Abstract
Advances in bladder cancer (BCa) treatment have been hampered by the lack of predictive biomarkers and targeted therapies. Here, we demonstrate that loss of the tumor suppressor NUMB promotes aggressive bladder tumorigenesis and worsens disease outcomes. Retrospective cohort studies show that NUMB-loss correlates with poor prognosis in post-cystectomy muscle-invasive BCa patients and increased risk of muscle invasion progression in non-muscle invasive BCa patients. In mouse models, targeted Numb ablation induces spontaneous tumorigenesis and sensitizes the urothelium to carcinogenic insults, accelerating tumor onset and progression. Integrative transcriptomic and functional analyses in mouse and human BCa models reveal that upregulation of YAP transcriptional activity via a RHOA/ROCK-dependent pathway is a hallmark of NUMB-deficient BCa. Pharmacological or genetic inhibition of this molecular pathway selectively inhibits proliferation and invasion of NUMB-deficient BCa cells in 3D-Matrigel organoids. Thus, NUMB-loss could serve as a biomarker for identifying high-risk patients who may benefit from targeted anti-RHOA/ROCK/YAP therapies.
Subject terms: Bladder cancer, RHO signalling, Targeted therapies, Prognostic markers
The transition from non-muscle-invasive bladder cancer (NMIBC) to muscle-invasive bladder cancer (MIBC) is associated with poorer patient prognosis. Here, the authors investigate NUMB loss as a driver of the NMIBC-to-MIBC transition and identify a RHOA/ROCK/YAP signaling pathway as a therapeutic vulnerability.
Introduction
Bladder cancer (BCa) ranks among the most common neoplasms in industrialized countries1. Most BCa manifest as urothelial carcinomas, originating from the transitional epithelium lining the inner surface of the bladder2–4. Clinical staging of these tumors depends on the extent of invasion into the bladder wall and their grade of cellular differentiation2,4. Tumors that have penetrated the superficial layers, infiltrating the detrusor muscle and beyond, are classified as muscle-invasive BCa (MIBC), and constitute ~25% of all BCa. Despite aggressive treatments, such as radical cystectomy and adjuvant platinum-based chemotherapy, MIBC has an overall poor prognosis, characterized by a high metastatic potential and a 5-year overall survival of ~50%. The aggressive treatments are also associated with high morbidity, severe side effects and a diminished quality of life4–6.
Conversely, tumors confined to the inner lining of the bladder are classified as non-muscle invasive BCa (NMIBC), representing ~75% of newly diagnosed BCa cases2,7. NMIBCs are highly heterogeneous, comprising tumors of different stages and grades: i) Ta papillary tumors, confined to the mucosa and typically low-grade/well-differentiated; ii) T1 tumors with submucosal invasion, representing a heterogeneous group that can present as either low- or high-grade/poorly differentiated lesions; and iii) carcinoma in situ (CIS; Tis in the TNM classification), typically presenting as flat and high-grade dysplasia lesions4. Although 5-year survival rates for NMIBC are favorable ( > 90%), a significant proportion of patients experience disease recurrence ( ~ 50-70%) and progression to MIBC disease ( ~ 20-30%)8. Notably, NMIBC patients who progress to MIBC often face a worse prognosis compared to patients with primary MIBC9. This, coupled with the lack of improvement in MIBC mortality rates, makes the transition from NMIBC to MIBC a life-threatening event. Therefore, high-risk NMIBC patients must undergo lifelong cystoscopic surveillance with bladder-sparing transurethral resection (TUR), often followed by adjuvant intravesical instillations of Bacillus Calmette-Guérin (BCG) or bladder perfusion chemotherapy to eradicate residual disease and reduce the frequency of recurrence and progression4,6,10. This treatment regimen makes BCa the most expensive cancer to treat11.
The considerable heterogeneity of NMIBC presents challenges in its clinical management, exacerbated by the inadequacy of current staging criteria which heavily rely on clinicopathological characteristics. Incorrect staging can lead to understaging and subsequent undertreatment, negatively affecting survival, or overtreatment with early cystectomy, associated with significant morbidity8. Moreover, despite the numerous molecular alterations described for this disease, targeted therapies to prevent disease progression are currently lacking2,12–15. Consequently, there is an urgent need for biomarkers of disease progression to guide clinical decision-making, particularly regarding the choice between conservative (surveillance and TUR) vs. aggressive treatments (early cystectomy plus adjuvant therapies). A deeper understanding of the biology underlying the NMIBC to MIBC transition is therefore essential for an improved personalized management of NMIBC patients3,8,10,13.
NUMB is a protein whose expression levels are variably altered in different types of human cancers. Both its underexpression and overexpression have been reported to play tumor-specific roles in tumorigenesis and cancer progression, and are associated with the acquisition of aggressive tumor phenotypes and adverse prognosis16–20. However, the relevance of NUMB alterations to BCa pathogenesis and disease progression has so far remained elusive.
In this work, we highlight a critical role of the loss of NUMB in bladder tumorigenesis and its potential clinical relevance as a prognostic and therapeutic biomarker in BCa. Through retrospective cohort studies, we demonstrate that a NUMB-deficient status is a prognostic biomarker for risk of MIBC progression in NMIBC patients, and for poor prognosis in MIBC patients. Furthermore, using a Numb-knockout (KO) mouse model, we show that Numb gene ablation is alone sufficient to drive spontaneous malignant transformation of the urothelium, causing the appearance of preneoplastic lesions, preinvasive tumors, and muscle-invasive tumors, and sensitizes the urothelium to carcinogen-induced malignant transformation, accelerating tumor onset and progression. By integrating functional and molecular studies in mouse and human BCa models, we show that NUMB loss leads to the upregulation of RHOA/ROCK signaling to the actin cytoskeleton. This, in turn, results in the downregulation of the Yes-associated protein-1 (YAP) inhibitory Hippo pathway, leading to enhanced YAP transcriptional activity. Through genetic and pharmacological inhibition studies in mouse and human BCa models, we demonstrate that hyperactivation of RHOA/ROCK/YAP signaling is responsible for the aggressive migratory/invasive phenotype of NUMB-deficient BCa cells, likely due to activation of an EMT program. These findings highlight the RHOA/ROCK/YAP circuitry as a therapeutic vulnerability in NUMB-deficient NMIBC patients that can be targeted pharmacologically to prevent progression to MIBC.
Results
NUMB loss is a hallmark of aggressive disease in human BCa patients
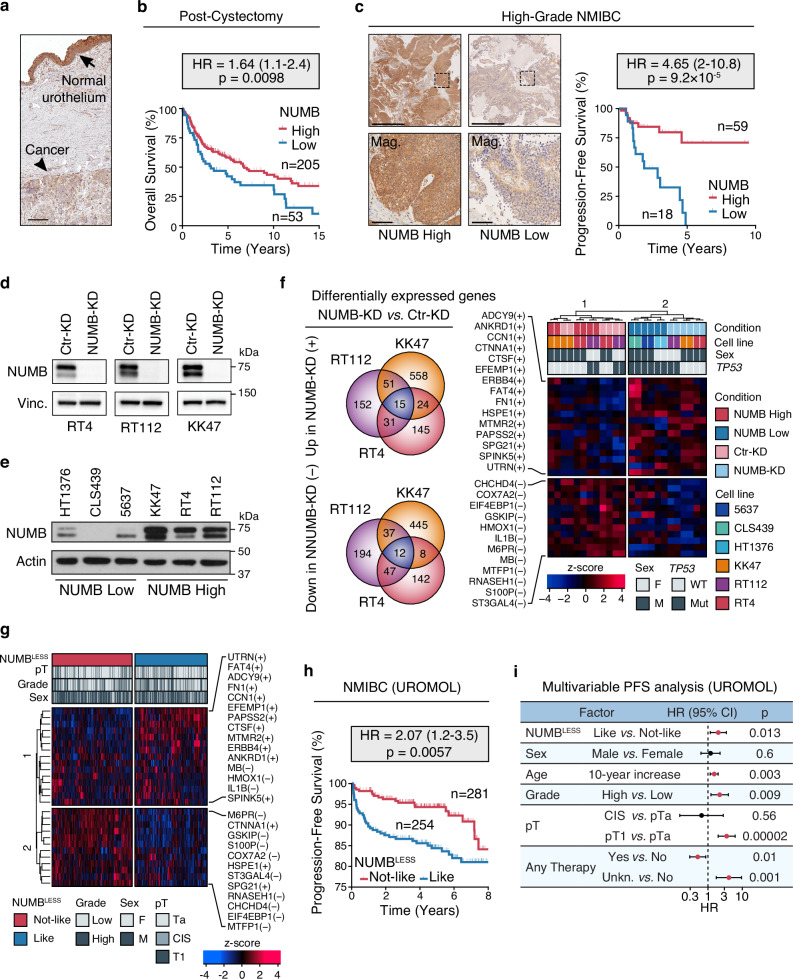
Alterations of NUMB expression, involving either overexpression or underexpression of the protein depending on the tumor type, have been previously implicated in tumorigenesis and disease progression16–20. To assess whether NUMB alterations are relevant to BCa, we performed a preliminary immunohistochemistry (IHC) analysis of formalin-fixed paraffin-embedded (FFPE) BCa samples obtained from patients who underwent radical cystectomy. We found that NUMB expression is frequently downregulated in the tumor lesion compared to the adjacent normal urothelium (Fig. 1a). To understand better the relevance of NUMB loss to the natural history of BCa, we surveyed a retrospective consecutive cohort of 356 MIBC patients (19.4% females, 80.6% males) who underwent radical cystectomy, assessing NUMB status by IHC on whole FFPE tumor sections. Complete clinicopathological follow-up (median, 5.73 years) was available for 258 patients. Approximately 80% of patients were categorized as NUMBHigh (73.9% females, 78.4% males) and ~20% as NUMBLow (26.1% females, 21.6% males). Notably, a NUMBLow status significantly correlated with clinical parameters of aggressive disease (e.g., tumor extension and vascular invasion) and positive lymph node status. It also predicted a higher rate of mortality, independently of standard clinical and pathological variables, including sex, age, tumor extension, lymph node status and vascular invasion (NUMBLow vs. NUMBHigh, HR = 1.64, CI = 1.1-2.4, p = 0.0098) (Fig. 1b, Supplementary Fig. 1a and Supplementary Table 1).
Fig. 1. NUMB loss is prognostic in human BCa and correlates with NMIBC progression.
a Representative IHC image of a NUMB-deficient MIBC (arrowhead) bordering the normal urothelium expressing NUMB (arrow). Bar, 400 μm. b Overall survival of post-cystectomy patients stratified by NUMB expression (NUMBHigh vs. NUMBLow) determined by IHC. HR, hazard ratio (95% confidence interval); p, two-sided log-rank test p-value; n, patient number; in this and all other relevant panels. c Left, Representative IHC images of NUMB expression in high-grade NUMBHigh and NUMBLow NMIBC. Boxed areas are magnified (Mag.) below. Bars, 1 mm; Mag. 100 µm. Right, progression-free survival of NMIBC patients stratified by NUMB expression. p, two-sided log-rank test p-value. d NUMB expression in RT4, RT112 and KK47 NIMBC cells knocked down for NUMB by siRNA (NUMB-KD) or mock siRNA in controls (Ctr-KD). Vinculin (Vinc.), loading control. Results are representative of two independent experiments. e NUMB expression in MIBC NUMBLow cell lines (HT1376, CLS439, 5637) and NMIBC NUMBHigh cell lines (KK47, RT4, RT112). Actin, loading control. Results are representative of two independent experiments. f Definition of the NUMBLESS signature. The Venn diagrams on the left show the intersection of genes up and down-regulated in the independent comparison of the NUMBHigh RT4, RT112 and KK47 NIMBC cells, NUMB-KD vs. Ctr-KD. Right, unsupervised hierarchical clustering of NUMBHigh and NUMBLow, and NUMB-KD and Ctr-KD BCa cell lines, based on the expression levels of the 15 genes consistently upregulated (+) and 12 genes consistently downregulated (-) (NUMBLESS signature). The color code scale indicates the z-score of log-normalized transcript abundance, in this and other relevant panels in the figure. g Unsupervised clustering of the 535 NMIBC patients from the UROMOL cohort, stratified by the NUMBLESS signature (NUMBLESS-Like vs. -Not-Like, corresponding to a NUMB-deficient or NUMB-proficient status, respectively). h Progression-free survival of NMIBC patients of the UROMOL cohort categorized as NUMBLESS-Like and NUMBLESS-Not-Like. p, two-sided log-rank test p-value. i. Multivariable progression-free survival (PFS) analysis of the association between the indicated factors and good (HR < 1) or poor (HR > 1) prognosis in the UROMOL cohort (n = 535). Significant associations are marked in red. HR, multivariable hazard ratios with error bars representing 95% confidence intervals (CI) by Cox proportional hazards model; p, two-sided Wald-test p-value. Source data are provided as Source Data file.
To investigate whether NUMB has clinical value in NMIBC, we analyzed a cohort of 77 high-grade NMIBC patients who either progressed or not to MIBC in a 4-month minimum follow-up period, as assessed by TUR (median follow-up 1.94 years; see Supplementary Table 2 for clinicopathological characteristics). Strikingly, in this cohort, a NUMBLow status predicted a worse progression-free survival with a higher rate of MIBC progression compared to NUMBHigh patients (HR = 4.65, CI = 2–10.8, p < 0.0001) (Fig. 1c). These retrospective clinical studies indicate that a NUMB-deficient status is a hallmark of aggressive disease and poor prognosis in BCa. In particular, in NMIBC patients, NUMB status appears to be a predictive biomarker of risk of progression to muscle-invasive disease independently of other clinical parameters, including sex, age and tumor extension (Supplementary Fig. 1b). These results highlight the potential use of NUMB status as a biomarker, which, in conjunction with clinicopathological parameters, could permit a more refined risk stratification of NMIBC patients.
Derivation of a clinically relevant gene signature characteristic of the NUMB-deficient state
NUMB dysfunction in cancer results from heterogeneous mechanisms, including exaggerated proteasomal degradation, isoform-specific transcriptional regulation, and post-translational modifications, as previously documented in breast cancer21–23. Therefore, we reasoned that a gene signature associated with a NUMB-deficient state could be a more precise tool than NUMB IHC, which assesses only protein loss, for identifying NMIBC patients at risk of progression to MIBC.
To obtain a comprehensive picture of the molecular alterations associated with NUMB loss in human BCa, we performed global transcriptomic profiling by bulk RNA-Seq of three NUMBHigh NMIBC cell lines: namely, RT4, RT112 and KK47, silenced or not for NUMB (NUMB-KD vs. Ctr-KD) (Fig. 1d). We also obtained the global transcriptomic profiles of three NUMBLow MIBC cell lines – CLS439, 5637 and HT1376 – for a comparative analysis (Fig. 1e). Notably, the NMIBC and MIBC cell lines selected for this analysis were derived from both male and female BCa patients (NMIBC, male: RT4 and KK47; female: RT112; MIBC, male: CLS439 and 5637; female: HT1376). The representation of both male and female BCa cell lines is important to extrapolate the results to the naturally occurring BCa disease. We initially performed an integrative comparative analysis of the transcriptomes of the three isogeneic NUMB-KD vs. Ctr-KD NMIBC cell lines to identify the most significantly deregulated genes (absolute Log2 fold-change > 1, FDR adjusted p < 0.01) displaying a consistent direction of regulation (up or down) relative to NUMB status across the three paired analyses. A minimal signature composed of 27 up/down-regulated genes, characteristic of the NUMB-defective condition, was identified and is hereafter referred to as the “NUMBLESS signature” (Fig. 1f). Remarkably, in an unsupervised clustering analysis, this signature correctly stratified the NUMBHigh (RT4, RT112 and KK47) and NUMBLow (CLS439, 1537 and HT1376) BCa cell lines into two distinct groups, independently of their respective male or female origin (Fig. 1f).
To test the clinical value of the NUMBLESS signature in NMIBC patients, we interrogated the transcriptomes of 535 NMIBC patients (females: n = 121, 22.6%; males: n = 414, 77.3%), with complete long-term clinicopathological follow-up (median follow-up, 5.12 months), who were included in the UROMOL study24 (see cohort description in Supplementary Table 3). The unsupervised clustering analysis of this cohort with the NUMBLESS signature identified two distinct groups of NMIBC patients who were classified as NUMBLESS-like (corresponding to a NUMB-deficient status) or NUMBLESS-not-like (corresponding to a NUMB-proficient status) (Fig. 1g). No statistically significant differences were observed in the distribution of these two groups among female and male patients (NUMBLESS-like: females, 51.2%, n = 62; males, 46.4%, n = 192; NUMBLESS-not-like: females, 48.8%, n = 59; males, 53.6%, n = 222; p = 0.35) indicating that NUMB loss is equally relevant to the male and female BCa disease. NUMBLESS-like NMIBC patients displayed a significantly higher risk of MIBC progression and shorter progression-free survival compared to NUMBLESS-not-like patients (HR = 2.07, CI = 1.2-3.5, p = 0.0057) (Fig. 1h). Notably, the ability of the NUMBLESS signature to predict risk of MIBC progression was maintained in a multivariable analysis adjusted for other clinical and pathological factors, including sex, age, grade, tumor extension and type of therapy (Fig. 1i). This finding, together with the results from the retrospective analysis of the NIMBC cohort by NUMB IHC (see Fig. 1c), strongly supports the value of assessing NUMB status, in addition to currently available clinical parameters, to predict the risk of MIBC progression in individual patients. In addition, the NUMBLESS 27-gene signature has a superior stratification power compared to IHC for identifying high-risk NUMB-deficient NMIBC patients ( ~ 50% NUMBLESS-like vs. 20% IHC NUMBLow). This finding suggests that the signature is capturing NUMB dysfunction associated not only with protein degradation but also other molecular mechanisms22,23.
Based on these findings, we investigated the relationship between the NUMBLESS signature and 14 BCa-specific transcriptomic signatures previously characterized for their clinical relevance25–38 (Supplementary Fig. 2a). We noted that there were only four overlapping genes (HMOX1, IL1B, EFEMP1, and ST3GAL4) between the NUMBLESS signature and the other signatures, with two of these genes (HMOX1 and IL1B) showing an opposite direction of regulation (Supplementary Fig. 2a). The NUMBLESS signature ranked as the third most potent signature, providing statistically significant prognostic information over the standard clinicopathological parameters in a multivariable analysis in which the different signatures were individually challenged against standard clinical variables to predict the risk of muscle invasion in the UROMOL cohort (Supplementary Fig. 2b). Remarkably, in a Lasso penalized Cox regression model analyzing the prognostic redundancy between the individual signatures, the NUMBLESS signature not only retained its prognostic value against all other signatures, but also displayed the highest prediction coefficient among the signatures that provided additional prognostic information beyond standard clinical variables (Supplementary Fig. 2c). These results suggest that the prognostic information provided by the NUMBLESS signature reflects specific aspects of BCa biology that appear to differ from those addressed by other BCa-specific transcriptomic predictors. This finding has important clinical implications, supporting the use of the NUMBLESS signature – either alone or in combination with other genomic signatures – for NMIBC patient stratification regarding risk of progression, in addition to standard clinicopathological parameters.
NUMB loss results in pathological regulation of YAP transcriptional activity and EMT in BCa
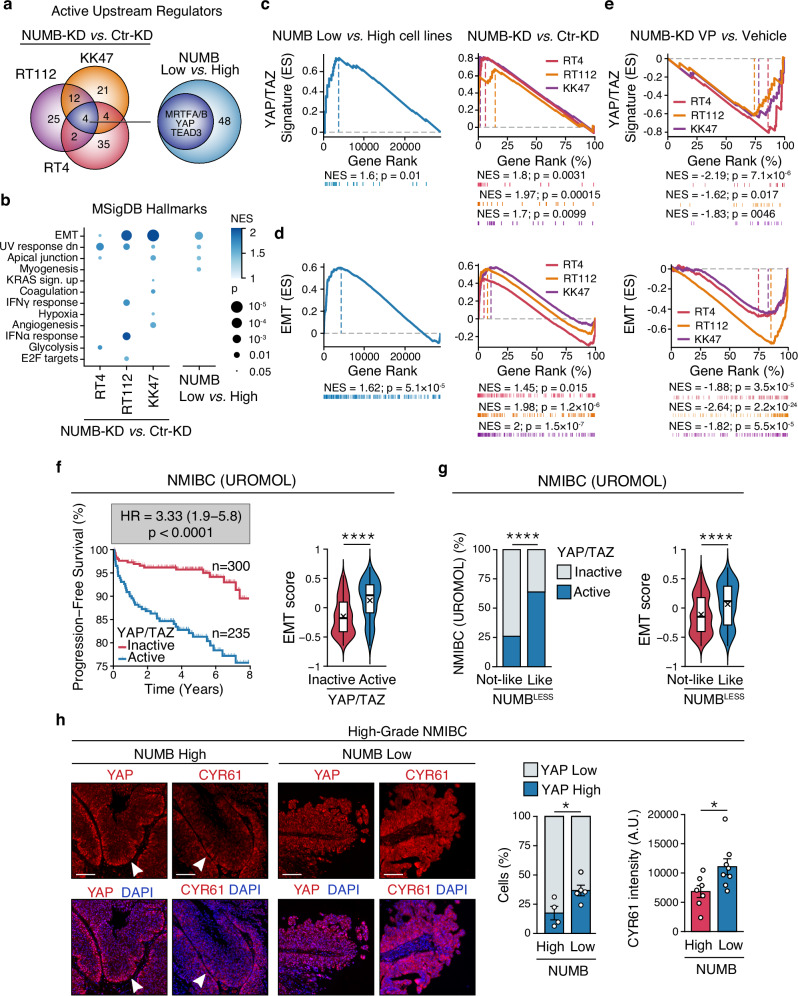
To investigate transcriptional programs associated with NUMB dysfunction in BCa, we used the IPA “Upstream Regulator Analysis” to compare the transcriptomes of the NUMBHigh NMIBC cell lines (RT4, RT112 and KK47) vs. NUMBLow MIBC cell lines (CLS439, 5637 and HT1376), and the paired NUMB-KD vs. Ctr-KD cell lines (RT4, RT112 and KK47). This analysis revealed common transcriptional regulators that are active in the NUMB-deficient condition (i.e., NUMBLow/NUMB-KD cells) relative to the NUMB-proficient condition (i.e., NUMBHigh/Ctr-KD cells). The most distinctive molecular traits associated with the NUMB-deficient condition were activation of upstream regulators of the YAP pathway (MRTFA/B, TEAD3 and YAP) and various EMT/phenotypic plasticity (e.g., TWIST1, SNAI1/2, ZEB1, CTNNB1, KLF4, SOX4) (Fig. 2a and Supplementary Fig. 3a).
Fig. 2. NUMB loss is associated with YAP activation and EMT.
a Venn diagrams show the activated transcriptional regulators identified in NUMB-KD vs. Ctr-KD RT4, RT112 and KK47 NIMBC cells (left) and in NUMBLow vs. NUMBHigh cell lines (right). The blue circle indicates the 4 common YAP/TAZ transcriptional regulators in the two paired comparisons. b Hallmark gene sets from MSigDB positively enriched by GSEA in NUMBLow vs. NUMBHigh cell lines and NUMB-KD vs. Ctr-KD RT4, RT112 and KK47 NIMBC cells. NES, Normalized Enriched Score; p, FDR adjusted two-sided permutation test p-value; n = 2. c, d Active YAP/TAZ (c) and EMT (d) gene signature by GSEA of the 3 NUMBLow vs. 3 NUMBHigh cell lines (left panels), and NUMB-KD vs. Ctr-KD RT4, RT112 and KK47 cells (right panels). ES, Enrichment Score; NES, Normalized Enrichment Score; p, two-sided permutation test p-value; n = 2. e GSEA showing downregulation of an active YAP/TAZ (top) and EMT (bottom) gene signature in the indicated NUMBHigh NUMB-KD cell lines treated with 500 nM verteporfin (VP) vs. vehicle for 24 h. ES, Enrichment Score; NES, Normalized Enrichment Score; p, two-sided permutation test p-value; n = 1. f Left, progression-free survival of the UROMOL patient cohort categorized as YAP/TAZ active or inactive by the 22-gene YAP/TAZ signature. HR, hazard ratio (95% confidence interval); p, two-sided log-rank test p-value; n, patient number. Right, box-violin plot showing the EMT score in the YAP/TAZ active and inactive groups. Boxplots are delimited by 25th and 75th percentiles and show the median (horizontal line) and the mean (X). The whiskers span from the smallest and largest data values within a 1.5 interquartile range. ****, p < 0.0001 by two-sided Welch’s t-test. g Left, association between the NUMBLESS and the 22-gene YAP/TAZ signatures in the UROMOL cohort (n = 535). ****, p < 0.0001, by two-sided Fisher’s exact test. Right, box-violin plot (see legend to Fig. 2f) showing the EMT score in the NUMBLESS-Like or NUMBLESS-Not-Like groups. ****, p < 0.0001 by two-sided Welch’s t-test. h Left, Immunofluorescence of YAP and CYR61 expression (red) and DAPI nuclear stain (blue) in high-grade NUMBHigh vs. NUMBLow NMIBC tumors. Bars, 100 μm. Right, quantification of the % of YAP-high vs. -low expressing cells in 4 NUMBHigh and 5 NUMBLow NIMBC, and CYR61 mean cytoplasmic intensity (arbitrary units, A.U.) of 7 NUMBHigh and 8 NUMBLow NIMBC. Values are expressed as mean ± SEM. *, p = 0.038 for YAP, p = 0.035 for CYR61, by two-sided Welch’s t-test for YAP and two-sided two-sample t-test for CYR61. Source data are provided as Source Data file.
In line with these results, GSEA analysis of the set of differentially expressed genes between the paired NUMB-deficient vs. NUMB-proficient cell models revealed the enrichment of a YAP/TAZ signature, composed of 22 transcriptional targets of the YAP pathway39, and the upregulation of an EMT signature (MSigDB “Hallmarks”, see Materials and Methods) associated with the NUMB-deficient condition (Fig. 2b–d, Supplementary Tables 4–7; see also Supplementary Data 1–4 for the complete list of differentially regulated genes). Both these effects were strongly inhibited following treatment of NUMB-KD RT4, RT112 and KK47 cells with verteporfin (VP), a well-established inhibitor of YAP transcriptional activity that works by preventing the YAP-TEAD interaction40 (Fig. 2e). These results suggest that activation of EMT downstream of NUMB loss is epistatically controlled by YAP signaling hyperactivation.
Relevance of YAP/EMT activation downstream of NUMB dysfunction in BCa patients
Considering the previously reported role of YAP activation in bladder tumorigenesis and its prognostic association with disease progression and response to immunotherapy in NMIBC patients41–43, we investigated the relevance of YAP/EMT activation downstream of NUMB dysfunction in NMIBC patients. We used the NUMBLESS signature, as well as the YAP and EMT signatures identified in the NUMB-KD vs. Ctr-KD cell models, to interrogate the UROMOL cohort. In keeping with previous findings43, we found that an active YAP status, assessed by the YAP/TAZ signature, is associated with an adverse disease course, predicting increased risk of muscle-invasion progression independently of clinicopathological parameters (Fig. 2f and Supplementary Fig. 3b, c), and with a positive response to BCG immunotherapy (Supplementary Fig. 3d). Notably, patients displaying high expression of the YAP signature also showed higher expression of the EMT signature (Fig. 2f). Moreover, a NUMBLESS-like status, which we showed is associated with increased risk of muscle invasion (see Fig. 1g–i), strongly correlated with an active YAP status and increased expression of the EMT signature (Fig. 2g). Similarly to the YAP signature, the NUMBLESS-like status also predicts a favorable response to BCG treatment (Supplementary Fig. 3e). Collectively, these results indicate that the link between NUMB dysfunction and YAP/EMT pathway upregulation is relevant to NMIBC patients. Further supporting this view, immunofluorescence (IF) analysis of high-grade NMIBC TUR samples showed a direct correlation between low expression of NUMB protein and increased nuclear accumulation of YAP and its direct transcriptional target, CYR61, indicative of activated YAP signaling44 (Fig. 2h).
NUMB deficiency drives malignant transformation of the mouse urothelium and accelerates carcinogen-induced bladder tumorigenesis
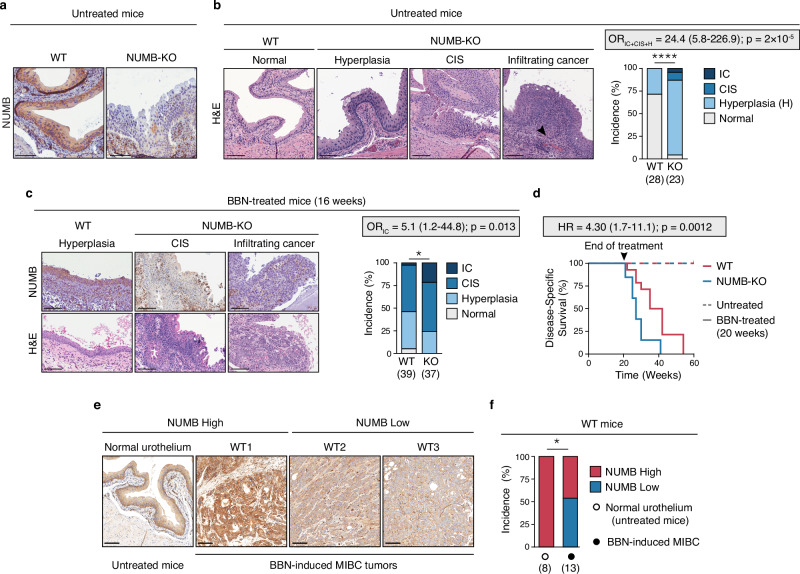
To investigate the impact of NUMB loss on the homeostasis of the normal urothelium, we used a transgenic NUMB-knockout (NUMB-KO) mouse model bearing targeted deletion of the Numb gene in the basal CK5+ layer (Fig. 3a). The NUMB-KO mouse was generated by crossing the Cre-loxP conditional NUMB-KO mouse (NUMBlox/lox) with the CK5-Cre mouse as previously described45,46. The rationale of this experimental strategy was based on evidence that, in both the mouse and human normal urothelium, NUMB is more highly expressed in basal (CK5 + /CK14 + ) cells compared to suprabasal intermediate (CK7 + ) and superficial umbrella (CK20 + ) cells (Supplementary Fig. 4a, b). We also reasoned that this model could have the potential to highlight key features of disease progression, considering previous evidence implicating basal, but not suprabasal, cells as the population-of-origin of aggressive BCa34,47,48.
Fig. 3. NUMB loss drives spontaneous malignant transformation of the normal urothelium and accelerates carcinogen-induced bladder tumorigenesis.
a NUMB expression in the urothelium of untreated adult WT (WT) and NUMB-KO (KO) mice. Bars, 100 μm. Images are representative of the bladders of 28 WT and 23 NUMB-KO mice examined. b Left, Representative H&E images of the normal WT urothelium, and preneoplastic (Hyperplasia) and neoplastic (carcinoma in situ, CIS; invasive cancer, IC) urothelial lesions from NUMB-KO mice. The black arrowhead indicates IC. Bars, 100 μm. Right, Incidence (%) of histological phenotypes in the urothelium of untreated, aged-matched 4- to 12-month old WT (n = 28) and NUMB-KO (n = 23) mice. ****, p = 0.00002 by two-sided Pearson’s Chi-squared test. Odds ratio (OR) with 95% CI and associated two-sided p-value by Fisher’s exact test is shown. c Aged-matched 8- to 16-week-old WT (n = 39) and NUMB-KO (n = 37) mice were exposed to 0.05% BBN in the drinking water for 16 weeks followed by a 2-week washout period. Bladder tissues were harvested and examined for histological changes (H&E) and NUMB expression (IHC). Left, Representative images of a preneoplastic (Hyperplasia) lesion from BBN-WT mice and neoplastic (CIS and IC) lesions from BBN-NUMB-KO mice. Right, Incidence (%) of histological phenotypes in WT vs. KO mice at the end of treatment. Bars, 100 μm. *, p = 0.025 by two-sided Pearson’s Chi-squared test; OR (95% CI) with associated two-sided p-value by Fisher’s exact test is shown. d Kaplan–Meier plot showing the disease-specific survival (%) of aged-matched 8- to 16-week-old WT (n = 14) and NUMB-KO (n = 13) mice treated for 20 weeks with BBN before switching to regular drinking water. Mice were sacrificed according to endpoints. Dashed-line, WT (n = 10) and NUMB-KO (n = 10) untreated mice. HR, (95% CI) with two-sided p-value by Log Rank test are shown. e MIBC lesions excised from BBN-WT mice (n = 13) were compared to the normal urothelium of untreated WT mice (n = 8) for NUMB expression by IHC. Shown are three MIBC lesions, one NUMBHigh (WT1) and two NUMBLow (WT2, WT3), and the normal NUMBHigh urothelium of untreated mice. Bars, 100 μm. f Quantification of the experiment in ‘e’ showing the incidence (%) of NUMBLow vs. NUMBHigh MIBC tumors induced by BBN treatment in WT mice. *, p = 0.018 by two-sided Fisher’s exact test. Source data are provided as Source Data file.
Comparative histology of the NUMB-KO vs. WT bladder mucosa of age-matched male mice revealed that the absence of NUMB in the urothelium leads to its early hyperplastic thickening, characterized by expansion of the basal layer, and the appearance of in situ (CIS) and overtly infiltrating neoplastic lesions (Fig. 3b and Supplementary Fig. 4c, d). These results suggest a direct involvement of NUMB loss in initiating early morphological alterations in the urothelium, which precedes the emergence of preneoplastic lesions with a marked propensity to progress to advanced tumor stages. Of note, the use of CK5-Cre/NUMB-KO male mice also allowed us to monitor alterations in the urothelium without having to consider the impact of preneopastic/neoplastic lesions resulting from NUMB loss in the mammary gland45. Indeed, a systematic histopathological survey of CK5+ tissues in these mice revealed no evident signs of aberrant morphology in any tissue, other than the bladder, including the mammary gland that was almost completely substituted by adipose tissue (Supplementary Fig. 5a and Supplementary Table 8).
To investigate whether NUMB loss could also have a cooperative role in bladder tumorigenesis, we examined the susceptibility of NUMB-KO vs. WT mice to prolonged exposure to the bladder-specific chemical carcinogen, N-butyl-N-(4-hydroxybutyl) nitrosamine (BBN) (Supplementary Fig. 5b). The BBN-induced BCa mouse model faithfully reproduces the histological features and genetic alterations observed in human BCa, including the progression from NMIBC to MIBC48–50. Male mice were also used for these experiments because of their increased susceptibility to BBN, developing earlier morphological alterations compared with female mice51. We found that, despite heterogeneity in the efficiency of Numb KO in the urothelium of individual NUMB-KO mice (see Supplementary Fig. 5c), they displayed increased susceptibility to BBN. Indeed, early tumor onset and a dramatically increased incidence of invasive carcinoma were observed in these mice compared to age-matched WT mice (OR = 5.1, CI = 1.2–44.8, p = 0.013) (Fig. 3c), accompanied by a significant reduction in the survival rate (HR = 4.30, CI = 1.7–11.1, p = 0.0012) (Fig. 3d). Remarkably, we also noted that ~50% of BBN-induced invasive tumors that developed in WT mice exhibited spontaneous loss of NUMB expression (Fig. 3e, f).
Together, these results highlight the potent pro-tumorigenic effects of NUMB loss in the urothelium: not only is NUMB loss per se sufficient to induce bladder tumorigenesis, it also expedites invasive tumorigenesis driven by other oncogenic insults. These results also suggest that loss of NUMB is a frequently selected event during the neoplastic transformation of the normal urothelium induced by a genotoxic insult, presumably as a means to alleviate its homeostatic tumor suppressor function.
Loss of NUMB induces aggressive biological phenotypes in BCa cells through YAP transcriptional hyperactivation
Based on the above results, we reasoned that the paired WT vs. NUMB-KO and BBN-WT vs. BBN-NUMB-KO mouse models represent an ideal preclinical setting to investigate the molecular mechanisms underlying the aggressive biological behavior of NUMB-deficient BCa, focusing on the NUMB-YAP interaction identified in established human BCa cell lines and in NMIBC patients. IHC analysis of endogenous YAP expression in the WT murine urothelium showed an evident YAP nuclear staining largely confined to the basal layer, similar to the expression pattern of the normal human urothelium (Supplementary Fig. 6a). Further analysis comparing the urothelium of WT vs. NUMB-KO mice by in situ IF with an antibody specific for active YAP revealed that in NUMB-KO mice, nuclear active YAP is not confined to the basal layer, as observed in WT mice, but is instead diffusely distributed within the hyperplastic urothelium (Supplementary Fig. 6b). Moreover, average active YAP levels were significantly higher in the NUMB-KO urothelium compared to WT (Supplementary Fig. 6b). The side-by-side comparison of both early preneoplastic/hyperplastic and non-infiltrating CIS lesions from BBN-treated NUMB-KO vs. WT mice revealed significantly higher active YAP levels in the NUMB-KO lesions (Supplementary Fig. 6c). This increased active YAP associated with NUMB loss was maintained in BBN-NUMB-KO vs. BBN-WT secondary tumors generated by transplantation of primary BBN-induced infiltrating tumors (Supplementary Fig. 6d, e).
Together, these findings indicate that the aberrant activation of YAP signaling is an early driving event in the stepwise process of spontaneous or carcinogen-induced urothelial transformation downstream of NUMB loss. These results also provide further evidence of the link between NUMB loss and YAP hyperactivation, underscoring the suitability of the paired NUMB-KO vs. WT and BBN-NUMB-KO vs. BBN-WT murine models to dissect the functional contribution of YAP hyperactivation to the biology of NUMB-deficient BCa.
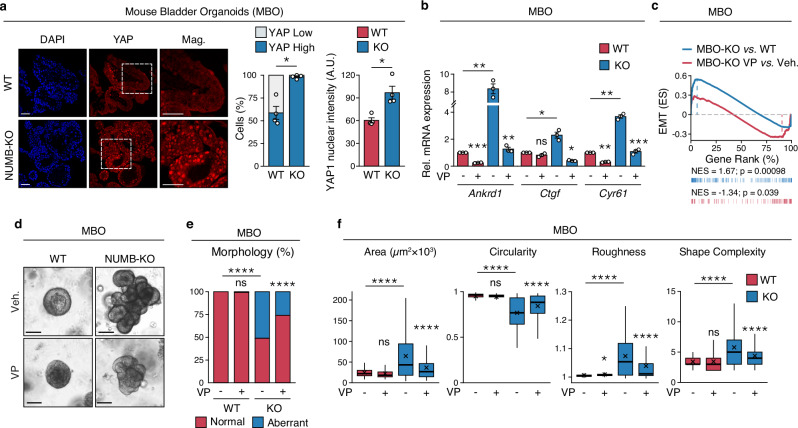
To investigate the phenotypic alterations linked to NUMB loss/YAP hyperactivation both in the normal urothelium and in bladder tumors, we took advantage of the ability of urothelial cells to originate self-organized 3D organotypic structures ex vivo in a reconstituted extracellular matrix (Matrigel). These structures (hereafter referred to as mouse bladder organoids, MBOs) are amenable to molecular and functional studies and therefore represent a highly tractable and pathophysiologically relevant experimental setting52. Male aged-matched, 4-6 month-old NUMB-KO and WT mice were used for the generation of MBOs.
An initial IF analysis of YAP expression in NUMB-KO vs. WT MBOs revealed that the absence of NUMB is associated with a diffuse distribution of cells exhibiting high intranuclear active YAP levels across the entire organoid population (Fig. 4a). This finding mirrors the results obtained in the in situ analysis of the NUMB-KO vs. WT bladder mucosa (Supplementary Fig. 6b). In keeping with the increased levels of active YAP, we observed higher expression levels of the YAP transcriptional targets, Ankrd1, Ctgf and Cyr61 (by RT-qPCR), as well as higher expression of the EMT/plasticity signature (by GSEA) in the molecular profiles of NUMB-KO vs. WT MBOs (Fig. 4b, c and Supplementary Table 9; see also Supplementary Data 5 for a complete list of differentially regulated gene). These alterations were reversed by the YAP inhibitor, VP (Fig. 4b, c), recapitulating the results observed in human NUMB-proficient RT4, RT112 and KK47 BCa cells silenced for NUMB (shown in Fig. 2e).
Fig. 4. NUMB loss induces invasive phenotypes and YAP signaling activation in normal urothelial cells.
a Left, Representative confocal IF images of endogenous YAP expression in FFPE sections of WT and NUMB-KO mouse bladder organoids (MBOs) grown in 3D-Matrigel. Nuclei are stained with DAPI. Bars, 100 μm; Mag., 50 μm. The % of YAP-positive cells (middle) and the mean nuclear YAP intensity (arbitrary units, A.U.) (right) are expressed as mean ± SEM, n = 4 fields/condition from one experiment, representative of two independent experiments. *, p = 0.011 (left), 0.017 (right) by two-sided Welch’s t-test. b RT-qPCR for the indicated YAP transcriptional targets in WT- and NUMB-KO MBO grown in 3D-Matrigel in the presence of vehicle (Veh) or 200 nM verteporfin (VP) for 48 hr. Graphs show the relative mean fold expression ± SEM from three independent experiments. p-values by FDR-adjusted two-sided one-sample t-test (vs. vehicle-treated MBO-WT): Ankrd1, MBO-WT VP, p = 0.00086 (***); MBO-KO Veh, p = 0.0083 (**); Ctgf, MBO-WT VP, p = 0.13 (ns, not significant); MBO-KO Veh, p = 0.041 (*); Cyr61, MBO-WT VP, p = 0.0017 (**); MBO-KO Veh, p = 0.0032 (**). p-values by two-sided Welch’s t-test (VP- vs. vehicle-treated MBO-KO samples): Ankrd1, p = 0.004 (**); Ctgf, p = 0.013 (*); Cyr61, p = 0.00011 (***). c GSEA enrichment plot of the EMT gene signature in RNA-seq data from WT- vs. NUMB-KO MBO cells (n = 6 for each condition) and NUMB-KO MBO treated with 200 nM VP vs. vehicle for 48 hr. (n = 1 for each condition). ES, Enrichment Score; NES, Normalized Enrichment Score; p, two-sided permutation test p-value. d Representative images of 3D-Matrigel WT- and NUMB-KO MBO grown in the presence of vehicle (Veh) or 25 nM verteporfin (VP) for 10 days. Bar, 100 μm. e Frequency of occurrence of normal and aberrant multiacinar morphology in MBO in ‘c’. MBO-WT+Veh, n = 219; MBO-WT + VP, n = 119; MBO-KO+Veh, n = 174; MBO-KO + VP, n = 189, obtained from three independent experiments. ****, p < 0.0001; ns, not significant (p = 0.35), relative to matching condition by FDR-adjusted pairwise two-sided Fisher’s t-test. f Analysis of morphometric parameters (Area, Circularity, Roughness, and Shape Complexity) of WT vs. NUMB-KO MBO treated as in ‘d’. Values are reported, in this and other relevant panels in the figure, as boxplots delimited by 25th and 75th percentiles and showing the median (horizontal line) and the mean (X). The whiskers span from the smallest and largest data values within a 1.5 interquartile range. MBO-WT+Veh, n = 219; MBO-WT + VP, n = 119; MBO-KO+Veh, n = 174; MBO-KO + VP, n = 189, obtained from three independent experiments. ****, p < 0.0001; *, p = 0.027; ns, not significant (p = 0.058 for Area, 0.15 for Circularity and 0.9 for Shape Complexity), relative to matching condition, by FDR-adjusted pairwise two-sided Welch’s t-test. Source data are provided as Source Data file.
A phenotypic inspection of the 3D-Matrigel cultures by brightfield microscopy revealed gross morphological differences between NUMB-KO vs. WT MBOs. WT-MBOs exhibited a typical rounded morphology and a smooth surface deprived of invasive protrusions (Fig. 4d). In contrast, Numb-KO MBOs appeared as rapidly growing, irregular, multilobular structures with numerous budding protrusions infiltrating the surrounding matrix, indicative of an invasive phenotype (Fig. 4d, e). To quantify these morphological differences, we assessed well-defined physical parameters which are informative of key biological processes that determine the final organoid morphology53–55 (see also Materials and Methods): i) overall surface area – cell growth and proliferation potential, ii) circularity/roundness – degree of differentiation/maturation and strength of cell-cell contacts, iii) roughness and shape complexity – cell invasion potential. From these analyses, we concluded that the absence of NUMB is associated with an aberrant morphogenetic program, characterized by hyperplasia (increased area), aberrant differentiation and loss of cell-to-cell cohesion (decreased circularity/roundness), as well as the emergence of an invasive phenotype (increased roughness/shape complexity) (Fig. 4d–f). Remarkably, VP strongly inhibited the hyperplastic and invasive phenotypes of NUMB-KO MBOs, resulting in smaller, more regularly shaped organoids, while having no effect on WT MBOs (Fig. 4d–f).
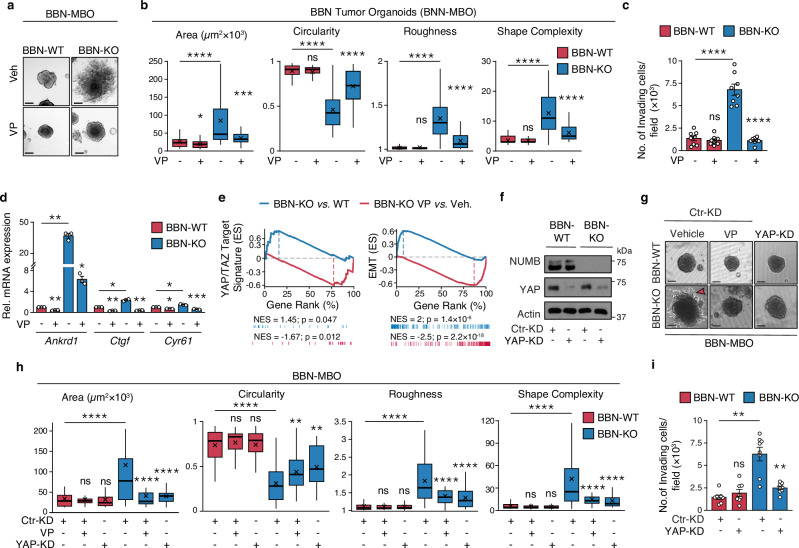
The molecular and phenotypic alterations observed in NUMB-KO vs. WT MBOs were fully recapitulated in clonally derived MBOs generated from BBN-induced tumors in WT and NUMB-KO mice. Indeed, BBN-NUMB-KO MBOs showed remarkably more aggressive tumor phenotypes compared to BBN-WT MBOs: i) a superior ability to grow and locally infiltrate the surrounding matrix with cellular protrusion and migratory cells outside their exterior borders (Fig. 5a, b); ii) a markedly increased propensity to invade and migrate through the extracellular matrix in the transwell Matrigel invasion assay (Fig. 5c). These aggressive phenotypes were reversed upon treatment with VP (Fig. 5a–c), pointing to their dependency on YAP transcriptional activity. This dependency was confirmed by a RT-qPCR analysis showing a strong inhibitory effect of VP on the transcription of the YAP targets genes, Ankrd1, Ctgf and Cyr61 in BBN-NUMB-KO MBOs, with no effect on BBN-WT MBOs (Fig. 5d).
Fig. 5. NUMB loss induces invasive phenotypes and YAP signaling activation in tumor urothelial cells.
a, b Morphology of 3D-Matrigel BBN-WT and BBN-KO tumor MBO exposed to 25 nM VP or vehicle for 7 days. Bar, 100 μm. Graphs in ‘b’ show the quantitation of the indicated morphometric parameters (see legend to Fig. 4f). BBN-WT+Veh, n = 47; BBN-WT + VP, n = 57; BBN-KO+Veh, n = 46; BBN-KO + VP, n = 45, obtained from three independent replicates. ****, p < 0.0001; ***, p = 0.0002; *, p = 0.023; ns, not significant (p = 0.42 for Circularity, 0.52 for Roughness and 0.87 for Shape Complexity), relative to matching condition by FDR-adjusted pairwise two-sided Welch’s t-test. c Transwell Matrigel invasion assay of BBN-WT and BBN-KO cells treated with VP (100 nM, 18 h) or vehicle. Number of invading cells/field expressed as the mean ± SEM of 8 microscope fields covering the entire migratory area, from two independent experiments. ****, p < 0.0001; not significant (ns), p = 0.54 by FDR-adjusted pairwise two-sided Welch’s t-test. d RT-qPCR for the indicated YAP transcriptional targets in BBN-WT vs. BBN-KO cells treated with VP (100 nM, 12 h) or vehicle. Graphs show the relative mean fold expression ± SEM from three independent experiments. p-values by FDR-adjusted two-sided one-sample t-test (vs. vehicle-treated BBN-WT): Ankrd1, BBN-WT VP, p = 0.0098 (**); BBN-KO vehicle, p = 0.0098 (**); Ctgf, BBN-WT VP, p = 0.0062 (**); BBN-KO vehicle, p = 0.011 (*); Cyr61, BBN-WT VP, p = 0.017 (*); BBN-KO vehicle, p = 0.017 (*). p-values by FDR-adjusted two-sided Welch’s t-test (VP- vs. vehicle-treated BBN-KO samples): Ankrd1, p = 0.01 (*); Ctgf, p = 0.005 (**); Cyr61, p = 0.00093 (***). e GSEA enrichment plot of the YAP (left) and EMT (right) gene signatures in RNA-seq data from BBN-KO vs. BBN-WT cells (n = 3) and BBN-KO cells treated with VP vs. vehicle (n = 1). ES, Enrichment Score; NES, Normalized Enrichment Score; p, two-sided permutation test p-value. f Efficiency of YAP silencing in BBN-WT and BBN-KO lentivirally silenced for YAP (YAP-KD) or control-silenced (Ctr-KD). Actin, loading control. Results are representative of two independent experiments. g Morphology of 3D-Matrigel BBN-MBO from cells in ‘f’. Ctr-KD BBN-WT and BBN-KO MBO were also treated with vehicle or 25 nM verteporfin (VP). Bar, 100 μm. The red arrowhead indicates invading protrusions. h Quantification of morphometric parameters of BBN-MBO in ‘g’ (see legend to Fig. 4f). BBN-WT Ctr-KD+Veh, n = 48; BBN-WT Ctr-KD + VP, n = 40; BBN-WT YAP-KD+Veh, n = 42; BBN-KO Ctr-KD+Veh, n = 59; BBN-KO Ctr-KD + VP, n = 35; BBN-KO YAP-KD+Veh, n = 33, obtained from two independent experiments. ****, p < 0.0001; **, p = 0.008 for BBN-KO Ctr-KD + VP, p = 0.001 for BBN-KO YAP-KD+Veh; not significant (ns) p-values are: for BBN-WT Ctr-KD + VP, p = 0.83 (Area), 0.47 (Circularity), 0.77 (Roughness) and 0.59 (Shape Complexity); for BBN-WT YAP-KD+Veh, p = 0.83 (Area), 0.89 (Circularity), 0.67 (Roughness) and 0.16 (Shape Complexity), vs. matching condition, by FDR-adjusted pairwise two-sided Welch’s t-test. i Transwell Matrigel invasion assay of BBN-WT and BBN-KO cells in ’f’. Number of invading cells/field 24 h after seeding, expressed as the mean ± SEM of 8 fields from two independent experiments. **, p = 0.001 for BBN-KO Ctr-KD cells, p = 0.003 for BBN-KO YAP-KD cells; ns, not significant (p = 0.31), vs. matching controls by FDR-adjusted pairwise two-sided Welch’s t-test.
These findings were mirrored in the global transcriptomic profiling analysis of BBN-NUMB-KO vs. BBN-WT MBOs by GSEA, which evidenced a significant enrichment of a YAP transcriptional target signature39 and an EMT signature (from MSigDB “Hallmarks”), both sensitive to VP treatment, in the NUMB-KO condition (Fig. 5e, Supplementary Table 10; see also Supplementary Data 6 for a complete list of differentially regulated genes). Note that the molecular alterations observed in the BBN-tumor model are in line with the results obtained in human NMIBC cell lines silenced for NUMB (see Fig. 2c–e), indicating that NUMB loss, via YAP hyperactivation, drives the acquisition of plasticity/EMT traits in BCa cells, which likely underlies their aggressive, invasive phenotypes.
To rule out possible off-target effects of VP, we checked whether genetic silencing of YAP phenocopies the inhibitory effects of VP treatment on the aggressive morphological traits of BBN-NUMB-KO tumor MBOs. Stable silencing of the Yap gene, using a lentiviral shRNA vector, inhibited the hyperproliferative and invasive/migratory phenotype of BBN-NUMB-KO MBOs, similarly to VP, but had no effect on BBN-WT MBOs (Fig. 5f–i).
To provide definitive evidence of the dependency of the molecular and functional phenotypes of BBN-NUMB-KO MBOs on NUMB loss, we restored NUMB expression in MBO-derived cells using a lentiviral NUMB-GFP vector. Ectopic NUMB expression inhibited YAP nuclear translocation, with evident YAP cytoplasmic retention (Supplementary Fig. 7a, b), and impaired YAP transcriptional activity (Supplementary Fig. 7c). These molecular changes were accompanied by a significant reversion of the aberrant morphological traits of BBN-NUMB-KO MBOs, resulting in BBN-WT-like morphological structures (Supplementary Fig. 7d).
Together, these data demonstrate that, through hyperactivation of YAP signaling, loss of NUMB, not only drives an aberrant morphological program in the normal mouse urothelium conducive to its overt malignant transformation, but also worsens the biological aggressiveness of already transformed BCa cells, enhancing their proliferative and invasive/migratory potential likely through YAP-dependent activation of an EMT program.
YAP nuclear localization and transcriptional activity is negatively controlled by the upstream activation of the Hippo cascade44,56,57. Core components of this pathway are the serine/threonine kinases, MST1 and LATS, whose sequential activation leads to the phosphorylation and subsequent cytoplasmic sequestration of YAP, ultimately repressing its nuclear translocation and transcriptional activity44,56,57. We therefore hypothesized that Hippo signaling could be repressed in NUMB-deficient BCa cells. By immunoblotting of cellular lysates from BBN-NUMB-KO vs. BBN-WT MBOs, we observed markedly reduced levels of the phosphorylated active forms of MST1 and LATS in BBN-NUMB-KO cells compared with BBN-WT cells, despite having similar levels of total protein (Supplementary Fig. 7e). This reduction in phosphorylated Hippo pathway kinases is in line with the evident reduction in inactive phosphorylated YAP in BBN-NUMB-KO vs. BBN-WT cells (Supplementary Fig. 7e). These results demonstrate that hyperactivation of the YAP pathway subsequent to NUMB loss is mechanistically linked to the downregulation of the upstream canonical Hippo signaling cascade.
YAP hyperactivation downstream of Numb loss is dependent on RHOA/ROCK signaling
The Hippo-YAP signaling pathway operates as a nexus that, in response to microenvironmental cues, controls multiple cellular and context-specific responses essential for tissue homeostasis, including proliferation, differentiation, cell plasticity and stemness56,58. A wide range of architectural and mechanical cues, transmitted through cell-cell junctions and cell-matrix adhesions, as well as multiple extracellular ligands/growth factors and downstream signaling pathways, can control this pathway through complex canonical and non-canonical arms57,59,60. In this context, both upstream and downstream events linked to actin dynamics and cytoskeletal organization, play a pivotal role in the regulation of Hippo-YAP signaling56–58,61. This is clearly demonstrated by the ability of the F-actin disrupting agent, latrunculin, to impede YAP activation in the context of both the canonical and non-canonical arms of Hippo-YAP signaling regulation59,62,63.
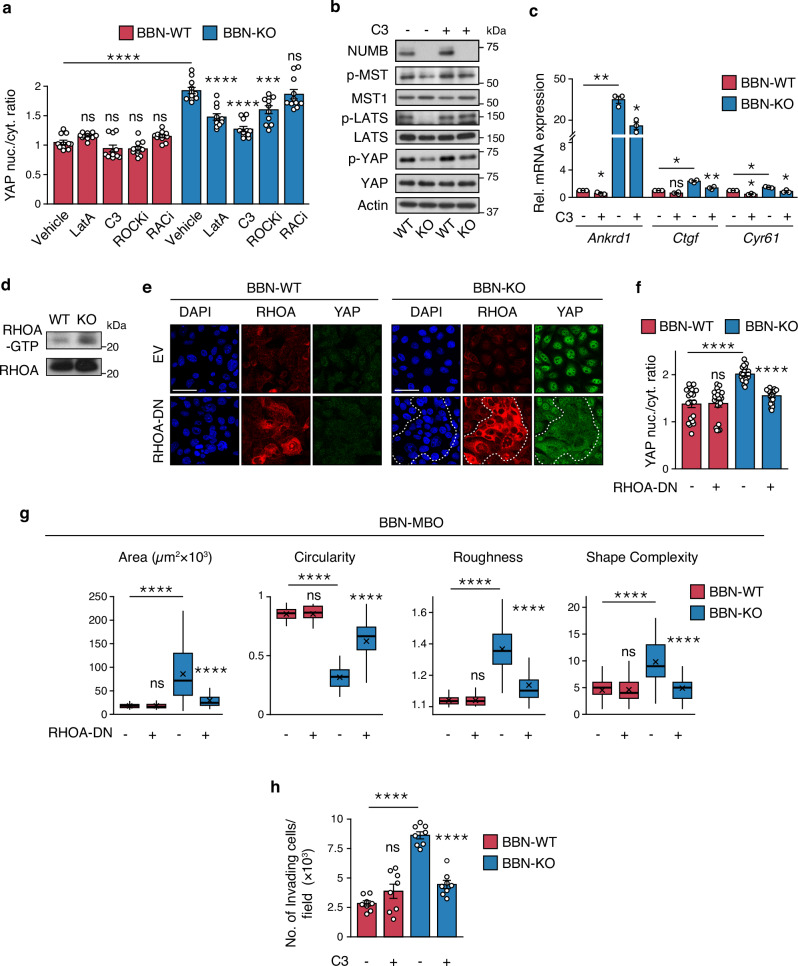
To gain a deeper understanding of the mechanism driving YAP hyperactivation following NUMB loss, we performed IF-based phenotypic screening of inhibitors of key regulators of actin dynamics and cytoskeleton organization in the BBN-NUMB-KO vs. WT MBO models. In particular, we focused on the small GTPases, RHOA and RAC1, guided by previous evidence indicating that loss of NUMB-mediated regulation of their activity results in alterations in actin dynamics and induces different cell motility phenotypes64–66. YAP intranuclear translocation vs. cytoplasmic retention was monitored in BBN-NUMB-KO vs. BBN-WT cells treated in 2D-culture conditions with inhibitors of RHOA (bacterial exoenzyme C3 transferase), RAC1 (RACi, NSC23766), RHO-associated protein kinase (ROCKi, Y-27632), and actin polymerization (latrunculin A, LatA)59. While RAC1 inhibition had no effect on YAP nuclear translocation in BBN-NUMB-KO cells, the inhibition of RHOA and its downstream effector ROCK reduced nuclear YAP levels similarly to LatA (Fig. 6a and Supplementary Fig. 8). In contrast, no significant effects were observed in BBN-WT cells (Fig. 6a and Supplementary Fig. 8). These results point to the involvement of the RHOA/ROCK actin regulatory pathway in the control of YAP signaling by NUMB.
Fig. 6. RHOA/ROCK-dependent actin remodeling is involved in YAP hyperactivation downstream of NUMB loss.
a Quantification of YAP nuclear/cytoplasmic ratio in BBN-WT and BBN-KO tumor cells treated with LatA (500 nM, 6 h), C3 transferase (3 µg/ml, 6 h), Y-27632 (ROCKi, 10 µM, 12 h), NSC-23766 (RACi, 10 µM, 12 h) or vehicle and co-stained for YAP, NUMB and DAPI. Graphs show the mean/field ± SEM, n = 11 fields/condition, from two independent experiments. ****, p < 0.0001; ***, p = 0.00064; not significant (ns) p-values are: p = 0.89, 0.91, 0.88 and 0.92, for BBN-WT cells treated with LatA, C3, ROCKi and RACi, respectively; p = 1 for BBN-KO cells treated with ROCKi, relative to matching controls by two-sided Tukey’s HSD test. Representative confocal images are shown in Supplementary Fig. 8. b Expression and phosphorylation status of the indicated Hippo pathway components in BBN-WT vs. BBN-KO cells, treated with C3 transferase (3 µg/ml, 6 h) or vehicle. Actin, loading control. Blots shown are representative of two independent experiments. c RT-qPCR for the indicated YAP transcriptional targets in BBN-WT vs. BBN-KO cells treated as in ‘b’. Graphs show the relative mean fold expression ± SEM from three independent experiments. p-values by FDR-adjusted two-sided one-sample t-test (vs. vehicle-treated BBN-WT): Ankrd1, BBN-WT C3, p = 0.043 (*); BBN-KO Veh, p = 0.0098 (**); Ctgf, BBN-WT C3, p = 0.068 (ns, not significant); BBN-KO Veh, p = 0.012 (*); Cyr61, BBN-WT C3; p = 0.017 (*); BBN-KO Veh, p = 0.017 (*). p-values by FDR-adjusted two-sided Welch’s t-test (C3- vs. vehicle-treated BBN-KO samples): Ankrd1, p = 0.01 (*); Ctgf, p = 0.006 (**); Cyr61, p = 0.027 (*). d Pull-down assay of activated RHOA in BBN-WT vs. BBN-KO cells. Blots are representative of two independent experiments. e Representative confocal fluorescence images of BBN-WT and BBN-KO tumor cells lentivirally transduced with RHOA-DN or empty vector (EV) and co-stained for RHOA, YAP and DAPI. Bars, 50 µm. Dashed line delineates a cluster of RHOA-DN overexpressing cells in BBN-KO cells. f Quantification of YAP nuclear/cytoplasmic ratio in BBN-WT- and BBN-KO cells treated as in ‘e’. Graphs show the mean/field ± SEM, n = 20 fields/condition, from two independent experiments. ****, p < 0.0001; ns, not significant (p = 1), relative to matching controls by two-sided Tukey’s HSD test. g Morphometric analysis of tumor BBN-MBO generated from cells described in ‘e’ (see legend to Fig. 4f). BBN-WT EV, n = 48; BBN-WT RHOA-DN, n = 48; BBN-KO EV, n = 37; BBN-KO RHOA-DN, n = 45, obtained from three independent experiments. ****, p < 0.0001; not significant (ns) p-values are: p = 1, 0.97, 0.91 and 0.82 for Area, Circularity, Roughness and Shape Complexity, respectively, relative to matching condition by FDR-adjusted pairwise two-sided Welch’s t-test. h Transwell Matrigel invasion assay of BBN-WT and BBN-KO cells treated with C3 transferase (3 µg/mL, 18 h) or vehicle. Number of invading cells/field expressed as the mean ± SEM of 8 microscope fields covering the migration area, from two independent experiments. ****, p < 0.0001; ns, not significant (p = 0.17), relative to matching controls by FDR-adjusted pairwise two-sided Welch’s t-test. Source data are provided as Source Data file.
Further investigation of the role of RHOA revealed that its inhibition by C3 transferase induced reactivation of the YAP-inhibitory Hippo pathway selectively in BBN-NUMB-KO vs. BBN-WT cells, as evidenced by the increased phosphorylation of MST1, LATS and YAP (Fig. 6b). In addition, C3 transferase reduced the expression of YAP transcriptional targets (Fig. 6c). Thus, it appears that active RHOA is required for YAP hyperactivation in the NUMB-KO condition. In line with this idea, we showed that BBN-NUMB-KO cells display higher levels of the active GTP-bound form of RHOA compared with BBN-WT cells (Fig. 6d). Moreover, expression of a dominant-negative RHOA mutant (T19N-RHOA) in BBN-NUMB-KO tumor cells inhibited YAP nuclear translocation (Fig. 6e, f) and reversed the phenotypes associated with biological aggressiveness, as witnessed in the morphological analysis of MBOs (Fig. 6g). In contrast, no effects of dominant-negative RHOA were observed in BBN-WT tumor cells (Fig. 6e–g). RHOA inhibition with C3 transferase also reduced the invasive/migratory potential of BBN-NUMB-KO tumor cells in the transwell Matrigel invasion assay, to levels similar to BBN-WT cells (Fig. 6h). Notably, none of the above phenotypes were affected by pharmacological (RACi, NSC-23766 inhibitor) or genetic (RAC1-T17N dominant-negative mutant) inhibition of RAC1 activity (Supplementary Fig. 9a–e).
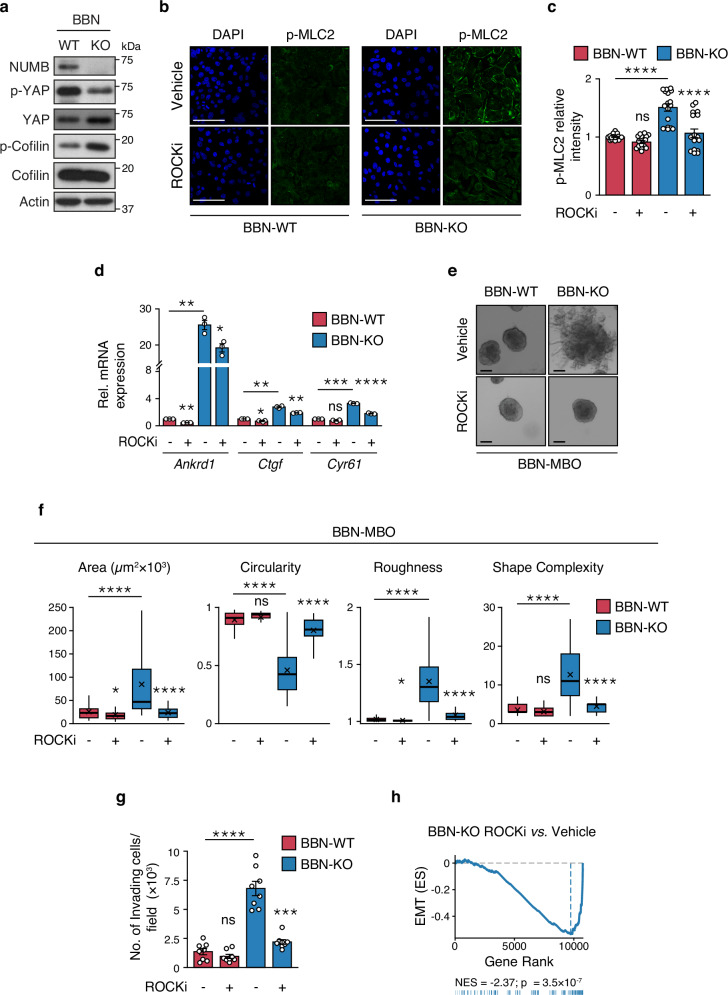
Next, we examined in more detail the involvement of the RHOA downstream effector, ROCK, which signals to the actin cytoskeleton and contractile machinery by regulating the phosphorylation status of two proteins: cofilin, a protein that in its phosphorylated form becomes inactive and loses its function in actin filament disassembly67; myosin light chain 2 (MLC2), a direct phosphorylation target of ROCK involved in the regulation of actomyosin contractility and stress fiber assembly/contraction68,69. We found increased levels of phosphorylated cofilin and MLC2 in BBN-NUMB-KO vs. BBN-WT cells, indicating that loss of NUMB is associated with higher basal ROCK activity (Fig. 7a–c). Using the ROCK inhibitor (ROCKi, Y-27632), we verified that the high levels of phosphorylated MLC2 (p-MLC2) in BBN-NUMB-KO cells were dependent on ROCK activity (Fig. 7b, c).
Fig. 7. ROCK inhibition prevents YAP hyperactivation induced by loss of NUMB.
a Immunoblot of total and phosphorylated cofilin and YAP in BBN-WT vs. BBN-KO cells. Actin, loading control. Data are representative of two independent experiments. b Representative confocal images of BBN-WT and BBN-KO cells treated with vehicle or ROCKi, Y-27632 (10 µM, 12 h) and co-stained for phosphorylated MLC2 (p-MLC2) and DAPI. Bar, 50 µm. c Quantification of the experiment in ‘b’. Graphs show the relative intensity of p-MLC2 in BBN-KO vs. BBN-WT cells expressed as the mean/field ± SEM, n = 18 random fields/condition from two independent experiments. ****, p < 0.0001; not significant (ns), p = 0.59, relative to matching controls by two-sided Tukey’s HSD test. d RT-qPCR of the indicated YAP transcriptional targets in BBN-KO vs. BBN-WT cells treated with ROCKi Y-27632 (10 µM, 8 h) or vehicle. Graphs show the relative mean fold expression ± SEM from three independent experiments. p-values by FDR-adjusted two-sided one-sample t-test (vs. vehicle-treated BBN-WT): Ankrd1, BBN-WT ROCKi, p = 0.0013 (**); BBN-KO Veh, p = 0.0038 (**); Ctgf, BBN-WT ROCKi, p = 0.041 (*); BBN-KO Veh, p = 0.0047 (**); Cyr61, BBN-WT ROCKi, ns, not significant (p = 0.081); BBN-KO Veh, p = 0.00079 (***). In ROCKi- vs. vehicle-treated BBN-KO samples, p-values by FDR-adjusted two-sided Welch’s t-test are: Ankrd1, p = 0.022 (*); Ctgf, p = 0.0089 (**); Cyr61, p = 0.000026 (****). e Representative images of tumor BBN-MBOs generated from BBN-WT and BBN-KO cells treated with ROCKi (Y-27632, 10 µM). Bars, 100 µm. f Morphometric analysis of the indicated parameters in BBN-MBO cell treated as in ‘f’ (see legend to Fig. 4f); BBN-WT+Vehicle, n = 47; BBN-WT+ROCKi, n = 65; BBN-KO+Vehicle, n = 46; BBN-KO+ROCKi, n = 58, obtained from three independent experiments. ****, p < 0.0001; *, p = 0.023, 0.024 for Area and Roughness measures, respectively; not significant (ns), p = 0.07, 0.074 for Circularity and Shape Complexity measures, respectively, relative to matching condition by FDR-adjusted pairwise two-sided Welch’s t-test. g Transwell Matrigel invasion assay of BBN-WT and BBN-KO cells treated with ROCKi Y-27632 (10 µM, 18 h) or vehicle. Number of invading cells/field expressed as the mean ± SEM of 8 random microscope fields covering the entire migration area, from two independent experiments. ****, p < 0.0001; ***, p = 0.00023; not significant (ns), p = 0.33, relative to matching controls by FDR-adjusted pairwise two-sided Welch’s t-test. h GSEA enrichment plot of the EMT gene signature in BBN-KO vs. WT cells and BBN-KO cells treated with ROCKi 10 μM vs. vehicle. n = 2. ES, Enrichment Score; NES, Normalized Enrichment Score; p, two-sided permutation test p-value. Source data are provided as Source Data file.
Finally, we demonstrated that ROCK has a key role in YAP signaling regulation downstream of NUMB, as evidenced by the reduction of YAP transcriptional activity in BBN-NUMB-KO MBOs treated with Y-27632 (Fig. 7d). In addition, ROCK inhibition reversed the aggressive phenotypes of BBN-NUMB-KO tumor cells, as witnessed in the analysis of morphological parameters measuring MBO growth and invasive/migratory potential (Fig. 7e–g). Most remarkably, the GSEA analysis of the transcriptomic profiles of BBN-NUMB-KO cells treated with Y-27632 showed that, like YAP inhibition with VP (see Fig. 5e), ROCK inhibition also results in EMT downregulation (Fig. 7h).
Together, these results point to the selective dependency of the aggressive invasive/migratory phenotype of BBN-NUMB-KO tumor cells on the aberrant activation of a RHOA/ROCK axis signaling to the actin cytoskeleton. These results also highlight the potential of ROCK as an actionable target to revert aberrant YAP and EMT activation, which likely underlie the biological aggressiveness of NUMB-deficient BCa.
Having shown in the BBN BCa model that RHOA/ROCK hyperactivation is responsible for the molecular alterations (YAP and EMT activation) linked to NUMB loss in BCa cells, leading to exacerbation of their aggressive phenotypes, we asked whether this circuitry could also account for the biological and phenotypic differences observed in NUMB-KO vs. WT MBOs derived from BBN-treatment naïve mice (characterized in Fig. 4). Supporting this hypothesis, treatment with Y-27632 reversed the aberrant morphology of NUMB-KO MBOs (Supplementary Fig. 9f–h) and markedly decreased YAP transcriptional activity (Supplementary Fig. 9i), while have no significant effects on WT MBOs. Y-27632 also prevented the enrichment of an EMT signature, as shown by GSEA analysis of the transcriptomes of Y-27632- vs. control-treated NUMB-KO MBOs (Fig. 7h). Therefore, ROCK inhibition by Y-27632 recapitulates the effects observed in NUMB-KO vs. WT MBO treated with the YAP inhibitor, VP (see Fig. 4b–d, f).
Dysregulation of RHOA/ROCK/YAP signaling underlies the invasive phenotype of NUMB-deficient human BCa cells
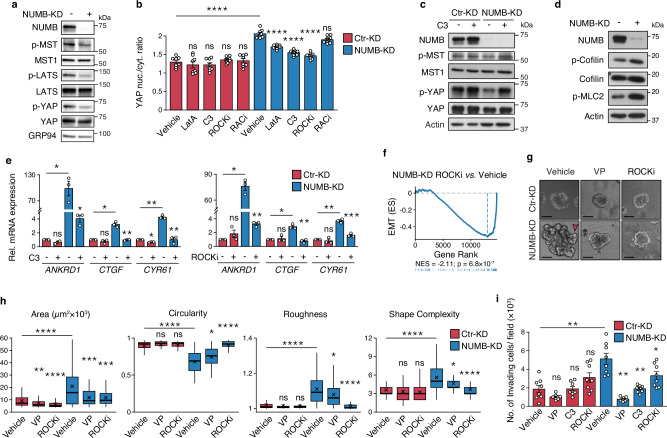
To investigate the relevance of RHOA/ROCK/YAP signaling to NUMB-deficient human BCa, we initially employed the human cell line, RT4, which represents a model of low aggressive, well-differentiated non-invasive luminal BCa of male origin70–72. The comparison of NUMB-KD vs. CTR-KD RT4 BCa cells confirmed the results obtained in the BBN-NUMB-KO vs. BBN-WT mouse tumor model, showing that the absence of NUMB leads to YAP hyperactivation through the downregulation of the canonical Hippo pathway, witnessed by the reduced phosphorylation levels of MST1, LATS, and YAP in NUMB-KD vs. CTR-KD RT4 cells (Fig. 8a). Multiple lines of evidence support the role of RHOA/ROCK signaling in mediating YAP hyperactivation in NUMB-deficient RT4 cells. Indeed, pharmacological inhibition of RHOA and ROCK, but not RAC1, reduced the nuclear/cytoplasmic YAP ratio selectively in NUMB-KD RT4 cells, mimicking the effect of the actin polymerization inhibitor, LatA (Fig. 8b and Supplementary Fig. 10a). This increase in cytoplasmic retention of YAP following RHOA and ROCK inhibition was consistent with evidence showing that: i) RHOA inhibition with C3 transferase induces increased levels of inactive phosphorylated YAP selectively in NUMB-KD RT4 cells, apparently linked to the restoration of the Hippo pathway as indicated by increased phospho-MST1/2 levels (Fig. 8c); ii) NUMB-KD vs. CTR-KD RT4 cells display higher constitutive levels of activated ROCK, as witnessed by higher levels of the phosphorylated forms of its downstream effectors, cofilin and MLC2 (Fig. 8d and Supplementary Fig. 10b, c); iii) inhibition of RHOA (C3 transferase) and ROCK (Y-27632) strongly inhibited the transcription of the YAP target genes, ANKRD1, CTGF and CYR6 (Fig. 8e) and EMT activation (Fig. 8f), similarly to the YAP transcriptional inhibitor, VP (Supplementary Fig. 10d; see also Fig. 2e for YAP and EMT pathway inhibition by VP in NUMB-KD RT4 cells). Therefore, it appears that the same molecular circuitry involving RHOA/ROCK, actin remodeling and Hippo signaling is required for the upregulation of YAP and EMT following NUMB loss in both mouse and human BCa cells.
Fig. 8. Loss of NUMB triggers RHOA/ROCK-dependent YAP hyperactivation in human RT4 BCa cells.
a Expression of NUMB and the indicated Hippo pathway components in control- (Ctr-KD) and NUMB-silenced (NUMB-KD) RT4 cells. GRP94, loading control. Data are representative of two independent experiments. b Quantification of YAP nuclear/cytoplasmic ratio in Ctr-KD and NUMB-KD RT4 cells treated with LatA (500 nM, 6 h), C3 transferase (3 µg/ml, 6 h), Y-27632 (ROCKi, 10 µM, 12 h), NSC-23766 (RACi, 10 µM, 12 h) or vehicle and co-stained for endogenous YAP and DAPI. Representative confocal images are shown in Supplementary Fig. 10a. Data are shown as the mean/field ± SEM, n = 8 fields/condition, from two independent experiments. ****, p < 0.0001; not significant (ns) p-values are: p = 0.98, 0.98, 1 and 1, for RT4 Ctr-KD cells treated with LatA, C3, ROCKi and RACi, respectively, p = 0.33 for RT4 NUMB-KD cells treated with RACi, relative to matching controls by two-sided Tukey’s HSD test. c Expression of NUMB, and total and phosphorylated YAP and MST1 in Ctr-KD vs. NUMB-KD RT4 cells treated with vehicle or C3 transferase (3 μg/mL, 6 h). Actin, loading control. Data are representative of two independent experiments. d Immunoblot of NUMB, total and phosphorylated cofilin, and phosphorylated MLC2 (p-MLC2), in Ctr-KD vs. NUMB-KD RT4 cells. Actin, loading control. Blots are representative of two independent experiments. e RT-qPCR of the indicated YAP transcriptional targets in NUMB-KD vs. Ctr-KD RT4 cells treated with C3 (3 µg/ml, 6 h) (left) or ROCKi Y-27632 (50 µM, 8 h) (right), or vehicle. Graphs show the relative mean fold expression ± SEM from three independent experiments. p-values by FDR-adjusted two-sided one-sample t-test (vs. vehicle-treated RT4 Ctr-KD): ANKRD1, Ctr-KD C3, not significant (ns, p = 0.076); NUMB-KD Veh, p = 0.042 (*); CTGF, Ctr-KD C3, ns (p = 0.36); NUMB-KD Veh, p = 0.03 (*); CYR61, Ctr-KD C3, p = 0.011 (*); NUMB-KD Veh, p = 0.007 (**) (left); ANKRD1, Ctr-KD ROCKi, ns (p = 0.263); NUMB-KD Veh, p = 0.013 (*); CTGF, Ctr-KD ROCKi, ns, (p = 0.7); NUMB-KD Veh, p = 0.034 (*); CYR61, Ctr-KD ROCKi, ns (p = 0.52); NUMB-KD Veh, p = 0.009 (**) (right). p-values by FDR-adjusted two-sided Welch’s t-test: C3- vs. vehicle-treated NUMB-KD (left), ANKRD1, p = 0.03 (*); CTGF, p = 0.0072 (**); CYR61, p = 0.0022 (**); ROCKi- vs. vehicle-treated NUMB-KD (right): ANKRD1, p = 0.0045 (**); CTGF, p = 0.0016 (**); CYR61, p = 0.00057 (***). f Enrichment of the EMT gene signature by GSEA in NUMB-KD RT4 cells treated with ROCKi vs. vehicle as in ‘e’. n = 1. ES, Enrichment Score; NES, Normalized Enrichment Score; p, two-sided permutation test p-value. g Morphology of organoids from stable Ctr-KD vs. NUMB-KD RT4 cells treated with vehicle (Veh), verteporfin (VP, 25 nM) or ROCKi Y-27632 (10 µM). The red arrow points to invasive protrusions in NUMB-KD RT4 cells. Bar, 100 µm. h Morphometric analysis of the experiment in ‘g’ (see legend to Fig. 4f). Ctr-KD+Vehicle, n = 122; Ctr-KD + VP, n = 77; Ctr-KD+ROCKi, n = 59; NUMB-KD+Vehicle, n = 68; NUMB + VP, n = 54; NUMB+ROCKi, n = 50, obtained from two independent experiments. ****, p < 0.0001; ***, p = 0.00032; **, p = 0.006; *, p = 0.024 (Circularity), 0.034 (Roughness), 0.034 (Shape Complexity); not significant (ns) p-values are: p = 0.2, 0.37 and 0.19, for Circularity, Roughness and Shape Complexity, respectively, for RT4 Ctr-KD cells treated with VP; p = 0.37, 0.17 and 0.12, for Circularity, Roughness and Shape Complexity, respectively, for RT4 Ctr-KD cells treated with ROCKi, relative to matching condition by FDR-adjusted pairwise two-sided Welch’s t-test. i Transwell Matrigel invasion assay of Ctr-KD and NUMB-KD RT4 cells treated with vehicle, VP (100 nM), C3 (3 µg/mL) and ROCKi Y-27632 (10 µM) for 48 h. Number of invading cells/field expressed as the mean ± SEM of 8 microscope fields from two independent experiments. **, p = 0.003, 0.002 and 0.003, for RT4 NUMB-KD cells treated with vehicle, VP and C3, respectively; *, p = 0.046; not significant (ns) p-values are: p = 0.091, 0.99 and 0.091, for RT4 Ctr-KD cells treated with VP, C3 and ROCKi, respectively, vs. matching condition by FDR-adjusted pairwise two-sided Welch’s t-test. Source data are provided as Source Data file.
Next, we verified whether this mechanism could be relevant to biological phenotypes induced by NUMB loss in human BCa, analyzing the effects of the different pathway inhibitors on the morphology and invasion/migration potential of 3D-Matrigel organoids generated from NUMB-KD vs. CTR-KD RT4 cells. Recapitulating the findings in the BBN tumor model, we found that the absence of NUMB exacerbates the biological aggressiveness of RT4 cells, as demonstrated by the increased organoid size (area), propensity to form invading protrusions (Fig. 8g, h), and invasive/migratory potential in the transwell Matrigel invasion assay of NUMB-KD RT4 cells compared to CTR-KD RT4 cells (Fig. 8i). These phenotypes were selectively reversed in NUMB-KD RT4 cells by pharmacological inhibition of YAP (VP) and RHOA/ROCK (C3 transferase/Y-27632) (Fig. 8g–i).
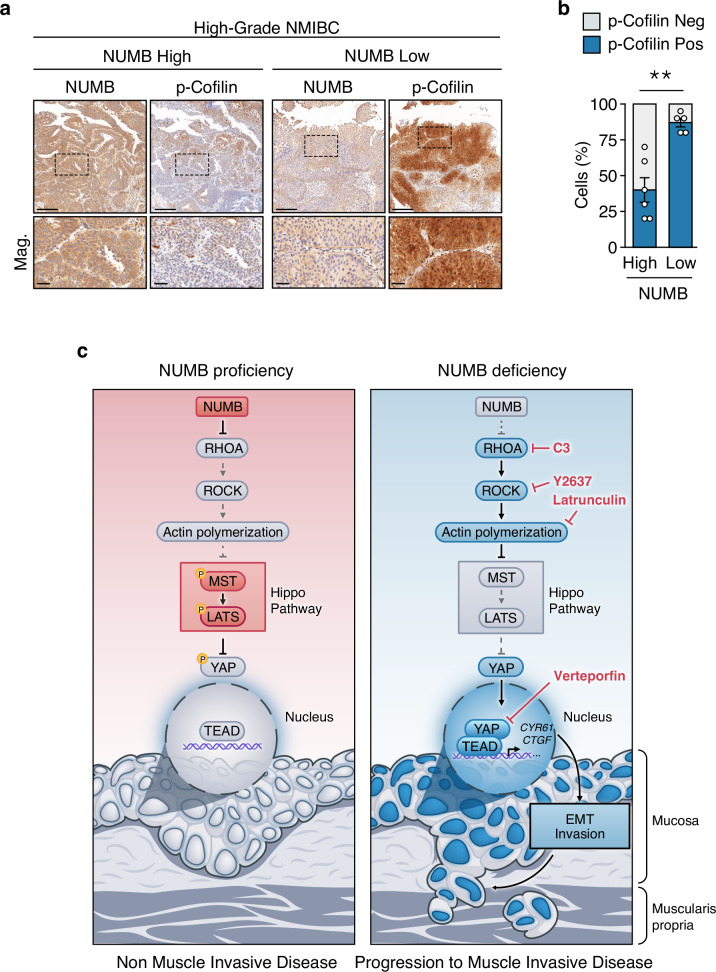
This set of molecular and functional findings obtained in the male RT4 BCa cell line was entirely recapitulated in the RT112 cell line originating from a low-aggressive, non-invasive human BCa of female origin70–72 (Supplementary Fig. 11a–h). Together, these data, obtained independently in a male (RT4) and a female (RT112) model of NUMBHigh low-aggressive human NMIBC further support the notion that NUMB loss drives the acquisition of biologically aggressive BCa phenotypes, establishing its relevance to the human BCa disease. Mechanistically, these data are further evidence that the aggressive biology of NUMB-deficient BCa cells depends on the aberrant activation of RHOA/ROCK signaling to the actin cytoskeleton, downstream of NUMB loss, which leads to suppression of the YAP-inhibitory Hippo pathway and consequently YAP hyperactivation. Remarkably, this molecular circuitry, functionally validated in the above human NMIBC cell lines, appears to be relevant to NMIBC patients, since we detected significantly higher levels of phosphorylated cofilin, indicative of increased ROCK activity, in NUMBLow NMIBC TUR samples compared with NUMBHigh samples (Fig. 9a, b). This finding is also in line with the increased nuclear accumulation and transcriptional activity of YAP observed in NMIBC patients with deficient NUMB status (see Fig. 2h).
Fig. 9. Loss of NUMB expression correlates with increased p-cofilin levels in NMIBC patients.
a Representative IHC staining of phosphorylated cofilin (p-Cofilin) and NUMB in NUMBHigh and NUMBLow high-grade NMIBC TUR specimens. Magnifications (Mag.) of the boxed areas are shown in the lower panels. Bars, 500 µm; Mag, 100 µm. b Quantification of the % of p-cofilin positive cells in 6 NUMBHigh vs. 5 NUMBLow high-grade NMIBC TUR specimens. Values are expressed as mean ± SEM. **, p = 0.0019 by two-sided Welch’s t-test. c Schematic representation of the molecular events influencing the Hippo/YAP pathway activation state in the bladder urothelium in NUMB-proficient or -deficient conditions. In NUMB-proficient conditions (NUMB proficiency), the presence of NUMB keeps in check RHOA/ROCK signaling to the actin machinery, leading to activation of the Hippo pathway, with ensuing YAP phosphorylation, cytoplasmic retention and transcriptional inactivation. In NUMB-deficient conditions (NUMB deficiency), the absence of NUMB leads to activation of RHOA/ROCK signaling to the actin machinery, which in turn suppresses the Hippo pathway, resulting in nuclear translocation of unphosphorylated YAP and ensuing transcription of its target genes via TEAD interaction. Through its transcriptional targets, CYR61 and CTGF, YAP induces EMT, which likely underlies the acquisition of mesenchymal/invasive traits responsible for the biological aggressiveness of NUMB-deficient BCa. Source data are provided as Source Data file.
Together, these findings point to the RHOA/ROCK/YAP signaling pathway as a potential actionable vulnerability for therapeutic intervention in NUMB-deficient NIMBC patients. The clinical relevance of this point is supported by our findings in the BBN tumor model and human BCa cell lines, which establish a previously uncharacterized mechanistic link between NUMB loss and EMT activation, dependent on the upregulation of the RHOA/ROCK/YAP signaling pathway. Indeed, the acquisition of mesenchymal/plasticity traits in tumor cells likely represents the main mechanism underlying local invasion and metastasis driving disease progression73,74, a scenario that is likely germane to the unique muscle-invasive propensity of NUMB-deficient NMIBC.
To gain a deeper insight into the possible mechanism underlying YAP-directed activation of EMT downstream of NUMB loss, we focused our attention on two YAP transcriptional targets, CYR61 and CTGF, which emerged in a list of upregulated genes in common between the 22-gene YAP signature and the EMT activation signature (Supplementary Fig. 12a), and have been previously mechanistically linked to EMT activation75,76. Simultaneous ablation of these two genes in BBN-NUMB-KO and BBN-WT MBO cells (Supplementary Fig. 12b) resulted in a marked transcriptional downregulation of EMT in BBN-NUMB-KO cells, as attested by GSEA of Cyr61/Ctgf-KD vs. Ctr-KD BBN-NUMB-KO cells (Supplementary Fig. 12c). These transcriptional changes were associated with profound inhibition of their invasive/migratory phenotype (Supplementary Fig. 12d). Similar results were obtained upon combined silencing of CYR61 and CTGF in NUMB-KD vs. CTR-KD RT4 cells (Supplementary Fig. 12e–g). Together, these results directly connect NUMB-loss driven induction of EMT and acquisition of invasive/migratory traits with YAP transcriptional activity, pointing to CYR61 and CTGF as key players in this process.
Discussion
NUMB is an evolutionarily conserved cell fate determinant and endocytic adaptor protein19, whose expression levels are variably altered in different types of human cancer, with both overexpression and underexpression being reported to have tumor type-specific roles in tumorigenesis and aggressive disease course. However, the best characterized function of NUMB remains that of a tumor suppressor whose loss has prognostic and pathogenetic relevance in various cancers, such as breast, prostate, brain, lung, and colon cancer16,21–23,45,66,77–79.
In this study, using a range of experimental systems, including established human BCa cell lines, in vivo mouse models and 3D-Matrigel organoid cultures, integrated with the analysis of BCa patient cohorts, we have established that NUMB behaves as a potent tumor suppressor in bladder tumorigenesis and its loss is a hallmark of a clinically aggressive BCa disease. This association with aggressive disease was evident from retrospective studies of longitudinal BCa patient cohorts, where a deficient NUMB status predicts worse overall survival in post-cystectomy MIBC patients and increased risk of muscle-invasion progression in NMIBC patients. Moreover, using genetically engineered mouse models, we uncovered that NUMB loss is causal in bladder tumorigenesis. Indeed, transgenic deletion of the Numb gene in the CK5-expressing basal layer of the mouse urothelium was alone sufficient to induce the formation of preneoplastic lesions, non-invasive tumors and muscle-invasive tumors. This finding aligns with the emerging view that intrinsically aggressive BCa with a propensity for muscle invasion originates from basal, rather than suprabasal, cells34,47,48,80. The development of a genetically engineered mouse model recapitulating the non-invasive to invasive BCa transition holds particular significance in the field of BCa, considering the relative paucity of in vivo models amenable to study the underlying biology of NMIBC to MIBC progression, which still remains largely unexplored81. Related to this point, evidence of a progressive range of lesions in our NUMB-KO mouse model supports the idea that NMIBC and MIBC represent different stages along a disease continuum. This finding is in keeping with recent experimental studies in mice34 and with evidence of a high degree of similarity between genetic alterations in NMIBC and MIBC patients14, which have challenged the historical view of NMIBC and MIBC as ab initio distinct pathobiological entities15.
In addition to NUMB loss-of-function being alone sufficient to drive spontaneous tumorigenesis, it can also cooperate with other oncogenic insults to accelerate tumor progression and fatal outcome, as demonstrated in NUMB-KO mice exposed to the chemical carcinogen, BBN. This compound induces bladder tumorigenesis in mice, mirroring the progression of naturally occurring human BCa disease48–50. By comparing the effects of BBN on WT and NUMB-KO mice, we could formally prove that the absence of NUMB exacerbates the biological aggressiveness of BCa cells by conferring a highly proliferative and invasive/migratory phenotype, as evidenced in the morphological comparison of BBN-NUMB-KO vs. BBN-WT tumor organoids and the transwell invasion/migratory assay.
These findings were entirely replicated in preclinical models of superficial non-invasive human BCa, the male RT4 and the female RT112 cell lines70–72, which invariably switched to an overtly invasive phenotype following silencing of NUMB expression. Together, these results point to NUMB loss as a molecular hallmark of aggressive BCa biology, which likely underlies the clinically aggressive disease course of human NUMB-deficient BCa.
At the mechanistic level, we demonstrated that the aggressive biological phenotypes conferred by NUMB loss are dependent on the functional upregulation of a RHOA/ROCK molecular circuitry, which epistatically induces Hippo cascade downregulation and ensuing YAP signaling hyperactivation (Fig. 9c). In the presence of functional NUMB, RHOA/ROCK signaling to the actin cytoskeleton is restrained, allowing an active Hippo pathway to inhibit the activity of YAP through its phosphorylation and cytoplasmic retention (Fig. 9c). In contrast, in the absence of NUMB, RHOA/ROCK signaling is upregulated, leading to suppression of the Hippo pathway via actin cytoskeleton remodeling and consequently increased nuclear translocation and transcriptional activity of YAP (Fig. 9c). These findings are in line with a recent report showing that NUMB-mediated inhibition of RHOA/ROCK activity is implicated in the regulation of migration and proliferation in colon cancer cells66. Therefore, the dysfunction of the NUMB/RHOA/ROCK/YAP regulatory axis identified in this study may have far-reaching implications in cancer biology beyond BCa. Mechanistically, our work also highlights a previously uncharacterized connection between NUMB loss and EMT/plasticity activation through upregulation of the RHOA/ROCK/YAP circuitry, which most likely accounts for the acquisition of invasive/migratory phenotypes observed in our functional in vitro mouse and human models, and is likely germane to the enhanced risk of NMIBC patients to progress to MIBC (Fig. 9c).
One open question from our study is how NUMB regulates RHOA/ROCK/YAP signaling. We previously showed that the endocytic/sorting function of NUMB is involved in the regulation of another small GTPase, RAC1. NUMB loss results in RAC1 activation, promoting the formation of specialized actin-based lamellipodia protrusions and cell motility phenotypes64,65. These events are associated with the relocalization of RAC1 to the plasma membrane through an EFA6B-ARF6-dependent endocytic recycling route, which is negatively regulated by NUMB through its direct interaction with the guanine nucleotide exchange factor (GEF) EFA6B65. Therefore, mirroring this mechanism, one possibility is that NUMB controls RHOA subcellular localization and activation by interacting with positive (GEFs) or negative (GAPs, GTPase-activating proteins; GDIs, guanine nucleotide dissociation inhibitors) regulators of RHOA activity; a hypothesis that would be in keeping with previous reports linking RHOA activation to its increased plasma membrane localization and decreased cytosolic distribution82,83.
On the other hand, our results selectively involving a RHOA-dependent, RAC1-independent, mechanism in NUMB loss-directed Hippo pathway downregulation/YAP activation, point to the involvement of actin cytoskeleton remodeling events specifically controlled by RHOA/ROCK, such as actomyosin contractility and stress fiber formation67,84. This hypothesis, supported by our evidence of the increased phosphorylation of MLC2 and cofilin in NUMB-deficient BCa cells, is consistent with reports that RHOA-induced stress fibers can suppress the Hippo pathway and activate YAP85,86. It has also been proposed that stress fibers can act as a scaffold for several Hippo pathway components, such as AMOT and MST1/287–90, while stress fiber/F-actin depolymerization activates MST1/290. It is also worth noting that NUMB has been reported to directly or indirectly interact with other actin cytoskeleton regulators, such as α-catenin, β-catenin, NF2/Merlin, AMOT and SPTAN1, which can influence the Hippo pathway’s MST1/2-LATS kinase cascade to inhibit YAP activity89,91,92. Therefore, while the definition of the exact mechanism(s) that connect NUMB loss to the activation of RHOA/ROCK/YAP circuitry warrants further investigation, it is apparent that NUMB loss has the potential to intersect multiple networks involved in actin cytoskeleton remodeling and cell geometry regulation, including tight junctions, adherens junctions and cell polarity complexes, as well as extracellular diffusible signals56,57,59–62, which could all contribute to the aggressive invasive phenotypes through RHOA/ROCK signaling.
The results of this study have potential clinical implications, in particular for the high-grade NMIBC disease that still represents a challenge in decision-making and clinical management due to the substantial inadequacy of currently available biomarkers for prediction of MIBC progression and the limited therapeutic options to prevent this life-threatening event. We submit that assessment of NUMB status, for instance through the use of the NUMBLESS signature, could be included in the development of new prognostic nomograms for individualized risk-adjusted treatment of NMIBC patients, in combination with other existing molecular signatures and standard clinicopathological parameters. More specifically, the NUMBLESS signature could provide the basis of a clinical tool to stratify NMIBC patients at high risk of MIBC progression, who might selectively benefit from innovative RHOA/ROCK/YAP targeted therapies. Indeed, our pharmacological inhibition studies in mouse and human 3D-Matrigel BCa models highlight the RHOA/ROCK/YAP axis as an actionable vulnerability to limit the aggressiveness of NUMB-deficient BCa cells through the inhibition of their proliferative and invasive/migratory phenotypes, which likely underlie the unique propensity for muscle invasion of NUMB-deficient NMIBCs (Fig. 9c).
Notably, several drugs that can interfere with the RHOA/ROCK/YAP circuitry are currently under investigation as potential anti-cancer treatments. Some of these drugs are already in clinical use, such as the YAP transcriptional activity inhibitor, VP40, which is employed in a broad range of ophthalmology conditions93,94, or the ROCK inhibitor, fasudil, currently in clinical trials for different types of vascular and neurodegenerative disorders95. However, predictive biomarkers of therapy response are still needed to aid their repurposing and expand their clinical indications, while minimizing their systemic toxicity. In the case of NMIBC, the translation of NUMB status into the clinical practice would provide a prognostic biomarker to identify those patients whose propensity to muscle-invasion depends specifically on RHOA/ROCK/YAP signaling hyperactivation, making them eligible for targeted anti-RHOA/ROCK/YAP treatments.
One aspect of our study that might apparently limit its relevance to human BCa is that the initial characterization of NUMB loss as a driver of spontaneous and carcinogen-induced tumorigenesis in vivo was based on the use of male mice. We note, however, that the molecular profiling studies that led to the identification of the NUMB loss/YAP/EMT connection and the derivation of the NUMBLESS signature included human BCa cell lines of both male and female origin. Furthermore, the two low-aggressive NMIBC models (male RT4 and female RT112), exhibited the same behaviors in the molecular and functional validation of the pathogenetic relevance of the NUMB loss/RHOA/ROCK/YAP axis. We also note that in our clinical studies the behavior of NUMB loss as a prognostic biomarker of adverse MIBC and NMIBC disease course appears to be independent of standard clinical variables, including sex, among others. This observation is particularly relevant to the potential use of the NUMBLESS signature as a prognostic classifier for NMIBC patients, considering that, in the UROMOL, NMIBC patients characterized by a NUMB dysfunction (NUMBLESS-like) were equally distributed among males and females. Considering that BCa is disproportionately diagnosed in men by a 4:1 ratio1, we submit that our findings are of potential relevance to naturally occurring BCa disease.
Methods
Ethical statement
This research complies with all relevant ethical regulations. All animal experiments were conducted with the approval of Italian Minister of Health (AUT. N. 125/2019-PR) and were performed in accordance with the Italian law (D.lgs. 26/2014), which enforces Dir. 2010/63/EU (Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the protection of animals used for scientific purposes) and EU 86/609 directive and under the control of the institutional organism for animal welfare and ethical approach to animals in experimental procedures (Cogentech OPBA).
Genetically engineered mouse models
Mice were housed in pathogen-free certified animal facilities at the Cogentech Mouse Facility located at IFOM (The AIRC Institute of Molecular Oncology, Milan, Italy). All mice have been maintained in a controlled environment, at 18–23 °C, 40–60% humidity, and with 12-h dark/12-h light cycles. The NUMB-KO mouse model was generated as previously described45. Briefly, mice were generated by crossing Numblox/lox mice96,97 with CK5-Cre mice98. Targeted deletion of the Numb allele was confirmed by PCR genotyping, as described previously96,98. Expression of the Cre-Recombinase allele was confirmed by PCR genotyping, using the following forward and reverse primers: CRE-FW: AACATGCTTCATCGTCGG; CRE-REV: TTCGGATCATCAGCTACACC. Male animals were used to monitor alterations in the urothelium without having to consider the impact of preneoplastic/neoplastic lesions resulting from NUMB loss in the mammary gland45. Histopathological evaluation of NUMB-KO and NUMB-WT mice was performed by board-certified pathologists (G. Renne and G. Bertalot), blinded to the genotype.
BBN-induced mouse model of bladder cancer and mouse pathology
Aged-matched 8- to 16-week-old male mice were given tap water containing 0.05% BBN (B8061, Merck Life Science Srl) for 16 or 20 weeks and afterward switched to regular drinking water until the end of the experiment. The control group received tap water throughout the entire experiment. Male mice were used for these experiments because of their increased susceptibility to BBN, developing earlier morphological alterations compared with female mice51. Mice treated for 16 weeks were sacrificed 2 weeks after the end of the BBN treatment (Fig. 3c and Supplementary Fig. 6c). Mice treated for 20 weeks were sacrificed upon reaching any of the predefined endpoints for high disease burden, including abdominal distension that impeded movement, loss of >15% of body weight, labored breathing, abnormal posture, hematuria (Fig. 3d). Tumor growth was measured by caliper every 3–4 days. Mice were euthanized once the primary tumor reached a maximal size of 1.2 cm in at least one dimension. Maximal tumor size was not exceeded in this study.
At sacrifice, bladders were removed and stored in 4% paraformaldehyde (PFA) for subsequent paraffin embedding and histological examination. The group assignment of bladder tissue sections was blinded to board-certified pathologists (G. Renne and G. Bertalot) for the objective histological evaluation and staging of BBN-induced lesions in the mouse urothelium. Carcinoma in situ (CIS) was defined as a flat tumor lesion confined to the superficial urothelial layer, with malignant urothelial cells displaying loss of cell polarity, cellular atypia, increased number of mitotic figures, and large irregular nuclei with a high nuclear/cytoplasmic ratio, according to standard pathology criteria.
Post-cystectomy MIBC patient cohort
Archival FFPE tissue specimens were collected from a retrospective consecutive cohort of 383 patients who underwent cystectomy for bladder cancer at the European Institute of Oncology (IEO, Milan, Italy) between 1999 and 2016, following approval by the Institutional Ethical Board. The clinicopathological characteristics of this cohort are reported in Supplementary Table 1. NUMB IHC staining was performed on whole FFPE sections from 356 tumor blocks with sufficient material. Staining intensity was scored on a scale from 0 to 3 (0, undetectable expression; 1, intermediate expression; 2-3, expression comparable to normal urothelial basal cells). The percentage of NUMB-positive cells was estimated in increments of 5% over the entire tumor area within the tissue section. Tumors were classified as NUMB-low if a tumor area larger than 70% exhibited an intensity score <1; otherwise, they were categorized as NUMB-high. Survival analysis was conducted using univariable and multivariable Cox proportional hazard models, with multivariable analysis adjusted for clinically relevant variables (age, sex, pT, pN, and vascular invasion). Of the 338 patients with available follow-up data, eighty patients with a pathological tumor stage of pT4 were excluded from the survival analysis to minimize the confounding effects linked to their generally very poor 5-year survival rate99.
NMIBC patient cohorts
Archival FFPE transurethral resection (TUR) specimens were collected from a longitudinal series of patients diagnosed with NMIBC at the European Institute of Oncology (IEO, Milan, Italy) and Policlinico Ospedali Riuniti di Foggia (Foggia, Italy), following standard operating procedures approved by the respective Institutional Ethical Boards. Patients with the first available high-grade NMIBC lesion were selected for the analyses, excluding those patients with a synchronous muscle-invasive disease. The primary outcome was time-to-progression to MIBC in a 4-month minimum follow-up period by TUR. Patients who did not progress were right-censored at the last recorded NMIBC lesion, cystectomy, or negative cystoscopy. Complete clinicopathological information of this NMIBC cohort, including age, sex, and tumor stage, are described in detail in Supplementary Table 2. Whole FFPE TUR sections from a total of 77 patients were IHC stained for NUMB. Assessment of NUMB status and univariable and multivariable analysis were performed as described above for the post-cystectomy MIBC cohorts.
Materials
Antibodies were obtained from the following sources: anti-NUMB mouse monoclonal antibody (clone Ab21, 1:1000 for IB) (#MABN2311 from Merck Life Science Srl); anti-NUMB rabbit monoclonal (clone C29G11) (#2756 from Cell Signaling Technologies, 1:500 for IF, 1:150 for IHC); anti-phospho MST1 (Thr183)/MST2(Thr180) (#49332 from Cell Signaling Technology, 1:1000 for IB); anti-MST1 (#3682 from Cell Signaling Technology, 1:1000 for IB); anti-phospho LATS1 (Thr1079) (#PA5-105442 from Invitrogen, 1:1000 for IB); anti-LATS1 (#3477 from Cell Signaling Technology, 1:1000 for IB); anti-phospho YAP (Ser127) (#4911 from Cell Signaling Technology, 1:1000 for IB); anti-YAP (63.7) (#sc-101199 from Santa Cruz, 1:1000 for IB, 1:200 for IF); anti-YAP (D8H1X) (#14074 from Cell Signaling Technology, 1:200 for IF); anti-active YAP (EPR19812) (#ab205270 from Abcam, 1:500 for IF); anti-phospho Myosin Light Chain 2 (Ser19) (p-MLC2) (#3671 from Cell Signaling Technology, 1:1000 for IB; 1:200 for IF); anti-phospho Cofilin-1 (Ser3) (F-11) (#sc-365882 from Santa Cruz, 1:1000 for IB, 1:300 for IF); anti-Cofilin (D3F9) (#5175 from Cell Signaling Technology, 1:1000 for IB); anti-RAC1 (clone 102/Rac1 (RUO) from BD Biosciences, 1:350 for IF); anti-RHOA (26C4) (#sc-418 from Santa Cruz, 1:400 for IF); anti-RHOA (#ARH03 from Cytoskeleton, 1:2000 for IB); anti-Actin (A4700 from Merck Life Science Srl, 1:10000 for IB); anti-Vinculin (#V9131 from Merck Life Science Srl, 1:10000 for IB); anti-CK5 (EP1601Y) (ab52635 from Abcam, 1:2000 for mice IHC, 1:200 for human IHC); anti-CK14 (LL002) (ab7800 from Abcam, 1:1000 for IF); anti-CK7 (RCK105) (ab9021 from Abcam, 1:500 for IF); anti-CK20 (ab118574 from Abcam, 1:200 for IF and IHC); anti-CYR61 (NB100-356 from Bio-techne Srl, 1:100 for IF); Bond Polymer Refine Detection Kit (#DC9800; Leica Biosystems); Bond Primary Ab Diluent (#AR9352; Leica Biosystems); goat anti-rabbit IgG (H + L)-HRP conjugate (#1706515 Bio-Rad, 1:2500 for IB); goat anti-mouse (H + L)-HRP conjugate (#1706516 Bio-Rad, 1:2500 for IB); Alexa Cy3- and 488-conjugated secondary antibodies from Jackson ImmunoResearch (1:500 for IF); Alexa Fluor 555 donkey anti-mouse IgG (1:400 for IF) and Alexa Fluor 488 donkey anti-rabbit IgG (1:500 for IF) secondary antibodies were obtained from Thermo Fisher; Mowiol-Dabco mounting medium (#81381, Merck Life Science Srl); SlowFade™ Gold Antifade glycerol-based Mountant (#S36936, Invitrogen); DAPI (#32670 from Merck Life Science Srl); Dispase from Stemcell Technologies; ACK Lysing Buffer (#A1049201, Gibco); Cell recovery solution (#354253, Corning); UltraPure low-melting-point (LMP) Agarose (#16520050, Thermo Fisher); Advanced-DMEM/F-12 was from Thermo Fisher; McCoy’s 5 A, RPMI and MEM mediums were from EuroClone; FGF10 (#100-26) and FGF7 (#100-19) from Peprotech; A83-01 from Merck Life Science Srl; B27 from Invitrogen (#A1895601); TrypLETM Express (#12604021) from Gibco; Matrigel growth factor–reduced basement membrane matrix (#356231) from Corning; fetal bovine serum (FBS) from Euroclone; ROCK inhibitor Y-27632 was purchased from Selleckchem (#S1049); Cell Permeable RHO Inhibitor (C3 Transferase) from Cytoskeleton Tebu-Bio (#027CT04-A); RAC1 inhibitor (NSC23766) from Merck Life Science Srl (#SML0952); Cell-permeable marine toxin latrunculin A (#L5163, from Merck Life Science Srl; verteporfin (#HY-B0146, from MedChemExpress and T3112, from TargetMol). N-butyl-N-(4-hydroxybutyl) nitrosamine (BBN) (#B8061, from Merck Life Science Srl).
Cells and culture procedures
RT4 and CLS439 cells were cultured in McCoy’s 5 A medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2mM L-Glutamine and 1% penicillin/streptomycin (P/S). RT112, KK47 and 5637 cells were cultured in RPMI medium supplemented with 10% heat-inactivated FBS, 2mM L-Glutamine and 1% P/S. HT1376 cells were cultured in MEM medium supplemented with 10% heat-inactivated FBS, 2mM L-Glutamine, 1% P/S and 0.1 mM of Non-Essential Amino Acids (NEAA). Primary murine cells were grown in advanced-DMEM/F-12 supplemented with 100 ng/mL FGF10, 25 ng/mL FGF7, 500 nM A83-01, and 2% B27. BBN-induced tumor cells were maintained in 1:1 mixture of DMEM and Ham’s F12 medium, supplemented with 2 mM L-Glutamine, 5 μg/ml insulin, 0.5 μg/ml hydrocortisone, 2% B27, 20 ng/ml EGF and bFGF, and 4 μg/ml heparin. To establish mouse bladder organoids (MBOs), we collected mouse bladders from 4-6 month-old untreated male mice and from BBN-induced tumor transplants, for tumoral-BBN-MBOs. Bladders from untreated male mice were enzymatically digested with 5 U/mL Dispase (#07913 from STEMCELL Technologies) for 30 minutes in 5% CO2 humidified atmosphere at 37°C and the entire superficial epithelium was gently scraped off with a scalpel and recovered by washing with DMEM/F12 medium. The epithelial structures were centrifuged at low speed (80 x g for 5 minutes) to remove fibroblasts and other non-epithelial contaminants and, after removal of the supernatant, the pellet was treated with ACK Lysing Buffer for 1 minute at RT to eliminate red blood cells. After centrifugation, the pellet containing the urothelial structures was dissociated to single cells by TrypLETM Express treatment. Single cell suspension was resuspended in pure Matrigel at density of 2000 cells/50 µL and plated to generate 3D-organoids as a single drop in a 24-well plate for 7-10 days in a total volume of 1 mL of culture medium. Cells from BBN-induced tumors were plated at 1000 cells/50 µL of pure Matrigel to establish 3D-BBN-organoids. Organoid cultures were incubated at 37°C in a humidified 5% CO2 atmosphere, with fresh medium changed every 7 days (4 days for drug treatment). Drug treatments started 3 days after seeding. The resulting 3D-organotypic structures were observed and photographed using an inverted phase-contrast microscope equipped with a digital camera.
Expression, hairpin constructs and engineering of vectors
NUMB knockdown in human bladder cancer cells was performed either by small interfering RNA (siRNA) or by lentiviral-driven expression of small hairpin RNA (shRNA). The efficiency of NUMB silencing was assessed by Western Blotting analysis or by qPCR. For siRNA experiments, cells were transfected with 5 nM of control siRNA (Ctr-KD) or NUMB siRNA (NUMB-KD), 10 nM CTGF and CYR61 human siRNAs using Lipofectamine TM RNAiMAX transfection reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. A second round of transfection was performed 48 hours after the first transfection, and cells were processed for the appropriate assay 48 hours after the second transfection. The targeted sequences used were as follows: NUMB siRNA, CAGCCACUGAACAAGCAGA and scrambled siRNA, AGACGAACAAGUCACCGAC21. CTGF siRNA (sc-39329), CYR61 siRNA (sc-39331) were purchased from Santa Cruz. pLKO.1-NUMB shRNA were purchased from Merck Life Science Srl (oligo identification: TRCN0000007226); a hairpin against Luciferase was used as control (TRCN0000072243). pLKO.1-Yap shRNA was obtained from Addgene (#166486) and used to knockdown YAP expression in BBN-induced cancer cells. Cyr61 shRNA Plasmid (m, SC-39332-SH) and Ctgf shRNA Plasmid (m, SC-39330-SH) were purchased from Santa Cruz. To express ectopic NUMB, a pLVX-NUMB-GFP construct was engineered by subcloning the NUMB-GFP fragment from a pEGFP-N1/NUMB construct (already available in the lab), into the EcoRI-SmaI sites of the pLVX-Puro vector (Clontech). DNA encoding full length human RAC1-DN (RAC1-T17N) was amplified by PCR from pRK5-myc/RAC1-N17 (#15904, Addgene) using the following primers: RAC1_T17N FW: TTTTTCTCGAGACGCGTATCGATTGAATTGGCCACC; RAC1_T17N RV: CTGCAGGAATTCTTACAACAGCAGGC; DNA encoding full-length human RHOA-DN (RHOA-T19N) was amplified by PCR from pSLIK/RHOA-T19N (# 84646, Addgene) using the following primers: RHOA_T19N, FW: TTTTTCTCGAGACGCGTATGGAGCAGAAGCTGATCTCCGAGG; RHOA_T19N, RV: GGATATCTGCAGAATTCACCGC. The resulting DNA fragments were inserted into the XhoI-EcoRI sites of pLVX-Puro. All constructs were sequence-verified. HEK293T were used for viral supernatants production and infections.
Quantitative real-time PCR
Total RNA was extracted with QIAzol™ Reagent and then purified using the RNeasy kit (Qiagen). For BBN NUMB-WT and NUMB-KO mouse BCa cells RNA reverse transcription was performed using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher) or iScript™ Reverse Transcription Supermix (BIO-RAD), according to the manufacturer’s instructions. For BBN NUMB-WT and NUMB-KO mouse BC cells, RT-qPCR was performed using the TaqMan™ Fast Advanced Cells-to-CT™ Kit (Thermo Fisher). For human bladder cancer cell lines, RT-qPCR was performed using the iQ™ SYBR® Green Supermix (BIO-RAD). The ΔCt method was used to calculate the mRNA levels of each target gene normalized against different housekeeping genes. The 2-ΔΔCt method was used to compare the mRNA levels of each target gene and the relative amplification value was plotted in the graph. TaqMan Gene Expression Assay IDs were as follow: Ankrd1 (Mm00496512_m1), Ctgf (Mm01192933_g1), Cyr61 (Mm00487498_m1), Gapdh (Mm99999915_g1), Tbp (mm00446973_m1). The following primer sequences were used for SYBR Green methodology: GAPDH FW: 5’-GTGGAAGGGCTCATGACCA-3’, RV: 5’-GGATGCAGGGATGATGTTCT-3’; TBP FW: 5’-AGTTCTGGGATTGTACCGCA-3’, RV: 5’- CATATTTTCTTGCTGCCAGTCTG-3’; NUMB FW: 5’- GTCCACATCAGTGGCAGACA-3’, RV: 5’- TCCAGTTTTTCCAAAGAAGCCT-3’; CTGF FW: 5’- CCAATGACAACGCCTCCTG-3’, RV: 5’-TGGTGCAGCCAGAAAGCTC-3’; CYR61 FW: 5’-AGCCTCGCATCCTATACAACC-3’, RV: 5’- TTCTTTCACAAGGCGGCACTC-3’; ANKRD1 FW: 5’-AGCGCCCGAGATAAGTTGCT-3’, RV: 5’-CACCAGATCCATCGGCGTCT-3’.
SDS-PAGE analysis
Cells were lysed in ice-cold RIPA buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, 1 mM beta-glycerophosphate, 1 mM Na3VO4, 1 µg/mL leupeptin (#9806, Cell Signaling Technology) supplemented with HaltTM Protease and Phosphatase Inhibitor Cocktail (#78440, Thermo Fisher Scientific). Samples were clarified by centrifugation and protein content was measured using the BCA protein assay kit (Thermo Scientific). Proteins (10-15 μg) were resolved on 4-15% SDS-PAGE gels and transferred onto nitrocellulose membranes (Bio-Rad Laboratories). Membranes were blocked in Tris-buffered saline (TBS) containing 5% non-fat dry milk and 0.1% Tween 20 (TBS-T), prior to incubation with primary antibodies overnight at 4 °C. The membranes were then washed with TBS-T followed by exposure to the appropriate horseradish peroxidase-conjugated secondary antibody for 1 hr at room temperature and visualized on Kodak X-ray film using the enhanced chemiluminescence (ECL) detection system (Bio-Rad Laboratories) or with ChemiDoc Imager followed by acquisition with Image Lab Software (Version 5.2.1). Uncropped and unprocessed scans of all the blots in the paper are available in the Source Data file.
RHO GTPase pull-down activation assay
Activated GTP-bound RHOA was measured using the RHOA/RAC1/CDC42 Activation Assay Combo Biochem KitTM (#BK030, Cytoskeleton, Inc, Tebu Bio Srl), following the manufacturer’s recommendations. Briefly, confluent cells were lysed in Cell Lysis Buffer (50 mM Tris pH 7.5, 10 mM MgCl2, 0.5 M NaCl and 2% Igepal) supplemented with Protease Inhibitor Cocktail (62 μg/mL Leupeptin, 62 μg/mL Pepstatin A, 14 mg/mL Benzamidine and 12 mg/mL tosyl-arginine methyl-ester) and incubated on ice for 20 minutes. Next, cell lysates were centrifuged at 16,000 x g for 15 minutes at 4 °C and supernatants were collected. For pull-down assay, 400 μg of protein lysate were added to a predetermined amount of rhotekin-RBD beads (25 μg). Twenty μg of cell lysate were used for the evaluation of total RHOA protein levels, detected using an antibody against RHOA.
Morphometric analysis of organoids and quantitation of invasive phenotypes in 3D-Matrigel
Phase-contrast images of organoids grown in Matrigel were acquired with EVOS FL Digital Inverted Fluorescence Microscope (Invitrogen, Massachusetts) at 4x magnification after 7-12 days. Images were then imported in Fiji software100 (Version 2.14.0) and manually segmented to obtain regions of interest (ROI) for each organoid. Organoids smaller than 70 μm in diameter or touching the image border, and cells adherent to the well plate were excluded from the analysis. For each ROI, the analysis of organoid morphology and invasive phenotype was performed by the extraction and quantitation of the following shape descriptors: area (μm²), a parameter that correlates with the overall organoid size, indicative of cellular proliferation rate; roughness (perimeter/convex perimeter), a parameter that describes the irregularity of the organoid boundary, indicative of cell invasiveness and migratory ability; circularity (4π*area/perimeter²), a general measure positively correlated with organoid maturation and inversely correlated with invasiveness; shape complexity (number of endpoint-pixels of the skeletonized shape), a measure of the complexity of the organoid shape, indicative of aberrant growth and correlated with the number of cells protruding from the edge55. Additionally, for MBOs from untreated mice, the occurrence of an irregular/multilobular morphology was manually annotated as aberrant, and the frequency of aberrant vs. normal morphology was reported.
Preparation of formalin-fixed/paraffin-embedded organoid samples for immunofluorescence studies
MBOs grown in Matrigel were harvested using ice-cold Cell Recovery Solution for 30 minutes, then pelleted by centrifugation at 200 x g for 5 minutes. The supernatant was discarded, and the pellet was resuspended in Cell Recovery Solution for an additional 30 minutes. After the complete removal of Matrigel, the organoids were washed in PBS and fixed in 4% PFA for 30 minutes at 4 °C under constant rotation. Following fixation, the organoids were pelleted to remove PFA, washed with PBS, and resuspended in 100 µL of 4% Low Melting Point Agarose solution. The agarose was allowed to solidify on ice and dehydrated in 70% ethanol overnight. Subsequently, the sample underwent standard procedures for paraffin embedding.
Immunofluorescence studies
For immunofluorescence (IF) staining of formalin-fixed paraffin-embedded (FFPE) samples (tissues and 3D-organoids), three µm-thick sections were deparaffinated with 100% xylene, rinsed in ethanol and re-hydrated in distilled water; antigen retrieval was performed by heating sections for 50 minutes at 95 °C in pH 6.0 citrate buffer solution; tissue sections were then washed in TBS-T for 5 minutes and permeabilized with 0,3% Triton X-100 diluted in PBS for 30 minutes at room temperature (RT). Next, slides were blocked with 5% BSA diluted in PBS with 0,1% Triton X-100 for 1 hour at RT. Finally, slides were incubated overnight at 4 °C with primary antibody appropriately diluted in blocking solution. The next day, sections were incubated with the proper secondary antibody and DAPI to counterstain the nuclei for 1 hour at room temperature before mounting using Mowiol solution. The same permeabilization, blocking, and staining procedure was used for IF on adherent cells. For the analysis of 3D-organoids, representative ROIs were acquired with a Leica SP5 Confocal Microscope. For human NMIBC TUR specimens, the whole slide was acquired with a NanoZoomer S60 Digital slide scanner (Hamamatsu Photonics K.K.) with a 20X/0.75 NA dry objective.
For the analysis of YAP on adherent cells (Figs. 6a, e, f, 8b, Supplementary Figs. 7a, b, 8, 9a,b, 10a, 11b,c), cells were co-stained for YAP and DAPI, along with any additional control markers of interest for the specific experiment and analyzed by Fiji image analysis software. The nucleus of each cell was segmented based on the DAPI channel and the mean YAP intensity was measured in an area comprising the nucleus itself and a 1 μm-thick cytoplasmic band surrounding the nucleus. The nucleus-to-cytoplasmic ratio was then computed for each cell and averaged across the different fields analyzed. For the analysis of YAP in mouse FFPE samples (Fig. 4a, Supplementary Fig. 6b–d), sections were co-stained for active (non-phosphorylated) YAP and DAPI. The nucleus of each cell was segmented based on the DAPI channel in Fiji followed by quantification of the mean active-YAP nuclear intensity. Cells with a nuclear intensity higher than 50 (arbitrary unit) were classified as YAP-High. Cells with a nuclear intensity lower than 50 (arbitrary unit) were classified as YAP-Low.
For the analysis of YAP in FFPE human NMIBC TUR samples (Fig. 2h), FFPE sections were co-stained for active YAP and DAPI and analyzed by QuPath101. Nuclei were automatically segmented on the DAPI channel using the deep-learning Stardist algorithm, based on an in-house trained model. The cytoplasmic cell area was represented as a 3 µm-thick band around the nucleus. Intensity of active YAP staining was measured in both nuclear and cytoplasmic cell compartments. For each patient, three independent representative regions of tumor tissue, each of average area equal to 1 mm2, were manually selected and analyzed for the distribution of cells with negative (nuclear intensity comparable to background cytoplasmic intensity) or positive (nuclear intensity 1.5-fold superior to background cytoplasmic intensity) active YAP nuclear expression. Detection measurements relative to each region were exported in R Studio (v. 2023.06.2). Filters were applied on the exported database to exclude non-reliable cell detections (objects with size lower than 15 µm2 and intensity lower than a custom threshold in the DAPI channel were discarded). The number of active YAP-High cells was then normalized to the total number of cells for each region and aggregated per patient to extract the mean percentage of YAP-High cells. For CYR61 IF staining of human FFPE NMIBC TUR specimens (Fig. 2h), antigen retrieval was performed with pH 9.0 Tris-EDTA buffer solution. Coverslip slides were mounted using the SlowFade™ Gold Antifade glycerol-based mountant and whole-slide widefield images were acquired using the Eclipse Ti2 microscope (Nikon Europe B.V.) with a 20X/0.8NA dry objective, and a sCMOS camera (Kinetix, Teledyne Photometrics). For each patient, three independent representative regions of tumor tissue, each of average area equal to 1 mm2, were manually selected and segmented using the same approach as described for YAP analysis. Then, the mean cytoplasmic intensity was averaged for each region and aggregated per patient. The quantitative analysis of p-MLC2 and p-Cofilin staining (Fig. 7b, c Supplementary Fig. 10b,c) was conducted in Fiji by computing the mean fluorescence intensity over the entire area of the field occupied by cells. The segmentation of this area was achieved by generating an image from the maximum intensity projection of all channels, following automatic brightness and contrast adjustment to scale the signals. A custom threshold was then established on this image to delineate the cell-occupied regions for intensity measurement.
Immunohistochemistry
Three-μm thick sections were prepared from FFPE tissue blocks, dried at 37 °C O/N and then processed with Bond-RX fully automated stainer system (Leica Biosystems) according to the following protocol. For phospho-Cofilin and NUMB staining, antigen retrieval was performed by heating sections in a microwave oven for 10 minutes on high power (∼900 watt) in citrate buffer solution pH 6.0 at 100 °C for 40 min. After the washing steps, peroxidase blocking was performed for 10 min using the Bond Polymer Refine Detection Kit (#DC9800, Leica Biosystems). Tissues were incubated for 30 min with the appropriate Ab diluted in Bond Primary Ab Diluent (#AR9352). Subsequently, tissues were incubated with post primary and polymer for 16 min, developed with DAB-chromogen for 10 min and counterstained with hematoxylin for 5 min. Slides were digitally scanned with the Aperio ScanScope. Digital images were processed with Adobe Photoshop CS3 (Adobe Inc.).
Transwell invasion assay
Transwell migration/invasion assays were performed using PET membrane inserts (BRAND® 24-well Cell Culture Insert, PET-membrane, #BR782711, Merck) covered with 20 μL of Matrigel:PBS (1:2), as described in the manufacturer’s protocol. Briefly, 1×105 cells were seeded in growth factor- or serum-free medium, depending on the cell lines, in the upper chamber of the transwell insert, and treated, when necessary, with the appropriate pharmacological inhibitors (added in the upper and lower chamber of the transwell). Complete medium supplemented with HGF (25 ng/mL) was added to the lower part of the transwell. After an appropriate incubation period ( ~ 18 hr for BBN-induced mouse tumor cells, 24 hr for RT112 cells, 48 hr for RT4 cells), the transwell inserts were removed and the cells were fixed in the transwell insert with 4% PFA for 15 minutes. Cells were then washed with water to remove the formaldehyde. Then, using a sterile cotton swap, cells which had not migrated through the membrane were scraped off the top of the transwell insert and stained by using DAPI + 0.1% Triton X-100 in PBS for 30 minutes. Images corresponding to the entire area of the transwell were collected with the Nikon Ti-2 microscopy using a 10x objective and the number of invaded cells was analyzed using the Fiji software.
RNA sequencing
Libraries were prepared using the TruSeq Stranded Total RNA Gold kit (Illumina) according to the manufacturer’s protocol. Sequencing was performed on an Illumina NovaSeq 6000 platform with paired-end reads of 50 bp. Sequenced raw reads were processed with NF-CORE RNA-Seq pipeline (https://github.com/nf-core/rnaseq, version 3.8.1), using the human GRCh38 and mouse GRCm39 as reference genomes. Transcript abundances were quantified using the Salmon pseudo-aligner. Genes abundance normalization and differential expression testing were performed with “DESeq2” (version 1.34). Log2 fold changes obtained were shrunken using the “apeglm” method to improve the stability of the estimates in the presence of a limited number of replicates. For samples with only one replicate, the only metric estimated was the log2 fold change, calculated by directly subtracting log1p(normalized counts) of each condition for genes expressed above a custom cutoff, defined based on the sequencing depth of each experiment. All samples for each condition tested were analyzed with principal component analysis and projected. When the batch effect was associated with a principal component that explained more than 20% of the variance, the batch of the experiment was added as a covariate in the statistical model. Genes with a Benjamini-Hochberg adjusted p-value < 0.05 and an absolute log2 fold change >0.5 were uploaded to the Ingenuity Pathway Analysis (IPA) software (Qiagen). IPA was used to infer upstream regulators from the RNA-Seq data by applying the “Upstream Regulator Analysis” tool. Transcription regulators with an activation Z-score >1.5 or <-1.5 and an overlap p-value < 0.05 were considered as significantly activated or inhibited, respectively. The association between gene expression modifications and changes in pathways and biological states was assessed with a Gene Set Enrichment Analysis (GSEA)6. The analysis was performed with the “fgsea” method implemented in the “ClusterProfiler” R package (version 4.2.2) and the MSigDB “Hallmarks”102 set was used as annotation. Genes were ranked based on their log2 fold change values. Genes with low counts or high dispersion that did not pass the DESeq2 independent filtering were not considered for the analysis. For the evaluation of the activity of YAP/TAZ pathway (not included in the “Hallmarks” set), a signature composed of 22 direct YAP/YAZ transcriptional targets39 was additionally used as annotation for the GSEA.
Definition of the NUMBLESS signature and clinical validation of its prognostic relevance to NMIBC
We established a gene signature reflective of NUMB deficiency in human BCa by identifying 15 consistently upregulated genes (Log2FC > 1, padj < 0.01) and 12 consistently downregulated genes (Log2FC < -1, padj < 0.01) following NUMB knockdown in the three NUMBHigh BCa cell lines (RTT4, RT112 and KK47). Raw gene abundances from the three NUMBHigh cell lines, silenced or not for NUMB, and the three NUMBLow cell lines (CLS439, HT1376 and 5637) were normalized using DESeq2. Normalized abundances were then converted to a log scale with the variance stabilizing transformations (implemented in DESeq2), before z-score computation. Using the expression levels of these 27 signature genes, we conducted unsupervised clustering of the three NUMBHigh and the three NUMBLow BCa cell lines with the Hierarchical Clustering algorithm from the complexHeatmap R library (version 2.13.4), employing a 1- Pearson’s correlation as the distance metric. We then utilized this 27-gene signature to cluster patients from the 535 NMIBC patients of the UROMOL cohort24. Raw gene abundances were normalized, transformed, and scaled as mentioned above. We then applied the Partitioning Around Medoids (PAM) algorithm for clustering, using the 1-Pearson’s correlation coefficient as the distance metric. The silhouette method used to determine the optimal number of clusters identified two distinct NMIBC tumor subtypes. The 27 genes in the signature were hierarchically clustered to confirm the consistent and coordinated modulation of genes upregulated and downregulated in clusters predicted as NUMBLESS “like” and “not-like”. The 22-gene YAP/TAZ transcriptional target signature was used to classify the NMIBC samples into YAP-active and YAP-inactive groups. The PAM algorithm was reapplied for clustering with the 1-Pearson’s distance metric. The differential propensity for MIBC progression among the two NMIBC subtypes, as defined by the NUMBLESS signature, was assessed using a univariable and multivariable Cox proportional hazards model. Multivariable analyses were adjusted for clinically relevant variables (age, gender, tumor grade, pT, history of intravesical therapy). Each sample was scored for Epithelial-to-Mesenchymal Transition (EMT) using the MSigDB “Hallmark EMT” gene set through Gene Set Variation Analysis (GSVA) implemented in the GSVA R library (version 1.51.17). We compared the NUMBLESS signature with 14 established prognostic BCa signatures by extracting genes from each signature and performing GSVA to generate continuous scores for each patient in the UROMOL cohort. For signatures that included genes with different directionalities, we calculated separate scores for genes positively and negatively associated with prognosis, then integrated these using a weighted average based on the number of genes. All scores were mean-centered and scaled to unit variance to obtain z-scores with comparable distribution. We assessed the clinical relevance of each signature by calculating the difference in the likelihood ratio chi-squared scores (LR-Δχ²) between the null Cox proportional hazards model, the base model including only clinicopathological variables (pT, Grade, Age, Sex), and the full model that also includes the gene signature. To analyze the individual contribution and redundancy of the signatures, we performed a Lasso penalized Cox regression incorporating both clinicopathological variables and all gene signatures.
Statistical analysis and reproducibility
PCR experiments were performed in biological triplicates with the exception of the control PCR experiments in Supplementary Figs. 9i, 10d, 12b, e in a single replicate. All the other experiments were repeated a minimum of two times with the exception of the RNA-Seq experiments confirming the reversion EMT signature enrichment under treatment with verteporfin, ROCK inhibitor Y-27632 or Cyr61/Ctgf-KD (Figs. 4c, 5e, 7h, 8f, Supplementary Figs. 9j, 11e, 12c,f). The number n of cells, animals, fields, patients, and independent experiments is reported in figures or in accompanying figure legends. Continuous data are represented as mean ± standard error (SEM) in all cases except for Figs. 2f, g, 4f, 5b,h, 6g, 7f, 8h, Supplementary Fig. 7d, 9d, h, 11f which are represented as boxplots. The boxplot is depicted with a box covering the interquartile range (IQR) from the 25th to the 75th percentile, the whiskers extend from the ends of the box to the smallest and largest data values within a 1.5 IQR. The horizontal line indicates the median, and the “X” indicates the mean. Means were compared using two sample Welch’s t-test, except for comparisons with the reference sample in RT-qPCR experiments (Figs. 4b, 5d, 6c, 7d, 8e, Supplementary Fig. 7c, 11d), where one-sample t-test was performed. When multiple conditions were present we performed a FDR adjusted pairwise Welch’s t-test following a significant Welch’s ANOVA except for Figs. 6a, f, 7c, 8b, Supplementary Fig. 6b,c, 9b, 11c which were analyzed with Tukey HSD test following a significant ANOVA. Data distribution was assumed to be normal although not formally tested. The Benjamini-Hochberg method was employed for FDR corrections in multiple comparison testing. When performing multiple comparisons, only comparisons of biological interest are shown in the graphs; complete results are available in the source data. Differences in proportions were tested with Fisher’s exact test in all 2×2 tables (Fig. 2g, Supplementary Tables 1-3) and with Pearson’s Chi-squared test in larger contingency tables. (Fig. 3b, c). Odds ratio and relative confidence intervals were calculated with Small Sample-Adjusted Unrestricted Maximum Likelihood Estimation and Normal Approximation (Wald) for Fig. 3b, c and with the Median-Unbiased Estimate and Mid-P Exact Confidence Intervals for Supplementary Tables 1–3. For survival analysis, the Cox proportional hazards model was employed as implemented in the “coxph” function of the “survival” R package (version 3.5) to compute hazard ratios (HR) and confidence intervals (CI) in both univariable and multivariable analyses. Global significance was tested using the log-rank test, while the significance of each variable was determined using the Wald test. To account for the batch effect between the two different rounds of staining of CYR61 (Fig. 2h), a linear mixed-effect model was adopted for the statistical analysis as implemented in the “lmerTest” R package (version 3.1). Marginal residuals of the model including only the random effect were used to plot batch-corrected intensity values (Arbitrary Units). All statistical tests were two-sided, except for Supplementary Fig. 3a. Significance was set at p < 0.05. Significance in figures is indicated as: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; ns, not significant. Exact p-values are reported in source data. All statistical analyses were performed using the R software (version 4.1.2). Details of each test are provided in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank the anonymous patients who donated their samples for research and the IEO Pharmacy for chemotherapy drugs. We thank A. Gobbi, M. Capillo, and the Mouse facility, and the Real-Time PCR and DNA Sequencing Service of Cogentech (Cogentech Srl, Milan) for mice handling and technical support; L. Rotta and the IEO Genomic Unit for RNA sequencing experiments; C. Luise, D. Ricca, M. Tillhon, F. Montani and the IEO Molecular and Digital Pathology Unit for technical assistance with tissue processing, staining and image acquisition; D. Disalvatore for help with statistical analyses; M. Monturano, G. Peruzzotti, the IEO Clinical Trial Office and the Retrospective Data Governance Board for assistance with clinical databases and ethical procedures; O. De Cobelli, F. Nolé, A. Mistretta, V.D. Matei, G. Cordima, G. Cozzi and the IEO Urology Program for help with clinical cohorts. M.G.F. was supported by a fellowship from the Fondazione Umberto Veronesi (FUV), and R.P. and D.C.R. were supported by an AIRC fellowship. This work was supported by: Associazione Italiana per la Ricerca sul Cancro (AIRC-IG 23049 to S.P., IG 23060 to P.P.D.F.); PSR-2020/2022 from the University of Milan to S.P.; The Italian Ministry of University and Scientific Research (MIUR-PRIN2017 and MIUR/PRIN2020 to S.P); The Italian Ministry of Health, RF-2016-02361540 e RF-2021-12373957 to P.P.D.F.; The Italian Ministry of Health with Ricerca Corrente and 5×1000 funds.
Author contributions
Conceptualization: D.T. and S.P. Methodology: R.P., D.C.R., M.G.F., R.B., F.R. performed the in vitro experiments; D.T. and R.B. performed the ex vivo experiments; F.A.T., F.S., G.R., N.F., G.B. and G.V. performed pathological analyses; G.J., R.B. and E.G. did IF and IHC stainings. Formal analysis: F.A.T., C.S. and S.R., imaging analysis; F.A.T., bioinformatic and statistical analysis on gene expression; F.A.T. and S.F. statistical analysis of patient data. Resources: F.S., G.R., N.F., P.P.D.F., G.V., G.P., G.M., S.P. Data curation: F.A.T., R.P, D.C.R., M.G.F., S.F, F.R. Investigation: F.A.T., R.P., D.C.R., M.G.F., D.T. Writing the original draft: D.T., S.P., F.A.T., R.H.G. Writing (review and editing): All Authors. Supervision: S.P. and D.T. Funding acquisition: S.P., D.T., P.P.D.F., G.P. D.T., and S.P. are the custodians of all the original documentation.
Peer review
Peer review information
Nature Communications thanks Cory Abate-Shen, Jianmin Zhang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The raw and processed RNA sequencing data generated in this study have been deposited in the GEO database under the accession code GSE256272. The clinicopathological data from the UROMOL cohort used in this study are available in the source data of the referenced publication24. Raw gene abundances were kindly provided by the authors. Human and mouse gene set annotations for pathway analysis were retrieved from the Molecular Signatures Database (MSigDB) [www.gsea-msigdb.org/gsea/msigdb/] with the “msigdbr” package (version 7.4.1) in R. Human GRCh38 and mouse GRCm39 reference genomes (primary genome assembly) used for the alignment of RNA sequencing data are available on GENCODE. Expression intensity data of CK5 and NUMB across diverse tissues were retrieved from the Human Protein Atlas [www.proteinatlas.org]. Materials and reagents are available upon request. Source data are provided with this paper.
Competing interests
G. Renne, G. Vago, D. Tosoni and S. Pece are co-inventors of a patent (EP24191940.6) related to this work, pending to the European Institute of Oncology and University of Milan. All the other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: F. A. Tucci, R. Pennisi.
These authors jointly supervised this work: D. Tosoni, S. Pece.
Contributor Information
D. Tosoni, Email: daniela.tosoni@ieo.it
S. Pece, Email: salvatore.pece@ieo.it
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54246-6.
References
- 1.Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin.71, 7–33 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Kaufman, D. S., Shipley, W. U. & Feldman, A. S. Bladder cancer. Lancet374, 239–249 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Robertson, A. G. et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell174, 1033 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamat, A. M. et al. Bladder cancer. Lancet388, 2796–2810 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Witjes, J. A. et al. European Association of Urology Guidelines on Muscle-invasive and Metastatic Bladder Cancer: Summary of the 2020 Guidelines. Eur. Urol.79, 82–104 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Powles, T. et al. Bladder cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol.33, 244–258 (2022). [DOI] [PubMed] [Google Scholar]
- 7.Dinney, C. P. et al. Focus on bladder cancer. Cancer Cell6, 111–116 (2004). [DOI] [PubMed] [Google Scholar]
- 8.van den Bosch, S. & Alfred Witjes, J. Long-term cancer-specific survival in patients with high-risk, non-muscle-invasive bladder cancer and tumour progression: a systematic review. Eur. Urol.60, 493–500 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Schrier, B. P., Hollander, M. P., van Rhijn, B. W., Kiemeney, L. A. & Witjes, J. A. Prognosis of muscle-invasive bladder cancer: difference between primary and progressive tumours and implications for therapy. Eur. Urol.45, 292–296 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Babjuk, M. et al. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder: update 2016. Eur. Urol.71, 447–461 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Svatek, R. S. et al. The economics of bladder cancer: costs and considerations of caring for this disease. Eur. Urol.66, 253–262 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Goebell, P. J. & Knowles, M. A. Bladder cancer or bladder cancers? Genetically distinct malignant conditions of the urothelium. Urol. Oncol.28, 409–428 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Hedegaard, J. et al. Comprehensive transcriptional analysis of early-stage urothelial carcinoma. Cancer Cell30, 27–42 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Pietzak, E. J. et al. Next-generation sequencing of nonmuscle invasive bladder cancer reveals potential biomarkers and rational therapeutic targets. Eur. Urol.72, 952–959 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu, X. R. Urothelial tumorigenesis: a tale of divergent pathways. Nat. Rev. Cancer5, 713–725 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Colaluca, I. N. et al. NUMB controls p53 tumour suppressor activity. Nature451, 76–80 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Yan, B. et al. Characterization of Numb expression in astrocytomas. Neuropathology28, 479–484 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Westhoff, B. et al. Alterations of the Notch pathway in lung cancer. Proc. Natl Acad. Sci. USA106, 22293–22298 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pece, S., Confalonieri, S., P, R. R. & Di Fiore, P. P. NUMB-ing down cancer by more than just a NOTCH. Biochim Biophys. Acta1815, 26–43 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Liu, Z. et al. Increased Numb protein expression predicts poor clinical outcomes in esophageal squamous cell carcinoma patients. Cancer Biol. Ther.19, 34–41 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pece, S. et al. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J. Cell Biol.167, 215–221 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colaluca, I. N. et al. A Numb-Mdm2 fuzzy complex reveals an isoform-specific involvement of Numb in breast cancer. J. Cell Biol.217, 745–762 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filippone, M. G. et al. Aberrant phosphorylation inactivates Numb in breast cancer causing expansion of the stem cell pool. J. Cell Biol.22110.1083/jcb.202112001 (2022). [DOI] [PMC free article] [PubMed]
- 24.Lindskrog, S. V. et al. An integrated multi-omics analysis identifies prognostic molecular subtypes of non-muscle-invasive bladder cancer. Nat. Commun.12, 2301 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dyrskjot, L. et al. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res64, 4040–4048 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Dyrskjot, L. et al. A molecular signature in superficial bladder carcinoma predicts clinical outcome. Clin. Cancer Res11, 4029–4036 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Kim, W. J. et al. Predictive value of progression-related gene classifier in primary non-muscle invasive bladder cancer. Mol. Cancer9, 3 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, W. J. et al. A four-gene signature predicts disease progression in muscle invasive bladder cancer. Mol. Med17, 478–485 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Heijden, A. G. et al. A five-gene expression signature to predict progression in T1G3 bladder cancer. Eur. J. Cancer64, 127–136 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Mo, Q. et al. Prognostic power of a tumor differentiation gene signature for bladder urothelial carcinomas. J. Natl Cancer Inst.110, 448–459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abudurexiti, M. et al. Development and external validation of a novel 12-gene signature for prediction of overall survival in muscle-invasive bladder cancer. Front Oncol.9, 856 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin, H., Zhang, C., Gou, X., He, W. & Gan, D. Identification of a 13‑mRNA signature for predicting disease progression and prognosis in patients with bladder cancer. Oncol. Rep.43, 379–394 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu, H. et al. Identification and validation of an individualized prognostic signature of bladder cancer based on seven immune related genes. Front Genet11, 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park, S. et al. Novel mouse models of bladder cancer identify a prognostic signature associated with risk of disease progression. Cancer Res81, 5161–5175 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tao, Y. et al. TP53-related signature for predicting prognosis and tumor microenvironment characteristics in bladder cancer: A multi-omics study. Front Genet13, 1057302 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang, F. et al. A 7-gene signature predicts the prognosis of patients with bladder cancer. BMC Urol.22, 8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, S. K. et al. A 23-gene prognostic index predicts progression and bacillus calmette-guerin response in non-muscle-invasive bladder cancer. Eur. Urol.85, 400–402 (2024). [DOI] [PubMed] [Google Scholar]
- 38.Xie, T. et al. Multi-cohort validation of Ascore: an anoikis-based prognostic signature for predicting disease progression and immunotherapy response in bladder cancer. Mol. Cancer23, 30 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, Y. et al. Comprehensive molecular characterization of the hippo signaling pathway in cancer. Cell Rep.25, 1304–1317 e1305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu-Chittenden, Y. et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev.26, 1300–1305 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, J. Y. et al. Overexpression of YAP 1 contributes to progressive features and poor prognosis of human urothelial carcinoma of the bladder. BMC Cancer13, 349 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gill, M. K. et al. A feed forward loop enforces YAP/TAZ signaling during tumorigenesis. Nat. Commun.9, 3510 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baek, S. W. et al. YAP1 activation is associated with the progression and response to immunotherapy of non-muscle invasive bladder cancer. EBioMedicine81, 104092 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zanconato, F., Cordenonsi, M. & Piccolo, S. YAP/TAZ at the Roots of Cancer. Cancer Cell29, 783–803 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tosoni, D. et al. The Numb/p53 circuitry couples replicative self-renewal and tumor suppression in mammary epithelial cells. J. Cell Biol.211, 845–862 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Filippone, M. G. et al. CDK12 promotes tumorigenesis but induces vulnerability to therapies inhibiting folate one-carbon metabolism in breast cancer. Nat. Commun.13, 2642 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin, K. et al. Cellular origin of bladder neoplasia and tissue dynamics of its progression to invasive carcinoma. Nat. Cell Biol.16, 469–478 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Batavia, J. et al. Bladder cancers arise from distinct urothelial sub-populations. Nat. Cell Biol.16, 982–991 (2014). 981-985. [DOI] [PubMed] [Google Scholar]
- 49.Nagao, M., Suzuki, E., Yasuo, K., Yahagi, T. & Seino, Y. Mutagenicity of N-butyl-N-(4-hydroxybutyl)nitrosamine, a bladder carcinogen, and related compounds. Cancer Res37, 399–407 (1977). [PubMed] [Google Scholar]
- 50.Fantini, D. et al. A Carcinogen-induced mouse model recapitulates the molecular alterations of human muscle invasive bladder cancer. Oncogene37, 1911–1925 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bertram, J. S. & Craig, A. W. Specific induction of bladder cancer in mice by butyl-(4-hydroxybutyl)-nitrosamine and the effects of hormonal modifications on the sex difference in response. Eur. J. Cancer (1965)8, 587–594 (1972). [DOI] [PubMed] [Google Scholar]
- 52.Mullenders, J. et al. Mouse and human urothelial cancer organoids: A tool for bladder cancer research. Proc. Natl Acad. Sci. USA116, 4567–4574 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moriconi, C. et al. INSIDIA: A FIJI Macro Delivering High-Throughput and High-Content Spheroid Invasion Analysis. Biotechnol. J.1210.1002/biot.201700140 (2017). [DOI] [PubMed]
- 54.Alinezhad, S. et al. Validation of Novel Biomarkers for Prostate Cancer Progression by the Combination of Bioinformatics, Clinical and Functional Studies. PLoS One11, e0155901 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harma, V. et al. Quantification of dynamic morphological drug responses in 3D organotypic cell cultures by automated image analysis. PLoS One9, e96426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Piccolo, S., Dupont, S. & Cordenonsi, M. The biology of YAP/TAZ: hippo signaling and beyond. Physiol. Rev.94, 1287–1312 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Panciera, T., Azzolin, L., Cordenonsi, M. & Piccolo, S. Mechanobiology of YAP and TAZ in physiology and disease. Nat. Rev. Mol. Cell Biol.18, 758–770 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Totaro, A., Panciera, T. & Piccolo, S. YAP/TAZ upstream signals and downstream responses. Nat. Cell Biol.20, 888–899 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dupont, S. et al. Role of YAP/TAZ in mechanotransduction. Nature474, 179–183 (2011). [DOI] [PubMed] [Google Scholar]
- 60.Meng, Z., Moroishi, T. & Guan, K. L. Mechanisms of Hippo pathway regulation. Genes Dev.30, 1–17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dupont, S. Role of YAP/TAZ in cell-matrix adhesion-mediated signalling and mechanotransduction. Exp. Cell Res343, 42–53 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Yu, F. X. et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell150, 780–791 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aragona, M. et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell154, 1047–1059 (2013). [DOI] [PubMed] [Google Scholar]
- 64.Lau, K. M. & McGlade, C. J. Numb is a negative regulator of HGF dependent cell scattering and Rac1 activation. Exp. Cell Res317, 539–551 (2011). [DOI] [PubMed] [Google Scholar]
- 65.Zobel, M. et al. A NUMB-EFA6B-ARF6 recycling route controls apically restricted cell protrusions and mesenchymal motility. J. Cell Biol.217, 3161–3182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang, Y. et al. Numb inhibits migration and promotes proliferation of colon cancer cells via RhoA/ROCK signaling pathway repression. Exp. Cell Res411, 113004 (2022). [DOI] [PubMed] [Google Scholar]
- 67.Ridley, A. J. Life at the leading edge. Cell145, 1012–1022 (2011). [DOI] [PubMed] [Google Scholar]
- 68.Kimura, K. et al. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science273, 245–248 (1996). [DOI] [PubMed] [Google Scholar]
- 69.Kureishi, Y. et al. Rho-associated kinase directly induces smooth muscle contraction through myosin light chain phosphorylation. J. Biol. Chem.272, 12257–12260 (1997). [DOI] [PubMed] [Google Scholar]
- 70.Masters, J. R. et al. Tissue culture model of transitional cell carcinoma: characterization of twenty-two human urothelial cell lines. Cancer Res46, 3630–3636 (1986). [PubMed] [Google Scholar]
- 71.Davies, G., Jiang, W. G. & Mason, M. D. Cell-cell adhesion molecules and their associated proteins in bladder cancer cells and their role in mitogen induced cell-cell dissociation and invasion. Anticancer Res19, 547–552 (1999). [PubMed] [Google Scholar]
- 72.Warrick, J. I. et al. FOXA1, GATA3 and PPARɣ cooperate to drive luminal subtype in bladder cancer: a molecular analysis of established human cell lines. Sci. Rep.6, 38531 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thiery, J. P., Acloque, H., Huang, R. Y. & Nieto, M. A. Epithelial-mesenchymal transitions in development and disease. Cell139, 871–890 (2009). [DOI] [PubMed] [Google Scholar]
- 74.Dongre, A. & Weinberg, R. A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol.20, 69–84 (2019). [DOI] [PubMed] [Google Scholar]
- 75.Haque, I. et al. Cyr61/CCN1 signaling is critical for epithelial-mesenchymal transition and stemness and promotes pancreatic carcinogenesis. Mol. Cancer10, 8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sonnylal, S. et al. Connective tissue growth factor causes EMT-like cell fate changes in vivo and in vitro. J. Cell Sci.126, 2164–2175 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tosoni, D. et al. Pre-clinical validation of a selective anti-cancer stem cell therapy for Numb-deficient human breast cancers. EMBO Mol. Med9, 655–671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shu, Y. et al. Loss of Numb promotes hepatic progenitor expansion and intrahepatic cholangiocarcinoma by enhancing Notch signaling. Cell Death Dis.12, 966 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Belle, V. A., McDermott, N., Meunier, A. & Marignol, L. NUMB inhibition of NOTCH signalling as a therapeutic target in prostate cancer. Nat. Rev. Urol.11, 499–507 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Papafotiou, G. et al. KRT14 marks a subpopulation of bladder basal cells with pivotal role in regeneration and tumorigenesis. Nat. Commun.7, 11914 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kobayashi, T., Owczarek, T. B., McKiernan, J. M. & Abate-Shen, C. Modelling bladder cancer in mice: opportunities and challenges. Nat. Rev. Cancer15, 42–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Laufs, U. & Liao, J. K. Post-transcriptional regulation of endothelial nitric oxide synthase mRNA stability by Rho GTPase. J. Biol. Chem.273, 24266–24271 (1998). [DOI] [PubMed] [Google Scholar]
- 83.Fujihara, H. et al. Inhibition of RhoA translocation and calcium sensitization by in vivo ADP-ribosylation with the chimeric toxin DC3B. Mol. Biol. Cell8, 2437–2447 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ridley, A. J. & Hall, A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell70, 389–399 (1992). [DOI] [PubMed] [Google Scholar]
- 85.Zhao, B. et al. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev.26, 54–68 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Feng, X. et al. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer Cell25, 831–845 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ernkvist, M. et al. p130-angiomotin associates to actin and controls endothelial cell shape. FEBS J.273, 2000–2011 (2006). [DOI] [PubMed] [Google Scholar]
- 88.Gagne, V. et al. Human angiomotin-like 1 associates with an angiomotin protein complex through its coiled-coil domain and induces the remodeling of the actin cytoskeleton. Cell Motil. Cytoskeleton66, 754–768 (2009). [DOI] [PubMed] [Google Scholar]
- 89.Mana-Capelli, S., Paramasivam, M., Dutta, S. & McCollum, D. Angiomotins link F-actin architecture to Hippo pathway signaling. Mol. Biol. Cell25, 1676–1685 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Densham, R. M. et al. MST kinases monitor actin cytoskeletal integrity and signal via c-Jun N-terminal kinase stress-activated kinase to regulate p21Waf1/Cip1 stability. Mol. Cell Biol.29, 6380–6390 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Murrell, M., Oakes, P. W., Lenz, M. & Gardel, M. L. Forcing cells into shape: the mechanics of actomyosin contractility. Nat. Rev. Mol. Cell Biol.16, 486–498 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Su, D. et al. SPTAN1/NUMB axis senses cell density to restrain cell growth and oncogenesis through Hippo signaling. J. Clin. Invest.13310.1172/JCI168888 (2023). [DOI] [PMC free article] [PubMed]
- 93.Huggett, M. T. et al. Phase I/II study of verteporfin photodynamic therapy in locally advanced pancreatic cancer. Br. J. Cancer110, 1698–1704 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Banerjee, S. M. et al. Combination of verteporfin-photodynamic therapy with 5-aza-2’-deoxycytidine enhances the anti-tumour immune response in triple negative breast cancer. Front Immunol.14, 1188087 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barcelo, J., Samain, R. & Sanz-Moreno, V. Preclinical to clinical utility of ROCK inhibitors in cancer. Trends Cancer9, 250–263 (2023). [DOI] [PubMed] [Google Scholar]
- 96.Zilian, O. et al. Multiple roles of mouse Numb in tuning developmental cell fates. Curr. Biol.11, 494–501 (2001). [DOI] [PubMed] [Google Scholar]
- 97.Wilson, A. et al. Normal hemopoiesis and lymphopoiesis in the combined absence of numb and numblike. J. Immunol.178, 6746–6751 (2007). [DOI] [PubMed] [Google Scholar]
- 98.Ramirez, A. et al. A keratin K5Cre transgenic line appropriate for tissue-specific or generalized Cre-mediated recombination. Genesis39, 52–57 (2004). [DOI] [PubMed] [Google Scholar]
- 99.Stein, J. P. et al. Radical cystectomy in the treatment of invasive bladder cancer: long-term results in 1,054 patients. J. Clin. Oncol.41, 3772–3781 (2023). [DOI] [PubMed] [Google Scholar]
- 100.Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bankhead, P. et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep.7, 16878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst.1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The raw and processed RNA sequencing data generated in this study have been deposited in the GEO database under the accession code GSE256272. The clinicopathological data from the UROMOL cohort used in this study are available in the source data of the referenced publication24. Raw gene abundances were kindly provided by the authors. Human and mouse gene set annotations for pathway analysis were retrieved from the Molecular Signatures Database (MSigDB) [www.gsea-msigdb.org/gsea/msigdb/] with the “msigdbr” package (version 7.4.1) in R. Human GRCh38 and mouse GRCm39 reference genomes (primary genome assembly) used for the alignment of RNA sequencing data are available on GENCODE. Expression intensity data of CK5 and NUMB across diverse tissues were retrieved from the Human Protein Atlas [www.proteinatlas.org]. Materials and reagents are available upon request. Source data are provided with this paper.