Abstract
To facilitate detection of glycoprotein K (gK) specified by herpes simplex virus, a 12-amino-acid epitope tag was inserted within gK domain III. Recombinant virus gKprotC-DIII, expressing the tagged gK, was isolated. This virus formed wild-type plaques and replicated as efficiently as the wild-type KOS virus in Vero cells. Anti-protein C MAb detected high-mannose and Golgi complex-dependent glycosylated gK within cells as well as on purified virions. The gK-null virus ΔgK (gK−/−) entered Vero cells substantially more slowly than the wild-type KOS (gK+/+), while ΔgK virus grown in complementing VK302 cells (gK−/+) entered with entry kinetics similar to those of the KOS virus.
Herpes simplex virus type 1 (HSV-1) specifies at least 12 glycoproteins, gB, gC, gD, gE, gG, gH, gI, gJ, gK, gL, gM, and gN, that are expressed during a productive viral infection. These glycoproteins function in pH-independent virus entry via fusion of the viral envelope with cellular membranes, cell-to-cell spread, egress of infectious virion particles, and virus-induced cell-to-cell fusion. (26, 37, 42, 43). Mutations that cause extensive virus-induced cell fusion, syncytia, can arise in at least four different regions of the viral genome, including the UL20 gene (1, 24), the UL24 gene (20, 39), the UL27 gene encoding glycoprotein B (gB) (5, 30), and the UL53 gene coding for glycoprotein K (gK) (2, 7, 32, 38). However, syncytial mutations (syn) in the UL53 (gK) gene are more frequently isolated than syncytium mutations in any other genes (2, 3, 7, 8, 32, 33, 35, 38).
HSV-1 gK is encoded by the UL53 open reading frame (ORF) (7, 25), and it has characteristics of a glycosylated membrane protein, including an N-terminal signal sequence, two potential sites for N-glycosylation 10 amino acids apart (7, 33), and several hydrophobic domains (7). Antisera generated in rabbits against gK-specific peptides indicated that gK exists as a single 40-kDa protein species in infected cells (17). In contrast, gK translated in vitro had an apparent molecular mass of 36 kDa, and N-linked glycosylation occurred in the first 112 residues of the protein, consistent with glycosylation at amino acids 48 and 58 (34). Initially, gK was predicted to have four transmembrane regions (7); however, experiments with in vitro-translated gK in the presence of microsomal membranes suggested that gK contained three instead of four membrane-spanning regions (27). This topological orientation placed all syn mutations within the proposed ectodomains of gK (10, 27).
Mutants that are deficient in gK expression have been isolated and characterized for several alphaherpesviruses. These studies have indicated that gK is important in virion morphogenesis and egress (10, 18, 21, 22, 28). Deletion of gK resulted in a small-plaque phenotype, reduced virus yield, and a restriction in the ability of virus to be translocated from the cytoplasm to the extracellular space. Moreover, at least for pseudorabies virus (PRV), there appeared to be a role for gK in preventing reinfection of cells (22).
Despite the established role of gK in membrane fusion, initial characterization of gK localization indicated that HSV-1 gK was retained within the endoplasmic reticulum (ER) and nuclear membranes (19). The inability of gK to be transported to the Golgi complex and, subsequently, to cell surfaces complicated the elucidation of the role gK could play in mediating cell-to-cell fusion. Furthermore, contrary to studies with other alphaherpesviruses, including PRV and varicella-zoster virus (VZV) (22, 28), HSV-1 gK was thought not to be a structural component of virion particles (19). The absence of gK from virions was difficult to reconcile with studies that had indicated that HSV-1 virions which specified mutations in gK exhibited delayed entry kinetics (31). Due to limitations in the ability of gK peptide antisera to detect HSV-1 gK, we generated recombinant viruses that specified protein C (protC) epitope tags within gK. Here we show that HSV-1 gK, like gK of other alphaherpesviruses, is a structural component of purified virions. Moreover, gK exists on purified virions and within cells as a Golgi complex-dependent glycosylated species.
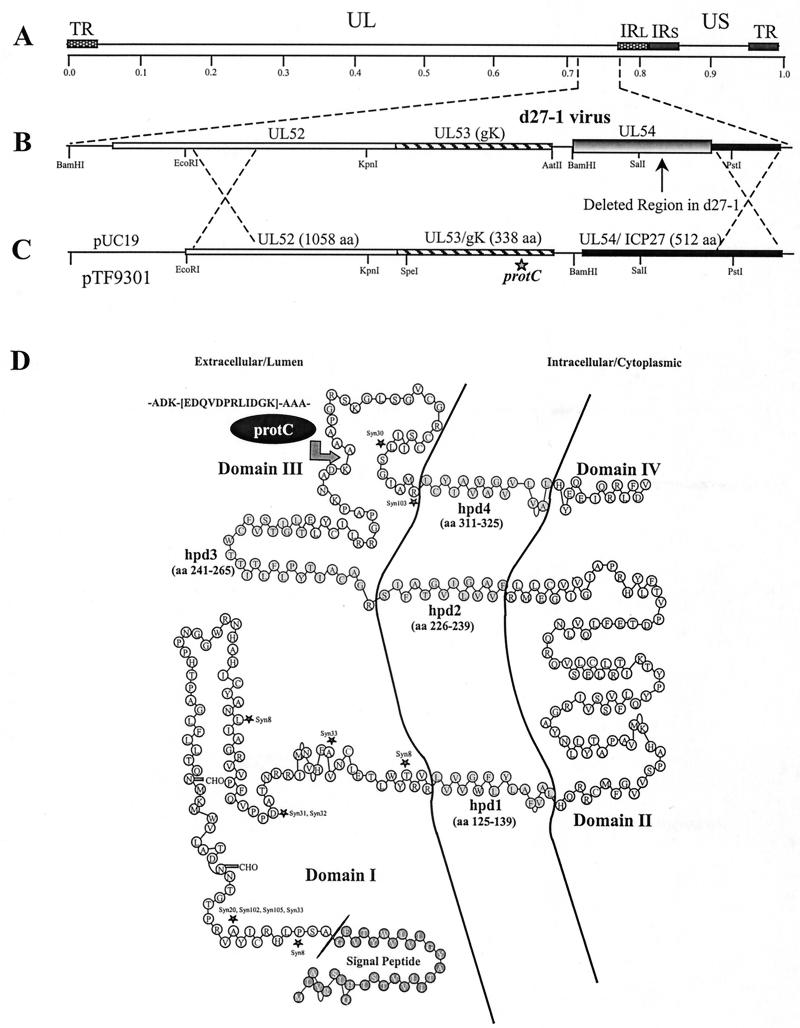
Construction and characterization of HSV-1 recombinant virus gKprotC-DIII expressing gK containing an in-frame insertion of a 12-amino-acid protC tag.
Over the past 20 years, different laboratories have attempted to generate monospecific antibodies and monoclonal antibodies (MAbs) against gK to facilitate its detection. To improve detection of gK, the 12-amino-acid protein C-derived epitope tag was inserted in-frame within gK domain III at a site predicted not to significantly affect the secondary structure of gK (Fig. 1D). The recombinant gK gene coding for the epitope-tagged gK was constructed using PCR-based splice-overlap extension methodology as described previously (6, 10, 11) and cloned into plasmid pSJ1723, generating plasmid pTF9301 (Fig. 1C). This plasmid contained gK-flanking sequences corresponding to the UL52 and UL54 (ICP27) genes to facilitate homologous recombination with viral DNA (21). Recombinant virus gKprotC-DIII was constructed by rescuing the mutant virus d27-1(KOS) (36), which has a lethal deletion within the UL54 gene specifying the immediate-early protein ICP27 as shown previously (9, 10, 21) (Fig. 1A to C). Putative recombinant virus isolates were plaque purified and tested by PCR and DNA sequencing for the presence of contaminating d27-1 virus and the engineered epitope-tagged gK (not shown).
FIG. 1.
Construction of recombinant virus gKprotC-DIII, specifying gK containing a protein C epitope tag. (A) The top line represents the prototypic arrangement of the HSV-1 genome with the unique long (UL) and unique short (US) regions flanked by the terminal repeat (TR) and internal repeat (IR) regions. (B) Shown below is the region of the mutant virus HSV-1 d27-1 genome (between map units 0.7 and 0.8) containing the UL52, UL53, and the partially deleted UL54 open reading frames (shaded and boxed regions of the UL54 gene pointed to by an arrow) with relevant restriction endonuclease sites. (C) Plasmid construct, pTF9301, containing the recombinant gK-protC gene and flanking UL52 and UL54 sequences used to generate recombinant virus gKprotC-DIII. (D) Schematic model of the predicted secondary structure of gK (10). The predicted putative hydrophobic domains (hpd) of gK that transverse the membrane are shown embedded within the membrane. The arrow points to the site of the protC epitope tag insertion. The primary structure of the epitope tag is shown. Known syncytial mutations are denoted by asterisks.
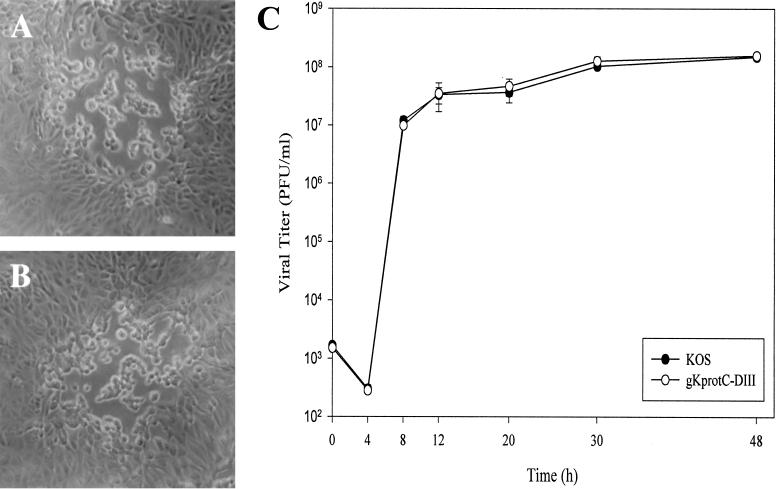
For analysis of virus spread and replication, Vero cells were infected with serial dilutions of wild-type HSV-1 (KOS) and gKprotC-DIII viruses, overlaid with medium containing 1% methyl cellulose, and incubated at 37°C for 48 h. Viral plaques were visualized by phase-contrast microscopy and photographed. Recombinant virus gKprotC-DIII produced viral plaques that were morphologically similar to KOS plaques (Fig. 2A and B).
FIG. 2.
Comparison of plaque morphology and replication characteristics of gKprotC-DIII and KOS viruses. Comparison of virus plaque morphologies formed on Vero cells at 48 h postinfection. (A) KOS. (B) gKprotC-DIII. (C) Time-dependent kinetics of infectious virus production after infection of Vero cells at an MOI of 5 and incubation at 37°C. The graph depicts one of three separate experiments with similar results. Each separate experiment was repeated in triplicate to obtain standard deviations.
For analysis of one-step growth kinetics, each virus at a multiplicity of infection (MOI) of 5 was adsorbed to approximately 8 × 105 Vero cells at 4°C for 1 h. Thereafter, warm medium was added, and virus was allowed to penetrate for 2 h at 37°C. Any remaining extracellular virus was inactivated by low-pH treatment (0.1 M glycine, pH 3.0). Cells and supernatants were harvested immediately (0 h) or after 4, 8, 12, 20, 30, or 48 h of incubation. Virus titers were determined by titration on Vero cells. The gKprotC-DIII virus replicated as efficiently as the KOS parental strain in Vero cells (Fig. 2C). These results indicated that insertion of the 12-amino-acid protC epitope tag within gK domain III did not adversely affect the structure and function of gK with regard to virus replication and cell-to-cell spread.
Detection and characterization of gK specified by gKprotC-DIII using anti-protC MAb.
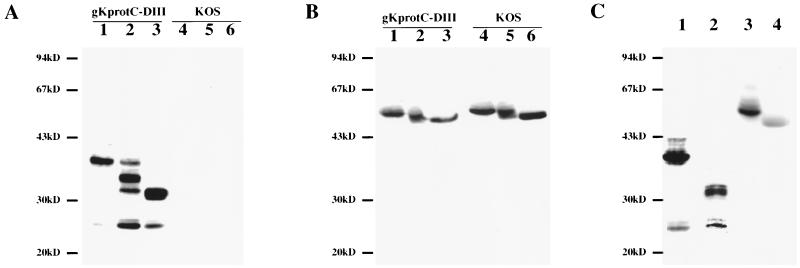
Subconfluent Vero cell monolayers were infected with either gKprotC-DIII or KOS at an MOI of 5. Forty-eight hours postinfection, cells were collected by low-speed centrifugation, washed with Tris-buffered saline (TBS), and lysed at room temperature for 15 min in mammalian protein extraction reagent (MPER) supplemented with a cocktail of protease inhibitors (Invitrogen-Life Technologies, Carlsbad, Calif.). Insoluble cell debris was pelleted, and samples were electrophoretically separated by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes (9). Blots were probed overnight with anti-protC MAb HPC-4 at 1:50 dilution (ATCC CRL HB-9892). Subsequently, blots were incubated for 1 h with a peroxidase-conjugated secondary antibody at a 1:50,000 dilution and visualized on x-ray film by chemiluminescence (Pierce Chemical, Rockford, Ill.) (9, 12). All antibody dilutions and buffer washes were performed in TBS supplemented with 0.135 M CaCl2 and 0.11 M MgCl2.
MAb HPC-4 detected gK protein species ranging from approximately 36 to 43 kDa, with the predominant species migrating with an apparent molecular mass of approximately 36 kDa. Additional minor protein species were detected with molecular masses ranging from 36 to 43 kDa, as well as a protein species with an apparent molecular mass of 25 kDa. In contrast, antibody HPC-4 did not react against extracts derived from KOS-infected cells (Fig. 3A). Similar to other herpesvirus glycoproteins, these results show that the protC-tagged gK exists as multiple protein species in virus-infected cell extracts.
FIG. 3.
Characterization of synthesis and processing of protC-tagged gK specified by gKprotC-DIII. Immunoblots of gKprotC-DIII- (lanes 1 to 3) and KOS (lanes 4 to 6)-infected cell extracts reacted with either anti-protC MAb HPC-4 (A) or anti-gD MAb 1103 (B). Cellular extracts were treated with Endo-H (lanes 2 and 5), PNGase-F (lanes 3 and 6), or mock treated (lanes 1 and 4). (C) Cellular extracts obtained from Vero cells infected with gKprotC-DIII in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of TM were probed with either anti-protC MAb (lanes 1 and 2) or anti-gD MAb (lanes 3 and 4).
The predicted molecular mass of the unglycosylated gK protein backbone (308 amino acids after signal peptide cleavage) is approximately 31 kDa; however, after addition of the protC tag, the molecular mass should be approximately 32.5 kDa. Thus, the gK-related species with molecular masses of 36 to 43 kDa represent glycosylated gK derivatives. The 25-kDa protein could be either a proteolytically processed peptide or a previously predicted gK-related protein, which may be produced by an alternate transcript utilizing an initiation codon within the UL53 ORF (29).
Previous investigations have shown that HSV-1 gK contained N-linked carbohydrate species (17, 34). To characterize the carbohydrate character of gK, Vero cell monolayers were infected with gKprotC-DIII at an MOI of 5 and incubated in the presence or absence of 5 μg of tunicamycin (TM) per ml. Cellular extracts were prepared at 48 h postinfection and analyzed by SDS-PAGE and immunoblot analysis using either anti-protC antibody, HPC-4, or anti-gD antibody 1103 (Rumbaugh-Goodwin Institute, Plantation, Fla.) (Fig. 3C). In the presence of TM, gK had an apparent molecular mass of approximately 32 kDa, which corresponded to the predicted molecular mass of the unglycosylated gK after removal of the signal peptide (Fig. 3C, lanes 1 and 2). As a control, a parallel blot was incubated with anti-gD antibody (1:2,000). As expected, TM reduced the apparent molecular mass of gD from 59 to 50 kDa (Fig. 3C, lanes 3 and 4) (41).
To further investigate the glycosylation of gK, infected cellular extracts were mock treated or incubated in the presence of either endoglycosidase H (Endo-H) or peptide:N-glycosidase F (PNGase F). N-linked carbohydrates were removed from proteins contained within infected cell lysates by a modified protocol of the manufacturer's instructions and essentially as described previously (22). Briefly, virally infected cells were lysed at 42°C for 1 h in MPER that contained a protease inhibitor cocktail, 0.5% SDS, and 1% β-mercaptoethanol. Samples were subsequently incubated for 18 h at 42°C either with 50 mM sodium citrate (pH 5.5) and 100 U of Endo-H (NEB Labs, Beverly, Mass.) for Endo-H digestions, with 50 mM sodium phosphate (pH 7.5), 1% NP-40, and 20 U of PNGase-F (NEB Labs) for PNGase-F digestions, or without enzyme prior to SDS-PAGE and Western blot analysis.
PNGase-F treatment, which cleaves off all N-linked carbohydrate chains from the protein backbone, reduced the apparent molecular mass of gK from 36 to 32 kDa (Fig. 3A, lane 3), in agreement with gK produced in the presence of TM (Fig. 3C, lane 2). Endo-H treatment, which is specific for digestion of high-mannose carbohydrate chains, resulted in the production of multiple protein species migrating with apparent molecular masses of 36, 34, 32, and 25 kDa. The 34-kDa band was the predominant species, indicating that the majority of gK expressed in infected cells contains high-mannose carbohydrate chains (Fig. 3A, lane 2). As controls for the PNGase F and Endo-H activities, the effect of these enzymes on gD specified by either gKprotC-DIII or KOS was also assessed. Both enzymes reduced the apparent molecular mass of gD similar to the reduction exhibited by gK, confirming that gD contains N-linked carbohydrates and that gD is present as both Endo-H-sensitive and Endo-H-resistant forms (Fig. 3B). These results are consistent with previous reports that suggested gK is expressed predominantly as a high-mannose glycosylated species (19). However, a portion of the expressed gK was Endo-H resistant even after an 18-h incubation with a high concentration of Endo-H, demonstrating that gK is transported to the Golgi apparatus and undergoes further processing of the high-mannose precursor carbohydrate moieties.
gK is a structural component of purified virions.
Previous reports suggested that HSV-1 gK might not be a structural component of virions (19). However, studies with PRV and VZV demonstrated that gK was present in purified virions. To ascertain whether gK was a structural component of HSV-1 virions, we investigated whether gK specified by gKprotC-DIII could be detected in purified virions.
KOS and gKprotC-DIII virions derived from infected Vero cell supernatants were purified by two consecutive sucrose density gradients as described previously (15, 16, 23). Specifically, roller bottles of Vero cells were infected at an MOI of 0.1 PFU/cell. Forty-eight hours postinfection, supernatants containing extracellular virus were harvested, and cellular debris was removed by centrifugation at 10,000 × g for 15 min. Virus was concentrated by pelleting through a 10% sucrose–TBS cushion at 100,000 × g for 1.5 h. Virus pellets were resuspended in TBS, layered onto a 60 to 10% continuous sucrose gradient, and sedimented at 100,000 × g for 2 h. Purified virus was collected from the light-scattering phase midway through the gradient, diluted threefold in TBS, and layered on a 10%–30%–60% sucrose–TBS discontinuous step gradient. Purified virion preparations were collected by side puncture at the 30%–60% interface, diluted in 30 ml of TBS, layered onto a 10% sucrose–TBS cushion, and centrifuged at 100,000 × g for 1.5 h to concentrate the virion preparations. Viral proteins were extracted and processed in MPER-protease inhibitor cocktail as described for cell lysates.
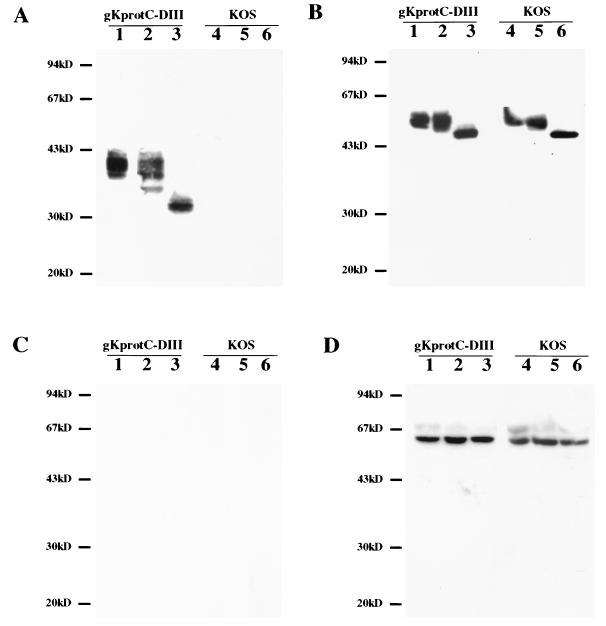
Antibody HPC-4 detected gK in gKprotC-DIII purified virions, while it did not react with KOS purified virions (Fig. 4A). Treatment of purified virions with PNGase-F caused the appearance of a gK species migrating as a 32-kDa band similar to that observed in PNGase-F-treated cellular extracts (compare Fig. 3A, lane 3, and Fig. 4A, lane 3). The 25-kDa species observed in cellular extracts was not detected in immunoblots of purified virion preparations. In contrast to the results obtained with Endo-H treatment of infected cellular extracts, the majority of gK detected in purified virions was resistant to enzymatic digestion, indicating that it contained peripheral sugars added at the Golgi apparatus (Fig. 4A, lane 2).
FIG. 4.
Detection and characterization of protC-tagged gK expressed on purified virions. Immunoblots of gKprotC-DIII (lanes 1 to 3) or KOS (lanes 4 to 6) purified virion preparations reacted with anti-protC MAb HPC-4 (A), anti-gD MAb 1103 (B), and anti-ICP27 MAb 1113 (C). Purified virion preparations were treated with Endo-H (lanes 2 and 5), PNGase-F (lanes 3 and 6), or mock treated (lanes 1 and 4). (D) Cellular extracts from Vero cells infected with either gKprotC-DIII (lanes 1 to 3) or KOS (lanes 4 to 6) were reacted with anti-ICP27 MAb. Cellular extracts were treated with Endo-H (lanes 2 and 5), PNGase-F (lanes 3 and 6), or mock treated (lanes 1 and 4).
For control purposes, parallel immunoblots were assayed for the presence of gD and ICP27. As expected, gD specified by KOS and gKprotC-DIII virions was found to contain N-linked, Golgi complex-dependent sugars (Fig. 4B). HSV-1 (UL54) ICP27 is a known nonstructural protein, which is expressed in infected cells but is not present in purified virions (45). As expected, ICP27 was detected in gKprotC-DIII- and KOS-infected cell extracts using anti-ICP27 antibody 1113 (Rumbaugh-Goodwin Institute); however, it was not present in purified virions (Fig. 4C and D).
HSV-1 gK enhances virion entry.
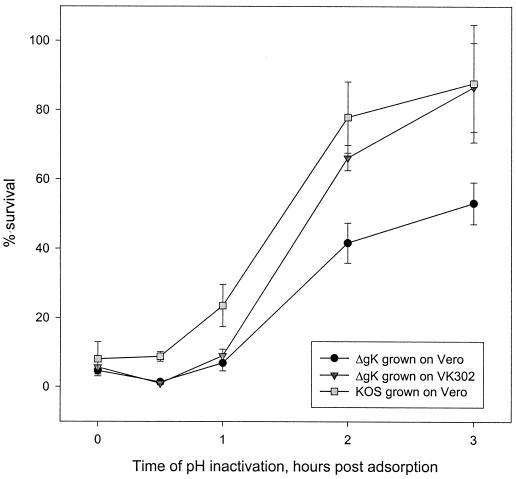
Based on the finding that gK was present in purified virions and the fact that syncytium mutations in gK alter virus entry kinetics (31) as well as cause substantial virus-induced membrane fusion (2, 8), we examined the penetration kinetics of the ΔgK virus in comparison to its parental strain, KOS. Subconfluent monolayers of VK302 cells in six-well culture dishes were infected at 4°C for 1 h with approximately 300 PFU of KOS or ΔgK grown in either Vero (gK−/−) or VK302 (gK−/+) cells. The inoculum was subsequently removed, warm medium (34°C) was added, and the cultures were shifted to 34°C to allow virus penetration. Immediately thereafter (0 h) and at 30, 60, 120, and 180 min, remaining extracellular virus was inactivated by treatment with low-pH buffer (0.1 M glycine, pH 3.0). Cells were washed three times with phosphate-buffered saline (PBS) and overlaid with 1% methyl cellulose in Dulbecco's modified Eagle's medium. Virus plaques were counted at 48 h postinfection, and the percentage of PFU surviving low-pH inactivation compared to PBS-treated controls was calculated. The kinetics of ΔgK (gK−/−) virion entry was substantially reduced in comparison to the KOS (gK+/+) strain. Furthermore, ΔgK virions produced in the complementing cell line, VK302 (gK−/+), which is transformed with the gK gene (18), entered substantially faster, exhibiting entry kinetics similar to that of the KOS virus (Fig. 5).
FIG. 5.
Penetration kinetics of KOS and ΔgK viruses into VK302 (Vero) cells. The penetration kinetics of KOS virus grown on Vero cells and ΔgK virus grown on either Vero or VK302 cells were obtained by determining the percentage of PFU surviving low-pH treatment relative to PBS-treated controls at different times postadsorption. Mean values and standard deviations of three independent experiments are shown.
Considering the importance of gK in membrane fusion phenomena, it is not surprising that gK was found to be glycosylated by the Golgi apparatus as well as being a structural component of the virion that functions to enhance virion entry. In this regard, gK localizes where it is capable of participating in multiple membrane fusion events during virion morphogenesis, virus-induced cell fusion, and virion penetration. ΔgK virions have a major defect in virion egress, which causes the accumulation of virions within vesicles in the cytoplasm of infected cells. The origin of these vesicles is not yet known; however, morphological data suggest that they may be derived from the Golgi complex. If this is true, it may mean that gK functions in a Golgi complex-dependent pathway involved in either reenvelopment at the Golgi complex according to the deenvelopment/reenvelopment model (4, 13, 14, 40, 44) or post-Golgi complex transport of virions to extracellular spaces.
It is important to note that lack of gK is not absolutely lethal for virus replication in cell culture, although it does reduce virus titers by more than 100-fold and substantially reduced the kinetics of virus entry into Vero cells. Importantly, experimental infections in mice using the eye route indicate that gK may be required for virus replication, spread, and neurovirulence in vivo (unpublished data). Therefore, it is possible that the true functions of gK may not be discernible in cell culture systems, such as Vero cells, that have been selected for the optimum replication of HSV virions. In this regard, more sensitive cell culture systems and experimental animal studies may be required to delineate the functions of gK.
Acknowledgments
This work was supported by a grant from the National Institute of Allergy and Infectious Diseases (AI43000) to K.G.K. We acknowledge support from the School of Veterinary Medicine, Louisiana State University.
REFERENCES
- 1.Baines J D, Ward P L, Campadelli-Fiume G, Roizman B. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J Virol. 1991;65:6414–6424. doi: 10.1128/jvi.65.12.6414-6424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bond V C, Person S. Fine structure physical map locations of alterations that affect cell fusion in herpes simplex virus type 1. Virology. 1984;132:368–376. doi: 10.1016/0042-6822(84)90042-4. [DOI] [PubMed] [Google Scholar]
- 3.Bond V C, Person S, Warner S C. The isolation and characterization of mutants of herpes simplex virus type 1 that induce cell fusion. J Gen Virol. 1982;61:245–254. doi: 10.1099/0022-1317-61-2-245. [DOI] [PubMed] [Google Scholar]
- 4.Browne H, Bell S, Minson T, Wilson D W. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J Virol. 1996;70:4311–4316. doi: 10.1128/jvi.70.7.4311-4316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bzik D J, Fox B A, DeLuca N A, Person S. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology. 1984;137:185–190. doi: 10.1016/0042-6822(84)90022-9. [DOI] [PubMed] [Google Scholar]
- 6.Chouljenko V, Jayachandra S, Rybachuk G, Kousoulas K G. Efficient long-PCR site-specific mutagenesis of a high GC template. BioTechniques. 1996;21:472–474. doi: 10.2144/96213st05. , 476–478, 480. [DOI] [PubMed] [Google Scholar]
- 7.Debroy C, Pederson N, Person S. Nucleotide sequence of a herpes simplex virus type 1 gene that causes cell fusion. Virology. 1985;145:36–48. doi: 10.1016/0042-6822(85)90199-0. [DOI] [PubMed] [Google Scholar]
- 8.Dolter K E, Ramaswamy R, Holland T C. Syncytial mutations in the herpes simplex virus type 1 gK (UL53) gene occur in two distinct domains. J Virol. 1994;68:8277–8281. doi: 10.1128/jvi.68.12.8277-8281.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foster T P, Chouljenko V N, Kousoulas K G. Functional characterization of the HveA homolog specified by African green monkey kidney cells with a herpes simplex virus expressing the green fluorescence protein. Virology. 1999;258:365–374. doi: 10.1006/viro.1999.9743. [DOI] [PubMed] [Google Scholar]
- 10.Foster T P, Kousoulas K G. Genetic analysis of the role of herpes simplex virus type 1 glycoprotein K in infectious virus production and egress. J Virol. 1999;73:8457–8468. doi: 10.1128/jvi.73.10.8457-8468.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster T P, Melancon J M, Kousoulas K G. An α-helical domain within the carboxyl terminus of herpes simplex virus type 1 (HSV-1) glycoprotein B (gB) is associated with cell fusion and resistance To heparin inhibition of cell fusion. Virology. 2001;287:18–29. doi: 10.1006/viro.2001.1004. [DOI] [PubMed] [Google Scholar]
- 12.Foster T P, Rybachuk G V, Kousoulas K G. Expression of the enhanced green fluorescent protein by herpes simplex virus type 1 (HSV-1) as an in vitro or in vivo marker for virus entry and replication. J Virol Methods. 1998;75:151–160. doi: 10.1016/s0166-0934(98)00107-4. [DOI] [PubMed] [Google Scholar]
- 13.Gershon A A, Sherman D L, Zhu Z, Gabel C A, Ambron R T, Gershon M D. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J Virol. 1994;68:6372–6390. doi: 10.1128/jvi.68.10.6372-6390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Granzow H, Weiland F, Jons A, Klupp B G, Karger A, Mettenleiter T C. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J Virol. 1997;71:2072–2082. doi: 10.1128/jvi.71.3.2072-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Handler C G, Cohen G H, Eisenberg R J. Cross-linking of glycoprotein oligomers during herpes simplex virus type 1 entry. J Virol. 1996;70:6076–6082. doi: 10.1128/jvi.70.9.6076-6082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Handler C G, Eisenberg R J, Cohen G H. Oligomeric structure of glycoproteins in herpes simplex virus type 1. J Virol. 1996;70:6067–6070. doi: 10.1128/jvi.70.9.6067-6070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutchinson L, Goldsmith K, Snoddy D, Ghosh H, Graham F L, Johnson D C. Identification and characterization of a novel herpes simplex virus glycoprotein, gK, involved in cell fusion. J Virol. 1992;66:5603–5609. doi: 10.1128/jvi.66.9.5603-5609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutchinson L, Johnson D C. Herpes simplex virus glycoprotein K promotes egress of virus particles. J Virol. 1995;69:5401–5413. doi: 10.1128/jvi.69.9.5401-5413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutchinson L, Roop-Beauchamp C, Johnson D C. Herpes simplex virus glycoprotein K is known to influence fusion of infected cells, yet is not on the cell surface. J Virol. 1995;69:4556–4563. doi: 10.1128/jvi.69.7.4556-4563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobson J G, Martin S L, Coen D M. A conserved open reading frame that overlaps the herpes simplex virus thymidine kinase gene is important for viral growth in cell culture. J Virol. 1989;63:1839–1843. doi: 10.1128/jvi.63.4.1839-1843.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jayachandra S, Baghian A, Kousoulas K G. Herpes simplex virus type 1 glycoprotein K is not essential for infectious virus production in actively replicating cells but is required for efficient envelopment and translocation of infectious virions from the cytoplasm to the extracellular space. J Virol. 1997;71:5012–5024. doi: 10.1128/jvi.71.7.5012-5024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klupp B G, Baumeister J, Dietz P, Granzow H, Mettenleiter T C. Pseudorabies virus glycoprotein gK is a virion structural component involved in virus release but is not required for entry. J Virol. 1998;72:1949–1958. doi: 10.1128/jvi.72.3.1949-1958.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kousoulas K G, Bzik D J, DeLuca N, Person S. The effect of ammonium chloride and tunicamycin on the glycoprotein content and infectivity of herpes simplex virus type 1. Virology. 1983;125:468–474. doi: 10.1016/0042-6822(83)90217-9. [DOI] [PubMed] [Google Scholar]
- 24.MacLean C A, Efstathiou S, Elliott M L, Jamieson F E, McGeoch D J. Investigation of herpes simplex virus type 1 genes encoding multiply inserted membrane proteins. J Gen Virol. 1991;72:897–906. doi: 10.1099/0022-1317-72-4-897. [DOI] [PubMed] [Google Scholar]
- 25.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 26.Mettenleiter T C. Aujeszky's disease (pseudorabies) virus: the virus and molecular pathogenesis—state of the art. Vet Res. 2000;31:99–115. doi: 10.1051/vetres:2000110. [DOI] [PubMed] [Google Scholar]
- 27.Mo C, Holland T C. Determination of the transmembrane topology of herpes simplex virus type 1 glycoprotein K. J Biol Chem. 1997;272:33305–33311. doi: 10.1074/jbc.272.52.33305. [DOI] [PubMed] [Google Scholar]
- 28.Mo C, Suen J, Sommer M, Arvin A. Characterization of varicella-zoster virus glycoprotein K (open reading frame 5) and its role in virus growth. J Virol. 1999;73:4197–4207. doi: 10.1128/jvi.73.5.4197-4207.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moyal M, Becker Y. Analysis of the transcription pattern of HSV-1 UL52 and UL53 genes. Virus Genes. 1994;9:15–21. doi: 10.1007/BF01703431. [DOI] [PubMed] [Google Scholar]
- 30.Pellett P E, Kousoulas K G, Pereira L, Roizman B. Anatomy of the herpes simplex virus 1 strain F glycoprotein B gene: primary sequence and predicted protein structure of the wild type and of MAb-resistant mutants. J Virol. 1985;53:243–253. doi: 10.1128/jvi.53.1.243-253.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pertel P E, Spear P G. Modified entry and syncytium formation by herpes simplex virus type 1 mutants selected for resistance to heparin inhibition. Virology. 1996;226:22–33. doi: 10.1006/viro.1996.0624. [DOI] [PubMed] [Google Scholar]
- 32.Pogue-Geile K L, Lee G T, Shapira S K, Spear P G. Fine mapping of mutations in the fusion-inducing MP strain of herpes simplex virus type 1. Virology. 1984;136:100–109. doi: 10.1016/0042-6822(84)90251-4. [DOI] [PubMed] [Google Scholar]
- 33.Pogue-Geile K L, Spear P G. The single base pair substitution responsible for the Syn phenotype of herpes simplex virus type 1, strain MP. Virology. 1987;157:67–74. doi: 10.1016/0042-6822(87)90314-x. [DOI] [PubMed] [Google Scholar]
- 34.Ramaswamy R, Holland T C. In vitro characterization of the HSV-1 UL53 gene product. Virology. 1992;186:579–587. doi: 10.1016/0042-6822(92)90024-j. [DOI] [PubMed] [Google Scholar]
- 35.Read G S, Person S, Keller P M. Genetic studies of cell fusion induced by herpes simplex virus type 1. J Virol. 1980;35:105–113. doi: 10.1128/jvi.35.1.105-113.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rice S A, Knipe D M. Genetic evidence for two distinct transactivation functions of the herpes simplex virus alpha protein ICP27. J Virol. 1990;64:1704–1715. doi: 10.1128/jvi.64.4.1704-1715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2231–2295. [Google Scholar]
- 38.Ruyechan W T, Morse L S, Knipe D M, Roizman B. Molecular genetics of herpes simplex virus. II. Mapping of the major viral glycoproteins and of the genetic loci specifying the social behavior of infected cells. J Virol. 1979;29:677–697. doi: 10.1128/jvi.29.2.677-697.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanders P G, Wilkie N M, Davison A J. Thymidine kinase deletion mutants of herpes simplex virus type 1. J Gen Virol. 1982;63:277–295. doi: 10.1099/0022-1317-63-2-277. [DOI] [PubMed] [Google Scholar]
- 40.Skepper J N, Whiteley A, Browne H, Minson A. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment-deenvelopment-reenvelopment pathway. J Virol. 2001;75:5697–5702. doi: 10.1128/JVI.75.12.5697-5702.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sodora D L, Cohen G H, Eisenberg R J. Influence of asparagine-linked oligosaccharides on antigenicity, processing, and cell surface expression of herpes simplex virus type 1 glycoprotein D. J Virol. 1989;63:5184–5193. doi: 10.1128/jvi.63.12.5184-5193.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spear P G. Entry of alphaherpesviruses into cells. Semin Virol. 1993;4:167–180. [Google Scholar]
- 43.Spear P G. Membrane fusion induced by herpes simplex virus. In: Bentz J, editor. Viral fusion mechanisms. Boca Raton, Fla: CRC Press; 1993. pp. 201–232. [Google Scholar]
- 44.Whealy M E, Robbins A K, Tufaro F, Enquist L W. A cellular function is required for pseudorabies virus envelope glycoprotein processing and virus egress. J Virol. 1992;66:3803–3810. doi: 10.1128/jvi.66.6.3803-3810.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yao F, Courtney R J. Association of ICP0 but not ICP27 with purified virions of herpes simplex virus type 1. J Virol. 1992;66:2709–2716. doi: 10.1128/jvi.66.5.2709-2716.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]