Abstract
Transporting epithelial cells in the gut and kidney rely on protocadherin-based apical adhesion complexes to organize microvilli that extend into luminal space. In these systems, CDHR2 and CDHR5 localize to the distal ends of microvilli, where they form an intermicrovillar adhesion complex (IMAC) that links the tips of these structures, promotes the formation of a well-ordered array of protrusions, and thus maximizes apical membrane surface area. Recently, we discovered that IMACs can also form between microvilli that extend from neighboring cells, across cell-cell junctions. As an additional point of physical contact between cells, transjunctional IMACs are well positioned to impact the integrity of canonical tight and adherens junctions that form more basolaterally. To begin to test this idea, we examined cell culture and mouse models that lacked CDHR2 expression and were unable to form IMACs. CDHR2 knockout perturbed cell and junction morphology, reduced key components from tight and adherens junctions, impaired barrier function, and increased the motility of single cells within established monolayers. These results support the hypothesis that, in addition to organizing apical microvilli, IMACs provide a layer of cell-cell contact that functions in parallel with canonical tight and adherens junctions to promote epithelial functions.
Epithelial microvilli are organized by CDHR2-based intermicrovillar adhesion links. These complexes connect the distal tips of adjacent protrusions that extend from the same cell, or from neighboring cells, across the junctional space.
Using multiple CDHR2 loss-of-function models, the authors establish that CDHR2 knockout disrupts the composition of canonical junctional complexes, which in turn leads to defects in epithelial monolayer physiology.
This study advances our understanding of mechanisms that stabilize cell junctions and further suggests that intermicrovillar adhesion links function in parallel with canonical junctional complexes to promote epithelial monolayer integrity and function.
INTRODUCTION
Cell junctions are essential for the integrity of epithelial tissues, forming physical connections between neighboring cells that provide structural support and control paracellular permeability (Horowitz et al., 2023). By partitioning apical and basolateral components in the plane of the plasma membrane, junctional complexes also play an essential role in the acquisition and maintenance of cell polarity, which in turn defines a functional axis at the cell and tissue scales (Buckley and St Johnston, 2022). Transporting epithelia found in the small intestine and kidney tubules are two examples of tissues that rely heavily on junctional complex formation (Garcia-Castillo et al., 2017). In this context, cell junctions are organized into stacked layers consisting of (from apical to basal) the tight junction (TJ), the adherens junction (AJ), desmosomes, gap junctions, and hemidesmosomes (Garcia et al., 2018). Loss of key components from these distinct layers results in broad ranging defects in cell and tissue architecture and function (Buckley and St Johnston, 2022; Horowitz et al., 2023).
At the level of the TJ, transmembrane proteins including junctional adhesion molecule, occludins, and claudins create adhesive strand-like structures that interact with TJ components on the opposing surface of a neighboring cell (Van Itallie and Anderson, 2014). Inside the cell, zonula occludens (ZO-1,-2,-3) act as scaffolding proteins to couple TJ adhesion molecules to F-actin and non-muscle myosin 2 (NM2), which provide peripheral support and generate mechanical tension (Horowitz et al., 2023). TJs are responsible for selectively controlling solute permeability in different tissue contexts, with specific complements of claudin isoforms creating distinct permeability profiles based on particle size and charge (Tsukita et al., 2019).
Immediately basal to TJs are AJs, which are defined by high levels of E-cadherin, a single-spanning transmembrane protein that drives strong homophilic adhesion between neighboring cells (Troyanovsky, 2023). Knockout (KO) of E-cadherin in mouse intestinal tissues demonstrated clear roles in intestinal morphogenesis and barrier function with blunted villi and increased permeability (Bondow et al., 2012). β-catenin is a core cytoplasmic component of the adherens junction, which binds to E-cadherin and through an interaction with α-catenin, forms a physical link to the underlying actin cytoskeleton (Tian et al., 2011; Valenta et al., 2012). Loss of E-cadherin releases β-catenin from adherens junctions, leading to activation of the Wnt signaling pathway, with downstream loss of the epithelial phenotype and enhanced cell migration, as observed in a broad range of cancers (Yap, 1998; Tian et al., 2011; van der Wal and van Amerongen, 2020).
While the factors alluded to above are well studied in the context of junctional biology, new roles and localizations for these components are still being discovered. For example, polarity proteins including CRB3A, PAR6β, and aPKC were recently localized along the length of apical microvilli, outside of the tight junction (Mangeol et al., 2022). Furthermore, Nectin-3, an adhesion protein classically categorized as part of the AJ, has now been identified in microvilli in human colonic tissue and cultured cells (Childress et al., 2023; Mangeol et al., 2024).
Outside of the large collection of adhesion molecules that accumulate in junctional complexes, other adhesive factors play key roles in shaping epithelial cells. Transporting cell types, like those found in the kidney and gut, leverage intermicrovillar adhesion complexes (IMACs) to drive apical brush border assembly (Crawley et al., 2014; Pinette et al., 2019). Here, protocadherins CDHR2 and CDHR5 target to the distal tips of protrusions, where they form heterophilic contacts that define the spacing between microvilli and maximize the number of protrusions that extend from the surface (Crawley et al., 2014; Pinette et al., 2019; Cencer et al., 2023). Unexpectedly, recent studies also established that CDHR2 and CDHR5 form complexes between the tips of microvilli that extend from neighboring cells, forming “transjunctional” IMACs that are extremely long-lived. These transjunctional complexes provide an anchoring mechanism for nascent microvilli and promote the long-term accumulation of protrusions on the apical surface (Cencer et al., 2023).
By providing an additional mode of physical contact between adjacent epithelial cells, transjunctional IMACs are well positioned to support the integrity of the more basolateral, canonical junctional complexes (e.g., TJs and AJs). To test this hypothesis, we examined multiple CDHR2 loss-of-function models and explored the possibility of junctional phenotypes in these systems. Here, we report that epithelial cell culture and mouse models lacking CDHR2 exhibit prominent defects in cell junctions, including a loss of established junctional complex components. These perturbations are accompanied by increased permeability and a highly motile phenotype. We propose that, under normal conditions, these defects are prevented by CDHR2-dependent formation of transjunctional IMACs. More generally, these data suggest that transjunctional IMACs form a previously unrecognized layer of contact between cells that promotes junctional complex stability.
RESULTS AND DISCUSSION
Loss of CDHR2 disrupts epithelial cell morphology
To characterize the potential impact of CDHR2 loss-of-function on canonical junctional complexes, we revisited our previously described CDHR2 KO LLC-PK1-CL4 (CL4) cell line (Cencer et al., 2023) and also developed a new KO model based on the intestinal epithelial CACO-2BBE cell line (Supplemental Figure S1A). Our previous study revealed that CDHR2 KO CL4 cells exhibit a striking loss of microvillar clustering and reduced accumulation of protrusions at cell margins (Cencer et al., 2023), as expected given the central function of this protocadherin in the IMAC (Crawley et al., 2014; Pinette et al., 2019). Analysis of CDHR2 KO CACO-2BBE cells revealed a similar phenotype, with a clear lack of microvilli clustering (Supplemental Figure S1B). As with CDHR2 KO CL4 cells (Cencer et al., 2023), levels of CDHR5 were also decreased (Supplemental Figure S1, C–E), a phenotype consistent with other IMAC loss-of-function models (Crawley et al., 2016; Weck et al., 2016; Li et al., 2017; Pinette et al., 2019; Choi et al., 2020).
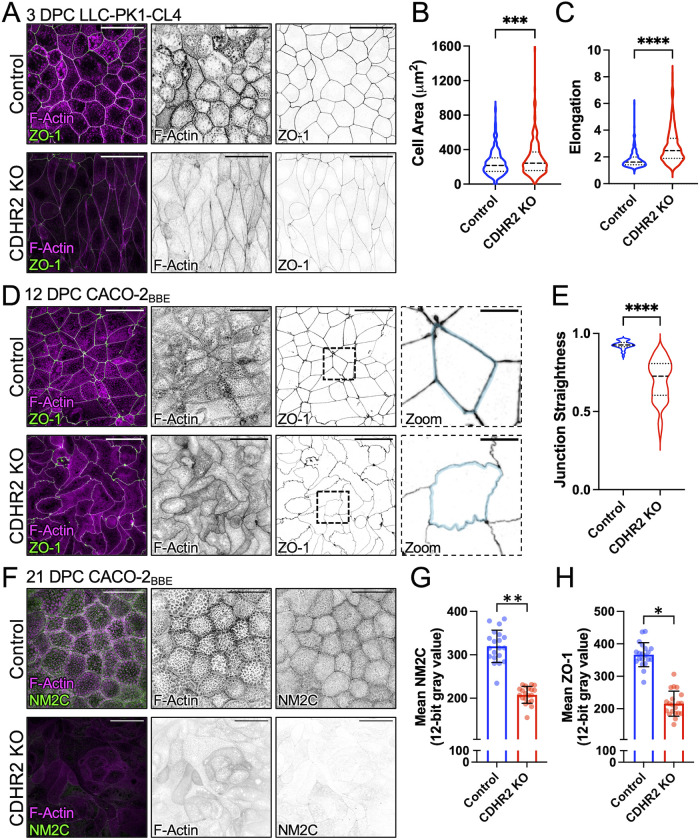
Further characterization of both CDHR2 KO cell lines at intermediate time points in differentiation, 3 days post confluence (DPC) for CL4 and 12 DPC for CACO-2BBE, revealed highly aberrant cell morphologies and junctional profiles. Confocal microscopy of CDHR2 KO CL4 cells stained for tight junction component ZO-1 revealed elongated cell shapes with larger overall areas when viewed en face (Figure 1, A–C). CDHR2 KO CACO-2BBE cells also displayed deformed junctions with a striking “ruffled” appearance (Figure 1D, Zoom). Straightness measurements revealed that junctions in this KO line were less linear, with a mean straightness of 0.70 ± 0.13 compared with Control junctions of 0.93 ± 0.03 (1 being a perfectly straight line; Figure 1E). Previous work established that junctional straightness is linked to the enrichment of NM2 (Van Itallie et al., 2009; Tokuda et al., 2014; Choi et al., 2016) and presumably high levels of tension in the contractile “belt” that encircles the cell at the level of junctional complexes. Interestingly, apical cell area (Figure 1, A and B) is also restricted by junctional tension, as reduced tension causes cells to spread (Diz-Munoz et al., 2013; Sumi et al., 2018). With these points in mind, we sought to determine whether the loss of CDHR2 affected non-muscle myosin-2C (NM2C), the most abundant NM2 variant in mature enterocytes (Ebrahim et al., 2013; Chinowsky et al., 2020). Staining for NM2C in fully differentiated (21 DPC) CDHR2 KO CACO-2BBE cells revealed significantly lower levels of NM2C signal (Figure 1, F and G), suggesting that junctional tension may be reduced in these cells. Together, these results indicate that CDHR2, which localizes primarily to the distal ends of brush border microvilli, has roles in maintaining normal epithelial cell and monolayer morphology.
FIGURE 1:
CDHR2 KO cells exhibit aberrant cell morphologies and decreased apical junction markers. (A) Three DPC Control and CDHR2 KO CL4 cells stained for F-actin (magenta) and ZO-1 (green). (B) Cell area measured in Control cells (n = 384) and CDHR2 KO cells (n = 259) from three experimental replicates. (C) Cell elongation (max Feret/min Feret ratio) measured from the cells analyzed in B. (D) A total of 12 DPC Control and CDHR2 KO CACO-2BBE cells stained for F-actin (magenta) and ZO-1 (green). Zooms show area in the dashed boxes, in this case highlighting straight versus ruffled junctions in Control and CDHR2 KO, respectively. (E) Junctional straightness (see Materials and Methods) from Control and KO cell junctional segments where 1.0 is a straight line; n = 62 segments from three experimental replicates per condition. (F) 21 DPC Control and CDHR2 KO CACO-2BBE cells stained for F-actin (magenta) and NM2C (green). (G) Mean NM2C and (H) ZO-1 intensities measured in CACO-2BBE cells from two experimental replicates, 10 fields per replicate, 20 fields total. Break in Y axis represents level of background fluorescence. In G and H, points represent individual image fields with mean ± SD. Significance levels from unpaired t tests calculated for experimental replicates are also shown above each plot (*p < 0.05, **p < 0.005). Scale bars: 40 µm (A, D, F), 10 µm (D, Zooms).
Loss of CDHR2 perturbs the composition of canonical junctional complexes
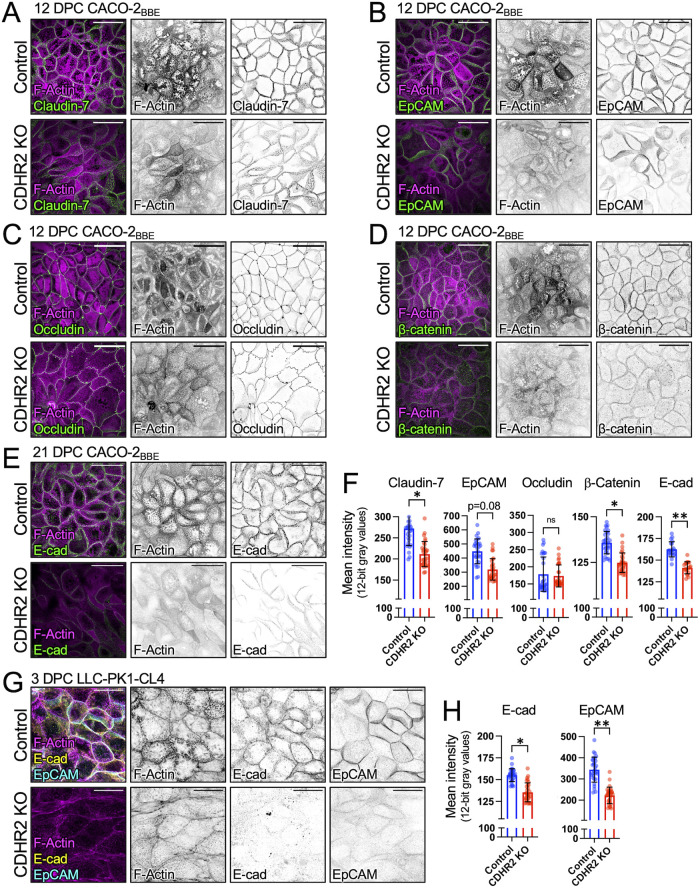
The abnormal cell morphology and perturbations in the circumferential actomyosin belt induced by loss of CDHR2 (Figure 1) could be due to defects in the canonical junctional complexes that mediate cell-cell contact. Indeed, confocal imaging of ZO-1 staining, which we used to delineate cell boundaries in monolayers, revealed reduced levels at TJs in CDHR2 KO CL4 and CACO-2BBE cells (Figures 1, A, D, and H). Claudin-7, another adhesive factor that directly contributes to TJs, was also significantly reduced at junctions, whereas epithelial cellular adhesion molecule (EpCAM) demonstrated a trend toward lower levels (p = 0.08), and occludin was not altered (Figure 2, A–C and F). As direct binding partners, claudin-7 and EpCAM contribute to TJ barrier maintenance through damage-induced cleavage of EpCAM and release of claudin-7 (Ladwein et al., 2005; Wu et al., 2013; Higashi et al., 2023). In addition, we observed a significant loss of canonical AJ markers from cell-cell contacts, including β-catenin and E-cadherin (Figure 2, D–F). CDHR2 KO CL4 cells exhibited parallel reductions in E-cadherin and EpCAM (Figure 2, G and H).
FIGURE 2:
CDHR2 KO cells exhibit defects in junctional composition. Confocal images of (A) 12 DPC Control and CDHR2 KO CACO-2BBE cells stained for F-actin (magenta) and Claudin-7 (green), (B) F-actin (magenta) and EpCAM (green), (C) F-actin (magenta) and Occludin (green), and (D) F-actin (magenta) and β-catenin (green). (E) Confocal images of 21 DPC Control and CDHR2 KO CACO-2BBE cells stained for F-actin (magenta) and E-cadherin (green). (F) Mean Control and CDHR2 KO CACO-2 BBE junctional protein fluorescence intensities (12-bit gray values) for Claudin-7 (n = 3), EpCAM (n = 3), Occludin (n = 2), β-catenin (n = 3), and E-cadherin (n = 2), where n = number of experimental replicates and each replicate consists of 10 image fields at 60X. (G) Confocal images of three DPC Control and CDHR2 KO CL4 cells stained for F-actin (magenta), E-cadherin (yellow), and EpCAM (cyan). (H) Mean Control and CDHR2 KO CL4 fluorescence intensities (12-bit gray values) for E-cadherin (n = 3) and EpCAM (n = 3), where n = number of experimental replicates and each replicate consists of 10 image fields at 60X. For plots in F and H, points represent individual 60X fields with mean ± SD. Significance levels from unpaired t tests calculated for experimental replicates are shown above each plot (*p < 0.05, **p < 0.005). Break in Y axes represents level of background fluorescence. Scale bars: 40 µm (A–E) and 20 µm (G).
To determine whether the reduced signals observed in our confocal datasets were due to lower expression levels in the absence of CDHR2, we generated whole cell lysates from both KO models and performed Western blots for a range of junctional components. Aside from EpCAM, which demonstrated markedly reduced expression in CDHR2 KO CL4 cells, most junctional factors were unchanged in their expression, suggesting that mislocalization rather than reduced expression was the basis for changes alluded to above (Supplemental Figure S2, A and B). Inspection of apical and basal sections and lateral views from confocal volumes did confirm mislocalization in some instances (Supplemental Figure S2, C and D). For example, in the case of ZO-1, basal sections through CDHR2 KO CACO-2BBE monolayers revealed punctate signal and aberrant stretches of lateral staining that were absent in Controls (Supplemental Figure S2C). Finally, to examine the possibility that loss of CDHR2 impaired apical-basal polarity establishment, we stained CDHR2 KO CACO-2BBE cells for Par3, a core polarity network component (Thompson, 2022). Inspection of those samples revealed that Par3 apical localization and junctional enrichment was unaffected by the loss of CDHR2 (Supplemental Figure S2, E and F), suggesting that polarity establishment remained intact.
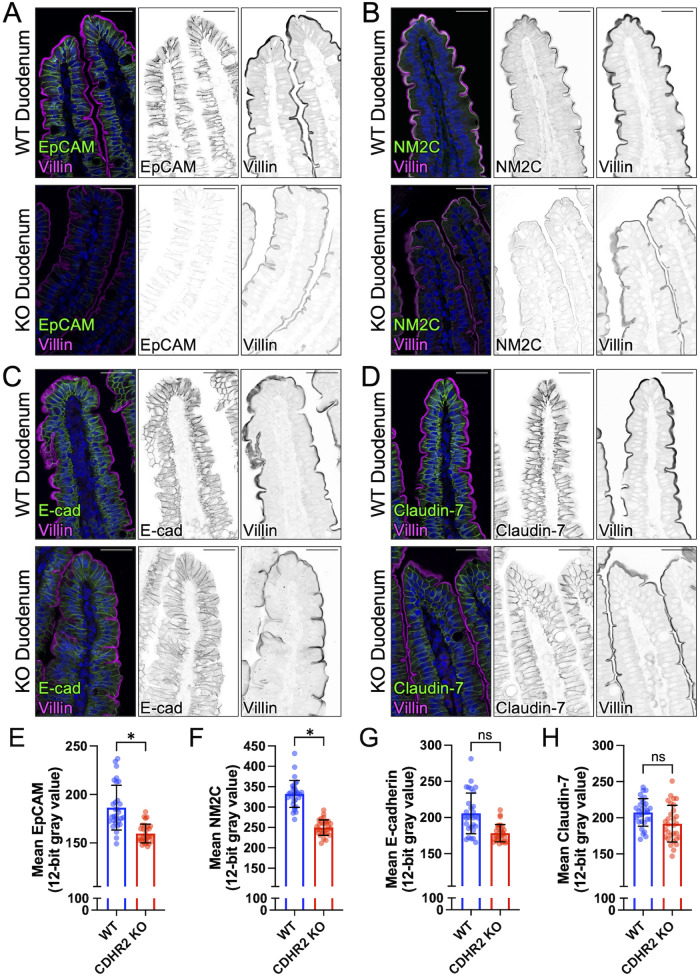
Our previous studies of the intestine-specific CDHR2 KO mouse focused on characterizing abnormalities in the apical brush border and revealed defects in microvillus structure and organization, as well as reduced IMAC enrichment at microvillar tips and decreased levels of apical solute transporters (Pinette et al., 2019). Based on our current findings, we revisited the CDHR2 KO mouse model to determine whether defects in junctional composition also manifest in vivo. Indeed, staining and confocal imaging of paraffin sections from CDHR2 KO mouse small intestine revealed a significant loss of multiple junctional components, including ZO-1 (Supplemental Figure S3, D and E), EpCAM and NM2C (Figure 3, A, B, E, and F), and a trend toward lower levels of E-cadherin and Claudin-7 (Figure 3 C, D, G, and H). CDHR2 KO intestinal tissue also exhibited lower levels of the microvillar actin bundler, villin (Supplemental Figure S3, D and E) as noted in our previous report (Pinette et al., 2019). Inspection of the full length of the small intestine revealed that these phenotypes were most prominent in the duodenum. In combination with our observations on CDHR2 KO CL4 and CACO-2BBE models, we conclude that the molecular composition of canonical junctional complexes is disrupted in epithelial cells lacking CDHR2.
FIGURE 3:
CDHR2 KO mouse duodenum exhibits reduced levels of some junctional components. Confocal images of wildtype (WT) and CDHR2 KO duodenal sections stained for villin (magenta) and the following markers in green: (A) EpCAM, (B) NM2C, (C) E-cadherin, and (D) Claudin-7. Mean intensities (12-bit gray values) measured for (E) EpCAM, (F) NM2C, (G) E-cadherin, and (H) Claudin-7. Nuclei are marked by DRAQ5 (blue). Intensity measurements for each marker were derived from two WT and CDHR2 KO littermate pairs and 15 image fields at 40X were analyzed from each animal. For plots in E–H, points represent individual 40X image fields with mean ± SD. Significance levels from unpaired t tests calculated for experimental replicates are shown above each plot (*p < 0.05). Break in Y axes represents level of background fluorescence. Scale bars: 40 µm (A–D).
CDHR2 KO cells exhibit a loss of junctional integrity and impaired wound healing
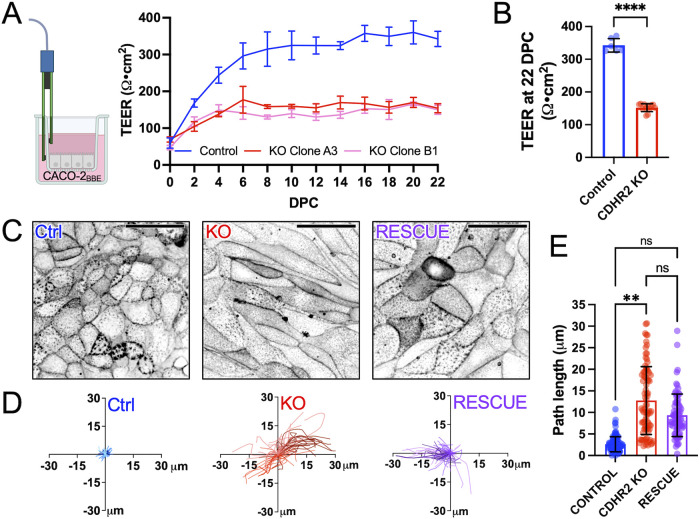
Based on the defects in junctional composition revealed by our quantitative immunostaining analysis, we next sought to determine whether junctional integrity and function were impacted by CDHR2 KO. Toward this end, we turned to measurements of transepithelial electrical resistance (TEER), an established approach for quantitatively characterizing TJ formation and barrier integrity (Srinivasan et al., 2015). Control and CDHR2 KO CACO-2BBE cells (two independently derived KO clones) were seeded on semipermeable Transwell filters and TEER was measured every 2 d starting one day postplating (Figure 4A). Measurements of TEER over the course of 22 d of differentiation revealed that, despite similar starting values, CDHR2 KO cells plateaued at a lower resistance compared with Controls, with 22 DPC averages of 152.1 ± 12.2 Ω·cm2 versus 342.6 ± 20.8 Ω·cm2, respectively (Figure 4, A and B). Moreover, whereas the Control cell TEER plateau fell within the range of resistance values reported in the literature for CACO-2 cells, the lower values measured for CDHR2 KO cultures indicate significant defects in TJ integrity (Narai et al., 1997; Srinivasan et al., 2015).
FIGURE 4:
CDHR2 KO monolayers exhibit decreased TEER and increased motility. (A) CACO cells were seeded on Transwell inserts and TEER was measured every other day postseeding to 22 DPC. (B) Mean TEER values from n = 6 Control and n = 12 CDHR2 KO (6 per CDHR2 KO clone) Transwells at 22 DPC: 342.6 ± 20.8 Ω·cm2 (Control) versus 152.1 ± 12.2 Ω·cm2 (CDHR2 KO clones). (C) Confocal images of live 3 DPC Control, CDHR2 KO, and Halo-CDHR2 Rescue CL4 cell monolayers labeled with membrane marker CellBriteSteady650. (D) Single-cell trajectories derived from Control, CDHR2 KO, and Halo-CDHR2 Rescue CL4 monolayers imaged for over 8 h. For each condition, three experimental replicates were imaged, and trajectories were calculated from 20 cells in each replicate (60 tracks in total per condition). (E) Total path lengths from trajectories shown in D; points represent individual trajectories shown with mean ± SD. Significance levels from pairwise comparisons using ANOVA calculated for experimental replicates are shown (**p < 0.005).
Movie S1.
related to Figure 4.
The stability of junctional complexes is important for defining the behavior of cells in the context of a monolayer (De Pascalis and Etienne-Manneville, 2017; Gupta and Yap, 2021). To examine how loss of CDHR2 might impact such dynamics, we performed live imaging of Control and CDHR2 KO CL4 monolayers. As a critical point of comparison, we also developed a CDHR2 KO line re-expressing a HALO-tagged variant of CDHR2 (Supplemental Figure S3, A–C). Importantly, the reduction in ZO-1 (and CDHR5, the heterophilic binding partner for CDHR2 in the IMAC) observed in CDHR2 KO cells was partially rescued upon CDHR2-HALO expression (Supplemental Figure S3, A–C), suggesting that the junctional perturbations alluded to above are due to the loss of CDHR2, rather than off-target effects. Control, CDHR2 KO, and Rescue CL4 monolayers were labeled with a plasma membrane dye (CellBriteSteady650) and subject to 8 h of confocal timelapse imaging (Figure 4C). Trajectory analysis of individual cells in the resulting movies revealed that control CL4 cells exhibited only a few microns of movement in this timeframe; in contrast, CDHR2 KO cells were highly motile, exchanging partners and covering up to tens of microns in the same duration (Figure 4, D and E). KO cells re-expressing CDHR2-HALO resulted in a partially rescued, intermediate phenotype as assessed by total trajectory path length (Figure 4E).
Based on the highly motile phenotype of CDHR2 KO cells, we also sought to determine whether collective migration of KO monolayers was impaired. For these experiments, we used chambers separated by a removeable barrier, which formed a wound-like gap on the surface once it was removed and allowed us to simultaneously observe the dynamics of both Control and CDHR2 KO cells in the same imaging field. Timelapse imaging revealed that Control cells exhibited well-ordered, collective migration toward the gap as expected (Supplemental Figure S3F). In contrast, CDHR2 KO cells demonstrated almost no collective migration in the same timeframe (Supplemental Figure S3F), and velocity analysis confirmed a significant reduction in advance of the leading edge for KO cells relative to Controls (Supplemental Figure S3G). CDHR2 KO cells remained motile within the plane of the monolayer, exhibiting uncoordinated trajectories (Supplemental Figure S3H) with average velocities that were similar to Control cells (0.11 ± 0.06 vs. 0.12 ± 0.09 µm/min, respectively). Together, these functional assays indicate that loss of junctional complex components in CDHR2 KO cells leads to decreased junctional integrity (indicated by reduced TEER), aberrant single-cell dynamics within intact monolayers, and impaired collective cell migration in wounded monolayers.
A role for intermicrovillar adhesion in stabilizing canonical junctional complexes
Given that transjunctional IMACs localize immediately above (i.e., apical to) TJs and AJs, we wondered if a loss of IMAC adhesion would impact the assembly and function of these canonical junctional complexes. To test this idea, we characterized CDHR2 KO phenotypes in a newly developed CACO-2BBE cell line and in a recently described CL4 line (Cencer et al., 2023). In both cell lines, CDHR2 KO led to defects in microvilli clustering, as expected (Crawley et al., 2014; Pinette et al., 2019). However, CDHR2 KO cells also presented with striking perturbations in cell and junction morphology (Figure 5). In the CL4 background, we observed significantly elongated cell profiles in confluent monolayers, a phenotype similar to previous reports in MDCK models lacking multiple junctional components (Choi et al., 2016). In the CACO-2BBE background, CDHR2 KO led to a slightly different phenotype, although one still indicative of defects in junctional integrity. In this case, loss of CDHR2 led to a decrease in the straightness of individual junctional segments. One interpretation of this result is that ruffled (nonlinear) junctions indicate a loss of mechanical tension, an idea consistent with the lower levels of NM2C that we observed at CDHR2 KO CACO-2BBE junctions. Our staining experiments and quantitative imaging also revealed that CDHR2 KO in both cell line backgrounds led to a loss of core components from TJs and AJs; we noted similar perturbations in intestinal tissues from CDHR2 KO mice. These striking changes in NM2 enrichment and junctional composition could be linked, as previous studies revealed a complex interplay between normal levels of NM2-dependent tension and proper accumulation of F-actin and other core components of cell-cell contacts. For example, E-cadherin–based cell-cell contacts are strengthened under tension because of extracellular domain conformational changes that lead to catch bonding (Rakshit et al., 2012). Other studies showed that tension also stabilizes the E-cadherin/β-catenin binding and interactions with F-actin (Buckley et al., 2014). At this point, however, our data do not allow us to determine whether the perturbations in junctional composition induced by CDHR2 KO are up or downstream of loss of NM2 and potentially reduced junctional tension.
FIGURE 5:
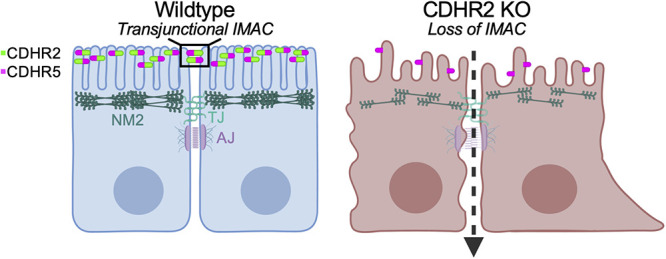
Loss of CDHR2-dependent intermicrovillar adhesion impairs basolateral junctions: a working model. Features of the CDHR2 KO phenotype include decreased microvillar clustering, aberrant cell morphology, reduced junctional tension, and a loss of key junctional proteins. These defects also give rise to increased junctional permeability and impaired collective cell migration during wound healing.
The changes in junctional morphology and composition induced by loss of CDHR2 were significant enough to disrupt the function of cell-cell contacts. Indeed, CDHR2 KO CACO-2BBE cells exhibited a significant reduction in TEER, indicating that TJ integrity is compromised in the absence of CDHR2 (Srinivasan et al., 2015; Yuan et al., 2020). Furthermore, we also noted that individual KO cells in established monolayers were highly motile and coordinated epithelial sheet motility in wound healing assays was severely impaired in in the absence of CDHR2. These observations are consistent with recent work from others, which established that normal levels of ZO-1 are required for coherent epithelial migration (Skamrahl et al., 2021).
The findings we present here are consistent with some of the earliest literature on CDHR2 (initially referred to as “protocadherin LKC”), which proposed a tumor suppressor role for this factor (Okazaki et al., 2002; Ose et al., 2009). Early work showed that CDHR2 is downregulated in a range of colon and liver cancer cell lines (Okazaki et al., 2002). Later investigations reported that reintroduction of CDHR2 into HTC116 colon carcinoma cells drove dramatic changes in cellular phenotype, including recruitment of β-catenin to cell-cell contacts, accelerated wound healing, and suppression of tumor growth in mice (Ose et al., 2009). These findings led the authors to propose that β-catenin signaling downstream of CDHR2 drives contact inhibition, and loss of this activity promotes tumor formation in epithelial tissues. Consistent with those ideas, we found that CDHR2 KO in CACO-2BBE demonstrated lower β-catenin levels at cell-cell contacts, a phenotype that has long been associated with aberrant epithelial cell motility and a loss of coordinated migration (Aman and Piotrowski, 2008).
Our group first discovered that CDHR2 participates in intermicrovillar adhesion on the apical surface of enterocytes in 2014 (Crawley et al., 2014). At that time, we were perplexed by the early studies invoking a tumor suppressor role in intestinal tissues, because they implied that CDHR2 was directly involved in β-catenin stabilization at junctional complexes (Okazaki et al., 2002; Ose et al., 2009). That model was difficult to reconcile with super-resolution images showing that CDHR2 instead targets specifically to the distal tips of brush border microvilli (Crawley et al., 2014). A resolution to this conundrum emerged with the discovery that CDHR2 forms IMACs between microvilli that extend from neighboring cells (Cencer et al., 2023). Based on the data we report here, we propose that these transjunctional IMACs form the most apical layer of physical contact between neighboring epithelial cells and promote the integrity of more basolateral adhesion complexes, including TJs and AJs. Future studies seeking to build on these results might focus on the role of transjunctional IMACs in response to challenges that arise in intestinal pathologies, such as Crohn's disease (VanDussen et al., 2018).
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Animal models
The CDHR2 KO mouse model was created and characterized by our laboratory as described in a previous study (Pinette et al., 2019). Animal experiments were carried out in accordance with Vanderbilt University Medical Center Institutional Animal Care and Use Committee guidelines under Institutional Animal Care and Use Committee Protocol M1600206-02.
Cell culture models
LLC-PK1-CL4 porcine kidney proximal tubule cells were grown in 1X high glucose DMEM/2 mM L-glutamine (Corning #10-013-CV) with an added 1% L-glutamine (Corning # 25-005-CI) and 10% FBS (R&D Systems). CACO-2BBE human colonic adenocarcinoma cells were grown in the same medium, with 20% FBS. Cells were incubated at 37°C and 5% CO2. Regular Mycoplasma testing was performed using the MycoStrip Mycoplasma Detection Kit (InvivoGen #rep-mys-50).
CRISPR/Cas9 genome editing
The CDHR2 CL4 KO cell line was created and validated in our previous study (Cencer et al., 2023). The CDHR2 CACO-2BBE KO cell line was generated and validated for the present study also as described previously, using the Lenti-CRISPR v2 system, with gRNA's designed to target exons 3 and 4 of human CDHR2 genomic sequence:
[Exon 3 FWD CACCGTAGGAACTTCGGGGCCACGT; Exon 3 REV AAACACGTGGCCCCGAAGTTCCTAC; Exon 4 FWD CACCGTCTTCCGCTACCAACCAGA; Exon 4 REV AAACTCTGGTTGGTAGCGGAAGAC] PCR/sequencing primers targeted regions surrounding Exon 3 or Exon 4, within which Cas9 was predicted to cut: [Exon 3 Fwd CAGCTATGGCTGTCCTGCTTC; Exon 3 Rev CAGTCAGAGACTGAAAGCGATGG; Exon 4 Fwd CTCCAAAGCTCTAGTCTGCACC; Exon 4 Rev CTCACCTGGATGTAGGGGTCG].
Rescue CL4 cell line generation
A validated CDHR2 KO CL4 cell clonal population was transfected with pHALO-N3-CDHR2 using FuGENE 6 (Promega #E2691) at a FuGENE:DNA (µl:µg) ratio of 3:1 following the reagent protocol in a T25 cell culture flask. The next day, cells were split to a T75 flask plus 1 mg/ml G418 sulfate for antibiotic selection. Cells were maintained in culture under G418 selection to create a stable cell line. To enrich for expressing cells, CDHR2 KO+HALO-CDHR2 rescue CL4 cells were incubated with 50 nM Janelia Fluor 635 dye for 1 h at 37°C to label cells, trypsinized, and sorted for midhigh expression as previously described by Vanderbilt University Medical Center's Flow Cytometry Shared Resource on a 5-Laser FACS Aria III system with a 100 µm sized nozzle (Cencer et al., 2023). Sorted cells were maintained under 1 mg/ml G418 selection.
Swiss roll preparation and paraffin embedding of intestinal tissue
The entire mouse small intestinal tube was excised and flushed with cold 1X PBS. Tissue was clamped at both ends with hemostats and fixed in the tube with room temperature (RT) 2% Paraformaldehyde for 15 min. After removing the hemostats, the intestinal tube was slid onto a metal cannula and cut lengthwise, down the entire length, with scissors. The flayed tissue was then rolled out, villi side up, onto a strip of parafilm. A hemostat was clamped onto the proximal end (duodenum) of the intestine and rolled, with the duodenum at the center of the roll. A 21 g needle was stuck through the roll and the hemostat was removed. The roll was then submerged in 10% neutral buffered formalin at RT for 48 h. After fixation, the needle was removed and the roll was cut in half, creating two thinner rolls, and each half was placed into a large tissue cassette and submerged back into the formalin. Cassettes were submitted to the Vanderbilt University Translational Pathology Shared Resource, embedded in paraffin wax, and sliced onto glass slides, at 10 µm thickness. Slides were stored at RT until staining.
Immunofluorescence staining of intestinal tissue
Using a Tissue-TekII manual slide staining set, slides were deparaffinized in Histo-Clear II (National Diagnostics) two times, 3 min each. Tissue was then rehydrated in a descending ethanol series (100%, 100%, 95%, 90%, 70%, 50%) 5 min each followed by washing in PBS 3 times, 3 min each. Slides were incubated in antigen retrieval buffer (10 mM Tris, 0.5 mM Ethyleneglycol‐bis(β‐aminoethyl)N,N,N′,N′‐tetraacetic Acid, pH 9.0) in coplin jars for 1 h using a rice cooker and then cooled to RT. Slides were washed three times, 3 min each in PBS and then blocked in 10% normal goat serum (NGS) for 1 h at RT. Primary antibody (diluted in 1% NGS) was added overnight at 4°C. The next day, slides were washed three times, 5 min each in PBS and secondary antibody (diluted in 1% NGS) was added for 1 h at RT in the dark. Slides were then washed three times, 5 min each in PBS followed by dehydration with an ascending ethanol series (50%, 70%, 90%, 95%, 100%, 100%) 5 min each. A coverslip was mounted with ProLong Gold. The following antibodies and dilutions were used for paraffin section staining: anti-ZO-1 (rabbit, Invitrogen #61-7300), 1:50; anti-Villin (mouse, Santa Cruz Biotechnology #SC-66022), 1:50; or anti-Villin (rabbit, Santa Cruz Biotechnology # SC-28283) 1:50; anti-E-cadherin (mouse, BD Biosciences #610182), 1:100; anti-EpCAM (rabbit, Invitrogen #PA5-19832), 1:100; anti-MYH14/NM2C (rabbit, Proteintech #20716-1-AP), 1:100; anti-Claudin-7 (rabbit, Invitrogen #34-9100), 1:100; Alexa Fluor F(ab’)2 fragment goat anti-rabbit 488 (Invitrogen #A11070), 1:1000; Alexa Fluor goat anti-mouse 568 (Invitrogen #A11019), 1:1000; Alexa Fluor F(ab’)2 fragment goat anti-rabbit 568 (Invitrogen #A21069), 1:1000; Alexa Fluor F(ab’)2 fragment goat anti-mouse 488 (Invitrogen #A11017), 1:1000. The secondary antibodies were spun down for 10 min at 4°C and 21 × g prior to using. DRAQ5 was used to label nuclei (Molecular Probes #62251); 1:500.
Fixing and immunofluorescent staining of cell culture models
CL4 and CACO-2BBE cells were grown to n days post-confluent (DPC) on acid-washed 22 × 22 mm #1.5H coverslips (Globe Scientific) in a 6-well plate to a polarization timepoint of 3 DPC and 12 or 21 DPC, respectively. Cells were fixed and stained as described previously (Cencer et al., 2023). The following antibodies and dilutions were used for cell staining: anti-PCLKC (CDHR2) (mouse, Abnova #H00054825-M01), 1:25; anti-CDHR5 (Rabbit, Sigma #HPA009173), 1:250; anti-ZO-1 clone R40.76 in CL4 (rat, EMD Millipore Sigma #MABT11), 1:100; anti-ZO-1 in CACO-2BBE (rabbit, Invitrogen #61-7300), 1:50; anti-E-cadherin (mouse, BD Biosciences #610182), 1:100; anti-EpCAM (rabbit, Invitrogen #PA5-19832), 1:100; anti-MYH14/NM2C (rabbit, Proteintech #20716-1-AP), 1:100; anti-Claudin-7 (rabbit, Invitrogen #34-9100), 1:100; anti-Beta-Catenin (rabbit, Invitrogen #71-2700), 1:100; anti-Occludin (mouse, Invitrogen #33-1500), 1:100; anti-Par3 (rabbit, EMD Millipore #07-330), 1:200; Alexa Fluor F(ab’)2 fragment goat anti-rabbit 488 (Invitrogen #A11070), 1:1000; Alexa Fluor goat anti-mouse 568 (Invitrogen #A11019), 1:1000; Alexa Fluor F(ab’)2 fragment goat anti-mouse 488 (Invitrogen #A11017) and goat anti-rabbit 568 (Invitrogen #A21069), 1:1000; Alexa Fluor goat anti-rat 647 (Invitrogen #A21247), 1:200; and Alexa Fluor Plus 405 Phalloidin (Invitrogen # A30104) or Alexa Fluor 647 Phalloidin (Invitrogen #A22287), 1:200 for actin staining. The secondary antibodies, not including phalloidin, were spun down for 10 min at 4°C and 21 × g prior to application.
Microscopy
Confocal microscopy was performed using a Nikon A1 microscope equipped with 488 nm, 568 nm, and 647 nm LASERs using an Apo TIRF 100x/1.49 NA or Apo TIRF 60x/1.49 NA TIRF oil immersion objective, or a Nikon Ti2 inverted light microscope equipped with a Yokogawa CSU-W1 spinning disk head, a Photometrics Prime 95B sCMOS camera, and four excitation LASERs (488, 568, 647, and 405 nm) and a 60X/1.49 NA TIRF oil immersion objective. For live monolayer imaging, Control, CDHR2 KO, or Halo-CDHR2 Rescue CL4 cells were seeded on 35 mm plasma cleaned glass-bottom dishes (CellVis #D35-20-1.5-N) and grown to 3 DPC. Prior to imaging, cells were rinsed with DPBS and incubated in 1:1000 CellBriteSteady650 membrane dye and 1:1000 kit enhancer (Biotium #30108) for 30 min at 37°C in Fluorobrite DMEM (Life Technologies #A1896701) +10% FBS, +1% L-glutamine. Monolayers were then imaged for 8 h at 4 min time intervals using the Yokogawa CSU-W1 spinning disk confocal, with a 647 nm excitation LASER using a Nikon Plan APO 40x SIL Silicone 1.25 NA objective lens.
TEER
Transwells (Greiner Bio-one #662641) were primed with 100 µl of cell culture media in the top chamber and 600 µl in the bottom chamber and left at 37°C for 15 min prior to cell seeding. Control and CDHR2 KO clone CACO-2BBE cells were counted, and 30,000 cells were seeded in each transwell with a total volume of 100 µl. A “blank” transwell was also maintained with media alone for background TEER measurements. A total of 24 h postseeding, the first TEER measurements were taken for the “0 DPC” timepoint. TEER was measured in ohms (Ω) using an EVOM3 epithelial voltohmmeter device (World Precision Instruments) equipped with a calibrated electrode (World Precision Instruments #STX2-PLUS). Prior to each TEER measurement, existing media was exchanged for fresh media, and cells were incubated for 4 h to ensure equal volume and equilibration in each well. The raw blank TEER value was subtracted from each monolayer TEER value and then multiplied by 0.33 cm2, the area of the transwell filter, to obtain the reported TEER value in Ω·cm2.
Wound healing assays
CL4 cells were seeded at a total of 30,000 cells per well in an Ibidi 2-chamber insert (Ibidi #80209) adhered to a 35 mm plasma cleaned glass-bottom dish (CellVis #D35-20-1.5-N). Cells were grown for 2 d or until they had just reached the edge of the chamber. The chamber media was aspirated, and the insert was carefully removed with forceps. Cells were gently rinsed twice with warm DPBS and fresh media was added to the dish. Cells were left in the incubator ∼8 h, to recover. Before imaging, cells were rinsed with DPBS and and incubated in 1:1000 CellBrite Steady 650 membrane dye and 1:1000 kit enhancer (Biotium #30108) for 30 min at 37°C. Dye was washed out and media replaced with Fluorobrite DMEM (Life Technologies #A1896701) +10% FBS, +1% L-glutamine. Cells were then imaged using a 10X Plan Apo 0.45 NA objective on a Yokogawa CSU-X1 spinning disk confocal microscope with a 647 nm excitation LASER for ∼24 h, or until 100% wound closure.
Western blotting
Control and CDHR2 KO cells were seeded in separate T25 cell culture flasks, one per condition. At 12 DPC (CACO) or 3 DPC (CL4), cells were rinsed in cold 1X DPBS and incubated, rocking, for 10 min in 100 µl cold lysis buffer (10 ml CelLytic-M Sigma #C2978; 4 mM Pefabloc Roche #11429868001; one pellet protease inhibitor Roche #05892791001). After lysis, cells were scraped and collected in prechilled Eppendorf tubes followed by centrifugation at 12,000 rpm for 10 min at 4°C. Supernatant protein concentration was measured via Bradford assay to ensure equal sample gel loading (“Blank” = Lysis Buffer alone). Samples were boiled for 5 min in 4X Laemmli Sample buffer (Bio-Rad #1610747) and 2-mercaptoethanol (Sigma #M3148). Samples were run on a NuPAGE 4-12% gradient gel (Invitrogen #NP0322BOX) with ∼20 µg total protein per well, with a protein ladder (Bio-Rad #1610373) at 110 V in fresh 1X MES SDS running buffer (NuPAGE #NP0002). Gels were transferred onto nitrocellulose membrane at 100 V for 1 h at 4°C in fresh transfer buffer (1X Tris-glycine; 20% methanol). The membrane was dyed with Ponceau and cut followed by blocking in 5% milk (Labscientific #M0841) in 1X PBS for 1 h at RT. Primary antibody (1:1000) was added overnight at 4°C in 3% milk and 0.1% Tween-20. Primary antibodies used are the same antibodies as previously noted for fixed cell immunofluorescence; in addition to anti-GAPDH for loading control using mouse (Santa Cruz Biotechnology #sc-32233) or rabbit (Cell Signaling Technology #14C10) antibody. The next day, membrane was washed 4 × 5 min each in PBST (1X PBS and 0.1% Tween-20) at RT. Secondary antibody (1:10,000) was added for 1 h at RT using the following: IRdye 800 donkey anti-rabbit (LI-COR #926-32213), IRdye 800 donkey anti-mouse (LI-COR #926–32212), IRDye 680 goat anti-rabbit (LI-COR #926-68071), and IRDye 680 donkey anti-mouse (LI-COR #926-68072). Note that GAPDH was labeled with 680 and all other proteins of interest with 800. Membrane was washed 3 × 5 min each in PBST and 1 × 5 min in PBS followed by imaging using the Odyssey CLx infrared scanner (LI-COR).
QUANTIFICATION AND STATISTICAL ANALYSIS
Cell shape measurements
60X image fields of 3 DPC Control and CDHR2 KO CL4 cells stained for ZO-1 were analyzed in Elements using a custom General Analysis 3 (GA3) pipeline. This involved first maximally projecting each image in Z followed by tight border segmentation. The binary was then inverted to highlight individual cells in the field and any “partial” cells touching the borders were eliminated from the selection. Cell area (µm2) and elongation were measured from the remaining binaries. Elongation is quantified by the software as a ratio of maximum feret length over minimum feret length. A value of greater than 1 indicates that the cell is stretched in one of its axes.
Cell junction straightness measurements
Randomly selected 60X image fields were maximum intensity projected in Z in FIJI. The ZO-1 channel was isolated and junctional segments (between two vertices) were cropped using the rectangle selection tool. The cropped segment was then binarized and dilated to segment the ZO-1 signal marking the cell junction. Next, the binary was skeletonized and the two-dimensional skeleton was analyzed. Branch length and Euclidian distance were exported from the measurements list to Excel. Junction straightness was calculated as the ratio of Euclidean distance over the branch length, with 1 being the most straight, as previously described in the literature (Sumi et al., 2018). Straightness ratios were calculated and displayed in Prism v.9.0 as violin plots.
Fluorescence intensity measurements
For marker intensity measurements in CL4 and CACO-2BBE cell culture models, 60X image fields were used as the basis for quantification for each condition (Control or CDHR2 KO). Raw images were maximum intensity projected in Z using FIJI and mean intensity of the entire field was measured in the channel of interest. The measured intensities were plotted and analyzed in Prism v. 9.0. For the quantification of staining levels in mouse intestinal tissues, 40X image fields were analyzed for mean intensity using a Nikon Elements GA3 analysis pipeline. Broadly, each raw image was maximum intensity projected in Z and the junctional marker channel was thresholded for intensity encompassing respective signal in Control mice. If necessary, to encompass all junctional signal, the thresholded Regions of Interest (ROI) were dilated and/or cleaned; the same thresholding parameters were applied to CDHR2 KO mouse images. Mean intensity readouts were exported from Elements and analyzed using Prism v. 9.0.
Single-cell tracking
Timelapse confocal volumes of Control, CDHR2 KO, Rescue CL4 monolayers were maximum intensity projected in Z using Nikon Elements, and circular ROIs were manually placed in the center of >20 cells per movie with parameters set to “ROIs change over time.” ROIs were manually moved every ∼10 frames to maintain a centrally located ROI position in each cell over time; the resulting full path of each cell was interpolated using Elements built-in ROI interpolation. Trajectories were plotted in Prism v. 9.0 for Control, CDHR2 KO, and Halo-CDHR2 rescue CL4 cell monolayers.
Wound healing analysis
Timelapse confocal volumes of wound healing assays were maximum intensity projected in Z using FIJI and divided in half with duplicate ROIs. For the Control side, 20 randomly placed horizontal lines were drawn along the entire vertical axis of the leading edge of the cell monolayer to the opposite end of the ROI (the wound “midline”). The length, in µm, was recorded in Excel along with the time, minutes, it took to reach the midline and velocity was calculated from µm/min. For the CDHR2 KO side, which exhibited far less movement toward the wound midline, 20 randomly placed horizontal lines were drawn from the monolayer edge to the furthest point of migration. Length of the resulting lines versus timepoint in the movie was recorded and velocity was calculated as µm/min.
Statistical analysis
All graphs were generated and statistical analyses performed using Prism (v.9.0, GraphPad). For statistical testing, we used unpaired parametric t tests for distributions that satisfied the normality and equal variance assumptions. Welch's correction was used in cases where datasets did not exhibit equal variance. Nonparametric t tests were used in cases where data failed to exhibit normality. Number of measurements and experimental replicates are indicated in figure legends.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank all members of the Tyska laboratory for their constructive feedback. We thank the Vanderbilt Genome Editing Resource (VGER) supported by the Cancer Center Support Grant P30 CA68485 for their expertise and assistance in generating the CDHR2 KO mouse. We also acknowledge the Translational Pathology Shared Resource supported by NCI/NIH Cancer Center Support Grant P30 CA068485 for their assistance in paraffin embedded tissue sectioning. FACS was performed in the VUMC Flow Cytometry Shared Resource supported by the Vanderbilt Ingram Cancer Center P30 CA68485, and the Vanderbilt Digestive Disease Research Center supported by P30 DK058404. This study was also supported by the NIH grants R01 DK095811 (M.J.T.), R01 DK125546 (M.J.T.), and R01 DK111949 (M.J.T.), and the Training Program in Developmental Biology T32 HD007502 (Christopher V. Wright).
Abbreviations used:
- AJ
adherens junction
- CL4
Clone 4
- DPC
days post confluence
- IMAC
intermicrovillar adhesion complex
- KO
knockout
- TEER
transepithelial electrical resistance
- TJ
tight junction.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E24-03-0113) on September 18, 2024.
REFERENCES
- Aman A, Piotrowski T (2008). Wnt/beta-catenin and Fgf signaling control collective cell migration by restricting chemokine receptor expression. Dev Cell 15, 749–761. [DOI] [PubMed] [Google Scholar]
- Bondow BJ, Faber ML, Wojta KJ, Walker EM, Battle MA (2012). E-cadherin is required for intestinal morphogenesis in the mouse. Dev Biol 371, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley CD, Tan J, Anderson KL, Hanein D, Volkmann N, Weis WI, Nelson WJ, Dunn AR (2014). Cell adhesion. The minimal cadherin-catenin complex binds to actin filaments under force. Science 346, 1254211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley CE, St Johnston D (2022). Apical-basal polarity and the control of epithelial form and function. Nat Rev Mol Cell Biol 23, 559–577. [DOI] [PubMed] [Google Scholar]
- Cencer CS, Silverman JB, Meenderink LM, Krystofiak ES, Millis BA, Tyska MJ (2023). Adhesion-based capture stabilizes nascent microvilli at epithelial cell junctions. Dev Cell 58, 2048–2062.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childress KO, Cencer CS, Tyska MJ, Lacy DB (2023). Nectin-3 and shed forms of CSPG4 can serve as epithelial cell receptors for Clostridioides difficile TcdB. mBio 14, e0185723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinowsky CR, Pinette JA, Meenderink LM, Lau KS, Tyska MJ (2020). Nonmuscle myosin-2 contractility-dependent actin turnover limits the length of epithelial microvilli. Mol Biol Cell 31, 2803–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi MS, Graves MJ, Matoo S, Storad ZA, El Sheikh Idris RA, Weck ML, Smith ZB, Tyska MJ, Crawley SW (2020). The small EF-hand protein CALML4 functions as a critical myosin light chain within the intermicrovillar adhesion complex. J Biol Chem 295, 9281–9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W, Acharya BR, Peyret G, Fardin MA, Mege RM, Ladoux B, Yap AS, Fanning AS, Peifer M (2016). Remodeling the zonula adherens in response to tension and the role of afadin in this response. J Cell Biol 213, 243–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley SW, Shifrin DA, Jr., Grega-Larson NE, McConnell RE, Benesh AE, Mao S, Zheng Y, Zheng QY, Nam KT, Millis BA, et al. (2014). Intestinal brush border assembly driven by protocadherin-based intermicrovillar adhesion. Cell 157, 433–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley SW, Weck ML, Grega-Larson NE, Shifrin DA, Jr., Tyska MJ (2016). ANKS4B is essential for intermicrovillar adhesion complex formation. Dev Cell 36, 190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pascalis C, Etienne-Manneville S (2017). Single and collective cell migration: The mechanics of adhesions. Mol Biol Cell 28, 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diz-Munoz A, Fletcher DA, Weiner OD (2013). Use the force: Membrane tension as an organizer of cell shape and motility. Trends Cell Biol 23, 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahim S, Fujita T, Millis BA, Kozin E, Ma X, Kawamoto S, Baird MA, Davidson M, Yonemura S, Hisa Y, et al. (2013). NMII forms a contractile transcellular sarcomeric network to regulate apical cell junctions and tissue geometry. Curr Biol 23, 731–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Nelson WJ, Chavez N (2018). Cell-cell junctions organize structural and signaling networks. Cold Spring Harb Perspect Biol 10, a029181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Castillo MD, Chinnapen DJ, Lencer WI (2017). Membrane transport across polarized epithelia. Cold Spring Harb Perspect Biol 9, a027912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Yap AS (2021). How adherens junctions move cells during collective migration. Fac Rev 10, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi T, Saito AC, Fukazawa Y, Furuse M, Higashi AY, Ono M, Chiba H (2023). EpCAM proteolysis and release of complexed claudin-7 repair and maintain the tight junction barrier. J Cell Biol 222, e202204079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz A, Chanez-Paredes SD, Haest X, Turner JR (2023). Paracellular permeability and tight junction regulation in gut health and disease. Nat Rev Gastroenterol Hepatol 20, 417–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladwein M, Pape UF, Schmidt DS, Schnolzer M, Fiedler S, Langbein L, Franke WW, Moldenhauer G, Zoller M (2005). The cell-cell adhesion molecule EpCAM interacts directly with the tight junction protein claudin-7. Exp Cell Res 309, 345–357. [DOI] [PubMed] [Google Scholar]
- Li J, He Y, Weck ML, Lu Q, Tyska MJ, Zhang M (2017). Structure of Myo7b/USH1C complex suggests a general PDZ domain binding mode by MyTH4-FERM myosins. Proc Natl Acad Sci USA 114, E3776–E3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeol P, Massey-Harroche D, Richard F, Concordet JP, Lenne PF, Le Bivic A (2022). Super-resolution imaging uncovers the nanoscopic segregation of polarity proteins in epithelia. Elife 11, e62087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeol P, Massey-Harroche D, Sebbagh M, Richard F, Le Bivic A, Lenne PF (2024). The zonula adherens matura redefines the apical junction of intestinal epithelia. Proc Natl Acad Sci USA 121, e2316722121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narai A, Arai S, Shimizu M (1997). Rapid decrease in transepithelial electrical resistance of human intestinal Caco-2 cell monolayers by cytotoxic membrane perturbents. Toxicol In Vitro 11, 347–354. [DOI] [PubMed] [Google Scholar]
- Okazaki N, Takahashi N, Kojima S, Masuho Y, Koga H (2002). Protocadherin LKC, a new candidate for a tumor suppressor of colon and liver cancers, its association with contact inhibition of cell proliferation. Carcinogenesis 23, 1139–1148. [DOI] [PubMed] [Google Scholar]
- Ose R, Yanagawa T, Ikeda S, Ohara O, Koga H (2009). PCDH24-induced contact inhibition involves downregulation of beta-catenin signaling. Mol Oncol 3, 54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinette JA, Mao S, Millis BA, Krystofiak ES, Faust JJ, Tyska MJ (2019). Brush border protocadherin CDHR2 promotes the elongation and maximized packing of microvilli in vivo. Mol Biol Cell 30, 108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakshit S, Zhang Y, Manibog K, Shafraz O, Sivasankar S (2012). Ideal, catch, and slip bonds in cadherin adhesion. Proc Natl Acad Sci USA 109, 18815–18820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skamrahl M, Pang H, Ferle M, Gottwald J, Rubeling A, Maraspini R, Honigmann A, Oswald TA, Janshoff A (2021). Tight junction ZO proteins maintain tissue fluidity, ensuring efficient collective cell migration. Adv Sci (Weinh) 8, e2100478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan B, Kolli AR, Esch MB, Abaci HE, Shuler ML, Hickman JJ (2015). TEER measurement techniques for in vitro barrier model systems. J Lab Autom 20, 107–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumi A, Hayes P, D'Angelo A, Colombelli J, Salbreux G, Dierkes K, Solon J (2018). Adherens junction length during tissue contraction is controlled by the mechanosensitive activity of actomyosin and junctional recycling. Dev Cell 47, 453–463.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BJ (2022). Par-3 family proteins in cell polarity & adhesion. FEBS J 289, 596–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Liu Z, Niu B, Zhang J, Tan TK, Lee SR, Zhao Y, Harris DC, Zheng G (2011). E-cadherin/beta-catenin complex and the epithelial barrier. J Biomed Biotechnol 2011, 567305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda S, Higashi T, Furuse M (2014). ZO-1 knockout by TALEN-mediated gene targeting in MDCK cells: Involvement of ZO-1 in the regulation of cytoskeleton and cell shape. PLoS One 9, e104994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyanovsky SM (2023). Adherens junction: The ensemble of specialized cadherin clusters. Trends Cell Biol 33, 374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita S, Tanaka H, Tamura A (2019). The claudins: From tight junctions to biological systems. Trends Biochem Sci 44, 141–152. [DOI] [PubMed] [Google Scholar]
- Valenta T, Hausmann G, Basler K (2012). The many faces and functions of beta-catenin. EMBO J 31, 2714–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wal T, van Amerongen R (2020). Walking the tight wire between cell adhesion and WNT signalling: A balancing act for beta-catenin. Open Biol 10, 200267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Itallie CM, Anderson JM (2014). Architecture of tight junctions and principles of molecular composition. Semin Cell Dev Biol 36, 157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Itallie CM, Fanning AS, Bridges A, Anderson JM (2009). ZO-1 stabilizes the tight junction solute barrier through coupling to the perijunctional cytoskeleton. Mol Biol Cell 20, 3930–3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDussen KL, Stojmirovic A, Li K, Liu TC, Kimes PK, Muegge BD, Simpson KF, Ciorba MA, Perrigoue JG, Friedman JR, et al. (2018). Abnormal small intestinal epithelial microvilli in patients with Crohn's disease. Gastroenterology 155, 815–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weck ML, Crawley SW, Stone CR, Tyska MJ (2016). Myosin-7b promotes distal tip localization of the intermicrovillar adhesion complex. Curr Biol 26, 2717–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CJ, Mannan P, Lu M, Udey MC (2013). Epithelial cell adhesion molecule (EpCAM) regulates claudin dynamics and tight junctions. J Biol Chem 288, 12253–12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap AS (1998). The morphogenetic role of cadherin cell adhesion molecules in human cancer: A thematic review. Cancer Invest 16, 252–261. [DOI] [PubMed] [Google Scholar]
- Yuan L, van der Mei HC, Busscher HJ, Peterson BW (2020). Two-stage interpretation of changes in TEER of intestinal epithelial layers protected by adhering bifidobacteria during E. coli challenges. Front Microbiol 11, 599555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.