Abstract
The accumulation of reactive oxygen species (ROS) significantly contributes to intervertebral disc degeneration (IDD), but the mechanisms behind this phenomenon remain unclear. This study revealed elevated ROS levels in the intervertebral discs (IVDs) of aged mice compared to those of younger mice. The local application of hydrogen peroxide (H2O2) near lumbar discs also induced ROS accumulation and IDD. Isobaric tags for relative and absolute quantitation (iTRAQ) analysis of discs from aged and H2O2-injected mice showed increased levels of YTH N6-methyladenosine RNA binding protein F2 (YTHDF2) and matrix metallopeptidase 1/3/7/9 (MMP1/3/7/9), along with decreased levels of forkhead box O3 (FOXO3) and TIMP1 (tissue inhibitor of metalloproteinases 1). Our experiments indicated that in nucleus pulposus (NP) cells and young mouse IVDs that were not exposed to ROS, FOXO3 recruited histone acetyltransferase CBP (CREB binding protein) and mediator complex subunit 1 (Med1) to activate TIMP1 expression, which inhibited MMP activity and prevented disc degeneration. However, ROS exposure activated YTHDF2 and promoted the degradation of m6A-modified FOXO3 mRNA, impairing FOXO3’s ability to activate TIMP1. This degradation exacerbated MMP activity and contributed to the degradation of the IVD extracellular matrix. Notably, administration of the YTHDF2 inhibitor DC-Y13-27 in older and H2O2-treated mice significantly enhanced FOXO3 and TIMP1 expression, reduced MMP activity, and mitigated IVD degeneration. Together, this study uncovers a novel ROS-regulated pathway in IDD, centered on the YTHDF2/FOXO3/TIMP1/MMPs axis, suggesting that targeting YTHDF2 may represent a promising therapeutic strategy for combating the progression of IDD.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-024-05503-w.
Keywords: Reactive oxygen species, Intervertebral disc degeneration, YTHDF2, FOXO3, TIMP1, MMP
Introduction
Intervertebral disc degeneration (IDD) is a prevalent musculoskeletal disorder that primarily presents as lower back pain and significantly contributes to disability and socioeconomic impact globally [1, 2]. Current clinical management approaches for IDD typically focus on pain relief in its early stages and surgical interventions in the advanced stages [1, 2]. Nonetheless, these strategies fail to address the underlying progression of the disease and are associated with various adverse effects, including gastrointestinal complications, increased risk of surgical revisions, mechanical failures, and further degeneration of adjacent discs [1, 2].
The incidence of IDD is largely attributed to the imbalance between anabolic and catabolic activities within the disc cells located in the nucleus pulposus (NP), annulus fibrosus (AF), and cartilaginous endplate (CEP) [3, 4]. These cells maintain the extracellular matrix (ECM), which mainly comprises collagen fibrils and proteoglycans [3, 4]. The progression of IDD is characterized by enhanced inflammation and an induction in degradative enzymes that lead to ECM breakdown and structural disc damage, including height reduction, fissuring of the AF, and loss of NP [5, 6]. Matrix metalloproteinases (MMPs) and a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTSs) are two major classes of enzymes that promote ECM degradation [7, 8]. MMPs and ADAMTSs degrade collagen and aggrecan, which are essential for disc structure and function. Their activities are typically regulated by tissue inhibitors of metalloproteinases (TIMPs) [7, 8]. In the pathogenesis of IDD, inflammatory cytokines like tumor necrosis factor alpha (TNF-α) and interleukin-1 beta (IL-1β) elevate MMP and ADAMTS expression but suppress TIMP production, creating an imbalance that favors ECM breakdown [7–9]. This disruption in ECM homeostasis accelerates the structural deterioration of the disc, underscoring the need for therapeutic strategies that restore this balance to potentially halt or reverse degeneration [7–9].
Emerging research highlights the significant role of reactive oxygen species (ROS) in the development of IDD. ROS, which mainly includes hydrogen peroxide (H2O2), hydroxyl radicals (•OH), and superoxide anion radicals (O2−•), are highly reactive molecules that induce oxidative stress, promoting inflammation, apoptosis, and cellular senescence in the disc [10–12]. Elevated ROS levels also downregulate essential ECM components, such as collagen type II and aggrecan, while upregulating catabolic agents like TNF-α, MMPs, and aggrecanases, thereby accelerating the IDD process [10–12]. Although it is established that the accumulation of ROS influences various biological processes associated with IDD, the underlying molecular mechanisms remain unclear.
N6-methyladenosine (m6A) represents the most prevalent internal modification of messenger RNA (mRNA) in higher eukaryotes and plays a crucial role in RNA metabolism by influencing mRNA stability, splicing, and translation [13–15]. The m6A modification is a dynamically reversible process regulated by three primary classes of proteins: “writers”, “erasers”, and “readers”. “Writers”, such as methyltransferase 3 (METTL3), METTL14, and Wilms tumor 1-associating protein (WTAP), are a group of adenosine methyltransferases that add m6A marks RNA, thereby enhancing its stability and localization [13–15]. In contrast, “erasers” like fat mass and obesity-associated protein (FTO) and AlkB family member 5 (ALKBH5), are RNA demethylases that remove m6A marks, influencing different biological processes [13–15]. “Readers” such as YT521-B homology domain proteins (YTHDC1-2, YTHDF1-3) and insulin-like growth factor 2 mRNA-binding proteins (IGF2BP1, 2, and 3), are m6A-binding proteins that recognize m6A sites and regulate mRNA stability, translation, and splicing [13–15]. This intricate network of enzymes plays a pivotal role in gene expression regulation and is implicated in various diseases, highlighting its significance in cellular regulation and disease pathology [13–15]. Recent studies have linked m6A modification to the pathogenesis of IDD. For instance, increased expression of ALKBH5 during IVD degeneration and NP cell (NPC) senescence has been observed, contributing to NPC senescence by demethylating DNMT3B transcripts, thereby enhancing DNMT3B expression through less YTHDF2 recognition [16]. Differential expression of m6A regulators such as RNA binding motif protein 15 (RBM15), YTHDC1, YTHDF3, heterogeneous nuclear ribonucleoproteins A2/B1 (HNRNPA2B1), and ALKBH5 has been noted in IDD models. Notably, YTHDC1 may affect the immune microenvironment of IDD through pathways involving Abelson tyrosine-protein kinase 1 (ABL1) and TXK tyrosine kinase (TXK) [17]. Despite these advances, the expression patterns and specific functions of these m6A regulators and their target genes in IDD remain to be fully elucidated.
While the role of ROS accumulation in the onset of IDD is crucial, the underlying mechanisms remain unclear. To investigate how ROS accumulation induces IDD, this study compared the whole-genome protein expression differences in the IVDs of young and aged mice, as well as in mice injected with PBS versus H2O2. Our findings revealed significant upregulation of YTHDF2 and MMP1/3/7/9, along with notable downregulation of the transcription factor FOXO3 (forkhead box O3) and TIMP1 in aged and H2O2-administrated mice. At both the cellular level and animal models, we demonstrated that ROS accumulation activated YTHDF2, leading to the degradation of FOXO3 mRNA. This degradation impaired FOXO3’s ability to transcriptionally activate TIMP1, resulting in increased expression of MMP1/3/7/9 and promoting the degradation of the IVD ECM, thereby causing IDD. This key discovery elucidates the molecular mechanism by which ROS accumulation modulates FOXO3 mRNA m6A modification and regulates the expression of downstream genes, providing novel insights into how ROS accumulation contributes to IDD.
Materials and methods
Animal experiments
C57BL/6 mice were obtained from the Beijing Vital River Laboratory Animal Technology Company (Beijing, China). Upon arrival, the mice were allowed a three-week acclimatization period in the laboratory environment before commencing any experimental interventions. They were housed under controlled conditions with a 12-hour light/dark cycle, a temperature of 22 °C, and 50–60% relative humidity. The mice had unrestricted access to standard rodent chow and water. To assess IDD in aged mice, a cohort (n = 8) was raised under the aforementioned standard conditions until they reached 52 weeks of age. These mice underwent magnetic resonance imaging (MRI) using a United-Imaging system (Shanghai, China; Model UMR770). For the experimental induction of IDD using H2O2, 8-week-old mice (22–25 g, n = 16) were randomly divided into two groups. Following anesthesia with 2% isoflurane (Sigma-Aldrich; Shanghai, China; #792632) at a flow rate of 0.6 L/min, one group (n = 8) received injections of 50 µL phosphate-buffered saline (PBS) (Thermo Fisher; Shanghai, China; #10010023), while the other group (n = 8) received 50 µL 2.5% H2O2 (Sigma-Aldrich; #H1009). These injections were administered subcutaneously near the lumbar discs every five days for 12 weeks. Post-experiment, MRI was conducted to evaluate the progression of IDD. To investigate the effect of the YTHDF2 inhibitor (DC-Y13-27) (MedChemExpress; Monmouth Junction, NJ, USA; #HY-154919) on IDD prevention, mice were randomly assigned to four groups. The treatments were administered as follows: the first group received 50 µL PBS, the second group received 50 µL PBS plus 5 mg/kg DC-Y13-27, the third group received 50 µL H2O2, and the fourth group received 50 µL H2O2 plus 5 mg/kg DC-Y13-27. These injections were administered subcutaneously near the lumbar discs every five days for 12 weeks. Following the treatment period, MRI assessments were conducted. Subsequently, IVDs from four groups were harvested for further analyses, including protein and RNA isolation, as well as histological evaluations.
Cell culture and transfection
Mouse AF (#CSC-C5389S) and NP (#CSC-C5390S) cells were acquired from Creative Bioarray (Shirley, NY, USA). These cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Thermo Fisher; #11965092) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher; #16000044) and 1% penicillin/streptomycin (Thermo Fisher; #15140122). Cultures were maintained in a humidified incubator (Thermo Fisher; #51033557) at 37 °C with 5% CO2. Upon reaching 80% confluence, cells were passaged using 0.25% trypsin-EDTA (Thermo Fisher; #25200072). For transfection experiments, cells were seeded at a density of 1 × 105 cells per well in 6-well plates and allowed to adhere overnight. Transfection was then performed using Lipofectamine 3000 reagent (Thermo Fisher; #L3000150), following the manufacturer’s protocol. The short hairpin RNAs (shRNAs) and plasmids utilized for transfection are listed in Tables S1 and S2. Transfection efficiency was evaluated 48 h post-transfection via real-time quantitative PCR (RT-qPCR) and Western blot analyses, enabling the assessment of gene silencing and protein expression levels.
Determination of ROS levels
To accurately measure ROS levels in the IVDs of C57BL/6 mice, the discs were carefully dissected and immediately immersed in ice-cold PBS to remove any residual blood or debris. The discs were then homogenized in a ice-cold buffer consisting of 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 10% glycerol, 0.1% SDS, and 5 mM sodium pyruvate, supplemented with a 1 × protease inhibitor cocktail (Thermo Fisher; #78429). The homogenate was then centrifuged at 13,000 g for 15 minutes, and the clear supernatant was collected for further analysis. ROS levels were detected using the peroxide-sensitive fluorescent probe 2’,7’-dichlorofluorescein diacetate (DCFH-DA) (Sigma-Aldrich; #D6883), following the manufacturer’s instructions. Fluorescence measurements were conducted on the Varioskan LUX microplate reader (Thermo Fisher; #VL0000D0) using an excitation wavelength of 485 nm and an emission wavelength of 530 nm. Quantification of ROS was accomplished by comparing the fluorescence intensities against a standard curve generated from known concentrations of H2O2. For intracellular ROS measurement, cells seeded in 6-well plates were washed twice with PBS and lysed in the same ice-cold lysis buffer used for the disc tissues. After centrifugation at 13,000 g for 15 min, the supernatant was incubated with DCFH-DA following the manufacturer’s instructions. Fluorescence was then recorded and quantified.
Total RNA isolation and RT-qPCR analysis
Mouse IVD tissues were homogenized and cultured cells were lysed in TRIzol Reagent (Thermo Fisher; #15596026). Following phase separation using chloroform (Sigma-Aldrich; #650498), RNA was precipitated with isopropanol (Sigma-Aldrich; #RTC000079), washed with 75% ethanol (Sigma-Aldrich; #E7023), and resuspended in RNase-free water. RNA integrity and concentration were assessed via agarose gel electrophoresis and NanoDrop spectrophotometer (Thermo Fisher; #ND-8000-GL). For RT-qPCR, 1 µg of total RNA was reverse transcribed into cDNA using the SuperScript III First-Strand Synthesis SuperMix (Thermo Fisher; #18080400), and qPCR was conducted using SYBR Green PCR Master Mix (Thermo Fisher; #4309155) with gene-specific primers (Table S3). The thermal cycling program consisted of an initial denaturation at 95 °C for 5 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Relative gene expression was calculated using the comparative cycle threshold (CT) (2ΔΔCT) method, normalized against beta-Actin.
Total protein isolation and Western blot analysis
To analyze protein expression in mouse IVD tissues and cultured cells, tissues were immediately frozen and homogenized in ice-cold radio-immunoprecipitation assay (RIPA) buffer supplemented with a 1 × protease inhibitor cocktail (Thermo Fisher; #78429). Cultured cells were washed twice with PBS before being directly lysed in the same RIPA buffer. The resulting lysates were incubated on ice for 30 min to ensure thorough lysis and then centrifuged at 14,000 g to remove insoluble material, allowing the collection of soluble proteins. Protein concentrations in the supernatants were determined using a NanoDrop spectrophotometer. Following quantification, proteins were denatured and separated by 10% SDS-PAGE gel electrophoresis. The separated proteins were then transferred onto polyvinylidene difluoride (PVDF) membranes (Thermo Fisher; #LC2000). For immunoblotting, membranes were blocked using 5% fat-free milk (Thermo Fisher; #50-488-785) to prevent non-specific binding and then incubated sequentially with specific primary and secondary antibodies (Table S4). After each antibody incubation, the membranes were thoroughly washed five times with PBST (PBS + 0.1% Tween-20) to remove unbound antibodies. Protein detection was carried out using an enhanced chemiluminescence (ECL) system (Thermo Fisher; #A38554), and the signals were captured with the ChemiDoc MP Imaging System (Bio-Rad, Shanghai, China). Quantification of protein signal intensities was performed using Image J software (National Institutes of Health, USA; version 1.54f), with values normalized against loading controls.
Isobaric tags for relative and absolute quantitation (iTRAQ) analysis
Mouse IVD tissues were harvested under sterile conditions and immediately immersed in liquid nitrogen to maintain protein integrity. These tissues were then homogenized in a lysis buffer containing 8 M urea (Sigma-Aldrich; #U5128), 2% 3-(3-cholamidopropyl) dimethylammonio-1-propanesulfonate (CHAPS) (Thermo Fisher; #28299), 30 mM HEPES (Thermo Fisher; #15630130), and 1 × concentration of protease inhibitor cocktail (Thermo Fisher; #78429). The homogenates were centrifuged at 14,000 g for 15 min, and the protein concentration in the supernatants was determined using a NanoDrop spectrophotometer. For protein processing, samples were reduced by treating them with 5 mM dithiothreitol (DTT) (Thermo Fisher; #R0861) at 56 °C for one hour, followed by alkylation with 15 mM iodoacetamide (Thermo Fisher; #A39271) in the dark at room temperature for 20 min. The protein solution was then diluted with 100 mM triethylammonium bicarbonate (TEAB) (Thermo Fisher; #90114) to reduce the urea concentration below 2 M. Overnight digestion was performed at 37 °C using trypsin (Thermo Fisher; #90057) at a trypsin-to-protein ratio of 1:50. After digestion, peptides were desalted using a Strata X C18 solid phase extraction (SPE) column (Phenomenex, CA, USA) and vacuum-dried. The peptides were reconstituted in 0.5 M TEAB and processed labelling with the iTRAQ Reagents Multiplex Kit (Sigma-Aldrich; #4352135). After a 2-hour incubation at room temperature, the peptides were pooled, desalted, and dried by vacuum centrifugation. Peptide fractionation was performed by reconstituting the dried mixture in a strong cation-exchange (SCX) loading buffer (0.1 M NH₄Cl and 10 mM K₂HPO₄, pH 7.0) and applying it to SCX chromatography (Phenomenex; #8B-SCX-100), where peptides were eluted using a salt gradient and collected in fractions. Each fraction was analyzed by liquid chromatography with tandem mass spectrometry (LC-MS/MS) on a high-resolution mass spectrometer (Thermo Fisher; #IQLAAEGAAPFADBMBHQ) in data-dependent acquisition mode. The MS/MS spectral data were processed using Mascot. Differentially expressed proteins (DEPs) were further subjected to Gene Ontology (GO) analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID) and the Protein ANalysis THrough Evolutionary Relationships (PANTHER) databases to determine GO terms in Biological Processes, Cellular Components, and Molecular Functions compared to a control dataset.
RNA stability assay
RNA stability was determined following a previous method [18]. Briefly, cells were treated with 2 µg/mL actinomycin D (Sigma-Aldrich; #A1410) to inhibit new RNA synthesis. Cells were collected at 0, 3, and 6 h post-treatment for RNA isolation. Total RNA was extracted, reverse transcribed into cDNA, and analyzed via RT-qPCR using specific primers (Table S3).
RNA immunoprecipitation (RIP)
The RIP assay was conducted using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Sigma-Aldrich; #17–700) following the manufacturer’s protocol. Briefly, cells were lysed in ice-cold RIPA buffer supplemented with 1 × protease inhibitor cocktail. After lysis, the cell extracts were centrifuged at 14,000 g for 15 min to separate the supernatant. The cleared lysate was then incubated with magnetic bead protein A/G (Thermo Fisher, #88802), which had been pre-conjugated with either 5 µg of control IgG antibody (Thermo Fisher, #02-6102) or anti-YTHDF2 antibody (Thermo Fisher, #MA5-47011). This mixture was rotated overnight at 4 °C to allow thorough antibody-antigen binding. The following day, RNA bound to the immunocomplexes was isolated using the QIAwave RNA Mini Kit (QIAGEN, Shanghai, China; #74534). The purified RNA was then subjected to RT-qPCR to quantify the relative expression levels of the FOXO3 gene with primers listed in Table S3.
Hematoxylin and Eosin (H&E) staining of IVDs
The H&E assay was performed to assess the histological structure of IVD tissues following a previously described method [19]. Briefly, IVDs were fixed with in 10% neutral buffered formalin (Sigma-Aldrich; #HT501128) for 48 h to preserve cellular integrity and structure. After fixation, the tissues were dehydrated through a graded ethanol series (70%, 80%, 95%, and 100%) (for pure ethanol, Sigma-Aldrich; #E7023), cleared in xylene (Sigma-Aldrich; #534056), and embedded in paraffin wax (Sigma-Aldrich; #76242) to ensure optimal tissue preservation for sectioning. Tissue sections were into 5 μm-thick slices using a microtome, deparaffinized in xylene, and rehydrated through a descending series of ethanol concentrations. The rehydrated sections were stained with the H&E staining kit (Abcam, Shanghai, China; #ab245880) according to the manufacturer’s protocol. The sections were then examined under a light microscope (Olympus Corporation; Tokyo, Japan; #BX53F) to observe the detailed cellular architecture and any pathological alterations within the IVD tissues. Histological images were independently evaluated by two observers using an established scoring system [20]. For the AF, the scoring criteria were as follows: 0 indicates normal; 1 signifies mildly serpentine without bulging beyond the endplate edge; 2 denotes moderately serpentine with slight bulging beyond the endplate edge; 3 describes severely serpentine with obvious bulging beyond the endplate edge; 4 characterizes severely serpentine and ruptured; 5 is classified as indistinct. For the NP, the scores are defined as: 0 for normal; 1 for condensed; 2 indicating the presence of chondrocyte-like cells with residual NP matrix; 3 for a widespread presence of chondrocyte-like cells; 4 describes mild replacement by fibrous cartilaginous tissue; 5 denotes moderate to severe replacement by fibrous cartilaginous tissue.
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was conducted following an established protocol [21]. Briefly, cells were fixed with 1% formaldehyde (Sigma-Aldrich; #47608) at room temperature for 15 min to cross-link proteins and DNA. The cross-linking reaction was stopped by the addition of 5 M glycine to a final concentration of 0.125 M. Cells were then rinsed with cold PBS buffer and lysed using ChIP-specific lysis buffer (Santa Cruz Biotechnology, Shanghai, China; #sc-45000) supplemented with a 1 × protease inhibitor cocktail. Chromatin in the lysates was sonicated to generate fragments ranging from 200 to 500 base pairs in length, and the efficiency of this shearing was confirmed via agarose gel electrophoresis. The chromatin fragments were incubated overnight at 4 °C with agarose beads conjugated to antibodies specific to YTHDF2, FOXO3, Med1, CBP, or control IgG (Table S4). To minimize nonspecific binding, the bead-bound complexes were washed five times with ChIP wash buffer (Santa Cruz Biotechnology; #sc-45002). The protein-DNA complexes were subsequently eluted using elution buffer (Santa Cruz Biotechnology; #sc-45003) and treated with Proteinase K (Thermo Fisher; #EO0491) to reverse the cross-links and digest proteins, thereby releasing the DNA. The DNA was purified using a DNA purification kit (ZYMO research, Beijing, China; #D5205) and finally, the purified DNA samples were analyzed by RT-qPCR with the primers listed in Table S5.
Immunoprecipitation (IP) and co-immunoprecipitation (co-IP) assays
For the IP assay, cells and IVDs were harvested and lysed in IP lysis buffer (Thermo Fisher; #87787) supplemented with protease and phosphatase inhibitors (Thermo Fisher; #A32959). The lysates were incubated on ice for 30 min and then centrifuged at 14,000 × g for 10 min at 4 °C to remove insoluble material. The clarified supernatant was incubated overnight at 4 °C with gentle agitation using anti-FOXO3 conjugated to protein A agarose beads (Thermo Fisher; #20334). The beads were washed four times with IP lysis buffer to remove non-specifically bound proteins. Proteins bound to the beads were eluted by boiling in SDS sample buffer (Bio-Rad, Shanghai, China; #1610747) for 5 min. The eluted proteins were separated by SDS-PAGE and visualized using the Pierce Silver Stain Kit (Thermo Fisher; # PI24612). Protein bands were excised, digested with trypsin, and analyzed by LC-MS/MS. Data analysis was performed using the Mascot software. For the co-IP assay, cells expressing various combinations of Flag-tagged and Myc-tagged vectors were lysed using IP lysis buffer. Immunoprecipitation was performed using anti-Flag-agarose (Thermo Fisher; #A36801) and anti-Myc-agarose (Sigma-Aldrich; #A7470) beads. The precipitated proteins were separated by 10% SDS-PAGE and analyzed by Western blotting with anti-Flag and anti-Myc antibodies to confirm the presence of and interaction between the tagged proteins.
Statistical analysis
All experiments were conducted independently and repeated three times. Data are presented as the mean ± standard deviation (SD). Statistical analyses were carried out using IBM SPSS (International Business Machines, Statistical Package for the Social Sciences) Statistics 21 (IBM SPSS Inc., USA). Group differences were evaluated via Analysis of Variance (ANOVA), followed by post hoc tests for multiple comparison analyses. In figures, significance levels are denoted as follows: P < 0.05 by one asterisk (*), P < 0.01 by two asterisks (**), and P < 0.001 by three asterisks (***).
Results
ROS accumulation accelerated the degeneration of lumbar IVDs in mice
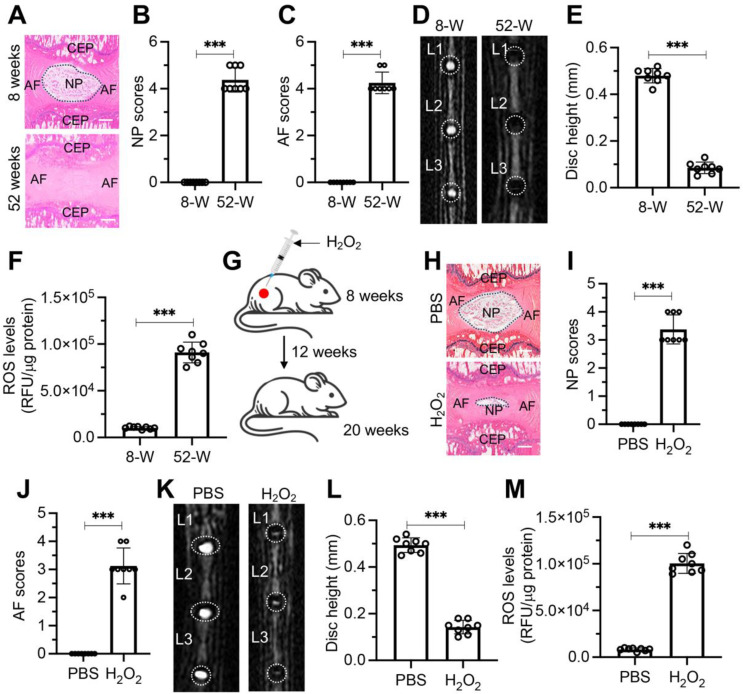
ROS have been linked to IVD degeneration [10–12]. To validate this association, we initially assessed the degenerative changes in the IVDs of mice at 8 weeks and 52 weeks of age. H&E staining showed no signs of degeneration in the IVDs of 8-week-old mice, whereas significant degeneration was observed in the IVDs of 52-week-old mice (Fig. 1A). Quantitative scoring of the H&E-stained sections revealed that the NP and AF tissues in 52-week-old mice had scores ranging from 3 to 5, indicating severe degeneration. In contrast, both NP and AF tissues in the 8-week-old mice received scores of 0 (Fig. 1B and C). MRI imaging of lumbar IVDs further supported these findings, showing no degeneration in 8-week-old mice, but severe degeneration in the 52-week-old group (Fig. 1D). Measurement of disc height indicated that the IVD disc height in 52-week-old mice was significantly lower than that in 8-week-old mice (Fig. 1E). Additionally, ROS levels in the lumbar IVDs of 52-week-old mice were approximately eight times higher than in the 8-week group (Fig. 1F).
Fig. 1.
ROS accumulation resulted in IDD in mice. (A) Representative H&E stained sections of L1/L2 IVDs from mice aged 8 weeks and 52 weeks. Scale bars = 100 μm. (B) Quantitative scoring of the NP in degenerative IVDs shown in (A). (C) Quantitative scoring of the AF in degenerative IVDs illustrated in (A). (D) Representative MRI scans of lumbar IVDs ranging from L1 to L3 in mice aged 8 weeks and 52 weeks. (E) Measurement of disc height in L1/L2 IVDs comparing 8-week-old and 52-week-old mice. (F) Levels of ROS in lumbar IVDs of mice at 8 weeks and 52 weeks of age. (G) Schematic representation of H2O2 injection procedure near lumbar discs in mice. (H) Representative H&E staining images of lumbar IVDs (L1/L2) from mice administered with PBS and H2O2. Scale bars = 100 μm. (I) Quantified NP scores of IVDs demonstrating degeneration in mice treated as described in (H). (J) Quantified AF scores of IVDs showing degenerative changes in mice treated as indicated in (H). (K) Representative MRI scans of lumbar IVDs (from L1 to L3) in mice treated with PBS and H2O2. (L) Disc height measurements in L1/L2 IVDs from PBS and H2O2-treated mice. (M) ROS levels in lumbar IVDs of mice treated with PBS and H2O2. ***P < 0.001
To demonstrate that elevated levels of ROS can directly promote IDD, we administered subcutaneous injections of 50 µL of 2.5% H2O2 or PBS (control) near the lumbar discs of mice every five days for 12 weeks (Fig. 1G). At the end of the experiment, H&E staining revealed no degeneration in the discs of PBS-injected mice, whereas severe degeneration was observed in the discs of H2O2-injected mice (Fig. 1H). The H&E-stained sections were scored, and both NP and AF tissues in the H2O2-injected mice received scores of 3 to 5, indicating severe degeneration. In contrast, the NP and AF tissues in the PBS-injected mice scored 0 (Fig. 1I and J). MRI imaging of the lumbar IVDs further confirmed these findings, showing no signs of degeneration in the PBS group but significant degeneration in the H2O2 group (Fig. 1K). IVD disc height in H2O2 group mice was dramatically decreased than that in the PBS group (Fig. 1L). ROS levels in the lumbar IVDs of the H₂O₂ group were approximately 12 times higher than in the control group (Fig. 1M). These results suggest that the accumulation of ROS can indeed accelerate the degeneration of IVDs.
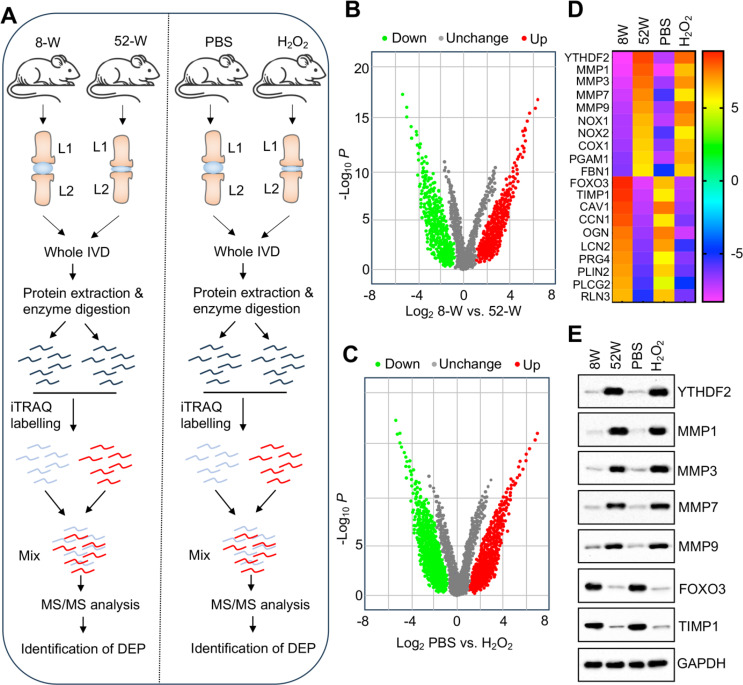
Identification of DEPs in IVDs from aged and H2O2-administrated mice
To investigate the molecular mechanisms by which ROS accumulation leads to IDD, we conducted iTRAQ protein profiling on two sets of IVDs: young vs. aged, and PBS- vs. H2O2-injected discs (n = 3 in each group) (Fig. 2A). The analysis revealed significant proteomic changes in the 52-week-old IVDs, identifying 1391 DEPs with fold changes of ≥ 1.5 and P ≤ 0.05 (Fig. 2B). Of these, 702 proteins were upregulated, while 689 were downregulated (Fig. 2B). Similarly, in the H2O2-injected mouse IVDs, 1216 proteins displayed significant alterations in expression, with 662 upregulated and 554 downregulated under the same criteria (Fig. 2C). Comparative analysis between the 52-week-old group and the H2O2 group revealed consistent upregulation of YTHDF2 and four matrix metalloproteinases (MMP1, MMP3, MMP7, and MMP9), alongside significant downregulation of the transcription factor FOXO3 and TIMP1 in both groups (Fig. 2D and Table S6). These consistent changes suggest that these proteins may play key roles in the pathogenesis of IDD induced by ROS accumulation.
Fig. 2.
Identification of DEPs in degenerative IVDs from aged and H2O2-injected mice. (A) Schematic of the experimental workflow for iTRAQ-based proteomic analysis conducted on degenerative IVDs from aged and H2O2-injected mice. (B and C) Volcano plots illustrating the log2 fold changes and statistical significance (− log10 p-value) of protein alterations. Red dots represent significantly upregulated proteins; green dots denote significantly downregulated proteins. (B) Comparison between 8-week-old and 52-week-old mice. (C) Comparison between PBS-treated and H2O2-injected mice. (D) Identification of proteins showing the most significant upregulation and downregulation in IVDs of 52-week-old mice and H2O2-injected mice. (E) Validation of protein expression levels for YTHDF2, FOXO3, TIMP1, MMP1, MMP3, MMP7, and MMP9 in IVDs from 8-week-old mice, 52-week-old mice, PBS-injected mice (20 weeks old), and H2O2-injected mice (20 weeks old). For each group, equal weights (0.05 g) of three independent IVDs were combined to generate homogenates, which were then analyzed via immunoblotting to assess protein levels
Furthermore, we performed Gene Ontology (GO) and Clusters of Orthologous Groups (COG) analyses on the DEPs. In the 52-week group, the upregulated proteins were primarily involved in biological processes such as cellular response to ROS, ECM organization, MMP activity, negative regulation of cell proliferation, and transcription activation (Fig. S1). In the H2O2-treated group, the upregulated proteins predominantly participated in biological processes including cellular response to ROS, ECM organization, MMP activity, and oxidative phosphorylation (Fig. S2). Additionally, analyses of the cellular components and molecular functions of these DEPs were performed, with results presented in Fig. S1 and S2.
To validate the iTRAQ analysis results, we assessed the protein expression levels of YTHDF2, MMP1, MMP3, MMP7, MMP9, FOXO3, and TIMP1 in the lumbar IVDs of four groups of mice. The results were consistent with the iTRAQ findings. Specifically, the protein levels of YTHDF2, MMP1, MMP3, MMP7, and MMP9 was significantly elevated in both 52-week-old mice and H2O2-injected mice, while the expression of FOXO3 and TIMP1 was significantly reduced in these groups (Fig. 2E).
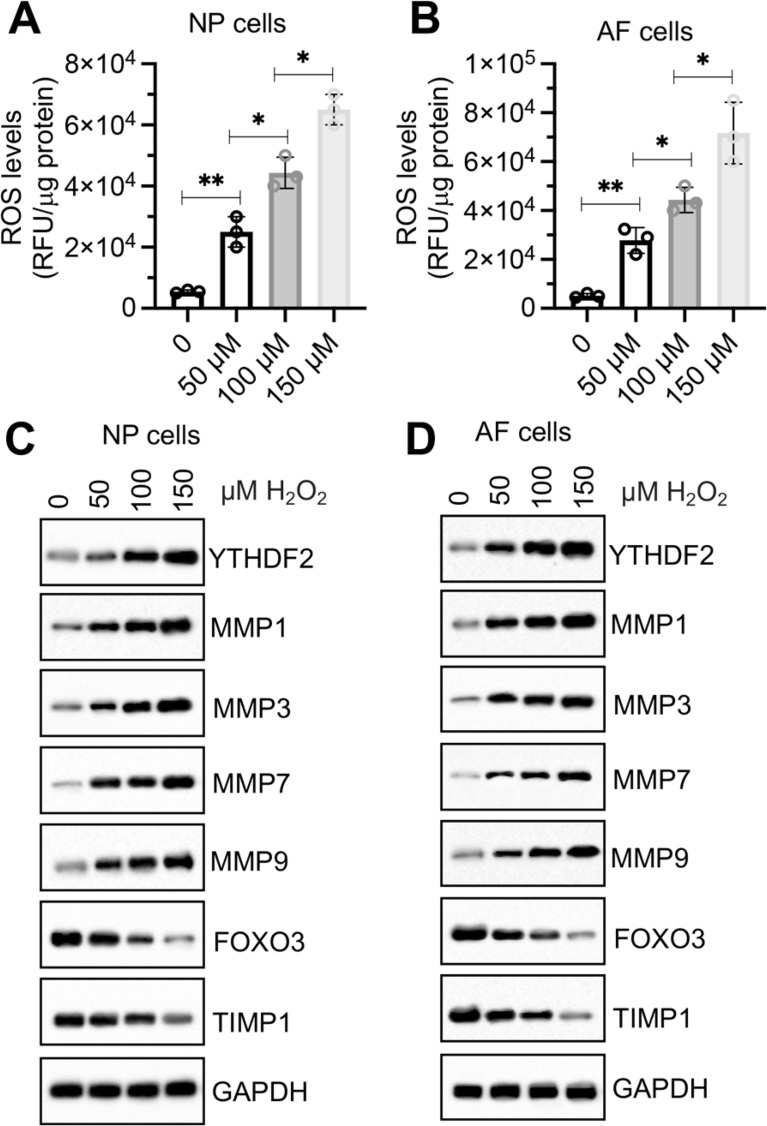
In vitro H2O2 treatment caused similar changes in protein levels of YTHDF2, FOXO3, TIMP1, and MMPs in NP and AF cells as observed in H2O2-injected mice
To verify that the observed changes in the expression of YTHDF2, FOXO3, TIMP1, MMP1, MMP3, MMP7, and MMP9 in aged and H2O2-injected mouse IVDs were induced by ROS, we treated mouse NP and AF cells with increasing concentrations of H2O2 (0, 50, 100, and 150 µM). Following H2O2 treatment, a rapid increase in intracellular ROS levels was observed in both NP and AF cells (Fig. 3A and B). Immunoblotting results revealed a dose-dependent increase in the protein levels of YTHDF2, MMP1, MMP3, MMP7, and MMP9 with increasing concentrations of H2O2 (Fig. 3C and D). Conversely, the expression levels of FOXO3 and TIMP1 decreased progressively with increasing H₂O₂ concentrations (Fig. 3C and D). These findings suggest that the expression of YTHDF2, FOXO3, TIMP1, MMP1, MMP3, MMP7, and MMP9 is directly influenced by ROS levels.
Fig. 3.
H2O2 dose-dependently changed the expression levels of YTHDF2/FOXO3/TIMP1/MMPs in NP and AF cells. (A and B) Changes in intracellular ROS levels in NP and AF cells after treatment with varying concentrations of H2O2 (0, 50, 100, and 150 µM) for 6 h. ROS levels were quantified using DCFH-DA staining. (A) NP cells. (B) AF cells. (C and D) Protein levels of YTHDF2, FOXO3, TIMP1, MMP1, MMP3, MMP7, and MMP9 in NP (C) and AF (D) cells following H2O2 treatment. *P < 0.05, **P < 0.01
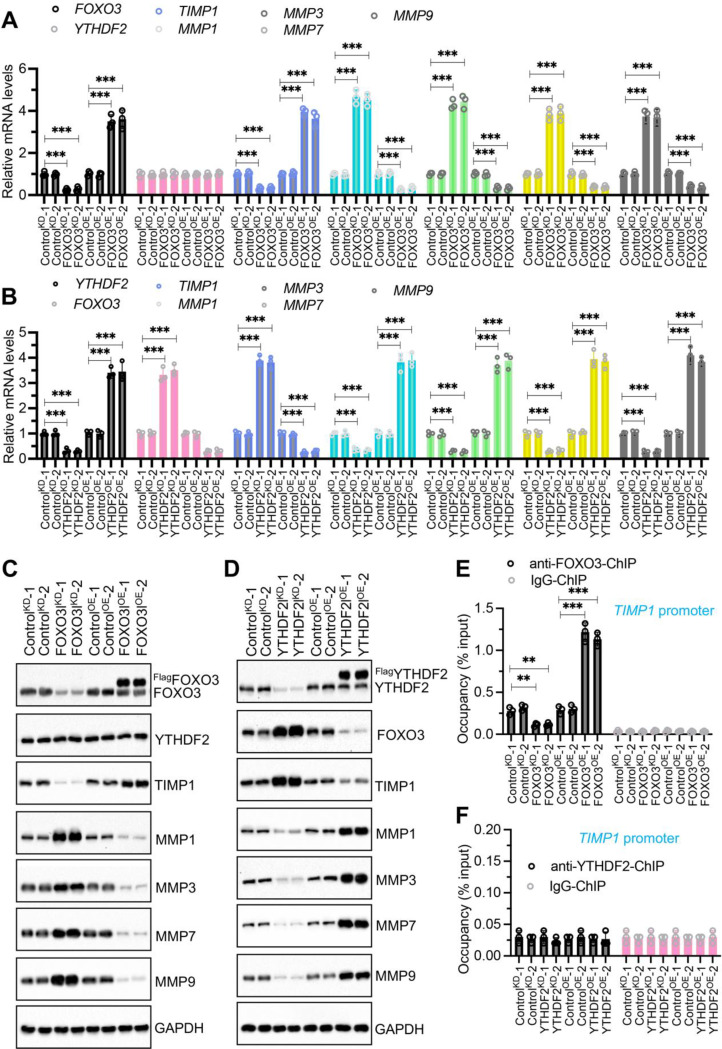
YTHDF2 was an upstream regulator of FOXO3, TIMP1, and MMP1/3/7/9
Among YTHDF2, FOXO3, TIMP1, MMP1, MMP3, MMP7, and MMP9, YTHDF2 is an m6A reader that selectively recognizes and binds to the m6A-modified RR(m6A)CH consensus sequence, whereas FOXO3 functions as a transcription factor. Thus, both YTHDF2 and FOXO3 have the potential to regulate the expression of TIMP1 and MMP1, MMP3, MMP7, and MMP9. To investigate whether YTHDF2 and FOXO3 are involved in regulating the expression of TIMP1 and MMP1/3/7/9, we generated knockdown (KD) and overexpression (OE) cell lines for YTHDF2 and FOXO3 in NP cells (Fig. S3). Using these cell lines, we measured the mRNA and protein levels of YTHDF2, FOXO3, TIMP1, MMP1, MMP3, MMP7, and MMP9. Our results indicated that altering FOXO3 levels did not affect the expression of YTHDF2 but did lead to changes in the expression of TIMP1 and MMP1/3/7/9 mRNA levels (Fig. 4A). Specifically, knockdown of FOXO3 resulted in decreased TIMP1 expression and increased expression of MMP1/3/7/9 (Fig. 4A). Conversely, overexpression of FOXO3 upregulated TIMP1 expression and reduced the levels of MMP1/3/7/9 (Fig. 4A). Unlike FOXO3, modulation of YTHDF2 levels affected the expression of FOXO3, TIMP1, and MMP1/3/7/9 (Fig. 4B). Knockdown of YTHDF2 upregulated FOXO3 and TIMP1 expression, but downregulated MMP1/3/7/9 (Fig. 4B). Overexpression of YTHDF2 had the opposite effect, decreasing FOXO3 and TIMP1 levels while increasing MMP1/3/7/9 expression (Fig. 4B). Immunoblot results confirmed that protein expression patterns of YTHDF2, FOXO3, TIMP1, MMP1, MMP3, MMP7, and MMP9 were consistent with their corresponding mRNA changes in both FOXO3KD/OE and YTHDF2KD/OE cell lines (Fig. 4C and D). These findings suggest that FOXO3 functions as an upstream regulator of TIMP1 and MMP1/3/7/9, while YTHDF2 acts upstream of FOXO3.
Fig. 4.
Impact of FOXO3 and YTHDF2 Knockdown and overexpression on the expression of TIMP1 and MMPs. (A) mRNA expression levels of YTHDF2, TIMP1, and MMPs in FOXO3KD and FOXO3OE cells. Quantitative PCR was performed on RNA from control knockdown (ControlKD-1 and ControlKD-2), FOXO3 knockdown (FOXO3KD-1 and FOXO3KD-2), control overexpression (ControlOE-1 and ControlOE-2), and FOXO3 overexpression (FOXO3OE-1 and FOXO3OE-2) cell lines (NP background) to assess levels of FOXO3, YTHDF2, TIMP1, MMP1, MMP3, MMP7, and MMP9. (B) mRNA expression levels of FOXO3, TIMP1, and MMPs in YTHDF2KD and YTHDF2OE cells. RNA from ControlKD-1/2, YTHDF2KD-1/2), ControlOE-1/2, and YTHDF2OE-1/2 cell lines (NP background) was analyzed to detect the expression levels of the same genes as in (A) by RT-qPCR. (C) Protein expression of YTHDF2, TIMP1, and MMPs in FOXO3KD and FOXO3OE cells. Protein samples from cells in (A) were analyzed by Western blot to detect levels of the indicated proteins. (D) Protein expression of FOXO3, TIMT1, and MMPs in YTHDF2KD and YTHDFOE cells. Protein samples from cells in (B) were subjected to Western blot analysis to detect levels of the indicated proteins. (E) The binding of FOXO3 on the promoter of TIMP1 by ChIP assay using cells in (A). (F) The binding of YTHDF2 on the promoter of TIMP1 by ChIP assay using cells in (B). **P < 0.01, ***P < 0.001
Additionally, we utilized ChIP assays to investigate whether YTHDF2 and FOXO3 could bind to the promoters of TIMP1 and MMP1/3/7/9. Our findings indicated that FOXO3 can bind to the TIMP1 promoter but not to the promoters of MMP1/3/7/9 (Fig. 4E and S4A-S4D). The binding of FOXO3 to the TIMP1 promoter was significantly reduced in FOXO3KD cells and notably increased in FOXO3OE cells (Fig. 4E). YTHDF2, however, did not show binding to the promoters of either TIMP1 or MMP1/3/7/9 (Fig. 4F and S4E-S4H). These results suggest that FOXO3 may regulate TIMP1 expression by directly binding to its promoter, and that TIMP1 subsequently influences the expression of MMP1/3/7/9. YTHDF2 appears to influence the expression of TIMP1 and MMP1/3/7/9 indirectly thorough a FOXO3-dependent mechanism.
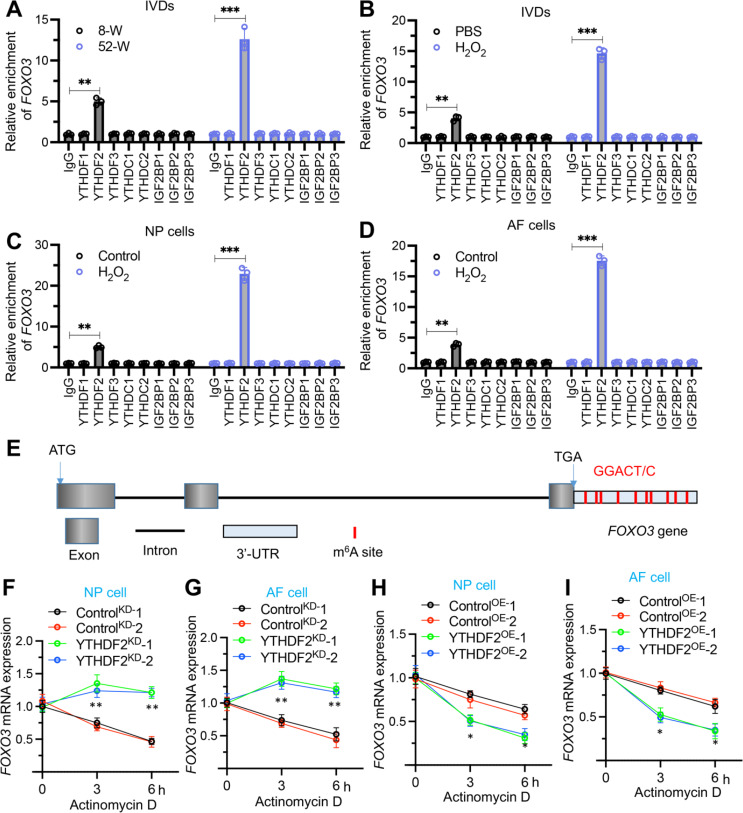
YTHDF2 was required for FOXO3 mRNA destabilization
Some m6A readers, such as YTHDF1, YTHDF2, and YTHDF3, are involved in m6A-mediated mRNA destabilization [13–15]. Given our observations of consistently inverse expression patterns between YTHDF2 and FOXO3 in aged IVDs, H2O2-injected IVDs, and H2O2-treated AF/NP cells, we hypothesized that YTHDF2 might function by negatively regulating the stability of FOXO3 mRNA. To test this hypothesis, we performed RIP assays on IVDs from 52-week-old mice (n = 8), H2O2-injected IVDs (n = 8), and H2O2-treated (0 and 150 µM) AF/NP cells to assess the binding of IgG (negative control), YTHDF1, YTHDF2, YTHDF3, YTHDC1, YTHDC2, IGF2BP1, IGF2BP2, and IGFBP3 to FOXO3 mRNA. The results revealed that only YTHDF2 was able to bind to FOXO3 mRNA in both mouse IVD and AF/NP cells, whereas other m6A readers did not bind to FOXO3 mRNA (Fig. 5A-5D). Notably, the binding capacity of YTHDF2 to FOXO3 mRNA was significantly enhanced in degenerated IVDs and in H2O2-treated cells compared to controls (Fig. 5A-5D). Additionally, nine GGACU motifs were identified within the 3’-UTR (untranslated region) of FOXO3 mRNA, which were potential binding sites for YTHDF2 (Fig. 5E). Further investigations into the effects of YTHDF2 knockdown and overexpression in AF and NP cellular backgrounds revealed that YTHDF2 deficiency significantly increased FOXO3 mRNA stability, while its overexpression dramatically reduced it (Fig. 5F and I). These findings suggest that YTHDF2 plays a negative regulatory role in controlling the stability of FOXO3 mRNA.
Fig. 5.
YTHDF2 was required for the stability of FOXO3 mRNA. (A) RIP assay of FOXO3 mRNA in L1/L2 IVDs from mice aged 8 weeks and 52 weeks, using antibodies against IgG, YTHDF1, YTHDF2, YTHDF3, YTHDC1, YTHDC2, IGFBP1, IGFBP2, and IGFBP3. (B) RIP assay of FOXO3 mRNA in L1/L2 IVDs from mice injected with PBS or H2O2, employing the same antibodies as in (A). (C) RIP assay of FOXO3 mRNA in NP cells treated with or without H2O2, using the same antibodies as in (A). (D) RIP assay of FOXO3 mRNA in AF cells treated with or without H2O2, utilizing antibodies as listed in (A). (E) Schematic representation of the m6A binding sites within the FOXO3 mRNA structure. (F-I) FOXO3 mRNA stability assessment. (F) FOXO3 mRNA decay in YTHDF2KD cells (NP background), measured after treatment with actinomycin D (2 µg/mL) at specified time points (0, 3, and 6 h), detected by RT-qPCR. (G) FOXO3 mRNA stability in YTHDF2KD cells (AF background) under similar conditions as in (F). (H) FOXO3 mRNA decay in YTHDF2OE cells (NP background). (I)FOXO3 mRNA stability in YTHDF2OE cells (AF background). *P < 0.05, **P < 0.01, ***P < 0.001
FOXO3 recruited histone lysine acetyltransferase CREBBP (CBP) and mediator complex subunit 1 (Med1) to regulate TIMP1 expression in the absence of ROS stimulation
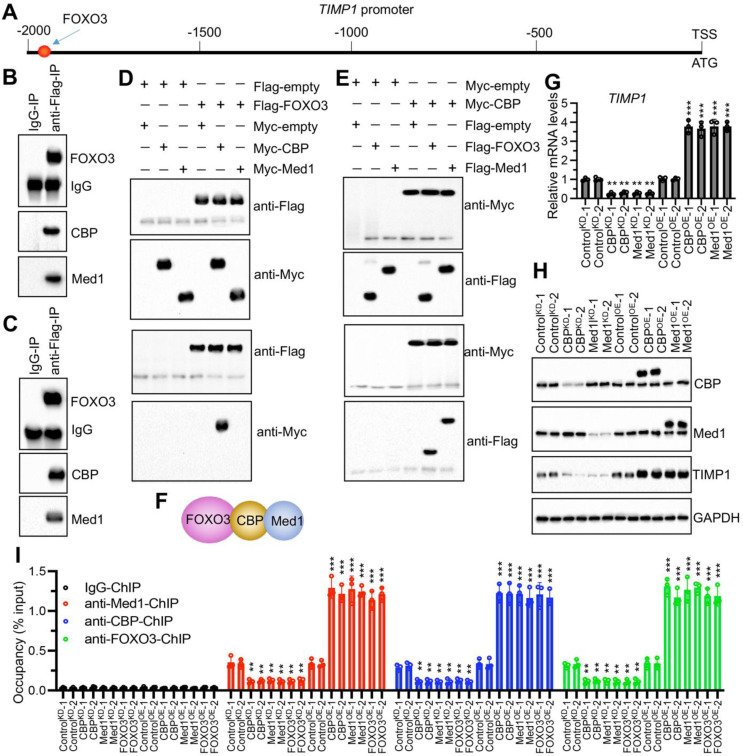
In our investigation detailed in Fig. 4, we demonstrated that FOXO3 bound to the promoter region of TIMP1. Analysis of the TIMP1 promoter (2000 bp upstream of the ATG start site) identified a specific FOXO3 binding site, GTAAACA, located at positions − 1967 to -1973 (Fig. 6A). In NP cells overexpressing FOXO3 (FOXO3OE-1), Flag-tagged FOXO3 was purified through IP assay, followed by LC-MS/MS analysis, which identified 112 proteins interacting with FOXO3 (Table S7). Notably, two transcriptional regulators—CBP and Med1—were found among these interactors (Table S7). Further IP assays in NP/AF cells overexpressing FOXO3 (FOXO3OE-1) confirmed that FOXO3 could pull down both CBP and Med1 (Fig. 6B and C). To dissect the assembly of the FOXO3, CBP, and Med1 complex, we transfected NP cells with Flag- and Myc-tagged constructs in pairwise combinations. Co-IP experiments demonstrated a direct interaction between FOXO3 and CBP but not between FOXO3 and Med1 (Fig. 6D). Conversely, CBP interacted directly with both FOXO3 and Med1 (Fig. 6E), indicating that CBP served as a pivotal bridge linking FOXO3 and Med1 (Fig. 6F).
Fig. 6.
FOXO3 recruited CBP and Med1 to assemble a complex in vivo and in vitro. (A) Identification of the FOXO3 binding site on the TIMP1 promoter. (B and C) FOXO3 interacted with CBP and Med1 in NP and AF cells overexpressing pCDNA3.1-Flag-FOXO3, assessed through immunoprecipitation assays using IgG-conjugated agarose A and anti-Flag resin. (B) In NP cells. (C) In AF cells. (D) Co-IP demonstrating direct interaction between FOXO3 and CBP, but not Med1. (E) Co-IP demonstrating direct interaction between CBP and FOXO3, as well as CBP and Med1. (F) Schematic diagram illustrating the assembly of the Med1-CBP-FOXO3 complex. (G) mRNA levels of TIMP1 in ControlKD, CBPKD, Med1KD, ControlOE, CBPOE, and Med1OE cells (NP background). (H) Protein levels of TIMP1 in the same cells as described in (G). (I) ChIP assays confirming the binding of the Med1-CBP-FOXO3 complex to the TIMP1 promoter, using antibodies against Med1, CBP, and FOXO3 in the same cells described in (G). **P < 0.01, ***P < 0.001
To further validate the function of this complex, both knockdown and overexpression cell lines of CBP and Med1 in NP background were generated (Fig. S5). Results indicated that depletion of either CBP or Med1 suppressed TIMP1 expression, while their overexpression led to an upregulation of TIMP1 (Fig. 6G and H). ChIP assays showed enrichment of CBP and Med1 on the TIMP1 promoter region. Disruption of any component of the FOXO3-CBP-Med1 complex decreased the enrichment of the others on the TIMP1 promoter, whereas overexpression of any complex member increased their collective enrichment (Fig. 6I).
Additionally, we examined the effects of oxidative stress by treating NP cells with H2O2. The results showed that H2O2 treatment notably reduced FOXO3 expression but did not significantly affect the expression levels of CBP or Med1 (Fig. S6A). However, the treatment significantly decreased the enrichment of the FOXO3-CBP-Med1 complex on the TIMP1 promoter (Fig. S6B). These findings elucidate the pivotal role of the FOXO3-CBP-Med1 complex in regulating TIMP1 expression in the absence of ROS stimulation and illustrate how ROS stimulation diminishes this complex’s binding to the TIMP1 promoter, leading to decreased TIMP1 expression.
Altered expression of the FOXO3-CBP-Med1 complex changed MMP1, MMP3, MMP7, and MMP9 levels
TIMP1 is an inhibitor of MMPs, and we observed an inverse expression trend between TIMP1 and MMP1/3/7/9 in degenerated IVDs and H2O2-treated cells. Given that the FOXO3-CBP-Med1 complex can regulate TIMP1 expression, we subsequently explored the effects of knocking down and overexpressing components of this complex on the expression of MMP1/3/7/9. Our findings indicated that knocking down any members of the FOXO3-CBP-Med1 complex led to upregulation of MMP1/3/7/9, while overexpressing these complex members suppressed MMP1/3/7/9 expression (Fig. S7A-S7D). Similar to FOXO3, neither CBP nor Med1 could bind directly to the promoter regions of MMP1/3/7/9 (Fig. S8A-S8D), suggesting that their regulatory effect on these MMPs was indirect.
Administration of YTHDF2 inhibitor in H2O2-treated cells upregulated TIMP1 expression but downregulated MMP1/3/7/9 expression
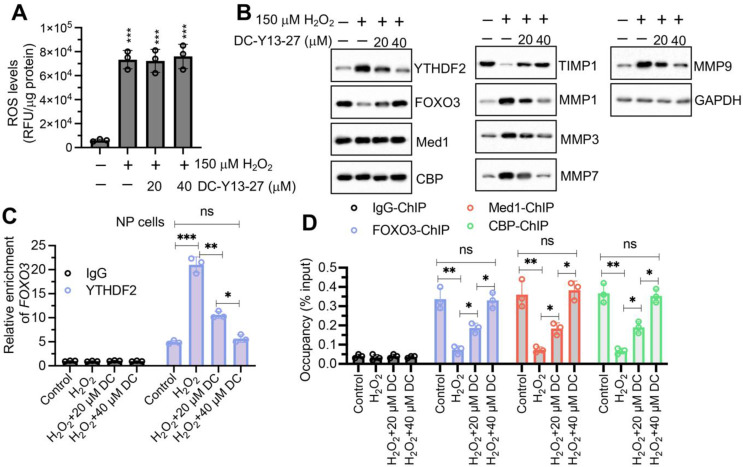
Since YTHDF2 was an upstream regulator of FOXO3, we hypothesized that inhibiting YTHDF2 might counteract the effects of ROS accumulation on TIMP1 and MMPs. To test this, we co-treated NP cells with the YTHDF2 inhibitor DC-Y13-27 (0, 20, and 40 µM) and H2O2 (0 and 150 µM). Following treatment, intracellular ROS levels were measured and found to be consistent with those observed with H2O2 treatment alone, indicating that DC-Y13-27 did not alter ROS levels in cells (Fig. 7A). Compared to treatment with H2O2 alone, the combined treatment with DC-Y13-27 and H2O2 consistently reduced YTHDF2 expression and resulted in increased expression of FOXO3 and TIMP1, along with decreased expression of MMP1, MMP3, MMP7, and MMP9 (Fig. 7B). RIP results demonstrated that treatment with DC-Y13-27 decreased the binding affinity of YTHDF2 to FOXO3 mRNA (Fig. 7C). Furthermore, the binding of the FOXO3-CBP-Med1 complex to the TIMP1 promoter was enhanced following treatment with DC-Y13-27 (Fig. 7D). These findings suggest that inhibition of YTHDF2 can reverse the ROS-mediated regulation of the FOXO3/TIMP1/MMP signaling axis.
Fig. 7.
DC-Y13-27 diminished the effect of H2O2 on the expression of FOXO3/TIMP1/MMPs in vitro. (A) Changes in intracellular ROS levels in NP cells following treatment with H2O2 (0 and 150 µM) and DC-Y13-27 (0, 20, and 40 µM) for 6 h. ROS levels were quantified using DCFH-DA staining. (B) Protein expression levels of YTHDF2, FOXO3, Med1, CBP, TIMP1, MMP1, MMP3, MMP7, and MMP9 in NP cells after treatment with H2O2 and DC-Y13-27. (C) RIP assay to detect FOXO3 mRNA association with YTHDF2 in NP cells treated with H2O2 and DC-Y13-27 (DC), using IgG and anti-YTHDF2 antibodies. (D) Assessment of the binding dynamics of the Med1-CBP-FOXO3 complex on the TIMP1 promoter in NP cells after treatment with H2O2 and DC-Y13-27 (DC). ns represents no significant difference. *P < 0.05, **P < 0.01, ***P < 0.001
Administration of DC-Y13-27 in aged and H2O2-injected mice attenuated the progression of IDD
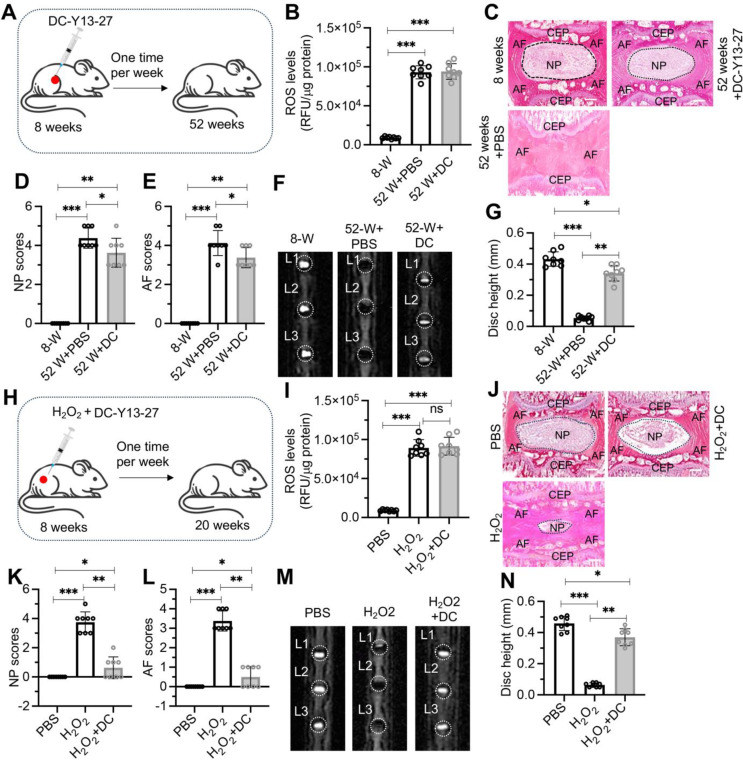
The effective suppression of MMPs by DC-Y13-27 in vitro under H2O2 treatment led us to evaluate its potential to inhibit the progression of IDD in mice. We initially randomized 8-week-old mice into two groups: one receiving weekly injections of PBS and the other receiving DC-Y13-27 injections (Fig. 8A). ROS levels in the IVDs of both the PBS and DC-Y13-27 groups (52-week-old mice) were significantly higher than in untreated 8-week-old mice (Fig. 8B). However, there was no notable difference in ROS levels between the PBS and DC-Y13-27 groups at the 52-week point (Fig. 8B). Mice injected with PBS exhibited severe degeneration at 52 weeks, whereas those treated with DC-Y13-27 showed only mild degeneration (Fig. 8C). Quantitative analysis of H&E staining, MRI scans, and disc height measurement confirmed that DC-Y13-27 significantly delayed IDD progression in mice (Fig. 8D and G).
Fig. 8.
Administration of DC-Y13-27 delayed IDD progression in aged and H2O2-injected mice. (A) Schematic representation of DC-Y13-27 administration in aged mice. (B) ROS levels in lumbar IVDs from young mice (8 weeks old), 52-week-old mice treated with PBS or DC-Y13-27 (DC). (C) Representative H&E stained sections of L1/L2 IVDs from young mice (8 weeks old) and 52-week-old mice treated with PBS or DC-Y13-27. Scale bars = 100 μm. (D) Quantitative scoring of the NP in degenerative IVDs shown in (C). (E) Quantitative scoring of the AF in degenerative IVDs illustrated in (C). (F) Representative MRI images of lumbar IVDs (L1 to L3) from young mice (8 weeks old) and 52-week-old mice treated with PBS or DC-Y13-27. (G) Measurement of disc height in L1/L2 IVDs. (H) Schematic representation of DC-Y13-27 and H2O2 injection procedure near lumbar discs in mice. (I) ROS levels in lumbar IVDs of mice treated with PBS, H2O2, and H2O2 + DC-Y13-27 (DC). (J) Representative H&E staining images of lumbar IVDs (L1/L2) from mice administered with PBS, H2O2, and H2O2 + DC-Y13-27 (DC). Scale bars = 100 μm. (K) Quantified NP scores of IVDs demonstrating degeneration in mice treated as described in (J). (L) Quantified AF scores of IVDs showing degenerative changes in mice treated as indicated in (J). (M) Representative MRI scans of lumbar IVDs (from L1 to L3) in mice treated with PBS, H2O2, and H2O2 + DC-Y13-27 (DC). (N) Disc height measurements in L1/L2 IVDs from mice treated with PBS, H2O2, and H2O2 + DC-Y13-27 (DC). ns represents no significant difference. *P < 0.05, **P < 0.01, ***P < 0.001
To further evaluate the effects of DC-Y13-27 under oxidative stress, we randomized another set of 8-week-old mice into three groups: PBS, H2O2, and a combination of H2O2 and DC-Y13-27, with injections administered weekly for 12 weeks (Fig. 8H). Results indicated that ROS levels in the IVDs of mice treated with DC-Y13-27 or H2O2 + DC-Y13-27 were significantly higher than those in the PBS group, yet there was no notable difference between the DC-Y13-27 and H2O2 + DC-Y13-27 groups (Fig. 8I). At 20 weeks, the PBS group showed no signs of IDD, the H2O2 group exhibited severe degeneration, while the H2O2 + DC-Y13-27 group displayed only mild degeneration (Fig. 8J). Quantitative analysis of H&E staining, MRI scans, and disc height measurement also supported these findings (Fig. 8K and N). Analysis of YTHDF2, FOXO3, TIMP1, Med1, CBP, and MMPs expression in these mouse groups revealed that DC-Y13-27 treatment significantly reduced the expression of YTHDF2 and MMP1/3/7/9, increased the expression of FOXO3 and TIMP1, and did not affect the expression of Med1 and CBP (Fig. S9). These results demonstrate that DC-Y13-27 effectively suppresses the expression of YTHDF2, thereby modulating downstream signaling pathways to mitigate IDD progression.
Discussion
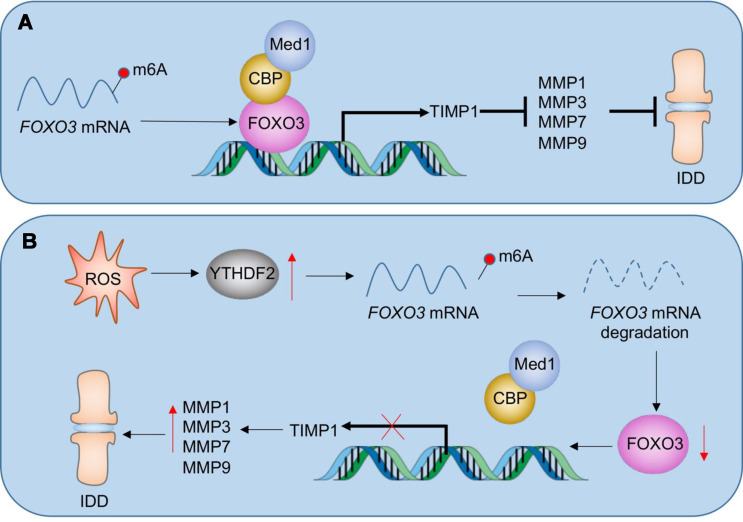
ROS plays a crucial role in the pathogenesis of IDD by inducing oxidative stress that damages cells and ECM, promoting inflammation, apoptosis, matrix degradation, and cellular senescence [10–12]. However, managing ROS in the process of IDD presents challenges due to the complexity of ROS sources and pathways, the dual effects of ROS as both damaging agents and cellular signaling molecules, difficulties in targeted therapeutic delivery to degenerating discs, and the lack of early detection methods [10–12]. Effective management of ROS in IDD requires a detailed understanding of their biological effects, innovative approaches to modulate oxidative stress, the development of targeted therapies, and improved diagnostic tools for early intervention [10–12]. In this study, we discovered that FOXO3 could form a transcriptional complex with CBP and Med1 in NP and AF cells to regulate the expression of TIMP1. TIMP1 protein, in turn, inhibited MMP1, MMP3, MMP7, and MMP9, thereby maintaining ECM homeostasis in NP and AF tissues (Fig. 9A). In aged mice and H2O2-injected mice, the accumulation of ROS led to YTHDF2 induction. Through both in vitro and in vivo experiments, we demonstrated that YTHDF2 destabilized FOXO3 mRNA. This destabilization prevented FOXO3 from forming a functional complex with CBP and Med1, thereby inhibiting TIMP1 expression. The downregulation of TIMP1 failed to suppress MMP1, MMP3, MMP7, and MMP9, resulting in their significant upregulation and consequent ECM degradation, ultimately promoting the development of IDD (Fig. 9B). Our findings reveal a ROS-mediated signaling pathway centered on the YTHDF2/FOXO3/TIMP1/MMPs axis, offering new insights into how ROS induces IDD. This pathway also provides promising therapeutic targets for future treatments aimed at combating IDD.
Fig. 9.
Schematic model of ROS-mediated pathway in the degeneration of IVDs. (A) Regulatory mechanism of the Med1-CBP-FOXO3 complex in healthy IVDs. FOXO3a recruits CBP and Med1 to form a transcriptional complex on the TIMP1 promoter, leading to TIMP1 activation. Increased TIMP1 levels inhibit the activity of matrix metalloproteinases (MMP1, MMP3, MMP7, and MMP9), thereby protecting the IVDs from degradation. (B) ROS accumulation-mediated pathway in IDD progression. Elevated ROS levels upregulate YTHDF2 expression, which destabilizes FOXO3 mRNA. The reduced stability of FOXO3 mRNA disrupts the formation of the functional FOXO3-CBP-Med1 complex, resulting in decreased TIMP1 expression. A lower TIMP1 level fails to regulate MMP activity, leading to the overexpression of MMP1, MMP3, MMP7, and MMP9, thereby accelerating ECM degradation and promoting IDD
Oxidative stress occurs when there is an imbalance between the production of ROS and the body’s ability to counteract or detoxify their harmful effects through neutralization by antioxidants [22, 23]. Elevated ROS level is a significant factor contributing to IDD [10–12]. During this biological process, RNA modifications—particularly m6A RNA methylation, the most common RNA modification in eukaryotic cells—serve as crucial post-transcriptional regulators influencing gene expression and RNA metabolism [13–15]. Importantly, m6A modifications can modulate cellular ROS levels, and oxidative stress, in turn, can regulate m6A RNA methylation dynamics [24, 25]. Utilizing high-performance liquid chromatography tandem mass spectrometry (HPLC-MS/MS), researchers have precisely quantified m6A modifications in purified yeast mRNA samples, revealing fluctuations in m6A levels in response to oxidative stress [26]. In human skin cells, such as HaCaT cells, ROS accumulation has been shown to upregulate methyltransferases including METTL3, METTL14, and WTAP [27]. Additionally, exposure to cadmium sulfate (CdSO4), a potent ROS inducer, in pancreatic β-cells significantly reduced m6A modifications, along with decreased mRNA levels of FTO and METTL3, in a dose-dependent manner [28]. Despite these advances, the specific roles and regulatory mechanisms of m6A modifying enzymes—writers, erasers, and readers—within the biological processes involved in IVD development and the pathogenesis of IDD remain underexplored. The intricate regulatory mechanisms involving ROS and m6A in these processes demand further investigation to better understand their implications in IDD. This study contributes to a more comprehensive understanding of how oxidative stress influences IDD pathogenesis through m6A-mediated RNA modifications. It highlights the complex interplay between oxidative stress and m6A modifications, providing key insights that are essential for developing targeted therapeutic strategies to slow or prevent IDD progression.
MMPs are a family of enzymes critically involved in the degradation of ECM and they play pivotal roles in the pathogenesis of IDD [8, 9]. Research in recent years has highlighted the complex expression patterns and regulatory mechanisms of MMPs in IDD, emphasizing their role in degrading essential disc matrix components such as collagen and proteoglycans [8, 9]. In particular, MMPs like MMP1, MMP3, MMP9, and MMP13 are significantly upregulated during the progression of IDD [8, 9]. This upregulation is often attributed to the increased levels of inflammatory cytokines such as IL-1β and TNF-α within the discs [29–32]. These cytokines stimulate disc cells to produce MMPs, which leads to ECM degradation and structure failure [29–31]. The molecular mechanisms underlying MMP activation in IDD involve several signaling pathways, including the MAPK, NF-κB, and Wnt/β-catenin pathways, which are activated by mechanical stress and cellular senescence [32–35]. Recently, Tseng et al. demonstrated that the accumulation of advanced glycation end products (AGEs) promotes the assembly of phosphorylated PPRC1 (pPPRC1) with two histone acetyltransferases, p300/CBP, and the transcription factor activator protein 1 (AP1) [36]. This assembly enhances the expression of 12 MMP genes (MMP1a/1b/3/7/9/10/12/13/16/19/23/28), contributing to the occurrence of IDD [36]. Counteracting the activity of MMPs, TIMPs play crucial roles in maintaining ECM integrity by inhibiting MMP activity [7, 37]. Disruption of this balance, such as decreased TIMP expression, can exacerbate ECM degradation in IDD [7, 37]. TIMPs, including TIMP1, TIMP2, TIMP3, and TIMP4, exhibit varying expression patterns during IDD progression, with a decrease in TIMP levels leading to enhanced ECM breakdown [7, 37]. However, the expression patterns of MMPs and TIMPs and their regulatory mechanisms in the process of ROS-induced IDD remain unknown. In this study, we investigated DEPs in IVDs from aged and H2O2-injected mice. We found significant downregulation of TIMP1 and upregulation of MMP1, MMP3, MMP7, and MMP9, but not of other TIMPs and MMPs. We demonstrated that the Med1-CBP-FOXO3 complex was responsible for the regulation of TIMP1 expression and indirectly regulated the expression of MMP1, MMP3, MMP7, and MMP9. To our knowledge, our study is the first to elucidate the mechanism by which ROS stimulation regulates the expression of TIMP1 and MMPs, offering new insights into the molecular basis of ROS-induced disc degeneration.
Current pharmacological treatments in IDD primarily focus on symptom relief rather than addressing the underlying causes [38]. One interesting finding in this study is the potential of targeting YTHDF2 for IDD therapy. Our study discovered that the YTHDF2 inhibitor DC-Y13-27 significantly slowed IDD progression, offering a promising new treatment option. However, given YTHDF2’s extensive range of target genes, its inhibition may lead to widespread gene expression alterations and unintended side effects, such as disruptions in cell proliferation, differentiation, and apoptosis across various tissues. Therefore, while DC-Y13-27 holds potential, its safety profile must be thoroughly evaluated to ensure that its therapeutic benefits outweigh any risks. In translating these findings into clinical practice, several potential challenges must be considered. First, the delivery of YTHDF2 inhibitors, such as DC-Y13-27, to IVDs presents significant obstacles. The IVD is a largely avascular structure, making targeted delivery of therapeutics difficult and reducing their effectiveness. Developing novel delivery methods, such as nanoparticles or localized injections, will be critical for ensuring the therapeutic agent reaches the degenerated disc in sufficient quantities. Second, the specificity of targeting the YTHDF2/FOXO3/TIMP1/MMP axis in humans raises concerns. YTHDF2 regulates a broad spectrum of mRNAs, and systemic inhibition may lead to off-target effects, affecting gene expression in other tissues, which could result in unintended consequences, such as impairments in cell proliferation or tissue regeneration. Therefore, optimizing the specificity of these inhibitors and evaluating their long-term safety profiles will be essential steps before moving toward clinical application.
In summary, our findings reveal a ROS-mediated signaling pathway in the pathogenesis of IDD involving the YTHDF2/FOXO3/TIMP1/MMPs axis. Disrupting this signaling pathway with the YTHDF2 inhibitor DC-Y13-27 has the potential to slow the progression of IDD, offering new insights and potential therapeutic targets for IDD treatment.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We extend our sincere gratitude to all the colleagues in the Department of Orthopedic Surgery at Zhejiang University Shaoxing Hospital for their invaluable support and assistance in this study.
Author contributions
Fei Wang: Writing– original draft, Methodology, Investigation, Data curation, Conceptualization. Yifeng Wang and Songou Zhang: Resources, Methodology, Investigation. Chun-Hung Hua: Resources, Methodology, Investigation. Mengyang Pu: Validation, Formal analysis. Ping Zhou: Writing– review & editing, Conceptualization, Methodology.
Funding
Not applicable.
Data availability
All analyzed data during this study are included in this published article (and its supplementary information files). The other materials are available from the corresponding author on reasonable request.
Declarations
Ethical approval
All animal procedures were conducted in strict accordance with a protocol (2020-032) reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of Shaoxing Hospital.
Consent to participate
Not applicable.
Consent to participate and publish
Not applicable.
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fei Wang, Yifeng Wang and Songou Zhang contributed equally to this work.
References
- 1.Oichi T, Taniguchi Y, Oshima Y, Tanaka S, Saito T (2020) Pathomechanism of intervertebral disc degeneration. JOR Spine 3:e1076. 10.1002/jsp2.1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiang Q, Zhao Y, Lin J, Jiang S, Li W (2022) The Nrf2 antioxidant defense system in intervertebral disc degeneration: molecular insights. Exp Mol Med 54:1067–1075. 10.1038/s12276-022-00829-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamali A, Ziadlou R, Lang G, Pfannkuche J, Cui S, Li Z et al (2021) Small molecule-based treatment approaches for intervertebral disc degeneration: current options and future directions. Theranostics 11:27–47. 10.7150/thno.48987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen F, Lei L, Chen S, Zhao Z, Huang Y, Jiang G et al (2024) Serglycin secreted by late-stage nucleus pulposus cells is a biomarker of intervertebral disc degeneration. Nat Commun 15:47. 10.1038/s41467-023-44313-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dou Y, Sun X, Ma X, Zhao X, Yang Q (2021) Intervertebral disk degeneration: the Microenvironment and tissue Engineering Strategies. Front Bioeng Biotechnol 9:592118. 10.3389/fbioe.2021.592118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bermudez-Lekerika P, Crump KB, Tseranidou S, Nuesch A, Kanelis E, Alminnawi A et al (2022) Immuno-Modulatory effects of intervertebral disc cells. Front Cell Dev Biol 10:924692. 10.3389/fcell.2022.924692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vo NV, Hartman RA, Yurube T, Jacobs LJ, Sowa GA, Kang JD (2013) Expression and regulation of metalloproteinases and their inhibitors in intervertebral disc aging and degeneration. Spine J 13:331–341. 10.1016/j.spinee.2012.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Risbud MV, Shapiro IM (2014) Role of cytokines in intervertebral disc degeneration: pain and disc content. Nat Rev Rheumatol 10:44–56. 10.1038/nrrheum.2013.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang WJ, Yu XH, Wang C, Yang W, He WS, Zhang SJ et al (2015) MMPs and ADAMTSs in intervertebral disc degeneration. Clin Chim Acta 448:238–246. 10.1016/j.cca.2015.06.023 [DOI] [PubMed] [Google Scholar]
- 10.Cheng F, Yang H, Cheng Y, Liu Y, Hai Y, Zhang Y (2022) The role of oxidative stress in intervertebral disc cellular senescence. Front Endocrinol (Lausanne) 13:1038171. 10.3389/fendo.2022.1038171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng C, Yang M, Lan M, Liu C, Zhang Y, Huang B et al (2017) ROS: crucial intermediators in the pathogenesis of intervertebral disc degeneration. Oxid Med Cell Longev 2017(5601593). 10.1155/2017/5601593 [DOI] [PMC free article] [PubMed]
- 12.Yang S, Lian G (2019) ROS and diseases: role in metabolism and energy supply. Mol Cell Biochem 467:1–12. 10.1007/s11010-019-03667-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, Yang C, Chen Y (2021) The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther 6:74. 10.1038/s41392-020-00450-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaccara S, Ries RJ, Jaffrey SR (2019) Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol 20:608–624. 10.1038/s41580-019-0168-5 [DOI] [PubMed] [Google Scholar]
- 15.Wiener D, Schwartz S (2021) The epitranscriptome beyond m6A. Nat Rev Genet 22:119–131. 10.1038/s41576-020-00295-8 [DOI] [PubMed] [Google Scholar]
- 16.Li G, Luo R, Zhang W, He S, Wang B, Liang H et al (2022) m6A hypomethylation of DNMT3B regulated by ALKBH5 promotes intervertebral disc degeneration via E4F1 deficiency. Clin Transl Med 12:e765. 10.1002/ctm2.765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu X, Shen L, Qu Y, Li D, Zhao X, Wei H, Yue S (2024) Experimental validation and comprehensive analysis of m6A methylation regulators in intervertebral disc degeneration subpopulation classification. Sci Rep 14:8417. 10.1038/s41598-024-58888-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu H, Lyu H, Jiang G, Chen D, Ruan S, Liu S et al (2022) ALKBH5-Mediated m6A demethylation of GLUT4 mRNA promotes glycolysis and resistance to HER2-Targeted therapy in breast Cancer. Cancer Res 82:3974–3986. 10.1158/0008-5472.CAN-22-0800 [DOI] [PubMed] [Google Scholar]
- 19.Huang L, Wang W, Xian Y, Liu L, Fan J, Liu H et al (2023) Rapidly in situ forming an injectable Chitosan/PEG hydrogel for intervertebral disc repair. Mater Today Bio 22:100752. 10.1016/j.mtbio.2023.100752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohnishi T, Sudo H, Iwasaki K, Tsujimoto T, Ito YM, Iwasaki N (2016) In vivo mouse intervertebral disc degeneration model based on a new histological classification. PLoS ONE 11:e0160486. 10.1371/journal.pone.0160486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tseng C, Han Y, Lv Z, Song Q, Wang K, Shen H, Chen Z (2023) Glucose-stimulated PGC-1alpha couples with CBP and Runx2 to mediate intervertebral disc degeneration through transactivation of ADAMTS4/5 in diet-induced obesity mice. Bone 167:116617. 10.1016/j.bone.2022.116617 [DOI] [PubMed] [Google Scholar]
- 22.Afzal S, Abdul Manap AS, Attiq A, Albokhadaim I, Kandeel M, Alhojaily SM (2023) From imbalance to impairment: the central role of reactive oxygen species in oxidative stress-induced disorders and therapeutic exploration. Front Pharmacol 14:1269581. 10.3389/fphar.2023.1269581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pizzino G, Irrera N, Cucinotta M, Pallio G, Mannino F, Arcoraci V et al (2017) Oxidative stress: Harms and benefits for Human Health. Oxid Med Cell Longev 2017(8416763). 10.1155/2017/8416763 [DOI] [PMC free article] [PubMed]
- 24.Yang B, Chen Q (2021) Cross-Talk between Oxidative Stress and m(6)A RNA Methylation in Cancer. Oxid Med Cell Longev 2021: 6545728. 10.1155/2021/6545728 [DOI] [PMC free article] [PubMed]
- 25.Ni Y, Zhang H, Chu L, Zhao Y (2023) m6A modification-association with oxidative stress and implications on Eye diseases. Antioxid (Basel) 12:510. 10.3390/antiox12020510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tardu M, Jones JD, Kennedy RT, Lin Q, Koutmou KS (2019) Identification and quantification of modified nucleosides in Saccharomyces cerevisiae mRNAs. ACS Chem Biol 14:1403–1409. 10.1021/acschembio.9b00369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen H, Zhao T, Sun D, Wu M, Zhang Z (2019) Changes of RNA N(6)-methyladenosine in the hormesis effect induced by arsenite on human keratinocyte cells. Toxicol Vitro 56:84–92. 10.1016/j.tiv.2019.01.010 [DOI] [PubMed] [Google Scholar]
- 28.Qu T, Mou Y, Dai J, Zhang X, Li M, Gu S, He Z (2021) Changes and relationship of N(6)-methyladenosine modification and long non-coding RNAs in oxidative damage induced by cadmium in pancreatic beta-cells. Toxicol Lett 343:56–66. 10.1016/j.toxlet.2021.02.014 [DOI] [PubMed] [Google Scholar]
- 29.Li Z, Yang H, Hai Y, Cheng Y (2023) Regulatory Effect of Inflammatory Mediators in Intervertebral Disc Degeneration. Mediators Inflamm 2023: 6210885. 10.1155/2023/6210885 [DOI] [PMC free article] [PubMed]
- 30.Li X, Wang X, Chen C, Zhang E, Zhang Y, Li H et al (2023) Accumulation of NCOA1 dependent on HERC3 deficiency transactivates matrix metallopeptidases and promotes extracellular matrix degradation in intervertebral disc degeneration. Life Sci 320:121555. 10.1016/j.lfs.2023.121555 [DOI] [PubMed] [Google Scholar]
- 31.Li X, Zhang J, Wang B, Chen C, Zhang E, Lv Z et al (2023) USP24-dependent stabilization of Runx2 recruits a p300/NCOA3 complex to transactivate ADAMTS genes and promote degeneration of intervertebral disc in chronic inflammation mice. Biol Direct 18:37. 10.1186/s13062-023-00395-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang HJ, Liao HY, Bai DY, Wang ZQ, Xie XW (2021) MAPK /ERK signaling pathway: a potential target for the treatment of intervertebral disc degeneration. Biomed Pharmacother 143:112170. 10.1016/j.biopha.2021.112170 [DOI] [PubMed] [Google Scholar]
- 33.He X, Hu W, Zhang Y, Chen M, Ding Y, Yang H et al (2023) Cellular senescence in skeletal disease: mechanisms and treatment. Cell Mol Biol Lett 28:88. 10.1186/s11658-023-00501-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhongyi S, Sai Z, Chao L, Jiwei T (2015) Effects of nuclear factor kappa B signaling pathway in human intervertebral disc degeneration. Spine (Phila Pa 1976) 40:224–232. 10.1097/BRS.0000000000000733 [DOI] [PubMed] [Google Scholar]
- 35.Zhang GZ, Liu MQ, Chen HW, Wu ZL, Gao YC, Ma ZJ et al (2021) NF-kappaB signalling pathways in nucleus pulposus cell function and intervertebral disc degeneration. Cell Prolif 54:e13057. 10.1111/cpr.13057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tseng C, Chen B, Han Y, Wang K, Song Q, Shen H et al (2024) Advanced glycation end products promote intervertebral disc degeneration by transactivation of matrix metallopeptidase genes. Osteoarthritis Cartilage 32:187–199. 10.1016/j.joca.2023.09.005 [DOI] [PubMed] [Google Scholar]
- 37.Arpino V, Brock M, Gill SE (2015) The role of TIMPs in regulation of extracellular matrix proteolysis. Matrix Biol 44–46:247–254. 10.1016/j.matbio.2015.03.005 [DOI] [PubMed] [Google Scholar]
- 38.Colella F, Garcia JP, Sorbona M, Lolli A, Antunes B, D’Atri D et al (2020) Drug delivery in intervertebral disc degeneration and osteoarthritis: selecting the optimal platform for the delivery of disease-modifying agents. J Control Release 328:985–999. 10.1016/j.jconrel.2020.08.041 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All analyzed data during this study are included in this published article (and its supplementary information files). The other materials are available from the corresponding author on reasonable request.