Abstract
Purpose
It is well known that individuals with hereditary retinoblastoma are at lifelong high risk for developing subsequent malignant neoplasms (SMN). However, the role that non-RB1 germline variants play in tumorigenesis and SMN risk has not yet been studied. The purpose of this study is to report the frequency and spectrum of non-RB1 germline cancer predisposing variants in individuals with retinoblastoma (RB).
Methods
Retrospective data collection from institutional electronic medical records of 94 individuals seen at our institution with personal history of retinoblastoma, who had undergone next-generation sequencing germline analysis.
Results
The prevalence of individuals with cancer predisposition was 57% (54/94). Of these individuals, 76% (41/54) had a pathogenic/likely pathogenic (P/LP) variant only in the RB1 gene, 9% (5/54) harbored a P/LP variant only in a non-RB1 gene, and 11% (6/54) had both. No difference was found between patients with and without non-RB1 variants when comparing demographic and clinical characteristics, including time to SMN. Variants were found in 7 different genes, with only 1 variant repeating 3 times.
Conclusion
In this small cohort of patients with retinoblastoma, non-RB1 variants did not appear to augment tumorigenesis or disease progression. Larger studies are required to determine associations between specific variants and development of SMN.
Keywords: Cancer, Genetics, Germline cancer susceptibility syndromes, Retinoblastoma, Secondary malignancies
Introduction
It is recognized that individuals with hereditary retinoblastoma (OMIM#180200) are at lifelong high risk for developing subsequent non-ocular malignancies.1, 2, 3, 4 Germline RB1 variants,4 radiation,2,5 systemic chemotherapy,1,5 and young age at treatment are important risk factors.6 Pan-cancer studies have shown that between 8% to 18% of individuals with retinoblastoma harbor pathogenic or likely pathogenic (P/LP) variants in cancer predisposition genes and that 34% to 59% of those variants may be unrelated to the primary cancer, depending on the study population.7, 8, 9 In fact, in an earlier report of individuals with retinoblastoma, we found 5 individuals who had pathogenic variants in cancer predisposing genes other than RB1.10 If and how these non-RB1 germline variants influence tumorigenesis, disease progression, and subsequent malignant neoplasms (SMN) has not yet been studied. The purpose of this study is to report the frequency and spectrum of non-RB1 germline cancer predisposing variants from individuals with hereditary (HR) and non-hereditary retinoblastoma (nHR) seen at our institution.
Materials and Methods
Retrospective data collection from institutional electronic medical records of 94 individuals seen at Memorial Sloan Kettering Cancer Center (MSKCC) with a personal history of retinoblastoma (RB), who had previously undergone next-generation sequencing (NGS) germline analysis using the MSK-IMPACT (Integrated Mutation Profiling of Actionable Cancer Targets) platform (MSKCC; NCT01775072).11 Testing was offered to all individuals undergoing enucleation and those who for clinical reasons were considered to benefit from broad germline testing. The study was approved by MSKCC’s Institutional Review Board/Privacy Board.
MSK-IMPACT is an FDA approved, hybridization capture-based NGS panel designed to identify known P/LP germline variants in 90 cancer-associated genes5, 6, 7,11 (the list of genes can be shared upon reasonable request). All the germline genes that are analyzed are clinically actionable. Germline variant classification follows the American College of Medical Genetics and Genomics guidelines.12 Unless otherwise specified, only P/LP germline variants are reported.
Statistical Analysis
Patients' characteristics among HR, nHR, RB1, and non-RB1 groups were analyzed using χ2 test, Fisher’s exact test, or Wilcoxon rank sum as appropriate.
Results
Description of the population
In this cohort of 94 individuals with retinoblastoma, 83 (88%) were ≤18 years and 11 (12%) were >18 years.
Participants’ demographics are presented in Table 1. There was an even distribution of individuals with HR (52%; n = 49) and nHR (48%; n = 45).
Table 1.
Demographics and Rate of non-RB1 variants in the HR and non-HR groups
| Characteristic | Hereditary, n = 49a | Non-hereditary, n = 45a | Overall, N = 94a |
|---|---|---|---|
| Sex | |||
| Female | 22 (45%) | 21 (47%) | 43 (46%) |
| Male | 27 (55%) | 24 (53%) | 51 (54%) |
| Age at time of study (years) | 7 (1-45)b | 6 (0-24)b | 7 (0-45)b |
| Age at RB diagnosis (years) | 1 (0-4)b | 1 (0-9)b | 1 (0-9)b |
| Unknown | 1 | 0 | 1 |
| Race description | |||
| Asian-Far East, Indian Subcontinent | 7 (16%) | 9 (21%) | 16 (18%) |
| Black or African American | 6 (14%) | 6 (14%) | 12 (14%) |
| Other | 4 (9%) | 4 (9%) | 8 (9%) |
| White | 27 (61%) | 24 (56%) | 51 (59%) |
| Unknown | 5 | 2 | 7 |
| Ethnicity description | |||
| Non-Spanish; Non-Hispanic | 40 (83%) | 35 (80%) | 75 (82%) |
| Spanish NOS; Hispanic NOS, Latino NOS | 8 (17%) | 9 (20%) | 17 (18%) |
| Unknown | 1 | 1 | 2 |
| Laterality | |||
| Bilateral | 41 (84%) | 0 (0%) | 41 (44%) |
| Trilateral | 1 (2%) | 0 (0%) | 1 (1%) |
| Unilateral | 7 (14%) | 45 (100%) | 52 (55%) |
| RB1 germline variant | |||
| Negative | 2 (4%) | 45 (100%) | 47 (50%) |
| Positive | 47 (96%) | 0 (0%) | 47 (50%) |
| Germline variant in gene other than RB1 | 6 12% (95%CI: 5%, 25%) |
5 11% (95%CI: 4%, 25%) |
P = .86c |
| CHEK2 | 1 | 0 | |
| ERCC3 | 0 | 1 | |
| FANCC | 0 | 1 | |
| MITF | 1 | 0 | |
| MSH3 | 1 | 1 | |
| MUTYH | 3 | 0 | |
| NTHL1 | 0 | 2 | |
| Second Germline variant in gene other than RB1:TSC2 c.4570-1G>A | 0 (0%) | 1 (2%) |
CI, confidence Interval; NOS, not otherwise specified.
n (%).
Median (Range).
Pearson’s χ2 test.
The HR group was defined by those with a germline RB1 variant and/or history of bilateral disease. This included 2 individuals with negative germline RB1 testing and bilateral disease, 7 with positive germline RB1 variants and unilateral disease, and 1 person with trilateral disease and a germline RB1 pathogenic variant. All others were defined within the nHR group.
Of note, hereditary groups refer to the presence or absence of P/LP variants in the RB1 gene. Two more groups are described based on the presence (non-RB1 variant group) or absence (control group) of P/LP variants in genes other than RB1.
Prevalence of non-RB1 P/LP variants
Eleven individuals (12%) had variants in a non-RB1 gene; 6 had HR and 5 had nHR. These represent 12% of all individuals with HR and 11% of individuals with nHR. The rate of non-RB1 variants was not significantly different between the 2 groups (Table 1).
Supplemental Table 1 compares clinical and demographic characteristics of patients with and without non-RB1 variants. No statistically significant difference was found between the 2 groups from the variables analyzed.
Development of secondary cancers
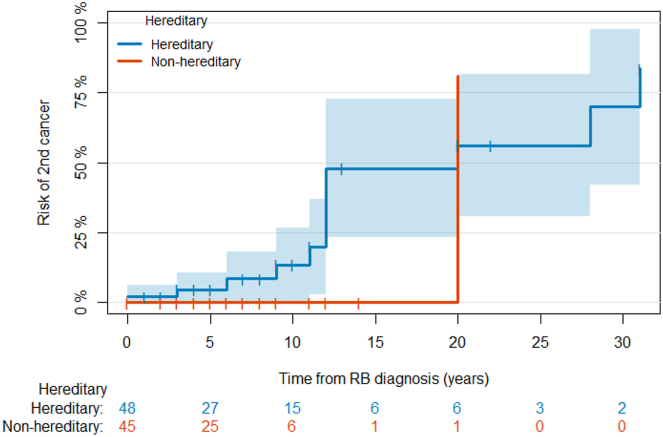
In an attempt to evaluate any impact on long-term outcomes, we looked for any association between having P/LP variants in RB1 and/or non-RB1 genes with the development of SMN (Figure 1). Only 13 individuals had developed a SMN at the time of our study, and 5 developed a third cancer. All individuals who developed SMN except 1 had HR; 3 individuals also had a non-RB1 variant.
Figure 1.
Cumulative incidence of second cancers. Because the age at second cancer is a censored observation, we used survival analysis to estimate the rate of SMN diagnosis in each of the groups and analyze any associations with presence of a non-RB1 variant. The cumulative incidence of SMN in the overall study population is presented in Figure 1. The cumulative incidence of SMN was 48% at 15 years in the hereditary group versus 0% in the non-hereditary group, although the difference was not statistically significant (P = .07). Likewise, there was no evidence of a significant difference in the time to second cancer between patients with and without non-RB1 variants in the overall study population nor within the subgroup of patients with hereditary disease. Tick marks represent censoring times. The subgroup of patients with non-hereditary disease was not analyzed because of the small number of second cancers in that group. Time to SMN was defined as the time from the age at RB diagnosis to the age at the first SMN diagnosis. Patients without SMN diagnoses were censored at the current age. Deaths before the occurrence of SMN were treated as competing risks. The cumulative incidence of SMN was estimated using an Aalen-Johansen estimator,13 and the curves were compared using a Gray’s test.14 Analyses were done using R v.3.6.3.
The mean age of SMN diagnosis was 13 years (12 and 24 years in the HR and non-HR groups, respectively).
Frequency of variants in RB1 and non-RB1 genes
The overall prevalence of individuals with cancer predisposition (individuals with HR and/or those with a non-RB1 variant) was 57% (n = 54), of which 11% (n = 6) had 2 different cancer predisposing variants, 42 (45%) had only an RB1 P/LP variant, and 5 (5%) had only a non-RB1 P/LP variant.
The frequencies of variants in RB1 and non-RB1 genes are presented in Table 1. Only 1 patient had reported variants in 2 non-RB1 genes: a LP variant in NTHL1 and a TSC2 variant, which was later reclassified as variant of uncertain significance.
Spectrum of variants in the non-RB1 group
A total of 9 non-RB1 heterozygous P/LP variants were found in 11 individuals; no one had biallelic variants. The variants were found in 7 different genes, MUTYH, MSH3, NTHL1, ERCC3, FANCC, CHEK2, and MITF, with 1 variant identified in each gene except for MSH3 and NTHL1, in which 2 different variants were found (Supplemental Table 3).
All non-RB1 variants in our study were unique, except for a MUTYH (NM_001128425) variant, c.1187G>A p.(Gly396Asp), which was found in 3 different individuals, all of whom had HR. Among them, 2 developed SMN (osteosarcoma and meningioma at 12 and 32 years, respectively). Only 1 other individual with a non-RB1 variant developed a SMN, a 23-year-old female with HR and a MSH3 variant who developed meningioma at age 20. None of the SMNs seemed to have any correlation with the respective non-RB1 gene identified on each individual, but they all have been previously reported as SMN in individuals with HR (Supplemental Tables 2 and 3).
Similarly, all RB1 variants were different when looking at the subgroup of individuals with HR in the non-RB1 variant group. The spectrum of RB1 variants within that group include 2 frameshift, 1 nonsense, 1 splice site, 1 exon-level deletion (all truncating variants), and 1 missense variant (believed to retain some residual activity and be associated with decreased penetrance15, 16, 17). All variants are spread along the RB1 gene from exon 4 to exon 20.18
Clinical, demographic, and molecular information of individuals with non-RB1 variants can be found in Supplemental Table 2.
Discussion
This represents the largest cohort of individuals with retinoblastoma in which the prevalence of germline non-RB1 P/LP variants has been reported to date. We found that 12% of individuals with retinoblastoma harbored a P/LP variant in a non-RB1 cancer predisposing gene, independent of their underlying germline predisposition to retinoblastoma. There was no significant difference of prevalence between individuals with HR and nHR (12% and 11%, respectively). Furthermore, our study has an ascertainment bias given by the nature of the IMPACT test, which was designed to be performed in individuals with available tumor sample; therefore, our study population is mainly composed by those who required enucleation (representing fewer than 5% of all individuals with retinoblastoma in our institution). No other treatment modalities or exposures were found to be overrepresented in our study population].
The overall prevalence of individuals with a cancer predisposition in our study is 57%, which, when compared with the prevalence of HR (52%), shows that a small but significant percentage (5%) of individuals with cancer predisposition can be missed if only RB1 directed testing is performed in persons with retinoblastoma. Considering that IMPACT only tests for actionable genes, some of these individuals and their families could be missing from surveillance interventions recommended based on each gene. These percentage is probably even higher given that (1) IMPACT only reports variants classified as P/LP and does not report variant of uncertain significance that may eventually be upgraded to pathogenic, and (2) testing was limited to the 90 gene version of IMPACT, but the current number of known cancer predisposition genes is significantly higher and continues to expand. Overall, this represents a significant number of persons being missed, considering that our cohort only represents 5% of the individuals with retinoblastoma seen at our institution. This supports the benefit of using broader gene panels for RB patients in the research setting, to facilitate for future studies to determine any gene variants that might be enriched in the RB population that could be considered for clinical testing in patients who would benefit from a diagnosis (ie, diagnosis would modify screening and surveillance recommendations).
We found that 11% of individuals with HR (6% of all individuals) harbored a second P/LP variant in a cancer predisposition gene. We have yet to determine the impact that non-RB1 variants can have in individuals with HR to understand if they confer an independently added risk of cancer or if they interact through common pathways and have disease modifying roles that could help predict disease outcomes and prognosis. We did not find any change in the mean age of retinoblastoma diagnosis when comparing individuals with and without non-RB1 variants that would suggest a modifying effect in the development of the primary tumor. Similarly, there was no significant difference between the same groups in the time to second cancer diagnosis among the overall study population nor within individuals with HR. However, these findings were not statistically significant, likely because of the low rate of second cancers in our study given a primarily pediatric population (mean age of 7 years), with second malignancies typically developing later in life (reported median range of age is 13-17 years).18, 19, 20, 21 Nonetheless, it should be noted that, among the 6 individuals with HR and a non-RB1 variant, 3 individuals are older than 10 years (all females) and all have developed a second cancer (Supplemental Table 3).22, 23, 24, 25, 26, 27, 28, 29
All of the genes in which a non-RB1 variant was identified have a different role in cell regulation. Only 1 is an oncogene (MITF), and the others are tumor suppressors, each involved in different mechanisms. Supplemental Table 2 provides information on each of the genes, including their role in carcinogenesis, and the known associated cancer risks. Although most of the genes in which variants were found are associated with autosomal recessive disease, these results provide valuable information for future family planning. Furthermore, screening recommendations also changed for some of the individuals (and their families) in whom the second variant was found in genes associated with autosomal dominant inheritance.
Given that RB1 not only initiates cancer in the retina but is also an important mechanism of cancer development in non-ocular cancers of later development,14,30, 31, 32, 33, 34 functional studies can help determine if germline variants in genes with specific roles within the cell could somehow play a role in the pathogenesis leading to the second somatic hit in RB1, facilitating the development of second cancers. A possible role of germline MUTYH variants in the pathogenesis of retinoblastoma has been recently suggested by Newman et al35 and Akdeniz et al,34 from results of functional studies from tumor samples of individuals with hereditary retinoblastoma and germline MUTYH variants, that suggest an important role of the loss of MUTYH function in the tumorigenesis of retinoblastoma.33,35 These findings along with the increased frequency of MUTYH variants in our cohort (found in half of the individuals with HR and a non-RB1 variant, with 2 of them who also developed a second cancer), could suggest a possible interaction between MUTYH and RB1 with an impact in tumorigenesis.
Further studies are required to identify those genes in which germline variants are more frequently found in persons with HR to determine good candidate genes for functional assessment, which might provide new insights into unknown biological mechanisms contributing to disease, which might help guide management and surveillance strategies in the future.
Conclusion
We found that 12% of individuals with a personal history of retinoblastoma harbored a P/LP variant in a non-RB1 cancer predisposing gene, and 6% of them had 2 different cancer predisposition syndromes. Although the size of our cohort limited the analysis in our study, there were no findings suggesting any association between having HR or non-HR and the presence of a non-RB1 variant and they likely represent 2 completely independent factors. However, further studies are required to determine possible associations with the presence of specific variants and the development of second cancers that could provide a better understanding of tumorigenesis in RB individuals and influence future management, screening, and surveillance recommendations.
Data Availability
All data relevant to the study are included in the article or uploaded as supplementary information. All data were deidentified for use in this study.
Conflict of Interest
All authors declare no conflicts of interest.
Acknowledgments
Funding
This research was funded in part through the NIH/NCI Cancer Center Support Grant P30 CA008748. The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Author Information
Conceptualization: A.M.R.B., M.F.W., D.M., J.H.F., M.F.B., D.H.A.; Data Curation: M.A.R., C.O., D.H.-L., D.M., M.F.B.; Formal Analysis: A.M.; Investigation: A.M.R.B., M.F.W., E.M.F., J.H.F., D.H.A.; Methodology: A.M.R.B., A.M., J.H.F., D.H.A.; Project Administration: A.M.R.B., M.A.R., C.O., D.H.A.; Resources: M.F.W., M.A.R., A.M., D.M., M.F.B., D.H.A.; Supervision: D.H.A.; Validation: A.M., D.M., J.H.F., M.F.B., D.H.A.; Visualization: A.M.R.B., A.M.; Writing-original draft: A.M.R.B., M.F.W., E.M.F., D.M., J.H.F., M.F.B., D.H.A.; Writing-review and editing: D.N.F., K.O.
ORCIDs
Ana Maria Rodriguez Barreto: http://orcid.org/0000-0002-9227-8979
Michael F. Walsh: http://orcid.org/0000-0001-5006-6778
Melissa A. Robbins: http://orcid.org/0000-0001-7216-2656
Audrey Mauguen: http://orcid.org/0000-0003-3236-6093
Elise M. Fiala: http://orcid.org/0000-0002-1626-5009
Cristina Olcese: http://orcid.org/0009-0009-4952-5477
Dianna Haggag-Lindgren: http://orcid.org/0000-0001-7867-2817
Diana Mandelker: http://orcid.org/0000-0003-4154-0567
Jasmine H. Francis: http://orcid.org/0000-0002-7637-3108
Michael F. Berger: http://orcid.org/0000-0003-3882-5000
Danielle Novetsky Friedman: http://orcid.org/0000-0003-4668-9176
Kenneth Offit: http://orcid.org/0000-0002-2180-2032
David H. Abramson: http://orcid.org/0000-0002-0118-6391
Ethics Declaration
Memorial Sloan Kettering Institutional Review Board (IRB) approval was obtained for the study, and all parents/guardians provided written consent for germline genetic testing under IRB # 12-245.
Footnotes
The Article Publishing Charge (APC) for this article was paid by the Memorial Sloan Kettering Cancer Center.
Additional Information
The online version of this article (https://doi.org/10.1016/j.gimo.2024.101836) contains supplemental material, which is available to authorized users.
Additional Information
Characteristics of patients with and without a non-RB1 variant
Supplementary Table 2. Clinical information of non-RB1 genes
Supplementary Table 3. Individuals with non-RB1 variants: clinical characteristics and variant information
References
- 1.Abramson D.H. Second nonocular cancers in retinoblastoma: a unified hypothesis. The Franceschetti lecture. Ophthalmic Genet. 1999;20(3):193–204. doi: 10.1076/opge.20.3.193.2284. [DOI] [PubMed] [Google Scholar]
- 2.Kleinerman R.A., Tucker M.A., Tarone R.E., et al. Risk of new cancers after radiotherapy in long-term survivors of retinoblastoma: an extended follow-up. J Clin Oncol. 2005;23(10):2272–2279. doi: 10.1200/JCO.2005.05.054. [DOI] [PubMed] [Google Scholar]
- 3.Abramson D.H., Melson M.R., Dunkel I.J., Frank C.M. Third (fourth and fifth) nonocular tumors in survivors of retinoblastoma. Ophthalmology. 2001;108(10):1868–1876. doi: 10.1016/s0161-6420(01)00713-8. [DOI] [PubMed] [Google Scholar]
- 4.Schefler A.C., Kleinerman R.A., Abramson D.H. Genes and environment: effects on the development of second malignancies in retinoblastoma survivors. Expert Rev Ophthalmol. 2008;3(1):51–61. doi: 10.1586/17469899.3.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong J.R., Morton L.M., Tucker M.A., et al. Risk of subsequent malignant neoplasms in long-term hereditary retinoblastoma survivors after chemotherapy and radiotherapy. J Clin Oncol. 2014;32(29):3284–3290. doi: 10.1200/JCO.2013.54.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abramson D.H., Frank C.M. Second nonocular tumors in survivors of bilateral retinoblastoma: a possible age effect on radiation-related risk. Ophthalmology. 1998;105(4):573–579. doi: 10.1016/S0161-6420(98)94006-4. discussion 579. [DOI] [PubMed] [Google Scholar]
- 7.Schrader K.A., Cheng D.T., Joseph V., et al. Germline variants in targeted tumor sequencing using matched normal DNA. JAMA Oncol. 2016;2(1):104–111. doi: 10.1001/jamaoncol.2015.5208. [published correction appears in JAMA Oncol. 2016;2(2):279] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiala E.M., Jayakumaran G., Mauguen A., et al. Prospective pan-cancer germline testing using MSK-Impact informs clinical translation in 751 patients with pediatric solid tumors. Nat Cancer. 2021;2:357–365. doi: 10.1038/s43018-021-00172-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zehir A., Benayed R., Shah R.H., et al. Variantal landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 individuals. Nat Med. 2017;23(6):703–713. doi: 10.1038/nm.4333. [published correction appears in Nat Med. 2017;23(8):1004] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Francis J.H., Richards A.L., Mandelker D.L., et al. Molecular changes in retinoblastoma beyond RB1: findings from next-generation sequencing. Cancers (Basel) 2021;13(1):149. doi: 10.3390/cancers13010149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng D.T., Mitchell T.N., Zehir A., et al. Memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets (MSK-Impact): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17(3):251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards S., Aziz N., Bale S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerds TA 2019. Prodlim: Product-Limit Estimation for Censored Event History Analysis. R Package Version 2019.11.13.
- 14.Gray B 2022. Cmprsk: Subdistribution Analysis of Competing Risks. R Package Version 2.2-11.
- 15.Levin A.M., Francis J.H., Abramson D.H. In: The Columbia Guide to Basic Elements of Eye Care. Casper D., Cioffi G., editors. Springer; 2019. Retinoblastoma. [DOI] [Google Scholar]
- 16.Sun H., Chang Y., Schweers B., et al. An E2F binding-deficient Rb1 protein partially rescues developmental defects associated with Rb1 nullizygosity. Mol Cell Biol. 2006;26(4):1527–1537. doi: 10.1128/MCB.26.4.1527-1537.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otterson G.A., Modi S., Nguyen K., Coxon A.B., Kaye F.J. Temperature-sensitive RB mutations linked to incomplete penetrance of familial retinoblastoma in 12 families. Am J Hum Genet. 1999;65(4):1040–1046. doi: 10.1086/302581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eloy P., Dehainault C., Sefta M., et al. A parent-of-origin effect impacts the phenotype in low penetrance retinoblastoma families segregating the c.1981C>T/p.Arg661Trp Mutation of RB1. PLoS Genet. 2016;12(2) doi: 10.1371/journal.pgen.1005888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Center for Biotechnology Information (NCBI) National Library of Medicine (US), National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/
- 20.Lohmann D.R., Gallie B.L. In: GeneReviews®. Adam M.P., Everman D.B., Mirzaa G.M., et al., editors. University of Washington, Seattle; 1993. Retinoblastoma; p. 2022. Accessed March 22, 2024. https://www.ncbi.nlm.nih.gov/books/NBK1452/ [Google Scholar]
- 21.Kleinerman R.A., Yu C.L., Little M.P., et al. Variation of second cancer risk by family history of retinoblastoma among long-term survivors. J Clin Oncol. 2012;30(9):950–957. doi: 10.1200/JCO.2011.37.0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kairupan C., Scott R.J. Base excision repair and the role of MUTYH. Hered Cancer Clin Pract. 2007;5(4):199–209. doi: 10.1186/1897-4287-5-4-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olkinuora A.P., Peltomäki P.T., Aaltonen L.A., Rajamäki K. From APC to the genetics of hereditary and familial colon cancer syndromes. Hum Mol Genet. 2021;30(R2):R206–R224. doi: 10.1093/hmg/ddab208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson D.J., Boyle A., Reinicke T., Woods B., Hsieh P. Benign tumors associated with heterozygous NTHL1 variant. Cureus. 2021;13(7) doi: 10.7759/cureus.16220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li N., Zethoven M., McInerny S., et al. Evaluation of the association of heterozygous germline variants in NTHL1 with breast cancer predisposition: an international multi-center study of 47,180 subjects. npj Breast Cancer. 2021;7(1):52. doi: 10.1038/s41523-021-00255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stradella A., Del Valle J., Rofes P., et al. ERCC3, a new ovarian cancer susceptibility gene? Eur J Cancer. 2020;141:1–8. doi: 10.1016/j.ejca.2020.09.023. [DOI] [PubMed] [Google Scholar]
- 27.Pan Z.W., Wang X.J., Chen T., et al. Deleterious Mutations in DNA Repair Gene FANCC Exist in BRCA1/2-Negative Chinese Familial Breast and/or Ovarian Cancer Patients. Front Oncol. 2019;9:169. doi: 10.3389/fonc.2019.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stolarova L., Kleiblova P., Janatova M., et al. CHEK2 germline variants in cancer predisposition: stalemate Rather than checkmate. Cells. 2020;9(12):2675. doi: 10.3390/cells9122675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bertolotto C., Lesueur F., Giuliano S., et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature. 2011;480(7375):94–98. doi: 10.1038/nature10539. [published correction appears in Nature. 2016;531(7592):126] [DOI] [PubMed] [Google Scholar]
- 30.Abramson D.H. Retinoblastoma: saving life with vision. Annu Rev Med. 2014;65:171–184. doi: 10.1146/annurev-med-061312-123455. [DOI] [PubMed] [Google Scholar]
- 31.Kleinerman R.A., Francis J.H., Abramson D.H. In: Albert and Jakobiec’s Principles and Practice of Ophthalmology. Albert D., Miller J., Azar D., Young L.H., editors. Springer; 2021. Second primary neoplasms in retinoblastoma: effect of gene and environment. [DOI] [Google Scholar]
- 32.Knudson A.G., Jr. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68(4):820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen M., Infante E., Brand R. In: GeneReviews®. Adam M.P., Everman D.B., Mirzaa G.M., et al., editors. University of Washington, Seattle; 1993. MUTYH polyposis; p. 2022. Accessed March 22, 2024. https://www.ncbi.nlm.nih.gov/books/NBK107219/ [PubMed] [Google Scholar]
- 34.Akdeniz D., Tuncer S.B., Kebudi R., et al. Investigation of new candidate genes in retinoblastoma using the TruSight One “clinical exome” gene panel. Mol Genet Genomic Med. 2019;7(8):e785. doi: 10.1002/mgg3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newman S., Nakitandwe J., Kesserwan C.A., et al. Genomes for kids: the scope of pathogenic mutations in pediatric cancer revealed by comprehensive DNA and RNA sequencing. Cancer Discov. 2021;11(12):3008–3027. doi: 10.1158/2159-8290.CD-20-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characteristics of patients with and without a non-RB1 variant
Supplementary Table 2. Clinical information of non-RB1 genes
Supplementary Table 3. Individuals with non-RB1 variants: clinical characteristics and variant information
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplementary information. All data were deidentified for use in this study.