Abstract
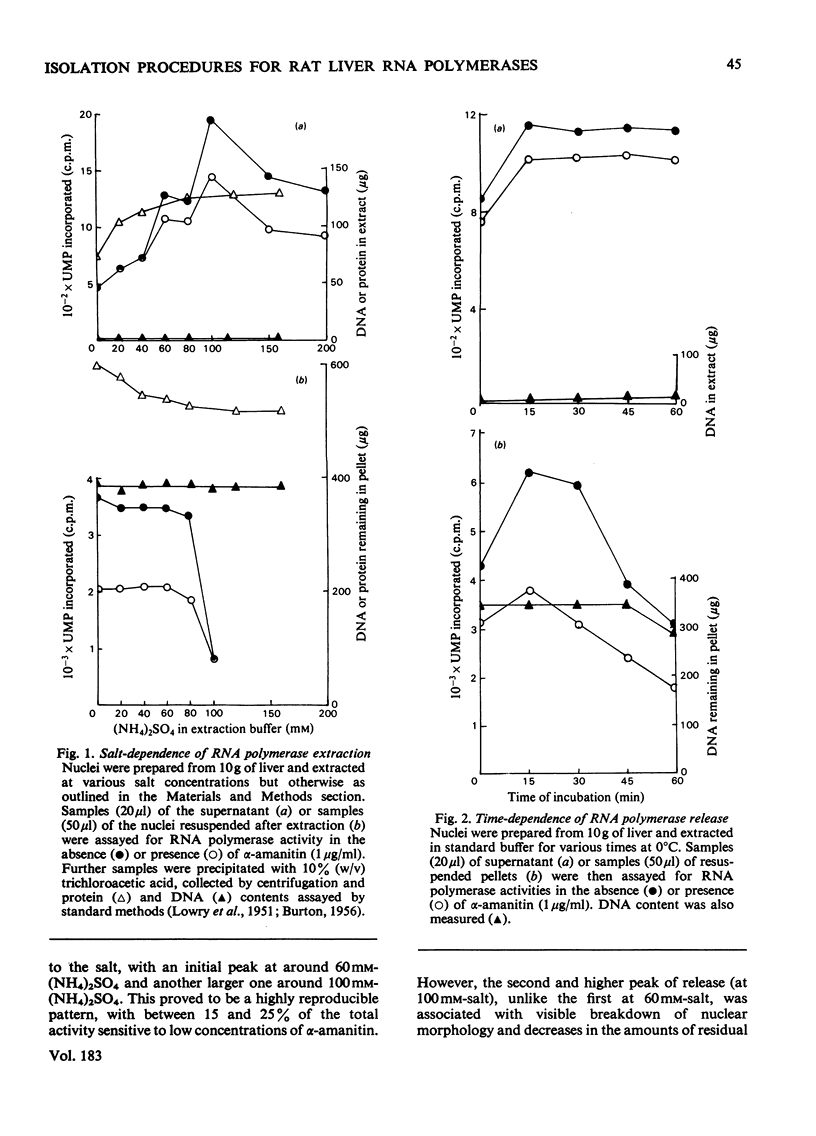
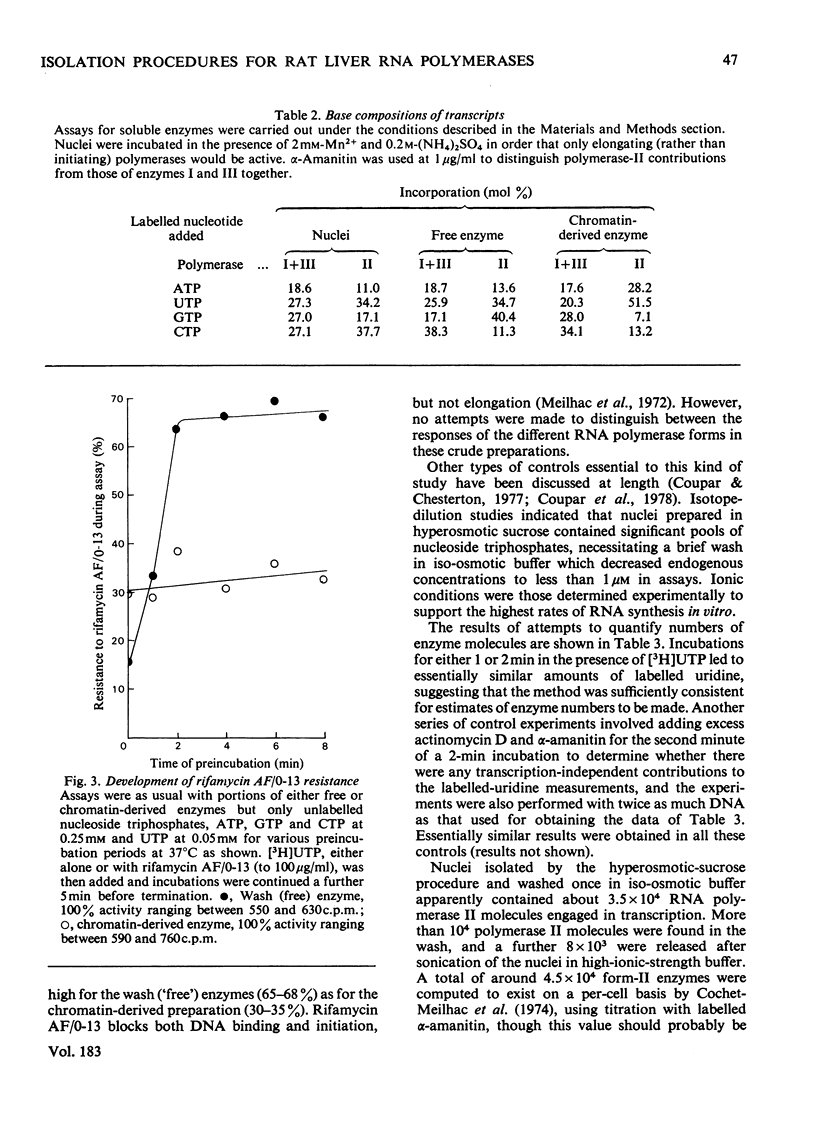
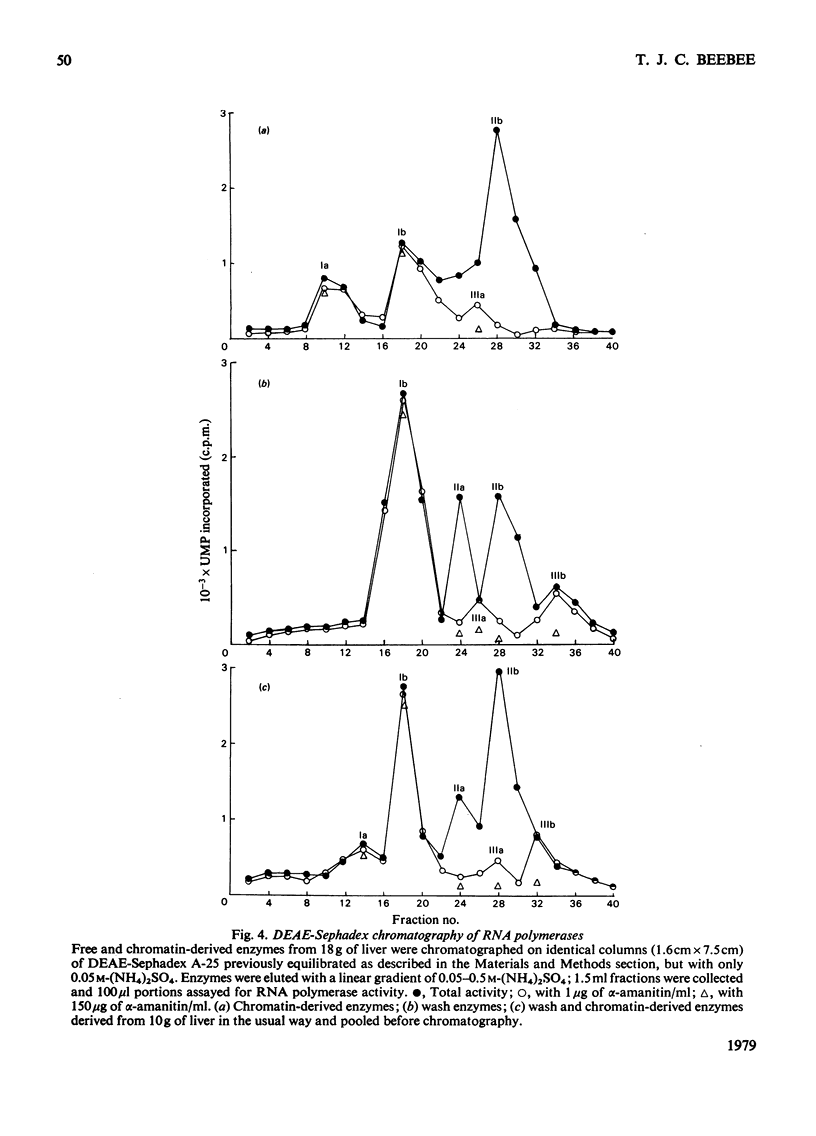
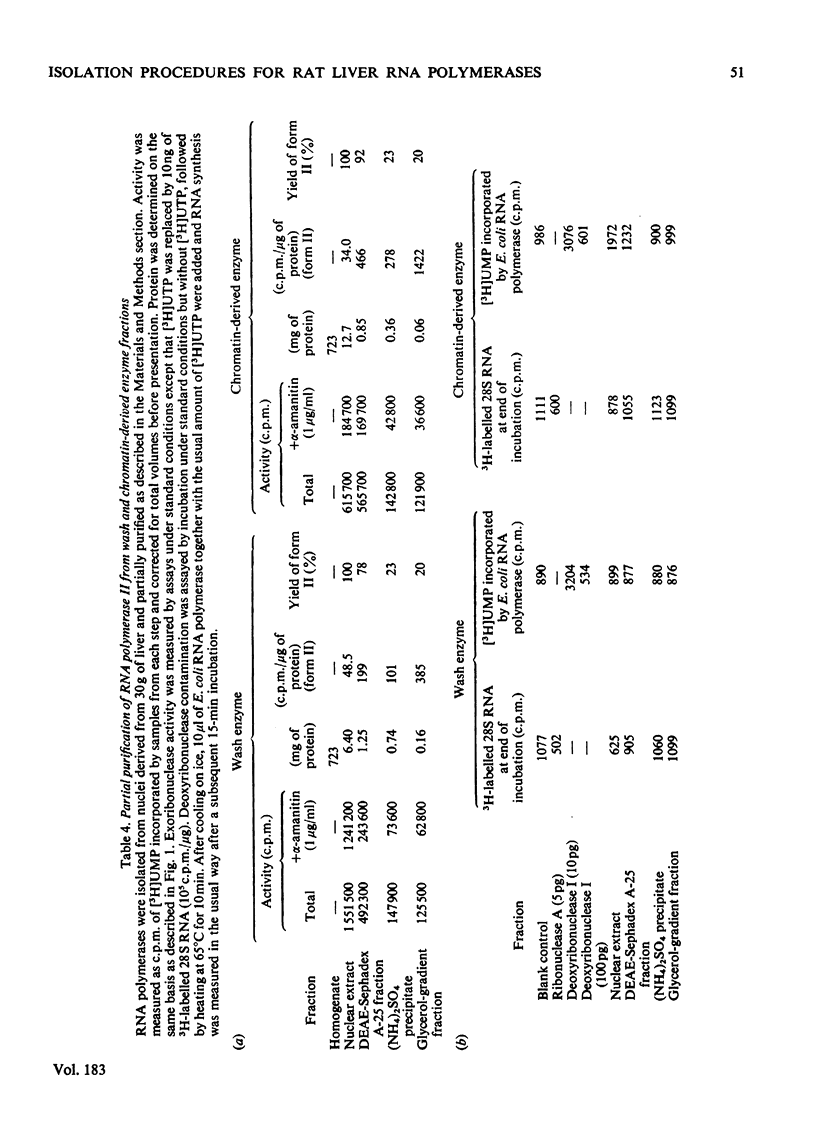
Nuclei were prepared from rat liver after homogenization of the tissue in hyperosmotic sucrose and RNA polymerases (EC 2.7.7.6) extracted by two methods applied sequentially. Optimal conditions for washing loosely bound enzymes out of nuclei were determined first, and involved short (10 min) incubations at 0 degrees C in the presence of 5 mM-Mg2+ and 60 mM-(NH4)2SO4. Subsequent sonication of the residual nuclear pellet after resuspension and lysis at high ionic strength resulted in further release of RNA polymerases. The primary wash yielded about 2 x 10(4) molecules of RNA polymerases I and III (altogether) and 1 x 10(4) molecules of form-II enzymes per original nucleus, whereas subsequent sonication released 2 x 10(4)-2.5 x 10(4) form-I and -III enzyme molecules (altogether) and a further 7 x 10(3)-8 x 10(3) form-II enzyme molecules, as measured by end-labelling of nascent RNA. RNA polymerase II was partially purified from both types of extracts and shown to initiate very poorly on high-molecular-weight homologous DNA irrespective of the source of the enzyme.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Austoker J. L., Beebee T. J., Chesterton C. J., Butterworth P. H. DNA-dependent RNA polymerase activity of Chinese hamster kidney cells sensitive to high concentrations of alpha-amanitin. Cell. 1974 Nov;3(3):227–234. doi: 10.1016/0092-8674(74)90136-6. [DOI] [PubMed] [Google Scholar]
- BURTON K. A study of the conditions and mechanism of the diphenylamine reaction for the colorimetric estimation of deoxyribonucleic acid. Biochem J. 1956 Feb;62(2):315–323. doi: 10.1042/bj0620315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebee T. J., Butterworth P. H. Template specificities of Xenopus laevis RNA polymerases. Selective transcription of ribosomal cistrons by RNA polymerase A. Eur J Biochem. 1974 Jun 15;45(2):395–406. doi: 10.1111/j.1432-1033.1974.tb03564.x. [DOI] [PubMed] [Google Scholar]
- Beebee T. J., Butterworth P. H. Transcription fidelity and structural integrity of isolated nucleoli. Eur J Biochem. 1977 Jul 15;77(2):341–348. doi: 10.1111/j.1432-1033.1977.tb11673.x. [DOI] [PubMed] [Google Scholar]
- Beebee T. J. The use of rat liver nucleoplasm for the characterization of heterogeneous nuclear ribonucleic acid synthesis in vitro. Biochem J. 1978 Dec 15;176(3):715–725. doi: 10.1042/bj1760715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler J. M., Iborra F., Sentenac A., Fromageot P. Structural studies on yeast RNA polymerases. Existence of common subunits in RNA polymerases A(I) and B(II). J Biol Chem. 1976 Mar 25;251(6):1712–1717. [PubMed] [Google Scholar]
- Burgess R. R., Travers A. A., Dunn J. J., Bautz E. K. Factor stimulating transcription by RNA polymerase. Nature. 1969 Jan 4;221(5175):43–46. doi: 10.1038/221043a0. [DOI] [PubMed] [Google Scholar]
- Chambon P. Eukaryotic nuclear RNA polymerases. Annu Rev Biochem. 1975;44:613–638. doi: 10.1146/annurev.bi.44.070175.003145. [DOI] [PubMed] [Google Scholar]
- Chesterton C. J., Butterworth P. H. Selective extraction of form I DNA dependent RNA polymerase from rat liver nuclei and its separation into two species. Eur J Biochem. 1971 Mar 11;19(2):232–241. doi: 10.1111/j.1432-1033.1971.tb01309.x. [DOI] [PubMed] [Google Scholar]
- Cochet-Meilhac M., Nuret P., Courvalin J. C., Chambon P. Animal DNA-dependent RNA polymerases. 12. Determination of the cellular number of RNA polymerase B molecules. Biochim Biophys Acta. 1974 Jun 27;353(2):185–192. doi: 10.1016/0005-2787(74)90183-x. [DOI] [PubMed] [Google Scholar]
- Coupar B. E., Chesterton C. J. The mechanism by which heparin stimulates transcription in isolated rat liver nuclei. Polyribonucleotide elongation rates and the number of transcribing RNA polymerase molecules present. Eur J Biochem. 1977 Oct 3;79(2):525–533. doi: 10.1111/j.1432-1033.1977.tb11837.x. [DOI] [PubMed] [Google Scholar]
- Coupar B. E., Davies J. A., Chesterton C. J. Quantification of hepatic transcribing RNA polymerase molecules, polyribonucleotide elongation rates and messenger RNA complexity in fed and fasted rats. Eur J Biochem. 1978 Mar 15;84(2):611–623. doi: 10.1111/j.1432-1033.1978.tb12204.x. [DOI] [PubMed] [Google Scholar]
- FURTH J. J., HO P. THE ENZYMATIC SYNTHESIS OF RIBONUCLEIC ACID IN ANIMAL TISSUE. I. THE DEOXYRIBONUCLEIC ACID-DIRECTED SYNTHESIS OF RIBONUCLEIC ACID AS CATALYZED BY AN ENZYME OBTAINED FROM BOVINE LYMPHOSARCOMA TISSUE. J Biol Chem. 1965 Jun;240:2602–2606. [PubMed] [Google Scholar]
- Fuhrman S. A., Gill G. N. Adrenocorticotropic hormone regulation of adrenal RNA polymerases. Stimulation of nuclear RNA polymerase III. Biochemistry. 1976 Dec 14;15(25):5520–5527. doi: 10.1021/bi00670a016. [DOI] [PubMed] [Google Scholar]
- Garel A., Axel R. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3966–3970. doi: 10.1073/pnas.73.11.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Bellard M., Oudet P., Chambon P. Isolation of high-molecular-weight DNA from mammalian cells. Eur J Biochem. 1973 Jul 2;36(1):32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- Guialis A., Beatty B. G., Ingles C. J., Crerar M. M. Regulation of RNA polymerase II activity in alpha-amanitin-resistant CHO hybrid cells. Cell. 1977 Jan;10(1):53–60. doi: 10.1016/0092-8674(77)90139-8. [DOI] [PubMed] [Google Scholar]
- Guilfoyle T. J., Key J. L. The subunit structures of soluble and chromatin-bound RNA polymerase II from soybean. Biochem Biophys Res Commun. 1977 Jan 10;74(1):308–313. doi: 10.1016/0006-291x(77)91409-7. [DOI] [PubMed] [Google Scholar]
- Jacob S. T., Sajdel E. M., Munro H. N. Altered characteristics of mammalian RNA polymerase following solubilization from nuclei. Biochem Biophys Res Commun. 1968 Sep 6;32(5):831–838. doi: 10.1016/0006-291x(68)90316-1. [DOI] [PubMed] [Google Scholar]
- Kellas B. L., Austoker J. L., Beebee T. J., Butterworth P. H. Forms AI and AII DNA-dependent RNA polymerases as components of two defined pools of polymerase activity in mammalian cells. Eur J Biochem. 1977 Feb;72(3):583–594. doi: 10.1111/j.1432-1033.1977.tb11281.x. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lampert A., Feigelson P. A short lived polypeptide component of one of two discrete functional pools of hepatic nuclear alpha-amanitin resistant RNA polymerases. Biochem Biophys Res Commun. 1974 Jun 18;58(4):1030–1038. doi: 10.1016/s0006-291x(74)80247-0. [DOI] [PubMed] [Google Scholar]
- Lin Y. C., Rose K. M., Jacob S. T. Evidence for the nuclear origin of RNA polymerases identified in the cytosol: release of enzymes from the nuclei isolated in isotonic sucrose. Biochem Biophys Res Commun. 1976 Sep 7;72(1):114–120. doi: 10.1016/0006-291x(76)90968-2. [DOI] [PubMed] [Google Scholar]
- Meilhac M., Tysper Z., Chambon P. Animal DNA-dependent RNA polymerases. 4. Studies on inhibition by rifamycin derivatives. Eur J Biochem. 1972 Jul 13;28(2):291–300. doi: 10.1111/j.1432-1033.1972.tb01913.x. [DOI] [PubMed] [Google Scholar]
- Roeder R. G. Multiple forms of deoxyribonucleic acid-dependent ribonucleic acid polymerase in Xenopus laevis. Isolation and partial characterization. J Biol Chem. 1974 Jan 10;249(1):241–248. [PubMed] [Google Scholar]
- Roeder R. G., Rutter W. J. Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature. 1969 Oct 18;224(5216):234–237. doi: 10.1038/224234a0. [DOI] [PubMed] [Google Scholar]
- Roeder R. G., Rutter W. J. Specific nucleolar and nucleoplasmic RNA polymerases. Proc Natl Acad Sci U S A. 1970 Mar;65(3):675–682. doi: 10.1073/pnas.65.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz L. B., Roeder R. G. Purification and subunit structure of deoxyribonucleic acid-dependent ribonucleic acid polymerase I from the mouse myeloma, MOPC 315. J Biol Chem. 1974 Sep 25;249(18):5898–5906. [PubMed] [Google Scholar]
- Schwartz L. B., Roeder R. G. Purification and subunit structure of deoxyribonucleic acid-dependent ribonucleic acid polymerase II from the mouse plasmacytoma, MOPC 315. J Biol Chem. 1975 May 10;250(9):3221–3228. [PubMed] [Google Scholar]
- Ueno K., Sekimizu K., Mizuno D., Natori S. Antibody against a stimulatory factor of RNA polymerase II inhibits nuclear RNA synthesis. Nature. 1979 Jan 11;277(5692):145–146. doi: 10.1038/277145a0. [DOI] [PubMed] [Google Scholar]
- Van Keulen H., Planta R. J., Retèl J. Structure and transcription specificity of yeast RNA polymerase A. Biochim Biophys Acta. 1975 Jun 16;395(2):179–190. doi: 10.1016/0005-2787(75)90157-4. [DOI] [PubMed] [Google Scholar]
- Widnell C. C., Tata J. R. Studies on the stimulation by ammonium sulphate of the DNA-dependent RNA polymerase of isolated rat-liver nuclei. Biochim Biophys Acta. 1966 Sep;123(3):478–492. doi: 10.1016/0005-2787(66)90216-4. [DOI] [PubMed] [Google Scholar]
- Yu F. L. An improved method for the quantitative isolation of rat liver nuclear RNA polymerases. Biochim Biophys Acta. 1975 Jul 7;395(3):329–336. doi: 10.1016/0005-2787(75)90204-x. [DOI] [PubMed] [Google Scholar]
- Yu F. L. Increased levels of rat hepatic nuclear free and engaged RNA polymerase activities during liver regeneration. Biochem Biophys Res Commun. 1975 Jan 2;64(3):1107–1115. doi: 10.1016/0006-291x(75)90161-8. [DOI] [PubMed] [Google Scholar]
- Yu F. L. Two functional states of the RNA polymerases in the rat hepatic nuclear and nucleolar fractions. Nature. 1974 Sep 27;251(5473):344–346. doi: 10.1038/251344a0. [DOI] [PubMed] [Google Scholar]


