Abstract
Background
The high invasiveness of phytophagous insects is related to their adaptability to various environments, that can be influenced by their associated microbial community. Microbial symbionts are known to play a key role in the biology, ecology, and evolution of phytophagous insects, but their abundance and diversity are suggested to be influenced by environmental stressors. In this work, using 16 S rRNA metabarcoding we aim to verify (1) if laboratory rearing affects microbial symbiont communities of Zeugodacus cucurbitae females, a cosmopolitan pest of cucurbitaceous crops (2) if temperature, diet quality, and antibiotic treatments affect microbial symbiont communities of both laboratory and wild populations, and (3) if changes in microbial symbiont communities due to temperature, diet and antibiotic affect longevity and fecundity of Z. cucurbitae.
Results
The results showed that microbial diversity, particularly the β-diversity was significantly affected by insect origin, temperature, diet quality, and antibiotic treatment. The alteration of gut microbial symbionts, specifically Enterobacteriaceae, was associated with low fecundity and longevity of Z. cucurbitae females feeding on optimal diet only. Fecundity reduction in antibiotic treated females was more pronounced when flies were fed on a poor diet without protein.
Conclusions
our study proves the relationship between gut microbiome and host fitness under thermal and diet fluctuation highlighting the importance of microbial community in the adaptation of Z. cucurbitae to environmental stress.
Clinical trial number
Not applicable.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-024-03673-y.
Keywords: Insect adaptation, Longevity, Fecundity, Microbial symbiont, Metabarcoding
Background
Biological invasions are recognized to pose substantial threats to species distributions, global biodiversity and ecosystem functioning [1, 2], leading to enormous economic and ecological costs [3, 4]. Insect pest invasions have dramatically increased in the past decades posing significant biosecurity threats to natural and agroecosystems [5, 6], and it is expected that the ranges of the top insect invaders are set to increase substantially in the future with climate change, land-use change, and globalization [5, 7]. Exotic species released from their native ecological area are forced to cope with new major environmental stressors and species that do not adjust to these changes are likely to suffer population declines and failure of establishment in the new environment.
Multiple factors and mechanisms have been proposed to explain the success or failure of biological invasions [8, 9]. Exotic species often respond to novel environments stressors with adaptive phenotypic divergence as a result of complex interactions between their genomes (local adaptation) and the environment (adaptive phenotypic plasticity). Phenotypic plasticity is the capacity of a given genotype to express different phenotypes according to the environment stressor they experience [10–12] over a very short timescale (intra-generational). Exotic pests with stronger adaptability to environmental conditions could easily persist under extreme biotic and abiotic stressors through adjusting their behavior, physiology, morphology, biochemistry, or life history [13, 14], facilitating the successful establishment of a species in unpredictable, heterogeneous or novel environments [15, 16].
Insect symbionts often accompany invading species to the new environment and may play a major role in facilitating pest invasions [17–19], because they can represent an important source of adaptability in insects toward new or stresfull environments [20]. Insect physiology, behaviour, biology, and ecology are influenced by individual variations in the microbial communities [21–23]. Insect symbionts consist mostly of primary and secondary that interact intimately with their insect host and can be located in different insect tissues like the gut, reproductive organs, and the bacteriome [24]. Obligate or primary symbionts are known to be essential for the survival and reproduction of their hosts and usually have an ancient stable host association through vertical transmission. Facultative or secondary symbionts are typically diverse and are not required for growth or reproduction but can affect adaptive host traits and can be horizontally transmitted and acquired from the environment [25, 26]. Once the transmission of microbes has occurred, different factors and environments in which the insects live affect the maintenance, diversity and assemblage of symbiont communities [20].
In phytophagous insects, diet is often suggested as one of the main external environmental factors that affect symbiont diversity via two ways [27–29]. The diet can act as a source of novel symbionts or affect the already existing symbionts via their nutritional properties [27, 29]. For example, host shifts and the ability to exploit new hosts have been associated with microbial community changes in many phytophagous insects [30–33]. So, the essential metabolic services provided by symbionts make plant tissues edible for phytophagous insects and promote host adaptation and expansion into previously uninhabitable environments [34, 35]. Environmental factors that can greatly affect insects’ microbiota diversity are temperature and humidity [36, 37]. Increase in temperature, as the major consequence of climate change, plays a key role in the life of insects and can lead to changes in microbial diversity [38, 39]. It has been demonstrated in Diptera including Drosophila [19, 40] and Tephritidae [41], but also in several other insect species, such as hemipteran stink bugs [42–44], damselflies [45], or Sarcophagidae [46, 47]. But, the impact of temperature stress on insect gut microbiomes is still being elucidated and the existing literature lends support to diverse, yet important interactions on the host. In this respect, microbiome variation may be considered as an adaptive trait, subject to natural selection, increasing fitness and enabling insect adaptation in changing environments [20, 48].
Zeugodacus cucurbitae (Coquillett) (Diptera: Tephritidae) is a major pest of Cucurbitaceae and fruits belonging to other families. It is an invasive species, originated from Central Asia and it has successfully established into tropical and sub-tropical areas of Africa, the Indian Ocean islands, Australia (Queensland) and the Pacific Islands, including Hawaii during the past century [49]. This tephritid species harboured a gut microbiome community that is in part transmitted vertically between generations [50, 51], and in part acquired from the environment, varying between individuals raised under similar conditions. This species was used as a model to investigate the role of gut microbial community in insect adaptability to diet quality and temperature stressors. Beyond bacterial community characterization, we addressed the following questions: (1) Do gut microbial symbiont communities vary between wild and laboratory populations of the same host species (Z. cucurbitae)? (2) Do temperature and diet quality affect microbial symbiont communities, and are laboratory populations more affected by these stressors than wild population? (3) Do changes in microbial symbiont communities due to temperature and food availability affect the longevity and fecundity of Z. cucurbitae? (4) Do manipulation of the microbiome by antibiotic treatment affect fitness of laboratory and wild strain of Z. cucurbitae?
Materials and methods
Flies rearing
For our experiments, we used laboratory and wild strains of Zeugodacus cucurbitae. The laboratory strain of Z. cucurbitae was obtained from laboratory stocks at the UMR PVBMT (La Réunion), which were reared from specimens originally obtained from wild cucurbit fruits on La Réunion island. Larvae were subsequently reared on uninfested zucchini (Cucurbita pepo) for generations F56-F61 under constant environmental conditions (25 ± 1 °C; 70% relative humidity; L:D 12:12 photoperiod supplemented with natural light to maintain twilight conditions). Adult flies of the wild population of Z. cucurbitae used, were collected in January 2020 from bitter gourd (Momordica charantia) in La Réunion Island. Infested fruits were brought to the laboratory and batches of 40–50 fruits from each fruit were placed in a box with sterile sand to allow pupation. Within one day after pupation, the pupae were collected and placed in rearing cages (30 × 30 × 30 cm) for emergence under constant laboratory conditions of 25 ± 1 °C, 65 ± 70% RH, and a photoperiod of L12:D12. Emerged wild and laboratory flies were reared on a protein: sugar mix (3:1) media and water was provided ad libitum for 3 days.
Experimental design and gut-microbial diversity
For each treatment group, 3-day-old adult flies were starved for at least 12 h. This period allowed the establishment of the natural microbiota in the gut [52]. For each tephritid species, thirty pairs of females and males were transferred to individual 1-L plastic vials to be used as our experimental unit for all assays, each vial represents one replication. Vials were kept in environmental chambers (Luminincube II, Analis, Belgium; MLR-350, Sanyo, Japan) under conditions of L12:D12 photoperiod, 70 ± 10% relative humidity.
Strain effect assay
To assess the difference on the gut microbial community between laboratory and wild strains of Z. cucurbitae, vials were kept at 25 °C ± 1 °C and adult flies were reared on an optimal diet containing sugar and protein (protein hydrolysate) at a ratio of 1:3. Six replicates were performed for each strain of Z. cucurbitae.
Temperature effect assay
To assess the effect of extremely high (35 °C) and low (10 °C) temperatures on the gut microbial community of laboratory and wild strains of Z. cucurbitae, experiments were conducted at the following constant temperatures: 10, 25, 35 °C (± 1 °C), with 25 °C representing the optimal temperature for the development of this tephritid species. Adult flies were reared on an optimal diet containing sugar and protein (protein hydrolysate) at a ratio of 1:3. Six replicates were performed for each temperature.
Diet quality assay
To assess the effect of diet quality on the gut microbial community of laboratory and wild strains of Z. cucurbitae, vials were kept in environmental chambers at 25 °C ± 1 °C. Adult flies were subjected to one of the three dietary treatments: (1) sugar and protein at a ratio of 1:3 (optimal diet), (2) no protein but sucrose solution at 10% (poor diet with restricted protein), (3) protein mix but no sucrose (poor diet with restricted carbohydrates). In the three dietary treatments, adult flies were given water ad libitum. Diets were provisioned twice a week in 5 mL sterile plastic cups and the treatment period lasted until 100% of the flies died. Six replicates were performed for each treatment group. Because adult flies were unable to survive more than 48 h when kept on poor diet with only protein, only the optimal diet and restricted carbohydrate diet (without protein source) experiments were included in the analysis.
Aposymbiotic conditions assay
Because we wanted to observe the effect of the gut microbiome on the fitness of Z. cucurbitae under diet quality and temperature stressors, experiments were repeated using aposymbiotic (diet with antibiotic) flies. In the case of aposymbiotic condition, a mixture of 200 µg/ml streptomycin, 45 µg/ml kanamycin, and 15 µg /ml of gentamycin antibiotics were added to the food consisting of protein: sugar mix (3:1) in the case of optimal diet (1) and to the sucrose (2) in the case of poor diet with restricted carbohydrate. The antibiotic mixture was used based on the result of published experiments on Tephritidae groups in order to reduce the diversity of gut bacteria as much as possible [53, 54]. In the case of symbiotic condition, flies were not exposed to the antibiotic treatments.
Fitness measures
The vials were monitored every two days in each treatment, the number of dead female flies was recorded, and dead ones were removed from the vials. Ten days after adult emergence, oviposition dishes were placed in each vial for females to lay eggs in. The oviposition devices were circular plastic cups with a volume of 5 mL containing a sheet of tissue paper sprayed with distilled water with 2 mL of mango concentrate. The opening of the plastic container was covered with a double layer of laboratory foil, which was then pierced 10 − 15 times with an entomological pin. Eggs laid by females were rinsed and counted. Female fecundity was estimated by counting the number of eggs produced by females in each vial during the treatment period.
For each Z. cucurbitae strain, female survival was analyzed using a general linear model (GLM) with a binomial error (logit link) as function of Z. cucurbitae strain, or temperature, or diet. To analyze female longevity, we compared the survivorship curves describing changes in number of female survivorship over time for the Z. cucurbitae strains, or temperature, or diet quality by calculating area under the disease progress curve (AUDPC) values, which were then analyzed using analysis of variance (ANOVA). The mean number of eggs laid per female and per day, were analyzed using a generalized linear model (GLM) with Poisson error (Log link) as a function of Z. cucurbitae strain, or temperature, or diet quality.
Gut dissection and DNA extraction
Live flies were starved for at least 8 h in order to clear the insect gut of allochthonous species before gut dissection [55]. At least three samples of tephritid females from each experimental treatment group were randomly selected for gut dissection at 10, 50, and 70% of fly mortality. Flies were collected and anaesthetized with carbon dioxide for 1 min. The flies were then surface sterilized by sequential immersion for 1 min in 70% ethanol, sterile distilled water, 0.05% sodium hypochlorite and lastly, sterile distilled water before individuals were placed on a sterile concave glass slide that had been surface treated by wiping with 70% ethanol and 0.05% sodium hypochlorite. The glass slide was placed on top of ice in a plastic Petri dish, then viewed under a stereomicroscope. Two pipette drops of sterile phosphate-buffered saline (PBS) were placed on top of the insect before dissection with sterile forceps. The individual surface-sterilized flies were dissected aseptically under laminar airflow. Dissection consisting of first removing the wings, the legs and the exoskeleton after softening by immersion in PBS for 1 min. The intact gut (excluding the Malpighian tubules) of the flies was then carefully removed and placed in a clean 1.5 mL microcentrifuge tube and immediately transferred to a freezer (-20 °C) for a maximum of 1 h. Samples were then stored at -80 °C until required.
Total genomic DNA was extracted from each gut homogenate using the DNeasy Blood and Tissue Kit (Qiagen), according to the manufacturer’s instructions. The quantity and quality of the extracted DNA was measured in each sample using a NanoDrop 1000 (Thermo Fisher Scientific, Wilmington, NC, United States). A quantity of 30 ng of DNA per sample was used as a template to generate amplicons corresponding to the V3-V4 hypervariable region of the bacterial 16 S rRNA gene. All PCR products were purified using Agencourt AMPure XP beads, dissolved in the elution buffer and finally labelled to complete library construction. Library size and concentration were obtained by Agilent 2100 Bioanalyzer. Qualified libraries were sequenced on HiSeq platform according to their insert size. The Illumina Hiseq 2500 sequencing was performed by BGI Genomics (Wuhan, China) (https://en.genomics.cn/).
Sequence data processing
Sequence data analysis was performed based on the DADA2 pipeline [56]. Paired forward and reverse reads from raw sequencing data files were trimmed and filtered by quality. Sequences with lengths greater than 200pb and mean quality value ≥ 20 were retained. These filtered files were subjected to several steps, starting with dereplication, followed by sequence variant inference and finally forward and reverse reads merging. A table of amplicon sequence variants (ASVs a higher analog of operational taxonomic units-OTUs) was created, recording the number of times each ASV was observed in each sample. Chimeras were removed. Taxonomic classification of the representative sequence for each ASV (What is the percentage of identity that is used in the classification? Given that it is based on my pipeline it probably is percentage of identity = 97% similarity, p-min‐ consensus = 0.51) was performed using the most recent SILVA taxonomic database (SILVA SSU ref 32 NR, September 2022). Chloroplast and mitochondrial sequences were removed using the R package decontam [57]. Prior to statistical analysis, a 0.025% relative abundance filter was applied based on [58] to remove spurious sequences.
The microbial community structure was characterized by measuring alpha-diversity (within samples) and beta-diversity (between samples). Alpha-diversity was estimated using the Abundance coverage estimator (ACE) to assess the ASV richness and the Inverse Simpson index to assess the ASV evenness. Differences in microbial α-diversity between groups was assessed by two-way ANOVAs. To ensure homoscedasticity, a log transformation was applied to the ACE and Inverse Simpson index. Microbial β-diversity variation between groups was examined by subjecting Bray-Curtis distances to a permutation analysis of variance (PERMANOVA, permutation = 1000) using the Adonis function of the VEGAN R package. Differences in β-diversity of microbial community between groups were visualized using a nonmetric multidimensional scaling (nMDS) analysis.
For each tephritid origin, we used ALDEx2 to test for a differential abundance of microbial genera (i.e., genera with relatively more sequences assigned to them) among adults reared at different temperatures and on different diets with and without antibiotic treatments. For the six most abundant genera, we constructed boxplots with ggplot2.
Relationship between tephritid ecology (survival and fitness) and gut microbial community
Multiple Factor Analysis (MFA) [59, 60] was used to study the relationship between gut microbial community of both tephritid species and adult longevity and female fecundity from laboratory and wild colonies reared under stressfull conditions (temperature or diet quality) using the function MFAshiny created within the package “Factoshiny” [61]. The MFA analysis consists of carrying out first principal component analysis (PCA) on each data set (gut microbial community, ecological data, stressfull conditions) which is then “normalized” by dividing all its elements by the square root of the first eigenvalue obtained from of its PCA. Then, the normalized data sets are merged to form a unique matrix and a second global PCA is performed on this matrix. The individual data sets are then projected onto the global analysis to analyze communalities and discrepancies [59, 60].
Results
Our dataset had a total of 216 Z. cucurbitae gut samples, which yielded 27.1 × 106 raw reads with an average of 129,142.7 (± 38,059.24) reads per gut sample. After filtering, demultiplexing, merging, and chimera removal, 21.0 × 106 reads were kept, then identified, and assigned to 826 ASVs. Rarefaction curves showed adequate saturation levels of sequencing across most samples (Supplementary figures S1). Taxonomic assignments of ASVs revealed that Z. cucurbitae gut microbiome comprised bacteria belonging to 25 unique phyla, 190 families and 400 genera.
Strains effect
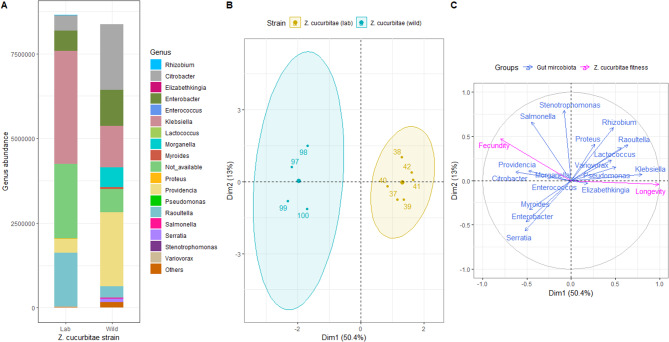
Bacteria belonging to the phylum Proteobacteria were dominant in females of both laboratory and wild strains of Z. cucurbitae (Supplementary figures S2). At the family levels, the most abundant taxa were Enterobacteriaceae, Enterococcaceae, and Flavobacteriaceae, where their relative abundance was different between laboratory and wild strain (Supplementary figures S3). At the genus levels (Fig. 1A), Klebsiella and Raoultella were the most abundant in the laboratory strain. Providencia, Citrobacter, Klebsiella, and Enterobacter were the most abundant genera in the wild strain. The gut-microbiota diversity was significantly more abundant in the wild strain than in the laboratory strain (Fig. 1A, anova, P < 0.001). No significant difference was observed between laboratory and wild strains using ASV richness (Table 1a, anova, P = 0.612) or Inverse Simpson alpha-diversity (Table 1a, anova, P = 0.605). A significant difference in the beta-diversity (Table 1b) was observed on the Bray-Curtis distances between laboratory and wild strains reared at 25 °C with optimal diet (PERMANOVA, F1,10 = 6.24; P = 0.008).
Fig. 1.
Variation in relative abundance (mean) at genus level (A) between laboratory and wild strains of Zeugodacus cucurbitae, and Multiple Factor Analysis (MFA) plots (individuals (B), and quantitative variables (C) maps) showing relationship between gut microbiome community and fitness (longevity and fecundity) among laboratory and wild Zeugodacus cucurbitae strains reared at 25 °C and with optimal diet
Table 1.
Analysis of variance on α diversity metrics (abundance coverage estimator and inverse Simpson index) (a) and perrmutational analysis of variance on Bray-Curtis distances (β diversity) (b) calculated for Zeugodacus cucurbitae strains, and for Zeugodacus cucuribitae laboratory and wild strain reared at different temperature (10, 25, and 35 °C) and with different diet quality (poor diet and optimal diet). Asterisks indicate different significant levels: p < 0.001 = ‘***’, p < 0.01 = ‘**’ p < 0.05 = ‘*’
| (a) | Abundance coverage estimator | Inverse Simpson index | |||||||
| Effect | Z. cucurbiate strain | df | Mean squares | Pseudo F | p Value | df | Mean squares | Pseudo F | p Value |
| Z. cucucrbitae strain | Z. cucurbitae | 1 | 0.009 | 0.274 | 0.612 | 1 | 0.026 | 0.285 | 0.605 |
| Temperature | Z. cucurbitae laboratory strain | 2 | 0.495 | 0.495 | 0.095 | 2 | 0.040 | 1.753 | 0.206 |
| Z. cucurbitae wild strain | 2 | 1.07 | 8.940 | 0.002 ** | 2 | 3.020 | 2.660 | 0.102 | |
| Diet | Z. cucurbitae laboratory strain | 1 | 0.25 | 1.939 | 0.193 | 1 | 0.379 | 3.318 | 0.098 |
| Z. cucurbitae wild strain | 1 | 0.007 | 0.041 | 0.842 | 1 | 0.043 | 0.346 | 0.569 | |
| (b) | Β-diversity index | ||||||||
| Effect | Z. cucurbiate strain | df | R² | Pseudo F | p Value | ||||
| Z. cucucrbitae strain | Z. cucurbitae | 1 | 0.38 | 6.247 | 0.008 ** | ||||
| Temperature | Z. cucurbitae laboratory strain | 2 | 0.48 | 6.99 | 0.001 *** | ||||
| Z. cucurbitae wild strain | 2 | 0.15 | 1.33 | 0.065 | |||||
| Diet | Z. cucurbitae laboratory strain | 1 | 0.21 | 2.641 | 0.017 * | ||||
| Z. cucurbitae wild strain | 1 | 0.16 | 1.97 | 0.008 ** | |||||
MFA analysis, with the first eigenvalue explaining 56.4% and the second one explaining 13.6% of the total variation, showed two distinct groups between laboratory (first group) and wild (second group) strains (Fig. 1B). Female longevity was positively associated with laboratory strain of Z. cucurbitae and was positively correlated with the abundance of Klebsiella and Raoultella gut-microbial genera (Fig. 1C). Female fecundity was positively associated with wild strain of Z. cucurbitae and was positively correlated with the abundance of Enterobacter and Citrobacter gut-microbial genera.
Temperature effect assay
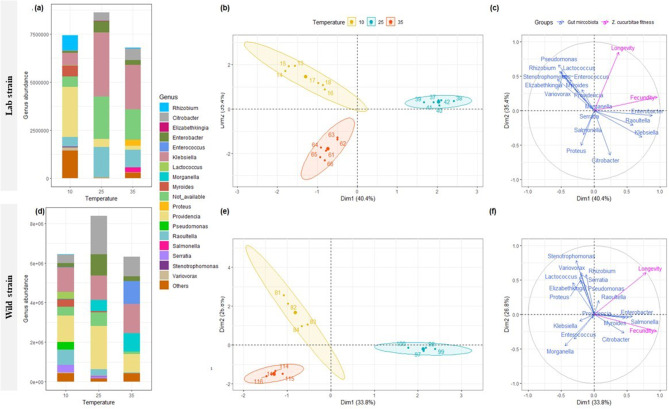
Gut-microbiota number was significantly more higher at 25 °C for wild and laboratory strains than at 10 and 35 °C (Fig. 2a and d, anova, P < 0.001 ). Bacteria belonging to the phylum Proteobacteria were dominant in females reared of both laboratory and wild strains for all temperatures (Supplementary figures S4). At the family level, the most abundant taxa were Enterobacteriaceae, Enterococcaceae, and Flavobacteriaceae, where their relative abundance was different between tested temperatures for laboratory and wild strains (Supplementary figures S5). At the genus level (Fig. 2a and d), Citrobacter and Enterobacter were the most abundant in the laboratory and wild strain reared at optimal temperature (25 °C) and decreased when females were reared at extremely low and high temperatures. ACE richness and Inverse Simpson alpha-diversity showed no significant difference (Table 1a) between temperatures for the laboratory strain, but significant differences were observed for the wild strain on ACE index (Anova, P = 0.002). A significant difference between temperatures in the beta-diversity was observed for the laboratory strain (PERMANOVA, F2,15 = 6.99; P < 0.001) but not for wild (PERMANOVA, F2,15 = 1.33; P = 0.056) strain.
Fig. 2.
Variation in relative abundance (mean) at genus level among tested temperature (10, 25, 35 °C) for laboratory (a) and wild (d) strains of Zeugodacus cucurbitae, and Multiple Factor Analysis (MFA) plots (individuals (b, e), and quantitative variables (c, f) maps) showing relationship between gut microbiome community and fitness (longevity and fecundity) for Zeugodacus cucurbitae laboratory and wild strains reared with optimal diet under different temperature
The first and second eigenvalues of MFA analysis explain 40.4 and 35.4%, and 33.8 and 26.8% of the total variation for laboratory and wild strains, respectively. MFA analysis (Fig. 2b + c, e + f) showed that females were split into three distinct groups according to their rearing temperatures. Female longevity and fecundity were positively correlated with the abundance of Enterobacter and Raoultella for both strains of Z. cucurbitae reared at 25 °C.
Diet effect assay
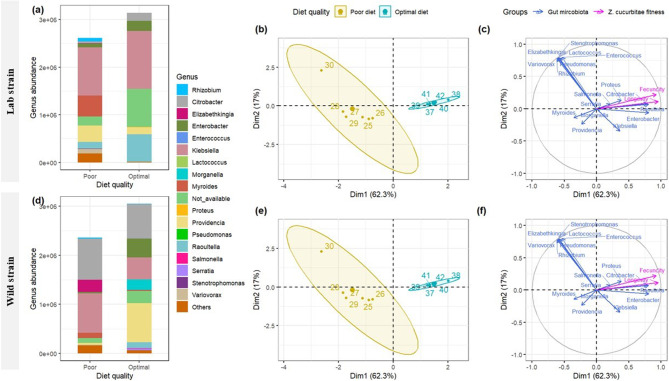
The gut-microbiota number was significantly higher in females reared with optimal diet than in females reared with poor diet (Fig. 3a and d, anova, P < 0.001). Bacteria belonging to the phylum Proteobacteria were dominant in females reared on the two diets of both laboratory and wild strains (Supplementary figures S6). At the family level, the most abundant taxa were Enterobacteriaceae, Enterococcaceae, and Flavobacteriaceae, where their relative abundance was different between tested diet quality for laboratory and wild strains of Z. cucurbitae (Supplementary figures S7). At the genus levels (Fig. 3a and d), the abundance of Enterobacter and Raoultella decrease and replaced by Myroides and Variovorax in famales of Z. cucurbitae (laboratory and wild strain) reared at poor diet ). ACE richness and Shannon alpha-diversity showed no significant difference (Table 1a, anova, P = 0.093; 0.842, respectively) between females reared with optimal or poor diet for laboratory and wild strains. A significant difference in the beta-diversity between diet conditions was observed between laboratory reared females (PERMANOVA, F1,10 = 2.64; P = 0.016) and wild (PERMANOVA, F1,10 = 1.97; P = 0.012) strains.
Fig. 3.
Variation in relative abundance (mean) at genus level for laboratory (a) and wild (d) strains of Zeugodacus cucurbitae females reared with optimal or poor diet at 25 °C, and Multiple Factor Analysis (MFA) plots (individuals (b, e), and quantitative variables (c, f) maps) showing relationship between gut microbiome community and fitness (longevity and fecundity) for Zeugodacus cucurbitae laboratory (b, c) and wild (e, f) strains reared under 25 °C and with different diet quality (poor diet and optimal diet)
The first eigenvalue explained respectively 62.3 and 55.4 of the total variation of the MFA analysis on the gut-microbiota community of laboratory and wild strains reared under optimal or poor diets. Z. cucurbitae populations were split into two distinct groups according to the diet quality (MFA, Fig. 3b and e). Females reared with an optimal diet represented the first group and females reared with a poor diet represented the second group. Female longevity and fecundity were positively associated with wild strain and laboratory strains of Z. cucurbitae reared on optimal diet and were positively correlated with the abundance of Raoultella, Enterobacter, and Citrobacter (MFA, Fig. 3c and f).
Fitness measures
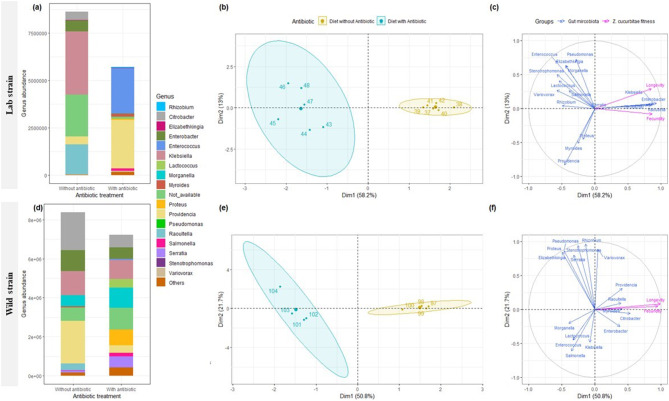
Bacteria belonging to the genus of Klebsiella and Raoultella are the most abundant genera in laboratory symbiotic strains that were disappeared under antibiotic treatments and replaced by other genera as Enterococcus and Providencia (Fig. 4a). Citrobacter and Providencia are the most abundant genera in wild symbiotic strain where their abundance was decreased under antibiotic treatment (Fig. 4b). This confirms that antibiotic treatments significantly reduced the beta-diversity of gut-microbiota of laboratory and wild strains (Table 2b, anova, P = 0.003, P = 0.023, respectively) but not the alpha-diversity (Table 2a). The same tendancy was observed when female flies were reared under temperature or diet stress and subjected to antibiotic treatment (Supplementary figures S8, S9). Analysis showed that antibiotic treatments significantly reduced the beta-diversity of gut-microbiota of females reared under temperature (Table 2b, anova, P < 0.001; P = 0.035) and diet stress (Table 2b, anova, P < 0.001; P < 0.001) of laboratory and wild strains, respectively.
Fig. 4.
Variation in relative abundance (mean) at genus level for lab (a) and wild (d) strains of Zeugodacus cucurbitae females reared with optimal and with or without antibiotic treatment at 25 °C for laboratory and wild strains, and Multiple Factor Analysis (MFA) plots (individuals (b, e), and quantitative variables (c, f) maps) showing relationship between gut microbiome community and fitness (longevity and fecundity) for Zeugodacus cucurbitae laboratory (b, c) and wild (e, f) strains reared at 25 °C with optimal diet with or without antibiotic treatment
Table 2.
Analysis of variance on α diversity metrics (abundance coverage estimator and inverse Simpson index) (a) and permutational analysis of variance on Bray-Curtis distances (β diversity) (b) calculated for Zeugodacus cucurbitae strain, and for Zeugodacus cucuribitae laboratory and wild strains reared with or without antibiotic under different temperature (10, 25, and 35 °C), or at different diet quality (poor diet and optimal diet). Asterisks indicate different significant levels: p < 0.001 = ‘***’, p < 0.01 = ‘**’ p < 0.05 = ‘*’
| (a) | Abundance coverage estimator | Inverse Simpson index | |||||||||||
| Effect | Z. cucurbiate strain | df | Mean squares | Pseudo F | p Value | df | Mean squares | Pseudo F | p Value | ||||
| Antibiotic | Laboratory strain | 1 | 0.018 | 0.131 | 0.724 | 1 | 2.501 | 22.356 | 0.001 *** | ||||
| Wild strain | 1 | 0.051 | 1.032 | 0.343 | 1 | 0.953 | 1.386 | 0.277 | |||||
| Temperature | Laboratory strain | 2 | 0.148 | 0.955 | 0.396 | 2 | 0.458 | 0.924 | 0.407 | ||||
| Antibiotic | 1 | 1.240 | 7.977 | 0.008 ** | 1 | 4.807 | 9.685 | 0.004 * | |||||
| Temperature x Antibiotic | 2 | 0.678 | 4.363 | 0.021 * | 2 | 0.442 | 0.892 | 0.420 | |||||
| Temperature | Wild strain | 2 | 0.180 | 1.421 | 0.258 | 2 | 1.043 | 0.689 | 0.510 | ||||
| Antibiotic | 1 | 1.645 | 12.96 | 0.002 ** | 1 | 6.069 | 4.007 | 0.055 | |||||
| Temperature x Antibiotic | 2 | 2.580 | 20.323 | 0.001 *** | 2 | 3.415 | 2.255 | 0.124 | |||||
| Diet | Laboratory strain | 1 | 0.790 | 5.312 | 0.032 * | 1 | 0.045 | 0.130 | 0.721 | ||||
| Antibiotic | 1 | 0.137 | 0.922 | 0.348 | 1 | 1.323 | 3.759 | 0.066 | |||||
| Diet x Antibiotic | 1 | 0.031 | 0.213 | 0.648 | 1 | 1.179 | 3.351 | 0.082 | |||||
| Diet | Wild strain | 1 | 0.005 | 0.004 | 0.985 | 1 | 0.741 | 2.706 | 0.115 | ||||
| Antibiotic | 1 | 0.339 | 2.669 | 0.117 | 1 | 0.275 | 1.007 | 0.327 | |||||
| Diet x Antibiotic | 1 | 0.018 | 0.149 | 0.703 | 1 | 0.454 | 1.658 | 0.212 | |||||
| (b) | Beta diversity index | ||||||||||||
| Effect | Z. cucurbiate strain | df | R² | Pseudo F | p Value | ||||||||
| Antibiotic | Laboratory strain | 1 | 0.511 | 10.465 | 0.003 ** | ||||||||
| Wild strain | 1 | 0.173 | 1.4677 | 0.0239 * | |||||||||
| Temperature | Laboratory strain | 2 | 0.178 | 5.128 | 0.001 *** | ||||||||
| Antibiotic | 1 | 0.180 | 10.347 | 0.001 *** | |||||||||
| Temperature x Antibiotic | 2 | 0.118 | 3.400 | 0.002 ** | |||||||||
| Temperature | Wild strain | 2 | 0.087 | 1.645 | 0.002 ** | ||||||||
| Antibiotic | 1 | 0.085 | 3.221 | 0.035 * | |||||||||
| Temperature x Antibiotic | 2 | 0.105 | 1.976 | 0.007 ** | |||||||||
| Diet | Laboratory strain | 1 | 0.071 | 2.390 | 0.023 * | ||||||||
| Antibiotic | 1 | 0.241 | 8.054 | 0.001 *** | |||||||||
| Diet x Antibiotic | 1 | 0.088 | 2.954 | 0.006 ** | |||||||||
| Diet | Wild strain | 1 | 0.071 | 2.390 | 0.020 * | ||||||||
| Antibiotic | 1 | 0.241 | 8.054 | 0.001 *** | |||||||||
| Diet x Antibiotic | 1 | 0.088 | 2.954 | 0.013 * | |||||||||
MFA analysis, with the first eigenvalue explaining 58.2 and 50.8% and the second one explaining 13 and 21.7% of the total variation, respectively, showed two distinct groups between symbiotic (first group) and aposymbiotic (second group) Z. cucurbitae (Fig. 4c + d, e + f). Female longevity and fecundity were positively associated with symbiotic Z. cucurbitae of both strains and were positively correlated with the abundance of Raoultella, Enterobacter, and Citrobacter.
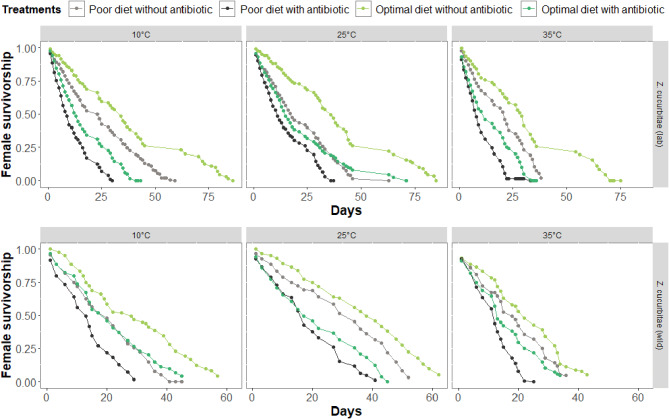
Female survival rates and longevity differed significantly (F1,3309 = 230.80; P < 0.001) between laboratory and wild strains (Fig. 5). For the laboratory strain, the survivorship of females significantly differed among temperatures (AUDPC; F2, 60 = 6.43, P = 0.002) and antibiotic (F1, 60 = 30.74, P < 0.001) treatments, but not significantly differed between diet quality (F1, 60 = 0.0001, P = 0.992). Only the interaction between the three parameters (temperature, antibiotic, and diet) was significant (F2, 60 = 10.64, P < 0.001). For the wild strain, Female survivorship was significantly different between antibiotic treated and non-treated (F1, 37 = 19.62, P < 0.001) populations, but not significantly different between temperatures (F2, 37 = 2.87, P = 0.069) nor between diets (F1, 37 = 0.053, P = 0.818). Only the interaction between temperatures and antibiotic treatment was significant (F2, 60 = 3.55, P = 0.038).
Fig. 5.
Females survivorship (%) and longevity (days) of laboratory (lab) and wild Zeugodacus cucurbitae strains under different temperatures (10, 25, and 35 °C), diets (optimal diet and poor diet) and antibiotic treatments (with antibiotic and without antibiotic)
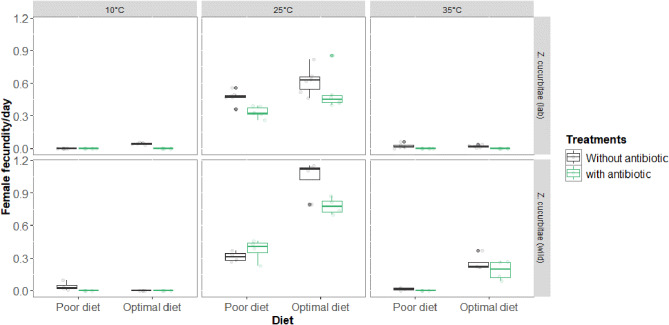
Female fecundities differed significantly (F1,3194 = 19.57; P < 0.001) between laboratory and wild strains (Fig. 6). For the laboratory strain, female fecundity differed significantly as a function of antibiotic treatments (Δdev 1, 2076= 39686; P < 0.001), diets (Δdev 1, 2075 = 35755; P < 0.001), and temperatures (Δdev 2, 2077 = 41664; P < 0.001). For wild strain, female fecundity differed significantly as a function of antibiotic treatments (Δdev 1, 1112= 28176; P < 0.001), diets (Δdev 1, 1111 = 23219; P < 0.001), and temperatures (Δdev 2, 1113 = 29266; P < 0.001).
Fig. 6.
Female daily fecundity (mean) of laboratory (lab) and wild Zeugodacus cucurbitae populations under different temperatures (10, 25, and 35 °C), diets (optimal diet and poor diet) and antibiotic treatments (with antibiotic and without antibiotic)
Discussion
Few studies have been performed to characterize and compare microbial symbiont communities of Z. cucurbitae from natural and laboratory rearing strains [62–65]. Most of them suggest that Z. cucurbitae microbial symbiont community is mainly composed by Proteobacteria phylum, with Enterobacteriaceae being the most abundant family [64, 65]. This is shown in our study for both wild and laboratory strains of Z. cucurbitae. Our study is in concordance with several other reports on the bacterial community of phytophagous insects, including Tephritidae [66].
Microbial symbiont communities of Z. cucurbitae were dominated by Klebsiella and Raoutella groups for laboratory strain and by Citrobacter and Providencia species for wild strain. The difference in microbial community composition may be due to diverse factors related to the environments in which insects live [67, 68], such as mass-rearing methods and host plant [69, 70]. As shown in previous studies in Tephritidae species [65, 71], the mass rearing under laboratory constant conditions that disconnect insects from their natural environment and natural diet can alter the structure of the microbial community of the laboratory strain of Z. cucurbitae, causing a reduction in bacterial symbiont richness and diversity, impacting all life stages [72, 73]. Also, given that the wild strain of Z. cucurbitae originated from bitter gourd and the laboratory strain is reared on zucchini, the difference could be due to host plant, particularly to the antibacterial action of bitter gourd. Similar results are found in another study on Z. cucurbitae originated from La Reunion island [31] showing that the majority of symbionts are usually acquired from the environment [74] The change in community composition and symbiont prevalence is a factor that could affect several aspects of host biology, physiology, and ecology [75–77]. We found that the high abundance of Raoultella and Klebsiella in laboratory strain were positively correlated with higher longevity, however, the high abundance of Citrobacter and Providencia were positively correlated with higher fecundity. This suggests that this difference provide ecological and fitness advantages for insects, which can help the host to adapt to a specific environment like mass rearing or natural environment. This study presented important findings for laboratory adaptation of this insect species.
Adaptation of phytophagous insects to stressors conditions is improved by the variation in gut microbial communities [20, 78]. Temperature is one of the important factors affecting both the host and its associated microbial symbionts [79]. We found that the abundance and the association of gut microbial communities varied greatly across temperatures. Particularly, the abundance of Klebsiella, Enterobacter, and Citrobacter in the gut of Z. cucurbitae was affected negatively by high and low temperatures. These results are similar to those for other insect groups [42, 67, 77, 80], suggesting that temperature has an important role in maintaining gut microbial symbionts in phytophagous insects.
At optimal temperature of 25 °C, the highest Z. cucurbitae fitness was correlated with the high abundance of Enterobacteraceae genera, as Enterobacter and Klebsiella for laboratory strain and with the abundance of Enterobacter and Citobacter for wild strain. It is known that thermal variation has a strong impact on host metabolism and physiology, and extreme temperatures threaten the host’s fitness [81–83], but also destabilize microbial symbiont communities that provide essential services to their host [39, 67, 84], leading to a survival and fecundity deterioration. We demonstrate that the gut microbial community does shift as results of increase or decrease of temperature, and in concert with adult fitness changes, suggesting that these changes may be linked to increase the resilience of this tephritid species to heat and cold temperature. The change in gut microbiome abundance and assemblage of insects and animals species according to seasonal change, particularly to temperature fluctuation, has been shown [85, 86]. This confirms that insects are able to establish vital associations with symbiotic bacteria, which influences their adaptation to changing environments [20]. Therefore, the dynamic change of gut microbial composition can be also an important manifestation of insect adaptation to the environment [87].
Zeugodacus cucurbitae females were reared at different diet quality, and results showed a significant decline in microbial symbiont diversity and abundance in poor artificial diet compared to optimal artificial diet, particularly in Enterobacter and Raoultella in both laboratory and wild strains. The decrease in gut microbial community under poor diet quality was accompanied by the colonization of other microbial species such as Variovorax and Myroides whose role in the fly’s biology is not fully known yet. Diet availability and quality have a significant impact on essential traits of host fitness and greatly influence the assemblage of microbial symbionts in the gut of phytophagous insects [28, 88, 89]. Diet quality can act as a selective constraint favouring the establishment of microbial symbiont communities that meet specific host requirements for nutritional services [89]. Microbial symbiont taxa that persist under highly stressful dietary conditions, such as Klebsiella, may be involved in helping Z. cucurbitae to overcome poor diet quality. Other studies from all around the globe have indicated Klebsiella to be an important component of tephritid fruit flies’ microbiome [90] that plays a major role in host nutrition [91–93].
We observed a change in host fitness according to the alteration of gut microbial community of Z. cucurbitae reared with poor diet which is well documented for Tephritidae [94–97]. This confirms that different diet quality may affects the composition of gut microbial community and thus influence the insect fitness [26, 28, 87, 98]. These microbial symbionts groups likely represent a substantial source of variation that could enhance adaptation to different diet quality and they enable insect hosts to inhabit otherwise unsuitable hosts, and thus either by being a direct food source (ingestion) or providing new metabolic pathways [27, 72, 99]. So, the possible role of these microbial groups in affecting the metabolic pathways of Z. cucurbitae need to be confirmed with targeted experimental support.
Symbiotic Z. cucurbitae females demonstrated significantly higher fecundity and longevity compared to adults treated with antibiotics, and these two measures are positively correlated with the abundance of Citrobacter, Enterobacter, and Raoultella belonging to Enterobacteriaceae family. Enterobacteriaceae has been suggested to be involved in biochemical pathways related to membrane transport and the metabolism of carbohydrates, amino acids, cofactors and lipids [100] and their suppression alter egg production [93] and ovarian development [101]. The alteration of gut microbial symbionts, specifically Enterobacteriaceae, was associated with low fecundity and longevity of Z. cucurbitae females feeding in the optimal diet. Fecundity reduction in antibiotic treated females was more pronounced when feeding on a poor diet without protein. The contribution of bacteria to fecundity has previously been demonstrated for Tephritidae species [93, 102, 103] and particularly this effect was diet-dependent [102]. This suggests that gut microbial symbionts can play important role to insect biology and particularly in enhancing female fecundity, either being a direct food source (ingestion) or providing new metabolic pathways [27, 72, 99]. Others studies showed that the ingestion of Klebsiella oxytoca satisfied the tephritid female requirements for egg maturation [99], supporting the idea of a functional contribution of microbial community to the host. It is also possible that the prevalence of specific microbial symbionts can be favoured by differences in the nutritional components of the two diets that require different types of bacteria in order to be processed [104], suggesting that beneficial microbial communities can increase adaptation of their insect host to unsuitable host plant range.
Conclusions
Our findings increase our knowledge about the responses of insect–gut microbial symbionts association to diet availability and temperature fluctuations in a climate change context. It is becoming increasingly clear that environmental factors (temperature and diet) not only directly impact the fitness of phytophagous insects, but also affect the abundance, diversity and assemblage of gut-associated microbial community with indirect implications for the host’s fitness. We speculate that the effect of temperature and diet on the beta-diversity of gut microbiota may be explained by Anna Karenina Principle (AKP) that can promote tephritid adaptation at micro and macroevolutionary scales as suggested by [48]. Mainly because the gut-symbionts community can expand the abiotic and trophic niches of their insect host by improving its adaptability to unsuitable host plant range and new climatic niche, facilitating thus their insect host establishment in the invasive ecosystem. We conclude that thermal and diet variability could play a key role in the ecology and evolution of insect-gut-symbiont associations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Jim Payet and Serge Glénac for their assistance in rearing of adult flies and in finding the infested fruits for wild strain. We think Christophe Simiand for its assistance in DAN extraction. The authors greatly acknowledge the Plant Protection Platform (3P, IBISA). We are grateful for all the financial support provided for this work.
Author contributions
AH: Conceptualization, Methodology, Validation, Formal analysis, Data curation, Writing – original draft, Visualization; LM: Validation, Investigation, Data curation, Writing – review; WH: Validation, Formal analysis, Writing – original draft; MDM: Conceptualization, Methodology, Validation, Writing – original draft; MV: Conceptualization, Methodology, Validation, Writing – original draft; HD: Conceptualization, Methodology, Validation, Writing – review, Supervision, Project administration.
Funding
A.H. received the financial support of the Conseil Régional de La Réunion and the University de La Réunion through the postdoctoral grant. H.D. was funded by the European Union: European Agricultural Funds for Rural Development (EAFRD) and the Centre de Coopération Internationale en Recherche Agronomique pour le Développement (CIRAD). This study used the facilities provided by the Plant Protection Platform (3P, UMR PVBMT), Saint-Pierre, La Réunion, France.
Data availability
The 16S rRNA sequence of gut microbiome symbiont of Z. cucurbitae can be accessed in NLM NCBI under the accession number PQ307258 : PQ308071[accn] (https://www.ncbi.nlm.nih.gov/nuccore?term=PQ307258+%3A+PQ308071%5Baccn%5D&cmd=DetailsSearch), and available in the open acces CIRAD Dataverse via the link ||10.18167/DVN1/XFGIG5.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mollot G, Pantel JH, Romanuk TN. Chapter two - the effects of invasive species on the decline in species richness: a global meta-analysis. In: Bohan DA, Dumbrell AJ, Massol F, editors. Advances in ecological research. Academic; 2017. pp. 61–83.
- 2.Simberloff D, Martin J-L, Genovesi P, Maris V, Wardle DA, Aronson J, et al. Impacts of biological invasions: what’s what and the way forward. Trends Ecol Evol. 2013;28:58–66. [DOI] [PubMed] [Google Scholar]
- 3.Diagne C, Leroy B, Vaissière A-C, Gozlan RE, Roiz D, Jarić I, et al. High and rising economic costs of biological invasions worldwide. Nature. 2021;592:571–86. [DOI] [PubMed] [Google Scholar]
- 4.Hulme PE. Climate change and biological invasions: evidence, expectations, and response options. Biol Rev. 2017;92:1297–313. [DOI] [PubMed] [Google Scholar]
- 5.Seebens H, Bacher S, Blackburn TM, Capinha C, Dawson W, Dullinger S, et al. Projecting the continental accumulation of alien species through to 2050. Glob Change Biol. 2021;27:970–82. [DOI] [PubMed] [Google Scholar]
- 6.Vilà M, Hulme PE, editors. Impact of biological invasions on ecosystem services. Cham: Springer International Publishing; 2017. [Google Scholar]
- 7.Turner RM, Brockerhoff EG, Bertelsmeier C, Blake RE, Caton B, James A, et al. Worldwide border interceptions provide a window into human-mediated global insect movement. Ecol Appl. 2021;31:e02412. [DOI] [PubMed] [Google Scholar]
- 8.Daly EZ, Chabrerie O, Massol F, Facon B, Hess MCM, Tasiemski A, et al. A synthesis of biological invasion hypotheses associated with the introduction–naturalisation–invasion continuum. Oikos. 2023;2023:e09645. [Google Scholar]
- 9.Lemoine MM, Engl T, Kaltenpoth M. Microbial symbionts expanding or constraining abiotic niche space in insects. Curr Opin Insect Sci. 2020;39:14–20. [DOI] [PubMed] [Google Scholar]
- 10.Fusco G, Minelli A. Phenotypic plasticity in development and evolution: facts and concepts. Phil Trans R Soc B. 2010;365:547–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price TD, Qvarnström A, Irwin DE. The role of phenotypic plasticity in driving genetic evolution. Proc R Soc Lond B. 2003;270:1433–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sgrò CM, Terblanche JS, Hoffmann AA. What can plasticity contribute to insect responses to climate change? Annu Rev Entomol. 2016;61:433–51. [DOI] [PubMed] [Google Scholar]
- 13.Agrawal AA, Vala F, Sabelis MW. Induction of preference and performance after acclimation to novel hosts in a phytophagous spider mite. Adapt Plasticity? Am Nat. 2002;159:553–65. [DOI] [PubMed] [Google Scholar]
- 14.Whitman D, Agrawal A. What is phenotypic plasticity and why is it important? In: Whitman D, Ananthakrishnan T, editors. Phenotypic plasticity of insects. Science; 2009.
- 15.Jardeleza MG, Koch JB, Pearse IS, Ghalambor CK, Hufbauer RA. The roles of phenotypic plasticity and adaptation in morphology and performance of an invasive species in a novel environment. Ecol Entomol. 2022;47:25–37. [Google Scholar]
- 16.Manfredini F, Arbetman M, Toth AL. A potential role for phenotypic plasticity in invasions and declines of social insects. Front Ecol Evol. 2019;7:375. [Google Scholar]
- 17.Amsellem L, Brouat C, Duron O, Porter SS, Vilcinskas A, Facon B. Chapter three - importance of microorganisms to macroorganisms invasions: is the essential invisible to the eye? (the little Prince, A. De Saint-Exupéry, 1943). In: Bohan DA, Dumbrell AJ, Massol F, editors. Advances in ecological research. Academic; 2017. pp. 99–146.
- 18.Lefort MC, Glare TR, Bouchon D, Boyer S. How hindgut microbiota may shape sympatric speciation in an invasive phytophagous scarab. Entomol Exp Appl. 2023;171:556–63. [Google Scholar]
- 19.Moghadam NN, Thorshauge PM, Kristensen TN, De Jonge N, Bahrndorff S, Kjeldal H, et al. Strong responses of Drosophila melanogaster microbiota to developmental temperature. Fly. 2018;12:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lange C, Boyer S, Bezemer TM, Lefort M-C, Dhami MK, Biggs E, et al. Impact of intraspecific variation in insect microbiomes on host phenotype and evolution. ISME J. 2023;17:1798–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engl T, Kaltenpoth M. Influence of microbial symbionts on insect pheromones. Nat Prod Rep. 2018;35:386–97. [DOI] [PubMed] [Google Scholar]
- 22.Lee J-H, Lee K-A, Lee W-J. Chapter four - microbiota, gut physiology, and insect immunity. In: Ligoxygakis P, editor. Advances in insect physiology. Academic; 2017. pp. 111–38.
- 23.Mondal S, Somani J, Roy S, Babu A, Pandey AK. Insect microbial symbionts: Ecology, interactions, and biological significance. Microorganisms. 2023;11:2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Douglas AE. Multiorganismal insects: diversity and function of resident microorganisms. Annu Rev Entomol. 2015;60:17–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engel P, Moran NA. The gut microbiota of insects–diversity in structure and function. FEMS Microbiol Rev. 2013;37:699–735. [DOI] [PubMed] [Google Scholar]
- 26.Paniagua Voirol LR, Frago E, Kaltenpoth M, Hilker M, Fatouros NE. Bacterial symbionts in Lepidoptera: their diversity, transmission, and impact on the host. Front Microbiol. 2018;9:556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Douglas AE. The microbial dimension in insect nutritional ecology. Funct Ecol. 2009;23:38–47. [Google Scholar]
- 28.Leite-Mondin M, DiLegge MJ, Manter DK, Weir TL, Silva-Filho M, de Vivanco C. The gut microbiota composition of Trichoplusia ni is altered by diet and may influence its polyphagous behavior. Sci Rep. 2021;11:5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Zhang F, Lu X. Diversity and functional roles of the gut microbiota in lepidopteran insects. Microorganisms. 2022;10:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fei-Ying Y, Saqib HSA, Jun-Hui C, Ruan Q-Q, Vasseur L, Wei-Yi H, et al. Differential profiles of gut microbiota and metabolites associated with host shift of Plutella Xylostella. Int J Mol Sci. 2020;21:6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hendrycks W, Delatte H, Moquet L, Bourtzis K, Mullens N, De Meyer M, et al. Eating eggplants as a cucurbit feeder: dietary shifts affect the gut microbiome of the melon fly Zeugodacus cucurbitae (Diptera, Tephritidae). MicrobiologyOpen. 2022;11:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jose PA, Yuval B, Jurkevitch E. Maternal and host effects mediate the adaptive expansion and contraction of the microbiome during ontogeny in a holometabolous, polyphagous insect. Funct Ecol. 2023;37:929–46. [Google Scholar]
- 33.Tsuchida T, Koga R, Matsumoto S, Fukatsu T. Interspecific symbiont transfection confers a novel ecological trait to the recipient insect. Biol Lett. 2011;7:245–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frago E, Dicke M, Godfray C. Insect symbionts as hidden players in insect-plant interactions. Trends Ecol Evol. 2012;705–11. [DOI] [PubMed]
- 35.Moran NA, Tran P, Gerardo NM. Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl Environ Microbiol. 2005;71:8802–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magoga G, Brunetti M, Kajtoch L, Spada A, Montagna M. Biotic and abiotic factors affecting the microbiota of Chrysomelidae inhabiting wetland vegetation. Hydrobiologia. 2023;850:3797–812. [Google Scholar]
- 37.Yun J-H, Roh SW, Whon TW, Jung M-J, Kim M-S, Park D-S, et al. Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl Environ Microbiol. 2014;80:5254–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hammer TJ, Le E, Moran NA. Thermal niches of specialized gut symbionts: the case of social bees. Proc R Soc B. 2021;288:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mason CJ, Shikano I. Hotter days, stronger immunity? Exploring the impact of rising temperatures on insect gut health and microbial relationships. Curr Opin Insect Sci. 2023;59:101096. [DOI] [PubMed] [Google Scholar]
- 40.Jaramillo A, Castañeda LE. Gut microbiota of Drosophila subobscura contributes to its heat tolerance and is sensitive to transient thermal stress. Front Microbiol. 2021;12:654108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ren L, Zhang X, Yang F, Jocelin NF, Shang Y, Wang Q, et al. Effects of heat tolerance on the gut microbiota of Sarcophaga Peregrina (Diptera: Sarcophagidae) and impacts on the life history traits. Parasites Vectors. 2023;16:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prado SS, Hung KY, Daugherty MP, Almeida RPP. Indirect effects of temperature on stink bug fitness, via maintenance of gut-associated symbionts. Appl Environ Microbiol. 2010;76:1261–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kikuchi Y, Tada A, Musolin DL, Hari N, Hosokawa T, Fujisaki K, et al. Collapse of insect gut symbiosis under simulated climate change. mBio. 2016;7:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kashkouli M, Fathipour Y, Mehrabadi M. Potential management tactics for pistachio stink bugs, Brachynema Germari, Acrosternum heegeri and Acrosternum Arabicum (Hemiptera: Pentatomidae): high temperature and chemical surface sterilants leading to symbiont suppression. J Econ Entomol. 2019;112:244–54. [DOI] [PubMed] [Google Scholar]
- 45.Theys C, Verheyen J, Delnat V, Janssens L, Tüzün N, Stoks R. Thermal and latitudinal patterns in pace-of-life traits are partly mediated by the gut microbiome. Sci Total Environ. 2023;855:158829. [DOI] [PubMed] [Google Scholar]
- 46.Byrd JH, Butler JF. Effects of temperature on Sarcophaga Haemorrhoidalis (Diptera: Sarcophagidae) development. J Med Entomol. 1998;35:694–508. [DOI] [PubMed] [Google Scholar]
- 47.Grassberger M, Reiter C. Effect of temperature on development of Liopygia (= Sarcophaga) argyrostoma (Robineau-Desvoidy)(Diptera: Sarcophagidae) and its forensic implications. J Forensic Sci. 2002;47:1332–6. [PubMed] [Google Scholar]
- 48.Mullens N, Hendrycks W, Bakengesa J, Kabota S, Tairo J, Svardal H, et al. Anna Karenina as a promoter of microbial diversity in the cosmopolitan agricultural pest Zeugodacus cucurbitae (Diptera, Tephritidae). PLoS ONE. 2024;19:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Meyer M, Delatte H, Mwatawala M, Quilici S, Vayssières J-F, Virgilio M. A review of the current knowledge on Zeugodacus cucurbitae (Coquillett)(Diptera, Tephritidae) in Africa, with a list of species included in Zeugodacus. ZooKeys. 2015;539. [DOI] [PMC free article] [PubMed]
- 50.Hassan B, Siddiqui JA, Xu Y. Vertically transmitted gut bacteria and nutrition influence the immunity and fitness of Bactrocera dorsalis Larvae. Front Microbiol. 2020;11:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang A, Yao Z, Zheng W, Zhang H. Bacterial communities in the gut and reproductive organs of Bactrocera minax (Diptera: Tephritidae) based on 454 pyrosequencing. PLoS ONE. 2014;9:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lauzon CR. Symbiotic relationships of tephritids. Insect Symbiosis. 2003;2:115–29. [Google Scholar]
- 53.Koskinioti P, Ras E, Augustinos AA, Tsiamis G, Beukeboom LW, Caceres C, et al. The effects of geographic origin and antibiotic treatment on the gut symbiotic communities of Bactrocera oleae populations. Entomol Exp Appl. 2019;167:197–208. [Google Scholar]
- 54.Rashid MA, Andongma AA, Dong Y-C, Ren X-M, Niu C-Y. Effect of gut bacteria on fitness of the Chinese citrus fly, Bactrocera minax (Diptera: Tephritidae). Symbiosis. 2018;76:63–9. [Google Scholar]
- 55.Andongma AA, Wan L, Dong Y-C, Wang Y-L, He J, Niu C-Y. Assessment of the bacteria community structure across life stages of the Chinese Citrus fly, Bactrocera minax (Diptera: Tephritidae). BMC Microbiol. 2019;19:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reitmeier S, Hitch TC, Treichel N, Fikas N, Hausmann B, Ramer-Tait AE, et al. Handling of spurious sequences affects the outcome of high-throughput 16S rRNA gene amplicon profiling. ISME Commun. 2021;1:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abdi H, Williams LJ, Valentin D. Multiple factor analysis: principal component analysis for multitable and multiblock data sets. WIREs Comput Stats. 2013;5:149–79. [Google Scholar]
- 60.Abdi H, Valentin D. Multiple factor analysis (MFA). Encyclopedia Meas Stat. 2007;657–63.
- 61.Vaissie P, Monge A, Husson F. Factoshiny: perform factorial analysis from FactoMineR with a shiny application. R Package Version. 2021;2.
- 62.Asimakis ED, Khan M, Stathopoulou P, Caceres C, Bourtzis K, Tsiamis G. The effect of diet and radiation on the bacterial symbiome of the melon fly, Zeugodacus cucurbitae (Coquillett). BMC Biotechnol. 2019;19:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bavithra ML, Siddanna PG, Abburi R, Reddy PK, Manjula K. Isolation and characterization of bacterial gut symbionts from irradiated, wild and lab reared males of melon fly, Zeugodacus cucurbitae (Coquillett). J Entomol Res Soc. 2023;25:351–62. [Google Scholar]
- 64.Choudhary JS, Naaz N, Prabhakar CS, Das B, Singh AK, Bhatt BP. High taxonomic and functional diversity of bacterial communities associated with melon fly, Zeugodacus cucurbitae (Diptera: Tephritidae). Curr Microbiol. 2021;78:611–23. [DOI] [PubMed] [Google Scholar]
- 65.Hadapad AB, Shettigar SKG, Hire RS. Bacterial communities in the gut of wild and mass-reared Zeugodacus cucurbitae and Bactrocera dorsalis revealed by metagenomic sequencing. BMC Microbiol. 2019;19:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hafsi A, Delatte H. Enterobactereaceae symbiont as facilitators of biological invasion: review of Tephritidae fruit flies. Biol Invasions. 2023;25:991–1006. [Google Scholar]
- 67.Iltis C, Tougeron K, Hance T, Louâpre P, Foray V. A perspective on insect–microbe holobionts facing thermal fluctuations in a climate-change context. Environ Microbiol. 2022;24:18–29. [DOI] [PubMed] [Google Scholar]
- 68.Lange-Enyedi NT, Borsodi AK, Németh P, Czuppon G, Kovács I, Leél-\HOssy S, et al. Habitat-related variability in the morphological and taxonomic diversity of microbial communities in two Hungarian epigenic karst caves. FEMS Microbiol Ecol. 2023;99:1–17. [DOI] [PubMed] [Google Scholar]
- 69.Jones AG, Mason CJ, Felton GW, Hoover K. Host plant and population source drive diversity of microbial gut communities in two polyphagous insects. Sci Rep. 2019;9:2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Z-W, Luo J-Y, Men Y, Liu Z-H, Zheng Z-K, Wang Y-H, et al. Different roles of host and habitat in determining the microbial communities of plant-feeding true bugs. Microbiome. 2023;11:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Augustinos AA, Tsiamis G, Cáceres C, Abd-Alla AM, Bourtzis K. Taxonomy, diet, and developmental stage contribute to the structuring of gut-associated bacterial communities in tephritid pest species. Front Microbiol. 2019;10:463894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deutscher AT, Chapman TA, Shuttleworth LA, Riegler M, Reynolds OL. Tephritid-microbial interactions to enhance fruit fly performance in sterile insect technique programs. BMC Microbiol. 2019;19:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jose PA, Ben-Yosef M, Lahuatte P, Causton CE, Heimpel GE, Jurkevitch E, et al. Shifting microbiomes complement life stage transitions and diet of the bird parasite Philornis Downsi from the Galapagos Islands. Environ Microbiol. 2021;23:5014–29. [DOI] [PubMed] [Google Scholar]
- 74.Behar A, Ben-Yosef M, Lauzon CR, Yuval B, Jurkevich E, Bourtzis K. Structure and function of the bacterial community associated with the Mediterranean fruit fly. Insect Symbiosis. 2008;3:251–71. [Google Scholar]
- 75.Kikuchi Y, Tada A, Musolin DL, Hari N, Hosokawa T, Fujisaki K, et al. Collapse of insect gut symbiosis under simulated climate change. mBio. 2016;7:e01578–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lv W-X, Cheng P, Lei J-J, Peng H, Zang C-H, Lou Z-W, et al. Interactions between the gut micro-community and transcriptome of Culex pipiens pallens under low-temperature stress. Parasites Vectors. 2023;16:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tougeron K, Iltis C. Impact of heat stress on the fitness outcomes of symbiotic infection in aphids: a meta-analysis. Proc R Soc B. 2022;289:20212660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sapkota R, Nakatsu CH, Scharf ME. Regulation of host phenotypic plasticity by gut symbiont communities in the eastern subterranean termite (Reticulitermes flavipes). J Exp Biol. 2021;224:jeb242553. [DOI] [PubMed] [Google Scholar]
- 79.Corbin C, Heyworth ER, Ferrari J, Hurst GD. Heritable symbionts in a world of varying temperature. Heredity. 2017;118:10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ayyasamy A, Kempraj V, Pagadala Damodaram KJ. Endosymbiotic bacteria aid to overcome temperature induced stress in the oriental fruit fly, Bactrocera dorsalis. Microb Ecol. 2021;82:783–92. [DOI] [PubMed] [Google Scholar]
- 81.Colinet H, Sinclair BJ, Vernon P, Renault D. Insects in fluctuating thermal environments. Annu Rev Entomol. 2015;60:123–40. [DOI] [PubMed] [Google Scholar]
- 82.Neven LG. Physiological responses of insects to heat. Postharvest Biol Technol. 2000;21:103–11. [Google Scholar]
- 83.Weaving H, Terblanche JS, Pottier P, English S. Meta-analysis reveals weak but pervasive plasticity in insect thermal limits. Nat Commun. 2022;13:5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Greenspan SE, Migliorini GH, Lyra ML, Pontes MR, Carvalho T, Ribeiro LP, et al. Warming drives ecological community changes linked to host-associated microbiome dysbiosis. Nat Clim Change. 2020;10:1057–61. [Google Scholar]
- 85.Ferguson LV, Dhakal P, Lebenzon JE, Heinrichs DE, Bucking C, Sinclair BJ. Seasonal shifts in the insect gut microbiome are concurrent with changes in cold tolerance and immunity. Func Ecol. 2018;32:2357–68. [Google Scholar]
- 86.Hou Z, Dong Y, Shi F, Xu Y, Ge S, Tao J, et al. Seasonal shifts in cold tolerance and the composition of the gut microbiome of Dendroctonus valens LeConte occur concurrently. Forests. 2021;12:888. [Google Scholar]
- 87.Tian Z, Chen L, Chen G, Wang J, Ma C, Zhang Y, et al. Effect of host shift on the gut microbes of Bactrocera cucurbitae (Coquillett) (Diptera: Tephritidae). Front Microbiol. 2023;14:1264788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Luo J, Cheng Y, Guo L, Wang A, Lu M, Xu L. Variation of gut microbiota caused by an imbalance diet is detrimental to bugs’ survival. Sci Total Environ. 2021;771:144880. [DOI] [PubMed] [Google Scholar]
- 89.Malacrinò A. Host species identity shapes the diversity and structure of insect microbiota. Mol Ecol. 2022;31:723–35. [DOI] [PubMed] [Google Scholar]
- 90.Raza MF, Yao Z, Bai S, Cai Z, Zhang H. Tephritidae fruit fly gut microbiome diversity, function and potential for applications. Bull Entomol Res. 2020;110:423–37. [DOI] [PubMed] [Google Scholar]
- 91.Azis K, Zerva I, Melidis P, Caceres C, Bourtzis K, Ntougias S. Biochemical and nutritional characterization of the medfly gut symbiont Enterobacter sp. AA26 for its use as probiotics in sterile insect technique applications. BMC Biotechnol. 2019;19:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Behar A, Yuval B, Jurkevitch E. Enterobacteria-mediated nitrogen fixation in natural populations of the fruit fly Ceratitis capitata. Mol Ecol. 2005;14:2637–43. [DOI] [PubMed] [Google Scholar]
- 93.Ben-Yosef M, Pasternak Z, Jurkevitch E, Yuval B. Symbiotic bacteria enable olive flies (Bactrocera oleae) to exploit intractable sources of nitrogen. J Evol Biol. 2014;27:2695–705. [DOI] [PubMed] [Google Scholar]
- 94.Chen E-H, Hou Q-L, Wei D-D, Jiang H-B, Wang J-J. Phenotypic plasticity, trade-offs and gene expression changes accompanying dietary restriction and switches in Bactrocera dorsalis (Hendel)(Diptera: Tephritidae). Sci Rep. 2017;7:1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Goane L, Salgueiro J, Pereyra PM, Arce OE, Ruiz MJ, Nussenbaum AL, et al. Antibiotic treatment reduces fecundity and nutrient content in females of Anastrepha fraterculus (Diptera: Tephritidae) in a diet dependent way. J Insect Physiol. 2022;139:104396. [DOI] [PubMed] [Google Scholar]
- 96.Goane L, Pereyra PM, Castro F, Ruiz MJ, Juárez ML, Segura DF, et al. Yeast derivatives and wheat germ in the adult diet modulates fecundity in a tephritid pest. Bull Entomol Res. 2019;109:178–90. [DOI] [PubMed] [Google Scholar]
- 97.Hafsi A, Facon B, Ravigné V, Chiroleu F, Quilici S, Chermiti B, et al. Host plant range of a fruit fly community (Diptera: Tephritidae): does fruit composition influence larval performance? BMC Ecol. 2016;16:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wong AC-N, Wang Q-P, Morimoto J, Senior AM, Lihoreau M, Neely GG, et al. Gut microbiota modifies olfactory-guided microbial preferences and foraging decisions in Drosophila. Curr Biol. 2017;27:2397–404. [DOI] [PubMed] [Google Scholar]
- 99.Drew RAI, Courtice AC, Teakle DS. Bacteria as a natural source of food for adult fruit flies (Diptera: Tephritidae). Oecologia. 1983;60:279–84. [DOI] [PubMed] [Google Scholar]
- 100.Ventura C, Briones-Roblero CI, Hernández E, Rivera-Orduña FN, Zúñiga G. Comparative analysis of the gut bacterial community of four Anastrepha fruit flies (Diptera: Tephritidae) based on pyrosequencing. Curr Microbiol. 2018;75:966–76. [DOI] [PubMed] [Google Scholar]
- 101.Noman MS, Shi G, Liu L, Li Z. Diversity of bacteria in different life stages and their impact on the development and reproduction of Zeugodacus tau (Diptera: Tephritidae). Insect Sci. 2021;28:363–76. [DOI] [PubMed] [Google Scholar]
- 102.Ben-Yosef M, Aharon Y, Jurkevitch E, Yuval B. Give us the tools and we will do the job: symbiotic bacteria affect olive fly fitness in a diet-dependent fashion. Proc R Soc B. 2010;277:1545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dimou I, Rempoulakis P, Economopoulos AP. Olive fruit fly Bactrocera (Dacus) oleae (Rossi) (Diptera: Tephritidae) adult rearing diet without antibiotic. J App Entomol. 2010;134:72–9. [Google Scholar]
- 104.Huang K, Wang J, Huang J, Zhang S, Vogler AP, Liu Q, et al. Host phylogeny and diet shape gut microbial communities within bamboo-feeding insects. Front Microbiol. 2021;12:633075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA sequence of gut microbiome symbiont of Z. cucurbitae can be accessed in NLM NCBI under the accession number PQ307258 : PQ308071[accn] (https://www.ncbi.nlm.nih.gov/nuccore?term=PQ307258+%3A+PQ308071%5Baccn%5D&cmd=DetailsSearch), and available in the open acces CIRAD Dataverse via the link ||10.18167/DVN1/XFGIG5.