Abstract
The human herpes simplex virus (HSV) protein VP16 induces formation of a transcriptional regulatory complex with two cellular factors—the POU homeodomain transcription factor Oct-1 and the cell proliferation factor HCF-1—to activate viral immediate-early-gene transcription. Although the cellular role of Oct-1 in transcription is relatively well understood, the cellular role of HCF-1 in cell proliferation is enigmatic. HCF-1 and the related protein HCF-2 form an HCF protein family in humans that is related to a Caenorhabditis elegans homolog called CeHCF. In this study, we show that all three proteins can promote VP16-induced-complex formation, indicating that VP16 targets a highly conserved function of HCF proteins. The resulting VP16-induced complexes, however, display different transcriptional activities. In contrast to HCF-1 and CeHCF, HCF-2 fails to support VP16 activation of transcription effectively. These results suggest that, along with HCF-1, HCF-2 could have a role, albeit probably a different role, in HSV infection. CeHCF can mimic HCF-1 for both association with viral and cellular proteins and transcriptional activation, suggesting that the function(s) of HCF-1 targeted by VP16 has been highly conserved throughout metazoan evolution.
When herpes simplex virus (HSV) infects a cell, the virion protein VP16 initiates a cascade of viral gene transcription that leads to productive lytic infection. VP16 (also known as Vmw65 and α-TIF) initiates viral gene transcription by directing formation of a multiprotein transcriptional regulatory complex—the VP16-induced complex—on HSV immediate-early promoters with two cellular proteins: Oct-1, a POU domain transcription factor, and HCF-1, a protein involved in cell proliferation (for reviews see references 6, 21, and 24).
HCF-1 (also known as C1, VCAF, and CFF) is an unusual protein. It is translated as a large polypeptide of 2,035 amino acids which undergoes proteolysis at a series of centrally located 26-amino-acid repeats, called HCF-1PRO repeats; the resulting amino (HCF-1N)- and carboxy (HCF-1C)-terminal polypeptides are stable and remain noncovalently associated (10, 25, 27) through the activity of two pairs of amino- and carboxy-terminal self-association sequences (SAS) (29). At its amino terminus, HCF-1 contains six sequence repeats that are related to a sequence repeat found in the Drosophila protein Kelch (1, 32) and are thus referred to as HCF-1KEL repeats; these repeats form a β-propeller structure which is responsible for binding VP16 and is sufficient to stabilize the VP16-induced complex (12, 28).
HCF-1 is known to be involved in cell proliferation independent of viral infection because, in a mutant hamster cell line called tsBN67, a proline-to-serine point mutation at position 134 (P134S) in the third HCF-1KEL repeat causes a stable temperature-induced cell proliferation arrest (5). This same mutation also impairs HCF-1 association with VP16 and stabilization of the VP16-induced complex (5, 28), suggesting that VP16 targets an activity involved in cell proliferation when it associates with HCF-1. Although a mutation in the HCF-1KEL repeat region causes cell proliferation arrest, this region alone is not sufficient to rescue cell proliferation—a neighboring basic region is also required (28).
In addition to VP16, the HCF-1KEL repeat region interacts with two human basic leucine zipper proteins, LZIP (also known as Luman) and Zhangfei (4, 16, 17, 19). LZIP and Zhangfei have the same tetrapeptide HCF-1 binding sequence, (D/E)HXY, as VP16 and, like VP16, fail to associate with the tsBN67 mutant HCF-1. These results suggest that VP16 mimics cellular proteins in its interaction with HCF-1 and that the role of HCF-1 in viral proliferation may reflect its role in cellular proliferation, although the precise mechanisms may differ in detail (20).
Consistent with an important role in cell proliferation, elements of HCF-1 are highly conserved in metazoans. For example, extracts from the worm Caenorhabditis elegans and the insects Drosophila and Spodoptera are able to fulfill the requirement for HCF-1 in VP16-induced-complex formation, implying the presence of functional HCF-1 homologs in those organisms (9, 26). Indeed, C. elegans contains a functional HCF-1 homolog called CeHCF that can support VP16-induced-complex formation (15). Human HCF-1, however, differs in some respects from CeHCF: although the amino- and carboxy-terminal regions of HCF-1 are highly conserved in CeHCF, the middle of the protein with its HCF-1PRO repeats is missing in CeHCF. Thus, CeHCF is much smaller than HCF-1 (782 versus 2,035 amino acids) and does not undergo proteolytic processing (15, 31).
Subsequently, a second human HCF protein called HCF-2 was discovered; like CeHCF, it is smaller than HCF-1, and the amino- and carboxy-terminal regions, but not the middle of the protein, are conserved (7). Unlike HCF-1 and CeHCF, however, the interaction of HCF-2 with VP16 is reportedly weak, which has led to the conclusion that HCF-2 is unlikely to play a role in transcriptional activation by VP16 or in HSV infection (7).
To better understand the relationship of HCF-1 and HCF-2 to each other and to CeHCF, we have compared all three proteins directly in both in vitro and in vivo assays. We have found, unexpectedly, that all three full-length HCF proteins can associate with VP16 and stabilize the VP16-induced complex effectively. Unlike HCF-1 and CeHCF, however, HCF-2 fails to support VP16-induced transcriptional activation effectively in vivo. These and other results suggest that, since humans diverged from worms during evolution, the HCF family has grown and diversified in humans.
MATERIALS AND METHODS
Constructs for HCF protein expression in mammalian cells.
The mammalian expression vector pCGN (23) was used for the expression of N-terminal hemagglutinin (HA)-HCF fusion proteins (pCGN series). The plasmids used in this study, pCGNHCF-1ΔRep, pCGNHCF-1N1011, pCGNHCF-1N1011/P134S, pCGNHCF-1N380, and pCGNHCF-1N380/P134S, are described in reference 28. pCGNCeHCFFL and pCGNCeHCFN395 are described in reference 15. pCGNHCF-2FL and pCGNHCF-2N373 are described in reference 7 and were kind gifts from A. Wilson (New York University).
HCF chimeras.
pCGNHCF2N356/HCF-1N364–1011 was made as follows. A cDNA sequence corresponding to HCF-2 amino acids 2 to 356 was amplified by PCR with primers containing XbaI (5′) and BsaI (3′) restriction enzyme recognition sites (5′, GCTCTAGAGCGGCTCCCAGCCTCCTC; 3′, GCGGGTCTCGGTGGTTTCTCAGTATC; the restriction sites are underlined), using pCGNHCF-2N373 as a template. The HCF-1 cDNA sequence corresponding to amino acids 364 to 1011 was amplified by PCR from pCGNHCF-1N1011 with a primer containing a 5′ BsaI restriction site and a 3′ primer corresponding to sequences 3′ of the HCF-1N1011 coding sequences and 3′ BamHI cloning site (5′, GCGGGTCTCGCCACCACCCCCAGCCCG; 3′, CAATCAAGGGTCCCCAAACTC). These two PCR products were digested with the appropriate restriction enzymes and ligated together with a pCGN vector linearized by digestion with XbaI and BamHI.
pCGNCeHCFN378/HCF-1N364–1011 was made in a two-step procedure as follows. First, pCGNCeHCFN395 was digested with BamHI and the cohesive end was flush ended by treatment with DNA polymerase I large (Klenow) fragment. The linear molecule was subsequently digested with XbaI. The resulting 1,185-bp XbaI-BamHI CeHCFN395 fragment was inserted into the pCGNHCF-1N1011 vector between the XbaI and PmlI restriction sites, creating pCGNCeHCFN395–HCF-1N160–1011. The coding sequences between CeHCF amino acid 378 and HCF-1 amino acid 363 were deleted by oligonucleotide-directed mutagenesis (11, 33) with the oligonucleotide CTTCTGGATACTATTTTACCACCACCCCCAGCCCG (CeHCF sequences are underlined).
COS cell transfection and electrophoretic mobility retardation assay.
COS cells were transfected by electroporation with a Bio-Rad Genepulser with an extender set at 200 mV and 960 μF. Twenty-four hours after transfection, the cells were washed and collected in ice-cold phosphate-buffered saline (PBS) and lysed in extraction buffer (300 mM NaCl, 100 mM Tris-HCl [pH 8.0], 0.2 mM EDTA, 10% glycerol, 0.1% NP-40, 1 mM phenylmethylsulfonyl fluoride). The extracts were normalized to the level of HCF protein after immunoblot analysis with the anti-HA mouse monoclonal antibody 12CA5. Electrophoretic mobility retardation assays using the normalized extracts were done as previously described (25). Briefly, extracts containing equal amounts of HCF were mixed with 10 ng of VP16ΔC, 5 ng of Oct-1 POU domain, and radiolabled DNA probe containing an (OCTA+)TAATGARAT VP16-responsive sequence from the HSV ICP0 promoter. Both VP16ΔC and Oct-1 POU were synthesized in Escherichia coli fused to glutathione S-transferase, purified by affinity to glutathione, and eluted by cleavage with thrombin. The binding mixtures were incubated at 30°C for 30 min, and the resulting VP16-induced complexes were resolved by electrophoresis through a 4% acrylamide gel as described previously (25).
Transfection and in vivo transcription assay.
Transfection of a subclone of tsBN67 cells called tsBN67HR1 (30) by calcium phosphate coprecipitation, preparation of RNA, and assay of in vivo transcription by RNase protection were done as previously described (5). Briefly, cells grown at 33.5°C were seeded at 1.2 × 106/10-cm-diameter dish. After 24 h at 33.5°C, they were transfected with 1 μg of the selected HCF expression plasmid (pCGN series), 100 ng of the internal reference plasmid pα4X(A+C), and 2 μg of the VP16-responsive β-globin-related reporter plasmid pU2/β6XTAAT, with or without 80 ng of the wild-type VP16 expression plasmid pCGNVP16. pUC119 was used as carrier DNA to normalize the total amount of DNA to 20 μg. At 24 h posttransfection, the cells were washed with PBS containing 2 mM EGTA and further incubated at 33.5°C for 12 h, after which the cells were harvested and cytoplasmic RNA was prepared by NP-40 lysis. Each sample was hybridized with radiolabled antisense β-globin (β134) and α-globin (α98) RNA probes and treated with RNases A and T1. Protected RNAs were separated on a 6% denaturing polyacrylamide gel in 0.5× Tris-borate-EDTA. Signal intensities were quantified using a Fuji BAS1000 phosphorimager. One-third of the collected cells were used for protein extraction, as described for COS cells above, and for anti-HA immunoblotting to visualize the expression levels of HCF and VP16.
tsBN67 cell rescue.
Rescue of the tsBN67HR1 temperature-sensitive cell proliferation defect was performed similarly to the method previously described (5; P. Reilly and W. Herr, unpublished results). tsBN67HR1 cells were seeded at 1.5 × 105/10-cm-diameter dish and incubated at 33.5°C for 24 h. Transfection by calcium phosphate coprecipitation was performed with 2 μg of pCGN HCF expression constructs, 4 μg of pBabe-puro (a puromycin resistance marker), 1 μg of pBabe-lacZ (a β-galactosidase expression construct to test the transfection efficiency), and 13 μg of pUC119 carrier DNA. Twenty-four hours after transfection, the cells were washed with 1× PBS–2 mM EGTA and incubated for a further 24 h before passage and temperature shift as follows. Cells from each 10-cm-diameter dish were split onto three 6-cm-diameter dishes for long-term incubation with puromycin at the nonpermissive 40°C temperature to measure rescue of cell proliferation (N) and protein synthesis after extract preparation (E) and for incubation at the permissive 33.5°C temperature to measure transfection efficiency (P). After 2 days of selection, the E plates were washed and the cells were collected with ice-cold PBS. The cell pellets were resuspended in Laemmli sample buffer and boiled for 5 min before being subjected to gel electrophoresis for immunoblot analysis. The N and P plates were maintained for 14 and 10 days, respectively, with refeeding at 3-day intervals, and then washed with PBS and fixed with 4% formaldehyde, and colonies were stained with 0.01% crystal violet solution.
Yeast two-hybrid assay.
The reporter Saccharomyces cerevisiae strain YGH1 (MATα ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3 gal4-542 gal80-538 LYS::GAL1UAS-gal1TATA-HIS3 URA3::gal1-lacZ) has GAL4-responsive HIS3 and lacZ genes. Yeasts were grown in synthetic complete media supplemented as indicated in Results. YGH1 was first transformed with pGBT9-HCF plasmids encoding GAL4 DNA-binding domain (DBD) residues 1 to 94 fused to various HCF coding sequences as C-terminal fusion proteins. pGBT9, pGBT9HCF-1N380, and pGBT9HCF-1N380/P134S are described by Freiman and Herr (4). pGBT9CeHCFN395 and pGBT9CeHCFFL contain C. elegans HCF amino acids 2 to 395 and 2 to 782, respectively, cloned between the XbaI-BamHI sites of pGBT9. pGBT9-transformed yeast was selected by growth in the absence of tryptophan. Yeast containing the pGBT9HCF series was transformed a second time with the pGADGH series, in which a GAL4 transcriptional activation domain (AD) is fused to VP16ΔC (pGADGHVP16ΔC), VP16ΔCE361A (pGADGHVP16ΔCE361A), LZIP (pGADGHLZIP), and SNF-4 (pGADGHSNF-4) as described previously (4, 14; R. Freiman and W. Herr, unpublished results). Transformed yeasts were selected by their ability to grow in the absence of tryptophan and leucine, and the interaction between the DBD fusion proteins and the AD fusion proteins was monitored by growth in the absence of tryptophan, leucine, and histidine.
RESULTS
To compare HCF-1, HCF-2, and CeHCF, we performed a pairwise comparison of their sequences and assessed their activities in three different assays: (i) VP16-induced-complex formation in vitro, (ii) VP16-induced transcriptional activation in vivo, and (iii) rescue of the tsBN67 cell proliferation defect.
Human and C. elegans HCF protein similarity.
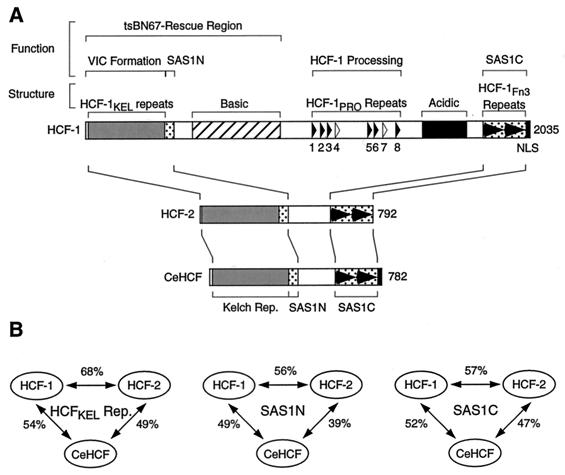
Figure 1A shows a schematic of the human HCF-1 and HCF-2 and the worm CeHCF proteins. Both HCF-2 and CeHCF lack similarity to the central region of HCF-1 containing the basic, HCF-1PRO repeat, and acidic regions. The amino-terminal HCF-1KEL repeat and a pair of HCF-1 subunit self-association sequences, called SAS1N for the amino-terminal SAS element and SAS1C for the carboxy-terminal SAS element (Fig. 1), are conserved in both HCF-2 and CeHCF (7, 15, 29). Additionally, CeHCF, but not HCF-2 (7), has sequence similarity to a carboxy-terminal nuclear localization signal (NLS) present in HCF-1 (13).
FIG. 1.
HCF proteins share sequence similarity in the amino- and carboxy-terminal regions. (A) Schematic diagram of the human HCF proteins HCF-1 and HCF-2 and the C. elegans HCF protein CeHCF. Above the diagram of HCF-1 are shown the positions of (i) functional regions of HCF-1 (e.g., VP16-induced complex [VIC] formation and HCF-1 subunit association [SAS1N and SAS1C]) and (ii) structural features of HCF-1 (e.g., HCF-1KEL repeats and basic region). The solid and open triangles indicate active and inactive HCF-1PRO repeats. The tandem solid arrowheads indicate fibronectin type 3 (Fn3) repeats. Below the diagram of HCF-1 are shown the schematic structures of HCF-2 and CeHCF. Regions of similarity are aligned by the lines connecting the diagrams. (B) Sequence identity of HCF proteins. The percentage of identical amino acid residues between each pair of proteins is shown. The sequence used in each comparison was as follows: HCFKEL repeats, HCF-1 (amino acids [aa] 18–360), HCF-2 (aa 8–353), and CeHCF (aa 29–375); SAS1N, HCF-1 (aa 361–401), HCF-2 (aa 354–393), and CeHCF (aa 376–416); and SAS1C, HCF-1 (aa 1812–2002), HCF-2 (aa 598–784), and CeHCF (aa 553–749).
Figure 1B shows the percent amino acid identity for each pairwise comparison among HCF-1, HCF-2, and CeHCF for the HCF-1KEL repeat, SAS1N, and SAS1C regions. The pairwise comparison shows that these three regions are all highly related to each other in these three proteins. HCF-1 and HCF-2, however, are more closely related to each other than to CeHCF, suggesting that the human HCF-1 and HCF-2 genes have resulted from a gene duplication after the divergence of worms and humans. The progenitor to the human and worm HCF proteins may have had a structure more similar to the present structure of HCF-2 and CeHCF. Although in overall structure CeHCF resembles HCF-2 more than HCF-1 (Fig. 1A), CeHCF is more closely related to HCF-1 than HCF-2 at the amino acid sequence level in each of the three conserved regions (Fig. 1B), suggesting that the functions that have been conserved in CeHCF may more closely resemble those of human HCF-1 than those of human HCF-2.
As previously noted (29), the sequences of the HCF-1 SAS elements are conserved in HCF-2 and CeHCF, even though these proteins are not known to be proteolytically processed. This observation has suggested that these regions have one or more functions other than “self-association” or that self-association is important irrespective of HCF proteolysis (29). The most highly conserved region, however, among all three proteins is the HCFKEL repeat region (Fig. 1B), which is responsible for association with VP16. Thus, when VP16 associates with HCF-1, it apparently targets the most conserved domain in the HCF protein family.
All three HCF proteins can support VP16-induced-complex formation.
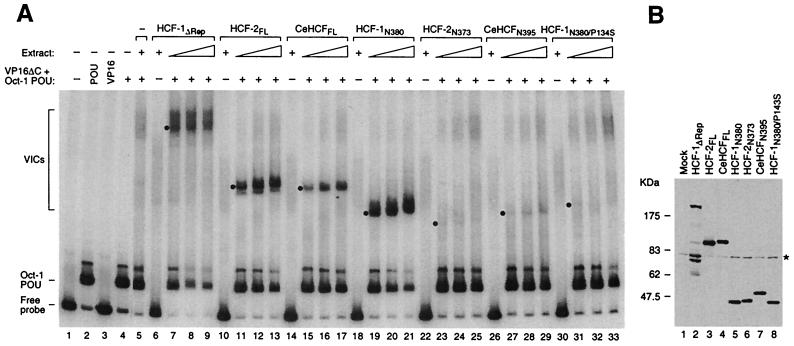
Figure 2 shows a comparison of the VP16-induced-complex formation activity of HCF-1, HCF-2, and CeHCF proteins in an electrophoretic mobility retardation assay with the Oct-1 DNA-binding POU domain and VP16 lacking the carboxyl transcriptional AD (VP16ΔC), both synthesized in E. coli, and a labeled HSV (OCTA+)TAATGARAT VP16 response element-containing DNA probe. We compared the activities of (i) full-length CeHCF and HCF-2; (ii) full-length HCF-1 lacking the HCF-1PRO repeats, which is smaller than wild-type HCF-1 and is not processed, resulting in better synthesis and easier normalization; and (iii) the HCF-1–VP16 interaction domain (residues 2 to 380) and corresponding regions of CeHCF (15) and HCF-2 (7). The HCF proteins, all containing an HA epitope tag at the amino terminus, were synthesized in monkey COS cells by transient expression (see Materials and Methods), and whole-cell extracts were prepared. The normalized levels of HCF protein in each extract used for complex formation are shown in Fig. 2B.
FIG. 2.
All three HCF proteins can stabilize the VP16-induced complex. (A) VP16-induced-complex formation. COS cell extracts containing different HCF proteins were analyzed for VP16-induced-complex formation activity with VP16ΔC, the Oct-1 POU domain, and radiolabeled VP16 response element DNA probe, as described in Materials and Methods. Protein-DNA complexes were resolved in an electrophoretic mobility retardation assay. The positions of the free probe, the Oct-1 POU domain-bound probe, and the VP16-induced complexes (VICs) formed by different HCF proteins are indicated on the left. Lane 1, probe alone; lane 2, probe with Oct-1 POU domain; lane 3, probe with VP16ΔC. Lanes 4, 5, 7 to 9, 11 to 13, 15 to 17, 19 to 21, 23 to 25, 27 to 29, and 31 to 33 contain the Oct-1 POU domain and VP16ΔC. Lane 5 contains in addition unprogrammed COS cell extract, and lanes 6 to 33 contain the COS cell extract with HCF protein as indicated. Each set of three titration lanes contains a twofold titration, and the non-VP16-containing sample contains the most concentrated extract. The position of each VP16-induced complex is indicated with a dot. +, present; −, absent. (B) Immunoblot analysis of the COS cell extracts used in the electrophoretic mobility retardation assay. Extracts containing HA-tagged HCF proteins were resolved by sodium dodecyl sulfate–8% polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and blotted with the 12CA5 anti-HA antibody. The asterisk on the right indicates a nonspecific band. The multiple species smaller than 175 kDa in lane 2 are unrelated to HCF-1 but instead reflect 12CA5-specific cross-reacting cellular proteins that appear only in this sample because it contains the most cell extract after sample normalization.
Figure 2A shows the result of the electrophoretic mobility retardation assay. Addition of the Oct-1 POU domain alone to the DNA probe results in an Oct-1–POU domain-specific complex (lane 2). Addition of VP16ΔC to the probe, either alone or in combination with the Oct-1 POU domain, has no evident effect (lanes 3 and 4). Addition of mock-transfected COS cell extract to VP16ΔC and the Oct-1 POU domain results in a low level of slowly migrating complexes (VP16-induced complexes) formed by endogenous HCF-1 (lane 5). Comparison of VP16-induced complex formation with that of the HCF-1ΔRep and full-length HCF-2 and CeHCF proteins shows that the levels of complex formation with HCF-2 are similar to those with HCF-1ΔRep, both of which are higher than with the CeHCF protein (lanes 7 to 9, 11 to 13, and 15 to 17). These results show that all three proteins can support VP16-induced-complex formation, although HCF-2 is more effective than CeHCF in this regard.
We were surprised to find that HCF-2 can effectively stabilize the VP16-induced complex, because Johnson et al. (7) showed that the region of HCF-2 (HCF-2N373) corresponding to the HCF-1 VP16 interaction domain (HCF-1N380), which in HCF-1 is sufficient for VP16-induced complex formation, does not stabilize the VP16-induced complex. We have shown previously, however, that the region of CeHCF (CeHCFN395) corresponding to the HCF-1 VP16 interaction domain has unexpectedly weak VP16-induced-complex formation activity (15). We therefore directly compared the VP16-induced-complex formation activities of the HCF-2N373 construct used by Johnson et al. (7) and the CeHCFN395 proteins. Consistent with the previously reported results, both HCF-2N373 and CeHCFN395 had very weak VP16-induced-complex formation activity compared to the HCF-1N380 construct (Fig. 2A, compare lanes 23 to 25 and 27 to 29 with lanes 19 to 21). Indeed, the HCF-2N373 and CeHCFN395 constructs did not support VP16-induced-complex formation significantly better than the corresponding mutant HCF-1N380/P134S molecule (5). We do not know why the HCF-2N373 and CeHCFN395 constructs do not effectively stabilize the VP16-induced complex, but this inactivity is reflected in the inability of these two proteins to bind VP16 in a coimmunoprecipitation assay (data not shown). Because the wild-type full-length HCF-2 and CeHCF proteins can stabilize the VP16-induced complex effectively, we conclude that stabilization of the VP16-induced complex is a conserved function of these two human and one C. elegans proteins.
HCF-1 and CeHCF, but not HCF-2, can promote VP16-induced transcription effectively.
Because all three HCF proteins can interact with VP16 and stabilize the VP16-induced complex, we next asked whether the VP16-induced complexes formed by these three different HCFs can promote VP16-induced transcriptional activation in vivo. To compare the abilities of different HCF proteins to activate transcription, we developed an assay in which ectopically expressed HCF proteins were tested for transcriptional activation of a β-globin promoter-reporter construct containing tandem VP16 response elements in the presence of full-length VP16. To circumvent the transcriptional activity of endogenous HCF-1, we used tsBN67 cells at permissive temperature; under these conditions, the cells proliferate normally but VP16 displays relatively little transcriptional activity compared to wild-type BHK21 hamster cells (5, 28). The set of HCF protein expression vectors used for the VP16-induced complex formation assay shown in Fig. 2 were transiently transfected both with and without a VP16 expression vector and the experimental β-globin and internal control α-globin reporter constructs. Additionally, the activities of the wild-type and tsBN67 mutant HCF-1N subunit (HCF-1N1011 and HCF-1N1011/P134S) were assayed. Levels of reporter mRNA synthesis were measured by RNase protection analysis. The levels of HCF and VP16 protein synthesis were monitored by immunoblot analysis as shown in Fig. 3C. The relative protein expression levels of HCF proteins are indicated above each lane. The levels of VP16 synthesis were similar in all samples containing VP16.
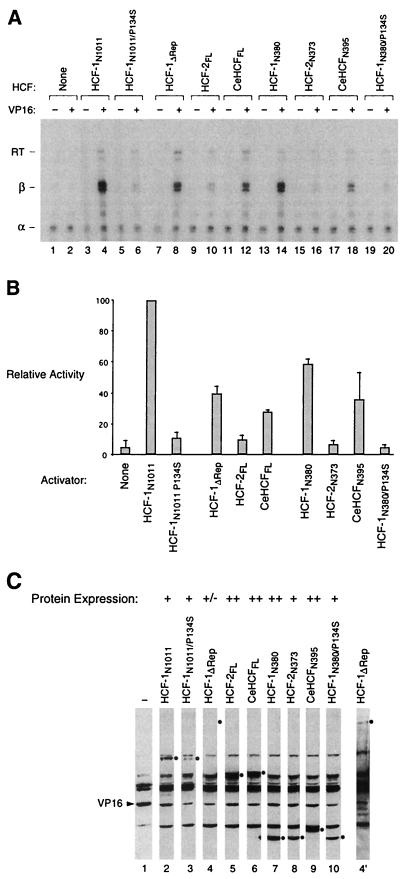
FIG. 3.
VP16 activates transcription in association with HCF-1 and CeHCF but only weakly in association with HCF-2. (A) Transcriptional activation by VP16 in tsBN67 cells grown at 33.5°C. The cells were transfected with an HCF expression construct, a VP16 expression construct, and reporter constructs, and the resulting β-globin and α-globin reporter RNAs were probed by RNase protection analysis as described in Materials and Methods. Odd-numbered samples contained no cotransfected VP16 expression vector. The samples shown here were normalized to the level of internal control α-globin transcript. The positions of the RNase-protected fragments corresponding to α-globin (α), correctly initiated β-globin (β), and read-through (RT) β-globin transcripts are indicated on the left. +, present; −, absent. (B) Quantitation of relative β-globin transcript levels. The intensity of each band corresponding to the β-globin transcript in the samples containing VP16 was measured by phosphorimager analysis. The transcript levels are shown relative to the HCF-1N1011 sample (panel A, lane 4). The results represent the average of two complete experiments; the CeHCFN395 was uncharacteristically high in the experiment not shown here (high error bars), and its apparently higher activity than CeHCFFL shown here is not a true representation of its activity. (C) Immunoblot analysis of the tsBN67 cell extracts used in the in vivo transcription assay shown in panel A. Extracts were resolved by sodium dodecyl sulfate–8% polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane, and the HA-tagged HCF and VP16 proteins were detected with the 12CA5 anti-HA antibody. Only the VP16-containing samples are shown. The position of each relevant HCF species is indicated with a black dot to the right of each lane. A long exposure of lane 4 is shown on the right (lane 4′). Relative levels of HCF-protein synthesis are indicated above each lane. Lane 1, no ectopic HCF protein synthesized.
Figure 3A shows a representative result of such an assay of HCF protein activity. Figure 3B shows the averaged quantitated results of two experiments. Without HCF expression vector transfection (lanes 1 and 2), the presence of VP16 results in little increase in reporter expression, probably owing in part to the relative inactivity of the endogenous mutant HCF-1 protein. Consistent with a low level of endogenous tsBN67 mutant HCF-1 activity, transfection of the tsBN67 mutant HCF-1N1011/P134S expression vector (lanes 5 and 6) resulted in little increased VP16 transcriptional activity. The most active HCF construct assayed was the HCF-1N1011 construct (lane 4). The transcriptional activity of HCF-1ΔRep (lane 8) was two- to threefold lower than that of HCF-1N1011 (lane 4), perhaps owing in part to lower levels of protein synthesis (Fig. 3B). In contrast to these two HCF-1 expression constructs, full-length HCF-2 displays much lower activity (compare lane 10 with lanes 4, 6, and 8), even though it was synthesized at high levels, suggesting that, although it can form a VP16-induced complex effectively (Fig. 2), the resulting complex is not as transcriptionally competent as its HCF-1 counterpart.
In contrast to HCF-2, CeHCF displays significant activity (lane 12) even though it originates from a distantly related organism and does not form a VP16-induced complex as effectively as the human HCF-2 protein (Fig. 2). This result suggests that, once formed, a VP16-induced complex containing CeHCF is more transcriptionally active in mammalian cells than one containing HCF-2. The pattern of differential transcriptional activity observed with the full-length proteins was also observed with the HCF-1N380, HCF-2N373, CeHCFN395, and HCF-1N380/P134S HCFKEL domain proteins (lanes 14, 16, 18, and 20).
Both HCF-2 and CeHCF fail to rescue the tsBN67 cell proliferation defect.
The third assay in which we compared the activities of HCF-1, HCF-2, and CeHCF was rescue of the temperature-sensitive tsBN67 cell proliferation defect caused by the P134S missense mutation in HCF-1 (5). This cell proliferation defect can be rescued by both full-length HCF-1 and the full-length HCF-1N subunit (28). Previous studies with full-length CeHCF and HCF-2 have provided contrasting results: CeHCF (15), but not HCF-2 (7), could rescue the tsBN67 cell proliferation defect. The activity of CeHCF was unexpected because it lacks the HCF-1-specific basic region, which is required for HCF-1 rescue of the tsBN67 defect (Fig. 1) (28). We have since discovered, however, that the original tsBN67 cell population (5) contains a minor population of proliferation revertants that are able to grow at the nonpermissive temperature of 40°C (Reilly and Herr, unpublished).
This finding raised the possibility that the unexpected rescue of tsBN67 cell proliferation by the CeHCF protein might have resulted not from the CeHCF protein but rather from growth of revertant tsBN67 cell colonies in the rescue assay. To test this hypothesis, we used a subclone of tsBN67 cells called tsBN67HR1 (30), which is devoid of revertant cells. Figure 4 shows the results of such an experiment. Cell proliferation was monitored in a colony formation assay in which tsBN67HR1 cells were cotransfected with an HCF expression vector (Fig. 4B) and a puromycin resistance marker, and the colonies were assayed after growth in the presence of puromycin at 33.5°C for 10 days to check for transfection efficiency or at 40°C for 14 days to assay rescue of proliferation at nonpermissive temperature (see Materials and Methods). Growth at the permissive temperature of 33.5°C showed that the transfection efficiencies were similar in all samples (data not shown).
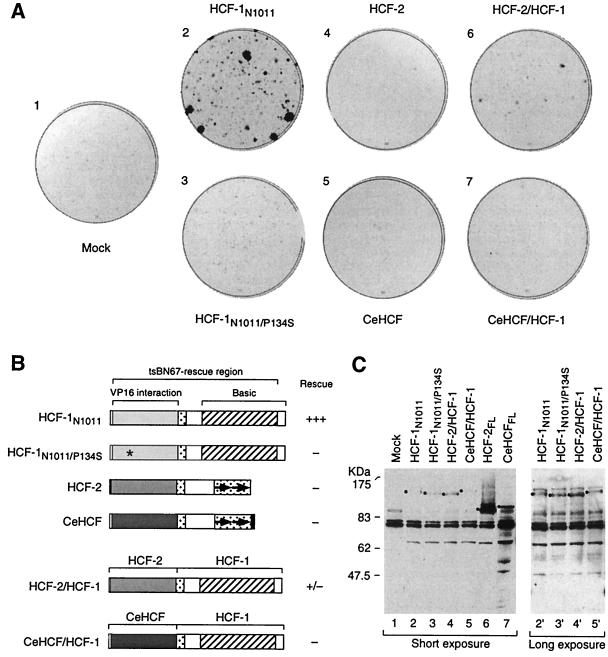
FIG. 4.
The HCF-2KEL repeat region can participate in rescue of the tsBN67 cell proliferation defect. (A) Colony formation assay. tsBN67 cells were transfected with different HCF expression plasmids as indicated. After transfection, the plates were incubated at 40°C for 14 days to permit colony formation. The colonies were visualized by staining them with crystal violet. (B) Schematic diagrams of HCF proteins analyzed and relative levels of rescue of tsBN67 cell proliferation at 40°C. See the legend to Fig. 1A for a description of the diagrams. Asterisk, P134S tsBN67 point mutation. (C) Levels of HCF proteins in transfected tsBN67 cells at nonpermissive temperature. Transfected cells (E plates [see Materials and Methods]) were collected 48 h after transfection and temperature shift to the nonpermissive temperature (40°C) and were used to make protein extracts. The extracts were resolved by sodium dodecyl sulfate–8% polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and probed with the 12CA5 anti-HA epitope tag antibody to monitor the levels of HA-tagged HCF protein synthesis. The position of each relevant band is indicated with a black dot. A long exposure of the immunoblot is shown for lanes 2 to 5 (lanes 2′ to 5′).
As expected, cells transfected with the HCF-1N1011 construct (Fig. 4A, plate 2), but not with the empty vector (plate 1) or mutant HCF-1N1011/P134S expression vector (plate 3), generated colonies of tsBN67 cells at 40°C. As previously described (7), HCF-2 failed to rescue the tsBN67 cell proliferation defect (plate 4), even though it was synthesized at levels higher than HCF-1N1011 (Fig. 4C, compare lanes 2 and 6). In contrast to our previous study (15), however, CeHCF also failed to rescue the tsBN67 defect in tsBN67HR1 cells (Fig. 4A, plate 5), even though it also was present at higher levels than the HCF-1N1011 protein (compare lanes 2 and 7; Fig. 4C). We therefore conclude that, contrary to our earlier findings (15), CeHCF, like human HCF-2, does not complement the tsBN67 HCF-1 defect. The apparent rescue by CeHCF in the earlier experiments may have resulted from higher transfection efficiency with the CeHCF samples, which was not monitored in those experiments (15).
As mentioned above, HCF-2 and CeHCF lack sequences corresponding to the basic region of HCF-1, which in HCF-1 are necessary for rescue of the tsBN67 cell proliferation defect (28). We therefore investigated whether either the HCF-2 or CeHCF HCFKEL repeat region can functionally replace the HCF-1 HCFKEL repeat region for rescue of the tsBN67 cell proliferation defect by exchanging the HCF-1KEL repeats (residues 1 to 363) for the corresponding HCF-2 (residues 1 to 356) or CeHCF (residues 1 to 378) sequences to create HCF-2/HCF-1 and CeHCF/HCF-1 chimeric proteins (Fig. 4B). These chimeric proteins were synthesized at levels similar to those of HCF-1N1011 (Fig. 4C, long exposure of lanes 2′, 4′, and 5′). As shown in Fig. 4A, the HCF-2/HCF-1 chimera, but not the CeHCF/HCF-1 chimera, could rescue the tsBN67 cell proliferation defect to some extent (compare plates 6 and 7 with plate 1). Although not as effective as HCF-1N1011 (plate 2), the activity of the HCF-2/HCF-1 chimera implies that the HCF-2KEL repeat region has retained some of the cellular function(s) of the HCF-1KEL repeat region. The reason(s) for the failure of the CeHCF/HCF-1 chimera to rescue the tsBN67 cell proliferation defect is not clear. The reason could be an inherent inability of CeHCF to promote mammalian-cell proliferation, or the activity or stability of the CeHCFKEL repeat region may be temperature sensitive, because the temperature required for the tsBN67 cell proliferation rescue assay (40°C) is 20°C higher than the optimal temperature for C. elegans growth (20°C).
CeHCF interacts with LZIP.
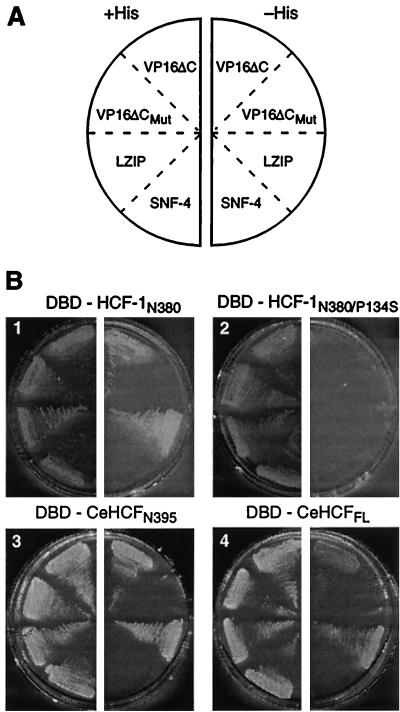
The studies described above show that, whereas CeHCF is functional in the two viral assays used—VP16-induced-complex formation and transcriptional activation—it is not active in the one cellular assay used—rescue of tsBN67 cell proliferation. The results of the rescue of tsBN67 cell proliferation, however, are problematic, as mentioned above. To test CeHCF function in a more defined molecular assay of cellular function, we tested the ability of CeHCF to associate with the human leucine zipper protein LZIP in an S. cerevisiae two-hybrid assay (3). In that assay, we compared the abilities of the HCF-1 and CeHCF proteins fused to the GAL4 DBD and LZIP or VP16ΔC fused to the GAL4 transcriptional AD to activate transcription of the HIS3 gene and thus support growth of HIS3− yeast in the absence of histidine.
Figure 5 shows the result of this experiment. Four GAL4 DBD fusions to the HCF-1N380, HCF-1N380/P134S, CeHCFN395, and CeHCFFL proteins were paired with four GAL4 AD fusions to (i) the VP16ΔC protein; (ii) the VP16ΔCE361A mutant (VP16ΔCMut), which fails to associate with HCF-1 (14); (iii) the human LZIP protein; and (iv) the irrelevant yeast protein SNF-4. As expected (4, 17), HCF-1N380 (plate 1), but not HCF-1N380/P134S (plate 2), interacted with the GAL4 AD-VP16ΔC and the GAL4 AD-LZIP proteins, but both failed to interact with the GAL4 AD-VP16ΔCMut and SNF-4 proteins. Like wild-type HCF-1N380, the GAL4 DBD-CeHCFN395 (plate 3), and GAL4 DBD-CeHCFFL (plate 4) proteins interacted with VP16ΔC and LZIP but not the VP16ΔCMut and SNF-4 fusion proteins. Thus, full-length CeHCF, as well as the CeHCFKEL repeat region, can associate with LZIP, suggesting that, like HCF-1, CeHCF possesses the ability to associate with both a human protein—LZIP—and the protein of a human viral pathogen—VP16. For unexplained reasons, HCF-2 failed to associate with either VP16 or LZIP in this yeast two-hybrid assay, even though it was synthesized at the same levels as CeHCF (data not shown). Nevertheless, the results shown here suggest that the association of HCF-1 and CeHCF with both LZIP and VP16 reflects the conservation of an important activity during metazoan evolution.
FIG. 5.
C. elegans HCF interacts with human LZIP in a yeast two-hybrid assay. A yeast GAL1-HIS3 reporter strain was transformed with expression plasmids encoding different HCF proteins fused to the GAL4 DBD together with an expression plasmid encoding either one of two known HCF-1 binding proteins (VP16 and LZIP) or nonbinding proteins (VP16ΔC and SNF-4) fused to a GAL4 AD. (A) Key for GAL4-AD fusion protein samples shown in panel B. (B) Yeast two-hybrid assays. The DBD fusion protein used is indicated above each plate. Successful growth with histidine (+His; left half of each plate) shows that each expression plasmid has no lethal effect. The interaction between a DBD fusion protein and an AD fusion protein is demonstrated by successful growth in the absence of histidine (−His; right half of each plate).
DISCUSSION
We have characterized and compared three HCF proteins in their interaction with the viral protein VP16, both in VP16-induced-complex formation and transcriptional activation and in their activities in mammalian cells. Our results show that all three HCF proteins—the human HCF-1 and HCF-2 proteins and the C. elegans CeHCF protein—can associate with VP16 and form a stable VP16-induced complex with Oct-1 (Fig. 2). The resulting VP16-induced complexes, however, have different activities for transcriptional activation (Fig. 3), raising the possibility that the two human HCF proteins can have opposing effects on how VP16 influences the outcome of viral infection. In contrast to HCF-2, but like HCF-1, CeHCF can both associate with VP16 and induce transcriptional activation by VP16 in mammalian cells, even though CeHCF does not normally interact with VP16, a key regulator of a human pathogen. This result suggests that HCF-1 and CeHCF possess a shared and conserved cellular activity that is used by VP16 to form a transcriptionally active VP16-induced complex. In contrast to our previous findings (15), however, CeHCF fails to complement the tsBN67 cell proliferation defect, although this failure may reflect more about the normal growth temperatures of humans and worms than about an inherent difference in cellular HCF function in these different species.
VP16 discriminates between HCF-1 and HCF-2 but not at the level of VP16-induced-complex formation.
Our results show that both HCF-1 and HCF-2 can stabilize the VP16-induced complex effectively. We did not expect this result, because Johnson et al. (7) had shown that the region of HCF-2 (HCF-2N373) analogous to a minimal region of HCF-1 (HCF-1N380) that stabilizes the VP16-induced complex (28) is unable to stabilize the VP16-induced complex effectively. We have reproduced this result, but when we assayed the full-length HCF-2 protein, it did stabilize the VP16-induced complex effectively (Fig. 2). We do not know the reason for the inactivity of the truncated HCF-2N373 protein. We note, however, that the HCFKEL repeats only extend to residue 360 in HCF-1 and residue 353 in HCF-2. Perhaps the 20 carboxy-terminal non-HCFKEL repeat SAS1N residues in the HCF-2N373 construct, but not in the HCF-1N380 construct, interfere with VP16-induced complex formation. Alternatively, the carboxy-terminal SAS1C region of HCF-2 may stabilize the VP16-induced complex, as has been described previously under certain conditions for HCF-1 (12).
Whatever the reason for the failure of the HCF-2N373 protein to stabilize the VP16-induced complex, the ability of the full-length wild-type HCF-2 protein to stabilize the VP16-induced complex has important implications for our understanding of the interplay of HCF-1 and HCF-2 with VP16 in human cells. The activity of the wild-type HCF-2 protein suggests that HCF-2 association with VP16 may influence the outcome of HSV infection, by either promoting or inhibiting lytic or latent infection.
Surprisingly, although HCF-2 can stabilize the VP16-induced complex effectively, it fails to support activation of transcription by VP16. Thus, HCF-1 and HCF-2 have dramatically different activities as a result of their association with VP16, in one instance (HCF-1) activating transcription and in the other instance (HCF-2) failing to do so effectively (Fig. 3). We do not know the reason for the difference in transcriptional activity by HCF-1 and HCF-2. One possibility is that HCF-2 lacks a carboxy-terminal NLS found in HCF-1 (Fig. 1) and the localization of HCF-2 is variable after transient overexpression (7). We do not favor this hypothesis, however, because HCF-1, which is a chromatin-bound protein, does not require the NLS for either nuclear localization or chromatin association (30) or for transcriptional activation with VP16 in either yeast (28) or mammalian cells (Fig. 3, HCF-1N1011). Furthermore, when stably synthesized in HeLa cells without overexpression, HCF-2 is also associated with nuclear chromatin (J. Wysocka and W. Herr, unpublished results). We therefore favor the hypothesis that an HCF-2-containing VP16-induced complex is able to bind appropriately to a VP16-inducible promoter but that the complex is inactive either because HCF-2 lacks an essential activity present in HCF-1 or because it possesses an inhibitory activity not present in HCF-1. For example, cyclin-dependent kinase activity is required for transcriptional activation by the VP16-induced complex (8). Perhaps an HCF-2-containing VP16-induced complex is not able to respond to cyclin-dependent kinase activity. Whatever the reason, the relative inability of HCF-2 to activate transcription compared to HCF-1 suggests that HCF-2 might inhibit VP16 function and thus perhaps either inhibit lytic infection or promote latent infection by HSV.
HCF-2 may also serve as an inhibitor of HCF-1 function in the uninfected cell. Johnson et al. (7) showed that full-length wild-type HCF-2 can inhibit the ability of wild-type HCF-1 to rescue the temperature-sensitive tsBN67 cell proliferation defect. Because we have shown here that HCF-1 and HCF-2 can associate similarly with VP16, it is plausible that HCF-2 can also associate similarly with cellular targets of HCF-1 but, as with VP16, display different activities with the shared targets.
The differences in HCF-1 and HCF-2 activity may result from differences in the regions conserved between the two proteins, such as the HCFKEL repeat region, or in the sequences that are not conserved between the two proteins, such as those corresponding to the HCF-1-specific basic region that are essential for tsBN67 cell rescue by HCF-1 (28). Indeed, both of these possibilities may play a role in the inhibition of HCF-1 rescue of the tsBN67 cell proliferation defect by HCF-2, because when the HCF-2 Kelch repeat region is fused to the HCF-1 SAS1N and basic regions, the chimeric HCF-2/HCF-1 protein displays an ability to rescue the tsBN67 cell proliferation defect that is intermediate between those of HCF-1 and HCF-2 (Fig. 4). The finding that the HCF-2KEL repeat region cannot functionally replace the HCF-1KEL repeat region to wild-type levels suggests that other functions of the HCF-1KEL repeat region in addition to association with cellular VP16-like binding activities are required to promote cell proliferation, as suggested previously by Mahajan and Wilson (20). In summary, the family of mammalian HCF-1 and HCF-2 proteins may represent a pair of regulators of cell proliferation that counteract each other's activities.
C. elegans HCF shares properties of human HCF-1 and HCF-2.
In contrast to human cells, C. elegans has only one evident HCF-like protein (15). The overall structure of this protein more closely resembles human HCF-2 (Fig. 1A), but in some features CeHCF more closely resembles human HCF-1, including amino acid sequence similarity (Fig. 1B) and VP16 transcriptional activation (Fig. 3). Like both HCF-1 and HCF-2, however, it is able to associate effectively with VP16. Because VP16 is an activator of the lytic pathway of a human viral pathogen not known to exist in worms, we believe that the ability of CeHCF to associate with VP16 is due to a cellular protein-protein interaction that has been conserved between humans and worms and that is mimicked by VP16. Underscoring the significance of this observation is the finding that, in contrast to HCF proteins, the other cellular component of the VP16-induced complex—Oct-1—has not conserved its ability to associate with VP16 even in a species as closely related to humans as mice, much less in invertebrates (2, 22).
A potential conserved cellular target of HCF proteins is LZIP, which, like VP16, can associate with both HCF-1 and CeHCF. Unlike in Drosophila, however, which contains the LZIP-like protein BBF-2/dCREB-A (4, 17), there is no evident LZIP homolog in worms (S. Lee and W. Herr, unpublished results). Furthermore, there is no known C. elegans homolog of the second human HCF-1-interacting protein called Zhangfei (19). One possible explanation for the lack of evident conservation of LZIP and Zhangfei homologs in C. elegans is that the family of HCFKEL repeat-interacting proteins in human cells is larger than presently known. Consistent with this hypothesis, and in contrast to LZIP (18) and Zhangfei (19), HCF-1 is an abundant human protein, and it is entirely tethered to chromatin through its VP16 interaction domain, suggesting that other human proteins are also involved in tethering HCF-1 to chromatin (30). We suggest that one or more of these other proteins have homologs in C. elegans that are involved in shared activities of HCF-1 and CeHCF.
The hybrid nature of CeHCF compared to the human HCF-1 and HCF-2 proteins suggests that it performs functions in the worm that in human cells are performed by either HCF-1 or HCF-2. Determination of those functions should help elucidate the function of the human HCF proteins, one of which at least—HCF-1—plays a critical role in human cell proliferation and HSV pathogenesis.
ACKNOWLEDGMENTS
We thank Patrick Reilly for suggesting that the rescue of tsBN67 cell proliferation by CeHCF may result from revertant cells in the tsBN67 cell population and for help with the tsBN67 rescue assay; A. Wilson for HCF-2 constructs; G. Hannon, M. Hengartner, N. Hernandez, and G. Thomsen for discussions and guidance; R. Freiman for early studies of HCF-1 activation of VP16-induced transcription in tsBN67 cells; J. Wysocka for communication of unpublished results; and N. Hernandez, E. Julien, C. Schmitt, A. Stenlund, and J. Wysocka for comments on the manuscript.
These studies were supported by PHS grants GM54598 and CA13106 and Cold Spring Harbor Laboratory funds.
REFERENCES
- 1.Adams J, Kelso R, Cooley L. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 2000;10:17–24. doi: 10.1016/s0962-8924(99)01673-6. [DOI] [PubMed] [Google Scholar]
- 2.Cleary M A, Stern S, Tanaka M, Herr W. Differential positive control by Oct-1 and Oct-2: activation of a transcriptionally silent motif through Oct-1 and VP16 corecruitment. Genes Dev. 1993;7:72–83. doi: 10.1101/gad.7.1.72. [DOI] [PubMed] [Google Scholar]
- 3.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 4.Freiman R N, Herr W. Viral mimicry: common mode of association with HCF by VP16 and the cellular protein LZIP. Genes Dev. 1997;11:3122–3127. doi: 10.1101/gad.11.23.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goto H, Motomura S, Wilson A C, Freiman R N, Nakabeppu Y, Fukushima K, Fujishima M, Herr W, Nishimoto T. A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 interaction. Genes Dev. 1997;11:726–732. doi: 10.1101/gad.11.6.726. [DOI] [PubMed] [Google Scholar]
- 6.Herr W. The herpes simplex virus VP16-induced complex: mechanisms of combinatorial transcriptional regulation. Cold Spring Harbor Symp Quant Biol. 1998;63:599–607. doi: 10.1101/sqb.1998.63.599. [DOI] [PubMed] [Google Scholar]
- 7.Johnson K M, Mahajan S S, Wilson A C. Herpes simplex virus transactivator VP16 discriminates between HCF-1 and a novel family member, HCF-2. J Virol. 1999;73:3930–3940. doi: 10.1128/jvi.73.5.3930-3940.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jordan R, Schang L, Schaffer P A. Transactivation of herpes simplex virus type 1 immediate-early gene expression by virion-associated factors is blocked by an inhibitor of cyclin-dependent protein kinases. J Virol. 1999;73:8843–8847. doi: 10.1128/jvi.73.10.8843-8847.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kristie T M, LeBowitz J H, Sharp P A. The octamer-binding proteins form multi-protein-DNA complexes with HSV αTIF regulatory protein. EMBO J. 1989;8:4229–4238. doi: 10.1002/j.1460-2075.1989.tb08608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kristie T M, Pomerantz J L, Twomey T C, Parent S A, Sharp P A. The cellular C1 factor of the herpes simplex virus enhancer complex is a family of polypeptides. J Biol Chem. 1995;270:4387–4394. doi: 10.1074/jbc.270.9.4387. [DOI] [PubMed] [Google Scholar]
- 11.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 12.LaBoissière S, Walker S, O'Hare P. Concerted activity of host cell factor subregions in promoting stable VP16 complex assembly and preventing interference by the acidic activation domain. Mol Cell Biol. 1997;17:7108–7118. doi: 10.1128/mcb.17.12.7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LaBoissière S, Hughes T, O'Hare P. HCF-dependent nuclear import of VP16. EMBO J. 1999;18:480–489. doi: 10.1093/emboj/18.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai J-S, Herr W. Interdigitated residues within a small region of VP16 interact with Oct-1, HCF, and DNA. Mol Cell Biol. 1997;17:3937–3946. doi: 10.1128/mcb.17.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Hengartner M O, Herr W. Selected elements of herpes simplex virus accessory factor HCF are highly conserved in Caenorhabditis elegans. Mol Cell Biol. 1999;19:909–915. doi: 10.1128/mcb.19.1.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu R, Yang P, O'Hare P, Misra V. Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol Cell Biol. 1997;17:5117–5126. doi: 10.1128/mcb.17.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu R, Yang P, Padmakumar S, Misra V. The herpesvirus transactivator VP16 mimics a human basic leucine zipper protein, Luman, in its interaction with HCF. J Virol. 1998;72:6291–6297. doi: 10.1128/jvi.72.8.6291-6297.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu R, Misra V. Potential role for Luman, the cellular homologue of herpes simplex virus VP16 (α gene trans-inducing factor), in herpesvirus latency. J Virol. 2000;74:934–943. doi: 10.1128/jvi.74.2.934-943.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu R, Misra V. Zhangfei: a second cellular protein interacts with herpes simplex virus accessory factor HCF in a manner similar to Luman and VP16. Nucleic Acids Res. 2000;28:2446–2454. doi: 10.1093/nar/28.12.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahajan S S, Wilson A C. Mutations in host cell factor 1 separate its role in cell proliferation from recruitment of VP16 and LZIP. Mol Cell Biol. 2000;20:919–928. doi: 10.1128/mcb.20.3.919-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Hare P. The virion transactivator of herpes simplex virus. Semin Virol. 1993;4:145–155. [Google Scholar]
- 22.Suzuki N, Peter W, Ciesiolka T, Gruss P, Scholer H R. Mouse Oct-1 contains a composite homeodomain of human Oct-1 and Oct-2. Nucleic Acids Res. 1993;21:245–252. doi: 10.1093/nar/21.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka M, Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 24.Thompson C C, McKnight S L. Anatomy of an enhancer. Trends Genet. 1992;8:232–236. [Google Scholar]
- 25.Wilson A C, LaMarco K, Peterson M G, Herr W. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell. 1993;74:115–125. doi: 10.1016/0092-8674(93)90299-6. [DOI] [PubMed] [Google Scholar]
- 26.Wilson A C, Cleary M A, Lai J-S, LaMarco K, Peterson M G, Herr W. Combinational control of transcription: the herpes simplex virus VP16-induced complex. Cold Spring Harbor Symp Quant Biol. 1993;18:167–178. doi: 10.1101/sqb.1993.058.01.021. [DOI] [PubMed] [Google Scholar]
- 27.Wilson A C, Peterson M G, Herr W. The HCF repeat is an unusual proteolytic cleavage signal. Genes Dev. 1995;9:2445–2458. doi: 10.1101/gad.9.20.2445. [DOI] [PubMed] [Google Scholar]
- 28.Wilson A C, Freiman R N, Goto H, Nishimoto T, Herr W. VP16 targets an amino-terminal domain of HCF involved in cell cycle progression. Mol Cell Biol. 1997;17:6139–6146. doi: 10.1128/mcb.17.10.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson A C, Boutros M, Johnson K M, Herr W. HCF-1 amino- and carboxy-terminal subunit association through two separate sets of interaction modules: involvement of fibronectin type 3 repeats. Mol Cell Biol. 2000;20:6721–6730. doi: 10.1128/mcb.20.18.6721-6730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wysocka J, Reilly P T, Herr W. Loss of HCF-1 chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol Cell Biol. 2001;21:3820–3829. doi: 10.1128/MCB.21.11.3820-3829.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wysocka J, Liu Y, Kobayashi R, Herr W. Developmental and cell-cycle regulation of Caenorhabditis elegans HCF phosphorylation. Biochemistry. 2001;40:5786–5794. doi: 10.1021/bi010086o. [DOI] [PubMed] [Google Scholar]
- 32.Xue F, Cooley L. kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell. 1993;72:681–693. doi: 10.1016/0092-8674(93)90397-9. [DOI] [PubMed] [Google Scholar]
- 33.Zoller M J, Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]