Abstract
The microRNA156 (miR156) has been widely studied in plants, however, the characterization of the miR156 family of genes in wheat and their expression patterns under abiotic stress are not completely clear. In this study, a total of 20 miR156 family members, referred to as tae-miR156a to tae-miR156t, were identified in wheat with their loci mapped to various chromosomes. These members were divided into five subgroups: miR156a/b/c/d/e/f, miR156g/h/i, miR156j/k, miR156l/m/n/o/p/q, and miR156r/s/t. They were highly conserved during evolution. The prediction of cis-elements in the tae-MIR156(s) promoter region revealed that the tae-MIR156(s) had diverse cis-acting elements; of these, 15 tae-MIR156(s) and 6 tae-MIR156(s) were found to be drought-responsive elements and cold-responsive elements, respectively. And the prediction target genes of tae-miR156(s) are mainly SPL transcription factor genes. Expression analysis based on quantitative real-time polymerase chain reaction (qRT‒PCR) showed that miR156(s) have different expression levels in the various wheat tissues, and the subgroups’ response to abiotic stress varied. Among them, miR156g/h/i were strongly induced in the root of cold and heat stress, and miR156a/b/c/d/e/f were significantly increased in roots after drought stress, whereas miR156r/s/t were highly inhibited in leaves and roots after salt stress. These findings imply that tae-miR156(s) are involved in stress response in wheat, and they provide new fundamental knowledge for further analysis of the function of miR156 and its regulatory mechanism in response to abiotic stress.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05899-4.
Keywords: MicroRNA, Wheat, MiR156, Expression, Abiotic stress
Background
MicroRNAs (miRNAs) are small non-coding RNAs of approximately 18–24 nucleotides in length that are now increasingly recognized as regulators of gene expression at the post-transcriptional level and have the ability to impact many biological processes [1]. MicroRNA156 (miR156) is widely found in monocots, dicots, and lower ferns and mosses [2–5]. A growing body of evidence shows that miR156 plays a significant role in regulating plant fitness, biomass, and yield [6]. Plant miRNAs from the same family share a high degree of sequence similarity, but they may have diverse roles in different plant species [7, 8]. For example, in rice (Oryza sativa), the deletion of miR156d/e/f/g/h/i suppressed ineffective tillering, increased plant height, grain length, and grain weight, improved stem resistance, and exhibited ideal plant morphology without affecting seed dormancy, whereas the miR156a/b/c/k/l mutant significantly improved seed dormancy and inhibited spike germination [9]. During symbiotic nodulation in legumes, miR156 family members were variably expressed, and miR156b inhibited nodulation by negatively regulating the expression of GmSPL9d (Soybean SQUAMOSA promoter-binding protein-like 9d) [10]. Overexpression of tae-miR156 resulted in increased tiller number and severe defects in spikelet generation for bread wheat (Triticum aestivum L.) [11]. MiR156 could impact fruit size and yield by regulating inflorescence architecture in tomatoes (Solanum lycopersicum L.) [12], as well as plant architecture and tuber development through starch accumulation in potatoes (Solanum tuberosum L.) [13], and somatic embryogenesis in citrus (Citrus reticulata Blanco) [14]. These findings indicated that miR156 plays a significant role in modifying plant growth and development.
Plants are constantly exposed to adverse conditions such as extreme temperatures, high salinity, and drought. These abiotic stresses are major factors limiting the geographical distribution of plants and their corresponding crop yields [15]. However, to reduce the adverse effects of these abiotic stresses, plants have plasticity in their defense mechanisms, enabling them to tolerate and survive stressful conditions [16]. For example, miR156 negatively impacted cold tolerance and positively regulated drought tolerance, while the overexpression of miR156a weakened salt resistance in apple (Malus pumila Mill.) [17–19]. MiR156 also exhibited a positive effect in Nicotiana tabacum subjected to cold stress [20]. Overexpression of miR156k resulted in lower tolerance to cold stress in rice [21]. Decreased expression of miR156 affected sugarcane (Saccharum officinarum L.) metabolism and growth after low-temperature treatment and enhanced cold tolerance by negatively regulating squamosa promoter binding protein-like (SPL) transcription factor [22]. Overexpression of miR156 could also improve drought and heat stress tolerance in alfalfa (Medicago sativa) [23, 24] while ahy-miR156 down-regulated target genes to enhance the level of drought resistance in groundnut legume (Arachis hypogaea) [25]. Newly published studies have also shown that in apple calli and Arabidopsis, MdmiR156n could regulate drought tolerance by inducing flavonoid accumulation and promoting reactive oxygen species (ROS) scavenging [26]. The miR156/SPL9 pathway suppressed anthocyanin production and significantly improved Arabidopsis durability to salt and drought stress [27]. Heterogeneous expression of Osa-MIR156bc increased abiotic stress resistance and forage quality in alfalfa [28]. In brief, miR156 exhibits diverse actions in various species and is vital for responding to unfavorable stress [29, 30]. However, not much is known about how miR156 is regulated and the role it plays in wheat under abiotic stress.
Wheat is an important staple crop globally and provides approximately 20% of the global dietary energy [31]. Although wheat is grown in large areas, its total production remains the lowest amongst cereals and rice. This is mainly a consequence of abiotic stressors such as drought, salt, and high temperatures which are the main contributors to losses in wheat production [32]. Bread wheat is a hexaploid whose genome size is estimated to be about 17 Gb. This large and complex genome poses a great challenge for the mining and application of stress-resistance genes as well as the cultivation and improvement of resistant varieties of wheat. MicroRNAs are short non-coding RNAs that have generated much interest in biological research due to their role as major regulators of gene expression at the post-transcriptional level. One key miRNA identified in plants is miR156 with its functions and molecular mechanisms well described for rice, Arabidopsis, and other plants. Although the presence of miR156 in wheat has been reported, the specific regulatory functions and mechanisms that involve miR156 have not been fully resolved and verified due to the large and complex wheat genome. Nevertheless, current advances in genome sequencing and chromosomal fine mapping can facilitate investigations into and validation of the roles of miR156 in wheat (Triticum aestivum L.). To this end, we analyzed the chromosomal localization, evolution, sequence conservation, cis‑acting regulatory elements, target genes, spatiotemporal expression patterns, and response to different abiotic stresses for miR156(s) in wheat. The results of this study provide comprehensive information to understand tae-miR156(s) and lay the foundation for functional research on abiotic stresses.
Methods
Identification and chromosomal localization of miR156(s) in wheat
The precursor sequences, mature sequences, and chromosomal location information of miR156(s) for wheat (Triticum aestivum L.) were downloaded from the PmiREN (Plant miRNA ENcyclopedia) database (https://www.pmiren.com/) [33].
Phylogenetic analysis of the tae-MIR156(s) in wheat
To construct the phylogenetic tree of plant miR156(s), we downloaded the precursor sequences of miR156(s) for wheat, rice, Zea mays, and Arabidopsis from the PmiREN database and performed multiple sequence alignments using the MEGA 11 software [34]. The neighbor-joining method was used to construct the phylogenetic tree with the bootstrap value set to 1000 [35].
Prediction of cis‑acting regulatory elements and target genes
The 2000 bp upstream sequences of tae-MIR156(s) and their cis-elements were predicted using the PlantCARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [36], and targets of tae-miR156(s) were predicted using the PsRNATarget (https://www.zhaolab.org/psRNATarget/) based on a T. aestivum cDNA library (EnsemblPlant, v.43) [37].
Plants and treatments
The hexaploid common wheat (Triticum aestivum L.) cultivar ‘Fielder’, an American, soft, white, pastry-type wheat, known for its amenability to Agrobacterium tumefaciens-mediated transformation and genome editing [38], was used for expression analysis and stress treatment. For spatial–temporal expression analysis, Fielder seeds (spring wheat) were directly sown in pots (30 cm height × 27 cm diameter) filled with a peat moss substrate (Pindstrup, Denmark) and mixed fertilizers (Osmocote Extract, Heerlen, the Netherlands) in a growth chamber set at 22 °C, a photoperiod regime of 16 h light/8 h darkness, and 45% humidity, different tissue samples were collected from Fielder plants at different developmental stages based on Zadoks’ scale [39], there were three independent biological replicates for each sample.
For abiotic stress expression analysis, Fielder seeds of the same size with full grains were selected and surface-sterilized in 1% sodium hypochlorite for 15 min, rinsed in distilled water, and germinated at room temperature [40], after which the seeds were placed in black plant hydroponic box with 96 holes in hydroponics containing Hoagland's nutrient solution, placed in an artificial climate chamber (22 °C light for 16 h, 18 °C dark for 8 h) for cultivation, and the nutrient solution was changed every two days. After normal growth of 14 days, they were subjected to stress treatments (cold, heat, salt, drought) separately. For cold and heat treatments, the temperature of the incubator was set at 4℃ and 42℃ [41], respectively. Drought was established with PEG6000 (20%) and NaCl (200 mM) solution was used to mimic salt stress [42, 43]. The leaves and roots were collected as experimental materials after 0 h (means no treatment), 3 h, 6 h, 12 h, 24 h, and 48 h. The collected samples were immediately frozen in liquid nitrogen and stored at − 80 °C for subsequent analysis. The leaves and roots of 15 seedlings were mixed as one sample, respectively. There were three independent biological replicates for each sample.
RNA extraction and qRT-PCR
The miRcute Plant miRNA Isolation Kit (Tiangen, Beijing, China) was used to extract miRNA from Fielder seedlings, roots, stems, and other tissues according to the manufacturer's instructions. The miRNA 1st Strand cDNA Synthesis Kit (by stem-loop) (Vazyme, Nanjing, China) was used for reverse transcription of the extracted RNA into cDNA. Quantification of miR156(s) expression by qRT-PCR was performed using a miRNA Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China). The wheat U6 gene was used as the internal reference. The 2−ΔΔCT method was used to calculate expression levels. All assays were performed in three independent experiments. Primers used in this study are listed in Supporting Information Table S1.
Statistical analysis
Statistical analyses were performed using GraphPad Prism9 (https://www.graphpad.com/scientific-software/prism/) by one-way ANOVA. All data are presented as mean ± SD. Differences between the means were compared using the Tukey's test. Lowercase letters represent significant differences at P < 0.05.
Results
Chromosome mapping and conserved analysis of miR156 in wheat
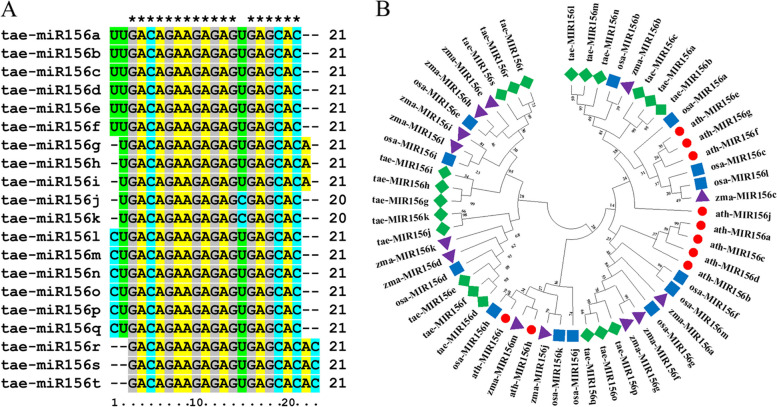
Based on the PmiREN database, a total of 20 wheat tae-MIR156 genes were identified. Among them, tae-MIR156r/s/t were localized on chromosomes 2A, 2B, and 2D, respectively. Tae-MIR156a/l, tae-MIR156b/m, and tae-MIR156c/n were localized on chromosomes 3A, 3B, and 3D, respectively. Tae-MIR156j/o, tae-MIR156k/p, and tae-MIR156q were localized on chromosomes 5A, 5B, and 5D, respectively. Tae-MIR156d/g, tae-MIR156e/h, and tae-MIR156f/i were localized on chromosomes 6A, 6B, and 6D, respectively (Table 1). Among these tae-MIR156, only tae-MIR156j and tae-MIR156k produced mature sequences of 20 nucleotides in length, while the rest produced mature sequences of 21 nucleotides. The mature miR156 members miR156a/b/c/d/e/f, miR156g/h/i, miR156j/k, miR156l/m/n/o/p/q, and miR156r/s/t have identical sequences (Fig. 1A). MiRNAs originate from primary precursor transcripts (pre-miRNAs) containing hairpin structures. However, we noted that the sequences and lengths of the pre-miRNA156(s) were highly varied, with the maximum sequence conservation noted for pre-miRNAs located in the stem section of the hairpin structures where mature miR156(s) are produced (Fig. S1). Phylogenetic analysis of miR156 family members in wheat, rice, maize, and Arabidopsis was performed. We noted that the different plant species have markedly different family numbers (Fig. 1B). Wheat has the largest number of miR156 genes (20 miR156s), while the other species have c. 10 miR156 family members. The phylogenetic analysis revealed that the miR156 family is conserved among Oryza sativa, Zea mays, and Arabidopsis thaliana (Fig. 1B).
Table 1.
Information on the members of the miR156 family in wheat
| miRNA | miRNA locus | Gene location | Mature sequence |
|---|---|---|---|
| tae-miR156a | tae-MIR156a | Chr3A: 76,564,147–76,564,167 ( +) | UUGACAGAAGAGAGUGAGCAC |
| tae-miR156b | tae-MIR156b | Chr3B: 109,363,904–109363924 (-) | UUGACAGAAGAGAGUGAGCAC |
| tae-miR156c | tae-MIR156c | Chr3D: 65,610,352–65610372 (-) | UUGACAGAAGAGAGUGAGCAC |
| tae-miR156d | tae-MIR156d | Chr6A: 127,182,601–127182621 ( +) | UUGACAGAAGAGAGUGAGCAC |
| tae-miR156e | tae-MIR156e | Chr6B: 191,531,743–191,531,763 ( +) | UUGACAGAAGAGAGUGAGCAC |
| tae-miR156f | tae-MIR156f | Chr6D: 104,978,824–104978844 ( +) | UUGACAGAAGAGAGUGAGCAC |
| tae-miR156g | tae-MIR156g | Chr6A: 437,637,057–437637077 (-) | UGACAGAAGAGAGUGAGCACA |
| tae-miR156h | tae-MIR156h | Chr6B: 448,349,738–448,349,758 ( +) | UGACAGAAGAGAGUGAGCACA |
| tae-miR156i | tae-MIR156i | Chr6D: 287,888,860–287888880 ( +) | UGACAGAAGAGAGUGAGCACA |
| tae-miR156j | tae-MIR156j | Chr5A: 75,281,037–75281056 (-) | UGACAGAAGAGAGCGAGCAC |
| tae-miR156k | tae-MIR156k | Chr5B: 51,122,100–51122119 (-) | UGACAGAAGAGAGCGAGCAC |
| tae-miR156l | tae-MIR156l | Chr3A: 76,564,340–76564360 ( +) | CUGACAGAAGAGAGUGAGCAC |
| tae-miR156m | tae-MIR156m | Chr3B: 109,363,710–109363730 (-) | CUGACAGAAGAGAGUGAGCAC |
| tae-miR156n | tae-MIR156n | Chr3D: 65,610,164–65610184 (-) | CUGACAGAAGAGAGUGAGCAC |
| tae-miR156o | tae-MIR156o | Chr5A: 440,504,399–440504419 (-) | CUGACAGAAGAGAGUGAGCAC |
| tae-miR156p | tae-MIR156p | Chr5B: 398,367,514–398,367,534 (-) | CUGACAGAAGAGAGUGAGCAC |
| tae-miR156q | tae-MIR156q | Chr5D: 339,371,652–339,371,672 (-) | CUGACAGAAGAGAGUGAGCAC |
| tae-miR156r | tae-MIR156r | Chr2A: 599,784,458–599,784,478 (-) | GACAGAAGAGAGUGAGCACAC |
| tae-miR156s | tae-MIR156s | Chr2B: 537,835,523–537,835,543 ( +) | GACAGAAGAGAGUGAGCACAC |
| tae-miR156t | tae-MIR156t | Chr2D: 456,504,616–456504636 (-) | GACAGAAGAGAGUGAGCACAC |
Fig. 1.
Sequence alignment and phylogenetic relationship analysis. A All 20 sequences of the wheat miR156 family members were aligned using the MEGA 11 software. Asterisks represent conserved nucleotides in all mature miRNAs. B Phylogenetic analysis of 56 precursor sequences of miR156 family members from four plant species. The pre-miRNA sequences of ten miR156 family members in Arabidopsis thaliana (red circle), 13 in Oryza sativa (blue square), 13 in Zea mays (purple triangle), and 20 in wheat (green square) were used for the alignment, and the phylogenetic neighbor-joining tree was reconstructed using MEGA 11 phylogenetic analysis software. Bootstrap values (percentages of 1000 replicates) are indicated on the nodes
Analysis of cis-acting elements in tae-MIR156(s) promoter regions in wheat
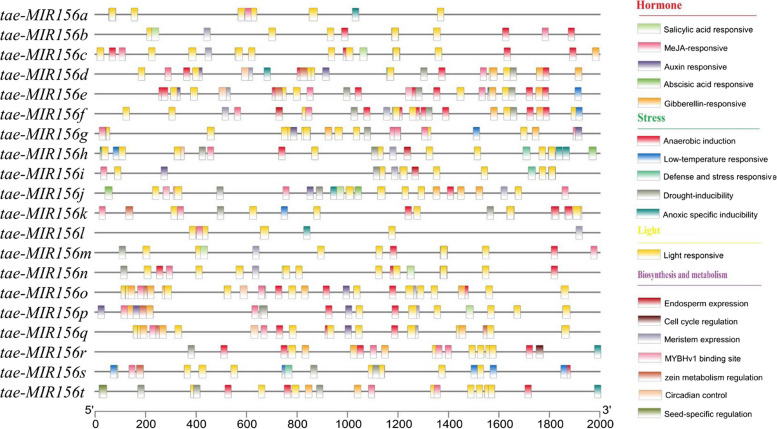
Plant gene promoter regions contain several critical cis-acting elements that are key to the regulation of gene expression [44]. The identification and analysis of the cis-acting elements of miRNA promoters will aid our understanding of the molecular mechanism by which miRNA regulates gene expression in plants. Therefore, to identify the function of tae-MIR156(s), 2000 bp upstream regions of tae-MIR156(s) in wheat were used as putative promoter regions for the prediction of cis-acting elements. Of the 20 tae-MIR156(s) promoters, all contained the core promoter element TATA-box and the common CAAT-box elements (Table S2). In addition, 44 specific promoter cis-acting elements were identified and divided into the following four categories: hormone-responsive, stress-responsive, light-responsive, and biosynthesis and metabolism related cis-acting elements (Fig. 2). Among them, the light-responsive category was shared among the 20 tae-MIR156(s), and five stress-response elements, namely, ‘anaerobic induction element’, ‘low-temperature responsive element’, ‘defense and stress responsive element’, ‘drought inducibility element’, and ‘anoxic specific inducibility element’, were identified. Among these stress response elements, a total of 15 tae-MIR156(s)(tae-MIR156d/e/f/g/h/i/j/k/m/n/o/p/r/s/t) were identified as possessing drought-responsive elements (MBS), and a total of 6 tae-MIR156(s)(tae-MIR156e/f/g/h/k/s) were identified as having low-temperature response elements (LTR).
Fig. 2.
Promoter analysis of tae-MIR156(s) in wheat
Tissue specific expression analysis of miR156 in wheat
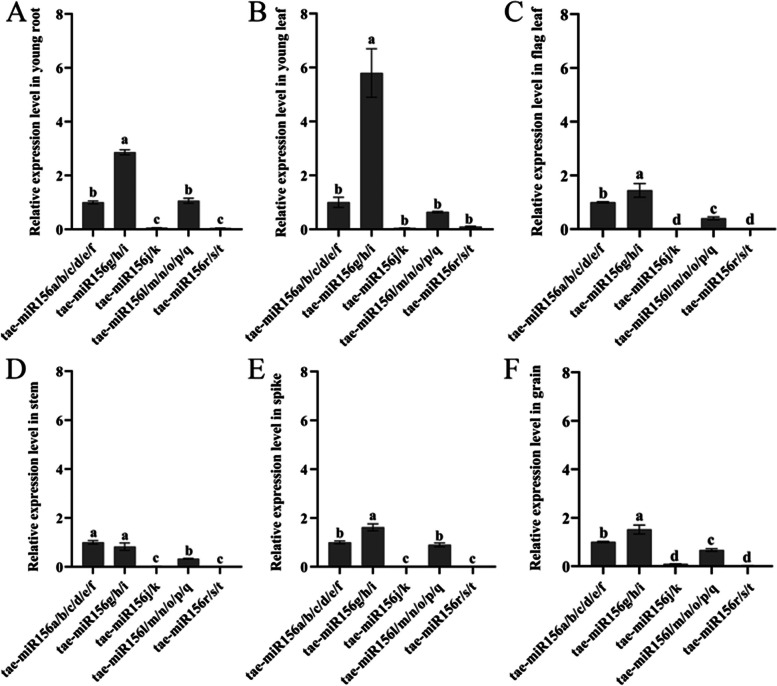
Constitutive expression of miR156 was previously reported to prolong the juvenile stage in Arabidopsis thaliana, rice, and tobacco, and influence resistance to abiotic stress [45–48]. Therefore, we analyzed spatial and temporal expression patterns of wheat tae-miR156(s) in different tissues using quantitative real-time PCR (qRT-PCR) and found that tae-miR156(s) were constitutively expressed in diverse wheat tissues at different developmental stages. Notably, tae-miR156g/h/i showed the highest expression levels in young roots and young leaves, while tae-miR156a/b/c/d/e/f exhibited the highest expression in stems at the heading stage (when the ear emerges from the flag leaf sheath). Interestingly, the expression of miR156j/k and miR156r/s/t in these tissues was limited (Fig. 3A-F). As the sequences of mature miR156(s) (miR156a/b/c/d/e/f, miR156g/h/i, miR156j/k, miR156l/m/n/o/p/q, and miR156r/s/t) were almost identical, the expression of these miR156(s) was the sum of the identical mature miR156(s). These results suggest that although miR156(s) from the same family are relatively conserved among themselves, they are expressed at different levels in various wheat tissues.
Fig. 3.
Spatial and temporal expression patterns of tae-miR156(s) in wheat. Expression of tae-miR156(s) in (A) young root and leaf (B) at seeding stage (Zadok’s growth stage, ZGS13); C flag leaf; D stem (ZGS37); E young spikes of 4 cm in length (ZGS37); (F) developing grains (ZGS77). Data are the mean of three replicates ± SD. Lowercase letters represent significant differences at P < 0.05
Expression pattern of tae-miR156(s) in response to abiotic Stress
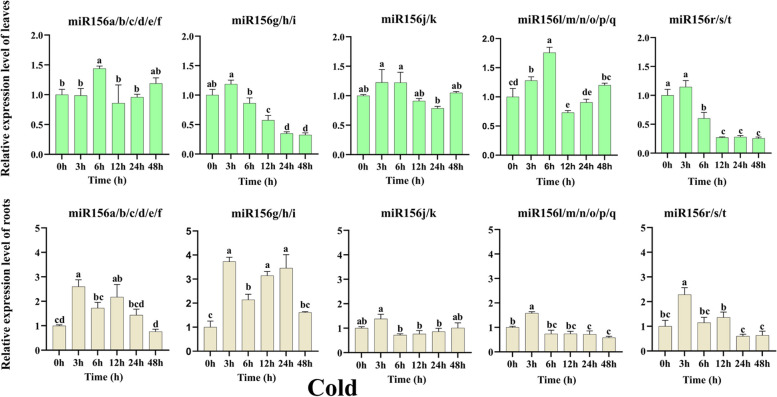
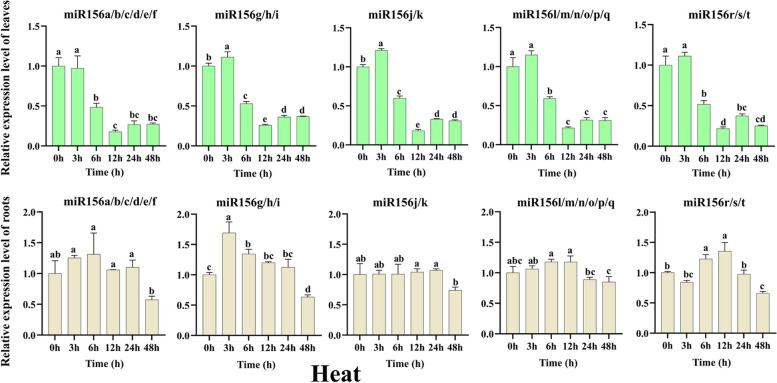
Roots and leaves are the initial organs that sense the signals of environmental stresses. To further assess the functions of tae-miR156(s), we analyzed their expression levels in response to cold, heat, drought, and salt treatments. In leaves of wheat subjected to cold treatment (Fig. 4), it was observed that expression levels of miR156a/b/c/d/e/f and miR156l/m/n/o/p/q showed similar patterns following cold stress. The expression of both subgroups increased significantly at 6 h (p < 0.05) and then decreased significantly at 12 h (p < 0.05), followed by another increase in expression levels over the subsequent treatment times. The most significant increase in expression levels was detected for miR156l/m/n/o/p/q, at 1.76 times higher than that of the untreated control after 6 h of cold treatment (p < 0.05). The expression levels of miR156g/h/i and miR156r/s/t increased after cold treatment for 3 h and significantly decreased during the rest of the period, with the greatest decrease at 48 h. miR156r/s/t showed the most significant decrease in expression level, which was 0.26 times that of the untreated control after 48 h of cold treatment. Compared to untreated leaves, the expression levels of miR156j/k increased after 3 h, 6 h, and 48 h of cold stress and decreased at 12 h and 24 h. In cold-treated root tissues, expression levels of all tae-miR156(s) were significantly increased after 3 h and then significantly decreased after 6 h. The upregulation of miR156g/h/i expression levels was the most significant, which was 3.72 times that of untreated, while miR156a/b/c/d/e/f, miR156j/k, miR156l/m/n/o/p/q, miR156r/s/t expression levels were 2.60, 1.38, 1.58, and 2.28 times higher than that of untreated control, respectively. The expression levels of miR156a/b/c/d/e/f were down-regulated at 48 h and increased at other time points, while that of miR156g/h/i were significantly up-regulated at all time points compared to the untreated control. However, the expression levels of miR156j/k, miR156l/m/n/o/p/q, and miR156r/s/t were significantly up-regulated (p < 0.05) after 3 h of cold treatment and there was no significant change in expression levels at the other time points (Fig. 4). These results indicated that cold stress affects the expression levels of tae-miIR156(s) in leaves and roots, but miR156g/h/i tends to be more sensitive in cold stressed root tissues.
Fig. 4.
The relative expression levels of tae-miR156(s) in leaves and roots after cold treatment as determined by qRT-PCR analysis. Data are the mean of three replicates ± SD. Lowercase letters represent significant differences at P < 0.05
Under heat treatment (Fig. 5), expression of all tae-miR156(s) was significantly down-regulated (P < 0.05) at 6 h in leaves, and the down-regulation was most pronounced at 12 h. Interestingly, except miR156a/b/c/d/e/f which demonstrated down-regulation in gene expression, the expression levels of other miR156(s) were higher after 3 h of heat treatment in leaves compared to the control. In roots subjected to heat treatment, the expression levels of miR156a/b/c/d/e/f, miR156g/h/i, and miR156j/k increased at 3 h, 6 h, 12 h, and 24 h but decreased significantly at 48 h compared to the untreated root samples. The most significant increase in expression was detected for miR156g/h/i, which was 1.70 times higher than that of the untreated control after 3 h of heat treatment. The most significant decrease in expression levels was observed for miR156a/b/c/d/e/f, which was 0.53 times that of the untreated control after 48 h of heat treatment. The expression of miR156l/m/n/o/p/q increased first and then decreased, but there was no significant difference between treatment and control. The expression of miR156r/s/t initially decreased, then increased to its highest level at 12 h after heat treatment but subsequently decreased at later time points. These results indicated that heat stress significantly affects the expression levels of tae-miR156(s).
Fig. 5.
The relative expression levels of tae-miR156(s) in leaves and roots after heat treatment as determined by qRT-PCR analysis. Data are the means of three replicates ± SD. Lowercase letters represent significant differences at P < 0.05
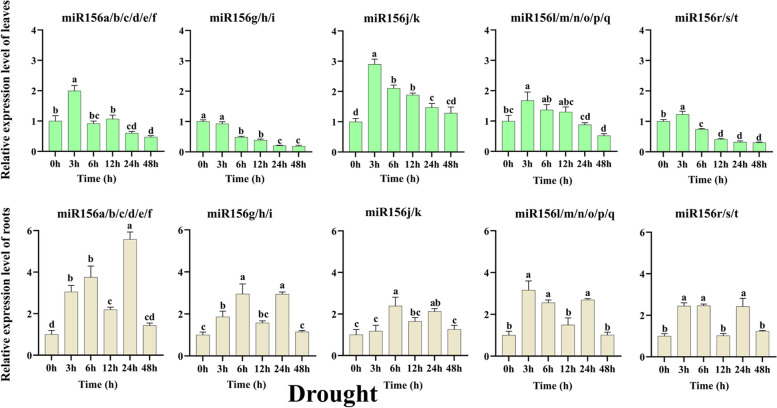
Under drought treatment (Fig. 6), the expression levels of miR156a/b/c/d/e/f, miR156j/k, miR156l/m/n/o/p/q, and miR156r/s/t increased significantly after 3 h, most significantly in miR156j/k, which was 2.90 times higher than that of the no treatment control. On the other hand, expression levels of miR156g/h/i decreased over all time points after drought treatment and were most significant after 48 h of treatment. In root tissues subjected to drought treatment, expression levels of all miR156(s) increased at different periods of treatment compared to untreated roots. The expression levels of miR156a/b/c/d/e/f were most significantly elevated after 24 h of treatment at 5.58 times higher than that of untreated. Expression levels of miR156g/h/i and miR156j/k were most significantly elevated after 6 h of treatment, at 2.95 and 2.40 times higher than that of untreated, respectively. miR156l/m/n/o/p/q and miR156r/s/t expression levels were significantly elevated after 3 h of treatment, which were 3.16 and 2.45 times higher than untreated control before treatment, respectively. These results indicate that all members of the wheat miR156 family are involved in the drought stress response, but the root tissues were more affected compared with the leaves.
Fig. 6.
The relative expression levels of tae-miR156(s) in leaves and roots after drought treatments by qRT-PCR analysis. Data are the mean of three replicates ± SD. Lowercase letters represent significant differences at P < 0.05
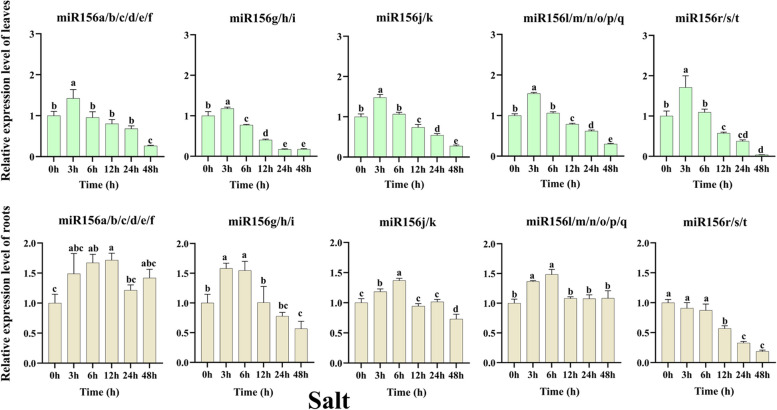
Under salt treatment (Fig. 7), all tae-miR156(s) showed a similar expression pattern with a significant increase after 3 h, followed by a decrease up to 48 h compared to untreated leaves. The most significant overexpression and suppression in expression levels were noted for miR156r/s/t, with expression levels 1.71-fold and 0.03-fold higher than untreated after 3 h and 48 h of treatment, respectively. Interestingly, in the root tissues under salt stress treatment, expression levels of miR156r/s/t decreased over the entire treatment period, but the most significant decrease was at 48 h, which was 0.19-fold of untreated. On the contrary, expression of miR156a/b/c/d/e/f increased over different periods compared to untreated, and the most significant increase was at 12 h, at 1.72 times higher than that of untreated. While the expression level of miR156g/h/i increased most significantly after 3 h of treatment (fold change of 1.58 times that of untreated), expression levels of miR156j/k and miR156l/m/n/o/p/q increased most significantly after 6 h of treatment at 1.37 and 1.48 times that of pre-treatment, respectively. These results indicated that the expression of miR156(s) was different in leaves and roots under the influence of salt stress.
Fig. 7.
The relative expression levels of tae-miR156(s) in leaves and roots after salt treatment as quantified by qRT-PCR analysis. Data are the mean of three replicates ± SD. Lowercase letters represent significant differences at P < 0.05
Prediction of tae-miR156(s) target genes
MicroRNA156 plays a key role in plant growth, development, and response to stress through the precise regulation of its target genes [6]. To further explore the function of miR156 in wheat, the possible targets of tae-miR156(s) were predicted by PsRNATarget [37]. Of them, tae-miR156a/b/c/d/e/f had the highest number of presented target genes (56) (Table S3), followed by tae-miR156g/h/i (39) (Table S4), tae-miR156j/k (37) (Table S5), tae-miR156l/m/n/o/p/q (34) (Table S6), and miR156r/s/t (32) (Table S7). However, the majority of each tae-miR156's targets were from the SPLs family, with 27 genes (Table 2), revealing that SPL transcription factors are essential in wheat. In addition, the genes encoding Beta-galactosidase, Heparanase, Pectate lyase, Serine protease, SNF1 protein kinase, Xyloglucan endotransglucosylase hydrolase, Beta-carotene hydroxylase, Kinesin-related protein, Pentatricopeptide repeat-containing protein, Mitochondrial carrier-like protein, Chaperone protein DnaJ, Metal tolerance protein, Transcriptional corepressor SEUSS, Mitochondrial carrier-like protein, Auxilin-related protein 1, F-box protein family, Peptide chain release factor 1, Exocyst complex component, transcription factor GTE10/8 and so on were also putative targets. The results indicated that tae-miR156(s) may interact with these possible target genes to regulate various biological processes in wheat.
Table 2.
Prediction of tae-miR156(s) target genes in wheat
| Number | Gene name | Gene locus | Direction | Binding sequences | Function |
|---|---|---|---|---|---|
| I | miR156a/b/c/d/e/f | 3'−5' | CACGAGUGAGAGAAGACAGUU | ||
| II | miR156g/h/i | 3'−5' | ACACGAGUGAGAGAAGACAGU | ||
| III | miR156j/k | 3'−5' | CACGAGUGAGAGAAGACAGU | ||
| IV | miR156l/m/n/o/p/q | 3'−5' | CACGAGUGAGAGAAGACAGUC | ||
| V | miR156r/s/t | 3'−5' | CACACGAGUGAGAGAAGACAG | ||
| 1 | TaSPL2-A | TraesCS3A02G432500 | 5'−3' | UGUGCUCUCUCUCUUCUGUCA | Cleavage |
| 2 | TaSPL2-B | TraesCS3B02G468400 | 5'−3' | UGUGCUCUCUCUCUUCUGUCA | Cleavage |
| 3 | TaSPL2-D | TraesCS3D02G425800 | 5'−3' | UGUGCUCUCUCUCUUCUGUCA | Cleavage |
| 4 | TaSPL3-A | TraesCS6A02G110100 | 5'−3' | CAUGCUCUCUCUCUUCUGUCA | Cleavage |
| 5 | TaSPL3-B | TraesCS6B02G138400 | 5'−3' | CAUGCUCUCUCUCUUCUGUCA | Cleavage |
| 6 | TaSPL3-D | TraesCS6D02G098500 | 5'−3' | CAUGCUCUCUCUCUUCUGUCA | Cleavage |
| 7 | TaSPL4-A | TraesCS6A02G155300 | 5'−3' | CGUGCUCUCUCUCUUCUGUCA | Cleavage |
| 8 | TaSPL4-B | TraesCS6B02G183400 | 5'−3' | CGUGCUCUCUCUCUUCUGUCA | Cleavage |
| 9 | TaSPL4-D | TraesCS6D02G145200 | 5'−3' | CAUGCUCUCUCUCUUCUGUCA | Cleavage |
| 10 | TaSPL7-A | TraesCS2A02G413900 | 5'−3' | GGUGCUCUCUCUCUUCUGUCA | Cleavage |
| 11 | TaSPL7-B | TraesCS2B02G432700 | 5'−3' | GGUGCUCUCUCUCUUCUGUCA | Cleavage |
| 12 | TaSPL7-D | TraesCS2D02G410700 | 5'−3' | GGUGCUCUCUCUCUUCUGUCA | Cleavage |
| 13 | TaSPL13-A | TraesCS2A02G232400 | 5'−3' | CAUGCUCCCUCUCUUCUGUCA | Cleavage |
| 14 | TaSPL13-B | TraesCS2B02G250900 | 5'−3' | CAUGCUCCCUCUCUUCUGUCA | Cleavage |
| 15 | TaSPL13-D | TraesCS2D02G232800 | 5'−3' | CAUGCUCCCUCUCUUCUGUCA | Cleavage |
| 16 | TaSPL14-A | TraesCS7A02G246500 | 5'−3' | UGUGCUCUCUCUCUUCUGUCA | Cleavage |
| 17 | TaSPL14-B | TraesCS7B02G144900 | 5'−3' | UGUGCUCUCUCUCUUCUGUCA | Cleavage |
| 18 | TaSPL14-D | TraesCS7D02G245200 | 5'−3' | UGUGCUCUCUCUCUUCUGUCA | Cleavage |
| 19 | TaSPL16-A | TraesCS7A02G260500 | 5'−3' | UGUGCUCUCUCUCUUCUGUCA | Cleavage |
| 20 | TaSPL16-B | TraesCS7B02G158500 | 5'−3' | UGUGCUCUCUCUCUUCUGUCA | Cleavage |
| 21 | TaSPL16-D | TraesCS7D02G261500 | 5'−3' | UGUGCUCUCUCUCUUCUGUCA | Cleavage |
| 22 | TaSPL17-A | TraesCS5A02G265900 | 5'−3' | UGUGCUCUCUCUCUUCUGUCA | Cleavage |
| 23 | TaSPL17-B | TraesCS5B02G265600 | 5'−3' | UGUGCUCUCUCUCUUCUGUCA | Cleavage |
| 24 | TaSPL17-D | TraesCS5D02G273900 | 5'−3' | UGUGCUCUCUCUCUUCUGUCA | Cleavage |
| 25 | TaSPL18-A | TraesCS5A02G286700 | 5'−3' | UGUGCUCUCUCUCUUCUGUCA | Cleavage |
| 26 | TaSPL18-B | TraesCS5B02G286000 | 5'−3' | UGUGCUCUCUCUCUUCUGUCA | Cleavage |
| 27 | TaSPL18-D | TraesCS5D02G294400 | 5'−3' | UGUGCUCUCUCUCUUCUGUCA | Cleavage |
Discussion
The miR156 gene family is widely distributed in plants, such as the rice miR156 family which has 12 members (miR156a to miR156l) [49], the Arabidopsis thaliana miR156 family with 10 members (miR156a to miR156j) [50], and the soybean miR156 family, comprising of 25 members (miR156a to miR156y) [51]. While many studies have reported on the function of miR156(s) in other plant species, the large and complex wheat genome has prevented similar studies in wheat. Nevertheless, current whole genome sequencing and assembly capabilities provide a platform to further explore the characteristics and functions of wheat MIR156 gene family members. In this study, we identified 20 MIR156 genes in wheat which are located on different chromosomes (Table 1), and have been largely conserved during the evolution (Fig. 1). However, they exhibited distinct spatiotemporal expression patterns at various stages of development (Fig. 3), demonstrating that they may have diverse regulatory functions. Whilst it has previously been demonstrated that MIR156a/b/c controlled the architecture of bread wheat [11], other miRNA members from the same family are proposed to have diverse functions [9, 52]. As the roles of tae-miR156(s) in other aspects of wheat have not been well described, the expression patterns of tae-miR156(s) described in our study provide a reference for future functional studies in wheat.
In many plants, miR156(s) respond to a wide range of abiotic stresses, showing complex and diverse functions and expression patterns, which are important to ensure plant adaptation to environmental stresses. And cis-acting elements play a crucial function in the regulation of gene expression. Previous studies have shown that promoters of MIR156(s) in apple and tea plants (Camellia sinensis (L.) O. Kuntze) harbor drought responsive elements known as MBS. Notably, during drought stress, the expression of csn-miR156f-2-5p was down-regulated in tea plants, but the expression of mdn-miR156ab was up-regulated in apples, indicating that various species had different expression patterns under drought stress [44, 53]. In our study, a total of 15 tae-MIR156(s) promoters possess drought-responsive elements (Fig. 2), implying that tae-miR156(s) may play a significant role in drought stress. In addition, tae-MIR156e/f/g/h/k/s contain low-temperature response elements (LTR), suggesting that they may be involved in cold stress (Fig. 2).
Abiotic stress affects miR156 expression in various plants. In Arabidopsis and sugarcane, low-temperature stress decreased miR156 expression and extended the nutritional growth phase, resulting in delayed growth metabolism [22, 45]. In young spikes of common wheat exposed to 48 h of cold treatment, miR156(s) were down-regulated [54] On the other hand, overexpression of OsmiR156k inhibited seedling growth under cold stress at the early development stage [21]. In our investigation, only expression of miR156g/h/i and miR156r/s/t were down-regulated in young leaves after 48 h of cold treatment when compared to untreated leaves, but in young root tissues, miR156g/h/i were up-regulated after 48 h of cold treatment. Almost all tae-miR156(s) were highly expressed following 3 h of cold stress treatment, with the most substantial up-regulation occurring in root tissues. These results proposed that tae-miR156(s) expression differed between leaves and roots during cold stress treatment (Fig. 4). Notably, the expression of tae-miR156g/h/i was up-regulated in root tissues at different time points compared to that of roots not exposed to the low temperature (Fig. 4), indicating that in wheat, tae-miR156g/h/i may be the most sensitive to cold stress of all miR156 members.
It has been demonstrated that without carbon dioxide fertilization, effective adaptation, and genetic improvement, global yields of wheat, rice, maize, and soybeans declined by an average of 6.0%, 3.2%, 7.0%, and 3.2%, respectively, for every 1 degree Celsius increase in global average temperature [55]. MiR156(s) were also involved in heat stress response in a variety of plants. In Arabidopsis seedlings, expression levels of miR156c, miR156d, and miR156h were up-regulated under heat stress proposing that overexpressing miR156 could enhance tolerance to heat stress [56]. Overexpression of miR156 also increased alfalfa's resistance to heat stress by boosting anthocyanin and chlorophyll accumulation [23]. As miR156 and its target genes are generally conserved in plants, it was proposed that the function of miR156 in heat stress memory may also be conserved in plants [57]. In our study, expression of miR156g/h/i, miR156j/k, miR156l/m/n/o/p/q, and miR156j/k were up-regulated in leaves after 3 h of heat treatment. Nonetheless, expression levels of all tae-miR156(s) in the leaves decreased sharply after 6 h of heat treatment and continued to decrease to a minimum level after 12 h (Fig. 5), suggesting that tae-miR156(s) have a similar response to heat stress. In contrast, in heat-treated roots, the expression of miR156g/h/i were most significantly up-regulated after 3 h of treatment compared with the other miR156s (Fig. 5). This suggested that among all the miR156 members in wheat, tae-miR156g/h/i were most likely implicated in resistance to heat stress.
The expression levels of miR156 may change in the presence of drought or high salt environments, allowing plants to adjust their growth strategies and improve drought and salt tolerance by regulating the expression of target genes. Under drought stress, the expression levels of miR156 were up-regulated in Brachypodium distachyon [58], maize (Zea mays) [59], tomato (Solanum lycopersicum) [60], and apple (Malus domestica) [26], but suppressed in rice [61], Chinese white poplar (Populus tomentosa) [62], and rape (Brassica napus) [63]. In addition, overexpression of miR156 enhanced tolerance to drought stress in alfalfa [64], tomato [60], and apple [26]. In this study, only miR156g/h/i expression in leaves was suppressed over all drought stress treatment time points while the expression levels of the other seventeen miR156(s) significantly increased after 3 h of drought treatment. And tae-miR156(s) were strongly induced by drought stress in root tissues compared to leaves (Fig. 6). These findings show that miR156 is differentially expressed in roots and leaves, implying that they serve separate functions during drought stress in wheat. Similar phenomena have been observed in plants such as cotton [65]. Of note, the expression levels of miR156a/b/c/d/e/f were significantly elevated after 24 h of treatment at 5.58 times higher than that of untreated (Fig. 6), showing that tae-miR156a/b/c/d/e/f may be the most susceptible to drought stress of all miR156 members in wheat.
For crops, salinity is one of the major abiotic stresses that often leads to reduced yields [66]. Under salt stress, the expression levels of miR156 in plants such as rice [67] and Arabidopsis [68] were up-regulated but the opposite was reported for expression in maize [69] and cotton [65]. Overexpression of miR156 reduced salt tolerance in apple seedlings [17]. MicroRNA156 positively regulates the physiological responses of Alfalfa under salinity stress [70] and manipulating the expression of ZmmiR156 in tobacco could enhance the salt tolerance of transgenic plants without affecting plant structure [48]. These studies demonstrated that miR156 was also involved in the response to salt stress. Previous genomics-based analyses showed that miR156(s) expression levels were down-regulated in the roots of salt-tolerant wheat cultivars after 24 h of salt stress treatment [71]. Meanwhile, in our study, only the expression of miR156g/h/i and miR156r/s/t declined after 24 h of salt stress in the wheat plant roots, suggesting a differential response of diverse tae-miR156 to salt stress. In contrast, all tae-miR156(s) showed a similar expression profile in leaves under salt treatment, with a significant increase at 3 h, followed by a decrease, and a minimum at 48 h compared with the untreated at the corresponding time point, with the most significant fluctuation in expression observed for miR156r/s/t. Moreover, the expression level of tae-miR156r/s/t in root tissues was inhibited in salt-treated root tissues (Fig. 7). These results indicate that tae-miR156r/s/t may be more sensitive to salt stress.
MicroRNA156 has been reported to be a "superstar microRNA", involved in a variety of biological processes and stress responses in plants [72]. Under stress conditions, miR156 is induced to maintain the juvenile state for a longer period, while under favorable conditions, miR156 is inhibited to accelerate developmental transitions [73]. These studies suggest that the expression pattern of miR156 is critical to plant growth and development and response to stress. Hence, the expression of miR156 can be manipulated for genetic improvement of plant resistance to various abiotic stresses [48]. Previous transcriptome analysis revealed that wheat miR156 was implicated in stress response, but no specific members were identified [40, 71, 74]. Although miR156 is highly conserved throughout plants, its function varies among members of the same family, demonstrating functional differentiation within the family [8, 9]. Therefore, it is crucial to identify which miRNA family members are most vulnerable to unfavorable stress.
The SPL transcription factors are well known to play conserved roles in regulating diverse developmental processes and responses to biotic and abiotic stresses in various plant species [6, 75, 76]. And miR156 targets a large number of SPL genes across a wide range of plant species, indicating that the miR156/SPLs regulatory module has evolved to regulate diverse processes in the plant kingdom [6]. Among the 17 Arabidopsis SPLs, 11 are miR156 targets [77]. In rice, there are 19 OsSPL genes, of which 11 SPL genes are predicted targets of miR156 [49]. The wheat genome has 56 TaSPL genes [78, 79]. In this study, 27 SPLs target genes were predicted of miR156 (Table 2). Among these TaSPLs target genes, TaSPL3, TaSPL13, TaSPL14, and TaSPL17 have been revealed to be targets of miR156, which may interact with miR156 to regulate plant architecture and improve agronomic traits in wheat [11, 79–82]. However, the interactions between wheat miR156 and other SPL target genes, as well as the molecular mechanisms of the miR156/SPLs pathway in regulating abiotic stresses still need to be further explored in wheat.
Conclusions
In this study, 20 miR156 family members (miR156a to miR156t) were identified in wheat. Although they are relatively conserved, the cis-elements of promoters and expression patterns were noted to be different. In addition, tae-miR156(s) were involved in abiotic stress response in wheat, and these miR156(s) showed different degrees of response in leaf and root tissues. Specifically, miR156g/h/i, miR156a/b/c/d/e/f, and miR156r/s/t may be the most effective molecular targets for inducing wheat stress resistance. Taken together, the miR156(s) despite originating from the same family, are involved in different responses to adverse conditions and may play a role in one of the most critical defense systems for wheat biotic stress tolerance. This lays the foundation for revealing the precise biological activities and molecular basis of adversity stress and will help to accelerate the breeding of stress-tolerant wheat varieties.
Supplementary Information
Acknowledgements
We thank the useful technical assistance provided by our colleagues in the laboratory. We also thank the rigorous evaluation of the manuscript made by the editors and reviewers.
Authors’ contributions
The experimental design and execution of this study were led by SR and HS. SR and JL (Juan Lin) were primarily responsible for conducting the experiments and participated in data analysis. TL, YW, CX, LM, and WL also contributed to experimental operations and data analysis. Following the completion of the experiments, SR drafted the initial manuscript. CC, JL (Jie Lu), CM, and HS provided in-depth improvements to the manuscript. All authors have read and agreed to the final version of the manuscript.
Funding
The work was supported by a grant from The Agriculture Research System (CARS-03), Anhui Provincial and Ministerial Collaborative Innovation Center for Food Crops (APMCIC-FC), and Jiangsu Collaborative Innovation Center for Modern Crop Production (JCIC-MCP).
Data availability
All the supporting data are included within the article and its additional files.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shuang Ruan, Juan Lin and Tiantian Li contributed equally to this work.
References
- 1.Wei X, Ke H, Wen A, Gao B, Shi J, Feng Y. Structural basis of microRNA processing by Dicer-like 1. Nat Plants. 2021;7:1389–96. [DOI] [PubMed] [Google Scholar]
- 2.Arazi T, Talmor Neiman M, Stav R, Riese M, Huijser P, Baulcombe DC. Cloning and characterization of microRNAs from moss. Plant J. 2005;43:837–48. [DOI] [PubMed] [Google Scholar]
- 3.Floyd SK, Bowman JL. Ancient microRNA target sequences in plants. Nature. 2004;428:485–6. [DOI] [PubMed] [Google Scholar]
- 4.Griffiths-Jones S, Grocock RJ, Van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao Y, Guo G, Ni Z, Sunkar R, Du J, Zhu JK, Sun Q. Cloning and characterization of microRNAs from wheat (Triticum aestivum L.). Genome Biol. 2007;8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, Wang HY. The miR156/SPL module, a regulatory hub and versatile toolbox, gears up crops for enhanced agronomic traits. Mol Plant. 2015;8:677–88. [DOI] [PubMed] [Google Scholar]
- 7.Lu YD, Gan QH, Chi XY, Qin S. Roles of microRNA in plant defense and virus offense interaction. Plant Cell Rep. 2008;27:1571–9. [DOI] [PubMed] [Google Scholar]
- 8.Du Q, Zhao M, Gao W, Sun S, Li WX. microRNA/micro RNA* complementarity is important for the regulation pattern of NFYA5 by miR169 under dehydration shock in Arabidopsis. Plant J. 2017;91:22–33. [DOI] [PubMed] [Google Scholar]
- 9.Miao C, Wang Z, Zhang L, Yao J, Hua K, Liu X, Shi H, Zhu JK. The grain yield modulator miR156 regulates seed dormancy through the gibberellin pathway in rice. Nat Commun. 2019;10:3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yun J, Sun Z, Jiang Q, Wang Y, Wang C, Luo Y, Zhang F, Li X. The miR156b-GmSPL9d module modulates nodulation by targeting multiple core nodulation genes in soybean. New Phytol. 2022;233:1881–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Cheng X, Liu P, Sun J. miR156-targeted SBP-box transcription factors interact with DWARF53 to regulate TEOSINTE BRANCHED1 and BARREN STALK1 expression in bread wheat. Plant Physiol. 2017;174:1931–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui L, Zheng F, Wang J, Zhang C, Xiao F, Ye J, Li C, Ye Z, Zhang J. miR156a-targeted SBP-Box transcription factor SlSPL13 regulates inflorescence morphogenesis by directly activating SFT in tomato. Plant Biotechnol J. 2020;18:1670–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhogale S, Mahajan AS, Natarajan B, Rajabhoj M, Thulasiram HV, Banerjee AK. MicroRNA156: A Potential Graft-Transmissible MicroRNA That Modulates Plant Architecture and Tuberization in Solanum tuberosum ssp andigena. Plant Physiol. 2014;164:1011–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng MQ, Lu MD, Long JM, Yin ZP, Jiang N, Wang PB, Liu Y, Guo WW, Wu XM. miR156 regulates somatic embryogenesis by modulating starch accumulation in citrus. J Exp Bot. 2022;73:6170–85. [DOI] [PubMed] [Google Scholar]
- 15.Song X, Li Y, Cao X, Qi Y. MicroRNAs and their regulatory roles in plant–environment interactions. Annual Rev Plant Biol. 2019;70:489–525. [DOI] [PubMed] [Google Scholar]
- 16.Megha S, Basu U, Kav NN. Regulation of low temperature stress in plants by microRNAs. Plant Cell Environ. 2018;41:1–15. [DOI] [PubMed] [Google Scholar]
- 17.Ma Y, Xue H, Zhang F, Jiang Q, Yang S, Yue P, Wang F, Zhang Y, Li L, He P. The miR156/SPL module regulates apple salt stress tolerance by activating MdWRKY100 expression. Plant Biotechnol J. 2021;19:311–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen X, Song Y, Ping Y, He J, Xie Y, Ma F, Li X, Guan Q. The RNA-binding protein MdHYL1 modulates cold tolerance and disease resistance in apple. Plant Physiol. 2023;192:2143–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niu C, Li H, Jiang L, Yan M, Li C, Geng D, Xie Y, Yan Y, Shen X, Chen P. Genome-wide identification of drought-responsive microRNAs in two sets of Malus from interspecific hybrid progenies. Horti Res. 2019;6:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y, Zhang X, Su Y, Zou J, Wang Z, Xu L, Que Y. miRNA alteration is an important mechanism in sugarcane response to low-temperature environment. BMC Genom. 2017;18:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui N, Sun X, Sun M, Jia B, Duanmu H, Lv D, Duan X, Zhu Y. Overexpression of OsmiR156k leads to reduced tolerance to cold stress in rice (Oryza Sativa). Mol Breeding. 2015;35:214. [Google Scholar]
- 22.Zhu PJ, Song QQ, Tan QL, Cheng Q, Li JH, Pang XH, Zhou QG, Lü P, Ou KW, Lu YF. Bioinformatics analysis of microRNAs and prediction of target genes associated with cold tolerance in sugarcane. Guihaia. 2022;42:1840–53.
- 23.Matthews C, Arshad M, Hannoufa A. Alfalfa response to heat stress is modulated by microRNA156. Physiol Plantarum. 2019;165:830–42. [DOI] [PubMed] [Google Scholar]
- 24.Arshad M, Feyissa BA, Amyot L, Aung B, Hannoufa A. MicroRNA156 improves drought stress tolerance in alfalfa (Medicago sativa) by silencing SPL13. Plant Sci. 2017;258:122–36. [DOI] [PubMed] [Google Scholar]
- 25.Chen H, Yang Q, Chen K, Zhao S, Zhang C, Pan R, Cai T, Deng Y, Wang X, Chen Y, et al. Integrated microRNA and transcriptome profiling reveals a miRNA-mediated regulatory network of embryo abortion under calcium deficiency in peanut (Arachis hypogaea L.). BMC Genom. 2019;20:392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen G, Wang Y, Liu X, Duan S, Jiang S, Zhu J, Zhang Y, Hou H. The MdmiR156n Regulates Drought Tolerance and Flavonoid Synthesis in Apple Calli and Arabidopsis. Int J Mol Sci. 2023;24:6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui LG, Shan JX, Shi M, Gao JP, Lin HX. The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 2015;82:901–901. [DOI] [PubMed] [Google Scholar]
- 28.Wang K, Liu Y, Teng F, Cen H, Yan J, Lin S, Li D, Zhang W. Heterogeneous expression of Osa-MIR156bc increases abiotic stress resistance and forage quality of alfalfa. Crop J. 2021;9:1135–44. [Google Scholar]
- 29.Yuan J, Wang X, Qu S, Shen T, Li M, Zhu L. The roles of miR156 in abiotic and biotic stresses in plants. Plant Physiol Bio. 2023;204:108150. [DOI] [PubMed] [Google Scholar]
- 30.Jerome Jeyakumar JM, Ali A, Wang WM, Thiruvengadam M. Characterizing the role of the miR156-SPL network in plant development and stress response. Plants. 2020;9:1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shewry PR. Wheat Journal of Exp Bot. 2009;60:1537–53. [DOI] [PubMed] [Google Scholar]
- 32.Abhinandan K, Skori L, Stanic M, Hickerson NM, Jamshed M, Samuel MA. Abiotic stress signaling in wheat an inclusive overview of hormonal interactions during abiotic stress responses in wheat. Front Plant Sci. 2018;9:734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo Z, Kuang Z, Wang Y, Zhao Y, Tao Y, Cheng C, Yang J, Lu X, Hao C, Wang T. PmiREN: a comprehensive encyclopedia of plant miRNAs. Nucleic Acids Res. 2020;48:1114–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38:3022–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai X, Zhuang Z, Zhao PX. psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018;46:49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato K, Abe F, Mascher M, Haberer G, Gundlach H, Spannagl M, Shirasawa K, Isobe S. Chromosome-scale genome assembly of the transformation-amenable common wheat cultivar ‘Fielder.’ DNA Res. 2021;28:dsab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zadoks JC, Chang TT, Konzak CF. A decimal code for the growth stages of cereals. Weed Res. 1974;14:415–21. [Google Scholar]
- 40.Qin D, Wu H, Peng H, Yao Y, Ni Z, Li Z, Zhou C, Sun Q. Heat stress-responsive transcriptome analysis in heat susceptible and tolerant wheat (Triticum aestivum L.) by using Wheat Genome Array. BMC Genom. 2008;9:432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao J, Qin Z, Cui G, Chen Z, Cheng X, Peng H, Yao Y, Hu Z, Guo W, Ni Z. Natural variation of STKc_GSK3 kinase TaSG-D1 contributes to heat stress tolerance in Indian dwarf wheat. Nat Commun. 2024;15:2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu TF, Xu ZS, Guo JK, Wang YX, Abernathy B, Fu JD, Chen X, Zhou YB, Chen M, Ye XG. Improved drought tolerance in wheat plants overexpressing a synthetic bacterial cold shock protein gene SeCspA. Sci Rep. 2017;7:44050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang M, Cheng J, Wu J, Chen J, Liu D, Wang C, Ma S, Guo W, Li G, Di D. Variation in TaSPL6-D confers salinity tolerance in bread wheat by activating TaHKT1; 5-D while preserving yield-related traits. Nat Genet. 2024;56:1257–69. [DOI] [PubMed] [Google Scholar]
- 44.Wen S, Zhou C, Tian C, Yang N, Zhang C, Zheng A, Chen Y, Lai Z, Guo Y. Identification and Validation of the miR156 Family Involved in Drought Responses and Tolerance in Tea Plants (Camellia sinensis (L.) O. Kuntze). Plants. 2024;13:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gang Wu, R Scott Poethig. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133:3539–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang JW, Park MY, Wang LJ, Koo Y, Chen XY, Weigel D, Poethig RS. miRNA control of vegetative phase change in trees. PLoS Genet. 2011;7:e1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cui LG, Shan JX, Shi M, Gao JP, Lin HX. The miR156-SPL 9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 2014;80:1108–17. [DOI] [PubMed] [Google Scholar]
- 48.Kang T, Yu CY, Liu Y, Song WM, Bao Y, Guo XT, Li B, Zhang HX. Subtly manipulated expression of ZmmiR156 in tobacco improves drought and salt tolerance without changing the architecture of transgenic plants. Front Plant Sci. 2020;10:1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie K, Wu C, Xiong L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 2006;142:280–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang C, Wang Q, Zhu X, Cui M, Jia H, Zhang W, Tang W, Leng X, Shen W. Characterization on the conservation and diversification of miRNA156 gene family from lower to higher plant species based on phylogenetic analysis at the whole genomic level. Funct Integr Genomics. 2019;19:933–52. [DOI] [PubMed] [Google Scholar]
- 51.Ding X, Ruan H, Yu L, Li Q, Song Q, Yang S, Gai J. miR156b from soybean CMS line modulates floral organ development. J Plant Biol. 2020;63:141–53. [Google Scholar]
- 52.Du Q, Zhao M, Gao W, Sun S, Li WX. microRNA/microRNA* complementarity is important for the regulation pattern of NFYA5 by miR169 under dehydration shock in Arabidopsis. Plant J. 2017;91:22. [DOI] [PubMed] [Google Scholar]
- 53.Chen F, Xiang Z, Bingyang D, Yuqin X, Yanyan W, Yueting S, Xin Z, Chao W, Yang L, TianHong L. MicroRNA156ab regulates apple plant growth and drought tolerance by targeting transcription factor MsSPL13. Plant Physiol. 2023;192:1836–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song G, Zhang R, Zhang S, Li Y, Gao J, Han X, Chen M, Wang J, Li W, Li G. Response of microRNAs to cold treatment in the young spikes of common wheat. BMC Genom. 2017;18:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao C, Liu B, Piao S, Wang X, Lobell DB, Huang Y, Huang M, Yao Y, Bassu S, Ciais P. Temperature increase reduces global yields of major crops in four independent estimates. Proc Natl Acad Sci. 2017;114:9326–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stief A, Altmann S, Hoffmann K, Pant BD, Scheible WR, Bäurle I. Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell. 2014;26:1792–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao J, He Q, Jin B. Regulation of non-coding RNAs in heat stress responses of plants. Front Plant Sci. 2016;7:1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bertolini E, Verelst W, Horner DS, Gianfranceschi L, Piccolo V, Inzé D, Pè ME, Mica E. Addressing the role of microRNAs in reprogramming leaf growth during drought stress in Brachypodium distachyon. Mol Plant. 2013;6:423–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang Q, Lv H, Li Q, Zhang X, Li L, Xu J, Wu F, Wang Q, Feng X, Lu Y. Characteristics of microRNAs and target genes in maize root under drought stress. Int J Mol Sci. 2022;23:4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Visentin I, Pagliarani C, Deva E, Caracci A, Turečková V, Novák O, Lovisolo C, Schubert A, Cardinale F. A novel strigolactone-miR156 module controls stomatal behaviour during drought recovery. Plant Cell Environ. 2020;43:1613–24. [DOI] [PubMed] [Google Scholar]
- 61.Zhou L, Liu Y, Liu Z, Kong D, Duan M, Luo L. Genome-wide identification and analysis of drought responsive microRNAs in Oryza sativa. J Exp Bot. 2010;61:4157–68. [DOI] [PubMed] [Google Scholar]
- 62.Ren Y, Chen L, Zhang Y, Kang X, Zhang Z, Wang Y. Identification of novel and conserved Populus tomentosa microRNA as components of a response to water stress. Funct Integr Genomic. 2012;12:327–39. [DOI] [PubMed] [Google Scholar]
- 63.Jian H, Wang J, Wang T, Wei L, Li J, Liu L. Identification of rapeseed microRNAs involved in early stage seed germination under salt and drought stresses. Front Plant Sci. 2016;7:658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arshad M, Gruber MY, Hannoufa A. Transcriptome analysis of microRNA156 overexpression alfalfa roots under drought stress. Sci Rep. 2018;8:9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang M, Wang Q, Zhang B. Response of miRNAs and their targets to salt and drought stresses in cotton (Gossypium hirsutum L.). Gene. 2013;530:26–32. [DOI] [PubMed] [Google Scholar]
- 66.Van Zelm E, Zhang Y, Testerink C. Salt tolerance mechanisms of plants. Annu Rev Plant Biol. 2020;71:403–33. [DOI] [PubMed] [Google Scholar]
- 67.Aycan M, Nahar L, Baslam M, Mitsui T. B-type response regulator hst1 controls salinity tolerance in rice by regulating transcription factors and antioxidant mechanisms. Plant Physiol Biochem. 2023;196:542–55. [DOI] [PubMed] [Google Scholar]
- 68.Liu HH, Tian X, Li YJ, Wu CA, Zheng CC. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA. 2008;14:836–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ding D, Zhang L, Wang H, Liu Z, Zhang Z, Zheng Y. Differential expression of miRNAs in response to salt stress in maize roots. Ann Bot. 2009;103:29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arshad M, Gruber MY, Wall K, Hannoufa A. An insight into microRNA156 role in salinity stress responses of alfalfa. Front Plant Sci. 2017;8:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zeeshan M, Qiu CW, Naz S, Cao F, Wu F. Genome-wide discovery of miRNAs with differential expression patterns in responses to salinity in the two contrasting wheat cultivars. Int J Mol Sci. 2021;22:12556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Y, Luo Z, Zhao X, Cao H, Wang L, Liu S, Wang C, Liu M, Wang L, Liu Z. Superstar microRNA, miR156, involved in plant biological processes and stress response. A review Sci Hortic. 2023;316:112010. [Google Scholar]
- 73.Cao C, Long R, Zhang T, Kang J, Wang Z, Wang P, Sun H, Yu J, Yang Q. Genome-wide identification of microRNAs in response to salt/alkali stress in Medicago truncatula through high-throughput sequencing. Int J Mol Sci. 2018;19:4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pandey R, Joshi G, Bhardwaj AR, Agarwal M, Katiyar AS. A comprehensive genome-wide study on tissue-specific and abiotic stress-specific miRNAs in Triticum aestivum. PLoS ONE. 2014;9:e95800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang J, Wei H, Hou M, Chen L, Zou T, Ding H, Jing Y, Zhang X, Zhao Y, Liu Q. ZmSPL13 and ZmSPL29 act together to promote vegetative and reproductive transition in maize. New Phytol. 2023;239:1505–20. [DOI] [PubMed] [Google Scholar]
- 76.Yin H, Hong G, Li L, Zhang X, Kong Y, Sun Z, Li J, Chen J, He Y. miR156/SPL9 regulates reactive oxygen species accumulation and immune response in Arabidopsis thaliana. Phytopathology. 2019;109:632–42. [DOI] [PubMed] [Google Scholar]
- 77.Addo Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol. 2008;18:758–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li L, Shi F, Wang G, Guan Y, Zhang Y, Chen M, Chang J, Yang G, He G, Wang Y. Conservation and divergence of SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) gene family between wheat and rice. Int J Mol Sci. 2022;23:2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gupta A, Hua L, Zhang Z, Yang B, Li W. CRISPR-induced miRNA156-recognition element mutations in TaSPL13 improve multiple agronomic traits in wheat. Plant Biotechnol J. 2023;21:536–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li L, Shi F, Wang Y, Yu X, Zhi J, Guan Y, Zhao H, Chang J, Chen M, Yang G. TaSPL13 regulates inflorescence architecture and development in transgenic wheat (Triticum aestivum L.). Plant Sci. 2020;296:110516. [DOI] [PubMed] [Google Scholar]
- 81.Cao J, Liu K, Song W, Zhang J, Yao Y, Xin M, Hu Z, Peng H, Ni Z, Sun Q. Pleiotropic function of the SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE gene TaSPL14 in wheat plant architecture. Planta. 2021;253:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cao L, Li T, Geng S, Zhang Y, Pan Y, Zhang X, Wang F, Hao C. TaSPL14–7A is a conserved regulator controlling plant architecture and yield traits in common wheat (Triticum aestivum L.). Front Plant Sci. 2023;14:1178624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the supporting data are included within the article and its additional files.