Abstract
Purpose
Fluorescence in situ hybridization (FISH) is the current gold standard assay that provides information related to risk stratification and therapeutic selection for individuals with plasma cell neoplasms. The differential hybridization of FISH probe sets in association with individuals’ genetic ancestry has not been previously reported.
Methods
This retrospective study included 1224 bone marrow samples from individuals who had an abnormal plasma cell proliferative disorder FISH result and a concurrent conventional G-banded chromosome study. DNA from bone marrow samples obtained from the G-banded chromosome study was genotyped, and a biogeographical ancestry prediction was carried out.
Results
Using a cohort of individuals with a plasma cell neoplasm, we identified reduced hybridization of a chromosome 15 centromere FISH probe (D15Z4). Metaphase FISH studies of cells with 2 copies of chromosome 15 demonstrated a failure of the D15Z4 FISH probe to hybridize to one chromosome 15 centromere, revealing a false-positive monosomy 15 FISH result in some individuals. Surprisingly, individuals with a monosomy 15 FISH result had a median African ancestry of 77.2% (95% CI 74.1%-80.3%), compared with a median African ancestry of 2.2% (95% CI 2.0%-2.5%) in the non–monosomy 15 cohort (P value = 9.4 × 10−10). Thus, individuals with African ancestry had an 8.02-fold (95% CI 3.73-17.25) increased probability of having a false-positive monosomy 15 result (P value = 9.92 × 10−8).
Conclusion
This study emphasizes a concern regarding the reliability of diagnostic genomic tools and their application in interpreting genetic testing results in diverse patient populations. We discuss alternative methodologies to better represent different ancestry groups in clinical diagnostic testing.
Keywords: African ancestry, D15Z4, Fluorescence in situ hybridization, Monosomy 15, Plasma cell neoplasm
Introduction
Multiple myeloma (MM) is a plasma cell neoplasm (PCN) that accounts for approximately 10% of all hematologic malignancies.1 Approximately 35,000 new cases were reported in the United States in 2021, accounting for 2% of all cancer deaths.2 MM is primarily a disease of adults with a median age of 65 years at the time of diagnosis. Similar to other malignancies in the United States such as prostate and stomach cancer,3 MM disproportionately affects Black/African American (AA) individuals. The age-adjusted prevalence and incidence of both the premalignant plasma cell condition, monoclonal gammopathy of undetermined significance, and MM are approximately 2- to 3-fold higher among AA compared with non-Hispanic White (NHW) individuals.4 The disparity in incidence is surmised to arise from complex interactions of socioeconomic, environmental, behavioral, and biologic factors.5 Although AA individuals account for 20% of the population that is diagnosed with MM in the United States, only approximately 5% of MM clinical trials6 and approximately 2% of translational research studies include AA individuals, demonstrating a quantitative underrepresentation of AA individuals in biomedical research.7,8 In addition, recent genomic studies have revealed a reduced quality and quantity of sequence data, resulting in the underdetection of genomic variants and inflation of the tumor mutation burden that is associated with African ancestry.9,10 These findings prompted us to investigate whether clinical diagnostic testing approaches that use genomic data display differential performance between AA and NHW individuals.
Fluorescence in situ hybridization (FISH) is the current gold standard clinical assay that uses DNA probes to detect genomic abnormalities of prognostic and therapeutic significance in MM.1 The favorable hyperdiploid MM subtype, found in approximately 50% of MM cases, is characterized by gains of at least 2 of the following odd-numbered chromosomes: 3, 5, 7, 9, 11, 15, or 19. Hyperdiploidy is typically detected using FISH probes targeting the centromere region of the gained chromosome.11 After chromosome 9 (42%), the most commonly observed trisomy involves chromosome 15 (37%). Gains of chromosome 15, either as trisomy (3 total copies) or tetrasomy (4 total copies) in a diploid cell, occur in nearly 40% of MM cases,12,13 whereas monosomy 15 is rarely observed outside of the hyperhaploid MM subtype.14,15 When comparing the enumeration of chromosome 15 by FISH using the alpha satellite D15Z4 probe with that obtained from Mate Pair Sequencing (MPseq),16 we discovered that a subset of PCN cases had a false-positive monosomy 15 FISH result. Herein, we describe the incidence and nature of this discrepant FISH result by examining the presence of monosomy 15 in 1224 individuals with a monoclonal gammopathy in association with biogeographical African ancestry.
Materials and Methods
Individual samples
This retrospective study included 1224 bone marrow (BM) samples from individuals who had an abnormal plasma cell proliferative disorder FISH result and a concurrent conventional G-banded chromosome study. All BM samples were referred to the Mayo Clinic Genomics Laboratory as part of routine clinical testing. Of these samples, 898 were described previously.17,18 This cohort was enriched to be representative of a diverse population (Diversity cohort, Figure 1). We also performed a search of the Mayo Clinic Genomics database of all cases found to be abnormal by the plasma cell FISH test from August 2015 to December 2019. Overall, this larger retrospective cohort included 7968 cases (Full Mayo cohort, Figure 1). The use of clinical data and residual BM samples was approved by the Mayo Clinic Institutional Review Board.
Figure 1.
Cohort description of both the diversity cohort (n = 1224) and the full Mayo cohort (n = 7968). Number and percentage of cases with either a monosomy 15 or no monosomy 15 FISH result. The number and percentage of cases with a monosomy 15 FISH result present in only the PC population, in all cells, including the PC and non-PC population or unknown if the PC/non-PC distribution could not be determined are indicated. Cases were classified as having a “polymorphism or variation” when the monosomy 15 FISH result was found in both the PC and non-PC population and classified as “somatic” when the monosomy 15 FISH result was found in only the PC population. FISH, fluorescence in situ hybridization; PC, plasma cell.
FISH to detect primary and secondary MM abnormalities
FISH of immunoglobulin kappa and lambda light chain (cIg)–stained positive plasma cell (PC) studies were performed as previously described17, 18, 19, 20 using the following probes described in Supplemental Materials. The chromosome 15 centromere probe (Vysis CEP 15 D15Z4) was used for chromosome 15 enumeration. At least 50 cIg-positive cells (MM tumor and normal PCs) were scored. Cells that were cIg negative represent either PCs that do not express light chains and all other cell types in the BM. An abnormal monosomy 15 FISH result required at least 10 of 50 (20%) cIg-positive cells to be considered abnormal.
Biogeographical ancestry genotyping
DNA extraction and genotyping was performed as previously described.17,18 Briefly, DNA from BM samples obtained from the G-banded chromosome study was genotyped using the Axiom Precision Medicine Research Array (Thermo Fisher Scientific) at the DNA Diagnostics Center. A biogeographical ancestry prediction was carried out using the Geographic Population Structure Origins tool21 (Supplemental Figure 1). Geographic Population Structure Origins tool estimates individual genetic ancestry in relation to 36 admixture proportions representing geographic regions and detects 10 African population components (African Bushmen, African Pygmies, Bantu Africa and the Niger-Congo Areas, Hadza, Madagascar, Nile Valley Peoples, Northwestern Africa, Southern Ethiopia, the Kalahari, and West Africa). Ancestry Painter and StructuRly were used to visualize admixture proportions.22,23 Ancestry designations were based on geographic origin and regionalization.23,24 Complete ancestry data are included in Supplemental Data.
Statistical analysis
A logistic regression model was performed to evaluate the association between high and low African ancestry and monosomy 15. The outcome variable was dichotomized as “1” for the presence and “0” for the absence of monosomy 15. African ancestry, derived from the addition of the 10 African-related gene pools (range from 0 to 1), was assigned the value of “1” for individuals with ≥80% African ancestry and “0” for those with <80% African ancestry, as performed in 2 studies.17,18 The adjusted covariates included age (continuous); sex (“1” male, “0” female); and presence of hyperdiploidy, t(11;14), t(14;16), t(14;20), t(4;14), and t(6;14) (with assigned values of “1” for yes and “0” for no). Odds ratio and 95% CIs were used to measure the strength and degree of certainty of the association. We tested normality of a continuous variable with Shapiro-Wilk method. Correlation tests were performed with t test for continuous variables following normal distribution and with Mann-Whitney U test for numeric variables deviating from normality. For the individual African ancestry estimations, the D'Agostino-Pearson test was used to assess the normality distribution of the data and selection of the statistical test. Furthermore, we estimated 95% CI for a median from skewed distribution with 1000 bootstraps (replace = F), each resampling 80% of the samples. A χ2 test was used for discrete frequency variables. All tests were two-sided, and P value < .05 was considered significant. Each test was run in either R (version 4.0.3) or GraphPad Prism version 9.2.0. Of the 1175 samples with a negative result for monosomy 15, 12.2% are from patients with ≥80% African ancestry. African ancestry accounted for 13.2% of the samples, and the prevalence of monosomy 15 in individuals with <80% African ancestry was approximately 3% (30 of 1062). Thus, a sample size of 1030 would ensure an 85% power to detect an odds ratio of 3.00 for African ancestry in the monosomy 15 samples at α = 0.05 level.25
Results
Discrepancy in the enumeration of chromosome 15 by FISH
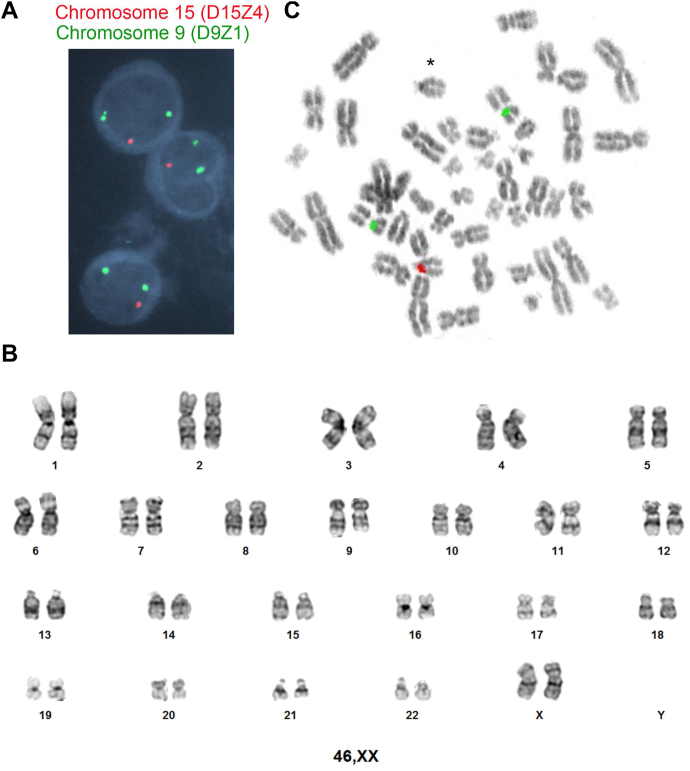
We previously identified a discrepancy involving the enumeration of chromosome 15 between FISH and MPseq in 6 of 70 MM tumors (8.6%).16 This discrepancy involved a monosomy 15 FISH result (1 copy of the D15Z4 FISH probe detected) (Figure 2A) with either 2 or 3 copies of chromosome 15 identified by MPseq (Supplemental Figure 2). In 1 instance, a normal chromosome 15 FISH result was observed, whereas 3 copies of chromosome 15 were identified by MPseq (case 52, Supplemental Figure 2). Of the 4 cases with available self-reported patient race data, 3 were from individuals who were Black/AA.
Figure 2.
Enumeration of chromosome 15 using the D15Z4 FISH probe. A. Interphase FISH analysis indicating monosomy 15 (1 red signal and 2 green signals per cell). Centromere 15 (D15Z4) FISH probe signals in red and centromere 9 (D9Z1) FISH probe signals in green. B. Banded analysis from a representative chromosome study demonstrating 2 chromosome 15s. C. Metaphase FISH of a cell with 2 chromosome 15s using the D15Z4 FISH probe (red) and the D9Z1 FISH probe (green).
To further investigate the incidence and nature of this discrepancy, we analyzed 1224 cases from individuals with a monoclonal gammopathy with available FISH and genomic ancestry data.17,18 In this cohort, monosomy 15 was identified in PCs by FISH in 49 cases (4.0%) (Table 1). We next asked whether the monosomy 15 cells were restricted to the PCs or found in other cells of the BM aspirate. Of the 42 cases with available light chain counterstain data, 3 (7.1%) cases had monosomy 15 present only in the PCs, whereas 39 (92.9%) had monosomy 15 in both the PC and non-PC populations (Table 2, Figure 1). Of the 39 cases, 100% of the analyzed non-PCs had evidence of monosomy 15, demonstrating that the monosomy 15 FISH result was not restricted to the PC (tumor) population and was seen in all analyzed cells within the BM. This finding differs from recurrent MM-specific genomic abnormalities, which are restricted to the PC population and absent in non-PCs.19,20
Table 1.
Demographics and clinical variables
| Variable | Monosomy 15, n = 49 (4.0%), Mean ± SD or n (%) | No Monosomy 15, n = 1175 (96.0%), Mean ± SD or n (%) | P Value |
|---|---|---|---|
| Age | 65.3 ± 12.2 | 63.5 ± 11.3 | .33a |
| Sex | |||
| Male | 25 (51%) | 663 (56%) | |
| Female | 24 (49%) | 512 (44%) | .39b |
| Median African ancestryc | 77.2% (95% CI 74.1%-80.3%) | 2.2% (95% CI 2.0%-2.5%) | 9.4 × 10−10d |
| African ancestrye | |||
| Yes (>80%) | 19 (39%) | 143 (12%) | |
| No (<80%) | 30 (61%) | 1032 (88%) | 2.34 × 10−7b |
Derived from t test.
Derived from χ2 test.
The data deviate from normality (P < .0001); thus, 1000 bootstraps with replace = F was used to derive 95% CI.
Derived from Mann-Whitney U test.
Derived from 10 top African related genes, coded as 1 if value ≥ 0.8 and 0 otherwise.
Table 2.
Detection of monosomy 15 in tumor and nontumor cells
| Variable | Monosomy 15 in Tumor (PCs) and Nontumor Cells, n = 39 | Monosomy 15 in Tumor Cells (PCs) Only, n = 3 |
|---|---|---|
| Average % of monosomy 15 in tumor cells | 98% (45%-100%) | 78% (41%-100%) |
| Average % of monosomy 15 in nontumor cells | 100% | 0% |
Seven cases had an unknown status of multiple myeloma tumor vs nontumor cells. Nontumor cells: 100% of the cells had monosomy 15.
PC, plasma cell.
To evaluate whether the monosomy 15 FISH result was a false positive, we analyzed the chromosome 15s from the conventional chromosome study. A chromosome study was available in 37 of the 39 cases. Although a single case had a loss of one chromosome 15 in 3 of 20 metaphase cells, all 20 metaphase cells from the remaining 36 cases (720 total metaphase cells) had 2 normal chromosome 15s, suggesting that the FISH result may represent a false-positive monosomy 15 FISH finding (Figure 2B). Metaphase FISH analysis of a cell with 2 copies of chromosome 15 demonstrated a failure of the D15Z4 FISH probe to hybridize to 1 chromosome 15 centromere (Figure 2C), demonstrating a false-positive monosomy 15 FISH result. This hybridization failure was observed in all 12 metaphase cells evaluated from 3 unique patients who had a monosomy 15 FISH result. Given that the false-positive monosomy 15 result is found in all non-PCs suggests a genetic variation reducing the hybridization of the D15Z4 FISH probe in some individuals.
A false-positive monosomy 15 FISH result is associated with African ancestry
Given our previous observation that the false-positive monosomy 15 result may be associated with individuals of African ancestry, we next sought to evaluate the frequency of African ancestry in the cohort of 49 individuals with monosomy 15 compared with the cohort of 1175 individuals without monosomy 15. Individuals with monosomy 15 had a median African ancestry of 77.2% (95% CI 74.1%-80.3%), compared with a median African ancestry of 2.2% (95% CI 2.0%-2.5%) in the non–monosomy 15 cohort (P value = 9.4 × 10−10) (Table 1, Figure 3A). Of individuals with monosomy 15, 39% had ≥80% African ancestry, whereas only 12% of the non-monosomy 15 cohort had ≥80% African ancestry (P value = 9.3 × 10−7) (Table 1, Figure 3B). Individuals with African ancestry had an 8.02-fold (95% CI 3.73-17.25) increased probability of having a false-positive monosomy 15 result (P value = 9.92 × 10−8). Of individuals with ≥80% or ≥50% African ancestry, 12% had a false-positive monosomy 15 result. Comparison of the distribution of African ancestry revealed a significantly lower contribution of Nile Valley Peoples and increased contribution of Northwestern African ancestry in African individuals with monosomy 15 compared with individuals without monosomy 15 (Supplemental Figure 3). None of the other African population components (African Bushmen, African Pygmies, Bantu Africa and the Niger-Congo Areas, Hadza, Madagascar, Southern Ethiopia, the Kalahari, and West Africa) resulted in a significant difference between individuals with monosomy 15 compared with the non–monosomy 15 cohort.
Figure 3.
False-positive monosomy 15 is associated with African ancestry. A. Violin plot showing the percentage of African ancestry in the monosomy 15 and no monosomy 15 cohorts. B. Pie charts of the monosomy 15 and no monosomy 15 cohort reflecting individuals with 80% to 100% African ancestry in red, 50% to 79% African ancestry in black, and 0% to 49% African ancestry in white. C. Allele frequency of 8 deletions across 5 superpopulations. The deletions are extracted from the 1000 Genomes Project phase 3 release, together covering 205 kb on hg38. Individual deletions are color coded, with circle size representing the size of a deletion. Presence and absence of a deletion is denoted with plus and minus sign, respectively, on the right panel. The start position and size of each deletion on CHM13v2.0/hs1 reference are based on liftover from hg38. Deletions not identified in each of the superpopulations were also plotted, shown with zero percent allele frequency. AFR, African; AMR, Admixed American; EAS, East Asian; EUR, European; SAS, South Asian.
To evaluate a potential mechanism for the false-positive monosomy 15 FISH result, we analyzed genomic data from The 1000 Genomes Project, which has mapped structural variants26 using genome sequencing data from 2504 individuals.27 The 15p11.1-q11.1 region targeted by the D15Z4 FISH probe was analyzed, from which a total of 12 structural variants were identified, including 8 deletions, 3 duplications, and a single ALU element. After liftover of coordinates from hg38 to Telomere-to-Telomere (T2T-CHM13v2.0/hs1) reference that includes gapless assembly of chromosome 15,28 the 8 deletions occurred in a 205-kb region, with sizes between 855 and 90,445 bp. By blastn search of the 171-bp centromeric alpha satellite sequence against the hs1 sequences corresponding to the 8 deletions, we found that 66% to 98% of the first 6 deletions were covered by this repeat. Notably, 6 of the deletions were identified in African, vs only 2 to 4 in the other 4 superpopulations (Figure 3C). Additionally, the 6 deletions in African together cover 118.7 kb, 4 times the total size (29.6 kb) of the 3 deletions in the European population. Thus, the deletions in the D15Z4-spanning region preferentially occur in the African population.
True monosomy 15 is a rare finding in monoclonal gammopathies
To determine the overall frequency of a true monosomy 15 FISH result, we searched the Mayo Clinic Genomics database for all cases evaluated by the PC FISH test from August 2015 to December 2019. Of the 7968 cases with an abnormal PC FISH result, monosomy 15 was detected in 95 cases (1.2%). Of the 95 cases with evidence of monosomy 15, evaluation of both PCs and non-PCs was possible in 93 cases. Of these, 32 of 93 (34.4%) had a monosomy 15 FISH result restricted to the PCs representing a somatic event in the PC clone. In the remaining 61 of 93 (65.6%) cases, a monosomy 15 was found in both the PCs and non-PCs, likely representing the false-positive monosomy 15 FISH reported here. Thus, the incidence of true somatic monosomy 15 in the Mayo Clinic cohort is approximately 0.4% (32 of 7968) (Figure 1). Therefore, evidence of a monosomy 15, particularly if also detected in most of the non-PC population cells, should raise concern for a possible false-positive monosomy 15 result, a finding highly associated with individuals with predominant African ancestry.
Discussion
Here, we describe the incidental observation of a false-positive monosomy 15 associated with African ancestry. To our knowledge, this is the first report of a genetic ancestry bias in the hybridization of a commonly used FISH probe in the study of hematological neoplasms.
Accessibility of genomic tools, including next-generation sequencing technology, has pushed precision medicine to the forefront of companion diagnostics and therapeutics. However, the clinical utility and impact of these technologies rely on currently used reference sequences, which have been reported to miss approximately 300 Mb in contigs from populations of African descent.29 This underrepresentation of African ancestry–specific sequences in the reference genome poses challenge in the interpretation of genetic sequencing related to both hereditary disease and cancer.30,31 For instance, lower sequencing depth was observed in African genomes compared with NHW populations in The Cancer Genome Atlas, resulting in the underdetection of variants and inflation of tumor mutational burden identified in AA patients with MM.9,10 Our observation of reduced hybridization of a commercially available FISH probe in association with African ancestry is a demonstration of the underrepresentation of African ancestry in the context of genomic tool development. Because research efforts and databases have largely focused on patients who self-report as NHW,32 a Eurocentric focus in research and medicine may contribute to health disparities.33
A false-positive FISH result for the chromosome 15 centromere increases the risk for an erroneous underreporting of trisomy 15 and/or overreporting of monosomy 15 in individuals with predominant African ancestry. Accurate reporting of cytogenetic abnormalities has implications for the prognosis and treatment management in patients with MM. For example, hyperdiploid MM with trisomy 15 and other odd-numbered chromosomes (3, 5, 7, 9, 11, or 19) is considered a favorable prognostic factor. In addition, individuals with high-risk cytogenetics with at least 1 trisomy had been reported to have improved overall survival compared with high-risk patients without a concurrent trisomy.13 Thus, underreporting trisomy 15 in this context could have prognostic implications for individuals with MM. In contrast to the high incidence of monosomies 13, 14, 16, and 17, the incidence of an apparent true monosomy 15 is approximately 0.4% in our overall Mayo Clinic cohort. Therefore, the identification of a monosomy 15 FISH result in both non-PCs and PCs together, combined with the rarity of true monosomy 15, raises a concern for a chromosome 15 centromere variant. Furthermore, the observation of increased t(11;14) translocations (Supplemental Results) in association with monosomy 15 could be due to the association of the chromosome 15 centromere variant with SNP rs9344, found to be increased in association with African ancestry and with t(11;14).17,34 Furthermore, although this study used a cohort of patients with a PCN, this finding was not restricted to this disease entity because a false-positive monosomy 15 result was also observed in other specimen types (Supplemental Results).
Clinical Laboratory Improvement Amendments requires the use of standard controls for validation of FISH probes within a clinical laboratory. However, these guidelines do not specify genetic ancestry of the control population.35 The incorporation of diverse populations may provide data informing hybridization variability of FISH probes in association with individuals’ ancestry. Alternatively, different FISH probes that do not target centromeric regions may be used. However, it is important to ensure that other structural variations will not interfere with their performance. Alternative approaches, such as flow cytometry,36 genome chromosomal microarray analysis, or next-generation sequencing,16,37 may provide improvements over FISH in the detection of hyperdiploid MM. In fact, our clinical laboratory has recently converted our plasma cell FISH assay from using centromeric FISH probes to performing flow cytometry to evaluate ploidy (unpublished data).36 This adjustment to the use of flow cytometry to evaluate ploidy will circumvent the issue of the chromosome 15 FISH probe. Alternatively, newer centromere FISH probes can be developed leveraging updated reference sequence data from the T2T Consortium,38,39 which recently reported the complete, gapless genomic sequence data, including centromeric regions of all chromosomes. In addition, the T2T Initiative combined with the Human Pangenome Reference Consortium can be used to understand centromeric variations in different populations. This information could possibly provide improved resources for genomic test development with equal representation of global populations.
A limitation of this study is the lack of complete chromosome 15 ploidy sequencing data for all samples restricting our ability to identify cases that had a normal FISH result from those that were trisomy 15 but harbored the centromeric variation. Thus, in addition to over-reporting monosomy 15, we suspect there is an underreporting of trisomy 15 using the D15Z4 FISH probe in individuals with African ancestry. This finding demonstrates that currently used routine cytogenetic tools, namely the D15Z4 FISH probe, may not have uniform hybridization within the chromosome 15 centromere, specifically affecting individuals of African ancestry. A second limitation of this study is the absence of outcome data for individuals with and without trisomy 15 within this diverse cohort. However, previous studies have demonstrated that patients with hyperdiploidy, including those with trisomy 15, are associated with a more favorable outcome compared with patients with high-risk abnormalities including t(4;14), t(14;16), t(14;20), and deletion 17p.12,13 Our data suggest that there exist genomic variations of the chromosome 15 centromere that results in variable D15Z4 FISH probe interaction.40 Whether other centromere FISH probes display similar hybridization bias warrants future investigation. Evaluation of genomic events using unbiased approaches, such as genome sequencing, along with a pangenome reference may circumvent some of these limitations.
Data Availability
The complete ancestry data sets for this study can be found in the Supplemental Data.
Conflict of Interest
Shaji Kumar served as a consultant for Celgene, Takeda, Amgen, Janssen, and Bristol-Myers Squibb and received research funding from Celgene, Takeda, Novartis, Amgen, AbbVie, Janssen, and Bristol-Myers Squibb. Linda B. Baughn serves as consultant for Genentech. Eran Elhaik is a consultant to DNA Diagnostics Center, and Michael Baird is the CSO of DNA Diagnostics Center. All other authors declare no conflicts of interest.
Acknowledgments
The authors thank the staff within the Mayo Clinic Genomics Laboratory for performing additional analysis that supports this work.
Funding
This work was supported by the National Cancer Institute of the National Institutes of Health (grant number P50CA186781) from the Mayo Clinic Multiple Myeloma Specialized Program of Research Excellence. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors also acknowledge support by the Mayo Clinic Department of Laboratory Medicine and Pathology and support of the Marion Schwartz Career Development Award in Multiple Myeloma.
Author Information
Conceptualization: L.B.B.; Data Curation: L.B.B., A.K., H.T., N.S., H.Y., S.T., J.S., R.M., E.E.; Formal Analysis: L.B.B., A.K., H.T., N.S., H.Y., S.T., J.S., R.M., E.E.; Funding Acquisition: L.B.B.; Investigation: L.B.B., A.K., H.T., N.S., H.Y., S.T., J.S., R.M., E.E.; Methodology: L.B.B., A.K., H.T., N.S., H.Y., S.T., J.S., R.M., E.E.; Project Administration: L.B.B.; Resources: L.B.B.; Software: L.B.B., A.K., H.T., N.S., H.Y., S.T., J.S., R.M., E.E.; Supervision: L.B.B.; Validation: L.B.B., A.K., H.T., N.S., H.Y., S.T., J.S., R.M., E.E.; Visualization: L.B.B., A.K., H.T., N.S., H.Y., S.T., J.S., R.M., E.E.; Writing-original draft: A.K., L.B.B.; Writing-review and editing: L.B.B., A.K., H.T., N.S., H.Y., S.T., J.S., R.M., E.E., C.Z.M., P.T.G., J.F.P., R.P.K., P.L.B., C.V., S.V.R., S.K., Y.W.A.
Ethics Declaration
The use of clinical data and residual bone marrow samples was approved with a waiver of consent by the Mayo Clinic Institutional Review Board. This research study involves no more than minimal risk to patients. No direct patient contact occurred; thus, the waiver did not affect the welfare of patients.
Footnotes
The Article Publishing Charge (APC) for this article was paid by Linda B. Baughn.
Alaa Koleilat and Hongwei Tang contributed equally to this work.
Additional Information
The online version of this article (https://doi.org/10.1016/j.gimo.2023.100816) contains supplementary material, which is available to authorized users.
Additional Information
References
- 1.Rajkumar S.V. Multiple myeloma: 2022 update on diagnosis, risk stratification, and management. Am J Hematol. 2022;97(8):1086–1107. doi: 10.1002/ajh.26590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 3.Özdemir B.C., Dotto G.P. Racial differences in cancer susceptibility and survival: more than the color of the skin? Trends Cancer. 2017;3(3):181–197. doi: 10.1016/j.trecan.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greenberg A.J., Vachon C.M., Rajkumar S.V. Disparities in the prevalence, pathogenesis and progression of monoclonal gammopathy of undetermined significance and multiple myeloma between blacks and whites. Leukemia. 2012;26(4):609–614. doi: 10.1038/leu.2011.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kyle R.A., Gertz M.A., Witzig T.E., et al. Review of 1027 patients with newly diagnosed multiple myeloma. Mayo Clin Proc. 2003;78(1):21–33. doi: 10.4065/78.1.21. [DOI] [PubMed] [Google Scholar]
- 6.Bhatnagar V., Gormley N., Kazandjian D., et al. FDA analysis of racial demographics in multiple myeloma trials. Blood. 2017;130(suppl 1) 4352-4352. [Google Scholar]
- 7.Bentley A.R., Callier S.L., Rotimi C.N. Evaluating the promise of inclusion of African ancestry populations in genomics. NPJ Genom Med. 2020;5:5. doi: 10.1038/s41525-019-0111-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gormley N., Fashoyin-Aje L., Locke T., et al. Recommendations on eliminating racial disparities in multiple myeloma therapies: a step toward achieving equity in healthcare. Blood Cancer Discov. 2021;2(2):119–124. doi: 10.1158/2643-3230.BCD-20-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asmann Y.W., Parikh K., Bergsagel P.L., et al. Inflation of tumor mutation burden by tumor-only sequencing in under-represented groups. NPJ Precis Oncol. 2021;5(1):22. doi: 10.1038/s41698-021-00164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wickland D.P., Sherman M.E., Radisky D.C., Mansfield A.S., Asmann Y.W. Lower exome sequencing coverage of ancestrally African patients in the cancer genome atlas. J Natl Cancer Inst. 2022;114(8):1192–1199. doi: 10.1093/jnci/djac054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pugh T.J., Fink J.M., Lu X., et al. Assessing genome-wide copy number aberrations and copy-neutral loss-of-heterozygosity as best practice: an evidence-based review from the Cancer Genomics Consortium working group for plasma cell disorders. Cancer Genet. 2018;228-229:184–196. doi: 10.1016/j.cancergen.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Chretien M.L., Corre J., Lauwers-Cances V., et al. Understanding the role of hyperdiploidy in myeloma prognosis: which trisomies really matter? Blood. 2015;126(25):2713–2719. doi: 10.1182/blood-2015-06-650242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar S., Fonseca R., Ketterling R.P., et al. Trisomies in multiple myeloma: impact on survival in patients with high-risk cytogenetics. Blood. 2012;119(9):2100–2105. doi: 10.1182/blood-2011-11-390658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterson J.F., Rowsey R.A., Marcou C.A., et al. Hyperhaploid plasma cell myeloma characterized by poor outcome and monosomy 17 with frequently co-occurring TP53 mutations. Blood Cancer J. 2019;9(3):20. doi: 10.1038/s41408-019-0182-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sawyer J.R., Tian E., Shaughnessy J.D., Jr., et al. Hyperhaploidy is a novel high-risk cytogenetic subgroup in multiple myeloma. Leukemia. 2017;31(3):637–644. doi: 10.1038/leu.2016.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smadbeck J., Peterson J.F., Pearce K.E., et al. Mate pair sequencing outperforms fluorescence in situ hybridization in the genomic characterization of multiple myeloma. Blood Cancer J. 2019;9(12):103. doi: 10.1038/s41408-019-0255-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baughn L.B., Li Z., Pearce K., et al. The CCND1 c.870G risk allele is enriched in individuals of African ancestry with plasma cell dyscrasias. Blood Cancer J. 2020;10(3):39. doi: 10.1038/s41408-020-0294-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baughn L.B., Pearce K., Larson D., et al. Differences in genomic abnormalities among African individuals with monoclonal gammopathies using calculated ancestry. Blood Cancer J. 2018;8(10):96. doi: 10.1038/s41408-018-0132-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahmann G.J., Jalal S.M., Juneau A.L., et al. A novel three-color, clone-specific fluorescence in situ hybridization procedure for monoclonal gammopathies. Cancer Genet Cytogenet. 1998;101(1):7–11. doi: 10.1016/s0165-4608(97)00058-7. [DOI] [PubMed] [Google Scholar]
- 20.Fonseca R., Barlogie B., Bataille R., et al. Genetics and cytogenetics of multiple myeloma: a workshop report. Cancer Res. 2004;64(4):1546–1558. doi: 10.1158/0008-5472.can-03-2876. [DOI] [PubMed] [Google Scholar]
- 21.Elhaik E., Tatarinova T., Chebotarev D., et al. Geographic population structure analysis of worldwide human populations infers their biogeographical origins. Nat Commun. 2014;5:3513. doi: 10.1038/ncomms4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Criscuolo N.G., Angelini C. StructuRly: a novel shiny app to produce comprehensive, detailed and interactive plots for population genetic analysis. PLoS One. 2020;15(2) doi: 10.1371/journal.pone.0229330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng Q., Lu D., Xu S. AncestryPainter: a graphic program for displaying ancestry composition of populations and individuals. Genomics Proteomics Bioinformatics. 2018;16(5):382–385. doi: 10.1016/j.gpb.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flanagin A., Frey T., Christiansen S.L., Bauchner H. The reporting of race and ethnicity in medical and science journals: comments invited. JAMA. 2021;325(11):1049–1052. doi: 10.1001/jama.2021.2104. [DOI] [PubMed] [Google Scholar]
- 25.Demidenko E. Sample size and optimal design for logistic regression with binary interaction. Stat Med. 2008;27(1):36–46. doi: 10.1002/sim.2980. [DOI] [PubMed] [Google Scholar]
- 26.Sudmant P.H., Rausch T., Gardner E.J., et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526(7571):75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.1000 Genomes Project Consortium. Auton A., Brooks L.D., Durbin R.M., et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nurk S., Koren S., Rhie A., et al. The complete sequence of a human genome. Science. 2022;376(6588):44–53. doi: 10.1126/science.abj6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherman R.M., Forman J., Antonescu V., et al. Assembly of a pan-genome from deep sequencing of 910 humans of African descent. Nat Genet. 2019;51(1):30–35. doi: 10.1038/s41588-018-0273-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halperin R.F., Carpten J.D., Manojlovic Z., et al. A method to reduce ancestry related germline false positives in tumor only somatic variant calling. BMC Med Genomics. 2017;10(1):61. doi: 10.1186/s12920-017-0296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manrai A.K., Funke B.H., Rehm H.L., et al. Genetic misdiagnoses and the potential for health disparities. N Engl J Med. 2016;375(7):655–665. doi: 10.1056/NEJMsa1507092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carress H., Lawson D.J., Elhaik E. Population genetic considerations for using biobanks as international resources in the pandemic era and beyond. BMC Genomics. 2021;22(1):351. doi: 10.1186/s12864-021-07618-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.West K.M., Blacksher E., Burke W. Genomics, health disparities, and missed opportunities for the Nation’s research agenda. JAMA. 2017;317(18):1831–1832. doi: 10.1001/jama.2017.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weinhold N., Johnson D.C., Chubb D., et al. The CCND1 c.870G>A polymorphism is a risk factor for t(11;14)(q13;q32) multiple myeloma. Nat Genet. 2013;45(5):522–525. doi: 10.1038/ng.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolff D.J., Bagg A., Cooley L.D., et al. Guidance for fluorescence in situ hybridization testing in hematologic disorders. J Mol Diagn. 2007;9(2):134–143. doi: 10.2353/jmoldx.2007.060128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sidana S., Jevremovic D., Ketterling R.P., et al. Rapid assessment of hyperdiploidy in plasma cell disorders using a novel multi-parametric flow cytometry method. Am J Hematol. 2019;94(4):424–430. doi: 10.1002/ajh.25391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bolli N., Genuardi E., Ziccheddu B., Martello M., Oliva S., Terragna C. Next-generation sequencing for clinical management of multiple myeloma: ready for prime time? Front Oncol. 2020;10:189. doi: 10.3389/fonc.2020.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aganezov S., Yan S.M., Soto D.C., et al. A complete reference genome improves analysis of human genetic variation. Science. 2022;376(6588) doi: 10.1126/science.abl3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altemose N., Logsdon G.A., Bzikadze A.V., et al. Complete genomic and epigenetic maps of human centromeres. Science. 2022;376(6588) doi: 10.1126/science.abl4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Keefe C.L., Matera A.G. Alpha satellite DNA variant-specific oligoprobes differing by a single base can distinguish chromosome 15 homologs. Genome Res. 2000;10(9):1342–1350. doi: 10.1101/gr.10.9.1342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete ancestry data sets for this study can be found in the Supplemental Data.