Abstract
Cell stresses occur in a wide variety of settings: in disease, during industrial processes, and as part of normal day‐to‐day rhythms. Adaptation to these stresses requires cells to alter their proteome. Cells modify the proteins they synthesize to aid proteome adaptation. Changes in both mRNA transcription and translation contribute to altered protein synthesis. Here, we discuss the changes in translational mechanisms that occur following the onset of stress, and the impact these have on stress adaptation.
Keywords: mRNA, proteostasis, signalling, stress, translation
When challenged by stress conditions such as disease, cells undergo proteome adaptation and modify the proteins they synthesize to facilitate this process. Changes in both mRNA transcription and translation contribute to altered protein synthesis. Here, we discuss the changes in translational mechanisms that occur following stress conditions, and the impact that these have on stress adaptation.
Abbreviations
- ADP
adenosine di‐phosphate
- AMP
adenosine mono‐phosphate
- AMPK
AMP‐activated protein Kinase
- ATP
adenosine tri‐phosphate
- CDS
coding sequence
- eEF
eukaryotic elongation factor
- eIF
eukaryotic initiation factor
- ER
endoplasmic reticulum
- eRF
eukaryotic release factor
- GDP
guanosine di‐phosphate
- GEF
guanine nucleotide exchange factor
- GTP
guanosine tri‐phosphate
- HRI
haem‐regulated eIF2α KinaseGcn2: general control non‐depressible 2
- IRES
internal ribosome entry site
- ISR
integrated stress response
- ORF
open reading frame
- PABP
poly(A)‐binding protein
- PB
P‐body
- PERK
PKR‐like ER Kinase
- PIC
pre‐initiation complex
- PKA
protein kinase A
- PKR
double‐stranded RNA‐dependent protein kinase
- RBP
RNA‐binding protein
- RiBi
ribosome biogenesis
- ROS
reactive oxygen species
- SG
stress granule
- TC
ternary complex
- TORC1
target of rapamycin complex 1, and its mammalian counterpart mTORC1
- uORF
upstream open reading frame
- UPR
unfolded protein response
- UTR
un‐translated region
Proteome adaptation upon stress
Environmental fluctuations cause all cells to be frequently subjected to multiple stresses in varying combinations and levels of severity. Common stresses include changes in temperature, osmolarity, reactive oxygen species and nutrient availability. To survive and proliferate, cells must adapt to and counter stresses as they arise. Altering the cellular proteome (Fig. 1) is key for stress adaptation across life [1, 2]. Extensive work has established roles for stress‐induced changes to the cellular mRNA pool across a wide variety of stresses, including in animals [3, 4, 5], plants [6, 7, 8] and fungi [9, 10, 11]. Protein production is also changed through regulation at the translational level. Alterations (or a lack thereof) in mRNA levels and translation efficiency affect protein production [12, 13]. Protein levels are further regulated by degradation, which can be induced for specific proteins upon stress [14, 15]. In this review, we discuss how mechanisms of eukaryotic translation change with stress, and the effect on mRNA translation.
Fig. 1.
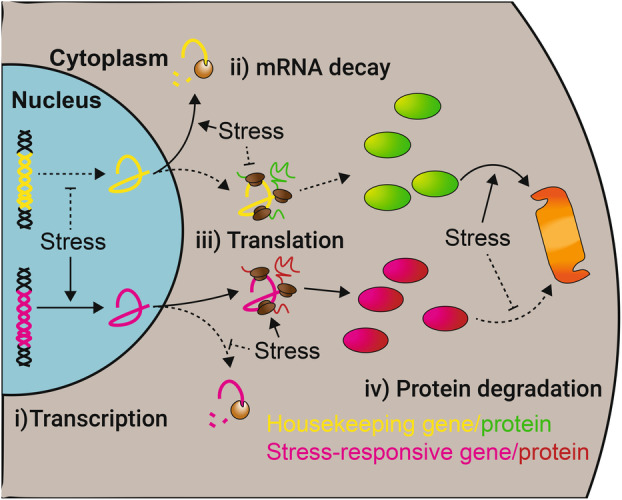
Mechanisms of proteome adaptation upon stress. (i) Transcription is altered upon stress. Production of stress‐responsive mRNAs is increased, while housekeeping mRNA production is decreased. (ii) mRNA decay further contributes to transcriptome adaptation. (iii) The altered transcriptome is subject to further regulation at the level of translation to specifically enhance production of stress‐responsive proteins. (iv) Protein degradation further aids proteome adaptation by selectively removing proteins. Dashed lines show where stresses inhibit processes. Housekeeping genes mRNAs are indicated in yellow while the proteins are in green. Stress‐responsive genes mRNAs are indicated in pink while the proteins are in red.
Translation in unstressed conditions
Canonical translation (Fig. 2) is widely described, with initiation considered the major rate‐limiting step [16, 17]. To initiate translation (Fig. 2A), a ternary complex (TC) consisting of initiator‐methionine (i‐met) tRNA, eIF2 and GTP is formed. This binds to the 40S small ribosomal subunit through eIF3 and eIF5. eIF1 and eIF1A also bind to the 40S to form the 43S pre‐initiation complex (PIC). The PIC binds the eIF4F cap‐binding complex (composed of eIF4G, eIF4E and eIF4A) on the 5′cap of an available mRNA to form the 48S initiation complex. This interaction is mediated through eIF3 and eIF5 binding to eIF4G, while ATP‐bound eIF4A allows formation of an open complex [18, 19, 20]. The 48S complex initiates scanning along the mRNA in this open conformation, with the eIF4A helicase unwinding the mRNA secondary structures, facilitated by its interaction with eIF4B [21]. Whether the eIF4F complex remains bound to the cap (or alternatively if eIF4E disconnects from the other eIF4F components while remaining bound to the cap) during scanning is an open question. Notable, but relatively small, 40S mRNA footprints indicative of queueing occurring during scanning are observed, suggesting that this disconnection from the cap can occur, allowing multiple PIC complexes in the mRNA 5′ UTR [22, 23]. Binding of eIF4E to the PIC increases scanning activity of the eIF4A helicase, implying a benefit for maintenance of the connection between the complete eIF4F complex and PIC [24]. The exact scanning mechanism may vary according to factors including mRNA secondary structure and modifications, although this remains to be determined. Once a suitable start codon is identified via codon‐matching with the i‐met tRNA, the TC's GTP is hydrolysed to GDP, and eIF2 is released. The 48S shifts from an open to a closed conformation, locking i‐met tRNA and mRNA together at the peptidyl transferase (P‐) site [25]. Initiation factors are replaced by the 60S large ribosomal subunit to form the 80S ribosome via an intermediate complex containing eIF5B [26].
Fig. 2.
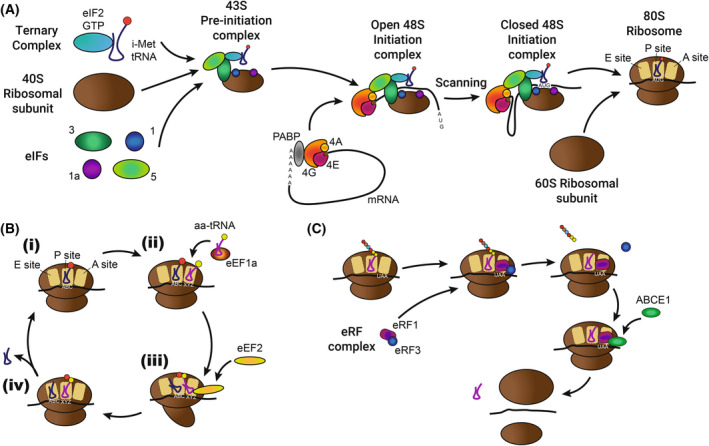
Canonical mechanism of translation. (A) Translation Initiation. The ternary complex, 40S ribosomal subunit and eIF3, eIF5, eIF1, eIF1a form the 43S pre‐initiation complex (PIC). The eIF4F complex, composed of eIF4A, eIF4G and eIF4E, binds the mRNA cap and poly‐A binding protein (PABP) to circularize the mRNA. The 43S PIC binds the eIF4F complex and scans along the mRNA until the initiator methionine (i‐met) tRNA of the ternary complex recognizes an AUG codon. The 60S ribosomal subunit is then recruited to form the 80S ribosome with the i‐met tRNA in the peptidyl transferase (P)‐site. (B) Translation elongation. (i) Following initiation or a round of translation, a ribosome has one tRNA in the P‐site. (ii) A codon‐matched amino‐acylated (aa‐)tRNA is recruited to the acceptor (A)‐site by eEF1a. (iii–iv) mRNA translocation occurs, mediated by eEF2, moving the tRNAs from the P‐ to the Exit (E)‐site, and A‐ to P‐site, adding one amino acid to the peptide chain. The E‐site tRNA is released and the process repeats from (i) until a stop codon is reached. (C) Translation termination. Once a stop codon is reached, it is recognized by the eRF1/3 complex, which causes the peptide chain to be released. The ribosome is then split for recycling.
During translation elongation (Fig. 2B), codon‐matched charged tRNAs, delivered by eEF1A, bind the mRNA in the aminoacyl (A‐) site of the ribosome [27]. mRNA translocation occurs via 40S conformational changes, assisted by eEF2, which move the tRNA from the A‐site to the P‐site [28, 29]. The tRNA‐conjugated amino acid is added to the C‐terminal end of the peptide chain and translocation occurs again to move the free tRNA to the exit (E‐) site, where it is ejected. A new tRNA can then bind in the A‐site to restart this process.
When a stop codon is reached, there are no codon‐matched tRNAs, so translation termination and ribosome disassembly occur (Fig. 2C). The eukaryotic release factor complex (composed of eRF1 and eRF3‐GTP) binds in the A‐site and terminates protein synthesis by hydrolysing the bond between the final tRNA and its amino acid [30, 31, 32]. Subsequent release of eRF3 and binding of an ABC‐ATPase protein and ATP hydrolysis splits the 80S back into 40S and 60S subunits, which are recycled for subsequent rounds of translation [33, 34]. Poly(A)‐binding protein (PABP) binds eIF4G, eIF4B and the 3′ mRNA poly(A) tail to: circularize the mRNA; stabilize the interaction of eIF4E with the 5′cap and boost translation termination efficiency to boost ribosome occupancy and translation [35, 36, 37, 38]. Closed loop formation is particularly biased towards shorter mRNAs [39]. Further, PABP can interact with both 40S and 60S ribosomal subunits, potentially limiting their diffusion away from the mRNA and promoting re‐initiation [40].
Many of these mechanisms are altered in either efficiency or character upon stress to change the rates and selection of mRNA for translation. Below, we discuss how changes in mRNA availability and mechanisms of translation affect the pool of translating mRNAs and protein production. In Box 1 and Fig. 3, we highlight some techniques and approaches which can be used to assess translation so researchers new to the field can get an overview of available methodologies.
Box 1. Techniques to monitor translation.
Translation is a highly dynamic, multi‐step process, readouts of which can be impacted by other processes (e.g. protein degradation). A further complicating factor is that there is no fixed rate of translation: much like transcription, translation occurs in bursts with periods of high and low activity [41, 42]. A large variety of techniques can be employed to measure translation; from the whole translatome to individual mRNAs. These range from comparing protein and mRNA levels to determine how much protein is produced per mRNA, while recognizing and controlling for the contribution of protein degradation (Fig. 3A) [43], incorporation of radioactive amino acids, chemically modified amino acids, or aminoacyl tRNA analogues into newly synthesized peptide chains (Fig. 3B) [44, 45, 46, 47, 48], investigation of mRNA–ribosome interactions (Fig. 3C) [49, 50, 51], and live or fixed cell microscopy of specific mRNA translation (Fig. 3D) [41, 52, 53]. Each technique has advantages and disadvantages for determining mRNA translation efficiency, and each requires a healthy degree of scepticism and appropriate controls when interpreting results due to several additional factors which can impact the observations. Researchers wishing to explore translation for the first time are advised to look through the methods highlighted here to help identify the one(s) most appropriate for their particular question.
Fig. 3.
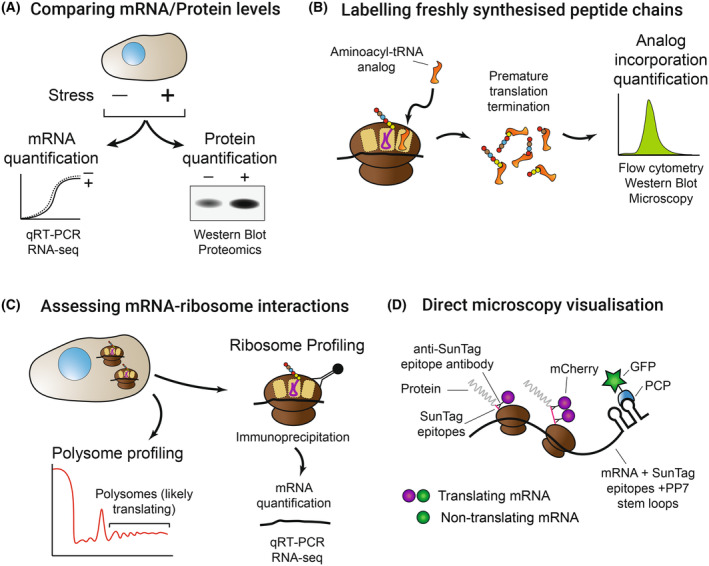
Methods to measure translation. (A) Comparing mRNA and protein levels to look for changes in translation of either one specific protein or multiple proteins using genomic and proteomic techniques. Protein degradation must be accounted for. (B) Labelling freshly synthesized peptide chains with aminoacyl‐tRNA analogues or labelled amino acids and quantifying the incorporation. (C) Assessing mRNA–ribosome interactions, either of specific mRNAs or transcriptome wide. (D) Direct microscopy visualization of translation of specific mRNAs through technologies such as SunTag, shown, which uses colocalization analysis of nascent chain (labelled with a fluorescent anti‐SunTag nanobody) and mRNA (labelled with green‐fluorescent protein tagged PP7 coat protein, PCP‐GFP).
Effect of stresses on mRNA availability
Stress‐induced formation of RNA‐containing granules
For translation to occur, both the translation machinery and mRNA must be available. Several stresses cause rapid sequestration of these components into stress granules (SGs: mRNA, translation initiation complexes and RNA binding proteins (RBPs)) and P‐bodies (PBs: mRNA and RBPs such as those involved in mRNA decay) mediated by liquid–liquid phase separation of RBPs [54, 55]. While SGs formed upon different stresses share a name and common components, there are significant compositional differences. SGs formed following eIF4A inhibition have relatively low levels of mRNAs, eIF4G and eIF3B compared to SGs formed following sodium arsenite treatment [56]. The importance of stress‐induced eIF2α phosphorylation to SG formation is also context dependent [56]. Care must, therefore, be taken when extrapolating SG function between stresses. Although most mRNAs in SGs are translationally repressed, stress‐activated (ATF4) and inhibited (5′TOP containing RPL32) mRNAs can be translated within these structures, with only a moderate decrease in the translationally active proportion compared to cytosolic mRNAs [57]. Phase‐separated granules supporting translation in non‐stressful conditions have also been widely reported across eukaryotes [58, 59, 60, 61, 62, 63]. Microscopy resolution remains a limiting factor for identifying small and/or more diffuse stress granules over the cytosolic background. While fundamental questions are being re‐opened about the nature of SGs as a translationally inactive mRNA storage compartment, further investigations are required to determine whether, and to what extent, other mRNAs are translated within SGs.
In contrast to SGs, PBs are devoid of translating mRNA and translation factors, instead containing proteins associated with mRNA decay and translational repression [57, 64]. Despite this, PBs do not seem to be sites of general mRNA degradation, and mRNA decay occurs when PB formation is prevented [65, 66]. Indeed, some mRNAs are stabilized upon stresses which induce PB formation: possibly by PBs sequestering mRNA degrading enzymes [66, 67, 68]. This has larger effects on some mRNAs (e.g. nonsense‐mediated decay‐regulated mRNAs) than others. Degradation of other mRNAs is regulated by alternative splicing, notably the HAC1 mRNA in S. cerevisiae (XBP1 in mammals) following ER stress, alternative splicing of which prevents degradation, allowing translation and unfolded protein response (UPR) initiation [69, 70]. Other mRNAs are similarly regulated [71, 72, 73]. Together with alterations at the transcriptional level, altered mRNA decay and splicing change the pool of mRNAs available for translation. For some mRNAs, this may be important to offset changes to translation efficiency, while for others it is the primary way of regulating protein output [74].
Stress regulation of mRNA–RNA‐binding protein interactions
mRNAs interact extensively with RNA‐binding proteins (RBPs), which can regulate mRNA translational capacity [75]. This includes, but is not limited to, factors involved in translation initiation, many of which have reduced mRNA association following stress [76, 77]. Other proteins known to affect translation, such as the S. cerevisiae translational repressor Puf3, have altered overall levels bound to mRNA following stress [76]. In most cases, additional studies to determine the specific mRNAs these proteins differentially interact with are yet to be performed. In follow‐up studies which have been performed, work has focused on either identifying mRNAs bound to a particular protein or using genetic manipulation followed by assessment of protein levels [78, 79]. Other stress‐regulated RBPs include metabolic proteins, such as thymidylate synthase and iron regulatory proteins, some of which regulate levels of certain proteins, including their own, either through regulation of mRNA stability or translation initiation [80, 81]. While many RBPs contain defined RNA recognition motifs, many do not. It is becoming more apparent that proteins without canonical RNA recognition motifs can interact with diverse mRNAs through phase separation, either in SGs/PBs or other similar bodies, and are thus able to manipulate their translation and sequestration into these structures [63, 82, 83, 84]. The molecular properties of the mRNAs which facilitate this remain unclear, although mRNA is more effective than other types of RNA at interacting with phase‐separated bodies, perhaps indicating an importance for both length and a relative lack of secondary structure [84].
Additional RNA‐containing structures involved in translation can have altered interactions with proteins which affect their function. This includes different ribosomal protein paralogues (discussed in the ‘translation elongation’ section and reviewed in [85]), and tRNAs. To deliver amino acids to the ribosome, tRNAs require aminoacylating. Under stress conditions, methionine‐tRNA‐synthetase loses specificity and mis‐charges non‐Met tRNAs with methionine, which can be used in translation. This occurs across species, possibly to aid in countering increased ROS [86, 87, 88]. Under certain stress conditions, tRNAs can be bound and cleaved by angiogenin, removing them from the tRNA pool [89]. Altered interactions between RNAs, tRNA and RBPs following stress have major impacts on translation.
Effect of stresses on translation initiation
Translation initiation is regulated by altered cellular signalling upon stress: principally through changing translation initiation component availability. The major stress‐regulated signalling pathways involved in altered translation upon stress are highly evolutionarily conserved. These include the integrated stress response (ISR) kinases, TORC1 (target of rapamycin complex 1 and its mammalian counterpart mTORC1 – collectively referred to here as TORC1), AMP‐activated protein kinase (AMPK) and protein kinase A (PKA). The regulation of these kinases is multi‐layered with substantial interplay (Fig. 4A). While we discuss the roles of these kinases individually, their interplay is key to producing complex and highly regulated changes to cellular translation. For example, both TORC1 inhibition and ISR activation are necessary for the expression of Nanog‐291 and Snail‐85 mRNAs upon stress simulation in breast cancer cells [90]. Depending upon the mRNA and its specific properties, translation can either be up‐ or downregulated by the same altered signalling, allowing a consolidated translational response to the stress. This response can change over the lifetime of the stress with different signalling outputs apparent in cells under acute and chronic stress [91, 92, 93, 94]. In the following sections, we detail these and other changes to the mechanisms and location of translation initiation.
Fig. 4.
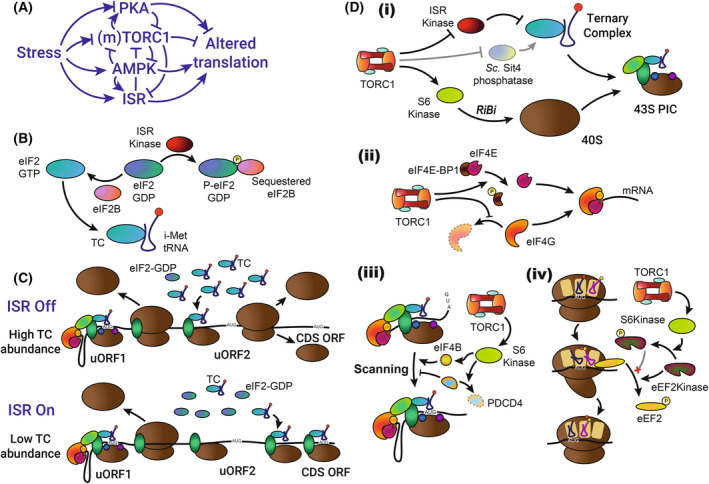
Changes to translation upon stress. (A) Stress impacts signalling from various kinases to alter translation. Arrowheads indicate activation, while barred lines indicate inhibition. ISR is the integrated stress response. (B) Inhibition of ternary complex formation by the ISR. ISR kinases phosphorylate eIF2α to sequester the eIF2 GEF and eIF2B, preventing GDP to GTP transition and re‐binding of initiator tRNA. (C) Regulation of ATF4/GCN4 translation by upstream open reading frames (ORFs) is dependent on ISR signalling. Following initiation and termination from uORF1, the restriction of ternary complex (TC) availability under conditions of ISR activation prevents premature re‐initiation at uORF2. This allows translation initiation from the start codon of the coding sequence (CDS) ORF. In mouse ATF4 mRNA, uORF2 overlaps the CDS ORF. For GCN4 mRNA, the uORF1 represented here is analogous to the first ORF pair (uORFs1&2) while uORF2 is analogous to the second ORF pair (uORFs3&4). (D) Impacts of the stress‐inhibited kinase TORC1 signalling on translation. (i) TORC1 inhibition restricts 43S pre‐initiation complex formation through ISR activation (see Fig. 4B) and reduced ribosome biogenesis (RiBi) through decreased S6Kinase activation. In S. cerevisiae, the TORC1‐inhibited phosphatase Sit4 can counter the ISR kinases. (ii) TORC1 inhibition restricts availability of the eIF4F components eIF4G (via increased degradation) and eIF4E (through eIF4E‐BP1 sequestration). (iii) TORC1 inhibition, and reduced S6Kinase activation, decreases eIF4B activation of eIF4A and prevents degradation of the eIF4A inhibitor PDCD4. (iv) Inhibition of TORC1, and resulting S6Kinase inhibition, allows eEF2Kinase to inhibit the ability of eEF2 in mRNA translocation.
Integrated stress response
The ISR is mediated by kinases which reduce, but do not eliminate, ternary complex (TC) assembly through inhibiting the ability of eIF2 to bind GTP [95]. The mechanisms of ISR activation are varied. In mammalian cells, the ISR is triggered by the activation of any one of four kinases (PERK, PKR, HRI, Gcn2) which are descended from the ancestral Gcn2, the only form found in yeast [96, 97]. PERK is activated by ER stress and hypoxia [98, 99, 100]. PKR is activated by double‐stranded RNA and interferon signalling (notably during viral infection), and possibly ER and oxidative stresses, although the evidence is circumstantial [101, 102, 103, 104]. HRI is activated by heat, osmotic, oxidative, haem deficiency, mitochondrial and proteotoxic stresses [99, 105, 106, 107, 108, 109, 110, 111]. Gcn2 is regulated by amino acid availability, interaction with the P‐stalk of non‐translating ribosomes, cytoskeletal perturbation, infection, TORC1 regulation of downstream phosphatases and phosphorylation by AMPK, in addition to oxidative, and possibly other stresses, in yeasts [112, 113, 114, 115, 116, 117, 118, 119, 120]. In the most well‐described mechanism of Gcn2 activation, Gcn2 interacts with Gcn1 and the yeast‐specific Gcn20, which recruit free tRNAs to Gcn2 at the ribosomal A‐site: a reaction that is enhanced under conditions where the availability of an alternative binding partner is limited (Yin1/IMPACT for Gcn1, eEF1α for Gcn2) [119, 121, 122, 123]. Gcn2 has additionally been described as a TORC1 inhibitor through regulating translation of Sestrin2 (which interferes with TORC1 localization), eIF2α phosphorylation and potentially by directly phosphorylating TORC1 itself [124, 125, 126].
Activated ISR kinases phosphorylate the eIF2 complex subunit eIF2α at Serine 51. This phosphorylation creates an alternate binding site for the eIF2 guanine nucleotide exchange factor (GEF) eIF2B, sequestering it and preventing GDP to GTP exchange on eIF2 [127]. GDP‐bound eIF2 has much lower affinity for the i‐Met tRNA and thus TC formation is inhibited (Fig. 4B) [128]. Reduced TC availability rapidly decreases translation initiation, allowing the cell time for transcriptome modification for stress‐adaptive translation. It additionally acts to decrease production of proteins which are liable to exacerbate stresses, such as the mitochondrial complex I protein NDUFAF2: the decrease of which limits oxidative stress [129]. Activation of the ISR during parasite infection can lead to reduced host‐cell nutrient usage, thereby enhancing pathogen growth, while for other infections the ISR inhibits pathogen multiplication [120, 130].
While translation initiation is decreased by eIF2α phosphorylation, TC levels are reduced rather than absent, allowing some initiation to occur. By modulating the level of ISR activation, cells can fine‐tune TC availability as appropriate for the conditions [131]. Altered TC availability does not uniformly reduce translation, and thereby changes the proteome composition in cells with activated ISR. Altered translation in response to ISR activation is best described for the upstream ORF (uORF)‐regulated transcription factors ATF4 and its yeast equivalent Gcn4. In mouse ATF4 mRNA, there are two uORFs: one distal from the start codon, and one proximal which encodes a peptide overlapping the start codon (human ATF4 mRNA contains three uORFs: two distal and one proximal). GCN4 has four uORFs, which can be thought of as occurring in pairs (1 and 2, 3 and 4) with similar functionality to those in ATF4 mRNA [97].
GCN4/ATF4 mRNA translation (Fig. 4C) initiates at the distal uORF(s) through canonical translation mechanisms and swiftly terminates. Following translation termination, a subset of the 40S ribosomes remains bound to the mRNA, aided by an interaction between a sequence‐specific part of the mRNA and eIF3, which can remain bound to the 40S for the initial ~ 75 amino acids [132, 133]. These 40S complexes resume scanning and reacquire a TC, which can interact with eIF3, to initiate at a downstream site [134]. When the TC is more abundant, this occurs rapidly, and translation is initiated at the proximal uORFs. Following termination at these uORFs, the 40S does not maintain its mRNA interaction and no further reinitiation occurs. When the TC is less abundant, it takes longer to acquire a new TC. Scanning consequently proceeds past the uORFs and initiates at the GCN4/ATF4 start codon. Spacing between the uORFs is critical for this regulation [135]. Delivery of i‐Met tRNA can be performed upon TC formation inhibition by the eIF2D/DENR complex, in the absence of which ATF4 expression is drastically reduced in Drosophila fat cells [136]. In contrast, eIF2D recycles 40S subunits and inhibits reinitiation in S. cerevisiae, although this role may be sequence dependent [137]. It is important to acknowledge that not all uORFs are involved in translational regulation: recent evidence indicates that only a small, albeit important, subset of uORFs regulate translation [138]. Other mRNAs with only one inhibitory uORF are more likely to have this uORF bypassed under stress [139]. Specific translation of Gcn4/ATF4 and other transcription factors allows transcriptional rewiring prior to the resumption of translation from a newly stress adapted transcriptome, contributing to stress adaptation.
Intriguingly, despite the TC's importance for translation initiation, inducing eIF2α phosphorylation in the absence of stress does not necessarily inhibit translation [140, 141]. Analysis of a mutant which sustains bulk translation at normal rates upon TORC1 inhibition despite high levels of phosphorylated eIF2α implicated defective regulation of eIF4G, which has been recently linked to isoform‐specific stress adaptive translation [141, 142]. Furthermore, other studies have shown that upon prolonged stress ATF4 expression can decrease while eIF2α phosphorylation remains high, potentially due to a reduction in the number of ribosomes [91, 92]. Underscoring the interplay between the different signalling elements discussed here, ATF4 protein expression can also be induced by activating TORC1 in a poorly understood process dependent on eIF4E availability and involving mRNA stabilization and increased uORF skipping independently of TC availability, although the transcriptional targets are distinct [143, 144]. Countering the ISR are the PP1 phosphatases which are active against P‐eIF2α when associated with either the constitutively expressed CReP (also known as PPP1R15B) regulatory subunit, or the ISR‐induced GADD34 (also known as PPP1R15A) regulatory subunit and G‐actin, which helps stabilize the complex [145, 146, 147]. In this way, the ISR acts to limit its own activation. Therefore, while great importance is ascribed to ISR activation, it is important to remember that this always occurs within a network of multiple interacting regulators (Fig. 4A).
Target of rapamycin complex 1
TORC1 regulates various downstream signalling pathways, or branches, which collectively promote anabolic, and inhibit catabolic, processes when TORC1 is active [148]. These branches include autophagy, S6Kinases and PP2A phosphatases, regulation of which are highly conserved from yeast to humans, demonstrating a central role in eukaryotic biology.
TORC1 signalling couples environmental status and nutrient availability with anabolic functions essential for cell growth, including translation. TORC1 activity towards either of the two most investigated downstream effectors, S6Kinase and eIF4E‐BP1, is inhibited by various stresses, including oxidative, envelope, hypoxic, osmotic, nutrient, heat, metabolic and ER stresses [9, 12, 149, 150, 151, 152, 153, 154, 155, 156, 157]. S6Kinase signalling can also be activated by mild oxidative and ER stresses [149, 158]. In mild stress conditions of nutrient limitation, akin to those found in the wild, S. cerevisiae TORC1 activity becomes oscillatory [93]. Other TORC1 effectors are regulated by some, but not all, of these stresses [9]. TORC1 activity is, thus, highly responsive to changing cellular conditions.
The majority of TORC1 is activated in the presence of amino acids through recruitment to the lysosome/vacuole by a GTPase complex. Once there, TORC1 is activated by the Rheb small GTPase, which is responsive to environmental status, particularly the presence of growth factors in mammalian cells. The link between Rheb and TORC1 has been lost in S. cerevisiae (although not S. pombe) and various other lower organisms, indicating that environmental status can be sensed through alternative mechanisms such as AMPK activation [159, 160, 161]. No Rheb homologue in plants is known, although a putative RhebGEF with the expected distribution and effect on growth has been identified [162]. While most TORC1 is at the lysosome/vacuole, a minority is found elsewhere in the cell, with these pools likely to have their own distinct functional effects and downstream signalling [163, 164, 165]. Little is known about how these non‐lysosomal/vacuolar pools are regulated and respond to stress, although Rheb localized to alternative membranes and the nucleoplasm is likely involved [165, 166, 167].
TORC1 inhibition restricts subunit availability for the PIC and eIF4F complexes, as well as scanning and elongation (Fig. 4D). TORC1 inhibition activates the ISR by Gcn2 dephosphorylation through the PP6C phosphatase in mammalian cells, and an unidentified phosphatase in S. cerevisiae [114, 115]. ISR activation, in turn, can inhibit TORC1 [124, 125, 126]. The S. cerevisiae PP2A phosphatase Sit4, which is activated by TORC1 inhibition, can counter ISR kinases to increase TC availability [113]. Further restriction of PIC formation (Fig. 4Di) is mediated by the S6Kinase branch of TORC1 signalling controlling the ribosome biogenesis (RiBi) transcriptional programme [168]. S6Kinases are named for their ability to phosphorylate ribosomal protein S6 (Rps6). In mammalian cells, Rps6 phosphorylation itself, likely in the nucleus, is important for RiBi, although in S. cerevisiae, this regulation occurs primarily through the S6Kinase Sch9 regulating transcriptional repressors, with the alternate S6Kinase Ypk3 being responsible for the majority of Rps6 phosphorylation [169, 170, 171, 172, 173]. Phosphorylated Rps6 in the 40S may aid in translation initiation, with a potential bias for shorter and 5′TOP mRNAs [174].
Availability of the eIF4F cap‐binding complex, composed of eIF4A, eIF4E and eIF4G, is regulated by TORC1 signalling (Fig. 4Dii). eIF4E, the cap‐binding subunit, is alternately bound by eIF4E‐binding proteins, most notably eIF4E‐BP1 in mammalian cells. TORC1 directly phosphorylates eIF4E‐BP1, preventing this binding from occurring and allowing eIF4F complex formation [175]. When TORC1 activity is reduced, eIF4E‐BP1 phosphorylation is reduced, allowing it to bind and sequester eIF4E, limiting translation initiation. Intriguingly, unlike the majority of TORC1 signalling events, the evolutionary history of eIF4E binding protein phosphorylation remains unclear. eIF4E binding proteins in yeasts and nematodes are not thought to be regulated by TORC1, unlike in flies, mammalian cells and amoebae [176, 177, 178, 179]. However, a recent report found that the S. cerevisiae phosphorylation of the eIF4E‐binding protein Caf20 is regulated by TORC1, although this does not cause eIF4E sequestration [180]. TORC1 activity controls the phosphorylation state of the eIF4F member eIF4G, which helps recruit the PIC to mRNA [44, 181]. S. cerevisiae eIF4G levels are controlled by TORC1 through autophagy‐mediated degradation, while mammalian eIF4G is degraded in a caspase‐dependent manner in conditions where TORC1 is inhibited, although whether the caspases act on eIF4G directly, or through promotion of autophagy remains unknown [141, 182, 183]. Intriguingly, the eIF4F complex also physically interacts with TORC1 and can facilitate reduced TORC1 activity in amino‐acid‐starved conditions [184, 185].
TORC1 promotes scanning and elongation via S6Kinase activation (Fig. 4Diii–iv). S6Kinase enhances phosphorylation of eIF4B, which then promotes eIF4A helicase activity and translational scanning [186]. S6Kinase further promotes degradation of the eIF4A inhibitor PDCD4 [187]. eEF2Kinase is phosphorylated and inactivated by S6Kinase, preventing it from inhibiting eEF2 [188]. Although this would be expected to mostly impact elongation, eEF2 activity is also important for ribosome recycling and translation initiation [189]. S6Kinase inhibition is equally important for decreasing translation in stressed cells: a phospho‐mimetic form of the S. cerevisiae S6Kinase Sch9 (simulating the ‘TORC1 active’ state) largely prevents translation downregulation, assessed by polysome profiling, upon TORC1 inhibition [171]. While TORC1 activity has a major role in regulating eIF4F‐dependent translation initiation, the effect of TORC1 inhibition, and its signalling branches, on translation in response to most stresses remains to be determined.
AMP‐activated protein kinase
AMPK is activated in response to stresses which reduce cellular ability to produce ATP (including hypoxia, amino acid starvation and TORC1 inhibition) by binding AMP/ADP [190, 191]. AMPK activation affects translation by both inhibiting TORC1, its downstream S6Kinase, and activating the ISR [113, 155, 156, 159]. AMPK activation by exercise is associated with eIF4E‐BP1 dephosphorylation, causing a change in the translation of certain mRNAs, although it remains unproven whether this is AMPK dependent and whether the pathway is mediated through TORC1 activity modulation or a parallel mechanism [192]. AMPK further promotes eEF2 phosphorylation, inhibiting translation elongation and ribosome recycling, by both activating eEF2Kinase and sequestering the eEF2Kinase phosphatase PP6C, which is intriguingly activated by TORC1 inhibition [114, 189, 193, 194].
Protein kinase A
PKA is regulated by intracellular levels of cAMP, becoming activated when cAMP binds the inhibitory regulatory subunits, releasing the catalytic subunits [195, 196]. Glucose stimulates cAMP production by boosting ATP generation, leading to PKA activation in S. cerevisiae [196, 197, 198]. While glucose induces mammalian PKA activity towards some substrates, glucose withdrawal can also stimulate PKA [199, 200, 201]. Glucose withdrawal causes both autophagic degradation of a PKA inhibitory subunit and ER stress, another PKA‐activating signal [200, 202, 203]. PKA interacts with other stress‐regulated signalling: TORC1 relieves PKA inhibition in S. cerevisiae, while in mammalian cells, PKA inhibits TORC1 and AMPK signalling [199, 204, 205, 206]. TORC1 and PKA have substantially overlapping downstream target proteins outlining a similar function in promoting cellular anabolism [207]. PKA phosphorylates eIF4B, likely promoting eIF4A activity and thus translation initiation [208]. Counterintuitively, the S. cerevisiae PKA catalytic subunits Tpk2 and Tpk3 are required for translation inhibition upon glucose starvation, and affect translation of certain mRNAs following heat stress: possibly by recruiting specific eIF4G subunits, which can affect stress‐adaptive translation, to the eIF4F complex [142, 209, 210, 211]. The PKA response to stress is multi‐faceted, with a deeper knowledge of the subsequent impacts upon translation still a work in progress. The multitude of downstream targets makes this work exceedingly challenging.
eIF4F‐independent translation initiation
Although ribosome recruitment via the eIF4F complex is the dominant mode of ribosome docking onto mRNA, alternatives exist. These alternatives become particularly relevant following stress when many components of the eIF4F complex are depleted either through degradation (eIF4G), sequestration (eIF4E) or dissociation (eIF4A) [141, 175, 212, 213].
Following viral infection cells shut down translation initiation to try to restrict viral amplification. To circumvent this, many viral mRNAs contain internal ribosome entry sites (IRESs), mRNA secondary structures which can recruit ribosomes in the absence of eIF4F bound to an mRNA 5′ cap [17, 214]. In at least some instances, IRES binding of eIF3 is involved in this recruitment, while other IRES sequences can recruit eIF4F components [215]. Interestingly, an IRES does not have to be upstream of the start AUG codon to promote translation: ribosome recruitment to the 3′UTR can drive increased translation by facilitating ribosome delivery to the 5′UTR [216]. There are additionally hundreds of known eukaryotic IRESs, although care must be taken before concluding that any sequence has IRES activity [217, 218].
While IRESs frequently contain large structural elements, they do not have to be structured. PABPs, which bind to both eIF4G and A‐rich sequences, can bind to unstructured A‐rich 5′UTRs in mRNAs and recruit ribosomes for increased translation upon stress [219, 220]. As different eIF4G proteins are differentially involved in stress‐adaptive translation from mammalian to yeast cells, it is possible that isoform‐specific differences in the PABP binding region could regulate IRES translation of these targets [142]. A‐rich mRNA sequences can also be methylated upon stress by METTL3, allowing binding of ABCF1 to the mRNA in the 5′UTR away from the cap. ABCF1 likely recruits eIF4G for cap‐independent initiation [221]. IRES sequences can circumvent eIF4F‐dependent translation initiation downregulation (thus maintaining translation) or upregulate translation of IRES‐containing mRNAs following stress [222]. IRESs can even facilitate translation of different protein isoforms [223].
eIF3, in addition to the ability to promote IRES‐regulated translation, can bind the mRNA cap and recruit ribosomes for eIF3‐dependent translation through the eIF3d subunit [224]. This method of ribosome recruitment becomes a major route for translation initiation for cells under various stresses [225, 226, 227]. Whether there is a bias of eIF3d binding towards stress‐adaptive mRNAs remains unknown. Recruitment of ribosomes to mRNA in such a distinct manner is likely to contribute to different translational efficiency of mRNAs following stress.
Stress‐induced translation from alternative start codons
While we have so far discussed translation initiation at AUG sites, initiation can also occur at alternative start codons, particularly at similar sequences like CUG [228]. Translation initiation site mapping indicates that up to 50% of translation initiation occurs at non‐AUG sites. Although it is possible that many of these sites are either initiation errors or artefacts of the experimental conditions, many likely encode short ORFs with regulatory functions [229]. A recent excellent review on this topic is available elsewhere [230]. Translation from non‐AUG start codons can be enhanced upon stress, including from two examples upstream of uORF1 on GCN4 mRNA, which may help promote Gcn4 expression [231]. In mammalian cells, the mitochondrial ribosome protein L18 can be translated from a downstream CUG codon upon stress to generate a cytosolic isoform, which helps promote heat shock protein translation [232]. Translation from non‐AUG initiation sites, therefore, may facilitate translational adaptation to stress.
Translation of stress‐adaptive mRNAs at translation hot spots
To boost translation upon stress, defined mRNAs can be recruited to translational platforms, including at regions of dense actin [1, 63, 82, 233, 234, 235]. Several proteins involved with these platforms have a high propensity for phase separation. Stress conditions which promote mRNA relocalization to SGs/PBs may similarly promote localization of certain mRNAs to these regions to increase stress‐responsive translation initiation. Similar phase‐separation‐driven promotion of the translation of translation factors is observed in unstressed conditions [59]. Such an increase in translation could be mediated by local activation of mTORC1 allowing increased local availability of the eIF4E cap‐binding protein for longer bursts of active translation, although this remains speculative [41, 236]. Evidence of whether these hotspots occur in mammalian cells under stress, and how extensively they are used in S. cerevisiae remains to be established.
Effect of stresses on translation elongation
For efficient elongation, there must be sufficient codon‐matched aminoacylated tRNAs alongside space in the A‐site for them to bind. eEF2 is important for translocation of the ribosome, thus vacating the A‐site for the subsequent round of tRNA binding. eEF2 activity is inhibited by phosphorylation from eEF2Kinase which has low activity under unstressed conditions. eEF2Kinase activity is constrained by phosphorylation from the TORC1‐activated S6Kinase (Fig. 4Div) [188, 237]. Upon stress, eEF2Kinase is no longer repressed but is activated by AMPK [194]. eEF2Kinase then phosphorylates and inhibits eEF2. This inhibition is countered by the TORC1‐repressed phosphatase PP6C, which activated AMPK sequesters [114, 193]. This interplay allows fine‐tuning of eEF2 activity according to the type and severity of stress.
tRNAs can be modified under stress conditions to allow for greater wobble at the third base, mitigating against reduced tRNA availability and promoting translation of select transcripts [238, 239]. For some tRNAs, this is insufficient: upon oxidative stress there is a reduction in Trp tRNA availability, leading to ribosome stalling and collisions at the single Trp UGG codon [240]. The increase in uncharged Trp tRNAs additionally leads to ISR activation, affecting translation initiation. Increased collisions could lead to erroneous translation through frameshifting, but this is mitigated by ribosome‐bound factors such as Slf1 [241].
Several ribosomal proteins have paralogues, resulting in numerous possible ‘flavours’ of ribosome, which have different preferences for mRNAs to translate [85]. Such a role has been reported in different tissues, and within single cells [242, 243]. Modulation of ribosome heterogeneity can be controlled by post‐translational modifications affecting ribosomal proteins or rRNA, with impacts upon mRNA translation [244, 245, 246]. This can be at the level of elongation rates through codon selection or through initial mRNA binding, including through IRESs [243, 246, 247]. Switching of the prevalent ribosome ‘flavours’ can occur in stressed yeast cells, and is seemingly more pronounced in the translating pool of ribosomes than the non‐translating pool [248]. Stresses, therefore, affect the type of ribosomes and their interaction with tRNAs to favour elongation of specific mRNAs upon stress.
Effect of stresses on translation termination
Reducing translation termination is a possible further mechanism to increase the abundance of stalled ribosomes and decrease translation. Large‐scale proteomic studies identified no changes in levels of the release factor complex proteins eRF1 and eRF3, or the ribosome recycling protein ABCE1 following UPR induction in mammalian cells [249, 250]. eRF1, eRF3 and ABCE1 remain largely cytosolic following stress, although a fraction of eRF1 and eRF3 have been observed in yeast PBs, and all three are marginally associated with mammalian SGs [251, 252, 253, 254]. Termination factor recruitment to SGs upon oxidative stress is coincident with an increase in stop codon readthrough and free 80S ribosomes, suggestive of a defect in translation termination and subsequent ribosome splitting [142, 254]. Supporting a stress‐induced reduction in ribosome recycling, there is an increase in free, dormant, 80S ribosomes following TORC1 inhibition, potentially mediated by Stm1 in yeast and SREBP1 in mammalian cells [255]. How these changes to termination protein availability affect protein translation needs further investigation through targeted experiments to increase their availability upon stress.
Future perspectives
Knowledge of the what and how of stress‐induced changes in translation is a critical challenge to our understanding of organismal homeostasis. This broad area, covering alterations in the translation efficiency of an individual mRNA, larger scale proteome realignments and the underlying molecular mechanisms remains a field ripe for discovery. Changes to these processes during the course of ageing and disease will likely have clinical relevance and are an important area for future research. Several kinases are known to play a role in translation, but determining the role that changes in their activity play upon stress to affect translation remains, beyond the well‐characterized ISR, largely unclear. Recent technological advances in RBP identification are likely to aid the identification of further important pathways, while the growing awareness of translational hotspots will add more detail to that currently known. The large spread of technical approaches available will power exciting discoveries for many years to come.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
TW: Writing – original draft, reviewing and editing; Figure preparation. AR: Writing – reviewing and editing.
Acknowledgements
The authors were funded by the MRC (grant #MC_UU_00018/8 to AR) and gratefully acknowledge the rest of the Rousseau lab, in particular Flavie Soubigou, and the MRC‐PPU for their support in putting together this review. We further acknowledge the British Society for Cell Biology, the Biochemical Society and the organizers of the Dynamic Cell V conference for giving us the platform which led to this review.
References
- 1. Williams TD & Rousseau A (2022) Actin dynamics in protein homeostasis. Biosci Rep 42, BSR20210848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Guo MS & Gross CA (2014) Stress‐induced remodeling of the bacterial proteome. Curr Biol 24, R424–R434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stankiewicz AM, Jaszczyk A, Goscik J & Juszczak GR (2022) Stress and the brain transcriptome: identifying commonalities and clusters in standardized data from published experiments. Prog Neuropsychopharmacol Biol Psychiatry 119, 110558. [DOI] [PubMed] [Google Scholar]
- 4. Singh A, Kandi AR, Jayaprakashappa D, Thuery G, Purohit DJ, Huelsmeier J, Singh R, Pothapragada SS, Ramaswami M & Bakthavachalu B (2022) The transcriptional response to oxidative stress is independent of stress‐granule formation. Mol Biol Cell 33, ar25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jovic K, Sterken MG, Grilli J, Bevers RPJ, Rodriguez M, Riksen JAG, Allesina S, Kammenga JE & Snoek LB (2017) Temporal dynamics of gene expression in heat‐stressed Caenorhabditis elegans . PLoS One 12, e0189445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li X, Li M, Zhou B, Yang Y, Wei Q & Zhang J (2019) Transcriptome analysis provides insights into the stress response crosstalk in apple (Malus × domestica) subjected to drought, cold and high salinity. Sci Rep 9, 9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Azameti MK, Ranjan A, Singh PK, Gaikwad K, Singh AK, Dalal M, Arora A, Rai V & Padaria JC (2022) Transcriptome profiling reveals the genes and pathways involved in thermo‐tolerance in wheat (Triticum aestivum L.) genotype Raj 3765. Sci Rep 12, 14831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou J, Chen S, Shi W, David‐Schwartz R, Li S, Yang F & Lin Z (2021) Transcriptome profiling reveals the effects of drought tolerance in Giant Juncao. BMC Plant Biol 21, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hughes Hallett JE, Luo X & Capaldi AP (2014) State transitions in the TORC1 signaling pathway and information processing in Saccharomyces cerevisiae . Genetics 198, 773–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gasch AP, Spellman PT, Kao CM, Carmel‐Harel O, Eisen MB, Storz G, Botstein D & Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11, 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Biswas P & Ghosh S (2015) Global transcriptomic profiling of Schizosaccharomyces pombe in response to nitrosative stress. Gene 558, 241–253. [DOI] [PubMed] [Google Scholar]
- 12. Rousseau A & Bertolotti A (2016) An evolutionarily conserved pathway controls proteasome homeostasis. Nature 536, 184–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Buccitelli C & Selbach M (2020) mRNAs, proteins and the emerging principles of gene expression control. Nat Rev Genet 21, 630–644. [DOI] [PubMed] [Google Scholar]
- 14. Nomura W, Futamata R & Inoue Y (2021) Role of RhoGAP Rgd1 in Pkc1 signaling‐related actin repolarization under heat shock stress in Saccharomyces cerevisiae . Biochim Biophys Acta Gen Sub 1865, 129853. [DOI] [PubMed] [Google Scholar]
- 15. Tao G‐Z, Rott LS, Lowe AW & Omary MB (2002) Hyposmotic stress induces cell growth arrest via proteasome activation and cyclin/cyclin‐dependent kinase degradation. J Biol Chem 277, 19295–19303. [DOI] [PubMed] [Google Scholar]
- 16. Aitken CE & Lorsch JR (2012) A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol 19, 568–576. [DOI] [PubMed] [Google Scholar]
- 17. Jackson RJ, Hellen CU & Pestova TV (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11, 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Singh CR, Watanabe R, Chowdhury W, Hiraishi H, Murai MJ, Yamamoto Y, Miles D, Ikeda Y, Asano M & Asano K (2012) Sequential eukaryotic translation initiation factor 5 (eIF5) binding to the charged disordered segments of eIF4G and eIF2β stabilizes the 48S preinitiation complex and promotes its shift to the initiation mode. Mol Cell Biol 32, 3978–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Villa N, Do A, Hershey JWB & Fraser CS (2013) Human eukaryotic initiation factor 4G (eIF4G) protein binds to eIF3c, ‐d, and ‐e to promote mRNA recruitment to the ribosome. J Biol Chem 288, 32932–32940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sokabe M & Fraser CS (2017) A helicase‐independent activity of eIF4A in promoting mRNA recruitment to the human ribosome. Proc Natl Acad Sci USA 114, 6304–6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harms U, Andreou AZ, Gubaev A & Klostermeier D (2014) eIF4B, eIF4G and RNA regulate eIF4A activity in translation initiation by modulating the eIF4A conformational cycle. Nucleic Acids Res 42, 7911–7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Duncan CDS & Mata J (2022) Translation‐complex profiling of fission yeast cells reveals dynamic rearrangements of scanning ribosomal subunits upon nutritional stress. Nucleic Acids Res 50, 13011–13025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wagner S, Herrmannová A, Hronová V, Gunišová S, Sen ND, Hannan RD, Hinnebusch AG, Shirokikh NE, Preiss T & Valášek LS (2020) Selective translation complex profiling reveals staged initiation and co‐translational assembly of initiation factor complexes. Mol Cell 79, 546–560.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Feoktistova K, Tuvshintogs E, Do A & Fraser CS (2013) Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc Natl Acad Sci USA 110, 13339–13344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Llácer JL, Hussain T, Marler L, Aitken CE, Thakur A, Lorsch JR, Hinnebusch AG & Ramakrishnan V (2015) Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol Cell 59, 399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang J, Wang J, Shin B‐S, Kim J‐R, Dever TE, Puglisi JD & Fernández IS (2020) Structural basis for the transition from translation initiation to elongation by an 80S‐eIF5B complex. Nat Commun 11, 5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schuller AP & Green R (2018) Roadblocks and resolutions in eukaryotic translation. Nat Rev Mol Cell Biol 19, 526–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Djumagulov M, Demeshkina N, Jenner L, Rozov A, Yusupov M & Yusupova G (2021) Accuracy mechanism of eukaryotic ribosome translocation. Nature 600, 543–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kaul G, Pattan G & Rafeequi T (2011) Eukaryotic elongation factor‐2 (eEF2): its regulation and peptide chain elongation. Cell Biochem Funct 29, 227–234. [DOI] [PubMed] [Google Scholar]
- 30. Shao S, Murray J, Brown A, Taunton J, Ramakrishnan V & Hegde RS (2016) Decoding mammalian ribosome‐mRNA states by translational GTPase complexes. Cell 167, 1229–1240.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eliseev B, Kryuchkova P, Alkalaeva E & Frolova L (2010) A single amino acid change of translation termination factor eRF1 switches between bipotent and omnipotent stop‐codon specificity. Nucleic Acids Res 39, 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Frolova L, Le Goff X, Zhouravleva G, Davydova E, Philippe M & Kisselev L (1996) Eukaryotic polypeptide chain release factor eRF3 is an eRF1‐ and ribosome‐dependent guanosine triphosphatase. RNA 2, 334–341. [PMC free article] [PubMed] [Google Scholar]
- 33. Shoemaker CJ & Green R (2011) Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci USA 108, E1392–E1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, Armache J‐P, Sieber H, Ungewickell C, Berninghausen O, Daberkow I et al. (2012) Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature 482, 501–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cheng S & Gallie DR (2007) eIF4G, eIFiso4G, and eIF4B bind the poly(a)‐binding protein through overlapping sites within the RNA recognition motif domains. J Biol Chem 282, 25247–25258. [DOI] [PubMed] [Google Scholar]
- 36. Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou MN & Sonenberg N (2005) Mammalian poly(a)‐binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev 19, 104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ivanov A, Mikhailova T, Eliseev B, Yeramala L, Sokolova E, Susorov D, Shuvalov A, Schaffitzel C & Alkalaeva E (2016) PABP enhances release factor recruitment and stop codon recognition during translation termination. Nucleic Acids Res 44, 7766–7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Costello J, Castelli LM, Rowe W, Kershaw CJ, Talavera D, Mohammad‐Qureshi SS, Sims PF, Grant CM, Pavitt GD, Hubbard SJ et al. (2015) Global mRNA selection mechanisms for translation initiation. Genome Biol 16, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thompson MK & Gilbert WV (2017) mRNA length‐sensing in eukaryotic translation: reconsidering the “closed loop” and its implications for translational control. Curr Genet 63, 613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Machida K, Shigeta T, Yamamoto Y, Ito T, Svitkin Y, Sonenberg N & Imataka H (2018) Dynamic interaction of poly(a)‐binding protein with the ribosome. Sci Rep 8, 17435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Livingston NM, Kwon J, Valera O, Saba JA, Sinha NK, Reddy P, Nelson B, Wolfe C, Ha T, Green R et al. (2023) Bursting translation on single mRNAs in live cells. Mol Cell 83, 2276–2289.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tunnacliffe E & Chubb JR (2020) What is a transcriptional burst? Trends Genet 36, 288–297. [DOI] [PubMed] [Google Scholar]
- 43. Edfors F, Danielsson F, Hallström BM, Käll L, Lundberg E, Pontén F, Forsström B & Uhlén M (2016) Gene‐specific correlation of RNA and protein levels in human cells and tissues. Mol Syst Biol 12, 883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mak T, Jones AW & Nurse P (2021) The TOR‐dependent phosphoproteome and regulation of cellular protein synthesis. EMBO J 40, e107911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schmidt EK, Clavarino G, Ceppi M & Pierre P (2009) SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 6, 275–277. [DOI] [PubMed] [Google Scholar]
- 46. Argüello RJ, Reverendo M, Mendes A, Camosseto V, Torres AG, Ribas de Pouplana L, van de Pavert SA, Gatti E & Pierre P (2018) SunRiSE – measuring translation elongation at single‐cell resolution by means of flow cytometry. J Cell Sci 131, jcs214346. [DOI] [PubMed] [Google Scholar]
- 47. Aviner R (2020) The science of puromycin: from studies of ribosome function to applications in biotechnology. Computat Struct Biotechnol J 18, 1074–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Klann K, Tascher G & Münch C (2020) Functional translatome proteomics reveal converging and dose‐dependent regulation by mTORC1 and eIF2α. Mol Cell 77, 913–925.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Brar GA & Weissman JS (2015) Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat Rev Mol Cell Biol 16, 651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chassé H, Boulben S, Costache V, Cormier P & Morales J (2017) Analysis of translation using polysome profiling. Nucleic Acids Res 45, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cacioppo R & Lindon C (2023) Immunoprecipitation of reporter nascent chains from active ribosomes to study translation efficiency. Bio Protoc 13, e4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Boersma S, Khuperkar D, Verhagen BMP, Sonneveld S, Grimm JB, Lavis LD & Tanenbaum ME (2019) Multi‐color single‐molecule imaging uncovers extensive heterogeneity in mRNA decoding. Cell 178, 458–472.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS & Vale RD (2014) A protein‐tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Riggs CL, Kedersha N, Ivanov P & Anderson P (2020) Mammalian stress granules and P bodies at a glance. J Cell Sci 133, jcs242487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Marcelo A, Koppenol R, de Almeida LP, Matos CA & Nóbrega C (2021) Stress granules, RNA‐binding proteins and polyglutamine diseases: too much aggregation? Cell Death Dis 12, 592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Aulas A, Fay MM, Lyons SM, Achorn CA, Kedersha N, Anderson P & Ivanov P (2017) Stress‐specific differences in assembly and composition of stress granules and related foci. J Cell Sci 130, 927–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mateju D, Eichenberger B, Voigt F, Eglinger J, Roth G & Chao JA (2020) Single‐molecule imaging reveals translation of mRNAs localized to stress granules. Cell 183, 1801–1812.e13. [DOI] [PubMed] [Google Scholar]
- 58. Lui J, Castelli LM, Pizzinga M, Simpson CE, Hoyle NP, Bailey KL, Campbell SG & Ashe MP (2014) Granules harboring translationally active mRNAs provide a platform for P‐body formation following stress. Cell Rep 9, 944–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pizzinga M, Bates C, Lui J, Forte G, Morales‐Polanco F, Linney E, Knotkova B, Wilson B, Solari CA, Berchowitz LE et al. (2019) Translation factor mRNA granules direct protein synthetic capacity to regions of polarized growth. J Cell Biol 218, 1564–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Morales‐Polanco F, Bates C, Lui J, Casson J, Solari CA, Pizzinga M, Forte G, Griffin C, Garner KEL, Burt HE et al. (2021) Core fermentation (CoFe) granules focus coordinated glycolytic mRNA localization and translation to fuel glucose fermentation. iScience 24, 102069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Luo Y, Pratihar S, Horste EH, Mitschka S, Mey ASJS, Al‐Hashimi HM & Mayr C (2023) mRNA interactions with disordered regions control protein activity. bioRxiv 2023.02.18.529068. 10.1101/2023.02.18.529068 [PREPRINT] [DOI]
- 62. Ma W & Mayr C (2018) A membraneless organelle associated with the endoplasmic reticulum enables 3′UTR‐mediated protein‐protein interactions. Cell 175, 1492–1506.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Williams TD, Winaya AR, Joshua I & Rousseau A (2023) Proteasome assembly chaperone translation upon stress requires Ede1 phase separation at the plasma membrane. iScience 27, 108732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Luo Y, Na Z & Slavoff SA (2018) P‐bodies: composition, properties, and functions. Biochemistry 57, 2424–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Eulalio A, Behm‐Ansmant I, Schweizer D & Izaurralde E (2007) P‐body formation is a consequence, not the cause, of RNA‐mediated gene silencing. Mol Cell Biol 27, 3970–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Horvathova I, Voigt F, Kotrys AV, Zhan Y, Artus‐Revel CG, Eglinger J, Stadler MB, Giorgetti L & Chao JA (2017) The dynamics of mRNA turnover revealed by single‐molecule imaging in single cells. Mol Cell 68, 615–625.e9. [DOI] [PubMed] [Google Scholar]
- 67. Brothers WR, Fakim H, Kajjo S & Fabian MR (2022) P‐bodies directly regulate MARF1‐mediated mRNA decay in human cells. Nucleic Acids Res 50, 7623–7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Goetz AE & Wilkinson M (2017) Stress and the nonsense‐mediated RNA decay pathway. Cell Mol Life Sci 74, 3509–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Xia X (2019) Translation control of HAC1 by regulation of splicing in Saccharomyces cerevisiae . Int J Mol Sci 20, 2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cherry PD, Peach SE & Hesselberth JR (2019) Multiple decay events target HAC1 mRNA during splicing to regulate the unfolded protein response. eLife 8, e42262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Laloum T, Martín G & Duque P (2018) Alternative splicing control of abiotic stress responses. Trends Plant Sci 23, 140–150. [DOI] [PubMed] [Google Scholar]
- 72. Zhang X, Zhang X, Yuan J & Li F (2023) The responses of alternative splicing during heat stress in the Pacific white shrimp Litopenaeus vannamei . Genes (Basel) 14, 1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kucherenko MM & Shcherbata HR (2018) miRNA targeting and alternative splicing in the stress response – events hosted by membrane‐less compartments. J Cell Sci 131, jcs202002. [DOI] [PubMed] [Google Scholar]
- 74. Blevins WR, Tavella T, Moro SG, Blasco‐Moreno B, Closa‐Mosquera A, Díez J, Carey LB & Albà MM (2019) Extensive post‐transcriptional buffering of gene expression in the response to severe oxidative stress in baker's yeast. Sci Rep 9, 11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Crawford RA & Pavitt GD (2019) Translational regulation in response to stress in Saccharomyces cerevisiae . Yeast 36, 5–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Matia‐González AM, Jabre I, Laing EE & Gerber AP (2021) Oxidative stress induces coordinated remodeling of RNA‐enzyme interactions. iScience 24, 102753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shchepachev V, Bresson S, Spanos C, Petfalski E, Fischer L, Rappsilber J & Tollervey D (2019) Defining the RNA interactome by total RNA‐associated protein purification. Mol Syst Biol 15, e8689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Panasenko OO, Somasekharan SP, Villanyi Z, Zagatti M, Bezrukov F, Rashpa R, Cornut J, Iqbal J, Longis M, Carl SH et al. (2019) Co‐translational assembly of proteasome subunits in NOT1‐containing assemblysomes. Nat Struct Mol Biol 26, 110–120. [DOI] [PubMed] [Google Scholar]
- 79. Ho JJD, Balukoff NC, Theodoridis PR, Wang M, Krieger JR, Schatz JH & Lee S (2020) A network of RNA‐binding proteins controls translation efficiency to activate anaerobic metabolism. Nat Commun 11, 2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wilkinson N & Pantopoulos K (2014) The IRP/IRE system in vivo: insights from mouse models. Front Pharmacol 5, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Liu J, Schmitz JC, Lin X, Tai N, Yan W, Farrell M, Bailly M, Chen T & Chu E (2002) Thymidylate synthase as a translational regulator of cellular gene expression. Biochim Biophys Acta 1587, 174–182. [DOI] [PubMed] [Google Scholar]
- 82. Nagaoka K, Udagawa T & Richter JD (2012) CPEB‐mediated ZO‐1 mRNA localization is required for epithelial tight‐junction assembly and cell polarity. Nat Commun 3, 675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Duran‐Arqué B, Cañete M, Castellazzi CL, Bartomeu A, Ferrer‐Caelles A, Reina O, Caballé A, Gay M, Arauz‐Garofalo G, Belloc E et al. (2022) Comparative analyses of vertebrate CPEB proteins define two subfamilies with coordinated yet distinct functions in post‐transcriptional gene regulation. Genome Biol 23, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Fay MM & Anderson PJ (2018) The role of RNA in biological phase separations. J Mol Biol 430, 4685–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bates C, Hubbard SJ & Ashe MP (2018) Ribosomal flavours: an acquired taste for specific mRNAs? Biochem Soc Trans 46, 1529–1539. [DOI] [PubMed] [Google Scholar]
- 86. Netzer N, Goodenbour JM, David A, Dittmar KA, Jones RB, Schneider JR, Boone D, Eves EM, Rosner MR, Gibbs JS et al. (2009) Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature 462, 522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wiltrout E, Goodenbour JM, Fréchin M & Pan T (2012) Misacylation of tRNA with methionine in Saccharomyces cerevisiae . Nucleic Acids Res 40, 10494–10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Rubio Gomez MA & Ibba M (2020) Aminoacyl‐tRNA synthetases. RNA 26, 910–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Saikia M & Hatzoglou M (2015) The many virtues of tRNA‐derived stress‐induced RNAs (tiRNAs): discovering novel mechanisms of stress response and effect on human health. J Biol Chem 290, 29761–29768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Jewer M, Lee L, Leibovitch M, Zhang G, Liu J, Findlay SD, Vincent KM, Tandoc K, Dieters‐Castator D, Quail DF et al. (2020) Translational control of breast cancer plasticity. Nat Commun 11, 2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Payea MJ, Dar SA, Anerillas C, Martindale JL, Belair C, Munk R, Malla S, Fan J, Piao Y, Yang X et al. (2023) Senescence suppresses the integrated stress response and activates a stress‐enhanced secretory phenotype. bioRxiv 2023.04.12.536613. 10.1101/2023.04.12.536613 [PREPRINT] [DOI] [PMC free article] [PubMed]
- 92. Mesclon F, Lambert‐Langlais S, Carraro V, Parry L, Hainault I, Jousse C, Maurin AC, Bruhat A, Fafournoux P & Averous J (2017) Decreased ATF4 expression as a mechanism of acquired resistance to long‐term amino acid limitation in cancer cells. Oncotarget 8, 27440–27453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. O'Neill JS, Hoyle NP, Robertson JB, Edgar RS, Beale AD, Peak‐Chew SY, Day J, Costa ASH, Frezza C & Causton HC (2020) Eukaryotic cell biology is temporally coordinated to support the energetic demands of protein homeostasis. Nat Commun 11, 4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Klein P, Kallenberger SM, Roth H, Roth K, Ly‐Hartig TBN, Magg V, Aleš J, Talemi SR, Qiang Y, Wolf S et al. (2022) Temporal control of the integrated stress response by a stochastic molecular switch. Sci Adv 8, eabk2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wek RC (2018) Role of eIF2α kinases in translational control and adaptation to cellular stress. Cold Spring Harb Perspect Biol 10, a032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Costa‐Mattioli M & Walter P (2020) The integrated stress response: from mechanism to disease. Science 368, eaat5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Hinnebusch AG (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59, 407–450. [DOI] [PubMed] [Google Scholar]
- 98. Harding HP, Zhang Y & Ron D (1999) Protein translation and folding are coupled by an endoplasmic‐reticulum‐resident kinase. Nature 397, 271–274. [DOI] [PubMed] [Google Scholar]
- 99. McEwen E, Kedersha N, Song B, Scheuner D, Gilks N, Han A, Chen J‐J, Anderson P & Kaufman RJ (2005) Heme‐regulated inhibitor kinase‐mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J Biol Chem 280, 16925–16933. [DOI] [PubMed] [Google Scholar]
- 100. Koumenis C, Naczki C, Koritzinsky M, Rastani S, Diehl A, Sonenberg N, Koromilas A & Wouters BG (2002) Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2alpha. Mol Cell Biol 22, 7405–7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. García MA, Meurs EF & Esteban M (2007) The dsRNA protein kinase PKR: virus and cell control. Biochimie 89, 799–811. [DOI] [PubMed] [Google Scholar]
- 102. García MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C & Esteban M (2006) Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev 70, 1032–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ito T, Yang M & May WS (1999) RAX, a cellular activator for double‐stranded RNA‐dependent protein kinase during stress signaling. J Biol Chem 274, 15427–15432. [DOI] [PubMed] [Google Scholar]
- 104. Pindel A & Sadler A (2011) The role of protein kinase R in the interferon response. J Interferon Cytokine Res 31, 59–70. [DOI] [PubMed] [Google Scholar]
- 105. Suragani RN, Zachariah RS, Velazquez JG, Liu S, Sun CW, Townes TM & Chen JJ (2012) Heme‐regulated eIF2α kinase activated Atf4 signaling pathway in oxidative stress and erythropoiesis. Blood 119, 5276–5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Girardin SE, Cuziol C, Philpott DJ & Arnoult D (2021) The eIF2α kinase HRI in innate immunity, proteostasis, and mitochondrial stress. FEBS J 288, 3094–3107. [DOI] [PubMed] [Google Scholar]
- 107. Yerlikaya A, Kimball SR & Stanley BA (2008) Phosphorylation of eIF2alpha in response to 26S proteasome inhibition is mediated by the haem‐regulated inhibitor (HRI) kinase. Biochem J 412, 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Lu L, Han AP & Chen JJ (2001) Translation initiation control by heme‐regulated eukaryotic initiation factor 2alpha kinase in erythroid cells under cytoplasmic stresses. Mol Cell Biol 21, 7971–7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Chen J‐J (2006) Regulation of protein synthesis by the heme‐regulated eIF2α kinase: relevance to anemias. Blood 109, 2693–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Fessler E, Eckl E‐M, Schmitt S, Mancilla IA, Meyer‐Bender MF, Hanf M, Philippou‐Massier J, Krebs S, Zischka H & Jae LT (2020) A pathway coordinated by DELE1 relays mitochondrial stress to the cytosol. Nature 579, 433–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Guo X, Aviles G, Liu Y, Tian R, Unger BA, Lin Y‐HT, Wiita AP, Xu K, Correia MA & Kampmann M (2020) Mitochondrial stress is relayed to the cytosol by an OMA1–DELE1–HRI pathway. Nature 579, 427–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Anda S, Zach R & Grallert B (2017) Activation of Gcn2 in response to different stresses. PLoS One 12, e0182143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Cherkasova V, Qiu H & Hinnebusch AG (2010) Snf1 promotes phosphorylation of the alpha subunit of eukaryotic translation initiation factor 2 by activating Gcn2 and inhibiting phosphatases Glc7 and Sit4. Mol Cell Biol 30, 2862–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wengrod J, Wang D, Weiss S, Zhong H, Osman I & Gardner LB (2015) Phosphorylation of eIF2α triggered by mTORC1 inhibition and PP6C activation is required for autophagy and is aberrant in PP6C‐mutated melanoma. Sci Signal 8, ra27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Cherkasova VA & Hinnebusch AG (2003) Translational control by TOR and TAP42 through dephosphorylation of eIF2alpha kinase GCN2. Genes Dev 17, 859–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Inglis AJ, Masson GR, Shao S, Perisic O, McLaughlin SH, Hegde RS & Williams RL (2019) Activation of GCN2 by the ribosomal P‐stalk. Proc Natl Acad Sci USA 116, 4946–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Dong J, Qiu H, Garcia‐Barrio M, Anderson J & Hinnebusch AG (2000) Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA‐binding domain. Mol Cell 6, 269–279. [DOI] [PubMed] [Google Scholar]
- 118. Harding HP, Ordonez A, Allen F, Parts L, Inglis AJ, Williams RL & Ron D (2019) The ribosomal P‐stalk couples amino acid starvation to GCN2 activation in mammalian cells. eLife 8, e50149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Silva RC, Sattlegger E & Castilho BA (2016) Perturbations in actin dynamics reconfigure protein complexes that modulate GCN2 activity and promote an eIF2 response. J Cell Sci 129, 4521–4533. [DOI] [PubMed] [Google Scholar]
- 120. Augusto L, Amin PH, Wek RC & Sullivan WJ Jr (2019) Regulation of arginine transport by GCN2 eIF2 kinase is important for replication of the intracellular parasite toxoplasma gondii. PLoS Pathog 15, e1007746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Masson GR (2019) Towards a model of GCN2 activation. Biochem Soc Trans 47, 1481–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Sattlegger E, Barbosa JARG, Moraes MCS, Martins RM, Hinnebusch AG & Castilho BA (2011) Gcn1 and actin binding to Yih1: implications for activation of the eIF2 kinase GCN2. J Biol Chem 286, 10341–10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Visweswaraiah J, Lageix S, Castilho BA, Izotova L, Kinzy TG, Hinnebusch AG & Sattlegger E (2011) Evidence that eukaryotic translation elongation factor 1A (eEF1A) binds the Gcn2 protein C terminus and inhibits Gcn2 activity. J Biol Chem 286, 36568–36579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Yuan W, Guo S, Gao J, Zhong M, Yan G, Wu W, Chao Y & Jiang Y (2017) General control nonderepressible 2 (GCN2) kinase inhibits target of rapamycin complex 1 in response to amino acid starvation in Saccharomyces cerevisiae. J Biol Chem 292, 2660–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Ye J, Palm W, Peng M, King B, Lindsten T, Li MO, Koumenis C & Thompson CB (2015) GCN2 sustains mTORC1 suppression upon amino acid deprivation by inducing Sestrin2. Genes Dev 29, 2331–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Averous J, Lambert‐Langlais S, Mesclon F, Carraro V, Parry L, Jousse C, Bruhat A, Maurin AC, Pierre P, Proud CG et al. (2016) GCN2 contributes to mTORC1 inhibition by leucine deprivation through an ATF4 independent mechanism. Sci Rep 6, 27698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Kenner LR, Anand AA, Nguyen HC, Myasnikov AG, Klose CJ, McGeever LA, Tsai JC, Miller‐Vedam LE, Walter P & Frost A (2019) eIF2B‐catalyzed nucleotide exchange and phosphoregulation by the integrated stress response. Science 364, 491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Kapp LD & Lorsch JR (2004) GTP‐dependent recognition of the methionine moiety on initiator tRNA by translation factor eIF2. J Mol Biol 335, 923–936. [DOI] [PubMed] [Google Scholar]
- 129. Zhang G, Wang X, Li C, Li Q, An YA, Luo X, Deng Y, Gillette TG, Scherer PE & Wang ZV (2021) Integrated stress response couples mitochondrial protein translation with oxidative stress control. Circulation 144, 1500–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Rodrigues L, Graça RSF & Carneiro LAM (2018) Integrated stress responses to bacterial pathogenesis patterns. Front Immunol 9, 1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Stonyte V, Boye E & Grallert B (2018) Regulation of global translation during the cell cycle. J Cell Sci 131, jcs220327. [DOI] [PubMed] [Google Scholar]
- 132. Szamecz B, Rutkai E, Cuchalová L, Munzarová V, Herrmannová A, Nielsen KH, Burela L, Hinnebusch AG & Valásek L (2008) eIF3a cooperates with sequences 5′ of uORF1 to promote resumption of scanning by post‐termination ribosomes for reinitiation on GCN4 mRNA. Genes Dev 22, 2414–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Lin Y, Li F, Huang L, Polte C, Duan H, Fang J, Sun L, Xing X, Tian G, Cheng Y et al. (2020) eIF3 associates with 80S ribosomes to promote translation elongation, mitochondrial homeostasis, and muscle health. Mol Cell 79, 575–587.e7. [DOI] [PubMed] [Google Scholar]
- 134. des Georges A, Dhote V, Kuhn L, Hellen CUT, Pestova TV, Frank J & Hashem Y (2015) Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature 525, 491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. You T, Stansfield I, Romano MC, Brown AJP & Coghill GM (2011) Analysing GCN4 translational control in yeast by stochastic chemical kinetics modelling and simulation. BMC Syst Biol 5, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Vasudevan D, Neuman SD, Yang A, Lough L, Brown B, Bashirullah A, Cardozo T & Ryoo HD (2020) Translational induction of ATF4 during integrated stress response requires noncanonical initiation factors eIF2D and DENR. Nat Commun 11, 4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Young DJ, Makeeva DS, Zhang F, Anisimova AS, Stolboushkina EA, Ghobakhlou F, Shatsky IN, Dmitriev SE, Hinnebusch AG & Guydosh NR (2018) Tma64/eIF2D, Tma20/MCT‐1, and Tma22/DENR recycle post‐termination 40S subunits in vivo. Mol Cell 71, 761–774.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Moro SG, Hermans C, Ruiz‐Orera J & Albà MM (2021) Impact of uORFs in mediating regulation of translation in stress conditions. BMC Mol Cell Biol 22, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Young SK, Palam LR, Wu C, Sachs MS & Wek RC (2016) Ribosome elongation stall directs gene‐specific translation in the integrated stress response. J Biol Chem 291, 6546–6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Zhigailov AV, Alexandrova AM, Nizkorodova AS, Stanbekova GE, Kryldakov RV, Karpova OV, Polimbetova NS, Halford NG & Iskakov BK (2020) Evidence that phosphorylation of the α‐subunit of eIF2 does not essentially inhibit mRNA translation in wheat germ cell‐free system. Front Plant Sci 11, 936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Black A, Williams TD, Soubigou F, Joshua IM, Zhou H, Lamoliatte F & Rousseau A (2023) The ribosome‐associated chaperone Zuo1 controls translation upon TORC1 inhibition. EMBO J 42, e113240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Cunningham J, Sfakianos AP, Kritsiligkou P, Kershaw CJ, Whitmarsh AJ, Hubbard SJ, Ashe MP & Grant CM (2023) Paralogous translation factors target distinct mRNAs to differentially regulate tolerance to oxidative stress in yeast. Nucleic Acids Res 51, 8820–8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Torrence ME, MacArthur MR, Hosios AM, Valvezan AJ, Asara JM, Mitchell JR & Manning BD (2021) The mTORC1‐mediated activation of ATF4 promotes protein and glutathione synthesis downstream of growth signals. eLife 10, e63326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Park Y, Reyna‐Neyra A, Philippe L & Thoreen CC (2017) mTORC1 balances cellular amino acid supply with demand for protein synthesis through post‐transcriptional control of ATF4. Cell Rep 19, 1083–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Choy MS, Yusoff P, Lee IC, Newton JC, Goh CW, Page R, Shenolikar S & Peti W (2015) Structural and functional analysis of the GADD34:PP1 eIF2α phosphatase. Cell Rep 11, 1885–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Novoa I, Zeng H, Harding HP & Ron D (2001) Feedback inhibition of the unfolded protein response by GADD34‐mediated dephosphorylation of eIF2alpha. J Cell Biol 153, 1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Chambers JE, Dalton LE, Clarke HJ, Malzer E, Dominicus CS, Patel V, Moorhead G, Ron D & Marciniak SJ (2015) Actin dynamics tune the integrated stress response by regulating eukaryotic initiation factor 2α dephosphorylation. eLife 4, e04872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Morozumi Y & Shiozaki K (2021) Conserved and divergent mechanisms that control TORC1 in yeasts and mammals. Genes (Basel) 12, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Deldicque L, Bertrand L, Patton A, Francaux M & Baar K (2011) ER stress induces anabolic resistance in muscle cells through PKB‐induced blockade of mTORC1. PLoS One 6, e20993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Takahara T & Maeda T (2012) Transient sequestration of TORC1 into stress granules during heat stress. Mol Cell 47, 242–252. [DOI] [PubMed] [Google Scholar]
- 151. Sun J, Conn CS, Han Y, Yeung V & Qian S‐B (2011) PI3K‐mTORC1 attenuates stress response by inhibiting cap‐independent Hsp70 translation. J Biol Chem 286, 6791–6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Yan G, Lai Y & Jiang Y (2012) The TOR complex 1 is a direct target of Rho1 GTPase. Mol Cell 45, 743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Yokota H, Gomi K & Shintani T (2017) Induction of autophagy by phosphate starvation in an Atg11‐dependent manner in Saccharomyces cerevisiae . Biochem Biophys Res Commun 483, 522–527. [DOI] [PubMed] [Google Scholar]
- 154. Su KH & Dai C (2017) mTORC1 senses stresses: coupling stress to proteostasis. Bioessays 39, doi: 10.1002/bies.201600268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE & Shaw RJ (2008) AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 30, 214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Hughes Hallett JE, Luo X & Capaldi AP (2015) Snf1/AMPK promotes the formation of Kog1/raptor‐bodies to increase the activation threshold of TORC1 in budding yeast. eLife 4, e09181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Cam H, Easton JB, High A & Houghton PJ (2010) mTORC1 signaling under hypoxic conditions is controlled by ATM‐dependent phosphorylation of HIF‐1α. Mol Cell 40, 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Sfakianos AP, Mellor LE, Pang YF, Kritsiligkou P, Needs H, Abou‐Hamdan H, Désaubry L, Poulin GB, Ashe MP & Whitmarsh AJ (2018) The mTOR‐S6 kinase pathway promotes stress granule assembly. Cell Death Differ 25, 1766–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Caligaris M, Nicastro R, Hu Z, Tripodi F, Hummel JE, Pillet B, Deprez M‐A, Winderickx J, Rospert S, Coccetti P et al. (2023) Snf1/AMPK fine‐tunes TORC1 signaling in response to glucose starvation. eLife 12, e84319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. van Dam TJ, Zwartkruis FJ, Bos JL & Snel B (2011) Evolution of the TOR pathway. J Mol Evol 73, 209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. De Virgilio C & Loewith R (2006) Cell growth control: little eukaryotes make big contributions. Oncogene 25, 6392–6415. [DOI] [PubMed] [Google Scholar]
- 162. Berkowitz O, Jost R, Pollmann S & Masle J (2008) Characterization of TCTP, the translationally controlled tumor protein, from Arabidopsis thaliana . Plant Cell 20, 3430–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Hatakeyama R, Péli‐Gulli MP, Hu Z, Jaquenoud M, Garcia Osuna GM, Sardu A, Dengjel J & De Virgilio C (2019) Spatially distinct pools of TORC1 balance protein homeostasis. Mol Cell 73, 325–338.e8. [DOI] [PubMed] [Google Scholar]
- 164. Laribee RN & Weisman R (2020) Nuclear functions of TOR: impact on transcription and the epigenome. Genes (Basel) 11, 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Makhoul C & Gleeson PA (2021) Regulation of mTORC1 activity by the Golgi apparatus. Fac Rev 10, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Hanker AB, Mitin N, Wilder RS, Henske EP, Tamanoi F, Cox AD & Der CJ (2010) Differential requirement of CAAX‐mediated posttranslational processing for Rheb localization and signaling. Oncogene 29, 380–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Zhong Y, Zhou X, Guan KL & Zhang J (2022) Rheb regulates nuclear mTORC1 activity independent of farnesylation. Cell Chem Biol 29, 1037–1045.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Chauvin C, Koka V, Nouschi A, Mieulet V, Hoareau‐Aveilla C, Dreazen A, Cagnard N, Carpentier W, Kiss T, Meyuhas O et al. (2014) Ribosomal protein S6 kinase activity controls the ribosome biogenesis transcriptional program. Oncogene 33, 474–483. [DOI] [PubMed] [Google Scholar]
- 169. Huber A, French SL, Tekotte H, Yerlikaya S, Stahl M, Perepelkina MP, Tyers M, Rougemont J, Beyer AL & Loewith R (2011) Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. EMBO J 30, 3052–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Reinhard C, Fernandez A, Lamb NJ & Thomas G (1994) Nuclear localization of p85s6k: functional requirement for entry into S phase. EMBO J 13, 1557–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H et al. (2007) Sch9 is a major target of TORC1 in Saccharomyces cerevisiae . Mol Cell 26, 663–674. [DOI] [PubMed] [Google Scholar]
- 172. González A, Shimobayashi M, Eisenberg T, Merle DA, Pendl T, Hall MN & Moustafa T (2015) TORC1 promotes phosphorylation of ribosomal protein S6 via the AGC kinase Ypk3 in Saccharomyces cerevisiae . PLoS One 10, e0120250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Yerlikaya S, Meusburger M, Kumari R, Huber A, Anrather D, Costanzo M, Boone C, Ammerer G, Baranov PV & Loewith R (2016) TORC1 and TORC2 work together to regulate ribosomal protein S6 phosphorylation in Saccharomyces cerevisiae . Mol Biol Cell 27, 397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Bohlen J, Roiuk M & Teleman AA (2021) Phosphorylation of ribosomal protein S6 differentially affects mRNA translation based on ORF length. Nucleic Acids Res 49, 13062–13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Romagnoli A, D'Agostino M, Ardiccioni C, Maracci C, Motta S, La Teana A & Di Marino D (2021) Control of the eIF4E activity: structural insights and pharmacological implications. Cell Mol Life Sci 78, 6869–6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Beauchamp EM & Platanias LC (2013) The evolution of the TOR pathway and its role in cancer. Oncogene 32, 3923–3932. [DOI] [PubMed] [Google Scholar]
- 177. Sewell AK, Poss ZC, Ebmeier CC, Jacobsen JR, Old WM & Han M (2022) The TORC1 phosphoproteome in C. elegans reveals roles in transcription and autophagy. iScience 25, 104186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Otsubo Y, Nakashima A, Yamamoto M & Yamashita A (2017) TORC1‐dependent phosphorylation targets in fission yeast. Biomolecules 7, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Jaiswal P & Kimmel AR (2019) mTORC1/AMPK responses define a core gene set for developmental cell fate switching. BMC Biol 17, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Kamada Y, Ando R, Izawa S & Matsuura A (2023) Yeast tor complex 1 phosphorylates eIF4E‐binding protein, Caf20. Genes Cells 28, 789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Raught B, Gingras AC, Gygi SP, Imataka H, Morino S, Gradi A, Aebersold R & Sonenberg N (2000) Serum‐stimulated, rapamycin‐sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. EMBO J 19, 434–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Tsapras P & Nezis IP (2017) Caspase involvement in autophagy. Cell Death Differ 24, 1369–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Clemens MJ, Bushell M & Morley SJ (1998) Degradation of eukaryotic polypeptide chain initiation factor (eIF) 4G in response to induction of apoptosis in human lymphoma cell lines. Oncogene 17, 2921–2931. [DOI] [PubMed] [Google Scholar]
- 184. Tsokanos F‐F, Albert M‐A, Demetriades C, Spirohn K, Boutros M & Teleman AA (2016) eIF4A inactivates TORC1 in response to amino acid starvation. EMBO J 35, 1058–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Liedmann S, Liu X, Guy CS, Crawford JC, Rodriguez DA, Kuzuoğlu‐Öztürk D, Guo A, Verbist KC, Temirov J, Chen MJ et al. (2022) Localization of a TORC1‐eIF4F translation complex during CD8+ T cell activation drives divergent cell fate. Mol Cell 82, 2401–2414.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Raught B, Peiretti F, Gingras AC, Livingstone M, Shahbazian D, Mayeur GL, Polakiewicz RD, Sonenberg N & Hershey JW (2004) Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J 23, 1761–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE & Pagano M (2006) S6K1‐ and betaTRCP‐mediated degradation of PDCD4 promotes protein translation and cell growth. Science 314, 467–471. [DOI] [PubMed] [Google Scholar]
- 188. Wang X, Li W, Williams M, Terada N, Alessi DR & Proud CG (2001) Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J 20, 4370–4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Smith PR, Loerch S, Kunder N, Stanowick AD, Lou T‐F & Campbell ZT (2021) Functionally distinct roles for eEF2K in the control of ribosome availability and p‐body abundance. Nat Commun 12, 6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Xiao B, Sanders MJ, Underwood E, Heath R, Mayer FV, Carmena D, Jing C, Walker PA, Eccleston JF, Haire LF et al. (2011) Structure of mammalian AMPK and its regulation by ADP. Nature 472, 230–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Wang S, Song P & Zou MH (2012) AMP‐activated protein kinase, stress responses and cardiovascular diseases. Clin Sci 122, 555–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Reiter AK, Bolster DR, Crozier SJ, Kimball SR & Jefferson LS (2008) AMPK represses TOP mRNA translation but not global protein synthesis in liver. Biochem Biophys Res Commun 374, 345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Zhou Q, Hao B, Cao X, Gao L, Yu Z, Zhao Y, Zhu M, Zhong G, Chi F, Dai X et al. (2022) Energy sensor AMPK gamma regulates translation via phosphatase PPP6C independent of AMPK alpha. Mol Cell 82, 4700–4711.e12. [DOI] [PubMed] [Google Scholar]
- 194. Browne GJ, Finn SG & Proud CG (2004) Stimulation of the AMP‐activated protein kinase leads to activation of eukaryotic elongation factor 2 kinase and to its phosphorylation at a novel site, serine 398. J Biol Chem 279, 12220–12231. [DOI] [PubMed] [Google Scholar]
- 195. Walker‐Gray R, Stengel F & Gold MG (2017) Mechanisms for restraining cAMP‐dependent protein kinase revealed by subunit quantitation and cross‐linking approaches. Proc Natl Acad Sci USA 114, 10414–10419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Tamaki H (2007) Glucose‐stimulated cAMP‐protein kinase a pathway in yeast Saccharomyces cerevisiae . J Biosci Bioeng 104, 245–250. [DOI] [PubMed] [Google Scholar]
- 197. Hahn DK, Tusell JR, Sprang SR & Chu X (2015) Catalytic mechanism of mammalian adenylyl cyclase: a computational investigation. Biochemistry 54, 6252–6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Ramos LS, Zippin JH, Kamenetsky M, Buck J & Levin LR (2008) Glucose and GLP‐1 stimulate cAMP production via distinct adenylyl Cyclases in INS‐1E Insulinoma cells. J Gen Physiol 132, 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Ferretti AC, Tonucci FM, Hidalgo F, Almada E, Larocca MC & Favre C (2016) AMPK and PKA interaction in the regulation of survival of liver cancer cells subjected to glucose starvation. Oncotarget 7, 17815–17828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Deng Z, Li X, Blanca Ramirez M, Purtell K, Choi I, Lu J‐H, Yu Q & Yue Z (2021) Selective autophagy of AKAP11 activates cAMP/PKA to fuel mitochondrial metabolism and tumor cell growth. Proc Natl Acad Sci USA 118, e2020215118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Briaud I, Lingohr MK, Dickson LM, Wrede CE & Rhodes CJ (2003) Differential activation mechanisms of Erk‐1/2 and p70S6K by glucose in pancreatic β‐cells. Diabetes 52, 974–983. [DOI] [PubMed] [Google Scholar]
- 202. Hong Q, Zhang Y, Lin W, Wang W, Yuan Y, Lin J, Xie Z, Li X & Meng Y (2022) Negative feedback of the cAMP/PKA pathway regulates the effects of endoplasmic reticulum stress‐induced NLRP3 Inflammasome activation on type II alveolar epithelial cell Pyroptosis as a novel mechanism of BLM‐induced pulmonary fibrosis. J Immunol Res 2022, 2291877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Ding B, Parmigiani A, Divakaruni AS, Archer K, Murphy AN & Budanov AV (2016) Sestrin2 is induced by glucose starvation via the unfolded protein response and protects cells from non‐canonical necroptotic cell death. Sci Rep 6, 22538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Soulard A, Cremonesi A, Moes S, Schütz F, Jenö P & Hall MN (2010) The rapamycin‐sensitive phosphoproteome reveals that TOR controls protein kinase a toward some but not all substrates. Mol Biol Cell 21, 3475–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Jewell JL, Fu V, Hong AW, Yu FX, Meng D, Melick CH, Wang H, Lam WM, Yuan HX, Taylor SS et al. (2019) GPCR signaling inhibits mTORC1 via PKA phosphorylation of raptor. eLife 8, e43038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Aw DKL, Sinha RA, Xie SY & Yen PM (2014) Differential AMPK phosphorylation by glucagon and metformin regulates insulin signaling in human hepatic cells. Biochem Biophys Res Commun 447, 569–573. [DOI] [PubMed] [Google Scholar]
- 207. Plank M (2022) Interaction of TOR and PKA signaling in S. cerevisiae . Biomolecules 12, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Chan GKL, Maisel S, Hwang YC, Pascual BC, Wolber RRB, Vu P, Patra KC, Bouhaddou M, Kenerson HL, Lim HC et al. (2023) Oncogenic PKA signaling increases c‐MYC protein expression through multiple targetable mechanisms. eLife 12, e69521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Ashe MP, De Long SK & Sachs AB (2000) Glucose depletion rapidly inhibits translation initiation in yeast. Mol Biol Cell 11, 833–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Tudisca V, Simpson C, Castelli L, Lui J, Hoyle N, Moreno S, Ashe M & Portela P (2012) PKA isoforms coordinate mRNA fate during nutrient starvation. J Cell Sci 125, 5221–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Barraza CE, Solari CA, Marcovich I, Kershaw C, Galello F, Rossi S, Ashe MP & Portela P (2017) The role of PKA in the translational response to heat stress in Saccharomyces cerevisiae . PLoS One 12, e0185416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Castelli LM, Lui J, Campbell SG, Rowe W, Zeef LA, Holmes LE, Hoyle NP, Bone J, Selley JN, Sims PF et al. (2011) Glucose depletion inhibits translation initiation via eIF4A loss and subsequent 48S preinitiation complex accumulation, while the pentose phosphate pathway is coordinately up‐regulated. Mol Biol Cell 22, 3379–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Berset C, Trachsel H & Altmann M (1998) The TOR (target of rapamycin) signal transduction pathway regulates the stability of translation initiation factor eIF4G in the yeast Saccharomyces cerevisiae . Proc Natl Acad Sci USA 95, 4264–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Komar AA & Hatzoglou M (2011) Cellular IRES‐mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle 10, 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI & Hellen CU (2001) Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci USA 98, 7029–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Paek KY, Hong KY, Ryu I, Park SM, Keum SJ, Kwon OS & Jang SK (2015) Translation initiation mediated by RNA looping. Proc Natl Acad Sci USA 112, 1041–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Zhao J, Li Y, Wang C, Zhang H, Zhang H, Jiang B, Guo X & Song X (2020) IRESbase: a comprehensive database of experimentally validated internal ribosome entry sites. Genomics Proteomics Bioinformatics 18, 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Yang Y & Wang Z (2019) IRES‐mediated cap‐independent translation, a path leading to hidden proteome. J Mol Cell Biol 11, 911–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Gilbert WV, Zhou K, Butler TK & Doudna JA (2007) Cap‐independent translation is required for starvation‐induced differentiation in yeast. Science 317, 1224–1227. [DOI] [PubMed] [Google Scholar]
- 220. Rachfall N, Heinemeyer I, Morgenstern B, Valerius O & Braus GH (2011) 5′TRU: identification and analysis of translationally regulative 5′untranslated regions in amino acid starved yeast cells. Mol Cell Proteomics 10, M110.003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Coots RA, Liu X‐M, Mao Y, Dong L, Zhou J, Wan J, Zhang X & Qian S‐B (2017) m6A facilitates eIF4F‐independent mRNA translation. Mol Cell 68, 504–514.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Koch A, Aguilera L, Morisaki T, Munsky B & Stasevich TJ (2020) Quantifying the dynamics of IRES and cap translation with single‐molecule resolution in live cells. Nat Struct Mol Biol 27, 1095–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Komar AA, Lesnik T, Cullin C, Merrick WC, Trachsel H & Altmann M (2003) Internal initiation drives the synthesis of Ure2 protein lacking the prion domain and affects [URE3] propagation in yeast cells. EMBO J 22, 1199–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Lee ASY, Kranzusch PJ, Doudna JA & Cate JHD (2016) eIF3d is an mRNA cap‐binding protein that is required for specialized translation initiation. Nature 536, 96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Guan B‐J, van Hoef V, Jobava R, Elroy‐Stein O, Valasek LS, Cargnello M, Gao X‐H, Krokowski D, Merrick WC, Kimball SR et al. (2017) A unique ISR program determines cellular responses to chronic stress. Mol Cell 68, 885–900.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Thompson L, Depledge DP, Burgess HM & Mohr I (2022) An eIF3d‐dependent switch regulates HCMV replication by remodeling the infected cell translation landscape to mimic chronic ER stress. Cell Rep. 39, 110767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Lamper AM, Fleming RH, Ladd KM & Lee ASY (2020) A phosphorylation‐regulated eIF3d translation switch mediates cellular adaptation to metabolic stress. Science (New York, NY) 370, 853–856. [DOI] [PubMed] [Google Scholar]
- 228. Lee S, Liu B, Lee S, Huang SX, Shen B & Qian SB (2012) Global mapping of translation initiation sites in mammalian cells at single‐nucleotide resolution. Proc Natl Acad Sci USA 109, E2424–E2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Cao X & Slavoff SA (2020) Non‐AUG start codons: expanding and regulating the small and alternative ORFeome. Exp Cell Res 391, 111973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Wright BW, Yi Z, Weissman JS & Chen J (2022) The dark proteome: translation from noncanonical open reading frames. Trends Cell Biol 32, 243–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Ingolia NT, Ghaemmaghami S, Newman JR & Weissman JS (2009) Genome‐wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Zhang X, Gao X, Coots RA, Conn CS, Liu B & Qian SB (2015) Translational control of the cytosolic stress response by mitochondrial ribosomal protein L18. Nat Struct Mol Biol 22, 404–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Williams T, Cacioppo R, Agrotis A, Black A, Zhou H & Rousseau A (2022) Actin remodelling controls proteasome homeostasis upon stress. Nat Cell Biol 24, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Katz ZB, Wells AL, Park HY, Wu B, Shenoy SM & Singer RH (2012) β‐actin mRNA compartmentalization enhances focal adhesion stability and directs cell migration. Genes Dev 26, 1885–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Casolari JM, Thompson MA, Salzman J, Champion LM, Moerner WE & Brown PO (2012) Widespread mRNA association with cytoskeletal motor proteins and identification and dynamics of myosin‐associated mRNAs in S. cerevisiae . PLoS One 7, e31912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Rabanal‐Ruiz Y, Byron A, Wirth A, Madsen R, Sedlackova L, Hewitt G, Nelson G, Stingele J, Wills JC, Zhang T et al. (2021) mTORC1 activity is supported by spatial association with focal adhesions. J Cell Biol 220, e202004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Ryazanov AG, Shestakova EA & Natapov PG (1988) Phosphorylation of elongation factor 2 by EF‐2 kinase affects rate of translation. Nature 334, 170–173. [DOI] [PubMed] [Google Scholar]
- 238. Chan C, Pham P, Dedon PC & Begley TJ (2018) Lifestyle modifications: coordinating the tRNA epitranscriptome with codon bias to adapt translation during stress responses. Genome Biol 19, 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Gu C, Begley TJ & Dedon PC (2014) tRNA modifications regulate translation during cellular stress. FEBS Lett 588, 4287–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Rubio A, Ghosh S, Mülleder M, Ralser M & Mata J (2021) Ribosome profiling reveals ribosome stalling on tryptophan codons and ribosome queuing upon oxidative stress in fission yeast. Nucleic Acids Res 49, 383–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Jennings MD, Srivastava P, Kershaw CJ, Talavera D, Grant CM & Pavitt GD (2023) Interaction of the La‐related protein Slf1 with colliding ribosomes maintains translation of oxidative‐stress responsive mRNAs. Nucleic Acids Res 51, 5755–5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T & Barna M (2011) Ribosome‐mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145, 383–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Shi Z, Fujii K, Kovary KM, Genuth NR, Röst HL, Teruel MN & Barna M (2017) Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome‐wide. Mol Cell 67, 71–83.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Erales J, Marchand V, Panthu B, Gillot S, Belin S, Ghayad SE, Garcia M, Laforêts F, Marcel V, Baudin‐Baillieu A et al. (2017) Evidence for rRNA 2′‐O‐methylation plasticity: control of intrinsic translational capabilities of human ribosomes. Proc Natl Acad Sci USA 114, 12934–12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto PE & Fox PL (2003) Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript‐specific translational control. Cell 115, 187–198. [DOI] [PubMed] [Google Scholar]
- 246. Jansson MD, Häfner SJ, Altinel K, Tehler D, Krogh N, Jakobsen E, Andersen JV, Andersen KL, Schoof EM, Ménard P et al. (2021) Regulation of translation by site‐specific ribosomal RNA methylation. Nat Struct Mol Biol 28, 889–899. [DOI] [PubMed] [Google Scholar]
- 247. Jack K, Bellodi C, Landry DM, Niederer RO, Meskauskas A, Musalgaonkar S, Kopmar N, Krasnykh O, Dean AM, Thompson SR et al. (2011) rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol Cell 44, 660–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Ghulam MM, Catala M & Abou Elela S (2020) Differential expression of duplicated ribosomal protein genes modifies ribosome composition in response to stress. Nucleic Acids Res 48, 1954–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Reich S, Nguyen CDL, Has C, Steltgens S, Soni H, Coman C, Freyberg M, Bichler A, Seifert N, Conrad D et al. (2020) A multi‐omics analysis reveals the unfolded protein response regulon and stress‐induced resistance to folate‐based antimetabolites. Nat Commun 11, 2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Cheng Z, Rendleman J & Vogel C (2016) Time‐course proteomics dataset monitoring HeLa cells subjected to DTT induced endoplasmic reticulum stress. Data Brief 8, 1168–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Youn J‐Y, Dyakov BJA, Zhang J, Knight JDR, Vernon RM, Forman‐Kay JD & Gingras A‐C (2019) Properties of stress granule and P‐body proteomes. Mol Cell 76, 286–294. [DOI] [PubMed] [Google Scholar]
- 252. Buchan JR, Muhlrad D & Parker R (2008) P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol 183, 441–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A & Parker R (2016) ATPase‐modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Makeeva DS, Riggs CL, Burakov AV, Ivanov PA, Kushchenko AS, Bykov DA, Popenko VI, Prassolov VS, Ivanov PV & Dmitriev SE (2023) Relocalization of translation termination and ribosome recycling factors to stress granules coincides with elevated stop‐codon readthrough and reinitiation rates upon oxidative stress. Cells 12, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Shetty S, Hofstetter J, Battaglioni S, Ritz D & Hall MN (2023) TORC1 phosphorylates and inhibits the ribosome preservation factor Stm1 to activate dormant ribosomes. EMBO J 42, e112344. [DOI] [PMC free article] [PubMed] [Google Scholar]


