Summary
Epileptic encephalopathies are severe epilepsy syndromes characterized by early onset and progressive cerebral dysfunction. A nonsense variant in the DALR anticodon binding domain containing 3 (DALRD3) gene has been implicated in epileptic encephalopathy, but no other disease-associated variants in DALRD3 have been described. In human cells, the DALRD3 protein forms a complex with the METTL2 methyltransferase to generate the 3-methylcytosine (m3C) modification in specific arginine tRNAs. Here, we identify an individual with a homozygous missense variant in DALRD3 who displays developmental delay, cognitive deficiencies, and multifocal epilepsy. The missense variant substitutes an arginine residue to cysteine (R517C) within the DALR domain of the DALRD3 protein that is required for binding tRNAs. Cells derived from the individual homozygous for the DALRD3-R517C variant exhibit reduced levels of m3C modification in arginine tRNAs, indicating that the R517C variant impairs DALRD3 function. Notably, the DALRD3-R517C protein displays reduced association with METTL2 and loss of interaction with substrate tRNAs. Our results uncover another loss-of-function variant in DALRD3 linked to epileptic encephalopathy disorders. Importantly, these findings underscore DALRD3-dependent tRNA modification as a key contributor to proper brain development and function.
Keywords: DALRD3, METTL2, 3-methylcytosine, m3C, epilepsy, epileptic encephalopathy, tRNA, neurodevelopment
This study identifies a homozygous missense variant in the DALRD3 gene of an individual with developmental and epileptic encephalopathy. The variant causes loss of function by perturbing DALRD3 protein levels and molecular interactions required for a specific tRNA modification. These findings provide evidence that DALRD3-dependent tRNA modification is required for proper neurodevelopment.
Introduction
The post-transcriptional modification of tRNA has emerged as a critical modulator of biological processes ranging from gene expression to development.1,2 In particular, tRNA modifications play critical roles in tRNA structure, stability, and function in protein synthesis.3,4,5 Defects in tRNA modification have emerged as the cause of many types of human diseases, highlighting the critical role of tRNA modification in human health and physiology.6,7,8 Notably, the brain appears to be sensitive to perturbations in tRNA modifications, as evidenced by the numerous neurological and neurodevelopmental disorders linked to deficiencies in tRNA modification.9,10
In mammalian cells, a subset of arginine tRNAs contain the 3-methylcytosine (m3C) modification at position 32 of the anticodon loop.11,12 The m3C modification is hypothesized to stabilize the folding of the anticodon loop and has been shown to influence mitochondrial tRNA structure in human cells.13,14 Thus, the m3C modification could affect the function of tRNAs in translation and protein expression (reviewed in Bohnsack et al.15). In human cells, the METTL2 methyltransferase forms a complex with the DALR anticodon binding domain containing 3 (DALRD3) protein to methylate tRNA-Arg-UCU and tRNA-Arg-CCU.16 The DALRD3 protein recognizes specific arginine-tRNA isoacceptors to target them for methylation by the METTL2 methyltransferase. Human cells deficient in DALRD3 exhibit nearly complete loss of m3C in tRNA-Arg-UCU and Arg-CCU, demonstrating that DALRD3 plays a key role in m3C formation in specific arginine tRNAs.16
We have previously identified two sibling individuals with a homozygous nonsense variant in exon 9 of the DALRD3 gene (rs1163930676, NM_001276405.1:c.1251C > A [p.(Tyr417∗)]).16 The C-to-A transversion introduces a premature stop codon in the DALRD3 mRNA and results in the loss of DALRD3 protein expression. The two individuals were born from consanguineous parents who are heterozygous for the nonsense variant. The parents are healthy and do not exhibit any detectable pathologies. In contrast, the homozygous siblings display a collection of clinical symptoms classified as severe developmental and epileptic encephalopathy disorder.17,18 Cells from the affected siblings exhibit a substantial reduction in m3C modification in tRNA-Arg-CCU and tRNA-Arg-UCU. These findings suggest a crucial biological role for DALRD3-dependent tRNA modification in development and nervous system function. However, the DALRD3 nonsense allele represents the only case of a DALRD3 variant that has been implicated in a neurodevelopmental disorder.
Here, we identify and characterize a DALRD3 missense variant in an individual with developmental delay and epileptic encephalopathy. The person described here is characterized by a similar set of symptoms as the previously described individuals with nonsense variants in DALRD3. We demonstrate that fibroblast cells from the affected individual are greatly reduced in m3C modification in arginine tRNAs. Moreover, we demonstrate that the amino acid substitution caused by the missense variant abrogates the ability of DALRD3 protein to interact with substrate tRNAs. Our study reveals a loss-of-function variant in DALRD3 linked to epileptic encephalopathy and further substantiates a role for DALRD3 protein function in ensuring proper neurodevelopment and neurological function.
Material and methods
Materials and methods can be found in the supplemental information.
Results
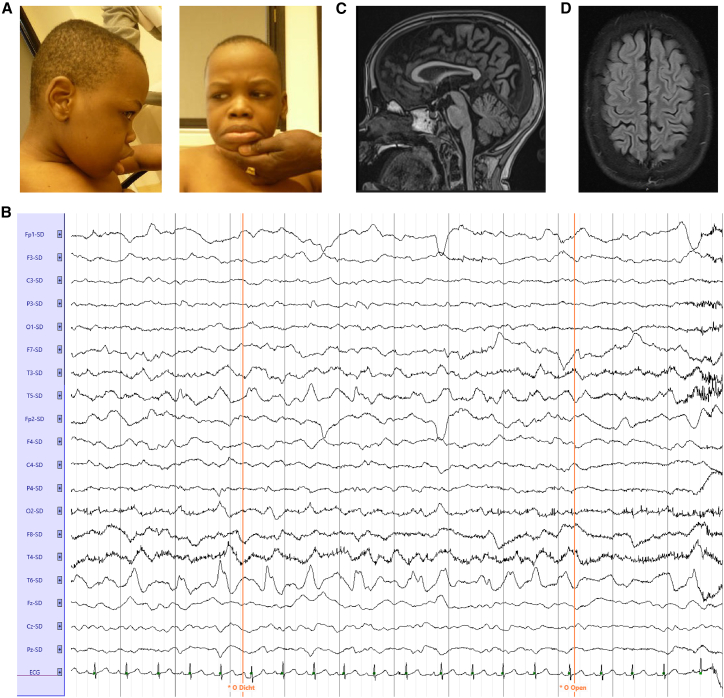
Based upon a match using the GeneMatcher platform,19 we describe an individual exhibiting severe developmental delay and multi-focal epilepsy associated with a variant in DALRD3 (Table 1, individual 1). The individual is a male of Sudanese ancestry who was born at term via cesarian section due to a previous cesarian section in the mother. Seizures started at age 4 years after a febrile episode followed by regression of psychomotor skills. The individual was referred to a pediatric neurologist again at 15 years of age for analysis because of progression of his symptoms. At neurological examination the affected individual displayed a dropped head and reduced facial expressivity (Figure 1A). In addition, he had progressive immobility, limited speech consisting of one-word expressions, increased muscle tone in his arms, subtle myoclonus, and ataxia. The individual has always had at least several epileptic seizures per month despite treatment with anti-epileptic drugs at therapeutic doses (see the supplemental note in the supplemental information for the full clinical description).
Table 1.
Clinical phenotype of individuals with homozygous variants in DALRD3
| Individual | 1 (this study) | 2 (Lentini et al.16) | 3 (Lentini et al.16) |
|---|---|---|---|
| ID | UMCG 2163660 | 19DG0509 | 19DG0510 |
| Gender | male | male | female |
| Genotype | NM_001009996.2: c.1549C>T: | NM_001276405.1:c.1251C>A: | NM_001276405.1:c.1251C>A: |
| ClinVar | VCV002234459.2 | VCV000918077.6 | VCV000918077.6 |
| ClinGen allele | CA2389547 | CA352710392 | CA352710392 |
| GnomAD | 3-49015671-G-A | 3-49016236-G-T | 3-49016236-G-T |
| Protein | p.(Arg517Cys) | p.(Tyr417∗) | p.(Tyr417∗) |
| Perinatal history | uncomplicated pregnancy, born term by cesarean section due to previous caesarean section | normal spontaneous vaginal delivery with history of placental insufficiency and oligohydramnios | Full-term product of caesarean section due to breech presentation, oligohydramnios, and placental insufficiency |
| Weight at birth (kg) | unknown | 2.25 (−2.2 SD) | 2.5 (fourth centile) |
| Developmental delay | severea | severe | severe |
| Motor | progressively immobile,b can walk some steps unaided | immobile | immobile |
| Speech | speaks very few wordsc | nonverbal | nonverbal |
| Seizures | seizures started at age 4 years after febrile episoded; thereafter regression of psychomotor skills; focal clonic seizures with impaired awareness to bilateral seizures,e,f often in clusters up to 10, during the night/early morning; at age 15 years, interictal subtle myoclonus of the hands and fingersg | seizures started at age 7 months in the form of myoclonic jerks which remains frequent and poorly controlled by antiepileptic medications | at age 6 months epilepsy ensued, initially as brief episodes of flexion tonic spasm of head followed by myoclonic seizures; unlike the sibling brother, the epilepsy of affected individual 2 is reasonably controlled by antiepileptic medications |
| EEG | intermittent focal epileptic phenomena in temporal regions,h slow background activity without normal differentiation,i high-amplitude slow waves over frontotemporal regions | independent multifocal epileptic discharges predominantly over the anterior head region bilaterally as well as over the right temporal and right parietal regions | markedly high voltage and slow background for age along with slow generalized polyspike and wave activity |
| Tone | axial hypotonic,j extremities slightly hypertonick; initially higherl and later low tendon reflexesm with extensor plantar responses; some ataxia of gait and handsn,o | axial and peripheral hypotonia with dystonic-like movement and generalized muscle wasting | central and peripheral hypotonia with dystonic-like movements and generalized muscle wasting |
| Microcephaly | no | no | yes |
| Brain finding | mild diffuse parenchymal volume lossp; normal aspect of white matter, basal ganglia, thalamus, and brain stem; arteriovenous malformation in right cerebellar hemisphereq; otherwise normal cerebellum; thickened skull by broad diploic spacer | mild diffuse brain parenchymal volume loss with diffuse paucity of the myelin within the brain parenchyma | normal topographical and morphological appearance of the infratentorial and supratentorial structures |
| Audiology assessment | N/A | moderate to severe conductive hearing loss in left ear and mild conductive hearing loss in right ear | N/A |
| Dysmorphism | none; prepubertal at age 13 yearss | subtle facial dysmorphia and small left ear | microcephaly with subtle facial dysmorphia |
| Other | marked lethargy and somnolencet,u; social interaction present, enjoys baby play; short stature (−3.7 SD) with delayed bone agev | severe gastroesophageal reflux disease necessitating gastronomy tube placement and fundoplication at age 4 years; no visual tracking or social smile | vomiting and choking on first day of life, mild congenital heart disease that resolved spontaneously, ectopic right kidney, bilateral optic disc pallor |
HP:0011344.
HP:0002505.
HP:0001344.
HP:0002373.
HP:0002266.
HP:0002069.
HP:0001336.
HP:0010857.
HP:0010845.
HP:0008936.
HP:0002509.
HP:0001347.
HP:0001315.
HP:0002066.
HP:0002070.
HP:0002283.
HP:0100026.
HP:0000929.
HP:0000823.
HP:0001254.
HP:0100786.
HP:0004322.
Figure 1.
Identification of an individual exhibiting epileptic encephalopathy and brain pathologies
(A) Affected individual 2163660 containing a homozygous c.1549C>T (NM_001009996.3) variant in DALRD3.
(B) EEG trace; Laplacian montage with eyes closed and opened.
(C) MRI of the affected individual; sagittal T1 Magnetization Prepared Rapid Gradient Echo imaging.
(D) MRI of the affected individual; transversal T2 Turbo Spin Echo imaging.
Electroencephalogram (EEG) testing revealed slow background activity without normal differentiation, intermittent focal epileptic phenomena in temporal regions, and high-amplitude slow waves over frontotemporal regions (Figures 1B and S1). No microcephaly was detected, but MRI showed a relatively thin corpus callosum, widening of sulci and the ventricular system as a sign of parenchymal loss, and an arteriovenous malformation in the right cerebellar hemisphere (Figures 1C, 1D, and S2). Based upon the pattern and onset of symptoms, the individual matches clinical conditions classified as developmental and epileptic encephalopathy (DEE 86 [MIM: 618910]).17
We used exome sequencing of the affected individual followed by our previously described strategy of variant selection and classification to identify homozygous variants with potential pathogenicity (Table S1).20 The candidate genes were then filtered by published association with epilepsy, which yielded a variant of unknown significance in the DALRD3 gene (ClinVar: VCV002234459.2). The variant is a homozygous nucleotide substitution resulting in a C-to-T transition in the last exon of the DALRD3 gene (rs140081609; MIM: 618904; Figures 2A and S3). Sanger sequencing confirmed the homozygous nucleotide substitution in the affected individual but not in a healthy control individual (Figure S4). The missense variant causes a substitution in the encoded DALRD3 protein of the canonical transcript (NM_001009996.3: c.1549C>T [p.(Arg517Cys)]). Translation of this transcript is expected to produce a protein containing an arginine-to-cysteine substitution at position 517 (R517C) in the DALR tRNA anticodon-binding domain (Figure 2A, protein). The parents of the affected individual are consanguineous and heterozygous carriers of the DALRD3 variant (Figures 2B and S5). Both parents are healthy with normal development and no detectable epilepsy. The DALRD3 variant was not detected in the individual’s older brother, who is healthy, developed normally, and did not exhibit epilepsy. The genotype of the sister is unknown, but she is healthy and exhibits normal development. These studies suggest that the R517C missense variant segregates with the disease in the family in an autosomal-recessive manner, being heterozygous in the parents, absent in the healthy sibling, and homozygous in the affected individual.
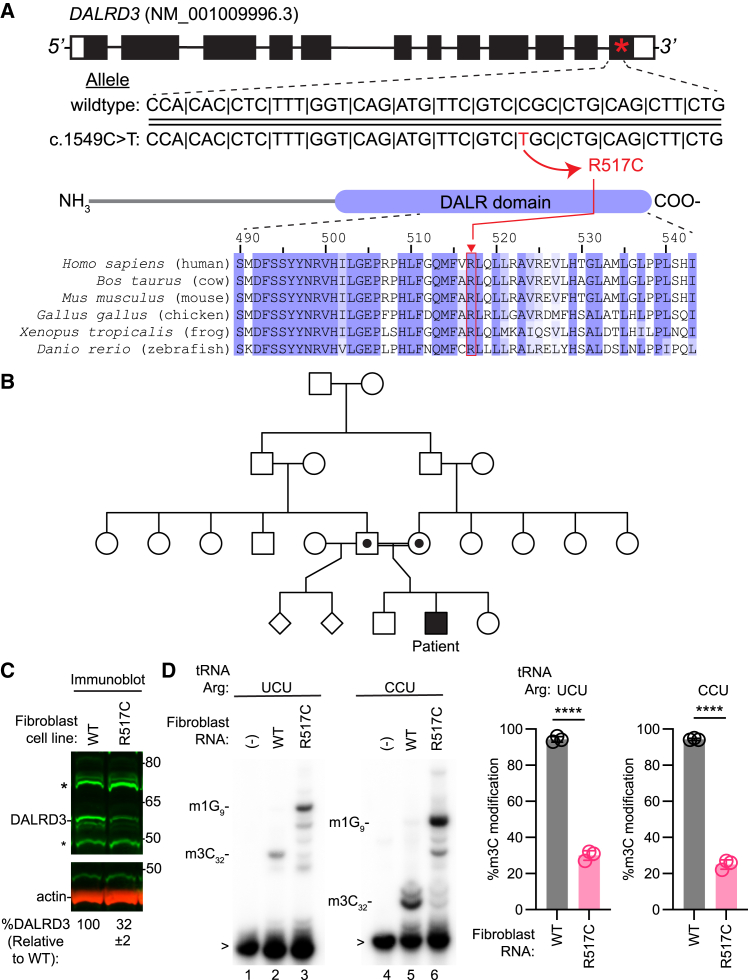
Figure 2.
Identification of a missense variant in the DALRD3 gene that impacts m3C modification status in arginine tRNAs
(A) DALRD3 exon and intron structure with encoded protein shown below. An asterisk represents the location of the variant in the mRNA (NM_001009996.3). WT and c.1549C>T alleles are shown, with the variant shown in red. The schematic depicts the domain structure of the DALRD3 protein and alignment of the region encompassing the R517C variant.
(B) Pedigree of the family harboring the missense variant in the DALRD3 gene.
(C) Immunoblot of lysates prepared from WT or R517 fibroblast cell lines. %DALRD3 represents the amount of DALRD3 protein relative to the WT fibroblast cell line and was quantified from three independent samples. ±, standard deviation from the mean.
(D) Primer extension analysis of tRNA-Arg-UCU and Arg-CCU extracted from WT or R517 fibroblast cell lines. (−) indicates that no RNA was added to the RT reaction. m3C32, 3-methylcytosine. m1G9, 1-methylguanine; >, labeled probe. Bar graphs represent quantification of m3C formation in tRNA-Arg-UCU or CCU by primer extension. n = 3. Error bars represent standard deviation from the mean. Significance was determined using an unpaired t test with two-tailed p value. ∗∗∗∗p ≤ 0.0001.
The DALRD3 c.1549C>T (p.(Arg517Cys)) variant has been detected with an allele frequency of <0.00001 among 1,613,950 alleles screened, with 14 heterozygotes and 0 homozygotes identified thus far (gnomAD v.4.1.0, SNV: 3-49015671-G-A). The group frequency for this allele among individuals of African or African American ancestry is 0.00004 among 74,866 alleles tested. The genomic constraint of the surrounding 1 kb of sequence is 0.82, while the loss-of-function intolerant score for DALRD3 is 0.
Sequence alignment reveals that the R517 residue of human DALRD3 protein is conserved in all known vertebrate DALRD3 orthologs from mammals to fish (Figure 2A). Based upon the high conservation in amino acid identity at this position, the nonsynonymous substitution caused by the R517C variant could disrupt the folding and function of the DARLD3 protein. Consistent with this hypothesis, the R517 variant is expected to be deleterious by multiple pathogenicity prediction algorithms (Figure S6).
To characterize the biological effects of the R517C variant on DALRD3 protein, we isolated fibroblast cells from the affected individual via skin biopsy (referred to as R517C fibroblast cells). We compared protein levels in the R517C fibroblast cells to fibroblast cells isolated from a healthy, age-matched control individual homozygous for the wild-type DALRD3 alleles. Based upon immunoblotting, the levels of DALRD3 protein in the R517C fibroblast cells were greatly reduced compared to fibroblast cells from the healthy wild-type individual (Figure 2C). These results suggest that the R517C variant alters the structure of DALRD3, leading to reduced cellular stability and increased degradation.
We next tested the impact of the DALRD3-R517 variant on m3C modification in arginine tRNAs of fibroblast cells from the affected individual. We monitored m3C modification using a primer extension assay in which the presence of m3C leads to a reverse transcriptase (RT) block at position 32 of arginine tRNAs, while the lack of m3C allows for readthrough and generation of an extended product to the next RT block. Using this assay, we find that the m3C modification is greatly reduced in R517C fibroblast cells compared to control cells from a healthy individual (Figure 2D, quantified in bar graphs). These results demonstrate that R517C fibroblasts exhibit a deficiency in the m3C modification in specific tRNA-Arg isoacceptors. The deficit in m3C modification in the arginine tRNAs of R517C fibroblast cells provides evidence that the R517C variant causes partial loss of function.
These results indicate that the R517C variant in the DALRD3 protein impacts m3C modification in human cells. To elucidate the molecular defects associated with the R517C variant, we investigated the interaction between DALRD3 and METTL2. We co-transfected HEK293T cells with a plasmid expressing Streptactin (Strep)-tagged METTL2 along with empty vector or plasmids expressing a FLAG-tagged version of the wild type (WT) or the R517C DALRD3 variant (Figure 3A). Expression of METTL2, DALRD3-WT, or the DALRD3-R517C variant was confirmed by immunoblotting for the Strep tag or FLAG tag (Figure 3B, input, lanes 1–3). We note that the DALRD3-R517C variant accumulated to lower levels than the WT DALRD3 protein in multiple independent transfections. The reduced levels of DALRD3-R517C protein compared to WT-DALRD3 in this system are consistent with the decreased amount of DALRD3 protein in the R517C fibroblast cells from the affected individual.
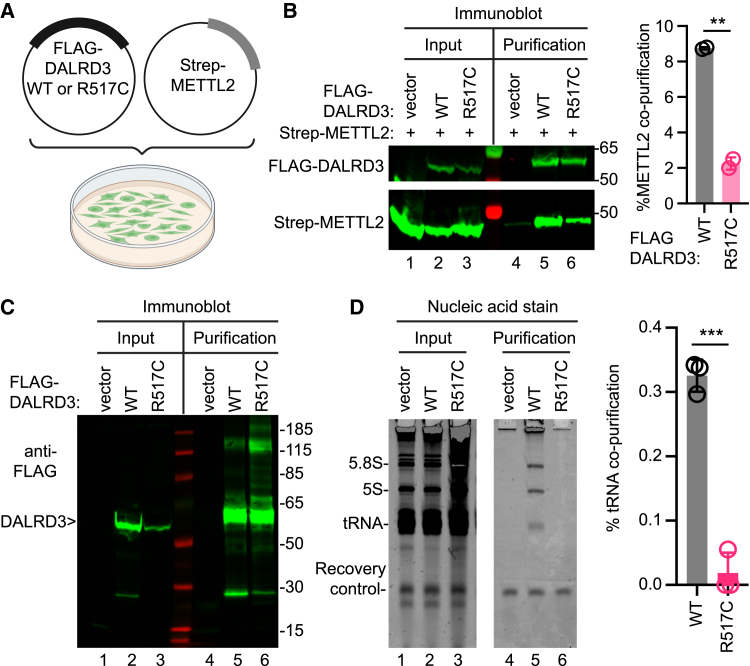
Figure 3.
The DALRD3-R517C variant exhibits reduced co-precipitation with METTL2 and RNAs
(A) Co-transfection setup with plasmids encoding FLAG-DALRD3 and Strep-METTL2.
(B) Immunoblot of input lysates and anti-FLAG purifications from HEK293T cells transfected with the indicated plasmids. The immunoblot was probed with anti-FLAG and anti-Strep antibodies. Input represents 4% of the total sample used for purification. Purification represents 50% of the purified sample. Bar graph represents quantification of METTL2 co-purifying with DALRD3-WT or the R517C variant. The percentage of co-purifying METTL2 represents the amount of METTL2 in the purified DALRD3 sample that was recovered from the total input.
(C) Immunoblot of FLAG-DALRD3 purified from HEK293T cells. The immunoblot was probed with anti-FLAG antibodies. Input represents 4% of the total sample used for purification. Purification represents 20% of the total purified sample.
(D) Nucleic acid stain of RNAs extracted from the indicated input or purified samples after denaturing PAGE. The migration patterns of 5.8S rRNA (∼150 nt), 5S rRNA (∼120 nt), and tRNAs (∼70–80 nt) are denoted. Input represents 2% of total extracts used for purification. The bar graph represents quantification of tRNA co-purifying with DALRD3-WT or the R517C variant. The percentage of tRNA co-purification represents the amount of tRNA in the purified DALRD3 sample that was recovered from the total input.
For bar graphs in (B) and (D), error bars represent standard deviation from the mean; significance was determined using an unpaired t test with two-tailed p value. ∗∗p = 0.0015 for (B). ∗∗∗p = 0.0002 for (D).
The FLAG-DALRD3 fusion proteins were then purified on anti-FLAG antibody resin, and recovery of FLAG-tagged DALRD3 was confirmed (Figure 3B, purification, FLAG-DALRD3, lanes 5 and 6). We detected an enrichment of METTL2 that copurified with DALRD3 above the background binding in the control purification (Figure 3B, purification, Strep-METTL2, compare lane 4 to lane 5). The amount of co-purifying METTL2 was decreased by ∼4-fold with the DALRD3-R517C variant compared to DALRD3-WT after normalization to the amount of DALRD3 protein purified (Figure 3B, Strep-METTL2, compare lanes 5 and 6, quantified in the bar graph). This result suggests that the DALRD3-R517C variant exhibits less stable interaction with METTL2 compared to DALRD3-WT.
Based upon this finding, we next tested the interaction of DALRD3 with tRNAs. Like above, we transfected HEK293T cells with empty vector or plasmids expressing FLAG-DALRD3-WT or the DALRD3-R517C variant. Expression and purification of DALRD3-WT or the DALRD3-R517C variant was confirmed by immunoblotting for the FLAG tag (Figure 3C). We examined the RNA species that co-purified with DALRD3 by denaturing PAGE followed by nucleic acid staining. A recovery control RNA was included during RNA extraction to normalize for differences in recovery efficiency. While purification from vector-transfected cell lysates exhibited only background binding to tRNAs, the purified DALRD3-WT sample contained several co-purifying RNA species that correspond in size to tRNAs along with 5S and 5.8S rRNA (Figure 3D, compare lanes 4 and 5). This pattern of co-purifying RNAs is consistent with our previous observation that DALRD3 exhibits a stable interaction with rRNA and tRNAs that are targets for m3C modification.16 In contrast to DALRD3-WT, the amount of co-purifying tRNAs with the DALRD3-R517C variant was reduced to nearly background levels (Figure 3D, quantified in the bar graph). Overall, these results reveal that the R517C substitution perturbs binding of DALRD3 to METTL2 and tRNAs.
Discussion
Altogether, we identify a loss-of-function variant in DALRD3 that substantiates the link between DALRD3-dependent tRNA modification and neurological function. Moreover, these results uncover the molecular basis for the deficit in tRNA modifications caused by the DALRD3 missense variant. In particular, the decreased amount of DALRD3-R517C protein in the cell could contribute to a reduction in METTL2 and tRNA binding that impairs tRNA modification. Moreover, this result suggests that misfolding and loss of protein-RNA interactions promote degradation of the DALRD3-R517C protein by the ubiquitin-proteasome system. Interestingly, global analysis of the human ubiquitin-modified proteome has identified a ubiquitination site on lysine residue 28 of DALRD3 that is responsive to proteasome inhibition.21 Thus, the levels of DALRD3 protein could potentially be restored in cells of affected individuals through proteasome inhibition.
The R517C substitution could also perturb the binding of METTL2 and tRNA substrates by affecting the folding of DALRD3 domains necessary for protein-RNA interactions. To gain insight into how the R517C substitution could affect DALRD3 folding, we mapped the R517 residue onto a predicted DALRD3 structure generated through AlphaFold.22,23 DALRD3 is predicted to fold into an N-terminal region unique to DALRD3 homologs and a C-terminal DALR anticodon binding domain (Figure S7A, N-terminal region in gray, DALR domain in violet). The C-terminal DALR domain of DALRD3 is predicted to form an all α-helical bundle that is characteristic of DALR domains found in arginyl tRNA synthetases.24,25 Based upon the AlphaFold model, the R517 residue lies within the last α helix of the all-α-helical bundle of the DALR domain, with the positively charged side chain partially exposed to solvent (Figures S7A and S7B, R517 residue in red). The change from an amphipathic arginine residue to a hydrophobic cysteine residue could alter the overall folding of the α-helical bundle of DALRD3 that impacts interactions with METTL2 and tRNAs. Moreover, the change from a positively charged arginine side chain to the uncharged cysteine side chain in DALRD3 could disrupt electrostatic or hydrogen bond interactions with METTL2 and arginine tRNAs.
The individual with the R517C variant in DALRD3 exhibits many of the phenotypes displayed by two previously characterized individuals with a homozygous nonsense variant in DALRD3.16 However, the onset of seizures in the individual described here was later in life compared to the two previously characterized individuals (4 years versus 6–7 months). Moreover, the individual described here exhibited less severe immobility and speech deficiencies compared to the two individuals with a homozygous nonsense variant. We also note that fibroblast cells from the individual with the R517C missense variant retain ∼25% of the m3C modification in arginine tRNAs, while cells with the DALRD3 frameshift variant exhibit nearly complete loss of m3C modification in arginine tRNAs.16 Thus, the reduced severity of clinical symptoms associated with the R517C variant is consistent with the partial loss-of-function phenotype compared to the nearly complete loss of function in the DALRD3 frameshift variant. Our findings set the stage for determining the biological pathways dependent upon DALRD3-dependent tRNA modifications that could be modulated in individuals with epileptic encephalopathy. Furthermore, our studies suggest that m3C modification status could serve as a functional assay to assist in the classification of uncharacterized DALRD3 variants.
The different clinical presentations and ages of onset for individuals with DALRD3 variants suggests that additional genes are likely to contribute to disease pathology. It is also possible that loss of DALRD3 protein function is independent of the observed phenotypes in families with DALRD3 variants. Thus, the identification of additional individuals with DALRD3 variants linked to disease will be the focus of future studies. Furthermore, the future generation of DALRD3-deficient animal models will provide a system for testing the role of DALRD3 function in neurodevelopment and brain function.
Data and code availability
The published article includes all data generated or analyzed during this study.
Acknowledgments
We thank the individuals and their family for participation in this study. We thank the Fu Lab for comments on this manuscript. This work was funded by NIH (USA) R01 GR532955 (to D.F.).
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2024.100377.
Web resources
ClinGen: https://clinicalgenome.org
ClinVar: https://www.ncbi.nlm.nih.gov/clinvar/
dbSNP: https://www.ncbi.nlm.nih.gov/snp/
gnomAD: https://gnomad.broadinstitute.org
gnomAD gene: https://gnomad.broadinstitute.org/gene/ENSG00000178149?dataset=gnomad_r4
gnomAD variant: https://gnomad.broadinstitute.org/variant/3-49015671-G-A?dataset=gnomad_r4
OMIM: https://www.omim.org
Supplemental information
References
- 1.Zhang W., Foo M., Eren A.M., Pan T. tRNA modification dynamics from individual organisms to metaepitranscriptomics of microbiomes. Mol. Cell. 2022;82:891–906. doi: 10.1016/j.molcel.2021.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suzuki T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 2021;22:375–392. doi: 10.1038/s41580-021-00342-0. [DOI] [PubMed] [Google Scholar]
- 3.McCown P.J., Ruszkowska A., Kunkler C.N., Breger K., Hulewicz J.P., Wang M.C., Springer N.A., Brown J.A. Naturally occurring modified ribonucleosides. Wiley Interdiscip. Rev. RNA. 2020;11 doi: 10.1002/wrna.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chujo T., Tomizawa K. Human transfer RNA modopathies: diseases caused by aberrations in transfer RNA modifications. FEBS J. 2021;288:7096–7122. doi: 10.1111/febs.15736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith T.J., Giles R.N., Koutmou K.S. Anticodon stem-loop tRNA modifications influence codon decoding and frame maintenance during translation. Semin. Cell Dev. Biol. 2024;154:105–113. doi: 10.1016/j.semcdb.2023.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pereira M., Francisco S., Varanda A.S., Santos M., Santos M.A.S., Soares A.R. Impact of tRNA Modifications and tRNA-Modifying Enzymes on Proteostasis and Human Disease. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19123738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orellana E.A., Siegal E., Gregory R.I. tRNA dysregulation and disease. Nat. Rev. Genet. 2022;23:651–664. doi: 10.1038/s41576-022-00501-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delaunay S., Helm M., Frye M. RNA modifications in physiology and disease: towards clinical applications. Nat. Rev. Genet. 2024;25:104–122. doi: 10.1038/s41576-023-00645-2. [DOI] [PubMed] [Google Scholar]
- 9.Blaze J., Akbarian S. The tRNA regulome in neurodevelopmental and neuropsychiatric disease. Mol. Psychiatr. 2022;27:3204–3213. doi: 10.1038/s41380-022-01585-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramos J., Fu D. The emerging impact of tRNA modifications in the brain and nervous system. Biochim. Biophys. Acta. Gene Regul. Mech. 2019;1862:412–428. doi: 10.1016/j.bbagrm.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Arimbasseri A.G., Iben J., Wei F.Y., Rijal K., Tomizawa K., Hafner M., Maraia R.J. Evolving specificity of tRNA 3-methyl-cytidine-32 (m3C32) modification: a subset of tRNAsSer requires N6-isopentenylation of A37. RNA. 2016;22:1400–1410. doi: 10.1261/rna.056259.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu L., Liu X., Sheng N., Oo K.S., Liang J., Chionh Y.H., Xu J., Ye F., Gao Y.G., Dedon P.C., Fu X.Y. Three distinct 3-methylcytidine (m(3)C) methyltransferases modify tRNA and mRNA in mice and humans. J. Biol. Chem. 2017;292:14695–14703. doi: 10.1074/jbc.M117.798298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lentini J.M., Bargabos R., Chen C., Fu D. Methyltransferase METTL8 is required for 3-methylcytosine modification in human mitochondrial tRNAs. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2022.101788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleiber N., Lemus-Diaz N., Stiller C., Heinrichs M., Mai M.M.Q., Hackert P., Richter-Dennerlein R., Höbartner C., Bohnsack K.E., Bohnsack M.T. The RNA methyltransferase METTL8 installs m(3)C(32) in mitochondrial tRNAs(Thr/Ser(UCN)) to optimise tRNA structure and mitochondrial translation. Nat. Commun. 2022;13:209. doi: 10.1038/s41467-021-27905-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bohnsack K.E., Kleiber N., Lemus-Diaz N., Bohnsack M.T. Roles and dynamics of 3-methylcytidine in cellular RNAs. Trends Biochem. Sci. 2022;47:596–608. doi: 10.1016/j.tibs.2022.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Lentini J.M., Alsaif H.S., Faqeih E., Alkuraya F.S., Fu D. DALRD3 encodes a protein mutated in epileptic encephalopathy that targets arginine tRNAs for 3-methylcytosine modification. Nat. Commun. 2020;11:2510. doi: 10.1038/s41467-020-16321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheffer I.E., Berkovic S., Capovilla G., Connolly M.B., French J., Guilhoto L., Hirsch E., Jain S., Mathern G.W., Moshé S.L., et al. ILAE classification of the epilepsies: Position paper of the ILAE Commission for Classification and Terminology. Epilepsia. 2017;58:512–521. doi: 10.1111/epi.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheffer I.E., Zuberi S., Mefford H.C., Guerrini R., McTague A. Developmental and epileptic encephalopathies. Nat. Rev. Dis. Prim. 2024;10:61. doi: 10.1038/s41572-024-00546-6. [DOI] [PubMed] [Google Scholar]
- 19.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghorbani F., Alimohamed M.Z., Vilacha J.F., Van Dijk K.K., De Boer-Bergsma J., Fokkens M.R., Lemmink H., Sijmons R.H., Sikkema-Raddatz B., Groves M.R., et al. Feasibility of Follow-Up Studies and Reclassification in Spinocerebellar Ataxia Gene Variants of Unknown Significance. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.782685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim W., Bennett E.J., Huttlin E.L., Guo A., Li J., Possemato A., Sowa M.E., Rad R., Rush J., Comb M.J., et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., Yuan D., Stroe O., Wood G., Laydon A., et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50:D439–D444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolf Y.I., Aravind L., Grishin N.V., Koonin E.V. Evolution of aminoacyl-tRNA synthetases--analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 1999;9:689–710. [PubMed] [Google Scholar]
- 25.Delagoutte B., Moras D., Cavarelli J. tRNA aminoacylation by arginyl-tRNA synthetase: induced conformations during substrates binding. EMBO J. 2000;19:5599–5610. doi: 10.1093/emboj/19.21.5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all data generated or analyzed during this study.