Abstract
Alternative polyadenylation (APA) is emerging as a post-transcriptional regulatory mechanism, similar as that of alternative splicing (AS), and plays a prominent role in regulating gene expression and increasing the complexity of the transcriptome and proteome. We use polyadenylation selected long-read isoform sequencing to obtain full-length transcript sequences in porcine muscles at five developmental stages. We identify numerous novel transcripts unannotated in the existing pig genome, including transcripts mapping to known and unknown gene loci, and widespread transcript diversity in porcine muscles. The top 100 most isoformic genes are mainly enriched in Gene Ontology terms related to muscle growth and development. It is revealed that intron retention/exon inclusion and the usage of distal polyadenylation site (PAS) are associated with ageing through analyzing changes of AS and PAS during muscle development. We also identify developmental changes in major transcripts and major PASs. Furthermore, genes/transcripts important for muscle development are identified. The results confirm the importance of AS and APA in pig muscles, substantially increasing transcriptional diversity and showing an important mechanism underlying gene regulation in muscles.
Subject terms: Developmental biology, Molecular biology
The integrative analysis of Iso-Seq and RNA-Seq reveals changes in alternative splicing and polyadenylation at five developmental stages of muscles in pigs.
Introduction
Skeletal muscle, the source of most meat, provides high-quality protein for human beings and is related closely to livestock economic traits. The growth and development of skeletal muscle determines directly the quality and quantity of meat in livestock production, and thereafter the economic benefits to the farm and producer. Although non-genetic factors such as management, environmental background, and nutrition can affect the formation and development of skeletal muscle, the genetic factors play crucial roles. It is a key priority to characterize the molecular mechanisms underlying the muscle growth and development, which will provide basis for genetic improvement of muscle trait. Studies have shown that muscle growth and development is precisely orchestrated by various transcription factors including myogenic factor 51, myocyte enhancer factor 22, and paired-homeobox family3. Recently, it was revealed that noncoding RNAs, including long noncoding RNA, microRNA, and circular RNA are indispensable in the regulation of the process4–6. However, it is far from to fully describe the mechanisms.
Alternative splicing (AS) and alternative polyadenylation (APA) extensively exist in eukaryotes and play important roles in post-transcriptional regulatory process. Both of them can produce multiple transcripts, called isoforms, from a single pre-mature mRNA. They are particularly prevalent in human beings: >95% of multi-exon genes undergo AS and >70% of protein-coding genes harbor >1 functional polyadenylation site (PAS)7–9. The production of alternative transcripts by AS and/or APA represents an important mechanism of spatiotemporal gene regulation, and contributes substantially to the diversification of transcriptome and proteome. Thereafter, there is increasing interest in the role of AS and APA in physiological and pathological processes in humans.
AS and APA, highly regulated during growth and development, appear to be particularly important and prevalent in muscles10–12, where they impact differentiation, development, regeneration, and numerous key functions13,14. Tropomyosin isoforms localize differently along actomyosin bundles and function non-redundantly in skeletal muscle15. Myocyte-specific enhancer factor 2D, a key transcription factor controlling cell differentiation and organogenesis, undergoes a switch of major isoforms during myogenesis16. Several exons of the bridging integrator-1 are regulated by AS in different developmental stages, tissues, and pathological conditions, and the splicing dysregulation of exon 11 results in T-tubule defects and muscle weakness17–19. APA determines muscle stem cell fate and muscle function through regulating the levels of transcription factor Pax320. Alteration of APA pattern caused by RBFOX2 depletion regulates mRNA levels and/or isoform expression of a series of genes, including mitochondrial and contractile genes, in rat myoblasts21.
Polyadenylation selected long-read isoform sequencing (Iso-Seq) developed by Pacific Biosciences (PacBio) can obtain full-length transcripts directly, providing a useful tool for high-throughput analyzing AS and APA isoforms in eukaryotes. Combined with short-read RNA-sequencing (RNA-Seq) method, Iso-Seq has been used to identify novel isoforms, and to explore the role of isoforms in tissues including muscles22–24. However, landscape and regulation of AS and APA during muscle growth and development remain to be identified. Here, the dynamic changes of AS and APA isoforms in porcine muscles at five developmental stages were profiled with PacBio Iso-Seq in combination with RNA-Seq. The results confirm the importance of AS and APA in muscles, substantially increasing transcriptional diversity and showing an important mechanism underlying gene regulation in muscles.
Results
Construction of an Iso-Seq muscle transcriptome
A total of 179.2 G of Iso-Seq data, including 2,057,277 circular consensus sequences (CCS), were obtained from muscles at five developmental stages with an average of 35.8 G per sample. On average, 314,705.6 full-length non-chimeric (FLNC) reads were identified per sample, producing an average of 137,713.8 consensus isoforms with a mean length of 2007.7 bp. After quality control and filtering by SQANTI3, a total of 688,352, accounting for 99.97%, high-quality consensus isoforms were obtained and used for further analysis (Supplementary Table 1). The resulting genome annotation files were given in Supplementary Data 1–5.
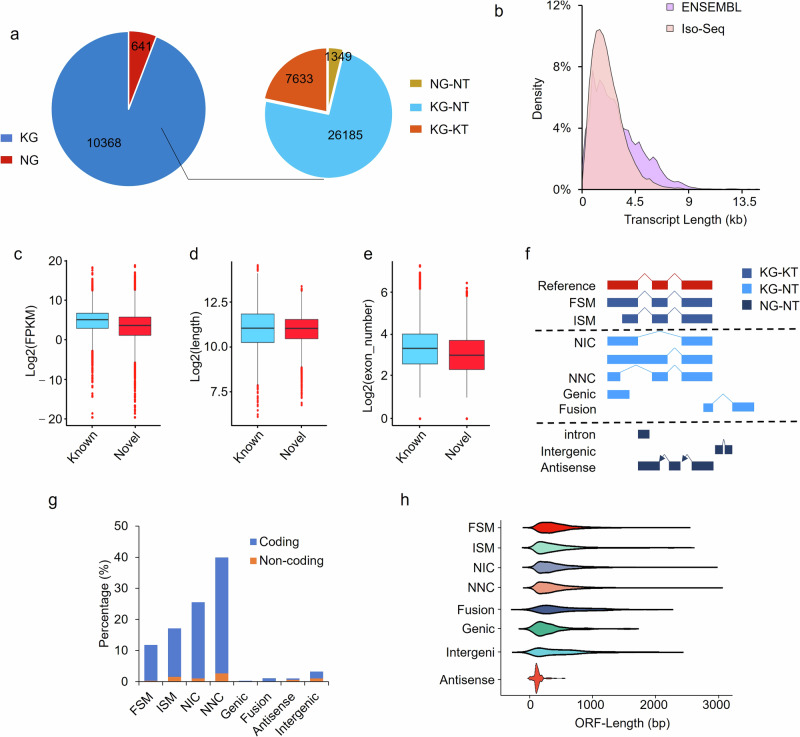
By integrating the transcripts of five stages, we recovered an Iso-Seq muscle transcriptome with SQANTI3 (Supplementary Data 6). After aligning to the reference genome (S. scrofa 11.1_release109) with Cupcake software, it was found that the transcriptome was composed of 35,167 transcripts corresponding to 11,009 gene loci of which 10,368 were known (mean length = 2.96 kb, SD = 1.8 kb, accounting for 94.18%) and 641 were novel (mean length = 1.73 kb, SD = 1.15 kb, accounting for 5.82%) (Table 1, Fig. 1a, Supplementary Table 2). These PB transcripts were mainly concentrated in 2 ~ 3 kb in length, consistent to those from the reference genome (Fig. 1b). All downstream analyses were based on the subset of Cupcake-filtered transcripts unless indicated.
Table 1.
Overview of the whole-transcriptome Iso-Seq datasets
| Total | A | B | C | D | E | |
|---|---|---|---|---|---|---|
| Unique genes | 11,009 | 6227 | 7151 | 7324 | 6998 | 6374 |
| Annotated genes | 10,368(94.18%) | 6047(97.11%) | 6930(96.91%) | 7120(97.21%) | 6824(97.51%) | 6216(97.52%) |
| Novel genes | 641(5.82%) | 180(2.89%) | 221(3.09%) | 204(2.79%) | 174(2.49%) | 158(2.48%) |
| Protein-coding gene | 10,768(97.81%) | 6097(97.91%) | 7006(97.97%) | 7196(98.25%) | 6882(98.34%) | 6241(97.91%) |
| Non-protein-coding gene | 241(2.19%) | 130(2.09%) | 145(2.03%) | 128(1.75%) | 116(1.66%) | 133(2.09%) |
| Isoforms | 35,167 | 10643 | 13741 | 13511 | 13357 | 11030 |
| Genes with >1 isoform | 6668(60.57%) | 2224(20.9%) | 2983(21.71%) | 2895(21.43%) | 2843(21.28%) | 2221(20.14%) |
| Genes with ≥10 isoforms | 461(4.19%) | 35(0.33%) | 55(0.4%) | 58(0.43%) | 64(0.48%) | 46(0.42%) |
| Known transcripts (%) | 7633(21.71%) | 3494(32.83%) | 4150(30.2%) | 4301(31.83%) | 4120(30.85%) | 3517(31.89%) |
| Novel transcripts | 27,534(78.29%) | 7149(67.17%) | 9591(69.8%) | 9210(68.17%) | 9237(69.15%) | 7513(68.11%) |
A, sample A (7-d-old pigs); B, sample B (30-d-old pigs); C, sample C (60-d-old pigs); D, sample D (90-d-old pigs); E, sample E (210-d-old pigs).
Fig. 1. Characterization of novel transcripts by Iso-Seq.
a Classification of genes identified. NG, novel genes. KG, known genes. NT, novel transcripts. KT, known transcripts. b Length distribution of transcripts. Differences in the abundance (c), length (d) and exon number (e) between novel and known transcripts identified by Cupcake program. f SQANTI3 program classifies the transcripts into nine categories. FSM, full-splice match. ISM, incomplete splice match. NIC, novel in catalog. NNC, novel not in catalog. g In-depth classification of Iso-Seq transcripts by SQANTI3 program. h Length distribution of open reading frames of transcripts among categories.
Novel transcripts were detected for a notable proportion of annotated genes in the pig muscles
7633 (21.71%) out of the 35,167 full-length transcripts were known in the reference genome, and the remaining 27,534 transcripts were novel with 26,185 (74.46%) from annotated genes (Fig. 1a, Table1). Many known genes were identified to have novel transcripts by the Iso-Seq in pig muscles. The muscle-specific creatine kinase gene, essential for maintaining energy homeostasis of muscle cell25,26, generates 67 transcripts including 2 known and 65 novels. The slow myosin binding protein-C gene, involved in the assembly and stabilization of muscle thick filaments27, produces 58 transcripts including 5 knowns and 53 novels. It was noted that, for some genes, only novel transcripts were obtained by the Iso-Seq although there were known transcripts in the reference genome. For example, a total of 15 transcripts were identified in uroporphyrinogen III synthase gene among five developmental stages, but none was known in the reference genome in which 3 transcripts were deposited.
99.19% of annotated transcripts were predicted as protein-coding, a little more than that of novel transcripts from annotated genes (98.16%); while 81.91% of transcripts from novel genes were predicted as protein-coding, far less than that from known genes. Compared to known transcripts, novel ones were generally less abundant (Mann-Whitney-U test: Z = –29.753, p < 0.001; Fig. 1c), and presumably harder to detect. Novel transcripts were also shorter (Z = –4.612, p < 0.001), and had less exons (Z = –20.803, p < 0.001; Fig. 1d, e).
To further classify the transcripts identified, SQANTI3 was used to process the muscle transcriptome constructed. A significant proportion of transcripts, accounting for 28.93%, were identified as annotated transcripts, including complete full splice match (FSM: 11.80%) and incomplete splice match (ISM: 17.13%) to existing annotations in reference genome. The majority, accounting for 66.82%, was characterized as novel transcripts from known genes. Most of these novel transcripts contained either known donor and acceptor splice sites, classified as novel in catalog (NIC: 25.52%), or at least one novel donor or acceptor site, classified as novel not in catalog (NNC: 39.91%). Also, there were a small number of fusions (1.1%) and genic transcripts (0.29%) in the group novel transcripts from known genes. Additionally, a very small number of transcripts (4.25%), classified as antisense and intergenic, were identified as novel transcripts from novel genes (Fig. 1f, g, Supplementary Table 3). No differences were found in length distribution of ORFs predicted among FSM, ISM, NIC, and NNC transcripts, while much differences were found between them and novel transcripts from novel genes, i.e. antisense and intergenic transcripts (Fig. 1h).
A widespread transcript diversity was detected in the pig muscle transcriptome
There were 10,882 and 19,627 multi-exon genes in the pig muscle transcriptome and reference genome, respectively, among which 61.28% and 51.53% underwent AS, producing 4.62 and 3.36 transcripts per gene, respectively, on average, indicating the sensitivity of Iso-Seq for identification of transcript diversity. Nebulin (NEB), a giant protein that winds around the actin filaments in the skeletal muscle sarcomere and plays important roles in force generation28, displays the greatest transcript diversity with 209 isoforms including 5 knowns and 204 novels. Gene Ontology (GO) analysis showed that the top 100 most isoformic genes were mainly involved in terms related to muscle growth and development such as Myofibril assembly, Muscle cell development, Contractile fiber, and Actin filament binding (Fig. 2a).
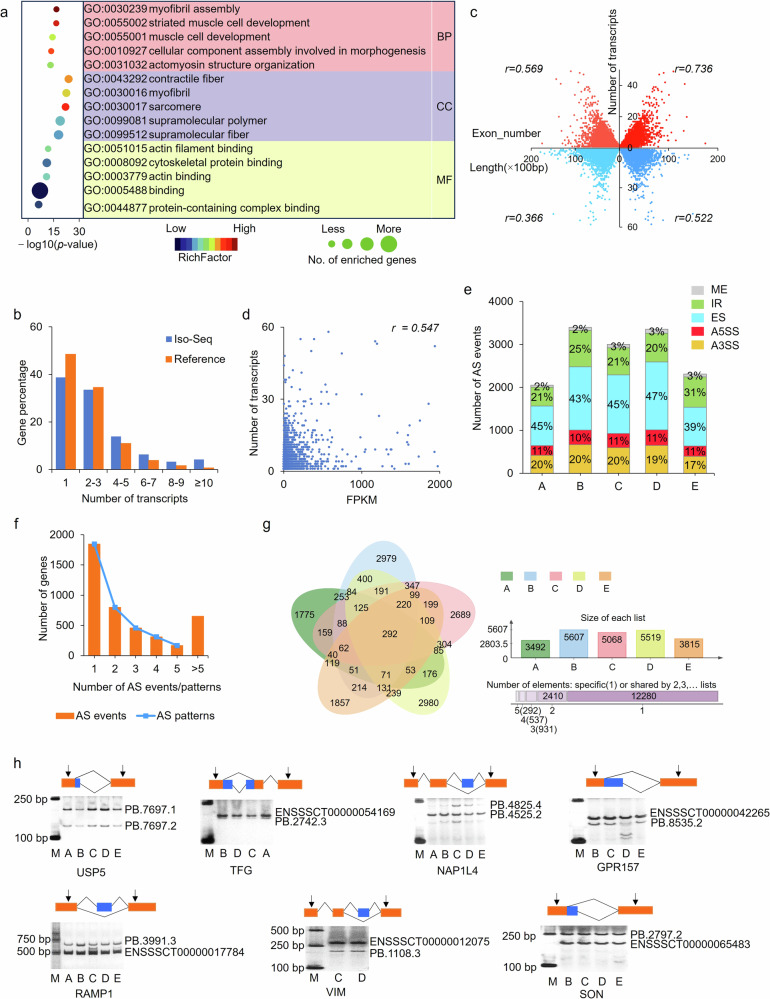
Fig. 2. Widespread transcript diversity was identified by Iso-Seq.
a GO terms enriched by the top 100 most isoformic genes. Top five terms are given in each category. b Distribution of number of transcripts among multi-exon genes. c Correlation analysis of number of transcripts with the number of exon and length of gene. d The Spearman Correlation analysis of the number of transcripts and FPKM of gene expression level. e Distribution of alternative splicing (AS) events among five developmental stages. f Distribution of AS events/patterns among genes. g Venn diagram of transcript among five developmental stages. h Validation of transcript diversity with RT-PCR method. In the legend, A, sample A (7-d-old pigs); B, sample B (30-d-old pigs); C, sample C (60-d-old pigs); D, sample D (90-d-old pigs); E, sample E (210-d-old pigs), the same as below.
The number of genes decreased with the number of transcripts increasing among multi-exon genes. The proportion of genes with isoforms ≥4 was much higher than that in reference genome, with the most differences in genes with transcripts ≥10 (Fig. 2b, Supplementary Table 4). Positive correlations exist between the number of transcripts and the number of exons or gene length, and the relationships were stronger among highly expressed genes: both the Spearman correlations increased when transcripts with Fragments Per Kilobase of transcript per Million (FPKM) < 10 were filtered out (Fig. 2c), reflecting the additional sensitivity for detecting transcripts of highly expressed genes. There also a moderate positive correlation between the number of transcript isoforms and gene expression level (Fig. 2d).
A total of 15,393 AS events including exon skipping (ES), intron retention (IR), alternative 5’ splicing site (A5SS), alternative 3’ splicing site (A3SS) and mutually exclusive exons (MEE) were obtained by Iso-Seq in pig muscles. These AS events were involved in a large number of genes (n = 4234), named AS-genes (Supplementary Table 5). ES and IR were the most prevalent events (ES: n = 6627 (43.05%), associated with 2828 genes, 66.79% of all AS-genes; IR: n = 3991 (25.93%), associated with 1569 genes, 38.8% of all AS-genes). There were little differences in the proportion of each AS type among five stages except for a significant increase in that of IR in 210-d-old pigs (Fig. 2e). Among them, 2388 genes (56.4%) had ≥ 2 AS events, and 1936 genes (45.73%) were characterized by more than one pattern of AS out of which 77 even contained all five types (Fig. 2f).
The AS-genes produced a total of 16,450 transcripts, i.e. AS-transcripts. Only 292 transcripts, corresponding to 177 genes, were shared by all five developmental stages (Fig. 2g). Most of AS-transcripts, n = 12,280 (74.65%), were specific to one period detected, corresponding to 3336 genes. Although significant differences existed in the number of AS-genes and AS-transcripts detected (4234 vs 16,450) among all the five periods, the number of AS-genes was similar to that of AS-transcripts in each of the five periods. These indicated that period-specific AS-transcripts might be a main reason regulating muscle growth and development. A total of seven AS genes were selected for validation with RT-PCR method, and expected products were obtained, confirming the accuracy of Iso-Seq in identifying AS and novel transcripts (Fig. 2h and Supplementary Fig. 1).
Illumina RNA-Seq reveals numerous differential alternative splicing events
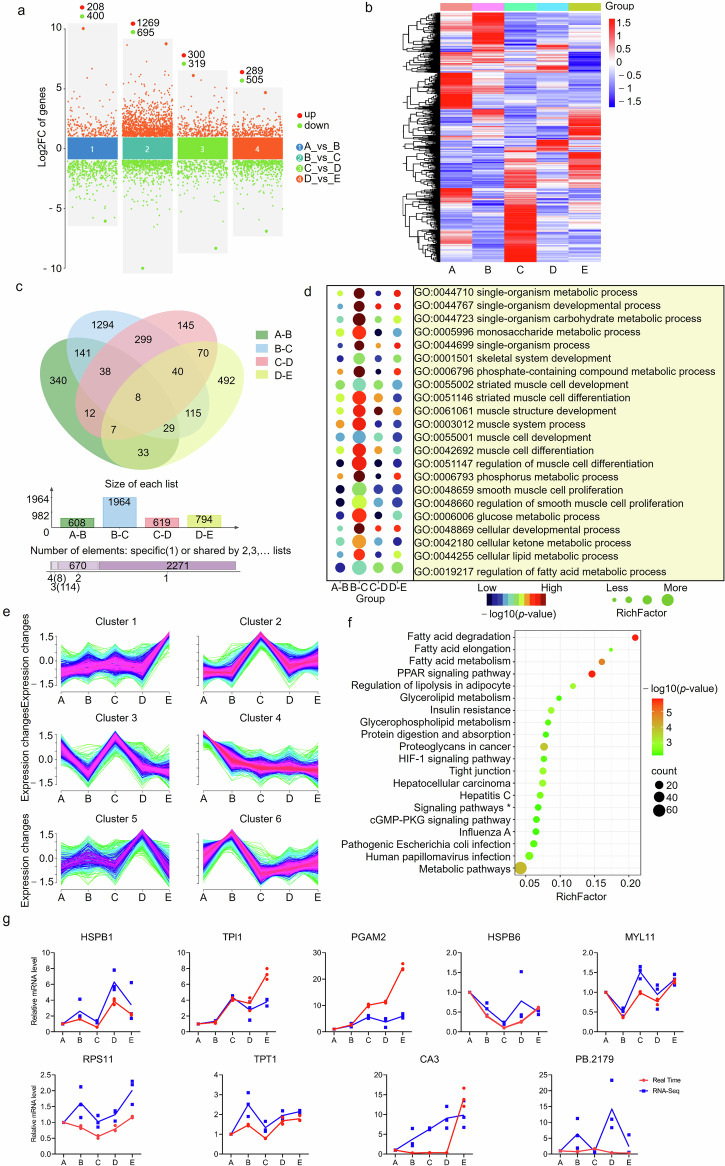
The gene expression profile was analyzed in muscles from five developmental stages with RNA-Seq, and the data overview was given in Supplementary Table 3. After pairwise comparison of adjacent periods, a total of 3063 differentially expressed genes (DEGs) were identified (Fig. 3a, b, Supplementary Table 6). There were more DEGs in pairwise group 30_vs_60 d than those in other three comparison groups. Majority of the DEGs were differentially expressed in only one comparison group, and only 8 DEGs were shared by all comparison groups in including delta like non-canonical Notch ligand 1, coiled-coil domain containing 69, protein kinase cGMP-dependent 1, actin binding Rho activating protein, LON peptidase N-terminal domain and ring finger 3, ENSSSCG00000063387, heat shock protein family A member 1 like, and nuclear receptor subfamily 4 group A member 3 (Fig. 3c). DEGs were enriched significantly in GO terms related to muscle growth in each comparison group (p < 0.05) (Fig. 3d).
Fig. 3. Characterization of differentially expressed genes (DEGs) in pairwise groups of adjacent periods.
a Multi-volcano plot of DEGs identified in the four comparison groups. b Heatmap of all DEGs identified in the four comparison groups. c, Venn diagram of DEGs. d Significantly enriched GO terms related to muscle growth (p < 0.05). e Clustering of all DEGs based on the expression patterns at five developmental stages. f KEGG pathways enriched by DEGs in cluster 4. *Signaling pathways regulating pluripotency of stem cells. g Real-time PCR validation of RNA-Seq data. Values are expressed as the mean of n = 3 repeats.
To explore the expression changes during skeletal muscle development, the 3063 DEGs were subjected to sequential analysis, and six clusters were obtained (Fig. 3e). The expression of 577 genes in cluster 4 showed a stable tendency to decline from 7 d to 210 d. The gene set were enriched in various GO terms related to metabolic and development process. It was interesting that the 577 genes were enriched significantly in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways associated with fat deposition (p < 0.05). Among top 20 pathways significantly enriched, nine were involved in fat accumulation including Fatty acid degradation, PPAR signaling pathway, Fatty acid metabolism, Regulation of lipolysis in adipocyte, Fatty acid elongation, HIF-1 signaling pathway, cGMP-PKG signaling pathway and Glycerolipid metabolism (Fig. 3f). Real-time PCR showed that seven out of nine DEGs randomly selected had similar expression changes among five samples, confirming the reliability of RNA-Seq (Fig. 3g and Supplementary Data 7).
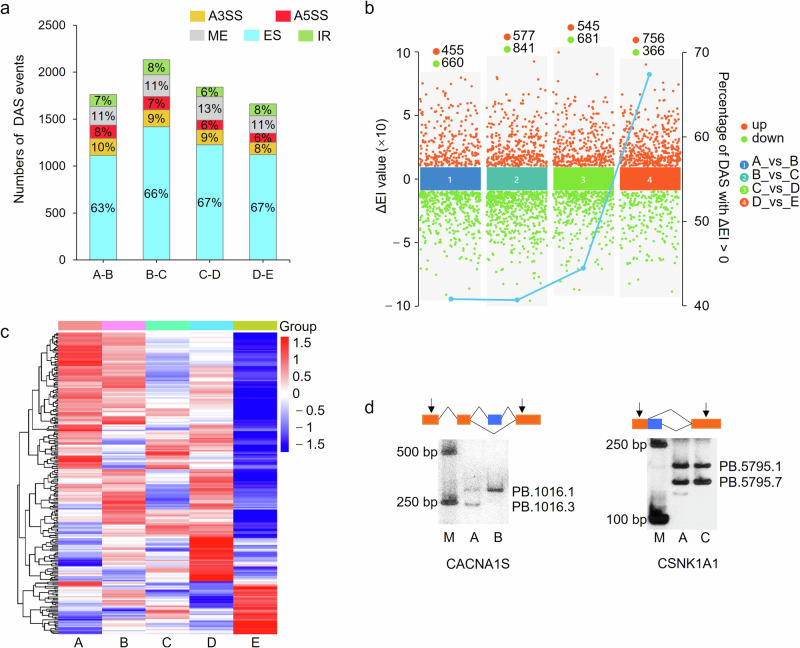
A total of 7399 unique differential AS (DAS) events were identified, corresponding to 2667 genes, in the four comparison groups of adjacent periods. Consistent with the distribution of DEGs, groups 30_vs_60 d had the most DAS events among the four pairwise comparisons (Fig. 4a, Supplementary Table 7). The composition of DAS events was similar among the four comparison groups. In each pairwise group differential ES ( > 60%) was the most frequently identified, indicating the dynamic changes in ES events might be closely related to muscle growth and development. We thereafter focused on ES-DAS events in subsequent analysis and found that △EI of them was increased significantly in groups 210-d-old compared to 90-d-old pigs (Fig. 4b), indicating that exon inclusion was generated with higher frequencies in 210-d-old pigs. Among all the pairwise comparisons, only group 90_vs_210 d had a proportion of events with △EI > 0 to all the events >50%, reached 67.46%.
Fig. 4. Identification of differential alternative splicing (DAS) events.
a Distribution of five types of DAS events. b The changes of △EI in exon skipping-DAS events. c Heatmap of genes encoding spliceosomal components in different samples. d RT-PCR validation of DAS events.
Among the DEGs in group 90_vs_210 d, 223 were related to splicesome components as annotated by Genecard (accessed on September 6th, 2023). Expression levels of most of these genes were generally lower in 210-d-old pigs than in other developmental stages (Fig. 4c), indicating that depressed expression of genes involved in splicesomal assembly might be partially responsible for increased exon inclusion in the development of muscles. Two DAS events were selected and validated by RT-PCR (Fig. 4d and Supplementary Fig. 2).
Integrated analysis of Iso-Seq and illumina RNA-Seq identifies abundant genes with alternative major transcripts
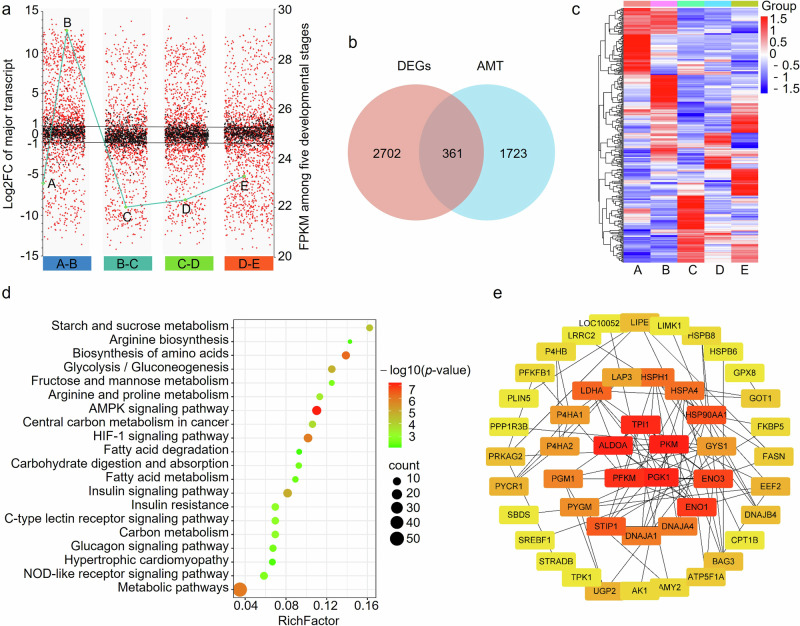
Next, we focused on the transcript with the most expression level, major transcript (MT), of each gene. The expression level of MTs seemed to be constant in muscles from 7- to 210-d-old pigs except for that at 30-d-old pigs (Fig. 5a). It was found that MT of the same gene was changed with muscle development, and the phenomenon was named alternative MT (AMT). AMT occurred extensively, and a total of 2084 genes, accounting for 49.17% of AS-genes, were identified to have AMT among periods detected (Supplementary Table 8). Generally, the expression level of genes with AMT was altered much greater than those with MT unchanged in each comparison groups (Fig. 5a).
Fig. 5. Identification of genes with alternative major transcripts (AMT).
a Expression changes of major transcripts. Genes with alternative major transcripts are shown with red and those with major transcript unchanged are shown with black between the comparison groups. b, Venn diagram of DEGs and genes with AMT. c, Heatmap of differentially expressed (DE)-AMT genes. d KEGG pathways significantly enriched by DE-AMT genes. e Protein-protein interaction analysis of DE-AMT. The minimum interaction score was set as 0.7, and the color of the rectangles indicates the degree of importance of the protein.
Among the 2084 genes with AMT, 361 were identified as differentially expressed, named DE-AMT genes, in at least one comparison group (Fig. 5b, c, Supplementary Table 8). The DE-AMT genes produce 3205 transcripts among which 980 can be identified as MT in at least one stage. Among the 361 genes, 103 were identified to have conserved domain by NCBI database, and 75 of them (72.81%) change the domain in AMT (Supplementary Table 8), indicating a potential for changes in the function of the polypeptide compared to the canonical one. These DE-AMT genes were annotated into various GO terms of all three functional categories, and mainly enriched in cellular process, metabolic process, and biological regulation in BP category. KEGG pathways associated with fat deposition, glycometabolism, and amino acid biosynthesis and metabolism were significantly enriched by the DE-AMT genes (p < 0.05) (Fig. 5d).
To explore the interaction among the DE-AMT genes, Protein-Protein Interaction (PPI) analysis was performed and genes with a score > 0.7 were visualized with Cytoscape. The network constructed was consisted of 48 nodes and 90 edges. Triosephosphate isomerase 1, pyruvate kinase, muscle, phosphoglycerate kinase 1, phosphofructokinase, muscle and fructose-bisphosphate aldolase A proteins had the most edges, indicating crucial roles of them for the muscle development (Fig. 5e).
A global increase in distal PAS usage was found during the development of muscle
APA can enhance transcriptome complexity by producing transcripts with difference in the 3’ ends. As the PASs of mRNAs were well obtained in the FLNC reads, we described the global polyadenylation events. We first made clear that there was weak correlation (r = 0.2799 ~ 0.3625) between the number of AS and PASs through Pearson analysis at five developmental stages, indicating that AS and APA are relatively independent.
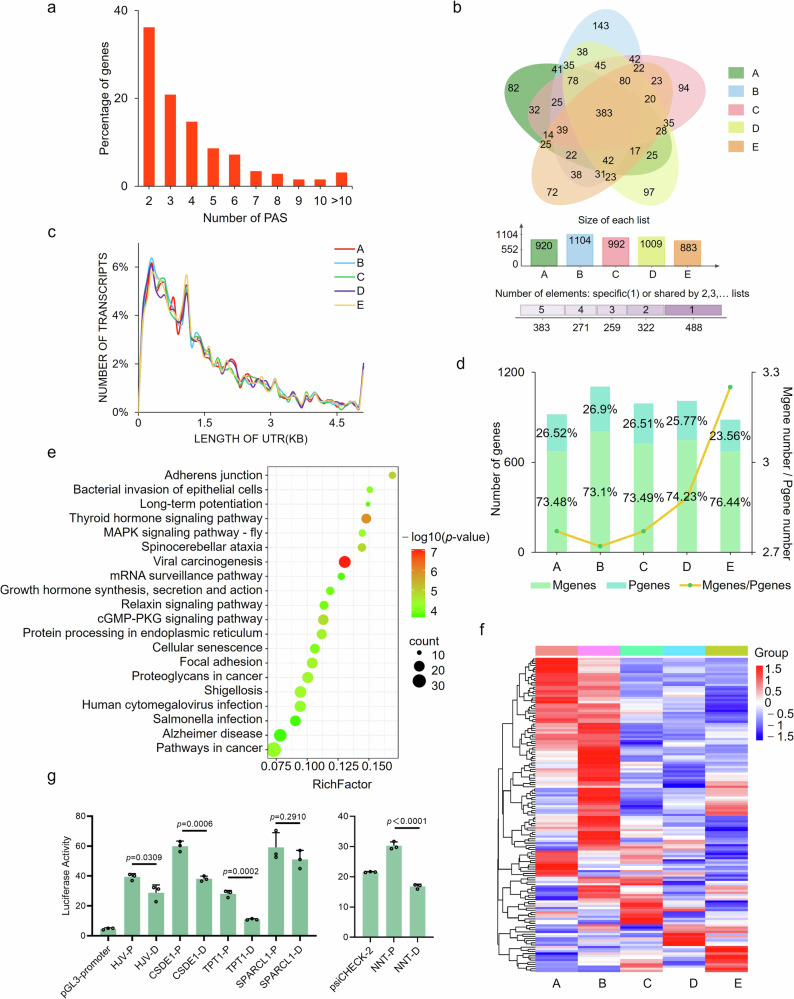
A total of 11,152 unique PASs, corresponding to 4025 genes, supported with an average of 10.26 reads per PAS, were predicted by TAPIS. To improve the accuracy of the data, we intersected the TAPIS data and those obtained by calculating the PAS location of each transcript. As a result, 1723 genes were found to have APA with an average of 4.03 per gene among five samples, and most of them produce 2 PASs (Fig. 6a, Supplementary Table 9). The most PASs, 53, were found in NEB gene. Among the 1723 genes 383 showed APA in all five periods, and 488 genes showed APA in only one period although most of them, 416, can express in multiple stages (Fig. 6b). The length of 3’ UTR of APA transcripts concentrated in 200 to 300 bp, and no significant differences were found among five periods (Fig. 6c). There were the most genes displaying APA, 1104, in pigs at 30-d-old among five developmental stages (Fig. 6d).
Fig. 6. Analysis of alternative polyadenylation (APA) in muscles at five developmental stages.
a Distribution of APA genes. b Venn diagram of APA genes among five developmental stages. c Length distribution of 3’ UTR of APA transcripts. d Distribution of PASs of APA transcripts. e KEGG pathways enriched by Mgenes at 210-d-old pigs. f Heatmap of Mgenes involved in the Fig. 6e. g Luciferase reporter analysis with proximal (P) and distal (D) PASs. Values are expressed as the mean ± SD of n = 3 independent samples.
To examine the usage of PAS in muscles, the expression ratio of the distal PAS isoform (M) to the proximal PAS isoform (m) was calculated for each gene. Over 70% of genes have M/m value > 1, i.e. Mgenes, much more than those with M/m value < 1 (Pgenes) in all five developmental stages (Fig. 6d, Supplementary Table 9), indicating that the distal PAS isoforms were generally more abundant. Notably, we observed a progressive increase in the usage of distal PAS during muscle development: the ratio of the number of Mgenes to that of Pgenes was positively correlated with the developmental periods (Pearson correlation = 0.83, Fig. 6d), indicating that with muscle development more genes select isoforms with distal PAS as major isoforms. The Mgenes in pigs at 210-d-old were mainly involved in KEGG pathways related to cellular senescence or aging such as Thyroid hormone29, Adherens junction30, Protein processing in endoplasmic reticulum31, Alzheimer disease, and Cellular senescence (Fig. 6e). It is interesting that genes involved in the Fig. 6e have a tendency to decrease with muscle development (Fig. 6f). Transcript isoforms with distal PASs are more likely to be degraded by microRNAs than those having proximal PASs, which might be the reason why the increases in the usage of distal PAS accompanied with decreases in gene expression during developmental stages. Luciferase reporter analysis showed that firefly luciferase genes with distal PASs, i.e. long 3’ UTRs, expressed lower than those with proximal PASs (Fig. 6g and Supplementary Data 8), verifying that 3’ UTRs lengthing have a role to inhibit the expression of genes compared to the shorter ones. These results suggested that a global increase of distal PAS usage, generating long 3’ UTR isoforms and thus decreasing the expression of genes, were associated with aging, and that 3’ UTR lengthening might act as a novel mechanism regulating growth and development of muscles.
Analyses on PAS alteration reveal genes important for muscle development
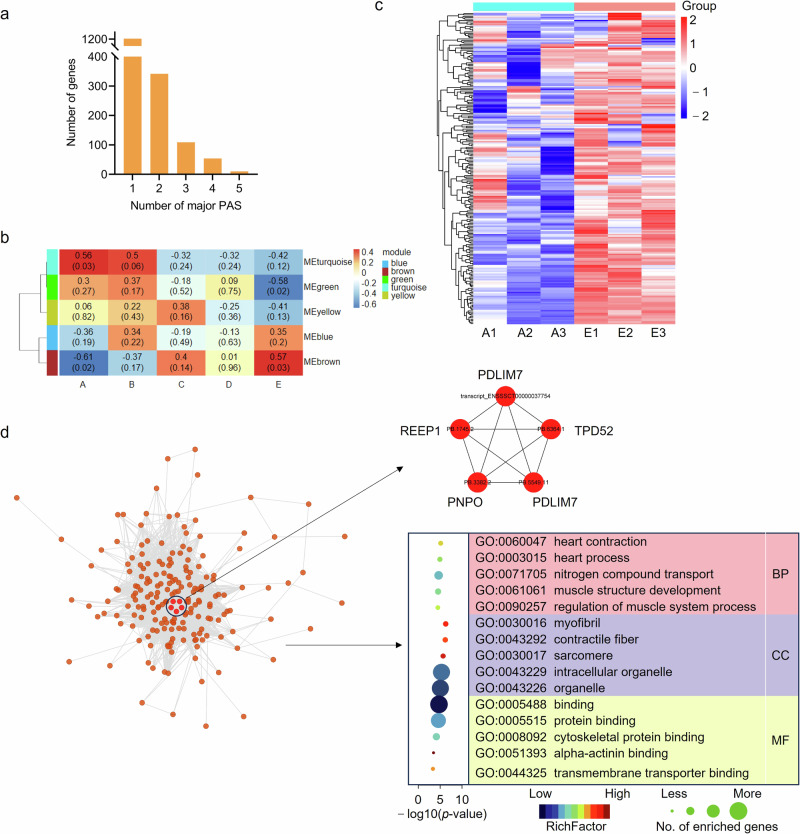
PAS with the highest usage was defined as the major PAS in each of the five tissues. Among the five developmental stages, 70.11% of multi-PAS genes, 1208, have constant major PAS, and the remaining ones, 515, change major PAS in at least one period among which 10 genes have unique major PAS in each of the stages (Fig. 7a, Supplementary Table 10). The changes of PAS were thought to be time-related32. To identify genes that might be significantly related to muscle development in pigs, Weighted Correlation Network Analysis (WGCNA) was conducted to screen out modules of highly interconnected genes. A total of five modules was obtained by using the total of 2129 transcripts from the 515 genes (Fig. 7b).
Fig. 7. Identification of PAS genes important for muscle development.
a Statistics of number of major PAS in genes identified among five developmental stages. b Module-trait association analysis. Each row represents a module, and column represents a trait; Blue and red color represent negative and positive correlation, respectively. c, Heatmap of transcripts clustered in Module brown. d Co-expression network characterizes hub transcripts in Module brown, and the following GO analysis shows the top 5 terms enriched by transcripts in the network.
Based on the connectivity, these modules were classified into two groups. Module brown, composed of 221 transcripts, corresponding to 128 genes, was the most associated with ages at 7-d-old and 210-d-old among the five modules (Supplementary Table 10). Notably, the expression level of all these transcripts was increased in pigs at 210-d-old compared to 7-d-old (Fig. 7c). A network was then built using the transcripts with a co-expression correlation > 0.05 in module brown. A total of 196 transcripts, corresponding to 118 genes, were included in the network among which pyridoxamine 5’-phosphate oxidase, PDZ and LIM Domain Protein, tumor protein D52, and receptor accessory protein 1 were identified as hub genes. These 118 genes were mainly involved in regulation of muscle system process, muscle structure development and muscle system process as revealed by GO analysis (Fig. 7d).
Discussion
Here, we used PacBio Iso-Seq combined with RNA-Seq to identify full-length transcripts and generate detailed maps of AS and APA in porcine muscles at five developmental stages. Considerable RNA isoform diversity, including novel transcripts not annotated in the pig genome, are characterized among expressed genes in muscles. Of note, full-length transcripts are detected from previously unannotated genes. Although global patterns of transcript diversity seem to be similar, we find developmental changes in RNA isoform diversity as AMT and alternative PAS exist among five stages. Additionally, we show that distal PAS usage of genes is associated with aging, indicating the importance of 3’ UTR lengthening in the growth and development of muscles. The results confirm the importance of AS and APA in muscles and highlight their roles on the growth and development of muscles.
To the best of our knowledge, we present the most detailed full-length transcriptome and comprehensive characterization of transcript diversity in porcine muscles yet undertaken. We firstly highlight that the current porcine database is incomplete and that novel isoforms are likely to exist for a notable proportion of genes expressed. There are not only examples of novel exons but also entire genes not annotated presently in pig genome. We show that the novel transcripts generally express with humble level, and that certain transcripts express in a period-dependent manner, even within the same detected genes. Here, we only detected five developmental stages after birth, more period-specific transcripts should exist in other stages, especially in embryonal stage. We also speculate that some isoforms might be breed- and tissue-specific. Additionally, it is worth noting that the sequencing depth did not achieve a saturation here, which might result in some transcript isoforms undiscovered. Thereafter, the reference annotation is even more incomplete than this study shows, and more sequencing could improve it further. However, a moderate positive correlation (r = 0.547) was found between the number of transcript isoforms and gene expression level. Genes with high expression level are prone to incorrect splicing, resulting in mis-assembled transcripts33. Thus, by-products of high gene expression and inaccurate regulation might exist among transcript isoforms obtained although RT-PCR verification of AS transcripts/events indicated the reliablity of Iso-Seq data. Transcript diversity caused by AS has been described in pigs in multiple tissues including muscles22,23,34, while studies on APA are limited. We make clear that multiple PASs are likely to exist for a large proportion of genes, and that the number (>4 per gene on average) is relatively higher than expected, indicating APA is an important mechanism underlying transcript diversity and transcriptome complexity. These might be one reason why substantial numbers of novel transcripts are identified here. Nevertheless, the results refresh the pig genome.
Secondly, we show the extent to which AS and APA events contribute to transcript diversity in porcine muscles. In particular, ES is a relatively common event compared to other AS events, followed by IR event, in all of the five developmental stages detected, different from previous studies on mixed skeletal muscles composed of longissimus thoracis and semitendinosus in which IR is more frequently identified than ES23. While, the other AS patterns, A3SS, A5SS and MEE, have the same ranking between the two studies. Here, only longissimus thoracis is used. The results might reflect the differences between longissimus thoracis and semitendinosus muscles. Strikingly, IR event increases substantially in pigs at 210-d-old compared to other four developmental stages. Through calculating △EI, we confirm that exon inclusion is increased in pigs at 210-d-old compared to the younger ones. Studies in C. elegans, mice and humans revealed that IR increases with an age-associated pattern and is involved in lifespan and Alzheimer’s disease35–37. Several lines of evidences implicate the intron-retained transcripts are likely to be less stable than those fully spliced ones38–40. IR is also showed to reduce the levels of transcripts through both nuclear sequestration and turnover and nonsense-mediated mRNA decay of intron-retained transcripts41. Here, we find exon inclusion increases with reduction of the expression level of splicesome component, partially explaining the reason of the increased exon inclusion in old pigs. Thereafter, IR events might play a role in developmental regulation of gene expression by degrading transcripts that are not or less required for the physiology of skeletal muscles in old pigs, which might be achieved by reducing the expression of splicesome component.
Thirdly, we reveal major developmental changes in isoform abundance in skeletal muscles, and highlight an essential role of transcript diversity in regulating muscle growth and development. There are numerous DAS events, alteration of major transcript and/or major PAS during the growth and development of skeletal muscle. It is widely known that alternatively spliced transcripts not only regulate the level of canonical mRNA but also can produce proteins with different functions42,43. Dysregulation of AS level has been related to diseases in humans including autism44,45, muscle fiber atrophy46, etc. APA results in the production of transcripts having different 3’ UTRs9,47,48. It is best known that 3’ UTRs regulate diverse fates of mRNAs such as degradation, nuclear export, localization, and translation. They can also determine the fate of proteins through regulating protein-protein interactions49. Additionally, if APA locates in the internal exon or intron, it might change the coding sequences, resulting in generation of multiple polypeptides from the same gene50. Thereafter, APA and AS represent an important mechanism of spatiotemporal gene regulation, which makes them an important tool for regulating phenotypic diversity. Here, we characterize 7399 DAS events, 2084 genes with AMT, 1723 genes with APA, as well as the MT usage profile among five developmental stages, indicating the importance of transcript diversity and providing data for further revealing the differential role of MTs in muscles. Concurrently, PAS usage profile and the changes in major PAS are identified among five stages in muscles. To our best knowledge this is the first study on changes of PAS in muscles which is the basis for revealing the role of APA during muscle development. Importantly, we make clear that the major PAS changes with aging, highlighting usage of distal PAS might represent a novel mechanism underlying muscle growth and development. Additionally, we show that genes related to muscle development tend to have more transcripts, and that a large number of period-specific transcripts are detected during muscle development. All these supports the importance of transcript diversity for muscle development.
In conclusion, we characterize developmental transcriptome landscape of muscles in pigs using long-read combined with short-read sequencing technology. Our data verify the importance of AS and APA in muscles, dramatically increasing transcript diversity and displaying an essential mechanism underlying gene regulation in muscles. Specifically, we characterize a lot of novel transcripts unannotated previously, and the global polyadenylation events, showing the reference annotation is far from complete. We also make clear the changes in MT and major PAS during the muscle development. Importantly, we highlight that a large proportion of AS-genes change MT with ages and that 3’ UTR lengthening of genes is associated with aging. Additionally, genes/transcripts important for muscle development are identified. The results not only refresh the pig genome but contribute to further revealing mechanisms underpinning growth and development of skeletal muscles.
Methods
Animals, tissues and RNA extraction
Min pigs, a Chinese local pig breed, were used here. A total of 15 male pigs, obtained from the Institute of Animal Husbandry, Heilongjiang Academy of Agricultural Sciences, Harbin, China, were used. The longissimus dorsi muscles were collected from pigs at 7-, 30-, 60-, 90- and 210-d-old, each with three individuals. Total RNA was isolated with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The purity and integrity of RNA was assessed with NanoDrop 2000 (Thermo Scientific, Wilmington, DE, USA) and Agilent 2100 (Agilent, Santa Clara, CA, USA). RNA concentration was measured using Qubit 3.0 Flurometer (Life Technologies, Carlsbad, CA, USA). All animal treatments were approved by the Laboratory Animal Welfare and Ethics Committee of Northeast Agricultural University. We have complied with all relevant ethical regulations for animal use.
Iso-Seq library construction and sequencing
Equal amount of qualified RNA from muscles of three individuals at the same developmental stages were pooled together to construct Iso-Seq library. Iso-Seq library preparation and sequencing was performed by Biomarker Technologies (Wuhan, China). Briefly, the full-length cDNA was synthesized with NEBNext Single Cell/Low Input cDNA Synthesis & Amplification Module (New England Biolabs, MA, USA). PCR amplification was performed with PrimeSTAR GXL DNA Polymerase (TaKaRa, Dalian, China). The PCR products were purified with AMPure PB magnetic beads (PacBio, Menlo Park, CA, USA), and measured with Qubit 3.0 (Life Technologies) and Agilent 2100 (Agilent) for concentration and size control. Finally, the PacBio Binding kit was used to bind the primer and Polymerase to the library. After purified with AMPure PB beads (PacBio), the library was sequenced on the sequel II platform (PacBio).
Raw read processing
The raw subreads were processed with the Iso-Seq3 pipeline (v3.4.0; https://github.com/PacificBiosciences/IsoSeq) to obtain FLNC reads. Briefly, polished CCS were generated from the raw subreads with CCS reads generator (--min-rq 0.9 --min-passes 3 -j 6 --min-length 200). Among the CCS reads those containing 5’/3’ cDNA primers and poly(A) tails were determined as full-length sequences. cDNA primers and poly(A) tails were removed from full-length reads with Lima (v2.1.0) and IsoSeq3 (v3.4.0) refine, respectively. The FLNC reads were filtered, clustered, and polished with IsoSeq3 (v3.4.0). The FLNC reads from the same transcript were clustered into a unique consensus isoform with SMRTLink software (v10.1). The high-quality consensus isoforms (accuracy >99%) were aligned to the reference genome (S. scrofa 11.1_release109) with minimap2 (v2.20-r1061). cDNA_Cupcake (v28.0.0) was then used to filter out reads with identity <0.9 and coverage <0.85, and to merge reads with differences only on the 5’ end. Owing to the intrinsic characteristics of Iso-Seq, the 5’ extremity might be incomplete, we considered the sequences with difference in 5’ end as the same isoform, and retained the longest one. The Iso-Seq data was annotated with the reference genome (S. scrofa 11.1_release109). Owing to the incomplete features of reference genome, there are Iso-Seq sequences longer than the reference ones, either in 3’-, 5’-end, or in both. To confirm these longer sequences, RNA-seq data were first used to extend the data of reference genome with StringTie software (v2.1.6, -eB -G). Briefly, if there were multiple overlapped RNA-seq reads mapped to the ends of the reference sequences, the reads were spliced to extend the reference sequences. Among serials of reads containing endmost nucleotides, the reads with the shortest 5’ end were selected to determine the start site of the sequences, while those with the longest 3’ end were used to determine the stop site. Then the optimized reference sequences were used to confirm the longer sequences obtained by Iso-Seq data. To improve the accuracy of alignment, redundant reads were further filtered out from each sample with SQANTI3 (v5.1.1) filter script to obtain five individual annotation file. At the same time an integrative annotation file of five samples were constructed with the following methods: the data of each sample were merged after cDNA_Cupcake (v28.0.0) filtering, and then redundant sequences caused by merging were removed wtih cDNA_Cupcake (v28.0.0) again, followed by SQANTI3 (v5.1.1) filtering. BUSCO (v3.0.2) was used to assess the integrity of the transcriptome. Protein-coding potential was predicted with SQANTI3 (v5.1.1).
Illumina RNA-Seq library construction and sequencing
The same muscles used for Iso-Seq were subjected to RNA-Seq. A total of 15 libraries (three for each period, five periods in total) were constructed and sequenced separately. Briefly, the libraries were prepared with the Hieff NGS Ultima Dual-mode mRNA Library Prep Kit for Illumina (Yeasen, Shanghai, China) according to the manufacturer’s instructions. Purification was performed with Hieff NGS DNA selection Beads (Yeasen) and AMPure XP system (Beckman Coulter, Beverly, USA). PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer (Yeasen). Library quality was assessed on the Agilent 2100 (Agilent). The libraries were sequenced with PE150 mode on Illumina NovaSeq6000 platform (Illumina, San Diego, CA, USA).
Transcript quantification
RNA-Seq data were used to quantify gene/isoform expression. Clean reads of each sample obtained by RNA-Seq were mapped individually to the reference genome (S. scrofa 11.1_release109) using HISAT2 (HISAT2 2.0.4, --dta -p 6 --max-intronlen 5000000). The uniquely mapped reads were assembled using StringTie software (v2.2.1, --merge -F 0.1 -T 0.1) in a reference-based approach. Then the data was used to quantify gene/isoform expression obtained by Iso-Seq. DEGs were identified between adjacent developmental stages according to read count values by using DESeq2 1.30.1 (default: test = “Wald”, fitType = “parametric”). In each comparison groups, genes with Counts Per Million (CPM) ≥ 1 in at least three replicates of one developmental stage were kept, and the selection criteria for DEGs was set as absolute log2 fold-change (FC) ≥ 1 and FDR < 0.05. Meanwhile, the expression level of gene/transcript was measured with FPKM method provided by StringTie. Gene clustering was performed with Mfuzz (http://mfuzz.sysbiolab.eu) according to the expression level. Scatter map was constructed with the OmicShare tools (https://www.omicshare.com/tools) to describe DEGs among multi-groups. Heatmap and venn diagram were plotted with online tools (http://www.bioinformatics.com.cn/). GO and KEGG were performed as described previously51. PPI analysis was performed with string (https://cn.string-db.org/, v12.0) and genes with a score > 0.7 were visualized with cytoscape (v3.9.1).
Alternative splicing analysis
Astalavista (v3.2)52 was used to analyze the basic AS types, including ES, IR, A5SS, A3SS and MEE, of each sample. The AS events were compared between adjacent groups by using rMATS (v4.0.2)53 to identify DAS. Factually, each of the basic AS patterns can be regarded as inclusion/exclusion of exon. The number of reads uniquely mapped to transcript containing the alternative exon is defined as IncLevel. The differences between adjacent samples (△EI = IncLevel2 – IncLevel1) were calculated. The rMATS (v4.0.2) statistical is used to measure the p-value of the differences by likelihood-ratio test. The p-values were then corrected by Benjamini Hochberg. In current analysis, the default threshold for rMATS screening is |Δψ | > c (c = 0.0001), i.e. p-value between mean ψ values of two samples larger than the threshold c.
Alternative polyadenylation analysis
The FLNC reads were further processed with TAPIS pipeline (v1.1.3) (https://bitbucket.org/comp_bio/tapis/overview) to obtain APA sites. At the same time the PAS of each transcript was first identified, and the number of PAS of the gene was calculated. Genes with > 1 PAS by both TAPIS and the location calculation methods were identified as APA genes, and used for further analysis. Among transcripts of APA genes, half of the difference between the farthest and nearest PAS positions was considered to be the middle position, and transcripts with PAS positions farther than the middle position were defined as distal PAS isoform (M) and the others were proximal PAS isoform (P). We calculated expression ratio of the distal PAS isoform (M) to the proximal PAS isoform (m), and genes with M/m value > 1 were identified as Mgenes, opposite genes were Pgenes (M/m value < 1). To analyze the role of genes with alternative major PAS among developmental stages, WGCNA was performed with ImageGP (http://www.ehbio.com/ImageGP/), and the soft-thresholding power of 12 and min module size of 85 was used.
PCRs and statistical analysis
Reverse transcription (RT) was performed with the PrimeScriptTM RT Reagent Kit (TaKaRa) according to the manufacturer’s instructions. Real-time quantitative PCR (qPCR) was carried out with ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) as described previously, and 2─ΔΔCt method was used to analyze the data54. PCR was performed with PrimeSTAR® HS (Premix) (TaKaRa), and the protocol was set as suggested. The PCR products were electrophoresed on 1.2% agarose gel or 8% polyacrylamide gel. The primer information was shown in Supplementary Table 11. The differences between two groups were compared with unpaired t-test by using GraphPad Prism 9.0.
Dual luciferase reporter analysis
PK-15 cells (RRID: CVCL_2160) were cultured as described previously in ref. 55. Genes with APA including cold shock domain containing E1 (CSDE1), hemojuvelin (HJV), translationally-controlled 1 (TPT1), SPARC like 1 (SPARCL1) and nicotinamide nucleotide transhydrogenase (NNT) were selected randomly, and the 3’ UTR was amplified with RT-PCR as described above. The products of CSDE1, HJV, TPT1 and SPARCL1 were inserted into pGL3-promoter (Promega) at XbaI site using ClonExpress® II One Step Cloning Kit (Vazyme). The 3’ UTR of NNT were cloned into psiCHECKTM-2 (Promega) at NheI site using ClonExpress® II One Step Cloning Kit (Vazyme). Each of the reporter genes constructed with pGL3-promoter backbone were contransfected with Renilla luciferase reporter (pRL-TK) (Promega), used as an inner control, into PK-15 cells with Lipofectamine 2000 (Invitrogen). Reporter genes of NNT were transfected singly with the same method. At 48 h post-transfection, the cells were collected and analyzed using Dual Luciferase Reporter Gene Assay Kit (Yeasen). The primer information was shown in Supplementary Table 11. The relative luciferase activities were calculated.
Statistics and reproducibility
Statistical analysis and data visualization in this study were performed by using SPSS (IBM SPSS Statistics 23) and GraphPad Prism 9.0 software. The differences in length, FPKM and exon number between novel and known transcripts were analyzed with Mann-Whitney U test. The correlations were described by Spearman’s Rank Correlation Coefficient between the number of transcripts and the number of exons or gene length. The value of relative luciferase activities was expressed as the mean ± standard deviation (SD) (n = 3 independent samples in each group), and compared with unpaired t-test. All the tests were performed in two-sided, and p or FDR < 0.05 were considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary File
Acknowledgements
This research was funded by the National Natural Science Foundation of China (32172696), Heilongjiang Provincial Natural Science Foundation Program (Joint Guidance, LH2022C095), China Agriculture Research System of MOF and MARA (CARS35).
Author contributions
X.Y. and D.L. funding acquisition and writing-reviewing. X.Y. conceptualization. Y. S. and Y.P. performed data analysis and generated all figures. X.W. and R.Z. completed experiments. L.W., M.T. and X.H. collected the samples. X.Y., Y.S. and Y.P. wrote and edited the manuscript. All authors contributed to the article and approved the submitted version.
Peer review
Peer review information
Communications Biology thanks Foissac Sylvan, Xinnxing Dong and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Kaliya Georgieva [A peer review file is available.].
Data availability
The datasets presented in this study have been deposited to the National Genomics Data Center (NGDC, https://ngdc.cncb.ac.cn/) with the dataset accession number CRA013591. Software and resources used for analysis and visualization are described in each method section. Supplementary Data 1–6 are provided in Supplementary Data. Numerical source data of Figs. 3g and 6g are provided in Supplementary Data named Supplementary Data 7 and 8. Supplementary Tables 1–11 are provided in Supplementary Data named Supplementary Data 9. Supplementary fig. 1 and 2 are provided in the Supplemental information pdf file.
Code availability
In this study all software used is open access, and the parameters were clearly delineated in the Methods section. If the detailed parameters of the software are not specified, the default parameters suggested by the developers were used.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yuanlu Sun, Yu Pang.
Contributor Information
Di Liu, Email: liudi1963@163.com.
Xiuqin Yang, Email: xiuqinyang@neau.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-07332-w.
References
- 1.Ott, M. O., Bober, E., Lyons, G., Arnold, H. & Buckingham, M. Early expression of the myogenic regulatory gene, myf-5, in precursor cells of skeletal muscle in the mouse embryo. Development111, 1097–1107 (1991). [DOI] [PubMed] [Google Scholar]
- 2.Black, B. L. & Olson, E. N. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol.14, 167–196 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Bhagavati, S., Song, X. & Siddiqui, M. A. RNAi inhibition of Pax3/7 expression leads to markedly decreased expression of muscle determination genes. Mol. Cell Biochem.302, 257–262 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Harding, R. L. & Velleman, S. G. MicroRNA regulation of myogenic satellite cell proliferation and differentiation. Mol. Cell Biochem.412, 181–195 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Li, L. et al. MyoD-induced circular RNA CDR1as promotes myogenic differentiation of skeletal muscle satellite cells. Biochim. Biophys. Acta Gene Regul. Mech.1862, 807–821 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Song, C. et al. Linc-smad7 promotes myoblast differentiation and muscle regeneration via sponging miR-125b. Epigenetics13, 591–604 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan, Q., Shai, O., Lee, L. J., Frey, B. J. & Blencowe, B. J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet.40, 1413–1415 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Wahl, M. C., Will, C. L. & Lührmann, R. The spliceosome: design principles of a dynamic RNP machine. Cell136, 701–718 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Hoque, M. et al. Analysis of alternative cleavage and polyadenylation by 3’ region extraction and deep sequencing. Nat. Methods10, 133–139 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merkin, J., Russell, C., Chen, P. & Burge, C. B. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science338, 1593–1599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Consortium, G. T. Ex Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science348, 648–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Söllner, J. F. et al. An RNA-Seq atlas of gene expression in mouse and rat normal tissues. Sci. Data4, 170185 (2017). [DOI] [PMC free article] [PubMed]
- 13.Vicente-García, C., Hernández-Camacho, J. D. & Carvajal, J. J. Regulation of myogenic gene expression. Exp. Cell Res.419, 113299 (2022). [DOI] [PubMed] [Google Scholar]
- 14.Lim, W. F. & Rinaldi, C. RNA Transcript Diversity in Neuromuscular Research. J. Neuromuscul. Dis.10, 473–482 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tojkander, S. et al. A molecular pathway for myosin II recruitment to stress fibers. Curr. Biol.21, 539–550 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Potthoff, M. J. & Olson, E. N. MEF2: a central regulator of diverse developmental programs. Development134, 4131–4140 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Lee, E. et al. Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science297, 1193–1196 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Pineda-Lucena, A. et al. A structure-based model of the c-Myc/Bin1 protein interaction shows alternative splicing of Bin1 and c-Myc phosphorylation are key binding determinants. J. Mol. Bio.351, 182–194 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Fugier, C. et al. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat. Med.17, 720–725 (2011). [DOI] [PubMed] [Google Scholar]
- 20.de Morree, A. et al. Alternative polyadenylation of Pax3 controls muscle stem cell fate and muscle function. Science366, 734–738 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao, J. et al. RBFOX2 is critical for maintaining alternative polyadenylation patterns and mitochondrial health in rat myoblasts. Cell Rep.37, 109910 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beiki, H. et al. Improved annotation of the domestic pig genome through integration of Iso-Seq and RNA-seq data. BMC Genomics20, 344 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hao, W. et al. Characterization of Alternative Splicing Events in Porcine Skeletal Muscles with Different Intramuscular Fat Contents. Biomolecules12, 154 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, Z. et al. Long- and short-read RNA sequencing from five reproductive organs of boar. Sci. Data10, 678 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maciejewska-Skrendo, A., Cięszczyk, P., Chycki, J., Sawczuk, M. & Smółka, W. Genetic Markers Associated with Power Athlete Status. J. Hum. Kinet.68, 17–36 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmetov, I. I., Hall, E. C. R., Semenova, E. A., Pranckevičienė, E. & Ginevičienė, V. Advances in sports genomics. Adv. Clin. Chem.107, 215–263 (2022). [DOI] [PubMed] [Google Scholar]
- 27.Ackermann, M. A. & Kontrogianni-Konstantopoulos, A. Myosin binding protein-C slow: a multifaceted family of proteins with a complex expression profile in fast and slow twitch skeletal muscles. Front Physiol.4, 391 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, Z. et al. Structures from intact myofibrils reveal mechanism of thin filament regulation through nebulin. Science375, eabn1934 (2022). [DOI] [PubMed] [Google Scholar]
- 29.Gauthier, B. R. et al. Thyroid hormones in diabetes, cancer, and aging. Aging Cell19, e13260 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang, F., Flavahan, S. & Flavahan, N. A. Impaired activity of adherens junctions contributes to endothelial dilator dysfunction in ageing rat arteries. J. Physiol.595, 5143–5158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martínez, G., Duran-Aniotz, C., Cabral-Miranda, F., Vivar, J. P. & Hetz, C. Endoplasmic reticulum proteostasis impairment in aging. Aging Cell16, 615–623 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu, W. et al. Dynamic landscape of alternative polyadenylation during retinal development. Cell Mol. Life Sci.74, 1721–1739 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu, C. & Zhang, J. Alternative polyadenylation of mammalian transcripts is generally deleterious, not adaptive. Cell Syst.7, 734–742 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, P. et al. Transcriptomic analysis of testis and epididymis tissues from Banna mini-pig inbred line boars with single-molecule long-read sequencing. Biol. Reprod.108, 465–478 (2023). [DOI] [PubMed] [Google Scholar]
- 35.Heintz, C. et al. Splicing factor 1 modulates dietary restriction and TORC1 pathway longevity in C. elegans. Nature41, 102–106 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tabrez, S. S., Sharma, R. D., Jain, V., Siddiqui, A. A. & Mukhopadhyay, A. Differential alternative splicing coupled to nonsense-mediated decay of mRNA ensures dietary restriction-induced longevity. Nat. Commun.8, 306 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adusumalli, S., Ngian, Z. K., Lin, W. Q., Benoukraf, T. & Ong, C. T. Increased intron retention is a post-transcriptional signature associated with progressive aging and Alzheimer’s disease. Aging Cell18, e12928 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yap, K., Lim, Z. Q., Khandelia, P., Friedman, B. & Makeyev, E. V. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev.26, 1209–1223 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong, J. J. et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell154, 583–595 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Ni, T. et al. Global intron retention mediated gene regulation during CD4+ T cell activation. Nucleic Acids Res.44, 6817–6829 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Braunschweig, U. et al. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res.24, 1774–8176 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eksi, R. et al. Systematically differentiating functions for alternatively spliced isoforms through integrating RNA-seq data. PLoS Comput. Biol.9, e1003314 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang, X. et al. Widespread Expansion of Protein Interaction Capabilities by Alternative Splicing. Cell164, 805–817 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voineagu, I. et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature474, 380–384 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stamova, B. S. et al. Evidence for differential alternative splicing in blood of young boys with autism spectrum disorders. Mol. Autism4, 30 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Solovyeva, E. M. et al. New insights into molecular changes in skeletal muscle aging and disease: Differential alternative splicing and senescence. Mech. Ageing Dev.197, 111510 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Tian, B., Hu, J., Zhang, H. & Lutz, C. S. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res.33, 201–212 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, H., Hu, J., Recce, M. & Tian, B. PolyA_DB: a database for mammalian mRNA polyadenylation. Nucleic Acids Res.33, D116–D120 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mayr, C. Regulation by 3’-Untranslated Regions. Annu. Rev. Genet51, 171–194 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Tian, B. & Manley, J. L. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol.18, 18–30 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun, Y. et al. Genome-wide characterization of lncRNAs and mRNAs in muscles with differential intramuscular fat contents. Front Vet. Sci.9, 982258 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Foissac, S. & Sammeth, M. ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res.35, W297–W299 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen, S. et al. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA111, E5593–E5601 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Yang, X. et al. Transcriptional Regulation Associated with Subcutaneous Adipogenesis in Porcine ACSL1 Gene. Biomolecules13, 1057 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary File
Data Availability Statement
The datasets presented in this study have been deposited to the National Genomics Data Center (NGDC, https://ngdc.cncb.ac.cn/) with the dataset accession number CRA013591. Software and resources used for analysis and visualization are described in each method section. Supplementary Data 1–6 are provided in Supplementary Data. Numerical source data of Figs. 3g and 6g are provided in Supplementary Data named Supplementary Data 7 and 8. Supplementary Tables 1–11 are provided in Supplementary Data named Supplementary Data 9. Supplementary fig. 1 and 2 are provided in the Supplemental information pdf file.
In this study all software used is open access, and the parameters were clearly delineated in the Methods section. If the detailed parameters of the software are not specified, the default parameters suggested by the developers were used.