Abstract
Fibroblast heterogeneity is increasingly recognised across cancer conditions. Given their important contribution to disease progression, mapping fibroblasts’ heterogeneity is critical to devise effective anti-cancer therapies. Cancer-associated fibroblasts (CAFs) represent the most abundant cell population in pancreatic ductal adenocarcinoma (PDAC). Whether CAF phenotypes are differently specified by PDAC cell lineages remains to be elucidated. Here, we reveal an important role for the MAPK signalling pathway in defining PDAC CAF phenotypes. We show that epithelial MAPK activity promotes the myofibroblastic differentiation of CAFs by sustaining the expression and secretion of TGF-β1. We integrate single-cell profiling of post-perturbation transcriptional responses from mouse models with cellular and spatial profiles of human tissues to define a MAPKhigh CAF (mapCAF) phenotype. We show that this phenotype associates with basal-like tumour cells and reduced frequency of CD8+ T cells. In addition to elevated MAPK activity, this mapCAF phenotype is characterized by TGF-β signalling, hypoxia responsive signatures, and immunoregulatory gene programs. Furthermore, the mapCAF signature is enriched in myofibroblastic CAFs from various cancer conditions and correlates with reduced response to immune checkpoint inhibition in melanoma. Altogether, our data expand our knowledge on CAF phenotype heterogeneity and reveal a potential strategy for targeting myofibroblastic CAFs in vivo.
Subject terms: Pancreatic cancer, Cancer microenvironment
Phenotypic and functional heterogeneity of cancer associated fibroblasts (CAFs) has been reported in pancreatic ductal adenocarcinoma (PDAC). Here the authors show that epithelial MAPK activity promotes myofibroblastic differentiation of CAFs. Furthermore, the epithelial basal-like subtype is associated with a CAF phenotype characterized by elevated MAPK activity and TGF-β signalling, associated with T cell exclusion in PDAC.
Introduction
Fibroblasts functionally contribute to disease progression and therapy response in solid tumours1,2. The diversity of phenotypes and functions displayed by cancer-associated fibroblasts (CAFs) within the tumour microenvironment (TME) is dependent on their cellular origin, spatial localisation and disease context2,3. In pancreatic ductal adenocarcinoma (PDAC), different subtypes and specialised functions of CAFs have been reported4–10. Myofibroblastic CAFs (myCAFs) and inflammatory CAFs (iCAFs) are two distinct states consistently reported in the PDAC TME4,6,8 for which mechanisms of cancer-induced rewiring have been provided4. The failure of agnostic targeting of PDAC CAFs in preclinical and clinical settings11–14 demonstrates our current insufficient understanding of fibroblast heterogeneity to properly guide therapeutic strategies. Here, we hypothesised that different neoplastic cell states contribute to PDAC fibroblast heterogeneity. Different transcriptional states of PDAC malignant cells have been described15–19, which result from the integration of cell intrinsic and extrinsic inputs20–22. Transcriptional infidelity to the pancreatic endoderm and acquisition of exogenous gene programmes (e.g., squamous/basal-like) is invariably associated with a more aggressive biological behaviour of cancer cells23. However, we have a limited understanding of whether specific CAF rewiring is induced by different neoplastic cell states. We argued that the identification of a CAF transcriptional phenotype specifically associated with a biologically aggressive cancer subtype may improve patients’ stratification based on the risk, lead to a better prediction of therapeutic responses and potentially guide effective personalised therapies. Activating mutations of KRAS are nearly universal in PDAC24 and result in the hyperactivation of the RAS/MEK/ERK (Mitogen-activated protein kinase, MAPK) pathway, which plays a central role in tumour initiation and maintenance25,26. Increased dosage of oncogenic KRAS is observed in basal-like/squamous subtypes17,27,28, yet classical PDAC cells have been reported to display superior sensitivity to the inhibition of MAPK29. Targeting of MAPK, alone or in combination, has demonstrated to be ineffective in PDAC30,31. However, the significance of MAPK activity in the PDAC TME has never been explored so far.
In this work, we uncover an important role for the MAPK signalling pathway in the definition of PDAC CAF phenotypes. We show that epithelial MAPK activity promotes the myofibroblastic differentiation of CAFs by sustaining the expression and secretion of TGF-β1 by malignant cells. Additionally, we demonstrate that hyperactivation of MAPK signalling occurs in myCAFs (MAPKhigh CAFs, mapCAFs) populating basal-like tumour niches with reduced frequency of CD8+ T cells. In addition to elevated MAPK activity, this mapCAF phenotype is characterised by TGF-β signalling, hypoxia responsive signatures and immunoregulatory gene programmes. In silico analyses confirm this phenotype in other solid tumours and further show that mapCAFs are indicative of primary resistance to immunotherapy in metastatic melanoma.
Results
Basal-like PDAC cells bear cancer-associated fibroblasts with elevated MAPK activity
To identify molecular features predictive of cellular dependency on MAPK signalling, we treated 12 human cell lines with increasing doses of the MEK1/2 inhibitor trametinib (MEKi). Next, we evaluated post-perturbation biochemical and transcriptomic responses with sub-IC50 doses of MEKi (Supplementary Fig. 1a–c). The cell lines approximated the two main PDAC subtypes (i.e., classical and basal-like) to varying extents (Supplementary Fig. 1b, c). Overall, the treatment did not affect cell lineage (Supplementary Fig. 1b, c). The reduction in cell fitness following short-term MEKi was not predicted by differential p-ERK1/2 phosphorylation, a proxy for pathway inhibition, nor by the amplitude of drug-induced transcriptomic changes (Supplementary Fig. 1d–f). Then, we used the RNA-seq data from the 12 MEKi treated cell lines to derive a transcriptomic signature of MEK1/2 inhibition (epithelial MEKi, eMEKi) based on downregulated genes only (Supplementary Data 1). To isolate the drug-induced transcriptional response, we employed batch correction to remove the major source of intrinsic variability in the RNA-seq data, i.e., cell lines’ identities (Supplementary Fig. 1g). The eMEKi signature levels did not correlate with drug sensitivity, biochemical pathway activation (i.e., p-ERK1/2 levels), or the ‘basalness’ score (see ‘Methods’) (Supplementary Fig. 1h, i). Conversely, eMEKi showed positive correlation, although not reaching statistical significance, with another transcriptional signature of MAPK activation (MAPK_Biocarta) (Supplementary Fig. 1j). The latter showed a better correlation with pathway activation, as assessed by levels of p-ERK1/2 in unperturbed condition (Supplementary Fig. 1h, k). To confirm this result, we used an established cell line (PaTu 8988S) where the basal-like programme is induced by RNAi silencing of the classical driver GATA632. In keeping with our observations, downregulation of GATA6 did not result in significant changes in MAPK fluxes nor in sensitivity to MEKi (Supplementary Fig. 1l–n).
Therefore, in our array of cell culture, MAPK activity does not discriminate basal-like vs classical PDAC cells nor identify cell lines with differential sensitivity to pathway inhibition.
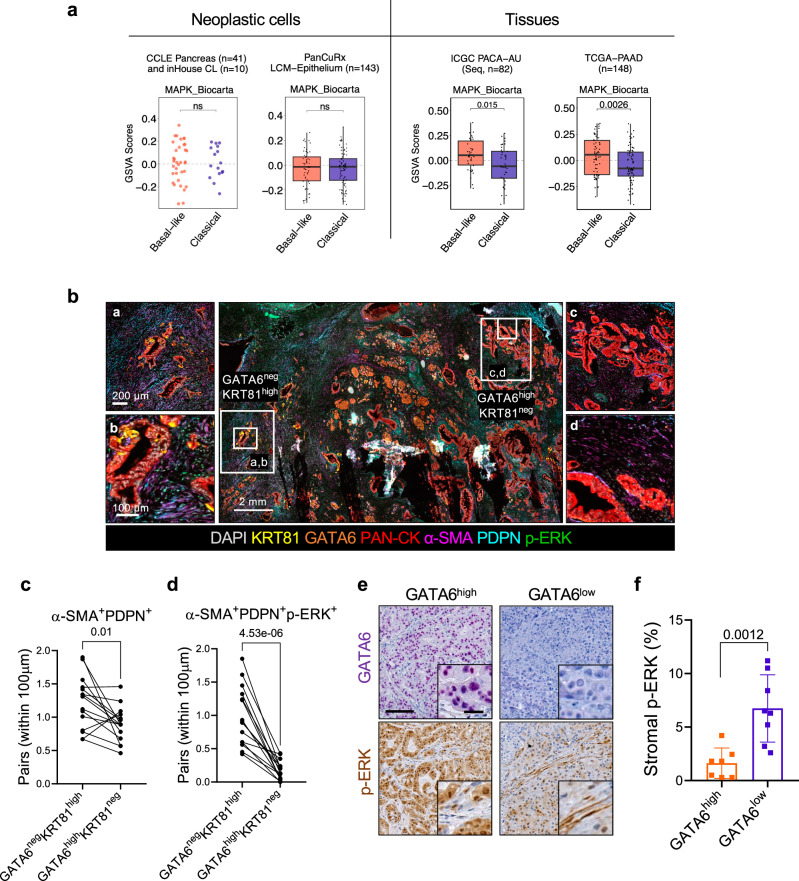
Next, we used the same transcriptional signature of MAPK activation (MAPK_Biocarta) to infer differential activity of the pathway in an extended set of human cell lines. We explored the transcriptomic data available from the Cancer Cell Line Encyclopaedia (CCLE) (n = 41)33 and those available in our laboratory (n = 10) to classify cell lines as either basal-like or classical19 (see ‘Methods’, Supplementary Data 2). In this dataset, the MAPK transcriptional signature could not discriminate classical from basal-like cell lines (Fig. 1a). Considering that in vitro models do not fully represent in vivo cell states, we next explored transcriptomic data from PDAC tissues. High levels of the MAPK signature were enriched in the basal-like PDAC of The Cancer Genome Atlas (TCGA)16 and the International Cancer Genome Consortium (ICGC)15 cohorts, but not in the PanCuRx17 cohort, which contains RNA-seq data from microdissected epithelia (Fig. 1a). Based on those results, we reasoned of a potential contribution of non-malignant cells to the elevated MAPK activation inferred in the basal-like tissues of the two cohorts. We focused on CAFs and tumour-associated macrophages as they represent the most abundant cell types in the PDAC TME. Independently of the neoplastic cell content of the tissues, MAPK transcriptional signatures were generally positively correlated with the levels of stromal genes such as Podoplanin (PDPN), Actin alpha 2 (ACTA2) and Fibroblast activation protein (FAP) (Supplementary Fig. 2a). Next, we applied a multiplex immunofluorescence (mIF) to 15 human PDAC tissues where neoplastic cells were classified as either basal-like or classical with well-established tissue markers15,34–37 (Fig. 1b). First, we found that every tumour tissue showed co-existence of classical (GATA6high KRT81neg) and basal-like (GATA6neg KRT81high) cells (Fig. 1b and Supplementary Fig. 2b). An increased abundance of (myCAFs, PDPN+α-SMA+) was observed in proximity of basal-like cells (Fig. 1b, c). In classical tumour regions38,39, p-ERK+ CAFs were rarely detected while the opposite was found in KRT81high expressing tumour regions (Fig. 1b). Spatial analysis showed that significantly more PDPN+ α-SMA+p-ERK+ CAFs can be found in proximity of basal-like cells (Fig. 1b, d). Stromal p-ERK1/2 signal was also significantly higher in GATA6low tissues from patient-derived xenografts (PDXs, n = 15) (Fig. 1e, f).
Fig. 1. Basal-like PDAC cells bear cancer-associated fibroblasts with elevated MAPK activity.
a Scatter dot plot and boxplots of GSVA scores obtained for the MAPK Biocarta gene set (MSigDB) stratified by Moffitt subtypes19. From left to right: CCLE 33and in house cell lines, PanCuRx17, ICGC15 and TCGA16. p values by Wilcoxon test (two-sided). TCGA Classical: Min −0.42, Max 0.35, a median of −0.08, with first quartile (Q1) at −0.15, third quartile (Q3) at 0.08, an interquartile range (IQR) of 0.23, lower whisker (LW) −0.42, upper whisker (UW) 0.35. TCGA Basal-like: Min −0.35, Max 0.35, Med 0.05, Q1 −0.14, Q3 0.19, IQR 0.33, LW −0.35, UW 0.35. ICGC Classical: Min −0.44, Max 0.27, Med −0.06, Q1 −0.18, Q3 0.09, IQR 0.27, LW −0.44, UW 0.27. ICGC Basal-like: Min −0.28, Max 0.38, Med 0.05, Q1 −0.05, Q3 0.19, IQR 0.24, LW −0.28, UW 0.38. PanCuRx Classical: Min −0.31, Max 0.31, Med −0.01, Q1 −0.12, Q3 0.05, IQR 0.17, LW −0.31, UW 0.30. PanCuRx Basal-like: Min −0.30, Max 0.26, Med −0.01, Q1 −0.12, Q3 0.07, IQR 0.19, LW −0.30, UW 0.26. b Representative images of multiplex IF performed on FFPE of human PDAC tissues. The panels represent different areas within the same tumour displaying different molecular phenotypes. GATA6negKRT81high, basal-like; GATA6highKRT81neg, classical. Scale bars as indicated. See also Supplementary Fig. 2b. c Paired dot plot showing quantification of α−SMA+PDPN+CAFs with a distance below 100 µm to classical and basal-like cells (n = 15 tissues). p values as determined by paired t-test (two-sided). d Paired dot plot showing the quantification of α−SMA+PDPN+p-ERK+ CAFs with a distance below 100 µm to classical and basal-like cells (n = 15 tissues). p values as determined by paired t-test (two-sided). e Representative images of immunohistochemistry for GATA6 and p-ERK on tissues from two patient derived xenografts. Scale bar, 200 μm. Inserts showed a magnification of selected areas (Scalebar, 25 μm). f Quantification of (e) showing the percentage of stromal p-ERK signal. p values as determined by Mann-Whitney test (two-sided). Results presented as mean values ± SD. n = 7 biological replicates GATA6high; n = 8 biological replicates GATA6low.
Pancreatic stellate cells (PSCs) are known precursors of CAFs40–42. We serum-starved mouse PSCs (mPSCs) and then briefly exposed them to the conditioned media from 10 human cell lines displaying varying degree of basalness before evaluating p-ERK. Although no clear trend was observed for increased nuclear p-ERK levels when mPSCs were exposed to conditioned media from more basal-like cultures, statistically significant differences were detected when mPSCs were treated with conditioned media from isogenic cell lines either proficient or deficient in GATA6 expression (Supplementary Fig. 2c–f). This result suggests that while other mechanisms might be at play, the phenotype of nuclear p-ERK accumulation in fibroblasts can be partially recreated through direct modulation of the neoplastic cell state. Collectively, these data support a mechanistic link of basal-like PDAC cancer cells driving elevated activity of MAPK in CAFs in vivo.
scRNA-seq of mouse PDAC tumours treated with MEKi reveals quantitative and qualitative changes in cell subsets
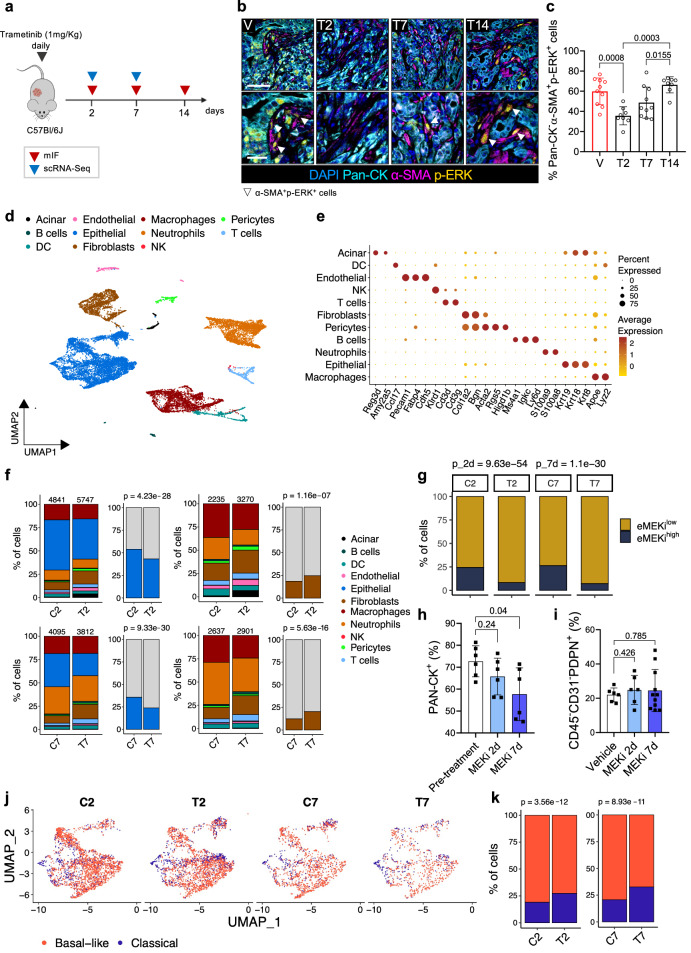
To investigate the role of MAPK in the definition of stromal phenotypes, we performed a multidimensional analysis of tissues from mouse basal-like PDAC (Fig. 2a). First, we generated and characterised a mouse model based on the orthotopic transplantation of a quasi-mesenchymal KPC (KrasLSL-G12D/wt; Tp53LSL-R172H/wt; Pdx1-Cre)43 derived cell line that in vivo produces cancer tissues aligning with the human basal-like PDAC phenotype44. As reported previously, this transplantation-based model shows accelerated tumour growth kinetics44–46. Tumour-bearing mice treated daily with 1 mg/kg of MEKi showed reduced activation of MAPK signalling (Supplementary Fig. 3a, b). Multiplex IF of tissues from tumour-bearing mice treated over the course of 14 days showed the presence of p-ERK+ CAFs in untreated tumours, the reduction of p-ERK signal in both the epithelial and stromal compartments at 2 days following treatment, and the rapid pathway rewiring particularly in the stromal compartment starting at 7 days of treatment (Fig. 2b, c and Supplementary Fig. 3c). In keeping with that, the reactivation of the MAPK pathway occurred in vitro within hours from the treatment of mPSCs (Supplementary Fig. 3d).
Fig. 2. scRNA-seq of mouse PDAC tumours treated with MEKi reveals quantitative and qualitative changes in cell subsets.
a Schematic representation of the in vivo experimental setting. Mouse and cell cartoon schematic from Malinova et al.23 b Images of multiplex immunofluorescence of tumour tissues from mice treated with: vehicle (C), MEKi for 2 (T2), 7 (T7) and 14 (T14) days. Scalebars, 50 µm (main), 25 μm (insets). c Quantification of b showing the percentage of Pan-CK–α-SMA+p-ERK+ cells. p values by Mann-Whitney test (two-sided). n ≥ 8 mice/condition. Results as mean values ± SD. d UMAP plot displaying unsupervised clustering of viable cells from 12 mouse tumours samples annotated in 11 different clusters, colour-coded by cell type. e Bubble plot showing selected cell type-specific markers across clusters. Dot size indicates cell percentage; colour intensity shows average expression. f Barplots show cell type percentages in samples from vehicle (C) or MEKi (T) treated mice (2 or 7 days). Numbers indicate total cells per sample. Malignant cells (blue) are shown relative to non-epithelial populations; fibroblasts (brown) are shown relative to other TME cells (grey). p values by χ² test (two-sided). g Barplot showing percentage of cells of the epithelial cluster stratified based on the expression level of the eMEKi signature in vehicle (C2, C7) or MEKi (T2, T7) treated samples for 2 or 7 days. p values by χ² test (two-sided). h Barplot showing the quantification of pan-Cytokeratin+ cells in tissues MEKi-treated tumour-bearing mice. p values by Mann-Whitney test (two-sided). n = 6 mice. Results as mean values ± SD. i FACS analysis of CAFs (PDPN+ cells) in tumour-bearing mice treated with MEKi for 2 or 7 days, shown as a percentage of total viable cells. p values by Mann-Whitney test (two-sided). n ≥ 6 mice/condition. Results as mean values ± SD. j UMAP plots of epithelial cluster cells from vehicle (C2–C7) and MEKi (T2–T7) treated mice, classified as classical or basal-like per Moffitt’s classification19. k Barplots showing percentage of epithelial cells from mice treated with vehicle or MEKi (2 or 7 days), classified by Moffitt’s subtypes19. p values by χ² test (two-sided).
Given the kinetics of the MAPK pathway rewiring, we then performed scRNA-seq on fresh tissues from tumour-bearing mice treated for 2 and 7 days with MEKi along with their untreated controls. We profiled a total of 18,495 cells across 12 tumours (6 untreated and 6 treated, see ‘Methods’) and recovered an overall cellular composition similar to that expected from PDAC tissues (Fig. 2d, e). Unsupervised clustering of single cell data identified 11 major cell types (Fig. 2d). Further annotation of the identified cell subsets was performed post hoc by using known gene signatures (Fig. 2e). The main cluster was almost exclusively composed by malignant epithelial cells which were confirmed by inferred copy-number alterations (CNA)47 (Supplementary Fig. 3e, f). Other cell types included non-malignant epithelial cells (i.e., acinar cells), immune and stromal cell types (Fig. 2d, e). The treatment was associated with significant changes in cell composition and cell states. We observed a significant reduction in the proportion of malignant epithelial cells both at 2 and 7 days of treatment (Fig. 2f). Consistent with the pharmacological treatment and proteomic analysis, the frequency of cells expressing high levels of the eMEKi transcriptional signature was significantly reduced in the epithelial compartment of treated tumours (Fig. 2g). In the non-malignant compartment, we observed a significantly higher fraction of CAFs in the treated groups (Fig. 2f). To validate our findings, we performed cytofluorimetric and mIF analyses in additional transplantation-based models of mouse PDAC subjected to MEKi. Tissues were obtained from syngrafts orthotopically transplanted with cell lines derived from KrasLSL-G12D/wt; Tp53fl/fl; Ptf1awt/Cre background mice48. Consistent with our in-silico findings, the fraction of epithelial cells (PanCK+, mIF; CD45–CD31–PDPN–, FACS) in the tissues decreased with the treatment (Fig. 2h and Supplementary Fig. 3g). However, flow cytometric analysis of the same tissues revealed no changes in the frequency of PDPN+ cells after treatment (Fig. 2i and Supplementary Fig. 3h), suggesting a context (i.e., cell line) dependent effect of MEKi on quantitative changes in the stromal compartment. MEKi was also associated with changes in the proportion of basal-like and classical neoplastic cells (Fig. 2j, k). However, molecular subtyping on epithelial pseudo-bulk showed no significant changes in cell state, which is in line with the results obtained on treated human PDAC cell lines (Supplementary Fig. 1b).
MAPK inhibition induces changes in the proportion of myCAFs and iCAFs
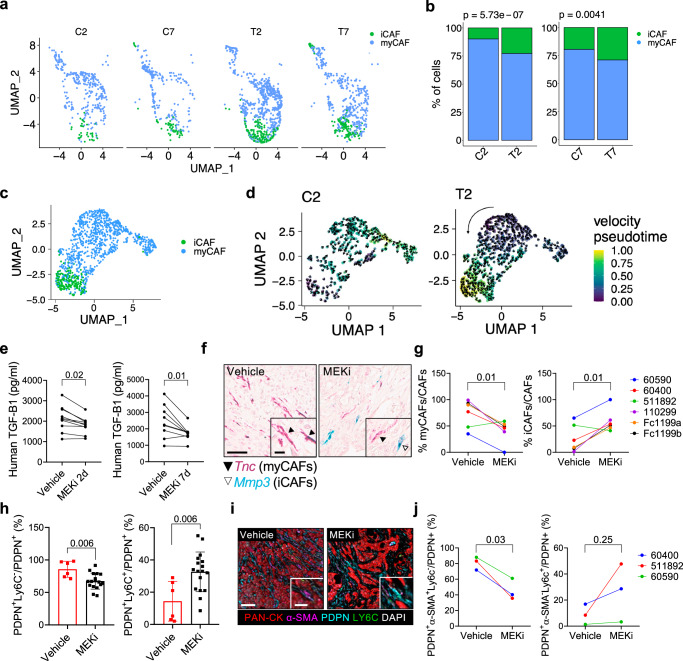
Analysis of fibroblast clusters within our scRNA-seq dataset identified 10 CAF subpopulations (Supplementary Fig. 4a, b), which were represented in both untreated and treated mice. However, only cells from untreated mice were considered for further characterisation of these subpopulations (Supplementary Fig. 4c, d and Supplementary Data 3). Cluster 6 was identified as ‘cycling’ fibroblasts based on expression of mitotic cell signatures (Supplementary Fig 4c, d and Supplementary Data 3). Clusters 7 and 8 were identified as ‘contaminant’ since they expressed markers of epithelial cells undergoing EMT and of cells of the innate immunity, respectively (Supplementary Fig 4c, d and Supplementary Data 3). Clusters 0 and 1 were identified as distinct groups with overlapping signatures of iCAFs, as the established mouse iCAF signature6 was enriched in these two clusters (Supplementary Fig. 4c, d and Supplementary Fig. 4e, f). Clusters 2–5 were identified as primarily composed by myCAFs6 (Supplementary Fig 4c–f), while Cluster 9 was characterised by the expression of apCAF markers6 (e.g., Cd74) and composed by rare cells (n = 50) (Supplementary Fig 4c–f). Considering their frequency across stages (C2, C7; Fig. 3a, b) and conditions (C vs T; Fig. 3a, b), we focused on the most abundant subpopulations iCAFs and myCAFs. In our model, progression was associated with an increase frequency of iCAFs, which we linked to the increased expression of Il1a and Il1b in malignant cells (Supplementary Fig. 4g).
Fig. 3. MAPK inhibition alters the myCAFs/iCAFs ratio in mouse PDAC.
a UMAP plot of fibroblast cluster from mice treated with vehicle (C2, C7) or MEKi (T2, T7) for 2 or 7 days, classified as myCAFs or iCAFs according to Elyada’s classification6. b Barplots show myCAFs and iCAFs percentages. p values determined by χ² test (two-sided). c UMAP plot of the fibroblast cluster obtained by the integration of vehicle- or MEKi-treated mice for 2 days, colour-coded by Elyada’s subtypes6. d UMAP showing velocity (arrows) and pseudotime (colour) for each cell of the fibroblast cluster (annotated in Fig. 3c) from mice treated with either vehicle (C2) or MEKi (T2) for 2 days. Black arrow indicates overall velocity direction. e Paired dot plot of human TGF-β1 levels in conditioned media from human cancer cell lines treated with MEKi for 2 (n = 10 cell lines) or 7 (n = 9 cell lines) days. p values by paired t-test (two-sided). f Representative in situ hybridisation images showing Tnc (red; myCAFs) and Mmp3 (green; iCAFs) expression in PDAC tissues from vehicle- or MEKi-treated mice. Scale bar, 60 μm (main), 15 μm (insets). See also Supplementary Fig. 4j, k. g Paired dot plot displaying the percentage of iCAFs and myCAFs in tissues from 6 tumour-bearing mice shown in (f). p values determined by paired t-test (two-sided). n = 6 biological replicates. h Flow cytometry of myCAFs (PDPN+LY6C−) and iCAFs (PDPN+LY6C+) as percentage of total CAFs (PDPN+ cells). Each value refers to an individual tumour-bearing mouse. Results presented as mean values ± SD. p values determined by Mann-Whitney test (two-sided). See also Supplementary Fig. 4m. n = 6 mice Vehicle; n = 17 mice MEKi. i Representative images of multiplex IF of tissues from vehicle- or MEKi-treated mice. Scale bar, 100 μm (main), 50 μm (insets). j Paired dot plot shows myCAFs (PDPN+α−SMA+LY6C− cells) and iCAFs (PDPN+α−SMA−LY6C+ cells) percentages relative to all CAFs. Each dot represents the average value derived from multiple mice, when available. p values by paired t-test (two-sided). n = 3 biological replicates.
As observed for the malignant compartment, the treatment induced quantitative and qualitative changes in CAFs. We observed a significant reduction in the proportion of myCAFs after treatment, both at 2 and 7 days of MEKi, which was associated with an increase in the number of iCAFs (Fig. 3a, b) in the fibroblast compartment. Next, we used the velociraptor toolkit49 to infer pseudotemporal trajectory from scRNA-seq data50. In untreated tumours, no dominant pseudotrajectory could be identified (Fig. 3c, d). Conversely, a dominant pseudotrajectory from myCAFs to iCAFs was inferred in treated tumours (Fig. 3d). Furthermore, malignant cells from MEKi treated tumours expressed lower levels of Tgfb1 both at T2 and T7, while expressing higher levels of Il1b only at 7 days of treatment (Supplementary Fig. 4h). Consistently, MEKi significantly reduced the secretion of TGF-β1 in a panel of human cancer cell lines (Fig. 3e). Conversely, direct MAPK inhibition of plastic or TGF-β1 activated mPSCs did not significantly alter the expression of myCAF or iCAF markers (Supplementary Fig. 4i).
To orthogonally validate these findings, we first explored scRNA-seq from untreated tumours to identify reliable markers of myCAF and iCAF subsets. We found that Tnc and Mmp3 were highly expressed in cell subsets showing myCAF and iCAF phenotypes, respectively (Supplementary Fig. 4j). In situ RNA hybridisation analysis (ISH) of mouse PanIN from autochthonous KC (KrasLSL-G12D/wt; Pdx1-Cre) models driven by oncogenic KrasG12D 51 revealed expression of Tnc in proximity of epithelial cells while abundant stromal cells expressing Mmp3 were found surrounding each lesion (Supplementary Fig. 4k). In cancer tissues from autochthonous KPC models driven by mutations of Kras and Trp5343, both Tnc and Mmp3 expressing cells showed a spatial segregation consistent with that observed for myCAF and iCAF phenotypes in this model4,6,8 (Supplementary Fig. 4k). Next, we performed RNA ISH on tissues from our mouse cohort, including KPC-transplantation based models, which are representative of either classical (60400, 110299) or basal-like (FC1199a/b, 60590, 511892) human PDAC (Supplementary Fig. 4l). Consistent with the scRNA-seq data, the treatment induced statistically significant changes in the proportion of myCAFs and iCAFs (Fig. 3f, g). Specifically, after short-term MEKi, there was a drastic reduction in the number of Tnc expressing cells with a concomitant increase in Mmp3 expressing cells (Fig. 3f, g). These changes occurred independently of the epithelial cell lineage and were confirmed by flow cytometric analysis (myCAFs, PDPN+LY6C−; iCAFs, PDPN+LY6C+; Fig. 3h and Supplementary Fig. 4m) and multiplex IF (myCAFs, PDPN+α-SMA+LY6C−; iCAFs, PDPN+α-SMA−LY6C+; Fig. 3i, j).
Altogether, our data indicates that epithelial MAPK activity promotes the myCAF phenotype in vivo.
MAPK inhibition alters ligand-receptor interactions between malignant and non-malignant cells in mouse PDAC
To understand how MAPK inhibition affects cell-to-cell communication in mouse PDAC, we used CellChat52, a computational tool that infers interactions between different cell types based on the identification of ligand-receptor (LR) pairs from scRNA-seq data52. Our analysis focused on the most prevalent cell types in both treated and untreated tumours, i.e., malignant cells, fibroblasts, macrophages and neutrophils (Fig. 2f). We did not analyse LR pairs at the level of malignant and stromal subtypes as the low prevalence of certain subtypes and the similar LR expression among them (Supplementary Fig. 5a) would have compromised statistical power. When comparing treated and untreated tumours, a systematic investigation of catalogued LR pairs between any two cell types revealed that the largest variation in the number of interactions occurred between fibroblasts and other cell types (Supplementary Fig. 5b, c). Moreover, the largest variation in interaction strength was observed for CAFs signalling to malignant cells (Supplementary Fig. 5d). Focusing on malignant cell-fibroblast communication (Supplementary Data 4), we found that enriched LR pairs were associated with cell response to extracellular matrix components, chemotaxis and the response to angiogenic factors (Supplementary Fig. 5e). The treatment-depleted LR pairs were associated with the EGFR pathway in both malignant cells and fibroblasts, integrin-mediated responses in fibroblasts and axon-guidance pathway in both malignant cells and fibroblasts (Supplementary Fig. 5e, f). These results suggest that the interactions between CAFs and malignant cells mostly rely on ECM-integrin signalling which is disrupted upon MEKi.
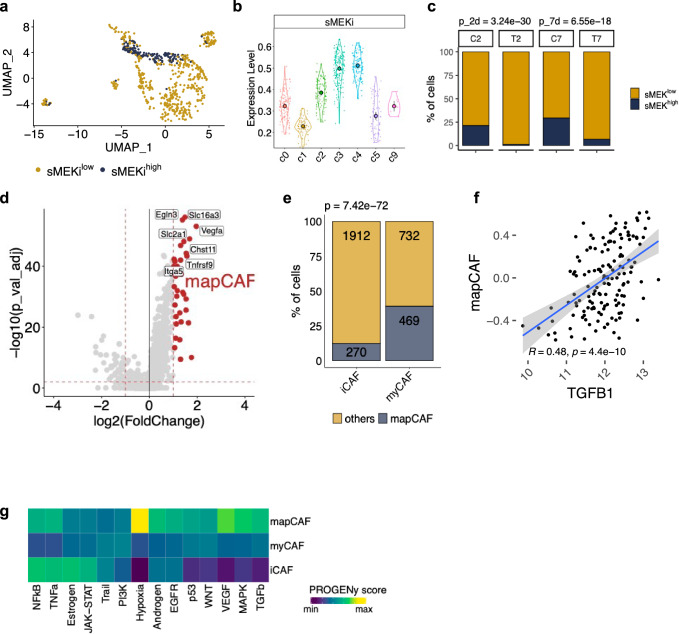
A MAPK CAF signature identifies a subcluster of myCAFs in mouse PDAC
To identify subsets of CAFs with elevated activity of MAPK in our scRNA-seq data, we derived a transcriptomic signature based on genes significantly downregulated by 2 days of MEKi in the CAF compartment (stromal MEK inhibition, sMEKi) (Supplementary Fig. 5g, and Supplementary Data 5). This timepoint corresponded to the abrogation of p-ERK1/2 signal in the stromal compartment (Fig. 2b, c). The expression of high levels of the sMEKi signature (n = 169 genes, see ‘Methods’) was enriched in myCAFs of the c3-c4 clusters (Supplementary Fig. 4c, d and Fig. 4a, b). Consistent with the dynamics of MAPK inhibition in our model (Fig. 2b, c), the proportion of CAFs expressing high level of the sMEKi signature decreased after 2 days of treatment and then increased at 7 days (Fig. 4c). Based on these results, we identified a MAPKhigh CAF (mapCAF, n = 38 genes, see ‘Methods’) transcriptional signature (Fig. 4d) to infer the presence of this phenotype in the scRNA-seq data from cancer tissues of autochthonous KPC mice6. In this dataset, we consistently found that mapCAFs were mostly myCAFs (Fig. 4e). In accordance with this, the mapCAF signature positively correlated with TGFB1 in the TCGA dataset16(Fig. 4f) and could be induced in vitro by treating mPSCs with TGF-β1 (Supplementary Fig. 5h).
Fig. 4. A MAPKhigh signature identifies a subcluster of myCAFs in mouse PDAC.
a UMAP plot of cells from the fibroblast cluster stratified according to sMEKi signature from mice treated with vehicle. b Violin plots showing the enrichment of cells (n = 742 cells) expressing high level of the sMEKi signature in myofibroblastic clusters. Data are presented as mean values and 95% confidence interval (CI). See Supplementary Fig. 4c, d. c Barplot representing the percentage of cells of the fibroblast cluster displaying either high or low levels of the sMEKi signature in vehicles (C) and in MEKi (T) treated samples. p values determined by χ2 (two-sided). d Volcano plot representing the differentially expressed genes from the comparison between fibroblasts displaying high vs low MAPK transcriptional activity (based on sMEKi levels). The red dots are some of the genes defining the mapCAF signature (n = 38). e Barplot displaying the frequency of cells expressing the mapCAF signature in the myCAF and iCAF clusters from Elyada et al.6 p values determined by χ² test (two-sided). f Scatter plot showing the positive correlation between TGFB1 expression and the mapCAF GSVA score for samples of the TCGA (n = 148 sample) cohort16. P value from r correlation test. Grey area represents 95% CI. g Heatmap showing expression of pathway-responsive genes in specific CAF phenotypes as assessed by PROGENy analysis101.
Then, we used PROGENy53 to infer pathway activity in each CAF subset from untreated tumours. In line with previous reports4–6,8,54, the activation of TGF-β was higher in myCAFs, while iCAFs were enriched for inflammation-associated signalling pathways (Fig. 4g). mapCAFs were characterised by elevated TGF-β and MAPK activity, the enrichment of hypoxia-driven programmes and activity of inflammatory pathways (Fig. 4g). Gene-set enrichment analysis of differentially expressed genes between CAFs with high and low MAPK activity confirmed metabolic reprogramming, as well as activation of inflammation-associated transcriptional programmes in mapCAFs (Supplementary Fig. 5i). We next applied decoupleR55,56 to infer transcription factor activity from scRNA-seq data and found that hypoxia- (e.g., Hif1a, Epas1), MAPK- (e.g., Etv4, Fosb) and TGF-β-related (e.g., Runx2) transcription factors were the main drivers of the mapCAF phenotype (Supplementary Fig. 5j).
The mapCAF phenotype is associated to basal-like human tumours
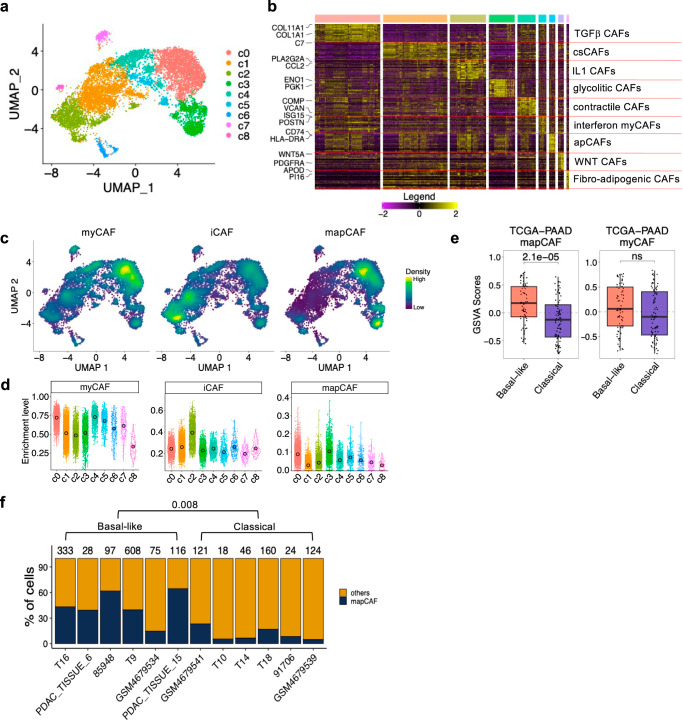
To translate our findings from mouse models to human PDAC tumours, we re-analysed the scRNA-seq dataset of human PDAC that we already investigated in Lupo et al.44. This dataset encompasses 126,530 cells from 63 patients. Subclustering in dimensionality-reduced space of the fibroblast compartment (n = 10,481 cells from 61 patients) revealed 9 CAF subpopulations (Fig. 5a, b). Cluster 2 was identified as IL1 dependent CAFs and accordingly the iCAF signature from Elyada6 was particularly enriched in this cluster (Fig. 5c, d). The expression of the meCAF marker PLA2G2A57 was also high in this cluster (Fig. 5a, b). Accordingly, the meCAF signature57 showed enrichment in c2 (Supplementary Fig. 6a, b). Cluster 1 was a distinct subcluster of iCAFs that expressed components of the complement system; therefore, they were identified as complement secreting CAFs (csCAF58) (Fig. 5a, b). Considering the relative levels of myCAF and iCAF signatures expression as well as their distinctive gene expression programmes (Supplementary Data 6), clusters 0 and clusters 3–5, were identified as myCAFs (Fig. 5c, d). Cluster 0 in particular was identified as TGF-β driven CAFs and accordingly showed enrichment of the LRCC15+ CAF signature (Supplementary Fig. 6a, b). Cluster 5 was characterised by the upregulation of IFN signalling responses while preserving its myofibroblastic identity and was therefore identified as interferon myCAFs (Fig. 5a, b and Supplementary Data 6). Cluster 6 was identified as apCAFs and indeed showed increased expression of CD74 and HLA_DRA (Fig. 5a, b). Clusters 7 and 8 formed somewhat distinct clusters and were identified as WNT CAFs and fibro-adipogenic CAFs, respectively (Fig. 5a, b).
Fig. 5. The human mapCAF signature identifies basal-like tumours.
a UMAP plot of fibroblast compartment after subclustering. Different cell type clusters are colour coded. b Heatmap showing the relative average expression of the most enriched genes for each cluster. Only two representative genes per cluster are reported. Clusters are colour coded as in UMAP plot of (a) and were annotated by markers and pathway enrichment analysis using GSEA93 by comparing cells within a cluster to all other cells in the dataset. See also Supplementary Data 6. c Density plot showing enrichment of human myCAF, iCAF and mapCAF signatures in the scRNA-seq dataset d Violin plots showing enrichment of the human myCAF, iCAF and mapCAF signatures in the clusters shown in (a). Data are presented as mean values and 95% CI e Boxplot of the GSVA scores for the human mapCAF and myCAF signatures in samples of TCGA (n = 148 sample) cohort16 stratified by Moffitt’s subtypes19. p values by Wilcoxon test (two-sided). mapCAF Classical: Min −0.73, Max 0.65, Med −0.12, Q1 −0.43, Q3 0.15, IQR 0.58, LW −0.73, UW 0.65. Basal-like: Min −0.55, Max 0.73, Med 0.17, Q1 −0.07, Q3 0.47, IQR 0.54, LW −0.55, UW 0.73. myCAF Classical: Min −0.84, Max 0.84, Med −0.10, Q1 −0.46, Q3 0.40, IQR 0.87, LW −0.84, UW 0.84. Basal-like: Min −0.79, Max 0.77, Med 0.06, Q1 −0.28, Q3 0.50, IQR 0.78, LW −0.79, UW 0.77. f Barplot showing the percentage of cells of the fibroblast cluster stratified according to the expression of mapCAF signature in PDAC samples displaying prevalent basal-like (n = 6) or classical (n = 6) epithelial cells as shown in Supplementary Fig. 6g. p values determined by Wilcoxon test (two-sided).
To identify CAFs displaying elevated MAPK activity, we mapped the sMEKi signature on the human CAF subclusters. Based on this, we defined a human mapCAF signature (n = 22, Supplementary Data 7) contrasting CAFs with high vs low MAPK transcriptional activity (Supplementary Fig. 6c). We exclusively retained genes which showed low to no expression in the malignant compartment (Supplementary Fig. 6d–f). Although to a different extent, mapCAF signature levels were enriched in CAFs clusters (c0–c3) which corresponded to the c3–c4 of the mouse subclusters (Supplementary Fig. 4c, d and Fig. 4a, b). The Hallmark_Hypoxia signature was enriched in the c2 iCAF clusters but also in myofibroblastic clusters, particularly c3 (Supplementary Fig. 6a, b). These results agree with prior observations from Mello and colleagues59 who showed that although hypoxia signalling is a defining feature of iCAFs, myCAFs remain the most abundant subpopulation in hypoxic tumour regions and a subset of myCAFs displays elevated levels of the Hallmark_Hypoxia signature59. Then, we inferred the mapCAF phenotype in the bulk RNA-seq of the TCGA16. In this dataset, the mapCAF, but not the human myCAF signature6, discriminated basal-like from classical tumours (Fig. 5e). To further confirm our observation, we isolated cases showing predominance of either basal-like or classical malignant cells from the scRNA-seq dataset (Supplementary Fig. 6g). We found that the mapCAF signature was significantly enriched in the stroma of human tumours displaying a greater proportion of basal-like cells (Fig. 5f). The epithelial cells coupled to the mapCAF phenotype showed enhanced glycolytic activity based on expression data (Supplementary Fig. 6h). This is in sharp contrast with the meCAF population, which is reported to couple with epithelial cells relying on oxidative phosphorylation57.
Spatial distribution of transcriptionally defined molecular subtypes
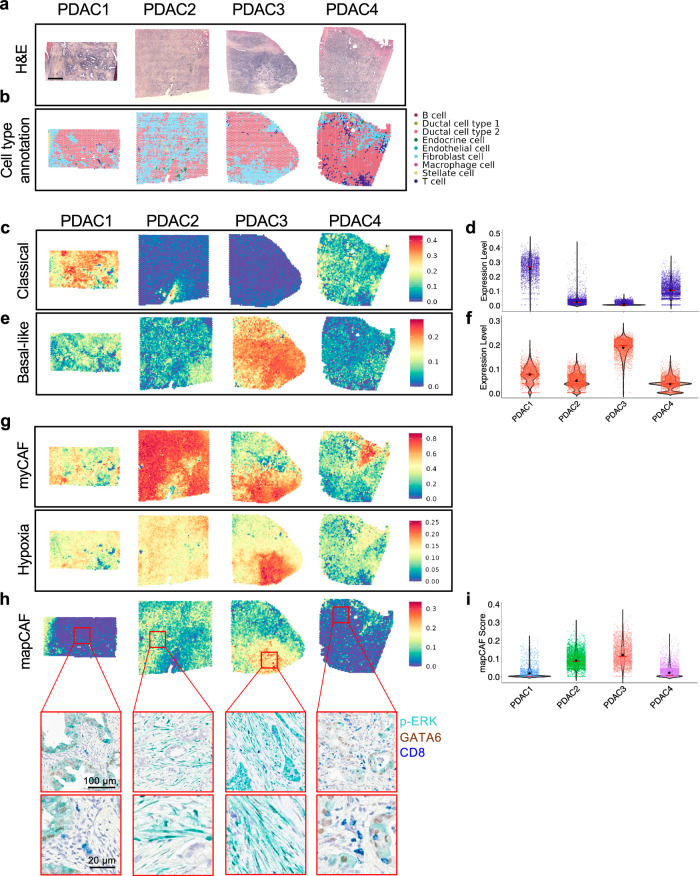
To understand how gene expression programmes are spatially organised in human PDAC, we employed Visium spatial transcriptomics (ST) on formalin-fixed paraffin embedded sections from 4 primary PDAC. The tumours exhibited different degrees of differentiation of the malignant epithelium and stromal cell content (Supplementary Fig. 7a). The number of Visium spots with data varied across the 4 tumours and ranged between 2234 and 4118 per section, also reflecting the different sizes of the investigated tumour tissues. The median number of genes detected per spot was 3724. To identify distinct cell populations, we deconvolved the spots data using scRNA-seq signatures from Peng et al.60 where tumour cells are indicated as Ductal cell type 2. We identified 9 major cell populations (Supplementary Fig. 7b). The spatial distribution of the identified cell types as well as the prediction of areas with high prevalence of either neoplastic or non-neoplastic cells, largely mirrored the histopathological evaluation of the sections by two expert pathologists (A.S., C.L.) (Fig. 6a, b). Next, we mapped the expression of malignant and stromal gene programmes, which have been identified through either bulk19 or single-cell6 RNA-seq of human PDAC tissues. First, we calculated the module score for Moffitt epithelial signatures19, i.e., classical and basal-like (Fig. 6c–f). This analysis revealed variability in the expression of the two epithelial programmes both between and within patients (Fig. 6c–f). Across each section, we found variability in the relative level of each signature indicating either the co-existence of subtypes or the presence of hybrid cells. However, given the size of the spots (55 µm diameter), it is also possible that the results are influenced by mixed gene expression signals due to more than one cell type contained in each spot. Considering the relative signal of each epithelial subtype, the whole slide estimate of the Moffitt gene programmes identified PDAC1 and PDAC4 as classical tumours, while PDAC2 and PDAC3 were defined as basal-like (Fig. 6c–f). This agreed with the expression of the basal-like marker S100A2 assessed on the Visium slides or with immunohistochemistry on serial sections (Supplementary Fig. 7c, d).
Fig. 6. Spatial mapping of epithelial and stromal gene programmes reveals specific association between p-ERK+CAFs and the mapCAF signature.
a Haematoxylin and Eosin (H&E) staining of 4 primary PDAC tumours selected for VISIUM spatial transcriptomics (ST). Scale bar, 1 mm. b Spatial visualisation of the cell types deconvolved using scRNA-seq data from Peng et al.60. See also Supplementary Fig. 7a, b. c Spatial visualisation of gene module score for the classical PDAC epithelial signature19. d Distribution of module scores for classical PDAC signature in each section. The red line indicates the median value, while the black dot the average expression value. All pairwise comparisons are statistically significant by Wilcoxon test (two-sided). e Spatial visualisation of gene module score for the basal-like PDAC epithelial signature19. f Distribution of module scores for basal-like PDAC signature in each section. The red line indicates the median value, while the black dot the average expression value. All pairwise comparisons are statistically significant by Wilcoxon test (two-sided). g Spatial visualisation of gene module score for the myCAFs and the Hypoxia_Hallmark signature. h Spatial visualisation of gene module score for the mapCAF signature. The red boxes indicate the corresponding regions on the multiplex IHC slide for p-ERK, GATA6 and CD8, conducted on a consecutive section of PDAC tissues. Scale bars as shown in figure. Conducted n = 1. i Distribution of module scores for the mapCAF signature in each section. The red line indicates the median value, while the black dot the average expression value. All pairwise comparisons are statistically significant by Wilcoxon test (two-sided).
Mapping of the CAF signatures6 revealed that myCAFs were the most abundant CAF subpopulation in each tumour section (Fig. 6g, and Supplementary Fig. 7e, f). In keeping with scRNA-seq data, the Hallmark_Hypoxia signature was associated with both iCAF and myCAF programmes (Fig. 6g, and Supplementary Fig. 7e, f).
Finally, we mapped the human mapCAF signature and calculated its module score for each section. Consistent with prior findings, the mapCAF signature score was higher in basal-like than in classical tumours and mapped in areas with predominance of fibroblasts (Fig. 6h, i). Next, we performed multiplex immunohistochemistry to show that the mapCAF programme captured areas displaying elevated stromal p-ERK1/2 staining (Fig. 6h, i and Supplementary Fig. 7g). Since gene expression programmes of mapCAFs suggested immunoregulatory functions, we also looked at the intra-tumoral frequency of cytotoxic T cells (CD8+ T cells) and observed that they were rare in mapCAFs occupied areas of the tumours (Fig. 6h).
mapCAFs are associated with T cell exclusion in PDAC and predict poor response to immunotherapy in melanoma
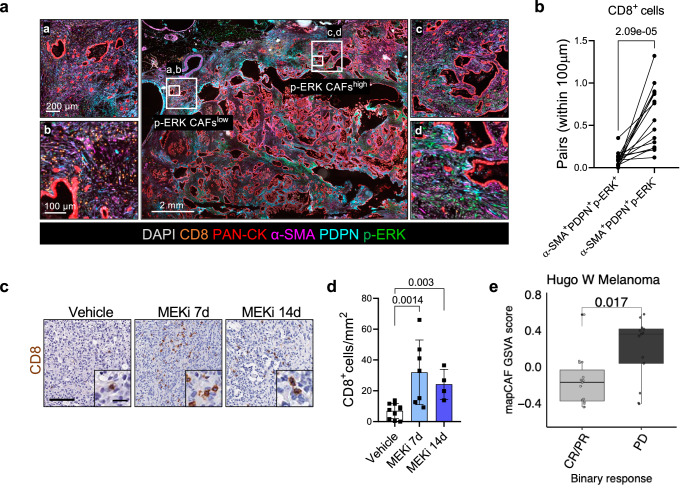
To confirm our initial observation on the reduced CD8+ T cell frequency in mapCAFs rich subTME, we performed mIF on additional human PDAC tumours (n = 15). Spatial analysis showed significantly less CD8+ T cells in proximity of PDPN+α-SMA+p-ERK+ CAFs (Fig. 7a, b and Supplementary Fig. 8a). The increased abundance of CD8+ T cells in proximity of p-ERK− CAFs only partially associated with increased vessel density in these areas (Supplementary Fig. 8b, c). In our mouse model, short-term inhibition of MAPK led to increased intra-tumoral density of CD8+ T cells with a kinetic that is compatible with MAPK rewiring in the stromal compartment (Fig. 7c, d and Supplementary Fig. 8d).
Fig. 7. mapCAFs are associated with T cell depleted sub-tumour microenvironment.
a Representative image of multiplex IF performed on FFPE of human PDAC tissues. The panels represent different areas within the same tumour displaying either low or high density of p-ERK+ CAFs. Scale bars as indicated. See also Supplementary Fig. 8a. b Paired dot plot showing the quantification of CD8+ T cells with a distance below 100 µm to p-ERK+ or p-ERK− CAFs (n = 15 tissues) shown in (a). p values as determined by Mann-Whitney test (two-sided). c Immunohistochemical staining for CD8 of tumour tissues from tumour-bearing mice treated with either vehicle or MEKi for 7 and 14 days. Scale bar, 100 µm. Inserts showed a magnification of selected areas (Scale bar, 20 μm). d Quantification shown as total number of CD8+ T cells per mm2 per sample. p values as determined by Mann-Whitney test (two-sided). n ≥ 4 mice/condition. Results presented as mean values ± SD. e Boxplots showing mapCAF GSVA score in melanoma69 (n = 28 sample). Samples are separated by binary drug response. CR complete response, PR partial response, SD stable disease, PD progressive disease. p value as determined by Wilcoxon test (two-sided). Hugo CR/PR: Min −0.45, Max 0.57, Med −0.17, Q1 −0.38, Q3 −0.04, IQR 0.34, LW −0.45, UW 0.06. PD: Min −0.41, Max 0.58, Med 0.36, Q1 0.03, Q3 0.41, IQR 0.38, LW −0.41, UW 0.58.
Next, we performed a single-cell pan-cancer analysis across 7 tumour types61 displaying myCAF and iCAF phenotypes in their TME62–68. The mapCAF signature was enriched in fibroblasts vs other cell types in all cancer indications (Supplementary Fig. 8e). Within the fibroblasts compartment, the mapCAF phenotype was significantly enriched in myCAFs (Supplementary Fig. 8e). Then, we sought to test whether the presence of mapCAFs correlated with the response to cancer immunotherapy by exploring data from two immuno-reactive tumour entities. We found that the mapCAF signature was enriched in patients with worse response to immunotherapy (stable and progressive disease) in patients with malignant melanoma in the cohort from Hugo et al.69 but not in patients with bladder cancer (BLCA) from the IMvigor210 trial70 (Fig. 7e and Supplementary Fig. 8f).
Overall, those data suggest that the mapCAF phenotype associates with a CD8+ T depleted microenvironment and could predict poor response to immunotherapy in immune-reactive tumours.
Discussion
Here, we disclosed an important role for the MAPK signalling pathway in the definition of PDAC CAF phenotypes. Our data suggest that epithelial MAPK signalling promotes the myofibroblastic differentiation of CAFs. Furthermore, we show that hyperactivation of MAPK signalling occurs in myCAFs populating basal-like tumour niches with reduced CD8+ T cells density.
Understanding how inhibition of the MAPK pathway affects malignant and non-malignant cell populations in PDAC is critical considering the recent advent of direct KRAS inhibitors which can specifically target mutant KRAS driven MAPK activity71. It is increasingly recognised that heterogeneity exists within CAFs subpopulation, with myCAFs exerting both tumour restraining and tumour promoting functions8,10,13,14,72. Thus, strategies to target the heterogeneous phenotypes and functions of CAFs in the PDAC TME is gaining increased attention.
In addition to elevated MAPK activity, the MAPKhigh CAF (mapCAF) phenotype we identified is characterised by TGF-β signalling, hypoxia responsive signatures and immunoregulatory gene programmes. In silico analyses confirmed this phenotype in other solid tumours and further showed that mapCAFs are indicative of primary resistance to immunotherapy in melanoma.
In our mouse model, short-term inhibition of MAPK result in the reduction of the mapCAF phenotype and increased intra-tumoral CD8+ T cell density. While it remains unclear how mapCAF would exert their immunosuppressive function, our data provide a strong basis for future mechanistic studies. Our findings expand on the heterogeneous phenotypes that can be found in the PDAC TME and provide new insights on a stromal phenotype specifically shaped by basal-like cells.
Signalling pathway activities and cell dependency on a given pathway are often successfully inferred from gene expression data53,73. Here, we used pathway mapping analysis and context-dependent pathway response signatures to infer MAPK activity in heterogeneous expression data from models and patients’ samples. Our data show that gene expression data can serve as predictor for the activation of the MAPK pathway.
However, in human cancer cell lines elevated MAPK transcriptional activity did not predict sensitivity to MEK1/2 inhibition nor discriminate between cell lines representative of the basal-like or the classical subtype. In vitro, a transiently disabled MAPK pathway was not associated with significant changes of cancer cell states. Our results are in line with previous observation from Miyabayashi and colleagues showing that RAS signalling hyperactivation is not fundamental for the definition of the basal-like/squamous subtype in the epithelial compartment27. We have previously shown that querying bulk transcriptional datasets from tissues with different neoplastic cell content is a viable strategy to localise the cellular compartment contributing to gene expression differences between molecular subtypes44,74. Using the same approach, we show that a MAPK transcriptional signature was particularly elevated in basal-like tumours from the TCGA and the ICGC cohort16, while it could not discriminate basal-like from classical in RNA-seq from microdissected epithelia. The inferred hyperactivation of MAPK in the stromal compartment of tumours classified as basal-like significantly correlated with increased density of fibroblasts displaying nuclear p-ERK, a known surrogate marker for MAPK activation75. Different neoplastic cell states often co-exist in human PDAC tissues17,21,76,77. Therefore, molecular subtyping based on bulk sequencing data might mask transcriptional heterogeneity17,22. Immunophenotyping of heterogeneous human tumours clearly showed that p-ERK+ CAFs were significantly enriched in basal-like niches. These p-ERK+ CAFs paired with basal-like cells also in mouse PDAC tumours as well as in heterospecies models. Conversely, this phenotype could not be reliably replicated in vitro. Topological constraints within in vivo tissue niches, along with other relevant biochemical or metabolic cues, might be responsible for the emergence of this phenotype.
To gain further insights into this CAF phenotype, we, therefore, integrated single-cell profiling of post-perturbation transcriptional responses from mouse models with cellular and spatial profiles of human tissues. Our mouse model aligned with the human basal-like PDAC44 and showed rapid kinetics of pathway rewiring in the stromal compartment following treatment. Short-term perturbations are often used to capture primary transcriptional response to a specific stimulus53. Contrasting the two conditions with known differential pathway activity, we obtained a stromal MAPK transcriptional signature (sMEKi) which we used to identify CAFs displaying MAPK hyperactivation. High levels of sMEKi were restricted to myCAF subpopulations in our mouse model. Our model preserved the CAFs heterogeneity reported in mouse and human tumours4,6,8, the expected myCAFs/iCAFs ratio in the PDAC TME1,5 and showed presence of p-ERK+ CAFs, as expected for a basal-like model. A MAPKhigh CAF gene signature (mapCAF) was consistently enriched in the myCAF compartment in an autochthonous model of PDAC6. Accordingly, the mouse mapCAF phenotype is enriched for ECM related genes/TGF-β driven programmes and could be induced, in vitro, by treating mouse PSCs with TGF-β1. Regulatory-network inference along with gene expression programmes analysis show that the mapCAF phenotype is mainly driven by Hypoxia and MAPK activity, while displaying expression of inflammatory-related pathways.
In line with prior works5,54, we observed both similarities and differences with the mouse models when translating our findings into the human setting. We used scRNA-seq data comprising 126,530 cells from 63 patients. While we found evidence for fibroblasts identified as inflammatory or myofibroblastic, overlapping gene expression programmes between iCAF and myCAF subsets suggest a higher heterogeneity of functions for human CAFs compared to mouse CAFs. For example, we found a cluster of interferon-responsive CAFs that, at the same time, have a strong myofibroblastic identity. Previously described meCAFs mapped within iCAF clusters. Similar to the mouse data, CAFs displaying elevated MAPK transcriptional activity were mostly myCAFs. This result is coherent with the spatial localisation of p-ERK+ CAFs in human tissues and with previous evidence that locate myCAFs in close proximity to cancer cells8. Integrating spatial with cellular profiles, we found that the mapCAFs tightly associate with basal-like cells to form communities depleted of CD8+ T cells.
Previous studies have shown that similar fibroblast lineages and phenotypes can be observed in different cancers1,65,78. We explored pan-cancer scRNA-seq data61 to find that the mapCAF signature is enriched in stromal cells from many cancer conditions and particularly in myCAFs79–81. Consistent with a potential immunoregulatory function, mapCAFs identify primary resistance to immunotherapy in metastatic melanoma69. Inferred dynamics in scRNA-seq data and orthogonal validation through in situ as well as cytofluorimetric analyses showed that MAPK inhibition leads to a reduced myCAFs/iCAFs ratio into the mouse PDAC TME. Changes into the myCAFs/iCAFs ratio were observed following MEKi in a large spectrum of transplantation-based models and regardless of the molecular subtype of the malignant compartment.
The myCAF and iCAF phenotypes are driven by a well-established antagonistic interaction between TGF-β and IL14, with TGF-β promoting myCAFs differentiation through the downregulation of IL1 receptor. In our mouse models, MEKi affected the relative abundance of myCAFs and iCAFs by favoring an inflammatory stroma. In scRNA-seq data, we found that MEKi was associated with reduced expression of TGF-β which paralleled the reduced secretion of the cytokine by human cancer cell lines upon MAPK inhibition. At the same time, direct MEKi of activated mPSCs did not affect expression of myCAF or iCAF markers. Overall, our data suggests that the changes in the fibroblast compartment are contributed by effective MAPK inhibition in malignant cells.
Altogether, our study shows that epithelial MAPK activity promotes the myCAF phenotype while MAPK hyperactivation in the stromal compartment is a distinctive feature of stromal cells in basal-like tumour niches. Inhibition of MAPK signalling using a potent MEK1/2 inhibitor has important consequences on stroma remodelling with changes in myCAFs to iCAFs ratio. Finally, our data show that the presence of these MAPK-activated myCAFs may represent a feature to predict responses to immunotherapy in PDAC and other malignancies, warranting further consideration for targeting.
Methods
Human samples
Human PDAC tissues used in this study were obtained from surgical resections of patients treated at the University and Hospital Trust of Verona (Azienda Ospedaliera Universitaria Integrata, AOUI). Written informed consent was acquired from patients before specimens’ acquisition. The FFPE samples used for staining were retrieved from the ARC-Net Biobank and were collected under the protocol number 1885 approved by the local Ethics Committee (Comitato Etico Azienda Ospedaliera Universitaria Integrata) to A.S. (Prot. 52070, Prog. 1885). Tissues from surgical resection used for the generation of primary cultures were collected under the protocol number 1911 approved by the local Ethics Committee (Comitato Etico Azienda Ospedaliera Universitaria Integrata) to V.C. (Prot. n 61413, Prog 1911 on 19/09/2018). All experiments were conducted in accordance with relevant guidelines and regulations. The Essen cohort is a retrospective study carried out according to the recommendations of the local ethics committee of the Medical Faculty of the University of Duisburg-Essen. Patients who had undergone pancreatic resection with a final histopathologic diagnosis of human PDAC between March 2006 and February 2016 was used (Approval no: 17-7340-BO). Clinical data were not collected for this study, and sex/gender was not considered in the study design.
Cell lines
We used 5 mouse PDAC cell lines, 1 mouse PSC line and 17 human PDAC cell lines. The mouse PDAC cell line FC1199 was generated from tumour of KPC mice (KrasG12D/+; p53R172H/+; Pdx1-Cre)43. FC1199 were provided by the Tuveson laboratory (Cold Spring Harbor Laboratory, NY, USA) and was cultured in DMEM supplemented with 10% foetal bovine serum (FBS) and 1% Penicillin-Streptomycin (Pen-Strep). Primary murine PDAC cell lines 60400, 60590, 511892 and 110299 were derived from corresponding tumour pieces of KPC mice (60400, 60590 and 511892: Ptf1awt/Cre;Kraswt/LSL-G12D;p53fl/fl 48; 110299: Ptf1awt/Cre;Kraswt/LSL-G12D;p53LSL-R172H/fl 82) and were cultured in DMEM high-glucose medium supplemented with 10% FBS. The mouse PSC line (mPSC4) has been established from WT C57BL/6J mice8. mPSC4 was provided by the Tuveson laboratory (Cold Spring Harbor Laboratory, NY, USA) and cultivated in DMEM containing 5% FBS and 1% Penicillin/Streptomycin. The human PDAC cell lines HPAF-II, PANC-1 and AsPC1 were obtained from ATCC (catalogue numbers CRL-1997, CRL-1469, CRL-1682). The Suit-2, Hs766T, Colo 357 and BxPC3 cell lines were generously supplied by Prof. Aldo Scarpa from the University of Verona. The MIA PaCa-2 line was provided by Prof. Vincenzo Bronte, also at the University of Verona. hF2, hT1, and hM1 were kindly provided by Dr. David A. Tuveson from Cold Spring Harbor Laboratory (USA). Patu 8988S were kindly provided by Dr. Francisco X. Real (CNIO, Madrid). All human PDAC cell lines were cultivated in DMEM supplemented with 10% FBS and 1% Pen-Strep. Human primary PDAC monolayer cell lines (VR2-2D, VR6-2D, VR9-2D, VR20-2D and VR23-2D) were established by digesting tissue samples and directly plating them onto tissue culture vessels to initiate monolayer cultures. Cell lines have been then cultured in the following medium: Advanced DMEM/F12 (Gibco), supplemented with HEPES (1X, Gibco), Glutamax™ (1X, Gibco), Primocin™ (1 mg/mL, Invivogen), mouse Epidermal Growth Factor (50 ng/mL, Gibco), Dexamethasone (3 nM, Sigma) and 5% Foetal Bovine Serum (FBS, Gibco). Cell lines were routinely screened for Mycoplasma contamination using MycoAlert Mycoplasma Detection Kit (Lonza).
Generation of mouse models
In this study we used both isograft and xenograft models. Six- to eight- weeks old C57Bl/6J (B6J) (Strain #:000664) and NSG (NOD.Cg-Prkdcscid;Il2rgtm1Wjl) (Strain #:005557) mice were purchased from Charles River Laboratory (Milan). All animal experiments regarding transplanted mice were conducted in accordance with procedures approved by CIRSAL at University of Verona (approved project 655/2017-PR) or were approved by the Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen (LANUV) under the license number 81-02.04.2020.A316. Animal care procedures and protocols were as prescribed in the national (Tierschutzgesetz) and European (Directive 2010/63/EU) laws and regulations as well as European Federation of Animal Science Associations (FELASA) http://www.felasa.eu. KC (KrasG12D/wt; Pdx1-Cre) and KPC (KrasLSL-G12D/wt; p53LSL-R172H/wt; Pdx1-Cre) mice were used as spontaneous model for pre-invasive lesions and PDAC, respectively43,51. Isograft models were generated with KPC-derived cell lines. For the generation of isograft based on KPC-derived cell line (FC1199), 2.5 × 105 cells were resuspended in 50 µL of a 2:3 mixture of Matrigel® (Corning) and cold PBS (Gibco) and injected into the pancreatic tail region using BD micro-fine insulin syringes (30-gauge). A successful injection was confirmed by the appearance of a bubble at the injection site without leakage. Tumour growth was monitored through weekly manual palpation beginning 7 days post-transplantation, followed by high-contrast ultrasound imaging using the Vevo 2100 System with an MS250 scanhead (13–24 MHz, Visual Sonics). For the generation of isograft based on KPC-derived cell lines (60400, 60590, 511892 and 110299), 5.0 × 103 cells were resuspended in 30 µL of a 1:1 dilution of Matrigel® (Corning) and cold plain medium and injected into the pancreas of B6J mice using insulin syringes (BD micro-fine 30 Gauge) under the guidance of Ultrasonic imaging. The injection was considered successful by the appearance of encapsulated cell suspension ball without signs of leakage. Mice were euthanized at designated time points, and pancreas was collected for further analysis. Patient-derived xenografts (PDX#1-15) were generated by subcutaneous implantation of a patient’s tumour fragment in the left flank of anaesthetised NSG mice. Tumour growth was measured twice weekly until they reached the volume of 1 cm3. Then, tumours were harvested, cut into small fragments (3 mm3), and transplanted subcutaneously into the left flank of anaesthetised NSG to generate the 2nd generation of xenograft. The same procedure was followed to generate the 3rd generation which was used for treatments. Mice were maintained under sterile and controlled conditions (22 °C, 50% relative humidity, 12 h light–dark cycle, autoclaved food and bedding, acidified drinking water). The maximal tumour size allowed by ethic committees (CIRSAL: diameter = 1.2 cm; LANUV: volume = 1000 mm3) was never exceeded in this study.
In vivo drug treatments
Isografts and xenografts were treated with Trametinib (MEKi) as indicated. Only female C57BL/6J mice were used for experiments with FC1199 derived grafts. Both male and female C57BL/6J mice were used for generating grafts from the other KPC cell lines. Sex was not considered in the study design. Before treatment, tumour masses were measured, and mice were randomised. KPC-derived isografts were treated with Trametinib dissolved in a solution of 0.5% hydroxypropyl-methylcellulose, 0.2% tween 80 and ddH2O (pH 8) with a final concentration of 1 mg/kg for daily oral administration. Isografts based on KPC-derived cell lines (60400, 60590, 511892 and 110299) were treated with Trametinib dissolved in a solution of 1% Kolliphor® EL, 1% PEG400 and ddH2O with a final concentration of 1 mg/kg for daily oral administration. Monitoring of tumour growth was performed as described in the section ‘Generation of mouse models’. Pancreas was collected for downstream analysis. For models orthotopically transplanted with 60400, 60590, 511892 and 110299 cell lines, tissue biopsies were collected from the same tumours under ultrasound guidance before treatment started or at different time points after treatment started as indicated.
In vitro drug treatments
Cells and organoids were treated with Trametinib as indicated. Trametinib (Selleck) was dissolved in DMSO, whose final concentration was less than 0.1% (v/v). For each cell culture, the IC50 concentration was determined by the luminescence ATP-based assay CellTiter-Glo (Promega, G9683) following the manufacturer’s instructions. Briefly, 1 × 103 cells were plated on white 96-well plate in 100 µL of culture medium. After 24 h cells were treated with increasing doses of Trametinib, and viability was measured at endpoint (72 h) using a microplate reader (BioTek, Synergy 2 Multi-mode Microplate Reader). For measurement of signalling pathways following pharmacological treatment, individual cell lines were counted and seeded in a 6 well plate or a 100 cm2 dish. After reaching 40% of the confluence, cells were treated for the time reported in each experiment. PDAC cell lines and mPSCs were treated with sub-IC50 doses of Trametinib.
Immunohistochemistry
The following primary antibodies were used on mouse PDAC tissues: p-ERK (#4376, 1:200, cl. 20G11, lot. 21, Cell Signaling Technology), GATA6 (#ab175349, lot. GR3447918-1, Abcam), S100A2 ([EPR5392], #109494, Abcam) and CD8 (CD8a (4SM15) (14-0808-82, Invitrogen)). Quantification of stromal p-ERK was performed by HALO software with the classifier function. After cell type segmentation performed by the software, p-ERK+ cells in the stroma regions are quantified as % of positive cells out of total cells in the whole tumour section. Quantification of CD8 was performed by counting the number of CD8 positive T cells per mm2. At least 5 individual areas per case and a minimum of 4 mice/arm were evaluated. Chromogenic multiplex IHC analysis was performed using Leica Biosystems BOND RX/RXm according to the manufacturer’s instructions using the following antibodies: p-ERK (#9101, lot.32, Cell Signaling Technology), GATA6 (#AF1700, lot. KWT0523031, Bio-techne) and CD8 (C8/144B, lot. 20042547, DAKO).
Multiplex immunofluorescence
mIF was carried out using the Opal multiplex system (Akoya Biosciences, MA, USA) following the manufacturer’s instructions. The following antibodies have been used: pan-Keratin (#ab6401, cl. C11, Abcam), p-ERK (#4376, 1:200, cl. 20G11, lot. 21, Cell Signaling Technology), α-SMA (ab5694, 1:200, GR3263275-13, Abcam), PDPN (ab236529, 1:200, cl. EPR22182, lot. GR3330154-1, Abcam), GATA6 (#ab175349, lot. GR3447918-1, Abcam), KRT81 (#sc100929, cl. 36-Z, lot. I3021, Santa Cruz Biotechnology), CD8 (#ab101500, 1:200, cl. SP16, lot. 1036318-32, Abcam), CD34 (NCL-L-END, 1:200, cl. QBEND/10, lot. 6088521, Leica) and LY6C (#ab15627, 1:100, cl. ER-MP20, lot. GR3261661-18, Abcam). Sections were counterstained with DAPI (Vector lab). Using a Zeiss Axio Scanner Z.1 (Carl Zeiss AG, Germany) at 10x objective magnification, slides were scanned and digitalised. For the mouse PDAC tissues from Fig. 2b, the following antibodies have been used: pan-Keratin (#4279, C11, Cell Signaling Technology), p-ERK (#9101, Cell Signaling Technology) and α-SMA (#ab5694, lot. GR3183259-39, Abcam). Sections were counterstained with DAPI (Sigma Aldrich). Images were acquired with Leica TCS SP5 laser scanning confocal (Leica) and digitalised by the Leica Application Suite X (LAS X) software.
Spatial imaging analysis
Quantification of individual and/or co-expressing markers in the multiplexed immunofluorescence images was performed using HALO image analysis software (Indica Labs, NM, USA). A gate for region of interest (ROI) was manually drawn on the image of each slide to exclude the non-tumour regions, areas containing tissue folds and stain artifacts. Tissue segmentation with classifier was performed to select for all tumour and stroma regions within the ROI. Nuclear detection based on DAPI staining was performed. Signal intensity for positivity of each marker was manually adjusted and the signal threshold for positivity was the same for all tissues within the cohort. After quantification of each marker and various phenotypes (co-expression of markers), close proximity analysis was performed to measure the close interaction between two target cell subsets. The threshold to pair up two cells was set to 100 µm.
Immunofluorescence on mPSCs
5 × 103 mPSCs were plated in a cell culture chamber slide. After 48 h, cells were serum starved for 6 h and treated for 10 min with media collected from cancer cell lines after 15 h of conditioning in serum starvation. After treatment, the IF was performed following the suggested protocol for p-ERK1/2 antibody (#9101, 1:200, lot. 32, Cell Signaling Technology). Nuclei were counterstained with DAPI. Quantification was performed by counting cells with nuclear signal for p-ERK per FOV divided by the total number of cells counted in the field (minimum 10 FOV/condition).
qRT-PCR on mPSCs
To perform qRT-PCR, mPSCs were cultured in both 2D and 3D formats. For plastic-activated mPSCs, 6 × 104 cells were plated in a 6-well plate. After 48 h, cells were treated with a sub-IC50 dose of Trametinib for 6 h. For TGF-β activated mPSCs, 8 × 104 cells were embedded in 50 µL Matrigel® domes. After 24 h, cells were pre-treated with 20 ng/mL TGF-β1 for 48 h, followed by treatment with a sub-IC50 dose of Trametinib for 6 h. RNA was isolated utilising the Trizol® Reagent (Life Technologies). Following DNase treatment, 1 µg of RNA was reverse transcribed into cDNA using the SensiFAST® cDNA Synthesis Kit (meridian BIOSCIENCE) in a total volume of 20 µL, following to the manufacturer’s guidelines. The resulting samples were then diluted to achieve a final concentration of 10 ng/µL. TaqMan assays were conducted in triplicate, employing 20 ng of cDNA along with the following TaqMan® probes (TaqMan® Gene Expression Assay): Il6 (Mm00446190_m1), Cxcl1 (Mm04207460_m1), Acta2 (Mm01546133_m1) and Ctgf (Mm01192933_g1). Hprt (Mm03024075_m1) was used as the reference gene. The relative quantification of gene expression was calculated using the ΔΔCt method with the Sequence Detection Systems Software, Version 1.9.1 (Applied Biosystems).
In situ hybridisation
The in-situ hybridisation (ISH) was performed on 4 μm section of mouse tissues. Briefly, slides were deparaffinized in xylene for 10 min followed by 100% ethanol for 2 min. After drying, slides were first incubated for 10 min with RNAscope® Hydrogen Peroxide (Advanced Cell Diagnostics) and then for 15 min at 99 °C with RNAscope® 1X Retrieval Reagents (Advanced Cell Diagnostics). After dehydration in 100% ethanol, slides were dried and incubated at 40 °C for 20 min with RNAscope® Protease Plus (Advanced Cell Diagnostics). The RNAscope® Probes (Mm-Mmp3 and Mm-Tnc-C2, Advanced Cell Diagnostics) were added to the slides following RNAscope® 2.5 Duplex Detection Reagents kit’s instructions. Positive control probe 2.5 Duplex Positive Control Probe-Mm and 2-plex Negative Control Probe (Advanced Cell Diagnostics) were used as positive and negative control, respectively. Quantification was performed by counting the total number of spindle-like cells with signal for Tnc or Mmp3 in each tumour section.
Immunoblotting
Protein lysates were obtained from cells using Lysis Buffer (Cell Signaling Technology) supplemented with phosphatases and proteases inhibitors (PhosSTOP™ and cOmplete (TM) Mini Protease Inhibitor Co, Roche). After electrophoretic separation, proteins were transferred on a PVDF membrane and incubated with the following antibodies: p-ERK (#9101, 1:2000, lot.32, Cell Signaling Technology), total ERK (#9102, 1:1000, lot. 26, Cell Signaling Technology), p-AKT (#4060, (Ser473) (D9E) XP®, lot.2, Cell Signaling Technology), total AKT (#9272, lot. 28, Cell Signaling Technology), p-S6 (#D57.2.2E/#4858, lot. 11, Cell Signaling Technology), total S6 (#2317, cl. 54D2, Cell Signaling Technology). Vinculin (#4650, 1:1000, lot. 5, Cell Signaling Technology) and GAPDH (#5174, (D16H11) XP®, 1:3000, lot. 9, Cell Signaling Technology) were used as loading controls. The signal was quantified with ImageJ, by measuring the integrated density of each band.
ELISA
Cell culture media were collected after 48 h of conditioning, centrifuged to remove particulates and stored at –80 °C. The ELISA assay was conducted using the Human TGF-β1 Quantikine® ELISA (DB100B, R&D System) according to the manufacturer’s instructions.
FACS analysis
Freshly isolated cells from KPC-derived orthotopic tumours (1 × 106) resuspended in 1 mL DPBS were first stained with BD Horizon™ Fixable Viability Stain 440UV (#566332, BD Biosciences), incubated at room temperature in the dark for 15 min and washed twice with FACS buffer (DPBS with 2% FBS). Upon the addition of 50 μL of BD Horizon Brilliant Stain Buffer (#563794, BD Biosciences), the cells were incubated with the following fluorochrome-conjugated antibodies: CD45 BV421 (#563890, 0.2 mg/ml, cl. 30-F11, lot. 4036259, BD Biosciences), CD31 PE-Cy7 (#561410, 0.2 mg/ml, cl. 390, lot. 3327027, BD Biosciences), PDPN (AF488 #127406, 0.2 mg/ml, cl. 8.1.1, lot. B355066, Biolegend) and LY6C APC (#128016, 0.2 mg/ml, cl. HK1.4, lot. 3115596, Biolegend) at 4 °C in the dark for 45 min in a final volume of 100 μL. After a final wash with FACS buffer, samples were acquired with BD FACSDiscover™ S8 Cell Sorter (BD Biosciences). Data analysis was conducted with the FlowJo software v10.10 (BD Biosciences).
RNA sequencing and data processing
RNA was extracted from cell lines using TRIzol (Life Technologies), followed by column-based purification with the PureLink RNA Mini Kit (Ambion). The quality of purified RNA samples was determined using a Bioanalyzer 2100 (Agilent) with RNA 6000 Nano Kit. RNAs with RNA Integrity Number (RIN) values greater than 7.5 were used to generate sequencing libraries using the TruSeq Stranded Total RNA Kit (Illumina) following manufacturer’s instructions. Libraries were prepared from TrueSeq Stranded RNA Kit (Illumina) and sequenced on Illumina instruments. After quality control and adaptor trimming, PSC reads were aligned to the GRCm38 genome using Salmon v1.4.083,84 Cell lines reads were aligned to the GRCh38 genome using STAR v2.785. RSEM transcripts’ quantification86 was imported in R through tximport package v4.0 and raw counts were normalised using the R/Biocondutor package DESeq2 v1.30.084. Differentially expression analysis has been performed using DESeq284. The eMEKi signature was derived by filtering the results of differential gene expression performed with DESeq284 between cell lines untreated and treated with MEKi for 2 days, filtering according to padj < 0.01 and log2FoldChange < –1. GSVA R package v1.38.287 was used to calculate the main PDAC transcriptomics subtypes gene set scores19. Subtyping of cell lines and tissue samples was based on the highest GSVA score between the basal and classical gene programmes.
Statistical analysis and data mining
Four transcriptomic datasets from either cell lines or cancer tissues were used for data mining15–17,33. Samples belonging to the ICGC15 and the TCGA-PAAD datasets16 were restricted to 82 and 148, respectively, by selecting only true PDAC cases. Two additional datasets were included for testing prediction capability of mapCAFs. Malignant melanoma FPKM from Hugo et al.69 were downloaded from GEO (GSE78220). BLCA RNAseq counts from the IMvigor210 trial70 were accessed through the R package easierData. GraphPad Prism was used for graphical representation of data. Statistical tests were performed with R or GraphPad Prism and are reported in each figure legend.
Mouse single cell RNA sequencing
Sample preparation and sequencing
Single cell RNA sequencing was performed on digested PDAC tissues from mice treated with either vehicle or MEKi for 2 or 7 days. For the digestion, tumour samples were collected in Splitting Medium (AdDMEM/F12 medium supplemented with HEPES (10 mM), Glutamax and Pen/Strep) supplemented with 0.1% BSA and RhoKi (10.5 μM). After washing with PBS, specimens were cut in small pieces (1 mm3) and incubated for 20 min in a tube rotator at 37 °C in warm Digestion Medium (PBS 1X, 2 mg/mL Dispase I, 1.25 mg/mL Collagenase Type II, 100 μg/mL DNAse I, and 0,05% FBS) supplemented with RhoKi. The cell suspension was pipetted and incubated on ice to let the larger tissue clumps settle to the bottom of the tube. The surnatant was collected, spun down and the pellet was resuspended in Splitting Medium supplemented with 0.1% BSA and 10 mg/ml Soybean trypsin inhibitor and stored on ice (Fraction 1). Then, the larger undigested clumps were digested again for 10 min, and every step previously described was repeated twice until the collection of Fraction 2 and 3. After digestion, Fractions 1, 2 and 3 were combined, filtered (40 μm nylon cell strainer) and centrifuged. The pellet was then resuspended in ACK lysing buffer supplemented with DNAse I to remove red blood cells from the sample and spun down. Cells were washed with PBS supplemented with 10% FBS and 1 × 104 cells (concentration 1.000 cells/ul) were submitted for sequencing. To generate single cell GEMs, cellular suspensions from 3 mice/condition were loaded on a GemCode Single Cell Instrument (10x Chromium System) and libraries were generated with GemCode Single Cell 3’ Gel Bead and Library Kit v3 (10x Genomics). After barcoding, GEMs were broken, and cDNA was cleaned with with DynaBeads MyOne Silane Beads. cDNA was amplified, cleaned with the AMPure beads and the quality was checked using Fragment Analyzer HS NGS Assay. Libraries were quantified by quantitative PCR (qPCR) (KAPA Biosystems Library Quantification Kit for Illumina platforms) and the sequencing was performed on NextSeq500 (Illumina) with 75 paired-end kit.
Data processing
Binary base call (BCL) files were processed with the 10X proprietary software Cell Ranger88, with default and recommended parameters. FASTQs files were aligned to reference transcriptome GRCm38 by count pipeline. Counts matrices for all samples were imported with Seurat89. Cells with low quality were filtered from counts matrices (200 < n° of genes x cell < 9000 & %mitochondrial gene count <25%). Vehicle and treatment datasets were integrated using Seurat integration pipeline89, clustering analysis were run on integrated dataset with Seurat FindCluster89 function using a resolution of 1. Annotation of the dataset were performed looking at the expression of well-known cell type markers. Copy number analysis was performed on epithelial compartment with InferCNV R package90 using as reference the non-epithelial cells.
Subtyping and enrichment analysis
Subtyping of epithelial cells was performed using signatures from Moffitt et al.19, Bailey et al.15 and Collisson et al.18. Subtyping of fibroblasts was performed using signatures from Elyada et al.6. To assess cells subtype, an enrichment score was assigned for each gene set to each cell using UCell R package91, the cell subtype was assigned based on maximum score achieved by a cell for a specific gene set. Other tested signatures were obtained through the msigdb packages92,93. The related single cell enrichment score was computed with UCell package91.
Analysis of the fibroblast subcluster
To assess the purity of the CAF cluster, we extrapolated the fibroblast cells from the integrated atlas and performed subclustering with FindCluster function of Seurat89 using a resolution of 0.5. Resolution value was decided after evaluation with clustree94. The unique features of the fibroblast subclusters were evaluated by marker genes expression. Of the 10 subclusters, 7 were annotated as CAFs (c0 = 442 cells, c1 = 398 cells, c2 = 357 cells, c3 = 282 cells, c4 = 209 cells, c5 = 149 cells, c9 = 50 cells) due to expression of both subtype specific signature genes6 and panCAF genes (Pdpn, Fap, Acta2, Pdgfra). One subcluster was annotated as cycling (140 cells) and the two remaining subclusters were annotated as EMT-like (61 cells) and Myeloid-like (53 cells) based on the high expression level of Ptprc, Pecam1 and Epcam for EMT-like cells and Itgam, Ptprc and Adgre1 for Myeloid-like cells. The 3 non-CAF clusters were filtered out before the subsequent analysis. To perform velocity analysis, the matrices of spliced and unspliced RNA counts were obtained from raw data using velocyto pipeline50 under default parameters. The resulting matrices were uploaded in Seurat objects selecting cells from the previously annotated CAF subclusters. The objects were integrated with Seurat integration pipeline by timepoints, regressing out cell cycle effect. Computation of velocity and velocity pseudotime values was performed through velociraptor R package49, using default parameters and precomputed PCA values. The sMEKi signature was derived by filtering the results of differential gene expression performed with Seurat function FindMarkers89 between vehicle and MEKi treated CAF subclusters of the 2 days timepoint, filtering according to p_val_adj < 0.05 and avg_log2FC < −0.25. Signature enrichment score was computed on fibroblast compartment using UCell R package91. CAFs of the vehicle samples were classified as sMEKhigh CAF when their signature enrichment score was above the third quartile of the score’s distribution. The mapCAF signature was instead obtained contrasting the transcriptome of sMEKhigh CAF vs those showing low level of the signature. Only genes showing p_val_adj < 0.05 and avg_log2FC > 1 were selected. Enrichment pathway analysis on the mapCAFs and other CAF subclasses was performed with both PROGENy53 and fgsea package. As input was used the ordered gene list from differential gene expression analysis performed with FindMarkers89 function and pathways from HALLMARK, REACTOME and GO collections from msigDB92,93. Transcription factor analysis was performed with decoupleR package56. Ligand receptor analysis was performed with CellChat R package52, first individually on vehicles and treatments data, then performing differential communication analysis between them.
Additional mouse scRNA-seq data
Single cell RNA-seq normalised counts of fibroblasts enriched dataset from Elyada et al.6 were downloaded from GEO (GSE129455). The data were imported and managed with Seurat89. mapCAF signature enrichment score was computed on dataset cells with UCell package91. Subtyping of the fibroblasts cluster was performed as described in the subtyping section of our mouse single cell RNA sequencing.
Human single cell RNA sequencing datasets
ScRNA-Seq from human PDAC tissues17,60,95,96 were downloaded from NGDC (GSA: CRA001160), GEO (Accession #GSE154778 and #GSE155698) and EGA (accession EGAS00001002543) together with annotation metadata, when provided. The dataset Peng et al.60 (primary PDAC = 24, ncells = 41964), Lin et al.95 (primary PDAC = 10, ncells = 7752), Chan-Seng-Yue et al.17 (primary PDAC = 13, ncells = 33970) and Steele et al.96 (primary PDAC = 16, ncells = 42844) were first preprocessed individually using Seurat V4.0.197 for quality control and filtering (percent_mt_max = 20, nFeature_min = 500, nCount_min = 500, nCount_max = 50,000), then integration was performed through harmony98 using default parameters and dataset metadata as grouping variable. The integrated dataset was annotated through singleR package using as reference the preloaded dataset HPCA from the celldex package. Epithelial cells were re-classified as ductal, acinar, or endocrine using known gene signatures99. Identification of tumour cells in the ductal cluster, was performed with copycat CNV analysis at sample level, using as normal reference non epithelial cells. Fibroblasts were re-classified using known gene signatures99.We then performed subclustering on the CAFs from the integrated atlas with FindCluster function of Seurat89 using a resolution of 0.2. Resolution value was decided after evaluation with clustree94. The unique features of the subclusters were evaluated by marker genes expression. Pan-cancer dataset from Luo et al.61 (primary tumour = 148, ncells = 494,610) was downloaded from Gene expression Omnibus (accession No. GSE210347) with annotation metadata6,19. All signature enrichment scores on human single cell datasets were computed with UCell package91. Subtyping of the epithelial and fibroblast clusters was performed as described in the subtyping section of mouse single cell RNA sequencing. For the definition of the human mapCAF phenotype, we first mapped the mouse sMEKi signature to human orthologs100. The human sMEKi was then used to identify CAFs with elevated expression of the signature which were then contrasted to all other CAFs to identify the human mapCAF signature. For that, we retained only genes showing p_val_adj < 0.05 and avg_log2FC > 1 with low to little expression in the malignant compartment of Peng et al.60.
Spatial transcriptomics
Five sections (thickness = 5 μm) from each sample were used to assess RNA quality (RIN) with 2100 Bioanalyzer (Agilent, USA); all samples showed DV200 > 30%. Spatial transcriptomics was performed with Visium Spatial Gene Expression for FFPE v2 (Manual protocol). One section from each patient was scored to fit 6.6 mm Visium capture areas and processed according to manufacturer protocols, with no modifications. Visium library quality was assessed with Tapestation (Agilent) and sequenced on Nextseq 2000 (Illumina). Binary base call (BCL) files were processed with the 10X proprietary software Space Ranger. V3.0.0 with default and recommended parameters. FASTQs files were aligned to reference transcriptome GRCh38 by count pipeline. All samples showed high QC parameters with a mean sequencing saturation around 60%. Counts matrices for all samples were imported with Seurat97 and integrated with RPCA method for batch effect correction. For annotation of cell types, we used single cell signatures from Peng et al.60 using the Seurat ‘anchor’-based integration workflow97.Signatures enrichment on the spatial dataset was performed using AddModuleScore function, with UCell method91.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We gratefully acknowledge the Centro Piattaforme Tecnologiche (CPT—University of Verona, Verona, Italy) for granting access to the genomic facility of University of Verona. We also acknowledge the GSTEP, Multiplex spatial profiling facility and Bioinformatics Research Core Facility. We would like to thank Dr. Samantha Bersani for performing chromogenic multiplex IHC. We would like to thank Dr. T. Jacks, Dr. D. Tuveson, Dr. J. Jonkers, Dr. A. Berns, Dr. H. Nakhai and Dr. R.M. Schmid for providing transgenic animals. We would like to thank Dr. Francisco X. Real (CNIO, Madrid), Prof. Vincenzo Bronte, Prof. Aldo Scarpa (University of Verona), and Dr. David A. Tuveson (Cold Spring Harbor Laboratory, NY, USA) for providing cell lines . We thank Dr. T. Ebel (Zentrum f. Pathologie Essen-Mitte) for kind support. We gratefully acknowledge the support from the West German Biobank (Essen, Germany) and Life & Brain GmbH (Bonn, Germany). AIRC Start up Grant No. 18178, AIRC IG No. 28801, EU MSCA project PRECODE (grant No: 861196) and National Cancer Institute (NCI, HHSN26100008); V.C. AIRC 29528; L.V. AIRC 25286-AIRC 29692; E.F. AIRC 28054-AIRC 29829; M.B. AIRC MFAG No 23681; C.C. AIRC MFAG No 29224; G.P. German Cancer Consortium (DKTK), Deutsche Forschungsgemeinschaft (DFG, German Research Foundation through 405344257/SI1549/3-2, 421166016 (SI1549/4–1), 450917483 (GRK2762/1)), German Federal Ministry of Education and Research (BMBF; 01KD2206A/SATURN3), European Union Seventh Framework Programme for research, technological development and demonstration (FP7/CAM‐PaC) under grant agreement no° 602783 and the research network CANcer TARgeting (CANTAR) of the Ministry of Culture and Science of the State of North Rhine‐Westphalia (MKW NRW); J.S. This study was conducted with the support of the Ontario Institute for Cancer Research through funding provided by the Government of Ontario. The funding agencies had no role in the collection, analysis and interpretation of data or in the writing of the manuscript. Disclaimer: This communication reflects the views of the authors, and the Joint Undertaking is not liable for any use that may be made of the information contained herein.
Author contributions
L.V., D.P., P.F.C., J.S., and V.C. designed the research; L.V., P.F.C., R.F., C.V., D.F., E.F., F.L., S.D.A., C.C., G.P., D.B., and M.B. performed experiments; D.P., P.D., D.R., A.A., and C.N. analysed omics data and generated displays; P.M., R.T.L., C.L., and A.S. collected and characterised human tissue samples; G.B. supervised in vitro experiments with mPSCs; V.C. and J.S. acquired funding; L.V., D.P., J.S., and V.C. wrote the manuscript; P.F.C., J.S., and V.C. supervised the study. All authors approved the final version of the manuscript.
Peer review
Peer review information
Nature Communications thanks Matthew Cribb, Christopher Hanley and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
RNA-seq data generated in this study have been deposited in the GEO database under accession code: GSE246457. scRNA-seq data generated in this study have been deposited in the GEO database under accession code: GSE246458. Spatial transcriptomics data generated in this study have been deposited in the GEO database under accession code: GSE274665. The publicly available RNA-seq data from CCLE used in this study were downloaded from DepMap portal[https://depmap.org/portal/]. The publicly available RNA-seq data from TCGA used in this study were downloaded from firebrowse [http://firebrowse.org/?cohort=PAAD]. The publicly available RNA-seq data from ICGC used in this study were downloaded from ICGC data portal, now migrated in ICGC 25 K data. The publicly available RNA-seq data from PanCuRx used in this study are available upon request in the EGA database under accession code EGAS00001002543. The publicly available RNA-seq data from ImVigor trial70 used in this study are available within easierData R package. The publicly available mouse sc-RNA-seq data used in this study are available in the GEO database under accession code GSE129455. The publicly available human sc-RNA-seq data used in this study are available in: GEO database under accession codes GSE15477895, GSE15569896, GSE21034761; in GSA database under accession code CRA00116060 and in EGA database upon request under accession code EGAS0000100254317. The remaining data are available within the Article, Supplementary Information or Source Data file. Source data are provided with this paper.
Competing interests
J.S. receives honoraria as consultant or for continuing medical education presentations from AstraZeneca, Bayer, Boehringer Ingelheim, Bristol-Myers Squibb, Immunocore, MSD Sharp Dohme, Novartis, Roche/Genentech and Servier. His institution receives research funding from Abalos Therapeutics, Boehringer Ingelheim, Bristol-Myers Squibb, Celgene, Eisbach Bio and Roche/Genentech; he holds ownership in FAPI Holding (<3%), all outside the submitted work. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lisa Veghini, Davide Pasini, Rui Fang.
These authors jointly supervised this work: Phyllis F. Cheung, Jens T. Siveke, Vincenzo Corbo.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54975-8.
References
- 1.Buechler, M. B. et al. Cross-tissue organization of the fibroblast lineage. Nature593, 575–579 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Chhabra, Y. & Weeraratna, A. T. Fibroblasts in cancer: unity in heterogeneity. Cell186, 1580–1609 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grunwald, B. T. et al. Spatially confined sub-tumor microenvironments in pancreatic cancer. Cell184, 5577–5592.e5518 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Biffi, G. et al. IL1-induced JAK/STAT signaling is antagonized by TGFbeta to shape CAF heterogeneity in pancreatic ductal adenocarcinoma. Cancer Discov.9, 282–301 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dominguez, C. X. et al. Single-cell RNA sequencing reveals stromal evolution into LRRC15(+) myofibroblasts as a determinant of patient response to cancer immunotherapy. Cancer Discov.10, 232–253 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Elyada, E. et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov.9, 1102–1123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hutton, C. et al. Single-cell analysis defines a pancreatic fibroblast lineage that supports anti-tumor immunity. Cancer Cell39, 1227–1244.e1220 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohlund, D. et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med.214, 579–596 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, Y. et al. Type I collagen deletion in alphaSMA(+) myofibroblasts augments immune suppression and accelerates progression of pancreatic cancer. Cancer Cell39, 548–565.e546 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhattacharjee, S. et al. Tumor restriction by type I collagen opposes tumor-promoting effects of cancer-associated fibroblasts. J. Clin. Investig.131, e146987 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim, E. J. et al. Pilot clinical trial of hedgehog pathway inhibitor GDC-0449 (vismodegib) in combination with gemcitabine in patients with metastatic pancreatic adenocarcinoma. Clin. Cancer Res.20, 5937–5945 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ko, A. H. et al. A phase I study of FOLFIRINOX plus IPI-926, a Hedgehog pathway inhibitor, for advanced pancreatic adenocarcinoma. Pancreas45, 370–375 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozdemir, B. C. et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell25, 719–734 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhim, A. D. et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell25, 735–747 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bailey, P. et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature531, 47–52 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas Research Network. Electronic address, a. a. d. h. e. & cancer genome Atlas research, N. Integrated genomic characterization of pancreatic ductal Adenocarcinoma. Cancer Cell32, 185–203.e113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan-Seng-Yue, M. et al. Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nat. Genet.52, 231–240 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Collisson, E. A. et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med.17, 500–503 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moffitt, R. A. et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet.47, 1168–1178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Candido, J. B. et al. CSF1R(+) macrophages sustain pancreatic tumor growth through T cell suppression and maintenance of key gene programs that define the squamous subtype. Cell Rep.23, 1448–1460 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang, W. L. et al. Single-nucleus and spatial transcriptome profiling of pancreatic cancer identifies multicellular dynamics associated with neoadjuvant treatment. Nat. Genet.54, 1178–1191 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raghavan, S. et al. Microenvironment drives cell state, plasticity, and drug response in pancreatic cancer. Cell184, 6119–6137.e6126 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malinova, A., Veghini, L., Real, F. X. & Corbo, V. Cell lineage infidelity in PDAC progression and therapy resistance. Front. Cell Dev. Biol.9, 795251 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waddell, N. et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature518, 495–501 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins, M. A. et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J. Clin. Investig.122, 639–653 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collisson, E. A. et al. A central role for RAF–>MEK–>ERK signaling in the genesis of pancreatic ductal Adenocarcinoma. Cancer Discov.2, 685–693 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyabayashi, K. et al. Intraductal transplantation models of human pancreatic ductal Adenocarcinoma reveal progressive transition of molecular subtypes. Cancer Discov.10, 1566–1589 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mueller, S. et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature554, 62–68 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Falcomata, C. et al. Selective multi-kinase inhibition sensitizes mesenchymal pancreatic cancer to immune checkpoint blockade by remodeling the tumor microenvironment. Nat. Cancer3, 318–336 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung, V. et al. Effect of selumetinib and MK-2206 vs oxaliplatin and fluorouracil in patients with metastatic pancreatic cancer after prior therapy: SWOG S1115 study randomized clinical trial. JAMA Oncol.3, 516–522 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tolcher, A. W. et al. Antitumor activity in RAS-driven tumors by blocking AKT and MEK. Clin. Cancer Res.21, 739–748 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kloesch, B. et al. A GATA6-centred gene regulatory network involving HNFs and DeltaNp63 controls plasticity and immune escape in pancreatic cancer. Gut71, 766–777 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barretina, J. et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature483, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Andres, M. P. et al. GATA4 and GATA6 loss-of-expression is associated with extinction of the classical programme and poor outcome in pancreatic ductal adenocarcinoma. Gut72, 535–548 (2023). [DOI] [PubMed] [Google Scholar]
- 35.Kaufmann, O., Fietze, E., Mengs, J. & Dietel, M. Value of p63 and cytokeratin 5/6 as immunohistochemical markers for the differential diagnosis of poorly differentiated and undifferentiated carcinomas. Am. J. Clin. Pathol.116, 823–830 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Martens, S. et al. Discovery and 3D imaging of a novel DeltaNp63-expressing basal cell type in human pancreatic ducts with implications in disease. Gut71, 2030–2042 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Somerville, T. D. D. et al. TP63-mediated enhancer reprogramming drives the squamous subtype of pancreatic ductal Adenocarcinoma. Cell Rep.25, 1741–1755.e1747 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muckenhuber, A. et al. Pancreatic ductal adenocarcinoma subtyping using the biomarkers hepatocyte nuclear Factor-1A and Cytokeratin-81 correlates with outcome and treatment response. Clin. Cancer Res.24, 351–359 (2018). [DOI] [PubMed] [Google Scholar]
- 39.O’Kane, G. M. et al. GATA6 expression distinguishes classical and basal-like subtypes in advanced pancreatic cancer. Clin. Cancer Res.26, 4901–4910 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Apte, M. V., Pirola, R. C. & Wilson, J. S. Pancreatic stellate cells: a starring role in normal and diseased pancreas. Front. Physiol.3, 344 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Helms, E. J. et al. Mesenchymal lineage heterogeneity underlies nonredundant functions of pancreatic cancer-associated fibroblasts. Cancer Discov.12, 484–501 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohlund, D., Elyada, E. & Tuveson, D. Fibroblast heterogeneity in the cancer wound. J. Exp. Med.211, 1503–1523 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hingorani, S. R. et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell7, 469–483 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Lupo, F. et al. Axon guidance cue SEMA3A promotes the aggressive phenotype of basal-like PDAC. Gut73, 1321–1335 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boj, S. F. et al. Organoid models of human and mouse ductal pancreatic cancer. Cell160, 324–338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Filippini, D. et al. Immunoevolution of mouse pancreatic organoid isografts from preinvasive to metastatic disease. Sci. Rep.9, 12286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patel, A. P. et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science344, 1396–1401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mazur, P. K. et al. Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nat. Med.21, 1163–1171 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rue-Albrecht K., L. A., Soneson C., Stadler M. velociraptor: Toolkit for Single-Cell Velocity. (2023).
- 50.La Manno, G. et al. RNA velocity of single cells. Nature560, 494–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hingorani, S. R. et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell4, 437–450 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun.12, 1088 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schubert, M. et al. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun.9, 20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krishnamurty, A. T. et al. LRRC15(+) myofibroblasts dictate the stromal setpoint to suppress tumour immunity. Nature611, 148–154 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garcia-Alonso, L., Holland, C. H., Ibrahim, M. M., Turei, D. & Saez-Rodriguez, J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res.29, 1363–1375 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Badia, I. M. P. et al. decoupleR: ensemble of computational methods to infer biological activities from omics data. Bioinform. Adv.2, vbac016 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, Y. et al. Single-cell analysis of pancreatic ductal adenocarcinoma identifies a novel fibroblast subtype associated with poor prognosis but better immunotherapy response. Cell Discov.7, 36 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen, K. et al. Single-cell RNA-seq reveals dynamic change in tumor microenvironment during pancreatic ductal adenocarcinoma malignant progression. EBioMedicine66, 103315 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mello, A. M. et al. Hypoxia promotes an inflammatory phenotype of fibroblasts in pancreatic cancer. Oncogenesis11, 56 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peng, J. et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res.29, 725–738 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luo, H. et al. Pan-cancer single-cell analysis reveals the heterogeneity and plasticity of cancer-associated fibroblasts in the tumor microenvironment. Nat. Commun.13, 6619 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen, S. et al. Single-cell analysis reveals transcriptomic remodellings in distinct cell types that contribute to human prostate cancer progression. Nat. Cell Biol.23, 87–98 (2021). [DOI] [PubMed] [Google Scholar]
- 63.Chen, Z. et al. Single-cell RNA sequencing highlights the role of inflammatory cancer-associated fibroblasts in bladder urothelial carcinoma. Nat. Commun.11, 5077 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Givel, A. M. et al. miR200-regulated CXCL12beta promotes fibroblast heterogeneity and immunosuppression in ovarian cancers. Nat. Commun.9, 1056 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kieffer, Y. et al. Single-cell analysis reveals fibroblast clusters linked to immunotherapy resistance in cancer. Cancer Discov.10, 1330–1351 (2020). [DOI] [PubMed] [Google Scholar]
- 66.Mi, Z., Kuo, M. C. & Kuo, P. C. RNA aptamer targeting of adam8 in cancer growth and metastasis. Cancers15, 3254 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mosa, M. H. et al. A Wnt-induced phenotypic switch in cancer-associated fibroblasts inhibits EMT in colorectal cancer. Cancer Res.80, 5569–5582 (2020). [DOI] [PubMed] [Google Scholar]
- 68.Puram, S. V. et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell171, 1611–1624.e1624 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hugo, W. et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell165, 35–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rosenberg, J. E. et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet387, 1909–1920 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hallin, J. et al. Anti-tumor efficacy of a potent and selective non-covalent KRAS(G12D) inhibitor. Nat. Med.28, 2171–2182 (2022). [DOI] [PubMed] [Google Scholar]
- 72.Mucciolo, G. et al. EGFR-activated myofibroblasts promote metastasis of pancreatic cancer. Cancer Cell42, 101–118.e111 (2024). [DOI] [PubMed] [Google Scholar]
- 73.Szalai, B. & Saez-Rodriguez, J. Why do pathway methods work better than they should? FEBS Lett.594, 4189–4200 (2020). [DOI] [PubMed] [Google Scholar]
- 74.D’Agosto, S. et al. Loss of FGFR4 promotes the malignant phenotype of PDAC. Oncogene41, 4371–4384 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, W. & Liu, H. T. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res.12, 9–18 (2002). [DOI] [PubMed] [Google Scholar]
- 76.Juiz, N. et al. Basal-like and classical cells coexist in pancreatic cancer revealed by single-cell analysis on biopsy-derived pancreatic cancer organoids from the classical subtype. FASEB J.34, 12214–12228 (2020). [DOI] [PubMed] [Google Scholar]
- 77.Krieger, T. G. et al. Single-cell analysis of patient-derived PDAC organoids reveals cell state heterogeneity and a conserved developmental hierarchy. Nat. Commun.12, 5826 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Galbo, P. M. Jr., Zang, X. & Zheng, D. Molecular features of cancer-associated fibroblast subtypes and their implication on cancer pathogenesis, prognosis, and immunotherapy resistance. Clin. Cancer Res.27, 2636–2647 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cancer Genome Atlas Research, N. Comprehensive molecular profiling of lung adenocarcinoma. Nature511, 543–550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cancer Genome Atlas Research, N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature507, 315–322 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Robertson, A. G. et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell32, 204–220.e215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Duewell, P. et al. RIG-I-like helicases induce immunogenic cell death of pancreatic cancer cells and sensitize tumors toward killing by CD8(+) T cells. Cell Death Differ.21, 1825–1837 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinform.12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hanzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform.14, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun.8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stuart, T. et al. Comprehensive Integration of single-cell data. Cell177, 1888–1902.e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science352, 189–196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Andreatta, M. & Carmona, S. J. UCell: robust and scalable single-cell gene signature scoring. Comput. Struct. Biotechnol. J.19, 3796–3798 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zappia, L. & Oshlack, A. Clustering trees: a visualization for evaluating clusterings at multiple resolutions. Gigascience7, giy083 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lin, W. et al. Single-cell transcriptome analysis of tumor and stromal compartments of pancreatic ductal adenocarcinoma primary tumors and metastatic lesions. Genome Med.12, 80 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Steele, N. G. et al. Multimodal mapping of the tumor and peripheral blood immune landscape in human pancreatic cancer. Nat. Cancer1, 1097–1112 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell184, 3573–3587.e3529 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Franzen, O., Gan, L. M. & Bjorkegren, J. L. M. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database2019, baz046 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Durinck, S., Spellman, P. T., Birney, E. & Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc.4, 1184–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gulati, G. S. et al. Single-cell transcriptional diversity is a hallmark of developmental potential. Science367, 405–411 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
RNA-seq data generated in this study have been deposited in the GEO database under accession code: GSE246457. scRNA-seq data generated in this study have been deposited in the GEO database under accession code: GSE246458. Spatial transcriptomics data generated in this study have been deposited in the GEO database under accession code: GSE274665. The publicly available RNA-seq data from CCLE used in this study were downloaded from DepMap portal[https://depmap.org/portal/]. The publicly available RNA-seq data from TCGA used in this study were downloaded from firebrowse [http://firebrowse.org/?cohort=PAAD]. The publicly available RNA-seq data from ICGC used in this study were downloaded from ICGC data portal, now migrated in ICGC 25 K data. The publicly available RNA-seq data from PanCuRx used in this study are available upon request in the EGA database under accession code EGAS00001002543. The publicly available RNA-seq data from ImVigor trial70 used in this study are available within easierData R package. The publicly available mouse sc-RNA-seq data used in this study are available in the GEO database under accession code GSE129455. The publicly available human sc-RNA-seq data used in this study are available in: GEO database under accession codes GSE15477895, GSE15569896, GSE21034761; in GSA database under accession code CRA00116060 and in EGA database upon request under accession code EGAS0000100254317. The remaining data are available within the Article, Supplementary Information or Source Data file. Source data are provided with this paper.