Abstract
Background and introduction
Two and half percent of the Indian population suffer from gallbladder cancer (GBC). The primary factors that lead GBC are associated with mutation of several protooncogenes such as EGFR, ERBB2, Myc, and CCND1 along with dysregulation of several tumor suppressor genes such as SMAD4 and CDKN2A. Bacterial infection caused by S.typhi and H.pylori are also hypothesized to be potential factors driving GBC.
Aims
This study aims to investigate the molecular mechanisms driving the progression of gallbladder adenocarcinoma in Eastern Indian patients. We specifically focussed on analyzing the mutational status of the KRAS gene, examining the amplification of the ERBB2/Her2-neu gene, and evaluating the expression patterns of six dysregulated genes (CCND1, MYC, EGFR, ERBB2/Her2-neu, CDKN2A, SMAD4). Additionally, we assessed the expression status of TGF-beta, the association between bacterial infections (S. Typhi and H. pylori) and GBC, and the impact of single nucleotide polymorphisms in ERBB2/Her2-neu and CCND1 genes within this population.
Methods
Sixty-seven samples from GBC-diagnosed patients, 26 other unrelated GBC samples for validation cohort, and 68 gallstone tissue samples were collected for this study. Genomic DNA from normal as well as tumor tissues were isolated, exon 2 and exon 3 of KRAS gene were amplified along, DNA sequenced and analyzed. KRAS codon 12 mutation was detected by allele specific PCR (ASPCR) method. Amplification of UreC A (coding for urease subunit α), VacA (coding for Vacuolating cytotoxin A) and CagA genes (coding for cytotoxin-associated gene A) in H.pylori were amplified using PCR. Similarly, FlicC (coding for flagellin gene C) in S.typhi was amplified using PCR. The ERBB2/Her2-neu SNP I655V, and CCND1 SNP A870G were analyzed using PCR followed by RFLP. Expression studies of CCND1, Myc, CDKN2A, ERBB2/Her2-neu, EGFR, and SMAD4 genes were measured in GBC tumor tissues by sybr green quantitative RT PCR.
Results
The oncogenes (EGFR and ERBB2/Her2-neu) were statistically significantly overexpressed and the tumor suppressor gene (SMAD4) downregulated in our GBC tumor patient samples. The EGFR and SMAD4 genes were negatively correlated (r = -0.01) in GBC patients and the data is statistically significant and validated through IHC technique. A significant downregulation of TGF-beta had also been observed. Lower frequency (i.e. 11.5%) of KRAS mutation in GBC tumor was observed.
Conclusions
EGFR and SMAD4 expression were found to be negatively correlated in GBC tissue samples. ERBB2 overexpression/amplification was observed in 30% of the GBC samples. We also found a low percentage of GBC samples to show KRAS codon 12 mutation in Indian GBC patient population, as had been previously documented in pancreatic cancers.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12876-024-03485-4.
Keywords: Gallbladder cancer, KRAS mutation, ERBB2 amplification, EGFR-SMAD4 expression correlation, Bacterial infection
Introduction
Gallbladder cancer (GBC) is very frequently reported among the biliary tract malignancies. GBC is often delineated at an advanced stage and has poor prognosis mainly presenting obvious symptoms [1]. GBC have reportedly higher incidences in places like Chile, North India, Korea, Japan and New Mexico of United States. According to GLOBOCAN 2024, the number of new cases in India is 25,999, having a percentage of 2.47 and current risk is 0.24. The numbers of deaths in India are 19,676, having a percentage of 2.74 and current risk is 0.18. GBC ranks 14th among newly reported cancer cases in India and holds a rank of 13th in the number of deaths. The 5-year prevalence rate in India is 31,357, having a proportion of 2.32 (GLOBOCAN, 2018). GBC also has a higher female incidence rate compared to men. The several risk factors concerning GBC include demographic elements, history of gallbladder disease, and environmental exposures [2]. The risk factorial conditions for GBC include chronic gallstones, gallbladder polyps, cholangitis related infections (e.g., Salmonella enterica typhi and H. pylori prevalent in India and Bangladesh), porcelain gallbladder, Mirizzi’s syndrome, and bile reflux [3]. Environmental factors like diet, high capsaicin ingestion, aflatoxins, and vitamin deficiencies have been disputably linked to GBC pathogenesis [4]. In various studies reported, S. typhi was found to colonize the gallbladder resulting in an asymptomatic chronic infection [5]. Various studies revealed various species of H. pylori in tissue and bile samples of the gallbladder. First hypothesis of a potential association between H. pylori infection and gallstone formation was given by [6]. Chile, Bolivia, India, Pakistan, Japan and Korea, which are S. typhi endemic zones have revealed that about 90% of chronically infected carriers are also gallstone patients, which can later develop GBC [5]. The advancement of gallbladder lesion into GBC includes staging such as metaplasia, dysplasia, carcinoma-in-situ and finally invasive carcinoma [4]. Subsequently, the lesion assembles mutations of KRAS. This genetic aberration is believed to be one of the principal reasons driving the lesion to develop into an invasive carcinoma [7]. Various studies report the spectrum of KRAS point mutation to be 3–40% [8]. KRAS mutation at codon-12 is reported to be 8% [9]. Various somatic mutations and amplifications have been observed in the ErbB signalling pathway, mainly consisting of the ERBB/Her-neu and or ERBB2/Her2-neu. ErbB pathway genes are important growth factor receptor genes frequently involved in multiple cancers including GBC [4]. The single nucleotide polymorphism (SNPs) in the human ERBB2/Her2-neu was identified in the transmembrane coding region of the gene at codon 655, encoding isoleucine or valine. In presence of “Val” allele may enhance dimerization of ERBB2/Her2-neu, resulting in increased autophosphorylation, tyrosine kinase activation and subsequently leading to cell transformation. CyclinD1 (CCND1) is a member of D-type cyclin proteins, involved in the regulation of cell cycle progression from G1 to S phase. The common site of SNP has been found at 870 position or codon 242 located in the common splice donor region of exon 4 site with increased expression. The “A” allele produces an alternative transcript, transcript-b, a bigger transcript which does not get spliced at exon 4 intron 4 boundary and has longer half-life, whereas “G” allele produces normal splicing of the exon 5, transcript-a. Transcript-b/A allele is associated with several cancers including colorectal, squamous cell carcinoma of the oesophagus, lung, SSC of head and neck, bladder and cervix [10]. Oncogenes, including EGFR, ERBB2/Her2-neu, Myc, and CCND1 are mostly dysregulated in GBC as have been reported from several Next Generation Sequencing (NGS) studies. Along with this loss of tumor suppressor genes (TSGs) and loss of heterozygosity (LOH) at polymorphic loci are also hallmarks of many different tumors including GBC. Previous studies also reported SMAD4 and CDKN2A to be the most frequently dysregulated TSGs in GBC [11]. Collectively, both TGF-beta signalling and SWI/SNF complex are correlated with MYC expression, and the MYC alteration is mutually exclusive with alterations in TGF and SWI/SNF complex, SMAD4 is the important gene in TGF-beta signalling pathway and acts as TSG in the GBC progression [12]. During the development of dysplasia, intermediate changes were observed including allelic loss at several chromosomal positions, CDKN2A, TSG particularly on the q arm of chromosome 9. GBC continues to pose a challenge in certain geographic locations such as in latin America and Asia including Arab countries [13]. Although it is still unknown, why certain ethnic groups have a higher predisposition to GBC than others, such as in northern and eastern India, where GBC has an extremely high incidence [14, 15]. Our hypothesis sought to establish the existence of huge variance of GBC incidences in different geographical locations across the world. The main reason is due to different kind of genomic alterations developed in GBC patients. The data reported by Kolkata cancer registry noted GBC to be the third most frequent malignancy in eastern Indian females [16]. The KRAS mutation frequency, ERBB2 amplification, and dysregulation of CDKN2A, EGFR, SMAD4, cMYC, CCND1, and ERBB2/Her2-neu genes varies with ethnicity and geographical location of GBC patient population. In addition to that S.typhi and H.pylori infection status also varies in different patient population of GBC. We hypothesized KRAS mutation in the early events in the GBC development. In Indian patient population, very less studies reported regarding these genomic alterations in previous decades, however there is no reports from Eastern Indian region of India.
In our present study, we have investigated on detecting the mutational proportion of KRAS gene in GBC and gallstone disease along with detecting the expression and amplification pattern of ERBB2/Her2-neu gene, with expression pattern of six mostly dysregulated genes, expression status of TGF-beta and status of bacterial infection such as S.Typhi and H.pylori, two SNPs of ERBB2/Her2-neu and CCND1 gene in Indian context in the population of West Bengal in Eastern Indian region.
Patient information and methods
Patients samples collection in the current study
All the tissues investigated in our study were obtained from patients from Kolkata megacity regions and state of West Bengal (one of the Eastern States in India) who had surgical resection between May 2013 to January 2022 at Medical College and Hospital, Kolkata, SSKM Hospital and IPGME&R, Kolkata. Written informed consent for gene expression analysis was received from all the patients before surgery and all the experiments were done according to the “Ethical guidelines for Biomedical Research on Human Participants” published by Indian Council of Medical Research (ICMR) (2006) and was approved by the Institutional Ethics Committee of Indian Statistical Institute (ISI). A systemic 2–5 year follow up was done for overall survival (OS). Tumor and adjacent normal tissue samples were collected during surgery and stored in RNA later solution (Sigma-Aldrich Co. LLC). Histopathological examination and TNM staging were done according to the 8th edition guidelines by American Joint Committee on Cancer (AJCC), by all the associated clinician. We had investigated the tumor purity through histopathological evaluation, and it was found to be > 80% in all samples (10 × imagery by using Leica DM 1000, camera: Leica EC3). We have generated a score for each sample regarding tumor cell percentage by randomly taking 8 images for each slide. The inclusion and exclusion criteria of the patients were considered prior to specimen collection and consenting.
Inclusion criteria of patient samples
The inclusion criteria included collection of only clinically diagnosed GBC samples within the age-limit 30–75 years and should positively be sero-negative and also capable of undergoing surgery procedure.
Exclusion criteria of the patient samples
The exclusion criteria were designed to target GBC subtype, which is a specific subtype of GBC. Hence periampullary adenocarcinomas were excluded and also GBC samples with sero -positivity were excluded. Some other exclusion criterias included were patients with age greater than 75 years and physically and mentally handicapped patients.
Patient Sample Collection Details: A total of 70 gallstone disease (GSD) tissues collected only from Medical College and Hospital, Kolkata, were included in the study. Demographic and clinico-pathological data were also collected during the tissue sample collection. Another 67 patients in a different discovery set including clinically and histologically diagnosed with GBC from SSKM and IPGME&R, and Medical College and Hospital, Kolkata, were included in the study. 5 ml of peripheral blood, GBC tumor tissue and adjacent normal tissue were collected. Excluding two above sets of patients, 20 independent GBC patients’ validation set was also included in detecting the frequency of KRAS codon 12 mutation, expression status of 6 mentioned genes, and ERBB2/Her2-neu amplification to confirm the previous findings in the present study. All tumor samples were collected at the primary stage and not treated with any drugs or chemotherapeutic agents. All patients underwent surgical resection and were staged according to the TNM staging system of the Union for International Cancer Control (UICC). Tumors were graded according to the World Health Organization (WHO) guidelines. Demographic and clinicopathological parameters such as age, sex, food habits, tobacco, smoking, alcohol habits, jaundice, diabetes pancreatitis, family history of cancer, gallstone history, tumor sizes, grades, stages, and survival data were recorded for all patients from the medical records and through follow up. Finally, informed consent from each patient or their relatives was taken during specimen collection.
Genomic DNA extraction from tissue samples
Genomic DNA was isolated from both the normal as well as the tumor tissue samples. Qiagen DNeasy Blood and Tissue kit, from Qiagen Inc. Germany (spin-column protocol) was used for isolation of DNA from tissue samples. The genomic DNA from blood was isolated by QiAmp DNA blood minikit using manufacturer protocol. At first, a day prior to isolation, tissue samples were transferred from -80 °C. DNA concentration was measured before PCR experiments.
PCR reaction for amplifying UrecA, CagA, VacA and FlicC was performed
Detailed protocol and respective primer sequences are described in Supplementary Tables 1 and 2.
KRAS Exon 2 and 3 PCR reaction was performed
Respective primer sequences and protocol are described in Supplementary Tables 1 and 2.
PCR amplification of ERBB2/Her2-neu Ile655Val and CCND1 A870G
ERBB2/Her2-neu Allele specific primers were designed against the SNP Ile655Val. Then (Restriction Fragment Length Polymorphism) RFLP was performed to screen the samples. The 655 “Val” allele gives rise to a restriction site for the enzyme BamH1 that is absent in 655 “Ile” allele. Using genomic DNA a 148 bp PCR product was generated. This was digested with BamH1 for 3 h at 55 °C, resulting in 116 bp and 32 bp fragments for “Val” at position 655 and 148 bp fragments for an “A is Ile” at this position. Digested products were separated by gel electrophoresis on 3.0% agarose gels. CCND1: Allele specific primer sequences were designed against the SNP A870G. Then RFLP was performed to screen the samples. The 870 “G” allele gives rise to a restriction site for the enzyme NciI that is absent in 870 “A” alleles. Using genomic DNA a 199-bp PCR product was generated. This was digested with NciI for 4 h at 37 °C, resulting in 176 bp and 23 bp fragments for an “A” at position 870 and in 141 bp, 35 bp, and 23 bp fragments for a “G” at this position. Digested products were separated by gel electrophoresis on 2.0% agarose gels. Detailed protocol and primer sequences are also described in Supplementary Tables 1 and 2.
Verification and validation of somatic mutation in KRAS gene by allele specific PCR
Allele specific polymerase chain reaction (ASPCR) primers were designed against mutant sequence and PCR were performed to amplify DNA strands in the corresponding mutant samples and visualized in 1% agarose gel. The primers (S2) were designed such that the last base of the forward primer changed as complementary to mutant allele. The mutations analyzed by this method were KRAS:p.G12A, KRAS:p.G12V, KRAS:p.G12D, KRAS:p.G12R, KRAS:p.and Q61H. Both the tumor and corresponding normal samples were screened by the allele specific primers as well as wild type primers to confirm the specific mutation. Detailed PCR protocol and primer sequences are described in Supplementary Tables 1 and 2.
KRAS 12th codon mutation detection by two-step enriched-semi nested PCR
The presence of mutation in codon 12 of KRAS was detected using a Two-Step Enriched- Semi Nested PCR by following the protocol described in supplemental method section.
ERBB2/Her2-neu amplification in GBC tumors
Copy number analysis for ERBB2/Her2-neu was done on all 93 samples using TaqMan Copy Number Assay (Hs00817646_cn) (Applied Biosystems, CA, USA). The PCR condition as follows: 10 min in 95 °C followed by 40 cycles of denaturation for 15 s at 95 °C, and annealing for 1 min at 60 °C. RNaseP (Applied Biosciences, CA, USA) (20X, VIC dye) used as a reference control with 2 copies in the human genome. Target and reference assays that were used for copy number calculation were derived from the mean of duplicate. Relative quantification determined as 2−ΔΔct was calculated for each of the samples to identify copy number change. Above twofold change was identified as amplified samples. The frozen Bio-Rad 2 × iQ™ SYBR® Green supermix (BioRad, USA)/LightCycler® 480 SYBR Green I Master (Roche Life Sciences, Germany), was used. In a reaction tube, master mix for each gene was prepared using (5 μl of SYBR® Green supermix, 0.3 μl of forward primer and 0.3 μl of reverse primer and 2.4 μl of nuclease-free water for each well) multiplied by total number of wells in a PCR microplate. Then 8 μl of the prepared master mix was added to each well in a 96 –well PCR microplate, followed by the addition of 2 μl of cDNA of each tissue sample into each well. Then, the microplate was run on Applied BIOsystem 7900 HT Real Time PCR machine with the thermal cycler programmed as follows:
Protocol for expression measurement and analysis of GAPDH, ACTB, CCND1, MYC, CDKN2A, ERBB2/Her2-neu, EGFR and SMAD4 genes in GBC tumor tissues
The expression of GAPDH, ACTB, CCND1, MYC, CDKN2A, ERBB2/Her2-neu, EGFR, and SMAD4 were analyzed in the Real Time PCR.All primer sequences and detailed PCR protocols for respective loci are mentioned in Supplementary Tables 1 and 2.
Relative gene expression and fold change analysis of tumor normal paired samples
The relative expression of six genes namely CCND1, MYC, EGFR, ERBB2/Her2-neu, CDKN2A, and SMAD4 were studied in GBC tumor with adjacent normal tissue samples. Two genes, namely GAPDH and ACTB were used as internal control (or, reference genes) to normalize the expression of the target gene to compensate for any difference in the amount of sample tissue.
Estimation of differential expression of genes in tumor and adjacent normal tissues
Target and reference gene Ct values were obtained. GAPDH and ACTB were used as reference control for normalization of target genes. Relative expression of targeted genes determined as 2−ΔΔct was calculated for each of the samples to identify fold change. More than twofold change was identified as dysregulation (overexpressed/under expressed) for the respective genes.
DNA sequencing and sequencing data analysis
Detailed methods described in supplementary method section.
Statistical analysis
Distribution of fold change differences and analysis of differential expression of genes in tumor and normal groups
Distributions of 2−Δct values after normalization with GAPDH for the respective genes were checked by Anderson–Darling test in R for both matched tumor and adjacent normal groups (n = 38) for the discovery group and (n = 14) for the replicative group. Wilcoxon signed rank test was used to measure any significant differences (p ≤ 0.05) of 2−Δct values between two groups for all genes. Fold change differences between tumor and normal group of respective genes represented by box plots (ggplot2 package in R Studio). Similar tests were done for unpaired samples, where total 68 tumors and 38 adjacent normal groups in the discovery set as well as 26 tumors and 26 adjacent normal groups in the replicative group were compared for respective genes.
Distribution of fold change differences and analysis of differential amplification in tumor and normal groups
Distributions of 2−Δct values after normalisation with RNaseP for the respective genes were checked by Anderson–Darling test in R package (ggplot2, R Studio) for both matched tumor and normal groups (n = 38). Wilcoxon signed rank test was used to measure any significant differences (p ≤ 0.05) of 2−Δctvalues between two groups for all genes. Fold change differences between tumor and normal group of respective genes represented by box plots (ggplot2 package in R).
Correlation analysis
Correlation analysis was done between gene expression and clinicopathological parameters using Pearson’s correlation in SPSS software (Version 16.0, Harvard University, MA, USA). This was done only for the tumor and normal paired samples. Gene expression fold change and clinicopathological variables data were converted in binary (0,1) format and analysed.
Survival analysis
Overall Survival (OS) analysis was done by Kaplan–Meier estimator using SPSS Inc. (Version 6.0, Harvard University, MA, USA). OS was calculated from the date of pathological diagnosis to the date of death or the date of the last confirmed contact. Survival curves were generated using the Kaplan–Meier method and assessed for statistically significant differences (p < 0.05) via the log rank test. Overall survival was also compared between the GBC patients group, and separately between ERBB2/Her2-neu amplified for 102 GBC samples and differential expression status of EGFR, ERBB2/Her2-neu, CCND1, Myc, CDKN2A, and SMAD4 with patient OS were done with 49 GBC patient samples to study the effect of genetic changes on patient survival.
Immunohistochemical (IHC) analysis of gallbladder tissue samples
For IHC staining, 5 um tissue sections were prepared in slides from formalin fixed, paraffin embedded GBC tissue blocks. Routine hematoxylin and eosin stain were carried out for all sections to ascertain histological features. Further, in separate experiments, all sections were taken for immunostaining following antigen retrieval in citrate buffer (pH 6.0) at 90 °C for 30 min. Primary monoclonal antibody anti-ERBB2/Her2-neu (Cell Signalling Technology Inc., US, cat. No. 2165, dilution 1:300), anti-EGFR (ABclonal Inc, MA, USA, cat. no.A5657,dilution 1:100) and anti-SMAD4 (ABclonal Inc, MA, USA, cat. no.A11351, dilution 1:100) and anti-rabbit HRP conjugated secondary antibody (Cell Signalling Technology Inc., US, cat. No. 7074, dilution 1:1000) were used for immunohistochemical localization of proteins followed by nuclear counterstaining by hematoxylin. Mounting of stained slides were done by DPX mount media and observed under Bright field microscope (Leica). Multiple areas of slide images were captured and analyzed with Image J software (version 1.54) for relative quantitative intensity. To quantify the intensity of immunostaining, we employed ImageJ Fiji software to determine the mean grey area values of deconvoluted images, which were subsequently represented graphically.
Results
Patient characteristics
At first, we had 38 paired (tumor and normal tissues), and 30 unpaired (only tumor tissues) tissue samples of GBC patients collected from multi hospitals of Kolkata City. We had another 14 paired and 6 unpaired samples of GBC for independent validation cohort. Out of these 68 GBC samples recruited in our study, 51 (75%) were female patients and 17 (25%) male patients, the mean age was found to be about 55. While only 7.4% (5 of them) had a smoking habit, and 5.9% (4 of them) had an alcohol habit, an overwhelming 82.4% (56 of them) recorded a gallstone history. The site of GBC lesion was found mostly in its fundus (41%), and neck (32.2%), followed by body (20.7%), head (4.6%) and intra luminal (1.5%). According to 7th edition of America Joint Committee on Cancer (AJCC) nomenclature on the stages (pathological) of the tumors were; out of total 68 patient samples, 25 (36.4%) had a stage IIB gallbladder tumor, 19 (28.1%) had a stage IIIB tumor, 8 (11.6%) had a stage IIIA tumor, while only 14 (20.4%) had a stage IV tumor (Table 1).
Table 1.
Characteristics of demography and clinico-pathological parameters of total patients
| Total Patients Recruited in the study | n = 68 |
|---|---|
| Demography and Clinicopathological Characteristics | |
| Age(mean) | 54.62 |
| Smoking habit | 7.4% |
| Alcohol habit | 5.9% |
| Gall Stone | 82.4% |
| Tumor Classification (7th AJCC) | |
| Stage I | 2.5% |
| Stage IIB | 36.4% |
| Stage IIIA | 11.6% |
| Stage IIIB | 28.1% |
| Stage IVA | 12.3% |
| Stage IVB | 9.1% |
| Tumor Differentiation | |
| Well differentiated | 18.2% |
| Moderately differentiated | 36.4% |
| Poorly differentiated | 22.7% |
| Unidentified | 15.2% |
| Lymph Node | |
| Present | 56.5% |
| Absent | 43.5% |
| Site of Lesions | |
| Body | 20.7% |
| Fundus | 41% |
| Neck | 32.2% |
| Head | 4.6% |
| Intraluminal | 1.5% |
Low frequency of KRAS codon 12 mutations detected in GBC cases
In biliary tract carcinomas, a low to high frequency of KRAS mutation had been noted. KRAS is known to be a commonly mutated driver gene in pancreatic and colonic cancer. We have selected 87 GSD samples and 68 GBC tissue samples for KRAS mutation detection at codon 12 position. We have performed PCR followed by RFLP based detection method at codon 12 position. We have observed three G12A, three G12V, and one G12D mutation among 87 GBC samples, but no KRAS codon 12 mutation was observed in GSD samples. In this study, we found a relatively lower frequency (i.e. 11.5%) of KRAS mutation in GBC tumors. To revalidate this lower frequency, we adopted two independent mutation validation approaches- first verified by ASPCR for G12A, G12R, G12V, and G12C (Fig. 1) and then KRAS 12th codon mutations were additionally validated by Two-Step Enriched-Nested PCR. We have done Sanger sequencing of 65 samples in exon 2 and exon 4 of KRAS gene (Supplementary Fig. 1). We did not observe any codon 13 mutation and codon 61 mutations in GBC patient samples.
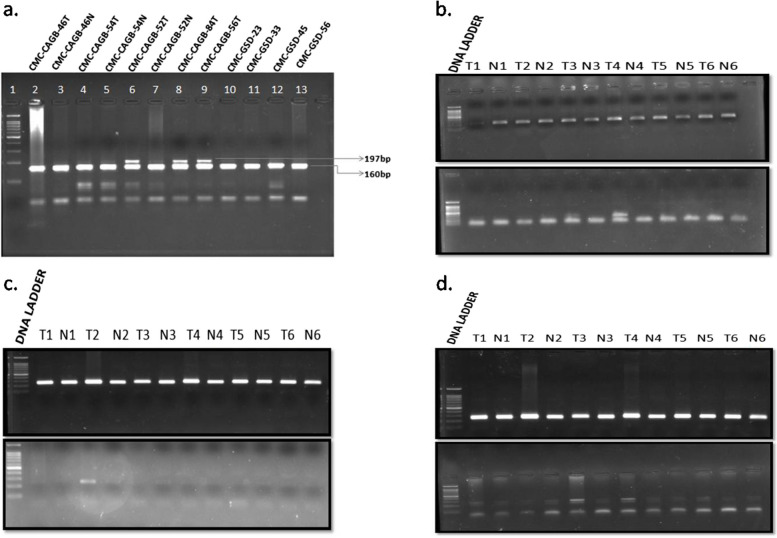
Fig. 1.
a The BstN1 enzyme digests PCR products, run in 2% agarose gel (stained with EtBr). Lane 1: 100 bp ladder; Lane 2 to 7 has GBC tumor and adjacent normal paired samples respectively; Lane 8 to 9 has positive control for codon 12th mutation of GBC tumor samples;. Lane 10–13 has Gallstone disease samples. A clear and strong band near 200 bp observed in lane 6, 8, and 9 suggesting presence of 197 bp fragment that differentiates the positive samples (codon 12 mutation of KRAS) with rest of the samples containing 160 bp band. b Detection of KRAS G12A mutations by allele specific PCR: Lane 1 represents 100 bp DNA ladder. Lane 2 to 12 represents 6 tumor normal paired samples. In the upper panel PCR done with primer corresponds to wild type allele for KRAS G12A mutation. A single band observed in all the samples due to the presence of normal allele in all the samples. In the lower panel PCR done with allele specific mutant primer for KRAS G12A mutation. A strong band is observed in the 4th tumor sample (T4) but not in the corresponding normal sample (N4). Samples T1, T2, and T3 do not contain the respective bands. T5 and T6 are negative controls so the bands are not present. c Detection of KRAS G12D mutations by allele specific PCR: Lane 1 represents 100 bp DNA ladder. Lane 2 to 12 represents 6 tumor normal paired samples. In the upper panel PCR done with primer corresponds to wild type allele for KRAS G12D mutation. A single band observed in all the samples due to the presence of normal allele in all the samples. In the lower panel PCR done with allele specific mutant primer for KRAS G12D mutation. A band is observed in the 2nd tumor sample (T2) but not in the corresponding normal sample (N2). Samples T1, T3, and T4 do not contain the respective bands. T5 and T6 are negative controls so the bands are not present. d Detection of KRAS G12V mutations by allele specific PCR: Lane 1 represents 100 bp DNA ladder. Lane 2 to 12 represents 6 tumor normal paired samples. In the upper panel PCR done with primer corresponds to wild type allele for KRAS G12V mutation. A single band observed in all the samples due to the presence of normal allele in all the samples. In the lower panel PCR done with allele specific mutant primer for KRAS G12V mutation. Double bands are observed in 3rd (T3), 4th (T4) tumor samples but not in the corresponding normal sample (N3 or N4). Samples T1, T2 does not contain the respective bands. T5 and T6 are negative controls so the bands are not present
Expression pattern of CCND1, Myc and CDKN2A genes in GBC patient samples
In our study, the relative expression of 6 most differentially regulated genes previously identified from Next Generation Sequencing (NGS) studies namely CCND1, MYC, EGFR, and ERBB2/Her2-neu, oncogenes and two tumor suppressor genes CDKN2A, and SMAD4 were studied in gallbladder tumor with respect to their adjacent normal tissue samples. It was found that in 38 paired tumor tissues, CCND1 was overexpressed in 42.1% (n = 16) (Fig. 2a), MYC was overexpressed in 42.1% (n = 16) of tumor samples (Fig. 2b), EGFR was overexpressed in 55.2% (n = 21) (Fig. 2c) and ERBB2/Her2-neu was overexpressed in 50% (n = 19) of tumor samples (Fig. 3a). On the other hand, SMAD4 was found to be downregulated in 42.1% (n = 16) (Fig. 3b), while CDKN2A was found to be downregulated in 55.2% (n = 21) of tumor samples (Fig. 3c). All normalization was done with respect to GAPDH and ACTB of internal control gene expression.
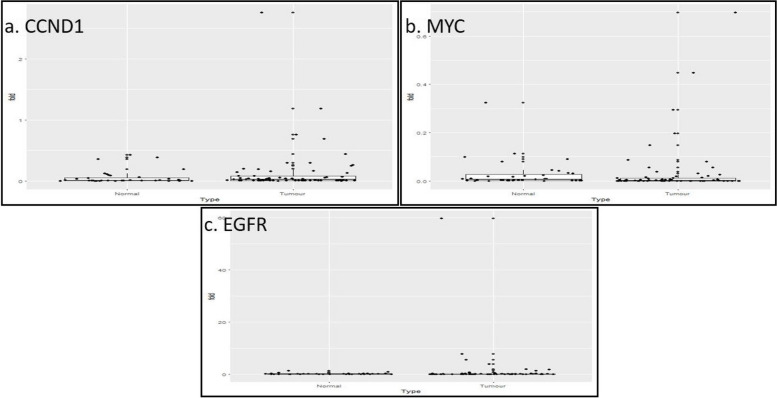
Fig. 2.
a Gene expression of CCND1 in 68 tumor and 38 adjacent normal tissue samples. Data normalized with internal control gene GAPDH. The expressions of CCND1 in tumor samples were compared with adjacent normal samples by Anderson–Darling statistical test. The twofold is the cut- off of the dysregulation of the gene with respect to the adjacent normal tissues. b Gene expression of MYC in 68 tumor and 38 adjacent normal tissue samples. Data normalized with internal control gene GAPDH. The expressions of MYC in tumor samples were compared with normal samples by Anderson–Darling statistical test. The twofold is the cut- off of the dysregulation of the gene with respect to the normal tissues. c Gene expression of EGFR in 68 tumor and 38 adjacent normal tissue samples. Data normalized with internal control gene GAPDH. The expressions of EGFR in tumor samples were compared with normal samples by Anderson–Darling statistical test. The twofold is the cut- off of the dysregulation of the gene with respect to the adjacent normal tissues
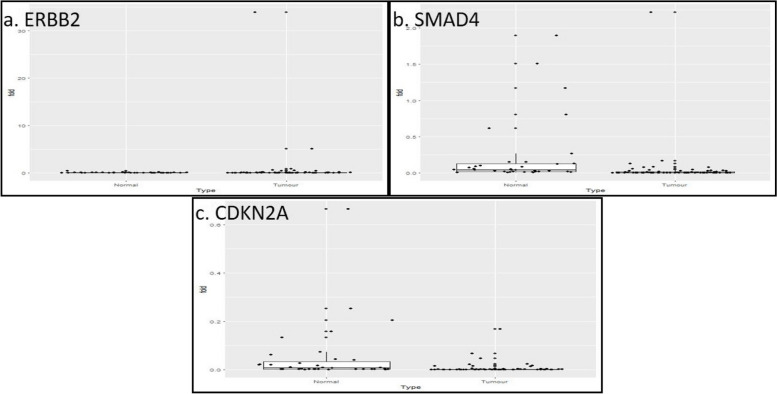
Fig. 3.
a Gene expression of ERBB2/Her2-neu in 68 tumor and 38 adjacent normal tissue samples. Data normalized with internal control gene GAPDH. The expressions of ERBB2/Her2-neu in tumor samples were compared with normal samples by Anderson–Darling statistical test. The twofold is the cut- off of the dysregulation of the gene with respect to the normal tissues. b Gene expression of SMAD4 in 68 tumor and 38 adjacent normal tissue samples. Data normalized with internal control gene GAPDH. The expressions of SMAD4 in tumor samples were compared with normal samples by Anderson–Darling statistical test. The twofold is the cut- off of the dysregulation of the gene with respect to the adjacent normal tissues. c Gene expression of CDKN2A in 68 tumor and 38 adjacent normal tissue samples. Data normalized with internal control gene GAPDH. The expression of CDKN2A in tumor samples was compared with adjacent normal samples by Anderson–Darling statistical test. The twofold is the cut- off of the dysregulation of the gene with respect to the normal tissues
Next, the distribution curve of the Ct values of each gene in normal tissue samples was constructed and the p-values of the distribution were found to be 0.013 for ACTB, 0.010 for GAPDH, 0.003 for CCND1, 0.0017 for MYC, 0.017 for CDKN2A, thus the hypothesis of an underlying normal distribution was rejected at 0.05 significance level (data not shown). As the distribution of the Ct values was found to be not normal, test was performed to determine the significance of differential expression of the genes in 38 paired tumor samples with respect to adjacent normal. Upon GAPDH normalization, CCND1 (p = 0.50) and MYC (p = 0.07) overexpression were found to be statistically non-significant. On the other hand, CDKN2A (p = 0.003) was found to be significantly downregulated. Then, again after pooling 30 unpaired tumor samples, when the test was performed on 68 tumor samples with 38 normal control, CCND1 and MYC overexpression were found to be statistically nonsignificant. CDKN2A (p = 0.003) downregulation was found to be statistically significant (Fig. 4a) and all data observed from this analysis were consistent with the previous finding of the same study.
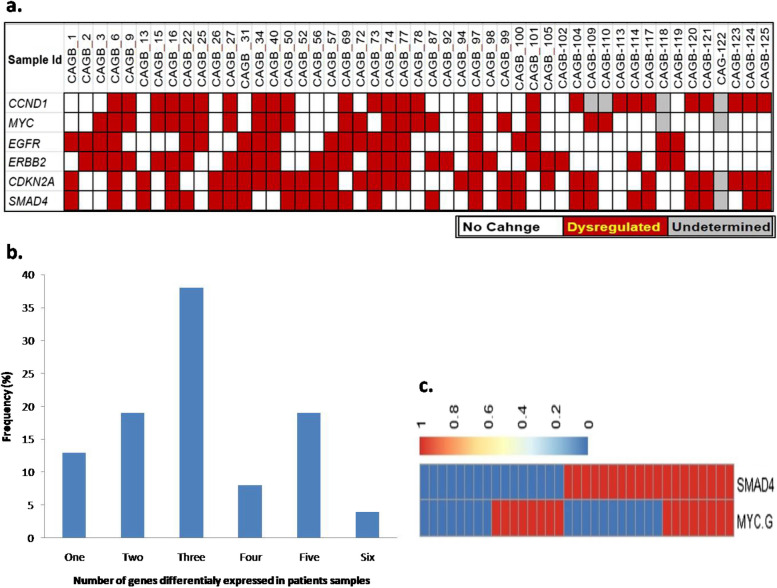
Fig. 4.
a Fold change status of studied 6 genes in all the patients. The red boxes indicate above twofold change of gene expression in tumor tissues compared to respective adjacent normal pair samples. White boxes indicate no change (< twofold) in the tumor tissues compared to adjacent normal pairs and grey boxes indicate un-determined values. b Cumulative grade of gene expression across samples. X axis denotes the combined alteration of genes and Y axis denotes frequency of types of alterations. c Combination of SMAD4 and MYC gene expression alteration in patient samples
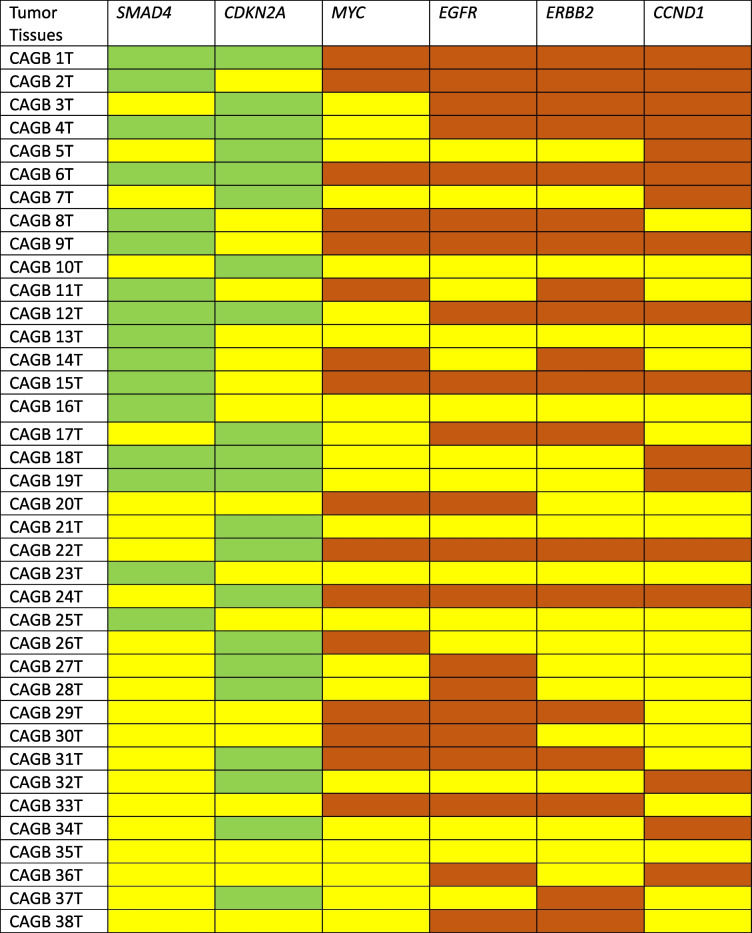
Among the 38 paired GBC patients, out of the 6 genes, any 1 studied showed altered expression in 13% (n = 5), any 2 showed altered expression in 19% (n = 7), any 3 showed altered expression in 37% (n = 14), any 4 in 8% (n = 3), and any 5 showed altered expression in 18% (n = 7) of tumor samples. And, all the 6 genes studied showed altered expression in only 5% (n = 2) of tumor samples (Fig. 4b). Next, SMAD4 downregulation and MYC overexpression was found to be mutually exclusive (i.e., if one shows altered expression, the other doesn't) in a majority, i.e., 20 (54.2%) of the paired 38 tumor samples, while both showed altered expression in only 8 (20%) of paired tumor samples (Fig. 4c). Out of the paired 38 tumor tissues both SMAD4 and CDKN2A downregulated in 6 samples. Among these samples, all 4 oncogenes (EGFR, ERBB2/Her2-neu, MYC and CCND1) showed upregulation in 2 samples, and any 3 of the oncogenes also showed upregulation in 2 samples. Another 10 samples showed downregulation of only SMAD4. Out of these 10 samples, all 4 oncogenes showed overexpression in 3 samples, while any 3 got overexpressed in 1 sample and any 2 got overexpressed in 2 samples. While another 16 samples showed only CDKN2A downregulation. Out of these 16 samples, all 4 of the oncogenes were upregulated in 2 samples, any 3 in 2 samples and any 2 in just one sample (Table 2).
Table 2.
Gene expression status of 6 genes in GBC patient samples
Here, yellow box indicates no change, green box indicate downregulation and red box indicate overexpression/upregulation of the respective genes
Expression pattern of ERBB2/Her2-neu gene and validation in GBC tissue samples
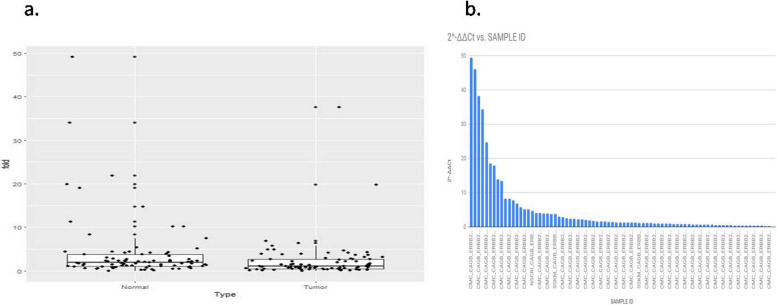
ERBB2/Her2-neu was found to be overexpressed in 50% (n = 19) of tumor samples (Fig. 3a). Next, the distribution curve of the Ct values in normal tissue samples was constructed through and the p-values of the distribution were found to be 0.013 for ACTB, 0.010 for GAPDH, 0.0000005 for ERBB2/Her2-neu, thus the hypothesis of an underlying normal distribution was rejected at 0.05 significance level (data not shown). Upon GAPDH normalization, ERBB2/Her2-neu (p = 0.00001) overexpression were found to be statistically significant. Then, after pooling 30 unpaired tumor samples, when the test was performed on 68 tumor samples with 38 normal control, similarly, oncogene ERBB2/Her2-neu (p = 0.00001) overexpression was found to be statistically significant. ERBB2/Her2-neu amplification was identified in 30% of GBC samples (Fig. 5). Among the ERBB2/Her2-neu amplified patients, in addition, when fold change between tumor and normal group were compared, a very significant difference was observed (p = 0.03). Ideally the amplification results are validated by other methods like, in situ hybridization and immunohistochemistry (IHC), we have performed by IHC method.
Fig. 5.
a Amplification of ERBB2/Her2-neu gene in Gallbladder adenocarcinoma samples. b Distribution of ERBB2/Her2-neu gene amplification in Gall bladder adenocarcinoma by Taqman copy number assay. The bar denotes the fold change in ERBB2/Her2-neu expression with respect to sample ID. X- axis denotes the sample ID of the patient cohort and the y-axis represents the fold change in expression of ERBB2/Her2-neu gene in the patient cohort
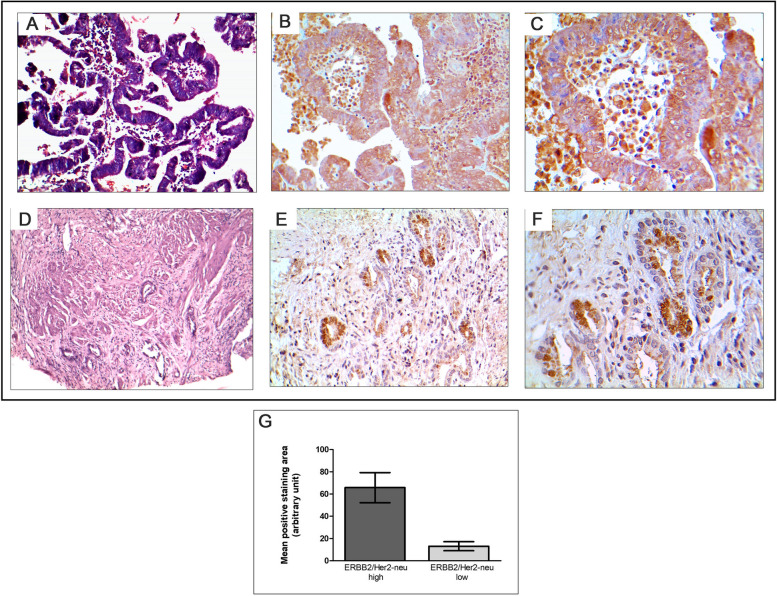
In our study, we conducted immunohistochemical staining to validate the gene expression pattern of ERBB2/Her2-neu in GBC. We selected 15 tumor tissues with both high and low expressions for ERBB2/Her2-neu and performed immunohistochemical localization of the respective proteins. Upon detailed analysis at different microscopic magnifications, we observed that in tissues with high ERBB2/Her2-neu expression, the staining was specific to tumor cells. This observation strongly suggests a tumor area-specific expression pattern of ERBB2/Her2-neu (Fig. 6).
Fig. 6.
Immunohistological localization of ERBB2/Her2-neu expression in gallbladder patient tissues. A Tumor tissue in H&E (magnification × 100); (B) ERBB2/Her2-neu high expression (magnification × 200); (C) ERBB2/Her2-neu high expression (magnification × 400); (D) H&E (magnification × 100); (E) ERBB2/Her2-neu low expression (magnification × 200); (F) ERBB2/Her2-neu low expression (magnification × 400) (G) Graphical representation of ERBB2 expression quantitated from positive staining intensity in tumor tissues
Expression pattern of EGFR, SMAD4 genes and validation in GBC tissue samples
Similarly, EGFR was found to be overexpressed in 55.2% (n = 21) of tumor samples (Fig. 2c). On the other hand, SMAD4 was found to be down regulated in 41.2% (n = 16) of tumor samples (Fig. 3b). Next, the distribution curve of the Ct values in normal tissue samples was constructed and the p-values of the distribution were found to be 0.013 for ACTB, 0.010 for GAPDH, 0.05 for EGFR, and 0.18 for SMAD4, thus the hypothesis of an underlying normal distribution was rejected at 0.05 significance level (data not shown). Then, again after pooling 30 unpaired tumor samples, when the test was performed on 68 tumor samples with 38 normal control, similarly, oncogene EGFR (p = 0.0005) overexpression and tumor suppressor gene SMAD4 (p = 0.009) down regulation was found to be statistically significant (Fig. 3a).
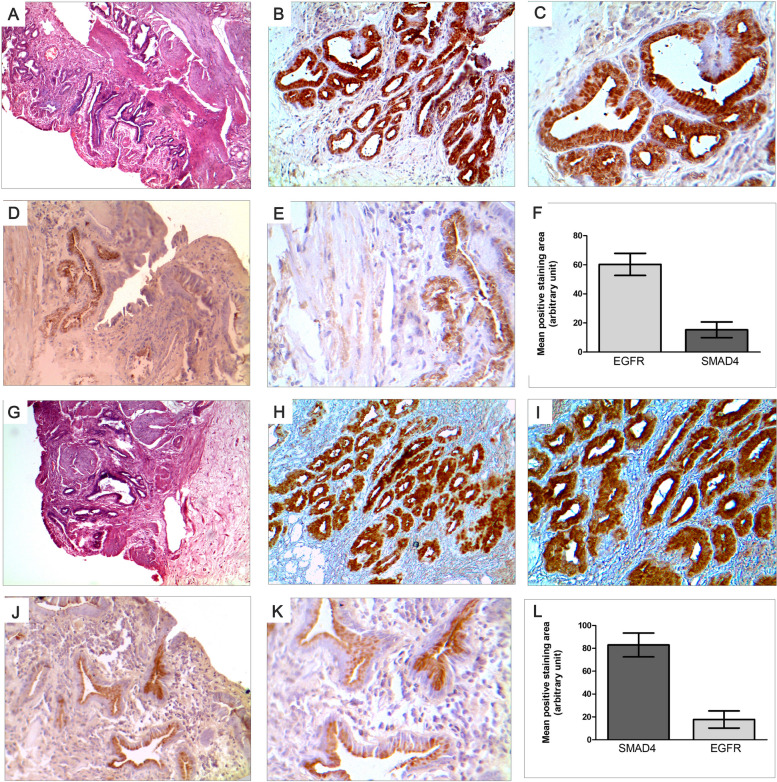
In our study, we conducted immunohistochemical staining to validate the gene expression patterns of EGFR and SMAD4 in GBC. There was a significant negative correlation between the expression of EGFR and SMAD4 in the majority of samples in consistence with the results obtained through RTPCR technique. We selected 15 tumor tissues with both high and low expressions for these genes and performed immunohistochemical localization of the respective proteins. Our investigation revealed an intriguing relationship between EGFR and SMAD4 expression. Tumor tissues exhibiting higher positive staining for EGFR demonstrated decreased SMAD4 levels, and conversely, tissues with lower EGFR levels had increased SMAD4 levels (Fig. 7). This corroborated the gene expression patterns observed in our earlier results.
Fig. 7.
Negative correlation of EGFR and SMAD4 expression in gallbladder patient tissues indicated by immunohistochemical localization in tumor areas (A) H&E staining of EGFR high expressing tumor tissues (magnification × 100); (B) EGFR high expression in same tissue (magnification × 200); (C) EGFR high expression (magnification × 400); (D) SMAD4 low expression in same tissue (magnification × 200); (E) SMAD4 low expression in same tissue (magnification × 400); (F) Representation of high expression of EGFR and low expression of SMAD4 as quantitated from positive staining intensity in tumor tissues: (G) H&E staining of SMAD4 low expressing tumor tissues (magnification × 100); (H) SMAD4 low expression in same tissue (magnification × 200); (I) SMAD4 low expression in same tissue (magnification × 400); (J) EGFR high expression in same tissue(magnification × 200); (K) EGFR high expression in same tissue(magnification × 400); (L) Representation of high expression of SMAD4 and low expression of EGFR as quantitated from positive staining intensity in tumor tissues
Detection of correlation between clinicopathological parameters and gene expression status in gallbladder adenocarcinoma
In our study, 38 sets of tumor and normal paired samples of GBC were considered to identify any correlation between clinicopathological data and expressions of several previously mentioned genes. Next, Pearson correlation test was performed to measure the strength of linear association between two variables and the p value determined to test the significance of the correlation coefficient (at significance level p ≤ 0.05). It has been found in our patients that there is a significant positive correlation between the presence of affected lymph nodes and overexpression oncogene ERBB2/Her2-neu (r = 0.00). The overexpression of CCND1 and MYC genes have a strong correlation with increase in stages of GBC (r = 0.00, and r = 0.00 respectively). It is also found that there is a significant positive correlation (r = 0.01) between the overexpression of the oncogenes, i.e. there is a strong incidence that CCND1 and Myc get overexpressed in same tumor samples. Also, it is found that there is a significant positive linear correlation (r = 0.00) between the expression of both the tumor suppressor genes CDKN2A and SMAD4 i.e. there is a strong incidence that CDKN2A and SMAD4 get downregulated in same samples. Our analysis also show a significant positive correlation between smoking and overexpression of CCND1 and MYC genes (r = 0.00 and r = 0.00 respectively); a positive correlation between stage and overexpression of CCND1 and MYC genes (r = 0.00 and r = 0.00 respectively). Similarly, our analysis also pointed out a significant negative correlation between EGFR and SMAD4 expression (r = -0.01) (Table 3). We did not find any significant correlation between gene expression status and gender or tumor stage.
Table 3.
Multivariate correlation analysis concerning genes of interest and associated clinico-pathological parameters
| Correlations | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex | Age | Alcohol | Smoking | GallStone | LymphNode | Stage | Grade | CCND1 | MYC | EGFR | ERBB2 | CDKN2A | SMAD4 | ||
| Sex | Pearson Correlation | 1 | .159 | .416a | .716a | -.141 | .025 | 1.000a | .159 | .416a | .716a | -.141 | .025 | -.038 | -.083 |
| Sig. (2-tailed) | .320 | .006 | .000 | .372 | .877 | .000 | .320 | .006 | .000 | .372 | .877 | .809 | .599 | ||
| N | 43 | 41 | 43 | 43 | 42 | 40 | 43 | 41 | 43 | 43 | 42 | 40 | 42 | 42 | |
| Age | Pearson Correlation | .159 | 1 | .111 | .087 | .479a | .013 | .159 | 1.000a | .111 | .087 | .479a | .013 | -.129 | -.250 |
| Sig. (2-tailed) | .320 | .488 | .588 | .002 | .937 | .320 | .000 | .488 | .588 | .002 | .937 | .427 | .120 | ||
| N | 41 | 41 | 41 | 41 | 40 | 39 | 41 | 41 | 41 | 41 | 40 | 39 | 40 | 40 | |
| Alcohol | Pearson Correlation | .416a | .111 | 1 | .497a | -.062 | .271 | .416a | .111 | 1.000a | .497a | -.062 | .271 | -.179 | -.106 |
| Sig. (2-tailed) | .006 | .488 | .001 | .696 | .091 | .006 | .488 | .000 | .001 | .696 | .091 | .256 | .505 | ||
| N | 43 | 41 | 43 | 43 | 42 | 40 | 43 | 41 | 43 | 43 | 42 | 40 | 42 | 42 | |
| Smoking | Pearson Correlation | .716a | .087 | .497a | 1 | .050 | -.207 | .716a | .087 | .497a | 1.000a | .050 | -.207 | -.050 | -.139 |
| Sig. (2-tailed) | .000 | .588 | .001 | .753 | .200 | .000 | .588 | .001 | .000 | .753 | .200 | .753 | .381 | ||
| N | 43 | 41 | 43 | 43 | 42 | 40 | 43 | 41 | 43 | 43 | 42 | 40 | 42 | 42 | |
| GallStone | Pearson Correlation | -.141 | .479a | -.062 | .050 | 1 | -.047 | -.141 | .479a | -.062 | .050 | 1.000a | -.047 | -.258 | -.375b |
| Sig. (2-tailed) | .372 | .002 | .696 | .753 | .777 | .372 | .002 | .696 | .753 | .000 | .777 | .104 | .016 | ||
| N | 42 | 40 | 42 | 42 | 42 | 39 | 42 | 40 | 42 | 42 | 42 | 39 | 41 | 41 | |
| LymphNode | Pearson Correlation | .025 | .013 | .271 | -.207 | -.047 | 1 | .025 | .013 | .271 | -.207 | -.047 | 1.000a | .272 | .127 |
| Sig. (2-tailed) | .877 | .937 | .091 | .200 | .777 | .877 | .937 | .091 | .200 | .777 | .000 | .094 | .442 | ||
| N | 40 | 39 | 40 | 40 | 39 | 40 | 40 | 39 | 40 | 40 | 39 | 40 | 39 | 39 | |
| Stage | Pearson Correlation | 1.000a | .159 | .416a | .716a | -.141 | .025 | 1 | .159 | .416a | .716a | -.141 | .025 | -.038 | -.083 |
| Sig. (2-tailed) | .000 | .320 | .006 | .000 | .372 | .877 | .320 | .006 | .000 | .372 | .877 | .809 | .599 | ||
| N | 43 | 41 | 43 | 43 | 42 | 40 | 43 | 41 | 43 | 43 | 42 | 40 | 42 | 42 | |
| Grade | Pearson Correlation | .159 | 1.000a | .111 | .087 | .479a | .013 | .159 | 1 | .111 | .087 | .479a | .013 | -.129 | -.250 |
| Sig. (2-tailed) | .320 | .000 | .488 | .588 | .002 | .937 | .320 | .488 | .588 | .002 | .937 | .427 | .120 | ||
| N | 41 | 41 | 41 | 41 | 40 | 39 | 41 | 41 | 41 | 41 | 40 | 39 | 40 | 40 | |
| CCND1 | Pearson Correlation | .416a | .111 | 1.000a | .497a | -.062 | .271 | .416a | .111 | 1 | .497a | -.062 | .271 | -.179 | -.106 |
| Sig. (2-tailed) | .006 | .488 | .000 | .001 | .696 | .091 | .006 | .488 | .001 | .696 | .091 | .256 | .505 | ||
| N | 43 | 41 | 43 | 43 | 42 | 40 | 43 | 41 | 43 | 43 | 42 | 40 | 42 | 42 | |
| MYC | Pearson Correlation | .716a | .087 | .497a | 1.000a | .050 | -.207 | .716a | .087 | .497a | 1 | .050 | -.207 | -.050 | -.139 |
| Sig. (2-tailed) | .000 | .588 | .001 | .000 | .753 | .200 | .000 | .588 | .001 | .753 | .200 | .753 | .381 | ||
| N | 43 | 41 | 43 | 43 | 42 | 40 | 43 | 41 | 43 | 43 | 42 | 40 | 42 | 42 | |
| EGFR | Pearson Correlation | -.141 | .479a | -.062 | .050 | 1.000a | -.047 | -.141 | .479a | -.062 | .050 | 1 | -.047 | -.258 | -.375b |
| Sig. (2-tailed) | .372 | .002 | .696 | .753 | .000 | .777 | .372 | .002 | .696 | .753 | .777 | .104 | .016 | ||
| N | 42 | 40 | 42 | 42 | 42 | 39 | 42 | 40 | 42 | 42 | 42 | 39 | 41 | 41 | |
| ERBB2 | Pearson Correlation | .025 | .013 | .271 | -.207 | -.047 | 1.000a | .025 | .013 | .271 | -.207 | -.047 | 1 | .272 | .127 |
| Sig. (2-tailed) | .877 | .937 | .091 | .200 | .777 | .000 | .877 | .937 | .091 | .200 | .777 | .094 | .442 | ||
| N | 40 | 39 | 40 | 40 | 39 | 40 | 40 | 39 | 40 | 40 | 39 | 40 | 39 | 39 | |
| CDKN2A | Pearson Correlation | -.038 | -.129 | -.179 | -.050 | -.258 | .272 | -.038 | -.129 | -.179 | -.050 | -.258 | .272 | 1 | .583a |
| Sig. (2-tailed) | .809 | .427 | .256 | .753 | .104 | .094 | .809 | .427 | .256 | .753 | .104 | .094 | .000 | ||
| N | 42 | 40 | 42 | 42 | 41 | 39 | 42 | 40 | 42 | 42 | 41 | 39 | 42 | 42 | |
| SMAD4 | Pearson Correlation | -.083 | -.250 | -.106 | -.139 | -.375b | .127 | -.083 | -.250 | -.106 | -.139 | -.375b | .127 | .583a | 1 |
| Sig. (2-tailed) | .599 | .120 | .505 | .381 | .016 | .442 | .599 | .120 | .505 | .381 | -.016 | .442 | .000 | ||
| N | 42 | 40 | 42 | 42 | 41 | 39 | 42 | 40 | 42 | 42 | 41 | 39 | 42 | 42 | |
aCorrelation is significant at the 0.01 level (2-tailed)
bCorrelation is significant at the 0.05 level (2-tailed)
Bacterial infection and correlation to the GBC disease
Our samples were also subjected to thorough screening for any bacterial DNA that could have a correlation with the GBC. This was hypothesized because every lower body cancer has such correlations and as suggested by a study, they have found correlation between S.typhi and H.pylori infections and GBC [17].
We found that in our tissue samples there were no strains of H.pylori when screened by an Urec A (a housekeeping gene of H.pylori) figure (Supplementary Fig. 2a). So, we screened for the CAG (Supplementary Fig. 2b) and VAC genes which are responsible for its pathogenicity. Only one tissue sample was VAC and CAG positive (Supplementary Fig. 3b). We have also screened 68 gallstone diseased tissues, for CAG A, Urec A, and VAC genes. Gallstone disease (GSD) is known as an early precancerous lesion of GBC. Hence, we may argue that H.pylori infection is not essential to develop GBC in Eastern Indian patients.
We also screened for S.typhi bacterial DNA in our tissue samples to see if this infection has any correlation to GBC in our region as suggested in many case–control endemic studies. When we screened our tissue samples for the FLIC gene responsible for making flagellin in S.typhi all tissue samples were FLIC negative (Supplementary Fig. 3a). We have also screened 68 gallstone diseased tissues for FLIC genes. As an essential component for S.typhi was not found, so we did not pursue further for its rRNA specific amplification.
Association of CCND1 870 A/G and ERBB2/Her2-neu codon 655 Ile/Val polymorphisms with GBC and control samples
The restriction analysis at CCND1 G870A and ERBB2/Her2-neu Ile655Val polymorphisms were done with 67 GBC patient DNA samples. The frequency of “AG” genotype was highest with respect to the other two genotypes in CCND1 870 A/G SNP (Supplementary Fig. 4a and 4b). The Ile/Ile genotype frequency was higher than the other two genotypes in ERBB2/Her2-neu Ile655Val SNP (Supplementary Fig. 5a and 5b). We have analyzed the correlation and survival data from patient samples separately with these SNPs, but we did not observe any significant correlation between survival and risk genotype of ERBB2/Her2-neu and CCND1 SNPs (Supplementary Table 3).
Survival analysis of gene expression and clinicopathological data of GBC tumor
We have included differential expression status of EGFR, ERBB2/Her2-neu, Myc, CCND1, SMAD4 and CDKN2A in a 50 paired tumor and adjacent normal sample set. On the other side we have included overall survivals of each and individual accounted patient. Unfortunately, we did not observe any statistically significant patient OS with gene expression status (Supplementary Fig. 6).
KRAS codon 12 Mutation detection, ERBB2/Her2-neu amplification, EGFR, ERBB2/Her2-neu, CCND1, Myc, SMAD4, and CDKN2A and TGF- β gene expression status in independent validation cohort of GBC tumors
We also have included another validation set of 26 unrelated/independent paired samples (tumor and normal tissues). Out of the 26 patients’ gallbladder tumor samples recruited in our study, 18 (69.2%) were female patients and 8 (30.8%) were male patients, mean age was found to be about 52. While 4 (15.4%) had a smoking habit, and 2(7.69%) had alcohol habit, an overwhelming 22 (84.6%) recorded a gallstone history. The site of GBC lesion was found mostly in it is neck (38.5%), and fundus (26.9%) followed by body (15.4%) and head (11.5%). Among the total 26 patient samples, 3 (11.5%) had a stage I gallbladder tumor, 6 (23.1%) had stage IIB, 2 (7.7%) had stage IIIA tumor, 6 (23.1%) had stage IIIB tumor and 9 (34.6%) had stage IVB tumor (Table 4).
Table 4.
Characteristics of demography and clinicopathological parameters of total patients in validation cohort
| Total Patients Recruited in the study | n = 26 |
|---|---|
| Demography and Clinicopathological Characteristics | |
| Age(mean) | 53.58 |
| Smoking habit | 15.4% |
| Alcohol habit | 7.69% |
| Gall Stone | 84.6% |
| Tumor Classification (7th AJCC) | |
| Stage I | 11.5% |
| Stage IIB | 23.1% |
| Stage IIIA | 7.7% |
| Stage IIIB | 23.1% |
| Stage IVB | 34.6% |
| Tumor Differentiation | |
| Well differentiated | 19.2% |
| Moderately differentiated | 38.5% |
| Poorly differentiated | 38.5% |
| Unidentified | 3.8% |
| Lymph Node | |
| Present | 53.84% |
| Absent | 46.16% |
| Site of Lesions | |
| Body | 15.4% |
| Fundus | 26.9% |
| Neck | 38.5% |
| Head | 11.5% |
| Intraluminal | 7.7% |
In our validation study cohort, we have done the most of the experiments that we have done in the previous section of our study. We first reidentified KRAS codon 12 mutation by PCR RFLP method and observed 10% KRAS codon 12 mutation in 26 GBC patients. ERBB2/Her2-neu amplification was observed in 46% (n = 12) when we compared tumor samples with adjacent normal tissue (Supplementary Tables 4 and 5). Test was also performed to determine the significance of differential expression of EGFR, ERBB2/Her2-neu, MYC, CCND1, SMAD4 and CDKN2A genes in validation cohort upon ACTB normalization and the results obtained are consistent with our previous findings. A significant ERBB2 overexpression was observed and a strikingly high negative/inverse correlation was observed between EGFR and SMAD4 expression, which is statistically significant.
Several previous studies demonstrated TGF-β downregulation increased the proliferation, and invasive ability of cancer cells, especially in pancreatic and colorectal cancers [18]. We have tried to analyze the trend in GBC tissue samples as well. Interestingly, in the 26 tissue samples in our validation cohort, we have also found a significant TGF-β downregulation in GBC tissues (data not shown).
Discussion
Cancer is a complex genetic disease characterized by cumulative genetic and epigenetic alterations that lead to activation of oncogenes and inactivation of tumor suppressor genes. GBC is no exception to this, with recent molecular genetic studies revealing involvement of specific proto-oncogenes and tumor suppressor genes in its development and progression [19, 20]
The ErbB signalling pathway is composed of EGFR, ERBB2, ERBB3, and ERBB4 receptors and their downstream genes. The EGFR (also known as ERBB1), and ERBB2 proteins have been overexpressed in GBC, and this pathway is associated with cell adhesion, differentiation, apoptosis, division and migration and has been linked to cancer initiation and progression among several tumor types. Expression of ERBB2/Her2-neu is mostly absent and/or mild in dysplasia or adenoma, however study show that advanced stages (stages II to IV) are closely associated with higher levels of ERBB2/Her2-neu. The EGFR overexpression vary between 6% to 70.7%, whereas ERBB2/Her2-neu overexpression has been reported to occur in 15.7%—63.6% cases in GBC from different studies [21]. Consistent with this above model in the present study, we also observed overexpression of ERBB1 and ERBB2/Her2-neu in our patient cohort and the overexpression pattern were statistically significant when we compared patient group with respect to normal control (EGFR/p = 0.01; and ERBB2/p = 0.02). Furthermore, few NGS studies also demonstrated EGFR point mutations, ERBB2/Her2-neu amplification and/or protein overexpression might be involved in the development of GBC. Thus, we have focused in our study to find the amplification status of ERBB2/Her2-neu in our GBC patients. A previous study even stated that ERBB2/Her2-neu gene amplification in GBC similar to that found in breast cancer [22]. In our study ERBB2/Her2-neu amplification was identified in 30% (17 out of 57) in GBC samples that supports previous findings. We have seen ERBB2/Her2-neu amplification to be statistically significant in GBC tumor type when compared with adjacent normal tissues (p = 0.03). Besides this above story, several studies have analyzed the protein expressions by immunostaining and mutation pattern of ERBB2/Her2-neu in GBC and resulted in (3–13%) and (4–24%) respectively [23]. The role of SMAD4 in modulating EGFR expression is a critical aspect. SMAD4 acts as a suppressor of EGFR expression. Loss of SMAD4 can result in increased EGFR expression, contributing to tumorigenesis [24]. SMAD4 functions as a suppressor of EGFR, and when SMAD4 is lost or downregulated, EGFR expression tends to rise in other GI tract cancers [25]. This is due to the loss of SMAD4's inhibitory effect on EGFR [26]. In our GBC cohort, expression analysis of ERBB2, EGFR, and SMAD4, led to some intriguing findings. One significant observation was the inverse correlation between EGFR and SMAD4 gene expression levels in our cohort of Indian GBC samples, which is in consistence with the typical trend reported in previous literature of other GI tract cancers like in pancreatic adenocarcinoma cells [26]. The results through RTPCR technique, corroborated with findings through IHC studies of EGFR and SMAD4 expression in GBC tissues, which was subsequently validated though another gene expression studies from validation cohort. This suggests similar genetic or molecular factors operating in the Indian population in GBC, which has barely been documented before. Our analysis also highlighted a link between ERBB2 expression and SMAD4 levels. When ERBB2 expression was high, SMAD4 expression also showed a corresponding increase. This intriguing relationship suggests potential crosstalk between the ERBB2 and SMAD4 pathways in GBC among Indian patients. Although such findings were documented in other GI tract cancers in different parts of the world, we confirm the same mechanisms operating in GBC in Indian patients. However, when the expression status of all the 6 genes were taken together, it is not possible to make a concrete prediction as to whether simultaneous overexpression of 2, 3 or 4 genes have any synergistic or antagonistic relation with downregulation of CDKN2A and SMAD4 taken together/independently for east Indian patient population.
Our study highlights the need for further research to unravel the intricate molecular mechanisms governing EGFR and SMAD4 expression in GBC, particularly among Indian patients. These findings can have significant implications for understanding the pathogenesis of GBC and may contribute to the development of targeted therapies tailored to specific populations.
Past NGS studies on GBC showed that the expression of EGFR, ERBB2/Her2-neu, CCND1, Myc, SMAD4, and CDKN2A were dysregulated most frequently. In a study, Feng et.al. 2011, pointed out the role of CCND1/CDK4/p16 pathway in GBC [27]. They also proposed that expression of CCND1 increased along with the progression of the disease [28]. Another study found that the expression rates of abnormal cyclin D1 were observed in 68.3% GBC and 57.1% gallbladder adenoma and these were significantly higher than those found in chronic cholecystitis (7.1%). Specimens with Cyclin D1 overexpression showed a high incidence of lymphatic permeation, venous permeation, lymph node metastasis and was frequently observed in adenocarcinomas and even in adenomas, but not in any specimen of normal epithelium or adenomyoma. This results strongly suggests that increased cyclin D1 probably play a critical role in the transformation of gallbladder epithelium cells in GBC. Our study also revealed that CCND1 expression was statistically significantly higher in GBC patient cohort with respect to their adjacent normal counterpart (p = 0.04). Besides this, overexpression of cyclin D1 proteins was detected by immunostaining in 41% of GBC.
Disruption of cell cycle is universal in tumors, and the most common abnormalities of this type involve the RB-CDK-INK4A pathway. Cyclin dependent kinases (CDKs) are in turn regulated by CDK inhibitors such as INK4A (also known as p16INK4 and encoded by CDKN2A). In addition to the overexpression of cyclin D1 gene described previously in GBC, a modest number of studies indicate that CDKN2A might have an important role in GBC associated with gallstone [29]. Deletions at the CDKN2A region (9p21) have been reported in half of GBC, suggesting that the loss of tumor suppressor activity of p16 may play a role in the early onset of preneoplastic lesions. Inactivation of CDKN2A in GBC can occur by deletion, mutation, methylation, homozygous deletions, and protein expression but the mechanism of inactivation is still not well understood. Combined data from whole exome and targeted sequencing studies, explored that only 5.9% of the GBC patients showed CDKN2A mutations which is very low in GBC suggesting that CDKN2A inactivating mutation frequency is lower than other inactivating mechanisms such as homozygous deletions, LOH, promoter hypermethylation and mRNA expression. Our study also explained the lower expression of CDKN2A, in gallbladder tumors with respect to their normal epithelium tissues measured by mRNA expression. Overall, CDKN2A expression was not statistically significant in 68 tumor tissues (p = 0.07). SMAD4 inactivation and alterations in transforming growth factor (TGF) beta family receptors were frequently observed in GBC and other biliary tract malignancies [12]. TGF- β suppresses tumor formation by blocking cell cycle progression, although this tumor-suppressive function is often lost in pancreatic adenocarcinoma cells by inactivation of SMAD4, which acts as a signalling mediator in the TGF- β signalling pathway [30, 31]. Thus, downregulation of TGF- β means loss of its tumor suppressive effect, which is corroborated in our studies, where we have observed decreased expression of TGF- β in GBC tissue samples.
Genetic studies have shed light on the role of specific genes in GBC development. KRAS, a proto-oncogene, has been implicated in several signal transduction pathways and associated pathways in the late stages of GBC malignancy. Mutations in codon 12 of the KRAS gene have been reported in GBC tissue, although the timing of these mutations in GBC pathogenesis remains under investigation. KRAS mutations exhibit significant variability across different populations, with Eastern Asian patients showing higher frequencies compared to Western populations [32]. Whereas, three NGS studies from whole-exome sequencing studies reported 7.8 -30% of KRAS mutations in their studies [33]. Most of the KRAS mutation has been aggregated on codon 12, the second nucleotide of codon 12 attributed to a G to A and G to C transition. GBC associated with an anomalous pancreatobiliary duct junction (APBDJ) has a high KRAS mutation rate (50–83%) at codon 12 in early stages, along with that overall the KRAS mutation rates vary from 10–67% in different studies [34]. But our study showed lower percentage (11.5%) of KRAS mutation. Kumari et al. also reported from Indian study that 1 out of 49 patients has KRAS mutation [35]. The observed lower KRAS mutation rate in Indian GBC patients, in contrast to higher rates in other populations, challenges the notion of KRAS as a predominant driver in Indian GBC.
The ErbB signalling pathway, involving receptors like EGFR and ERBB2, has also been linked to GBC. The expression levels of EGFR and ERBB2 vary with GBC stages, and studies have reported overexpression patterns from early to advanced stages of GBC and involved in cancer initiation and progression [22, 36]. Chronic bacterial cholangitis, primarily due to Salmonella and Helicobacter infections, also increases the risk of biliary tract malignancy. In our study, we did not find evidence of H. pylori or S. typhi infection in GBC patients. Gallstone disease shows remarkable geographical variations, being more common in certain regions like the UK, USA, and Europe and less common in Africa, China, and Japan [37]. In India, gallstone disease is more prevalent in the northern and eastern parts of the country, particularly among younger women. This regional variation is also reflected in our study, where a majority of patients were female from eastern zone of India [38].
Our findings demonstrated that the genotype and allele frequencies of ERBB2/Her2-neu Ile655Val polymorphism among GBC was not significantly different, we also found that “Val” allele or carriers of “Val” allele or (Ile/Val + Val/Val) genotype were not significantly associated with GBC patient in our cohort. Similarly, we also found that genotype and allele frequencies of the CCND1 G870A polymorphism were not significantly different in the same patient cohort. Our data showed that ERBB2/Her2-neu Ile655Val, and CCND1 A870A gene polymorphisms may not be potential markers for GBC prognosis at least in a hospital based Indian population.
Alterations in genes such as EGFR, SMAD4, and others in gallbladder cancer in this study could potentially be linked to arsenic or heavy metal contamination, as indicated by recent studies conducted in regions with heavy metal exposure. Arsenic, a prevalent heavy metal contaminant in groundwater, has been associated with various malignancies, including gall bladder. Studies conducted in areas like the Middle Ganga Plain in Bihar, India, have revealed significant arsenic contamination in tube wells, surpassing safety thresholds [39].Investigations in countries with high arsenic exposure, such as Chile, have demonstrated higher arsenic concentrations in gall bladder tissue samples compared to non-cancerous gallbladder tissues. These findings suggest a potential association between arsenic exposure and the molecular alterations observed in gall bladder, including changes in EGFR, SMAD4, and other genes implicated in cancer development and progression [40].
Conclusion
In conclusion, combining the data, Epidemiology and clinical profile of GBC in our study was similar to the findings in previous literature. The sample data shows high prevalence of GBC among women compared to men. Approximately, 82% of GBC patients had a gallstone history as is consistent with other studies conducted in India on GBC. In our study, we found a negative correlation between EGFR and SMAD4 expression levels in Indian GBC samples, in consistent with typical negative correlation reported in previous studies in other GI tract cancers. We also found ERBB2/Her2-neu amplification/overexpression in 30% of the GBC samples, which is consistent with previous studies in this field. The study also demonstrates SMAD4 downregulation and MYC overexpression to be mutually exclusive in majority of the GBC samples. We also found a low percentage (11.5%) of GBC samples to show KRAS codon 12 mutation, indicating a lower KRAS mutation frequency among Indian GBC patients. However, no GSD tissue samples showed KRAS mutation, indicating KRAS mutation has got no role in early gall bladder dysplasia to cancer progression. We also found a significant TGF- β downregulation in GBC tissue samples.
Supplementary Information
Acknowledgements
This work was supported by the grant sanctioned for N.S. by the Government of West Bengal, Department of Higher Education, Science and Technology and Biotechnology. The grant number RD-37-2016 dated 14/03/2018. N.S. also funded a scheme Research Internship Programme in Biotechnology Based Science and Education (RISE) fellowship to PI for M.Sc. Summer students from the Government of West Bengal, Department of Higher Education, Science and Technology and Biotechnology. N.S. was thankful to Dr. Shalini Dutta, from Indian Statistical Institute, Prof. Santasabuj Das, and Prof. Asish Mukhopadhyay, from National Institute for Cholera and Enteric Diseases, Kolkata for providing DNA of H.pylori strain and S.typhi positive Strains and primers. N.S. acknowledged to Prof. Bidyut Roy, Indian Statistical Institute for valuable discussions for data analysis for this manuscript. N.S. also thankful to all the patients for their participation in this study. Dr. Gourab Saha, Mr. Riju Ghosh, Miss. Anwesha Das, and Miss. Asmita Biswas, from Human Genetics Unit, Indian Statistical Institute, also helped in patient’s data, specimen collection and research work are gratefully acknowledged. N.S. thanks to Master Saraswan Sikdar.
Financial and competing interests’ disclosure
The work was supported also by the Department of Higher, Education, Science & Technology and Biotechnology, Government of West Bengal, India. (BT/P/Budget/RD-37/2016). The corresponding author N.S. is supported by the funding from the Department of Biotechnology, Government of India (RLS/BT/Re-entry/05/2012) The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial’ interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. No writing assistant was utilized in the production of the manuscript.
Abbreviations
- GBC
Gallbladder Cancer
- GSD
GallStone Disease
- RFLP
Restriction Fragment Length Polymorphism
- SNP
ERBB2: Single Nucleotide Polymorphism
- ISI
Indian Statistical Institute
- IRB
Institutional Review Board
- ICMR
Indian Council of Medical Research
- AJCC
American Joint Committee on Cancer
- UICC
Union for International Cancer Control
- WHO
World Health Organization
- ASPCR
Allele Specific PCR
- APBDJ
Anomalous Pancreatobiliary Duct Junction
Authors’ contributions
S.C. K.G. S.P. San.G. and P.C. performed the experiments and analyzed the data. S.S., V.S., J.M., and A.M., collected specimen and clinicopathological data from patients. San.G. and B.B helped in immunohistochemistry work. B.K.C., Shi.G., and S.D. are clinicians who collaborated in this project and have done clinical analysis, and are involved in the clinical part of the manuscript writing. S.C. and San.G are involved in manuscript writing. N.S. designed the experiments, performed the experiments, collected specimens, involved in data analysis, manuscript writing and editing, data interpretation and corresponds to the manuscript.
Funding
We do not have funding availability for the publication.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The studies involving human participants were reviewed and approved by the Institutional Research Ethics Review Board of every hospitals and institutions involved and the IRB certificate is attached. The two state hospitals included are Medical College & Hospital, Kolkata, SSKM & I.P.G.M.E.&R Hospital, Kolkata. The study was approved by the Institutional Ethics Committee of Indian Statistical Institute (ISI). The patients/participants provided their written informed consent to participate in this study. All our methods followed the guidelines of the Helsinki declaration.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Goetze TO. Gallbladder carcinoma: Prognostic factors and therapeutic options. World J Gastroenterol. 2015;21:12211–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lowenfels AB, Lindström CG, Conway MJ, et al. Gallstones and risk of gallbladder cancer. J Natl Cancer Inst. 1985;75:77–80. [PubMed] [Google Scholar]
- 3.Zhu AX, Hong TS, Hezel AF, et al. Current management of gallbladder carcinoma. Oncologist. 2010;15:168–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nemunaitis JM, Brown-Glabeman U, Soares H, et al. Gallbladder cancer: Review of a rare orphan gastrointestinal cancer with a focus on populations of New Mexico. BMC Cancer. 2018;18(1):665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Domenico EG Di, Cavallo I, Pontone M, et al. Biofilm Producing Salmonella Typhi: Chronic Colonization and Development of Gallbladder Cancer. Int J Mol Sci 2017;18 [DOI] [PMC free article] [PubMed]
- 6.Cen L, Pan J, Zhou B, et al. Helicobacter Pylori infection of the gallbladder and the risk of chronic cholecystitis and cholelithiasis: A systematic review and meta-analysis. Helicobacter 2018;23 [DOI] [PubMed]
- 7.Mishra SK, Kumari N, Krishnani N. Molecular pathogenesis of gallbladder cancer: An update. Mutat Res. 2019;816–818:111674. [DOI] [PubMed] [Google Scholar]
- 8.Marks EI, Yee NS. Molecular genetics and targeted therapeutics in biliary tract carcinoma. World J Gastroenterol. 2016;22:1335–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma A, Sharma KL, Gupta A, et al. Gallbladder cancer epidemiology, pathogenesis and molecular genetics: Recent update. World J Gastroenterol. 2017;23:3978–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naidu MSK, Suryakar AN, Swami SC, et al. Oxidative stress and antioxidant status in cervical cancer patients. Indian J Clin Biochem. 2007;22:140–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldin RD, Roa JC. Gallbladder cancer: a morphological and molecular update. Histopathology. 2009;55:218–29. [DOI] [PubMed] [Google Scholar]
- 12.Dardare J, Witz A, Merlin J-L, et al. SMAD4 and the TGFβ Pathway in Patients with Pancreatic Ductal Adenocarcinoma. Int J Mol Sci. 2020;21:3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alqahtani SA, Alghamdi IG. Epidemiology of gallbladder cancer in Saudi Arabia. Cancer Manag Res. 2020;12:9527–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Acharya MR, Patkar S, Parray A, et al. Management of gallbladder cancer in India. Chin Clin Oncol. 2019;8:35. [DOI] [PubMed] [Google Scholar]
- 15.Singh S, Bhatnagar S, Lohia N, et al. Epidemiological and Geographical Profile of Gall Bladder Cancer Patients from a Hospital-based Registry of Northern Gangetic Plains. Asian Pac J Environ Cancer. 2022;5:3–10. [Google Scholar]
- 16.Sen U, Sankaranarayanan R, Mandal S, et al. Cancer patterns in eastern India: the first report of the Kolkata cancer registry. Int J Cancer. 2002;100:86–91. [DOI] [PubMed] [Google Scholar]
- 17.Maurya SK, Tewari M, Mishra RR, et al. Genetic aberrations in gallbladder cancer. Surg Oncol. 2012;21:37–43. [DOI] [PubMed] [Google Scholar]
- 18.Matsushita M, Matsuzaki K, Date M, et al. Down-regulation of TGF-beta receptors in human colorectal cancer: implications for cancer development. Br J Cancer. 1999;80:194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pérez-Moreno P, Riquelme I, García P, et al. Environmental and Lifestyle Risk Factors in the Carcinogenesis of Gallbladder Cancer. J Pers Med 2022;12 [DOI] [PMC free article] [PubMed]
- 20.Huang J, Lucero-Prisno DE, Zhang L, et al. Updated epidemiology of gastrointestinal cancers in East Asia. Nat Rev Gastroenterol Hepatol. 2023;20:271–87. [DOI] [PubMed] [Google Scholar]
- 21.Verma P, Gupta P, Gupta N, et al. HER2/ERBB2 overexpression in advanced gallbladder carcinoma: comprehensive evaluation by immunocytochemistry and fluorescence in situ hybridisation on fine-needle aspiration cytology samples. J Clin Pathol 2023; [DOI] [PubMed]
- 22.Roa I, de Toro G, Schalper K, et al. Overexpression of the HER2/neu Gene: A New Therapeutic Possibility for Patients With Advanced Gallbladder Cancer. Gastrointest Cancer Res. 2014;7:42–8. [PMC free article] [PubMed] [Google Scholar]
- 23.Li M, Liu F, Zhang F, et al. Genomic ERBB2/ERBB3 mutations promote PD-L1-mediated immune escape in gallbladder cancer: a whole-exome sequencing analysis. Gut. 2019;68:1024–33. [DOI] [PubMed] [Google Scholar]
- 24.Moz S, Basso D, Bozzato D, et al. SMAD4 loss enables EGF, TGFβ1 and S100A8/A9 induced activation of critical pathways to invasion in human pancreatic adenocarcinoma cells. Oncotarget. 2016;7:69927–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao S, Wang Y, Cao L, et al. Expression of oncogenic K-ras and loss of Smad4 cooperate to induce the expression of EGFR and to promote invasion of immortalized human pancreas ductal cells. Int J Cancer. 2010;127:2076–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y-W, Hsiao P-J, Weng C-C, et al. SMAD4 Loss triggers the phenotypic changes of pancreatic ductal adenocarcinoma cells. BMC Cancer. 2014;14:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng Z, Chen J, Wei H, et al. Feng et al. Exp Mol Pathol 2011;91:569–77. [DOI] [PubMed]
- 28.Doval DC, Azam S, Sinha R, et al. Expression of epidermal growth factor receptor, p53, Bcl2, vascular endothelial growth factor, cyclooxygenase-2, cyclin D1, human epidermal receptor-2 and Ki-67: Association with clinicopathological profiles and outcomes in gallbladder carcinoma. J Carcinog. 2014;13:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roa JC, Vo Q, Araya JC, et al. [Inactivation of CDKN2A gene (p16) in gallbladder carcinoma]. Rev Med Chil. 2004;132:1369–76. [PubMed]
- 30.Ahmed S, Bradshaw A-D, Gera S, et al. The TGF-β/Smad4 Signaling Pathway in Pancreatic Carcinogenesis and Its Clinical Significance. J Clin Med 2017;6 [DOI] [PMC free article] [PubMed]
- 31.Massagué J. TGFβ in Cancer. Cell. 2008;134:215–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yokoyama M, Ohnishi H, Ohtsuka K, et al. KRAS Mutation as a Potential Prognostic Biomarker of Biliary Tract Cancers. Jpn Clin Med. 2016;7:33–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li M, Zhang Z, Li X, et al. Whole-exome and targeted gene sequencing of gallbladder carcinoma identifies recurrent mutations in the ErbB pathway. Nat Genet. 2014;46:872–6. [DOI] [PubMed] [Google Scholar]
- 34.Iyer P, Shrikhande SV, Ranjan M, et al. ERBB2 and KRAS alterations mediate response to EGFR inhibitors in early stage gallbladder cancer. Int J Cancer. 2019;144:2008–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumari N, Corless CL, Warrick A, et al. Mutation profiling in gallbladder cancer in Indian population. Indian J Pathol Microbiol. 2014;57:9–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abeer I, Khan S, Hasan Mohd J, et al. EGFR and HER2neu Expression in Gall Bladder Carcinoma and Its Association with Clinicopathological Parameters - An Institutional Experience from North India. Asian Pac J Cancer Biol. 2021;6:57–65. [Google Scholar]
- 37.Sikdar N. The Geographical, Ethnic Variations and Risk factors of Gallbladder Carcinoma: A Worldwide View. J Investigative Genom 2016;3
- 38.Kapoor VK, McMichael AJ. Gallbladder cancer: an “Indian” disease. Natl Med J India. 2003;16:209–13. [PubMed] [Google Scholar]
- 39.Chakraborti D, Mukherjee SC, Pati S, et al. Arsenic groundwater contamination in Middle Ganga Plain, Bihar, India: a future danger? Environ Health Perspect. 2003;111:1194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shridhar K, Krishnatreya M, Sarkar S, et al. Chronic Exposure to Drinking Water Arsenic and Gallbladder Cancer Risk: Preliminary Evidence from Endemic Regions of India. Cancer Epidemiol Biomarkers Prev. 2023;32:406–14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.