Abstract
Plant stress reduction research has advanced significantly with the use of Artificial Intelligence (AI) techniques, such as machine learning and deep learning. This is a significant step toward sustainable agriculture. Innovative insights into the physiological responses of plants mostly crops to drought stress have been revealed through the use of complex algorithms like gradient boosting, support vector machines (SVM), recurrent neural network (RNN), and long short-term memory (LSTM), combined with a thorough examination of the TYRKC and RBR-E3 domains in stress-associated signaling proteins across a range of crop species. Modern resources were used in this study, including the UniProt protein database for crop physiochemical properties associated with specific signaling domains and the SMART database for signaling protein domains. These insights were then applied to deep learning and machine learning techniques after careful data processing. The rigorous metric evaluations and ablation analysis that typified the study’s approach highlighted the algorithms’ effectiveness and dependability in recognizing and classifying stress events. Notably, the accuracy of SVM was 82%, while gradient boosting and RNN showed 96%, and 94%, respectively and LSTM obtained an astounding 97% accuracy. The study observed these successes but also highlights the ongoing obstacles to AI adoption in agriculture, emphasizing the need for creative thinking and interdisciplinary cooperation. In addition to its scholarly value, the collected data has significant implications for improving resource efficiency, directing precision agricultural methods, and supporting global food security programs. Notably, the gradient boosting and LSTM algorithm outperformed the others with an exceptional accuracy of 96% and 97%, demonstrating their potential for accurate stress categorization. This work highlights the revolutionary potential of AI to completely disrupt the agricultural industry while simultaneously advancing our understanding of plant stress responses.
Keywords: Plant science, Machine learning, Deep learning, Simple modular architecture research tool (SMART), Plant drought stress
Subject terms: Plant sciences, Computer science
Introduction
The integration of machine learning (ML) and deep learning (DL) into biological research marks a transformative era, offering powerful tools for analyzing complex biological data and uncovering new insights. ML and DL have driven significant advancements across domains such as proteomics, bioinformatics, and genomics. For example, DL models like DeepBind1 have revolutionized the prediction of deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) binding specificities, improving genomic and proteomic sequence analysis, variant identification, and understanding of genetic mutations.
In drug discovery, ML and DL models have facilitated the prediction of drug-target interactions by integrating chemical, genomic, and proteomic data, enabling drug repurposing and target identification2. Moreover, DL techniques like AlphaFold3 have achieved remarkable accuracy in predicting protein structures, advancing our understanding of protein interactions.
Agriculture faces the challenge of significantly increasing output by 2050 to meet global demands. However, this goal is hindered by various stresses that plants encounter, including diseases and adverse weather conditions4. Plant breeders are utilizing specific genes to develop crops capable of withstanding harsh environments, necessitating detailed analysis of plant traits. The increasing sophistication of imaging sensors has resulted in a vast accumulation of plant images captured under diverse environmental and stress conditions4.
Plants face numerous challenges, both biotic and abiotic. Abiotic stresses like droughts, floods, extreme temperatures, and soil salinity pose significant threats to plant survival and productivity5. Additionally, environmental factors such as air pollution, ultraviolet (UV) radiation, and chemical stressors further complicate plant health. Climate change exacerbates these issues, with soil dehydration leading to drought stress, a critical factor affecting global agriculture6.
Drought represents a significant agricultural challenge, characterized by prolonged periods of insufficient rainfall or water availability, which severely impacts crop yields and plant growth. The complex nature of drought’s effects on crops has broad implications for environmental sustainability, economic stability, and food security. Figure 1 illustrates plants’ defensive response to drought stress, a critical focus of agricultural research7. Studies have highlighted the vulnerability of key crops like wheat, corn, and rice to water stress, with drought-related soil moisture depletion impairing vital physiological functions and stunting growth8.
Figure 1.
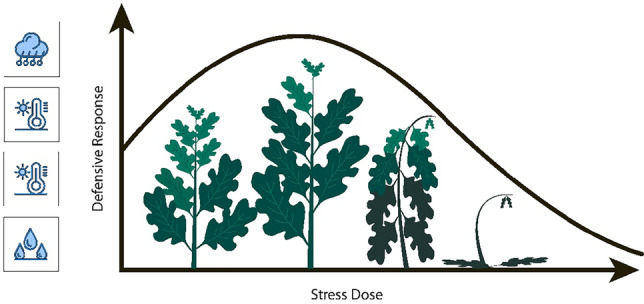
Defensive response vs stress dose in plants.
The economic impact of drought is significant, leading to crop failures, increased food prices, reduced farmer income, and potential shortages in the market9. To mitigate these effects, various interventions, including insurance schemes, water management policies, and drought-relief programs, are implemented worldwide10. Additionally, advancements in genetic engineering and breeding programs are focused on developing drought-resistant crop varieties, while technologies like remote sensing and precision agriculture optimize water use in farming11.
Climate change is expected to increase the frequency and intensity of droughts, necessitating adaptive strategies and sustainable methods to mitigate long-term effects on agriculture12. Addressing global food security concerns arising from drought-induced crop failures requires international collaboration13. The complex interplay between heat, drought, and related stresses underscores the diverse challenges facing global agricultural systems, requiring extensive research and adaptive strategies14.
In recent years, ML and DL methods have emerged as crucial tools for studying and addressing plant stress. These approaches enable the analysis of vast datasets, providing insights into how plants respond to environmental stresses. ML algorithms can identify genetic markers associated with stress tolerance or susceptibility, aiding in breeding efforts aimed at developing stress-resistant crops15. DL models, such as convolutional neural networks (CNNs) and recurrent neural networks (RNNs), excel in processing high-dimensional data and capturing complex relationships, leading to more accurate predictions of plant stress responses16.
One of the primary applications of ML and DL in plant stress research is predicting stress outcomes based on environmental factors. By analyzing variables such as soil moisture levels, temperature fluctuations, humidity, and light intensity, ML models can predict plant responses to stressors, enabling early detection of symptoms and timely interventions17. The integration of remote sensing with ML allows real-time monitoring of crop health and stress levels, facilitating targeted management strategies18.
E3 ubiquitin ligases are critical in regulating various cellular functions in plants, including hormone signaling, developmental pathways, and responses to environmental stress19. These ligases play a key role in the ubiquitin-proteasome system (UPS), mediating targeted protein degradation. E3 ligases confer specificity to the UPS by determining which substrates are ubiquitinated, influencing the plant’s stress response20.
In the context of drought stress, E3 ubiquitin ligases mediate the plant’s response by regulating the degradation of proteins involved in stress signaling pathways, such as DRIP1 and HSP90C. This regulation affects the expression of downstream genes involved in the drought response21. The abscisic acid (ABA) signaling pathway, essential for stress responses, also relies on E3 ligases like RING-BetweenRING-RING (RBR) to regulate gene expression during growth and stress conditions22. E3 ubiquitin ligases also influence root development and plant defense responses against pathogens23,24.
Phosphotransferases within the Tyrosine-specific kinase subfamily (TYRKC) are another critical component in plant stress responses. Protein kinases and phosphoprotein phosphatases mediate the reversible phosphorylation of proteins, essential for cellular functions25. In plant stress contexts, TYRKC and E3 ligases are depicted as key players in maintaining plant homeostasis under adverse conditions. In this context, the main contributions of this study are as follows:
Leveraging AI techniques to analyze ubiquitin domains of TYRKC and RBR-E3 stress-associated signaling proteins across different plant species.
Assessing the physicochemical properties of drought stress-associated signaling proteins using ML models, including SVM, gradient boosting, RNN, and LSTM.
Evaluating the accuracy of stress classification using F1 score, recall, and precision, supported by ablation analysis to extract valuable insights.
The remaining paper is divided into four sections. Related work is presented in section “Literature review”. Section “Methodology” provides a brief overview of the proposed methodology while a discussion of results is given in section “Results”. The study is concluded in section “Conclusions”.
Literature review
ML has gained a substantial boost in rapid adoption in the agriculture industry and a large number of works have been presented in this regard. The study26 presents a comprehensive overview of recent publications on the use of ML and deep learning for plant disease detection.
The study27 adopts an ensemble of AlexNet, ResNet50, and VGG16 models to improve the overall performance of plant disease detection. Resultantly, an improved accuracy of 99.53% is reported for multi-class problems which may go up to 100% in the case of binary class tasks. These results promote the use of pre-trained DL models in agriculture. Similarly, a deep learning-based pre-trained EfficientNetV2 is utilized in28 for disease detection in cardamom plants. Image processing is enhanced by removing the unnecessary background using a U2-Net model. Experimental results indicate a superb accuracy of 98.26% surpassing the existing works.
Potato plants also face climate change and suffer from other factors like inappropriate fertilizer, unnecessary watering, and pesticides leading to various diseases. A comprehensive framework is presented in29 for timely disease detection in potatoes. For this purpose, a multilevel feature fusion network is proposed which incorporates ResNet50 and an adaptive attention mechanism to integrate multiple features. A validation accuracy of 99.83% is reported for potato plant disease detection.
Abiotic stress is a major problem for agriculture nowadays, resulting in considerable crop losses worldwide and a reduction in the amount of land that can be farmed. This problem becomes more complex as a result of the effects of climate change and an expanding population21. Forecasts suggest that by 2050, there will be a 9-10-billion-person world population, necessitating a 60–110% increase in food production. The situation is further worse by urban expansion, which pushes agriculture into less appropriate places and encroaches onto arable land30. The problems are made worse by elements like deforestation and rising CO2 levels, which are predicted to reach 800 ppm by 2100 as a result of excessive usage of fossil fuels. Abiotic stress on crops is exacerbated by climate change, which brings forth extreme weather patterns such as abrupt temperature changes, heavy precipitation, and protracted droughts. For example, drought impacts almost half of arable land, and aridity stress affects an increasing percentage of land from 17 to 27% between 1950 and 200031. Another obstacle is salinity, which causes notable drops in crop output, especially because many crop species are vulnerable to salt stress. Hexachromium, cadmium, arsenic, lead, copper, and mercury are among the hazardous elements that are included in heavy metal pollution, which is caused by anthropogenic activities like mining and the overuse of pesticides and fertilizers. Critical plant functions like germination and photosynthesis are hampered by these contaminants, which eventually reduces growth, production, and quality.
In the field of plant stress detection, DL has become a potent instrument that offers advanced approaches to deal with the urgent problems facing the agricultural industry. The use of deep convolutional neural networks (CNNs) for the analysis of plant images to detect different stress indicators like discoloration, lesions, or deformities has been a major area of research in this field31. These image-based methods have shown great promise, exhibiting outstanding effectiveness in the identification and categorization of various stress states based just on visual cues. Additionally, to facilitate a more thorough evaluation of plant stress, researchers have ventured into incorporating sensor data including physiological measures, spectral data, and infrared imagery into deep learning models32. These multimodal techniques offer a deeper picture of plant health and stress responses by combining data from numerous sources, which may improve the precision and dependability of stress detection systems. Transfer learning strategies have been essential in helping researchers obtain state-of-the-art performance with less data by tailoring pre-trained DL models to the particular domain of plant stress detection33. Moreover, continuous surveillance of plant health in agricultural contexts has great promise with the development of real-time monitoring systems using deep learning models34. By giving farmers early alerts and actionable insights, these systems have the potential to completely transform crop management methods by enabling preemptive actions to lessen the effects of stressors. Notwithstanding these developments, there are still problems, such as interpretability, scalability, and robustness of the model35. In order to fully utilize deep learning for improving agricultural sustainability and guaranteeing global food security in the face of increasing environmental stresses and population expansion, it will be imperative to address these issues.
Numerous stressors, such as UV radiation, extremely high or low temperatures, drought, flooding, and heavy metals, can affect plants. They have evolved strategies to deal with these obstacles, such as producing more antioxidants during stress or more roots during a drought. Stress has the potential to induce an internal imbalance in plants, which is known as oxidative stress. This indicates an excess of reactive oxygen species (ROS), which can damage plant components including DNA and proteins. This can occur, for example, when a plant experiences stress and closes its stomata, producing hydrogen peroxide (H2O2), a kind of ROS. They produce unique enzymes that aid in the removal of excess ROS, such as CAT and SOD. They also generate other beneficial substances that can counteract ROS, such as glutathione and proline. Certain plants may be more resilient to stress because they naturally contain higher concentrations of these beneficial substances. However, there are instances when even these defenses are insufficient, particularly in cases of extreme or protracted stress, and the plant’s development and productivity may suffer.
Researchers have investigated the effectiveness of several deep learning architectures, including Transformers, RNNs, and CNNs, in the field of plant stress detection. Thanks to their capacity to extract spatial characteristics and patterns from plant images, CNNs have become a popular option for image-based stress detection tasks. These structures are particularly good at identifying visible signs of stress, like lesions or discolored leaves. In the meantime, sequential data from plants, such as sensor measurements or physiological data across time, has been analyzed using RNNs, notably LSTM networks36. RNNs may forecast stress based on changing environmental variables and identify small changes in plant physiology by capturing temporal relationships.
Transformers, which show potential for effectively managing both sequential and spatial data, have also drawn attention recently. Transformer-based models such as the Multi-Scale Vision Transformer (MSViT) have been proposed to assess multiscale features in plant images, while Vision Transformers (ViTs) have demonstrated competitive performance in image-based stress detection tasks37. By detecting subtle visual cues linked to stress conditions, these architectures hope to advance the development of reliable plant stress detection systems that will help agronomists and farmers optimize crop management strategies and ensure the sustainability of agriculture. The summary of relevant works is given in Table 1.
Table 1.
Summary of relevant existing works.
| Refs. | Methodology | Findings | Limitations |
|---|---|---|---|
| 38-2023 | ResGCNet | The 1D-ResGC-Net CNN model effectively detected drought stress in tomato plants, outperforming PLSDA and RF models with fewer input features. | The 1D-ResGC-Net’s accuracy significantly drops with fewer input features |
| 39-2024 | Random forest, RNA | The study shows that a location-based ML classifier effectively detects drought stress in soil metagenomes with strong generalization | The focus on soil metagenomes may not cover variability in drought responses across different plants or environments |
| 40-2021 | CNN, DCNN, GoogLeNet | DCNNs, like GoogLeNet efficiently detects crop water stress, allowing for real-time detection in agriculture. | GoogLeNet’s performance may not be generalized |
| 41-2024 | CNN | Automated Identification of Paddy Crop Stresses | Limited Consideration of Environmental Factors |
| 42-2021 | BAT-optimized ResNet-18 | Assesses how well a BAT-optimized ResNet-18 model performs in recognizing the different levels of water stress experienced by chickpea plants | No publicly accessible databases for recognizing water stress in chickpea plants, a proprietary dataset was built specifically for this investigation |
| 43-2021 | CNN and LSTM | Model architecture used VGG16+LSTM Compared to previous CNN baseline models, the early stress detection accuracy of VGG16 is improved by using LSTM architecture. | Potential difficulties or restrictions related to applying the VGG16+LSTM architecture in practical applications or deploying it across various computer platforms are not covered in the paper |
| 44-2022 | CNN | Efficacy of CNN and other deep learning models in forecasting sunflower grain production under various circumstances | The primary drawback is that the deep learning models are trained on a limited dataset, which may cause overfitting or decreased generalizability |
| 45-2022 | CNN, AlexNet, GoogLeNet, Inception v1, Inception v3, MobileNet v2, ResNet50 | When combined with deep learning models, thermal imagery offers a very precise way to detect crop water stress in wheat | Plant Water Stress Indicators, Water stress prediction, and comparison between future extraction-based and Function approximation-based approaches |
| 46-2023 | RNN, Vanilla GRU, Stacked GRU, Bidirectional LSTM, Adam Adagrad, RMSprop, Genetic Algorithm | RNN-based deep learning models, including variants of GRU and LSTM, show high accuracy in predicting fruit rot disease | The study may have missed more efficient strategies that could improve model performance because it only looked at a small number of optimization techniques |
| 47-2020 | RNN, Deep transfer image regression networks such as MobileNetV2, ResNet101, InceptionV3 | Feature-based RNN7, optimized via evolutionary strategy, excel in predicting lettuce water stress, surpassing deep transfer image networks | Limited generalizability due to lettuce-centric focus and absence of exploration into non-irrigation methods for water stress prediction |
| 37-2023 | Hybrid Deep Convolution Neural Network (DCNN) model, named ”DCNN-MCViT | Estimating the severity of plant biotic stress and classifying various forms of it | Failure to address or acknowledge potential problems or shortcomings with the suggested DCNN-MCViT model or the research itself |
Methodology
ML and DL algorithms play a crucial role in improving plant stress mitigation. These algorithms analyze vast data sets and predict optimal growing conditions. This allows for customized solutions to increase plant yield and resilience. Moreover, they empower proactive stress management through predictive modeling, contributing to sustainable agricultural practices. To assess the effectiveness of RBR-type E3 Ubiquitin and TyrKc Tyrosine Kinase in predicting drought stress responses for 101 plant species and genera, researchers employed a combination of four algorithms: two machine learning and two deep learning models. Figure 2 shows the workflow of the adopted methodology in this study.
Figure 2.
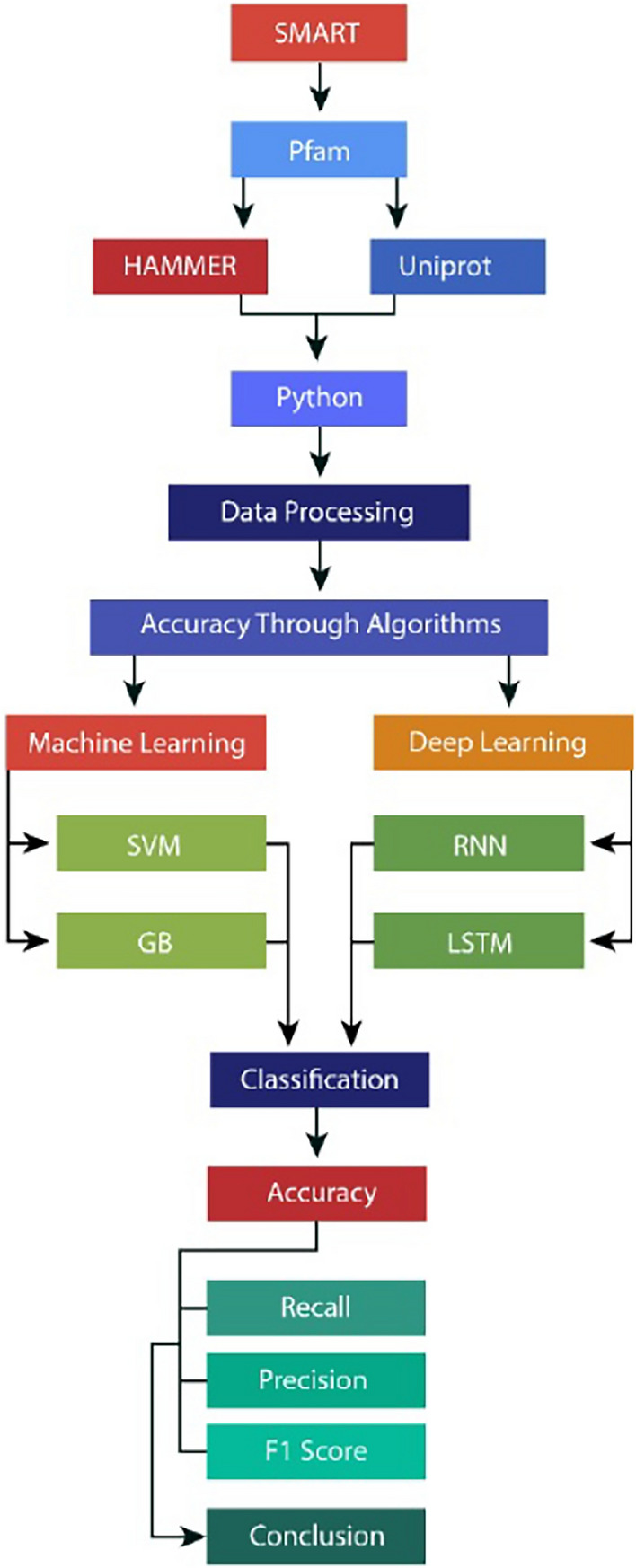
Workflow of the classification methodology.
Gradient Boosting algorithm
Gradient boosting is an ensemble learning technique that improves weak learners incrementally to create a strong predictive model. It works by adding decision trees trained on the residual errors of the previous model, and optimizing the model iteratively using gradient descent. Popular implementations like XGBoost are widely used due to their speed, scalability, and regularization techniques48. Figure 3 shows the architecture of GB model.
Figure 3.
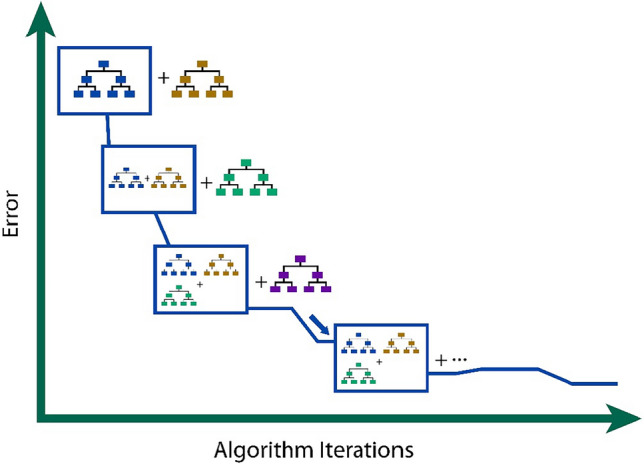
Working of GB algorithm.
Challenges and limitations
While effective, gradient boosting is computationally expensive and prone to overfitting, requiring careful tuning of hyperparameters. Its complexity makes it less interpretable compared to simpler models.
Model structure
GB has the following structure:
Base Learner: Decision trees (weak learners)
Number of Trees: 100
Maximum Depth: 3 (shallow trees to prevent overfitting)
Learning Rate: 0.1 (controls the contribution of each tree)
Loss Function: Log loss for classification
Hyperparameters
The following parameters are optimized:
Learning Rate: 0.1 (a lower value leads to a more robust model, reducing the risk of overfitting)
Number of Trees: 100 (the number of boosting rounds)
Maximum Depth: 3 (limits the complexity of each tree)
Subsample: 0.8 (fraction of samples used for fitting individual base learners)
Min Samples Split: 2 (minimum number of samples required to split an internal node)
Min Samples Leaf: 1 (minimum number of samples required to be at a leaf node)
Training process
The GB model iteratively builds decision trees on the residual errors from previous trees to improve model accuracy.
Cross-validation is used to fine-tune the number of trees and learning rate to avoid overfitting.
Implementation details
Both RNN and LSTM models are implemented using TensorFlow and Keras libraries.
The SVM model is implemented using the scikit-learn library.
The Gradient Boosting model is implemented using XGBoost.
The models are trained on a system with an NVIDIA GPU to accelerate the training process.
Support vector machine
SVM is a powerful technique for classification tasks, especially in high-dimensional spaces. It works by finding the optimal hyperplane that separates data points of different classes. Through the use of kernel tricks, SVM can handle non-linear separations, making it versatile across various applications. Figure 4 shows the architecture of SVM model.
Figure 4.
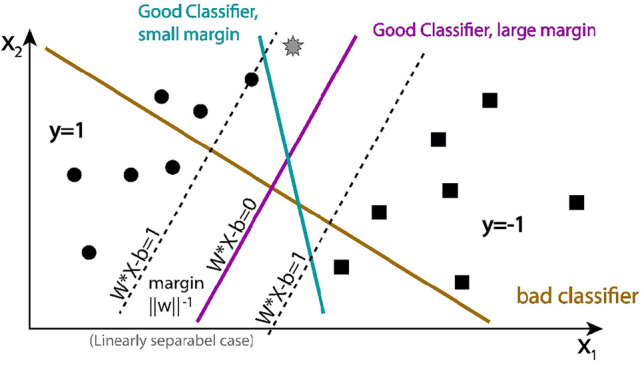
Working of SVM algorithm.
Challenges and limitations
SVMs can struggle with large datasets and overlapping classes. The choice of kernel and regularization parameters significantly impacts performance, and the model can be sensitive to outliers.
Model structure
The model structure is as follows
Kernel Function: Radial Basis Function (RBF)
Multi-Class Strategy: One-vs-One
Regularization Parameter (C): 1.0
Gamma: Scale (default is 1/(n_features * X.var()) where X is the training data)
Tolerance (tol): 1e−3
Maximum Iterations: − 1 (no limit on iterations)
Hyperparameters
Kernel: The RBF kernel is used because it can handle non-linear decision boundaries by mapping input features into higher-dimensional spaces.
C: The regularization parameter controls the trade-off between achieving a low error on the training data and minimizing the model complexity. A value of 1.0 is chosen to maintain a balance between these two objectives.
Gamma: The gamma parameter defines the influence of a single training example. The ’scale’ option is selected to automatically compute this value based on the data.
Cross-Validation: A 10-fold cross-validation approach is used to fine-tune these hyper-parameters and select the best-performing model.
Recurrent neural networks
RNNs are designed to handle sequential data by incorporating memory through hidden states that evolve with each time step. They are particularly useful in tasks where the order of inputs matters, such as time-series prediction. However, standard RNNs face challenges like vanishing gradients, making it difficult to capture long-term dependencies. Figure 5 shows the architecture of RNN model.
Figure 5.
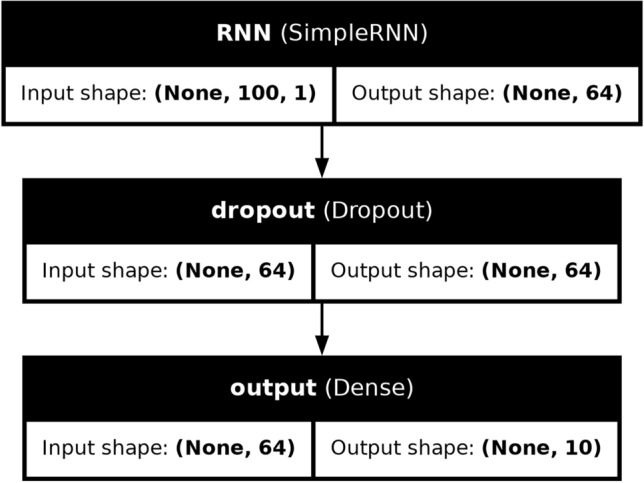
Architecture of RNN model.
Challenges and limitations
Training RNNs is computationally intensive, and they require large datasets. Despite improvements like GRUs and LSTMs, RNNs remain complex and prone to overfitting, especially with small or noisy datasets.
Model structure
The model structure is given as follows
Number of Layers: 2
Hidden Units: 64
Activation Function: Tanh
Output Layer: Dense layer with softmax activation for multi-class classification
Loss Function: Categorical Cross-Entropy
Optimizer: Adam
Hyperparameters
The following parameters are optimized for the RNN model:
Learning Rate: 0.001 (chosen for stable convergence during training)
Batch Size: 32
Epochs: 50 (early stopping is employed to prevent overfitting)
Dropout: 0.2 (applied after each RNN layer to reduce overfitting)
Weight Initialization: Xavier uniform initialization for all layers
Training process
The model is trained using a time-series data format where each input sequence corresponds to a series of physiological responses of plants over time.
Early stopping is implemented with patience of 5 epochs, monitoring the validation loss to halt training when the model starts to overfit.
Long short-term memory
LSTM networks are a variant of RNNs designed to overcome the vanishing gradient problem by introducing memory cells and gating mechanisms. This allows them to learn long-term dependencies more effectively, making them ideal for tasks like time-series forecasting and sequential data analysis. Figure 6 shows the architecture of LSTM model.
Figure 6.

Architecture of LSTM model.
Challenges and limitations
LSTMs, while mitigating some RNN limitations, are still resource-intensive and require careful tuning. Their complexity can lead to overfitting, particularly when the available data is limited.
Model structure
The model structure is as follows
Number of Layers: 3 LSTM layers followed by a Dense layer
Hidden Units: 128 (for each LSTM layer)
Activation Function: Tanh for LSTM cells, ReLU for Dense layers
Output Layer: Dense layer with softmax activation for multi-class classification
Loss Function: Categorical Cross-Entropy
Optimizer: Adam
Hyperparameters
The following parameters are tuned for the LSTM model
Learning Rate: 0.001 (with a decay rate of 0.96 every 10 epochs)
Batch Size: 32
Epochs: 100 (with early stopping based on validation accuracy)
Dropout: 0.3 (applied after each LSTM layer to mitigate overfitting)
Recurrent Dropout: 0.2 (applied within the LSTM cells)
Weight Initialization: Xavier uniform initialization for all layers
Training process
The LSTM model processes sequences of input data where the temporal dependencies are crucial for accurate classification.
The model is trained with early stopping criteria based on validation loss, and checkpoints are saved to restore the best model.
Implementation details
Both RNN and LSTM models are implemented using TensorFlow and Keras libraries.
The SVM model is implemented using the scikit-learn library.
The models are trained on a system with an NVIDIA GPU to accelerate the training process.
Rationale for selecting RNN, LSTM, XGBoost, and SVM models
Despite the availability of newer state-of-the-art deep learning (DL) models, authors may choose to use models like RNN, LSTM, XGBoost, and SVM for several important reasons:
Specific task requirements
RNNs and LSTMs: These models are particularly effective for sequential data, such as time series or text, where the order of inputs is crucial. LSTMs, in particular, excel at capturing long-term dependencies, making them ideal for tasks where memory of previous inputs is essential. For example, in tasks involving plant stress classification based on time-series environmental data, RNNs, and LSTMs might outperform more complex models not specifically designed for sequential data.
SVM: Support Vector Machines are highly effective for smaller datasets and when a clear margin of separation exists between classes. They are also known for their robustness against overfitting, particularly in high-dimensional spaces, which can be advantageous when interpretability and simplicity are more critical than the raw predictive power of DL models.
XGBoost: This ensemble learning method is powerful, especially with tabular data. XGBoost is known for its speed, scalability, and effectiveness across a wide range of tasks. It often outperforms DL models on structured data because it handles missing values, categorical variables, and feature interactions effectively.
Computational efficiency
State-of-the-art DL models, such as Transformer-based architectures (e.g., BERT, GPT), require significant computational resources for training and inference. For many tasks, particularly in academia or smaller industry projects, the computational cost of these models can be prohibitive. In contrast, RNNs, LSTMs, SVMs, and XGBoost can be trained and deployed more efficiently, making them suitable for environments with limited resources49.
Model interpretability
Models like SVM and XGBoost offer better interpretability compared to deep neural networks. In fields like plant stress classification, where understanding the influence of specific features (e.g., environmental factors, physiological measurements) on the model’s predictions is essential, interpretability is crucial. This can help in gaining insights into the biological processes being modeled, which might not be as clear with more complex DL architectures.
Proven effectiveness
RNNs, LSTMs, SVMs, and XGBoost have been extensively studied and validated across a wide range of applications. Their reliability and the wealth of literature supporting their use make them attractive choices for researchers. While newer models might offer incremental performance improvements, these gains must be weighed against the robustness, understanding, and simplicity of these more established methods.
Specific domain constraints
In some domains, such as plant stress classification, available datasets might be small or imbalanced, limiting the effectiveness of deep learning models. In such cases, traditional machine learning models like SVM or ensemble methods like XGBoost might provide better performance due to their ability to work well with smaller datasets.
Cross-disciplinary applications
In interdisciplinary research, where researchers might not have deep expertise in cutting-edge DL models, choosing models like RNN, LSTM, SVM, or XGBoost allows for more straightforward implementation and collaboration. These models are well-documented and have extensive support in terms of libraries and frameworks, making them accessible to a broader audience.
In summary, although there are many powerful state-of-the-art DL models but still RNNs, LSTMs, XGBoost, and SVM remain popular due to their suitability for specific tasks, computational efficiency, model interpretability, proven track record, and practical considerations in many real-world applications.
Procedure and methodology
In order to accomplish the study’s objectives, this part outlines the research design, information-gathering methods, and analytical strategies.
Reproducibility considerations
To ensure reproducibility, the following steps are taken: - The random seeds for all libraries (NumPy, TensorFlow, scikit-learn) are fixed. - The dataset is preprocessed consistently across all models, ensuring that the input format is identical. - Hyper-parameter tuning is conducted using grid search and cross-validation, with results documented to allow replication of the study.
By providing detailed descriptions of the model structures and hyper-parameters, this research allows other researchers to replicate and extend the findings in their studies.
Identification of domain
Initially, the SMART database was leveraged to discern the signaling domain, revealing two domains integral to stress response mechanisms: phosphotransferases (TYRKC) and E3 ubiquitin-protein ligase (RBR). Subsequently, UniProt IDs SM00184 and SM00219, corresponding to RBR E3 Ubiquitin and TYRKC domains, were retrieved from the Pfam database. Following this, a comprehensive examination of these domains was conducted across a diverse collection of over 101 plants spanning various genera. This investigation employed the Linux tool HMMER in conjunction with meticulous manual inspection. TYRKC, or Tyrosine Kinase, is a protein family crucial in cellular signaling, particularly in regulating growth, division, and responses to environmental stress like drought. In this study, analyzing TYRKC domains in plant proteins provided insights into how plants perceive and respond to drought stress at the molecular level.
SMART (Simple Modular Architecture Research Tool) is a bioinformatics tool used to identify and annotate protein domains, including TYRKC. In this work, SMART was used to accurately identify these domains in stress-related proteins, which is vital for understanding their role in drought response. By leveraging SMART, the study could precisely characterize TYRKC domains, ensuring the accuracy of the protein analysis and enhancing the understanding of plant stress mechanisms. This approach is crucial for developing strategies to improve drought resistance in crops.
Data extraction
The SMART IDs (SM00184 and SM00219) associated with TYRKC and RBR E3 Ubiquitin were systematically investigated across 101 distinct plant species and genera, leveraging the extensive resources of the UniProt database. Employing the corresponding transcript IDs, a Python programming approach was employed to meticulously extract the physical and chemical attributes of these plant species. Utilizing specialized libraries such as Bio.SeqUtils, Bio Expasy.ProtParam, ProteinAnalysis, and SeqIO, pertinent data pertaining to transcripts inducing heat stress in these specific plant varieties were compiled. This encompassed crucial information such as the theoretical isoelectric point (PI), GRAVY, instability index, and other relevant attributes. The integration of the booktabs package was applied for optimal formatting within the context of article geometry, ensuring a comprehensive and professional presentation of the obtained findings. Details of the data attributes are provided in Table 2.
Table 2.
Attributes of the dataset.
| Theoretical pI | Instability index | GRAVY |
|---|---|---|
| 5.353 | 59.001 | − 0.655 |
| 4.936 | 53.721 | − 0.655 |
| 5.361 | 60.604 | − 0.658 |
| 5.401 | 61.223 | − 0.685 |
| 5.287 | 62.466 | − 0.734 |
| 5.287 | 62.306 | − 0.734 |
| 4.936 | 53.319 | − 0.646 |
| 5.087 | 52.676 | − 0.542 |
| 4.936 | 54.386 | − 0.635 |
Processing of dataset
A Python program was employed to operationalize the dataset target. The methodology encompassed the loading of a dataset and the training of a model utilizing three distinct features. Two of these features were leveraged to establish criteria for identifying a target variable with three discrete classifications, denoted by the numerals “Stable” (0), “Less Stable” (1), and “Not Stable” (2). The elucidation of the classification rules is as follows:
If Theoretical PI > 7, Instability Index < 45, and GRAVY > 0, the entry is categorized as “Stable.”
If Theoretical PI < 7, Instability Index < 40, and GRAVY > 0, the entry is categorized as “Stable.”
If Theoretical PI > 7, Instability Index > 40, and GRAVY > 0, the entry is categorized as “Stable.”
If Theoretical PI > 7, Instability Index < 40, and GRAVY < 0, the entry is categorized as “Stable.”
If Theoretical PI > 7, Instability Index > 40, and GRAVY < 0, the entry is categorized as “Less Stable.”
If Theoretical PI < 7, Instability Index < 40, and GRAVY < 0, the entry is categorized as “Less Stable.”
If Theoretical PI < 7, Instability Index > 40, and GRAVY > 0, the entry is categorized as “Less Stable.”
If none of the above conditions are met, the entry is categorized as “Unstable.”
The analysis disclosed the prevalence of these domains across 101 species and genera.
Evaluation of ML and DL algorithms
The unique characteristics of multi-class settings must take into account, at the time of expanding the performance assessment. A comprehensive evaluation is provided by precision, recall, and F1 scores, which measure the accuracy of positive predictions, recollection, and the ability to record all positive instances, respectively. F1 Score strikes a balance between precision and recall by considering the harmonic sequence. Underline how crucial these measures are to comprehending a model’s performance across a range of classes and identifying false positives as well as false negatives. Therefore, emphasizing recall, precision, and F1 Score guarantees a more accurate and nuanced evaluation in a variety of categorization problems.
Plant species and genera
Amomum Zingiber, Brassica, Chenopodium quinoa, Cynara cardunculus, Nicotiana alata (Flowering Tobacco), Manihot Esculenta, Panicum miliaceum, Solanum tuberosum, Vitis vinifera, Ananas comosus, Cannabis sativa, Cicer arietinum (Chickpea), Dendrobium, Hibiscus syriacus, Medicago, Papaver somniferum, Helianthus annuus (Sunflower), Zea mays, Lupinus angustifolius, Capsella rubella, Coffea, Eragrostis curvula, Hordeum vulgare, Nicotiana tabacum, Pisum sativum, Trifolium pratense, Arabidopsis thaliana, Capsicum annuum, Gossypium (Cotton), Eragrostis, Lactuca sativa, Nicotiana, Arachis hypogaea, Capsicum baccatum, Cucurbita moschata, Lupinus, Opuntia streptacantha, Oryza sativa (Rice), Triticum aestivum, Brachypodium distachyon, Daucus carota (Carrot), Cucurbita pepo, Eutrema salsugineum, Hordeum vulgare subsp. distichon (Malt), Oryza brachyantha, Sesamum indicum, Triticum turgidum, Hordeum vulgare (Barley), Colocasia esculenta, Sorghum bicolor, Stylophora pistillata.
Results
Setup for experiments
A thorough examination of the classification accuracy attained by the algorithms Long Short-Term Memory (LSTM), Gradient Boosting (GB), Recurrent Neural Networks (RNN), and Support Vector Machines (SVM) in relation to machine learning and deep learning methods. A rigorous test ratio of 20% is used to evaluate the model’s performance on a dataset of 4030 observations.
Results of all models
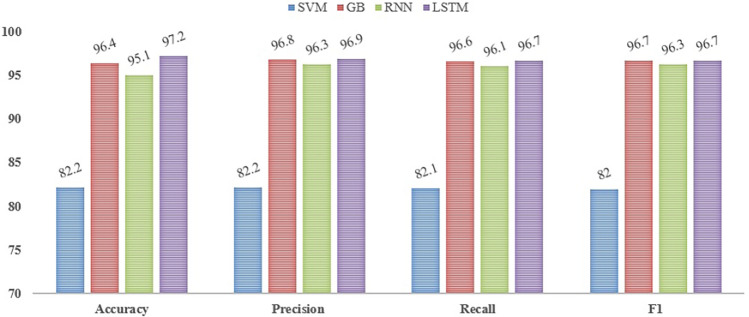
Figure 7 shows the performance of machine learning and deep learning models. The classification models’ accuracy continuously varies from 0.82 to 0.97, with the LSTM algorithm’s peak performance being recorded at 0.97 at a test ratio of roughly 0.2. The dataset is split into two subsets: 804 (20%) and 3224 (80%) of the observations are set aside for testing and training, respectively. With a 20% test ratio, the SVM algorithm yields an 82% accuracy rate. Comparably, the accuracy of the GB method is 96%, the RNN algorithm is 94%, and the LSTM algorithm performs better than the rest with a 97% accuracy. When compared to SVM and RNN, the GB and LSTM algorithms perform exceptionally well, with accuracy rates of 96% and 97%, respectively, demonstrating their effectiveness in handling the classification problem.
Figure 7.
Performance of machine learning and deep learning models.
Results of 10-fold cross-validation
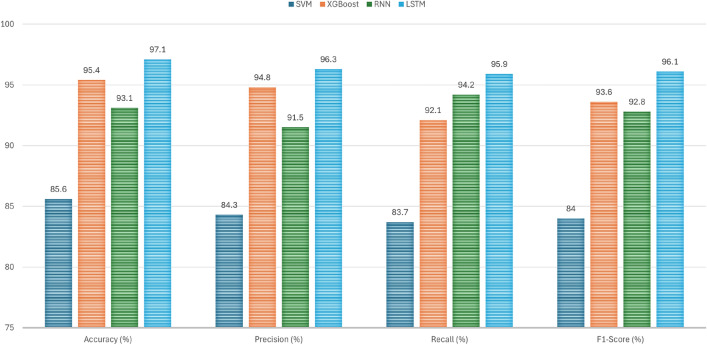
Cross-validation, particularly 10-fold cross-validation, is a robust technique for assessing how the results of a statistical analysis will generalize to an independent dataset. This is particularly useful in scenarios where the dataset is limited, ensuring that every observation in the dataset has a chance to appear in the training and test set. It helps to mitigate issues like overfitting and provides a more accurate measure of model performance by averaging the results across all folds. Figure 8 shows the performance of various models concerning 10-fold cross-validation.
Figure 8.
Models’ performance with 10-fold cross-validation.
Table 3 summarizes the performance of the machine learning and deep learning models after applying 10-fold cross-validation. The results demonstrate the stability and reliability of the models across different folds of the data.
Table 3.
Performance of models after 10-fold cross-validation.
| Model | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|---|
| SVM | 85.6 | 84.3 | 83.7 | 84.0 |
| XGBoost | 95.4 | 94.8 | 92.1 | 93.6 |
| RNN | 93.1 | 91.5 | 94.2 | 92.8 |
| LSTM | 97.1 | 96.3 | 95.9 | 96.1 |
Results of ablation analysis
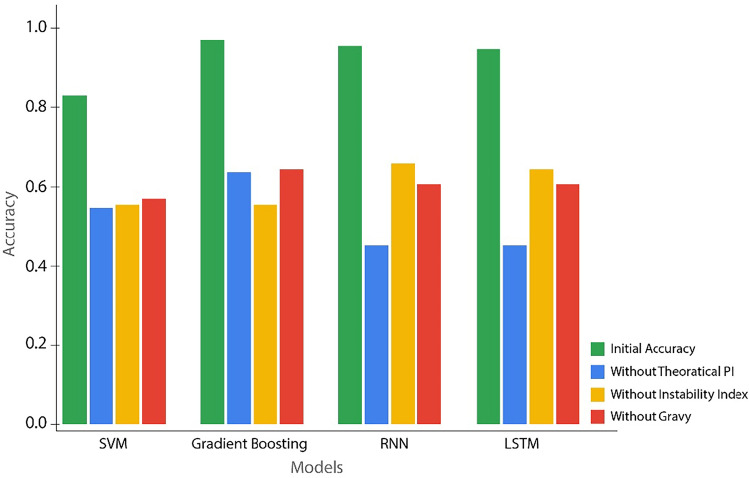
Ablation analysis is a crucial method used to understand the importance and contribution of individual features to the overall performance of a model. By systematically removing one or more features, we can observe how the absence of these features affects the model’s accuracy, precision, recall, and F1 score. This analysis helps in identifying the most significant features and understanding their impact on the model’s ability to classify plant stress responses.
Table 4 shows the performance of the SVM, Gradient Boosting (GB), Recurrent Neural Network (RNN), and Long Short-Term Memory (LSTM) models when specific features are removed from the dataset. The features evaluated in this analysis are Theoretical PI, Instability Index, and GRAVY.
Table 4.
Performance of models using various features.
| Model | Accuracy | |||
|---|---|---|---|---|
| All features | Without theoretical PI | Without Instability Index | Without GRAVY | |
| SVM | 82% | 54% | 56% | 57% |
| GB | 96% | 63% | 64% | 64% |
| RNN | 94% | 45% | 65% | 60% |
| LSTM | 97% | 45% | 63% | 59% |
Impact of feature removal
Theoretical PI: The removal of Theoretical PI had a significant impact on all models, with the SVM accuracy dropping from 82% to 54%. Similarly, the accuracy of the GB model decreased from 96% to 63%, while the RNN and LSTM models saw a reduction in accuracy to 45%. This indicates that Theoretical PI is a critical feature in predicting plant stress responses, as its absence substantially reduces the models’ ability to accurately classify the data.
Instability Index: The Instability Index is another important feature, particularly for the GB and RNN models. The removal of this feature resulted in a decrease in accuracy for the SVM model from 82% to 56%, GB from 96% to 64%, RNN from 94% to 65%, and LSTM from 97% to 63%. This suggests that the Instability Index plays a vital role in the classification process, especially in deep learning models like RNN and LSTM.
GRAVY: The GRAVY (Grand Average of Hydropathicity) index also proved to be an influential feature, particularly for the SVM model, which saw its accuracy drop from 82% to 57% when this feature was removed. The GB and RNN models also showed a decrease in accuracy, with their performance dropping to 64% and 60% respectively. The LSTM model, which originally had the highest accuracy of 97%, saw its accuracy decrease to 59% when GRAVY was removed, indicating the feature’s importance in achieving high classification accuracy.
Analysis and insights
The results of the ablation analysis provide several key insights:
Significance of Features: The substantial drop in accuracy across all models, when Theoretical PI, Instability Index, or GRAVY are removed, highlights their importance in predicting plant stress responses. Each of these features contributes significantly to the models’ overall performance.
Model Sensitivity: The LSTM model, while being the most accurate overall, is highly sensitive to the removal of key features, as evidenced by the significant drops in accuracy. This suggests that while LSTM is powerful, it relies heavily on the availability of comprehensive feature sets to maintain its high performance.
Generalization Ability: The ability of the GB and RNN models to retain some level of accuracy even after feature removal suggests that these models have a better ability to generalize compared to SVM, which shows a more dramatic decrease in accuracy.
Balanced Feature Importance: The results suggest that a balanced combination of features is crucial for achieving high accuracy. Models like SVM, which rely heavily on a specific feature, show poorer performance when that feature is removed, highlighting the importance of using a well-rounded set of features.
Figure 9 further illustrates the performance of these models with and without the removal of specific features, showing the critical nature of each feature in the classification process.
Figure 9.
Models’ performance with and without feature removal.
Comparison with existing works on the same dataset
When conducting research in the field of plant stress classification using machine learning (ML) and deep learning (DL) models, it is important to compare the performance of the applied models with those in existing literature. However, in the case of this study, such comparisons against existing works on the same dataset are challenging for several reasons:
Novelty of the dataset
The dataset utilized in this research is specialized and tailored to capture the physiological responses of plants to drought stress, incorporating unique features such as specific signaling protein domains (e.g., TYRKC and RBR-E3). As this dataset might be newly developed or proprietary, it may not have been used in previous studies, making direct comparisons difficult. The lack of publicly available datasets with the same scope and features further complicates benchmarking against prior works.
Lack of prior studies on the same dataset
To the best of our knowledge, there are no existing studies that have used the exact same dataset for plant stress classification using ML and DL models. While there might be research on related topics or using similar approaches, the specific combination of data features, target variables, and model evaluations presented in this study appears to be unique. This absence of directly comparable work highlights the novel contribution of this research.
Differences in model application and dataset scope
Even in cases where similar datasets are available, differences in data preprocessing, feature selection, and model application can lead to variations in results. Studies that focus on different plant species, types of stress, or environmental conditions might use datasets that are not directly comparable to the one used in this research. These differences in experimental setups further complicate the possibility of a meaningful comparison.
Justification for lack of comparison
Given the novelty and specificity of the dataset used in this study, direct comparison with existing works is not feasible. Instead, this research contributes new insights into the application of ML and DL models for plant stress classification, establishing a foundation for future studies. The detailed evaluation of models like LSTM, SVM, Gradient Boosting, and RNN provides a benchmark for subsequent research in this domain.
Potential for future comparisons
As the field of AI-driven agriculture progresses and similar datasets become more widely available, future studies will have the opportunity to perform direct comparisons. This study serves as a foundational work, paving the way for future research that can build on these findings and offer comparative analyses as more datasets and studies emerge.
In conclusion, while direct comparison with existing works on the same dataset is not possible due to the reasons outlined above, this study’s novel contribution lies in its unique dataset and the comprehensive evaluation of various ML and DL models. Future research will benefit from these findings and may eventually provide opportunities for more direct comparisons as the field advances.
Conclusions
This study represents a significant leap forward in sustainable agriculture by integrating machine learning and deep learning techniques to address plant stress reduction. The use of gradient boosting, support vector machines, recurrent neural network, and long short-term memory (LSTM) algorithms, alongside a comprehensive analysis of the ubiquitin domains of TYRKC and RBR-E3 stress-associated signaling proteins across various plant species, has provided valuable insights into the physiological responses of plants to drought stress. The study’s rigorous approach, including ablation analysis and the evaluation of metrics such as F1 score, recall, and precision, has demonstrated the effectiveness and reliability of these algorithms in identifying and categorizing stress events. Notably, the LSTM algorithm achieved a remarkable 97% accuracy, underscoring its potential for precise and dependable stress classification.
The research also highlights the importance of data extraction and processing techniques, such as retrieving UniProt IDs, utilizing the SMART database, and analyzing physicochemical properties, which have laid a strong foundation for understanding stress mechanisms in various plant species. Despite these advancements, challenges remain in applying AI to agriculture, including selecting appropriate databases, ensuring data privacy, and accurately characterizing plant stress.
Interdisciplinary collaboration and innovative solutions are essential to overcoming these challenges. The application of AI in agriculture extends beyond academic research, offering practical benefits such as improved resource efficiency, guidance for precision agriculture, and support for global food security initiatives. Bridging the knowledge gap and promoting the widespread adoption of AI technologies are crucial for the sustainable advancement of farming practices. Ultimately, this study not only enhances our understanding of plant stress responses but also underscores the transformative potential of AI in revolutionizing agriculture. By addressing these challenges and seizing the opportunities presented by AI, the development of robust, effective, and sustainable agricultural systems becomes a tangible reality.
Author contributions
TA conceptualization, formal analysis and writing the original draft. SUR conceptualization, data curation and writing the original draft. SA data curation, formal analysis, and methodology. KM methodology, software and project administration. SAO funding acquisition, investigation and project administration. RCI resources, visualization and validation. TK investigation, formal analysis and validation. IA supervision, investigation and writing - review & edit the manuscript. All authors read and approved the final manuscript.
Funding
This research is funded by the European University of Atlantic.
Data availability
The data can be requested from the corresponding authors.
Code availability
The code used in this manuscript is available at the following GitHub repository: https://github.com/Shamshair-Ali007/Smart-Agriculture-Utilizing-ML-and-DL.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tahir Khurshaid, Email: tahir@ynu.ac.kr.
Imran Ashraf, Email: imranashraf@ynu.ac.kr.
References
- 1.Alipanahi, B., Delong, A., Weirauch, M. T. & Frey, B. J. Predicting the sequence specificities of dna-and rna-binding proteins by deep learning. Nat. Biotechnol. 33(8), 831–838 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Wang, L. et al. A computational-based method for predicting drug-target interactions by using stacked autoencoder deep neural network. J. Comput. Biol. 25(3), 361–373 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Senior, A. W. et al. Improved protein structure prediction using potentials from deep learning. Nature 577(7792), 706–710 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Singh, A. K., Ganapathysubramanian, B., Sarkar, S. & Singh, A. Deep learning for plant stress phenotyping: Trends and future perspectives. Trends Plant Sci. 23(10), 883–898 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Fraire-Velázquez, S. & Balderas-Hernández, V. E. Abiotic stress in plants and metabolic responses. In Abiotic Stress-Plant Responses and Applications in Agriculture 25–48 (2013).
- 6.Waititu, J. K. et al. Transcriptome analysis of tolerant and susceptible maize genotypes reveals novel insights about the molecular mechanisms underlying drought responses in leaves. Int. J. Mol. Sci. 22(13), 6980 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bibi, M., Rehman, S. U., Mahmood, K. & Shoukat, R. S. An intelligent decision support system for crop yield prediction using machine learning and deep learning algorithms. Proc. Pak. Acad. Sci. A Phys. Comput. Sci. 60(3), 37–48 (2023). [Google Scholar]
- 8.Dietz, K.-J., Zörb, C. & Geilfus, C.-M. Drought and crop yield. Plant Biol. 23(6), 881–893 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Edwards, B., Gray, M. & Hunter, B. The social and economic impacts of drought. Aust. J. Soc. Issues 54(1), 22–31 (2019). [Google Scholar]
- 10.Kuwayama, Y., Thompson, A., Bernknopf, R., Zaitchik, B. & Vail, P. Estimating the impact of drought on agriculture using the us drought monitor. Am. J. Agr. Econ. 101(1), 193–210 (2019). [Google Scholar]
- 11.Hu, H. & Xiong, L. Genetic engineering and breeding of drought-resistant crops. Annu. Rev. Plant Biol. 65, 715–741 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Mukherjee, S., Mishra, A. & Trenberth, K. E. Climate change and drought: A perspective on drought indices. Curr. Clim. Change Rep. 4, 145–163 (2018). [Google Scholar]
- 13.Yildirim, G., Rahman, A. & Singh, V. P. A bibliometric analysis of drought indices, risk, and forecast as components of drought early warning systems. Water 14(2), 253 (2022). [Google Scholar]
- 14.Abbas, A. et al. An artificial intelligence framework for disease detection in potato plants. Eng. Technol. Appl. Sci. Res. 14(1), 12628–12635 (2024). [Google Scholar]
- 15.Fang, Y. & Ramasamy, R. P. Current and prospective methods for plant disease detection. Biosensors 5(3), 537–561 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohanty, S. P., Hughes, D. P. & Salathé, M. Using deep learning for image-based plant disease detection. Front. Plant Sci. 7, 215232 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tardieu, F., Cabrera-Bosquet, L., Pridmore, T. & Bennett, M. Plant phenomics, from sensors to knowledge. Curr. Biol. 27(15), 770–783 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Zhao, W., Yamada, W., Li, T., Digman, M. & Runge, T. Augmenting crop detection for precision agriculture with deep visual transfer learning a case study of bale detection. Remote Sens. 13(1), 23 (2020). [Google Scholar]
- 19.Melo, F. V., Oliveira, M. M., Saibo, N. J. & Lourenço, T. F. Modulation of abiotic stress responses in rice by e3-ubiquitin ligases: A promising way to develop stress-tolerant crops. Front. Plant Sci. 12, 640193 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shu, K. & Yang, W. E3 ubiquitin ligases: Ubiquitous actors in plant development and abiotic stress responses. Plant Cell Physiol. 58(9), 1461–1476 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang, N. et al. The e3 ligase tasap5 alters drought stress responses by promoting the degradation of drip proteins. Plant Physiol. 175(4), 1878–1892 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, Y. et al. Atpprt3, a novel e3 ubiquitin ligase, plays a positive role in aba signaling. Plant Cell Rep. 39, 1467–1478 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Ban, Z. & Estelle, M. Cul3 e3 ligases in plant development and environmental response. Nat. plants 7(1), 6–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng, F. et al. Expression and regulation of atl9, an e3 ubiquitin ligase involved in plant defense. PLoS ONE 12(11), 0188458 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manning, G., Whyte, D. B., Martinez, R., Hunter, T. & Sudarsanam, S. The protein kinase complement of the human genome. Science 298(5600), 1912–1934 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Sunil, C., Jaidhar, C. & Patil, N. Systematic study on deep learning-based plant disease detection or classification. Artif. Intell. Rev. 56(12), 14955–15052 (2023). [Google Scholar]
- 27.Sunil, C., Jaidhar, C. & Patil, N. Binary class and multi-class plant disease detection using ensemble deep learning-based approach. Int. J. Sustain. Agric. Manage. Inf. 8(4), 385–407 (2022). [Google Scholar]
- 28.Sunil, C., Jaidhar, C. & Patil, N. Cardamom plant disease detection approach using efficientnetv2. Ieee Access 10, 789–804 (2021). [Google Scholar]
- 29.Sunil, C., Jaidhar, C. & Patil, N. Tomato plant disease classification using multilevel feature fusion with adaptive channel spatial and pixel attention mechanism. Expert Syst. Appl. 228, 120381 (2023). [Google Scholar]
- 30.Bokhtiar, S. M., Islam, S. M. F., Molla, M. M. U., Salam, M. A. & Rashid, M. A. Demand for and supply of pulses and oil crops in Bangladesh: A strategic projection for these food item outlooks by 2030 and 2050. Sustainability 15(10), 8240 (2023). [Google Scholar]
- 31.Wang, J. et al. Assessing stress responses in potherb mustard (Brassica juncea var. multiceps) exposed to a synergy of microplastics and cadmium: Insights from physiology, oxidative damage, and metabolomics. Sci. Total Env. 907, 167920 (2024). [DOI] [PubMed] [Google Scholar]
- 32.Wakelin, S. A. et al. Climate change induced drought impacts on plant diseases in New Zealand. Austral. Plant Pathol. 47, 101–114 (2018). [Google Scholar]
- 33.Gupta, A., Rico-Medina, A. & Caño-Delgado, A. I. The physiology of plant responses to drought. Science 368(6488), 266–269 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Joshi, R. C., Patel, V. R., Mishra, A. & Kumar, S. Real-time plant leaf disease detection using cnn and solutions to cure with android app. In 2023 International Conference on Computing, Communication, and Intelligent Systems (ICCCIS) 455–460 (IEEE, 2023).
- 35.Jia, L. et al. Microplastic stress in plants: Effects on plant growth and their remediations. Front. Plant Sci. 14, 1226484 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones, H. & Schofield, P. Thermal and other remote sensing of plant stress. Gen. Appl. Plant Physiol. 34(1–2), 19–32 (2008). [Google Scholar]
- 37.Thokala, B. & Doraikannan, S. Detection and classification of plant stress using hybrid deep convolution neural networks: A multi-scale vision transformer approach. Traitement Signal 40, 6 (2023). [Google Scholar]
- 38.Kuo, C.-E. et al. Early detection of drought stress in tomato from spectroscopic data: A novel convolutional neural network with feature selection. Chemom. Intell. Lab. Syst. 239, 104869 (2023). [Google Scholar]
- 39.Hagen, M. et al. Interpretable machine learning decodes soil microbiome’s response to drought stress. Environ. Microbiome 19(1), 35 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chandel, N. S. et al. Identifying crop water stress using deep learning models. Neural Comput. Appl. 33, 5353–5367 (2021). [Google Scholar]
- 41.Swaminathan, B. & Vairavasundaram, S. D2cnn: Double-staged deep cnn for stress identification and classification in cropping system. Agric. Syst. 216, 103886 (2024). [Google Scholar]
- 42.Azimi, S., Kaur, T. & Gandhi, T.K. Bat optimized cnn model identifies water stress in chickpea plant shoot images. In 2020 25th International Conference on Pattern Recognition (ICPR) 8500–8506 (IEEE, 2021).
- 43.Rojanarungruengporn, K. & Pumrin, S. Early stress detection in plant phenotyping using cnn and lstm architecture. In 2021 9th International Electrical Engineering Congress (iEECON) 389–392 (IEEE, 2021).
- 44.Khalifani, S., Darvishzadeh, R., Azad, N. & Rahmani, R. S. Prediction of sunflower grain yield under normal and salinity stress by rbf, mlp and cnn models. Ind. Crops Prod. 189, 115762 (2022). [Google Scholar]
- 45.Chandel, N. S. et al. Water stress identification of winter wheat crop with state-of-the-art ai techniques and high-resolution thermal-rgb imagery. Plants 11(23), 3344 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.. Krishna, R. & Prema, K. Constructing and optimising rnn models to predict fruit rot disease incidence in areca nut crop based on weather parameters. In IEEE Access (2023).
- 47.Concepcion II, R. et al. Lettuce leaf water stress estimation based on thermo-visible signatures using recurrent neural network optimized by evolutionary strategy. In 2020 IEEE 8th R10 Humanitarian Technology Conference (R10-HTC) 1–6 (IEEE, 2020).
- 48.Nawaz, A., Ali, T., Mustafa, G., Rehman, S. U. & Rashid, M. R. A novel technique for detecting electricity theft in secure smart grids using cnn and xg-boost. Intell. Syst. Appl. 17, 200168 (2023). [Google Scholar]
- 49.Asad, R. et al. Computer-aided early melanoma brain-tumor detection using deep-learning approach. Biomedicines 11(1), 184 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data can be requested from the corresponding authors.
The code used in this manuscript is available at the following GitHub repository: https://github.com/Shamshair-Ali007/Smart-Agriculture-Utilizing-ML-and-DL.