Abstract
Few studies have analyzed the role of the lung microbiome in the diagnosis of pulmonary coinfection in ventilated ICU COVID-19 patients. We characterized the lung microbiota in COVID-19 patients with severe pneumonia on invasive mechanical ventilation using full-length 16S rRNA gene sequencing and established its relationship with coinfections, mortality, and the need for mechanical ventilation for more than 7 days. This study included 67 COVID-19 ICU patients. DNA extracted from mini-bronchoalveolar lavage fluid and endotracheal aspirates was amplified by PCR with specific 16S primers (27F and 1492R). General and relative bacterial abundance analysis was also conducted. Alpha diversity was measured by the Shannon and Simpson indices, and differences in the microbiota were established using beta diversity. A linear discriminant analysis (LDA) effect size algorithm was implemented to describe biomarkers. Streptococcus spp. represented 51% of the overall microbial abundance. There were no differences in alpha diversity between the analyzed variables. There was variation in bacterial composition between samples that had positive and negative cultures. The genera Veillonella sp., Granulicatella sp., Enterococcus sp. and Lactiplantibacillus sp., with LDA scores > 2, were biomarkers associated with negative cultures. Rothia sp., with an LDA score > 2, was a biomarker associated with mortality.
Subject terms: Microbiology, Diseases, Medical research, Molecular medicine
Introduction
COVID-19 can present with no symptoms, or with disease ranging from mild respiratory illness to severe lung injury, systemic inflammation, and ultimately death1. Mostafa et al.2 suggested that there is dysbiosis of the lung microbiome in patients with COVID-19 and that the microbiome participates in the development of critical illness by influencing both homeostasis and proinflammatory conditions.
Associations between alterations in the respiratory tract microbiome in severe COVID-19 with bacterial coinfection and mortality remain a matter of controversy. There are many confounding variables that affect the conclusions of the different studies such as differences in the analysis of the oropharyngeal microbiome vs lung microbiome, previous use of antibiotics, mechanical ventilation and length of hospitalization among others.
Kullberg et al.3 found that in 114 patients with COVID-19 and acute respiratory distress syndrome (ARDS) on mechanical ventilation, there was an association between increased bacterial and fungal loads in the lung microbiota and a decreased probability of release from invasive mechanical ventilation (IMV) and increased mortality. They included patients who were hospitalized longer than 48 h, reflecting an analysis of the nosocomial microbiota with possible pulmonary superinfection and not of the community microbiota with pulmonary coinfection.
Merenstein et al.4 analyzed 56 studies that investigated the lung microbiota in patients with COVID-19, 31 of which performed sequencing with amplification of the 16S rRNA gene; of the latter, only three took samples from the lower respiratory tract (LRT) of patients hospitalized in the intensive care unit (ICU). Only one study evaluated the contribution of the 16S/18S rRNA gene to the diagnosis of pulmonary coinfections by endotracheal aspiration in patients on IMV5. The sample included 34 patients whose mean number of lung samples taken was greater than 48 h prior to admission to the ICU [2.7 (0–16) days], and most patients (59%) had received antibiotics prior to respiratory sample collection, which interferes with the results.
We hypothesized that the best setting for studying these associations was critically ill patients, who had not been hospitalized for more than 48 h (reflecting community-acquired infection), and with lung microbiome studies. The main objective of this study was to determine whether the bacterial taxonomy through full-length 16S rRNA gene sequencing was positively related to lung coinfection based on the positivity of respiratory cultures and/or molecular tests of the Biofire® FilmArray® pneumonia panel (FA-PNEU) and with mortality and remaining on IMV on day 7 of the ICU stay. We included patients with COVID-19 on invasive mechanical ventilation (IMV) who were hospitalized for less than 48 h and who had not received antibiotics.
Results
The clinical characteristics of the 67 patients are described in Table 1. Twenty-two (32.4%) patients died, 59 (88.1%) patients were on IMV on day 7 of their ICU stay, and 22.5% and 30.9% of the cultures and FA-PNEU were positive, respectively.
Table 1.
Demographic characteristics of the 67 patients with severe COVID-19 pneumonia admitted to Intensive Care Units in Colombia.
| Characteristic (n = 67) | Frequency (%) |
|---|---|
| Men | 38 (55.9) |
| Age (Me: P25–P75) | 58 (46–66) |
| Hypertension | 27 (39.7) |
| Diabetes | 16 (23.5) |
| Chronic kidney disease | 1 (1.5) |
| Rheumatological disease | 0 (0) |
| Neoplasia | 3 (4.4) |
| Chronic obstructive pulmonary disease | 1 (1.5) |
| HIV | 1 (1.5) |
| Heart failure | 0 (0) |
| Cirrhosis | 1 (1.5) |
| Positive lung culture < 48 h | 15 (22.5) |
| Positive FA-PNEUM < 48 h | 17 (30.9) |
| Deads | 22 (32.4) |
| Ventilated patients on 7th day | 59 (88.1) |
| Pulmonary embolism | 9 (13.2) |
| SOFA score (Me: P25 – P75) | 2 (2–5) |
| APACHE II score (Me: P25 – P75) | 9 (6–12) |
| Lung superinfection | 28 (41.2) |
| Treatment | |
| Dexamethasone | 28 (41.7) |
| Methylprednisolone | 39 (58.2) |
| Intermediate dose or anticoagulation | 46 (68.6) |
| Tocilizumab | 1 (1.5) |
| Empirical antibiotics | 37 (55.2) |
| Ceftriaxone | 17 (30.9) |
| Cefepime | 12 (17.9) |
| Ampicillin/sulbactam | 9 (13.2) |
Me Median, P25–P75 25th and 75th percentiles, % percentage, HIV human immunodeficiency virus, APACHE II Chronic health disease classification system II, SOFA sequential organ failure assessment, FA-PNEUM Biofire® FilmArray® pneumonia panel.
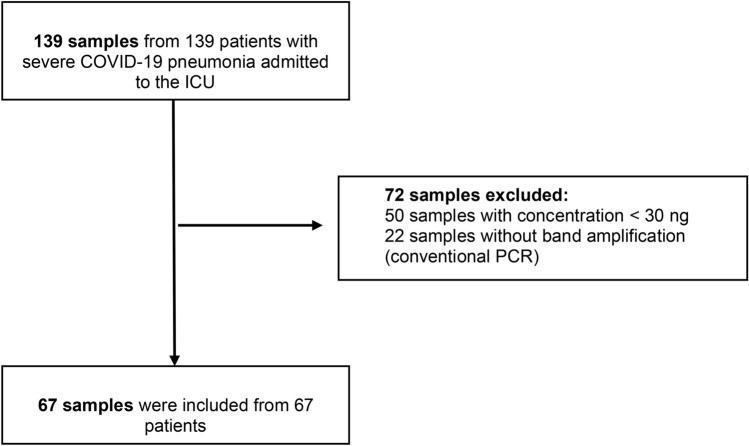
Supplementary Fig. 1 describes the overall library size of the 67 samples. A total of 2009 ASVs were filtered, leaving 279. A total of 8,509,922 reads were obtained, with a median of 127,562 reads (p25–75, 7765–457,678 reads).
The rarefaction curve in supplementary Fig. 2 displays that only four samples reach the plateau, those samples contain the higher read count in supplementary Fig. 1. In addition, in supplementary Table 1, the average Good’s coverage displays a minimum value of 76.47, a maximum of 100, a median 99.6 and interquartile range of 4.07. This suggests that it is necessary to sequence some samples in a high depth (Fig. 1).
Fig. 1.
Study flow chart.
Visual exploration
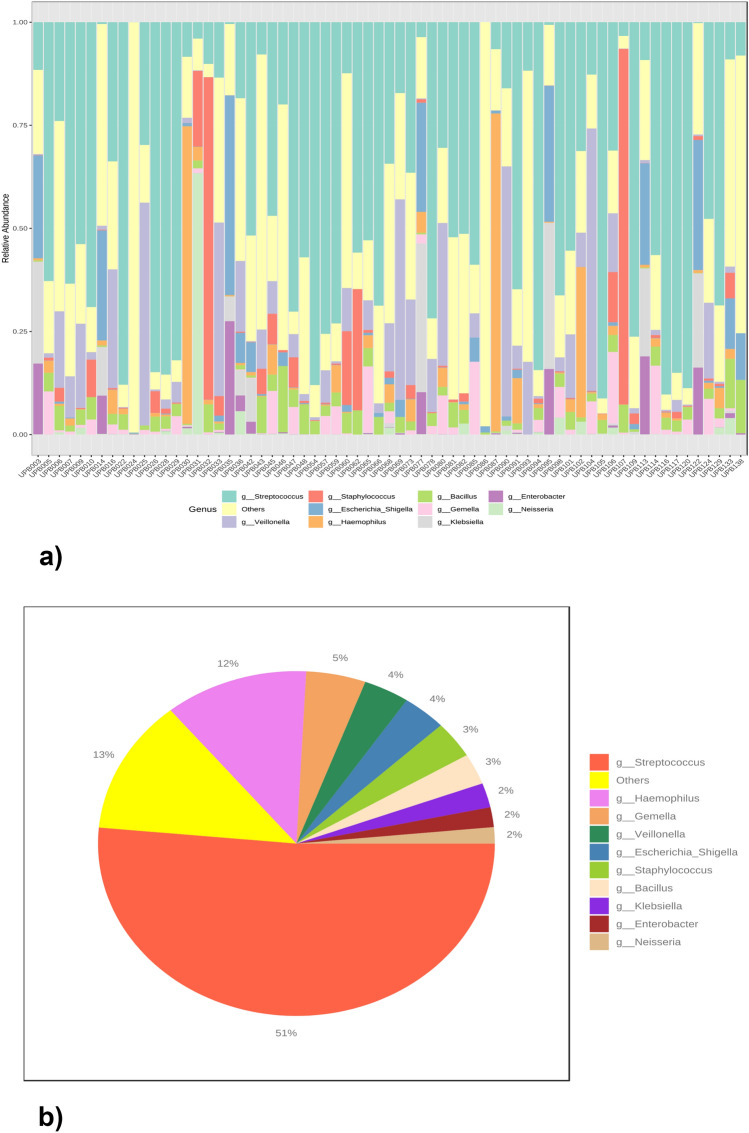
Figure 2a shows the relative abundance of the top ten taxa in all the samples. The three most common genera are Streptococcus, Veillonella and Staphylococcus. Figure 2b shows the general abundance profile, that is, the sum of each of the genera in all the samples, for the ten main taxa. Streptococcus accounted for 51%, and Haemophilus accounted for 12%.
Fig. 2.
(a) Relative bacterial abundance of the 67 lung samples. (b) General bacterial abundance.
We performed taxonomic bar graphs organized to make comparisons with the results related to pulmonary coinfection, that is, with positive and negative cultures and FA-PNEU. Supplementary Figs. 3 and 4.
Bacterial community profile
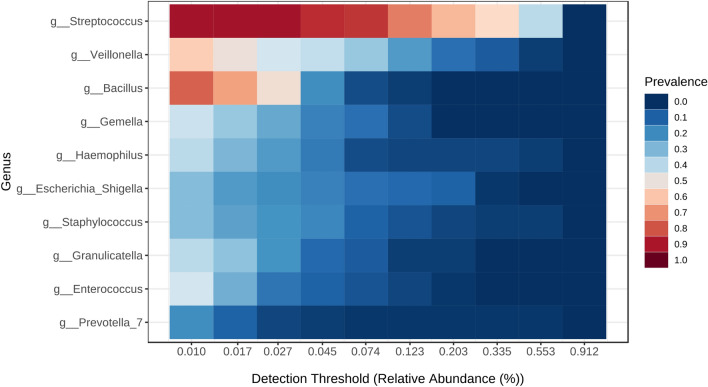
The heat profiles of the core microbiota are shown in Fig. 3. The genus Streptococcus was present in 90% of the samples with a minimum relative abundance of 2.7%, and Bacillus was present in 80% of the samples with a minimum relative abundance of 1%.
Fig. 3.
Core microbiota heat map.
The richness that determines the number of unique ASVs found in each sample at the genus level is shown in supplementary Fig. 5. There were no differences in richness between groups for the four variables: mortality (a), being ventilated on day 7 (b), and the positivity or not of the cultures (c) and the FA-PNEU (d).
We also did not find differences in α diversity, measured with the Shannon and Simpson indices, between the four variables. Supplementary Fig. 6.
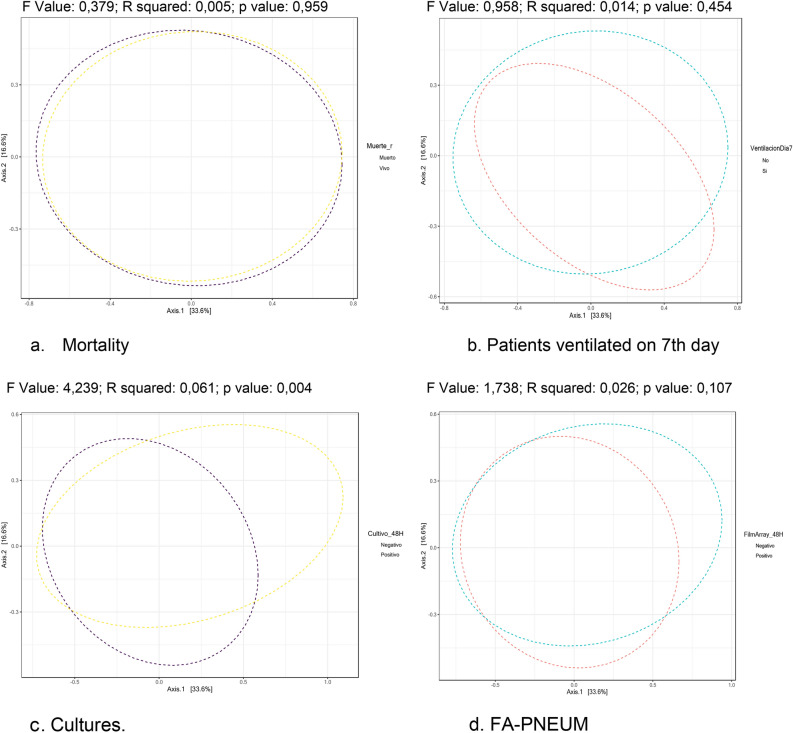
The β diversity for each of the four variables, mortality, mechanical ventilation on day 7, cultures and FA-PNEU, is shown in Fig. 4. There was a significant difference (F value: 4.2393; R squared: 0.061227; p value: 0.004) only between the bacterial communities when the culture was positive or negative.
Fig. 4.
Diversity β in each of the four variables: mortality (a), mechanical ventilation on day 7 (b), cultures (c) and AF-PNEU (d) in the 67 lung samples from the 67 patients with severe COVID-19 pneumonia. 19 admitted to Intensive Care Units in Colombia. There is only statistical significance between the bacterial communities with the cultures variable. Abbreviations: p p value with the PERNANOVA statistical method, R coefficient of determination, F F test, ratio of two variances, FA-PNEU Biofire® FilmArray® Pneumonia Panel.
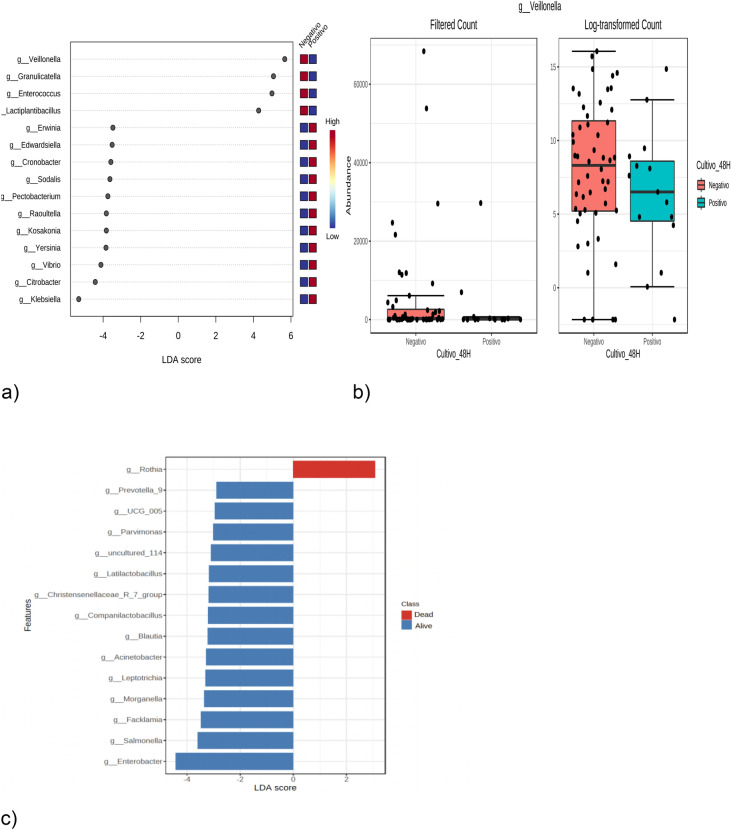
Using LefSe, we found that only the culture and mortality variables were significantly different between the groups. Figure 5a shows the LEfSe at the sex level based on the culture results (positive or negative). At the genus level, the presence of Veillonella, Granulicatella, Enterococcus and Lactiplantibacillus, with an LDA score > 2, are biomarkers of negative cultures; in other words, the patients who had that taxon in their lung samples were potentially protected from bacterial coinfection. Figure 5b shows the unifactor statistical comparison at the genus level with Veillonella and the culture variable (positive or negative); Veillonella with p = 0.0289, is associated with negative culture. Figure 5c shows the effect size of the linear discriminant analysis (LEfSe) at the genus level based on the mortality variable (living or dead); the presence of Rothia, with an LDA score > 2, and the presence of Rothia, with an LDA score > 2, were associated with mortality. In other words, patients who had Rothia in their lung samples were more likely to die.
Fig. 5.
(a) LEfSe at the gender level based on the culture results (positive or negative), in the 67 lung samples of the 67 patients with COVID-19 pneumonia admitted to Care Units Intensives in Colombia. (b) Unifactor statistical comparison, at the genus level with Veillonella and the culture variable (positive or negative). (c) Effect size of the linear discriminant analysis (LEfSe) at the genus level based on the mortality variable (living or dead). Abbreviations: LDA linear discriminant analysis, g genus, High high probability, Low low probability.
Correlations between metataxonomy and serum cytokine levels
Only three of the 9 evaluated cytokines were correlated with any microbial genus. Supplementary Table 2. Interferon gamma has a weak positive correlation with Fusobacterium; that is, the higher the concentration of interferon gamma, the greater the abundance of this bacterial genus. Interleukin 2 (IL-2) has a weak negative correlation with the genus Erysipelatoclostridium, and IL 1-beta has a weak positive correlation with Fusobacterium. None of the 7 most frequently isolated microorganisms according to sequencing were correlated with cytokines.
Discussion
The most important findings of the present study are as follows: first, the genus Streptococcus spp. has the highest overall abundance; second, there are differences in bacterial composition between samples that had positive culture results and negative cultures; third, the presence of Veillonella sp., Granulicatella sp., Enterococcus sp. and Lactiplantibacillus sp. are biomarkers of negative cultures; and fourth, the presence of Rothia is a biomarker of mortality.
Rothia sp. is associated with the development of ARDS in patients with COVID-19 pneumonia6. Han et al.7 performed a metatranscriptomic study in BAL fluid from 19 patients with COVID-19 and 23 healthy controls and found a correlation between Rothia mucilaginosa and SARS-CoV-2, suggesting that it plays a role in the host’s inflammatory response. We did not find a correlation between Rothia and IL-1β; we previously described that patients with IL-1β levels < 1365 pg/mL had increased mortality. The levels of only three cytokines, IFN-γ, GM-CSF and IL-17A, were correlated with the levels of some genera; however, we did not find studies on COVID-19 with similar findings.
Veillonella sp., Granulicatella sp. and other opportunistic oral pathogens have been detected in the BAL fluid of patients with COVID-198. Kumar D et al.9 reported that Veillonella was a biomarker for survival in COVID-19 pneumonia patients and improved the inflammatory response in these patients. Sulaiman et al.10 in an observational cohort of 142 critically ill adults with COVID-19 who underwent bronchoscopy, analyzed the lower respiratory tract microbiome using metagenomics and metatranscriptomics. Poor clinical outcome was associated with lower airway enrichment with an oral commensal (Mycoplasma salivarium).
Meng et al.11 performed metatranscriptomic sequencing in 72 patients with severe COVID-19 pneumonia and 57 patients who had already recovered, finding that Veillonella rodentium was a marker of recovery. We found that the presence of Veillonella in the lung samples of patients was a biomarker of negative cultures. Whether this lower probability of bacterial coinfection is a possible explanation for the higher probability of recovery is a possible hypothesis. We did not find similar studies in severe COVID-19 pneumonia that correlated the genera Veillonella sp., Granulicatella sp., Enterococcus sp. and Lactiplantibacillus sp. by metataxonomy with negative cultures.
The main limitation of the study was that the samples had a low concentration of the isolated DNA. The lung samples from these patients with severe COVID-19 pneumonia in the IMV were scant, bloody, and contained a large amount of mucus, so we performed pretreatment to break up and inactivate the mucus. Other limitations were as follows: first, we did not have control samples from ventilated patients without pneumonia or from patients with pneumonia not related to COVID-9, which could serve as a reference for the community microbiota and be able to determine whether there was dysbiosis. Second, there was variability in the number of reads obtained from the samples, some with few reads and others with many reads. Third, metataxonomy does not provide species-level resolution for comparisons with cultures and the FA-PNEU.
The main strength of our research is the scarcity of studies in patients with severe community-acquired pneumonia due to COVID-19 in the ICU at VMI, in which lung samples were taken to analyze the correlation between sequencing with amplification of the 16S rRNA gene with cultures and FA-PNEU. Thomsen et al.5 evaluated the diagnosis of respiratory coinfections with the analysis of 16S/18S rRNA gene amplicons in 34 patients with severe COVID-19. They detected potential pathogens in four patients (12%) with the 16S gene, seven patients (21%) using conventional cultures, and one patient (3%) using a molecular respiratory panel. Fourteen patients had not received antibiotics, and four microorganisms (3%) Haemophilus influenzae and 1 Fusobacterium necrophorum) were detected in the 16S gene. They concluded that metataxonomy complements conventional microbial diagnosis.
Lloréns-Rico et al.12 studied 21 patients with V4 amplification of the 16S gene in BAL fluid samples and demonstrated that the length of stay in the ICU, IMV and the use of antibiotics explained the greatest variation within the lung microbiota. We did not find differences in the composition of the bacterial communities between the samples of patients who were or were not on IMV for more than 7 days or in mortality. Merenstein et al.13 analyzed V1-V2 of the 16S rRNA gene in endotracheal aspirates from 24 patients with COVID-19 on IMV, with a median of 4 days of hospitalization. Six patients had taxa dominated by Staphylococcus, and three were a prominent minority; only 3 patients had Staphylococcus aureus identified by culture, suggesting that sequencing was more sensitive than culture. In our study, Staphylococcus was detected by taxonomy in 58 of the 67 lung samples (86.56%).
Another strength is that the patients did not receive previous antibiotics, and their samples were taken in the first 12 h of IMV, which reduces the probability of alteration of the bacterial composition. Castilhos et al.14 demonstrated that the use of antibiotics leads to greater dysbiosis in critically ill patients with COVID-19. IMV alters the microbiome, allowing the growth of opportunistic pathogens4.
We used Oxford Nanopore technology with a GridION (ONT) device, and the articles used Illumina MiSeq technology. Sequence reads are longer with Nanopore, and its accuracy, per base, is lower than that of MiSeq (95% vs. 99.9%)15, so we would be biased in performing comparisons. Heikema et al.16 compared both technologies in 59 nasal swab samples in which Nanopore presented problems in the detection of bacteria of the genus Corynebacterium. Another difference is that we sequenced the complete 16S rRNA gene and not the individual hypervariable regions, which improved the precision of the measurements of bacterial diversity17.
Conclusion
We found that the Streptococcus genus had the highest overall abundance. The presence of Veillonella, Granulicatella, Enterococcus and Lactiplantibacillus are biomarkers of negative cultures, and the presence of Rothia is a biomarker of mortality.
Materials and methods
This was a case‒control study nested within a cohort of 139 patients included in the original study18. The patients were > 18 years of age and admitted to nine ICUs in Medellín, Colombia, with severe COVID-19 infection on mechanical ventilation. To meet the coinfection criteria, patients could not have been hospitalized for more than 48 h at the time of LRT sampling. Patients who had received any dose of empiric antimicrobial therapy were excluded. However, the treating physicians were free to initiate antibiotics after taking the respiratory samples. The study was conducted between March 1 and July 30, 2021. Respiratory therapists in each ICU collected samples from the lower respiratory tract on the first day of intubation with mini-bronchoalveolar lavage fluid [mini-BAL] or endotracheal aspirate [ETA]. Of the total volume of samples, 5–10 ml (mL) was distributed for BioFire® FilmArray® Pneumonia Panel (FA-PNEU) testing, 5–10 ml for conventional culturing, and another 5–10 ml for lung microbiota analysis with the extraction of deoxyribonucleic acid (DNA). The samples were not processed when a quality level of 0 or 1 was detected by the Murray/Washington criteria, based on the number of squamous cells and neutrophils per field19. Venous blood samples were collected upon admission to the ICU, in order to measure cytokine levels. Patients had a median of 9 days of symptom onset from day 1 of stay.
A total of 139 samples were pretreated with 300 mL of isopropanol, 20 mL of 10 M NaOH, 179.1 mL of water and 0.9 mL of Tween-20 (volume of 500 mL) to break up mucus and inactivate respiratory samples. The protocol used was as follows: in a 15 ml conical tube, 400 μl of sample and 1200 μl of pretreatment reagent were added, the tube was vortexed for 30 to 60 s, incubated at room temperature for a maximum of 2 h and vortexed again. A total of 450 μl of the mixture was removed, placed in 1.5 ml vials and centrifuged at 13,000 rpm for 1 min. Finally, 400 μl was removed without disturbing the generated pellet. The automated extraction protocol was then carried out with the MagMAX DNA Multi-Sample Ultra kit (Applied Biosystems, San Francisco, USA) using the KingFisher Flex System following the manufacturer’s instructions for a final elution of 40 μl of genetic material. Qubit quantification was performed by fluorometric DNA quantification (Qubit dsDNA HS Assay kit, for Qubit 3.0, Life Technologies). Of the 139 samples, 89 had an optimal concentration (30 ng).
Quality filtering was performed with conventional PCR using the universal primers 27F and 1492R, with the objective of verifying the amplification of the band corresponding to the 16S rRNA gene. A total of 67 samples were considered optimal, and the 16S rRNA gene was sequenced using Oxford Nanopore technology on a GridION (ONT) device. The flow chart of the study is shown in Fig. 1. Four negative controls (no DNA template control) were included in the analyses to control for contamination of the reagents. According to the taxonomy classification of non-template controls and patients, we cannot be sure that the 16S sequences detected belong to pathogenic or environmental species. In addition, the classification was limited to genus level.
16S DNA sequencing
DNA was amplified by PCR using specific 16S primers (27F and 1492R) containing 5’ tags facilitating ligase-free ligation of rapid sequencing adapters (https://store.nanoporetech.com/16s-barcoding-kit-1-24.html).
The 16S Barcoding Kit 1-24 SQK-16S024 (Oxford, Nanopore, Florida, USA) was used for library preparation and sequencing following the protocol recommended by the manufacturer. One microliter of the purified library was quantified using a Qubit 3.0 (Life Technologies, USA) following the manufacturer’s instructions. Subsequently, all the libraries were loaded into an R9.4 flow cell (Oxford Nanopore, UK) and run in the GrdION Mk1 system (Oxford Nanopore, UK).
Predictor variables
The main objective was to determine whether the bacterial taxonomy was positively related to the FA-PNEU and conventional culture results. Other objectives were to determine the relationships of the metataxonomy with mortality and remaining on IMV on day 7 of the ICU stay. We hypothesize that bacterial DNA richness, α diversity, and bacterial community composition would serve as predictors of the outcomes defined by these four variables.
Additionally, we examined the relationship of bacterial taxonomy with the serum levels of eight cytokines (pg/ml) taken at ICU admission: IL-1β, IL-2, IL-6, IL-10, IL-12p70, IL-18, IFN-γ and TNFα. The Human ProcartaPlex TM Multiplex Immunoassay Mix & Match of 10-Plex system based on magnetic beads was used to detect the serum biomarker PPX-10-MX323G4 (Invitrogen, Whatman, Massachusetts, United States). The 10 cytokines were analyzed using the LuminexR MAGPIX R System (Thermo Fisher Scientific, Luminex Corporation 12,212 Technology Blvd. Austin, Texas 78,727)20.
Bioinformatics analysis
The generation of raw data in fastq format was performed with MinKNOW software. The adapters and chimeras were removed (porechop v:0.2.4 and fastp v0.23.4), leaving only reads between 1000 and 2000 base pairs with quality scores > 9. Taxonomic classification was carried out with Kraken v2.1.3 and the SILVA v138.1 database.
We used the open access analysis platform MicrobiomeAnalyst (https://www.microbiomeanalyst.ca) with 4 variables: mortality (alive-dead), mechanical ventilation on day 7 (yes–no), culture (positive–negative) and FA-PNEU (positive–negative). Filtering of the ASVs was performed, excluding those with a low count of 3 reads in less than 20% of the samples. Cumulative sum scaling (CSS) was used for data normalization.
We performed a visual exploration by calculating the relative abundance of all samples and an overall abundance profile using a pie chart. A profile of the bacterial community was generated through the following steps: one, a heatmap of the core microbiota at the genus taxonomic level, with a minimum sample prevalence of 20% and a relative abundance of 0.01%; two, the calculation of microbial richness at the genus level, with the use of the Mann‒Whitney test as a statistical method to compare the richness between the four variables; three, the α diversity measured with the Shannon and Simpson indices, with the use of the Mann‒Whitney test as a statistical method to compare the indices between the four variables; and four, the differences in the composition of the bacterial community, with β diversity using the main coordinate analysis, with the Bray‒Curtis index at the genus level, with the PERMANOVA statistical method.
We used the linear discriminant analysis effect size (LEfSe) algorithm to identify and interpret sex-related biomarkers for each of the four variables, with the Kruskal‒Wallis rank sum test (p value of 0.1) and univariate statistical comparisons with the t test, with a p value < 0.05 indicating statistical significance.
Statistical analyses
The median and interquartile range were calculated for the continuous variables, and the categorical variables are presented as frequencies and percentages. To compare the concentrations of serum cytokines with those of microorganisms identified by metataxonomy, the Spearman correlation coefficient was used, with a predefined value greater or less than 0.6 and a p < 0.05. The data were analyzed using R version 4.2.3 software.
Supplementary Information
Acknowledgements
The authors acknowledge the institutions participating in the study with the Respective collaborators: Clínica Universitaria Bolivariana, Francisco Molina; Clínica El Rosario Tesoro, Álvaro Ochoa; Clínica CardioVid, Juan David Uribe; Clínica Sagrado Corazón, Nelson Fonseca; Clínica Las Américas Auna, Bladimir Gil; Clínica Medellín, Juan Echeverry; Hospital La María, Marco González; Hospital Manuel Uribe Ángel, Victoria Ángel; and Hospital Pablo Tobón Uribe, Gisella de la Rosa.
Abbreviations
- COVID-19
Coronavirus disease 2019
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- LDA
Linear discriminant analysis
- ARDS
Acute respiratory distress syndrome
- IMV
Invasive mechanical ventilation
- LRT
Lower respiratory tract
- ICU
Intensive care unit
- FA-PNEU
Biofire® FilmArray® pneumonia panel
- DNA
Deoxyribonucleic acid
- CSS
Cumulative sum scaling
- LEfSe
Discriminant analysis effect size
- GM-CSF
Human granulocyte and macrophage colony-stimulating factor
- IFN-γ
Interferon gamma
Author contributions
FJM, LEB, JPI, LEC, LL, LV and AT designed the study; collected, compiled, analyzed and interpreted the data; and wrote the manuscript. AJA, IM, LSP, JU, KC, and JPH and QM performed laboratory analyses, interpreted the data and wrote the manuscript. RLA and LF interpreted the data and wrote the manuscript. All authors approved the final version of the manuscript.
Funding
This study was funded by Minciencias, Colombia, 121084468048.
Data availability
The datasets generated during and analysed during the current study are available in the National Center for Biotechnology Information Repository. The BioProject accession number is PRJNA1129550. The SRA records will be accessible with the following link: https://www.ncbi.nlm.nih.gov/sra/PRJNA1129550.
Declarations
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
This study was approved by the ethics committee of the Universidad Pontificia Bolivariana and by the committees of the clinics and hospitals that participated in the study. Written informed consent was obtained from the participants or their legal representatives.
Consent for publication
Not applicable.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-81738-8.
References
- 1.Chen, N. et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet395, 507–513 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mostafa, H. H. et al. Metagenomic next-generation sequencing of nasopharyngeal specimens collected from confirmed and suspect COVID-19 patients. mBio11, e01969-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kullberg, R. F. J. et al. Lung microbiota of critically Ill patients with COVID-19 are associated with nonresolving acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med.206, 846–856 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merenstein, C., Bushman, F. D. & Collman, R. G. Alterations in the respiratory tract microbiome in COVID-19: Current observations and potential significance. Microbiome10(1), 165 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomsen, K. et al. Extensive microbiological respiratory tract specimen characterization in critically ill COVID-19 patients. APMIS129, 431–437 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Battaglini, D. et al. The role of dysbiosis in critically Ill patients with COVID-19 and acute respiratory distress syndrome. Front. Med. (Lausanne)8, 671714 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han, Y., Jia, Z., Shi, J., Wang, W. & He, K. The active lung microbiota landscape of COVID-19 patients through the metatranscriptome data analysis. Bioimpacts12, 139–146 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao, L. et al. Oral microbiome and SARS-CoV-2: Beware of lung co-infection. Front. Microbiol.31(11), 1840 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar, D. et al. Nasopharyngeal microbiome of COVID-19 patients revealed a distinct bacterial profile in deceased and recovered individuals. Microb. Pathog.173, 105829 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sulaiman, I. et al. Microbial signatures in the lower airways of mechanically ventilated COVID-19 patients associated with poor clinical outcome. Nat. Microbiol.6, 1245–1258 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng, H. et al. Respiratory immune status and microbiome in recovered COVID-19 patients revealed by metatranscriptomic analyses. Front. Cell Infect. Microbiol.12, 1011672 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lloréns-Rico, V. et al. Clinical practices underlie COVID-19 patient respiratory microbiome composition and its interactions with the host. Nat. Commun.12, 6243 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merenstein, C. et al. Signatures of COVID-19 severity and immune response in the respiratory tract microbiome. Bio12, e0177721 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Castilhos, J. et al. Severe dysbiosis and specific haemophilus and neisseria signatures as hallmarks of the oropharyngeal microbiome in critically Ill coronavirus disease 2019 (COVID-19) patients. Clin Infect Dis75, e1063–e1071 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stevens, B. M., Creed, T. B., Reardon, C. L. & Manter, D. K. Comparison of oxford nanopore technologies and Illumina miseq sequencing with mock communities and agricultural soil. Sci. Rep.13, 9323 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heikema, A. P. et al. Comparison of illumina versus nanopore 16S rRNA gene sequencing of the human nasal microbiota. Genes11, 1105 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.del Rosario, R. M. & del Carmen, M. M. Identification of bacteria through 16S rRNA sequencing: Principles, methods and applications in clinical microbiology. Enferm. Infecc. Microbiol. Clin.22(4), 238–45 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Molina, F. J. et al. Diagnostic concordance between BioFire® FilmArray® pneumonia panel and culture in patients with COVID-19 pneumonia admitted to intensive care units: The experience of the third wave in eight hospitals in Colombia. Crit. Care26, 130 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alvarez-Lerma, F. et al. Reccomendations for the diagnosis of pneumonia associated with mechanical ventilation. Enferm. Infecc. Microbiol. Clin.19, 479–487 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Molina, F. J. et al. Cytokine levels as predictors of mortality in critically ill patients with severe COVID-19 pneumonia: Case-control study nested within a cohort in Colombia. Front. Med.9, 1005636 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and analysed during the current study are available in the National Center for Biotechnology Information Repository. The BioProject accession number is PRJNA1129550. The SRA records will be accessible with the following link: https://www.ncbi.nlm.nih.gov/sra/PRJNA1129550.