Abstract
Background:
Microbial dysbiosis has been reported to contribute to development of neurodegenerative diseases, however, there is a need to identify causative/prognostic indicators.
Objectives:
To comparatively analyze gut microbiome composition in symptomatic LBD (dementia/mild cognitive impairment), iRBD, and cohabiting controls without LBD or iRBD.
Methods:
16S rRNA amplicon sequencing was performed in 38 cases (27 LBD, 11 iRBD) and 39 cohabitant controls. 19 non-cohabitant healthy controls (HCs) were also included to contrast differences between cohabitant cases and controls.
Results:
Microbiome composition of cohabitant controls and LBD and iRBD cases were strikingly similar. No differences were observed between LBD, and iRBD only showed reduced Bacteroides, compared with cohabitant controls. There were several taxonomic differences in gut microbiome composition between non-cohabitant HCs and cases.
Conclusions:
Minimal microbiome differences were observed between iRBD or LBD cases and cohabitant controls. These findings underscore the importance of using cohabiting controls in future gut microbiome studies.
1. Introduction
Gastrointestinal (GI) dysfunction is common in all forms of Lewy body disease (LBD). Gut microbiome research in LBD has primarily focused on Parkinson’s disease (PD) and lacked cohabitant control groups or data drawn from North American populations, with few studies on isolated REM sleep behavior disorder (iRBD), which in the majority of cases represents a prodromal phase of LBD, and cognitive manifestations of LBD such as dementia with Lewy bodies (DLB) and mild cognitive impairment due to Lewy body disease (MCI-LB) [1-6]. Prior studies have found a decreased relative abundance of short-chain fatty-acid (SCFA)-producing taxa in PD and DLB, which could drive increased intestinal permeability and inflammation [1,4-7]. Thus, understanding gut microbiome changes associated with LBD may be important for both diagnostics and therapeutics development.
In this study, we leveraged fecal microbiota taxonomic composition data and diversity metrics from patients with cognitive manifestations of LBD and its putative prodromal state (iRBD) and cohabitant controls.
2. Methods
2.1. Participants
The study was approved by the Mayo Clinic Institutional Review Board. Informed consent was obtained from all participants. Participants included 27 LBD cases (12 MCI-LB and 15 DLB) and 11 polysomno-graphically proven iRBD cases with a consensus diagnosis from sleep/behavioral neurology subspeciality neurologists and neuropsychologists based on established criteria and recruited from the Mayo Clinic Alzheimer’s Disease Research Center and North American Prodromal Synucleinopathy Study. A cohabitant control cohort (n = 39), consisting of the participants’ cohabitants without LBD or iRBD, was also included. To help parse differences, and for comparisons with previous studies, an established healthy cohort (HC; not cohabitants) was also included, comprising 19 robustly screened participants in the University of Minnesota Microbiota Therapeutics stool donor program [8]. Average age of HC cohort was 28 years of age with 38 % female.
2.2. Sample collection and preparation
Fecal samples were collected using standard stool specimen collection kits at the time of clinical assessment. DNA was extracted from fecal samples using the DNeasy PowerSoil Pro kit on an automated QIAcube platform with the inhibitor removal technology (IRT) protocol by the University of Minnesota Genomics Center (UMGC). PCR amplification was done using the 515F/806R primer set, targeting the V4 region of the 16S rRNA gene.
2.3. Sequencing and bioinformatics
Dual-indexed sequencing was conducted on the MiSeq platform (Illumina, Inc., San Diego, CA, USA) at a read length of 301 nucleotides (nt) by UMGC. Sequence data were processed using mothur (v.1.41.1) as previously described [9]. Reads were trimmed to 170 nt to remove low-quality regions, paired-end joined, screened for high quality, and aligned against the SILVA database (v.138_1). Chimeras were removed with UCHIME (v.4.2.40). Amplicon sequence variants (ASVs) were classified against the version 18 release from the Ribosomal Database Project. Samples were normalized to 8500 reads. Raw sequencing data were uploaded to the Sequence Read Archive under accession number SRP434593.
2.4. Statistical analyses
Continuous data are presented as medians with interquartile ranges (IQR) and categorical data as frequencies due to non-normally distributed data. Continuous and categorical data were compared with Kruskal-Wallis and Fisher exact tests, respectively. Alpha- and beta-diversity analyses were performed using mothur. Beta-diversity was calculated using Bray-Curtis distances and visualized through ordination by principal coordinate analysis (PCoA). Dissimilarity in community composition between study groups was evaluated using analysis of similarity (ANOSIM). A Bonferroni corrected significance of p < 0.0083 was used for individual taxa comparisons.
3. Results
3.1. Study participant demographics
Median age at stool sample collection was similar between iRBD, LBD, and cohabitant controls. Consistent with typical iRBD/LBD demographics, male sex was overwhelmingly represented in iRBD and LBD cohorts; and thus cohabitant controls were primarily female. Clinical and demographic characteristics are shown in Table 1.
Table 1.
Demographic and clinical characteristics of the study cohorts.
| Cohabitant controls (n = 39) |
iRBD (n = 11) |
LBD (n = 27) | p-value | |
|---|---|---|---|---|
| Age at stool sample, yrs (IQR) | 63.0 (10.5) | 68.0 (6.5) | 68.0 (8.5) | NS |
| Male, n (%) | 5 (12.8 %) | 9 (81.8 %) | 22 (81.5 %) | <0.001 |
| MCI/DLB | NA | NA | 12/15 | |
| iRBD, n (%) | NA | 11 (100.0 %) | 25 (92.6 %) | NS |
| Age RBD onset (IQR) | NA | 60.0 (20.4) | 57.53 (9.4) | NS |
| Years RBD Sx (IQR) | NA | 7.0 (12.5) | 9.06 (11.8) | NS |
| Cognitive Sx, n (%) | NA | 2 (18.2 %) | 27 (100 %) | <0.001 |
| Parkinsonism, n (%) | NA | 1 (9.1 %)* | 19 (70.4 %) | <0.001 |
| Hallucinations, n (%) | NA | 0 | 9 (33.3 %) | 0.028 |
| Fluctuations, n (%) | NA | 0 | 12 (44.4 %) | 0.008 |
| Constipation, n (%) | NA | 5 (45.5 %) | 22 (84.6 %) | 0.01 |
| Global CDR (IQR) | NA | 0.00 (0.5) | 0.50 (0.5) | <0.001 |
| CDR-SB (IQR) | NA | 0.00 (0.5) | 3.00 (3.0) | <0.001 |
| MoCA (IQR) | NA | 26.00 (4.5) | 21.50 (9.5) | 0.01 |
| MDS-UPDRS-III (IQR) | NA | 0.00 (2.0) | 19.00 (22.5) | 0.003 |
| Alpha Diversity | ||||
| Shannon Index (IQR) | 3.72 (3.54–3.88) | 3.87 (3.60–3.89) | 3.70 (3.34–4.05) | NS |
Data are presented as Medians and interquartile range (IQR); iRBD = isolated REM sleep behavior disorder; LBD = Lewy body disease; MCI = mild cognitive impairment; DLB = dementia with Lewy bodies; Sx = Symptoms; CDR = Clinical Dementia Rating; SB = sum of boxes; MDS-UPDRS-III = Movement Disorders Society Unified Parkinson’s Disease Rating Scale Motor Subscale Part III; MoCA = Montreal Cognitive Assessment; NA = not applicable; NS = not significant.
Parkinsonism not significant enough to be considered phenoconverted.
3.2. Differences in relative abundances of bacterial taxa
Alpha-diversity of cohabitant controls was not different from iRBD or LBD cases (Table 1). HC alpha-diversity was greater than LBD cases and cohabitant controls, but not iRBD cases. Beta-diversity did not differ between cohabitant controls, iRBD and LBD (ANOSIM R = 0.001; p = 0.5) but HCs differed significantly from all groups (p < 0.001 for all comparisons).
No bacterial genera differed between cohabitant controls and patients with LBD (Fig. 1A-F). Patients with iRBD only had lower abundance of a single taxa (Bacteroides) compared to cohabitant controls (Fig. 1A). The addition of HCs successfully helped parse differences between patients and cohabitant controls (Fig. 1A-F). Compared to HCs, patients with LBD had lower abundances of Lachnospiraceae, Faecalibacterium, and Alistipes (Fig. 1D, E, 1F), but these taxa did not differ between HCs and cohabitant controls. In addition to lower abundances of Bacteroides (Fig. 1A), patients with iRBD were also found to have decreased Alistipes (Fig. 1F) compared to HCs. No differences in Alistipes were observed between HCs and cohabitant controls. Compared to HCs, the abundance of Blautia was increased in patients with LBD, iRBD, and cohabiting controls. Finally, an increased relative abundance of Collinsella was observed in both patients with LBD and iRBD compared to HCs (Fig. 1C), but these differences failed to reach Bonferroni-adjusted significance (p < 0.0083).
Fig. 1.
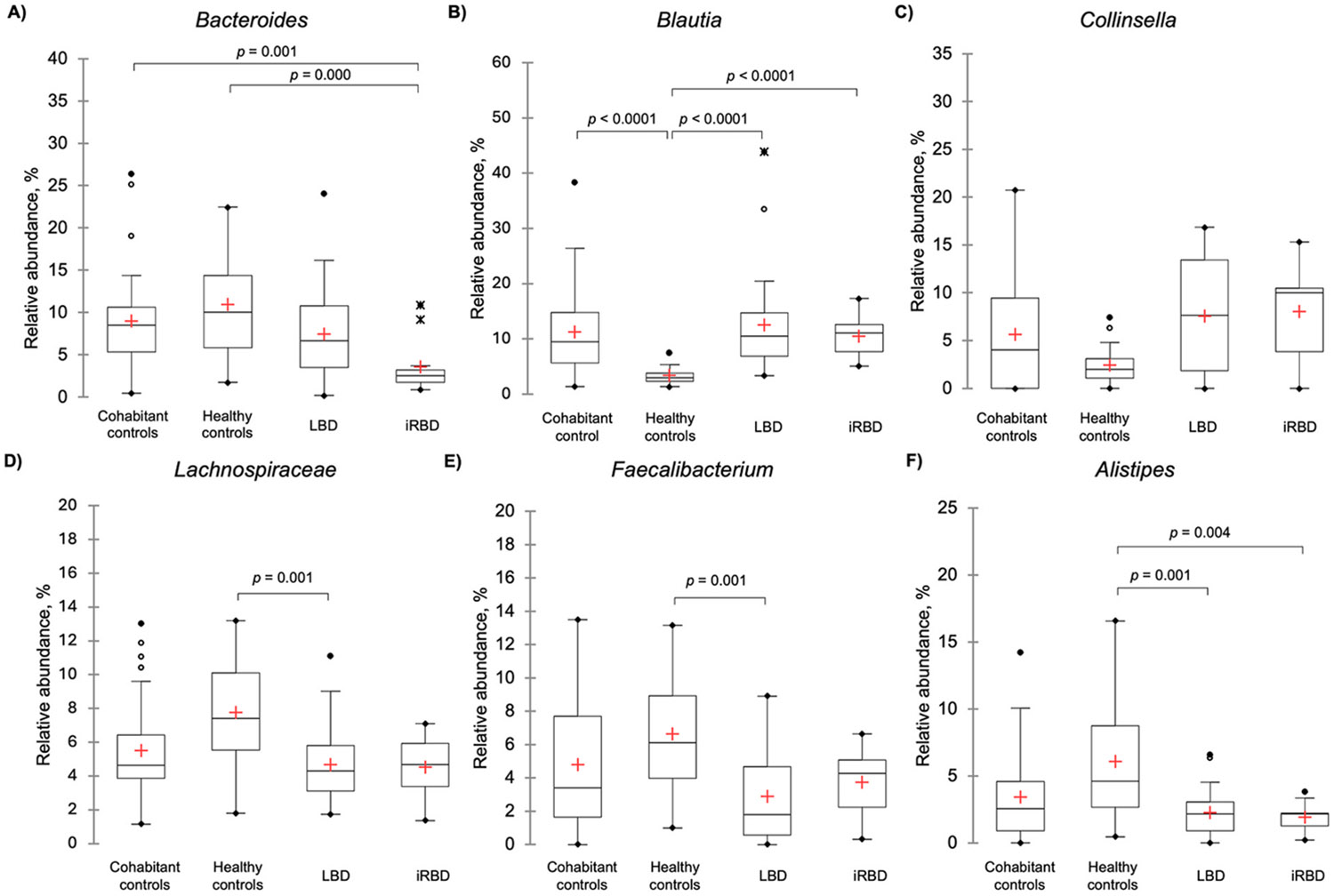
Comparisons of relative abundance distributions of taxa between Lewy body disease, isolated REM sleep behavior disorder, cohabitant controls and healthy controls
Legend - Box-plots of relative abundance distributions of taxa found to be significantly different between study groups along with Collinsella which lost significance after Bonferroni correction. The red cross represents the mean. The solid black lines in ascending order represent the minimum, quartile 1, median, quartile 3, and maximum. Data points more than 1.5 times the IQR away from the first or third quartiles are considered outliers and are marked by hollow, solid, and X points.
4. Discussion
Our data underscores the importance of using matched cohabitant controls in microbiome research in LBD and iRBD. No bacterial genera differed between cohabitant controls and patients with LBD and only a single taxa (Bacteroides) was found to differ between cohabitant controls and patients with iRBD. These lack of differences between disease states and cohabitant controls are in line with recent work with cohabitant controls in a pulmonary arterial hypertension patient population [10] and represents an important consideration in microbiome research more broadly. Inclusion of a HC cohort, identified microbial differences in the LBD spectrum in patients from the United States with iRBD and the cognitive impairment-predominant phenotypes of LBD (MCI-LB, DLB) which are similar to those reported in studies from other countries that did not rely exclusively on matched cohabitant controls and have primarily focused on PD [3,5,6]. Specifically, we found a lower abundance of SCFA producers Faecalibacterium and Lachnospiraceae in LBD and non-significantly increased Collinsella in both LBD and iRBD when compared with HCs, similar to prior reports [3,5,6]. This supports the possibility of shared dysbiotic changes across PD, DLB and iRBD. However, rigorous studies across the spectrum of these diseases are needed to refine our understanding of the significance of these changes, particularly in the absence of significant differences when exclusively comparing cohabitant controls.
We found substantially similar microbiome compositions between iRBD, LBD, and their cohabitant controls compared to HCs, consistent with the premise of microbial sharing with the cohabitants with and without disease [10,11]. This underscores the importance of incorporating matched cohabitant controls in future LBD microbiome research, and may shed light on whether previously reported apparent microbiome composition differences between LBD populations and controls may have instead been ascribable to lack of including, or limiting analysis to, matched cohabiting controls rather than disease-specific group differences [1,2,4-7,12]
Limitations of this study include a relatively small number of participants, a homogeneous predominantly Caucasian population, and limited available clinical data regarding cohabitant controls, although none had known iRBD or LBD diagnoses. Associations are limited to the high-level taxonomic information provided by 16S rRNA amplicon sequencing which limits the direct association of microbial function and disease state. We were unable to age match our HCs to cases and cohabitants controls, which may have influenced some differences between cases and HC. However, if age was the primary driver of our findings, we would expect similar differences between HC and cohabitant controls which were not present, thus age difference does not fully explain differences between HC and iRBD or LBD. Finally, our cohort was limited to cognitive manifestations of LBD and its proposed prodromal state, iRBD. Future studies evaluating microbiome changes across the spectrum of synucleinopathies (PD, iRBD, DLB, multiple system atrophy, pure autonomic failure) are warranted.
Beyond a single taxa in iRBD, we didn’t observe differences in microbiome composition between cases and cohabitant controls in our study. While further evaluation of potential microbiome influences on LBD pathophysiology is warranted, future research should be premised largely around cohabitant controls due to shared microbiome features. Future research should also incorporate more robust approaches such as shotgun metagenomics and metabolomics to further delineate microbiome-related mechanistic differences in LBD, and further probe differences between LBD spectrum disorders and cohabiting controls.
Funding
Study funded by NIH P30AG062677 (B. Boeve, R. Petersen), U01NS100620 (B. Boeve, K. Kantarci), R34AG056639 (B. Boeve, E. St Louis), U19AG071754 (B. Boeve, E. St Louis), the University of Minnesota Data Science Initiative (L. Teigen), the Mangurian Foundation for Lewy body disease research (B. Boeve), The Little Family Foundation (B. Boeve, O. Ross), the Turner Foundation (B. Boeve), Ted Turner and Family (O. Ross) and the Robert H. and Clarice Smith and Abigail van Buren Alzheimer Disease Research Program (R. Petersen).
Footnotes
CRediT authorship contribution statement
Levi M. Teigen: Writing – review & editing, Writing – original draft, Funding acquisition, Formal analysis, Conceptualization. Stuart J. McCarter: Writing – review & editing, Writing – original draft, Formal analysis, Data curation, Conceptualization. Zachary Ziegert: Writing – review & editing, Writing – original draft, Formal analysis. Christopher Staley: Writing – review & editing, Writing – original draft, Visualization, Funding acquisition, Formal analysis. Kiera M. Grant: Writing – review & editing, Data curation. Vinod K. Gupta: Writing – review & editing, Formal analysis. Xiaowei Zhao: Writing – review & editing, Formal analysis. Erik K. St Louis: Writing – review & editing, Conceptualization. Kejal Kantarci: Writing – review & editing. Val J. Lowe: Writing – review & editing. Leah K. Forsberg: Writing – review & editing. Rodolfo Savica: Writing – review & editing. Vijay K. Ramanan: Writing – review & editing. David T. Jones: Writing – review & editing. Ronald C. Petersen: Writing – review & editing. Jaeyun Sung: Writing – review & editing, Formal analysis. Alexander Khoruts: Writing – review & editing, Formal analysis, Conceptualization. Bradley F. Boeve: Writing – review & editing, Funding acquisition, Conceptualization. Owen A. Ross: Writing – review & editing, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Chen SJ, Chen CC, Liao HY, et al. , Association of fecal and plasma levels of short-chain fatty acids with gut microbiota and clinical severity in patients with Parkinson disease, Neurology 98 (8) (Feb 22 2022) e848–e858, 10.1212/WNL.0000000000013225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Heintz-Buschart A, Pandey U, Wicke T, et al. , The nasal and gut microbiome in Parkinson’s disease and idiopathic rapid eye movement sleep behavior disorder, Mov. Disord 33 (1) (Jan 2018) 88–98, 10.1002/mds.27105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Huang B, Chau SWH, Liu Y, et al. , Gut microbiome dysbiosis across early Parkinson’s disease, REM sleep behavior disorder and their first-degree relatives, Nat. Commun 14 (1) (May 2 2023) 2501, 10.1038/s41467-023-38248-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Nishiwaki H, Hamaguchi T, Ito M, et al. , Short-chain fatty acid-producing gut microbiota is decreased in Parkinson’s disease but not in rapid-eye-movement sleep behavior disorder, mSystems 5 (6) (Dec 8 2020), 10.1128/mSystems.00797-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nishiwaki H, Ito M, Ishida T, et al. , Meta-analysis of gut dysbiosis in Parkinson’s disease, Mov. Disord 35 (9) (Sep 2020) 1626–1635, 10.1002/mds.28119. [DOI] [PubMed] [Google Scholar]
- [6].Nishiwaki H, Ueyama J, Kashihara K, et al. , Gut microbiota in dementia with Lewy bodies, NPJ Parkinsons Dis 8 (1) (Dec 9 2022) 169, 10.1038/S41531-022-00428-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ryman S, Vakhtin AA, Richardson SP, Lin HC, Microbiome-gut-brain dysfunction in prodromal and symptomatic Lewy body diseases, J. Neurol 270 (2) (Feb 2023) 746–758, 10.1007/s00415-022-11461-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hamilton MJ, Weingarden AR, Sadowsky MJ, Khoruts A, Standardized frozen preparation for transplantation of fecal microbiota for recurrent Clostridium difficile infection, Am. J. Gastroenterol 107 (5) (May 2012) 761–767, 10.1038/ajg.2011.482. [DOI] [PubMed] [Google Scholar]
- [9].Staley C, Kaiser T, Vaughn BP, et al. , Predicting recurrence of Clostridium difficile infection following encapsulated fecal microbiota transplantation, Microbiome 6 (1) (Sep 18 2018) 166, 10.1186/s40168-018-0549-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Moutsoglou DM, Tatah J, Prisco SZ, et al. , Pulmonary arterial hypertension patients have a proinflammatory gut microbiome and altered circulating microbial metabolites, Am. J. Respir. Crit. Care Med 207 (6) (Mar 15 2023) 740–756, 10.1164/rccm.202203-0490OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Song SJ, Lauber C, Costello EK, et al. , Cohabiting family members share microbiota with one another and with their dogs, Elife 2 (Apr 16 2013) e00458, 10.7554/eLife.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Boertien JM, Murtomaki K, Pereira PAB, et al. , Fecal microbiome alterations in treatment-naive de novo Parkinson’s disease, NPJ Parkinsons Dis 8 (1) (Oct 10 2022) 129, 10.1038/s41531-022-00395-8. [DOI] [PMC free article] [PubMed] [Google Scholar]


