Abstract
Purpose
Spina bifida (SB) arises from complex genetic interactions that converge to interfere with neural tube closure. Understanding the precise patterns conferring SB risk requires a deep exploration of the genomic networks and molecular pathways that govern neurulation. This study aims to delineate genome-wide regulatory signatures underlying SB pathophysiology,
Methods
An untargeted, genome-wide approach was used to interrogate regulatory regions for rare single-nucleotide and copy-number variants (rSNVs and rCNVs, respectively) predicted to affect gene expression, comparing results from SB patients with healthy controls. Qualifying variants were subjected to a deep learning prioritization framework to identify the most functionally relevant variants, as well as the likely target genes affected by these rare regulatory variants.
Results
This ensemble of computational tools identified rSNVs in specific transcription factor binding sites (TFBSs) that distinguish SB cases from controls. rSNV enrichment was found in specific TFBSs, especially CCCTC-binding factor binding sites. These TFBSs were subjected to a deep learning prioritization framework to identify the most functionally relevant variants, as well as the likely target genes affected by these rSNVs. The functional pathways or modules implicated by these regulated genes serve protein transport, cilia assembly, and central nervous system development. Moreover, the detected rare copy-number variants in SB cases are positioned to disrupt gene regulatory networks and alter 3-dimensional genomic architectures, including brain-specific enhancers and topologically associated domain boundaries of relevant cell types.
Conclusion
Our study provides a resource for identifying and interpreting genomic regulatory DNA variant contributions to human SB genetic predisposition.
Keywords: Deep learning, Intergenic variants, Neural tube defects, Topologically associating domains (TADs), Transcription factor binding sites (TFBS)
Introduction
In the first 5 weeks of human gestation, a flat plate emerges of neuroepithelial cells that proliferate, become polarized and migrate in a process of convergence-extension, apically constrict, and fold into a neural tube that forms the brain and spinal cord. Failure of this neurulation leads to neural tube defects (NTDs). Rostral NTDs expose brain (anencephaly) and are lethal in utero or at term, whereas spina bifida (SB, aka myelomeningocele) involves more caudal vertebrae, spinal cord, and/or nerve roots and is survivable. Despite folic acid fortification efforts and recent advances in fetal repair surgery, SB remains a debilitating birth defect of the spinal cord with brain comorbidity and carries a prevalence of 3.5 to 5.3 cases per 10,000 live births worldwide.1 Even with surgical intervention, NTDs carry a heavy clinical, financial, and public health cost. Analyzing genomic data of human NTD patients is advancing the field beyond protein coding variant studies of disease pathophysiology. Achieving a precision medicine inspired approach to NTD risk assessment requires a deeper exploration of the gene regulatory networks and transcriptional programs that govern the early developmental processes of neural tube formation and closure. To test the hypothesis that significant contributions to NTD risk are not solely attributable to likely gene-disrupting (LGD) DNA variants, we have begun to interrogate not only exomes but also the elements of the genome regulating gene expression.
Although the potential role of intergenic variation in NTD causation remains to be clarified, noncoding variants are known to account for greater than 90% of the statistically significant findings from genome-wide association studies, including across a range of neurological disorders.2 Recent studies provide evidence to support that regulatory noncoding variation plays a significant role in numerous neurodevelopmental disorders and structural birth defects.3,4 Specific regulatory changes, including rare variants that disrupt transcription factor binding sites (TFBS), defined as 6 to 12 nucleotide stretches of DNA with specific motifs to which transcription factors bind within enhancers have been shown to contribute to congenital malformations, as well as neurodevelopmental disorders.5 Rare genetic variants at TFBS have not only been associated with genetic phenotypes but have also been directly linked with altering local methylation profiles.6 We therefore sought to methodically interrogate the regulatory genome in our SB patient cohorts and to offer an untargeted approach avoiding candidate gene searches and ascertainment bias.
An inherent challenge to interpreting regulatory variation stems from our expanding nonlinear view of nucleotide sequences. TFBS are not necessarily regulating the nearest neighbor transcription unit but often affect transcription of genes at great distances from the site. Recent advanced methods for investigation of the 3-dimensional genome have reinforced the notion that gene regulation is inherently associated with chromatin topology and cellular function.7 This requires computational interrogation of rare regulatory variants and assessment of their potential role in genomic organization from a 3-dimensional DNA perspective.
Topologically associating domains (TADs) are a feature of genomic organization in which chromosomes fold into domains with preferential interactions. It has been shown that contacts between enhancers and promoters are largely restricted within TADs and that TAD features, including their boundaries, are strongly conserved in mammals.8 Advanced high-throughput chromosome conformation capture—or Hi-C—technologies are now detecting high-resolution finer domains and sub-TADs, highlighting an enrichment of chromatin marks, as well as important features, such as interaction sites for the CCCTC-binding factor (CTCF).9 There is an increasing amount of evidence suggesting that TADs represent a functional subdivision of genomic organization in which enhancer-promoter contacts are spatially restricted and that TAD boundary disruption may lead to aberrant transcriptional signatures, which may be predicted to be pathogenic.10
Here, we establish a framework to identify and prioritize these regulatory variants within the genetic architecture of NTD risk, which builds upon several recent genome-wide studies. Wolujewicz et al11 previously detected a rare copy-number variant (rCNV) burden in human SB cases affecting exonic regions of the genome and Aguiar-Pulido et al12 highlighted an approach to distinguish SNV signatures and predict SB risk based on rare LGD variants. Here, we investigate the contribution of rare SNVs (rSNVs) and rCNVs in the regulatory genomes of SB cases compared with ancestry-matched controls, supporting the hypothesis that rare regulatory variants may have functional pathophysiological roles in the genetic susceptibility to SB.
Materials and Methods
Study population and genome sequencing
This case-control study was conducted by integrating population cohorts from the United States, as well as from Qatar. Genomic DNA was extracted from deidentified infant blood spot and venipuncture samples collected in the United States and participants in the national Spina Bifida Clinic at the Hamad Medical Corporation, Qatar. The human participant research study protocol was approved by Institutional Review Boards in the United States (Weill Cornell Medical College-NY) and in the Middle Eastern population receiving their health care in Qatar (Hamad Medical Corporation and Weill Cornell Medical College-Qatar). Consent documentation was provided in both English and Arabic. DNA extraction was completed using the Pure-gene DNA Extraction Kit from Qiagen, and the input amounts of DNA ranged from 200 to 500 ng for infant blood spots and 2 to 3 μg from venipuncture samples. Genome sequencing was conducted on all DNA samples using Illumina v.3 chemistries on HiSeq 2500 instruments to obtain short paired-end reads of 2 × 100 base pairs (bp). After passing all quality control measures, our case participants comprised 149 SB cases who presented with nonsyndromic myelomeningocele, and our control cohort included 149 unrelated individuals ancestry matched to the case population. After population structure analyses using PLINK13 genomic distances were calculated from 9 gene pools followed by subsequent optimization of case-control pairings as previously described.12
Alignment and quality control
All sequences, whether from our collection or the control sequences obtained from existing databases,14, 15, 16 were joint genotyped to assure consistent alignment and variant calls. Sequencing reads in the form of FASTQ files were aligned to the hg38 reference genome using Burrows-Wheeler Aligner.17 SAMtools18 was used on individual bam files to run quality control measures such as mapping quality and to assess read depth uniformity. Read depth statistics were calculated both in SAMtools and using the Genome Analysis Tool Kit.19 The median insert size for samples included in the analysis was 413 bp.
SNV detection and annotation pipeline
Variant calling was performed with Genome Analysis Tool Kit 4 Best Practices, and joint genotyping was carried out on the entire integrated cohort comprising 149 cases and 149 controls. Subsequently, variant quality score recalibration was performed on the variant call sets to model the data set profile and filter out potential variant artifacts. Variant quality was evaluated and only variants that included a “PASS” in the filter column were retained and annotated utilizing Ensembl Variant Effect Predictor v.95.20
CNV detection and annotation pipeline
For CNV calling and annotations, high-confidence custom pipelines were deployed as previously detailed.11 Each variant detection pipeline was deployed independently on each case and control in our study using individual sample bam files, as well as the additional input of unannotated SNV calls. An optimized consensus filtering approach was performed on the raw CNV outputs, and only high-confidence CNV calls were retained for further analysis. Annotation of detected CNVs was performed using AnnotSV.21 CNVs were considered rare if they occurred with less than 1% minor allele frequency according to population genetic databases (gnomAD22 and DGV23). Visual validation and inspection of genomic variants was conducted using samplot24 and Integrative Genomics Viewer.25
Brain specific TFBS annotations and motif analysis
We leveraged tissue-specific TFBS regions as determined by Funk et al.26 In brief, deoxyribonuclease treatment followed by nucleotide sequencing footprinting provided binding site predictions for transcription factors genome wide, and by analyzing a compendium of ENCODE deoxyribonuclease treatment followed by nucleotide sequencing experiments, the chromosomal loci and tissue specificity predictions for 1515 human transcription factors for 27 tissue types were determined. We subset the rare variants detected in our cohort with the variants that overlapped brain-tissue-specific TFBS regions and that had Hmm-based IdeNtification of Transcription factor (HINT) scores >200 on the flat files of the tissue-specific open chromatin footprints. TFmotifView27 was used for further analysis, including the motif-specific enrichments and visualizations of TFBS-rSNVs in our case and control cohorts.
Deep learning prioritization
After intersecting our rare variant call sets with brain derived TFBS regions, we subjected our TFBS-rSNVs to a deep learning prioritization framework using DeepSEA.28 DeepSEA was trained on 919 cell-type-specific epigenomic features, allowing its interpretation for any cell type. For the TFBS-rSNVs in our call set, DeepSEA predictions were obtained using the online tool (http://deepsea.princeton.edu/job/analysis/create). We assembled VCF files comprising 370,556 rare TFBS-rSNVs, which were provided as input to DeepSEA and submitted in 10 batches. The functional significance score distribution that was associated with each of our detected TFBS-rSNVs were computed as the product of the geometric mean E-value across chromatin features and the geometric mean E-value of the evolutionary conservation scores. We further binned these functional significance scores into quartiles representing benign, uncertain, and likely expression-modifying noncoding variation.
Target gene prediction and enrichment
To determine the target genes potentially transcription modified and/or aberrantly expressed by rare binding site disruption, we leveraged the GeneHancer29 database, which includes chromosome conformation capture and expression quantitative trait loci (eQTL) data to identify enhancer regions in a gene-specific manner. We intersected the relevant brain-specific footprints and TFBS in our annotation data sets with the double elite enhancer regions from GeneHancer, which maintain a more stringent level of enhancer gene pairing, to assign a target gene for each of our TFBS-rSNVs. For the detected TFBS-rSNVs in our SB cases that have been classified as likely to alter gene expression (LAGE)—with a functional significance score in the top quartile—we evaluated the predicted target genes that were overrepresented in the SB cases compared with predicted likely altered target gene expression from the matched controls. These genes were used to further characterize the biological signaling processes and pathways predicted to be perturbed by TFBS-rSNVs underlying SB.
Functional module analysis
Brain-specific functional module prediction was conducted utilizing the approach outlined by Krishnan et al.30 We identified brain-specific functional modules by using the overrepresented predicted target genes in our SB cases as input in the Functional Module Detection query at https://humanbase.flatironinstitute.org. Each gene list was clustered using a shared nearest neighbor-based community-finding algorithm to identify distinct modules of tightly connected genes, and the resulting modules were then tested for functional enrichment using genes annotated to Gene Ontology (GO) biological process terms. The associated Q values for each term were calculated using one-sided Fisher’s exact tests and subsequent Benjamini-Hochberg corrections to account for multiple testing.
GO and pathway analyses
Gene set enrichment and overrepresentation analyses were performed using WebGestalt,31 EnrichR,32 and MonaGO.33 Genes or terms were ranked based on the adjusted P value (Benjamini-Hochberg), and significantly affected gene sets were selected based on an adjusted P value of <.05. Ingenuity Pathway Analysis and GeneAnalytics34 were used to investigate the functional consequences of our enriched and implicated gene sets.
TAD coordinates and boundaries
As a high-quality data set for relevant cell type genomic organization derived using Hi-C interaction frequency data, we utilized the TAD boundary designations from Dixon et al,8 who mapped genome-wide chromatin interactions in human embryonic stem cells and 4 human embryonic-stem-cell-derived lineages. To improve the resolution and incorporate other cell type information in our analyses, we further used preciseTAD,35 which uses a transfer learning model for TAD boundary prediction and resolves the boundary to the base-pair level.
Results
An integrative approach for rare regulatory variation
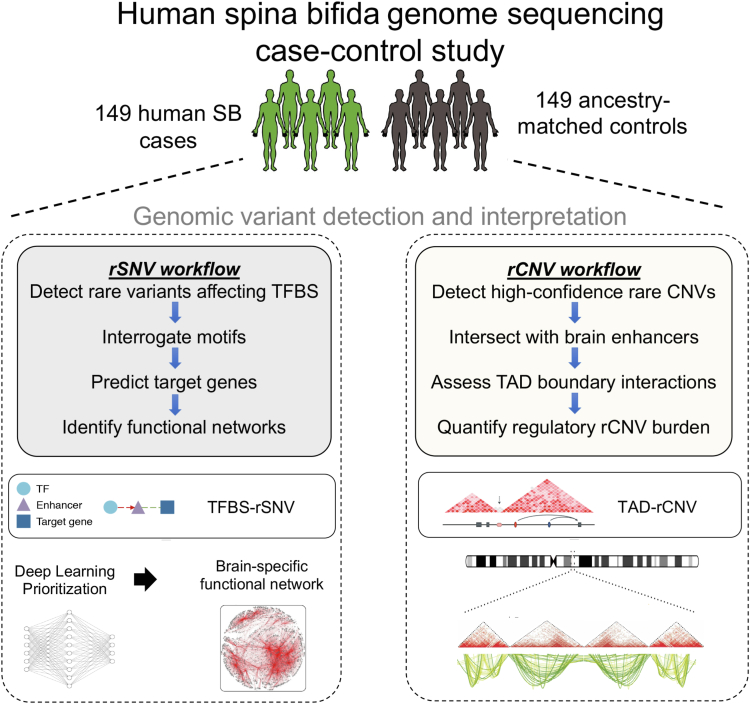
All 298 samples passed initial preprocessing and quality control checks (see Materials and Methods), and our optimization of case-control ancestry pairing was based on a mixed admixture model to control stratification as previously described.12 The methodology and approach used in this study of regulatory element contributions to SB risk leverages genome sequencing data from our study cohorts, as well as a tissue-specific TFBS atlas26 and high-resolution Hi-C and TAD maps (Figure 1). We restricted our analyses to rare variation that was represented in population databases at a threshold under 1% minor allele frequency. We defined our subset of detected rSNVs that overlap brain-specific transcription factor binding sites as TFBS-rSNVs. Similarly, rCNVs that overlap hESC TAD boundaries were termed TAD-rCNVs. Given the early developmental dynamics of neural tube closure in the neuroepithelium, we utilized TAD boundaries and chromatin related information from well-characterized hESC lines. We postulate that stem cell data sets delineating embryonic chromatin topology are better suited to our study than brain tissue because neural tube closure occurs before neuronal differentiation.
Figure 1.
Regulatorygenome sequencinganalyses. Rare single-nucleotide variants that overlap brain-specific transcription factor binding sites were interrogated and subjected to deep learning analyses. Predicted gene targets were analyzed on the level of pathways as well as brain-specific functional networks. Rare copy-number variants were analyzed for their proximity to human embryonic stem cell topologically associating domain boundaries.
TFBS motifs are enriched in SB cases by rare single-nucleotide variants
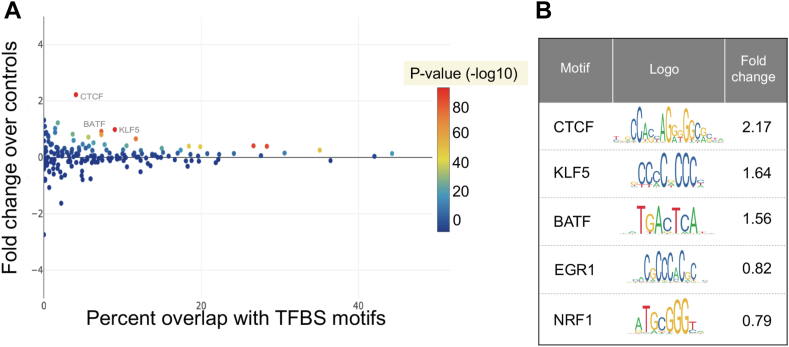
Our analysis uncovered 186,060 TFBS-rSNVs in SB cases and 184,496 in controls. There was no significant burden between our study cohorts (P = .639), and no recurrent mutational hot spots were uncovered in this motif-independent analysis. After cross-filtering between our SB case and control cohorts to exclude redundant variants, we quantified enrichments of individual motifs within each cohort. This was calculated as a fold change of motif frequency in SB cases vs controls, with an associated hypergeometric P value (Figure 2A). We display the binding motifs that were the most statistically significant and enriched in our SB cases in Figure 2B. Among the implicated transcription factor motifs, the top 3 in terms of fold-change enrichment were CTCF, KLF5, and BATF. CTCF motifs were disrupted by rare variants in SB cases vs controls—as assessed by our detected TFBS-rSNVs—at a ratio of 2.17:1. KLF5 and BATF motif disruption were detected in SB cases at 1.64-fold and 1.56-fold above matched controls, respectively.
Figure 2.
Transcription factor binding site motifs affected by rare single-nucleotide variants in spina bifida cases showing statistically significant enrichment over matched controls. A. Scatterplot displaying the significance of transcription factor motif occurrences in genomic regions of spina bifida cases compared with control regions. B. Top 5 transcription factor motifs with the corresponding fold-change enrichment and logo representation for the most significant motif enrichments in cases vs controls.
The detected TFBS-rSNVs in our cohorts were further subjected to a deep-learning-based algorithmic framework, DeepSEA, to computationally predict functional effects on chromatin and ultimately prioritize our identified rare regulatory variants. DeepSEA’s algorithmic framework, which has been trained on large-scale chromatin profiling data, provides functional significance scores that are computed on the basis of chromatin effect predictions, evolutionary-derived information, and can accurately predict over 2000 chromatin features. When applied to our set of TFBS-rSNVs, we were able to generate deep learning derived functional significance scores for our TFBS-rSNVs. The TFBS-rSNVs were subsequently binned for each cohort by quartiles according to their functional significance scores, which is analogous to assigning predicted interpretations or classifications of benign, uncertain, and LAGE.
Furthermore, by leveraging aggregated chromosome conformation capture assay and eQTL information from GeneHancer, we were able to predict the gene targets of our detected TFBS-rSNVs; that is, which genes may be transcriptionally dysregulated because of the disruption of transcription factor binding to its site. If they were not in the promoter region of a gene, TFBS-rSNV coordinates were intersected with high-confidence enhancer regions to predict transcriptionally associated target genes of the detected TFBS-rSNVs. The target genes were sought from our predicted LAGE TFBS-rSNVs in SB cases, and the comparison of the chromosomes harboring these gene targets for both expected and observed distributions are displayed in Supplemental Figure 1. Potential hot spots and chromosomal enrichments of target genes are evident for SB cases vs controls (P = 1.6E−07). These predicted target genes that were overrepresented in SB cases vs matched controls were used to pinpoint which pathways or functional modules may be ultimately affected by these rare variants (Figure 3). The target genes most prevalent in SB cases, encompassing 107 genes in total, are listed in Supplemental Table 1.
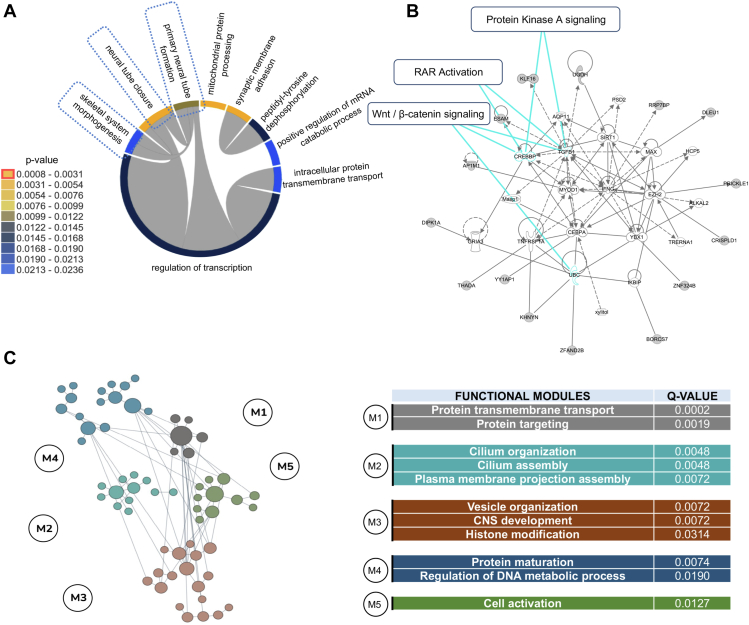
Figure 3.
Enhancer gene targets from transcription factor binding sites (TFBS)-rSNVs point to regulatory machinery and pathways affected in spina bifida (SB). A. Gene Ontology analyses on our overrepresented target gene set in SB cases include biological processes of neural tube closure and skeletal system morphogenesis, as well as gene subsets that are also associated as transcriptional regulators. B. Ingenuity Pathway Analysis of our overrepresented target genes. C. Brain-specific functional module prediction of our SB overrepresented target genes.
Figure 3A displays dynamic clustering of GO terms related to biological processes for our predicted gene targets of LAGE variants that are overrepresented in SB cases. The top statistically significant gene set ontologies include neural tube closure, transcriptional regulation, neural tube formation, and skeletal system morphogenesis, which point to relevant NTD pathophysiology. Importantly, we did not observe statistical significance with any of the putative target gene sets implicated in controls due to rare noncoding variants. Figure 3B illustrates the core pathway modules perturbed by our predicted TFBS-rSNVs. Among the major pathways are Wnt/ß-catenin signaling, retinoic acid receptors (RAR), and protein kinase A signaling. Further machine learning analysis facilitated prioritization of these variants and genes to ultimately obtain core networks potentially affected in SB cases. This analysis used a shared k-nearest neighbor approach of our overrepresented gene target set to point to brain-specific functional modules, which yielded 5 statistically significant modules encompassing protein transmembrane transport, cilia organization, and central nervous system development (Figure 3C). Representative Q values for each term were calculated using one-sided Fisher’s exact tests and Benjamini-Hochberg corrections to account for multiple tests. By comparing the LAGE target genes from our SB cases with those from controls, we were able to assess and predict which protein classes may be most affected by TFBS-rSNVs, as well as the potential imbalances between cases and controls. Ultimately, this analysis identified gene-specific transcriptional regulators and RNA metabolism proteins that were significantly more affected in SB cases by LAGE TFBS-rSNVs than in controls (Supplemental Figure 2).
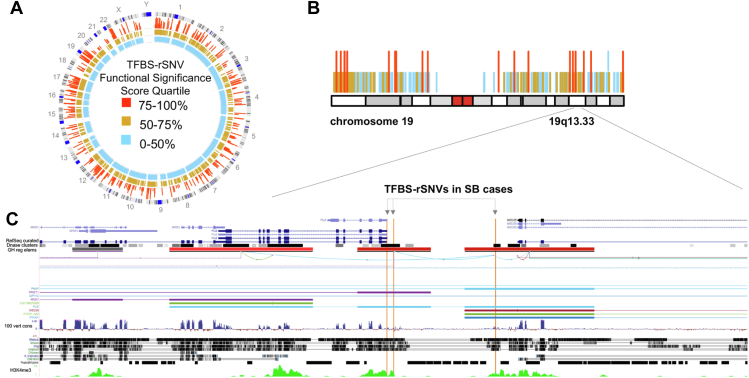
Visual representations of the regulatory variation predicted using our computational approach are shown in Figure 4. Figure 4A illustrates—at the genome-wide level—the TFBS-rSNVs that we detected, as well as their functional significance predictions using DeepSEA. The distribution of these functional scores on chromosome 19 in Figure 4B highlights these variants and their functional impact on the chromosomal level. This example illustrates a cluster of regulatory variants found at this chromosome. Figure 4C displays a higher resolution and nucleotide-level perspective of the LAGE regulatory variation detected near the FUZ gene, which encodes the Fuzzy Planar Cell Polarity protein. The high-impact TFBS-rSNVs, which are positioned to affect CTCF and SP1 binding sites, are displayed with vertical bars, as well as corresponding genomic tracks, including conservation metrics and epigenetic marks. We did not observe LGD variants overlapping FUZ exons in our SB cases; however, several high-impact TFBS-rSNVs predicted to modulate FUZ were identified in our SB cases.
Figure 4.
Genomic view of representative high-impact TFBS-rSNV in spina bifida cases. A. Circos plot depicting the TFBS-rSNVs detected in our cohort along with the deep learning prioritization scores represented as disease impact scores. B. Chromosome 19 and its TFBS-rSNVs are shown. C. Gene browser level visualization from the University of California Santa Cruz Genome Browser depicts rare regulatory variation, as well as conservation and epigenetic marks.
Rare CNVs localize to relevant functional regulatory elements
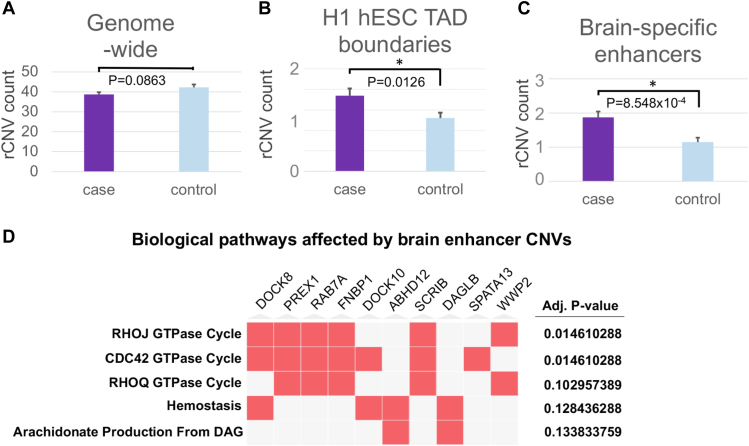
Additionally, we analyzed rCNVs in our cohort, following a high-confidence ensemble approach and pipeline previously described.11 In brief, we deployed a suite of 5 structural variant calling algorithms and used a consensus-based workflow leveraging the read depth, split-read, and read pair information in an optimized fashion. This approach was benchmarked on both real and simulated genomes to maximize both recall and precision for genome-wide deletions and duplications that are at least 1 kb in size. On a genome-wide level, the tally of rare CNVs was not significantly different between SB cases and controls (P = .0863) (Figure 5A). However, when we focused on rCNVs with relevant regulatory features—particularly rCNVs that overlap with TAD domain boundaries and brain-specific enhancer elements—we found statistically significant associations (P = .0126 for TAD boundaries and P = 8.548 × 10−4, for brain enhancer elements) (Figure 5B and C). The TAD boundary enrichment seen in our SB cases was also supported using other relevant TAD boundary coordinates, including H9 hESCs (P = .0145), a progenitor cell type known for their neuronal differentiation capability. We further tested the association between the rCNVs observed in our cohort with TAD boundary elements of SK-N-SH, a human neuroblastoma cell line that exhibits dysregulated neural crest cell differentiation. This association also proved to be significant (P = .0164) and is consistent with the relative stability of TAD boundaries in different cell types
Figure 5.
Rare copy-number variants (<0.01 MAF) localize preferentially to functional regulatory elements in spina bifida. A. A global analysis across the genome did not suggest a statistically significant burden of rare copy-number variation. B and C. Human embryonic stem cell topologically associating domain boundaries and brain-specific enhancer regions, however, did suggest a significant localization pattern. D. Biological pathways affected by rare brain enhancer copy-number variants.
Moreover, using a recently derived multiomic data set for differentially active enhancers during human brain development with clinical relevance,36 we also observed a significant increase in the rCNVs in SB cases that overlapped these putative critical regions compared with controls (P = .0053) (Supplemental Figure 3). This orthogonal data set further supports the brain enhancer association we found in our SB cases. We did not see a similar effect among rare SNVs. That is, the total number of rare SNVs overlapping brain enhancer regions were not significantly different between SB and controls in our study, on average 3807 rare SNVs per genome in SB cases and 3748 rare SNVs per genome in controls (P = .140) (Supplemental Figure 4).
We further investigated the target genes affected by rCNVs that overlap brain enhancer regions in our SB cases by using the same target gene prediction framework as in our TFBS-rSNV analyses in which 107 target genes were identified. This rCNV target gene set, provided in Supplemental Table 2, points to 2 statistically significant biological pathways: RHOJ guanine trinucleotide phosphatase cycle and CDC42 guanine trinucleotide phosphatase cycle. These are known regulators in biological pathways that affect cytoskeletal architecture, gene expression, and progression of the cell cycle. Within the significant pathways identified, genes predicted to be affected by the rCNVs include DOCK8, PREX1, RAB7A, FNBP1, DOCK10, SCRIB, SPATA13, and WWP2. The total set of 226 genes predicted to be affected by enhancer-associated rCNVs shows an enrichment in protein-protein interactions (P = 1.09 × 10−6), suggesting a potential disruption of functional networks on the protein level (Supplemental Figure 5). When restricting our analyses of rCNVs to those located within genomic promoter regions, we detected a statistically significant enrichment in the annotations that include methylation-dependent chromatin silencing and covalent chromatin modifications (Supplemental Figure 6), further underscoring the potential contribution of DNA topology and accessibility.
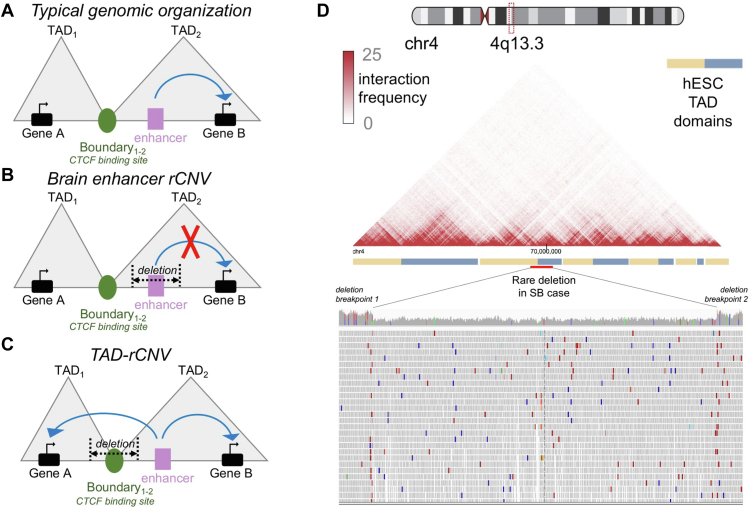
Figure 6 illustrates typical wild-type genomic organization with regard to TAD insulation features (ie, CTCF binding), as well as aberrant organization leading to potential miswiring of enhancer-promoter contacts (Figure 6A-C). Typically, enhancer-promoter contacts are spatially restricted to TAD compartments, as shown in Figure 6A, in which an enhancer modulates the expression only of a gene within the same TAD. We have identified statistically significant enrichments in our SB cases in which rCNVs overlap brain-specific enhancer regions. This scenario is depicted in Figure 6B and shows a plausible mechanism of genomic regulatory variation that can have transcriptional consequences within the particular domain. Figure 6C illustrates another scenario that we observed in our SB cases in which a rCNV disrupts a relevant TAD boundary element, which is often demarcated by CTCF binding. This event may interfere with the insulation properties at the boundaries and further promote ectopic enhancer-promoter contacts across TAD regions. A representative example of this phenomenon is depicted in Figure 6D, which includes a Hi-C matrix and empirically derived interaction frequencies across chromatin regions in hESCs. A rare 30-kb deletion overlaps the TAD boundaries as shown and may fuse these adjacent TADs because of the loss of CTCF insulation. This would promote ectopic regulatory crosstalk among the enhancers and promoters within each TAD and provide a putative mechanism for transcriptional dysfunction underlying SB.
Figure 6.
Topologically associating domain-rare copy-number variants are positioned to perturb 3-dimensional regulatory genomic interactions in spina bifida cases. A-C. Schematic depicting miswiring of enhancer-promoter contacts due to enhancer and boundary element variants. D. Hi-C interaction frequency map of human embryonic stem cell topologically associating domains and shown alongside a rare deletion represented in Integrative Genomics Viewer that was detected in an spina bifida case.
Discussion
In this report, we devised a computational genome-wide approach to interrogate the potential effects that rare variants (both SNVs and CNVs) may have on the regulatory genome of human SB cases. Although rare variation across the entire genome was found equally in both cases and controls, SB cases displayed significant enrichment of rSNVs at the level of known transcription factor binding motifs. Within the set of statistically enriched motifs in SB cases, CTCF, KLF5, and BATF were the most significant. Of particular interest, KLF5 is a crucial transcription factor that controls the expression of multiple downstream target genes and can regulate cell stemness and differentiation, proliferation, and apoptosis.37 In addition, CTCF motifs delineate insulation marks in the 3-dimensional genome and indicate sub-TAD boundaries. CTCF, as a chromatin factor, is increasingly studied in the context of neurological disorders.38 An enrichment in CTCF motif disruption among SB cases compared with controls suggests a potential contribution from a dysregulated sub-TAD genomic organization. TAD fusions and neoTADs can form to contribute to genetic disorders and have been linked with other structural birth defects, such as limb malformations, including brachydactyly and F-syndrome.39 Further implicating the contribution to SB of perturbed 3D genomic organization, our quantitative assessment of TAD-rCNVs detected an enrichment of observed rare CNVs overlapping hESC TAD boundaries in SB cases, consistent with the notion that rCNVs in regulatory motifs can contribute to SB pathophysiology. Our data suggest that a 3-dimensional gene regulatory perspective will inform the understanding of rare CNVs that fall near TAD boundaries.
Based on our untargeted genome-wide approach, the estimated LAGE TFBS-rSNVs in SB cases are predicted to modulate the expression of genes belonging to pathways involved in neural tube closure and associated biological signaling. Several of the putative target genes found here, such as planar cell polarity protein, PRICKLE1, and transcription factor, MAX, were previously associated with increased NTD risk.12 Furthermore, our regulatory pathway analyses from predicted TFBS-rSNV targets converges with the genes and pathway analyses we observed in our likely gene-disrupting (LGD) studies. Several novel genes may warrant further investigation, such as transcription factor, PREX1, for which Prex1−/− mice have recently been shown to display autism spectrum-like features.40
Because regulatory dynamics are often cell type specific and temporally derived, there are inherent limitations to this study. To mitigate these limitations and not overreach in our attempt to link regulatory variants with target genes, we restricted our analyses to only high-confidence enhancers. We also used genomic viewers to visually validate each variant (rSNV and rCNV) that was computationally predicted to alter gene expression. It is likely that emerging tools (eg, Sei, Enformer, EUGENe, and GraphReg)—that will leverage larger training sets of chromatin features and diverse cell types to predict variant impact on gene expression—will allow for the expansion and reinforcement of the SB associations with the genomic regulatory elements observed here.
Multiomic investigations utilizing not only genome sequencing but also RNA-sequencing and Hi-C will be needed to independently correlate aberrant chromatin changes and genomic organization with SB genomic risk. Future functional studies probing the regulatory networks underlying SB will include genetic editing of in vitro stem cell and in vivo animal models to further test this computational interrogation. The integration of these computational and functional assays will no doubt further advance the accuracy of a personalized approach to genetic counseling with regard to SB and other neural tube defect recurrence risks and leverage knowledge of the variant composition in SB affected individuals to inform developmental prognosis and optimize treatment.
Data Availability
Data pertaining to specific variants generated during the downstream analyses, which support the findings of this study, are available upon request to the corresponding author (M.E.R.). Deidentified data will be made available upon request.
Conflict of Interest
Dr Finnell participated in TeratOmic Consulting LLC, a consulting company no longer in existence. Additionally, Dr Finnell serves on the editorial board for the journal Reproductive and Developmental Medicine and receives travel funds to attend editorial board meetings.
Acknowledgments
Material in the present manuscript contains similarities to the doctoral dissertation of P.W. but has not previously been published in a peer-reviewed journal. The authors thank the California Department of Public Health Maternal Child and Adolescent Health Division for providing data. The findings and conclusions herein are the authors’ and do not necessarily represent the official position of the California Department of Public Health.
Funding
This project was supported by the National Institutes of Health P01HD067244 and R01HD111089 (M.E.R., R.H.F.), Weill Cornell, and Qatar Foundation (NPRP4-149-3-049; the Biomedical Research Program at Weill Cornell Medicine-Qatar) (M.E.R., K.S.)
Author Contributions
Conceptualization: P.W., V.A-P., O.E., M.E.R.; Data Curation (including subject enrollment): P.W., M.E.R., R.H.F., G.T., K.S.; Formal Analysis: P.W., V.A-P., O.E., R.H.F., M.E.R.; Funding Acquisition: M.E.R., R.H.F., K.S.; Investigation: all co-authors; Methodology: P.W., V.A-P., O.E., M.E.R.; Project Administration: M.E.R.; Resources: M.E.R., O.E., K.S., R.H.F.; Software: P.W., V.A-P.; Supervision: P.W., V.A-P., O.E., M.E.R., K.S.; Validation: P.W., V.A-P., O.E., G.T.; Visualization: P.W., V.A-P., M.E.R., O.E.; Writing-original draft: P.W., V.A-P., M.E.R.; Writing-review and editing: P.W., V.A-P., M.E.R., R.H.F., O.E., and all co-authors.
Ethics Declaration
Recruitment protocols were approved by Institutional Review Boards (IRBs) in the US (state of California and Stanford University, the University of Texas at Austin, Weill Cornell Medical College-NY, IRB# 1106011749, specific to this study), and the Middle Eastern population receiving their health care in Qatar (Hamad Medical Corporation and Weill Cornell Medical College-Qatar IRB # 15-00065, specific to this study), including informed consent documentation provided in both English and Arabic.
Footnotes
The Article Publishing Charge (APC) for this article was paid by Margaret Elizabeth Ross.
Additional Information
The online version of this article (https://doi.org/10.1016/j.gimo.2024.101894) contains supplemental material, which is available to authorized users.
Contributor Information
Paul Wolujewicz, Email: paw2010@med.cornell.edu.
M. Elizabeth Ross, Email: mer2005@med.cornell.edu.
Additional Information
References
- 1.Crider K.S., Qi Y.P., Yeung L.F., et al. Folic acid and the prevention of birth defects: 30 years of opportunity and controversies. Annu Rev Nutr. 2022;42:423–452. doi: 10.1146/annurev-nutr-043020-091647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schipper M., Posthuma D. Demystifying non-coding GWAS variants: an overview of computational tools and methods. Hum Mol Genet. 2022;31(R1):R73–R83. doi: 10.1093/hmg/ddac198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner T.N., Eichler E.E. The role of de novo noncoding regulatory mutations in neurodevelopmental disorders. Trends Neurosci. 2019;42(2):115–127. doi: 10.1016/j.tins.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lupo P.J., Mitchell L.E., Jenkins M.M. Genome-wide association studies of structural birth defects: a review and commentary. Birth Defects Res. 2019;111(18):1329–1342. doi: 10.1002/bdr2.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Postma A.V., Bezzina C.R., Christoffels V.M. Genetics of congenital heart disease: the contribution of the noncoding regulatory genome. J Hum Genet. 2016;61(1):13–19. doi: 10.1038/jhg.2015.98. [DOI] [PubMed] [Google Scholar]
- 6.Martin-Trujillo A., Patel N., Richter F., et al. Rare genetic variation at transcription factor binding sites modulates local DNA methylation profiles. PLoS Genet. 2020;16(11) doi: 10.1371/journal.pgen.1009189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moyon L., Berthelot C., Louis A., Nguyen N.T.T., Roest Crollius H. Classification of non-coding variants with high pathogenic impact. PLoS Genet. 2022;18(4) doi: 10.1371/journal.pgen.1010191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon J.R., Selvaraj S., Yue F., et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vietri Rudan M., Barrington C., Henderson S., et al. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015;10(8):1297–1309. doi: 10.1016/j.celrep.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lupiáñez D.G., Kraft K., Heinrich V., et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161(5):1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolujewicz P., Aguiar-Pulido V., AbdelAleem A., et al. Genome-wide investigation identifies a rare copy-number variant burden associated with human spina bifida. Genet Med. 2021;23(7):1211–1218. doi: 10.1038/s41436-021-01126-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aguiar-Pulido V., Wolujewicz P., Martinez-Fundichely A., et al. Systems biology analysis of human genomes points to key pathways conferring spina bifida risk. Proc Natl Acad Sci U S A. 2021;118(51) doi: 10.1073/pnas.2106844118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.ICGC/TCGA Pan-cancer Analysis of Whole Genomes Consortium Pan-cancer analysis of whole genomes. Nature. 2020;578(7793):82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Croen L.A., Shaw G.M., Jensvold N.G., Harris J.A. Birth defects monitoring in California: a resource for epidemiological research. Paediatr Perinat Epidemiol. 1991;5(4):423–427. doi: 10.1111/j.1365-3016.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 16.Kumar P., Al-Shafai M., Al Muftah W.A., et al. Evaluation of SNP calling using single and multiple-sample calling algorithms by validation against array base genotyping and Mendelian inheritance. BMC Res Notes. 2014;7:747. doi: 10.1186/1756-0500-7-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H., Handsaker B., Wysoker A., et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43(1110):11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLaren W., Gil L., Hunt S.E., et al. The Ensembl variant effect predictor. Genome Biol. 2016;17(1):122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geoffroy V., Herenger Y., Kress A., et al. AnnotSV: an integrated tool for structural variations annotation. Bioinformatics. 2018;34(20):3572–3574. doi: 10.1093/bioinformatics/bty304. [DOI] [PubMed] [Google Scholar]
- 22.Collins R.L., Brand H., Karczewski K.J., et al. A structural variation reference for medical and population genetics. Nature. 2020;581(7809):444–451. doi: 10.1038/s41586-020-2287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacDonald J.R., Ziman R., Yuen R.K.C., Feuk L., Scherer S.W. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42(Database issue):D986–D992. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belyeu J.R., Chowdhury M., Brown J., et al. Samplot: a platform for structural variant visual validation and automated filtering. Genome Biol. 2021;22(1):161. doi: 10.1186/s13059-021-02380-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson J.T., Thorvaldsdóttir H., Winckler W., et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Funk C.C., Casella A.M., Jung S., et al. Atlas of transcription factor binding sites from ENCODE DNase hypersensitivity data across 27 tissue types. Cell Rep. 2020;32(7) doi: 10.1016/j.celrep.2020.108029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leporcq C., Spill Y., Balaramane D., Toussaint C., Weber M., Bardet A.F. TFmotifView: a webserver for the visualization of transcription factor motifs in genomic regions. Nucleic Acids Res. 2020;48(W1):W208–W217. doi: 10.1093/nar/gkaa252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou J., Troyanskaya O.G. Predicting effects of noncoding variants with deep learning-based sequence model. Nat Methods. 2015;12(10):931–934. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fishilevich S., Nudel R., Rappaport N., et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford) 2017;2017:bax028. doi: 10.1093/database/bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krishnan A., Zhang R., Yao V., et al. Genome-wide prediction and functional characterization of the genetic basis of autism spectrum disorder. Nat Neurosci. 2016;19(11):1454–1462. doi: 10.1038/nn.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47(W1):W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuleshov M.V., Jones M.R., Rouillard A.D., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xin Z., Cai Y., Dang L.T., et al. MonaGO: a novel gene ontology enrichment analysis visualisation system. BMC Bioinformatics. 2022;23(1):69. doi: 10.1186/s12859-022-04594-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ben-Ari Fuchs S., Lieder I., Stelzer G., et al. GeneAnalytics: an integrative gene set analysis tool for next generation sequencing, RNAseq and microarray data. OMICS. 2016;20(3):139–151. doi: 10.1089/omi.2015.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stilianoudakis S.C., Marshall M.A., Dozmorov M.G. preciseTAD: a transfer learning framework for 3D domain boundary prediction at base-pair resolution. Bioinformatics. 2022;38(3):621–630. doi: 10.1093/bioinformatics/btab743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yousefi S., Deng R., Lanko K., et al. Comprehensive multi-omics integration identifies differentially active enhancers during human brain development with clinical relevance. Genome Med. 2021;13(1):162. doi: 10.1186/s13073-021-00980-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo Y., Chen C. The roles and regulation of the KLF5 transcription factor in cancers. Cancer Sci. 2021;112(6):2097–2117. doi: 10.1111/cas.14910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janowski M., Milewska M., Zare P., Pękowska A. Chromatin alterations in neurological disorders and strategies of (epi)genome rescue. Pharmaceuticals (Basel) 2021;14(8):765. doi: 10.3390/ph14080765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang F., Lupski J.R. Non-coding genetic variants in human disease. Hum Mol Genet. 2015;24(R1):R102–R110. doi: 10.1093/hmg/ddv259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo D., Yang X., Shi L. Rho GTPase regulators and effectors in autism spectrum disorders: animal models and insights for therapeutics. Cells. 2020;9(4):835. doi: 10.3390/cells9040835. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data pertaining to specific variants generated during the downstream analyses, which support the findings of this study, are available upon request to the corresponding author (M.E.R.). Deidentified data will be made available upon request.