Abstract
Stress granules (SGs) are macromolecular assemblies that form under cellular stress. Formation of these membraneless organelles is driven by the condensation of RNA and RNA-binding proteins such as G3BPs. G3BPs form SGs following stress-induced translational arrest. Three G3BP paralogues (G3BP1, G3BP2A, and G3BP2B) have been identified in vertebrates. However, the contribution of different G3BP paralogues to SG formation and gene expression changes is incompletely understood. Here, we probed the functions of G3BPs by identifying important residues for SG assembly at their N-terminal domain such as V11. This conserved amino acid is required for formation of the G3BP–Caprin-1 complex, hence promoting SG assembly. Total RNA sequencing and ribosome profiling revealed that a G3BPV11A mutant leads to changes in mRNA levels and ribosome engagement during the integrated stress response (ISR). Moreover, we found that G3BP2B preferentially forms SGs and promotes changes in mRNA expression under endoplasmic reticulum (ER) stress. Furthermore, our work is a resource for researchers to study gene expression changes under cellular stress. Together, this work suggests that perturbing protein–protein interactions mediated by G3BPs affect SG assembly and gene expression during the ISR, and such functions are differentially regulated by G3BP paralogues under ER stress.
INTRODUCTION
Stress granules (SGs) are membraneless organelles composed of ribonucleoprotein (RNP) complexes that condense into macromolecular assemblies upon cellular stress (Kedersha et al., 1999). Association of untranslated mRNAs and the enrichment of proteins from the translational machinery into SGs has led to the model that these RNP condensates inhibit mRNA expression and translation during the stress response (Kedersha et al., 2002; Buchan and Parker, 2009). For instance, SG assembly leads to suppression of Csnk2a1 mRNA translation preventing axonal growth postinjury (Sahoo et al., 2018, 2020). In yeast, Ded1p condensation into SGs during heat shock promotes the translation of stress-induced transcripts, while suppressing the translation of housekeeping genes (Iserman et al., 2020). However, it has been reported that SGs are dispensable for stress-induced global translational arrest (Kedersha et al., 2016). Moreover, active translation of mRNA molecules has been identified inside SGs (Mateju et al., 2020), leading to uncertainty around the function of SGs in mRNA expression and their relevance during the stress response.
SGs assemble by the condensation of RNAs and RNA-binding proteins such as Ras-GTPase–activating protein (SH3 domain)-binding proteins (G3BPs). G3BPs are common components and crucial SG nucleators under multiple cellular stresses. Knockout (KO) of G3BPs inhibits the formation of stress granules under oxidative and endoplasmic reticulum (ER) stress (Kedersha et al., 2016; Yang et al., 2020). Previous work revealed that G3BP1 participates in protein interaction networks that drive SG assembly via phase separation of core SG proteins (Guillén-Boixet et al., 2020; Sanders et al., 2020; Yang et al., 2020). Multivalency with RNA is increased via binding of G3BP1 with proteins such as UBAP2L and Caprin-1, promoting condensation of SGs. The G3BP1–Caprin-1 interaction is mediated by the nuclear transport factor 2 like domain (NTF2L) of G3BP1 (Schulte et al., 2023). Thus, studying this interaction will expand our understanding of the role of the NTF2L domain on SG formation and uncover insights of G3BP's functions during the stress response.
Three G3BP paralogues have been identified in vertebrates: G3BP1 and the two splice isoforms encoded by G3BP2: G3BP2A, and G3BP2B (Kennedy et al., 2002). Aside from SG assembly, G3BPs are involved in RNA metabolism and in the response against viral infection by either promoting or inhibiting the replication of virus (Alam and Kennedy, 2019). Furthermore, they interact with key cellular pathways during the progression of certain cancers and neurodegenerative diseases (Kang et al., 2021; Mukhopadhyay and Zhou, 2023). The function of G3BP paralogues in SG assembly has been considered predominantly redundant (Yang et al., 2020). For this reason, most research has been focused on understanding the function of G3BP1. However, there is growing evidence suggesting that G3BP paralogues may have different roles in certain biological contexts, such as regulation of mTOR signaling and the response against poliovirus infection (Dougherty et al., 2015; Prentzell et al., 2021).
SGs assemble upon activation of stress response programs such as the integrated stress response (ISR). The ISR is mediated by stress-sensing kinases (PKR, PERK, HRI, and GCN2) that phosphorylate translation initiation factor eIF2α causing global changes in gene expression (Holcik and Sonenberg, 2005; Wek et al., 2006; Sonenberg and Hinnebusch, 2009). Previous work showed that stress granule assembly by G3BP1 differs across multiple ISR-dependent stressors (Aulas et al., 2017). This could suggest that G3BPs may have different roles across stress conditions. Moreover, the role of G3BPs in the regulation of gene expression during the ISR is not well understood.
In this study, we identified and investigated the function of the SG-deficient G3BP1V11A mutant. This mutated residue at the NTF2L domain does not disrupt dimerization of G3BP1, but rather its interaction with RNA-binding protein Caprin-1, leading to stress granule inhibition. To explore the role of G3BPs in mRNA expression during the stress response, we performed ribosome profiling (Ribo-seq), a technique that measures ribosome density on mRNAs (McGlincy and Ingolia, 2017), and total RNA sequencing (RNA-seq). We found that SG-deficient G3BP1V11A led to marginal changes in mRNA levels at steady state and under oxidative stress via arsenite (NaAs) treatment. To better understand differences between G3BP paralogues, we studied the function of G3BP1/2 on SG assembly upon activation of the ISR via oxidative or ER stress. By performing fluorescence microscopy and SG imaging, we found that G3BP paralogues assemble differently into SGs under ER stress. Under NaAs stress, all three G3BP paralogues robustly form SGs with similar properties. In contrast, while G3BP1 forms SGs poorly under thapsigargin (Tg) treatment, G3BP2B robustly assembles granules in cells. Moreover, we found that G3BP2 isoforms have a bigger impact on mRNA expression and translation compared with G3BP1 under ER stress. Finally, we found that G3BP2BV11A mutant disrupts G3BP2B SGs and leads to substantial changes in the expression of specific mRNAs both at steady state and under ER stress. Together, we found that mutating residue V11 on the NTF2L domain of G3BPs inhibits G3BP–Caprin-1 interaction, leading to a decrease in stress granule assembly and changes in gene expression during the ISR. Moreover, we found that G3BP paralogues are not functionally redundant under ER stress, due to differences in SG assembly and regulation of gene expression.
RESULTS
Key residues in the G3BP1 NTF2L domain are necessary for stress granule assembly
To better understand the role of stress granules during the stress response, we sought to block protein–protein interactions (PPIs) mediated by G3BPs that lead to SG assembly in cells. Previous work has shown that KO of endogenous G3BP1/2 inhibits the formation of stress granules under oxidative and ER stress (Kedersha et al., 2016; Yang et al., 2020). We validated G3BP KO suppresses SG assembly by IF against PABP foci, a common SG protein, in wild-type and G3BP1/2 KO cells under NaAs and Tg treatments, which cause oxidative and ER stress, respectively (Kedersha et al., 1999; Bertolotti et al., 2000; Kimball et al., 2003). As expected, we observed a significant decrease in the percentage of cells with PABP foci under both NaAs and Tg (Supplemental Figure S2). By performing live-cell imaging on G3BP1/2 KO cells expressing transgenic G3BP1 N-terminally tagged to monomeric EGFP and treated with 200 µM NaAs, we confirmed previous reports that truncating the intrinsically disordered regions of G3BP1, IDR2, and IDR3, disrupts stress granule assembly (Guillén-Boixet et al., 2020; Sanders et al., 2020; Yang et al., 2020) (Supplemental Figure S3, B–E). On the other hand, mutating the S149 residue located at the IDR1 of G3BP1 into phosphomimetic and phosphoincompetent amino acids did not decrease SG formation by G3BP1 under oxidative stress (Supplemental Figure S3, B–E), as reported previously (Panas et al., 2019).
G3BP1 IDRs are involved in multiple functions such as RNA binding (Yang et al., 2020). Therefore, the truncation of these domains can impair important functions of G3BPs outside of their role in SG formation, complicating the study of G3BP's function on mRNA expression. Instead, we sought to perturb PPIs mediated by G3BPs driving SG assembly by mutagenesis of amino acid residues in the NTF2L domain of G3BP1. The NTF2L domain is implicated in mediating interactions between G3BPs and other scaffold proteins, such as Caprin-1 and UBAP2L (Kedersha et al., 2016; Sanders et al., 2020; Yang et al., 2020; Schulte et al., 2023). In fact, the NTF2L domain's role in SG formation is exploited by viruses which outcompete interactions between G3BPs and host proteins to prevent stress granule assembly and improve viral replication (Panas et al., 2012, 2015; Schulte et al., 2016). We therefore generated 12 variants of G3BP1 including alanine substitutions at conserved amino acids (Figure 1, A and B). Within the NTF2L domain, these mutations were proximal to the G3BP1–Caprin-1 complex interface (Schulte et al., 2023) (Supplemental Figure S3F), and therefore potentially implicated in regulating this interaction during stress granule assembly.
FIGURE 1:
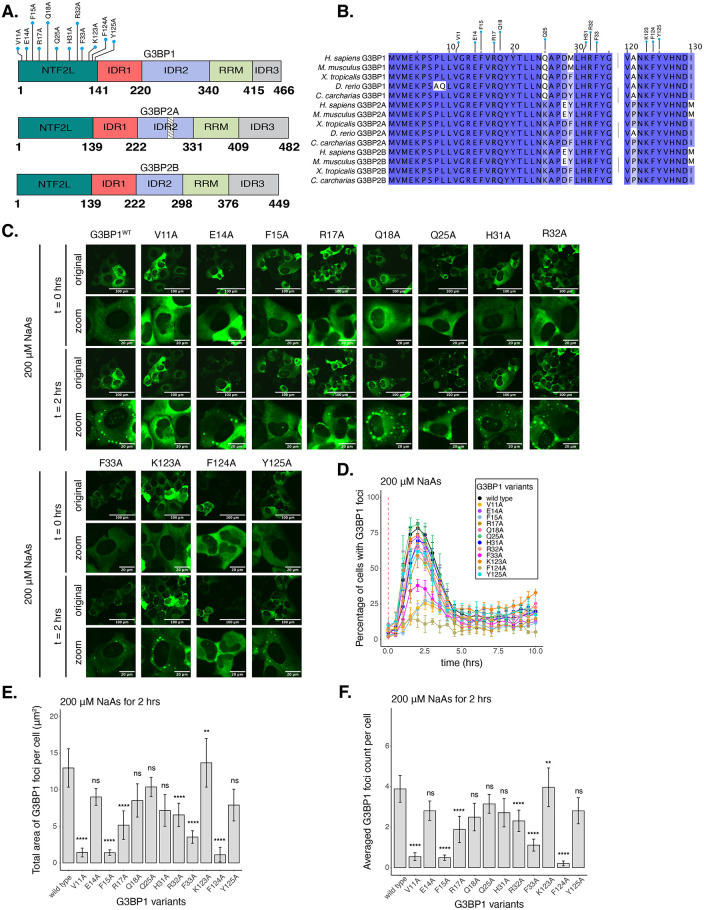
Select residues in the NTF2L domain are key for stress granule formation. (A) Schematic of G3BP1 (top) domains showing the location of NTF2L alanine substitutions. Schematics of G3BP2A (middle) and G3BP2B (bottom) are also shown. Striped box on G3BP2A shows the region that is spliced out of the G3BP2B IDR2. (B) Conservation of indicated G3BP NTF2L-domain regions across vertebrate species. (C) Images (20X objective) of cells expressing mEGFP-G3BP1 variants at t = 0 h and t = 2 h posttreatment with 200 µM NaAs. Puncta in the t = 2 h timepoint are stress granules. (D) Percentage of cells with G3BP1 foci. Vertical red dashed line shows when NaAs was added to cells. (E) Total area of G3BP1 foci per cell at 2 h under NaAs. (F) G3BP1 foci count per cell at 2 h under NaAs. Plots D–F are showing mean ± SEM across Nreplicates ≥ 3. P-values were calculated based on whole cell populations (ncells ≥ 100 per replicate) relative to G3BP1WT. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
To test the impact of NTF2L domain mutations on stress granules, we used live-cell microscopy (Figure 1C). We found that alanine substitutions of hydrophobic residues V11, F15, F33, or F124 caused a 50–80% decrease in the percentage of cells with G3BP1 foci at 2 h of 200 µM sodium arsenite treatment (Figure 1D). Foci area and count per cell were significantly decreased for these mutations (Figure 1, E and F), suggesting that these hydrophobic interactions play a crucial role in SG assembly. Mutation of charged residues R17 and R32 also significantly decreased G3BP1 foci, suggesting that they may mediate important molecular interactions, while most mutated polar residues such as E14, Q18, Q25, and Y125 did not significantly impact the assembly of G3BP1 granules under these conditions (Figure 1, D–F). Interestingly, K123A mutation caused a mild increase in G3BP1 foci area and count per cell (Figure 1, E and F). Prior work identified a mild effect of H31A (Schulte et al., 2023), which we also observed in our data (Figure 1, D–F). Expression of G3BP1 mutants did not deviate beyond 25% from the median expression of G3BP1WT, suggesting that changes in granule properties were not primarily due to differences in protein levels (Supplemental Figure S3G). Foci disassembled after 2 h of stress exposure, confirming the transient nature of G3BP1 granules under acute stress (Figure 1D; Supplemental Figure S3C).
Mutation of V11 in G3BP1 inhibits association with Caprin-1
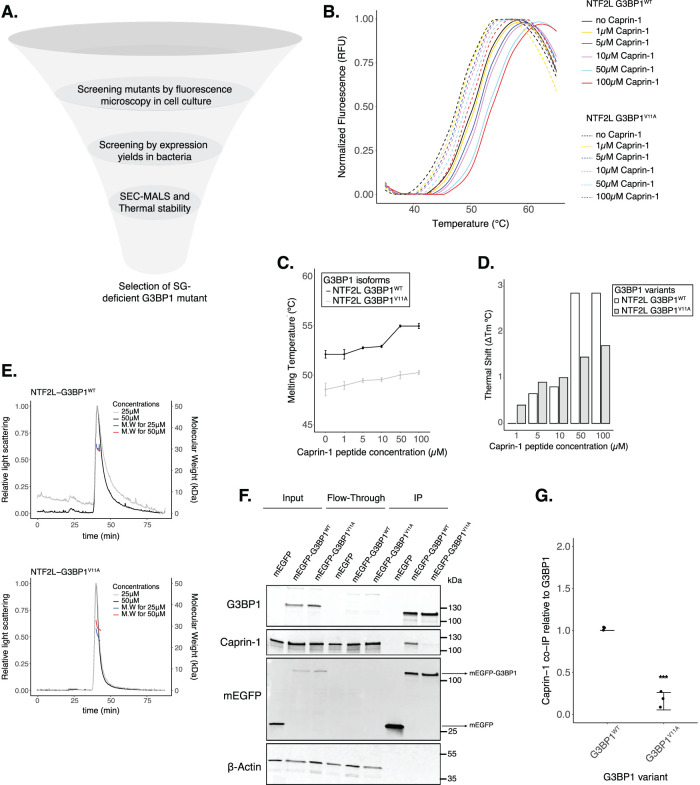
To narrow down SG-deficient mutants to study the function of G3BPs on mRNA expression, we characterized several mutants in vitro (Figure 2; Supplemental Figure S4). The NTF2L domain promotes SG assembly by forming a network with proteins such as Caprin-1 and UBAP2L, which increases the valency of interactions with RNA to drive RNP condensation upon translation arrest and RNA influx (Sanders et al., 2020; Yang et al., 2020). Recently, the complex between G3BP1 NTF2L domain and a Caprin-1 short linear motif was characterized as a critical mechanism for SG assembly (Schulte et al., 2023), giving us a better mechanistic understanding of this interaction. Most of the mutations we generated are located at the interface of the G3BP1—Caprin-1 complex (Supplemental Figure S3F) and are predicted to inhibit this interaction. However, substitution of hydrophobic amino acids found in this interface may also destabilize the entire NTF2L domain and generate a dysfunctional protein. To test this possibility, we expressed and purified recombinant NTF2L domain mutants and characterized protein stability (Supplemental Figure S4). Purification of G3BP1F15A and G3BP1F124A resulted in low yields due to low protein expression in contrast to G3BP1WT (Supplemental Figure S4, A–C). This suggests that F15A and F124A mutants may destabilize the NTF2L domain, thereby leading to decreased stress granules in cells. However, G3BP1V11A and G3BP1R32A expressed robustly and resulted in high protein yields. To quantitatively measure the stability of G3BP1 mutants, we performed DSF. Results revealed that G3BP1R32A, a mild SG-deficient mutant, did not significantly decrease the stability of G3BP1 in solution (Supplemental Figure S4, K and L). On the other hand, G3BP1V11A caused a thermal shift of 3.6°C relative to G3BP1WT (Figure 2, B and C), suggesting a mild decrease in stability by this mutant. However, we performed SEC-MALS to test dimerization of the NTF2L domain by G3BP1V11A, and we did not observe a disruption of G3BP1 dimers (Figure 2E). The NTF2L domain (1–139 residues) has a molecular weight of 15.9 kDa, which leads to a corresponding dimer of 30–32 kDa. G3BP1WT formed dimers of 30.5 (+/− 5.5%) kDa and 30.0 (+/− 2.1%) kDa at 25 µM and 50 µM, respectively. Similarly, G3BP1V11A formed dimers of 27.1 (+/− 2.1%) kDa and 32.1 (+/− 1.1%) kDa at 25 µM and 50 µM, respectively. This suggests that V11A mutation does not inhibit dimerization under the conditions tested.
FIGURE 2:
G3BP1V11A inhibits the G3BP1–Caprin-1 complex. (A) Diagram showing screening of G3BP1 mutants (image designed in biorender.com). (B) DSF melting curves for NTF2L domains of G3BP1WT (solid lines) and G3BP1V11A (dashed lines) supplemented with increasing Caprin-1 concentrations (1-5-10-50-100 µM). (C) Estimated melting temperatures (Tm) from curves on panel B. (D) Thermal shifts (ΔTm) relative to apo NTF2L-G3BP1 proteins. (E) SEC-MALS curves for G3BP1WT (upper) and G3BP1V11A (lower). (F) Western blot showing co-immunoprecipitation of the G3BP1–Caprin-1 complex in cells treated with 200 µM NaAs for 2 h. (G) Estimations of Caprin-1 band intensities from Western on panel F by normalizing on G3BP1 levels. Mean ± SD across Nreplicates = 3. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
To test binding of G3BP1 to a short linear motif of Caprin-1 (Schulte et al., 2023), we measured the thermal stability of the NTF2L domain in the presence of a Caprin-1 peptide (residues 356–386; 1–100 µM) via DSF (Figure 2, B–D). Caprin-1 induced a thermal shift of ∼3°C for G3BP1WT at 50 µM and 100 µM. However, Caprin-1 induced a thermal shift of 1–1.5°C for G3BP1V11A. V11 is surrounded by hydrophobic residues from the Caprin-1 peptide and within the NTF2L domain (Supplemental Figure S3F). Mutating V11 may lead to either mild misfolding of G3BP1 and/or inhibition of the G3BP1–Caprin-1 interaction. However, SEC-MALS data revealed that G3BP1V11A remains functional (Figure 2E). Overall, these data suggest that V11A might induce a moderate decrease in stability in vitro, but it still promotes dimerization of the NTF2L domain. Moreover, Caprin-1 interaction with wild-type protein is stronger than V11A, suggesting a decrease in G3BP1–Caprin-1 complex formation by V11A mutant.
To validate the inhibition of the G3BP1–Caprin-1 complex by G3BP1V11A in cell lysates, we performed a co-immunoprecipitation assay from cells treated with 200 µM NaAs for 2 h (Figure 2, F and G). Indeed, we observed a significant decrease in this complex with G3BP1V11A, suggesting that granule deficiency is caused by the loss of Caprin-1 interaction. Because we found G3BP1V11A to be a stable variant that cannot interact with Caprin-1, we decided to study the function of this mutant on mRNA expression and translation to better understand the role of stress granules during cellular stress.
SG-deficient G3BP1V11A marginally impacts mRNA levels under oxidative stress
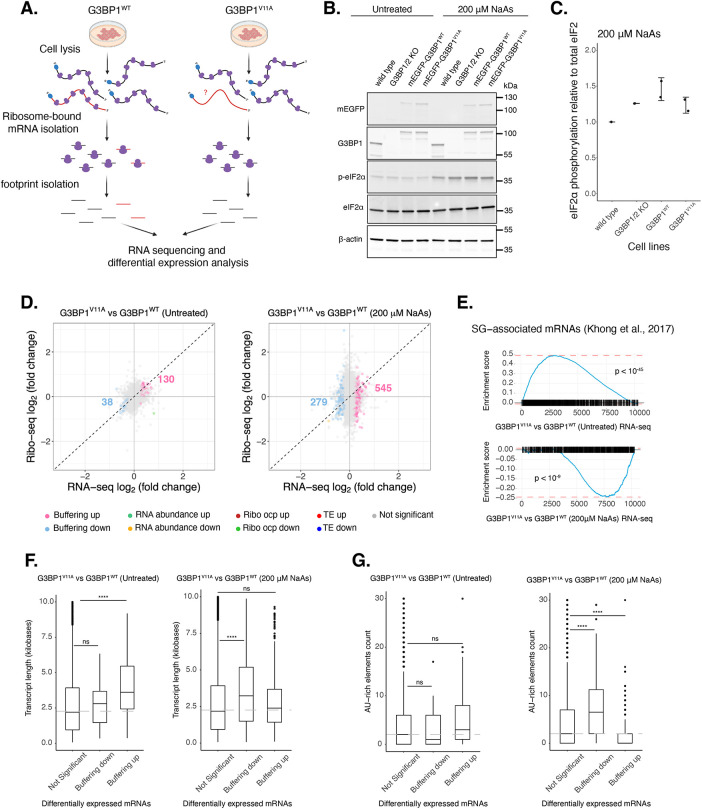
The role of stress granules during the ISR is not completely understood. To test the model that SG assembly leads to changes in gene expression, we measured global translation using Ribo-seq and total RNA sequencing in U-2OS G3BP1/2 KO cells expressing either transgenic G3BP1WT or G3BP1V11A under the ISR via oxidative stress (Figure 3). We first confirmed G3BP1 was expressed at comparable levels relative to wild-type cells by Western blot (Supplemental Figure S6, A and B). Then, we determined peak phosphorylation of eIF2α occurred around 1–2 h under oxidative stress (Supplemental Figure S6, C and D), and therefore harvested cells treated with 200 µM NaAs for 2 h. No difference in eIF2α phosphorylation was observed for cells expressing G3BP1WT and G3BP1V11A (Figure 3, B and C). Furthermore, no significant difference in the averaged translation efficiency of ISR canonical factors was observed between G3BP1 variants (Supplemental Figure S6, E and F), suggesting that stress granule deficiency does not affect ISR activation, as reported previously (Kedersha et al., 2016). To test reconstitution of G3BP1WT functionality relative to G3BPs in wild-type cells, we quantified SG assembly via IF by measuring PABP foci under NaAs (Supplemental Figure S6G). Approximately 40–50% of wild-type cells were positive for PABP foci at 2 h posttreatment. ∼25% of G3BP1WT expressing cells were positive for PABP foci (Supplemental Figure S6H). This suggests that G3BP1WT transgene partially reconstitute stress granule formation relative to endogenous G3BPs in wild-type cells. On the other hand, G3BP1/2 KO and G3BP1V11A substantially decreased the percentage of cells with PABP foci (Supplemental Figure S6H), as expected.
FIGURE 3:
G3BP1V11A leads to marginal changes in the expression of select mRNAs under arsenite stress. (A) Diagram showing Ribo-seq workflow (image designed in biorender.com). (B) Western blot showing levels of eIF2α phosphorylation across U-2OS wild-type cells, G3BP1/2 KO cells, and G3BP1/2 KO cells expressing either G3BP1WT or G3BP1V11A N-terminally tagged to mEGFP. Cells were treated either with water for control or 200 µM NaAs for 2 h. (C) Comparing levels of eIF2α phosphorylation under NaAs treatment across cell lines from Western on panel B. Mean ± SD across Nreplicates = 2. These lysates were used for Ribo-seq and total RNA-seq. (D) DGE plots for Ribo-seq and total RNA-seq. Cells expressing transgenic G3BP1V11A were compared with G3BP1WT cells at steady state (left) and under 200 µM NaAs for 2 h (right). Colored numbers on plots correspond to count of Buffering up (pink) and Buffering down (light blue) genes. (E) GSEA for SG-associated mRNAs in RNA-seq profiles at steady state (upper) and arsenite stress (lower) for G3BP1V11A versus G3BP1WT. (F) Box plots comparing transcript lengths for Not significant, Buffering down and Buffering up genes at steady state (left) and arsenite stress (right). (G) Box plots comparing count of AU-rich elements for Not significant, Buffering down and Buffering up genes at steady state (left) and arsenite stress (right).
G3BPs mediate RNA stability and expression under physiological and disease contexts (Alam and Kennedy, 2019; Laver et al., 2020; Kang et al., 2021). We determined both SG-independent and -dependent changes in mRNA levels and translation by comparing ribosome profiling and RNA-seq of G3BP1V11A with G3BP1WT at steady state and under 200 µM NaAs (Figure 3D). Genes with a significant Δ Ribo/RNA ratio were called TE down or TE up, if their translation efficiency decreased or increased, respectively. If changes in translation were due to corresponding changes in relative levels of RNA, genes were categorized RNA abundance up or RNA abundance down. On the other hand, genes that only changed at the level of RNA without significant changes in ribosome density were termed as Buffering up or Buffering down, if their relative abundances increased or decreased, respectively. We observed that V11A mutant mostly affected the levels of RNAs at steady state and under oxidative stress. Moreover, the number of Buffering up and Buffering down genes under stress increased 4-fold and 7-fold, respectively, relative to steady state. This suggests a bigger effect of V11A mutant on mRNA expression under a SG-dependent context.
G3BP1V11A impacts the levels of SG-associated mRNAs
Interestingly, a GSEA with previously identified SG-associated mRNAs (Khong et al., 2017) under NaAs showed that they are significantly activated (ES = 0.49, p < 10−45) by G3BP1V11A at steady state (Figure 3E). However, these genes were suppressed (ES = −0.24, p < 10−9) under arsenite stress. Furthermore, G3BP1V11A down-regulated the expression of significantly longer transcripts and mRNAs with higher counts of AU-rich elements (Figure 3, F and G). These two properties are common among mRNAs enriched in stress granules (Khong et al., 2017; Namkoong et al., 2018). This may suggest that G3BP1 may protect SG-associated mRNAs under NaAs in a SG-dependent manner. However, these data also demonstrate that V11A may affect the expression of SG-associated mRNAs independently of SGs at steady state.
G3BP paralogues form SGs differently under ER stress
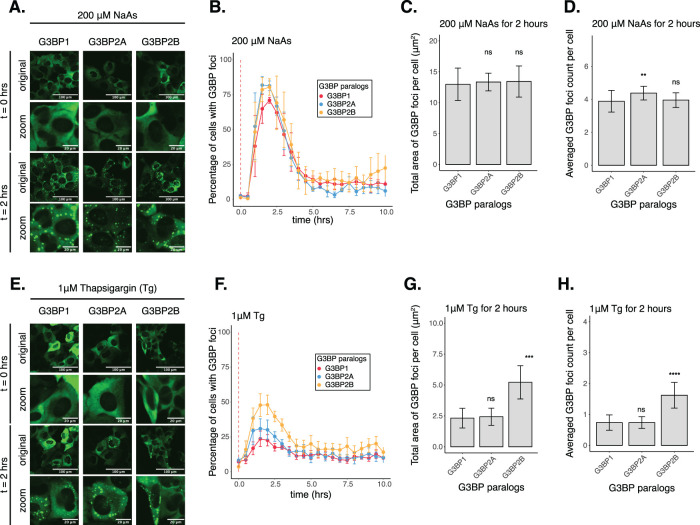
While our work thus far has focused on G3BP1, three paralogues of G3BPs have been identified in mammals (Kennedy et al., 2002). Even though G3BP paralogues are considered redundant in SG assembly, their functions have been found to differ in mTOR signaling regulation and the response against poliovirus infection (Dougherty et al., 2015; Prentzell et al., 2021). Furthermore, G3BP paralogues are differentially expressed across tissues and human diseases (Kennedy et al., 2002; Ali et al., 2021; Li et al., 2021). We therefore hypothesized that they may play differential roles in regulating translation during the ISR. To investigate this idea, we generated G3BP1/2 KO cell lines stably expressing G3BP1, G3BP2A, or G3BP2B fused to mEGFP. To test the role of G3BP paralogues under stress, we performed live-cell imaging on G3BP1/2 KO cells expressing transgenic G3BP paralogues N-terminal tagged with monomeric EGFP and treated with 200 µM NaAs to induce oxidative stress (Figure 4, A–D). G3BP paralogues exhibited minimal differences in the percentage of cells with G3BP foci (Figure 4B) and on the averaged count of foci per cell (Figure 4D). No significant differences were observed on the total area of G3BP foci per cell (Figure 4C). To validate our tagging strategy and confirm our results, we also performed live-cell imaging on G3BP1/2 KO cells expressing C-terminally tagged G3BP proteins (Supplemental Figure S7). These results suggest that G3BP1/2 paralogues are redundant under oxidative stress. Similar to G3BP1 granules, G3BP2 foci disassembled after 2 h of stress exposure, suggesting that G3BP2 SGs are transient in nature. Moreover, the percentage of cells with G3BP foci on Figure 4B was comparable to percentages measured via IF on wild-type cells (Supplemental Figure S9, A and B), suggesting functional reconstitution of transgenes relative to endogenous G3BPs.
FIGURE 4:
G3BP paralogues form stress granules differently under ER stress. (A) Images of cells expressing mEGFP-G3BP paralogues at t = 0 h and t = 2 h posttreatment with 200 µM NaAs. (B) Percentage of cells with G3BP foci. Vertical red dashed line shows when NaAs was added to cells. (C) Total area of G3BP foci per cell at 2 h under NaAs. (D) G3BP foci count per cell at 2 h under NaAs. (E) Images of cells expressing mEGFP-G3BP paralogues at t = 0 h and t = 2 h posttreatment with 1 µM Tg. (F) Percentage of cells with G3BP foci. Vertical red dashed line shows when Tg was added to cells. (G) Total area of G3BP foci per cell at 2 h under Tg. (H) G3BP foci count per cell at 2 h under Tg. Plots B–D and F–H are showing mean ± SEM across Nreplicates ≥ 3. P-values were calculated based on whole cell populations (ncells ≥ 100 per replicate) relative to G3BP1. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
Previous work indicates that properties of G3BP stress granules may differ across stress conditions (Aulas et al., 2017). Therefore, we also investigated the role of G3BPs in SG assembly under ER stress induced by 1 µM Tg (Figure 4, E–H; Supplemental Figure S8, A–D). The percentage of cells with G3BP1 foci under Tg was ∼30% lower than under arsenite stress (Supplemental Figure S8B). Stress granule total area and count were also smaller under Tg than arsenite (Supplemental Figure S8, C and D). Surprisingly, we found that G3BP2B-expressing cells formed more foci under ER stress than G3BP1 or G3BP2A (Figure 4F). We found that both the total area and count of G3BP2B foci per cell were significantly higher than G3BP1, while no significant differences were observed between G3BP1 and G3BP2A granules (Figure 4, G and H). This suggests that G3BP1 may not be the primary driver of SG formation under ER stress. This also suggests that G3BP2B may form functionally different granules under ER stress, and that the G3BP paralogues differ in their propensity to form SGs under different stressors.
Additionally, expression of G3BP2 isoforms did not deviate beyond 25% from the median expression of G3BP1 (Supplemental Figure S8, E and F), suggesting that changes in granule properties were not primarily due to differences in protein levels among different cell lines. Interestingly, endogenous G3BP1/2 paralogues in U-2OS wild-type cells did not form granules differently under either oxidative or ER stress, as revealed by IF (Supplemental Figure S9). This may suggest that G3BP2 cooperates with G3BP1 to form granules under ER stress by potentially forming G3BP heterodimers in cells, as reported previously (Matsuki et al., 2013).
G3BP1/2 paralogues regulate mRNA expression and translation differently under ER stress
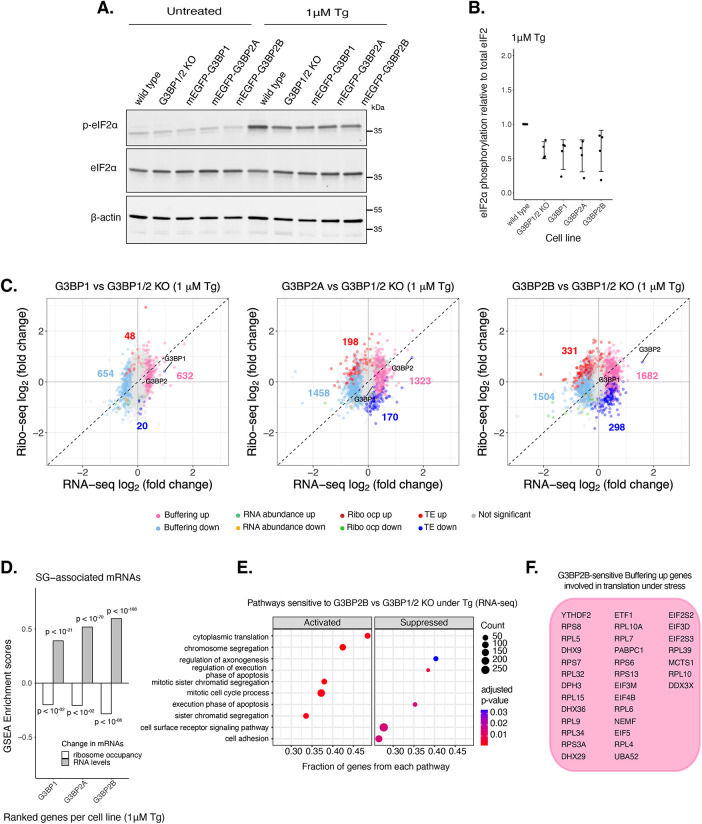
Given the differences in SG formation across paralogues, we hypothesized that they may play distinct roles in regulating mRNA expression and translation under ER stress. Therefore, we performed Ribo-seq and RNA-seq with G3BP1/2 KO cells stably expressing each transgenic G3BP paralogue at comparable levels as revealed by Western blot for mEGFP (Supplemental Figure S10, A and B). G3BP2A/B levels were approximately three to four times higher than endogenous G3BP2 levels in wild-type cells (Supplemental Figure S10D). This expression level was selected to match the levels of transgenic G3BP1 in G3BP1/2 KO cells to maintain a constant total amount of G3BP protein, which was similar to the total amount of all G3BP protein in wild-type cells as revealed by Ribo-seq TPMs (Supplemental Figure S10I). Phosphorylation of eIF2α under 1 µM Tg peaked around 1–2 h posttreatment (Supplemental Figure S12, A and B), therefore cells were harvested at 2 h for ribosome profiling. ISR activation was validated by the increased expression of ATF4, GADD34, and CHOP (Supplemental Figure S12C). Expression of different G3BPs does not affect activation of the ISR, as no significant differences in eIF2α phosphorylation or the average translation efficiency of ISR canonical factors were observed among G3BP paralogue cell lines (Figure 5, A and B; Supplemental Figure S12D).
FIGURE 5:
G3BP paralogues impact mRNA expression differently under ER stress. (A) Western blot showing levels of eIF2α phosphorylation across U-2OS wild-type cells, G3BP1/2 KO cells, and G3BP1/2 KO cells expressing either G3BP1, G3BP2A, or G3BP2B N-terminally tagged to mEGFP. Cells were treated either with DMSO for control or 1 µM Tg for 2 h. (B) Comparing levels of eIF2α phosphorylation under Tg treatment across cell lines from Western on panel A. mean ± SD across Nreplicates = 4. (C) DGE plots for Ribo-seq and total RNA-seq. Cells expressing either transgenic G3BP1, G3BP2A, or G3BP2B were compared with G3BP1/2 KO cells under 1 µM Tg for 2 h. Colored numbers correspond to count of Buffering down (light blue), Buffering up (pink), TE up (red), and TE down (blue) genes. (D) Enrichment scores for SG-associated mRNAs in RNA-seq (RNA levels) and Ribo-seq (ribosome occupancy) profiles for G3BP1, G3BP2A, and G3BP2B. (E) GSEA for activated and suppressed pathways by G3BP2B under 1 µM Tg. (F) List of G3BP2B-sensitive Buffering up genes that are involved in the “cytoplasmic translation” pathway identified in panel E.
By comparing each G3BP paralogue relative to G3BP1/2 KO cells, we found that the number of differentially translated mRNAs (Δ TE) sensitive to G3BP2A/B isoforms was higher than those sensitive to G3BP1, whereas G3BP2B has the biggest impact under ER stress (Figure 5C). Furthermore, there was a higher correlation of Δ TE genes between G3BP2A and G3BP2B profiles (0.81), than G3BP1 compared with either G3BP2 isoform (0.50 and 0.43, for 2A and 2B, respectively) (Supplemental Figure S13A). This implies that G3BP1/2 paralogues regulate differently the translation of transcripts under ER stress. We also found that buffered mRNA expression (Δ Buffering) is different among paralogues where G3BP2 isoforms affected the RNA levels of a higher number of transcripts compared with G3BP1 (Figure 5C). The correlation of RNA-seq profiles was higher between G3BP2 isoforms (0.84) than G3BP1 compared with either G3BP2A or G3BP2B (0.55 and 0.36, respectively) (Supplemental Figure S13B). This implies that G3BP2A/B isoforms also have a greater impact on RNA levels than G3BP1 under ER stress. Moreover, GSEA with previously identified SG-associated mRNAs under Tg (Namkoong et al., 2018) showed that G3BPs differentially suppressed their translation under ER stress (Figure 5D). G3BP2 isoforms also led to increased RNA levels of SG-associated mRNAs compared with G3BP1. This is in accordance with a higher number of SGs being formed by G3BP2 isoforms under ER stress (Figure 4, E–H). This result suggests that G3BP paralogues differentially suppress translation of SG-associated transcripts, while stabilizing their RNA levels, potentially via the formation of stress granules under ER stress.
G3BP2B increases the RNA levels of specific genes
GSEA revealed that G3BP2B significantly activated the expression of genes that may be involved in multiple pathways such as cell cycle regulation and mRNA translation (Figure 5, E and F). In fact, previously identified transcripts enriched in granules and known to be involved in cell growth and survival (Namkoong et al., 2018), such as RICTOR and BRCA1, were categorized as Buffering up G3BP2B-sensitive mRNAs (Supplemental Figure S13, C and D). BRCA1 did not significantly change for G3BP1; however, both BRCA1 and RICTOR were also identified as G3BP2A-sensitive Buffering up genes. Additionally, dead box helicase DDX3X, an essential gene involved in translation initiation and cell growth (Wilkins et al., 2024), was identified as a Buffering up gene sensitive to all G3BP paralogues (Supplemental Figure S13E). We performed differential gene expression (DGE) analysis of wild-type cells relative to G3BP1/2 KO cells under Tg (Supplemental Figure S13F). We observed that wild-type cells increased the expression of G3BP-sensitive genes such as RICTOR, DDX3X, and transcripts involved in translation. Hypergeometric analysis revealed a significant overlap between Buffering up and Buffering down genes sensitive to each transgenic G3BP and wild-type cells (Supplemental Figure S13G). This suggests that transgenic cell lines reconstitute the expression of activated genes in wild-type cells. These results highlight both the potential differences and similarities between G3BP paralogues on regulating translation and RNA levels of select transcripts under ER stress.
G3BP2BV11A leads to SG-independent and -dependent changes in mRNA levels
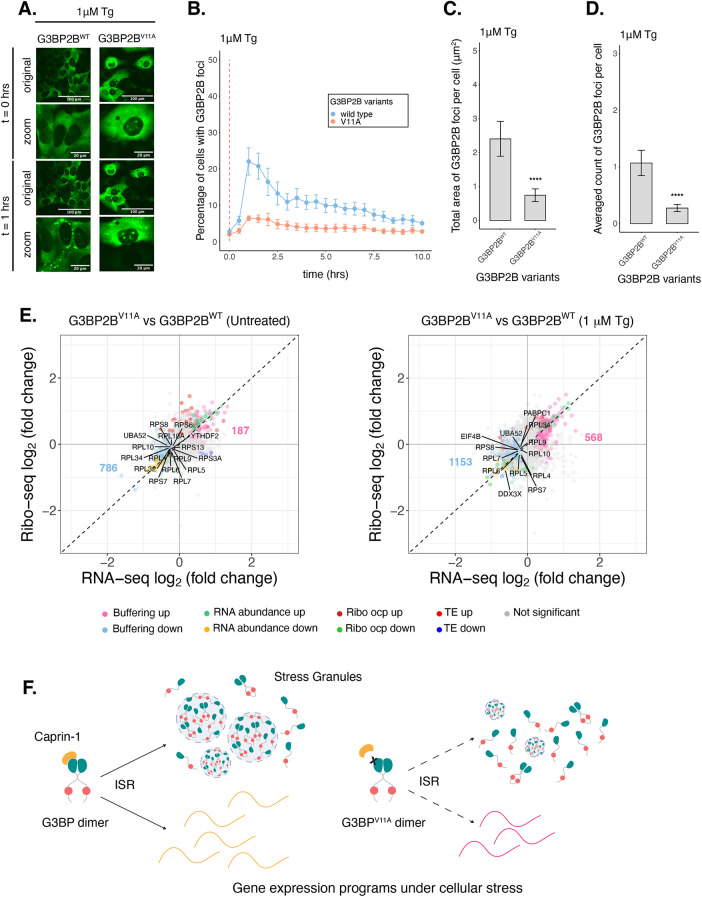
To determine the potential impact of G3BP2B-SGs on gene expression under ER stress, we mutated V11 on G3BP2B by site-directed mutagenesis, and performed live-cell imaging on G3BP1/2 KO cells expressing either G3BP2BWT or G3BP2BV11A N-terminally tagged with monomeric EGFP (Figure 6A). We found that G3BP2BV11A-expressing cells exhibited a reduction of the percentage of cells with G3BP2B foci under both oxidative and ER stress (Figure 6B; Supplemental Figure S15B). The total area and count of foci per cell was also significantly decreased (Figure 6, C and D; Supplemental Figure S15, C and D). This suggests that PPIs with the V11 residue are critical for G3BP2B SG formation during the stress response. Expression of G3BP2BV11A mutant did not deviate beyond 25% from the median expression of G3BP2BWT (Supplemental Figure S15E), suggesting that changes in granule properties were not primarily due to differences in protein levels.
FIGURE 6:
G3BP2BV11A leads to substantial changes in the expression of select mRNAs under Tg. (A) Images of cells expressing mEGFP-G3BP2B variants at t = 0 h and t = 1 h posttreatment with 1 µM Tg. (B) Percentage of cells with G3BP2B foci. Vertical red dashed line shows when Tg was added to cells. (C) Total area of G3BP2B foci per cell at 1 h under Tg. (D) G3BP2B foci count per cell at 1 h under Tg. Plots B–D are showing mean ± SEM across Nreplicates ≥ 3. P-values were calculated based on whole cell populations (ncells ≥ 100 per replicate). * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001. (E) DGE plots for Ribo-seq and total RNA-seq. Cells expressing G3BP2BV11A were compared with G3BP2BWT cells at steady state or under 1 µM Tg for 2 h. (F) Model showing the effect of G3BPV11A mutation on stress granule assembly and gene expression programs during the ISR.
We then performed Ribo-seq and RNA-seq with G3BP1/2 KO cells stably expressing either G3BP2BWT or G3BP2BV11A and harvested at steady state or after 2 h of Tg treatment (Figure 6E). To minimize artifacts caused by higher levels of transgenic G3BP2B, we developed cell lines expressing comparable levels of G3BP2B relative to wild-type cells as measured by Western blot (Supplemental Figure S15, F and G). eIF2α phosphorylation did not differ between cells expressing G3BP2BWT and G3BP2BV11A (Supplemental Figure S15H) and average translation efficiency of ISR canonical factors was similar across cell lines (Supplemental Figure S15, I and J). To test reconstitution of G3BP2BWT functionality relative to G3BPs in wild-type cells, we quantified SG assembly via IF by measuring PABP foci under Tg (Supplemental Figure S14E). Approximately >50% of wild-type cells were positive for PABP foci at 2 h posttreatment. ∼25% of G3BP2BWT expressing cells were positive for PABP foci (Supplemental Figure S14F). This suggests that G3BP2BWT transgene partially reconstitute stress granule formation relative to endogenous G3BPs in wild-type cells. On the other hand, G3BP1/2 KO and G3BP2BV11A substantially decreased the percentage of cells with PABP foci (Supplemental Figure S14F), as expected.
Comparing G3BP2BV11A with G3BP2BWT via DGE analysis revealed that V11A mutant affects mRNA expression under ER stress (Figure 6E). This may suggest that V11A regulates gene expression in a SG-dependent manner. However, we found that G3BP2BV11A also affected expression of transcripts at steady state. This implies that PPIs disrupted by this mutation have important roles for gene expression independently of stress granules. The number of Buffering up and Buffering down genes affected by G3BP2BV11A under stress were 3-fold and 1.5-fold higher, respectively, relative to genes differentially expressed at steady state. This suggests a bigger impact of G3BP2BV11A on gene expression during ER stress. Overall, these results suggest that G3BP2B regulates gene expression both dependently and independently from stress granule assembly.
G3BP2BV11A changes the expression of genes involved in global mRNA translation
DGE analysis revealed that G3BP2BV11A decreased gene expression of components of the translational machinery both at steady state and under ER stress, such as DDX3X (Figure 6E). This implies that PPIs regulated by G3BP2B are relevant for the expression of specific mRNAs. The ISR leads to global changes in gene expression programs to promote cell survival during cellular stress. This can lead to a redistribution of metabolic fates for specific mRNAs. We propose a hypothesis where G3BPs may stabilize RNA levels of essential transcripts during the ISR, such as genes involved in translation, to enhance protein synthesis during recovery from stress. To test this hypothesis, we measured global protein synthesis via an OPP detection assay before, during, and after Tg treatment on G3BP1/2 KO, G3BP2BWT, and G3BP2BV11A expressing cells (Supplemental Figure S16). As expected, we observed a substantial decrease (>30%) in OPP signal at 1 h Tg treatment relative to steady state for all cell lines, demonstrating global inhibition of protein synthesis during ER stress. Furthermore, we observed a mild increase in protein synthesis recovery rate at 4 h of recovery from stress in cells expressing G3BP2BWT relative to either G3BP1/2 KO or G3BP2BV11A cells (Supplemental Figure S16D). This supports a model where G3BP2B potentiates global protein synthesis during recovery from stress, presumably via the RNA stabilization of components from the translational machinery.
DISCUSSION
To perturb molecular interactions mediated by G3BPs, we introduced alanine substitutions along the NTF2L domain of G3BP1. By performing fluorescence microscopy and SG imaging, we found that hydrophobic residues V11, F15, F33, and F124 have substantial impact on granule assembly under arsenite stress (Figure 1). The NTF2L domain mediates PPIs with proteins such as USP10, Caprin-1, and UBAP2/2L (Sanders et al., 2020; Yang et al., 2020; Schulte et al., 2023). Macromolecules such as Caprin-1 act as bridges that facilitate SG assembly (Sanders et al., 2020). Binding between G3BP1 and Caprin-1 was characterized previously, where the NTF2L domain was shown to interact with Caprin-1 via a short linear motif (Schulte et al., 2023). NTF2L residues explored in this study are proximal to this interface. Hence, we found that G3BP1V11A mutant inhibits the G3BP1–Caprin-1 complex in cells under NaAs, without substantially affecting protein stability or its homodimerization in solution (Figure 2). However, we also contemplate the possibility that other G3BP PPIs such as the G3BP–UBAP2/2L complex may be perturbed by this mutant. Thus, we emphasize the need for more work to expand our understanding of this mutant and its relevance in mediating protein networks by G3BPs.
The role of stress granules on gene expression has remained uncertain. Here we found that stress granule deficient G3BP1V11A mutant leads to marginal changes in mRNA expression under NaAs and does not impact levels of ISR activation, confirming that the ISR is not regulated by G3BP expression or stress granule formation (Figure 3). We observed a significant down-regulation of SG-associated transcripts by G3BP1V11A, suggesting that stress granules may regulate RNA levels during stress. However, we also found that G3BP1V11A can impact mRNA expression at steady state albeit at a lesser degree compared with arsenite stress. This suggests that V11A mutant may be impacting SG-independent functions of G3BP1. Even though these data imply that SGs are directly involved in the regulation of mRNA levels during the ISR, we cannot rule out the possibility that SG-independent functions of G3BP1 may be contributing to changes in gene expression observed in our experiments.
In this work, we found that G3BP1/2 paralogues form SGs similarly under arsenite stress (Figure 4, A–D) while differentially under ER stress induced by thapsigargin treatment. Under Tg stress, G3BP2B formed foci in a higher percentage of cells, also leading to bigger granule sizes and numbers per cell compared with either G3BP1 or G3BP2A (Figure 4, E–H). These results imply that SGs form differently under different stress conditions, as reported previously (Aulas et al., 2017). Composition and architecture of G3BP1 granules has been previously studied by proteomics, RNA isolation, and super-resolution microscopy techniques (Jain et al., 2016; Khong et al., 2017; Youn et al., 2018; Seo and Kleiner, 2023). Our findings motivate performing similar studies focused on G3BP2 granules to better understand the differences and heterogeneity of these condensates from G3BP1 SGs.
Interestingly, we found that U-2OS cells, which naturally express G3BP1, G3BP2A, and G3BP2B, do not form granules differently under arsenite versus ER stress (Supplemental Figure S9). A previous study showed that G3BP1/2 can form heterodimers in cells (Matsuki et al., 2013). G3BP2, with a higher propensity to form condensates due to its increased multivalency (Guillén-Boixet et al., 2020), potentially dimerizes with G3BP1 and they cooperate to increase stress granule assembly. Our findings motivate future work to further understand possible cooperative behavior between G3BP paralogues and how this impacts cellular stress response.
Our results also suggest that G3BP1/2 paralogues may have different functions specifically under ER stress. We performed Ribo-seq to test the role of G3BPs on mRNA expression during ER stress, and we found that G3BP2B regulates the expression of more genes compared with either G3BP1 or G3BP2A (Figure 5C). These observations may be explained by a higher propensity of G3BP2B for SG-assembly. To test the role of G3BP2B SGs on mRNA expression, we performed Ribo-seq in cells expressing G3BP2BV11A, which led to deficiency in G3BP2B foci assembly under both oxidative and ER stress (Figure 6, A–D; Supplemental Figure S15, A–D). We found that G3BP2BV11A leads to substantial changes in gene expression under ER stress. G3BP2B-sensitive genes identified on Figure 5 were significantly down-regulated by V11A mutation. Some of these genes are part of the translational machinery, such as DDX3X and ribosomal proteins. This led us to hypothesize that G3BP2B may be involved in regulating protein synthesis to promote cell survival and recovery from stress. To test this model, we performed an OPP detection assay, and we observed a slight increase in the recovery of protein synthesis by G3BP2BWT relative to either G3BP2BV11A or G3BP1/2 KO cells at 4 h poststress. Overall, this could imply that G3BP2B granules play a role in regulating gene expression to promote recovery from stress. However, we also found that G3BP2BV11A affects gene expression at steady state. For this reason, our results also suggest that G3BP2BV11A may affect mRNA levels in a SG-independent manner. Furthermore, more follow-up experiments are needed to further validate changes in expression of specific genes by G3BPs.
Here, we propose that stress granules impact mRNA expression and translation during the ISR. However, there are alternate mechanisms that are potentially playing a direct role on gene expression. Mutating V11 may inhibit interaction networks between G3BPs and proteins critical for mRNA regulation both inside and outside of stress granules. Our work identifies a relationship between perturbations to G3BP PPIs that are critical for stress granule assembly and gene expression changes under stress. However, more biochemical work is needed to further characterize the mechanism of this G3BPV11A mutant and to better understand the extent of its function on stress granule formation and mRNA regulation.
In conclusion, we identified G3BPV11A to study the function of G3BP stress granules. Dysregulation of SGs is correlated to the progression of neurodevelopmental diseases, cancer, and viral infection (Alam and Kennedy, 2019; Wolozin and Ivanov, 2019; Mukhopadhyay and Zhou, 2023). Therefore, mutagenesis of V11 could be utilized to better understand the role of G3BP SGs in these contexts. Finally, we revealed that G3BPs lead to differential mRNA expression, translation, and SG formation, suggesting different roles of G3BP paralogues during the ISR.
MATERIALS and METHODS
Request a protocol through Bio-protocol
Plasmids
All G3BP paralogues (G3BP1, G3BP2A, and G3BP2B) cDNAs were cloned by Gibson assembly (NEB, E2611L) into either an mEGFP-C1 (Addgene, 54759) or mEGFP-N1 (Addgene, 54767) plasmid with Kanamycin resistance cassettes. Fusion proteins were interspaced with a glycine-serine (GS) linker of eight residues to improve folding and stability of flanking domains. G3BP1/2 mutants were generated by site directed mutagenesis. For lentivirus expression, these synthetic constructs were cloned by Gibson assembly into a lentivirus backbone with a SFFV promoter, and an ampicillin resistance cassette for bacterial selection. For the generation of cell lines with labeled nuclei, a H2B-mCherry construct cloned in a lentivirus backbone was kindly shared by Dr. Xiaokun Shu (University of California, San Francisco, USA). For in vitro reconstitution, NTF2 domain of G3BP1 variants was cloned into a pET28 MBP-TEV bacterial backbone (Addgene, 69929) with Gibson assembly.
Cell culture
Cells were grown in DMEM/Ham's F-12 media, supplemented with 10% FBS and 1x of a penicillin streptomycin solution (100x, 10,000 I.U. penicillin [per ml], 10,000 µg/ml streptomycin) and kept at 37°C, 5% CO2. Cells were tested for Mycoplasma and maintained at a low passage number.
Immunofluorescence
U-2OS wild-type and U2OS G3BP1/2 KO cells (Kedersha et al., 2016) were seeded at a 40–50% confluency in triplicate in a glass-bottom 96-well plate with #1.5 cover glass (Cellvis, P96-1.5H-N) in DMEM/Ham's F-12 media, supplemented with 10% FBS and 1x of a penicillin streptomycin solution (100x, 10,000 I.U. penicillin [per ml], 10,000 µg/ml streptomycin). Cells were incubated overnight at 37°C, 5% CO2. Then, they were treated with 100 µl of 200 µM sodium arsenite (LabChem, LC229001) or 1 µM thapsigargin (Millipore Sigma, 586005-1MG) for 2 h to induce oxidative and ER stress, respectively. Treated media was removed and cells were washed once with ice-cold 1X PBS. Cells were fixed in 4% paraformaldehyde (Electron Microscopy Sciences, 15710) for 10 min at room temperature. Samples were washed three times with ice-cold PBS, incubated with 0.1% Triton x-100 at 4°C for 10 min, and followed with three more washes with PBS at 4°C for 5 min. Blocking was performed under 1% BSA in PBS-T solution for 30 min at 4°C. Primary antibodies for PABP (Abcam, ab21060, 1:1000), G3BP1 (Proteintech, 13057-2-AP, 1:2000), and G3BP2 (Abcam, ab86135, 1:2000) were incubated in 1% BSA PBS-T overnight at 4°C. Then, samples were washed three times with ice-cold PBS for 5 min, and incubated with secondary antibody, Goat anti-Rabbit IgG Alexa Fluor 488 nm (Invitrogen, A11008, 1:2000) for 1 h at room temperature in the dark in 1% BSA PBS-T. Finally, samples were washed three more times in PBS for 5 min in the dark, and stained with 1x Hoechst solution (Bio-Rad, 1351304) at room temperature for 15 min in the dark. Images were acquired by capturing 5 fields of view per well using a 20x objective (Air objective; N.A. 0.75, W.D 1 mm), 100 ms of exposure time for 405 nm and 488 nm lasers at 25% power, on a Nikon CSU-W1/SoRa spinning disk confocal microscope.
Generation of stable cell lines
U-2OS G3BP1/2 KO cells (Kedersha et al., 2016) seeded in a 6-well plate were infected at a 60–70% confluency with lentivirus expressing mEGFP-GS-G3BP variants (G3BP1, G3BP2A, and G3BP2B, G3BP1V11A, G3BP2BV11A) and coincubated with 8 µg/ml polybrene (Millipore Sigma, TR-1003-G). Cells were spun down at 1000 × g, 30°C for 2 h. Then, infected cells were incubated at 37°C, 5% CO2 for 72 h. Infected cells were sorted twice to achieve mEGFP positive populations.
Live-cell imaging
U-2OS G3BP1/2 KO (Kedersha et al., 2016) cells at a 60–70% confluency were infected with lentivirus expressing H2B-mCherry plasmids obtained from the Shu lab at UCSF. Positive mCherry single-cell populations were sorted. U-2OS H2B-mCherry positive G3BP1/2 KO cells were seeded at a 40–50% confluency in a 96-well plate in DMEM/Ham's F-12 media, supplemented with 10% FBS and 1x of a penicillin streptomycin solution (100x, 10,000 I.U. penicillin [per ml], 10,000 µg/ml streptomycin). Cells were incubated overnight at 37°C, 5% CO2. Cells were infected with lentivirus expressing mEGFP-GS-G3BP variants and coincubated with 8 µg/ml polybrene. Four different concentrations of virus (1-5-10-50 µl) were titered per G3BP variant to achieve a similar range of G3BP expression levels between proteins. Cells were spun down at 1000 × g, 30°C for 2 h and incubated at 37°C, 5% CO2. After 72 h of infection, ∼5000 cells were pooled and seeded in a glass-bottom 96-well plate with #1.5 cover glass in triplicates and incubated overnight at 37°C, 5% CO2 in no phenol red DMEM (Mediatech 17-205-CV) supplemented with 10% FBS, 1x of a penicillin streptomycin solution (100x, 10,000 I.U. penicillin [per ml], 10,000 µg/ml streptomycin), autoclaved 25 mM HEPES buffer, and 1x Glutamax (Thermo Fisher Scientific, 35-050-061). G3BP condensation was induced with either 200 µM sodium arsenite or 1 µM thapsigargin. To account for G3BP condensation induced by blue light, cells were also treated with DMSO. Images were captured every 30 min for 10 h at a 20x magnification (Air objective; N.A. 0.75, W.D 1 mm), 100 ms of exposure time for 561 nm and 488 nm lasers, on a customized Ti inverted Nikon microscope with a Spectral Applied Research LMM5 laser merge module, and a Borealis modified Yokogawa CSU-X1 spinning disk head, as described previously (Stehbens et al., 2012).
O-propargyl puromycin protein synthesis assay
U-2OS cells at a 40–50% confluency were seeded in triplicate in a glassed-bottom 96-well plate with #1.5 cover glass (Cellvis, P96-1.5H-N) in DMEM/Ham's F-12 media, supplemented with 10% FBS and 1x of a penicillin streptomycin solution (100x, 10,000 I.U. penicillin [per ml], 10,000 µg/ml streptomycin). Cells were incubated overnight at 37°C, 5% CO2. Cells were treated with 1 µM thapsigargin for 1 h. For recovery, cells were replenished with fresh medium and incubated for 1–4 h. OPP assay was performed as recommended by manufacturer's instructions (Thermo Fisher Scientific, C10458). O-propargyl puromycin (OPP) analogue was detected by a click reaction with an Alexa Fluor 594 picolyl azide. Images were acquired by capturing 5 fields of view per well at a 20x magnification (Air objective; N.A. 0.75, W.D 1 mm), 100–200 ms of exposure time for the 405 nm, 488 nm, and 561 nm lasers at 25–50% power, on a Nikon CSU-W1/SoRa spinning disk confocal microscope.
Ribosome profiling library construction
Ribo-seq library preparation was performed as described previously (Venkataramanan et al., 2021). Briefly, cells at 80–90% confluency in a 15-cm dish were incubated with either 200 µM sodium arsenite or 1 µM thapsigargin for 2 h, then treated with 50 µg/ml cycloheximide (CHX) for 1 min, and harvested. Cells were also harvested after treatment with either water or DMSO, as controls. Then, harvested cells were washed gently with PBS containing 50 µg/ml CHX and lysed in ice-cold lysis buffer (20 mM Tris pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1% vol/vol Triton x-100, 1 mM DTT, 20-25 U/mL TURBO DNase [Thermo Fisher Scientific, AM2238], 100 µg/ml CHX). Cells were triturated with a 26-gauge needle. Lysate was recovered after spinning for 10 min, 20,000 × g, at 4°C. Lysates were treated with RNase I (Ambion, 100 U/µl) at room temperature for 45 min in slow agitation. Then, treated with SUPERase Inhibitor (Ambion, 20 U/µl) on ice. Monosomes were recovered by size exclusion chromatography (SEC; Illustra MicroSpin Columns, S-400 HR, VWR, 95017-619), and footprint RNA fragments were extracted from the flow-through using a Direct-zol kit (Zymo Research). Gel slices of RNA fragments with sizes between 26 and 34 nt were excised from a 15% polyacrylamide TBE-Urea gel (Thermo Fisher Scientific, EC68855BOX). Eluted RNA was treated with T4 PNK and preadenylated linker was ligated to the 3′ end using T4 RNA ligase 2 truncated KQ (NEB, M0373L). Linker-ligated RNA was reverse transcribed with Protoscript II (NEB, M0368L) for 30 min at 50°C. Template RNA was hydrolyzed with 1M NaOH for 20 min at 70°C. Gel slices of reverse transcribed cDNA around 105 nt long, were excised from 15% polyacrylamide TBE-Urea gel. Eluted cDNA was circularized with CircLigase II ssDNA ligase (Lucigen, CL9021K) for 1 h at 60°C. rRNA was depleted with biotinylated oligos (Ingolia et al., 2012). cDNA libraries were amplified with different reverse indexing primers per sample. Libraries were quantified and checked for quality using a Qubit fluorimeter and Bioanalyzer (Agilent) and sequenced on HiSeq 4000 or NovaSeqX sequencing systems.
RNA-seq library construction
RNA-seq library preparation was performed by extracting RNA from 25 µl of lysate with a Direct-zol kit. rRNA was depleted with a NEBNext rRNA Depletion Kit (NEB, E7400), and following manufacturer's instructions. Libraries were prepared with a NEBNext Ultra II Directional RNA Library Prep Kit of Illumina (E7760), and following manufacturer's instructions. cDNA libraries were amplified with multiplex oligos for Illumina sequencing (NEB, E6609S). Libraries were quantified and checked for quality using a Qubit fluorimeter and Bioanalyzer (Agilent) and sequenced on HiSeq 4000 or NovaSeqX sequencing systems (single end, 65 nt reads and 100 nt reads, respectively).
Western blot
U-2OS cells, at a 70–80% confluency, were harvested by scraping from a 6-well plate. Cells were lysed and rotated in RIPA buffer (20 mM Tris-HCl at pH 7.5–7.7, 150 mM NaCl, 1% NP-40, 0.5% DOC, 0.05% SDS) supplemented with a protease inhibitor cocktail (Sigma-Aldrich, 4693159001) for 30 min at 4°C. Samples were analyzed by SDS–PAGE electrophoresis. G3BP1 (Bethyl, A302-033A, 1:1000), G3BP2 (Abcam, ab86135, 1:1000), eIF2⍺ (Cell Signaling Technology, 9722S, 1:1000), p-eIF2⍺ (Abcam, ab32157, 1:1000), GFP (Thermo Fisher Scientific , A-11122, 1:1000), Caprin-1 (Proteintech, 15112-1-AP, 1:1000), β-actin (Abcam, ab184092, 1:5000) were detected by Western blot. Secondary antibody (LI-COR Biosciences, 926-32211, 1:10,000) was detected with a LI-COR Odyssey DLx imaging system.
Co-immunoprecipitation of G3BP1–Caprin-1 complex
U-2OS cells, at a 70–80% confluency, were treated with 200 µM NaAs for 2 h and harvested by scraping from a 10-cm plate. Cells were lysed in lysis buffer (20 mM Tris-HCl pH 7.7, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.5% NP-40, 10% glycerol) containing 1 U/µL RNAse I and a protease inhibitor cocktail. Cells were rotated for 30 min at 4ºC, and the lysate was cleared by centrifugation at 5000 rpm for 5 min at 4ºC. GFP-trap agarose beads (ChromoTek, gta-10) were equilibrated in ice-cold wash/dilution buffer (20 mM Tris-HCl pH 7.7, 150 mM NaCl, 5 mM MgCl2). Lysates were rotated end-over-end with the equilibrated beads for 2 h at 4ºC. After binding, beads were washed with a wash/dilution buffer twice. Beads were then resuspended in SDS buffer (5% β-mercaptoethanol, 30% Glycerol, 10% SDS, 250 mM Tris-Cl, pH 6.8) and analyzed by SDS–PAGE electrophoresis and Western blot.
Protein expression and purification
BL21 (DE3) Escherichia coli bacteria were transformed with NTF2L domain of G3BP1 variants and cultivated in agar plates at 37°C overnight. Colonies were selected by kanamycin resistance and expanded in a 1 l Terrific Broth culture media, supplemented with a potassium phosphate buffer (17 mM KH2PO4,72 mM K2HPO4) and incubated at 37°C while shaking at 200 rpm. Once the culture reached an optical density (OD) between 0.7 and 0.9, 1 mM IPTG was added to induce the expression of the MBP-NTF2L proteins at 18°C overnight. Bacterial pellets were collected by centrifugation for 30 min at 4°C and 4500 rpm. Pellets were resuspended in 40 ml lysis buffer (50 mM sodium phosphate pH 8.0, 250 mM sodium chloride, 30 mM imidazole, 20 µg/ml DNase I [Roche, #10104159001], 20 µg/ml RNase A [QIAGEN, #19101], one tablet EDTA-free protease inhibitor [Roche, #11836170001]). After pellets were fully resuspended, they were further lysed by sonication (3 s on, 3 s off, 25% amplitude). The sonicated lysates were then spun down for 30 min at 4°C and 40,000 rpm. The supernatant was filtered with 0.45 µm filters before loading onto a His-Trap HP column (Cytiva, #17524801) connected to an AKTA HPLC purification system. The column was equilibrated with buffer A (20 mM sodium phosphate at pH 8.0, 300 mM sodium chloride, 30 mM imidazole) and protein was eluted with a linear gradient of buffer B (20 mM sodium phosphate at pH 8.0, 300 mM sodium chloride, 500 mM imidazole). Eluted protein was cleaved with 1 mg TEV protease and 5 mM β-mercaptoethanol at 4°C overnight to remove histidine tags. Cleaved protein was purified by collecting the flowthrough on a His-Trap column. Then, collected protein was purified by SEC with a Superdex 75 Increase 10/300 GL column (Cytiva, #29148721) equilibrated with 10 mM Tris-Cl pH 8.0, 150 mM NaCl. Finally, MBP tags that coeluted with NTF2L proteins were purified by incubating with amylose resin (New England BioLabs, #E8021L) overnight at 4°C and collecting the flow through the following day. Collected protein was concentrated to 1 mg/ml using a 3 kDa concentrator, flash-frozen, and stored at −80°C.
Differential scanning fluorimetry
Differential scanning fluorimetry (DSF) was performed in a 96-well clear PCR plate (Bio-Rad, #MLL9601) with 2 µM recombinant NTF2L domain, 5x SYPRO orange (Sigma, #S5692) and filled to 20 µl with SEC buffer (10 mM Tris-Cl pH 8.0, 150 mM NaCl). A control well included SEC buffer alone with 5x SYPRO orange. The PCR plate was sealed and loaded to a RT-PCR (Bio-Rad, #1855201) machine where the plate was incubated at 25°C for 1 min followed by gradual temperature increases by 0.2°C increments until the instrument reached 95°C. Samples were excited at 535 nm and emission collected at 559 nm. The melting temperature (Tm) was calculated by taking the first derivative of the melting curve. We performed DSF to test binding of the NTF2L domain to a synthesized Caprin-1 peptide (GenScript) corresponding to residues 356–386. NTF2L apo protein at 2 µM was mixed with various concentrations of Caprin-1 peptide (1-5-10-50-100 µM), and melting temperatures were estimated as described above.
Size exclusion chromatography multi-angle light scattering
Size exclusion chromatography multi-angle light scattering (SEC-MALS) was performed at 4°C using a Superdex 75 Increase 10/300 GL column (Cytiva, #29148721) equilibrated with 10 mM Tris-Cl pH 8.0, 150 mM NaCl. The column was connected in series with a MALS detector (Wyatt Technology, miniDAWN) and a refractive index detector (Wyatt Technology, Optilab). We loaded 25 µM and 50 µM recombinant NTF2L domains onto the column. The data were analyzed using the Astra software (Wyatt Technology) to derive the molar mass of each sample.
Data analysis
Stress granule segmentation
Image analysis was performed by developing CellProfiler pipelines (Carpenter et al., 2006). Briefly, for immunofluorescence (IF) and live-cell imaging experiments, both nuclear and cytoplasmic signals were rescaled by dividing pixel intensities by a factor. Nuclei were segmented with the Otsu thresholding method. Single cells were segmented by propagation from the nuclear signal. PABP and G3BP foci were segmented by enhancing their speckle-like feature and applying either a manual or the Otsu thresholding method for IF and live-cell imaging experiments, respectively. Finally, potential segmentation artifacts were filtered based on eccentricity, where foci less than 0.875 were kept (Supplemental Figure S1). Further data processing and plotting was done in python and R.
Preprocessing and alignment of next-generation sequencing data
Next-generation sequencing data were processed as described previously (Calviello et al., 2021). Briefly, Ribo-seq footprint reads were trimmed of adapter sequences with cutadapt and filtered based on a 32 nt cutoff. Unique molecular identifier (UMI) sequences were collapsed and removed. Reads were aligned with bowtie2 2.4.1 and filtered against a collection of repeat RNAs including tRNA, rRNA, snRNA, srpRNA, among others (RepeatMasker). Filtered reads were mapped to the hg38 version of the human genome using STAR (Dobin et al., 2013) 2.7.5a with –sjdbOverhang set to 29. Quality control of mapped footprint reads was performed with R package Ribo-seQC (Calviello et al., 2019) and MultiQC tool (Ewels et al., 2016). Spearman correlations were calculated with ggstatsplot (Patil, 2021) R package. RNA-seq and Ribo-seq profiles had spearman correlation factors of 0.8–1.0 per condition, suggesting good reproducibility among replicates (Supplemental Figures S5, A and B; S11, A and B; S14, A and B). As expected, the frequency of fragment lengths for ribosome footprints showed peaks around ∼28 nt, and three nucleotide periodicities at open reading frames of transcripts (McGlincy and Ingolia, 2017) (Supplemental Figures S5, C and D; S11, C and D; S14, C and D). Deletions at both exons 2 of endogenous G3BP1 and G3BP2 genes (Kedersha et al., 2016) to create functional G3BP1/2 KOs were validated by RNA-seq and Ribo-seq (Supplemental Figure S5, E and F).
Differential gene expression analysis
log base 2 fold change (LFC) of differentially expressed genes was estimated with DESeq2 (Love et al., 2014). A “local” fit for both total RNA and ribosome footprint reads was applied in DESeq2. Only genes with total RNA baseMean and ribosome occupancy baseMean above 20 were considered. Genes with RNA LFC more or less than 0 and P adjusted values less than 0.01 were identified as Buffering up or Buffering down, respectively. Genes with Ribo LFC more or less than 0 and P adjusted values less than 0.05 were identified as Ribo ocp up or Ribo ocp down, respectively. Genes changing significantly (P adjusted less than 0.05) at both RNA and ribosome occupancy were identified as either RNA abundance up or RNA abundance down. Translation efficiency (TE) was estimated with Riborex package (Li et al., 2017). Genes with TE LFC more or less than 0, and P adjusted values less than 0.05, were identified as TE up or TE down, respectively.
TE and RNA correlations
Correlations of ΔTE and ΔRNA were plotted with R package ggplot2. Adjusted R2 coefficient was calculated with stat_poly_eq function and regression line was estimated with stat_poly_line function from the ggpmisc package.
Gene set enrichment analysis
Gene lists generated by DESeq2 were sorted in decreased order based on their RNA-seq LFC scores. Gene set enrichment analysis (GSEA) was performed with gseGO function from clusterProfiler R package and genome wide annotation for human “org.Hs.eg.db”. P values were adjusted with the “fdr” method and a cutoff of 0.05. GSEA was performed with 10,000 permutations and ontology defined by biological processes. For SG-associated mRNAs GSEA, gene lists generated by DESeq2 were sorted in decreased order based on their Ribo-seq LFC scores and analyzed with the fgsea function.
Venn diagrams
Venn diagrams were generated with VennDiagram R package. A hypergeometric test was performed with the venn.diagram function. Population size per test was calculated by counting the number of genes with transcript per millions (TPMs) above 1 in the RNA-seq read count datasets corresponding to the cell lines in question.
Estimation of transcript features
mRNA features such as transcript length and the count of AU-rich elements was performed as reported previously (Floor and Doudna, 2016). Briefly, whole-transcript regions were extracted by decomposing a GTF genome annotation file from GENCODE release 25 with python program https://github.com/stephenfloor/extract-transcript-regions on a high-performance computing cluster. Then, transcript length and count of AU-rich elements were calculated locally with python program transcriptome_properties.py found on https://github.com/stephenfloor/tripseq-analysis. Extracted features for G3BP1-sensitive Buffering down and Buffering up genes were analyzed in R.
ACKNOWLEDGMENTS
Sequencing was performed at the UCSF CAT, supported by UCSF PBBR, RRP IMIA, and NIH 1S10OD028511-01 grants. Read alignment was performed on the UCSF Wynton High Performance Computing Cluster. Microscopy was partly performed with a next-generation CSU-W1/SoRa spinning disk microscope system funded by NIH S10OD028611-01 grant. Flow cytometry and cell sorting was performed at the UCSF Parnassus Flow Cytometry CoLab. U-2OS G3BP1/2 KO cells were shared by Dr. Nancy Kedersha. Special thanks to all current and former members of the Floor lab, specially Srivats Venkataramanan and Yizhu Lin for providing feedback on RNA sequencing, data analysis, and initial conceptualization of this project. Furthermore, special thanks to Edmond Linossi from the Jura lab at UCSF for sharing his expertise in protein biochemistry. This project was funded by the National Institutes of Health F31GM143845 award (to J.M.L-L.) and R35GM149255 (to S.N.F.). S.N.F. is a Pew Scholar in the Biomedical Sciences, supported by The Pew Charitable Trusts.
Abbreviations used:
- DGE
differential gene expression
- DSF
differential scanning fluorimetry
- ER
endoplasmic reticulum
- GEO
gene expression omnibus
- GSEA
gene set enrichment
- ISR
integrated stress response
- KO
knockout
- NaAs
sodium arsenite
- OD
optical density
- PPI
protein-protein interaction
- RNP
ribonucleoprotein
- SEC-MALS
size exclusion chromotography with multi-angle light scattering
- SG
stress granules
- TE
translation efficiency
- Tg
thapsigargin
- TPM
transcript per million
- UMI
unique molecular identifier.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E24-02-0062) on October 2, 2024.
Sequencing data availability
Sequencing data are available at the Gene Expression Omnibus (GEO) accession numbers GSE254636, GSE254637, and GSE254638.
Pipelines availability
REFERENCES
- Alam U, Kennedy D (2019). Rasputin a decade on and more promiscuous than ever? A review of G3BPs. Biochim Biophys Acta Mol Cell Res 1866, 360–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali N, Prasad K, AlAsmari AF, Alharbi M, Rashid S, Kumar V (2021). Genomics-guided targeting of stress granule proteins G3BP1/2 to inhibit SARS-CoV-2 propagation. Int J Biol Macromol 190, 636–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulas A, Fay MM, Lyons SM, Achorn CA, Kedersha N, Anderson P, Ivanov P (2017). Stress-specific differences in assembly and composition of stress granules and related foci. J Cell Sci 130, 927–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D (2000). Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol 2, 326–332. [DOI] [PubMed] [Google Scholar]
- Buchan JR, Parker R (2009). Eukaryotic stress granules: The ins and outs of translation. Mol Cell 36, 932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calviello L, Sydow D, Harnett D, Ohler U (2019). Ribo-seQC: Comprehensive analysis of cytoplasmic and organellar ribosome profiling data. [Preprint]. Bioinformatics 10.1101/601468 [DOI] [Google Scholar]
- Calviello L, Venkataramanan S, Rogowski KJ, Wyler E, Wilkins K, Tejura M, Thai B, Krol J, Filipowicz W, Landthaler M, Floor SN (2021). DDX3 depletion represses translation of mRNAs with complex 5′ UTRs. Nucleic Acids Res 49, 5336–5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang I, Friman O, Guertin DA, Chang J, Lindquist RA, Moffat J, et al. (2006). CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty J, Tsai W-C, Lloyd R (2015). Multiple poliovirus proteins repress cytoplasmic RNA granules. Viruses 7, 6127–6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels P, Magnusson M, Lundin S, Käller M (2016). MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floor SN, Doudna JA (2016). Tunable protein synthesis by transcript isoforms in human cells. Elife 5, e10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillén-Boixet J, Kopach A, Holehouse AS, Wittmann S, Jahnel M, Schlüßler R, Kim K, Trussina IREA, Wang J, Mateju D, et al. (2020). RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181, 346–361.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N (2005). Translational control in stress and apoptosis. Nat Rev Mol Cell Biol 6, 318–327. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS (2012). The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc 7, 1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iserman C, Desroches Altamirano C, Jegers C, Friedrich U, Zarin T, Fritsch AW, Mittasch M, Domingues A, Hersemann L, Jahnel M, et al. (2020). Condensation of Ded1p promotes a translational switch from housekeeping to stress protein production. Cell 181, 818–831.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R (2016). ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang W, Wang Y, Yang W, Zhang J, Zheng H, Li D (2021). Research progress on the structure and function of G3BP. Front Immunol 12, 718548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Chen S, Gilks N, Li W, Miller IJ, Stahl J, Anderson P (2002). Evidence that ternary complex (eIF2-GTP-tRNA i Met )–deficient preinitiation complexes are core constituents of mammalian stress granules. Mol Biol Cell 13, 195–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P, Anderson P (2016). G3BP–Caprin1–USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol 212, e201508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P (1999). RNA-binding proteins Tia-1 and Tiar link the phosphorylation of Eif-2α to the assembly of mammalian stress granules. J Cell Biol 147, 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy D, French J, Guitard E, Ru K, Tocque B, Mattick J (2002). Characterization of G3BPs: Tissue specific expression, chromosomal localisation andrasGAP120 binding studies. J Cell Biochem 84, 173–187. [DOI] [PubMed] [Google Scholar]
- Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, Parker R (2017). The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol Cell 68, 808–820.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball SR, Horetsky RL, Ron D, Jefferson LS, Harding HP (2003). Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am J Physiol Cell Physiol 284, C273–C284. [DOI] [PubMed] [Google Scholar]
- Laver JD, Ly J, Winn AK, Karaiskakis A, Lin S, Nie K, Benic G, Jaberi-Lashkari N, Cao WX, Khademi A, et al. (2020). The RNA-binding protein rasputin/G3BP enhances the stability and translation of its target mRNAs. Cell Rep 30, 3353–3367.e7. [DOI] [PubMed] [Google Scholar]
- Li H, Lin P-H, Gupta P, Li X, Zhao SL, Zhou X, Li Z, Wei S, Xu L, Han R, et al. (2021). MG53 suppresses tumor progression and stress granule formation by modulating G3BP2 activity in non-small cell lung cancer. Mol Cancer 20, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wang W, Uren PJ, Penalva LOF, Smith AD (2017). Riborex: Fast and flexible identification of differential translation from Ribo-seq data. Bioinformatics 33, 1735–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateju D, Eichenberger B, Voigt F, Eglinger J, Roth G, Chao JA (2020). Single-molecule imaging reveals translation of mRNAs localized to stress granules. Cell 183, 1801–1812.e13. [DOI] [PubMed] [Google Scholar]
- Matsuki H, Takahashi M, Higuchi M, Makokha GN, Oie M, Fujii M (2013). Both G3BP1 and G3BP2 contribute to stress granule formation. Genes Cells 18, 135–146. [DOI] [PubMed] [Google Scholar]
- McGlincy NJ, Ingolia NT (2017). Transcriptome-wide measurement of translation by ribosome profiling. Methods 126, 112–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay C, Zhou P (2023). Role(s) of G3BPs in human pathogenesis. J Pharmacol Exp Ther 387, 100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namkoong S, Ho A, Woo YM, Kwak H, Lee JH (2018). Systematic characterization of stress-induced RNA granulation. Mol Cell 70, 175–187.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas MD, Kedersha N, Schulte T, Branca RM, Ivanov P, Anderson P (2019). Phosphorylation of G3BP1-S149 does not influence stress granule assembly. J Cell Biol 218, 2425–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas MD, Schulte T, Thaa B, Sandalova T, Kedersha N, Achour A, McInerney GM (2015). Viral and cellular proteins containing FGDF motifs bind G3BP to block stress granule formation. PLoS Pathog 11, e1004659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas MD, Varjak M, Lulla A, Eng E, K. M, A. KH, G. B, McInerney GM. (2012). Sequestration of G3BP coupled with efficient translation inhibits stress granules in Semliki Forest virus infection. Mol Biol Cell 23, 4701–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil I (2021). Visualizations with statistical details: The “ggstatsplot” approach. J Open Source Softw 6, 3167. [Google Scholar]
- Prentzell MT, Rehbein U, Cadena Sandoval M, De Meulemeester A-S, Baumeister R, Brohée L, Berdel B, Bockwoldt M, Carroll B, Chowdhury SR, et al. (2021). G3BPs tether the TSC complex to lysosomes and suppress mTORC1 signaling. Cell 184, 655–674.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo PK, Kar AN, Samra N, Terenzio M, Patel P, Lee SJ, Miller S, Thames E, Jones B, Kawaguchi R, et al. (2020). A Ca2+-dependent switch activates axonal casein kinase 2α translation and drives G3BP1 granule disassembly for axon regeneration. Curr Biol 30, 4882–4895.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo PK, Lee SJ, Jaiswal PB, Alber S, Kar AN, Miller-Randolph S, Taylor EE, Smith T, Singh B, Ho TS-Y, et al. (2018). Axonal G3BP1 stress granule protein limits axonal mRNA translation and nerve regeneration. Nat Commun 9, 3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders DW, Kedersha N, Lee DSW, Strom AR, Drake V, Riback JA, Bracha D, Eeftens JM, Iwanicki A, Wang A, et al. (2020). Competing protein-RNA interaction networks control multiphase intracellular organization. Cell 181, 306–324.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte T, Liu L, Panas MD, Thaa B, Dickson N, Götte B, Achour A, McInerney GM (2016). Combined structural, biochemical and cellular evidence demonstrates that both FGDF motifs in alphavirus nsP3 are required for efficient replication. Open Biol 6, 160078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte T, Panas MD, Han X, Williams L, Kedersha N, Fleck JS, Tan TJC, Dopico XC, Olsson A, Morro AM, et al. (2023). Caprin-1 binding to the critical stress granule protein G3BP1 is influenced by pH. Open Biol 13, 220369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo KW, Kleiner RE (2023). Profiling dynamic RNA–protein interactions using small-molecule-induced RNA editing. Nat Chem Biol 19, 1361–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG (2009). Regulation of translation initiation in Eukaryotes: Mechanisms and biological targets. Cell 136, 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehbens S, Pemble H, Murrow L, Wittmann T (2012). Imaging intracellular protein dynamics by spinning disk confocal microscopy. Methods Enzymol 504, 293–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataramanan S, Gadek M, Calviello L, Wilkins K, Floor SN (2021). DDX3X and DDX3Y are redundant in protein synthesis. RNA 27, 1577–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek RC, Jiang H-Y, Anthony TG (2006). Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans 34, 7–11. [DOI] [PubMed] [Google Scholar]
- Wilkins KC, Schroeder T, Gu S, Revalde JL, Floor SN (2024). A novel reporter for helicase activity in translation uncovers DDX3X interactions. RNA 30, 1041–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolozin B, Ivanov P (2019). Stress granules and neurodegeneration. Nat Rev Neurosci 20, 649–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Mathieu C, Kolaitis R-M, Zhang P, Messing J, Yurtsever U, Yang Z, Wu J, Li Y, Pan Q, et al. (2020). G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 181, 325–345.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn J-Y, Dunham WH, Hong SJ, Knight JDR, Bashkurov M, Chen GI, Bagci H, Rathod B, MacLeod G, Eng SWM, et al. (2018). High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol Cell 69, 517–532.e11. [DOI] [PubMed] [Google Scholar]