Abstract
Over the last three decades, human genetics has gone from dissecting high-penetrance Mendelian diseases to discovering the vast and complex genetic etiology of common human diseases. In tackling this complexity, scientists have discovered the importance of numerous genetic processes – most notably functional regulatory elements – in the development and progression of these diseases. Simultaneously, scientists have increasingly used multiplex assays of variant effect to systematically phenotype the cellular consequences of millions of genetic variants. In this article, we argue that the context of genetic variants – at all scales, from other genetic variants and gene regulation to cell biology to organismal environment – are critical components of how we can employ genomics to interpret these variants, and ultimately treat these diseases. We describe approaches to extend existing experimental assays and computational approaches to examine and quantify the importance of this context, including through causal analytic approaches. Having a unified understanding of the molecular, physiological, and environmental processes governing the interpretation of genetic variants is sorely needed for the field, and this perspective argues for feasible approaches by which the combined interpretation of cellular, animal, and epidemiological data can yield that knowledge.
Introduction
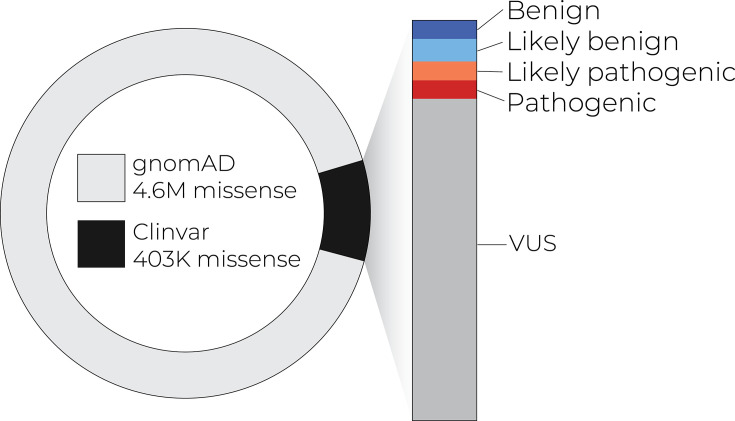
As a consequence of stunning technological advances – especially in DNA-sequencing – current databases hold hundreds of millions of human single-nucleotide variants, with nearly 5 million in the tiny portion of the genome that encodes protein sequence (Figure 1). Yet our knowledge of the functional effects of all this variation is vanishingly small: even for the changes that result in amino acid replacements, only about 2% have been clinically interpreted, and about 80% of those have been interpreted as ‘variants of uncertain significance’ (Fayer et al., 2021; Starita et al., 2017; Figure 1). Breaking through this interpretative bottleneck constitutes a central challenge for human genomics research.
Figure 1. Only a small number of coding variants have annotations that can guide diagnosis and treatment.
As exome and whole-genome sequencing becomes commonplace in the clinic, the number of variants of uncertain significance is likely to increase.
Multiplex assays of variant effect (MAVEs) experimentally assess the effects of single-nucleotide variants at scale. Here, a single open-reading frame, exon, or regulatory region is saturated with all possible single-nucleotide changes, and a single property is measured via coupling this property to the number of DNA sequence reads of each variant before and after a functional selection (Esposito et al., 2019; Findlay, 2021; Kinney and McCandlish, 2019; Starita et al., 2017). We and others have shown that the resulting functional data can reveal whether and how each variant alters function, and that the functional data empower the interpretation of variants of uncertain significance (Fayer et al., 2021; Scott et al., 2022; Starita et al., 2017; Sun et al., 2016; Tabet et al., 2022; Weile et al., 2021; Wu et al., 2021; Yang et al., 2017). For example, the integration of multiplex functional data for cancer-related genes led to the reinterpretation of ~70% of variants of uncertain significance in TP53, ~50% in BRCA1, and ~15% in PTEN (Fayer et al., 2021). Spurred on by these and other results, the first generation of MAVEs is being deployed widely (Da et al., 2021; Esposito et al., 2019; Fowler et al., 2021; Rubin et al., 2021), and comprehensive variant effect maps for easy-to-measure cellular properties, such as growth, are within reach for many clinically relevant human genes.
Despite this success, we are far from being able to reliably interpret the organismal effects of all human genetic variation, much less to use genetic information to accurately predict individual phenotype and disease risk. For one, MAVEs have not been extended to structural variation, copy number variation in repetitive DNA, and other large and complex variants that are likely numerous and highly impactful (Manolio et al., 2009; Miller et al., 2021; Mitra et al., 2021; Press et al., 2014; Press et al., 2019). To make such complex variation accessible for phenotyping in high throughput, new experimental and computational approaches are needed. Yet even for single-nucleotide variants for which MAVEs exist or can be envisioned, major challenges for accurate variant effect interpretation remain. Existing MAVEs generally do not account for genetic, environmental, or tissue/developmental context (Figure 2). Assessing and perturbing this context is essential for fully characterizing the effect of a genetic variant (Claussnitzer and Susztak, 2021). This lack of context constrains the utility of MAVEs for understanding how variants interact with genetic background, affect non cell-autonomous phenotypes, and alter organismal phenotype in interplay with environmental factors. Here, we explore the importance of each of these contexts for understanding single-nucleotide variants and describe a next generation of MAVEs, applicable to both coding and regulatory variants, and computational approaches that, together, can generate contextually informative variant effect maps and predict individual disease risk.
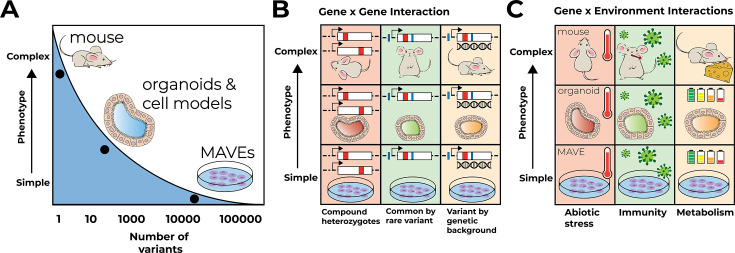
Figure 2. Multiplex assays of variant effect (MAVEs) in context.
(A) MAVEs in cell lines can assay many variants for simple phenotypes like cell growth. Models like organoids and mice allow for measuring complex multicellular phenotypes like proportions of cell types but are currently limited to assaying only a few variants at a time. (B) Gene–gene interactions are examined in different models at different levels of phenotype complexity. Gene–gene interactions suggested for prioritization include compound heterozygotes, combinations of common and rare variants in a given locus in cis and trans, and experiments testing variants on different genetic backgrounds. (C) Gene–environment interactions are examined at different levels of phenotype complexity. Three broad categories are suggested to model the complexity of environmental context in the laboratory: abiotic stress, challenges to immunity, and metabolism.
Variant effect mapping in genetic context
Onset, severity, and even incidence of disease can differ widely among carriers of a given disease-associated variant. This is particularly true for small and moderate effect variants that are associated with common diseases through genome-wide association studies (GWAS). The heritability explained by GWAS variants tends to be small, and these variants typically have little power to predict the disease risk of individuals (Eichler et al., 2010; Gibson, 2012; Khera et al., 2018; Manolio et al., 2009). This ‘missing heritability’ of common diseases and the differences in variant expressivity among patients with highly penetrant, rare Mendelian disorders are commonly attributed to uncharacterized non-additive genetic interactions, among other factors (Eichler et al., 2010; Gibson, 2012; Manolio et al., 2009).
Non-additive genetic interaction, or epistasis, describes the phenomenon that the combined effect of alleles at two or more loci deviates from the sum of their individual effects (Fisher, 1919; Mackay, 2001). The importance of non-additive genetic interactions, in particular for complex traits, has been the subject of a long-standing debate (Fisher, 1930; Hivert et al., 2021; Wade and Goodnight, 1998; Wright, 1931), and the importance of epistasis across the allele frequency spectrum is not well understood. Model organism research has identified many examples of non-additive genetic interactions affecting a wide variety of morphological and quantitative traits in fungi, animals, and plants (Costanzo et al., 2019; Forsberg et al., 2017; Mackay, 2014; Press et al., 2014). These genetic interactions often reflect functional relationships such as those among genes coding for subunits of a multimeric protein complex or proteins functioning in a common pathway (Costanzo et al., 2019; Miller and Piccolo, 2020; Narasimhan et al., 2017; Ni et al., 2017; Pamplona-Cunha et al., 2020). Quantitative genetics theory and empirical data for human and crop traits show that additive genetic models can explain over half of the total genetic contributions to complex traits (Darnell et al., 2022; Dudley, 2007; Hivert et al., 2021; Sheppard et al., 2021; Visscher et al., 2008). However, accounting for non-additive genetic effects, and particularly epistatic interactions, can lead to more accurate phenotype predictions (Carlborg et al., 2006; Forsberg et al., 2017; Lachowiec et al., 2015; Matsui et al., 2022). Methods that examine genetic ancestry and portability can also improve prediction and estimation in the presence of these effects (Brown, 2016; Park et al., 2018; Patel et al., 2022).
Because a large majority of disease-associated GWAS variants reside within or in linkage with accessible chromatin regions, complex diseases are assumed to arise through the additive action of many regulatory variants (Maurano et al., 2012; Meuleman et al., 2020). The enrichment of GWAS variants in or near regulatory regions, their large numbers, and their small contribution to heritability and disease risk were conceptualized in the ‘Omnigenic’ model (Boyle et al., 2017; Liu et al., 2019). This theoretical framework posits that ‘core genes’, which are functionally related to a phenotype of interest, carry common, small-effect, significantly trait-associated variants that together contribute little to heritability while the bulk of heritability is contributed by a huge number of common ‘peripheral’ variants with individually tiny effects that ultimately affect core gene expression. Peripheral variants are uniformly distributed over the genome, with each chromosome contributing to trait heritability according to its size, reminiscent of Fisher’s Infinitesimal model (Barton et al., 2017; Walsh and Lynch, 2018). The Omnigenic model is not universally accepted (Connally et al., 2022; Sohail et al., 2019), but it offers an explanation for the missing heritability conundrum and is supported by recent empirical studies (Sinnott-Armstrong et al., 2021; Smith et al., 2022). If most heritability is not located at genes or regulatory regions directly altering the phenotype of interest, how can we use experimental variant effect mapping in context to understand disease risk? In any case, there are several ways that experimental variant effect mapping in context can be applied to understand disease risk.
An obvious approach is systematically studying compound heterozygotes (Figure 2B). There are many recessive variants of unknown molecular or cellular functional consequence in disease-associated genes, even amongst the thousands of genes that play a role in highly penetrant Mendelian disorders. Examples abound of cases in which individuals carry different alleles of a gene that together – as a compound heterozygote – alter gene activity enough to cause disease. These alleles may act additively or non-additively, enabling valuable mechanistic insights on disease origins, a first crucial step toward future therapy. Another promising approach is assaying combinations of common, disease-associated GWAS variants with all possible other variants in the same gene and regulatory region (Figure 2B), a likely scenario by which the penetrance and expressivity of a common variant may be altered. To address both of these scenarios, variants of a given gene or regulatory region will need to be assessed in pairwise fashion. Executing such experiments in human cells across all possible variant pairs will require some innovation in variant barcoding, genome engineering, and sequencing strategies to unambiguously link both variants with a cell’s phenotype. However, the systematic evaluation of compound heterozygotes and of combinations of GWAS variants with rare variants affecting the same locus will yield actionable information for genetic counselors, physicians and patients.
Experimentally addressing the consequences of more complex genetic interactions (Figure 2B) will require sophisticated technological innovations and novel data analysis methods. For example, variant libraries could be introduced into many different cell lines, including induced pluripotent stem cells (iPSCs) derived from diverse individuals, thereby testing the consequences and interactions of many different genetic variants, including structural variants. Identifying causal additive or non-additive genetic interactions in this scenario will be a formidable challenge. Alternatively, variant libraries could be introduced into cells carrying programmed genetic perturbations such as gene deletion, knockdown, or overexpression. Most ambitious, variant libraries for a disease-associated core gene could be introduced into cell lines carrying common variants in other core genes for the same trait, testing some assumptions of the Omnigenic model empirically. If it were possible to conduct variant effect mapping in whole animals, genetic crosses and high-throughput phenotyping could be used to interrogate the phenotypes of many different combinations of variant libraries with each other, other variant libraries, or possible modifier loci.
Most importantly, these experimental efforts must go hand in hand with innovative theoretical studies to enable predictions of variant effects in genetic context. For example, combining existing variant effect maps of disease-associated genes with genome-wide polygenic risk scores is one approach to investigate the Omnigenic model. Such a modeling approach could leverage the growing resources of biobanks and human phenotype data to identify modifier loci and refine variant effect maps (Schiabor Barrett et al., 2022; Tabet et al., 2022). Similarly, taking advantage of the multitude of high-quality GWAS, one could systematically explore loci that are significantly associated with different disorders; these loci represent candidates for genetic modifiers that alter the penetrance and expressivity of other variants (Lehner et al., 2006; Queitsch et al., 2012). One such locus is known: the major histocompatibility complex (MHC), which encodes cell surface proteins that are essential for the adaptive immune system by virtue of their ability to ‘display’ a cell’s repertoire of contained peptides to the immune system. The fact that different MHC alleles display different sets of variant peptides offers avenues for MHC variation to alter the penetrance and expressivity of many other variants. Stratifying existing GWAS by outliers, for example, focusing on individuals with the most and least severe disease phenotypes, might also identify genetic modifiers of variant penetrance and expressivity, where they exist (Gibson, 2009; Lehner et al., 2006; Queitsch et al., 2012). Although there is limited evidence for dominance in human genotype–phenotype mapping and epistasis remains difficult to quantify genome-wide thus far, we anticipate that understanding epistatic effects at individual genes will yield valuable insight into the extent of these epistatic interactions and their phenotypic consequences.
Variant effect mapping in cell, tissue, and developmental contexts
Although variants in some genes affect all cell types similarly across development, variants more often exert their effect in specific tissues, and at specific stages of development. For example, pathogenic germline variants in BRCA1 and BRCA2 genes, which play a fundamental role in DNA repair, confer a greatly increased risk of some, but not all, types of cancer (Welcsh and King, 2001). Variants affecting metabolic or neurodevelopmental disorders like atypical pantothenate kinase-associated neurodegeneration and schizophrenia can show effects in specific tissues and/or developmental stages (Immonen et al., 2017; Kurian and Hayflick, 2013). Moreover, different variants in the same gene can affect different tissues and cause different diseases. For example, pathogenic germline variants in LMNA most often cause cardiomyopathy, but can also cause muscular dystrophy, lipodystrophy, or the premature aging syndrome progeria (Novelli and D’Apice, 2003). This complexity only expands with regulatory variants at a given locus, where enhancer elements are able to activate multiple genes (Hu and Tee, 2017), causing pleiotropic effects on tissue specificity. While recent approaches have dramatically improved the linkage of regulatory variants to target genes (Andersson et al., 2014; Dey et al., 2022; Gschwind et al., 2023; Nasser et al., 2021; Kundaje et al., 2015), this remains an area of substantial ongoing work. Even so, numerous existing studies have begun to tackle the application of MAVE technologies to regulatory regions (Gasperini et al., 2016; Gordon et al., 2020; Kircher et al., 2019; Klein et al., 2018; Morova et al., 2023; Vockley et al., 2015). These studies rely on the same assumptions as coding variant MAVEs, namely that there is a direct link to the phenotype being selected and that variants have variable effects on that phenotype in the cell type being screened.
Tissue- and developmental stage-specific effects cannot be addressed in human cell lines that lack the expression programs, proteomes, and cellular structures found in fully differentiated cells. However, most MAVEs have been executed in human cell lines and have focused on easily screenable phenotypes like cell growth, protein abundance, or protein–protein interactions. Recent efforts to develop MAVEs based on rich phenotypes like cell morphology or transcriptional programs have demonstrated the potential of investigating these more complex phenotypes (Hasle et al., 2020; Martin-Rufino et al., 2023; Ursu et al., 2022; Xu et al., 2023). Disentangling the relationships between phenotypes is challenging, primarily as a result of pleiotropy (Solovieff et al., 2013). However, MAVEs test the entire set of possible alleles in a gene or regulatory region, allowing us to determine the full spectrum of allele-specific effects on phenotypes that can be modeled in a scalable assay.
Moreover, researchers have begun to explore iPSC-derived differentiated cells to model variant effects (Bajpai et al., 2021). However, owing to the challenges in engineering iPSC genomes, MAVEs in these cells are in their infancy (Lv et al., 2018). A potentially more powerful approach would be to isolate specific primary cell populations from individual patients (Claussnitzer et al., 2015; Shifrut et al., 2018), introduce variant libraries, and ascertain variant effects on cellular phenotypes. Complementing these efforts with other approaches like base or prime editing (Erwood et al., 2022; Hanna et al., 2021) could lead to the characterization of large variant libraries in the correct cellular context. Successful proof-of-principle studies demonstrate the potential of this approach (Martin-Rufino et al., 2023). However, these approaches are predicated on knowing the correct cell type(s) for the phenotype of interest. This information is often not available, but deriving robust cell type–disease associations is the subject of significant ongoing work (Tabet et al., 2022; Jones et al., 2022). Recent computational methods have also opened the possibility of accurate estimation of cell types of action in silico (Jagadeesh et al., 2022; Yu et al., 2022).
However, even differentiated cells do not recapitulate cell–cell interactions found in human tissues, much less the complex ballet of interactions required for normal development (Dorrity et al., 2022; Saunders et al., 2022). To model cell–cell communication will require multicellular models. Organoids, especially those derived from patient cells, offer a possible solution as they can model various types of tissues (Hendriks et al., 2021; Meng et al., 2022). The main challenges here are developing genome engineering approaches that can yield a large set of clonal organoids while accounting for the sometimes large phenotypic variation among organoids of the same type generated with the same protocol; such efforts are in progress (Sockell et al., 2022).
Model organisms are another option. Multicellular model organisms from worms to mice have proven extraordinarily useful in probing the tissue and developmental effects of individual genetic variants. However, to map variant effects at large scale in a model organism, it must produce large numbers of offspring to allow for adequate coverage of variants and high-throughput phenotypic screens of developmental or behavioral traits; The ideal organism would offer strong conservation of genes and pathways affected in human disease. Model organism approaches are particularly powerful when paired with single-cell transcriptomic readouts, as was used to investigate the consequences of 23 genetic perturbations affecting the development of cell lineages in zebrafish (Danio rerio) (Saunders et al., 2022). Leveraging single-cell transcriptomics, large numbers of replicates, and sophisticated statistical analysis, this study ascertained the consequences of a particular perturbation on the variance in cell type abundance organism-wide and detected the perturbation-dependent effects on cell type composition relative to wild-type embryos. Theoretically, single-cell genomics could be applied to a large number of small animals, each expressing a single variant or variant combinations, to similarly determine the consequences of genetic variants on cell type composition, gene expression, chromatin accessibility, protein abundance, and other single-cell phenotypes.
However, to enable MAVEs in whole animals, new technology is needed. The biggest challenge is that variant libraries must be introduced into animals such that thousands or more variants are represented across many thousands of animals. Germline editing is preferred to generate distinct, clonal recombinant animals, avoiding mosaicism and easing phenotype interpretation. Considering currently available genetically tractable models and their offspring numbers, the few in which such technology development seems worthwhile include Caenorhabditis elegans, Drosophila melanogaster, and Danio rerio. These animal models lend themselves readily to ascertaining the phenotypes of compound heterozygotes and variant combinations in different genes through crosses. However, it remains a formidable, unsolved challenge to successfully introduce large variant libraries into an animal germline such that each genome contains just one variant, each variant is present in many animals for replication of variant effects, and each variant is expressed at the same level.
Variant effect mapping in environmental context
Environmental context (gene–environment interactions) plays a large role in variant penetrance and expressivity, particularly for variants associated with common diseases such as diabetes, asthma, depression, and cardiovascular disease. Their incidence has risen sharply in recent decades in the United States and elsewhere (Bingley and Gale, 1989; Cockram, 2000; Ebrahim et al., 2010; Gibson, 2009; Harsanyi et al., 2022; Rewers et al., 2018; Sørensen, 2000). Because human genetic makeup has not fundamentally changed in the last 50 years, changing environmental context has either altered the genetic contributions of a subset of polymorphisms or shifted the liability threshold for these disorders. Fundamental recent changes to environmental context include altered pathogen exposure and microbiomes through increased hygiene, refrigeration, and antibiotics; radical dietary shifts toward industrially produced food; and environmental stress through artificial light, house dust, and harmful chemicals. Although we posit that environmental context is more crucial than genetic or developmental context for understanding variant penetrance and expressivity, environment is also the hardest context to fully define and measure in the laboratory.
The challenges of exploring environmental context in humans are well illustrated by the TEDDY study (The Environmental Determinants of Diabetes in the Young) (Rewers et al., 2018). The study followed ~9000 children who carry high-risk alleles for type I diabetes for 15 years, collecting clinical metadata (e.g., diet, household exposures, medications, pre- and perinatal exposures, psychosocial stressors, among many others), and the results of many ‘omics’ analyses (e.g., whole-genome sequencing, metabolomics, microbiome, lipidomics, transcriptomics, proteomics). One challenge was participant dropout (26%) due to the high participation effort (Johnson et al., 2011; Johnson et al., 2014). Another was the relatively small number of children who ultimately developed type 1 diabetes (~300). These logistical challenges limited the study’s power to detect gene–environment interactions. The published results suggest little effect of many environmental factors such as breastfeeding (Hummel et al., 2021), early antibiotic treatment (Kemppainen et al., 2017), vaccinations (Elding Larsson et al., 2018), and maternal exposures (Johnson et al., 2021b; Johnson et al., 2021a; Silvis et al., 2019), while a few studies report evidence for possible risk factors (infection) (Lönnrot et al., 2017; Vehik et al., 2019) or interventions (vitamin D, probiotics) (Norris et al., 2018; Uusitalo et al., 2016).
While it may be difficult to pinpoint relevant gene–environment interactions across large human populations, these studies nevertheless hold much promise, in particular when considered together with the results of experimental variant effect maps. For example, a dense variant effect map for a particular gene or regulatory region produced in a single condition could be supplemented with human phenotype data for carriers of particular variants. These individuals will have experienced a large range of environmental conditions, beginning in utero and continuing after birth. To estimate their gene–environment interactions, one would have to account for (potentially non-additive) genetic background, which can be accomplished through sibling study designs, kinship analyses, or use of polygenic risk scores (Mostafavi et al., 2020). The ever increasing size (i.e., sample number) and sophistication (i.e., inclusion of multiple traits, ancestries, and environmental exposures) of today’s GWAS make such an approach imminently feasible, though the approach might still be insufficient for traits with large epistatic components in their genetic architecture. Large sibling and twin cohorts collected over many decades may also aid in computational efforts to decipher the effect size of gene–environment interactions and the variants most affected. As siblings and twins share (to varying degrees) both their genome and their environment, increasing concordance of disease incidence and severity would be expected for relatives carrying genetic variants associated with moderate or low disease risk in GWAS and implicated as pathogenic or likely pathogenic in functional assays. If gene–environment interactions have changed, family studies will allow extrapolation of gene–environment effect size, if not necessarily identification of responsible environmental factors. The same approach can be applied in reverse using migration studies and other approaches for causal effect estimation from the social sciences to further isolate gene–environment effects (Lea et al., 2023; Figure 3). This can include direct approaches for modeling gene–environment interactions (Kerin and Marchini, 2020; Marderstein et al., 2021; Moore et al., 2019), as well as integration of causal effect size estimation using quasi-experimental approaches (Barcellos et al., 2018; Barcellos et al., 2021; Davies et al., 2018; Plotnikov et al., 2020; Abdellaoui et al., 2019; Ebrahim et al., 2010; von Hinke and Sørensen, 2023).
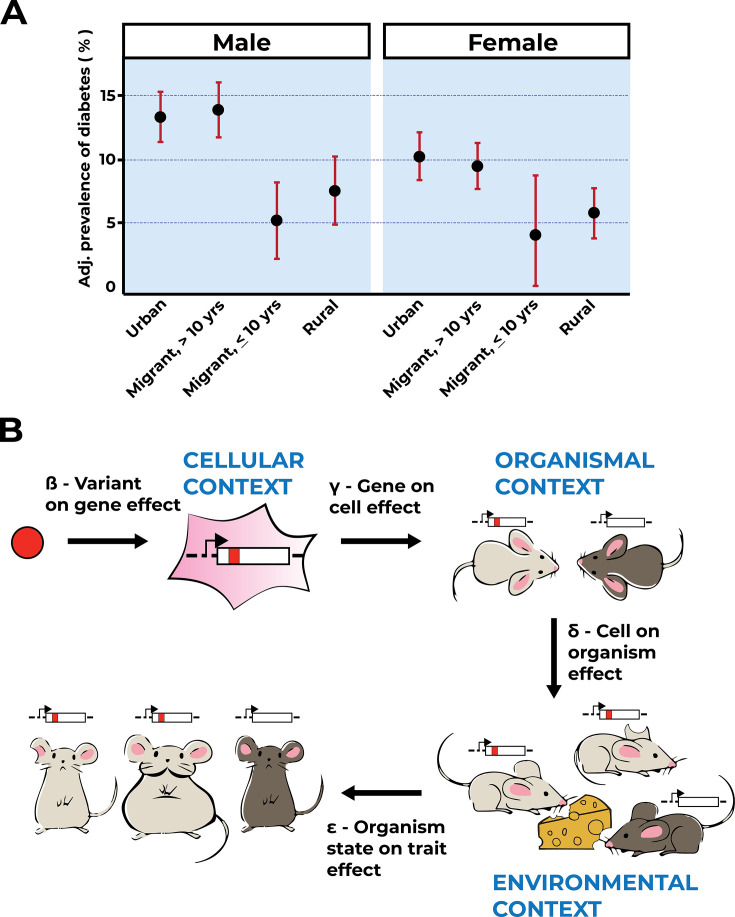
Figure 3. Environmental context is key to trait interpretation.
(A) Adapted from Figure 1 of Ebrahim et al., 2010, age-, factory-, and occupation-adjusted percent prevalence (95% CI) of diabetes by type of migrant and sex, Indian migration study 2005–2007. Diabetes is prevalent in urban residents and residents who migrated to urban areas and resided there for more than 10 years. (B) Gene–environment interactions will affect an organismal trait at the level of genes, cells, tissues, and whole organisms. Extending the Mostafavi et al., 2022 model to incorporate environmental context captures more relevant biology, and hence facilitates variant effect interpretation. As shown, a variant (red allele) affects a gene’s function within a particular cellular context. Cells affected by the red allele function exist within the context of the organism, here a light-gray mouse as compared to a dark-gray mouse that does not carry the red allele. Mice with or without the red allele are exposed to environmental factors, symbolized by the cheese as a dietary factor challenging metabolism. Note that one of the mice carrying the red variant is not exposed to this environmental challenge (light gray, clipped ear). In the example shown, environmental context determines the trait value (obesity, big light-gray mouse) for mice carrying the red variant.
Efforts to determine gene–environment interactions with MAVEs are nascent. An example is a recent collection of variant effect maps for MTHFR, encoding a key enzyme in folate metabolism. MTHFR deficiency can be severe, with diverse early-onset consequences of a massive accumulation of homocysteine in the blood, or relatively mild, with later-emerging thromboembolism. Milder MTHFR deficiency can be remediated for some patients by increasing folate levels. Still milder effects result from homozygosity of the common Ala222Val variant (30% global minor allele frequency), for which there is a risk of neural tube defects that is entirely remediated by sufficient dietary folate. Thus, the pathogenicity of variants is dependent on environmental context. This dependency of variant effect on the environmental context of folate levels (and on the genetic context of an A222V variant) was captured for nearly every MTHFR missense variant (Weile et al., 2021). Other examples include efforts to account for proteotoxic stress, for example, by evaluating transcription factor variants at elevated temperature or in response to chaperone inhibitors (Dorrity et al., 2018; Morton et al., 2020). None of these examples yet captures the full complexity of environmental context given that only a few environmental variables were each altered one at a time.
Addressing environmental context has to grapple with the overwhelming number of possible environmental factors that could alter a variant’s impact. For some genes, relevant environmental factors are known and these factors can be directly incorporated into multiplexed assays, as for MTHFR. Even for MTHFR, however, there is evidence that levels of riboflavin, as well as folate, can also remediate the effects of the common Ala222Val variant (Jarrett et al., 2022; McNulty et al., 2006), and the interactions across the broader set of MTHFR variants alone or in combination with folate and Ala222Val remain to be explored. More broadly, the profound changes in environmental context in the last 50–100 years suggest a focus on three broad categories: environmental factors altering metabolism, causing abiotic stress, and challenging the immune system. These broad categories can be modeled (with increasing degrees of difficulty) in the laboratory and studied both individually and in combination. Combinatorial studies are essential as responses to individual perturbations are often not predictive of responses to combinations, at least in model organisms (Ammeux et al., 2016). We envision environmental perturbations in cell lines, organoids, and even model animals carrying variant libraries (Figure 2C).
In addition to variant effect mapping across a range of external conditions, the internal conditions of the cells and organisms carrying variants can be drastically changed. The relative importance of external versus internal cellular environments is much debated (Billman, 2020). Here, an approach to induce mistranslation across the entire proteome for a given codon (Berg et al., 2019; Cozma et al., 2022) can manipulate internal cellular environments and allow their effects on individual variants to be measured. Similarly, one may consider manipulating splicing, protein folding, protein turnover, or mitochondrial function as general measures to affect cellular environment as it occurs in human aging (Johnson et al., 1999; Leutert et al., 2023). Analogously, pharmacological perturbation of key cellular processes could be applied to alter cell–cell interactions and niche formation to understand intra-organismal (i.e., organism-autonomous) effects of genetic variants.
Quantifying gene–environment effects on an organismal trait must consider prior knowledge of variant effects on gene function, the consequences of perturbed gene function on cellular function within the context of the whole organism, and an organism’s environmental exposures (Figure 3B). These multiple unknown factors are difficult to disentangle without acquiring knowledge of each component, but data from MAVEs enable us to do so systematically. Similar to pleiotropy (Solovieff et al., 2013), gene–environment interactions will affect an organismal trait at the level of genes, cells, tissues, and whole organisms (Figure 3B). Under the simplifying assumption that most genetic effects are due to variants acting on genes or their products, we can separate variant effects on a trait into two components, the contribution of that variant to the target gene and the effect of that gene on the trait of interest (Mostafavi et al., 2022). Incorporating environmental context into this model makes it substantially more complicated, but more reflective of biological reality, and hence more predictive of variant effects and disease risks (Figure 3B). The scale and highly controlled nature of MAVE-derived variant effects will make it possible to disentangle the contributions of environmental interactions from the two components as defined above.
Multiplex assays of variant effect in context to decipher mechanisms, improve disease risk predictions, and facilitate prevention and treatment
In the clinic, the individual matters. Each of us has our private set of common and rare variants and our unique environmental exposures that together determine our disease risk and prognosis. We argue that MAVEs in genetic, developmental, and environmental contexts will allow a systematic characterization of the relative importance of each context for variant effect as well as the characterization of variants particularly impacted by a given context or context combination. Moreover, as MAVEs that can measure phenotypes more complex than cell survival or cell fitness are deployed widely, we will begin to gather mechanistic insight on variant function at large scale and in response to multiple environmental perturbations. Current approaches to decipher biological mechanisms of specific variants through variant-to-function analyses remain laborious and highly specialized (Claussnitzer and Susztak, 2021; Tabet et al., 2022). As we must pursue mechanistic insights into variant effects to separate confounding from causal association, conducting MAVEs in context offers the promise of scale and systematic analysis. The more contextual multiplex variant functional data becomes available, the more modeling approaches that rely on vast datasets such as machine learning and artificial intelligence can be used to infer variant-specific and context-dependent biological mechanisms, thereby informing individualized predictions of disease risk for rare and common diseases and facilitating their prevention and treatment.
A key question is which genes and regulatory regions are most likely to yield the most useful information for precision medicine, and therefore should be the focus of MAVEs in context. A simple answer is to focus on the genes that contain many variants of uncertain significance and that are actionable with existing prevention and treatment options. This approach, however, will exclude many of the regulatory and coding GWAS variants associated with common disease for which context, in particular environmental context, will likely matter the most. If we are to understand complex common diseases, experimentalists and theoreticians will need to come together to innovate and develop strategies to study human variants at genome scale and in multiple contexts.
Acknowledgements
The authors thank S Gorjifard for figure design and execution, and T Templin for helpful feedback on this work. NS-A is funded by the Bruce G.Cochener Foundation through a Creativity Award and by a Fred Hutch Microbiome Research Initiative pilot grant. NS-A, SF, FR, LMS, CT, JV, DMF, and CQ are supported by NIH NHGRI 5RM1HG010461. CQ is supported by NIH NIGMS R35GM139532. FR was supported by a Canadian Institutes of Health Research Foundation Grant.
Appendix 1
Glossary of the terms used herein
Variant of uncertain significance
A genetic variant that has been identified but whose significance to disease etiology and health of an individual is not known. These variants cannot guide diagnosis or treatment.
Heritability
An attribute of a quantitative trait in a population that expresses how much of the observed total phenotypic variation is due to genetic variation. Narrow-sense heritability describes the fraction of phenotypic variation due to the additive effects of genes. Broad-sense heritability captures the fraction of phenotypic variation due to genetic values that may include non-additive effects.
Dominance
In human genetics, this usually refers to alleles that manifest their phenotype in the heterozygous state.
Epistasis
In human genetics, this usually refers to non-additive genetic interaction and describes the phenomenon that the combined effect of alleles at two or more loci deviates from the sum of their individual effects.
Pleiotropy
Describes the phenomenon in which a single gene is responsible for a number of distinct and seemingly unrelated phenotypic effects.
Liability
A term used to collectively describe all the genetic and environmental factors that contribute to the development of a complex disease or trait. A large number of factors are summed to yield a 'liability' curve with a threshold marking a binary outcome (e.g., unaffected or affected by a disease). Phenotypic outcome is determined by whether an individual’s liability is smaller or greater than the threshold.
Variant penetrance
Refers to the proportion of individuals carrying a particular genetic variant with the expected phenotype(s) commonly associated with that variant.
Variant expressivity
Refers to the range and severity of phenotypes associated with a given genetic variant in different genetic and environmental contexts.
Rare genetic variants
Defined as variants with a minor allele frequency of less than 0.01.
Common genetic variants
Defined as variants with a minor allele frequency of 0.01 or greater.
Mendelian disease
Disease that follows Mendelian patterns of inheritance due to alterations in one gene or regulatory region inherited from one (dominant disorder) or both parents (recessive disorder). Mendelian diseases are often highly penetrant; although many such disorders are individually rare, together, they affect an estimated 5% of the world’s population (Rahit and Tarailo-Graovac, 2020).
Common complex disease
Common complex diseases lack simple Mendelian patterns of inheritance. These disorders are thought to be a consequence of genetic variation in many loci and environmental factors, whose interplay remains little understood and difficult to study.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Nasa Sinnott-Armstrong, Email: nasa@fredhutch.org.
Douglas M Fowler, Email: dfowler@uw.edu.
Christine Queitsch, Email: queitsch@uw.edu.
Ziyue Gao, University of Pennsylvania, United States.
David E James, University of Sydney, Australia.
Funding Information
This paper was supported by the following grants:
National Institutes of Health 5RM1HG010461 to Nasa Sinnott-Armstrong, Stanley Fields, Frederick Roth, Lea M Starita, Cole Trapnell, Judit Villen, Douglas M Fowler, Christine Queitsch.
Bruce G. Cochener Foundation Creativity Award to Nasa Sinnott-Armstrong.
National Institutes of Health NIGMS R35GM139532 to Christine Queitsch.
Additional information
Competing interests
No competing interests declared.
is a scientific advisory board member, consultant, and/or cofounder of Algen Biotechnologies, Altius Therapeutics, and Scale Biosciences.
Author contributions
Conceptualization, Supervision, Funding acquisition, Investigation, Visualization, Methodology, Writing – original draft, Project administration, Writing – review and editing.
Supervision, Investigation, Project administration, Writing – review and editing.
Conceptualization, Supervision, Investigation, Writing – original draft, Project administration, Writing – review and editing.
Conceptualization, Supervision, Funding acquisition, Investigation, Project administration, Writing – review and editing.
Conceptualization, Supervision, Funding acquisition, Project administration, Writing – review and editing.
Conceptualization, Funding acquisition, Investigation, Writing – review and editing.
Conceptualization, Supervision, Funding acquisition, Investigation, Methodology, Writing – original draft, Project administration, Writing – review and editing.
Conceptualization, Supervision, Funding acquisition, Investigation, Visualization, Methodology, Writing – original draft, Project administration, Writing – review and editing.
References
- Abdellaoui A, Hugh-Jones D, Yengo L, Kemper KE, Nivard MG, Veul L, Holtz Y, Zietsch BP, Frayling TM, Wray NR, Yang J, Verweij KJH, Visscher PM. Genetic correlates of social stratification in Great Britain. Nature Human Behaviour. 2019;3:1332–1342. doi: 10.1038/s41562-019-0757-5. [DOI] [PubMed] [Google Scholar]
- Ammeux N, Housden BE, Georgiadis A, Hu Y, Perrimon N. Mapping signaling pathway cross-talk in Drosophila cells. PNAS. 2016;113:9940–9945. doi: 10.1073/pnas.1610432113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, Ntini E, Arner E, Valen E, Li K, Schwarzfischer L, Glatz D, Raithel J, Lilje B, Rapin N, Bagger FO, Jørgensen M, Andersen PR, Bertin N, Rackham O, Burroughs AM, Baillie JK, Ishizu Y, Shimizu Y, Furuhata E, Maeda S, Negishi Y, Mungall CJ, Meehan TF, Lassmann T, Itoh M, Kawaji H, Kondo N, Kawai J, Lennartsson A, Daub CO, Heutink P, Hume DA, Jensen TH, Suzuki H, Hayashizaki Y, Müller F, Forrest ARR, Carninci P, Rehli M, Sandelin A, The FANTOM Consortium An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajpai VK, Swigut T, Mohammed J, Tycko J, Naqvi S, Arreola M, Kim TC, Arora N, Pritchard JK, Bassik MC, Wysocka J. A genome-wide genetic screen uncovers novel determinants of human pigmentation. bioRxiv. 2021 doi: 10.1101/2021.09.29.462413. [DOI] [PMC free article] [PubMed]
- Barcellos SH, Carvalho LS, Turley P. Education Can reduce health disparities related to genetic risk of obesity: Evidence from a british reform. bioRxiv. 2018 doi: 10.1101/260463. [DOI] [PMC free article] [PubMed]
- Barcellos SH, Carvalho L, Turley P. The effect of education on the relationship between genetics, early-life disadvantages, and later-life SES. National Bureau of Economic Research. 2021;1:28750. doi: 10.3386/w28750. [DOI] [Google Scholar]
- Barton NH, Etheridge AM, Véber A. The infinitesimal model: definition, derivation, and implications. Theoretical Population Biology. 2017;118:50–73. doi: 10.1016/j.tpb.2017.06.001. [DOI] [PubMed] [Google Scholar]
- Berg MD, Zhu Y, Genereaux J, Ruiz BY, Rodriguez-Mias RA, Allan T, Bahcheli A, Villén J, Brandl CJ. Modulating mistranslation potential of tRNASer in Saccharomyces cerevisiae. Genetics. 2019;213:849–863. doi: 10.1534/genetics.119.302525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billman GE. Homeostasis: the underappreciated and far too often ignored central organizing principle of physiology. Frontiers in Physiology. 2020;11:200. doi: 10.3389/fphys.2020.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingley PJ, Gale EA. Rising incidence of IDDM in Europe. Diabetes Care. 1989;12:289–295. doi: 10.2337/diacare.12.4.289. [DOI] [PubMed] [Google Scholar]
- Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BC. Asian genetic epidemiology network type 2 diabetes consortium. American Journal of Human Genetics. 2016;99:76–88. doi: 10.1016/j.ajhg.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlborg O, Jacobsson L, Ahgren P, Siegel P, Andersson L. Epistasis and the release of genetic variation during long-term selection. Nature Genetics. 2006;38:418–420. doi: 10.1038/ng1761. [DOI] [PubMed] [Google Scholar]
- Claussnitzer M, Dankel SN, Kim KH, Quon G, Meuleman W, Haugen C, Glunk V, Sousa IS, Beaudry JL, Puviindran V, Abdennur NA, Liu J, Svensson PA, Hsu YH, Drucker DJ, Mellgren G, Hui CC, Hauner H, Kellis M. FTO obesity variant circuitry and adipocyte browning in humans. The New England Journal of Medicine. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussnitzer M, Susztak K. Gaining insight into metabolic diseases from human genetic discoveries. Trends in Genetics. 2021;37:1081–1094. doi: 10.1016/j.tig.2021.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockram CS. The epidemiology of diabetes mellitus in the Asia-Pacific region. Hong Kong Medical Journal = Xianggang Yi Xue Za Zhi / Hong Kong Academy of Medicine. 2000;6:43–52. [PubMed] [Google Scholar]
- Connally NJ, Nazeen S, Lee D, Shi H, Stamatoyannopoulos J, Chun S, Cotsapas C, Cassa CA, Sunyaev SR. The missing link between genetic association and regulatory function. eLife. 2022;11:e74970. doi: 10.7554/eLife.74970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Kuzmin E, van Leeuwen J, Mair B, Moffat J, Boone C, Andrews B. Global Genetic Networks and the Genotype-to-Phenotype Relationship. Cell. 2019;177:85–100. doi: 10.1016/j.cell.2019.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozma E, Rao M, Dusick M, Genereaux J, Rodriguez-Mias RA, Villén J, Brandl CJ, Berg MD. Probing the genetic code and impacts of mistranslation using trnaala anticodon variants. bioRxiv. 2022 doi: 10.1101/2022.11.23.517754. [DOI]
- Da K, Weile J, Kishore N, Nguyen M, Rubin AF, Fields S, Fowler DM, Roth FP. MaveRegistry: a collaboration platform for multiplexed assays of variant effect. Bioinformatics. 2021;37:3382–3383. doi: 10.1093/bioinformatics/btab215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell G, Smith SP, Udwin D, Ramachandran S, Crawford L. Partitioning tagged non-additive genetic effects in summary statistics provides evidence of pervasive epistasis in complex traits. bioRxiv. 2022 doi: 10.1101/2022.07.21.501001. [DOI]
- Davies NM, Dickson M, Davey Smith G, van den Berg GJ, Windmeijer F. The causal effects of education on health outcomes in the UK biobank. Nature Human Behaviour. 2018;2:117–125. doi: 10.1038/s41562-017-0279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey KK, Gazal S, van de Geijn B, Kim SS, Nasser J, Engreitz JM, Price AL. SNP-to-gene linking strategies reveal contributions of enhancer-related and candidate master-regulator genes to autoimmune disease. Cell Genomics. 2022;2:100145. doi: 10.1016/j.xgen.2022.100145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrity MW, Cuperus JT, Carlisle JA, Fields S, Queitsch C. Preferences in a trait decision determined by transcription factor variants. PNAS. 2018;115:E7997–E8006. doi: 10.1073/pnas.1805882115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrity MW, Saunders LM, Duran M, Srivatsan SR, Ewing B, Queitsch C, Shendure J, Raible DW, Kimelman D, Trapnell C. Proteostasis governs differential temperature sensitivity across embryonic cell types. bioRxiv. 2022 doi: 10.1101/2022.08.04.502669. [DOI] [PMC free article] [PubMed]
- Dudley JW. From means to QTL: the illinois long‐term selection experiment as a case study in quantitative genetics. Crop Science. 2007;47:S–20. doi: 10.2135/cropsci2007.04.0003IPBS. [DOI] [Google Scholar]
- Ebrahim S, Kinra S, Bowen L, Andersen E, Ben-Shlomo Y, Lyngdoh T, Ramakrishnan L, Ahuja RC, Joshi P, Das SM, Mohan M, Davey Smith G, Prabhakaran D, Reddy KS, Indian Migration Study group The effect of rural-to-urban migration on obesity and diabetes in India: a cross-sectional study. PLOS Medicine. 2010;7:e1000268. doi: 10.1371/journal.pmed.1000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH. Missing heritability and strategies for finding the underlying causes of complex disease. Nature Reviews Genetics. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elding Larsson H, Lynch KF, Lönnrot M, Haller MJ, Lernmark Å, Hagopian WA, She JX, Simell O, Toppari J, Ziegler AG, Akolkar B, Krischer JP, Rewers MJ, Hyöty H, TEDDY Study Group Pandemrix vaccination is not associated with increased risk of islet autoimmunity or type 1 diabetes in the TEDDY study children. Diabetologia. 2018;61:193–202. doi: 10.1007/s00125-017-4448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwood S, Bily TMI, Lequyer J, Yan J, Gulati N, Brewer RA, Zhou L, Pelletier L, Ivakine EA, Cohn RD. Saturation variant interpretation using CRISPR prime editing. Nature Biotechnology. 2022;40:885–895. doi: 10.1038/s41587-021-01201-1. [DOI] [PubMed] [Google Scholar]
- Esposito D, Weile J, Shendure J, Starita LM, Papenfuss AT, Roth FP, Fowler DM, Rubin AF. MaveDB: an open-source platform to distribute and interpret data from multiplexed assays of variant effect. Genome Biology. 2019;20:223. doi: 10.1186/s13059-019-1845-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayer S, Horton C, Dines JN, Rubin AF, Richardson ME, McGoldrick K, Hernandez F, Pesaran T, Karam R, Shirts BH, Fowler DM, Starita LM. Closing the gap: systematic integration of multiplexed functional data resolves variants of uncertain significance in BRCA1, TP53, and PTEN. American Journal of Human Genetics. 2021;108:2248–2258. doi: 10.1016/j.ajhg.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay GM. Linking genome variants to disease: scalable approaches to test the functional impact of human mutations. Human Molecular Genetics. 2021;30:R187–R197. doi: 10.1093/hmg/ddab219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. XV.—The correlation between relatives on the supposition of mendelian inheritance. Transactions of the Royal Society of Edinburgh. 1919;52:399–433. doi: 10.1017/S0080456800012163. [DOI] [Google Scholar]
- Fisher RA. The Genetical Theory of Natural Selection. Clarendon; 1930. [DOI] [Google Scholar]
- Forsberg SKG, Bloom JS, Sadhu MJ, Kruglyak L, Carlborg Ö. Accounting for genetic interactions improves modeling of individual quantitative trait phenotypes in yeast. Nature Genetics. 2017;49:497–503. doi: 10.1038/ng.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Hurles M, Adams DJ, Gloyn AL, Hahn WC. The atlas of variant effects (AVE) alliance: understanding genetic variation at nucleotide resolution. Version 4Zenodo. 2021 doi: 10.5281/zenodo.4989960. [DOI]
- Gasperini M, Starita L, Shendure J. The power of multiplexed functional analysis of genetic variants. Nature Protocols. 2016;11:1782–1787. doi: 10.1038/nprot.2016.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G. Decanalization and the origin of complex disease. Nature Reviews Genetics. 2009;10:134–140. doi: 10.1038/nrg2502. [DOI] [PubMed] [Google Scholar]
- Gibson G. Rare and common variants: twenty arguments. Nature Reviews Genetics. 2012;13:135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon MG, Inoue F, Martin B, Schubach M, Agarwal V, Whalen S, Feng S, Zhao J, Ashuach T, Ziffra R, Kreimer A, Georgakopoulos-Soares I, Yosef N, Ye CJ, Pollard KS, Shendure J, Kircher M, Ahituv N. lentiMPRA and MPRAflow for high-throughput functional characterization of gene regulatory elements. Nature Protocols. 2020;15:2387–2412. doi: 10.1038/s41596-020-0333-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gschwind AR, Mualim KS, Karbalayghareh A, Sheth MU, Dey KK, Jagoda E, Nurtdinov RN, Xi W, Tan AS, Jones H, Ma XR, Yao D, Nasser J, Avsec Ž, James BT, Shamim MS, Durand NC, Rao SSP, Mahajan R, Doughty BR, Andreeva K, Ulirsch JC, Fan K, Perez EM, Nguyen TC, Kelley DR, Finucane HK, Moore JE, Weng Z, Kellis M, Bassik MC, Price AL, Beer MA, Guigó R, Stamatoyannopoulos JA, Lieberman Aiden E, Greenleaf WJ, Leslie CS, Steinmetz LM, Kundaje A, Engreitz JM. An encyclopedia of enhancer-gene regulatory interactions in the human genome. bioRxiv. 2023 doi: 10.1101/2023.11.09.563812. [DOI]
- Hanna RE, Hegde M, Fagre CR, DeWeirdt PC, Sangree AK, Szegletes Z, Griffith A, Feeley MN, Sanson KR, Baidi Y, Koblan LW, Liu DR, Neal JT, Doench JG. Massively parallel assessment of human variants with base editor screens. Cell. 2021;184:1064–1080. doi: 10.1016/j.cell.2021.01.012. [DOI] [PubMed] [Google Scholar]
- Harsanyi S, Kupcova I, Danisovic L, Klein M. Selected biomarkers of depression: what are the effects of cytokines and inflammation? International Journal of Molecular Sciences. 2022;24:578. doi: 10.3390/ijms24010578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasle N, Cooke A, Srivatsan S, Huang H, Stephany JJ, Krieger Z, Jackson D, Tang W, Pendyala S, Monnat RJ, Trapnell C, Hatch EM, Fowler DM. High-throughput, microscope-based sorting to dissect cellular heterogeneity. Molecular Systems Biology. 2020;16:e9442. doi: 10.15252/msb.20209442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks D, Artegiani B, Hu H, Chuva de Sousa Lopes S, Clevers H. Establishment of human fetal hepatocyte organoids and CRISPR-Cas9-based gene knockin and knockout in organoid cultures from human liver. Nature Protocols. 2021;16:182–217. doi: 10.1038/s41596-020-00411-2. [DOI] [PubMed] [Google Scholar]
- Hivert V, Sidorenko J, Rohart F, Goddard ME, Yang J, Wray NR, Yengo L, Visscher PM. Estimation of non-additive genetic variance in human complex traits from a large sample of unrelated individuals. American Journal of Human Genetics. 2021;108:786–798. doi: 10.1016/j.ajhg.2021.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Tee WW. Enhancers and chromatin structures: regulatory hubs in gene expression and diseases. Bioscience Reports. 2017;37:BSR20160183. doi: 10.1042/BSR20160183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel S, Weiß A, Bonifacio E, Agardh D, Akolkar B, Aronsson CA, Hagopian WA, Koletzko S, Krischer JP, Lernmark Å, Lynch K, Norris JM, Rewers MJ, She JX, Toppari J, Uusitalo U, Vehik K, Virtanen SM, Beyerlein A, Ziegler AG, TEDDY Study Group Associations of breastfeeding with childhood autoimmunity, allergies, and overweight: The Environmental Determinants of Diabetes in the Young (TEDDY) study. The American Journal of Clinical Nutrition. 2021;114:134–142. doi: 10.1093/ajcn/nqab065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immonen J, Jääskeläinen E, Korpela H, Miettunen J. Age at onset and the outcomes of schizophrenia: A systematic review and meta-analysis. Early Intervention in Psychiatry. 2017;11:453–460. doi: 10.1111/eip.12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeesh KA, Dey KK, Montoro DT, Mohan R, Gazal S, Engreitz JM, Xavier RJ, Price AL, Regev A. Identifying disease-critical cell types and cellular processes by integrating single-cell RNA-sequencing and human genetics. Nature Genetics. 2022;54:1479–1492. doi: 10.1038/s41588-022-01187-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett H, McNulty H, Hughes CF, Pentieva K, Strain JJ, McCann A, McAnena L, Cunningham C, Molloy AM, Flynn A, Hopkins SM, Horigan G, O’Connor C, Walton J, McNulty BA, Gibney MJ, Lamers Y, Ward M. Vitamin B-6 and riboflavin, their metabolic interaction, and relationship with MTHFR genotype in adults aged 18-102 years. The American Journal of Clinical Nutrition. 2022;116:1767–1778. doi: 10.1093/ajcn/nqac240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson FB, Sinclair DA, Guarente L. Molecular biology of aging. Cell. 1999;96:291–302. doi: 10.1016/s0092-8674(00)80567-x. [DOI] [PubMed] [Google Scholar]
- Johnson SB, Lee HS, Baxter J, Lernmark B, Roth R, Simell T, TEDDY Study Group The Environmental Determinants of Diabetes in the Young (TEDDY) study: predictors of early study withdrawal among participants with no family history of type 1 diabetes. Pediatric Diabetes. 2011;12:165–171. doi: 10.1111/j.1399-5448.2010.00686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SB, Lynch KF, Lee HS, Smith L, Baxter J, Lernmark B, Roth R, Simell T, TEDDY Study Group At high risk for early withdrawal: using a cumulative risk model to increase retention in the first year of the TEDDY study. Journal of Clinical Epidemiology. 2014;67:609–611. doi: 10.1016/j.jclinepi.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SB, Lynch KF, Roth R, Lundgren M, Parikh HM, Akolkar B, Hagopian W, Krischer J, Rewers M, She JX, Toppari J, Ziegler AG, Lernmark Å, TEDDY Study Group First-appearing islet autoantibodies for type 1 diabetes in young children: maternal life events during pregnancy and the child’s genetic risk. Diabetologia. 2021a;64:591–602. doi: 10.1007/s00125-020-05344-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RK, Tamura R, Frank N, Uusitalo U, Yang J, Niinistö S, Andrén Aronsson C, Ziegler AG, Hagopian W, Rewers M, Toppari J, Akolkar B, Krischer J, Virtanen SM, Norris JM, TEDDY Study Group Maternal food consumption during late pregnancy and offspring risk of islet autoimmunity and type 1 diabetes. Diabetologia. 2021b;64:1604–1612. doi: 10.1007/s00125-021-05446-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RC, Karkanias J, Krasnow MA, Pisco AO, Quake SR, Salzman J, Yosef N, Bulthaup B, Brown P, Harper W, Hemenez M, Ponnusamy R, Salehi A, Sanagavarapu BA, Spallino E, Aaron KA, Concepcion W, Gardner JM, Kelly B, Neidlinger N, Wang Z, Crasta S, Kolluru S, Morri M, Tan SY, Travaglini KJ, Xu C, Alcántara-Hernández M, Almanzar N, Antony J, Beyersdorf B, Burhan D, Calcuttawala K, Carter MM, Chan CKF, Chang CA, Chang S, Colville A, Culver RN, Cvijović I, D’Amato G, Ezran C, Galdos FX, Gillich A, Goodyer WR, Hang Y, Hayashi A, Houshdaran S, Huang X, Irwin JC, Jang S, Juanico JV, Kershner AM, Kim S, Kiss B, Kong W, Kumar ME, Kuo AH, Li B, Loeb GB, Lu W-J, Mantri S, Markovic M, McAlpine PL, de Morree A, Mrouj K, Mukherjee S, Muser T, Neuhöfer P, Nguyen TD, Perez K, Puluca N, Qi Z, Rao P, Raquer-McKay H, Schaum N, Scott B, Seddighzadeh B, Segal J, Sen S, Sikandar S, Spencer SP, Steffes LC, Subramaniam VR, Swarup A, Swift M, Van Treuren W, Trimm E, Veizades S, Vijayakumar S, Vo KC, Vorperian SK, Wang W, Weinstein HNW, Winkler J, Wu TTH, Xie J, Yung AR, Zhang Y, Detweiler AM, Mekonen H, Neff NF, Sit RV, Tan M, Yan J, Bean GR, Charu V, Forgó E, Martin BA, Ozawa MG, Silva O, Toland A, Vemuri VNP, Afik S, Awayan K, Botvinnik OB, Byrne A, Chen M, Dehghannasiri R, Gayoso A, Granados AA, Li Q, Mahmoudabadi G, McGeever A, Olivieri JE, Park M, Ravikumar N, Stanley G, Tan W, Tarashansky AJ, Vanheusden R, Wang P, Wang S, Xing G, Dethlefsen L, Ezran C, Gillich A, Hang Y, Ho P-Y, Irwin JC, Jang S, Leylek R, Liu S, Maltzman JS, Metzger RJ, Phansalkar R, Sasagawa K, Sinha R, Song H, Swarup A, Trimm E, Veizades S, Wang B, Beachy PA, Clarke MF, Giudice LC, Huang FW, Huang KC, Idoyaga J, Kim SK, Kuo CS, Nguyen P, Rando TA, Red-Horse K, Reiter J, Relman DA, Sonnenburg JL, Wu A, Wu SM, Wyss-Coray T, Tabula Sapiens Consortium* The Tabula Sapiens: a multiple-organ, single-cell transcriptomic atlas of humans. Science. 2022;376:eabl4896. doi: 10.1126/science.abl4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemppainen KM, Vehik K, Lynch KF, Larsson HE, Canepa RJ, Simell V, Koletzko S, Liu E, Simell OG, Toppari J, Ziegler AG, Rewers MJ, Lernmark Å, Hagopian WA, She JX, Akolkar B, Schatz DA, Atkinson MA, Blaser MJ. Environmental determinants of diabetes in the young (TEDDY):association between early-life antibiotic use and the risk of islet or celiac disease autoimmunity. JAMA Pediatrics. 2017;171:1217–1225. doi: 10.1001/jamapediatrics.2017.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerin M, Marchini J. Inferring gene-by-environment interactions with a Bayesian whole-genome regression model. American Journal of Human Genetics. 2020;107:698–713. doi: 10.1016/j.ajhg.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, Kathiresan S. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nature Genetics. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney JB, McCandlish DM. Massively parallel assays and quantitative sequence-function relationships. Annual Review of Genomics and Human Genetics. 2019;20:99–127. doi: 10.1146/annurev-genom-083118-014845. [DOI] [PubMed] [Google Scholar]
- Kircher M, Xiong C, Martin B, Schubach M, Inoue F, Bell RJA, Costello JF, Shendure J, Ahituv N. Saturation mutagenesis of twenty disease-associated regulatory elements at single base-pair resolution. Nature Communications. 2019;10:3583. doi: 10.1038/s41467-019-11526-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JC, Keith A, Rice SJ, Shepherd C, Agarwal V, Loughlin J, Shendure J. Functional testing of thousands of osteoarthritis-associated variants for regulatory activity. bioRxiv. 2018 doi: 10.1101/379727. [DOI] [PMC free article] [PubMed]
- Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu Y-C, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh K-H, Feizi S, Karlic R, Kim A-R, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJM, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai L-H, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M, Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian MA, Hayflick SJ. Pantothenate kinase-associated neurodegeneration (PKAN) and PLA2G6-associated neurodegeneration (PLAN): review of two major neurodegeneration with brain iron accumulation (NBIA) phenotypes. International Review of Neurobiology. 2013;110:49–71. doi: 10.1016/B978-0-12-410502-7.00003-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachowiec J, Shen X, Queitsch C, Carlborg Ö. A genome-wide association analysis reveals epistatic cancellation of additive genetic variance for root length in Arabidopsis thaliana. PLOS Genetics. 2015;11:e1005541. doi: 10.1371/journal.pgen.1005541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea AJ, Caldas IV, Garske KM, Echwa J, Gurven M, Handley C, Kahumbu J, Kinyua P, Lotukoi F, Lopurudoi A, Lowasa S, Mallarino R, Martins D, Messer PW, Miano C, Muhoya B, Peng J, Phung T, Rabinowitz JD, Roichman A, Siford R, Stone A, Oill AT, Mathew S, Wilson MA, Ayroles JF. Adaptations to Water Stress and Pastoralism in the Turkana of Northwest Kenya. bioRxiv. 2023 doi: 10.1101/2023.01.17.524066. [DOI]
- Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nature Genetics. 2006;38:896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- Leutert M, Armstrong JO, Ollodart AR, Hess K, Muir M, Rodriguez-Mias RA, Kaeberlein M, Dunham M, Villén J. Multidimensional proteomics identifies molecular trajectories of cellular aging and rejuvenation. bioRxiv. 2023 doi: 10.1101/2023.03.09.531951. [DOI]
- Liu X, Li YI, Pritchard JK. Trans effects on gene expression can drive omnigenic inheritance. Cell. 2019;177:1022–1034. doi: 10.1016/j.cell.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lönnrot M, Lynch KF, Elding Larsson H, Lernmark Å, Rewers MJ, Törn C, Burkhardt BR, Briese T, Hagopian WA, She JX, Simell OG, Toppari J, Ziegler AG, Akolkar B, Krischer JP, Hyöty H, Group TS. Respiratory infections are temporally associated with initiation of type 1 diabetes autoimmunity: the TEDDY study. Diabetologia. 2017;60:1931–1940. doi: 10.1007/s00125-017-4365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv W, Qiao L, Petrenko N, Li W, Owens AT, McDermott-Roe C, Musunuru K. Functional annotation of TNNT2 variants of uncertain significance with genome-edited cardiomyocytes. Circulation. 2018;138:2852–2854. doi: 10.1161/CIRCULATIONAHA.118.035028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF. The genetic architecture of quantitative traits. Annual Review of Genetics. 2001;35:303–339. doi: 10.1146/annurev.genet.35.102401.090633. [DOI] [PubMed] [Google Scholar]
- Mackay TFC. Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nature Reviews Genetics. 2014;15:22–33. doi: 10.1038/nrg3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TFC, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marderstein AR, Davenport ER, Kulm S, Van Hout CV, Elemento O, Clark AG. Leveraging phenotypic variability to identify genetic interactions in human phenotypes. American Journal of Human Genetics. 2021;108:49–67. doi: 10.1016/j.ajhg.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Rufino JD, Castano N, Pang M, Grody EI, Joubran S, Caulier A, Wahlster L, Li T, Qiu X, Riera-Escandell AM, Newby GA, Al’Khafaji A, Chaudhary S, Black S, Weng C, Munson G, Liu DR, Wlodarski MW, Sims K, Oakley JH, Fasano RM, Xavier RJ, Lander ES, Klein DE, Sankaran VG. Massively parallel base editing to map variant effects in human hematopoiesis. Cell. 2023;186:2456–2474. doi: 10.1016/j.cell.2023.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Mullis MN, Roy KR, Hale JJ, Schell R, Levy SF, Ehrenreich IM. The interplay of additivity, dominance, and epistasis on fitness in a diploid yeast cross. Nature Communications. 2022;13:1463. doi: 10.1038/s41467-022-29111-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, Shafer A, Neri F, Lee K, Kutyavin T, Stehling-Sun S, Johnson AK, Canfield TK, Giste E, Diegel M, Bates D, Hansen RS, Neph S, Sabo PJ, Heimfeld S, Raubitschek A, Ziegler S, Cotsapas C, Sotoodehnia N, Glass I, Sunyaev SR, Kaul R, Stamatoyannopoulos JA. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty H, Dowey LRC, Strain JJ, Dunne A, Ward M, Molloy AM, McAnena LB, Hughes JP, Hannon-Fletcher M, Scott JM. Riboflavin lowers homocysteine in individuals homozygous for the MTHFR 677C->T polymorphism. Circulation. 2006;113:74–80. doi: 10.1161/CIRCULATIONAHA.105.580332. [DOI] [PubMed] [Google Scholar]
- Meng X, Yao D, Kelley KW, Reis N, Thete MV, Kulkarni S, Bassik MC, Pasca SP. CRISPR screens in 3D assembloids reveal disease genes associated with human interneuron development. bioRxiv. 2022 doi: 10.1101/2022.09.06.506845. [DOI]
- Meuleman W, Muratov A, Rynes E, Halow J, Lee K, Bates D, Diegel M, Dunn D, Neri F, Teodosiadis A, Reynolds A, Haugen E, Nelson J, Johnson A, Frerker M, Buckley M, Sandstrom R, Vierstra J, Kaul R, Stamatoyannopoulos J. Index and biological spectrum of human DNase I hypersensitive sites. Nature. 2020;584:244–251. doi: 10.1038/s41586-020-2559-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DB, Piccolo SR. Compound heterozygous variants in pediatric cancers: a systematic review. Frontiers in Genetics. 2020;11:493. doi: 10.3389/fgene.2020.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DE, Sulovari A, Wang T, Loucks H, Hoekzema K, Munson KM, Lewis AP, Fuerte EPA, Paschal CR, Walsh T, Thies J, Bennett JT, Glass I, Dipple KM, Patterson K, Bonkowski ES, Nelson Z, Squire A, Sikes M, Beckman E, Bennett RL, Earl D, Lee W, Allikmets R, Perlman SJ, Chow P, Hing AV, Wenger TL, Adam MP, Sun A, Lam C, Chang I, Zou X, Austin SL, Huggins E, Safi A, Iyengar AK, Reddy TE, Majoros WH, Allen AS, Crawford GE, Kishnani PS, University of Washington Center for Mendelian Genomics. King MC, Cherry T, Chong JX, Bamshad MJ, Nickerson DA, Mefford HC, Doherty D, Eichler EE. Targeted long-read sequencing identifies missing disease-causing variation. American Journal of Human Genetics. 2021;108:1436–1449. doi: 10.1016/j.ajhg.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra I, Huang B, Mousavi N, Ma N, Lamkin M, Yanicky R, Shleizer-Burko S, Lohmueller KE, Gymrek M. Patterns of de novo tandem repeat mutations and their role in autism. Nature. 2021;589:246–250. doi: 10.1038/s41586-020-03078-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R, Casale FP, Jan Bonder M, Horta D, Consortium B, Franke L, Barroso I, Stegle O. A linear mixed-model approach to study multivariate gene-environment interactions. Nature Genetics. 2019;51:180–186. doi: 10.1038/s41588-018-0271-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morova T, Ding Y, Huang CCF, Sar F, Schwarz T, Giambartolomei C, Baca SC, Grishin D, Hach F, Gusev A, Freedman ML, Pasaniuc B, Lack NA. Optimized high-throughput screening of non-coding variants identified from genome-wide association studies. Nucleic Acids Research. 2023;51:e18. doi: 10.1093/nar/gkac1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton EA, Dorrity MW, Zhou W, Fields S, Queitsch C. Transcriptional re-wiring by mutation of the yeast Hsf1 oligomerization domain. bioRxiv. 2020 doi: 10.1101/2020.05.23.112250. [DOI]
- Mostafavi H, Harpak A, Agarwal I, Conley D, Pritchard JK, Przeworski M. Variable prediction accuracy of polygenic scores within an ancestry group. eLife. 2020;9:e48376. doi: 10.7554/eLife.48376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafavi H, Spence JP, Naqvi S, Pritchard JK. Limited overlap of eQTLs and GWAS hits due to systematic differences in discovery. bioRxiv. 2022 doi: 10.1101/2022.05.07.491045. [DOI]
- Narasimhan VM, Rahbari R, Scally A, Wuster A, Mason D, Xue Y, Wright J, Trembath RC, Maher ER, van Heel DA, Auton A, Hurles ME, Tyler-Smith C, Durbin R. Estimating the human mutation rate from autozygous segments reveals population differences in human mutational processes. Nature Communications. 2017;8:303. doi: 10.1038/s41467-017-00323-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasser J, Bergman DT, Fulco CP, Guckelberger P, Doughty BR, Patwardhan TA, Jones TR, Nguyen TH, Ulirsch JC, Lekschas F, Mualim K, Natri HM, Weeks EM, Munson G, Kane M, Kang HY, Cui A, Ray JP, Eisenhaure TM, Collins RL, Dey K, Pfister H, Price AL, Epstein CB, Kundaje A, Xavier RJ, Daly MJ, Huang H, Finucane HK, Hacohen N, Lander ES, Engreitz JM. Genome-wide enhancer maps link risk variants to disease genes. Nature. 2021;593:238–243. doi: 10.1038/s41586-021-03446-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Y, Seballos S, Fletcher B, Romigh T, Yehia L, Mester J, Senter L, Niazi F, Saji M, Ringel MD, LaFramboise T, Eng C. Germline compound heterozygous poly-glutamine deletion in USF3 may be involved in predisposition to heritable and sporadic epithelial thyroid carcinoma. Human Molecular Genetics. 2017;26:243–257. doi: 10.1093/hmg/ddw382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris JM, Lee HS, Frederiksen B, Erlund I, Uusitalo U, Yang J, Lernmark Å, Simell O, Toppari J, Rewers M, Ziegler AG, She JX, Onengut-Gumuscu S, Chen WM, Rich SS, Sundvall J, Akolkar B, Krischer J, Virtanen SM, Hagopian W, TEDDY Study Group Plasma 25-Hydroxyvitamin D concentration and risk of islet autoimmunity. Diabetes. 2018;67:146–154. doi: 10.2337/db17-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novelli G, D’Apice MR. The strange case of the “lumper” lamin A/C gene and human premature ageing. Trends in Molecular Medicine. 2003;9:370–375. doi: 10.1016/s1471-4914(03)00162-x. [DOI] [PubMed] [Google Scholar]
- Pamplona-Cunha H, Medeiros MF, Sincero TCM, de C Back I, da Silva E. Compound heterozygous familial hypercholesterolemia caused by LDLR variants. Arquivos Brasileiros de Cardiologia. 2020;115:587–589. doi: 10.36660/abc.20190582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DS, Eskin I, Kang EY, Gamazon ER, Eng C, Gignoux CR, Galanter JM, Burchard E, Ye CJ, Aschard H, Eskin E, Halperin E, Zaitlen N. An ancestry-based approach for detecting interactions. Genetic Epidemiology. 2018;42:49–63. doi: 10.1002/gepi.22087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RA, Musharoff SA, Spence JP, Pimentel H, Tcheandjieu C, Mostafavi H, Sinnott-Armstrong N, Clarke SL, Smith CJ, Durda PP, Taylor KD, Tracy R, Liu Y, Johnson WC, Aguet F, Ardlie KG, Gabriel S, Smith J, Nickerson DA, Rich SS, Rotter JI, Tsao PS, Assimes TL, Pritchard JK, V.A. Million Veteran Program Genetic interactions drive heterogeneity in causal variant effect sizes for gene expression and complex traits. American Journal of Human Genetics. 2022;109:1286–1297. doi: 10.1016/j.ajhg.2022.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikov D, Williams C, Atan D, Davies NM, Ghorbani Mojarrad N, Guggenheim JA, Eye UB, Consortium V. Effect of education on myopia: evidence from the United Kingdom ROSLA 1972 reform. Investigative Ophthalmology & Visual Science. 2020;61:7. doi: 10.1167/iovs.61.11.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press MO, Carlson KD, Queitsch C. The overdue promise of short tandem repeat variation for heritability. Trends in Genetics. 2014;30:504–512. doi: 10.1016/j.tig.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press MO, Hall AN, Morton EA, Queitsch C. Substitutions are boring: some arguments about parallel mutations and high mutation rates. Trends in Genetics. 2019;35:253–264. doi: 10.1016/j.tig.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Carlson KD, Girirajan S. Lessons from model organisms: phenotypic robustness and missing heritability in complex disease. In PLOS Genetics. 2012;8:e1003041. doi: 10.1371/journal.pgen.1003041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahit K, Tarailo-Graovac M. Genetic modifiers and rare mendelian disease. Genes. 2020;11:239. doi: 10.3390/genes11030239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rewers M, Hyöty H, Lernmark Å, Hagopian W, She JX, Schatz D, Ziegler AG, Toppari J, Akolkar B, Krischer J, Group TS. The environmental determinants of diabetes in the young (TEDDY) study: 2018 update. Current Diabetes Reports. 2018;18:136. doi: 10.1007/s11892-018-1113-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin AF, Min JK, Rollins NJ, Da EY, Esposito D, Harrington M, Stone J, Bianchi AH, Dias M, Frazer J, Fu Y, Gallaher M, Li I, Moscatelli O, Ong JY, Rollins JE, Wakefield MJ, “Sunny” Ye S, Tam A, McEwen AE, Starita LM, Bryant VL, Marks DS, Fowler DM. MaveDB v2: a curated community database with over three million variant effects from multiplexed functional assays. bioRxiv. 2021 doi: 10.1101/2021.11.29.470445. [DOI]
- Saunders LM, Srivatsan SR, Duran M, Dorrity MW, Ewing B, Linbo T, Shendure J, Raible DW, Moens CB, Kimelman D, Trapnell C. Deep molecular, cellular and temporal phenotyping of developmental perturbations at whole organism scale. bioRxiv. 2022 doi: 10.1101/2022.08.04.502764. https://doi.org/10.1101/2022.08.04.502764 [DOI]
- Schiabor Barrett KM, Masnick M, Hatchell KE, Savatt JM, Banet N, Buchanan A, Willard HF. Clinical validation of genomic functional screen data: analysis of observed BRCA1 variants in an unselected population cohort. HGG Advances. 2022;3:100086. doi: 10.1016/j.xhgg.2022.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A, Hernandez F, Chamberlin A, Smith C, Karam R, Kitzman JO. Saturation-scale functional evidence supports clinical variant interpretation in Lynch syndrome. Genome Biology. 2022;23:266. doi: 10.1186/s13059-022-02839-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard B, Rappoport N, Loh PR, Sanders SJ, Zaitlen N, Dahl A. A model and test for coordinated polygenic epistasis in complex traits. PNAS. 2021;118:e1922305118. doi: 10.1073/pnas.1922305118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifrut E, Carnevale J, Tobin V, Roth TL, Woo JM, Bui CT, Li PJ, Diolaiti ME, Ashworth A, Marson A. Genome-wide CRISPR screens in primary human T cells reveal key regulators of immune function. Cell. 2018;175:1958–1971. doi: 10.1016/j.cell.2018.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvis K, Aronsson CA, Liu X, Uusitalo U, Yang J, Tamura R, Lernmark Å, Rewers M, Hagopian W, She JX, Simell O, Toppari J, Ziegler A, Akolkar B, Krischer J, Virtanen SM, Norris JM, TEDDY Study Group Maternal dietary supplement use and development of islet autoimmunity in the offspring: TEDDY study. Pediatric Diabetes. 2019;20:86–92. doi: 10.1111/pedi.12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnott-Armstrong N, Naqvi S, Rivas M, Pritchard JK. GWAS of three molecular traits highlights core genes and pathways alongside a highly polygenic background. eLife. 2021;10:e58615. doi: 10.7554/eLife.58615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJ, Sinnott-Armstrong N, Cichońska A, Julkunen H, Fauman EB, Würtz P, Pritchard JK. Integrative analysis of metabolite GWAS illuminates the molecular basis of pleiotropy and genetic correlation. eLife. 2022;11:e79348. doi: 10.7554/eLife.79348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sockell A, Wong W, Longwell S, Vu T, Karlsson K, Mokhtari D, Schaepe J, Lo YH, Cornelius V, Kuo C, Van Valen D, Curtis C, Fordyce PM. A microwell platform for high-throughput longitudinal phenotyping and selective retrieval of organoids. bioRxiv. 2022 doi: 10.1101/2022.11.01.514733. https://doi.org/10.1101/2022.11.01.514733 [DOI] [PubMed]
- Sohail M, Maier RM, Ganna A, Bloemendal A, Martin AR, Turchin MC, Chiang CW, Hirschhorn J, Daly MJ, Patterson N, Neale B, Mathieson I, Reich D, Sunyaev SR. Polygenic adaptation on height is overestimated due to uncorrected stratification in genome-wide association studies. eLife. 2019;8:e39702. doi: 10.7554/eLife.39702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. Pleiotropy in complex traits: challenges and strategies. Nature Reviews Genetics. 2013;14:483–495. doi: 10.1038/nrg3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen TI. The changing lifestyle in the world. Body weight and what else? Diabetes Care. 2000;23 Suppl 2:B1–B4. [PubMed] [Google Scholar]
- Starita LM, Ahituv N, Dunham MJ, Kitzman JO, Roth FP, Seelig G, Shendure J, Fowler DM. Variant interpretation: functional assays to the rescue. American Journal of Human Genetics. 2017;101:315–325. doi: 10.1016/j.ajhg.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Yang F, Tan G, Costanzo M, Oughtred R, Hirschman J, Theesfeld CL, Bansal P, Sahni N, Yi S, Yu A, Tyagi T, Tie C, Hill DE, Vidal M, Andrews BJ, Boone C, Dolinski K, Roth FP. An extended set of yeast-based functional assays accurately identifies human disease mutations. Genome Research. 2016;26:670–680. doi: 10.1101/gr.192526.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabet D, Parikh V, Mali P, Roth FP, Claussnitzer M. Scalable functional assays for the interpretation of human genetic variation. Annual Review of Genetics. 2022;56:441–465. doi: 10.1146/annurev-genet-072920-032107. [DOI] [PubMed] [Google Scholar]
- Ursu O, Neal JT, Shea E, Thakore PI, Jerby-Arnon L, Nguyen L, Dionne D, Diaz C, Bauman J, Mosaad MM, Fagre C, Lo A, McSharry M, Giacomelli AO, Ly SH, Rozenblatt-Rosen O, Hahn WC, Aguirre AJ, Berger AH, Regev A, Boehm JS. Massively parallel phenotyping of coding variants in cancer with Perturb-seq. Nature Biotechnology. 2022;40:896–905. doi: 10.1038/s41587-021-01160-7. [DOI] [PubMed] [Google Scholar]
- Uusitalo U, Liu X, Yang J, Aronsson CA, Hummel S, Butterworth M, Lernmark Å, Rewers M, Hagopian W, She JX, Simell O, Toppari J, Ziegler AG, Akolkar B, Krischer J, Norris JM, Virtanen SM, Group TS. Association of early exposure of probiotics and islet autoimmunity in the TEDDY study. JAMA Pediatrics. 2016;170:20–28. doi: 10.1001/jamapediatrics.2015.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vehik K, Lynch KF, Wong MC, Tian X, Ross MC, Gibbs RA, Ajami NJ, Petrosino JF, Rewers M, Toppari J, Ziegler AG, She JX, Lernmark A, Akolkar B, Hagopian WA, Schatz DA, Krischer JP, Hyöty H, Lloyd RE, Group TS. Prospective virome analyses in young children at increased genetic risk for type 1 diabetes. Nature Medicine. 2019;25:1865–1872. doi: 10.1038/s41591-019-0667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Hill WG, Wray NR. Heritability in the genomics era--concepts and misconceptions. Nature Reviews Genetics. 2008;9:255–266. doi: 10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- Vockley CM, Guo C, Majoros WH, Nodzenski M, Scholtens DM, Hayes MG, Lowe WL, Jr, Reddy TE. Massively parallel quantification of the regulatory effects of noncoding genetic variation in a human cohort. Genome Research. 2015;25:1206–1214. doi: 10.1101/gr.190090.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hinke S, Sørensen EN. The long-term effects of early-life pollution exposure: evidence from the London smog. Journal of Health Economics. 2023;92:102827. doi: 10.1016/j.jhealeco.2023.102827. [DOI] [PubMed] [Google Scholar]
- Wade MJ, Goodnight CJ. Perspective: the theories of fisher and wright in the context of metapopulations: when nature does many small experiments. Evolution; International Journal of Organic Evolution. 1998;52:1537–1553. doi: 10.1111/j.1558-5646.1998.tb02235.x. [DOI] [PubMed] [Google Scholar]
- Walsh B, Lynch M. In: Oxford Scholarship Online. Walsh B, Lynch M, editors. Oxford Academic; 2018. The infinitesimal model and its extensions; pp. 875–912. [DOI] [Google Scholar]
- Weile J, Kishore N, Sun S, Maaieh R, Verby M, Li R, Fotiadou I, Kitaygorodsky J, Wu Y, Holenstein A, Bürer C, Blomgren L, Yang S, Nussbaum R, Rozen R, Watkins D, Gebbia M, Kozich V, Garton M, Froese DS, Roth FP. Shifting landscapes of human MTHFR missense-variant effects. American Journal of Human Genetics. 2021;108:1283–1300. doi: 10.1016/j.ajhg.2021.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcsh PL, King MC. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Human Molecular Genetics. 2001;10:705–713. doi: 10.1093/hmg/10.7.705. [DOI] [PubMed] [Google Scholar]
- Wright S. Evolution in Mendelian Populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Li R, Sun S, Weile J, Roth FP. Improved pathogenicity prediction for rare human missense variants. American Journal of Human Genetics. 2021;108:1891–1906. doi: 10.1016/j.ajhg.2021.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Chen L, Sun M, Jean-Baptiste K, Cao D, Zhou X, Wong S, Xiao C, Liu T, Quijano V, Brandes N, Germino J, Ntranos V, Ye CJ, Aguet F, Farh KKH. Single cell sequencing as a general variant interpretation assay. bioRxiv. 2023 doi: 10.1101/2023.12.12.571130. https://doi.org/10.1101/2023.12.12.571130 [DOI]
- Yang F, Sun S, Tan G, Costanzo M, Hill DE, Vidal M, Andrews BJ, Boone C, Roth FP. Identifying pathogenicity of human variants via paralog-based yeast complementation. PLOS Genetics. 2017;13:e1006779. doi: 10.1371/journal.pgen.1006779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Cato LD, Weng C, Liggett LA, Jeon S, Xu K, Chiang CWK, Wiemels JL, Weissman JS, de Smith AJ, Sankaran VG. Variant to function mapping at single-cell resolution through network propagation. Nature Biotechnology. 2022;40:1644–1653. doi: 10.1038/s41587-022-01341-y. [DOI] [PMC free article] [PubMed] [Google Scholar]