Abstract
Yersinia pestis is the perilous pandemics that occurred in Asia and Europe. The bacterium has shown drug resistance that can cause the future pandemic and destroy the drug treatment against plague. As known, effective therapeutics such as designing potent vaccine that can aid world to protect against plague. The immunoinformatics approaches was implemented via different server. The 4 potent antigens (F1 capsule, LcrV, OmpA, and PH6) were listed as essential protein target for creating the multi-epitope vaccine. These targets were selected for designing multi-epitope vaccine that predicted the CTL and HTL epitopes. The vaccine construct included different linkers such as EAAAK, AAY, GPGPG, and SSL that an adjuvant (Beta defensin-3) inserted at N-terminal of vaccine. The computational physiochemical properties and other immunological analysis showed stable, soluble, antigen, non-allergen, and non-toxin. The molecular docking confirmed the stable binding and good interaction and the iMODS server showed the stable binding. Furthermore, computational immune simulation of multi-epitope vaccine showed that vaccine can stimulate adaptive and innate responses after second doses. In this study, the vaccine designed for Y. pestis that in future require immunological examination to unveil real efficiency.
Keywords: Reverse vaccinology, Computational immunology, Immunoinformatics, Multi-epitope vaccine
Highlights
-
•
Novel multi-epitope vaccine targeting Yersinia pestis, utilizing advanced immunoinformatics tools for epitope prediction.
-
•
The final vaccine construct consists of 16 HTL/CTL epitopes, strategically separated by linkers to enhance stability and efficacy, incorporating TAT peptide and PADRE sequence to boost antigenicity and delivery.
-
•
The vaccine showed high antigenicity, low allergenicity and toxicity, and favorable solubility for formulation.
1. Introduction
Plague is a vector-borne sickness disseminated by fleas that affects a broad range of wildlife rodents, which serve as natural reservoirs for the disease in a number of environments across the world [1]. The Gram-negative bacterium Yersinia pestis is responsible for plague. In 1894, the Swiss-French physician Alexandre Yersin, associated with the Pasteur Institute, identified the cause of the plague and contributed to understanding part of the transmission cycle involving the rat (Rattus rattus) and its ectoparasites (Xenopsylla cheopis) [2]. There are five major types of plague caused by an infection with Y.pestis: bubonic, septicemic, pneumonic, meningeal, and pharyngeal [3]. Septic or secondary pneumonic infection can occur in patients with bubonic plague. Pneumonic plague can then be transmitted from person to person by respiratory droplets produced by sufferers' sneezing and coughing [4]. Additionally, Y.pestis has been observed to induce skin ulcers at its point of entrance, including pustules, boils, petechiae, bruising, and gangrene [5]. Plague has shaped human history through numerous pandemics that originated in Central Asia and spread to Africa and Europe, ultimately affecting every continent over the past 150 years [6]. Plague is still present in Asia, Africa, and America in the twenty-first century [7]. It is important to remind the public that plague continues to pose a main health threat. Genomic analyses of Y. pestis reveal that the pathogen shares genetic similarities with Y. pseudotuberculosis. However, a series of gene gain and loss events have led to markedly different disease mechanisms, as well as distinct niche preferences and lifestyles [8]. Y. pestis exhibited resistance to ampicillin, chloramphenicol, kanamycin, streptomycin, spectinomycin, sulfonamides, tetracycline, and minocycline. Another strain, Y.pestis, was resistant to streptomycin but not to spectinomycin or other antibiotics, including those used in plague treatment (chloramphenicol and tetracycline) and prophylaxis (spectinomycin) (sulfonamides and tetracycline) [9,10]. One of the most severe infectious illnesses, which is caused by Yersinia pestis, has no perfect plague vaccine available for human usage, The fact that plague poses such a serious threat to people highlights the urgent need for more effective vaccinations. For human usage, there are no foolproof plague vaccinations. Furthermore, the multi-epitope vaccine has been shown to robustly enhance both humoral and cellular immune responses [11]. This enhanced immune response is attributed to the presence of T cell and B cell epitopes within the vaccine formulation. Additionally, the inclusion of adjuvants in the multi-epitope vaccine is anticipated to elicit sustained immune reactions and promote a heightened level of immunogenicity, thereby enhancing its efficacy in conferring protection against targeted pathogens [12]. The utilization of immunoinformatics methodologies has the potential to significantly expedite the research efforts of scientists in the discovery and development of cutting-edge vaccines to combat emerging and evolving diseases in the future. This innovative approach offers a promising avenue for streamlining the vaccine development process, ultimately leading to substantial time savings and improved efficiency in the field of immunology [13]. In this research, we are looking for an effective vaccine with a new generation that is adequate.
2. Method
2.1. Retrieval of protein sequences
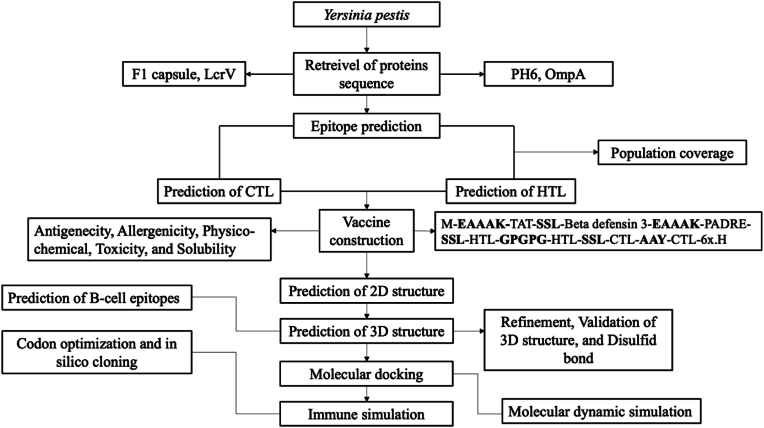
The FASTA sequences of F1 capsule (CAA43966.1), LcrV (AAA27641.1), OmpA (TPW82762.1), and PH6 (P3522) of Yersinia pestis were retrieved from NCBI and Uniprot. In addition, the analysis of antigen sequences was implemented by MEGA11 [14] to detect the conserved region of the protein. The comprehensive workflow of the study showed in Fig. 1.
Figrue 1.
The comprehensive workflow of designing multi-epitope vaccine against Yersinia pestis.
2.2. The extraction and assessment of experimental T-cell epitopes via immonoinformatics
The meticulous process of extracting the experimental T-cell epitopes was systematically executed, during which we meticulously filtered this extensive dataset through the lens of antigenicity to ensure the selection of the most promising epitope candidate, utilizing the sophisticated capabilities of the VaxiJen server, which is renowned for its ability to predict the antigenic potential of various epitopes with a high degree of accuracy and reliability.
2.3. Prediction of CTL epitopes and IL4 inducing
The role of presenting peptides from the Major Histocompatibility Complex class I (MHC-I) is of significant importance in the activation of the immune system, and as a result, the selection of Cytotoxic T Lymphocyte (CTL) epitopes plays an important role in the creation of vaccines. In order to identify potential CTL epitopes, a computational tool known as CTLpred was utilized. This tool, accessible at the link http://crdd.osdd.net/raghava/ctlpred/, was employed for the prediction of CTL epitopes specifically for the F1 capsule, LcrV, OmpA, and PH6 proteins. The method being discussed in this study demonstrates a remarkable capability to forecast CTL epitopes within the antigen sequence through the utilization of sophisticated Artificial Neural Network (ANN) and Support Vector Machine (SVM) algorithms. The integration of these computational models allows for a more nuanced and comprehensive analysis of potential epitope candidates, enhancing the accuracy and reliability of the predictions made. Furthermore, in order to anticipate the binding of major histocompatibility complex class I (MHC-I) peptides to prevalent mice alleles, the researchers leveraged the Immune Epitope Database (IEDB) tool (http://tools.iedb.org/mhci/), employing the default prediction methodology provided by the platform. This approach not only simplifies the prediction process but also ensures alignment and consistency with current literature and experimental findings, facilitating better comparability. The finalized epitopes and the analysis of IL4 was implemented by IL4Pred (https://webs.iiitd.edu.in/raghava/il4pred/design.php).
2.4. Prediction of HTL epitopes
MHC-II molecules have a significant role in modulating the adaptive immune response through the activation of CD4+ T-lymphocytes (HTL). In order to identify potential MHC-II peptide binders derived from the aforementioned antigens, we made use of the IEDB MHC-II epitope prediction tool. In this process, we explored various approaches for epitope prediction and specifically followed the guidelines provided by IEDB, which involved the integration of NN-align, SMM-align, CombLib, and Sturniolo algorithms, whenever a corresponding predictor was accessible for the particular molecule under investigation.
2.5. Construction of multi-epitope
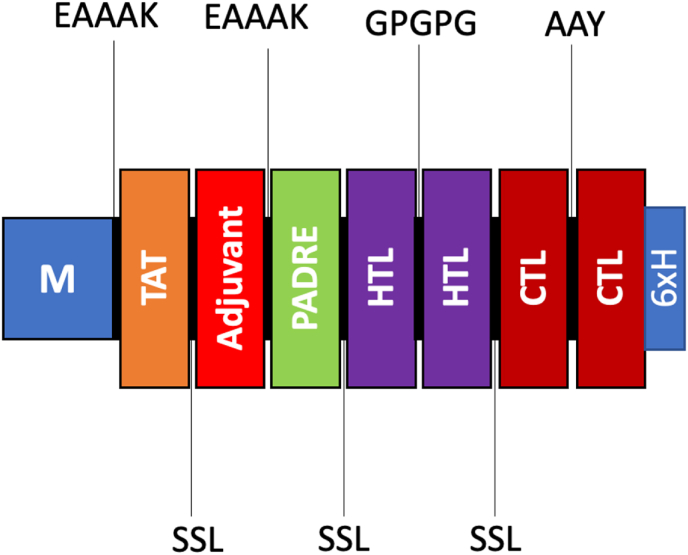
The epitopes that were predicted in this study were systematically collected using innovative linkers, including EAAAK, SSL, GPGPG, and AAY. The initiation of the vaccine sequence was marked by the presence of methionine, while the N-terminal of the vaccine was comprised of human Beta-defensin 3 (hBD3) and TAT sequences serving as a cell-penetrating peptide. Additionally, the N-terminal included TLR3 agonist that stimulated the excretion of pro-inflammatory cytokines in vitro. Subsequently, the PADRE sequence was introduced to the adjuvant through the utilization of the EAAAK linker. The HTL epitopes were combined utilizing the GPGPG linker, whereas the CTL epitopes were linked together through the AAY linker. In order to ensure the segregation of each component of the vaccine, SSL linkers were employed. Finally, a sequence of 6-Histidine residues was incorporated at the C-terminal end of the vaccine. The overall sequence structure contains M-EAAAK-TAT-SSL-Beta defensin 3-EAAAK-PADRE-SSL-HTL-GPGPG-HTL-SSL-CTL-AAY-CTL-6x.H.
2.6. Assessment of multi-epitope vaccine
The multi-epitope vaccine's antigenicity was predicted using the VaxiJen v2.0 program, with a threshold value of 0.456 being used [15]. In order to delve into the allergenicity aspect of the vaccine, an analysis was conducted using the AllerTOP v.2.0 tool [16]. In addition, a complete evaluation of the construct's chemical and physical properties was conducted using the ProtParam server (https://web.expasy.org/protparam/). This analysis included a thorough look at a number of factors, including the molecular weight, the in vitro and in vivo half-lives, the aliphatic index, the grand average of hydropathicity (GRAVY), the theoretical isoelectric point (pI), and the makeup of amino acids. The use of the protein-sol software enabled the analysis of the amino acid sequence, allowing key calculations to be performed for predicting the solubility of the multi-epitope vaccine. Through the application of solubility analysis, an evaluation was performed to ensure the integrity and quality of a given substance, thus highlighting the significance of this analytical approach in the context of biological research and vaccine development. The incorporation of such computational tools and methodologies in scientific investigations underscores the importance of leveraging technological advancements for enhancing the efficiency and accuracy of predictive analyses in the realm of biotechnology and pharmaceutical sciences [17]. SolPro (http://scratch.proteomics.ics.uci.edu) evaluates protein solubility. Compared to SolPro, an SVM-based tool used for predicting the solubility of protein sequences demonstrates around 74 % accuracy through tenfold cross-validation [18]. The ToxinPred tool finally predicts the toxicity levels of both the vaccine and its epitopes. This platform utilizes the Support Vector Machine (SVM) model in conjunction with a dataset consisting of 1805 toxic peptides (with ≤35 residues) to differentiate between toxic and non-toxic peptides [19]. In order to detect the homology of the human and mice proteins with vaccine, we used the Blastp to avoid the autoimmune reaction.
2.7. Prediction of 2D structure
All vaccine constructs underwent additional analysis by being input into the PSIPRED 4.0 server, (http://bioinf.cs.ucl.ac.uk/psipred/), in order to acquire information regarding their 2D structure. The PSIPRED 4.0 server executed predictions of protein secondary structures using position-specific scoring matrices, achieving an impressive prediction accuracy rate exceeding 84 %. This methodological approach proved to be highly effective in elucidating the structural characteristics of the vaccine constructs, thereby contributing valuable insights into their potential functionality and immunogenicity. The Self-Optimized Prediction Method with Alignment (SOPMA) server was utilized to anticipate the secondary structure of the multi-epitope vaccine. The SOPMA server predicted the alpha helix, extended strand, and random coil.
2.8. Prediction of 3D structure and validation
Robetta tool (https://robetta.bakerlab.org/), an online service dedicated to the prediction of protein structures, engages in a continuous process of assessing various parameters of a given prediction such as coverage, local accuracy, and completeness, among others. The final 3D structure of vaccines was effectively modeled through a collaborative social project undertaken by this platform. The process of protein folding within this server is underpinned by sophisticated techniques including deep learning methodologies, the employment of RoseTTAFold, TrRosetta, as well as an interactive submission interface which facilitates the integration of custom sequence alignments tailored for homology modeling, the imposition of constraints, incorporation of local fragments, and other elements necessary for the prediction process to be conducted swiftly and with a high degree of accuracy [20]. Subsequently, the most advantageous rough 3D representations of every vaccine sequence were entered into the GalaxyRefine module hosted on the GalaxyWEB server (https://galaxy.seoklab.org/), a platform capable of rectifying questionable loops or endpoints within the preliminary model configurations through an enhancement technique grounded on optimization, ultimately producing five enhanced versions of each initial model [21].
Model validation was conducted by utilizing the ProSA web server (https://prosa.services.came.sbg.ac.at/prosa.php), along with the SAVES v6.0 server (https://saves.mbi ucla.edu) [22]. Through the assessment of the Z-score, the ProSA tool undertakes an analysis to verify and validate both the global and local quality aspects of the model that has been predicted, thereby generating an overarching quality assessment metric for the specific input configuration under consideration. In instances where the Z-score deviates beyond the predefined threshold established for the native protein structure, there exists a heightened likelihood of encountering inaccuracies within the predicted structural arrangement. In addition, the analysis of Ramachandran plot and the ERRAT was carried out via SAVE v6.0 server, as well as facilitate the assessment and analysis of a diverse array of stereochemical parameters that are inherent to the protein structure.
2.9. Engineering of disulfide bond
Enhancing the stability and facilitating the analysis of protein interactions and dynamics are advantageous attributes of utilizing disulfide bond engineering. Disulfide by Design 2 (DbD2) (http://cptweb.cpt.wayne.edu/DbD2/index.php) quickly assesses residue pairs based on their geometry and proximity for potential disulfide bond formation, assuming they are mutated to cysteines within a given protein model. Disulfide engineering has been used in vaccine development to increase the protein vaccine candidates' thermal stability.
2.10. Prediction of B-cell epitopes
The ElliPro server (http://tools.iedb.org/ellipro) was used to predict conformational and linear B-cell epitopes after the 3D structure of the vaccine was predicted, refined, and validated. Based on the protein's structural characteristics, such as solvent accessibility and flexibility, this approach is excellent at locating B-cell conformational epitopes, which are usually longer than other kinds of epitopes [23]. The minimum score and maximum distance (in Angstrom) were set at 0.8 and 6, respectively, as the two main prediction parameters to guarantee the precision and effectiveness of the predicted epitopes in the context of vaccine development.
2.11. Molecular docking
The investigation involved in the molecular docking of the vaccine candidates with Toll-like receptor 3 (TLR3) was carried out utilizing the innovative ClusPro 2.0 server (https://cluspro.bu.edu/login.php). Since its inception in 2004, ClusPro has consistently stood out as one of the leading docking servers in the field, as evidenced by its performance in the Critical Assessment of Predicted Interactions (CAPRI) evaluations, showcasing a remarkable track record of accurate predictions for the docking of protein-protein complexes [24]. The utilization of ClusPro has been instrumental in advancing our understanding of molecular interactions and holds significant promise for further research in the domain of protein docking analysis. Additionally, the PDBsum server was utilized to examine the protein-protein interactions between the vaccine and TLR3 [25,26].
2.12. Molecular dynamic simulation
Normal mode analysis (NMA) was conducted to evaluate the conformational stability of the selected docked complex using the iMODS web server (http://imods.chaconlab.org). This tool visually illustrates the system's motion dynamics through arrows representing the mobility in the NMA [27]. The Eigenvalues derived from this analysis serve as indicators of the rigidity levels present within the molecular structures under investigation while deformability plots offer insights into the flexible or non-rigid regions of these structures. Additionally, the iMODS server offers valuable information such as B-factor values, variance calculations, covariance mapping, and the linking matrix associated with the desired structural model. By utilizing these functionalities, researchers can gain a comprehensive understanding of the structural dynamics and conformational changes within their docked complexes, ultimately enhancing their insights into molecular interactions and stability assessments.
2.13. Immune simulation
An in-silico immune simulation was performed using the C-ImmSim server (http://kraken.iac.rm.cnr.it/C-IMMSIM/index.php) to validate the immunogenicity and the immune response triggered by the selected vaccine [28]. This server operates by simulating the functions of the bone marrow, thymus, and lymph nodes in mammals, using the vaccine's FASTA format. The efficacy of the vaccine in triggering immune responses from various immune cells such as cytotoxic T cells, helper T-cells, B-cells, immunoglobulins, and cytokines was assessed within this simulation framework. The simulation process involved the administration of three injections spaced at 4-week intervals to mimic a realistic vaccination schedule. In this simulation, parameters for 1 and 45 time steps were configured, with each step representing an equivalent of 8 h in real-time scenarios. Consequently, the total number of steps in the simulation amounted to 1050, providing a comprehensive evaluation of the vaccine's impact on the immune system. During the simulation, all other parameters were kept at their default settings to maintain consistency and reliability in the results. This thorough and methodical approach enabled a comprehensive analysis of the vaccine's capacity to trigger immune responses, providing valuable insights into its potential effectiveness in practical applications.
2.14. Codon optimization and in silico cloning
The utilization of the Java Codon Adaptation Tool (JCat) (http://www.prodoric.de/JCat) was implemented in the process of codon optimization and reverse translation [29]. Subsequent to the optimization of codons within the E. coli strain K12, a specific vaccine cDNA sequence was formulated, which possesses the capability to facilitate highly efficient expression within this particular strain. The subsequent step in the sequence analysis involved the meticulous selection of various parameters aimed at circumventing the occurrence of rho-independent transcription terminators, prokaryotic ribosome binding sites, as well as restriction enzyme cleavage sites. Furthermore, the deliberate addition of restriction enzyme sites XhoI and BamHI was carried out at both the N-terminal and the C-terminal ends of the optimized codon sequence, with the intention of enabling seamless in-silico cloning within the pET-28a (+) vector through the utilization of the sophisticated SnapGene 6.2 software.
3. Result
3.1. Prediction of CTL and HTL epitopes
Cytotoxic T-lymphocyte (CTL) epitopes were chosen for their high scores, reflecting strong specificity and low sensitivity towards the adaptive immune recipient, as outlined in Table 1. The process involved identifying MHC-I peptides that have a high binding affinity to specific mice alleles, including those considered as reference alleles, which was carried out using the IEDB server. Furthermore, the IL4 epitope online server utilized a combination of Motif and SVM (Hybrid method) approach to predict IL4 epitopes. Our research team specifically configured this server to accurately predict IL4 triggering epitopes that could be vital in understanding the immune response mechanisms. This methodological approach guarantees a thorough analysis of epitopes capable of triggering specific immune responses, thus aiding the progress of vaccine development and immunotherapy strategies. The integration of computational tools and predictive models enhances the precision and efficiency of epitope selection, ultimately leading to a more targeted and effective immune response modulation in various health conditions. The IEDB MHC-II prediction platform has been chosen specifically for the purpose of identifying mice MHC-II alleles in Yersinia pestis antigens. The process of predicting Helper T-lymphocyte (HTL) epitopes was conducted by adhering to the percentile rank and ultimately selecting those with high scores. A total of 16 epitopes were carefully handpicked from the pool of predicted HTL epitopes. It is essential to note that a higher percentile rank signifies the potential for better binding interactions, as outlined in Table 2. This selection process underscores the importance of accuracy and efficiency in identifying epitopes that hold significant promise for further research and exploration. The utilization of advanced prediction platforms such as IEDB plays a crucial role in streamlining the identification of potential targets for immunological studies and vaccine development, ultimately contributing to advancements in the field of biomedicine.
Table 1.
Predicted final CTL epitopes and analysis of IL4 inducing.
| Antigen | CTL Epitope | IL4 inducer | Score | MHC I | Allele | Position |
|---|---|---|---|---|---|---|
| LcrV | ITDDIELLKK NKHLSSSGT LAELTAELK MNHHGDARS |
+ + + + |
1.00 1.00 0.99 0.99 |
NRVITDDIELLKKI QAEINKHLSSSGTI LAELTAELKIYSVI VDSMNHHGDARSKL |
H-2-Kk H-2-Db H-2-Kk H-2-Kk |
61–74 171–184 157–170 140–153 |
| F1 | FTSQDGNN ISSVIAIAL TASTTATAT ATGSQDFFV |
+ – + + |
0.95 0.99 0.97 0.95 |
YLTFTSQDGNNHQF KISSVIAIALFGTI AADLTASTTATATL VLATGSQDFFVRSI |
H-2-Db H-2-Db H-2-Kk H-2-Kk |
92–105 3–16 21–34 134–147 |
| PH6 | YMMSWVGID IPTGDSKGG INDDSPKDI QPGDVTMFT |
+ + – + |
1.00 0.99 0.99 0.98 |
YMMSWVGIDNNWYI RLIPTGDSKGGYMI YINDDSPKDIKDHL FAGKQPGDVTMFTL |
H-2-Db H-2-Ld H-2-Ld H-2-Ld |
109–122 84–97 121–134 59–72 |
| OmpA | YPVAQDLDV FGQEDAAAP EGQANPVTG GNTCDNVKP |
– – + + |
1.00 1.00 0.99 0.99 |
SYPVAQDLDVYTRL VSYRFGQEDAAAPI RGEGQANPVTGNTC TGNTCDNVKPRAAL |
H-2-Kd H-2-Kb H-2-Dd H-2-Ld |
106–119 193–206 305–318 314–327 |
Table 2.
Predicted final MHC-II and HTL epitopes.
| Antigen | MHC II | Position | Allele | Percentile rank |
|---|---|---|---|---|
| LcrV | ILEKMPQTTIQVDGSEKK KIYSVIQAEINK KRVKEFLESSPNTQWE TSRFNSAIEALNR |
215–232 165–176 99–114 295–307 |
H2-IAd H2-IAd H2-IAb H2-IAb |
0.6 1.1 0.3 2.8 |
| F1 | TTATATLVEPAR GGYKTGTTSTSV GKYTDAVTVTVS TLTYKEGAPITIMDNGN |
28–39 70–81 157–168 41–57 |
H2-IAd H2-IAb H2-IAb H2-IAb |
0.31 0.36 0.37 0.3 |
| PH6 | EIFAGKQPGDVT GMANASTVINSKD SKDVSGEVTVKQGNT IKDHLYVKAGTVLKP |
57–68 22–34 32–46 130–144 |
H2-IAb H2-IAd H2-IAd H2-IAd |
2.5 3.4 0.6 0.2 |
| OmpA | QEDAAAPIVAPTPAPAP ADRIGQPAPNLA DTGVSPLVALGAEYA VKGYKEVVTQPQA |
199–215 268–279 143–157 341–353 |
H2-IAb H2-IAb H2-IAd H2-IAb |
0.01 0.07 1.1 0.4 |
3.2. The assessment of experimental data
We extracted 370 MHC epitopes, which were filtered based on their antigenicity. The final evaluation identified 12 epitopes as the best candidates, determined through immunoinformatics approaches. The data supporting this analysis can be found in the supplementary material.
3.3. Construction of multi-epitope vaccine
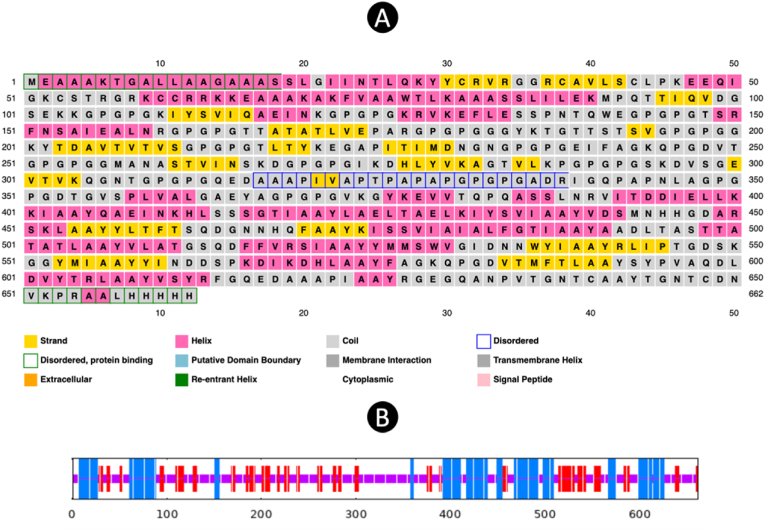
The vaccine construct is composed of a total of 16 HTL and 16 CTL epitopes, which have been strategically separated by linkers to avoid any potential conformational clash that may hinder the efficacy of the vaccine. Furthermore, the inclusion of the TAT peptide serves the purpose of facilitating improved vaccine delivery, while the incorporation of the PADRE sequence aims to enhance the antigenicity of the vaccine. This particular characteristic of PADRE highlights its capacity to function as a TLR agonist, elucidating its role as an adjuvant in numerous immunotherapeutic vaccines. The examination of the resultant vaccine product was conducted to identify a highly efficient candidate that could be further utilized in the development of future vaccines. The detailed representation of the construct can be observed in Fig. 2, providing a visual aid for better understanding and analysis of the vaccine design and components. This visual representation aids in comprehending the spatial arrangement of epitopes, linkers, and additional sequences within the vaccine construct, thereby contributing to the overall assessment of its potential effectiveness in inducing a robust immune response.
Fig. 2.
The diagram depicting the assembly of a multi-epitope vaccine comprising various linkers, TAT, adjuvant, PADRE, HTL epitopes, and CTL epitopes.
3.4. Evaluation of vaccine
Antigenicity, allergenicity, toxicity, and solubility were all considered in the evaluation of the vaccine's final sequence, and it successfully satisfied each requirement. When the VaxiJen v2.0 server was used to assess the vaccine's antigenicity, the results showed that the final vaccine design had a high level of antigenicity, with a score of 0.7114 and 0.7846, respectively, according to ANTIGENpro. Additionally, analyses conducted with the AllergenFP 1.0 and AllerTOP 2.0 servers determined that the final vaccine is non-allergenic and does not provoke allergic reactions upon administration. The SVM prediction mode in the ToxinPred server was utilized to analyze the complete final vaccine sequence, including the adjuvant sequence, all epitopes, linkers, fynomer sequence, and H5E tag, indicating that there were no toxic components present. The solubility of the vaccine was predicted using SolPro (0.9049) and the Protein-sol server (0.549), showing favorable solubility characteristics as detailed in Table 3. Solubility is an essential factor in vaccine development, influencing various aspects.
Table 3.
The comprehensive properties of multi-epitope vaccine.
| Property | Measurement |
|---|---|
| SOLpro | 0.9049 (Soluble) |
| Number of amino acids | 662aa |
| Protein-Sol | 0.549 |
| Molecular weight | 68131.85 Da |
| Formula | C3019H4737N831O932S17 |
| Theoretical pI | 8.77 (Basic) |
| Ext. coefficient | 71,670M−1 cm−1 |
| Instability index | 25.69(Stable) |
| Aliphatic index | 72.82 (Thermostable) |
| Half-Life | 30 h (mammalian reticulocytes, in vitro). >20 h (yeast, in vivo). >10 h (Escherichia coli, in vivo). |
| Antigenicity | ANTIGENpro (0.7114), VexiJen v2.0 (0.7846) Antigenic |
| Toxicity | Non-Toxin |
| Allergenicity | Non-Allergen |
| GRAVY | −0.229 (Hydrophilic) |
The analysis of the physicochemical characteristics of the vaccine revealed that the final configuration contains 662 residues and has a molecular weight of 68.13185 kDa. The theoretical isoelectric point (pI) of this structure is calculated to be 9.36, indicating its basic nature. This configuration includes 52 negatively charged residues, consisting of aspartic acid and glutamic acid, along with 60 positively charged residues made up of arginine and lysine. The net charge of the vaccine structure amounts to 8, a value that tends to diminish in an alkaline milieu, with a preference generally leaning towards higher positive charge values. The extinction coefficient for this structure is reported to be 71,670 M−1 cm−1, with absorption values at 280 nm of 1052 (0.1 %) (g/l), which includes all cysteine pairs in aqueous conditions. Moreover, the expected half-life of the vaccine in mammalian reticulocytes (in vitro) is estimated to be about 30 h, around 20 h in yeast (in vivo), and 10 h in E. coli (in vivo), highlighting the vaccine's resilience across various biological environments. Variations in the half-life of a vaccine among different organisms underscore its robustness and efficacy within distinct biological settings. A shorter half-life could necessitate more frequent doses, thereby impacting the dosing regimen and overall practicality. The ideal half-life is contingent upon factors such as the intrinsic nature of the vaccine, the specific target pathogen, and practical considerations surrounding administration. Predictions suggest that the vaccine structure, which has an instability index of 25.69 (below 40), demonstrates a high level of stability suitable for eliciting an immune response. The aliphatic index of the vaccine is found to be 72.82, indicating its thermostable properties; an increase in the aliphatic index enhances the protein's heat resistance. The average hydrophilicity of the vaccine, denoted by the GRAVY score of −0.229, suggests enhanced solubility, thereby facilitating interactions with blood and water, and streamlining the identification of the target. The thorough analysis of the physicochemical properties clearly confirms that the vaccine structure meets the necessary criteria for vaccine formulation.
3.5. Prediction of secondary structure
The prediction of the 2D of the vaccine was conducted through the utilization of PSI-blast-based secondary structure prediction method known as PSIPRED, revealing that the structure comprises 35.20 % helix, 15.86 % strand, and 48.94 % coil. In addition to this, the SOPMA tool was employed to provide further insights into the secondary structure, yielding values of 27.64 % for alpha-helix, 21.15 % for extended strand, and 51.21 % for random coil, all based on default parameters. A visual depiction illustrating the features of the secondary structure can be observed in the accompanying Fig. 3, allowing for a more comprehensive understanding of the composition. The considerable proportion of random coil highlighted in the graphical representation serves as an indication of the existence of epitopes distributed across various regions within the construct. This arrangement of epitopes within various structural elements indicates potential interaction sites with immune system components, thereby increasing the immunogenicity of the vaccine candidate. Consequently, the detailed analysis of the secondary structure aids in elucidating key characteristics that contribute to the overall efficacy and functionality of the vaccine design.
Fig. 3.
(A) and (B) The result of 2D structure that predicted by PSIPRED and SOPMA server.
3.6. Prediction of tertiary structure
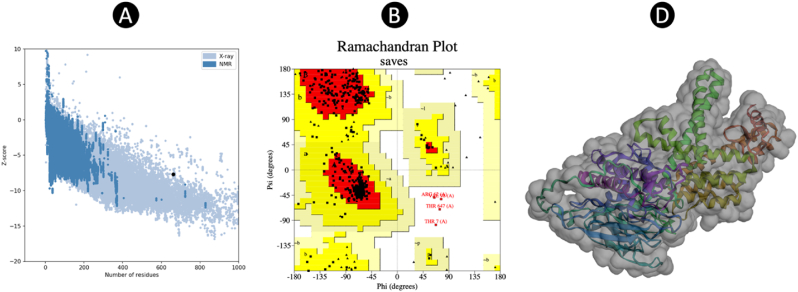
After generating the 3D vaccine model using the Robetta server, one model was identified as the most optimal predictive representation. Based on this model, the final 3D configuration of the vaccine was created. Following the refinement of the 3D structural model obtained from the modeling phase by the GalaxyRefine server, model 1 was selected as the best final vaccine model, evaluated against several criteria, including GDT-HA (0.9823), RMSD (0.311), MolProbity (1.125), clash score (17.2), and poor rotamers (0.8), in comparison to other refined models. The Z-score attributed to the vaccine structure within the PROSA diagram was noted to be −7.71 (Fig. 4), a proximity to the spectrum of native proteins of analogous magnitude, signifying a minimal margin of error and heightened precision of the simulation, thereby reinforcing the overall dependability of the projected model. Furthermore, the quality metric of 88.941 derived from the ERRAT server serves as an indicator of the optimal caliber of the protein model. The categorization of amino acids based on phi and psi angles using the Ramachandran plot through PROCHECK analysis on the SAVE server revealed that 91.8 % of the residues were located in the most favorable region, with an additional 7.5 % in the additional allowed category, and 0.0 % in the generously allowed category, while 0.8 % were found in the disallowed region. The outcomes ascertain that the total residuals residing in the desired sector aligned within the ideal value range, surpassing the 90 % threshold, thereby validating the trustworthiness of this specific model.
Fig. 4.
3D structure validation. (A) and (B) structure validation by ProSA and PROCHECK. (C) The final 3D structure after refinement.
3.7. Disulfide bond prediction
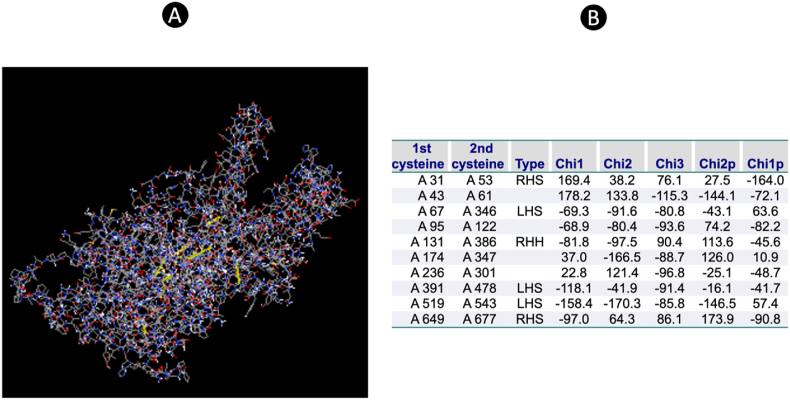
Regions in need of stabilization within the vaccine structure can greatly benefit from the implementation of disulfide engineering techniques. Through our analysis, we were able to pinpoint specific residues that are suitable for mutation into cysteine, which is essential for the formation of disulfide bonds. The examination of the energy value utilized for the screening of residues revealed that it fell below the critical threshold of 2.2, indicating the feasibility of the mutation process. Additionally, the chi3 value exhibited a wide range, spanning from −80.8 to +97, further supporting the potential for successful engineering. Our comprehensive analyses not only identified individual residues suitable for mutation but also highlighted specific residue pairs that could be targeted for the creation of disulfide bonds (Fig. 5). A total of 10 positions were identified as candidates for mutation to facilitate disulfide bond formation, and these findings have been visually represented in a detailed figure for reference and further analysis.
Fig. 5.
(A) The disulfide bond calculation that mutated in the 3D structure (Yellow line). (B) The position of disulfide bond in the 3D structure. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.8. Prediction of B-cell epitopes
Moreover, upon analysis using the ElliPro algorithm, it was discovered that the vaccine's structural composition unveiled a total of eight conformational B-cell epitopes, each possessing scores that varied between 0.566 and 0.811 as depicted in the accompanying Table 4. This particular observation serves to underscore the remarkable immunogenic potential exhibited by the vaccine, thereby enhancing its overall efficacy in eliciting an immune response. Furthermore, these conformational B-cell epitopes further complement the previously identified linear B-cell epitopes, which amounted to a total of 14 predicted epitopes, ultimately contributing to a more comprehensive understanding of the vaccine's antigenic profile.
Table 4.
The Linear and conformational B-cell epitopes were predicted by Ellipro.
| Start | Linear Epitopes | Number of residues | Score |
|---|---|---|---|
| 24–65 | NTLQKYYCRVRGGRCAVLSCLPKEEQIGKCSTRGRKCCRRKK | 42 | 0.81 |
| 594–662 | YPVAQDLDVYTRLAAYVSYRFGQEDAAAPIAAYRGEGQANPVTGNTCAAYTGNTCDNVKPRAALHHHHH | 69 | 0.804 |
| 297–346 | VSGEVTVKQGNTGPGPGQEDAAAPIVAPTPAPAPGPGPGADRIGQPAPNL | 50 | 0.779 |
| 403–435 | AAYQAEINKHLSSSGTIAAYLAELTAELKIYSV | 33 | 0.778 |
| 265–285 | NSKDGPGPGIKDHLYVKAGTV | 21 | 0.693 |
| 192–205 | TSVGPGPGGKYTDA | 14 | 0.672 |
| 230–257 | DNGNGPGPGEIFAGKQPGDVTGPGPGGM | 28 | 0.658 |
| 176–186 | ARGPGPGGGYK | 11 | 0.642 |
| 86–105 | LILEKMPQTTIQVDGSEKKG | 20 | 0.62 |
| 530–538 | VGIDNNWYI | 9 | 0.614 |
| 559–579 | YINDDSPKDIKDHLAAYFAGK | 21 | 0.599 |
| 1–8 | MEAAAKTG | 8 | 0.59 |
| 459–464 | TFTSQD | 6 | 0.569 |
| 162–166 | PGPGT | 5 | 0.543 |
| No. | Conformational epitopes | Number of residues | Score |
|---|---|---|---|
| 1 | A:A403, A:A404, A:Y405, A:Q406, A:A407, A:E408, A:I409, A:N410, A:K411, A:H412, A:L413, A:S414, A:S415, A:S416, A:G417, A:T418, A:I419, A:A420, A:A421, A:Y422, A:L423, A:A424, A:E425, A:L426, A:T427, A:A428, A:E429, A:L430, A:K431, A:I432 | 30 | 0.811 |
| 2 | A:Y594, A:P595, A:V596, A:A597, A:Q598, A:D599, A:L600, A:D601, A:V602, A:Y603, A:T604, A:R605, A:L606, A:A607, A:A608, A:Y609, A:V610, A:S611, A:Y612, A:R613, A:F614, A:G615, A:Q616, A:E617, A:D618, A:A619, A:A620, A:A621, A:P622, A:I623, A:A624, A:A625, A:Y626, A:R627, A:G628, A:E629, A:G630, A:Q631, A:A632, A:N633, A:P634, A:V635, A:T636, A:G637, A:N638, A:T639, A:C640, A:A641, A:A642, A:Y643, A:T644, A:G645, A:N646, A:T647, A:C648, A:D649, A:N650, A:V651, A:K652, A:P653, A:R654, A:A655, A:A656, A:L657, A:H658, A:H659, A:H660, A:H661, A:H662 | 69 | 0.804 |
| 3 | A:N24, A:T25, A:L26, A:Q27, A:K28, A:Y29, A:Y30, A:C31, A:R32, A:V33, A:R34, A:G35, A:G36, A:R37, A:C38, A:A39, A:V40, A:L41, A:S42, A:C43, A:L44, A:P45, A:K46, A:E47, A:E48, A:Q49, A:I50, A:G51, A:K52, A:C53, A:S54, A:T55, A:R56, A:G57, A:R58, A:K59, A:C60, A:C61, A:R62, A:R63, A:K64, A:K65, A:K121, A:A176, A:R177, A:G178, A:P179, A:G180, A:P181, A:G182, A:G183, A:G184, A:Y185, A:T192, A:S193, A:V194, A:G195, A:P196, A:G197, A:P198, A:G199, A:G200, A:K201, A:Y202, A:T203, A:D204, A:A205, A:E222, A:G223, A:A224, A:P225, A:I226, A:T227, A:I228, A:D230, A:N231, A:G232, A:N233, A:G234, A:P235, A:G236, A:P237, A:G238, A:E239, A:I240, A:F241, A:A242, A:G243, A:K244, A:Q245, A:P246, A:G247, A:D248, A:V249, A:T250, A:G251, A:P252, A:G253, A:P254, A:G255, A:G256, A:M257, A:A258, A:N265, A:S266, A:K267, A:D268, A:G269, A:P270, A:G271, A:P272, A:G273, A:I274, A:K275, A:D276, A:H277, A:L278, A:Y279, A:V280, A:K281, A:A282, A:G283, A:T284, A:V285, A:L286, A:P288, A:K295, A:V297, A:S298, A:G299, A:E300, A:V301, A:T302, A:V303, A:K304, A:Q305, A:G306, A:N307, A:T308, A:G309, A:P310, A:G311, A:P312, A:G313, A:Q314, A:E315, A:D316, A:A317, A:A318, A:A319, A:P320, A:I321, A:V322, A:A323, A:P324, A:T325, A:P326, A:A327, A:P328, A:A329, A:P330, A:G331, A:P332, A:G333, A:P334, A:G335, A:A336, A:D337, A:R338, A:I339, A:G340, A:Q341, A:P342, A:A343, A:P344, A:N345, A:L346, A:A347, A:G348, A:P349, A:G350, A:P351, A:G352, A:D353, A:P369 | 185 | 0.705 |
| 4 | A:Y490, A:A491, A:A492, A:D493, A:L494, A:T495, A:A496, A:S497, A:T498, A:T499, A:V530, A:G531, A:I532, A:D533, A:N534, A:N535, A:Y537, A:I538, A:Y559, A:I560, A:N561, A:D562, A:D563, A:S564, A:P565, A:K566, A:D567, A:I568, A:D570 | 29 | 0.634 |
| 5 | A:Q93, A:T94, A:T95, A:I96, A:Q97, A:V98, A:D99, A:G100, A:S101, A:E102, A:K103, A:K104, A:G105 | 13 | 0.608 |
| 6 | A:M1, A:E2, A:A3, A:A4, A:A5, A:K6, A:T7, A:G8, A:A9, A:L86, A:I87, A:L88, A:E89, A:K90, A:M91, A:P92, A:P162, A:G163, A:P164, A:G165, A:T166 | 21 | 0.586 |
| 7 | A:F460, A:T461, A:S462, A:Q463, A:D464 | 5 | 0.569 |
| 8 | A:H571, A:A573, A:A574, A:Y575, A:A577, A:G578, A:K579 | 7 | 0.566 |
3.9. Molecular docking
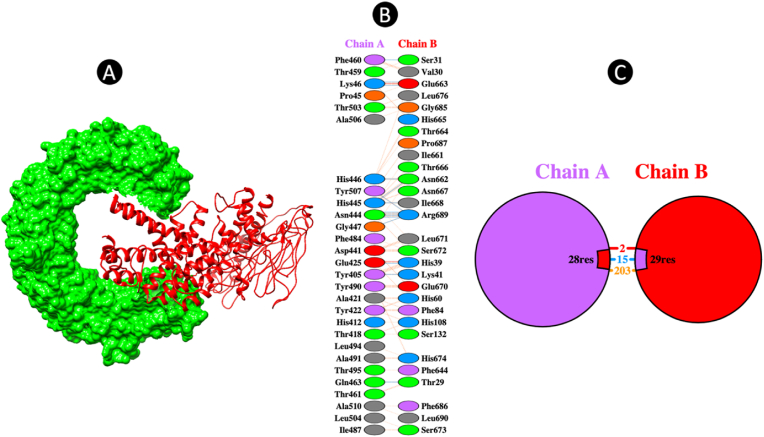
The ClusPro online server for protein-protein docking was used to carry out the molecular docking process between the refined vaccine model and the immune receptor TLR3, identified by the PDB ID: 2A0Z. The results generated by the server exhibited a total of 30 clusters for each of the docked complexes, with a ranking from 0 to 29 based on the composition of cluster members. Additionally, the outcomes included the weighted scores corresponding to the energies of the clusters. Among these clusters, Cluster 0 of the TLR-3-vaccine docked complex stood out as it comprised the highest number of members, specifically 29, and displayed the lowest energy value of −952.5. Due to these distinctive characteristics, Cluster 0 was singled out for further in-depth analysis and exploration to gain a comprehensive understanding of the interactions within the TLR-3-vaccine complex. This detailed analysis of Cluster 0 will yield important insights into the binding mechanisms and overall stability of the docked complex, significantly advancing research in protein-protein interactions and vaccine design. In the subsequent phase of the study, we employed the PDBsum tool to investigate the protein-protein interactions and identify the types of interactions present. The vaccine-TLR3 complex has 2 ion-bridges and 15 hydrogen bonds that confirmed the potential interaction (Fig. 6).
Fig. 6.
(A) The final docked complex between TLR3 and multi-epitope vaccine (B) and (C) The residue-residue interaction between TLR3 and vaccine (Chain A: TLR3, Chain B: Vaccine).
3.10. Molecular dynamic simulation
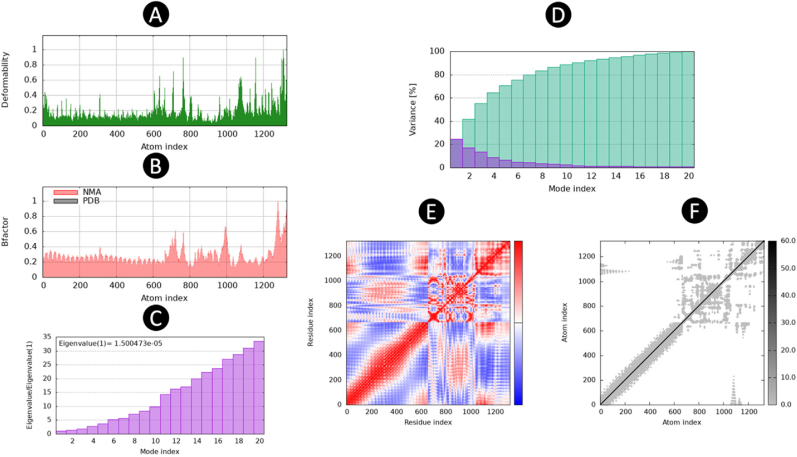
Furthermore, the stability analysis of the Normal Mode Analysis (NMA) for the intricate simulation led to the generation of a deformation graph that effectively highlights the peaks observed in Fig. 7A. The computed eigenvalue for the intricate system was determined to be 1.500473e − 05, which is visually represented in Fig. 7B. By examining the variance and B-factor graph in Fig. 7C, one can observe the cumulative variance (depicted in green) and individual variance (depicted in red), providing insight into the relationship within the docked complex. The covariance map, as illustrated in Fig. 7D, showcases the dynamic motion between pairs of residues within the complex. Here, the color scheme is utilized to signify the nature of motion; red corresponds to correlated motion, white indicates uncorrelated motion, and blue represents anti-correlated motion. Furthermore, the elastic map of the complex, presented in Fig. 7E, offers a visualization of the interatomic relationships. The darker gray regions in this map serve as indicators of stiffer regions within the complex, thus shedding light on its structural properties.
Fig. 7.
The results of molecular dynamics simulation of vaccine construct and TLR3 docked complex. (A) Deformability, (B) B-Factor, (C) eigenvalues, (D) variance, (E) co-variance map (correlated (red), uncorrelated (white), or anti-correlated (blue) motions), and (F) Atom index. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.11. Immune simulation
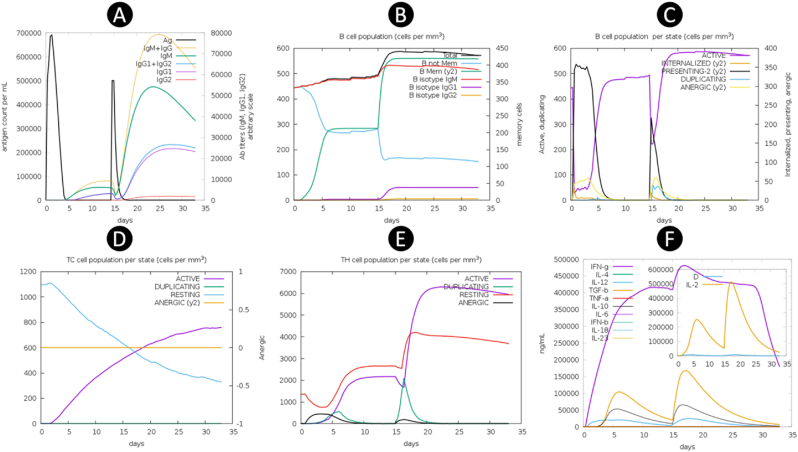
The immune simulation was conducted over a period of one month, allowing for a comprehensive analysis of immune responses. It was observed that following the administration of an additional booster dose of the vaccine, the immune responses exhibited a notable increase in strength as compared to the initial prime immunization. This enhancement in immune response was accompanied by a significant decrease in the antigen levels, while there was a substantial elevation in the levels of antibodies, specifically Immunoglobulin G (IgG) and Immunoglobulin M (IgM), being produced. Throughout the course of the simulation, fluctuations in the levels of immunoglobulins and antigens were observed, indicating a dynamic interplay within the immune system. Notably, the abundance of antigens reached its peak at each point of vaccine injection, as illustrated in Fig. 8A. The humoral response, characterized by the production of antibodies, was consistently triggered following each vaccine administration, with antibody levels remaining elevated in the weeks subsequent to the final injection. The immunization process effectively stimulated the activity of B cells, as depicted in Fig. 8B and C. Furthermore, the activation of CTLs was clearly evident, as shown in Fig. 8D. Analysis of HTLs counts revealed that the replication phase commenced immediately after each injection, as depicted in Fig. 8E. In the days following each injection, upon successful interaction with antigen-presenting cells, HTLs initiated the processes of replication and differentiation into memory cells. These HTLs were also essential in stimulating cytokine secretion and boosting the humoral response. Remarkably, each injection led to a significant increase in the levels of various cytokines, including Interferon-gamma (IFN-γ), Transforming Growth Factor Beta (TGF-β), Interleukin-10 (IL-10), and Interleukin-12 (IL-12), as illustrated in Fig. 8F.
Fig. 8.
Immune simulation results by C-ImmSim. (A) Antigen and various subtypes of immunoglobulin levels are visually depicted through distinct peaks of varying colors on the graph. The elevation in IgG production serves as a reflection of the intricate process involving the activation and expansion of primary, secondary, and tertiary immune responses following the introduction of the vaccine. This phenomenon showcases the dynamic nature of the immune system's ability to mount a multifaceted defense mechanism against specific antigens, highlighting the complexity and robustness of the human immune response. (B,C) B lymphocytes by total count and population per entity state (active, presenting, internalized, duplicating, or anergic). (C) Cytotoxic T cell response. (E) Helper T cell population. (F) The levels of cytokines and interleukins exhibit significant variations as they progress through distinct developmental phases, demonstrating dynamic fluctuations in their concentrations and biological activities. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.12. In silico cloning
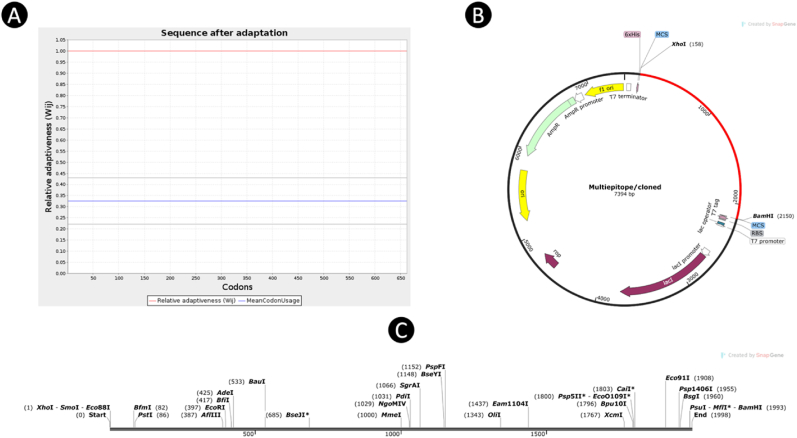
The codon adaptation of the designed vaccine construct was carried out using the JCat server, taking into account the codon usage patterns specific to the E. coli K12 strain. The target sequence had a total length of 1998 nucleotides, offering a substantial amount of genetic material for the vaccine construct. Through analysis, CAI of the refined sequence was determined to be 1, signifying a strong potential for high expression levels within the bacterial system under consideration. Additionally, the GC content of the sequence was estimated to be 54.63 %, well within the optimal range of 30–70 %, suggesting a good probability of successful expression in the E. coli K12 strain. Following the adaptation process, the vaccine sequence was seamlessly integrated into the pET28a (+) expression vector, precisely positioned between the XhoI and BamHI restriction enzymes (Fig. 9), facilitated by the utilization of the sophisticated SnapGene tool for enhanced accuracy and efficiency in the genetic engineering procedure.
Fig. 9.
(A) Codon adaptation; Codon utilization in the optimized gene shown. (B) Computational cloning of vaccine via engineering a recombinant plasmid. in order to clone the vaccine in pET21b (+) vector, XhoI and BamHI restriction sites were attached at the N-terminal and C-terminal of the improved codon. (C) The position of restriction of enzyme in vaccine sequences.
4. Discussion
Antibiotic controls have been traditionally utilized for managing infectious diseases; however, their efficacy has been diminishing, prompting the scientific community to explore alternative approaches such as the development of vaccines as potential replacements for antibiotics [30]. Initially, vaccinations were employed for controlling infectious diseases and played a crucial role in this regard. Nevertheless, the escalating reliance on vaccines in recent times has spurred scientists to investigate more efficient, inexpensive, and safer methods for crafting vaccines [31]. The conventional vaccine technology is associated with the risk of virulence resurgence and is not particularly cost-effective, leading to the emergence of a novel approach known as immune-informatics. This innovative technology necessitates a profound understanding of genomics and proteomics to facilitate the introduction of new vaccines into the marketplace. Immuninformatics focuses solely on a specific portion of the pathogen, known as the epitope, which serves to enhance the immune response [32]. The utilization of computational software, online servers, and databases characterizes immune informatics technology, which delivers high-quality outcomes at a reduced cost while ensuring enhanced safety and effectiveness. Numerous vaccine variants have been created based on epitope prediction strategies to combat various human pathogens such as SARS-CoV-2, Mers-CoV, Monkeypox virus, and Marburg virus [11,[33], [34], [35]]. The progress of a multi-epitope vaccine against Yersinia pestis, the causative agent of plague, represents a significant step forward in the field of vaccine design and immunotherapy. In pervious study, the design of multi-epitope vaccine against Yersinia pestis was implemented that created the vaccine based on 5 protein such as rstB, YPO2385, hmuR, flaA1, and psaB via different analysis [36]. Our study integrates advanced computational methods for epitope prediction of 4 proteins (F1 capsule, LcrV, OmpA, and PH6) that previous study showed that is potent target for designing vaccine [[37], [38], [39]] and vaccine construction, followed by extensive in silico evaluations to ensure a robust and effective vaccine candidate. The prediction and selection of CTL and HTL epitopes were conducted using sophisticated computational tools, ensuring high specificity and sensitivity. The use of the IEDB server for MHC-I and MHC-II binding affinity predictions, coupled with the IL4 epitope online server, allowed for the identification of epitopes with strong potential to trigger specific immune responses. By selecting epitopes with high binding affinities to reference mouse alleles, our approach provides a solid foundation for subsequent in vivo validation, highlighting the translational potential of our findings. The multi-epitope vaccine construction was carefully designed, incorporating 16 HTL and 16 CTL epitopes. These epitopes were strategically separated by linkers to avoid conformational clashes that could hinder vaccine efficacy. Additionally, the inclusion of the TAT peptide aimed to enhance vaccine delivery, while the PADRE sequence was included to boost antigenicity, acting as a TLR agonist and functioning as an adjuvant. The detailed structural representation of the vaccine construct, provides a clear understanding of the spatial arrangement and functional components, aiding in the overall assessment of the vaccine design. Comprehensive evaluation of the vaccine's properties was conducted to ensure its safety and effectiveness. Antigenicity assessments using VaxiJen v2.0 and ANTIGENpro servers indicated high antigenic potential, with scores of 0.7114 and 0.7846, respectively. Allergenicity and toxicity analyses using AllergenFP 1.0, AllerTOP 2.0, and ToxinPred servers confirmed that the vaccine is non-allergenic and non-toxic. Solubility predictions using SolPro and Protein-sol servers demonstrated favorable solubility characteristics, essential for efficient vaccine delivery and effectiveness. The physicochemical properties of the vaccine were meticulously analyzed, revealing a final configuration of 662 residues with a molecular weight of 68.13185 kDa and a theoretical isoelectric point (pI) of 9.36. The vaccine structure exhibited a high level of stability, as indicated by an instability index of 25.69, and favorable solubility, as suggested by a GRAVY score of −0.229. These properties are crucial for ensuring the vaccine's stability and solubility, facilitating effective interaction with the immune system. Secondary and tertiary structure predictions further validated the robustness of the vaccine design. The secondary structure, as predicted by PSIPRED and SOPMA tools, highlighted a significant proportion of random coils, suggesting potential sites for interactions with immune components. The tertiary structure, refined by the GalaxyRefine server, exhibited high accuracy and stability, with 91.8 % of residues positioned within the most favorable regions of the Ramachandran plot, as indicated by PROCHECK analysis. Disulfide bond engineering was identified as a critical step for enhancing the stability of the vaccine structure. Specific residues suitable for mutation to cysteine were pinpointed, facilitating the formation of disulfide bonds, which are crucial for structural stabilization. The prediction of B-cell epitopes using the ElliPro algorithm revealed a total of eight conformational B-cell epitopes, underscoring the vaccine's immunogenic potential. These conformational epitopes complement the previously identified linear B-cell epitopes, contributing to a comprehensive understanding of the vaccine's antigenic profile.Molecular docking studies using the ClusPro server demonstrated strong interactions between the vaccine and the TLR3 immune receptor, with Cluster 0 exhibiting the highest number of members and the lowest energy value. This interaction was further confirmed by PDBsum analysis, which identified ion-bridges and hydrogen bonds, indicative of a stable and effective vaccine-receptor complex. Molecular dynamics simulations offered valuable insights into the stability and dynamic behavior of the vaccine-receptor complex. The NMA deformation graph, eigenvalues, variance, B-factors, covariance map, and elastic map together underscored the structural integrity and potential flexibility of the complex, which are essential for effective immune activation. The immune simulation results demonstrated a strong immune response after vaccine administration, marked by significant increases in IgG and IgM antibody levels, along with the activation of B cells, CTLs, and HTLs. The observed cytokine profiles, which included elevated levels of IFN-γ, TGF-β, IL-10, and IL-12, further confirmed the vaccine's capacity to elicit a comprehensive immune response. In silico cloning of the vaccine construct using the JCat server and subsequent integration into the pET28a (+) expression vector confirmed the potential for high expression levels in the E. coli strain K12, supported by a CAI of 1 and a GC-content of 54.63 %. The multi-epitope vaccine against Yersinia pestis designed in this study demonstrates significant potential for inducing a robust and specific immune response. The integration of advanced computational tools for epitope prediction, vaccine construction, and in silico evaluations provides a comprehensive framework for vaccine development. Future experimental validation and in vivo studies are warranted to confirm these findings and pave the way for the development of an effective plague vaccine.
5. Conclusion
In essence, it is imperative to advance the creation of a next-generation, powerful, and effective vaccine candidate targeted at combating Yersinia pestis. The current approach in immunoinformatics allows for the development of a potential vaccine that can enhance the efficiency of the vaccine production process. The findings indicate that the multi-epitope vaccine stands out as a promising new vaccine option with the potential to trigger robust immune responses. Moreover, the utilization of a unified multi-epitope vaccine strategy in this study could have a significant impact on the enhancement of an effective vaccine against Yersinia pestis, thereby mitigating the risks of drug resistance and the emergence of new variants. Such advancements are crucial in the ongoing battle against infectious diseases.
Author contributions
M.J.S, H.H.A, R.A.K, L.B, L.V, G.V.S.P, M.C, W.M. T, M.A, M.J.J, and A.K.H conceptualized the study and contributed to drafting the manuscript. M.J.S and H.H.A performed the statistical analyses and prepared figures. H.H.Aand M.J.S finalized the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
Authors are grateful to the Researchers Supporting Project (ANUI2024M111), Alnoor University, Mosul, Iraq.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2024.101871.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
The data that has been used is confidential.
References
- 1.Stenseth N.C., Atshabar B.B., Begon M., Belmain S.R., Bertherat E., Carniel E., Gage K.L., Leirs H., Rahalison L. Plague: past, present, and future. PLoS Med. 2008;5:e3. doi: 10.1371/journal.pmed.0050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zietz B.P., Dunkelberg H. The history of the plague and the research on the causative agent Yersinia pestis. Int. J. Hyg Environ. Health. 2004;207:165–178. doi: 10.1078/1438-4639-00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perry R.D., Fetherston J.D. Yersinia pestis--etiologic agent of plague. Clin. Microbiol. Rev. 1997;10:35–66. doi: 10.1128/CMR.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H., Cui Y., Wang Z., Wang X., Guo Z., Yan Y., Li C., Cui B., Xiao X., Yang Y., Qi Z., Wang G., Wei B., Yu S., He D., Chen H., Chen G., Song Y., Yang R. A dog-associated primary pneumonic plague in Qinghai Province, China. Clin. Infect. Dis. an Off. Publ. Infect. Dis. Soc. Am. 2011;52:185–190. doi: 10.1093/cid/ciq107. [DOI] [PubMed] [Google Scholar]
- 5.R.C.A. Rocke T.E. vol. 1372. U.S. Geological Survey Circular; 2012. (Plague). [DOI] [Google Scholar]
- 6.Vogler A.J., Chan F., Nottingham R., Andersen G., Drees K., Beckstrom-Sternberg S.M., Wagner D.M., Chanteau S., Keim P. A decade of plague in Mahajanga, Madagascar: insights into the global maritime spread of pandemic plague. mBio. 2013;4 doi: 10.1128/mBio.00623-12. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plague around the world, 2010–2015 Relev. Epidemiol. Hebd. 2016;91:89–93. [PubMed] [Google Scholar]
- 8.McNally A., Thomson N.R., Reuter S., Wren B.W. “Add, stir and reduce”: Yersinia spp. as model bacteria for pathogen evolution. Nat. Rev. Microbiol. 2016;14:177–190. doi: 10.1038/nrmicro.2015.29. [DOI] [PubMed] [Google Scholar]
- 9.Galimand M., Guiyoule A., Gerbaud G., Rasoamanana B., Chanteau S., Carniel E., Courvalin P. Multidrug resistance in Yersinia pestis mediated by a transferable plasmid. N. Engl. J. Med. 1997;337:677–680. doi: 10.1056/NEJM199709043371004. [DOI] [PubMed] [Google Scholar]
- 10.Guiyoule A., Gerbaud G., Buchrieser C., Galimand M., Rahalison L., Chanteau S., Courvalin P., Carniel E. Transferable plasmid-mediated resistance to streptomycin in a clinical isolate of Yersinia pestis. Emerg. Infect. Dis. 2001;7:43–48. doi: 10.3201/eid0701.010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aram C., Alijanizadeh P., Saleki K., Karami L. Development of an ancestral DC and TLR4-inducing multi-epitope peptide vaccine against the spike protein of SARS-CoV and SARS-CoV-2 using the advanced immunoinformatics approaches. Biochem. Biophys. Reports. 2024;39 doi: 10.1016/j.bbrep.2024.101745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomar N., De R.K. Immunoinformatics: a brief review. Methods Mol. Biol. 2014;1184:23–55. doi: 10.1007/978-1-4939-1115-8_3. [DOI] [PubMed] [Google Scholar]
- 13.Oli A.N., Obialor W.O., Ositadimma M., Ifeanyichukwu, Odimegwu D.C., Okoyeh J.N., Emechebe G.O., Adejumo S.A., Ibeanu G.C. Immunoinformatics and vaccine development: an overview. ImmunoTargets Ther. 2020;9:13–30. doi: 10.2147/ITT.S241064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamura K., Stecher G., Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doytchinova I.A., Flower D.R. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinf. 2007;8:4. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dimitrov I., Bangov I., Flower D.R., Doytchinova I. AllerTOP v.2--a server for in silico prediction of allergens. J. Mol. Model. 2014;20:2278. doi: 10.1007/s00894-014-2278-5. [DOI] [PubMed] [Google Scholar]
- 17.Hebditch M., Carballo-Amador M.A., Charonis S., Curtis R., Warwicker J. Protein-Sol: a web tool for predicting protein solubility from sequence. Bioinformatics. 2017;33:3098–3100. doi: 10.1093/bioinformatics/btx345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magnan C.N., Randall A., Baldi P. SOLpro: accurate sequence-based prediction of protein solubility. Bioinformatics. 2009;25:2200–2207. doi: 10.1093/bioinformatics/btp386. [DOI] [PubMed] [Google Scholar]
- 19.Gupta S., Kapoor P., Chaudhary K., Gautam A., Kumar R., Raghava G.P.S. Peptide toxicity prediction. Methods Mol. Biol. 2015;1268:143–157. doi: 10.1007/978-1-4939-2285-7_7. [DOI] [PubMed] [Google Scholar]
- 20.Kim D.E., Chivian D., Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ko J., Park H., Heo L., Seok C. GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res. 2012;40:W294–W297. doi: 10.1093/nar/gks493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ponomarenko J., Bui H.-H., Li W., Fusseder N., Bourne P.E., Sette A., Peters B. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinf. 2008;9:514. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozakov D., Hall D.R., Xia B., Porter K.A., Padhorny D., Yueh C., Beglov D., Vajda S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017;12:255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laskowski R.A., Jabłońska J., Pravda L., Vařeková R.S., Thornton J.M. PDBsum: structural summaries of PDB entries. Protein Sci. 2018;27:129–134. doi: 10.1002/pro.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saleki K., Aram C., Alijanizadeh P., Khanmirzaei M.H., Vaziri Z., Ramzankhah M., Azadmehr A. Matrix metalloproteinase/Fas ligand (MMP/FasL) interaction dynamics in COVID-19: an emin silico em study and neuroimmune perspective. Heliyon. 2024;10 doi: 10.1016/j.heliyon.2024.e30898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.López-Blanco J.R., Aliaga J.I., Quintana-Ortí E.S., Chacón P. iMODS: internal coordinates normal mode analysis server. Nucleic Acids Res. 2014;42:W271–W276. doi: 10.1093/nar/gku339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rapin N., Lund O., Castiglione F. Immune system simulation online. Bioinformatics. 2011;27:2013–2014. doi: 10.1093/bioinformatics/btr335. [DOI] [PubMed] [Google Scholar]
- 29.Grote A., Hiller K., Scheer M., Münch R., Nörtemann B., Hempel D.C., Jahn D. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33:W526–W531. doi: 10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lei C., Kumar S. Yersinia pestis antibiotic resistance: a systematic review., Osong Public Heal. Res. Perspect. 2022;13:24–36. doi: 10.24171/j.phrp.2021.0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joon S., Singla R.K., Shen B. Vaccines and immunoinformatics for vaccine design. Adv. Exp. Med. Biol. 2022;1368:95–110. doi: 10.1007/978-981-16-8969-7_5. [DOI] [PubMed] [Google Scholar]
- 32.Oluwagbemi O.O., Oladipo E.K., Kolawole O.M., Oloke J.K., Adelusi T.I., Irewolede B.A., Dairo E.O., Ayeni A.E., Kolapo K.T., Akindiya O.E., Oluwasegun J.A., Oluwadara B.F., Fatumo S. Bioinformatics, computational informatics, and modeling approaches to the design of mRNA COVID-19 vaccine candidates. Computation. 2022;10 doi: 10.3390/computation10070117. [DOI] [Google Scholar]
- 33.Obaidullah A.J., Alanazi M.M., Alsaif N.A., Albassam H., Almehizia A.A., Alqahtani A.M., Mahmud S., Sami S.A., Bin Emran T. Immunoinformatics-guided design of a multi-epitope vaccine based on the structural proteins of severe acute respiratory syndrome coronavirus 2. RSC Adv. 2021;11:18103–18121. doi: 10.1039/D1RA02885E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suleman M., Rashid F., Ali S., Sher H., Luo S., Xie L., Xie Z. Immunoinformatic-based design of immune-boosting multiepitope subunit vaccines against monkeypox virus and validation through molecular dynamics and immune simulation. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1042997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sami S.A., Marma K.K.S., Mahmud S., Khan M.A.N., Albogami S., El-Shehawi A.M., Rakib A., Chakraborty A., Mohiuddin M., Dhama K., Uddin M.M.N., Hossain M.K., Tallei T.E., Bin Emran T. Designing of a multi-epitope vaccine against the structural proteins of Marburg virus exploiting the immunoinformatics approach. ACS Omega. 2021;6:32043–32071. doi: 10.1021/acsomega.1c04817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haq A.U., Khan A., Khan J., Irum S., Waheed Y., Ahmad S., Nizam-Uddin N., Albutti A., Zaman N., Hussain Z., Ali S.S., Waseem M., Kanwal F., Wei D.-Q., Wang Q. Annotation of potential vaccine targets and design of a multi-epitope subunit vaccine against Yersinia pestis through reverse vaccinology and validation through an agent-based modeling approach. Vaccines. 2021;9 doi: 10.3390/vaccines9111327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levy Y., Vagima Y., Tidhar A., Aftalion M., Gur D., Nili U., Chitlaru T., Zauberman A., Mamroud E. Targeting of the Yersinia pestis F1 capsular antigen by innate-like B1b cells mediates a rapid protective response against bubonic plague. Npj Vaccines. 2018;3:52. doi: 10.1038/s41541-018-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fields K.A., Nilles M.L., Cowan C., Straley S.C. Virulence role of V antigen of Yersinia pestis at the bacterial surface. Infect. Immun. 1999;67:5395–5408. doi: 10.1128/IAI.67.10.5395-5408.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Byvalov A.A., V Konyshev I., Uversky V.N., V Dentovskaya S., Anisimov A.P. Yersinia outer membrane vesicles as potential vaccine candidates in protecting against plague. Biomolecules. 2020;10 doi: 10.3390/biom10121694. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that has been used is confidential.