Abstract
Purpose
For patients with congenital heart disease (CHD), the most common birth defect, genetic evaluation is not universally accepted, and current practices are anecdotal. Here, we analyzed genetic evaluation practices across centers, determined diagnostic yield of testing, and identified phenotypic features associated with abnormal results.
Methods
This is a multicenter cross-sectional study of 5 large children’s hospitals, including 2899 children ≤14 months undergoing surgical repair for CHD from 2013 to 2016, followed by multivariate logistics regression analysis.
Results
Genetic testing occurred in 1607 of 2899 patients (55%). Testing rates differed highly between institutions (42%-78%, P < .001). Choice of testing modality also differed across institutions (ie, chromosomal microarray, 26%-67%, P < .001). Genetic testing was abnormal in 702 of 1607 patients (44%), and no major phenotypic feature drove diagnostic yield. Only 849 patients were seen by geneticists (29%), ranging across centers (15%-52%, P < .001). Geneticist consultation associated with increased genetic testing yield (odds ratio: 5.7, 95% CI 4.33-7.58, P < .001).
Conclusion
Genetics evaluation in CHD is diagnostically important but underused and highly variable, with high diagnostic rates across patient types, including in infants with presumed isolated CHD. These findings support recommendations for comprehensive testing and standardization of care.
Keywords: Cardiovascular genetics, Congenital heart disease, Genetic testing
Introduction
Congenital heart disease (CHD) is the most common congenital anomaly and is the leading cause of death due to birth defects.1 CHD frequently has a genetic etiology, with a genetic syndrome identified in up to 25% of patients with CHD.2, 3, 4, 5, 6, 7, 8, 9, 10 A scientific statement from the American Heart Association highlights the need for genetic testing in CHD to identify comorbidities, formulate prognosis, identify risk to other family members, and identify recurrence risk for both patients and family members.11,12 Obtaining a genetic diagnosis in the newborn period allows early implementation of health supervision and early intervention. In CHD, genetic testing results have been shown to inform prognosis, including poor respiratory13 and surgical outcomes,14, 15, 16, 17 and may increasingly drive management. As genetic testing increasingly drives clinical care in CHD, geneticist involvement is important to formulate a differential diagnosis, select optimal testing, interpret complex results, and optimize cost.
Recommendations for genetic testing are available for infants with birth defects, including CHD.18,19 Despite these guidelines, the degree to which practice varies between centers is unknown. In recent single-center studies of infants with CHD, genetic testing is underused,20,21 and there seems to be significant practice variability between centers, with reported genetic testing rates ranging from 25% to 87%.9,20, 21, 22, 23, 24, 25, 26 However, the interpretation of data from these studies is limited, given that they are single-center cohorts, focused on older technologies such as chromosome analysis, with different patient populations and inclusion criteria. Nevertheless, studies of CHD genetic testing report consistently high diagnostic yield (25%-36%).9,20, 21, 22, 23, 24 We hypothesize that underutilization and variability in genetic testing leads to missed genetic diagnoses that affect clinical care. To improve care, we need to assess and compare genetic evaluation practices.
This is a large multicenter review of genetic evaluation and testing practices in infants with CHD. Given the recommendation for widespread utilization of chromosome microarray analysis (CMA) in 2010,18 and a highly variable implementation of exome sequencing (ES) and genome sequencing (GS) in recent years, we chose a time frame from 2013 to 2016, a window with consistent recommendations and potentially less testing variability. We compare genetic testing rate and yield across institutions and patient subtypes, including cardiac lesion type and extracardiac features. This information will help drive recommendations to standardize and improve care in these critically ill patients.
Materials and Methods
Study population
We performed a multicenter retrospective analysis of genetic testing and evaluation practices and results. The Cytogenomics of Cardiovascular Malformations Consortium27 is a multisite alliance of geneticists and cardiologists contributing to a CHD registry, and the study sites included 5 children’s hospitals participating in the Consortium: Baylor College of Medicine, Indiana University School of Medicine, Nationwide Children’s Hospital, Children’s Healthcare of Atlanta at Emory University, and Medical College of Wisconsin.27 Cohort ascertainment used the Society of Thoracic Surgeons (STS) National Database to comprehensively identify patients with significant CHD.28 STS is the largest database in North America dealing with congenital cardiac malformations. STS consists of congenital heart surgery procedure records and captures all surgical procedures occurring at participating sites, including the 5 centers in this study. We included a total of 2899 children who underwent surgical repair for CHD at ≤14 months of age. The patients underwent surgical repair between January 1, 2013, and January 1, 2016 (a historic time period with consistent recommendations for genetic testing), and the data collection range was from November 1, 2011, to December 31, 2016. The study was approved by the institutional review board at each center.
Data collection
Patient medical records were comprehensively reviewed, and study data were entered into a Research Electronic Data Capture database hosted at Indiana University.29,30 Genetic testing categories include the following: chromosome analysis, CMA, fluorescence in situ hybridization (FISH) analysis, and molecular testing including single-gene sequencing, gene panels, and exome testing. Interpretation from the clinical testing laboratory was used, including both in-house and send-out testing results, and prenatal results were used if available. Testing was reported as “normal,” “abnormal,” or “not done/unknown.” Abnormal testing results were reviewed in more detail, when available, by a genetic counselor and further categorized as positive (pathogenic/likely pathogenic), or of undetermined significance (variants of uncertain significance, runs of homozygosity, incidental findings, unable to determine pathogenicity with available information, and variants initially reported being of uncertain significance but subsequently downgraded to likely benign or population-based variants). Additional congenital anomalies were determined from documentation within the medical record and classified by location, including the brain, ear, nose or throat, eye, chest, lung, diaphragm, congenital diaphragmatic hernia, gastrointestinal and abdominal wall, kidney, spleen, pancreas/annular pancreas, liver and gallbladder, anus, imperforate anus or anal atresia, genitourinary, ribs and vertebrae, limbs, lymphatic, lymphatic dysplasia, skin, arteriovenous malformation, and umbilical. During statistical analysis, categories with too few patients were grouped as “other.” All fields not obtained directly from the STS database were obtained from the medical record, using thorough review by a trained medical professional. If there was no indication after thorough chart review, the variable was classified as “not present, not done, or unknown.” Data obtained directly from the STS National Database included date of birth, sex, race, ethnicity, CHD diagnosed prenatally, multiple gestation, weight at birth, age at first surgery, cardiac diagnoses, primary procedure, number of cardiac surgeries, and length of hospitalization. The race and ethnicity data collected from the STS National Database used fixed categories, including an option of “other.” A subset of race and ethnicity data was verified within the electronic medical record, which is most often obtained from patient self-reporting during hospital registration, and similarly used fixed categories determined by each hospital, including an option of “other.” The “other” category for race and ethnicity was also used for comparison in data analysis when the numbers of those in some subgroups were too small for meaningful analyses.
Cardiac classification
We identified the fundamental diagnosis listed in the STS database for each patient. Based on this diagnosis information, we assigned an overall cardiac diagnosis type in the classification system developed by the National Birth Defects Prevention Study.31 This classification was done in a hierarchical fashion based on the approach used by prior epidemiological studies.32
Statistical analysis
The univariate comparisons of patients’ characteristics, testing rate, and testing yield across centers were conducted using Pearson’s χ2 test (all expected cell counts in contingency table ≥ 5) or Fisher exact test (any expected cell counts <5) for categorical variables and one-way analysis of variance for continuous variables. The effect (ie, odds ratio [OR]) of covariates on testing rate and testing yield was estimated via multivariate logistic regression models, including center, sex, race, family history of CHD, presence and type of major congenital malformation, geneticist consultation, history of intrauterine growth restriction (IUGR) or small for gestational age (SGA), CHD group, infant of diabetic mother, maternal infection (any), maternal teratogens (alcohol, illegal drugs, tobacco, and prescription opioids at any point during the pregnancy), prenatal diagnosis, and extracorporeal membrane oxygenation (ECMO). Survey of all participating centers indicated that genetic testing was not limited by insurance at any involved center; therefore, it was not used as a variable. For testing yield, the number of genetic tests (chromosome analysis, FISH, CMA, and molecular) were also adjusted. We reported nominal testing P values and determined statistical significance using Bonferroni adjustment to account for the multiple testing during univariate analyses. For multivariate analysis, we used Tukey’s honestly significance difference test to account for the post hoc pairwise comparison between centers. All analyses were conducted in R version 3.5.2 (R Foundation for Statistical Computing).
Results
Cohort description
There were 2899 patients ≤14 months of age who underwent surgical repair of CHD at 1 of the 5 tertiary care centers participating in the study. Patient characteristics across centers are provided, including maternal and environmental factors (focusing on factors implicated in CHD), clinical characteristics, extracardiac malformations, surgical data, need for ECMO, and vital status (Table 1, Supplemental Table 1). Within the entire cohort, 1590 (55%) were male, 2310 (80%) were born full term, and the average birth weight was 2.99 kg. Apparently isolated CHD occurred in 2362 (82%), whereas 537 (19%) had CHD plus additional congenital anomalies (multiple congenital anomalies [MCAs]). CHD types were classified as per previous studies (Supplemental Table 1).25,31,32 Prenatal diagnosis of CHD occurred in 1233 (43%). An average of 1.75 cardiac surgeries per patient were performed, and ECMO was required in 10%. Mortality, defined as death during the data collection window, occurred in 10%.
Table 1.
Patient characteristics
| No. (%) |
P Value for Comparison Across Centers | |
|---|---|---|
| N = 2899 | ||
| Sex | .396 | |
| Male | 1590 (54.8%) | |
| Female | 1308 (45.1%) | |
| Unknown | 1 | |
| Race | <.001 | |
| African American /Black | 576 (19.9%) | |
| Asian | 120 (4.1%) | |
| Other | 165 (5.7%) | |
| White | 2038 (70.3%) | |
| Weight at birth (kg) | .392 | |
| Mean (SD) | 2.99 (0.69) | |
| Gestational age | .676 | |
| Term (≥37 wk) | 2310 (79.7%) | |
| Late preterm (32-37 wk) | 507 (17.5%) | |
| Preterm (<32 wk) | 80 (2.8%) | |
| Unknown | 2 (0.1%) | |
| Additional congenital malformation | <.001 | |
| Yes | 537 (18.5%) | |
| No | 2362 (81.5%) | |
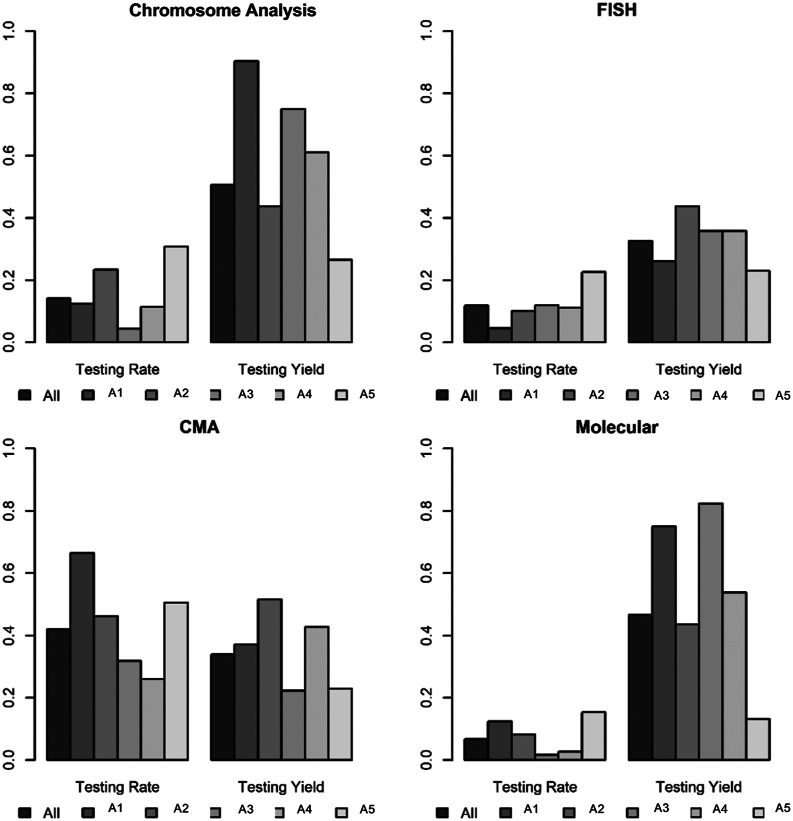
Participants included children ≤14 months of age who underwent surgical repair for congenital heart disease across 5 centers, A1-A5 (all large children’s hospitals participating in the Cytogenomics of Cardiovascular Malformations Consortium). The proportion of patients who had genetic testing (testing rate) and the proportion of patients whose genetic testing results were reported as abnormal (testing yield) were analyzed, across 4 major genetic testing methods: chromosome analysis, FISH analysis, CMA, and molecular testing. The proportion of patients who received testing, the choice of modality, and the yield of testing differed significantly across centers.
CMA, chromosomal microarray analysis; FISH, fluorescence in situ hybridization analysis.
Genetic testing
Of the 2899 patients with CHD included in the study, genetic testing was performed in 1607 (55%) (Table 2). In patients with apparently isolated CHD, genetic testing occurred in 1212 of 2362 patients (51%) and in patients with MCAs, genetic testing occurred in 395 of 537 (74%). CMA was the most frequent testing modality, performed in 1220 (42%), followed by chromosome analysis in 408 (14%) and FISH analysis in 344 (12%) (Figure 1, Table 3). Molecular testing was least frequent, occurring in 197 (7%).
Table 2.
Frequency of genetic testing completion and yields
| Centers | Total | A1 | A2 | A3 | A4 | A5 | P Value |
|---|---|---|---|---|---|---|---|
| All patients | N = 2899 | n = 503 | n = 478 | n = 1001 | n = 475 | n = 442 | |
| Testing | |||||||
| Yes | 1607 (55.4%) | 393 (78.1%) | 286 (59.8%) | 432 (43.2%) | 201 (42.3%) | 295 (66.7%) | <.001 |
| No | 1292 (44.6%) | 110 (21.9%) | 192 (40.2%) | 569 (56.8%) | 274 (57.7%) | 147 (33.3%) | |
| Abnormal | <.001 | ||||||
| Yes | 702 (43.7%) | 202 (51.4%) | 173 (60.5%) | 143 (33.1%) | 96 (47.8%) | 88 (29.8%) | |
| No | 905 (56.3%) | 191 (48.6%) | 113 (39.5%) | 289 (66.9%) | 105 (52.2%) | 207 (70.2%) | |
| Isolated CHD | N = 2362 (81.5%) | n = 418 | n = 373 | n = 832 | n = 380 | n = 359 | |
| Testing | |||||||
| Yes | 1212 (51.3%) | 316 (75.6%) | 209 (56.0%) | 315 (37.9%) | 145 (38.2%) | 227 (63.2%) | <.001 |
| No | 1150 (48.7%) | 102 (24.4%) | 164 (44.0%) | 517 (62.1%) | 235 (61.8%) | 132 (36.8%) | |
| Abnormal | |||||||
| Yes | 486 (40.1%) | 114 (45.6%) | 127 (60.8%) | 93 (29.5%) | 62 (42.8%) | 60 (26.4%) | <.001 |
| No | 726 (59.9%) | 172 (54.4%) | 82 (39.2%) | 222 (70.5%) | 83 (57.2%) | 167 (73.6%) | |
| CHD + MCA | N = 537 (18.5%) | n = 85 | n = 105 | n = 169 | n = 95 | n = 83 | |
| Testing | <.001 | ||||||
| Yes | 395 (73.6%) | 77 (90.6%) | 77 (73.3%) | 117 (69.2%) | 56 (58.9%) | 68 (81.9%) | |
| No | 142 (26.4%) | 8 (9.4%) | 28 (26.7%) | 52 (30.8%) | 39 (41.4%) | 15 (18.1%) | |
| Abnormal | |||||||
| Yes | 216 (54.7%) | 58 (75.3%) | 46 (59.7%) | 50 (42.7%) | 34 (60.7%) | 28 (41.2%) | <.001 |
| No | 179 (45.3%) | 19 (24.7%) | 31 (40.3%) | 67 (57.3%) | 22 (39.3%) | 40 (58.8%) |
CHD, congenital heart disease; MCA, multiple congenital anomaly.
Figure 1.
Testing rate and testing yield by testing modality across centers. CMA, chromosomal microarray analysis; FISH, fluorescence in situ hybridization analysis.
Table 3.
Testing rate and testing yield by cardiac lesion
| Any Tests | Chromosome Analysis |
FISH | CMA | Molecular | |
|---|---|---|---|---|---|
| All patients | |||||
| Testing | |||||
| Yes | 1607 (55.4%) | 408 (14.1%) | 344 (11.9%) | 1220 (42.1%) | 197 (6.8%) |
| No | 1292 (44.6%) | 2491 (85.9%) | 2555 (88.1%) | 1679 (57.9%) | 2702 (93.2%) |
| Abnormal | |||||
| Yes | 702 (43.7%) | 207 (50.7%) | 112 (32.6%) | 413 (33.9%) | 92 (46.7%) |
| No | 905 (56.3%) | 201 (49.3%) | 232 (67.4%) | 807 (66.1%) | 105 (53.5%) |
| Conotruncal defect | |||||
| Testing | |||||
| Yes | 599 (66.2%) | 89 (9.8%) | 179 (19.8%) | 464 (51.3%) | 71 (7.8%) |
| No | 306 (33.8%) | 816 (90.2%) | 726 (80.2%) | 441 (48.7%) | 834 (92.2%) |
| Abnormal | |||||
| Yes | 234 (39.1%) | 24 (27.0%) | 59 (33.0%) | 157 (33.8%) | 32 (45.1%) |
| No | 365 (60.9%) | 65 (73.0%) | 120 (67.0%) | 307 (66.2%) | 39 (54.9%) |
| LVOTO | |||||
| Testing | |||||
| Yes | 362 (58.9%) | 89 (14.5%) | 67 (10.9%) | 312 (50.7%) | 46 (7.5%) |
| No | 253 (41.1%) | 526 (85.5%) | 548 (89.1%) | 303 (49.3%) | 569 (92.5%) |
| Abnormal | |||||
| Yes | 128 (35.4%) | 27 (30.3%) | 15 (22.4%) | 99 (31.7%) | 18 (39.1%) |
| No | 234 (64.6%) | 62 (69.7%) | 52 (77.6%) | 213 (68.3%) | 28 (60.9%) |
| Septal defect | |||||
| Testing | |||||
| Yes | 168 (35.3%) | 60 (12.6%) | 25 (5.3%) | 112 (23.5%) | 23 (4.8%) |
| No | 308 (64.7%) | 416 (87.4%) | 451 (94.7%) | 364 (76.5%) | 453 (95.2%) |
| Abnormal | |||||
| Yes | 94 (56.0%) | 47 (78.1%) | 11 (44.0%) | 44 (39.3%) | 9 (39.1%) |
| No | 74 (44.0%) | 13 (21.7%) | 14 (56.0%) | 68 (60.7%) | 14 (60.9%) |
| AVSD | |||||
| Testing | |||||
| Yes | 166 (51.4%) | 111 (34.4%) | 19 (5.9%) | 63 (19.5%) | 12 (3.7%) |
| No | 157 (48.6%) | 212 (65.6%) | 304 (94.1%) | 260 (80.5%) | 311 (96.3%) |
| Abnormal | |||||
| Yes | 132 (79.5%) | 98 (88.3%) | 13 (68.4%) | 33 (52.4%) | 8 (66.7%) |
| No | 34 (20.5%) | 13 (11.7%) | 6 (31.6%) | 30 (47.6%) | 4 (33.3%) |
| RVOTO | |||||
| Testing | |||||
| Yes | 81 (57.9%) | 18 (12.9%) | 20 (14.3%) | 67 (47.9%) | 15 (10.7%) |
| No | 59 (42.1%) | 122 (87.1%) | 120 (85.7%) | 73 (52.1%) | 125 (89.3%) |
| Abnormal | |||||
| Yes | 26 (32.1%) | 1 (5.6%) | 3 (15.0%) | 20 (29.9%) | 6 (40.0%) |
| No | 55 (67.9%) | 17 (94.4%) | 17 (85.0%) | 47 (70.1%) | 9 (60.0%) |
| APVR | |||||
| Testing | |||||
| Yes | 60 (47.2%) | 9 (7.1%) | 9 (7.1%) | 58 (45.7%) | 2 (1.6%) |
| No | 67 (52.8%) | 118 (92.9%) | 118 (92.9%) | 69 (54.3%) | 125 (98.4%) |
| Abnormal | |||||
| Yes | 25 (41.7%) | 3 (33.3%) | 7 (77.8%) | 22 (37.9%) | 1 (50.0%) |
| No | 35 (58.3%) | 6 (66.7%) | 2 (22.2%) | 36 (62.1%) | 1 (50.0%) |
| Heterotaxy | |||||
| Testing | |||||
| Yes | 60 (75.9%) | 12 (15.2%) | 16 (20.3%) | 55 (69.6%) | 7 (8.9%) |
| No | 19 (24.1%) | 67 (84.8%) | 63 (79.7%) | 24 (30.4%) | 72 (91.1%) |
| Abnormal | |||||
| Yes | 19 (31.7%) | 0 (0%) | 0 (0%) | 15 (27.3%) | 6 (85.7%) |
| No | 41 (68.3%) | 12 (100%) | 16 (100%) | 40 (72.7%) | 1 (14.3%) |
| Arteriopathy | |||||
| Testing | |||||
| Yes | 27 (35.5%) | 1 (1.3%) | 4 (5.3%) | 17 (22.4%) | 6 (7.9%) |
| No | 49 (64.5%) | 75 (98.7%) | 72 (94.7%) | 59 (77.6%) | 70 (92.1%) |
| Abnormal | |||||
| Yes | 11 (40.7%) | 0 (0%) | 2 (50%) | 5 (29.4%) | 4 (66.7%) |
| No | 16 (59.3%) | 1 (100%) | 2 (50%) | 12 (70.6%) | 2 (33.3%) |
| Other | |||||
| Testing | |||||
| Yes | 84 (53.2%) | 19 (12.0%) | 5 (3.2%) | 72 (45.6%) | 16 (10.1%) |
| No | 74 (46.8%) | 139 (88.0%) | 153 (96.8%) | 86 (54.4%) | 142 (89.9%) |
| Abnormal | |||||
| Yes | 33 (39.3%) | 7 (36.8%) | 2 (40%) | 18 (25.0%) | 9 (56.2%) |
| No | 51 (60.7%) | 12 (63.2%) | 3 (60%) | 54 (75.0%) | 7 (43.8%) |
APVR, anomalous pulmonary venous return; AVSD, atrioventricular septal defect; CMA, chromosomal microarray analysis; FISH, fluorescence in situ hybridization analysis; LVOTO, left ventricle outflow tract obstruction; RVOTO, right ventricle outflow tract obstruction.
Genetic testing use is highly variable between institutions
The proportion of patients who had genetic testing differed significantly between institutions (42%-78%, P < .001) (Table 2). There were statistically significant differences in testing utilization across institutions, for both isolated CHD (38%-76%, P < .001) and in patients with MCAs (59%-91%, P < .001). The chosen genetic testing modality also differed by institution (Figure 1, Supplemental Table 2). The difference across institutions remained statistically significant after Bonferroni adjustment of multiple testing (ie, 14 tests in Table 2 and Supplemental Table 2). CMA, which was the most frequent test performed, also had the widest range in utilization across institutions (26%-67%, P < .001) (Supplemental Table 2). Chromosome analysis had the second wideset range (4%-31%, P < .001). FISH analysis (5%-23%, P < .001) and molecular testing (2%-15%, P < .001) showed similar variability across institutions. Prenatal genetic testing (which includes any prenatal genetic screening) occurred in 347 (12%), and utilization varied significantly across centers (rate 8%-21%, P < .001) (Supplemental Table 1).
Factors affecting genetic testing
Next, we determined the effects of multiple contributing factors to genetic testing rate. We conducted a multivariate analysis, evaluating whether the difference in testing rate across centers can be attributed to these factors, and to estimate their effects (OR) on genetic testing. The results (Table 4) show that several factors were associated with testing rate, but collectively they did not fully account for the differences across centers. Analyzing by the type of cardiac defect, compared with conotruncal defects (CTDs) (used as the reference because of long standing recommendations for genetic testing in patients with CTD),11 patients with septal defects were the least likely to receive genetic testing (OR 0.22, 95% CI 0.17-0.30, P < .001), followed by arteriopathy (OR 0.32, 95% CI 0.18-0.56, P < .001) and atrioventricular septal defect (AVSD) (OR 0.41, 95% CI 0.30-0.57, P < .001). Genetics consultation was associated with a significantly increased rate of genetic testing (OR 13.0, 95% CI 9.87-17.39, P < .001). Females were tested slightly more frequently than males (OR 1.28, 95% CI 1.07-1.53, P = .008), as were patients with a family history of CHD (OR 1.37, 95% CI 1.01-1.86, P = .047), infants whose CHD was diagnosed prenatally (OR 1.26, 95% CI 1.03-1.53, P = .02), infants with IUGR or SGA (OR 1.69, 95% CI 1.26-2.27, P < .001) and infants of diabetic mothers (OR 1.49, 95% CI 1.11-2.02, P = .009). There was no significant difference in testing rate based on race, maternal teratogen exposure, requirement for ECMO, or presence of MCA after adjusting for the effect of other factors.
Table 4.
Effects of contributing factors on testing rate and testing yield
| Factors Contributing to Testing Rate |
Factors Contributing to Testing Yield |
||||
|---|---|---|---|---|---|
| Odds Ratio (95% CI) | P Value | Odds Ratio (95% CI) | P Value | ||
| Institution | A4 | Reference | Reference | ||
| A3 | 1.92 (1.45-2.56) | <.001 | 0.99 (0.66-1.49) | .957 | |
| A2 | 3.42 (2.49-4.73) | <.001 | 2.79 (1.80-4.35) | <.001 | |
| A1 | 2.48 (1.77-3.49) | <.001 | 1.84 (1.23-2.76) | .003 | |
| A5 | 8.21 (5.86-11.6) | <.001 | 0.30 (0.19-0.47) | <.001 | |
| Sex | Male | Reference | Reference | ||
| Female | 1.28 (1.07-1.53) | .008 | 1.18 (0.93-1.49) | .172 | |
| Race | White | Reference | Reference | ||
| AA/Black | 0.83 (0.66-1.04) | .105 | 1.14 (0.82-1.57) | .431 | |
| Others | 0.96 (0.70-1.31) | .789 | 1.18 (0.80-1.74) | .395 | |
| Family history of CHD | No | Reference | Reference | ||
| Yes | 1.37 (1.01-1.86) | .047 | 0.54 (0.38-0.77) | <.001 | |
| Congenital malformation | No | Reference | Reference | .380 | |
| Yes | 1.33 (0.92-1.93) | .128 | 1.21 (0.80-1.83) | ||
| Type of malformations | Brain | Reference | Reference | ||
| ENT | 0.67 (0.26-1.73) | .414 | 1.83 (0.74-4.51) | .189 | |
| Limb | 0.51 (0.18-1.51) | .222 | 1.20 (0.46-3.18) | .707 | |
| Other | 0.72 (0.30-1.65) | .439 | 2.08 (0.92-4.77) | .080 | |
| Rib/vertebra | 0.65 (0.20-2.19) | .479 | 0.61 (0.20-1.82) | .370 | |
| Umbilical | 0.88 (0.28-2.84) | .834 | 1.41 (0.49-4.07) | .523 | |
| Infant of a diabetic mother | No | Reference | Reference | ||
| Yes | 1.49 (1.11-2.02) | .009 | 0.65 (0.45-0.93) | .021 | |
| Maternal teratogens | No | Reference | Reference | ||
| Yes | 1.22 (0.85-1.75) | .272 | 0.99 (0.65-1.50) | .961 | |
| CHD diagnosed prenatally | No | Reference | Reference | ||
| Yes | 1.26 (1.03-1.53) | .022 | 0.64 (0.50-0.83) | <.001 | |
| Prenatal genetic testing | No | Reference | Reference | ||
| Yes | 0.70 (0.50-0.98) | .039 | 2.48 (1.73-3.57) | <.001 | |
| Geneticist consultation | No | Reference | Reference | ||
| Yes | 13.0 (9.87-17.39) | <.001 | 5.71 (4.33-7.58) | <.001 | |
| History of IUGR or SGA | No | Reference | Reference | ||
| Yes | 1.69 (1.26-2.27) | <.001 | 1.2 (0.86-1.68) | .287 | |
| ECMO | No | Reference | Reference | ||
| Yes | 1.17 (0.87-1.59) | .298 | 0.7 (0.47-1.04) | .080 | |
| CHD groups | CTD | Reference | Reference | ||
| APVR | 0.46 (0.29-0.72) | <.001 | 1.02 (0.55-1.86) | .958 | |
| Arteriopathy | 0.32 (0.18-0.56) | <.001 | 1.20 (0.48-2.88) | .685 | |
| AVSD | 0.41 (0.30-0.57) | <.001 | 6.29 (3.99-10.15) | <.001 | |
| HTX | 1.05 (0.57-2.00) | .869 | 0.60 (0.30-1.14) | .127 | |
| LVOTO | 0.71 (0.56-0.91) | .008 | 0.76 (0.55-1.03) | .080 | |
| Other | 0.58 (0.39-0.87) | .008 | 0.97 (0.56-1.65) | .913 | |
| RVOTO | 0.59 (0.39-0.91) | .018 | 0.63 (0.35-1.10) | .110 | |
| Septal defect | 0.22 (0.17-0.30) | <.001 | 1.66 (1.11-2.49) | .014 | |
AA, African American; APVR, anomalous pulmonary venous return; AVSD, atrioventricular septal defect; CHD, congenital heart disease; CMA, chromosomal microarray analysis; CTD, conotruncal defect; ECMO, extracorporeal membrane oxygenation; ENT, ear, nose, throat; FISH, fluorescence in situ hybridization analysis; HTX, heterotaxy; IUGR, intrauterine growth retardation; LVOTO, left ventricle outflow tract obstruction; RVOTO, right ventricle outflow tract obstruction; SGA, small for gestational age.
Genetic testing results
We measured the yield of genetic testing by the proportion of patients with genetic testing results who were reported in the medical record as abnormal and found that 702 of 1607 (44%) tested patients had abnormal results (Table 2). Testing was abnormal in 486 of 1212 (40%) patients with isolated CHD and 216 of 395 (55%) patients with MCA. A summary of common genetic syndromes identified is provided (Supplemental Table 3) and includes trisomy 21 in 323, 22q11.2 deletion syndrome in 104, and Turner syndrome in 27.
CMA, which is the most frequent test performed, was abnormal in 413 of 1220 (34%) (Figure 1, Table 3), and similarly, FISH analysis was abnormal in 112 of 344 (33%). Molecular testing had the second highest yield (92 of 197 [47%]). Chromosome analysis had the highest yield and was abnormal in 207 of 408 patients tested (51%). Of these, 179 (87%) represent common aneuploidies (trisomy 21, trisomy 18, trisomy 13, or Turner syndrome). We also analyzed the data after excluding trisomy 21, trisomy 18, trisomy 13, Turner syndrome, and 22q11.2 deletion syndrome, given their prevalence, well-characterized features, and existing guidelines for genetic testing. After excluding these patients, overall testing yield remained high (31%, Supplemental Table 4), including in both patients with isolated CHD (26%) and patients with MCAs (45%). Not surprisingly, with exclusion of common aneuploidies, the yield of chromosome analysis dropped from 51% to 11% (24 of 217), and exclusion of 22q11.2 deletion syndrome resulted in a decreased yield of FISH analysis from 33% to 14% (36 of 257), whereas yield remained consistent for CMA (abnormal in 315 of 1117 [28%]) and molecular testing (abnormal in 84 of 187 [45%]) (Supplemental Table 5). These results indicate that genetic testing diagnostic yields remain high in patients with CHD even after excluding well-characterized syndromes associated with CHD, and the high yields are driven by molecular testing and CMA analysis.
We performed a more detailed review of available testing results and categorized abnormal results based on their likelihood of pathogenicity as positive (pathogenic/likely pathogenic) or of undetermined significance (Supplemental Table 3). Overall, there were 611 of 702 abnormal tests available for detailed review; 299 (49%) were positive, whereas 312 (51%) were of undetermined significance. When comparing individual testing modalities, there were 399 abnormal CMAs available for review; 206 (52%) were positive, whereas 193 (48%) were of undetermined significance. There were 134 abnormal molecular tests available for review; 55 (41%) were positive, whereas 79 (59%) were of undetermined significance. Therefore, compared with CMA, molecular testing had a slight but significantly lower rate of likely positive results (41% vs 52%, P = .03) and higher rate of results with undetermined significance (59% vs 48%, P = .03).
Factors affecting testing yield
Next, we determined the effects on genetic testing yield of multiple contributing factors. The difference of testing yield among centers remained statistically significant after adjusting for other factors via multivariate analysis (Table 4). We evaluated these factors for impact on diagnostic yield (Table 4). Although the testing yield was higher among patients with MCAs (55%) than among those with isolated CHD (40%), the difference was no longer statistically significant after adjusting for the effect of other factors (OR 1.2, 95% CI 0.80-1.83, P = .38) (Table 4). The results were similar after excluding common aneuploidies and 22q11.2 deletion syndrome (OR 1.29, 95% CI 0.8-2.08, P = .294) (Supplemental Table 6). We analyzed specific congenital malformation classes (ear, nose, and throat, brain, limb, etc) and found that none significantly increased the likelihood of abnormal genetic testing. We found no statistically significant impact on genetic testing yield by sex, race, IUGR or SGA, or a requirement for ECMO after adjusting for the effect of other factors.
In analysis of environmental perinatal risk factors, maternal diabetes decreased the likelihood of abnormal genetic testing (OR 0.65, 95% CI 0.45-0.93, P = .02), but other maternal teratogens had no effect on abnormal testing (OR 0.99, 95% CI 0.65-1.50, P = .96) (Table 4). Family history of CHD and maternal diabetes both increased testing rate and decreased testing yield.
Of 1607 tested patients, 447 (28%) were evaluated by multiple testing modalities, and the rate of multiple testing varies significantly by institution (P < .001) (Supplemental Table 7). The likelihood of abnormal testing increased with 2 tests (OR 1.57, 95% CI 1.16-2.13, P = .004) or with 3 or 4 tests (OR 2.36, 95% CI 1.40-3.99, P = .001).
We reviewed the 10 patients with 3 abnormal tests, which included the following: 6 patients with abnormal chromosome analysis, FISH, and CMA; 2 patients with abnormal chromosome analysis, CMA, and molecular testing; 1 patient with abnormal chromosome analysis, FISH, and molecular testing; and 1 patient with abnormal FISH, CMA, and molecular testing (Supplemental Table 8). Although duplicate abnormal results were similar, they were unique based on the nature of the testing modality.
Institution, cardiac lesion type, and geneticist consultation significantly affected the yield of genetic testing
There was a significant difference in the yield of genetic testing across institutions (P < .001) (Tables 2 and 4). Additionally, there was a significant difference in yield of genetic testing across cardiac lesion types (P < .001) (Tables 3 and 4). When comparing CHD lesions, AVSD (OR 6.3, 95% CI 3.99-10.15, P < .001), and septal defects (OR 1.66, 95% CI 1.11-2.49, P = .01) had significantly increased yield over CTD (Table 4). After excluding the patients with aneuploidies and 22q11.2 deletion syndrome, the yield of AVSD decreased but remained significantly higher than CTD (OR 2.04, 95% CI 1.08-3.83, P < .027), and the difference between septal defects and CTD was no longer significant (OR 1.03, 95% CI 0.6-1.76, P = .913) (Supplemental Table 6).
Whereas the overall rate of subspecialty consultation was high, with 2122 of 2899 (73%) patients being evaluated by a subspecialist, only 849 (29%) patients were evaluated by a medical geneticist. Geneticist evaluation rates differed significantly by institution (15%-52%, P < .001) (Supplemental Table 1). The genetics team member involved in initial evaluation was a geneticist in 782 patients and a genetic counselor in 329 patients, and the evaluation occurred at an average of 122 days of life. The genetics evaluation was performed inpatient in 633 patients and outpatient in 193 patients (with 23 undetermined). In patients with geneticist involvement, testing yield increased significantly (OR 5.7, 95% CI 4.33-7.58, P < .001) (Table 4). Patients who were evaluated by genetics had a significantly higher rate of MCAs (10% vs 39%, P < .00001). There was also a significant difference across centers in reported family history of CHD (mean 11%, range 3.2%-21%, P < .001) (Supplemental Table 1). Of note, center A5 had the highest rate of geneticist consultation (52%) and also the highest rate of reported familial CHD (21%).
Institution affected genetic testing rate and yield
Given that the institution had the greatest impact on genetic testing rate and yield, we conducted a post hoc pairwise comparison between centers to explore which centers were driving overall differences and to determine whether the P value is driven by differences between all 5 centers vs a large discrepancy between 1 center and the other 4 centers (Supplemental Table 9). Tukey’s honestly significance difference test was used to account for the multiple testing of 10 pairs of centers. The results suggest that the testing rates may have 4 levels that significantly differed from one another, whereas the testing yields may have 3 levels. The testing rates of centers significantly increased in the order of A4, A3, A2, and A5. Although the testing rate of center A1 lied between that of centers A3 and A2, the differences were not statistically significant after adjusting for multiple testing. For testing yield, there were no significant differences between A3 and A4 (P = .99) and between A2 and A5 (P = .17), suggesting 3 levels of testing rate in increasing order of A3/A4, A1, and A2/A5. Overall, these results highlight the variability in genetics evaluation between centers.
Discussion
To our knowledge, this is the first comprehensive multicenter review of genetic evaluation practices in CHD. The findings have several important clinical implications. In our cohort, genetic testing occurred in 55% and was driven by cardiac lesion type, congenital anomalies, and family history of CHD. Institution was the greatest overall driver of genetic testing rate. Testing modalities also varied by institution (eg, CMA, 26%-67%, P < .001), with important implications for obtaining a genetic diagnosis because the conditions detected are test specific. Our study population included patients born 6 years after the first American Heart Association consensus statement on the genetic basis of CHD and 3 years after the International Standard Cytogenomic Array Consortium recommended universal chromosome analysis or CMA in patients with congenital anomalies including CHD.18 Our results demonstrate these guidelines were not incorporated universally and testing remained inconsistent and highly variable, highlighting the need to standardize care.
In patients with genetic testing, close to half (44%) were abnormal. There were variable rates of testing by institution; yet, this did not correlate with yield, suggesting that more restricted testing is not reflective of clinical discernment. Interestingly, although the testing yield was higher among patients with MCA (55%) than among those with isolated CHD (40%), the difference was not statistically significant after adjusting for the effects of other contributing factors via multivariate analysis. The high yield in patients thought to be nonsyndromic further supports that clinical ascertainment of genetic conditions is limited in this patient population, and geneticist consultation may improve care. In the neonatal period, many patients with common syndromes present with an isolated heart defect or with subtle physical features difficult to ascertain without a geneticist’s input. Indeed, in our cohort, a majority of patients with the most commonly identified syndromes presented with an “isolated heart defect” during the study period (250 of 295 patients with trisomy 21, 65 of 88 patients with 22q11.2 deletion syndrome, and 15 of 24 patients with Turner syndrome.) A recent study by Shikany et al,25 in which all patients were evaluated by a geneticist, found a greater yield of genetic testing in patients with MCA and identified specific extracardiac features associated with abnormal genetic testing. However, another single-center study by Ahrens-Nicklas et al,24 again in which all patients were evaluated by a geneticist, mirrors our results, in that patients with MCA had a trend toward increased abnormal results that fails to reach significance. However, the Ahrens-Nicklas study demonstrated that dysmorphic facial features increase the likelihood of abnormal results. Notably, both studies identified a higher proportion of patients with MCA than our study (30% and 60% vs 19% in our study). Our results also indicate a higher rate of MCA in patients evaluated by a geneticist (10% vs 39%.) Overall, the results indicate genetic testing has a higher yield in the setting of MCA, but still has value in isolated heart defects. This is especially true given the difficulty assessing patient phenotypes in the newborn period.
Our genetic testing yield was 44%, and upon secondary review of available testing results (Supplemental Table 3), 49% of these were confirmed to be pathogenic or likely pathogenetic, with the remainder being of uncertain significance. The diagnostic yield identified in this study is in line with genetic testing yield in previous studies in CHD that range from 25% to 50%,21,24,25 with some variability based on how an abnormal genetic test was defined.7 Accounting for the higher proportion of abnormal testing in our cohort, our results reflect any testing reported as abnormal in the medical record, without distinguishing clinical significance. We report a breakdown of likely pathogenic results, vs those of uncertain significance, determined upon reanalysis of genetic testing results (Supplemental Table 3). Additionally, whereas other studies report the proportion of abnormal tests, our results report the proportion of patients with abnormal testing results, and 28% of tested patients have multiple genetic tests. In fact, up to 16% of patients with duplicate testing have multiple positive results. After removing patients with common aneuploidies and 22q11.2 deletion syndrome, the yield of genetic testing remained high (31%), but the yield of karyotype and FISH analysis decreased by more than half. The yield remained consistently high for CMA and molecular testing. These findings reinforce the selective use of karyotype when aneuploidy is suspected. During the years evaluated in the study, CMA was a high yield choice for first tier CHD testing. Molecular testing in this time frame was most frequently performed with panel testing and required a specific differential diagnosis. Expectedly, there was a significantly higher rate of results with undetermined significance, including variants of uncertain significance, in the molecular testing compared with CMA.
Consultation with genetics during the study was uncommon and varied highly by institution; yet, genetics involvement was associated with a significant increase in the yield of genetic testing. All institutions involved in this study are centers where expertise in cardiovascular genetics exists, and it is possible that the variability in testing rates and geneticist evaluation would be even more marked at other institutions. It is unclear whether geneticists were consulted on cases with a higher a priori risk of an identifiable underlying genetic cause or if clinical discernment led to higher diagnostic yield or both. We did note that 89% of molecular testing that was diagnostic was concomitant with geneticist evaluation. We also identified significant variability in reported family history of CHD across centers (3%-21%, Supplemental Table 1), and the center with the highest rate of geneticist involvement also has the highest rate of CHD family history, possibly reflecting that a careful family history was facilitated by geneticist involvement and that the finding correlates with the difference in genetics evaluation practices across centers. Previous studies demonstrate that geneticist involvement optimizes care in CHD21 and may help to overcome historic roadblocks to genetic testing and improve interpretation of results. There are many institution-specific barriers to genetic consultation and testing that may be contributing to variability. Access to a genetics provider is limiting factor, and the number of clinical geneticists currently on staff across institutions within the cohort varies from 6 to 17 (6, 9, 14, and 17, respectively). Although access to genetics providers may be a historic roadblock, technologic advancement, such as telemedicine, may improve access in the future.
Cardiac lesion type is a significant driver of genetic testing frequency, occurring least frequently in septal defects, followed by arteriopathy and AVSD; however, as compared with CTD defects, septal defects and AVSD are the only lesions to significantly increase likelihood of abnormal testing results (Table 4). Although it is somewhat unclear how interdependent the variables are for testing, AVSD has the greatest diagnostic yield, even outside of common aneuploidies. This finding has been demonstrated in multiple single-center studies,22,24 including Ahrens-Nicklas et al24 who demonstrate similarly increased diagnostic yield in septal defects, AVSD, and interrupted aortic arch but identify a decreased diagnostic yield in right ventricle outflow tract obstruction. In contrast, the study by Shikany et al25 identifies an increased yield in right ventricle outflow tract obstruction, and this was the only lesion which affected yield in their study. However, this was identified only in their reanalysis using a nonhierarchical CHD classification system.25 Our results reflect a hierarchical cardiac classification system. Future studies may need to use a more complex nonhierarchical CHD classification systems and explore larger sample sizes to identify differences in genetic testing yield based on cardiac lesions that will guide genetic testing.
We analyzed a comprehensive list of other factors that may influence genetic testing rate and yield and determined the difference across centers were not fully explained by the variation. There are significant differences between the centers, including CHD groups, ECMO use, average number of cardiac surgeries, teratogen exposure, infection, and congenital malformations, which may reflect differences in the baseline patient population across centers vs differences in clinical evaluation, reporting in the medical record or bias in ascertainment. We hypothesize that some of the difference is attributed to the variation in clinical evaluation practices across centers, including the variability in geneticist evaluation. For example, center A5 had the highest rate of geneticist consultation (52%) and also the highest rate of reported familial CHD (21%). Patients who were evaluated by a geneticist also had a significantly higher rate of MCA (10% vs 39%, P < .00001). The difference across centers in race/ethnicity likely reflects a difference in the baseline patient population across geographic areas but is difficult to interpret in the absence of reporting on the structure of the background population. By using the STS database, our data focus on critical CHD; therefore, our results do not include, and may be less applicable in, patients with CHD who do not require CHD repair in infancy. The study also does not capture patients who died before surgery, a population whose risk for genetic diagnosis is not insignificant.
In 2010, International Standard Cytogenomic Array Consortium released a consensus statement for patients with MCAs, recommending genetic testing.18 The guidelines recommended the use of chromosome analysis for patients with obvious chromosomal syndromes, a family history of chromosomal rearrangement, or a history of multiple miscarriages, and the use of CMA for all others. Our study time period from 2013 to 2016 should reflect these recommendations, with standardized and widespread use of chromosome analysis or CMA. However, our results show that both the testing rate and choice of modality were highly variable across centers. We chose this historical cohort in an attempt to focus on a time frame with consistent testing recommendations in this patient population, and the belief findings in this cohort have relevance to the current practice of genetics. Subsequent guidelines incorporate CMA into prenatal testing, including in cases of isolated structural anomalies.33,34 Our results demonstrate an increasing use of molecular testing, including ES. Recently ES and GS have demonstrated utility35,36 and cost-effectiveness,37 and in 2021, the American College of Medical Genetics and Genomics incorporated ES and GS as first-line testing in updated recommendations.19 Our results indicate a significantly higher rate of results with unknown significance in molecular testing compared with CMA. ES and GS interpretation can be nuanced, and its use in infants with CHD will likely necessitate greater geneticist involvement, adding an additional layer of complexity. Our results also highlight that adoption of straightforward recommendations such as universal CMA, were inconsistent and variable, likely affecting care. Going forward, incorporation of ES and GS should be accompanied by careful assessment of utilization and utility through increased quality improvement efforts.
To our knowledge, this is the first comprehensive multicenter review of genetic evaluation practices in CHD, and the results demonstrate the utility of genetic evaluation as well as the need to standardize practices. These findings should help drive recommendations and improve care in these critically ill patients.
Data Availability
Deidentified data sets are available upon request.
Conflict of Interest
The authors declare no conflicts of interest.
Acknowledgments
The authors thank the patients and their families for their contribution. The authors are also grateful to the clinical laboratories at respective sites.
Funding
This work was supported by the American Heart Association Transformational Award (grant number 19TPA34850054 [to S.M.W.]), the Indiana University Health-Indiana University School of Medicine (Strategic Research Initiative and Physician Scientist Initiative [to S.M.W.]), and National Institutes of Health (grant number 2P01HL134599 [to S.M.W. and M.D.D.] and grant number K08HL148508 [to M.D.D.]).
Author Information
Conceptualization: M.D.D., L.R.H., M.L., W.B., S.F.-B., V.G., G.C.G., B.M.H., S.R.L., K.L.M., A.M., D.K.M., C.N.M., S.B.W., B.J.L., S.M.W.; Data Curation: M.D.D., L.R.H., W.B., S.F.-B., V.G., G.C.G., B.M.H., S.R.L., K.L.M., A.M., D.K.M., C.N.M., S.B.W., B.J.L., S.M.W.; Formal Analysis: M.L.; Funding Acquisition: M.D.D., S.M.W.; Project Administration: L.R.H., S.M.W.; Supervision: S.M.W.; Visualization: M.L.; Writing-original draft: M.D.D.; Writing-review and editing: M.D.D., L.R.H., M.L., W.B., S.F.-B., V.G., G.C.G., B.M.H., S.R.L., K.L.M., A.M., D.K.M., C.N.M., S.B.W., B.J.L., S.M.W.
Ethics Declaration
Indiana University Institutional Review Board (IRB) was the main IRB and all institutions involved also received local IRB approval. A waiver of consent was approved by the IRB at all institutions involved for this minimal-risk study. The data were identifiable at each individual site, but all data were entered into the multicenter database deidentified, except for date of birth which was used for data integrity. Only summary results are presented within the research article, and all data presented within the article are deidentified.
Footnotes
The Article Publishing Charge (APC) for this article was paid by Stephanie M. Ware.
Additional Information
The online version of this article (https://doi.org/10.1016/j.gimo.2023.100814) contains supplementary material, which is available to authorized users.
Additional Information
References
- 1.Writing Group Members. Mozaffarian D., Benjamin E.J., et al. Executive summary: heart disease and stroke statistics—2016 update: a report from the American Heart Association. Circulation. 2016;133(4):447–454. doi: 10.1161/CIR.0000000000000366. [DOI] [PubMed] [Google Scholar]
- 2.Thienpont B., Mertens L., de Ravel T., et al. Submicroscopic chromosomal imbalances detected by array-CGH are a frequent cause of congenital heart defects in selected patients. Eur Heart J. 2007;28(22):2778–2784. doi: 10.1093/eurheartj/ehl560. [DOI] [PubMed] [Google Scholar]
- 3.Richards A.A., Santos L.J., Nichols H.A., et al. Cryptic chromosomal abnormalities identified in children with congenital heart disease. Pediatr Res. 2008;64(4):358–363. doi: 10.1203/PDR.0b013e31818095d0. [DOI] [PubMed] [Google Scholar]
- 4.Breckpot J., Thienpont B., Peeters H., et al. Array comparative genomic hybridization as a diagnostic tool for syndromic heart defects. J Pediatr. 2010;156(5):810–817.e4. doi: 10.1016/j.jpeds.2009.11.049. [DOI] [PubMed] [Google Scholar]
- 5.Goldmuntz E., Paluru P., Glessner J., et al. Microdeletions and microduplications in patients with congenital heart disease and multiple congenital anomalies. Congenit Heart Dis. 2011;6(6):592–602. doi: 10.1111/j.1747-0803.2011.00582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lalani S.R., Shaw C., Wang X., et al. Rare DNA copy number variants in cardiovascular malformations with extracardiac abnormalities. Eur J Hum Genet. 2013;21(2):173–181. doi: 10.1038/ejhg.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Syrmou A., Tzetis M., Fryssira H., et al. Array comparative genomic hybridization as a clinical diagnostic tool in syndromic and nonsyndromic congenital heart disease. Pediatr Res. 2013;73(6):772–776. doi: 10.1038/pr.2013.41. [DOI] [PubMed] [Google Scholar]
- 8.Van Der Bom T., Zomer A.C., Zwinderman A.H., Meijboom F.J., Bouma B.J., Mulder B.J. The changing epidemiology of congenital heart disease. Nat Rev Cardiol. 2011;8(1):50–60. doi: 10.1038/nrcardio.2010.166. [DOI] [PubMed] [Google Scholar]
- 9.Cowan J.R., Ware S.M. Genetics and genetic testing in congenital heart disease. Clin Perinatol. 2015;42(2):373–393. doi: 10.1016/j.clp.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 10.LaHaye S., Corsmeier D., Basu M., et al. Utilization of whole exome sequencing to identify causative mutations in familial congenital heart disease. Circ Cardiovasc Genet. 2016;9(4):320–329. doi: 10.1161/CIRCGENETICS.115.001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pierpont M.E., Basson C.T., Benson D.W., Jr., et al. Genetic basis for congenital heart defects: current knowledge: a scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation. 2007;115(23):3015–3038. doi: 10.1161/CIRCULATIONAHA.106.183056. [DOI] [PubMed] [Google Scholar]
- 12.Pierpont M.E., Brueckner M., Chung W.K., et al. Genetic basis for congenital heart disease: revisited: a scientific statement from the American Heart Association. Circulation. 2018;138(21):e653–e711. doi: 10.1161/CIR.0000000000000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harden B., Tian X., Giese R., et al. Increased postoperative respiratory complications in heterotaxy congenital heart disease patients with respiratory ciliary dysfunction. J Thorac Cardiovasc Surg. 2014;147(4):1291–1298.e2. doi: 10.1016/j.jtcvs.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 14.Carey A.S., Liang L., Edwards J., et al. The impact of CNVs on outcomes for infants with single ventricle heart defects. Circ Cardiovasc Genet. 2013;6(5):444–451. doi: 10.1161/CIRCGENETICS.113.000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim D.S., Kim J.H., Burt A.A., et al. Patient genotypes impact survival after surgery for isolated congenital heart disease. Ann Thorac Surg. 2014;98(1):104–110. doi: 10.1016/j.athoracsur.2014.03.017. discussion 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim D.S., Kim J.H., Burt A.A., et al. Burden of potentially pathologic copy number variants is higher in children with isolated congenital heart disease and significantly impairs covariate-adjusted transplant-free survival. J Thorac Cardiovasc Surg. 2016;151(4):1147–1151.e4. doi: 10.1016/j.jtcvs.2015.09.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landis B.J., Cooper D.S., Hinton R.B. CHD associated with syndromic diagnoses: peri-operative risk factors and early outcomes. Cardiol Young. 2016;26(1):30–52. doi: 10.1016/j.jtcvs.2015.09.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller D.T., Adam M.P., Aradhya S., et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86(5):749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manickam K., McClain M.R., Demmer L.A., et al. Exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability: an evidence-based clinical guideline of the American College of Medical Genetics and Genomics (ACMG) Genet Med. 2021;23(11):2029–2037. doi: 10.1038/s41436-021-01242-6. [DOI] [PubMed] [Google Scholar]
- 20.Connor J.A., Hinton R.B., Miller E.M., Sund K.L., Ruschman J.G., Ware S.M. Genetic testing practices in infants with congenital heart disease. Congenit Heart Dis. 2014;9(2):158–167. doi: 10.1111/chd.12112. [DOI] [PubMed] [Google Scholar]
- 21.Geddes G.C., Basel D., Frommelt P., Kinney A., Earing M. Genetic testing protocol reduces costs and increases rate of genetic diagnosis in infants with congenital heart disease. Pediatr Cardiol. 2017;38(7):1465–1470. doi: 10.1007/s00246-017-1685-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buckley J.R., Kavarana M.N., Chowdhury S.M., Scheurer M.A. Current practice and utility of chromosome microarray analysis in infants undergoing cardiac surgery. Congenit Heart Dis. 2015;10(3):E131–E138. doi: 10.1111/chd.12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker K., Sanchez-de-Toledo J., Munoz R., et al. Critical congenital heart disease—utility of routine screening for chromosomal and other extracardiac malformations. Congenit Heart Dis. 2012;7(2):145–150. doi: 10.1111/j.1747-0803.2011.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahrens-Nicklas R.C., Khan S., Garbarini J., et al. Utility of genetic evaluation in infants with congenital heart defects admitted to the cardiac intensive care unit. Am J Med Genet A. 2016;170(12):3090–3097. doi: 10.1002/ajmg.a.37891. [DOI] [PubMed] [Google Scholar]
- 25.Shikany A.R., Landis B.J., Parrott A., et al. A comprehensive clinical genetics approach to critical congenital heart disease in infancy. J Pediatr. 2020;227:231–238.e14. doi: 10.1016/j.jpeds.2020.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helm B.M., Landis B.J., Ware S.M. Genetic evaluation of inpatient neonatal and infantile congenital heart defects: new findings and review of the literature. Genes. 2021;12(8):1244. doi: 10.3390/genes12081244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinton R.B., McBride K.L., Bleyl S.B., et al. Rationale for the Cytogenomics of Cardiovascular Malformations Consortium: a phenotype intensive registry based approach. J Cardiovasc Dev Dis. 2015;2(2):76–92. doi: 10.3390/jcdd2020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grover F.L., Shahian D.M., Clark R.E., Edwards F.H. The STS national database. Ann Thorac Surg. 2014;97(suppl 1):S48–S54. doi: 10.1016/j.athoracsur.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 29.Harris P.A., Taylor R., Thielke R., Payne J., Gonzalez N., Conde J.G. Research Electronic Data Capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris P.A., Taylor R., Minor B.L., et al. The REDCap consortium: building an international community of software platform partners. J Biomed Inform. 2019;95 doi: 10.1016/j.jbi.2019.103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Botto L.D., Lin A.E., Riehle-Colarusso T., Malik S., Correa A., National Birth Defects Prevention Study Seeking causes: classifying and evaluating congenital heart defects in etiologic studies. Birth Defects Res A Clin Mol Teratol. 2007;79(10):714–727. doi: 10.1002/bdra.20403. [DOI] [PubMed] [Google Scholar]
- 32.Øyen N., Poulsen G., Boyd H.A., Wohlfahrt J., Jensen P.K., Melbye M. National time trends in congenital heart defects, Denmark, 1977-2005. Am Heart J. 2009;157(3):467–473.e1. doi: 10.1016/j.ahj.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 33.Committee on Genetics and the Society for Maternal-Fetal Medicine. Committee Opinion No.682: microarrays and next-generation sequencing technology: the use of advanced genetic diagnostic tools in obstetrics and gynecology. Obstet Gynecol. 2016;128(6):e262–e268. doi: 10.1097/AOG.0000000000001817. [DOI] [PubMed] [Google Scholar]
- 34.Society for Maternal-Fetal Medicine (SMFM) Norton M.E., Kuller J.A. The use of chromosomal microarray for prenatal diagnosis. Am J Obstet Gynecol. 2016;215(4):B2–B9. doi: 10.1016/j.ajog.2016.07.016. [DOI] [PubMed] [Google Scholar]
- 35.Reuter M.S., Chaturvedi R.R., Liston E., et al. The Cardiac Genome Clinic: implementing genome sequencing in pediatric heart disease. Genet Med. 2020;22(6):1015–1024. doi: 10.1038/s41436-020-0757-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sweeney N.M., Nahas S.A., Chowdhury S., et al. Rapid whole genome sequencing impacts care and resource utilization in infants with congenital heart disease. NPJ Genom Med. 2021;6(1):29. doi: 10.1038/s41525-021-00192-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stark Z., Schofield D., Martyn M., et al. Does genomic sequencing early in the diagnostic trajectory make a difference? A follow-up study of clinical outcomes and cost-effectiveness. Genet Med. 2019;21(1):173–180. doi: 10.1038/s41436-018-0006-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deidentified data sets are available upon request.