Abstract
Purpose
Wolf-Hirschhorn syndrome (WHS), a contiguous gene syndrome caused by heterozygous deletions of the distal short arm of chromosome 4 that includes NSD2, reportedly causes specific DNA methylation signatures in peripheral blood cells. However, the genomic loci responsible for these signatures have not been elucidated. The present study aims to define the loci underlying WHS-related DNA methylation signatures and explore the role of NSD2 in these signatures.
Methods
We conducted genome-wide methylation analysis of individuals with WHS or NSD2 variants using an array method. We studied genome-edited knockin mice and induced pluripotent stem cells to explore the function of NSD2 variants.
Results
Three undiagnosed cases with NSD2 variants showed WHS-related DNA methylation signatures. In patient-derived induced pluripotent stem cells and genome-edited knockin mice, these variants cause NSD2 loss of function, respectively. The p.Pro905Leu variant caused decreased Nsd2 protein levels and altered histone H3-lysine 36 dimethylation levels similarly to what was observed in Nsd2 knockout mice. Nsd2 knockout and p.Pro905Leu knockin mice exhibited common DNA methylation changes.
Conclusion
These results revealed that WHS-related DNA methylation signatures are dependent on NSD2 dysfunction and could be useful in identifying NSD2 variants of uncertain significance.
Keywords: DNA methylation signature, Histone H3-lysine 36 dimethylation, NSD2, Peripheral blood cells, Wolf-Hirschhorn syndrome
Introduction
Wolf-Hirschhorn syndrome (WHS, OMIM 194190), also known as 4p deletion syndrome, is a contiguous gene deletion syndrome caused by heterozygous loss of the short arm of chromosome 4 (4p). The critical region for phenotypes located on chromosome 4p16.3 contains 3 genes, namely, LETM1 (OMIM ∗604407), NSD2 (also named WHSC1, OMIM ∗602952), and NELFA (also named WHSC2, OMIM ∗606026).1,2 Single variants in the NSD2 gene can cause mild phenotypes of WHS.3, 4, 5, 6 This is known as Raunch-Steindl syndrome (RAUST) (OMIM 619695), which is caused by heterozygous NSD2 deficiency.6 The clinical features of RAUST are less severe dysmorphic facial features, less severe developmental disabilities in general than WHS, and absence of a seizure disorder. NSD2, a member of the Nuclear Receptor Binding SET Domain Protein (NSD) family, is a Histone-Lysine 36 (H3K36) N-Methyltransferase.7 NSD1 (OMIM ∗606681) variants cause Sotos syndrome 1 (Sotos, OMIM 117550) and a specific DNA methylation signature consequently.8 Many epigenome-related gene variants exhibit unique combinations of DNA methylation changes defined as “episignature.”9, 10, 11 NSD2 variants also exhibit DNA methylation signatures.12 However, the specific loci of DNA methylation changes for NSD2 variants remain unknown. In this study, we define WHS-related DNA methylation signatures and determine whether NSD2 loss of function established through functional assays of variants of uncertain significance (VUS) can be classified as WHS-related DNA methylation signature.
Materials and Methods
Patients
A total of 21 patients with del(4)(p16) (Supplemental Table 1), 3 patients with undiagnosed neurodevelopmental disorders with congenital anomalies (Supplemental Table 2), and 3 patients with NSD2 likely benign variants (Supplemental Table 3) were recruited. The deleted regions at chromosome 4 in 16 out of the 21 patients with del(4)(p16) have been previously described (Supplemental Table 1).13,14 Some of the patients were originally recruited to Initiative on Rare and Undiagnosed Diseases for diagnosis using exome sequencing.15 NSD2 variant nomenclature refers to the NC_000004.12(NM_001042424.3). Pathogenicity classification is based on the American College of Medical Genetics and Genomics/Association for Molecular Pathology guidelines.16
DNA methylation
Genomic DNA was obtained from the peripheral blood of the patients. DNA methylation data were obtained using the Illumina Infinium Methylation EPIC BeadChip (EPIC) array (Illumina). Control methylation data of EPIC were obtained from publicly available data sets taken from the Gene Expression Omnibus (GEO: GSE154566, GSE179759, and GSE166503).17, 18, 19 Overall, the 142 assays from unrelated healthy individuals from the general population without clinically apparent neurodevelopmental phenotypes were divided into 106 and 36 assays for the training and the testing set, respectively (Supplemental Table 4).
Identification of WHS-related methylation probes
WHS-related methylation in the training set was assessed by linear regression modeling using M-values, by applying logit transformation to the beta value. The speculated cell compositions,20 gender, and batch effects were entered into the linear regression model as separate confounders. WHS-related methylation was identified using the following threshold; difference in absolute beta from controls greater than 0.2 and with <Bonferroni-corrected P = 2.4e-16. Clustering of samples by DNA methylation data was performed using the heatmap.2 function from the gplots package. Distances were measured using the Euclidean formula. Hierarchical cluster analysis was performed using “ward.D.” Beta value variance between groups were compared using var.test. Support vector machine (SVM) modeling was performed to classify cases by WHS-related DNA methylation signature in a training set, using the e1071 R package (v.1.7-9).21 SVM decision values ranging between 0 and 1 in the testing set were converted to probability scores using Platt’s scaling.22
Functional assay of NSD2 VUS
Induced pluripotent stem cells (iPSCs) were generated from case 2’s and healthy control’s peripheral blood cells as previously described.23 To investigate the function of the case 3’s variant in NSD2, genome-edited mice carrying Nsd2 NM_001081102.2: c.2717C>T substitution (Nsd2WT/P906L) were generated using the CRISPR/Cas9 systems as previously described.24 Nsd2 knockout mice (Nsd2WT/−) were also generated by genome editing. The guide RNA sequences are listed in Supplemental Table 5. In mouse genome, NM_001081102.2:c.2717C>T NP_001074571.2:p.(Pro906Leu) is an orthologous substitution of human NSD2 NC_000004.12(NM_001042424.3):c.2714C>T NP_001035889.1:p.(Pro905Leu) harbored by case 3. Transient expression of NSD2 VUS in HeLa cells was performed using FuGENE 6 (Promega). pCMV-3xFLAG-NSD2 and pCMV-3xFLAG-NSD2 c.2714C>T plasmids were constructed by VectorBuilder; 24 hours after transfection, cells were lysed with AllPrep DNA/RNA Mini Kit (Qiagen) or Radio-Immunoprecipitation Assay (RIPA) buffer for assays as described below.
Mouse phenotypes
Body weights were assessed to compare the growth of each genotype mice. For the comparison of intra-uterine growth, body weight of wild type (n = 14), Nsd2WT/P906L (n = 21), and Nsd2 P906L/P906L (n = 7) at embryonic day (E)18.5 were compared in some littermates with relative ratio. Ones of wild type (n = 9), Nsd2WT/− (n = 13), and Nsd2 −/− (n = 7) were also compared similarly. For the comparison of postnatal growth, body weight of wild type (n = 18: male, n = 15: female), Nsd2WT/P906L (n = 17: male, n = 18: female), and Nsd2WT/− (n = 9: male, n = 10: female) at age 8 weeks were compared in each sex with relative ratio. Sample size estimation was performed with power.t.test of Rpackage.
DNA methylation in mouse tissue
Genomic DNA was extracted from mouse tissues using the AllPrep DNA/RNA Mini Kit, followed by bisulfite treatment using the Zymo EZ DNA Methylation-GoldTM kit (Zymo Research). Genome-wide DNA methylation was analyzed using the Infinium Mouse Methylation BeadChip (Illumina). Methylation data were acquired using the iScan system and processed using GenomeStudio 2.0 (Illumina). The background was corrected using the method provided by Illumina. We removed probes with detection P values > .01 in at least 1 sample and filtered probes located on the X, Y, and mitochondrial chromosomes. This yielded 265,725 autosomal probes from 22 assays. DNA methylation differences were assessed through linear regression modeling using beta values.
RNA-seq
Total RNA was isolated from the peripheral blood cells treated with RNA later, using the RiboPure-Blood Kit (Ambion). RNA amplification was performed using the SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (TaKaRa) and cDNA libraries were prepared using Nextera XT DNA Sample Preparation Kit (Illumina). Sequencing was performed using NovaSeq (Illumina). The data were aligned to GRCh38, and transcript count was performed using Dragen (Illumina) while referring to the GRCh38.35 gtf files. In mouse tissue samples, total RNA was isolated from thymocytes with biological duplicates of each genotype using the AllPrep DNA/RNA Mini Kit. RNA-seq libraries were prepared using the NEBNext UltraII Directional RNA Library Prep Kit for Illumina (New England BioLabs). Sequencing was performed using HiSeq X Ten (Illumina). The data were aligned to GRCm38 mm10 using HISAT2-2.1.0, and transcript count was performed using Cufflinks 2.2.1. Significant changes in transcript expression were calculated using Cuffdiff.25
Chromatin immunoprecipitation sequencing (ChIP-seq)
ChIP assay was performed with biological duplicates of each genotype using ChIP Reagents (Nippon Gene). Thymocyte-rich fraction26 (1 × 106) were fixed with 1% formaldehyde for 5 minutes. Cells were resuspended in SDS lysis buffer, and the lysate was sonicated using the S220 Focused-ultrasonicator (Covaris), to fragment the chromatin. The chromatin was purified through centrifugation and immunoprecipitated using Dynabeads Protein A (Veritas Life Sciences) conjugated with anti-H3K36me2 antibodies (ab176921; Abcam) in 1× RIPA (150 mM) buffer with protease inhibitors, for 2 hours at 4 °C. The chromatin-bound beads were washed sequentially with 150 mM and 500 mM 1× RIPA, and Tris-EDTA (TE) buffers. Next, the chromatin-bound beads were incubated in ChIP direct elution buffer with proteinase K (200 μg/ml), overnight at 65 °C. DNA was purified using AMPure XP beads (Beckman Coulter) as per manufacturer’s instructions. ChIP-seq libraries were prepared using NEBNext ChIP-Seq Library Prep Master Mix Set and Multiplex Oligos for Illumina. Sequencing was performed using HiSeq 2500 (Illumina). The data were aligned to the GRCm38 mm10 reference genome using Burrows-Wheeler Alignment tool (BWA) v.0.7.17. Multiple mapped reads and PCR duplicates were removed. Generated Bam files were processed using deepTools to generate a coverage track bigwig.27 The coverage was calculated as the number of reads per 100 bp bin using the reads per kilobase per million mapped reads normalization. Differences between the mice samples were filtered by 1 reads per kilobase per million mapped reads.
Western blot analysis
Western blot analysis was performed as previously described.28 Anti-Histone H3 K36me2 (ab9049), anti-H3 (ab1791), and anti-NSD2 (ab75359) antibodies were procured from Abcam; ab75359 is a monoclonal antibody to a fusion protein, corresponding to amino acids (a.a.) 1-647 of Human GST-NSD2. Anti-beta-Actin (M177-3) and Anti-DDDDK-tag (M185-3S) antibody for detection of FLAG were purchased from MBL. Mice thymocytes or iPSCs were lysed with RIPA buffer. Gel electrophoresis and membrane transfer were performed as per manufacturer’s protocol (BioRad).
Comparison of methylation signature probes of each syndrome
To compare the DNA methylation signature associated with NSD2 defect with that of NSD1 defect8 (GSE74432), DNMT3A defect of Tatton-Brown-Rahman syndrome29 (TBRS, GSE128801), and DNMT3B defect of immunodeficiency, centromeric region instability, facial anomalies syndrome 130 (ICF1, GSE95040), which were identified using the Illumina Infinium HumanMethylation450 BeadChip (450K) array, we first compared unrelated healthy individual beta values between EPIC and 450K arrays in the GEO database. Finally, we selected 299,928 common probes between 2 arrays with a difference of < |0.1| between the 142 EPIC healthy controls (described above) and 213 assays of the 450K healthy controls (GEO: GSE36064, GSE42861 (non-smokers aged ˂50 years), GSE74432).8,31,32 Among the 299,928 common probes, we filtered differentially methylated probes in NSD2 and NSD1 defect respectively, according to the threshold; mean beta values differed more than 0.15 in absolute from 355 controls with <Bonferroni-corrected P = .05 by regression model. NSD2 defects-specific probes were then selected with <Bonferroni-corrected P = .05 by comparing NSD2 defects and NSD1 defects.
Results
DNA methylation signature in WHS
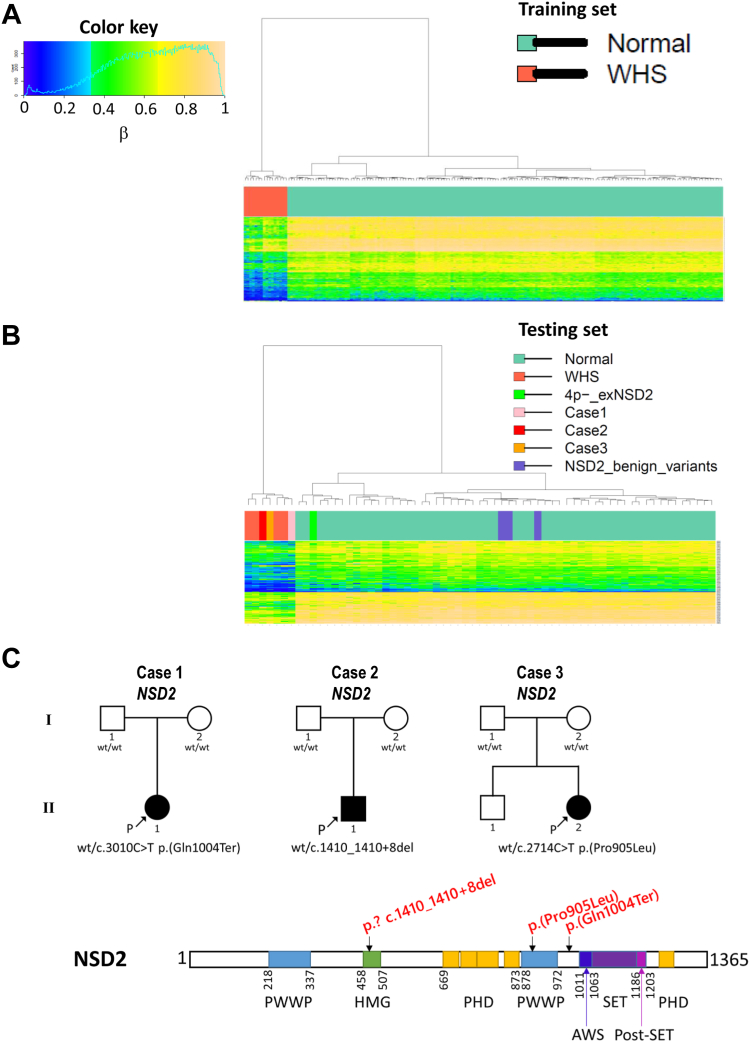
We selected a random 80% subset of all case subjects with WHS (n = 16, Supplemental Table 1) and controls (n = 106, Supplemental Table 4) as a training cohort for the purpose of DNA methylation feature selection and model training. Technical effects on the data (Supplemental Figure 1A) have been removed as an adjustment for confounders in the analysis. The remaining 20% was set aside as a testing data set to be used for the assessment of the performance of the classification model. We selected 280 probes that showed more than |0.2| methylation beta value difference compared with the controls. Although beta value variances of 122 out of the 280 probes were significantly greater in WHS, they were significantly different with Bonferroni-corrected P value <2.4e-16 after statistically adjusted for blood cell type compositions (Supplemental Table 6). The 280 probes distinguished methylation related to WHS from those in control samples through hierarchical clustering (Figure 1A). The 280 probes were confirmed to classify WHS cases from controls in the testing data set by clustering (Figure 1B) and SVM model (Supplemental Figure 1B). Leave-one-out cross-validation with SVM also classified all 20 cases as WHS by the beta values of the 280 probes (Supplemental Figure 1C). These results validated availability of the 280 probes as “WHS-related DNA methylation signatures.”
Figure 1.
DNA methylation pattern of WHS and undiagnosed patients with NSD2 variants. A. The 280 probes identified in the training set by regression analysis were clearly separated into 16 WHS and 106 control samples through hierarchical clustering. B. The 280 probes were validated to classify 4 WHS from 36 controls in the testing set. Three undiagnosed patients with NSD2 VUS were also classified into a WHS branch. Another 3 patients with likely benign NSD2 variants were classified into a control branch. C. Schematic illustration of variants of the 3 cases classified into a WHS branch in (B).
Speculation of pathogenicity of NSD2 variants with WHS-related DNA methylation signatures
An individual with deleted 4p16.2p15.31 that did not include NSD2 was classified in the cluster of controls (Figure 1B),and was similar to controls in the SVM model (Supplemental Figure 1A). These results suggest that WHS-related DNA methylation signatures could be caused by NSD2 loss-of-function variants. Therefore, we analyzed 6 cases harboring rare variants in NSD2. Among them, 3 cases showed WHS-related DNA methylation signature (Figure 1B). Case 1 harbors a de novo nonsense variant of upstream of NSD2 SET domain (c.3010C>T p.(Gln1004Ter)) (Figure 1C). Case 2 harbors a de novo 9 bp deletion spanning exon 5 to intron 5 of NSD2 (c.1410_1410+8del), which could cause abnormal splicing. Case 3 harbors a de novo missense variant in the highly conserved PWWP domain (for the proline-tryptophan-tryptophan-proline motif in the consensus amino acid sequence) of NSD2 (c.2714C>T p.(Pro905Leu)). These 3 variants were clearly clustered in the same branch as WHS in the testing set (Figure 1B). They were also verified with the SVM model and exhibited over 0.9 scores (Supplemental Figure 1B). Choufani et al33 reported that over 0.7 SVM scores show syndrome-specific DNA methylation signature. These results strongly suggest they cause the WHS-related DNA methylation signature. Therefore, we thought that these 3 undiagnosed cases could be NSD2 monogenic disorder.
Meanwhile, the control cluster included 3 cases with “likely benign” NSD2 variants (Figure 1B and Supplemental Table 3). These results strongly support that the WHS-related DNA methylation signatures isolated in this study could detect DNA methylation changes in undiagnosed cases with pathogenic NSD2 variants.
Pathogenicity of the NSD2 variants identified in undiagnosed patients
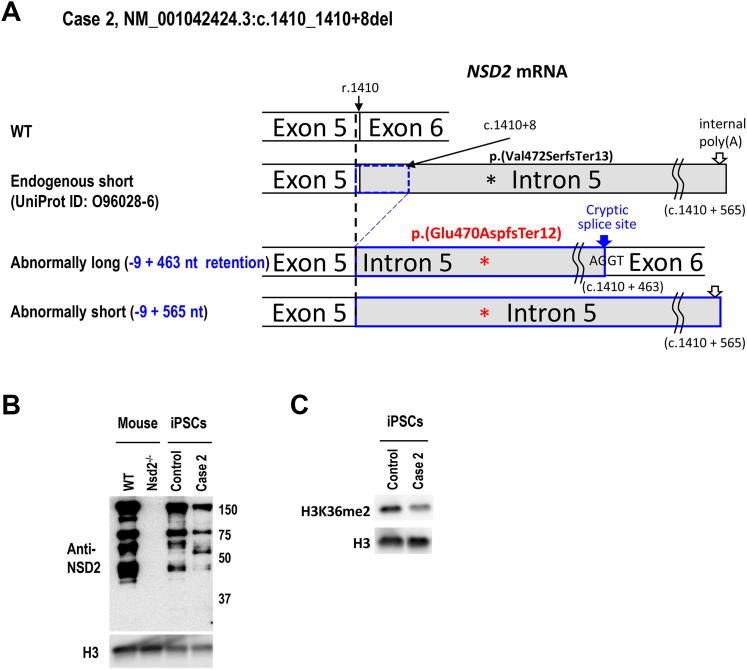
Because case 1 harbors a nonsense variant, it is clearly considered as a loss-of-function variant. Regarding case 2, RNA-seq of peripheral blood cells from this patient revealed the case 2-specific mRNA isoforms, which retain 5’ end of intron 5 because of the 1 bp deletion at the 3’ end of exon 5 and the 8 bp deletion at the 5’ end of intron 5 (Supplemental Figure 2). Further analysis identified a cryptic splice site at c.1410+464 in intron 5, which generated the patient-specific 464 bp intron 5 retention between exons 5 and 6 that was present in 35% of all NSD2 mRNA isoforms in the case 2’s peripheral blood cells (“Abnormally long” in Figure 2A). In addition, another case 2-specific isoform was observed that terminated at c.1410+565 (“Abnormally short” in Figure 2A). The last 21 nt of this isoform was “A,” indicating intrinsic poly(A). The transcript termination at this intrinsic poly(A) has been reported as ENST00000508355.5 in Ensembl release 110 July 2023 (“Endogenous short” in Figure 2A, Supplemental Figure 2B). The protein isoforms that could be translated from these 2 transcript isoforms are the same. The genomic location of the termination codon is the same as that used by the NSD2 protein isoform O96028-6 in UniProtKB (release 2023_05), ie, c.1410 + 43 to 45. The amino acid sequence of O96028-6 is p.(Val472SerfsTer13). That of the case2-specific protein isoform is NP_001035889.1:p.(Glu470AspfsTer12) because of the 9 bp deletion, which is predicted by Expasy to be 53.18 KDa.
Figure 2.
Effects of NSD2 variants in Case 2. A. Schematic description of NSD2 transcript from both alleles of case 2. B. NSD2 antibody (ab75359) was specific to proteins derived from the Nsd2 protein via immunoblotting because there was no band in Nsd2−/− brain. The decreased levels of NSD2 proteins (approx. 150 kDa of O96028-1 and 75 KDa of O96028-3) were confirmed in the case 2 iPSCs. In contrast, a case 2-specific protein was detected by ab75359 (approx. 55 kDa), which was close to the 53.18 kDa speculated molecular weight of NSD2 p.(Glu470AspfsTer12) by Expasy (https://www.expasy.org/). C. The modification levels of H3K36me2 were decreased in case 2 iPSCs.
Western blotting of NSD2 from the established case 2’s iPSCs showed a decrease in NSD2 protein, including 2 major functional NSD2 isoforms of 152 kDa (UniProt ID:O96028-1. 1365 a.a.) and 71 kDa (UniProt ID:O96028-3. 647 a.a.) (Figure 2B). Compared with control iPSCs, the 3rd and 4th bands at approximately 65 kDa and 45 kDa were rarely detected in case 2’s iPSCs. Seven NSD2 protein isoforms, produced by alternative splicing, are reported in UniProtKB (release 2023_05). Five of the 7 isoforms could be detected by the antibody that we used, eg, 69 kDa O96028-5, 53 kDa O96028-6, 30 kDa O96028-7, and the 2 isoforms described in above (O96028-1 and -3). To the best of our knowledge, it is unknown whether the 2 bands at around 65 and 45 kDa on western blotting represent processed NSD2 proteins or those generated by alternative splicing or posttranslational modification. Case 2-specific signal was clearly detected at approximately 55 kDa (Figure 2B). This is thought to be NSD2 NP_001035889.1:p.(Glu470AspfsTer12) translated from abnormal NSD2 mRNA isoforms from the pathogenic variant allele that somehow escaped nonsense-mediated decay. H3K36me2 was apparently reduced in case 2’s iPSCs (Figure 2C).
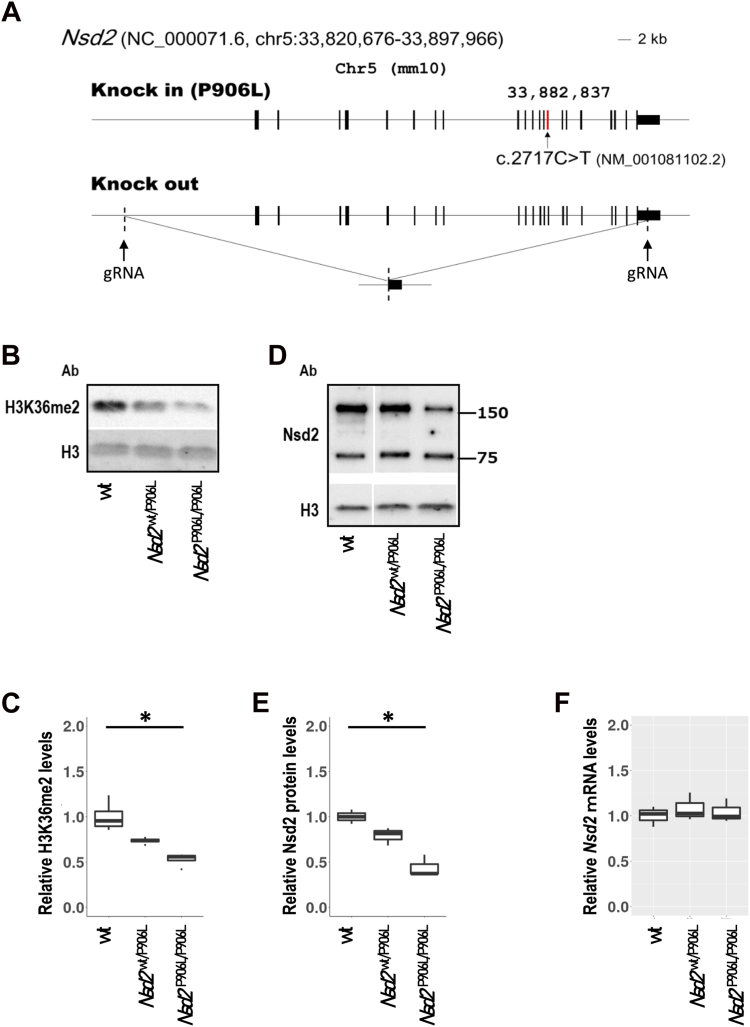
Because we could not get an informed consent on establishing case 3 iPSCs, we created knockin mice of Nsd2 NP_001074571.2:p.Pro906Leu corresponding to case 3’s NSD2 variant and Nsd2 null mutant mice (Nsd2−/−) (Figure 3A and Supplemental Figure 3A and B). Only few Nsd2 p.Pro906Leu homozygous mice (Nsd2P906L/P906L) survived and most Nsd2P906L/P906L died within a day after birth. All Nsd2 null mutant mice (Nsd2−/−) died within a day after birth. Bodyweights at embryonic day 18.5 showed that Nsd2P906L/P906L and Nsd2−/− mice were significantly lower than those of wild-type (WT) mice. At delivery, body weights of Nsd2WT/P906L and Nsd2WT/− neonates were normal, although they showed lighter body weight than that of WT mice at age 8 weeks (Supplemental Figure 3C). We analyzed thymocytes at age 10 weeks because thymocyte’s abnormality is reported in Nsd2 mutant mice34 and high expression of Nsd2 in thymus has been reported.35 Western blotting revealed significantly decreased H3K36me2 in the Nsd2P906L/P906L mice, compared with WT mice (Figure 3B and 3C, Supplemental Figure 4A). Nsd2 P906L mutant protein expression was significantly lower than Nsd2 WT protein (Figure 3D and E, Supplemental 4B), although Nsd2 mRNA remains as much as WT (Figure 3F). This is reconfirmed in transient expression of cDNA of FLAG-tagged NSD2 mutant protein, which suggests lower expression of NSD2 NP_001035889.1:p.(Pro905Leu) in case 3 (Supplemental Figure 3D). Decrease of signal at 152 kDa was confirmed without lower molecular weight signals indicating degradative products.
Figure 3.
Functional assay of the Pro905Leu variant of NSD2. A. Schematic representation of Nsd2 variant (knockin) and Nsd2 deletion (knockout) alleles generated by CRISPR/Cas9-mediated genome editing in this study. For the latter, the genomic loci targeted by CRISPR/Cas9 guide RNAs are indicated by arrows. B. Nsd2 p.Pro906Leu knock-in mice showed a decrease in H3K36me2 in thymocytes at E18.5. C. Student’s t test showed significant decrease of H3K36me2 in homozygous mice with P < .01 as indicated ∗. Biological replicates were n = 4. D. Nsd2 p.Pro906Leu homozygous knockin mice showed a decrease in Nsd2 protein. E. Student’s t test showed significant decrease of Nsd2 in homozygous knock-in mice with P < .01 as indicated ∗, (F) but not Nsd2 mRNA, in brains at E18.5 (n = 4).
These results revealed that the NSD2 variants in cases 1 to 3 are NSD2 loss-of-function variants.
Multi-omics analysis of Nsd2 mutant mice
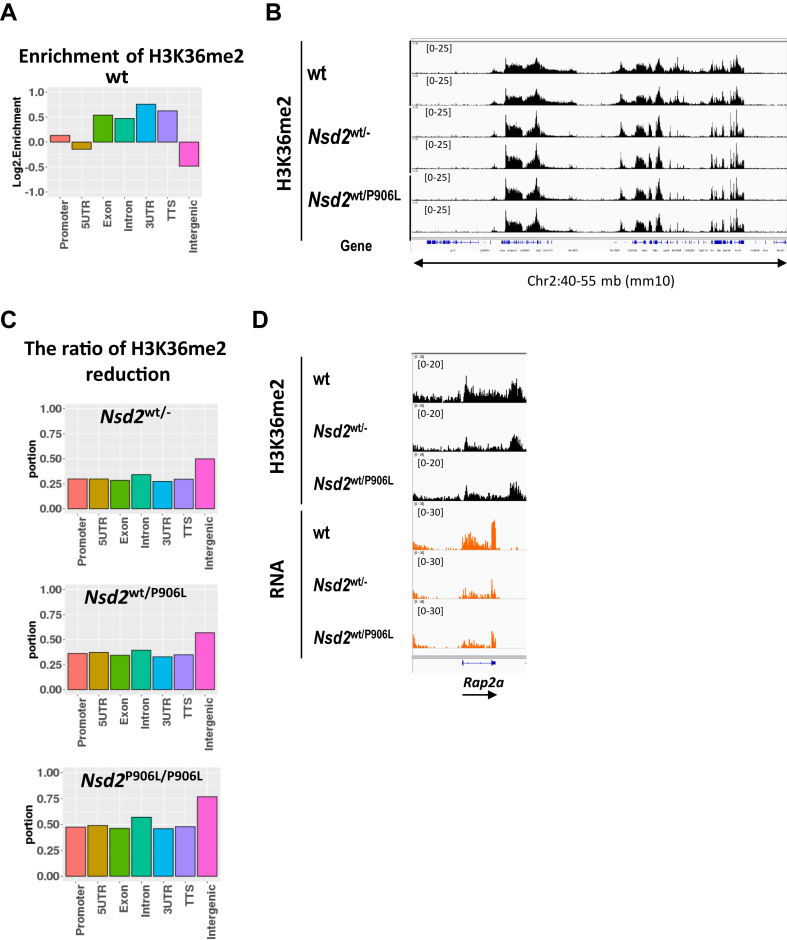
The distribution of H3K36me2 in WT thymocytes was enriched in the gene body and promoter region, but not in the transcription start site (TSS), as previously reported (Figure 4A and D).7 At the same time, the distribution of H3K36me2 in the intergenic region was not enriched compared with the whole genome, with a log2. enrichment of -0.484 (Figure 4A). To determine the effect of Nsd2 mutations on epigenetic regulation and gene expression, we performed a multi-omics analysis. H3K36me2 modification upstream of the TSS was correlated with gene expression as previously reported (Supplemental Figure 5A).7 Nsd2WT/− showed a genome-wide decrease in H3K36me2 (Figure 4B). Nsd2WT/P906L changed H3K36me2 similarly to Nsd2WT/− with a correlation coefficient of 0.99 (Figure 4B and Supplemental 4C). The ratio of the windows in which H3K36me2 reduction was confirmed in the mutant was highest in intergenic regions in all 3 genotypes (Figure 4C). These results strongly suggested that NSD2 p.(Pro905Leu) in case 3 are pathogenic variants. Significant downregulation of expression were observed in 56 genes in Nsd2WT/− (Table S7). Primarily (86%), they were regulated in the same direction of H3K36me2 changes in TSS (Supplemental Figure 5B).73% of significantly expression-changed genes associating with H3K36me2 changes in Nsd2WT/P906L were overlapped with those in Nsd2WT/− (Figure 4D, Supplemental 5B). Regarding to DNA methylation, genome-wide DNA hypomethylation was entirely observed in mutant mice (Supplemental Figure 5D and E). Concomitant changes with DNA hypo-methylation and H3K36me2 decrease in the intergenic region were most frequent changes in Nsd2 mutant mice (Supplemental Figure 5F). Enrichment of DNA methylation changes at the intergenic region was highest in the WHS-related DNA methylation signatures, too (Supplemental Figure 1D). DNA methylation changes in Nsd2WT/− with |>0.1| were not always associated with nearest genes expression changes, as generally confirmed (Supplemental Figure 5G).36 In addition, H3K36me2 downregulation at promoter by Nsd2 mutation were observed in barely expressed genes (Supplemental Tables 8-10).
Figure 4.
Multiomics analysis of NSD2-variants. A. Genome-wide H3K36me2 log 2 enrichment by genetic features in wild-type mice. B. Representative genomic tracks of H3K36me2 changes in each mice genotype. C. Ratio of H3K36me2-decreased windows in each genetic region by Nsd2 KO or knockin mice from wild-type mice. D. Representative tracks at Rap2a locus of H3K36me2 and gene expression in each mice genotype.
These results suggest that Nsd2 defects change both the expression of a part of genes and intergenic DNA methylation concordant with H3K36me2 changes. Disease-specific DNA methylation changes in blood cells are not always observed in causal genes of systemic phenotypes. It was the same as in Nsd2 mutant mice.
DNA methylation differences in patients with NSD2 loss-of-function variants and Sotos syndrome 1
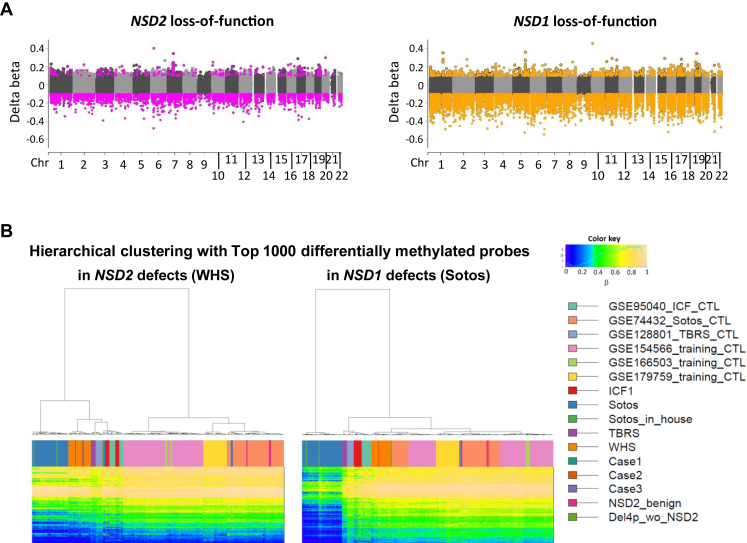
NSD2 and NSD1 are H3K36me2 methyltransferases. The loss of function of NSD2 and NSD1 genes could cause similar epigenetic abnormalities. Indeed, patients with Sotos syndrome 1 exhibited high scores for WHS based on the SVM model using WHS-related DNA methylation signature (Supplemental Figure 1A, Supplemental Table 11). However, as indicated by WHS and Sotos syndrome phenotypes, NSD2 defects and NSD1 defects influence development in different ways. Therefore, we attempted to identify DNA methylation changes in peripheral blood cells that would distinguish NSD2 defects and NSD1 defects and normal controls, using publicly available data. We identified 3402 and 15,327 differentially methylated probes in NSD2 and NSD1 defects, respectively; 2088 out of the 3402 probes overlapped with the 15,327 probes. More robust changes in NSD1 defects than in NSD2 defects were confirmed (Figure 5A). Only 20 probes clearly separated NSD2 defects from NSD1 defects and controls among the 3402 probes (Supplemental Figure 6A, Supplemental Table 12). Sotos, TBRS with heterozygous DNMT3A (OMIM ∗602769) loss of function (OMIM615879), ICF1 with homozygous DNMT3B (OMIM ∗602900) loss of function (OMIM 242860), and NSD2 defects are all characterized by genome-wide DNA hypomethylation.8,10,29,30 Hierarchical clustering using the top 1000 Value differentially methylated (DMP) probes in NSD2 defects showed a single dendrogram with only NSD2 defects. However, DNA methylation pattern of DMP probes in NSD2 defects was similar between 4 syndromes. Meanwhile, that of NSD1 defects is unique to NSD1 defects, which was clustered in a distinct branch (Figure 5B). Dimensionality reduction by t-distributed stochastic neighbor embedding (tSNE) of DMP probes in NSD2 defects also clustered NSD1 defects (Supplemental Figure 6B). This suggests that the major targets of the NSD2 defects are similar to those of NSD1 defects. Excluded 2 Sotos patients were the ones harboring the same variant in NSD1 at the end of the gene and also display slightly different DNA methylation changes compared with the other Sotos patients in the study by Choufani et al.8 Meanwhile, tSNE of DMP probes in NSD1 defects did not gather NSD2 defects in 1 place. Again, it showed distinct DNA methylation changes in NSD1 defects. However, TBRS clustered differently from their controls. This may indicate common molecular mechanisms between Sotos and TBRS. The 4 syndromes were clustered separately by tSNE of mixed top 100 P value DMP probes of each syndrome, which indicates that DNA methylation signatures specific to each syndrome exist (Supplemental Figure 6C).
Figure 5.
DNA methylation changes in NSD2 loss-of-function variants and other syndromes. A. Comparison of genome-wide DNA methylation changes between NSD2 and NSD1 loss-of-function variants in peripheral blood cells. Pink and orange dots indicate 3402 and 15,327 probes with significantly differential methylation in NSD2 and NSD1 loss-of-function variants, respectively, with Bonferroni-corrected P < .05 and |delta beta| > 0.15. B. Hierarchical clustering of individuals with NSD2 defects (WHS and Cases), NSD1 defects (Sotos and Sotos inHouse), DNMT3A defects (TBRS), DNMT3B defects (ICF1), and controls of each study by methylation levels of the top 1000 P values DMP probes in NSD2 or in NSD1 defects.
Discussion
The WHS-related DNA methylation signatures and the DNA methylation signature in NSD2 loss-of-function variants are almost the same. But the cases with NSD2 loss-of-function variants were not clinically diagnosed as WHS. Reportedly, NSD2 loss-of-function variants lead to a distinct, rather mild phenotype partially overlapping with WHS.3, 4, 5, 6 Overlapping phenotype could be related to common epigenetic dysfunctions similar to that in DNA methylation signatures.
H3K36me2 modifications were enriched in the genic region in WT mice; however, most decreased H3K36me2 modifications were observed in the intergenic region in adult Nsd2 heterozygous KO and knockin (ie, Nsd2WT/P906L) mice thymocytes. Intergenic H3K36me2 decrease was consistent with the findings by Weinberg et al37 from the mesenchymal stem cells in which Nsd1/2 were genetically ablated and from the embryonic stem cells in which Nsd1 was genetically ablated. Increased H3K36me2 was also detected in some genomic regions in Nsd2WT/− and Nsd2WT/P906L mice. This conflicting phenomenon was also reported in multiple myelomas with NSD2 variants7 or squamous cell carcinomas with NSD1 variants.38 It might be a result of the redundancy of other enzymes for H3K36me2 modification than NSD2 and/or the balance between reciprocal modification of H3K36me2 and H3K27me339 that also involves Histone H1.40,41 Most downregulated genes in Nsd2WT/− and Nsd2WT/P906L mice correlated with decreased H3K36me2 before TSS (Supplemental Figure 5B). NSD2 is reportedly located near TSS but avoids it42 and is involved in transcription.43 These genes could be involved in pathological conditions as direct targets of Nsd2 loss-of-function variants. Although there were no differentially methylated loci in Nsd2WT/− and Nsd2WT/P906L mice in a statistically significant manner, the trends of hyper-methylation around TSS and of general hypo-methylation across the entire gene body of significantly downregulated genes were observed (Supplemental Figure 5G). However, DNA methylation changes occurred at similar levels in the gene body of nonexpressed genes (Supplemental Figure 5G). These results highlight the difficulty in speculating gene expression changes through DNA methylation changes in the gene body. The effects of DNA methylation changes on gene expression could be related to tissue specificity of gene expression.44
H3K36me2 recruits DNMT3A.37,45,46 DNA hypomethylation were most often observed in the intergenic region along with H3K36me2 decrease caused by Nsd2 variants (Supplemental Figure 5F) as previously reported37 and in blood cells of patients with NSD2 defects (Supplemental Figure 1D). However, DNA methylation is regulated by other histone modifications, too.47 DNA methylation changes were observed not only in regions that H3K36me2 was regulated by Nsd2 variants but also in regions where H3K36me2 were undetected in all genotypes (Supplemental Figure 5D). It is possible that the DNA methylation signature identified herein, specific to NSD2 loss-of-function variants, might not be direct targets of H3K36me2 by NSD2. In contrast, episignature is reported in each single epigenetic regulator gene’s variants.11 This indicates common epigenetic and etiological backgrounds in patients with NSD2 loss-of-function variants: WHS and RAUST.
Our study revealed the DNA methylation signature in NSD2 loss-of-function variants and p.(Pro905Leu) is a loss-of-function variant. Opposite DNA methylation patterns between Sotos syndrome 1 and Hunter McAlpine syndrome with duplication of NSD1 (OMIM 601379) compared with controls have been previously reported.10,48 This indicates that opposite DNA methylation patterns at some of the 280 probes could be identified between NSD2 gain-of-function and loss-of-function variants, too. In addition, distinct DNA methylation signatures in Helsmoortel-van der Aa Syndrome (OMIM 615873) have been reported based on the loci of missense variants in ADNP gene (OMIM ∗611386).49 Regarding KAT6B (OMIM ∗605880), variants resulted in distinct DNA methylation signatures are associated with different syndromes, namely, Genitopatellar syndrome (OMIM 606170) and Say-Barber-Biesecker-Young-Simpson syndrome (OMIM 603736).10 Hence, DNA methylation patterns of the 280 probes could be criteria in diagnosing the pathogenicity of NSD2 VUS and to assess of function of NSD2 variants.
Data Availability
All DNA methylation array and sequencing data have been deposited in the Gene Expression Omnibus (GEO) at GSE174251 (human methylation array), GSE176070 (RNA-seq), GSE220890 (ChIP-seq), and GSE221621 (mouse methylation array).
Conflict of Interest
The authors declare no conflicts of interest.
Acknowledgments
English language editing services were provided by Editage.
Funding
This work has been funded by research grants from the Japan Agency for Medical Research and Development (AMED) (Practical Research Project for Rare/Intractable Diseases; ek0109489 to K.H., ek0109205 to T.K.), JSPS KAKENHI (21H02887 and 21K19584 to K.H.), National Center for Child Health and Development (NCCHD 2022A-3 to K.H.), Gunma university for the promotion of scientific research to K.H.
Author Information
Conceptualization: T.K., K.H.; Formal analysis: T.K.; Funding acquisition: T.K., K.H.; Investigation: T.K., S.K., Y.T., E.O., H.Kamura, K.Kojima, H.Kikuchi; Methodology: M.T., T.S., S.T., H.A., K.U.; Project administration: T.K., K.H.; Resources: O.M., M.K., T.I., Y.Y., K.W., H.O., K.S., S.M., N.O., Y.F., F.T., K.Kosaki; Supervision: K.H.; Validation: T.K., S.K.; Visualization: T.K., S.K.; Writing-original draft: T.K.; Writing-review & editing: K.N., K.H.
ORCIDs
Tomoko Kawai: https://orcid.org/0000-0001-8137-0334
Kenichiro Hata: https://orcid.org/0000-0002-7809-7637
Shuji Takada: https://orcid.org/0000-0002-9406-4683
Ethics Declaration
Approval for review and reporting these cases was given by all the relevant Japanese medical institutions: National Center for Child Health and Development, St. Marianna University School of Medicine, Gunma Children’s Medical Center, Keio University School of Medicine, Kitasato University School of Medicine, Shinshu University School of Medicine, Osaka Women’s and Children’s Hospital, Saitama Children’s Medical Center, and Central Hospital, Aichi Human Service Center. Written informed consent was obtained from the individuals’ parents for genetic testing. Human cells were collected post ethical approval from the Institutional Review Board of National Institute for Child Health and Development, Japan. Signed informed consent was obtained from donors or their parents, and the specimens were irreversibly de-identified. All experiments were performed according to the tenets of the Declaration of Helsinki. All protocols for animal experiments were approved by the Animal Care and Use Committee of the National Research Institute for Child Health and Development, Tokyo, Japan.
Footnotes
The Article Publishing Charge (APC) for this article was paid by Tomoko Kawai.
Additional Information
The online version of this article (https://doi.org/10.1016/j.gimo.2024.101838) contains supplemental material, which is available to authorized users.
Contributor Information
Tomoko Kawai, Email: kawai-tm@ncchd.go.jp.
Kenichiro Hata, Email: khata@gunma-u.ac.jp.
Additional Information
are provided in a pdf file.
are provided in a excel file.
References
- 1.Wright T.J., Ricke D.O., Denison K., et al. A transcript map of the newly defined 165 kb Wolf-Hirschhorn syndrome critical region. Hum Mol Genet. 1997;6(2):317–324. doi: 10.1093/hmg/6.2.317. [DOI] [PubMed] [Google Scholar]
- 2.Zollino M., Lecce R., Fischetto R., et al. Mapping the Wolf-Hirschhorn syndrome phenotype outside the currently accepted WHS critical region and defining a new critical region, WHSCR-2. Am J Hum Genet. 2003;72(3):590–597. doi: 10.1086/367925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Derar N., Al-Hassnan Z.N., Al-Owain M., et al. De novo truncating variants in WHSC1 recapitulate the Wolf–Hirschhorn (4p16.3 microdeletion) syndrome phenotype. Genet Med. 2019;21(1):185–188. doi: 10.1038/s41436-018-0014-8. [DOI] [PubMed] [Google Scholar]
- 4.Lozier E.R., Konovalov F.A., Kanivets I.V., et al. De novo nonsense mutation in WHSC1 (NSD2) in patient with intellectual disability and dysmorphic features. J Hum Genet. 2018;63(8):919–922. doi: 10.1038/s10038-018-0464-5. [DOI] [PubMed] [Google Scholar]
- 5.Boczek N.J., Lahner C.A., Nguyen T.M., et al. Developmental delay and failure to thrive associated with a loss-of-function variant in WHSC1 (NSD2) Am J Med Genet A. 2018;176(12):2798–2802. doi: 10.1002/ajmg.a.40498. [DOI] [PubMed] [Google Scholar]
- 6.Zanoni P., Steindl K., SenGupta D., et al. Loss-of-function and missense variants in NSD2 cause decreased methylation activity and are associated with a distinct developmental phenotype. Genet Med. 2021;23(8):1474–1483. doi: 10.1038/s41436-021-01158-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuo A.J., Cheung P., Chen K., et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell. 2011;44(4):609–620. doi: 10.1016/j.molcel.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choufani S., Cytrynbaum C., Chung B.H., et al. NSD1 mutations generate a genome-wide DNA methylation signature. Nat Commun. 2015;6 doi: 10.1038/ncomms10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aref-Eshghi E., Bend E.G., Colaiacovo S., et al. Diagnostic utility of genome-wide DNA methylation testing in genetically unsolved individuals with suspected hereditary conditions. Am J Hum Genet. 2019;104(4):685–700. doi: 10.1016/j.ajhg.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aref-Eshghi E., Kerkhof J., Pedro V.P., et al. Evaluation of DNA methylation episignatures for diagnosis and phenotype correlations in 42 Mendelian neurodevelopmental disorders. Am J Hum Genet. 2020;106(3):356–370. doi: 10.1016/j.ajhg.2020.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levy M.A., McConkey H., Kerkhof J., et al. Novel diagnostic DNA methylation episignatures expand and refine the epigenetic landscapes of Mendelian disorders. HGG Adv. 2022;3(1) doi: 10.1016/j.xhgg.2021.100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerkhof J., Squeo G.M., McConkey H., et al. DNA methylation episignature testing improves molecular diagnosis of Mendelian chromatinopathies. Genet Med. 2022;24(1):51–60. doi: 10.1016/j.gim.2021.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Shimizu K., Wakui K., Kosho T., et al. Microarray and FISH-based genotype-phenotype analysis of 22 Japanese patients with Wolf-Hirschhorn syndrome. Am J Med Genet A. 2014;164A(3):597–609. doi: 10.1002/ajmg.a.36308. [DOI] [PubMed] [Google Scholar]
- 14.Okamoto N., Ohmachi K., Shimada S., Shimojima K., Yamamoto T. 109 kb deletion of chromosome 4p16.3 in a patient with mild phenotype of Wolf–Hirschhorn syndrome. Am J Med Genet A. 2013;161A(6):1465–1469. doi: 10.1002/ajmg.a.35910. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi Y., Date H., Oi H., et al. Six years’ accomplishment of the Initiative on Rare and Undiagnosed Diseases: nationwide project in Japan to discover causes, mechanisms, and cures. J Hum Genet. 2022;67(9):505–513. doi: 10.1038/s10038-022-01025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards S., Aziz N., Bale S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kandaswamy R., Hannon E., Arseneault L., et al. DNA methylation signatures of adolescent victimization: analysis of a longitudinal monozygotic twin sample. Epigenetics. 2021;16(11):1169–1186. doi: 10.1080/15592294.2020.1853317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y., Long H., Wang S., et al. Genome-wide DNA methylation pattern in whole blood associated with primary intracerebral hemorrhage. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.702244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Acton R.J., Yuan W., Gao F., et al. The genomic loci of specific human tRNA genes exhibit ageing-related DNA hypermethylation. Nat Commun. 2021;12(1):2655. doi: 10.1038/s41467-021-22639-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haghshenas S., Bhai P., Aref-Eshghi E., Sadikovic B. Diagnostic utility of genome-wide DNA methylation analysis in Mendelian neurodevelopmental disorders. Int J Mol Sci. 2020;21(23):9303. doi: 10.3390/ijms21239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scholkopf B., Smola A.J., Williamson R.C., Bartlett P.L. New support vector algorithms. Neural Comput. 2000;12(5):1207–1245. doi: 10.1162/089976600300015565. [DOI] [PubMed] [Google Scholar]
- 22.Platt J.C. Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. Adv Large Margin Classifiers. 1999;10(3):61–74. [Google Scholar]
- 23.Sugawara T., Miura T., Kawasaki T., Umezawa A., Akutsu H. The hsa-miR-302 cluster controls ectodermal differentiation of human pluripotent stem cell via repression of DAZAP2. Regen Ther. 2020;15:1–9. doi: 10.1016/j.reth.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hara S., Terao M., Muramatsu A., Takada S. Efficient production and transmission of CRISPR/Cas9-mediated mutant alleles at the IG-DMR via generation of mosaic mice using a modified 2CC method. Sci Rep. 2019;9(1) doi: 10.1038/s41598-019-56676-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C., Roberts A., Goff L., et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baran-Gale J., Morgan M.D., Maio S., et al. Ageing compromises mouse thymus function and remodels epithelial cell differentiation. Elife. 2020;9 doi: 10.7554/eLife.56221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramírez F., Ryan D.P., Grüning B., et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44(W1):W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taniguchi K., Kawai T., Kitawaki J., et al. Epitranscriptomic profiling in human placenta: N6-methyladenosine modification at the 5′-untranslated region is related to fetal growth and preeclampsia. FASEB J. 2020;34(1):494–512. doi: 10.1096/fj.201900619RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeffries A.R., Maroofian R., Salter C.G., et al. Growth disrupting mutations in epigenetic regulatory molecules are associated with abnormalities of epigenetic aging. Genome Res. 2019;29(7):1057–1066. doi: 10.1101/gr.243584.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Velasco G., Grillo G., Touleimat N., et al. Comparative methylome analysis of ICF patients identifies heterochromatin loci that require ZBTB24, CDCA7 and HELLS for their methylated state. Hum Mol Genet. 2018;27(14):2409–2424. doi: 10.1093/hmg/ddy130. [DOI] [PubMed] [Google Scholar]
- 31.Alisch R.S., Barwick B.G., Chopra P., et al. Age-associated DNA methylation in pediatric populations. Genome Res. 2012;22(4):623–632. doi: 10.1101/gr.125187.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y., Aryee M.J., Padyukov L., et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol. 2013;31(2):142–147. doi: 10.1038/nbt.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choufani S., McNiven V., Cytrynbaum C., et al. An HNRNPK-specific DNA methylation signature makes sense of missense variants and expands the phenotypic spectrum of Au-Kline syndrome. Am J Hum Genet. 2022;109(10):1867–1884. doi: 10.1016/j.ajhg.2022.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campos-Sanchez E., Deleyto-Seldas N., Dominguez V., et al. Wolf-Hirschhorn syndrome Candidate 1 is necessary for correct hematopoietic and B cell development. Cell Rep. 2017;19(8):1586–1601. doi: 10.1016/j.celrep.2017.04.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chesi M., Nardini E., Lim R.S., Smith K.D., Kuehl W.M., Bergsagel P.L. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood. 1998;92(9):3025–3034. [PubMed] [Google Scholar]
- 36.Lister R., Pelizzola M., Dowen R.H., et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinberg D.N., Papillon-Cavanagh S., Chen H., et al. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature. 2019;573(7773):281–286. doi: 10.1038/s41586-019-1534-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farhangdoost N., Horth C., Hu B., et al. Chromatin dysregulation associated with NSD1 mutation in head and neck squamous cell carcinoma. Cell Rep. 2021;34(8) doi: 10.1016/j.celrep.2021.108769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Streubel G., Watson A., Jammula S.G., et al. The H3K36me2 methyltransferase Nsd1 demarcates PRC2-mediated H3K27me2 and H3K27me3 domains in embryonic stem cells. Mol Cell. 2018;70(2):371–379.e5. doi: 10.1016/j.molcel.2018.02.027. [DOI] [PubMed] [Google Scholar]
- 40.Yusufova N., Kloetgen A., Teater M., et al. Histone H1 loss drives lymphoma by disrupting 3D chromatin architecture. Nature. 2021;589(7841):299–305. doi: 10.1038/s41586-020-3017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willcockson M.A., Healton S.E., Weiss C.N., et al. H1 histones control the epigenetic landscape by local chromatin compaction. Nature. 2021;589(7841):293–298. doi: 10.1038/s41586-020-3032-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka H., Igata T., Etoh K., Koga T., Takebayashi S.I., Nakao M. The NSD2/WHSC1/MMSET methyltransferase prevents cellular senescence-associated epigenomic remodeling. Aging Cell. 2020;19(7) doi: 10.1111/acel.13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H., Farnung L., Dienemann C., Cramer P. Structure of H3K36-methylated nucleosome–PWWP complex reveals multivalent cross-gyre binding. Nat Struct Mol Biol. 2020;27(1):8–13. doi: 10.1038/s41594-019-0345-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blake L.E., Roux J., Hernando-Herraez I., et al. A comparison of gene expression and DNA methylation patterns across tissues and species. Genome Res. 2020;30(2):250–262. doi: 10.1101/gr.254904.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heyn P., Logan C.V., Fluteau A., et al. Gain-of-function DNMT3A mutations cause microcephalic dwarfism and hypermethylation of Polycomb-regulated regions. Nat Genet. 2019;51(1):96–105. doi: 10.1038/s41588-018-0274-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu W., Li J., Rong B., et al. DNMT3A reads and connects histone H3K36me2 to DNA methylation. Protein Cell. 2020;11(2):150–154. doi: 10.1007/s13238-019-00672-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weinberg D.N., Rosenbaum P., Chen X., et al. Two competing mechanisms of DNMT3A recruitment regulate the dynamics of de novo DNA methylation at PRC1-targeted CpG islands. Nat Genet. 2021;53(6):794–800. doi: 10.1038/s41588-021-00856-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peeters S., Declerck K., Thomas M., et al. DNA methylation profiling and genomic analysis in 20 children with short stature who were born small for gestational age. J Clin Endocrinol Metab. 2020;105(12) doi: 10.1210/clinem/dgaa465. [DOI] [PubMed] [Google Scholar]
- 49.Bend E.G., Aref-Eshghi E., Everman D.B., et al. Gene domain-specific DNA methylation episignatures highlight distinct molecular entities of ADNP syndrome. Clin Epigenetics. 2019;11(1):64. doi: 10.1186/s13148-019-0658-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
are provided in a pdf file.
are provided in a excel file.
Data Availability Statement
All DNA methylation array and sequencing data have been deposited in the Gene Expression Omnibus (GEO) at GSE174251 (human methylation array), GSE176070 (RNA-seq), GSE220890 (ChIP-seq), and GSE221621 (mouse methylation array).