Abstract
In mucosal barriers, tissue cells and leukocytes collaborate to form specialized niches that support host-microbiome symbiosis. Understanding the spatial organization of these barriers is crucial for elucidating the mechanisms underlying health and disease. The gingiva, a unique mucosal barrier with significant health implications, exhibits intricate tissue architecture and likely contains specialized immunological regions. Through spatial transcriptomic analysis, this study reveals distinct immunological characteristics between the buccal and palate regions of the murine gingiva, impacting natural alveolar bone loss. The microbiota primarily affects gingival immunity in the buccal region. Additionally, a significant influence of the microbiota on the junctional epithelium facing the oral biofilm offers new insights into neutrophil recruitment. The microbiota also regulates the proliferation and barrier-sealing function of the gingival epithelium. This underscores the presence of immunological niches in the gingiva, with the microbiota differentially influencing them, highlighting the high complexity of this oral mucosal barrier.
Subject terms: Dentistry, Next-generation sequencing
Introduction
Mucosal barriers play a crucial role in sustaining life by serving as the primary defense against various challenges, including those posed by the local microbiota. These barriers are comprised of epithelial layers, which can either be monolayered, as observed in the intestine and lungs or multi-layered, as seen in the oral cavity1,2. Each epithelium has a unique immunological network promoting symbiotic relationships with its microbiota2,3. This symbiosis is established through intricate interplay during the initial colonization of commensal microbiota after birth, shaping the barrier immunity throughout adulthood4–8.
In the oral mucosa, postnatal colonization is characterized by substantial fluctuations in the load and composition of the microbiome until weaning, a process essential for the proper development of physiological and immunological functions of the epithelium9. Unlike other mucosae, the oral mucosa contains a distinctive barrier—the gingiva. Formed around the only hard tissue exposed to the external environment—the teeth, the gingiva poses complex challenges to the immune system. These challenges involve coping with masticatory mechanical forces damaging the gingival epithelium, along with managing the formation of bacterial plaque on the tooth surface10,11. Several studies have demonstrated the involvement of the microbiota in recruiting and regulating the function of neutrophils within the gingival epithelium9,12,13. The microbiota also facilitates the recruitment of various leukocytes to the gingival epithelium, encompassing Langerhans cells (LCs), innate lymphoid cells (ILCs), CD8+ T cells, and γδT cells11,14,15. This underscores the close relationship between the microbiota and the gingival epithelium, suggesting the potential for a broader impact of commensal bacteria on this important oral barrier.
Spatial transcriptomic technologies have emerged as an effective approach to studying tissue architecture and function16–19. These methods have been applied to investigate the human gingiva, revealing distinctly specialized epithelial and stromal compartments with unique gene expression signatures in health and periodontitis20,21. In this context, the steady-state microbiome associated with the human oral mucosa displays regional differences22, indicating functional specialization of different epithelial regions based on their specific microbial challenges. This fosters the emergence of an updated perspective that challenges the traditional view of the oral epithelium as a passive physical structure merely housing leukocytes responsible for host defense. Instead, it acknowledges the active involvement of epithelial cells in surveilling the oral barrier, endowed with the ability to adapt and respond to various challenges in a region-specific manner2,23,24. To uncover new insights into how the microbiota impacts the gingiva, particularly the gingival epithelium, this study conducted spatial and single-cell transcriptomic analyses on gingival tissues from germ-free (GF) and specific pathogen-free (SPF) mice. The analysis reveals the inherent diversity within the gingival epithelium, identifying distinct immunological features between the buccal and palate epithelial regions of the gingiva that influence natural alveolar bone loss. Additionally, it unveils a role for the microbiota in influencing gingival epithelial properties, such as barrier sealing and the induction of immunological function in the junctional epithelium facing the dental biofilm. These findings highlight the existence of microbiota-dependent and independent mucosal niches in the gingiva, which are essential for maintaining local homeostasis.
Results
Characterization of gingival cell clusters through single-cell RNA sequencing
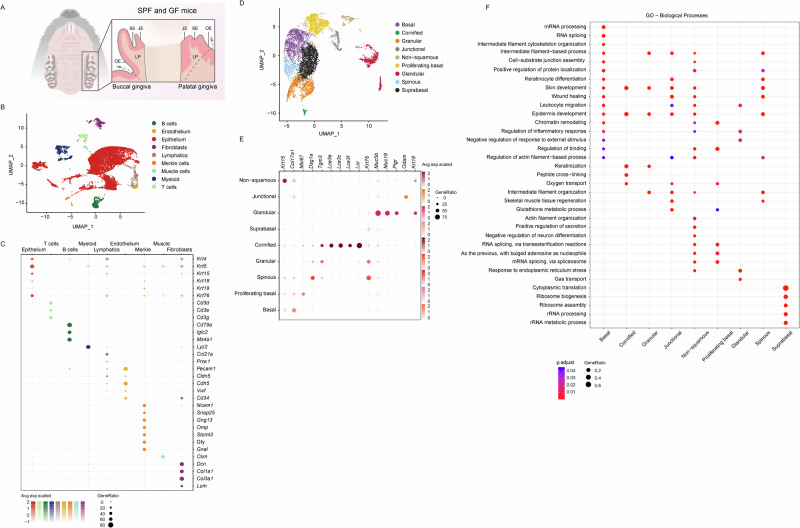
To investigate the type of cells residing in the gingiva, we excised and pooled maxillary gingival mucosal tissues surrounding the three molars from 8-week-old SPF and GF mice (Fig. 1A). In order to construct a comprehensive cellular map of the gingiva, we conducted single-cell RNA sequencing (scRNA-seq) and obtained expression profiles for SPF (16,810 cells) and GF (6,070 cells) mice. The integration of SPF with the GF data revealed nine transcriptionally distinct clusters (Fig. 1B). The categorization of these nine distinct clusters utilized canonical markers, as detailed in Fig. 1C. Epithelial cells, which represent the majority of cells, were discerned based on the expression of recognized epithelial markers, such as Krt4, Krt5, Krt15, Krt18, and Krt76. Other clusters included endothelial cells (Pecam1, Cldn5, Cdh5, Vwf, and Cd34), lymphatics (Ccl21a and Prox1), muscle cells (Ckm), stromal fibroblasts (Dcn, Col1a1, Col1a3, and Lum) and Merkel cells (Ncam1, Snap25, Gng13, Omp, Stoml3, Gfy, and Gnal). Additionally, diverse immune subsets were identified, including T cells (Cd3d/e/g), B cells (Cd79a, Iglc2, and Ms4a1), and myeloid cells (Lyz2) (Fig. 1C).
Fig. 1. Single-cell transcriptomic profiling of the gingival barrier.
A Schematic representation of the gingival tissues collected from 8-week-old murine molars. JE junctional epithelium, SE sulcular epithelium, OE oral epithelium, LP lamina propria. B UMAP visualization of major cell types identified in the gingiva via scRNA-seq. C Dot plots illustrate the expression of cluster-defining genes and the percentage of cells expressing each gene. Expression values are normalized and presented as scaled averages. D UMAP visualization of major cell types expressing epithelial-related genes. E Dot plots display the expression of epithelial cluster-defining genes and the percentage of cells expressing each gene. Expression values are normalized and presented as scaled averages. F Dot plots presenting the primary biological pathways enriched in each epithelial cluster and the percentage of genes expressing each pathway.
Subsequently, attention was directed towards the epithelial cluster, given its role as the outermost barrier in the gingiva. As depicted in Fig. 1D-E, the epithelial cluster was subdivided into nine subclusters based on previous transcriptomic data: the junctional epithelium (Odam) and basal epithelium (Col17a1, Krt15), with the latter further subdivided to proliferating basal epithelium that also expresses Mki67. Other epithelial clusters comprised the spinous (Dsg1a), granular (Tgm3), and cornified (Lor, Lce3e/f/c) epithelial layers. A subset of cells expressing Krt76 was identified in differentiating, non-proliferative cells, referred to as suprabasal epithelium, as reported in humans and mice25,26. The analysis also revealed two clusters expressing Krt18 segregated from clusters representing stratified epithelium. One of these clusters was generally termed non-squamous epithelium27–30. The expression of Muc5b/19 in addition to Krt18 by the other cluster, identified it as glandular epithelium31 of minor salivary glands located in the submucosa32. This cluster was further divided into two subclusters based on the expression of genes related to chemosensing, olfactory (Omp, Gnal, Gfy, Chga)33,34, and neuronal (Gng13, Stoml3)35,36 functions (data not shown).
Next, ClusterProfiler analysis was employed to identify unique Gene Ontology (GO) pathways for each epithelial subtype (Fig. 1F). As anticipated, most subclusters showed involvement in skin development, reflecting the resemblance between the epidermis and gingival epithelium. The basal and spinous epithelium were associated with keratinocyte differentiation, while the granular and cornified epithelium were linked to keratinization processes. The formers were also enriched with genes involved in wound healing, possibly due to mechanical damage caused by masticatory mechanical forces10. The wound-healing singling was also evident in the junctional epithelium located adjacent to the oral plaque, which was also linked to glutathione metabolism, indicating its participation in antioxidant defense and various cellular processes37. Immune-related pathways were identified in the basal, junctional, and non-squamous epithelia as well as the glandular epithelium, the latter was also associated with protein localization, secretion, and the regulation of inflammation and neuron differentiation. The suprabasal epithelial cluster appeared distinct, not aligning with skin development pathways. Rather, this cluster upregulated translation and ribosomal activity pathways, which in epithelial cells are important for achieving successful cell regeneration after injury38. Collectively, these findings highlight the gingival heterogeneity and the specialization of each epithelial region.
The buccal and palate epithelial regions of the gingiva exhibit unique structural and functional characteristics
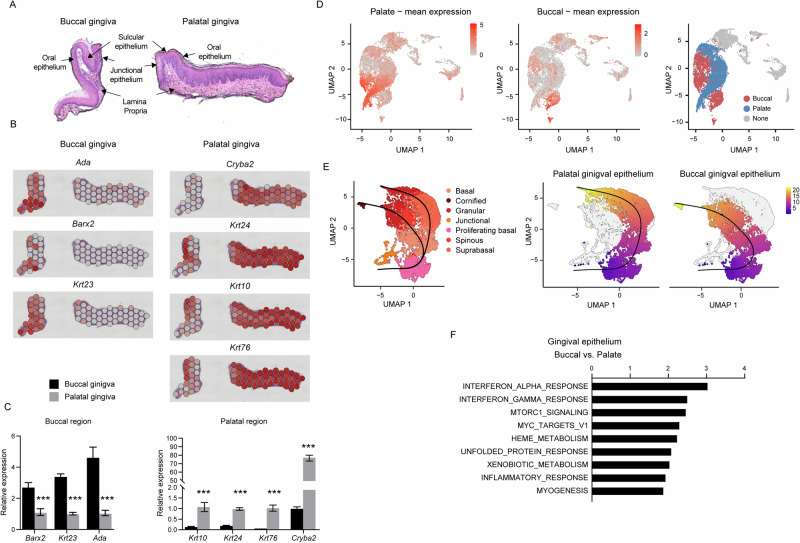
Considering that the gingiva is covered by epithelia originating from the buccal and palate oral mucosal tissues, our subsequent inquiry focused on whether these areas can be distinctly identified in the gingiva and if they exhibit diverse features. Leveraging Visium spatial analysis, we identified several genes with high expression in the buccal gingiva (Ada, Barx2, and Krt23) and the palatal gingiva (Cryba2, Krt24, Krt10, and Krt76), primarily localized within the spinous and granular clusters (Fig. 2A, B). The differential expression of these genes was validated through a quantitative PCR (qPCR) assay conducted on isolated buccal and palatal gingival regions (Fig. 2C). The mean expression of the noted genes further facilitated the identification of the buccal and palatal epithelial regions through transcriptomic analysis (Fig. 2D). Next, we continued to perform pseudotemporal trajectory analysis on the various epithelial subclusters. The analysis revealed that the proliferating basal cell cluster gives rise to both types of epithelia, and the pseudotemporal projection initiated to branch in the basal cell cluster (Fig. 2E). A similar trajectory of the buccal and palatal gingival epithelium was observed in SPF and GF samples when analyzed separately (data not shown). In contrast, the junctional epithelium was not linked to the proliferating basal cells, suggesting a distinct developmental origin. The buccal epithelial region includes the typically defined layers of basal, spinous, granular, and cornified cell clusters. The palate region, on the other hand, consists of distinct basal, suprabasal, and granular cell clusters. Notably, although both the buccal and palate regions are keratinized in mice, the analysis excludes the cornified cluster in the projection of the palate region, potentially stemming from differences in the level of keratinization in each tissue that affect the availability of the RNA transcripts. Next, we conducted a gene set enrichment analysis (GSEA) to identify differences in biological pathways between the buccal and palatal epithelial regions. As depicted in Fig. 2F, analysis of all clusters associated with each epithelium revealed the upregulation of immunological pathways in the buccal gingival epithelium in comparison to the palate region of the gingiva. This includes IFN-α, IFN-γ, and inflammatory responses that are known to affect local immunity11,39. Additionally, pathways related to cell growth and proliferation such as mTORC1 signaling, unfolded protein response, heme metabolism, and MYC target V1 were upregulated in the buccal region. An upregulation of the xenobiotic metabolism, reflecting microbiota-mediated modification of ingested compounds40, was observed in the buccal region. Supplementary Fig. 1 further demonstrated changes in biological pathways specific to each epithelial cluster in the buccal and palate regions of the gingival epithelium. Taken together, these data suggest that, despite originating from similar precursors, the buccal and palate gingival epithelium follow distinct developmental routes, resulting in structural and functional differences that can affect local immunity.
Fig. 2. The buccal and palatal gingiva exhibit distinct structural and functional features.
A Representative H&E image of a gingival cross-section from 8-week-old SPF mice utilized for spatial analysis, illustrating general tissue morphology and delineating the various tissue compartments. B Spatial feature plots display the expression of distinct genes that delineate the buccal or palatal regions of the gingiva. C Validation of the expression of the indicated genes in epithelial tissues isolated separately from the buccal and palatal regions of 8-week-old mice. Graphs represent transcript levels quantified via qPCR and normalized to the palatal epithelium, depicted as the mean ± SEM (n = 5). Representative data from two independent experiments. D UMAP plots illustrate the mean expression of genes associated with the buccal and palatal regions of the gingiva. A UMAP demonstrating the segregation of the epithelial cluster into the buccal and palatal regions is provided. E Differentiation trajectory of epithelial cells constructed using the slingshot R package. F GESA demonstrates differential pathways between epithelial clusters associated with the buccal and palatal regions of the gingiva.
Local immunity differs between the buccal and palate gingival regions, implicating natural alveolar bone loss
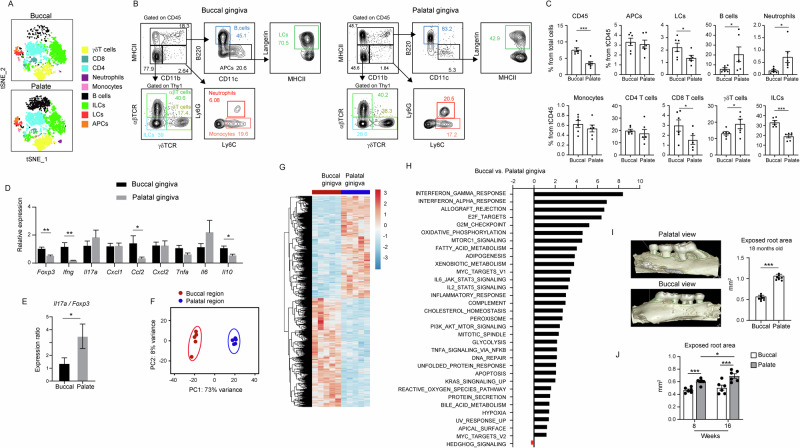
Since the transcriptomic analysis indicated variations in immunological pathways between the buccal and palate regions of the gingival epithelium, we aimed to address this aspect directly. To accomplish this, the gingival epithelium was collected from 8-week-old mice, and the buccal and palate regions were isolated and subjected to flow cytometry analysis separately. In Fig. 3A, tSNE plots illustrate the presence of various innate and adaptive subsets of leukocytes in each region. Further analysis revealed the presence of higher frequencies of total CD45+ leukocytes in the buccal epithelium compared to the palate (Fig. 3B, C). Additionally, while the buccal epithelium contains higher percentages of LCs and ILCs than the palate epithelium, the latter consists of elevated frequencies of B cells and neutrophils. To gain insight into the activity of the immune system in each region, mRNA was isolated from each region to quantify various immunologically related genes. As depicted in Fig. 3D, a reduction in the expression of Foxp3, Ifng, Ccl2, and Il10 was observed in the palate compared to the buccal gingiva epithelium. In addition, the ratio between Il17a and Foxp3, which signifies the immunological status of the oral mucosa39, was elevated in the palate region (Fig. 3E). Next, the buccal and palate regions of 8-week-old gingiva were excised separately, and the epithelium (containing epithelial cells and local leukocytes) was isolated and subjected to bulk RNA-seq analysis. Principal component analysis (PCA) and hierarchical clustering indicated a significant difference between these two regions of the gingival epithelium (Fig. 3F, G). As depicted in Fig. 3H, the epithelium of the buccal gingiva demonstrated elevated immunological signaling such as IFN-γ, IFN-α, IL-2/STAT5, IL6/JAK/STAT3, allograft rejection, inflammatory response, and TNF-α/NFκB pathways. Of note, the IFN-γ response, the most altered pathway in the analysis, was shown to be secreted by gingival CD4+ T cells, playing a crucial role in maintaining epithelial integrity11. Metabolic pathways such as oxidative phosphorylation, fatty acid metabolism, cholesterol homeostasis, and reactive oxygen species signaling were also increased in the buccal compared to the palate region of the gingiva. Interestingly, the buccal also demonstrated elevated levels of wound repair pathways, for instance, PI3K/AKT/MTOR signaling.
Fig. 3. Distinct physiological immune responses in the buccal and palatal gingiva impact natural alveolar bone loss.
A tSNE flow cytometry plot illustrating the principal leukocyte subsets present in the gingival epithelium of both buccal and palatal regions in 8-week-old SPF mice. Representative data from two independent experiments. B Flow cytometry plots and C corresponding graphs showcasing the frequencies of major leukocyte subsets within the epithelium of buccal and palatal gingiva. Data represented as mean frequencies ± SEM (n = 6). Representative outcomes from two independent experiments. D and E Relative expression of the noted genes in the buccal and palatal gingiva of 8-week-old SPF mice. Graphs depict transcript levels quantified via qPCR and normalized to the buccal region, presented as mean ± SEM (n = 5). Representative data from two independent experiments. F PCA of the most variable transcripts expressed by cells isolated from the buccal and palatal gingiva. G Hierarchical clustering of genes exhibiting differential expression in the epithelium of buccal and palatal gingiva isolated from 8-week-old mice. H Identification of significantly upregulated and downregulated gene pathways through GSEA among the epithelium of the distinct gingival regions (family-wise error rate [FWER] < 0.05). I Representative μCT images and corresponding graphs illustrating the exposed root area in the buccal and palatal regions of the maxilla collected from 18-month-old SPF mice (n = 8). J Graphs demonstrating the exposed root area in buccal and palatal regions of 8- and 16-week-old mice (n = 6). Representative data from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
Next, the high presence of neutrophils and increased Il17a/Foxp3 ratio along with the reduction in immunoregulatory (IL-2/STAT5, Foxp3, and Il10) and anti-osteoclastogenic (IFN-γ) pathways, proposed that the palate gingival region is more susceptible to immune-mediated natural alveolar bone loss. To address this issue directly, we conducted µCT scans on the maxilla of 18-month-old mice. As illustrated in Fig. 3I, the exposed tooth root area, representing the loss of alveolar bone, was higher in the palatal area compared to the buccal. Notably, elevated bone loss in the palatal region of the gingiva was already observed in 8-week-old mice, and this effect was further increased in 16-week-old mice (Fig. 3J). These findings suggest that, in addition to structural changes, there are distinct differences in mucosal immunity between the buccal and palate regions of the gingival epithelium, leading to a substantial impact on the adjacent alveolar bone.
The microbiota regulates the proliferation of gingival epithelial cells and cell-cell interactions
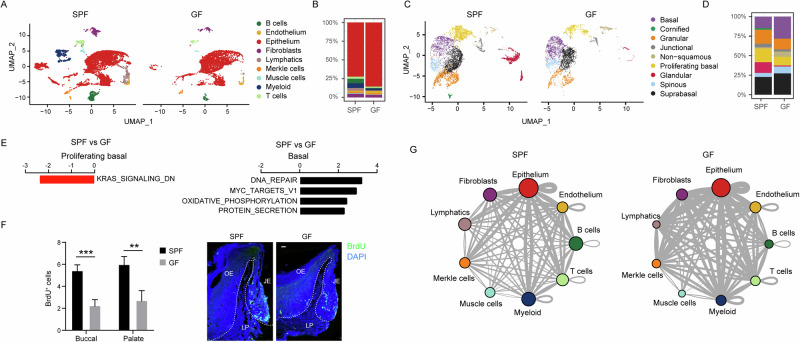
To evaluate the impact of the microbiota on the gingival epithelium, we conducted a comparative analysis of various cell clusters in 8-week-old GF and SPF mice. The analysis unveiled an augmented relative frequency of epithelial cells in the GF samples (Fig. 4A, B). Within the epithelial clusters, there was an increase in the relative fraction of the basal epithelial cells in GF mice, accompanied by a reduction in proliferating basal cells (Fig. 4C, D and Supplementary Fig. 2). Random subsampling of the SPF and GF samples to achieve equal cell numbers in each group showed that the overall cell composition and cluster structures remained consistent with those observed in the full dataset (Supplementary Fig. 3). This observation suggests that the microbiota plays a role in regulating epithelial turnover and proliferation kinetics. Supporting this notion, GESA indicated a decrease in KRAS downregulated genes within the cluster of proliferating basal cells—a signaling pathway crucial for cell proliferation and differentiation (Fig. 4E). Conversely, in the elevated basal cell cluster, an upregulation of pathways associated with cell growth was observed in GF mice. In line with this, the spinous and suprabasal epithelial clusters showed a relative increase in GF mice, although the GESA analysis did not reveal any changes in their cellular pathways in GF mice. To examine directly the impact of the microbiota on epithelial cell proliferation in vivo, we injected BrdU into SPF and GF mice, 5 h later gingival cross-sections were prepared for immunofluorescence analysis. As depicted in Fig. 4F, BrdU+ cells were mainly detected in the basal layer of the epithelium of both SPF and GF mice. However, the number of BrdU+ cells was significantly higher in the SPF epithelium compared to the GF epithelium, in both the buccal and palatal gingiva (Fig. 4F). This aligns with our observation that both regions originate from the same pool of proliferating basal cells. Next, a CellPhoneDB analysis was used to profile the communication among cell types in SPF and GF gingival tissues. The analysis revealed stronger cell-cell interactions in GF mice compared to SPF mice (Fig. 4G and Supplementary Fig. 4). This could be attributed to increased tissue damage in GF mice caused by masticatory forces, stemming from improper microbiota-dependent maturation of the gingival epithelium after birth9,11. These data demonstrate that exposure to the microbiota regulates cell-cell interaction in the gingiva and facilitates the proliferation rate of the gingival epithelium.
Fig. 4. Microbiota influence on gingival epithelial cell proliferation and function.
A and B UMAP representation of major cell types identified in the gingiva of 8-week-old SPF and GF mice via scRNA-seq. The graph illustrates the proportion of each cell type in both mouse groups. C and D UMAP visualization of various epithelial cell populations in the gingiva of SPF and GF mice. The graph denotes the proportion of each cell type in the respective mouse groups. E GESA reveals distinct pathways expressed in basal cells and proliferating basal cells within the gingiva of SPF and GF mice. F Representative images and a graph depict the counts + SEM of BrdU-positive cells in the buccal and palatal gingival epithelium of 8-week-old SPF and GF mice (n = 5) at the 5-h mark following BrdU injection. Representative data from two independent experiments. JE, junctional epithelium, OE oral epithelium, LP lamina propria. G Interaction network among the major cell types in SPF and GF mice, constructed by CellPhoneDB. Thicker lines indicate more interaction with other types of cells. **P < 0.01; ***P < 0.001.
The microbiota exerts a greater influence on immunological function in the buccal gingiva compared to the palatal gingiva
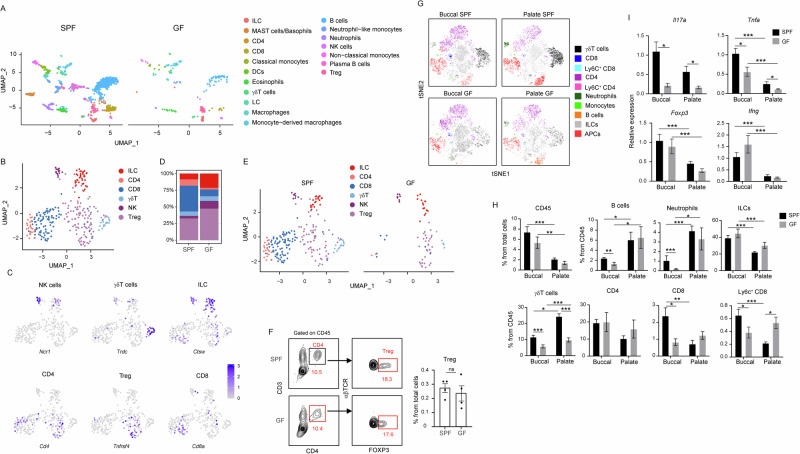
In addition to the influence of the microbiota on gingival epithelial cells, the analysis unveiled changes in the leukocyte clusters in the gingiva of 8-week-old GF mice (Fig. 5A). Turning our attention to T and innate lymphocytes, well-known for their role in modulating gingival immunity41, we initially characterized their various subsets. The analysis identified clusters of CD8+ (Cd8a, Cd8b1, Tcra) CD4+ (Cd4, Tcra) T cells, with a significant proportion of CD4+ T cells representing regulatory (Treg) cells (Cd4, Foxp3, Ctla4, Tnfsf4) (Fig. 5B, C). Additionally, γδT cells, specifically IL-17-producing Vγ6+ cells (Tcrd, Cxcr6, Cd163l1, Il17a), natural killer (NK) cells (Gzma, Nkg7, Ncr1, Klrk1), and innate lymphoid cells (ILC), predominantly ILC1 (Thy1, Ctsw, Ifng, Tnfsf10, Nkg7, Ncr1, Klrk1), were identified (Fig. 5B, C). The analysis further revealed that while CD8 and non-Treg CD4 cells were reduced in GF mice, the frequencies of ILCs and Treg cells were not reduced (Fig. 5D, E). The comparable frequencies of Treg cells from total cells were further verified by flow cytometry (Fig. 5F). Next, given the relatively low leukocyte numbers in the scRNA-seq data, we incorporated flow cytometry analysis of gingival epithelial tissues to support the investigation of the impact of the microbiota on local leukocytes. This approach also allowed us to concentrate on the epithelium of the buccal and palate gingival regions. As illustrated in Fig. 5G-H, the microbiota exhibited no significant influence on the frequencies of total CD45+ leukocytes in the gingival epithelium, although a trend of reduction can be observed in the buccal region. However, while the frequencies of B cells and neutrophils in the palate gingiva of GF and SPF mice were comparable, the levels of these cells in the buccal region were significantly diminished in GF mice. CD8 T cells, which are present in higher frequencies in the buccal gingiva, were reduced in the buccal gingiva of GF mice but left unaffected in the palate region. Ly6C expressing CD8 T cells, representing a small population of short-lived T cells42, were also reduced in the buccal region but upregulated in the palate gingiva of GF mice. The γδT cells were significantly reduced due to the absence of the microbiota in both regions of the gingival epithelium. Concerning CD4+ T cells, the overall population was unchanged in the epithelium of GF mice compared to SPF mice regardless of the specific region. To further dissect the immunological functions affected by the microbiota, we quantified the expression of key immunological genes in the gingival epithelium. Figure 5I demonstrates that expression of Il17a and Tnfa was upregulated by the microbiota in both regions of the gingival epithelium, while Tnfa expression was higher in the buccal than the palatal region regardless of the microbiota. Additionally, the levels of Foxp3 and Ifng were higher in the buccal compared to the palate gingival epithelium, and the microbiota had no impact on their expression levels. These findings underscore the capability of the microbiota to shape the immunological landscape of the gingival epithelium. Additionally, the data suggest a region-specific impact, emphasizing heightened responsiveness of the buccal epithelial region to the microbiota.
Fig. 5. Immune response of the buccal gingiva is more influenced by the microbiota in comparison to the palatal gingiva.
A UMAP representation of the various leukocytes identified in the gingiva of 8-week-old SPF and GF mice. B and C UMAP representation of lymphocytes (excluding B cells) in the gingiva, upon integration of SPF and GF data. A selected set of key genes related to the various lymphocytes are also presented. D and E UMAP representation of gingival leukocytes in SPF versus GF mice. The graph illustrates the proportion of each cell type in both mouse groups. F Flow cytometry plots and graphs show the frequencies + SEM of CD4+ and Foxp3+ CD4+ T cells (Treg) in the gingiva of 8-week-old SPF and GF mice (n = 5). Representative data from two independent experiments. G tSNE flow cytometry plots display the main subsets of leukocytes present in the gingival epithelium of the buccal and palatal regions of 8-week-old SPF and GF mice. Representative data from two independent experiments. H Graphs depict the frequencies of the main leukocyte subsets in the epithelium of the buccal and palatal gingiva of SPF and GF mice. Data are presented as the mean frequencies + SEM (n = 5). Representative results from two independent experiments. I Relative expression of the noted genes in the buccal and palatal gingiva of 8-week-old SPF and GF mice. Graphs present the transcript levels quantified by qPCR and normalized to the buccal region of SPF mice and depicted as the mean + SEM (n = 5). Representative data from two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
The microbiota shapes the structure and function of the junctional epithelium
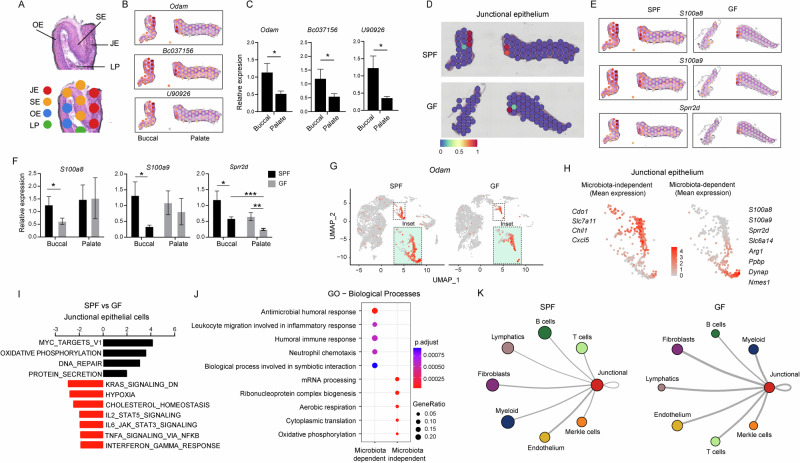
The junctional epithelium, a distinctive region of the gingiva positioned close to the dental plaque, prompted an exploration into the potential regulatory role of the microbiota on its structure and function. Through Visium spatial analysis, distinct gingival regions, comprising the junctional, sulcular, and oral epithelium, along with the underlying lamina propria, were delineated in both the buccal and palate regions of the gingiva (Fig. 6A). This identification relied on gene expression patterns, including Odam, Bc037156, and U90026, which were subsequently validated through qPCR (Fig. 6B, C). Notably, the expression of these genes was higher in the buccal than the palatal gingival epithelium. The visium analysis also indicates that the microbiota is crucial for the development of the junctional epithelium, since areas defined as the junctional epithelium were less visible, particularly in the buccal region (Fig. 6D). Indeed, the spatial expression of genes such as S100a8, S100a9, and Sprr2d that are enriched in the junctional epithelium were reduced in GF mice (Fig. 6E). qPCR also verified the reduction in the expression of the noted genes in GF mice and further demonstrated that while all three genes were reduced in the buccal region, Sprr2d was also reduced in the palate region (Fig. 6F). Utilizing the scRNAseq analysis, Odam expression, which identifies the junctional epithelium, was reduced in GF mice (Fig. 6G). Interestingly, two sub-clusters can be visualized in the junctional epithelium, while the lower cluster was considerably diminished in the GF mice (Fig. 6G, insets). This enables the identification of genes whose expression is microbiota-dependent (S100a8, S100a9, Sprr2d, Slc6a14, Arg1, Ppbp, Dynap, and Nmes1) and independent (Cdo1, Slc7a11, Chil1, and Cxcl5) within the junctional epithelium (Fig. 6H).
Fig. 6. The microbiota shapes the structure and function of the junctional epithelium.
A Representative H&E image of a gingival cross-section from an 8-week-old SPF mouse demonstrating the capability of the spatial analysis to discern distinct anatomical regions within the gingiva. B Spatial feature plots showing expression of distinct genes that define the junctional epithelium in the buccal or palatal regions of the gingiva. C Validation of the expression of the specified genes in epithelial tissues isolated from the buccal and palatal regions separately using qPCR (n = 5). Representative data from two independent experiments. D Spatial and scRNA-seq integration allows for demarcation of the junctional epithelium in SPF and GF gingiva. E Spatial layout showing expression of distinct genes defining the junctional epithelium in the buccal or palatal gingiva of SPF and GF mice. F qPCR validation of the expression of the specified genes in epithelial tissues isolated from the buccal and palatal regions of 8-week-old SPF and GF mice (n = 5). Representative data from two independent experiments. G UMAP layout depicting Odam expression to demarcate the junctional epithelium in SPF and GF mice (Inset, enlargement of the junctional epithelium). H UMAP plots demonstrate the mean expression of microbiota-dependent and independent genes as specified. I GESA demonstrates the distinctive pathways expressed by the junctional epithelium of SPF and GF mice. J Dot plots depicting the main biological pathways expressed by the microbiota-dependent and -independent clusters of the gingival epithelium. Expression values are normalized and scaled averages. K Interaction network of junctional epithelial cells with the other major cell types in the gingiva of SPF and GF mice, constructed by CellPhoneDB. Thicker lines indicate more interaction with other types of cells. *P < 0.05, **P < 0.01, ***P < 0.001.
Subsequently, GESA was conducted on epithelial cells from the junctional epithelium of SPF versus GF mice. As depicted in Fig. 6I, the analysis revealed notable changes in cell growth and proliferation pathways, coupled with the upregulation of oxidative phosphorylation and protein secretion pathways in GF mice. Conversely, there was a reduction in pathways related to hypoxia and cholesterol hemostasis in this group. These findings suggest a metabolic shift in epithelial cells due to the absence of microbiota, potentially impacting both epithelial renewal and integrity43. Additionally, the analysis revealed a reduction in immunological pathways such as IFN-γ, TNF-α, and IL-16 singling as well as IL2/STAT5 signaling associated with T regulatory cell development, all known to shape oral mucosal immunity44. Further exploration using GO analysis unveiled additional immunological pathways reduced in the junctional epithelium of GF mice. As depicted in Fig. 6J, microbiota-dependent immunological pathways in junctional epithelia cells involve antimicrobial humoral response, symbiotic interactions, processes related to migration of inflammatory cells, and neutrophil chemotaxis. A CellPhoneDB analysis further demonstrated stronger cell-cell interactions of the junctional epithelium with other cell types in GF mice than in SPF mice (Fig. 6K and Supplementary Fig. 5). Taken together, these findings demonstrate that the microbiota plays a crucial role in the development of the junctional epithelium, with a notable impact on the buccal region.
CXCL7, rather than CXCL1 or CXCL2, mediate microbiota-dependent neutrophil recruitment by the junctional epithelium
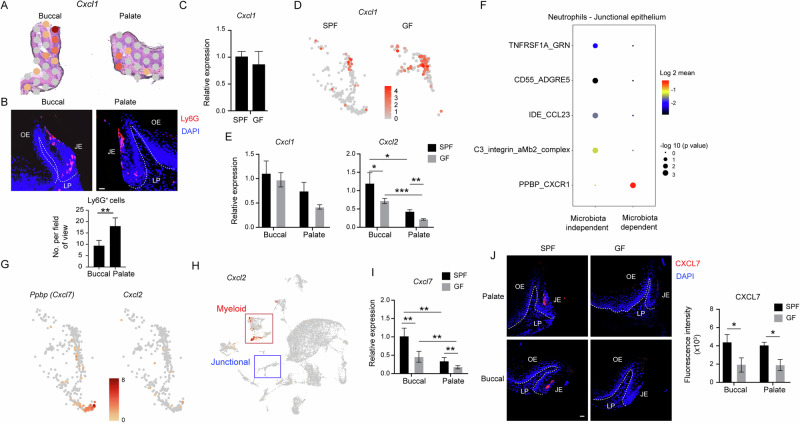
Concerning neutrophil chemotaxis, Cxcl1, a gene encoding the neutrophil-attracting chemokine CXCL1, exhibited preferential expression in the junctional epithelium (Fig. 7A). Accordingly, Ly6G-positive cells, indicative of neutrophils, were visualized by immunofluorescence staining in the junctional epithelium at the buccal and palatal regions, with a higher number of neutrophils detected on the palatal side (Fig. 7B). However, the microbiota did not influence the expression of Cxcl1 (Fig. 7C). Moreover, Cxcl1 expression was restricted to the upper subcluster of the junctional epithelium that is considered microbiota-independent (Fig. 7D). Next, the expression of neutrophils-attracting chemokines was examined in the buccal and plate regions of GF and SPF mice. As demonstrated in Fig. 7E, Cxcl1 expression was comparable in both regions. Cxcl2, on the other hand, was highly expressed in the buccal region, while the microbiota reduced the expression in both regions. To further investigate the interaction between neutrophils and the junctional epithelium, a receptor–ligand analysis was conducted. Figure 7F illustrates the most significant receptor–ligand interactions in the upper (microbiota-independent) and lower (microbiota-dependent) subclusters. Notably, the interaction between CXCR1 and PPBP (CXCL7), which is involved in neutrophil recruitment, was found to be influenced by the microbiota. Following this observation, the expression of Cxcl7 was predominantly confined to the lower cluster of the junctional epithelium (Fig. 7G). Conversely, Cxcl2 was almost absent in the junctional epithelium; instead, the primary cluster expressing this chemokine was the myeloid cell cluster (Fig. 7H). A qPCR analysis further demonstrated that Cxcl7 expression was affected by the microbiota in both the buccal and palate regions, while the expression in the buccal, which is more microbiota-dependent, was higher than the palate (Fig. 7I). Moreover, immunofluorescence staining confirmed the specific expression of CXCL7 protein in the junctional epithelium, which was diminished in GF mice (Fig. 7J). These results imply that junctional epithelial cells mediate neutrophil recruitment through CXCL1 independently of the microbiota, whereas CXCL7 mediates this process in a microbiota-dependent manner.
Fig. 7. Microbiota-dependent expression of CXCL7 in junctional epithelial cells.
A Spatial plot depicting the expression of Cxcl1 in the junctional epithelium of buccal and palatal gingiva in 8-week-old SPF and GF mice. B Immunofluorescence staining of buccal and palatal gingival cross-sections with antibodies against Ly6G (red) and DAPI (blue) for nuclear visualization. The accompanying bar graph shows the mean number of Ly6G+ cells + SEM (n = 4–5) in the palatal and buccal gingiva. Representative image from three independent experiments. Scale bar, 50 µm. C Expression levels of Cxcl1 + SEM in the gingival tissue isolated from 8-week-old SPF and GF mice (n = 5) using qPCR. D UMAP plots demonstrating the expression of Cxcl1 in the junctional epithelium clusters of SPF and GF mice. E Quantification of Cxcl1 and Cxcl2 expression in the buccal and palatal gingiva of 8-week-old SPF and GF mice. Graphs present the transcript levels quantified by qPCR and normalized to the buccal region depicted as the mean ± SEM (n = 5). Representative data from two independent experiments. F Selected receptor–ligand interactions between neutrophils and the microbiota-dependent and microbiota-independent clusters of the junctional epithelium. G UMAP plots signifying the expression of Cxcl7 and Cxcl2 in the junctional epithelium clusters of SPF and GF mice. H UMAP plots demonstrating the expression of Cxcl2 by major cell types in the gingiva of SPF mice. I Quantification of Cxcl7 expression in the buccal and palatal gingiva of 8-week-old SPF and GF mice. Graphs present the transcript levels quantified by qPCR and normalized to the buccal region depicted as the mean ± SEM (n = 5). J Immunofluorescence staining of buccal and palatal gingival cross-sections collected from 8-week-old SPF and GF mice with antibodies against CXCL7 (red) and DAPI (blue). Representative images and a graph display the immunofluorescence intensity as the mean ± SEM (n = 4–5). Scale bar, 50 µm. Representative data from two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
The microbiota enhances the sealing of the gingival epithelium and regulates the development of the glandular epithelium
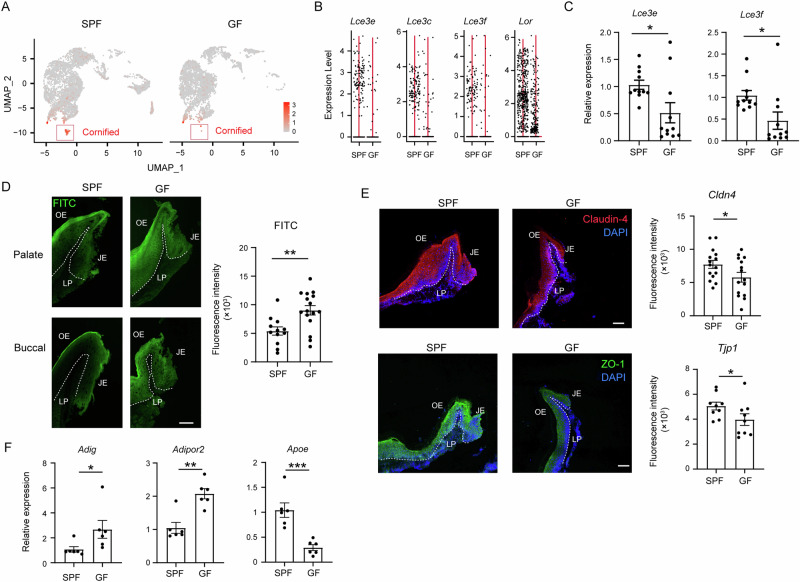
We have shown previously that the oral epithelium is sealed during the weaning period, represented, in part, by elevated expression of tight junction proteins9. We thus asked whether the microbiota affect the sealing process in the gingival epithelium. First, the scRNA-seq analysis shows that the cluster of cornified cells, which provides a permeability barrier45, was reduced in GF mice (Fig. 8A). As depicted in Fig. 8B, genes associated with the cornified cluster such as Lce3e, Lce3c, Lce3c, and Lor were downregulated due to the absence of the microbiota. The downregulation of some of these genes in GF mice was further confirmed by qPCR (Fig. 8C). To directly evaluate the influence of the microbiota on the permeability of the gingival epithelium, a FITC solution was applied to the gingiva, and the penetration of the fluorescent dye into the tissue was examined 30 min later. Cross-sections of the gingiva, prepared from both the buccal and palate sides, revealed a greater intensity of FITC labeling in the gingival epithelium of GF mice, highlighting the crucial role of the microbiota in epithelial sealing (Fig. 8D). Next, we assessed the expression of tight junction proteins, specifically claudin 4 and tight junction protein 1 (Tjp1, also known as ZO-1), through immunofluorescence staining. Consistent with the increased permeability observed in GF mice, the expression levels of these proteins were down-regulated in these mice, as depicted in Fig. 8E. In addition to tight junction proteins, cholesterol metabolism plays a crucial role in the development of the cornified layer46. To investigate this further, we conducted qPCR analysis to measure the expression levels of genes associated with cholesterol metabolism, including Adig, Adipor2, and Apoe. As illustrated in Fig. 8F, the expression levels of these genes were found to be decreased in the epithelium of GF mice compared to the SPF group. Besides the gingiva, the microbiota also governed the non-squamous and glandular epithelial clusters, as evidenced by the significant reduction in the latter in GF mice, while the non-squamous cells appeared to be augmented (Supplementary Fig. 6A). Pseudotemporal trajectory analysis revealed that glandular epithelial cells stem from non-squamous epithelial cells (Supplementary Fig. 6B), suggesting that the observed increase in non-squamous cells in GF mice is due to diminished differentiation capacity into glandular epithelial cells. This notion is supported by the GESA, which demonstrates heightened cellular and developmental pathways in non-squamous epithelial cells of SPF mice compared to GF mice (Supplementary Fig. 6C). Collectively, these findings imply that the microbiota plays a crucial role in sealing the gingival epithelium. The microbiota also governs the differentiation of glandular epithelial cells.
Fig. 8. Impaired gingival epithelial sealing in GF mice.
A UMAP plots delineating the cornified epithelial cells in SPF and GF mice. B Violin plots displaying the expression of the specified genes in the two groups of mice. C Validation of gene expression in gingival epithelial tissues from 8-week-old SPF and GF mice via qPCR. Graphs depict transcript levels quantified and normalized to the SPF epithelium, represented as mean ± SEM (n = 5). Representative data from two independent experiments. D Representative images and graph demonstrating FITC penetration into the gingival epithelium of 8-week-old SPF and GF mice, 30 min post-application of FITC solution on the gingiva. The bar graph presents fluorescence intensity quantification as mean ± SEM (n = 5). Scale bar, 50 µm. Representative data from two independent experiments. E Immunofluorescence of gingival cross-sections from 8-week-old SPF and GF mice stained with monoclonal antibodies directed against ZO-1 (green), claudin 4 (red), and Hoechst (blue) for nuclear visualization. The bar graph shows fluorescence intensity quantification as mean ± SEM (n = 5). Representative image from three independent experiments. Scale bar, 50 µm. F Expression levels of specified genes in the gingival epithelium of 8-week-old SPF and GF mice. Graphs depict transcript levels quantified by qPCR and normalized to SPF mice, presented as mean ± SEM (n = 5). Representative data from two independent experiments.
Discussion
While originating from similar precursors, the buccal and palatal regions of the gingival epithelium differentiate into distinct types of stratified epithelia with varying immunological activities. This divergence may be attributed to the respective interactions of each epithelium with the microbiota, given the less keratinized nature of the buccal epithelium, leading to more complex immunological signaling47. Consequently, the lack of microbiota results in reduced frequencies of several leukocytes in the buccal gingival epithelium (e.g., neutrophils, B cells, and CD8+ T cells), contrasting with the palate side where no such reduction occurred. Moreover, the buccal gingiva also contains higher levels of LCs than the palate region, the major antigen-presenting cells of stratified epithelia48. The development of LCs in the vicinity of the dental plaque is enhanced by the microbiota11,14, supporting the stronger interaction of the buccal gingiva with the microbiota. The elevated levels of CD8+ T cells and ILCs in the buccal gingival epithelium of SPF mice also align with this notion, as both cell types have been shown to play a role in preventing viral infection in the oral mucosa49,50. This implies that the buccal and palatal gingival epithelia constitute immunologically distinct regions, potentially influencing pathological processes within this oral barrier.
Under steady-state conditions, neutrophils are rarely present in the oral epithelium of adult mice, with the exception of the gingival epithelium where they are primarily located in the junctional epithelium9,13. The present study further substantiates this by showing the specific expression of neutrophil-attracting chemokines in this region, and by visualizing the neutrophils in the junctional epithelium of both the palate and buccal sides. Interestingly, despite the lower association of the palate gingiva with the microbiota compared to the buccal region, the palate exhibited higher frequencies of neutrophils. This observation may contribute to the increased kinetics of natural alveolar bone loss found in the palate region, as neutrophils have been implicated in this process51. The elevated neutrophil population in this region is likely associated with the increased levels of γδT cells, which are the predominant producers of the neutrophil-recruiting cytokine IL-17 in the gingiva and are involved in periodontal bone loss15,52. Moreover, neutrophils can enhance the survival of B cells expressing the receptor activator for nuclear factor κ B ligand (RANKL), which has the capacity to induce bone loss53,54. Indeed, our analysis revealed higher frequencies of B cells in the palate region of the gingiva. Since the frequencies of both neutrophils and B cells in the palate gingiva are not reduced in GF mice, this supports the view that natural bone loss is less influenced by microbiota and more by masticatory forces10,11. Regardless, the reasons for the higher abundance of neutrophils in the palate region remain unclear. This may arise from greater masticatory forces exerted on the palate side of the maxilla compared to the buccal side, which could influence epithelial function. Supporting this notion, masticatory forces have been reported to affect gingival epithelial cell growth and the wound-healing process55. Alternatively, the physiological process of neutrophil clearance from circulation to maintain neutrophil homeostasis, partially mediated by the gingiva56, could be more robust in the palate region of the gingiva.
This study unveiled a substantial impact of the microbiota on the junctional epithelium, with an entire subcluster of this region significantly reduced in GF mice. This reduction might be attributed to the incapacity of certain cell populations to develop in GF mice, aligning with a previous study reporting a decreased size of the junctional epithelium in GF mice compared to conventionalized mice57. In this regard, commensals are known as active participants in the development of the structure and function of host tissues, particularly in mucosal sites58. Alternatively, the diminishment could result from the downregulation of microbiota-dependent genes that segregate the junctional epithelium into two distinct subclusters. Regardless, the spatial analysis indicates that the junctional epithelium in the buccal region is more affected by the microbiota. Accordingly, neutrophils, primarily located in the junctional epithelium, were reduced only in the buccal region. This refines earlier observations indicating reduced neutrophils in the GF gingiva13, suggesting a region-specific reduction. However, the mechanism contributing to this differential process remains unclear. While Cxcl2 is downregulated in GF mice, in agreement with other reports13,59, this chemokine is predominantly expressed by myeloid cells such as neutrophils, LCs, and monocytes. These myeloid cells were reported to express CXCL260–62 and are present in or near the junctional epithelium and their numbers are reduced in GF mice11,13,14. Thus, another chemokine likely recruits neutrophils in a microbiota-dependent fashion. Indeed, the present study demonstrates that CXCL7 is expressed in epithelial cells of the junctional epithelium and is regulated by the microbiota. In this regard, CXCR1, the receptor of CXCL7 is expressed by murine gingival neutrophils and involved in periodontal bone loss63, while CXCL7 is reported to be associated with murine periodontitis64.
The analysis further reveals stronger cell–cell interactions in the gingiva of GF mice compared to the SPF group. We have previously demonstrated that exposure to the microbiota during the neonatal period is crucial for the proper maturation of the oral epithelium2,9 and for the recruitment of gingival leukocytes, particularly in the epithelium11,14,15. This process is essential for preparing the gingiva for the weaning period, during which masticatory forces are initiated for the first time in life. In the absence of microbiota, the gingiva is not perfectly sealed, as demonstrated in the present study, and its ability to cope with the damage caused to the epithelium by mastication is imperfect11. This might explain the stronger cell-cell interactions detected in the GF mice, as the gingiva requires extra activity to compensate for the impaired development of the epithelium during neonatal life.
The findings from the mouse model provide valuable insights into the human oral mucosa, as both species share key immunological and structural features in their mucosal barriers. Notably, CXCL7 is expressed in human gingiva and is upregulated during periodontitis, suggesting it may be similarly regulated by the microbiota as observed in mice65. Moreover, in both healthy individuals and periodontitis patients, the residual alveolar bone on the buccal side of the maxillary anterior teeth is thicker than on the lingual side, aligning with our observations in mice66. These findings highlight that the distinct immunological niches identified in murine gingiva reflect regional variations in human gingival immunity, reinforcing the utility of mouse models in uncovering mechanisms of oral homeostasis and disease that are likely translatable to humans.
In summary, through spatial and transcriptomic analysis, this study has elucidated unique developmental and immunological characteristics inherent to the buccal and palate regions of the gingiva. Additionally, it sheds light on the influence of the microbiota on the diverse niches constituting the gingival epithelium, thereby advancing our understanding of this crucial oral mucosal barrier.
Methods
Mice
C57BL/6 (B6) mice were bred and maintained in the central animal facility at the Hebrew University Faculty of Medicine (Jerusalem, Israel). The mice were maintained under SPF conditions and analyzed at various ages as described in the text. All animal protocols were approved by the Hebrew University Institutional Animal Care and Use Committee (IACUC). Germ-free (GF) B6 mice were maintained in sterile isolators at the Weizmann Institute of Science, and the GF studies were approved by the IACUC of the Weizmann Institute of Science. Both GF and SPF mice, 8 weeks of age unless otherwise specified, were used for all experiments.
Isolation of tissue leukocytes
The gingival tissues were excised. In some experiments, gingival tissues were incubated in 1 mL of 4 mg/ml Dispase in PBS + 0.04% BSA until fully distended for 30 min. The epithelium was carefully separated using forceps and a binocular microscope. Tissues were then minced and treated with a Collagenase type II (2 mg/mL; Worthington Biochemicals) and DNase I (1 mg/mL; Sigma) solution in PBS plus 2% FCS for 25 min at 37 °C in a shaker bath. A total of 20 μL of 0.5 M EDTA per 2 mL sample was added to the digested tissues and incubated for an additional 10 min. The cells were then washed, filtered with a 70-μM filter, stained with antibodies, run in Aurora (Cytek) flow cytometer, and further analyzed generating plots and tSNE plots using FlowJo software (BD Biosciences).
Antibodies
| Antibodies | Source | Identifier |
|---|---|---|
| Rat PE anti-mouse CD3 | BioLegend | Cat# 100206; RRID: AB_312663 |
| Armenian hamster FITC anti-mouse TCR γ/δ | BioLegend | Cat# 118106; RRID: AB_313830 |
| Mouse Pacific Blue anti-mouse CD45.2 | BioLegend | Cat# 109820; RRID: AB_492872 |
| Mouse PE anti-mouse, human Langerin (CD207) | BioLegend | Cat# 144204; RRID: AB_2561499 |
| Armenian hamster Brilliant Violet 605 anti-mouse TCR β chain | BioLegend | Cat# 109241; RRID: AB_2629563 |
| Rat PerCP/Cyanine 5.5 anti-mouse I-A/I-E | BioLegend | Cat# 107626; RRID: AB_ 2191071 |
| Rat APC anti-mouse Ly-6G | BioLegend | Cat# 127614; RRID: AB_2227348 |
| Rat Brilliant Violet 711 anti-mouse CD326 (EpCAM) | BioLegend | Cat# 118233; RRID: AB_2632775 |
| Rat PE/Cyanine7 anti-mouse Ly-6C | BioLegend | Cat# 128018; RRID: AB_1732082 |
| Armenian hamster Brilliant Violet anti-mouse CD69 | BioLegend | Cat# 104543; RRID: AB_2629640 |
| CD207 (Langerin) Monoclonal Antibody (eBioRMUL.2), eBioscience™ | Invitrogen | Cat# 14-2073-82; RRID: AB_493943 |
| Anti-gamma H2A.X (phosph S139) antibody [9F3] | abcam | Cat# ab26350 |
| Rat PE/Cyanine7 anti-mouse/human CD45R/B220 | BioLegend | Cat# 103222; RRID: AB_ 313005 |
| Rat Brilliant Violet 711™ anti-mouse Ly-6C | BioLegend | Cat# 128037; RRID: AB_ 2562630 |
| Rat PE/Cyanine7 anti-mouse/human CD11b | BioLegend | Cat# 101216; RRID: AB_312799 |
| Rat APC/Cy7 anti-mouse CD4 | BioLegend | Cat# 100414; RRID: AB_ 312699 |
| Rat PE/Cyanine5 anti-mouse CD8a | BioLegend | Cat# 100709; RRID: AB_312749 |
| Rat Brilliant Violet 650TM anti-mouse/human CD11b | BioLegend | Cat# 101259; RRID: AB_ 2566568 |
| Rat PE/DazzleTM 594 anti-mouse Ly-6G | BioLegend | Cat# 127647; RRID: AB_ 2566318 |
| Armenian hamster APC/Cyanine7 anti-mouse CD11c | BioLegend | Cat# 117324; RRID: AB_ 830649 |
| Rat anti-Mouse I-A/I-E BUV496 M5/114.15.2 | BD Biosciences | Cat# 750281; RRID: AB_ 2874472 |
| Rat BD741261 BUV563 anti-mouse CD103 | BD Biosciences | Cat# 741261; RRID: AB_ 2870808 |
| Rat Alexa Fluor® 488 anti-mouse FOXP3 | BioLegend | Cat# 126406; RRID: AB_ 1089113 |
| Rabbit Anti-CXCL7/PBP antibody | Abcam | Cat# ab206406 |
Immunofluorescence staining
For whole-mount staining, gingival tissues were incubated in 1 mL of 4 mg/ml Dispase in PBS + 2% FCS until fully distended for 30 min, and epithelium was carefully separated using forceps and binocular microscope. Tissues were then fixed in ice-cold 95% ethanol for 40 min. For frozen section staining, the mandibles were fixed overnight at 4 °C in 4% paraformaldehyde/PBS solution, decalcified for 2–3 weeks in EDTA, embedded in OCT, and cryo-sectioned into 10-μm-thick sections. The cross sections, as well as the whole tissues, were washed 3 times in PBS, blocked in blocking buffer (5% FCS, 0.1% Triton X-100 in PBS) for 1 h at room temperature, and incubated with a primary antibody overnight at 4 °C. Following three washing steps in PBS, the samples were incubated with a secondary antibody diluted 1:200 in blocking buffer for 2 h at room temperature, washed 3 times, stained with DAPI and mounted. For paraffin sections, the salivary glands were fixed overnight at 4 °C in 4% paraformaldehyde/PBS solution, and then the tissues were dehydrated using 70%, 80%, 90%, and 100% ethanol and then xylene to dissolve the alcohol. Next, the tissues were embedded in paraffin and micro-sectioned into 7 μM-thick sections. Slides were deparaffinized with xylene, and 100%, 95%, 80% and 70% ethanol washed 3 times with PBS, blocked in blocking buffer (PBS, 10% FCS, 10% BSA, 2% triton X-100) for 1.5 h at room temperature and incubated with primary anti-rat antibodies overnight at 4°C. Following 3 washing steps in PBS, the samples were incubated with secondary antibodies: Donkey anti-rabbit IgG (Invitrogen) or donkey anti-rat IgG (Invitrogen) diluted 1:200 in blocking buffer for 2 h at RT, washed 3 times, stained with DAPI and mounted. As a negative staining control, the primary antibody was omitted and replaced by blocking buffer. Signals were visualized and digital images were obtained using a Nikon TL microscope for the cross sections and a Nikon spinning disk confocal microscope for the wholemount tissues.
Micro-computed tomography (μCT) analysis
The hemi-maxillae were secured in an acrylic mold using orthodontic wax for stability during acts scans, which were conducted at 8 µm resolution using the μCT instrument Skyscan1272 (Bruker microCT, Kontich, Belgium). Air served as the scan medium to optimize the contrast between the sample and the background. 3D reconstruction and analysis of the μCT images were executed using Dragonfly software [Version 2022.2 for Windows; Object Research Systems (ORS) Inc., Montreal, Canada] with Tiff files.
RNA extraction and qPCR
For RNA isolation, the excised gingival tissues and tongues were homogenized in 500 µl TRI reagent (Sigma) using an electric homogenizer (IKA labortechnik) and RNA was extracted according to the manufacturer’s instructions. cDNA synthesis was performed using the qScript cDNA Synthesis Kit (Quanta-BioSciences). qPCR reactions (10 µL volume) were performed using Power SYBR Green PCR Master Mix (Quanta-BioSciences) and specific primers to the examined gene. The following reaction conditions were used: 10 min at 95 °C, 40 cycles of 15 s at 95 °C, and 60 s at 60 °C. The samples were normalized to GAPDH as control mRNA, by change in cycling threshold (ΔCT) method and calculated based on 2−ΔΔCT.
RNA-Seq differential expression analysis
In brief, 1–2 µg RNA was used for the library construction. For quality control of RNA yield and library synthesis products, the RNA ScreenTape and High Sensitivity D1000 ScreenTape kits (both from Agilent Technologies), Qubit® RNA HS Assay kit, and Qubit® DNA HS Assay kit (both from Invitrogen) were used for each specific step.; KAPA Stranded mRNA-Seq Kit Illumina® Platforms kit (Roche) was used for library preparations according to manufacture protocol, and the final DNA library was eluted in 30 µL of elution buffer. Libraries were normalized and pooled together. Multiplex sample pool was loaded in NovaSeq 6000 SP Reagent Kit v1.5 (100 cycles) (Illumina). Run conditions were in single end and 122 bp length and loaded on NovaSeq 6000 system machine (Illumina).” Binary Base Call (BCL) output files from a Novaseq600 machine were converted to FASTQ format raw reads, using BCL to FASTQ (bcl2fastq v2.20.0.422 Copyright (c) 2007–2017 Illumina, Inc.).
Raw reads were processed according to the QuantSeq User Guide recommendations, reads were trimmed at their 5’ end to remove the first 12 bases, then low quality and technical bases were removed from the 3’ end using cutadapt (version 1.12). Finally, low-quality reads, with more than 30 percent of the bases with quality below 20, were filtered out using the FASTX package (version 0.0.14). Processed reads were aligned against the mouse genome using TopHat (v2.1.1)67. The genome version was GRCm38, with annotations from Ensembl release 89. Htseq-count (version 0.6.0)68 was then used for quantification of raw counts per gene per sample, excluding short or otherwise unwanted gene types, such as rRNA or miRNA. Normalization and differential expression analysis were performed with the DESeq2 package (version 1.12.4)69. Genes with a sum of counts less than 10 over all samples were filtered out prior to normalization. Differential expression, comparing 8-weeks to 1-week-old mice, was calculated with default parameters, except not using the independent filtering algorithm. Statistical significance 6 thresholds were taken as an adjusted p-value (padj) < 0.1. Exact commands with the full parameters used can be found underthe GEO accession.
Gene set enrichment analysis (GSEA)
Whole differential expression data were subjected to gene set enrichment analysis using GSEA70. GSEA uses all differential expression data (cut-off independent) to determine whether a priori-defined sets of genes show statistically significant, concordant differences between two biological states. GSEA was run against the hallmark gene set collection from the molecular signatures database (MSigDB, v6.2, July 2018).
Epithelial permeability essay
20 mg of FITC (Sigma) was dissolved in 100 µl DMSO (sigma), and the solution was diluted in acetone (1:1). Mice were anesthetized, and 40 µl of the solution was carefully applied to the gingiva. The gingival tissues were excised and embedded in OCT for frozen block preparation, and coronal sections were generated. Images were obtained using confocal microscopy, and epithelial permeability was assessed based on FITC fluorescence intensity in cross sections as measured using FIJI image.
Single-cell RNA-seq (scRNA-seq) (10× Chromium)
Library preparation
The gingival epithelial tissues were collected from 4 SPF and 4 GF mice and processed as described above, except that EDTA was not used. The samples were then subjected to the Chromium Next GEM Single Cell 3ʹ GEM, Library & Gel Bead Kit v3.1 (10x Genomics, CA, USA) and libraries were prepared for sequencing following manufacturer company instructions. Sequencing was done using Illumina Nextseq500 platforms with the following sequencing conditions: 28 bp (Read1) and 54 bp (Read2).
Single-cell analysis methods
CellRanger pipeline71 (v6.0.1, 10x genomics) with default parameters was used for demultiplexing, alignment (mm10 reference genome, 2020-A version, downloaded from 10x website), filtering, barcode counting, and UMI counting. The Seurat R package72 (v4.0.4) was used for downstream analysis and visualization. Gene-cell matrices were filtered to remove cells with more than 25% of reads mapped to mitochondrial genes, less than 250 genes and more than 6000 genes, <500 UMIs, and more than 50,000 UMI. In addition, genes detected in fewer than 10 cells were excluded from the analysis. After implementing these quality control measures, a total of 4,161 GF cells, 10,300 SPF cells, and 16,835 genes were retained for further analysis. To remove doublets, cells with an unusually high number of genes/UMIs or those showing co-expression of specific marker genes were manually filtered. Next, the scDblFinder package in R was used to computationally identify doublets, marking 600 doublets in GF and 1515 in SPF samples. After manual filtering, 464 potential doublets in GF and 679 in SPF remained. These were evenly distributed across clusters, indicating that no specific cluster was disproportionately affected by doublets. To further evaluate the impact of the remaining doublets, they were entirely removed, and the analysis was rerun. The results confirmed a high degree of consistency with the original clusters.
The expression data was normalized using Seurat’s NormalizeData function, which normalizes the feature expression measurements for each cell by the total expression, multiplies this by a scale factor (10,000), and then log-transforms the results. The top 2000 highly variable genes were identified using Seurat’s FindVariableFeatures function with the ‘vst’ method. Potential sources of unspecific variation in the data were removed by regressing out the UMI count using linear models and scaling and centering the residuals as implemented in the function “ScaleData” of the Seurat package. Principal component analysis (PCA) was performed. 20 PCs were used for clustering and data reduction. Cell clusters were generated using Seurat’s unsupervised graph-based clustering functions “FindNeighbors” and “FindClusters” (resolution = 0.5). UMAP was generated using the RunUMAP on the projected principal component (PC) space. Using manual inspection, several cells/clusters classified as erythrocytes (based on the expression of Hbbs-bs, Hbbs-bt, Hba-a1, and Hba-a2 genes) were removed, and the above steps were re-done to generate new clusters and embeddings. The same steps were done also for specific sub-populations.
Seurat’s functions FeaturePlot and DimPlot were used for visualization. Seurat’s DotPlot and VlnPlot functions were used to visualize gene expression for each cluster. Plots were further formatted using custom R scripts with the packages ggplot273 and patchwork (https://CRAN.R-project.org/package=patchwork). Heatmaps were produced with Seurat’s function DoHeatmap or the R package pheatmap (https://cran.r-project.org/package=pheatmap). Marker genes for each cluster were identified by performing differential expression between a distinct cell cluster and the cells of the other clusters with the non-parametric Wilcoxon rank sum test (Seurat’s FindAllMarkers function). Cell types were assigned manually based on the expression of classic marker genes. DE analysis between specific populations was done using FindMarkers function with default parameters. Trajectory analysis for specific sub-populations was done using slingshot R package74. CellPhoneDB75 was used to find interactions between clusters. Results were plotted using the R package ktplots (10.5281/zenodo.5717922).
10X Visium spatial transcriptomics
Preparation of gingival tissues
The freshly obtained gingival tissues were snap-frozen on dry ice and embedded in a freezing and embedding compound at Optimal Cutting Temperature (OCT). The OCT-embedded tissues were cryosectioned at a 10 μm thickness and placed on Visium Spatial slides. One to two sections from each mouse were placed on each spatial slide, with the two spatial slides containing tissues from SPF mice and two spatial slides containing tissues from GF mice.
RNA quality assessment
The quality of RNA in the tissue blocks was assessed by calculating the RNA integrity number (RIN) using High Sensitivity RNA ScreenTape (Agilent) of freshly collected PCa tissue. Tissue sections with an RIN ≥ 7 were selected for placement on Visium Spatial slides.
Preparation of spatially barcoded arrays, staining and imaging
The Visium Spatial Gene Expression Slide & Reagent kit (10× Genomics, CA, USA) was used to generate spatially barcoded cDNA from tissue sections according to the manufacturer’s instructions. The Visium Spatial slide is a Codelink-activated microscope glass slide upon which poly(dT)VN oligonucleotides (IDT) are distributed. The arrays on the slide were designed so that 4992 (64 × 78) spots containing unique barcoded oligonucleotides with poly(dT)VN were printed in each 6.5 × 6.5 mm2 capture area. The diameter of each spot was 55 μm, and the center-to-center distance of two adjacent spots was 100 μm. The fiducial frame (red spots) was printed as a border around the capture areas to maintain orientation. The Visium Spatial Tissue Optimization Slide with PCa tissue sections was subjected to methanol fixation and hematoxylin–eosin (H&E) staining.
Tissue optimization and permeabilization
In the quality control experiment, the permeabilization conditions for the tissue sections were optimized prior to spatial barcoding experiments to maximize mRNA yields in tissue sections. The experiment was performed by utilizing the Visium Spatial Tissue Optimization Slide & Reagent kit (10X Genomics) according to the manufacturer’s instructions. The optimal permeation time (with the strongest fluorescence signal intensity and the lowest dispersion among tested times of 30, 24, 18, 12, 6, and 3 min) was determined by fluorescence imaging after fluorescence cDNA synthesis and tissue removal. The glass slides were scanned using an Olympus BX53 microscope (Olympus, Shinjuku, Tokyo, Japan), and signal intensities were investigated using the cellSens Dimension Software (Olympus) system. The cDNA fluorescence signals should be consistent with the structure of the tissue section shown by histology. This step was performed to ensure that all array sequences necessary for mRNA capture without a spatial barcode were present under the optimal permeabilization conditions. Then, a penetration operation was performed based on the optimal permeation time.
Reverse transcription, second-strand cDNA synthesis, and cDNA amplification
The polyadenylated mRNAs released from the overlying PCa cells were captured by primers (IDT) in the spots. Reverse transcription was performed to synthesize spatially barcoded, full-length cDNA (from polyadenylated mRNA on the slide) through the incubation of the permeabilized PCa sections with RT Master Mix reagents. Second-strand was synthesized by adding second-strand-mix to the PCa sections on the slide. The cDNAs were denatured and transferred from each capture area to the corresponding tube and then amplified via PCR to generate a sufficient mass of DNA for library construction.
Library construction of cDNA and sequencing
Libraries of tissue sections were generated according to the 10X Genomics Visium library preparation protocol. cDNA amplicon size was optimized via enzymatic fragmentation and size selection. P5, P7, i5 and i7 sample indexes and TruSeq Read 2 sequences were added by performing end repair, tailing, adaptor ligation, and PCR. The final libraries containing the P5 and P7 primers were used for Illumina amplification. After library construction, 150PE-mode sequencing was carried out on the Illumina NovaSeq600 platform (Illumina, CA, USA).
Visium analysis
Spaceranger pipeline (https://support.10xgenomics.com/spatial-gene-expression/software/pipelines/latest/what-is-space-ranger) (v1.3.1, 10X genomics) was used for alignment, tissue detection, fiducial detection, and barcode/UMI counting. Seurat R package was further used for visualization and exploration. Specifically, the functions SpatialDimPlot and SpatialFeaturePlot. Integration with single-cell data was done using Seurat’s integration workflow76. Specifically, both datasets were normalized using the SCTransform function and functions FindIntegrationAnchors and TransferData were used to identify anchors between the two datasets and crate a prediction assay with a score for each spot for each annotation in the single cell data. The annotation with the highest score was assigned for each spot.
Statistical analysis
Data are expressed as means ± SEM. Statistical tests were performed using unpaired t-test comparing two groups and one-way ANOVA comparing more than two groups. A P value of <0.05 was considered significant. Detailed information on the number of biological samples and animals used can be found in figure legends.
Supplementary information
Acknowledgements
We thank Yuval Nevo, Inbar Plaschkes, and Hadar Benyamini for the bioinformatic analysis and Dr. Simon Yona for technical support. This work was supported by the Israel Science Foundation grant 2272/20 (A.-H.H).
Author contributions
Conceptualization: Y.J., A.W., A.-H.H. Methodology: Y.J., A.S.-P., L.E.B. Investigation: Y.J., A.S.-P., Y.N., O.S., N.D., S.Y. Visualization: Y.J., R.N., S.Y. Funding acquisition: A.-H.H., A.W. Project administration: L.E.B., H.S. Supervision: A.-H.H., A.W., E.E.
Data availability
All data are available in the main text or the supplementary materials. The gingival Visium, scRNA-seq, and bulk RNA-seq data generated during this study are available at GEO/NCBI under accession numbers GSE269575, GSE269576, and GSE269171.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
12/31/2024
A Correction to this paper has been published: 10.1038/s41522-024-00636-z
Supplementary information
The online version contains supplementary material available at 10.1038/s41522-024-00625-2.
References
- 1.Peterson, L. W. & Artis, D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat. Rev. Immunol.14, 141–153 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Zubeidat, K. & Hovav, A. H. Shaped by the epithelium - postnatal immune mechanisms of oral homeostasis. Trends Immunol.42, 622–634 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Torow, N., Hand, T. W. & Hornef, M. W. Programmed and environmental determinants driving neonatal mucosal immune development. Immunity56, 485–499 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris-Tryon, T. A. & Grice, E. A. Microbiota and maintenance of skin barrier function. Science376, 940–945 (2022). [DOI] [PubMed] [Google Scholar]
- 5.Hooper, L. V., Littman, D. R. & Macpherson, A. J. Interactions between the microbiota and the immune system. Science336, 1268–1273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivanov, I. I. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell139, 485–498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naik, S. et al. Compartmentalized control of skin immunity by resident commensals. Science337, 1115–1119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soderholm, A. T. & Pedicord, V. A. Intestinal epithelial cells: at the interface of the microbiota and mucosal immunity. Immunology158, 267–280 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koren, N. et al. Maturation of the neonatal oral mucosa involves unique epithelium-microbiota interactions. Cell Host Microbe29, 197–209.e195 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Dutzan, N. et al. On-going Mechanical Damage from Mastication Drives Homeostatic Th17 Cell Responses at the Oral Barrier. Immunity46, 133–147 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaber, Y. et al. Langerhans cells shape postnatal oral homeostasis in a mechanical-force-dependent but microbiota and IL17-independent manner. Nat. Commun.14, 5628 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silva, L. M. et al. Fibrin is a critical regulator of neutrophil effector function at the oral mucosal barrier. Science374, eabl5450 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Zenobia, C. et al. Commensal bacteria-dependent select expression of CXCL2 contributes to periodontal tissue homeostasis. Cell Microbiol.15, 1419–1426 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capucha, T. et al. Sequential BMP7/TGF-beta1 signaling and microbiota instruct mucosal Langerhans cell differentiation. J. Exp. Med.215, 481–500 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilharm, A. et al. Mutual interplay between IL-17-producing gammadeltaT cells and microbiota orchestrates oral mucosal homeostasis. Proc. Natl Acad. Sci. USA116, 2652–2661 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, H. et al. Dissecting mammalian spermatogenesis using spatial transcriptomics. Cell Rep.37, 109915 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, S. et al. Spatially resolved transcriptomics reveals genes associated with the vulnerability of middle temporal gyrus in Alzheimer’s disease. Acta Neuropathol. Commun.10, 188 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Danan, C. H., Katada, K., Parham, L. R. & Hamilton, K. E. Spatial transcriptomics add a new dimension to our understanding of the gut. Am. J. Physiol. Gastrointest. Liver Physiol.324, G91–G98 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maynard, K. R. et al. Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex. Nat. Neurosci.24, 425–436 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caetano, A. J. et al. Spatially resolved transcriptomics reveals pro-inflammatory fibroblast involved in lymphocyte recruitment through CXCL8 and CXCL10. Elife12, e81525 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lundmark, A. et al. Gene expression profiling of periodontitis-affected gingival tissue by spatial transcriptomics. Sci. Rep.8, 9370 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mark Welch, J. L., Dewhirst, F. E. & Borisy, G. G. Biogeography of the oral microbiome: the site-specialist hypothesis. Annu. Rev. Microbiol.73, 335–358 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allaire, J. M. et al. The intestinal epithelium: central coordinator of mucosal immunity. Trends Immunol.39, 677–696 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Whitsett, J. A. & Alenghat, T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat. Immunol.16, 27–35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DiTommaso, T. et al. Keratin 76 is required for tight junction function and maintenance of the skin barrier. PLoS Genet.10, e1004706 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sequeira, I. et al. Immunomodulatory role of Keratin 76 in oral and gastric cancer. Nat. Commun.9, 3437 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dale, B. A., Salonen, J. & Jones, A. H. New approaches and concepts in the study of differentiation of oral epithelia. Crit. Rev. Oral Biol. Med.1, 167–190 (1990). [DOI] [PubMed] [Google Scholar]
- 28.Garzon, I. et al. In vitro and in vivo cytokeratin patterns of expression in bioengineered human periodontal mucosa. J. Periodontal Res.44, 588–597 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Omary, M. B. Intermediate filament proteins of digestive organs: physiology and pathophysiology. Am. J. Physiol. Gastrointest. Liver Physiol.312, G628–G634 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vercauteren Drubbel, A. & Beck, B. Single-cell transcriptomics uncovers the differentiation of a subset of murine esophageal progenitors into taste buds in vivo. Sci. Adv.9, eadd9135 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das, B. et al. Tissue distribution of murine Muc19/smgc gene products. J. Histochem. Cytochem.58, 141–156 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi, S., Passuti, N. & Delecrin, J. Interpretation and utility of traction radiography in scoliosis surgery. Analysis of patients treated with Cotrel–Dubousset instrumentation. Spine (Philos. PA 1976)22, 2542–2546 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Dibattista, M., Al Koborssy, D., Genovese, F. & Reisert, J. The functional relevance of olfactory marker protein in the vertebrate olfactory system: a never-ending story. Cell Tissue Res.383, 409–427 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Modlin, I. M. et al. Chromogranin A—biological function and clinical utility in neuro endocrine tumor disease. Ann. Surg. Oncol.17, 2427–2443 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Agostinelli, E. et al. A role for STOML3 in olfactory sensory transduction. eNeuro8, ENEURO.0565–20.2021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, Z. et al. Identification of new binding partners of the chemosensory signaling protein Ggamma13 expressed in taste and olfactory sensory cells. Front. Cell. Neurosci.6, 26 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, G., Fang, Y. Z., Yang, S., Lupton, J. R. & Turner, N. D. Glutathione metabolism and its implications for health. J. Nutr.134, 489–492 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Nguyen, T., Mills, J. C. & Cho, C. J. The coordinated management of ribosome and translation during injury and regeneration. Front. Cell Dev. Biol.11, 1186638 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nassar, M. et al. GAS6 is a key homeostatic immunological regulator of host-commensal interactions in the oral mucosa. Proc. Natl Acad. Sci. USA114, E337–E346 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collins, S. L. & Patterson, A. D. The gut microbiome: an orchestrator of xenobiotic metabolism. Acta Pharm. Sin. B10, 19–32 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moutsopoulos, N. M. & Konkel, J. E. Tissue-specific immunity at the oral mucosal barrier. Trends Immunol.39, 276–287 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeLong, J. H. et al. Cytokine- and TCR-mediated regulation of T cell expression of Ly6C and Sca-1. J. Immunol.200, 1761–1770 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rath, E. & Haller, D. Intestinal epithelial cell metabolism at the interface of microbial dysbiosis and tissue injury. Mucosal Immunol.15, 595–604 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park, J. Y., Chung, H., DiPalma, D. T., Tai, X. & Park, J. H. Immune quiescence in the oral mucosa is maintained by a uniquely large population of highly activated Foxp3(+) regulatory T cells. Mucosal Immunol.11, 1092–1102 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madison, K. C. Barrier function of the skin: “la raison d’etre” of the epidermis. J. Investig. Dermatol.121, 231–241 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Schmidt, R. et al. Modulation of cellular cholesterol and its effect on cornified envelope formation in cultured human epidermal keratinocytes. J. Investig. Dermatol.97, 771–775 (1991). [DOI] [PubMed] [Google Scholar]
- 47.Groeger, S. & Meyle, J. Oral mucosal epithelial cells. Front. Immunol.10, 208 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hovav, A. H. Mucosal and skin Langerhans cells—nurture calls. Trends Immunol.39, 788–800 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Shannon, J. P. et al. Group 1 innate lymphoid-cell-derived interferon-gamma maintains anti-viral vigilance in the mucosal epithelium. Immunity54, 276–290.e275 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stolley, J. M. et al. Depleting CD103+ resident memory T cells in vivo reveals immunostimulatory functions in oral mucosa. J. Exp. Med.220, e20221853 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hajishengallis, G., Moutsopoulos, N. M., Hajishengallis, E. & Chavakis, T. Immune and regulatory functions of neutrophils in inflammatory bone loss. Semin. Immunol.28, 146–158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barel, O. et al. gammadelta T cells differentially regulate bone loss in periodontitis models. J. Dent. Res.101, 428–436 (2022). [DOI] [PubMed] [Google Scholar]
- 53.Abe, T. et al. The B cell-stimulatory cytokines BLyS and APRIL are elevated in human periodontitis and are required for B cell-dependent bone loss in experimental murine periodontitis. J. Immunol.195, 1427–1435 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scapini, P. et al. Proinflammatory mediators elicit secretion of the intracellular B-lymphocyte stimulator pool (BLyS) that is stored in activated neutrophils: implications for inflammatory diseases. Blood105, 830–837 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Rouabhia, M., Rouabhia, D., Park, H. J., Giasson, L. & Zhang, Z. Effect of soft foods on primary human gingival epithelial cell growth and the wound healing process. Food Res. Int.100, 433–441 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Cortes-Vieyra, R., Rosales, C. & Uribe-Querol, E. Neutrophil functions in periodontal homeostasis. J. Immunol. Res.2016, 1396106 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsukamoto, Y. et al. Role of the junctional epithelium in periodontal innate defense and homeostasis. J. Periodontal Res.47, 750–757 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Macpherson, A. J. & Harris, N. L. Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol.4, 478–485 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Greer, A. et al. Site-specific neutrophil migration and CXCL2 expression in periodontal tissue. J. Dent. Res.95, 946–952 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Girbl, T. et al. Distinct compartmentalization of the chemokines CXCL1 and CXCL2 and the atypical receptor ACKR1 determine discrete stages of neutrophil diapedesis. Immunity49, 1062–1076.e1066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oulee, A. et al. Identification of genes encoding antimicrobial proteins in Langerhans cells. Front. Immunol.12, 695373 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rydstrom, A. & Wick, M. J. Monocyte and neutrophil recruitment during oral Salmonella infection is driven by MyD88-derived chemokines. Eur. J. Immunol.39, 3019–3030 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Huang, Y. et al. Blocking CXCR1/2 attenuates experimental periodontitis by suppressing neutrophils recruitment. Int Immunopharmacol.128, 111465 (2024). [DOI] [PubMed] [Google Scholar]
- 64.Shusterman, A. et al. The PF4/PPBP/CXCL5 gene cluster is associated with periodontitis. J. Dent. Res.96, 945–952 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zeng, W. J. et al. The expression levels of chemotaxis-related molecules CXC chemokine receptor 1, interleukin-8, and pro-platelet basic protein in gingival tissues. J. Dent. Sci.19, 58–63 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang, X. et al. The dimension and morphology of alveolar bone at maxillary anterior teeth in periodontitis: a retrospective analysis-using CBCT. Int. J. Oral Sci.12, 4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol.14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun.8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol.33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wickham H. ggplot2: Elegant Graphics for Data Analysis XVI, 260 (Springer-Verlag New York, 2016).
- 74.Street, K. et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genom.19, 477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Garcia-Alonso, L. et al. Single-cell roadmap of human gonadal development. Nature607, 540–547 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stuart, T. et al. Comprehensive integration of single-cell data. Cell177, 1888–1902.e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplementary materials. The gingival Visium, scRNA-seq, and bulk RNA-seq data generated during this study are available at GEO/NCBI under accession numbers GSE269575, GSE269576, and GSE269171.