Abstract
Background
Dendrobium officinale Kimura et Migo is a perennial epiphytic herb in traditional Chinese medicine, showing remarkable resistance to salt stress. Water-soluble sugars serve as important osmoprotectants and play crucial roles in plant stress responses. Previous studies have primarily focused on sugar metabolism in individual tissues under stress, resulting in a limited understanding of the regulatory differences between tissues and the relationship between sugar metabolism and transport.
Results
A variety of salt-responsive genes were identified through transcriptome analysis of D. officinale. GO and KEGG enrichment analyses revealed functional differences among the differentially expressed genes (DEGs) between leaves and roots. Expression analysis indicated that sugar metabolic genes and D. officinale Sugars Will Eventually be Exported Transporters (DoSWEETs) displayed distinct expression patterns in leaves and roots under salt stress. Most sugar metabolic genes were up-regulated in the leaves and down-regulated in the roots in response to salt, while DoSWEETs predominantly responded in the roots. Specifically, DoSWEET2a, 6a, 12a, 14, and 16 were confirmed via RT-qPCR. Additionally, positive correlations were observed between certain genes (scrK, INV, SUS) and DoSWEETs, with INV (LOC110096666) showing a strong positive correlation with all detected DoSWEETs in both leaves and roots.
Conclusions
Our findings not only illustrated the distinct responses of leaves and roots to salt stress, but also highlighted the relationship between sugar metabolic genes and DoSWEETs in adapting to such stress. This enhances our understanding of the differential responses of plant tissues to salt stress and identified candidate genes for salt-resistance breeding in D. officinale.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-024-11069-5.
Keywords: Transcriptome, Tissue-specific response, Sugar biosynthesis, Sugar transport
Background
Dendrobium officinale Kimura et Migo is a valued traditional Chinese medicine known for its high content of polysaccharides, alkaloids, flavonoids, and various bioactivities in its stem [1, 2]. Water-soluble polysaccharides, a medicinal ingredient isolated from D. officinale stems [3], have shown therapeutic effects, including immunity enhancement, gastric benefits, anti-inflammation properties, and anti-tumor activity [4, 5].
In plants, photosynthetic sucrose is loaded into and unloaded from the phloem by sugar transporters [6]. Subsequently, sucrose is hydrolyzed by sugar hydrolases or synthetases for reutilization and storage as metabolites in sink tissues, including stems and leaves [7]. Throughout its growth and development, D. officinale often experiences various abiotic stresses [8]. Salinity is a common abiotic stress factor that significantly affects plant growth, development, and productivity [9]. When plants are exposed to salinity stress, high salinity initially causes osmotic stress in the roots, leading to reduced water availability. This inhibition affects the growth of young leaves and reduces stomatal conductance in mature leaves. In the second phase, ionic stress inflicts considerable damage on plant growth and development by disrupting ion balance or causing excessive ion accumulation [9]. High salinity can also trigger various signal transduction pathways, including phytohormone-mediated and Ca2+ signaling pathways, to facilitate communication between roots and leaves [10–12]. This communication may lead to a decrease in photosynthetic efficiency [12] or promote the biosynthesis of secondary metabolites [13]. Water-soluble polysaccharides, recognized as a type of plant secondary metabolite [14], may function as osmoprotectants to help mitigate the challenges caused by salt stress [15]. However, the impact of salt stress on sugar metabolism and transport in both leaves and roots remains not fully understood.
Dendrobium has demonstrated remarkable tolerance to high salinity (15 dS m− 1) [16]. Benefiting from transcriptome analysis and the available D. officinale genome, the expression profiles of specific metabolite biosynthetic genes under salt stress have been elucidated [13, 17]. Salt stress significantly alters various metabolic pathways, including phenylalanine metabolism, flavonoid biosynthesis, and α-linolenic acid metabolism [13]. However, the impact of salt on sugar metabolism in D. officinale remains poorly understood. Sugars Will Eventually be Exported Transporters (SWEETs), a novel type of sugar transporters [18], exhibit bi-directional transport capabilities [7], and play roles in responses to various abiotic stresses [19–23]. Our previous study identified several members of the SWEET family in D. officinale and revealed that the expression of DoSWEETs was regulated by cold, drought, and, Methyl jasmonate (MeJA) treatment [24]. However, it is still unclear whether DoSWEETs respond to salt stress and how sugar metabolism and transport are regulated and coordinated under such conditions.
In this study, using online transcriptome data from leaves and roots under salt treatment [13], we analyzed the expression profiles and correlations of sugar metabolic genes and DoSWEETs employing multiple bioinformatic tools and resources, which were further verified by RT-qPCR. Our findings illustrated the differential regulation and potential cooperation of sugar metabolic genes and DoSWEETs in different tissues of D. officinale in response to salt stress. The new insight enhances our understanding of salt adaptation in D. officinale and provides potential candidates as molecular markers for breeding salt-resistant varieties.
Results
Gene expression comparison of salt-treated leaves and roots from D. Officinale
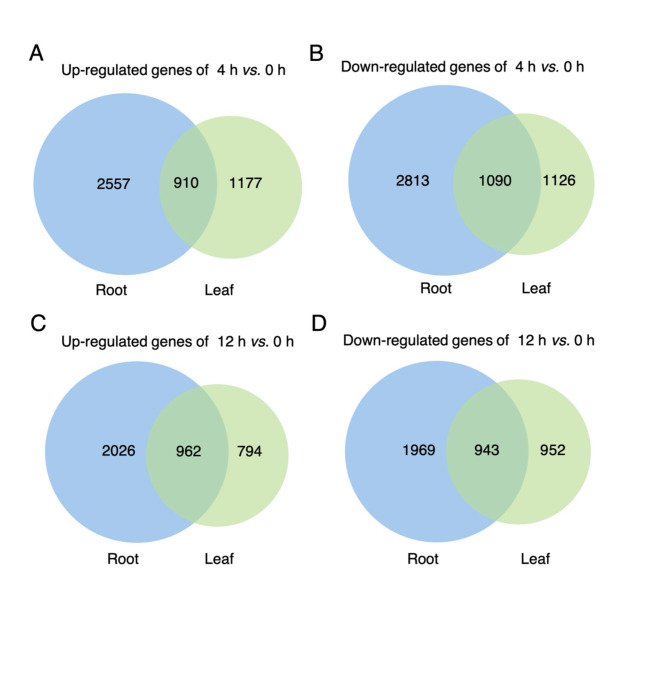
To identify the differences in gene regulation between the leaves and roots of D. officinale in response to salt stress, the relevant transcriptome datasets with accession number PRJNA715099 [13] were downloaded for bioinformatic analysis. TPM values of genes at different time points (0 h, 4 h, and 12 h) under salt treatment were obtained (Additional file 1: Table S1). In the roots, about 6,000 genes were significantly regulated by salt stress, defined by the criteria of |log2FoldChange| > 1 and p-value < 0.05. In comparison, the number of differentially expressed genes (DEGs) in the leaves was relatively lower, at around 4,000 genes (Fig. 1 and Additional file 1: Table S2). This suggested that salt stress had a more pronounced impact on gene regulation in the roots compared to the leaves.
Fig. 1.
Venn diagram of salt-responsive DEGs in leaves and roots. A, C Venn diagrams illustrating the up-regulated genes in leaves and roots for the comparisons of 4 h vs. 0 h (A) and 12 h vs. 0 h (C). B, D Venn diagram of the down-regulated genes in leaves and roots for the comparisons of 4 h vs. 0 h (B) and 12 h vs. 0 h (D)
Notably, the numbers of up-regulated and down-regulated genes in response to salt treatment showed little variation at both 4 h and 12 h in both leaves and roots (Fig. 1). Among the DEGs, over 900 genes were identified as responsive to salt treatment in both tissues (Fig. 1). Specifically, we identified 151 up-regulated DEGs (Additional file 2: Fig. S1A) and 69 down-regulated DEGs (Additional file 2: Fig. S1B) in the leaves across the three comparisons (4 h vs. 0 h, 12 h vs. 0 h, and 12 h vs. 4 h). In the roots, 109 up-regulated DEGs (Additional file 2: Fig. S1C) and 53 down-regulated DEGs (Additional file 2: Fig. S1D) were detected in the same comparisons. These observations revealed that approximately 200 genes were consistently up- or down-regulated in both leaves and roots under salt stress.
Functional comparison of DEGs of in salt-treated leaves and roots
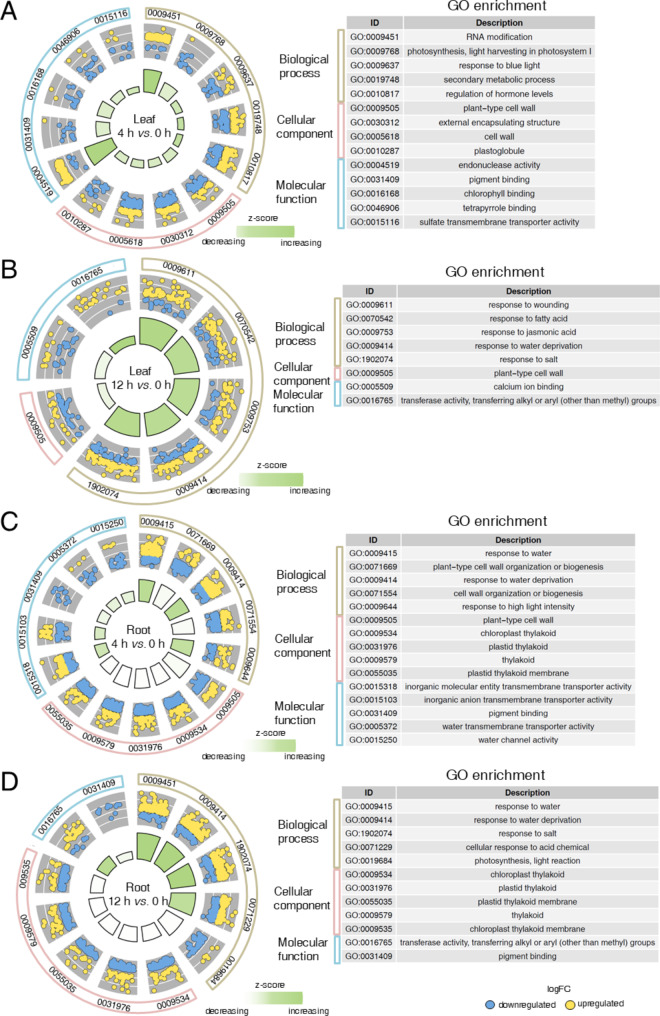
To investigate the potential functional differences between the DEGs in leaves and roots in response to salt stress, we conducted Gene Ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) enrichment analyses. The DEGs were assigned into three major GO categories, highlighting significant differences in the response patterns of leaves and roots (Fig. 2 and Additional file 1: Table S3). In the biological process category, a majority of the DEGs in leaves (4 h vs. 0 h) were associated with functions such as ‘RNA modification’, ‘photosynthesis’, ‘response to blue light’, ‘secondary metabolic process’, and ‘regulation of hormone level’ (Fig. 2A). In contrast, the most significantly enriched function among the DEGs in the roots was ‘response to water’, observed in both comparisons (4 h vs. 0 h and 12 h vs. 0 h) (Fig. 2C and D, and Additional file 1: Table S3). Moreover, functions related to ‘response to salt’ and ‘response to water deprivation’ were enriched among the DEGs in both leaves and roots (12 h vs. 0 h) (Fig. 2B and D). Interestingly, in the cellular component category, DEGs in the roots were more frequently associated with thylakoid-related functions, including ‘chloroplast thylakoid’, ‘plastid thylakoid’, ‘thylakoid’, ‘plastid thylakoid membrane’, and ‘chloroplast thylakoid membrane’ (Fig. 2C and D). While the DEGs in the leaves exhibited significant enrichment in the ‘plant-type cell wall’ category (Fig. 2A and B).
Fig. 2.
Top five GO enrichment circle diagrams for DEGs in response to salt in leaves and roots. A-B Go enrichment circle diagrams for DEGs in response to salt in the leaves for the comparisons of 4 h vs. 0 h (A) and 12 h vs. 0 h (B). C-D Go enrichment circle diagram for DEGs in response to salt in the roots for the comparisons of 4 h vs. 0 h (C) and 12 h vs. 0 h (D). Outside circle: classification of GO categories. Medium circle: enriched dots representing up-regulated genes (yellow) and down-regulated genes (blue) within each category. Inside circles: change trend of each category, represented by z-score
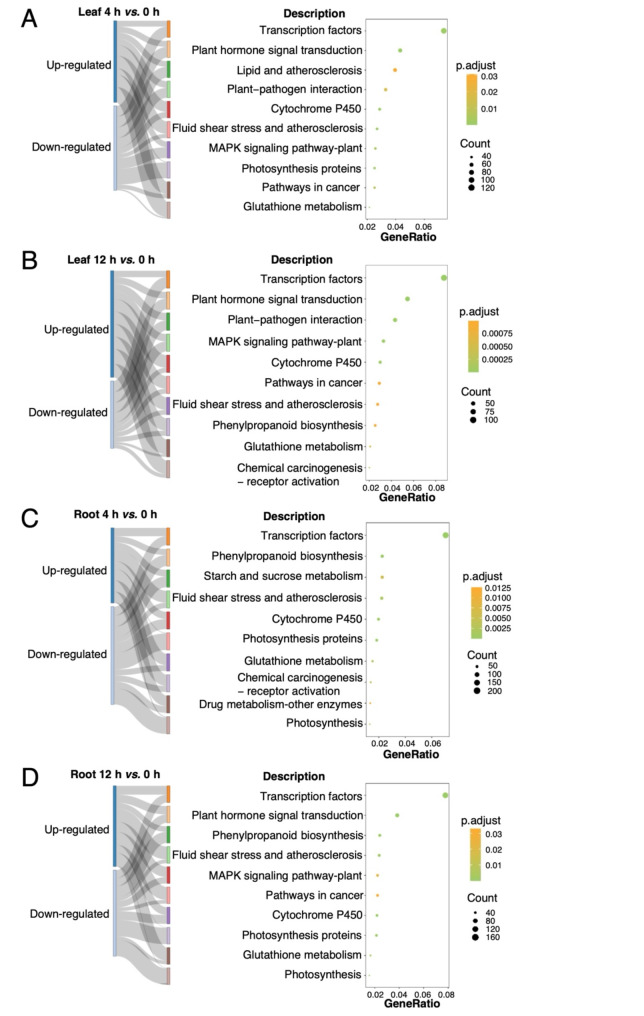
KEGG analysis revealed that the number of DEGs related to transcription factors was the highest in both leaves and roots under salt stress (Fig. 3 and Additional file 1: Table S4). Five pathways, including ‘cytochrome P450’, ‘glutathione metabolism’, ‘plant hormone signal transduction’, ‘MAPK signaling pathway-plant’, and ‘photosynthesis proteins’, were enriched across at least three comparison groups (Fig. 3 and Additional file 1: Table S4). Furthermore, DEGs in the roots (4 h vs. 0 h) were significantly associated with the ‘starch and sucrose metabolism’ pathway (Fig. 3C and Additional file 1: Table S4), indicating that sucrose might play a critical role in the rapidly response to salt stress in D. officinale roots. Additional analysis revealed that genes involved in the ‘glutathione metabolism’ pathway were more likely to be induced by salt stress, whereas those in the ‘photosynthesis proteins’ and ‘photosynthesis’ pathways predominantly consisted of down-regulated genes (Fig. 3).
Fig. 3.
Top ten significantly enriched KEGG pathways for DEGs in response to salt in leaves and roots. A-B Enriched pathways for up- and down-regulated genes from leaves, comparing 4 h vs. 0 h (A) and 12 h vs. 0 h (B). C-D Enriched pathways for up- and down-regulated genes from roots, comparing 4 h vs. 0 h (C) and 12 h vs. 0 h (D)
Differential responses of the sugar metabolic genes in leaves and roots under salt stress
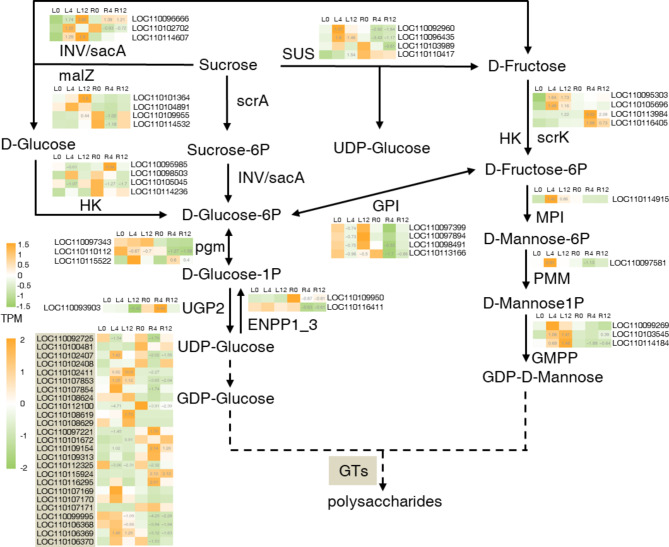
To analyze the response of sugar metabolic genes in D. officinale under salt stress, we focused on the expression of genes within the KEGG pathway ko00500 (starch and sucrose metabolism) and ko00051 (fructose and mannose metabolism). The selected genes included invertase (INV)/β-fructofuranosidase (sacA), α-glucosidase (malZ), phosphotransferase system (scrA), hexokinase (HK), phosphoglucomutase (pgm), UTP-glucose-1-phosphate uridylyltransferase 2 (UGP2), ectonucleotide pyrophosphatase/phosphodiesterase family member 1/3 (ENPP1_3), sucrose synthase (SUS), fructokinase (scrK), mannose-6-phosphate isomerase (MPI), phosphomannomutase (PMM), and mannose-1-phosphate guanylyltransferase (GMPP). We also analyzed the expression changes of cellulose synthase like genes (CSLs), which are a class of glycosyltransferases (GTs) involved in the biosynthesis of water-soluble polysaccharides in D. officinale [25, 26].
Under salt stress, the analyzed sugar metabolic genes were predominantly up-regulated in the leaves, with a ratio of 19/59 (up-regulated/total, log2FC > 1 and p < 0.05), while in the roots, they were primarily down-regulated, with a ratio of 20/59 (down-regulated/total, log2FC < -1 and p < 0.05). In contrast, six genes showed decreased expression in the leaves (log2FC < -1 and p < 0.05), whereas seven genes were induced in the roots (log2FC > 1 and p < 0.05) (Fig. 4). Notably, different regulatory patterns were observed for specific genes between leaves and roots. SUS, malZ, and INV/sacA were up-regulated in the leaves at 4–12 h under salt treatment, indicating an increase in sucrose hydrolysis during short-term salt stress. Conversely, while the expression of SUS in the roots significantly decreased in response to salt, malZ and INV/sacA exhibited an upward trend (Fig. 4). Similar different expression patterns were evident for genes encoding HK, PMM, GMPP, and CSL, all with |log2FC| > 1 and p < 0.05 (Fig. 4). These findings indicated that the expression of sugar metabolic genes was enhanced in the leaves but weakened in the roots in response to salt stress.
Fig. 4.
Expression profiles of sugar metabolic genes and DoSWEETs in response to salt stress. Each cell represents the mean TPM value derived from three biological replicates. The number in each cell represents the log2FC value compared to the control group (L0 or R0) with a significant level of p < 0.05. L0, L4, and L12 represent leaves treated with salt for 0 h, 4 h, and 12 h, respectively. R0, R4, and R12 represent roots treated with salt for 0 h, 4 h, and 12 h, respectively. Glycosyltransferases (GTs) are marked in light brown
Differential responses of DoSWEETs between leaves and roots under salt stress
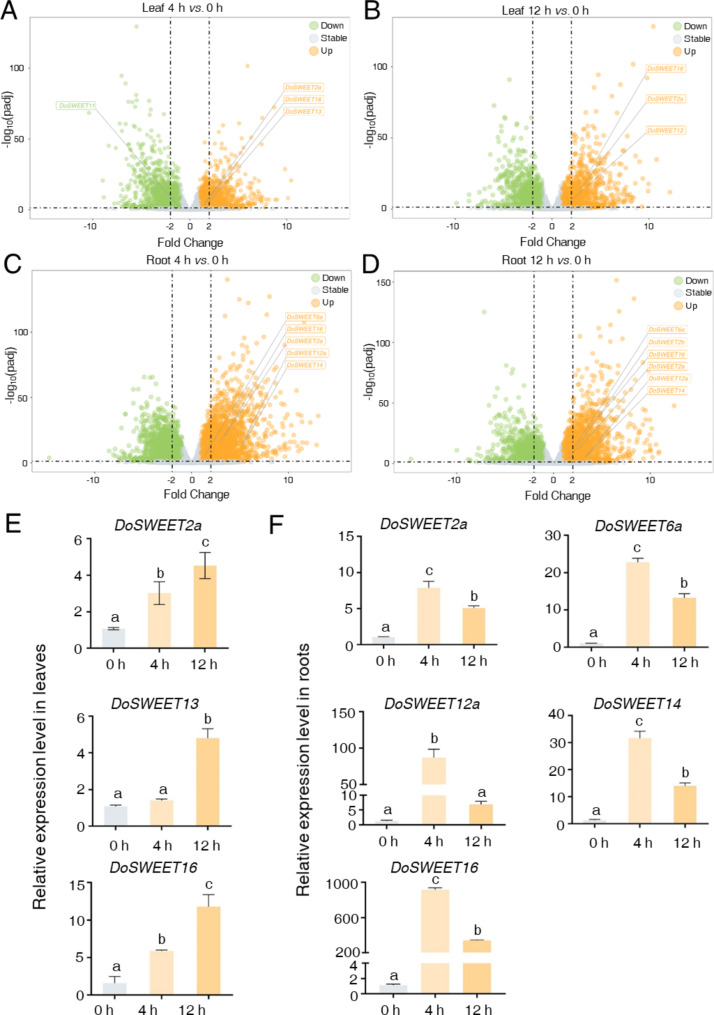
In addition to metabolic functions, the transport of water-soluble sugars plays a crucial role in plant resistance to salt stress. The SWEET family of sugar transporters in known to participate in responses to various abiotic stresses [19–23]. In our previous study, we identified 25 members of the SWEET family in D. officinale [24]. To further elucidate the roles of DoSWEETs in salt response, we analyzed their expression patterns (Additional file 1: Table S5). In the transcriptomic datasets examined in this study, nine DoSWEETs were detected, of which eight were significantly regulated by salt stress (Additional file 1: Table S6 and Additional file 2: Fig. S2). Among these eight salt-responsive DoSWEETs, only DoSWEET11 exhibited a downward trend in expression in the leaves following 4 h of treatment, the remaining DoSWEETs were all induced by salt stress (Fig. 5A and D). Notably, compared to the leaves, a greater number of DoSWEETs were induced in the roots under salt stress. DoSWEET2a and DoSWEET16 were up-regulated in both leaves and roots following salt treatment (Fig. 5A and D). Meanwhile, DoSWEET13 was uniquely induced in the leaves by salt stress, while DoSWEET6b, DoSWEET12a, and DoSWEET14 were only up-regulated in the roots (Fig. 5A and D). Results from RT-qPCR further confirmed the salt-responsive behavior of DoSWEETs (Fig. 5E and F). Importantly, a more rapid regulation of DoSWEETs was observed in the roots compared to the leaves under salt stress. Following a 4 h treatment, the expression changes of salt-responsive DoSWEETs in the leaves (DoSWEET2a, DoSWEET13, and DoSWEET16) were noticeably smaller than those detected in the roots (DoSWEET2a, DoSWEET6a, DoSWEET12a, DoSWEET13, DoSWEET14, and DoSWEET16) (Fig. 5E and F). In conclusion, the expression of DoSWEETs involved in sugar transport was enhanced in both leaves and roots after salt treatment. However, the roots exhibited a greater sensitivity to high salinity, showing a quicker and more substantial regulation in DoSWEETs expression than the leaves during short-term salt stress.
Fig. 5.
Volcano plot and RT-qPCR analysis of differentially expressed DoSWEETs in response to salt stress. A-B Volcano plots illustrating the differentially expressed DoSWEETs in the leaves for the comparison of 4 h vs. 0 h (A) and 12 h vs. 0 h (B). C-D Volcano plots showing the differentially expressed DoSWEETs in the roots for the comparison of 4 h vs. 0 h (C) and 12 h vs. 0 h (D). Green dots represent down-regulated genes (log2FC < -1 and padj < 0.05). Orange dots represent up-regulated genes (log2FC > 1 and padj < 0.05). Gray dots represent stable genes (|log2FC| <= 1 or padj > = 0.05). E-F Expression analysis of DoSWEETs in the leaves (E) and roots (F) following salt treatment assessed by RT-qPCR. Bars represent means ± SD (n = 3). Different letters above each column indicate significant differences as determined by Duncan’s multiple range test (p < 0.05)
Correlation of sugar metabolic genes and DoSWEETs under salt stress
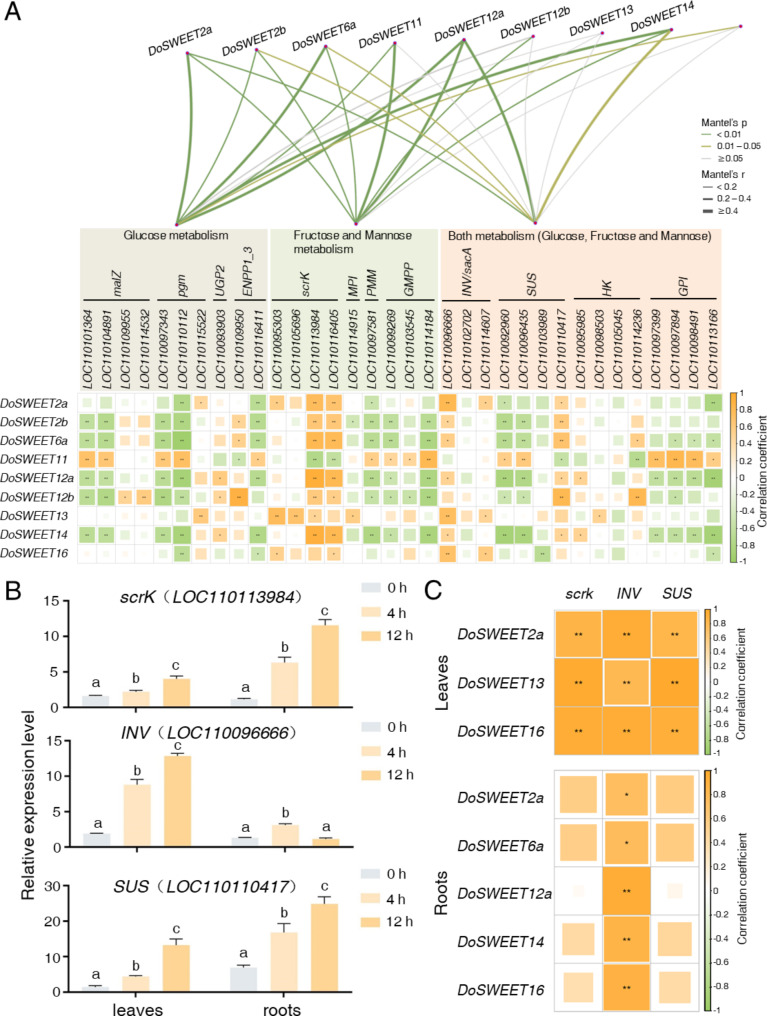
To elucidate the relationship between sugar metabolism and transport in response to salt stress, we analyzed the correlation between sugar metabolic genes and DoSWEETs. Based on the associated KEGG pathways, sugar metabolic genes were categorized into three groups: those involved in glucose metabolism, those in fructose and mannose metabolism, and genes implicated in both pathways (Fig. 6A, S3, and Additional file 1: Tables S7-S10). DoSWEET12a exhibited strong correlations with all three groups, with correlation coefficients of 0.64, 0.41, and 0.41, respectively (Fig. 6A and Additional file 1: Table S7). At the single gene level, scrK (LOC110113984 and LOC110116405), INV (LOC110096666), and SUS (LOC110110417) exhibited positive correlations with most DoSWEETs (Fig. 6A and Additional file 1: Table S8). RT-qPCR analysis revealed that the expression levels of scrK (LOC110113984), INV (LOC110096666), and SUS (LOC110110417) consistently increased in both leaves and roots during short-term salt stress, except for INV (LOC110096666) in the roots (Fig. 6B). Based on the confirmed expression patterns of DoSWEETs (Fig. 5E and F) and sugar metabolic genes (Fig. 6B), the correlation relationships were re-evaluated (Fig. 6C). In leaves, the up-regulated DoSWEET2a, DoSWEET13, and DoSWEET16 exhibited high positive correlations (correlation coefficient > 0.82) with scrK (LOC110113984), INV (LOC110096666), and SUS (LOC110110417). In roots, the up-regulated DoSWEET2a, DoSWEET6a, DoSWEET12a, DoSWEET14, and DoSWEET16 showed positive correlations with INV (LOC110096666) (Fig. 6C). These findings aligned closely with the transcriptome data (Additional file 2: Fig. S3, Additional file 1: Tables S9 and S10), suggesting potential cooperation between DoSWEETs and key sugar metabolic genes in modulating resistance to salt stress.
Fig. 6.
Correlation analysis of sugar metabolic genes and DoSWEETs. A Correlation analysis of sugar metabolic genes and DoSWEETs based on transcriptome data. Symbol in each cell indicates the significance levels (**p < 0.01; *p < 0.05). B Expression analysis of key sugar metabolic genes in response to salt treatment via RT-qPCR. Bars represent means ± SD (n = 3). Different letters above each column indicate significant differences based on Duncan’s multiple range test (p < 0.05). C Correlation analysis of sugar metabolic genes and DoSWEETs based on RT-qPCR data
Discussion
Salt stress is a significant abiotic factor that poses a major threat to the agriculture industry, adversely affecting plant growth and development, ultimately leading to reduced yields [12]. Under salt stress conditions, Na+ or Cl− can rapidly impact plant roots by triggering osmotic stress, or they can exert a gradual impact through ionic stress [9]. Consequently, high salinity diminishes photosynthetic rates in leaves and promotes the accumulation of water-soluble osmoprotectants, such as proline, glycine betaine, and sugars, in the roots, stems, and leaves [15, 27]. Dendrobium species belong to the largest family of angiosperm [28] and have evolved various mechanisms to adapt to diverse natural habitats. These plants characterized by thick roots that enable them to live epiphytically on rock surfaces or tree trunks [29]. They exhibit a high tolerance to salt stress [16]. Under salt stress conditions, specific biosynthesis pathways are activated, leading to the accumulation of secondary metabolites, including sugars, flavonoids, and alkaloids in the leaves of D. officinale [13]. Notably, water-soluble polysaccharides, key osmoprotectants involved in the response to salt stress [15], are also the most important medicinal ingredients in D. officinale. Polysaccharides are synthesized through the sugar metabolic pathway and are transported by sugar transporters [14, 24]. Thus, elucidating the differential regulation and potential interactions between sugar metabolic genes and sugar transporters in both leaves and roots is crucial for gaining comprehensive insights into the transcriptional regulation of D. officinale in response to salt stress.
Based on the expression profiles, a greater number of DEGs were observed in the roots in rapid salt-response compared to the leaves (Fig. 1), indicating tissue-specific responses at the transcriptional level. Among these numerous DEGs, approximately 200 showed consistent up- or down-regulation in response to salt treatment (Additional file 2: Fig. S1). This suggested that the majority of early salt-responsive genes were activated rapidly and displayed transient changes in expression. Notably, the consistently up-regulated DEGs in the leaves were significantly related to cytochrome P450, α-linolenic acid metabolism, and the jasmonic acid (JA) metabolic process (Additional file 1: Table S11). The α-linolenic acid serves as a precursor for JA biosynthesis and has been observed with increased content in salt-tolerant transgenic rapeseed [30]. Similarly, sugar beet, is a high-sugar-yielding crop known for storing amount of sugar in its roots, exhibited DEGs and differentially accumulated metabolites (DAMs) associated with starch and sucrose metabolism, α-linolenic acid metabolism, and plant hormone signal transduction pathways. When subjected to salt stress for 24 h [31]. These findings suggest that sugar metabolism and α-linolenic acid metabolism likely play critical roles in enhancing salt tolerance in D. officinale. The consistently up-regulated genes may serve as key regulatory components in this progress.
In the roots, the DEGs were significantly enriched in biological processes related to ‘response to water’, ‘response to water deprivation’, and ‘response to salt’ (Fig. 2C and D). The pattern aligns with the conditions observed in roots experiencing osmotic stress during the initial phase of salinity stress. Osmotic stress not only impairs the root’s ability to absorb water but also triggers water loss from leaves [15]. Consistent with the physiological change observed in model plants, DEGs in the leaves of D. officinale were significantly enriched in the category ‘response to water deprivation’ during late-stage salt stress (12 h) (Fig. 2B). Conversely, during early salt stress (4 h), the DEGs were associated with ‘photosynthesis’, ‘secondary metabolic process’, and ‘regulation of hormone levels’ (Fig. 2A). The findings suggest that the roots of D. officinale respond to salt stress by regulating water loss, while the leaves initially react to salt stress by down-regulating photosynthesis and activating secondary metabolic processes and hormone-mediated pathways. Previous reports have indicated that osmotic adjustment in the leaves helps protect Dendrobium plants from salt stress [16]. Osmoprotectants, including proline, sugars, and polyols produced through secondary metabolic processes, play a crucial role in osmotic adjustment in plants [32, 33]. In this study, we detected the sugar content, including sucrose, glucose, and fructose, in both leaves and root following salt treatment. Generally, the sugar contents in both tissues were found to be induced by salt stress (Additional file 2: Fig. S4). In the leaves, sucrose and glucose levels continued to rise after salt treatment, which was consisted with the result from metabolome [13]. Comparatively, the sugar content in the roots exhibited greater sensitivity to high salinity, showing a rapid and significant increase during early salt stress (4 h), followed by a decreased or stabilization at 12 h (Additional file 2: Fig. S). Therefore, the induction of secondary metabolic processes in salt-treated D. officinale may contribute to the plant’s adaptation to salt stress by generating osmoprotectants, such as water-soluble sugars.
Salt stress typically inhibits cell expansion and root growth in plants [34]. Hormonal signaling from the roots can regulate the growth of other plant tissues [35, 36]. In this study, the ‘plant-type cell wall’ emerged as the predominant cellular component among the DEGs in both roots and leaves under short-term salt stress (Fig. 2). The cell wall is crucial for maintaining plant growth and morphology. Osmotic stress caused by high salinity leads to water loss in the roots, disrupting the integrity of the cell-wall [37]. This elucidates the association of salt-responsive DEGs with the ‘plant-type cell wall’ cellular component. Additionally, the thylakoid membrane, which is abundant in ion channels and transporter [38], plays a crucial role in regulating pH and osmosis balance. This is consistent with our observations in the roots, as illustrated in Fig. 2.
Sugar metabolism is vital for plants responses to salt stress [39–42]. Water-soluble polysaccharides are abundant in the stems and leaves of D. officinale. Distinct patterns were noted in the responses of sugar metabolic genes in the leaves and roots under salt stress. The majority of DEGs related to sucrose hydrolysis, including SUS, malZ, and INV/sacA, were significantly induced in the leaves during salt stress. However, SUS expression markedly decreased in the roots when exposed to salt stress in transcriptome analysis (Fig. 4). SUS, which functions as a reversible enzyme, is crucial in plant sugar metabolism by hydrolyzing sucrose into D-fructose and UDP-glucose [43]. An increased level of SUS resulted in elevated D-fructose accumulation in the leaves of rice seedlings under salt stress [44], which was also observed in D. officinale in this study (Additional file 2: Fig. S4). Elevated fructose content helps alleviate osmotic stress and maintain osmotic balance, thus preventing further water loss [45].
Leaves serve as the primary source of sugar in D. officinale. In addition to sugar metabolism, its transportation is essential for plant adaptation to salt stress. We observed that several DoSWEETs were up-regulated in response to salt stress. Interestingly, roots exhibited a more rapid and pronounced regulation of DoSWEETs compared to leaves during short-term salt exposure (Fig. 5), which was consistent with the changes of sugar content (Additional file 2: Fig. S4). In the leaves, the expression of DoSWEET2a, DoSWEET13, and DoSWEET16 also gradually increased with treatment time (Fig. 5B). This suggests that sugars synthesized in the leaves may be transported to the roots to mitigate osmotic stress induced by high salinity.
Both sugar metabolism and transportation play crucial roles in plant responses to salt stress, but how do they interact? In the leaves of D. officinale, three salt-responsive DoSWEETs (DoSWEET2a, DoSWEET13, and DoSWEET16) exhibited a strong positive correlation with various enzymes involved in sugar metabolism (Additional file 2: Fig. S3A). Conversely, in the roots, the salt-responsive DoSWEETs (DoSWEET2a, DoSWEET6a, DoSWEET12a, DoSWEET14, and DoSWEET16) showed a negative association with most sugar metabolic enzymes (Additional file 2: Fig. S3B). These findings indicated that not only do the transcriptional regulations of sugar-related genes differed between leaves and roots in response to salt stress, but the relationships among these genes also displayed diverse patterns. In Arabidopsis thaliana (L.) Heynh., SWEET14 was up-regulated after salt treatment [46] and correlated with a gene involved in proline biosynthesis during early salt stress [47]. Our previous study identified DoSWEET13 and DoSWEET14 as potential paralog pairs in sugar transport, both closely associated with SUC2 [24], a predominant sucrose/proton symporter in phloem loading in A. thaliana [48]. In the current study, DoSWEET13 and DoSWEET14 exhibited a high correlation with pgm (LOC110115522) and INV (LOC110096666) in D. officinale roots (Additional file 2: Fig. S3B), indicating their potential function in sugar transport within the roots under salt stress. However, further experimental evidence is required to illustrate the underlying regulatory mechanisms of sugar-related genes in different tissues of D. officinale, as well as to investigate their functions and interactions in response to salt stress.
Conclusions
In this study, we analyzed the differential responses of tissues in D. officinale to salt stress, focusing on the interplay between sugar metabolic genes and DoSWEETs in adapting to high salinity. A total of eight DoSWEETs were found to respond to salt, exhibiting tissue-specific preferences. Notably, DoSWEET2a and DoSWEET16 showed a strong correlation with INV in both leaves and roots. These findings greatly advance our understanding of the mechanisms underlying salt adaptation in different plant tissues of D. officinale and highlight potential candidates, such as the key enzymes and the DoSWEETs, for breeding salt-resistant varieties by regulating the sugar metabolism and transport.
Methods
Transcriptome analysis of D. Officinale
Transcriptome datasets of D. officinale treated with 250 mM NaCl for 0, 4, and 12 h were obtained with the accession number PRJNA715099 (https://www.ncbi.nlm.nih.gov/bioproject/, accessed on December 15, 2023) [13]. Gene expression levels were quantified using Salmon 1.8.0 (accessed on April 14, 2022) [49]. Before quantification, a salmon index was built utilizing the RNA and genome sequences of D. officinale with the accession number PRJNA262478 (https://www.ncbi.nlm.nih.gov/bioproject/, accessed on December 15, 2023) [26, 50]. Clean data, filtered using Trimmomatic (accessed on April 14, 2022) [51], were then input into salmon 1.8.0 to calculate values of transcripts per million (TPM) using the quasi-mapping-based mode. To access DEGs in response to salt stress, the DESeq2 package [52] (accessed on April 14, 2022) was employed to analyze the salmon outputs, applying the criteria of |log2FoldChange| > 1 and p-value < 0.05. Visualization of the results was performed using the ggplot2 package (https://github.com/tidyverse/ggplot2, accessed on April 14, 2022).
Functional enrichment of DEGs in response to salt
GO enrichment analysis [53] was performed using the clusterProfiler package (accessed on April 14, 2022) [54], with pvalueCutoff = 0.05 and qvalueCutoff = 0.02. The top five most significant processes in each category (biological process, cellular component, and molecular function) were extracted for visualization using the GOplot package (accessed on February 27, 2024) [55].
KEGG enrichment analysis [56] was also conducted using the clusterProfiler package (accessed on April 14, 2022) [54], with the same pvalueCutoff and qvalueCutoff values. The top ten most significant pathways were visualized with the ggplot2 package (https://github.com/tidyverse/ggplot2, accessed on April 14, 2022). The numbers of up- and down-regulated genes in each pathway were visualized using the sankeyNetwok function from the networkD3 package (https://github.com/christophergandrud/networkD3, accessed on February 29, 2024).
Screening and expression profiling of the genes involved in sugar metabolism in D. Officinale
Initially, the protein sequences of genes in D. officinale genome were annotated using eggNOG-MAPPER (http://eggnog-mapper.embl.de, accessed on February 26, 2024) [57]. Genes associated with sugar metabolism were extracted based on their ORTHOLOGY numbers (K02810, K01193, K01187, K00695, K00844, K00847, K01810, K01835, K00963, K01809, K17497, K00966, and K01513) in KEGG pathways ko00500 (starch and sucrose metabolism) and ko00051 (fructose and mannose metabolism) (https://www.kegg.jp/, accessed on March 1, 2024). Cellulose synthase like genes (CSLs), classified as glycosyltransferase genes (GTs), are known to be involved in the biosynthesis of glucomannan polysaccharides in D. officinale [25, 26]. DoCSLs were screened using the following ORTHOLOGY numbers: K13680, K20887, K20924, and K20923.
The expression patterns of the above genes derived from the transcriptome data were visualized using the pheatmap package (https://github.com/raivokolde/pheatmap, accessed on April 14, 2022), based on the mean TPM values obtained from three biological replicates.
Expression correlation analysis between DoSWEETs and sugar metabolism related genes
To analyze the correlation between DoSWEETs and sugar metabolism-related genes, we utilized the Hmisc package (https://github.com/haxscramper/hmisc, accessed on April 14, 2022) to calculate the Pearson correlation coefficients. The results were subsequently visualized using the corrplot R package (https://github.com/taiyun/corrplot, accessed on April 14, 2022).
To further investigate the correlation between DoSWEETs and genes involved in different sugar metabolism pathways, the previously identified genes were categorized into three groups based on their catalytic functions: the glucose-related group, the fructose and mannose-related group, and the both-related group. Then we analyzed the correlation between DoSWEETs and these gene groups using the vegan package (https://github.com/vegandevs/vegan, accessed on March 13, 2024) and visualized the results using the linkET package (https://github.com/Hy4m/linkET, accessed on March 13, 2024).
Plant materials and salt treatment
The tissue-cultured D. officinale used in this study was identified and cultured by professor Yuehua Wang at Chengdu University. A voucher specimen of this material has been deposited in the herbarium of Institute of Botany, Chinese Academy of Sciences, with deposition number PE02032867 (D. officinale Kimura et Migo, identified by Zhanhe Ji).
Tissue-cultured D. officinale plantlets with roots were grown on Murashige and Skoog (MS) medium [58] at pH 5.8, maintained at a temperature of 24 ± 1 °C under a photoperiod of 16 h light /8 h dark. For salt treatment, the plantlets were transferred to the MS medium supplemented with 250 mM NaCl. Leaves and roots were harvested at 0 h, 4 h, and 12 h post salt treatment. All samples were rapidly frozen in liquid nitrogen and stored at -80 °C. Each experiment was conducted three times.
Detection of sucrose, D-glucose, and D-fructose
Leaves and roots were harvested from three tissue-cultured D. officinale plantlets after following salt treatment. Fresh samples for the detection of sucrose, D-glucose were weighed to 0.1 g and then crushed. For the detection of D-fructose, fresh samples were weighed to 0.05 g and extracted using the Plant Tissue Fructose Content Assay Kit (Solarbio Science & Technology Co., Ltd, Beijing, China). Sucrose was extracted using the Plant Sucrose Content Assay Kit (Solarbio Science & Technology Co., Ltd, Beijing, China), while glucose was extracted using the Glucose Content Assay Kit (Sangon Biotech Co., Ltd. Shanghai, China). The sugar content was measured using a spectrophotometer. All experiments were performed with three independent biological replicates.
RNA extraction and RT-qPCR
RNA was isolated form collected samples using the EASYspin Plus Complex Plant RNA (Aidlab Biotechnologies Co., Ltd., Beijing, China), followed by one-step TRUEscript 1st Strand cDNA Synthesis Kit (Aidlab Biotechnologies Co., Ltd., Beijing, China). RT-qPCR was performed using the 2× SYBR Green qPCR Mix (Low ROX) (Aidlab Biotechnologies Co., Ltd., Beijing, China) with QuantStudio 3 (ABI, California, USA) in a total reaction volume of 20 µL. The reaction program was set according to previously reported protocols [24]. DoGAPDH was used as the internal control [59]. The relative expression levels of the target genes were calculated using the 2−ΔΔCt method [60]. All experiments included three independent biological replicates. The primers used in this study are listed in Additional file 1: Table S12.
Statistical analysis
The results of RT-qPCR and sugar content were visualized using Graph-Pad Prism 9 (accessed on May 12, 2024). All experiments were conducted in triplicate. Statistical analysis to determine significant differences was performed using Duncan’s multiple range test within a one-way ANOVA framework in SPSS 25 (accessed on May 20, 2024).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We sincerely thank all researchers who unselfishly shared transcriptome data of salt stress and the genome of D. officinale.
Author contributions
Y.Z. and L.H. conceived and designed the study. L.H. collected the data and materials, performed data analyses, and drafted the manuscript. X.S., S.Y.W., J.Q.C., and K.X.L. performed the formal analyses. Y.Q.C., S.M.Y., C.Y.Y., and Y.X.S. contributed to the visualization of the results. Y.Z. revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by Sichuan Science and Technology Program (2024NSFSC1330) and the Key Laboratory of Medicinal and Edible Plant Resources Development of Sichuan Education Department, Chengdu University (No. 10Y202402).
Data availability
Data is provided within the manuscript or supplementary information files.
Declarations
Ethics approval and consent to participate
The tissue-cultured D. officinale was kept in Chengdu University and the permission to use has been obtained from Chengdu University. All experiments involving the plant and its material complied with relevant institutional, national, and international guidelines and legislation, and the IUCN Policy Statement on Research Involving Species at Risk of Extinction and the Convention on the Trade in Endangered Species of Wild Fauna and Flora.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ng TB, Liu J, Wong JH, Ye X, Wing Sze SC, Tong Y, Zhang KY. Review of research on Dendrobium, a prized folk medicine. Appl Microbiol Biotechnol. 2012;93(5):1795–803. [DOI] [PubMed] [Google Scholar]
- 2.Cao H, Ji YL, Li SC, Lu L, Tian M, Yang W, Li H. Extensive metabolic profiles of leaves and stems from the medicinal plant Dendrobium Officinale Kimura et Migo. Metabolites. 2019;9(10):215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo QL, Tang ZH, Zhang XF, Zhong YF, Yao SZ, Wang LS, Lin CW, Luo X. Chemical properties and antioxidant activity of a water-soluble polysaccharide from Dendrobium officinale. Int J Biol Macromol. 2016;89:219–27. [DOI] [PubMed] [Google Scholar]
- 4.Tang H, Zhao T, Sheng Y, Zheng T, Fu L, Zhang Y. DendrOfficinalecinale Kimura et Migo: a review on its ethnopharmacology, phytochemistry, pharmacology, and industrialization. Evid Based Complement Alternat Med. 2017;2017:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teixeira da Silva JA, Ng TB. The medicinal and pharmaceutical importance of Dendrobium species. Appl Microbiol Biotechnol. 2017;101(6):2227–39. [DOI] [PubMed] [Google Scholar]
- 6.Braun DM. Phloem loading and unloading of sucrose: what a long, strange trip from source to sink. Annu Rev Plant Biol. 2022;73:553–84. [DOI] [PubMed] [Google Scholar]
- 7.Eom JS, Chen LQ, Sosso D, Julius BT, Lin IW, Qu XQ, Braun DM, Frommer WB. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr Opin Plant Biol. 2015;25:53–62. [DOI] [PubMed] [Google Scholar]
- 8.Zeng X, Ling H, Chen X, Guo S. Genome-wide identification, phylogeny and function analysis of GRAS gene family in Dendrobium catenatum (Orchidaceae). Gene. 2019;705:5–15. [DOI] [PubMed] [Google Scholar]
- 9.Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59(1):651–81. [DOI] [PubMed] [Google Scholar]
- 10.Knight H, Trewavas AJ, Knight MR. Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J. 2003;12(5):1067–78. [DOI] [PubMed] [Google Scholar]
- 11.Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53(1):247–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaleem F, Shabir G, Aslam K, Rasul S, Manzoor H, Shah SM, Khan AR. An overview of the genetics of plant response to salt stress: present status and the way forward. Appl Biochem Biotechnol. 2018;186(2):306–34. [DOI] [PubMed] [Google Scholar]
- 13.Zhang M, Yu Z, Zeng D, Si C, Zhao C, Wang H, Li C, He C, Duan J. Transcriptome and metabolome reveal salt-stress responses of leaf tissues from Dendrobium officinale. Biomolecules. 2021;11(5):736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Si C, Zeng D, Yu Z, Teixeira da Silva JA, Duan J, He C, Zhang J. Transcriptomic and metabolomic analyses reveal the main metabolites in Dendrobium officinale leaves during the harvesting period. Plant Physiol Biochem. 2022;190:24–34. [DOI] [PubMed] [Google Scholar]
- 15.Gupta B, Huang B. Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular characterization. Int J Genomics. 2014;2014:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdullakasim S, Kongpaisan P, Thongjang P, Saradhuldhat P. Physiological responses of potted Dendrobium orchid to salinity stress. Hortic Environ Biote. 2018;59(4):491–8. [Google Scholar]
- 17.Zhang M, Liu N, Teixeira da Silva JA, Liu X, Deng R, Yao Y, Duan J, He C. Physiological and transcriptomic analysis uncovers salinity stress mechanisms in a facultative crassulacean acid metabolism plant Dendrobium officinale. Front Plant Sci. 2022;13:1028245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, Guo WJ, Kim JG, Underwood W, Chaudhuri B, et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468(7323):527–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klemens PAW, Patzke K, Deitmer J, Spinner L, Le Hir R, Bellini C, Bedu M, Chardon F, Krapp A, Neuhaus HE. Overexpression of the vacuolar sugar carrier AtSWEET16 modifies germination, growth, and stress tolerance in Arabidopsis. Plant Physiol. 2013;163(3):1338–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miao H, Sun P, Liu Q, Miao Y, Liu J, Zhang K, Hu W, Zhang J, Wang J, Wang Z, et al. Genome-wide analyses of SWEET family proteins reveal involvement in fruit development and abiotic/biotic stress responses in banana. Sci Rep. 2017;7(1):3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji J, Yang L, Fang Z, Zhang Y, Zhuang M, Lv H, Wang Y. Plant SWEET family of sugar transporters: structure, evolution and biological functions. Biomolecules. 2022;12(2):205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Yao L, Hao X, Li N, Qian W, Yue C, Ding C, Zeng J, Yang Y, Wang X. Tea plant SWEET transporters: expression profiling, sugar transport, and the involvement of CsSWEET16 in modifying cold tolerance in Arabidopsis. Plant Mol Biol. 2018;96(6):577–92. [DOI] [PubMed] [Google Scholar]
- 23.Le Hir R, Spinner L, Klemens PA, Chakraborti D, de Marco F, Vilaine F, Wolff N, Lemoine R, Porcheron B, Gery C, et al. Disruption of the sugar transporters AtSWEET11 and AtSWEET12 affects vascular development and freezing tolerance in Arabidopsis. Mol Plant. 2015;8(11):1687–90. [DOI] [PubMed] [Google Scholar]
- 24.Hao L, Shi X, Qin S, Dong J, Shi H, Wang Y, Zhang Y. Genome-wide identification, characterization and transcriptional profile of the SWEET gene family in Dendrobium officinale. BMC Genomics. 2023;24(1):378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He C, Zhang J, Liu X, Zeng S, Wu K, Yu Z, Wang X, Teixeira da Silva JA, Lin Z, Duan J. Identification of genes involved in biosynthesis of mannan polysaccharides in Dendrobium officinale by RNA-seq analysis. Plant Mol Biol. 2015;88(3):219–31. [DOI] [PubMed] [Google Scholar]
- 26.Zhang GQ, Xu Q, Bian C, Tsai WC, Yeh CM, Liu KW, Yoshida K, Zhang LS, Chang SB, Chen F, et al. The Dendrobium Catenatum Lindl. Genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci Rep. 2016;6(1):19029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hazen SP, Jaarsma R, de Vries RSM, de Boer AH. Effect of salt stress on growth, Na+ accumulation and proline metabolism in potato (Solanum tuberosum) cultivars. PLoS ONE. 2013;8(3):e60183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiang XG, Schuiteman A, Li DZ, Huang WC, Chung SW, Li JW, Zhou HL, Jin WT, Lai YJ, Li ZY, et al. Molecular systematics of Dendrobium (Orchidaceae, Dendrobieae) from mainland Asia based on plastid and nuclear sequences. Mol Phylogenet Evol. 2013;69(3):950–60. [DOI] [PubMed] [Google Scholar]
- 29.Niu Z, Zhu F, Fan Y, Li C, Zhang B, Zhu S, Hou Z, Wang M, Yang J, Xue Q, et al. The chromosome-level reference genome assembly for Dendrobium officinale and its utility of functional genomics research and molecular breeding study. Acta Pharm Sinica B. 2021;11(7):2080–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Y, Li D, Liu T, Liao M, Li Y, Zhang W, Liu Z, Chen M. Effect of overexpression of γ-tocopherol methyltransferase on α-tocopherol and fatty acid accumulation and tolerance to salt stress during seed germination in Brassica napus L. Int J Mol Sci. 2022;23(24):15933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui J, Li J, Dai C, Li L. Transcriptome and metabolome analyses revealed the response mechanism of sugar beet to salt stress of different durations. Int J Mol Sci. 2022;23(17):9599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Zelm E, Zhang Y, Testerink C. Salt tolerance mechanisms of plants. Annu Rev Plant Biol. 2020;71(1):403–33. [DOI] [PubMed] [Google Scholar]
- 33.Yang Y, Guo Y. Unraveling salt stress signaling in plants. J Integr Plant Biol. 2018;60(9):796–804. [DOI] [PubMed] [Google Scholar]
- 34.Geng Y, Wu R, Wee CW, Xie F, Wei X, Chan PMY, Tham C, Duan L, Dinneny JR. A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell. 2013;25(6):2132–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munns R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002;25(2):239–50. [DOI] [PubMed] [Google Scholar]
- 36.Fricke W, Akhiyarova G, Wei W, Alexandersson E, Miller A, Kjellbom PO, Richardson A, Wojciechowski T, Schreiber L, Veselov D, et al. The short-term growth response to salt of the developing barley leaf. J Exp Bot. 2006;57(5):1079–95. [DOI] [PubMed] [Google Scholar]
- 37.Feng W, Kita D, Peaucelle A, Cartwright HN, Doan V, Duan Q, Liu MC, Maman J, Steinhorst L, Schmitz-Thom I, et al. The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr Biol. 2018;28(5):666–e755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bose J, Munns R, Shabala S, Gilliham M, Pogson B, Tyerman SD. Chloroplast function and ion regulation in plants growing on saline soils: lessons from halophytes. J Exp Bot. 2017;68(12):3129–43. [DOI] [PubMed] [Google Scholar]
- 39.Li Z, Cheng B, Liu W, Feng G, Zhao J, Zhang L, Peng Y. Global metabolites reprogramming induced by spermine contributing to salt tolerance in creeping bentgrass. Int J Mol Sci. 2022;23(9):4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang W, Pang J, Zhang F, Sun L, Yang L, Fu T, Guo L, Siddique KHM. Salt–responsive transcriptome analysis of canola roots reveals candidate genes involved in the key metabolic pathway in response to salt stress. Sci Rep. 2022;12(1):1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Chu Y, Yao K, Shi C, Deng X, Lin J, Rennenberg H. Response of sugar metabolism in the cotyledons and roots of Ricinus communis subjected to salt stress. Plant Biol. 2022;25(1):62–71. [DOI] [PubMed] [Google Scholar]
- 42.Sarkar AK, Sadhukhan S. Imperative role of trehalose metabolism and trehalose-6‐phosphate signaling on salt stress responses in plants. Physiol Plant. 2022;174(1):e13647. [DOI] [PubMed] [Google Scholar]
- 43.Jiang Z, Zhang H, Gao S, Zhai H, He S, Zhao N, Liu Q. Genome-wide identification and expression analysis of the sucrose synthase gene family in sweet potato and its two diploid relatives. Int J Mol Sci. 2023;24(15):12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meng F, Feng N, Zheng D, Liu M, Zhang R, Huang X, Huang A, Chen Z. Exogenous hemin alleviates NaCl stress by promoting photosynthesis and carbon metabolism in rice seedlings. Sci Rep. 2023;13(1):3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao X, Wang Q, Ma X, Lang D, Guo Z, Zhang X. Physiological biochemistry-combined transcriptomic analysis reveals mechanism of Bacillus cereus G2 improved salt-stress tolerance of Glycyrrhiza Uralensis Fisch. Seedlings by balancing carbohydrate metabolism. Front Plant Sci. 2022;12:712363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sellami S, Le Hir R, Thorpe MR, Vilaine F, Wolff N, Brini F, Dinant S. Salinity effects on sugar homeostasis and vascular anatomy in the stem of the Arabidopsis thaliana inflorescence. Int J Mol Sci. 2019;20(13):3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Székely G, Ábrahám E, Cséplő Á, Rigó G, Zsigmond L, Csiszár J, Ayaydin F, Strizhov N, Jásik J, Schmelzer E, et al. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 2007;53(1):11–28. [DOI] [PubMed] [Google Scholar]
- 48.Srivastava AC, Dasgupta K, Ajieren E, Costilla G, McGarry RC, Ayre BG. Arabidopsis plants harbouring a mutation in AtSUC2, encoding the predominant sucrose/proton symporter necessary for efficient phloem transport, are able to complete their life cycle and produce viable seed. Ann Bot. 2009;104(6):1121–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14(4):417–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang GQ, Liu KW, Li Z, Lohaus R, Hsiao YY, Niu SC, Wang JY, Lin YC, Xu Q, Chen LJ, et al. The apostasia genome and the evolution of orchids. Nature. 2017;549(7672):379–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30(15):2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Love MI, Huber W, Anders S. Moderated estimation of Fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innov (Camb). 2021;2(3):100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walter W, Sanchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31(17):2912–4. [DOI] [PubMed] [Google Scholar]
- 56.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27(1):29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cantalapiedra CP, Hernandez-Plaza A, Letunic I, Bork P, Huerta-Cepas J. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol Biol Evol. 2021;38(12):5825–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15(3):473–97. [Google Scholar]
- 59.Jiang M, Xu SZ, Wen GS, Zhao CL. Validation of seven housekeeping genes as reference ones for qRT-PCR normalization in Dendrobium catenatum. RUSS J PLANT PHYSL+. 2017;64(4):497–508. [Google Scholar]
- 60.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is provided within the manuscript or supplementary information files.