi: 10.1016/j.jmb.2009.07.025. [DOI] [PubMed] [Google Scholar]
- 147.Toudic C., Vargas A., Xiao Y., St-Pierre G., Bannert N., Lafond J., Rassart É., Sato S., Barbeau B. Galectin-1 interacts with the human endogenous retroviral envelope protein syncytin-2 and potentiates trophoblast fusion in humans. Faseb. J. 2019;33:12873–12887. doi: 10.1096/fj.201900107R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gauster M., Maninger S., Siwetz M., Deutsch A., El-Heliebi A., Kolb-Lenz D., Hiden U., Desoye G., Herse F., Prokesch A. Downregulation of p53 drives autophagy during human trophoblast differentiation. Cell. Mol. Life Sci. 2018;75:1839–1855. doi: 10.1007/s00018-017-2695-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ballabio A., Bonifacino J.S. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 2020;21:101–118. doi: 10.1038/s41580-019-0185-4. [DOI] [PubMed] [Google Scholar]
- 150.Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P., et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gerri C., McCarthy A., Alanis-Lobato G., Demtschenko A., Bruneau A., Loubersac S., Fogarty N.M.E., Hampshire D., Elder K., Snell P., et al. Initiation of a conserved trophectoderm program in human, cow and mouse embryos. Nature. 2020;587:443–447. doi: 10.1038/s41586-020-2759-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Esbin M.N., Dahal L., Fan V.B., McKenna J., Yin E., Darzacq X., Tjian R. TFEB controls expression of human syncytins during cell–cell fusion. Genes Dev. 2024;38:718–737. doi: 10.1101/gad.351633.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zheng W., Zhang Y., Xu P., Wang Z., Shao X., Chen C., Cai H., Wang Y., Sun M.-a., Deng W., et al. TFEB safeguards trophoblast syncytialization in humans and mice. Proc. Natl. Acad. Sci. USA. 2024;121 doi: 10.1073/pnas.2404062121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cesana M., Tufano G., Panariello F., Zampelli N., Soldati C., Mutarelli M., Montefusco S., Grieco G., Sepe L.V., Rossi B., et al. TFEB controls syncytiotrophoblast formation and hormone production in placenta. Cell Death Differ. 2024;31:1–13. doi: 10.1038/s41418-024-01337-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Deretic V. Autophagy in inflammation, infection, and immunometabolism. Immunity. 2021;54:437–453. doi: 10.1016/j.immuni.2021.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Signorelli P., Avagliano L., Virgili E., Gagliostro V., Doi P., Braidotti P., Bulfamante G.P., Ghidoni R., Marconi A.M. Autophagy in term normal human placentas. Placenta. 2011;32:482–485. doi: 10.1016/j.placenta.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 157.Brickle A., Tran H.T., Lim R., Liong S., Lappas M. Autophagy, which is decreased in labouring fetal membranes, regulates IL-1β production via the inflammasome. Placenta. 2015;36:1393–1404. doi: 10.1016/j.placenta.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 158.Bawolak M.-T., François Marceau M. Vacuolar ATPase-mediated sequestration of local anesthetics in swollen macroautophagosomes. Can. J. Anesth. 2010;57:230. doi: 10.1007/s12630-009-9220-9. [DOI] [PubMed] [Google Scholar]
- 159.Avagliano L., Terraneo L., Virgili E., Martinelli C., Doi P., Samaja M., Bulfamante G.P., Marconi A.M. Autophagy in normal and abnormal early human pregnancies. Reprod. Sci. 2015;22:838–844. doi: 10.1177/1933719114565036. [DOI] [PubMed] [Google Scholar]
- 160.Doulaveris G., Orfanelli T., Benn K., Zervoudakis I., Skupski D., Witkin S.S. A polymorphism in an autophagy-related gene, ATG16L1, influences time to delivery in women with an unfavorable cervix who require labor induction. J. Perinat. Med. 2013;41:411–414. doi: 10.1515/jpm-2012-0278. [DOI] [PubMed] [Google Scholar]
- 161.Liassides C., Papadopoulos A., Siristatidis C., Damoraki G., Liassidou A., Chrelias C., Kassanos D., Giamarellos-Bourboulis E.J. Single nucleotide polymorphisms of Toll-like receptor-4 and of autophagy-related gene 16 like-1 gene for predisposition of premature delivery: A prospective study. Medicine. 2019;98 doi: 10.1097/MD.0000000000017313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wang L., Hu H., Morse A.N., Han X., Bao J., Yang J., Chen Y., Liu H. Activation of autophagy in human uterine myometrium during labor. Reprod. Sci. 2020;27:1665–1672. doi: 10.1007/s43032-020-00198-3. [DOI] [PubMed] [Google Scholar]
- 163.Chen L., Luo Y., Chen Y., Wang L., Wang X., Zhang G., Ji K., Liu H. Time Course Analysis of Transcriptome in Human Myometrium Depending on Labor Duration and Correlating With Postpartum Blood Loss. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Fu J., Warmflash A., Lutolf M.P. Stem-cell-based embryo models for fundamental research and translation. Nat. Mater. 2021;20:132–144. doi: 10.1038/s41563-020-00829-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Liu X., Polo J.M. Human blastoid as an in vitro model of human blastocysts. Curr. Opin. Genet. Dev. 2024;84 doi: 10.1016/j.gde.2023.102135. [DOI] [PubMed] [Google Scholar]
- 166.Wang L., Yan Z.-H., He T.-R., Liu H.-X., Li Y.-K., Niu Y.-L., Wang J.-J., De Felici M., Ge W., Shen W. In vitro oogenesis from murine premeiotic germ cells using a new three-dimensional culture system. Cell Death Discov. 2023;9:276. doi: 10.1038/s41420-023-01577-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kulibin A.Y., Malolina E.A. In vitro spermatogenesis: In search of fully defined conditions. Front. Cell Dev. Biol. 2023;11 doi: 10.3389/fcell.2023.1106111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Romualdez-Tan M.V. Modelling in vitro gametogenesis using induced pluripotent stem cells: a review. Cell Regen. 2023;12:33. doi: 10.1186/s13619-023-00176-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gonen N., Eozenou C., Mitter R., Elzaiat M., Stévant I., Aviram R., Bernardo A.S., Chervova A., Wankanit S., Frachon E., et al. In vitro cellular reprogramming to model gonad development and its disorders. Sci. Adv. 2023;9 doi: 10.1126/sciadv.abn9793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cherubini M., Erickson S., Haase K. Modelling the Human Placental Interface In Vitro-A Review. Micromachines. 2021;12 doi: 10.3390/mi12080884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Okae H., Toh H., Sato T., Hiura H., Takahashi S., Shirane K., Kabayama Y., Suyama M., Sasaki H., Arima T. Derivation of Human Trophoblast Stem Cells. Cell Stem Cell. 2018;22:50–63.e6. doi: 10.1016/j.stem.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 172.Nursalim Y.N.S., Groom K.M., Blenkiron C., Chamley L.W. A simple method to isolate term trophoblasts and maintain them in extended culture. Placenta. 2021;108:1–10. doi: 10.1016/j.placenta.2021.03.009. [DOI] [PubMed] [Google Scholar]
- 173.Horii M., Bui T., Touma O., Cho H.Y., Parast M.M. An Improved Two-Step Protocol for Trophoblast Differentiation of Human Pluripotent Stem Cells. Curr. Protoc. Stem Cell Biol. 2019;50:e96. doi: 10.1002/cpsc.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mischler A., Karakis V., Mahinthakumar J., Carberry C.K., San Miguel A., Rager J.E., Fry R.C., Rao B.M. Two distinct trophectoderm lineage stem cells from human pluripotent stem cells. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Karakis V., Jabeen M., Britt J.W., Cordiner A., Mischler A., Li F., San Miguel A., Rao B.M. Laminin switches terminal differentiation fate of human trophoblast stem cells under chemically defined culture conditions. J. Biol. Chem. 2023;299 doi: 10.1016/j.jbc.2023.104650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Haider S., Meinhardt G., Saleh L., Kunihs V., Gamperl M., Kaindl U., Ellinger A., Burkard T.R., Fiala C., Pollheimer J., et al. Self-Renewing Trophoblast Organoids Recapitulate the Developmental Program of the Early Human Placenta. Stem Cell Rep. 2018;11:537–551. doi: 10.1016/j.stemcr.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Turco M.Y., Gardner L., Kay R.G., Hamilton R.S., Prater M., Hollinshead M.S., McWhinnie A., Esposito L., Fernando R., Skelton H., et al. Trophoblast organoids as a model for maternal-fetal interactions during human placentation. Nature. 2018;564:263–267. doi: 10.1038/s41586-018-0753-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yang L., Semmes E.C., Ovies C., Megli C., Permar S., Gilner J.B., Coyne C.B. Innate immune signaling in trophoblast and decidua organoids defines differential antiviral defenses at the maternal-fetal interface. Elife. 2022;11 doi: 10.7554/eLife.79794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Karvas R.M., Khan S.A., Verma S., Yin Y., Kulkarni D., Dong C., Park K.M., Chew B., Sane E., Fischer L.A., et al. Stem-cell-derived trophoblast organoids model human placental development and susceptibility to emerging pathogens. Cell Stem Cell. 2022;29:810–825.e8. doi: 10.1016/j.stem.2022.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hori T., Okae H., Shibata S., Kobayashi N., Kobayashi E.H., Oike A., Sekiya A., Arima T., Kaji H. Trophoblast stem cell-based organoid models of the human placental barrier. Nat. Commun. 2024;15:962. doi: 10.1038/s41467-024-45279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Karakis V., Britt J.W., Jabeen M., Miguel A.S., Rao B.M. Derivation of human trophoblast stem cells from placentas at birth. bioRxiv. 2024 doi: 10.1101/2024.05.01.592064. Preprint at. [DOI] [Google Scholar]
- 182.Yang L., Liang P., Yang H., Coyne C.B. Trophoblast organoids with physiological polarity model placental structure and function. J. Cell Sci. 2024;137 doi: 10.1242/jcs.261528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Richardson L.S., K Kammala A., Costantine M.M., Fortunato S.J., Radnaa E., Kim S., Taylor R.N., Han A., Menon R. Testing of drugs using human feto-maternal interface organ-on-chips provide insights into pharmacokinetics and efficacy. Lab Chip. 2022;22:4574–4592. doi: 10.1039/d2lc00691j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Park J.Y., Mani S., Clair G., Olson H.M., Paurus V.L., Ansong C.K., Blundell C., Young R., Kanter J., Gordon S., et al. A microphysiological model of human trophoblast invasion during implantation. Nat. Commun. 2022;13:1252. doi: 10.1038/s41467-022-28663-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Pu Y., Gingrich J., Veiga-Lopez A. A 3-dimensional microfluidic platform for modeling human extravillous trophoblast invasion and toxicological screening. Lab Chip. 2021;21:546–557. doi: 10.1039/d0lc01013h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Li X., Kodithuwakku S.P., Chan R.W.S., Yeung W.S.B., Yao Y., Ng E.H.Y., Chiu P.C.N., Lee C.L. Three-dimensional culture models of human endometrium for studying trophoblast-endometrium interaction during implantation. Reprod. Biol. Endocrinol. 2022;20:120. doi: 10.1186/s12958-022-00973-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bentin-Ley U., Pedersen B., Lindenberg S., Larsen J.F., Hamberger L., Horn T. Isolation and culture of human endometrial cells in a three-dimensional culture system. J. Reprod. Fertil. 1994;101:327–332. doi: 10.1530/jrf.0.1010327. [DOI] [PubMed] [Google Scholar]
- 188.Wang H., Pilla F., Anderson S., Martínez-Escribano S., Herrer I., Moreno-Moya J.M., Musti S., Bocca S., Oehninger S., Horcajadas J.A. A novel model of human implantation: 3D endometrium-like culture system to study attachment of human trophoblast (Jar) cell spheroids. Mol. Hum. Reprod. 2012;18:33–43. doi: 10.1093/molehr/gar064. [DOI] [PubMed] [Google Scholar]
- 189.Arnold J.T., Lessey B.A., Seppälä M., Kaufman D.G. Effect of normal endometrial stroma on growth and differentiation in Ishikawa endometrial adenocarcinoma cells. Cancer Res. 2002;62:79–88. [PubMed] [Google Scholar]
- 190.Turco M.Y., Gardner L., Hughes J., Cindrova-Davies T., Gomez M.J., Farrell L., Hollinshead M., Marsh S.G.E., Brosens J.J., Critchley H.O., et al. Long-term, hormone-responsive organoid cultures of human endometrium in a chemically defined medium. Nat. Cell Biol. 2017;19:568–577. doi: 10.1038/ncb3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Meng C.X., Andersson K.L., Bentin-Ley U., Gemzell-Danielsson K., Lalitkumar P.G.L. Effect of levonorgestrel and mifepristone on endometrial receptivity markers in a three-dimensional human endometrial cell culture model. Fertil. Steril. 2009;91:256–264. doi: 10.1016/j.fertnstert.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 192.Alzamil L., Nikolakopoulou K., Turco M.Y. Organoid systems to study the human female reproductive tract and pregnancy. Cell Death Differ. 2021;28:35–51. doi: 10.1038/s41418-020-0565-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Muruganandan S., Fan X., Dhal S., Nayak N.R. Development of A 3D Tissue Slice Culture Model for the Study of Human Endometrial Repair and Regeneration. Biomolecules. 2020;10 doi: 10.3390/biom10010136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Buck V.U., Gellersen B., Leube R.E., Classen-Linke I. Interaction of human trophoblast cells with gland-like endometrial spheroids: a model system for trophoblast invasion. Hum. Reprod. 2015;30:906–916. doi: 10.1093/humrep/dev011. [DOI] [PubMed] [Google Scholar]
- 195.Hewitt S.C., Dickson M.J., Edwards N., Hampton K., Garantziotis S., DeMayo F.J. From cup to dish: how to make and use endometrial organoid and stromal cultures derived from menstrual fluid. Front. Endocrinol. 2023;14 doi: 10.3389/fendo.2023.1220622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Klionsky D.J., Abdel-Aziz A.K., Abdelfatah S., Abdellatif M., Abdoli A., Abel S., Abeliovich H., Abildgaard M.H., Abudu Y.P., Acevedo-Arozena A., et al. Guidelines for the use and interpretation of assays for monitoring autophagy 1. Autophagy. 2021;17:1–382. doi: 10.1080/15548627.2020.1797280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Mizushima N., Murphy L.O. Autophagy assays for biological discovery and therapeutic development. Trends Biochem. Sci. 2020;45:1080–1093. doi: 10.1016/j.tibs.2020.07.006. [DOI] [PubMed] [Google Scholar]
- 198.Ueno T., Komatsu M. Monitoring autophagy flux and activity: principles and applications. Bioessays. 2020;42 doi: 10.1002/bies.202000122. [DOI] [PubMed] [Google Scholar]
- 199.Yoshii S.R., Mizushima N. Monitoring and measuring autophagy. Int. J. Mol. Sci. 2017;18:1865. doi: 10.3390/ijms18091865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Dong C., Fu S., Karvas R.M., Chew B., Fischer L.A., Xing X., Harrison J.K., Popli P., Kommagani R., Wang T., et al. A genome-wide CRISPR-Cas9 knockout screen identifies essential and growth-restricting genes in human trophoblast stem cells. Nat. Commun. 2022;13:2548. doi: 10.1038/s41467-022-30207-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Shimizu T., Oike A., Kobayashi E.H., Sekiya A., Kobayashi N., Shibata S., Hamada H., Saito M., Yaegashi N., Suyama M., et al. CRISPR screening in human trophoblast stem cells reveals both shared and distinct aspects of human and mouse placental development. Proc. Natl. Acad. Sci. USA. 2023;120 doi: 10.1073/pnas.2311372120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Jauniaux E., Collins S., Burton G.J. Placenta accreta spectrum: pathophysiology and evidence-based anatomy for prenatal ultrasound imaging. Am. J. Obstet. Gynecol. 2018;218:75–87. doi: 10.1016/j.ajog.2017.05.067. [DOI] [PubMed] [Google Scholar]
- 203.Kashiwagi H., Mariya T., Umemoto M., Ogawa S., Hirohashi Y., Fujibe Y., Kubo T., Someya M., Baba T., Ishioka S., et al. Pregnancy-specific beta-1-glycoprotein 6 is a potential novel diagnostic biomarker of placenta accreta spectrum. Med. Mol. Morphol. 2024;57:35–44. doi: 10.1007/s00795-023-00371-y. [DOI] [PubMed] [Google Scholar]
- 204.Chen Y., Wang L., Bao J., Sha X., Cui L., Huang Q., Gu C., Li X., Liu H. Persistent hypoxia induced autophagy leading to invasiveness of trophoblasts in placenta accreta. J. Matern. Fetal Neonatal Med. 2021;34:1297–1303. doi: 10.1080/14767058.2019.1635582. [DOI] [PubMed] [Google Scholar]
- 205.Vogel J.P., Chawanpaiboon S., Moller A.-B., Watananirun K., Bonet M., Lumbiganon P. The global epidemiology of preterm birth. Best Pract. Res. Clin. Obstet. Gynaecol. 2018;52:3–12. doi: 10.1016/j.bpobgyn.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 206.Oaks B.M., Adu-Afarwuah S., Ashorn P., Lartey A., Laugero K.D., Okronipa H., Stewart C.P., Dewey K.G. Increased risk of preterm delivery with high cortisol during pregnancy is modified by fetal sex: a cohort study. BMC Pregnancy Childbirth. 2022;22:727. doi: 10.1186/s12884-022-05061-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Agrawal V., Jaiswal M.K., Mallers T., Katara G.K., Gilman-Sachs A., Beaman K.D., Hirsch E. Altered autophagic flux enhances inflammatory responses during inflammation-induced preterm labor. Sci. Rep. 2015;5:9410. doi: 10.1038/srep09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Menon R. Epithelial to mesenchymal transition (EMT) of feto-maternal reproductive tissues generates inflammation: a detrimental factor for preterm birth. BMB Rep. 2022;55:370–379. doi: 10.5483/BMBRep.2022.55.8.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Severino M.E.L., Richardson L., Kammala A.K., Radnaa E., Khanipov K., Dalmacio L.M.M., Mysorekar I.U., Kacerovsky M., Menon R. Autophagy determines distinct cell fates in human amnion and chorion cells. Autophagy Rep. 2024;3 doi: 10.1080/27694127.2024.2306086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Scifres C.M., Nelson D.M. Intrauterine growth restriction, human placental development and trophoblast cell death. J. Physiol. 2009;587:3453–3458. doi: 10.1113/jphysiol.2009.173252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Curtis S., Jones C.J.P., Garrod A., Hulme C.H., Heazell A.E.P. Identification of autophagic vacuoles and regulators of autophagy in villous trophoblast from normal term pregnancies and in fetal growth restriction. J. Matern. Fetal Neonatal Med. 2013;26:339–346. doi: 10.3109/14767058.2012.733764. [DOI] [PubMed] [Google Scholar]
- 212.Metzger B.E., Lowe L.P., Dyer A.R., Trimble E.R., Chaovarindr U., Coustan D.R., Hadden D.R., McCance D.R., Hod M., McIntyre H.D., et al. Hyperglycemia and adverse pregnancy outcomes. Obstet. Anesth. Digest. 2009;29:39–40. doi: 10.1097/01.aoa.0000344706.95925.dc. [DOI] [Google Scholar]
- 213.Avagliano L., Massa V., Terraneo L., Samaja M., Doi P., Bulfamante G.P., Marconi A.M. Gestational diabetes affects fetal autophagy. Placenta. 2017;55:90–93. doi: 10.1016/j.placenta.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 214.Lu L., Ma Y., Deng J., Xie J., Huang C. Lower ATG7 Levels are Associated with a Higher Risk of Gestational Diabetes Mellitus: A Cross-Sectional Study. Diabetes Metab. Syndr. Obes. 2022;15:2335–2343. doi: 10.2147/DMSO.S377041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Tsai K., Tullis B., Jensen T., Graff T., Reynolds P., Arroyo J. Differential expression of mTOR related molecules in the placenta from gestational diabetes mellitus (GDM), intrauterine growth restriction (IUGR) and preeclampsia patients. Reprod. Biol. 2021;21 doi: 10.1016/j.repbio.2021.100503. [DOI] [PubMed] [Google Scholar]
- 216.Bianchi C., Taricco E., Cardellicchio M., Mandò C., Massari M., Savasi V., Cetin I. The role of obesity and gestational diabetes on placental size and fetal oxygenation. Placenta. 2021;103:59–63. doi: 10.1016/j.placenta.2020.10.013. [DOI] [PubMed] [Google Scholar]
- 217.Dimitriadis E., Rolnik D.L., Zhou W., Estrada-Gutierrez G., Koga K., Francisco R.P.V., Whitehead C., Hyett J., da Silva Costa F., Nicolaides K., Menkhorst E. Pre-eclampsia. Nat. Rev. Dis. Prim. 2023;9:8. doi: 10.1038/s41572-023-00417-6. [DOI] [PubMed] [Google Scholar]
- 218.Oh S.-Y., Choi S.-J., Kim K.H., Cho E.Y., Kim J.-H., Roh C.-R. Autophagy-related proteins, LC3 and Beclin-1, in placentas from pregnancies complicated by preeclampsia. Reprod. Sci. 2008;15:912–920. doi: 10.1177/1933719108319159. [DOI] [PubMed] [Google Scholar]
- 219.Akaishi R., Yamada T., Nakabayashi K., Nishihara H., Furuta I., Kojima T., Morikawa M., Yamada T., Fujita N., Minakami H. Autophagy in the placenta of women with hypertensive disorders in pregnancy. Placenta. 2014;35:974–980. doi: 10.1016/j.placenta.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 220.Tranquilli A.L. Introduction to ISSHP new classification of preeclampsia. Pregnancy Hypertens. 2013;3:58–59. doi: 10.1016/j.preghy.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 221.Pan Y.-J., He L., Zhou S.-J., Zhang L.-J., Zhang A.-H., Zhao Y.-Y. Expression of urotensin II is associated with placental autophagy in patients with severe preeclampsia. J. Hum. Hypertens. 2018;32:759–769. doi: 10.1038/s41371-018-0083-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Cheng S., Huang Z., Banerjee S., Jash S., Buxbaum J.N., Sharma S. Evidence From Human Placenta, Endoplasmic Reticulum–Stressed Trophoblasts, and Transgenic Mice Links Transthyretin Proteinopathy to Preeclampsia. Hypertension. 2022;79:1738–1754. doi: 10.1161/HYPERTENSIONAHA.121.18916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Akcora Yildiz D., Irtegun Kandemir S., Agacayak E., Deveci E. Evaluation of protein levels of autophagy markers (Beclin 1 and SQSTM1/p62) and phosphorylation of cyclin E in the placenta of women with preeclampsia. Cell. Mol. Biol. 2017;63:51–55. doi: 10.14715/cmb/2017.63.12.12. [DOI] [PubMed] [Google Scholar]
- 224.Tong M., Cheng S.-b., Chen Q., DeSousa J., Stone P.R., James J.L., Chamley L.W., Sharma S. Aggregated transthyretin is specifically packaged into placental nano-vesicles in preeclampsia. Sci. Rep. 2017;7:6694. doi: 10.1038/s41598-017-07017-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Kalkunte S.S., Neubeck S., Norris W.E., Cheng S.-B., Kostadinov S., Vu Hoang D., Ahmed A., von Eggeling F., Shaikh Z., Padbury J., et al. Transthyretin is dysregulated in preeclampsia, and its native form prevents the onset of disease in a preclinical mouse model. Am. J. Pathol. 2013;183:1425–1436. doi: 10.1016/j.ajpath.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Landers K.A., McKinnon B.D., Li H., Subramaniam V.N., Mortimer R.H., Richard K. Carrier-mediated thyroid hormone transport into placenta by placental transthyretin. J. Clin. Endocrinol. Metab. 2009;94:2610–2616. doi: 10.1210/jc.2009-0048. [DOI] [PubMed] [Google Scholar]
- 227.McKinnon B., Li H., Richard K., Mortimer R. Synthesis of thyroid hormone binding proteins transthyretin and albumin by human trophoblast. J. Clin. Endocrinol. Metab. 2005;90:6714–6720. doi: 10.1210/jc.2005-0696. [DOI] [PubMed] [Google Scholar]
- 228.Buhimschi I.A., Nayeri U.A., Zhao G., Shook L.L., Pensalfini A., Funai E.F., Bernstein I.M., Glabe C.G., Buhimschi C.S. Protein misfolding, congophilia, oligomerization, and defective amyloid processing in preeclampsia. Sci. Transl. Med. 2014;6:245ra92. doi: 10.1126/scitranslmed.3008808. [DOI] [PubMed] [Google Scholar]
- 229.Yung H.W., Atkinson D., Campion-Smith T., Olovsson M., Charnock-Jones D.S., Burton G.J. Differential activation of placental unfolded protein response pathways implies heterogeneity in causation of early-and late-onset pre-eclampsia. J. Pathol. 2014;234:262–276. doi: 10.1002/path.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Alves C., Jenkins S.M., Rapp A. 2023. Spontaneous abortion NBK560521. [PubMed] [Google Scholar]
- 231.Cai H., Chen L., Zhang M., Xiang W., Su P. Low expression of MFN2 is associated with early unexplained miscarriage by regulating autophagy of trophoblast cells. Placenta. 2018;70:34–40. doi: 10.1016/j.placenta.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 232.Zhou F., Wang Y., Tan Y., Wu C., Chen Y. HMGB1 regulates lipopolysaccharide-induced cellular dysfunction in HTR8/SVneo cells: Implications for the role of HMGB1 in unexplained spontaneous miscarriage. Placenta. 2021;112:16–22. doi: 10.1016/j.placenta.2021.06.012. [DOI] [PubMed] [Google Scholar]
- 233.Eskelinen E.-L. The dual role of autophagy in cancer. Curr. Opin. Pharmacol. 2011;11:294–300. doi: 10.1016/j.coph.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 234.Singh S.S., Vats S., Chia A.Y.-Q., Tan T.Z., Deng S., Ong M.S., Arfuso F., Yap C.T., Goh B.C., Sethi G., et al. Dual role of autophagy in hallmarks of cancer. Oncogene. 2018;37:1142–1158. doi: 10.1038/s41388-017-0046-6. [DOI] [PubMed] [Google Scholar]
- 235.Yun C.W., Jeon J., Go G., Lee J.H., Lee S.H. The dual role of autophagy in cancer development and a therapeutic strategy for cancer by targeting autophagy. Int. J. Mol. Sci. 2020;22:179. doi: 10.3390/ijms22010179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Galluzzi L., Bravo-San Pedro J.M., Levine B., Green D.R., Kroemer G. Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat. Rev. Drug Discov. 2017;16:487–511. doi: 10.1038/nrd.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]