Summary
In humans, dietary patterns impact weight and metabolism differentially across individuals. To uncover genetic determinants for differential dietary effects, we subjected four genetically diverse mouse strains to humanized diets (American, Mediterranean, vegetarian, and vegan) with similar macronutrient composition, and performed body weight, metabolic parameter, and RNA-seq analysis. We observed pronounced diet- and strain-dependent effects on weight, and triglyceride and insulin levels. Differences in fat mass, adipose tissue, and skeletal muscle glucose uptake, and gene expression changes in most tissues were strain-dependent. In visceral adipose tissue, ∼400 genes responded to diet in a strain-dependent manner, many of them in metabolite transport and lipid metabolism pathways and several previously identified to modify diet effects in humans. Thus, genetic background profoundly impacts metabolism, though chosen dietary patterns modify the strong genetic effects. This study paves the way for future mechanistic investigations into strain-diet interactions in mice and translation to precision nutrition in humans.
Subject areas: Genetics, Transcriptomics, Model organism, Diet
Graphical abstract
Highlights
-
•
Genetics has a stronger effect on metabolic traits and gene expression than diet
-
•
Gene by environment interaction effects are most evident in visceral adipose tissue
-
•
Nutrient transporter genes show unique expression with diet and genetic background
-
•
Gene expression in C57BL/6J visceral adipose tissue is most unique on a vegan diet
Genetics; Transcriptomics; Model organism; Diet
Introduction
In the US, 73% of the adult population is obese (42%) or overweight (31%).1 Diet modification is considered key to weight management and prevention of complications of excess weight, prominently type 2 diabetes and cardiovascular disease.2,3 The most common dietary patterns that have been investigated for effects on weight loss and cardiometabolic disease are Mediterranean,4,5 vegetarian and vegan,6,7,8,9 low and very low fat,10 low and very low carbohydrate,10,11,12,13,14 Dietary Approaches to Stop Hypertension (DASH),15 and Paleo diets.16 Mediterranean, DASH, and vegetarian diets proved most beneficial for preventing and treating diabetes and cardiovascular disease.2,3 They support long-term weight loss, improve insulin sensitivity, lower circulating glucose and lipid levels,4,17 and are associated with advantageous cardiometabolic outcomes.18,19,20,21,22 Common to these favorable diets are large amounts of plant-based foods, including vegetables, fruits, nuts, seeds, whole grains, legumes, and beans. They are thus high in fiber, phytonutrients (including antioxidants and polyphenols), and omega-3 fatty acids, and low in saturated fatty acids.
Most investigated diets have modest long-term net effects on weight loss when the whole population is considered. However, they cause large differences in weight change among individuals. For example, subjects on isocaloric low-fat and low-carbohydrate-containing diets exhibited weight changes between 25 kg lost to 5 kg gained.10 Iso-caloric Mediterranean and Chinese plant-based diets in pre-diabetic patients also produced high variability in weight effects between individuals.23 These observations suggested that genetic background modifies an individual’s response to diets.10
Indeed, human studies show that the effects of diet on metabolic outcomes depend on the genetic background of the individual. Often the protective effects of diet on metabolism are only conferred to individuals with a certain allelic variant of a single nucleotide polymorphism (SNP). For example, the effects of the Mediterranean diet on body composition, circulating triglyceride, and cholesterol levels were modified by SNPs in the transcription factor 7-like 2 (TCF7L2),24,25,26,27 apo-lipoprotein E (APOE),28 and cholesteryl ester transfer protein (CETP),29,30 respectively. The effect of a Mediterranean/DASH combination diet on circulating lipids was altered by an SNP in the gene Caveolin 1 (CAV1),31,32 the impact of an Inuit diet on circulating fatty acids by an SNP in Carnitine Palmitoyltransferase 1A (CPT1A),33 and the effect of a high-fish diet on circulating triglycerides by an SNP in Glucokinase Regulatory Protein (GCKR).34,35 Protective effects of various dietary macronutrients on metabolism were modified by SNPs located in metabolically relevant genes, including Fat Mass and Obesity associated (FTO),36 Neuropeptide Y (NPY),37 Perilipin (PLIN),38,39,40 Fibroblast Growth Factor 21 (FGF21),41 Adiponectin (ADIPOQ),42 Melatonin Receptor 1B (MTNR1B),43 Fatty Acid Desaturase (FADS2),44,45,46 Hepatocyte Nuclear Factor 1α (HNF1A),47 3-Hydroxy-3-Methylglutaryl-CoenzymeA Reductase (HMGCR),48 and Apo-Lipoprotein A2 (APOA2).49 Finally, lower overall energy intake or caloric content had beneficial effects on metabolism that were modified by SNPs in Insulin Receptor Substrate 1 (IRS1),50 Uncoupling Protein 2 (UCP2),51 and Beta-2 Adrenergic Receptor (ADRB2) genes.52 Analysis of the UK Biobank showed that common SNPs near Solute Carrier Family 12 member 3 (SLC12A3), ATP Binding Cassette Subfamily A Member 6 (ABCA6), MLX Interacting Protein Like (MLXIPL), and the gap junction locus (GJB6-GJB2-GJA3) modified the positive impact of dietary fish oil on circulating cholesterol or triglyceride levels.53 Many SNPs also interact with diet to influence cardiovascular disease outcomes.54,55
While such gene-diet interactions have been recognized, human studies often encounter limitations including low subject numbers, minimal environmental control, and poor diet adherence.56,57 Such factors make it hard to identify subtle genotype-diet interaction effects.58 Smaller studies, referenced above, primarily identified SNPs that are in coding regions of genes, and these genes are likely to have large effect sizes. However, few studies have enough power to detect interactions between diet and common regulatory variants.53 Mouse populations with genetic diversity comparable to humans provide a renewable resource for controlled studies, the results of which can be tested in humans at a limited scale.59 While a uniform environment and adherence to provided diets reduce noise, mouse models also allow deeper tissue and cellular phenotyping. Studies with ∼100 genetically diverse mouse strains showed that, when challenged with high fat or high carbohydrate diets, genetic background causes disparate responses in body weight gain,59,60,61 atherosclerosis development,59,62 insulin resistance,59,63,64 circulating metabolites,61,64 glucose uptake,64 adipocyte size,64 and protein abundances.64 Interestingly, mouse genetic loci that favor body fat gain on a high fat high sucrose diet and loci associated with atherosclerosis overlapped with loci identified in humans.59,62,65 For example, mouse obesity loci near Npc1, Klf14, Atp2a1, Apob48r, Opcml, and Sorl1 have been associated with various obesity phenotypes, including body mass index (BMI) and waist circumference, in humans.65 Furthermore, many lipid metabolism genes and pathways in adipose tissue are conserved between human and mouse.66 These earlier studies demonstrate that the effect of dietary macronutrient content on metabolic parameters is significantly modified by genetic variation, and the genetic and cellular mechanisms identified in mice are likely conserved in humans.
Most studies in mice used high fat diets and did not test the more subtle effects of dietary nutrient sources. Recently, Barrington et al. studied the effects of humanized mouse diets on metabolic parameters and DNA methylation, thereby expanding mouse studies to include human diet patterns.67 Mouse diets representative of human American, Mediterranean, ketogenic, and Japanese diets were tested in four genetically diverse mouse strains. Significant interactions between diet and genetic background were observed for body fat, LDL cholesterol, glucose tolerance, and liver triglycerides. However, the diets varied widely in macronutrient composition with fat contents between 4.3% and 55.8%. Such differences could have at least in part driven the large dietary effects that were observed.
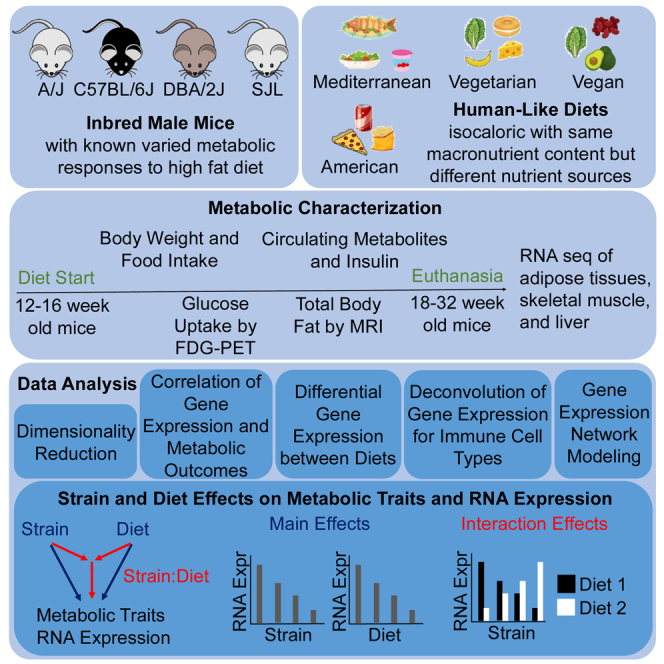
The goal of our study was to investigate the metabolic effects of different humanized mouse diets with equivalent macronutrient composition but differing macronutrient sources, and evaluate how genetic background modifies dietary effects on body weight and other metabolic parameters, as well as on gene transcription in metabolic tissues. We used common human diets that derive macronutrients predominantly from animals or plants (American, Mediterranean, vegetarian, and vegan) and studied four genetically diverse mouse strains (A/J, C57BL/6J, DBA/2J, and SJL). We observed clear strain-dependent differences in metabolic parameters and distinct strain-dependent effects for different diets suggesting an interplay between diets and genetic background.
Results
Design of mouse diets with human nutrient sources
Mediterranean, vegetarian, and vegan diets were chosen as established human diet patterns with beneficial metabolic effects.2,3,18,19,20,21,22 They were compared to a typical American diet pattern that is most often associated with negative health outcomes. To test responses to variations in dietary macronutrient sources only, macronutrient content was kept the same in all diets and adjusted to average consumption in the American population (35% fat, 50% carbohydrate, and 15% protein).68 The American diet contained the highest amount of animal products (red meats, dairy) with high amounts of saturated fatty acids, and refined easily absorbable carbohydrates. The Mediterranean diet consisted of more plant and less animal products. It included fish and was higher in fiber and monounsaturated fatty acids, and lower in saturated fatty acids when compared to the American diet. The vegetarian diet contained mostly plant products, and eggs and dairy as animal products, while the vegan diet consisted only of plant products. Vegetarian and vegan diets were high in fiber, unsaturated fatty acids, and low in cholesterol. Human versions of each diet were designed based on 7-day menu plans, and mouse chow was prepared using powdered versions of human food sources (Figures S1A–S1E; Tables S1 and S2).
Selection of mouse strains
We selected four genetically diverse inbred mouse strains, A/J, C57BL/6J, DBA/2J, and SJL,69 to test the contributions of genetic background on metabolic changes in response to the diets described above. Among the ∼100 mouse strains in the hybrid mouse diversity panel (HDMP), the four selected strains responded to a high fat diet with widely varying fat mass gain and changes in insulin sensitivity.59,63 SJL mice remained lean, while the other three strains dramatically increased body fat mass.64 A/J mice remained insulin sensitive, while C57BL/6J developed intermediate and DBA/2J severe insulin resistance.62 We thus expected that the metabolic responses of these four mouse strains would noticeably differ. Since male mice of selected strains showed more pronounced effects than females on fat mass and insulin sensitivity,63 important parameters for our study, we only used male mice.
Genetic background and diet interaction affect body weight gain and metabolic parameters
In an initial study, diets were fed to mice (n = 7–8 for each condition) starting at 14–15 weeks of age. Body weights and food intake were determined weekly, and blood samples for measuring glucose, insulin, triglycerides, and non-esterified fatty acids were taken before starting the diets and at 4, 8, and 16 weeks on the diets (Study 1, Figure S1F).
Body weight changes (study 1)
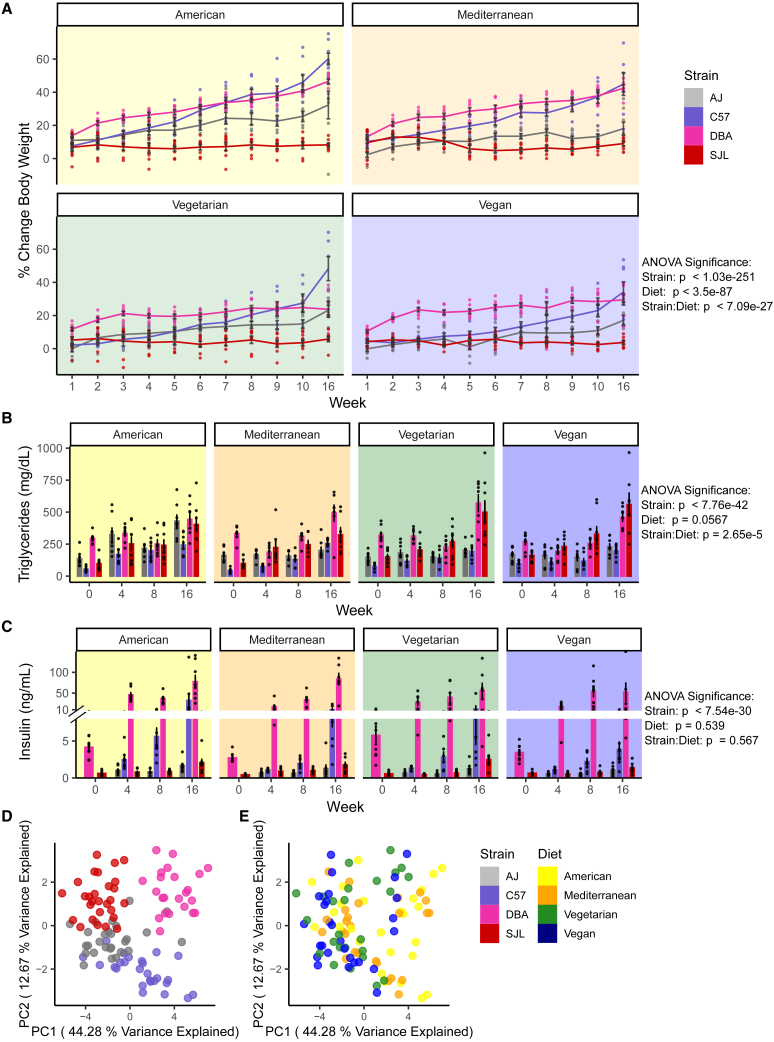
Overall, among the four strains analyzed, C57BL/6J and DBA/2J mice showed the highest percent change in body weight over the course of the study, with the degree of gain modified by diet. American and Mediterranean diets promoted the most weight gain (Figures 1A and S2A). C57BL/6J continued to gain weight over the 16 weeks on all diets while DBA/2J mice did so only on the American or Mediterranean diets. When fed vegetarian or vegan diets, DBA/2J weights plateaued around week 4. A/J mice gained the most weight on the American diet and gained relatively little on the other 3 diets. SJL mice did not gain weight on any of the four diets over the 16-week period. Variances in body weight gain between strains and diets could not be explained by divergent food intake. We observed no significant differences in caloric intake normalized to body weight between diet groups (Figure S2B). There were consistent differences between strains, with SJL mice eating more and DBA/2J mice eating less than average. However, these effects were inversely related to body weight changes in these strains.
Figure 1.
Genetic background and diet interaction affects body weight gain and metabolic parameters
(A) Percent body weight change from baseline over 16 weeks on diets (n = 7–8 mice for each strain and diet, 14 weeks old at week 0, male mice).
(B) Plasma or serum triglycerides at 0, 4, 8, and 16 weeks on diets (n = 7–8, 14 weeks old at week 0, male mice).
(C) Plasma or serum insulin at 0, 4, 8, and 16 weeks on diets (n = 7–8, 14 weeks old at week 0, male mice).
(D) Principal component analysis of all traits measured in study 1 using all samples, principal components PC1 and 2 shown with samples colored by genetic background.
(E) Principal component analysis of all traits measured in study 1 using all samples, PC1 and 2 shown with samples colored by diet.
Error bars represent mean ± SEM, p-values from three-way ANOVA performed considering the effects of Strain, Diet, Time, and all interaction effects (Table 1).
See also Figures S1–S4.
Since we were interested in unique dietary effects on different genetic backgrounds, we used Analysis of Variance (ANOVA) tests to determine the independent effects of the main variables, strain and diet, as well as the effects of interactions between both variables (strain:diet) on metabolic outcomes. ANOVA is a form of linear regression used to deconvolve the effects of multiple variables,70 where interaction effects are determined by accounting for the effects of the main variables (Graphical Abstract). Significant interactions occur when the effects of one variable depend on the state of the other, i.e., a particular diet prevents weight gain in some strains but not others. Using a three-way ANOVA to account for the effects of the main variables strain, diet, and time, we determined that there are significant effects of strain and diet, and strain:diet interaction effects on percent change in body weight (Table 1).
Table 1.
P-values derived from three-way ANOVA interaction tests for study 1 phenotypic data
| Parameter | Strain | Diet | Strain: Diet | Week | Strain: Week | Diet: Week | Strain: Diet: Week |
|---|---|---|---|---|---|---|---|
| % Change Body Weight | 1.0E-251 | 3.5E-87 | 2.0E-150 | 7.1E-27 | 4.4E-10 | 2.9E-100 | 1.1E-06 |
| Triglycerides | 7.8E-42 | 5.7E-02 | 8.5E-46 | 2.7E-05 | 8.7E-02 | 4.7E-12 | 5.1E-02 |
| Insulin | 7.5E-30 | 5.4E-01 | 2.9E-16 | 5.7E-01 | 4.2E-01 | 2.4E-15 | 3.1E-01 |
| Glucose | 7.8E-18 | 3.7E-01 | 2.0E-12 | 1.3E-01 | 4.7E-01 | 1.8E-12 | 4.1E-03 |
| NEFA | 2.2E-06 | 2.1E-04 | 1.9E-10 | 1.8E-04 | 8.2E-07 | 7.6E-10 | 5.7E-08 |
NEFA, non-esterified fatty acids.
Circulating insulin, triglycerides, glucose, and non-esterified fatty acids (study 1)
Among the four strains, DBA/2J mice had the highest triglyceride levels at baseline (Figures 1B and S3A). For all diets, these levels remained stable at earlier time points and increased to similar levels at 16 weeks. SJL mice had substantially raised triglycerides only after challenge with diets, and vegetarian and vegan diets caused the largest elevations over the 16 weeks on diets. In A/J mice, only the American diet triggered an increase in triglycerides. C57BL/6J mice had the lowest triglyceride levels at baseline and showed modest progressive increases over time on all diets, with American diet resulting in the largest increase at the earlier time points. For insulin, DBA/2J mice had, when compared to the other three strains, elevated baseline levels that further increased to a similar extent on all diets (Figures 1C and S3B). A/J and SJL mice had consistently low insulin levels that responded to all diets to a similarly small degree. Insulin levels of C57BL/6J were higher than those of A/J and SJL and showed pronounced differential increases in response to the four diets; vegan diet raised insulin levels the least overall and American diet the most at the earlier time points. Using a three-way ANOVA, we found significant effects due to strain and strain:diet interactions for triglycerides and insulin, as well as for glucose (Table 1; Figure S4). NEFA levels showed significant strain, diet, and strain:diet interaction effects (Table 1; Figure S4).
Principal component analysis of aggregate metabolic parameters (study 1)
We used principal component analysis (PCA) to explore the multi-dimensional metabolic phenotype data in two dimensions using linear combinations of the input variables. The axis that describes the most variance through the data, principal component 1 (PC1), separates DBA/2J and SJL data into clear groups, while the axis with second highest variance, principal component 2 (PC2), separates C57BL/6J from DBA/2J and SJL mice (Figure 1D). Neither axis groups the data by diet (Figure 1E). These results suggest that strain background explains more of the variability in the metabolic phenotypes than diet.
Genetic background impacts variations in total body fat and glucose uptake in visceral adipose tissue and skeletal muscle
The observed large differences in percent change in body weight and metabolic parameters prompted us to investigate the metabolic phenotype of the mice in more depth with additional approaches. We placed a second cohort of mice on diets starting at 12 weeks of age (n = 4 for each condition), collected blood samples at the start of the diet, and took weekly body weight and food intake measurements. Glucose uptake in adipose tissues and skeletal muscle was measured at 4 weeks on diets using 2-[18F] fluoro-2-deoxy-D-glucose positron emission tomography (FDG-PET). Total body fat mass was determined at 5 weeks on diets using magnetic resonance imaging (MRI). At euthanasia, after 6 weeks on diets, blood and tissues were collected for analysis of circulating metabolites and RNA-seq analysis, respectively (Study 2, Figure S1F).
Body weight changes and circulating insulin, triglycerides, and glucose (study 2)
Similar to the first study, DBA/2J mice gained weight rapidly, C57BL/6J and A/J had intermediate weight gain and SJL mice were protected from weight gain (Figure S5). We again observed that DBA/2J and C57BL/6J were most responsive to diet, gaining the most weight on the American and least on the vegan diet. Effects were less pronounced than in the first study, but using three-way ANOVA we show that the effects of strain and diet, and the interaction effects of strain:diet on percent change in body weight were all significant (Table 2). We also found that circulating triglyceride and insulin levels, at baseline and after 6 weeks on diets, were similar to the first study (Figure S5). Circulating insulin, triglycerides, and glucose were all significantly affected by strain, while strain:diet interactions had a significant effect on triglycerides and glucose (Table 2).
Table 2.
P-values derived from two- and three-way ANOVA interaction tests of study 2 phenotypic data
| Parameter | Strain | Diet | Strain: Diet | Week | Strain: Week | Diet: Week | Strain: Diet: Week |
|---|---|---|---|---|---|---|---|
| % Change Body Weight | 1.1E-45 | 2.9E-16 | 1.5E-04 | 3.6E-34 | 2.8E-11 | 4.3E-03 | 1.7E-01 |
| Triglycerides | 4.7E-24 | 3.5E-01 | 9.8E-03 | 4.0E-05 | 1.1E-02 | 1.7E-01 | 1.7E-01 |
| Insulin | 3.6E-07 | 3.5E-01 | 2.0E-01 | 2.6E-04 | 6.1E-05 | 1.7E-01 | 1.7E-01 |
| Glucose | 2.0E-13 | 3.5E-01 | 2.1E-02 | 5.1E-16 | 5.3E-03 | 1.7E-01 | 1.7E-01 |
| MRI-Total Fat | 2.9E-11 | 4.1E-01 | 5.6E-01 | – | – | – | – |
| FDG-PET-VAT | 1.3E-12 | 3.4E-01 | 9.6E-01 | – | – | – | – |
| FDG-PET-Quad | 1.4E-02 | 5.7E-01 | 5.6E-01 | – | – | – | – |
| FDG-PET-BAT | 1.3E-02 | 5.7E-01 | 5.6E-01 | – | – | – | – |
| VAT Wt/BW | 4.5E-09 | 2.1E-01 | 5.6E-01 | – | – | – | – |
| SAT Wt/BW | 3.0E-04 | 4.4E-01 | 7.6E-01 | – | – | – | – |
| BAT Wt/BW | 1.3E-16 | 1.9E-02 | 7.6E-01 | – | – | – | – |
| Liver Wt/BW | 1.7E-07 | 8.3E-01 | 7.3E-01 | – | – | – | – |
| Quad Wt/BW | 1.6E-11 | 4.0E-01 | 7.8E-01 | – | – | – | – |
| Gastro Wt/BW | 6.2E-19 | 1.3E-01 | 6.2E-01 | – | – | – | – |
| Pancr Wt/BW | 2.1E-03 | 6.8E-01 | 7.9E-01 | – | – | – | – |
| Brain Wt/BW | 9.5E-08 | 5.2E-01 | 7.6E-01 | – | – | – | – |
BAT, brown adipose tissue; FDG-PET, 2-[18F] fluoro-2-deoxy-D-glucose positron emission tomography; gastro, gastrocnemius muscle; MRI, magnetic resonance imaging; Pancr, pancreas; Quad, quadriceps muscle; SAT, subcutaneous adipose tissue; VAT, visceral adipose tissue; Wt, weight.
Body fat mass
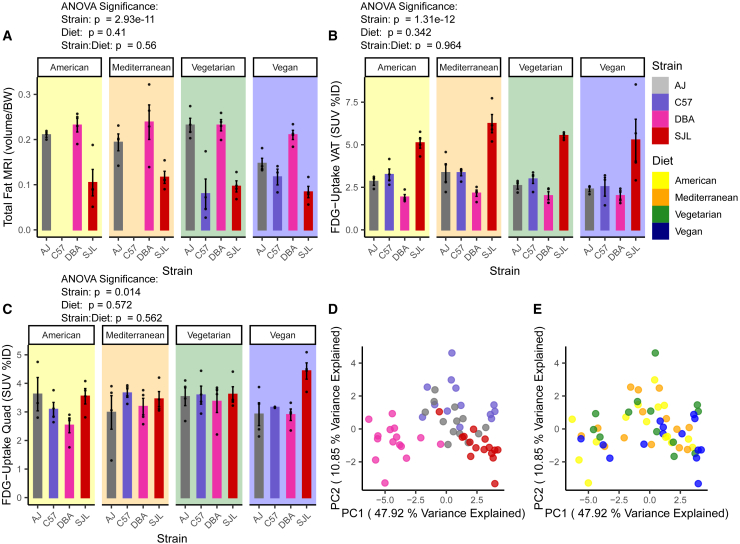
Using MRI, we observed that A/J and DBA/2J mice had the overall highest total body fat mass relative to body weight, except for A/J mice fed a vegan diet (Figure 2A). C57BL/6J and SJL mice had similarly low fat mass relative to body weight, at least on vegetarian and vegan diets. Due to technical issues, measurements for American and Mediterranean diets are missing for C57BL/6J. Two-way ANOVA revealed significant strain-dependent effects for relative total body fat mass (Table 2).
Figure 2.
Genetic background impacts variations in total body fat and glucose uptake in visceral adipose tissue and skeletal muscle
(A) Total body fat volume per gram body weight measured via magnetic resonance imaging (MRI) after 5 weeks on diets (n = 3–4, 17 weeks old, male mice). BW, body weight. Note, due to technical issues, data for C57BL/6J on American and Mediterranean diets are missing.
(B) Glucose uptake in visceral adipose tissue (VAT) measured via 2-[18F] fluoro-2-deoxy-D-glucose positron emission tomography (FDG-PET) imaging at 4 weeks on diet (n = 3–4, 16 weeks old, male mice). SUV%ID, standardized uptake values (SUV) normalized to injected dose per gram body weight.
(C) Glucose uptake in quadriceps (Quad) muscle measured via FDG-PET imaging at 4 weeks on diet (n = 3–4, 16 weeks old, male mice).
(D) Principal component analysis of all traits measured in study 2 using all samples, Principal components PC1 and 2 shown with samples colored by genetic background.
(E) Principal component analysis of all traits measured in study 2 using all samples, PC1 and 2 shown with samples colored by diet.
Error bars represent mean ± SEM, p-values from three-way ANOVA performed considering the effects of Strain, Diet, Time, and all interaction effects (Table 2).
See also Figures S5–S7.
Glucose uptake into adipose tissues and skeletal muscle
Using FDG-PET imaging, we measured glucose uptake in visceral adipose tissue (VAT), brown adipose tissue (BAT), and quadriceps (the largest skeletal muscle). We found that SJL VAT accumulates the most, DBA/2J VAT the least, and A/J and C57BL/6J VAT similar intermediate amounts of glucose with results independent of diet (Figure 2B). Glucose uptake for all strains and diets was similar in the quadriceps muscle, except for SJL uptake that was larger on the vegan diet (Figure 2C). In BAT, DBA/2J and A/J mice on the vegetarian diet took up more glucose than the other two strains (Figure S6). C57BL/6J on all diets except for Mediterranean took up the least amount of glucose in BAT. Two-way ANOVA showed significant strain-dependent effects for all tissues analyzed but no significant diet or strain:diet effects (Table 2).
Analysis of adipose tissues, skeletal muscle, and liver
After 6 weeks on diets, major metabolic organs were harvested and weighed, and tissue weight was normalized to body weight (Figure S7). VAT, subcutaneous adipose tissue (SAT), and BAT were consistently the largest in DBA/2J regardless of diet (Figures S7A–S7C). The other three strains had similar adipose tissue proportions. Quadriceps and gastrocnemius muscle weights relative to body weight were consistently the lowest for DBA/2J mice on all diets, while the other three strains had similar skeletal muscle weights (Figures S7E and S7F). Relative liver and pancreas weights were similar among diets and strains (Figures S7D and S7G), while relative brain weight was the lowest for DBA/2J mice on all diets (Figure S7H). A two-way ANOVA test showed significant strain-dependent effects for relative organ weights (Table 2). BAT weight was significantly affected by diet (Table 2), with the lowest relative weights on the vegan and vegetarian diets for all strains except A/J on the vegetarian diet (Figure S7C). Gastrocnemius and VAT were suggestive of a diet effect (p = 0.13 and p = 0.21, respectively, Table 2) with relative gastrocnemius weights trending higher on vegetarian and vegan diets for C57BL/6J and SJL mice (Figure S7F), and relative VAT weights trending lower on the vegan when compared to the American diet for all strains (Figure S7A). No organ weights differed significantly due to strain:diet interactions (Table 2).
Principal component analysis of aggregate metabolic parameters (study 2)
Using all phenotype data from study 2, we performed PCA and plotted the data for principal components 1 and 2 (PC1 and PC2), the axes of highest variance. We show that these separate the data based on strain but not diet (Figures 2D and 2E), suggesting, like in study 1, that strain explains more of the variability in the measured traits.
Genetic background and diet impact gene expression in adipose tissues, skeletal muscle, and liver
To determine which genes are expressed in response to diets with different genetic backgrounds, we isolated and sequenced mature RNA species from VAT, SAT, BAT, liver, and quadriceps muscle from two mice for each strain and diet group. We found that between ∼24,000 (Liver) and ∼31,000 (SAT) genes were expressed in each tissue.
Principal component analysis of tissue gene expression data
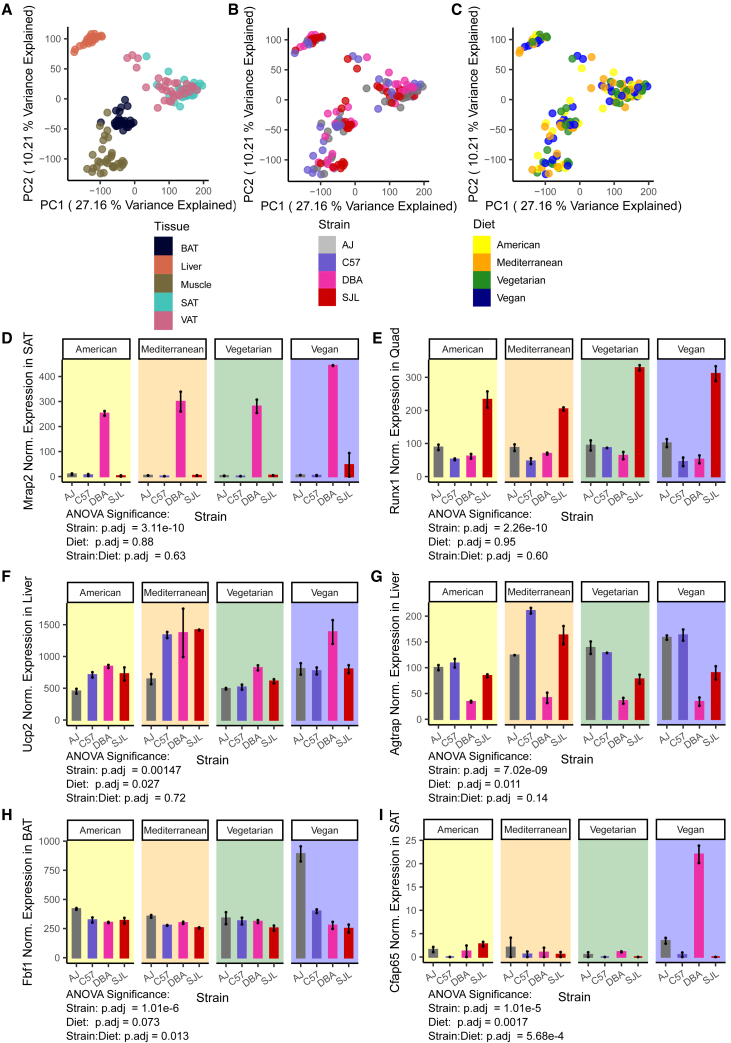
We used PCA to reduce the dimensionality of the gene expression data. When viewing all samples on principal components 1 and 2 (PC1 and PC2), we observed clear separation of samples for all tissues except for the two white adipose tissues (SAT and VAT) that strongly overlapped (Figure 3A). PC1 and PC2 did not separate data by strain or diet (Figures 3B and 3C). PC3 and PC4 explained a smaller portion of the variation in the data and separated the samples again by tissue, including notably dividing SAT and VAT samples, but did not split the data by strain or diet (Figures S8A–S8C). PC5 and PC6 finally separated the samples by strain (Figure S8E), but not tissue or diet (Figures S8D and S8F), and no principal components separated samples by diet. However, as would be expected, differences between tissue types accounted for the most variation in expressed genes. When separating the data by tissue and performing PCA separately for each tissue, we found that PC1 and PC2 separated samples reasonably well by strain for all tissues, but no principal components separated samples by diet (Figure S9). Thus, strain explained the variation in gene expression in examined tissues better than diet.
Figure 3.
Genetic background and diet impact gene expression in liver, quadriceps muscle, SAT, and BAT
(A) Principal component analysis of expressed RNA species using all samples, Principal components PC1 and 2 shown with samples colored by tissue.
(B) Principal component analysis of expressed RNA species using all samples, PC1 and 2 shown with samples colored by genetic background.
(C) Principal component analysis of expressed RNA species using all samples, PC1 and 2 shown with samples colored by diet.
(D) Expression of gene Mrap2 in subcutaneous adipose tissue (SAT), with significant strain-dependent effects. (n = 2, 18 weeks old, male mice).
(E) Expression of gene Runx1 in quadriceps (Quad) muscle, with significant strain-dependent effects (n = 2, 18 weeks old, male mice).
(F) Expression of gene Ucp2 in liver, with significant diet-dependent effects (n = 2, 18 weeks old, male mice).
(G) Expression of gene Agtrap in liver, with significant diet-dependent effects (n = 2, 18 weeks old, male mice).
(H) Expression of gene Fbf1 in brown adipose tissue (BAT), with significant strain:diet-dependent effects. (n = 2, 18 weeks old, male mice).
(I) Expression of gene Cfap65 in subcutaneous adipose tissue (SAT), with significant strain:diet-dependent effects (n = 2, 18 weeks old, male mice).
Error bars represent mean ± SEM, p-values from two-way ANOVA performed considering the effects of Strain, Diet, and interaction effects, p-values adjusted using FDR correction (n = 26,000–33,000) (Table S3). Gene expression plots show normalized counts.
See also Figures S8 and S9.
Effects of diets on gene expression in visceral adipose tissue
To characterize more subtle effects of diets, we performed differential expression analysis for each of the adipose tissues, quadriceps muscle, and liver comparing the American diet to each of the other three diets. We show data for visceral adipose tissue since it showed the most pronounced differences (Figure S10). Each diet comparison showed large effects on gene expression (Figures S10A, S10C, and S10E). KEGG pathway analysis of genes differentially expressed between diets suggested that when compared to the American diet, the Mediterranean diet upregulated genes involved in immune responses, the vegetarian diet promoted muscle-like expression patterns, and the vegan diet modified gene expression in multiple metabolic pathways (Figures S10B, S10D, and S10F).
Expression of strain or diet dependent genes in adipose tissues, skeletal muscle, and liver
For each tissue, we used a two-way ANOVA test to identify genes for which expression varied across samples due to effects of strain and diet, and strain:diet interactions. We identified between ∼8,300 (Quad) and ∼13,600 (SAT) genes with significant strain effects (Table S3). For example, Melanocortin 2 Receptor Accessory Protein 2 (Mrap2) expression in SAT (Figure 3D) and RUNX Family Transcription Factor 1 (Runx1) expression in quadriceps muscle (Figure 3E) were significantly affected by strain, with Mrap2 expression dramatically higher in DBA/2J and Runx1 expression highest in SJL mice when compared to the other strains. Far fewer genes were impacted by diet; we identified between 0 (BAT) and 50 (VAT) genes with significant diet effects (Table S3). After VAT, the liver showed the largest response to diet with 32 genes (Table S3). For example, Uncoupling Protein 2 (Ucp2) and Angiotensin II Receptor Associated Protein (Agtrap) expressions in the liver were significantly affected by diet (Figures 3F and 3G). Ucp2 expression was highest on the Mediterranean diet in all strains except A/J and on the vegan diet in DBA/2J (Figure 3F). Agtrap was highest in C57BL/6J and SJL on the Mediterranean diet and in A/J and C57BL/6J on the vegan diet (Figure 3G).
Strain:Diet interaction effects on gene expression in adipose tissues, skeletal muscle, and liver
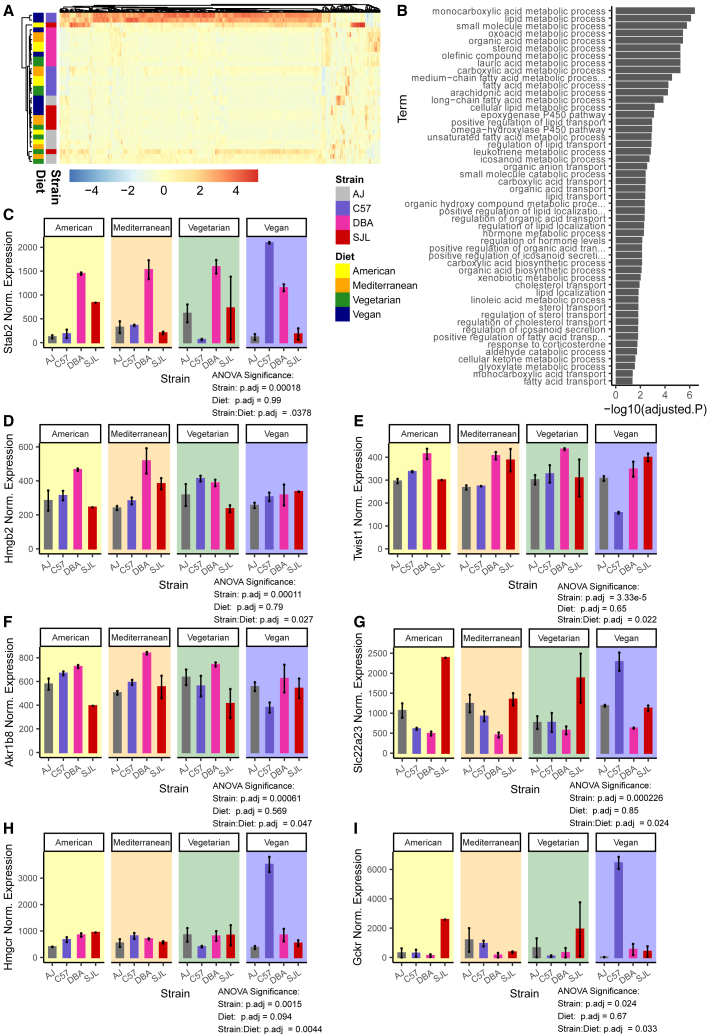
In most tissues, we identified a few genes with significant strain:diet interaction effects (Table S4). Fas Binding Factor 1 (Fbf1) was the only gene in BAT and had highest expression in A/J mice on a vegan diet (Figure 3H). In SAT there were six genes, including Cilia and Flagella Associated Protein 65 (Cfap65), that were highest expressed in DBA/2J mice on the vegan diet (Figure 3I). In the liver, five genes (including transcription factors Dmrtb1 and Pbx4, and a ribonuclease family member Rnase2a, all with currently unspecified functions), and in quadriceps muscle (Quad), two genes (with unknown functions) were dependent on strain:diet interaction effects (Table S4). In VAT, however, 421 genes had significant strain:diet interaction effects on gene expression (Figure 4A; Table S4).
Figure 4.
Gene expression response driven by strain:diet interaction effects in visceral adipose tissue correlates with metabolic responses
(A) Heatmap of 421 visceral adipose tissue genes that exhibit significant strain:diet-dependent effects. Color represents the normalized expression value across samples, normalization performed with DESeq2.
(B) Pathway analysis of 421 visceral adipose tissue genes that exhibit significant strain:diet-dependent effects. Significantly enriched pathways from the Gene Ontology-biological processes database. P-values adjusted using FDR correction for multiple tests.
(C) Expression of gene Stab2 in visceral adipose tissue, with significant strain:diet-dependent effects (n = 2, 18 weeks old, male).
(D) Expression of gene Hmgb2 in visceral adipose tissue, with significant strain:diet-dependent effects (n = 2, 18 weeks old, male).
(E) Expression of gene Twist1 in visceral adipose tissue, with significant strain:diet-dependent effects (n = 2, 18 weeks old, male).
(F) Expression of gene Akr1b8 in visceral adipose tissue, with significant strain:diet-dependent effects (n = 2, 18 weeks old, male).
(G) Expression of gene Slc22a23 in visceral adipose tissue, with significant strain:diet-dependent effects (n = 2, 18 weeks old, male).
(H) Expression of gene Hmgcr in visceral adipose tissue, with significant strain:diet-dependent effects (n = 2, 18 weeks old, male).
(I) Expression of gene Gckr in visceral adipose tissue, with significant strain:diet-dependent effects (n = 2, 18 weeks old, male).
Error bars represent mean ± SEM, p-values from two-way ANOVA performed considering the effects of Strain, Diet, and interaction effects, p-values adjusted using FDR correction (n = ∼31,000) (Table S4). Gene expression plots show normalized counts.
See also Figures S12–S20.
Diets and immune response in adipose tissues
It is well known that an American/Western diet promotes an inflammatory response in visceral adipose tissue, marked by the infiltration of different immune cells.71 To determine if strain or diet affected the composition of immune cells present in each adipose tissue, we deconvolved the bulk RNA sequencing data using a reference panel of gene expression in 16 immune cell types. We found that the three adipose tissues varied considerably in immune cell type proportions (Figures S11A–S11C). Strikingly, SAT tissue had overall low macrophage content compared to BAT and VAT. Significant strain-dependent effects, determined by two-way ANOVA, were observed for six immune cell types in VAT, for four in SAT, and three in BAT (Figure S11D). However, no significant diet effects were found, and only for B cell proportions in VAT was a strain:diet interaction effect observed (Figure S11D). In VAT, various types of macrophages and T cells differed significantly by strain, as determined by two-way ANOVA (Figures S11E and S11F).
Gene expression response driven by strain:diet interaction effects in visceral adipose tissue correlate with metabolic responses
Pathways in VAT affected by Strain:Diet interaction effects
To understand which pathways were affected by strain:diet interactions, we identified gene ontology (GO) biological process pathways and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways that were enriched within our set of 421 genes. We found that many fatty acid and sterol transport and metabolism pathways were highly enriched in our strain:diet responsive gene set (Figures 4B and S12).
Correlation of Strain:Diet dependent gene expression with other metabolic phenotypes
To reconcile gene expression patterns with observed phenotypes, we investigated which of the 421 differentially expressed genes were significantly correlated with metabolic traits (Table S5). To highlight some examples, VAT expression of Stabilin 2 (Stab2), a gene involved in hyaluronan transport, was significantly correlated with BAT weight normalized to body weight (R2 = 0.59, Padj = 0.037), and its levels are elevated in DBA/2J mice, as previously observed72 (Figure 4C). VAT expression of High-Mobility Group Box 2 (Hmgb2), a gene that regulates pre-adipocyte expansion,73 was significantly correlated with BAT weight normalized to body weight (R2 = 0.60, Padj = 0.032) and circulating insulin at week 0 (R2 = 0.63, Padj = 0.016) (Figure 4D). VAT expression of Twist Basic-Helix-Loop-Helix Transcription Factor 1 (Twist1), an early adipogenesis gene,74,75 was significantly correlated with SAT (R2 = 0.61, Padj = 0.025) and BAT weight normalized to body weight (R2 = 0.58, Padj = 0.045) (Figure 4E). Aldo-keto-reductase 1 B8 (Akr1b8) expression in VAT was highly correlated with all adipose tissue weights per body weight, total fat mass, and percent change in body weight at various time points (Table S5; Figure 4F). Akr1b8 is likely involved in the regulation of lipotoxicity.76,77 An additional ∼15 genes in VAT positively correlated with several parameters including percent change in body weight, relative adipose tissue weights, and/or insulin and triglyceride levels (Table S5; Figure S13). VAT expression of Solute Carrier Family 22, Member 23 (Slc22a23), an adipose norepinephrine importer,78 was significantly negatively correlated with VAT weight (R2 = −0.57, Padj = 0.048) and percent change in body weight at weeks 3, 4, 5 and 6 on diets (R2 = −0.65, −0.67, −0.68, −0.68; Padj = 0.01, 0.007, 0.005, 0.005) (Figure 4G). Nine other genes had significant negative correlations with percent change in body weight and/or relative adipose tissue weights (Table S5; Figure S14).
Expression of genes in C57BL/6J VAT on a vegan diet
Other VAT genes with strain:diet interactions were expressed specifically in some strains in response to the diets in a way that did not reflect observed phenotypes. This included the largest proportion of strain:diet interaction genes for which expression was highest in C57BL/6J vegan samples, notably Hmgcr (Figure 4H) and Gckr (Figure 4I). Other C57BL/6J VAT vegan-specific genes fell into broad categories, such as solute carriers and transporters (Figure S15). These encode transporters of metabolically relevant molecules (Slc2a2 - glucose, Slc17a8 - glutamate, Slc13a2 - citrate, Slc22a7 - prostaglandins, Slc22a1 - Na+/vitamin C, Slc30a1 - manganese, Slc35a2 - nucleotide sugars, Slc36a1 - apolar amino acids, Slc39a2 - zinc, Slc40a1 - iron, Slc66a1 - cationic amino acids, Kcnk10 – potassium). Other categories included ATP-binding cassette (ABC) transporter genes (Figure S16), cytochrome P450 genes (Figure S17), hydroxysteroid dehydrogenase genes (Figure S18), and serine protease inhibitor genes (Figure S19). Other notable C57BL/6J VAT vegan-specific genes include sterol and fatty acid metabolism genes such as fatty acid binding proteins (Fabp1 and Fabp2) and lipases (Lipc and Lipg), other genes involved in regulating metabolism such as 3-hydroxybutyrate dehydrogenase 1 (Bdh1), and genes involved in hormone signaling pathways including Insulin-Like Growth Factor Binding Protein 2 (Igfbp2) (Figure S20).
Effects of diet components on gene expression in visceral adipose tissue
To determine if specific diet components (Table S1) are potentially responsible for changes in gene expression, we used the statistical method partial least-squares regression (PLSR). Like PCA, this approach reduces high dimensional data to one or two component axes; however, PLSR is based on the shared variance structure underlying two linked datasets. We used PLSR to predict how the sources of each diet (Table S1) covary with and predict the expression of the 421 strain:diet dependent genes (Table S4). The results of this analysis are shown in Figure 5.
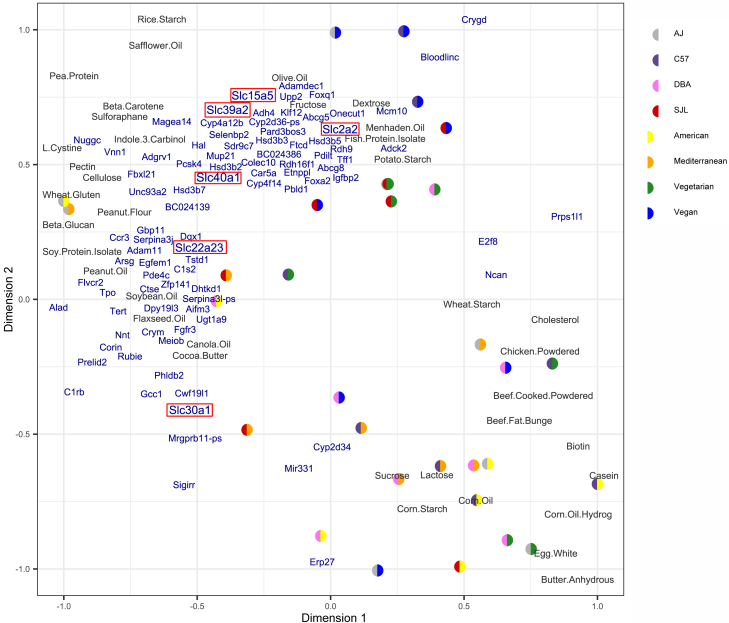
Figure 5.
Partial least squares regression (PLSR) aligns diet components and RNA expression in reduced-dimension space
PLSR used 38 diet components (Table S1) to predict the expression of the 421 strain:diet-dependent VAT genes (Table S4) in reduced-dimension space, based on the shared variance structures of the datasets. Briefly, X and Y were decomposed into loading vectors and latent variables, such that the covariance of the representations of X and Y in component space is maximized (see STAR Methods). The new axes represent scaled, centered versions of the X- and Y- loading vectors, called components, which are linear combinations of X and Y input variables, respectively. The position of X input variables (diet sources in black) and Y output variables (genes in blue) on the new component axes are defined as the calculated X- or Y- loading for each input variable. The position of the mice (colored by strain and diet) is defined by the latent variables; here we averaged the position calculated for X- and Y-spaces. In this shared space, the covariance between diet sources and gene expression is related to the cosine of the angle between them; items nearby each other are strongly related, items opposite each other along a component are negatively related, and items at 90-degree angles apart are not related to each other. Genes referenced in discussion are boxed in red.
PLSR works by decomposing the predictor dataset (diet sources) and outcomes dataset (gene expression) into loading vectors and latent variables, such that the covariance between them in component space is maximized. The new axes represent scaled, centered versions of loading vectors, called components, which are linear combinations of diet source or gene expression input variables. The position of diet source input variables and gene expression output variables on the new component axes are defined as the calculated loading vectors, while the position of the mice (colored by strain and diet in Figure 5) is defined by the latent variables. The genes and diet sources that covary are located near each other, and we hypothesize that these genes may be responsive to the nutrients located nearby (Figure 5). Genes and diet sources located opposite each other are negatively related, while variables at 90° from one another are not strongly related.
In the reduced components space, we find that the plot cleanly divides between animal diet sources and mice fed animal-based diets at the bottom right and plant diet sources and mice fed plant-based diets at the top and left (Figure 5). We observe that transporter genes Slc2a2 (glucose), Slc40a1 (iron exporter), Slc15a5 (unknown function), Slc22a23 (organic ion transporter), and Slc39a2 (zinc transporter) are associated with sugars in the diet. Another zinc transporter, Slc30a1, is opposite the sugar processing genes (Figure 5). These genes may directly contribute to the response to available plant or animal nutrient sources.
Co-expression networks identify additional Strain:Diet responsive genes
Finally, since we were underpowered to detect subtle strain:diet effects in the genome-wide ANOVA analysis (Table S3), we used co-expression network analysis to identify additional strain:diet responsive genes. We used the tool iterativeWGCNA79 to identify groups of genes with highly similar expression patterns, then used ANOVA to identify which modules contain genes whose expression varies by strain and diet, using an aggregate expression value per module to reduce the burden of multiple test corrections.
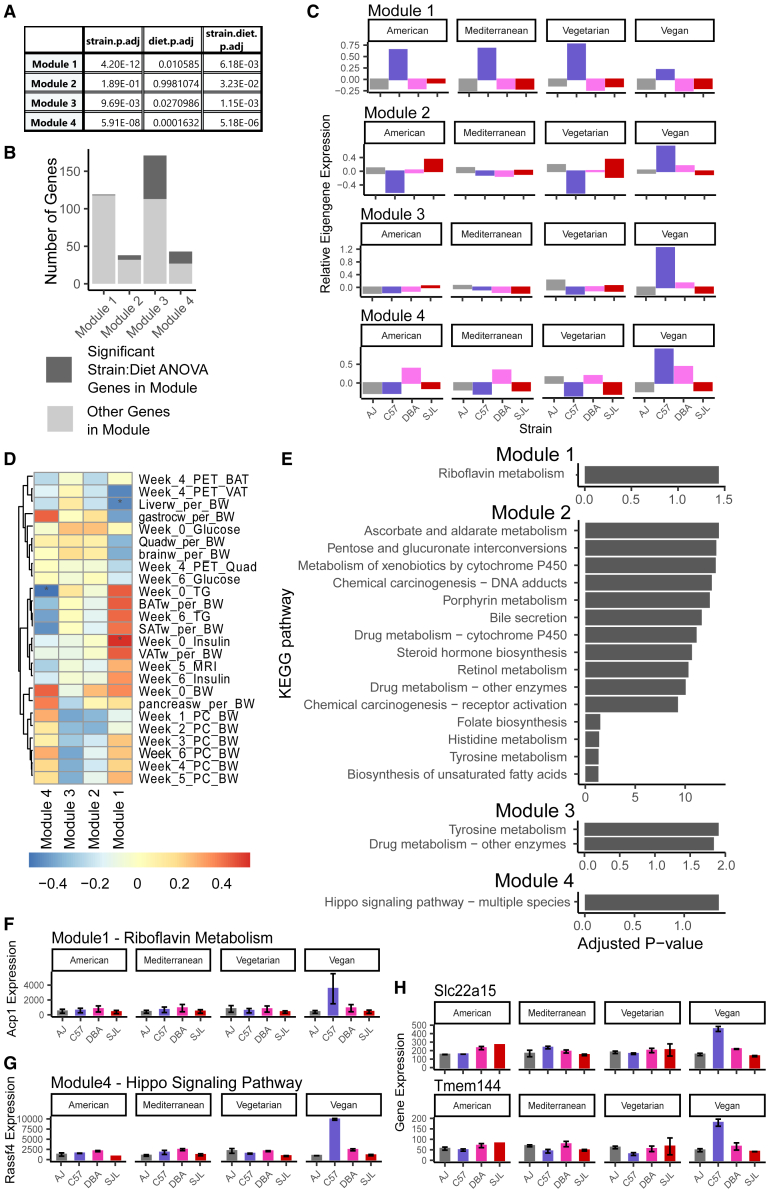
Considering all expressed genes in VAT (STAR Methods), we identified 131 modules of co-expressed genes, containing ∼15,000 genes in total. We defined the aggregate expression value using a metric called the eigengene, which is the first principal component (PC1) of the expression data of genes in the module. Using a two-way ANOVA by strain and diet, we identified four modules for which aggregate expression responded to strain:diet interactions (Figure 6A; Table S6). In these 4 modules, the expression of 285 genes varies by strain:diet interactions, including 78 new strain:diet genes that were not identified in the genome-wide ANOVA (Figure 6B). The aggregate expression in each module is similar to the 421 genes in Table S4; C57BL/6J mice on a vegan diet have significantly different expression than other groups (Figure 6C).
Figure 6.
Co-expression network analysis for visceral adipose tissue uncovers four modules for which genes vary by strain and diet
(A) Results of a two-way ANOVA determining if expression of eigengene (similar to normalized average expression of module genes (see STAR Methods)) in each module varies due to strain, diet, and strain:diet interaction effects. p-values adjusted using FDR correction (n = 131).
(B) The total number of genes in each of the selected modules, with the number of genes previously identified using ANOVA colored black (Table S3) and the number of genes discovered in this analysis only colored gray.
(C) Relative expression of module eigengenes in VAT.
(D) Pearson correlation of module eigengene expression with metabolic traits. BAT, brown adipose tissue; BW, bodyweight; gastroc, gastrocnemius; MRI, magnetic resonance imaging; PC, percent; PET, positron emission tomography; quad, quadriceps; SAT, subcutaneous adipose tissue; TG, triglycerides; VAT, visceral adipose tissue; w, tissue weight. p-values from Pearson correlation adjusted using FDR correction (n = 131).
(E) KEGG pathways that were enriched for module genes.
(F) Expression of module 1 gene Acid Phosphatase 1 (Acp1) in VAT. Acp1 is a gene in the KEGG-riboflavin metabolism pathway that is enriched in module 1.
(G) Expression of module 4 gene Ras Association Domain Family Member 4 (Rassf4) in VAT. Rassf4 is a gene in the KEGG- Hippo signaling pathway that is enriched in module 4.
(H) Expression of selected transporter genes from module 4 in VAT.
To consider the function of these strain:diet responsive genes, we calculated the correlation of metabolic traits with the module aggregate expression value. Two of the four modules' aggregate gene expression (modules 1 and 4) is significantly correlated with metabolic traits (Figure 6D). Module 1 expression was positively correlated with body weight traits but negatively with adipose tissue weights and circulating triglyceride levels. Module 4 expression was positively correlated with body weight, adipose tissue weights, and insulin and triglyceride levels, and was negatively correlated with muscle weight, liver weight, circulating glucose, and glucose uptake traits. Using enrichment testing, we found that all four modules were enriched for different KEGG signaling pathway genes (Figure 6E). Module 1 contained Acid Phosphatase 1 (Acp1), a gene in the riboflavin metabolism pathway (Figure 6F), and module 4 contained Ras Association Domain Family Member 4 (Rassf4), a gene in the Hippo signaling pathway (Figure 6G). Module 4 also contained the organic cation transporter gene Slc22a15 and putative transporter gene Tmem144/Slc35g7 (Figure 6H). While Slc22a15 was identified in the previous genome-wide ANOVA (Table S4), the co-expression analysis allowed us to prioritize Tmem144, a gene of unknown function.
Discussion
Our study shows that diet, genetic background, and the interaction between the two can all modify body weight gain and various metabolic parameters in mice. Among the four strains analyzed, DBA/2J mice had the most pronounced increases in body weight, stored the largest amount of fat, and had the highest insulin and triglyceride levels, with diet types modifying the extent of the changes. On the other hand, SJL had the lowest fat mass and low insulin levels, but relatively high triglyceride levels. While diet had little impact on fat mass or body weight in SJL mice, diet affected triglyceride and insulin levels. A/J mice had high fat mass relative to body weight with some diet-dependent effects; they increased body weight, triglyceride, and insulin levels most with the American diet, but showed little change in these parameters on the other three diets. C57BL/6J mice showed large body weight gains with different diet patterns modifying the extent of gains. Although the data were incomplete, C57BL/6J exhibited relatively low fat mass versus body weight, similar to SJL, except on the American diet (as judged by visceral adipose tissue weight at euthanasia). They also had the lowest triglyceride levels overall and intermediate insulin levels that were highest on the American and lowest on the vegan diet. Glucose uptake varied largely in a strain-dependent manner. When data for all metabolic parameters were combined in principle component analysis, we consistently observed clear separation by strain, but not by diet, suggesting that the genetic background exerted a much stronger effect on the metabolic phenotype than a diet.
In previous mouse studies that tested dietary effects on metabolic parameters in a panel of ∼100 genetically diverse mouse strains, including the four strains we used, mice were fed high fat high sucrose diets.59,62,63,65 Our diets contained a similar amount of fat (35 kcal%) as the ones used in these previous studies (32 kcal%).59,62,63,65 When comparing results with our American diet that had the highest sucrose content, similar to our results, large differences in body weight and insulin levels between A/J, C57BL/6J, DBA/2J, and SJL strains at baseline and in response to the high fat high sucrose diet were observed.59,62,63,65 Triglyceride levels were also consistent with data shown for DBA/2J, A/J, and C57BL/6J in the study by Parks et al.63 with DBA/2J highest, A/J intermediate, and C57BL/6J lowest levels among these three strains (no data shown for SJL). DBA/2J had decreased insulin-stimulated glucose uptake in isolated soleus muscle when compared to A/J.63 We found lower glucose uptake for DBA/2J when compared to A/J in quadriceps in vivo. With regard to body fat percentage, consistent with our data, DBA/2J and A/J had similarly high body fat percentages and SJL the lowest.59,62,65 Thus, consistent with these earlier studies, we capture a large effect of strain on body weight gain and insulin levels at baseline and in response to a high fat diet.
The earlier study that used humanized diets showed stronger diet effects than ours.67 Large differences in macronutrient composition between diets in the earlier study could have accounted for observed differences. Additionally, keeping fat, carbohydrate, and protein levels the same in our diets may have blunted dietary effects. In line with the average fat content in the human diet, our diets contained around twice the amount of fat when compared to regular mouse chow (17kcal%). Thus, the higher fat content could also have muted effects of macronutrient sources.
We observed larger, more significant differences in metabolic parameters in study 1 than study 2, though the directionality and pattern between strains and diets were largely conserved. This was likely due to the more intrusive tests performed during study 2 (FDG-PET and MRI) that elicited stress responses and introduced noise impacting weight gain and metabolic parameters. Due to limited resources, we also used only four mice per group in study 2 making it underpowered to detect smaller differences. Similarly, with n = 2 mice per group, RNA-seq did not have enough power to identify the full spectrum of strain:diet responsive genes; only large magnitude changes in gene expression were identified as significant. Furthermore, we only used male mice which limits our conclusions to this population. Despite these limitations, we were able to capture large-scale differences due to genetic background that were evident when visualizing the data in reduced dimensions. Further, using correlation analysis and co-expression networks, we were able to link the expression of diet and genetic background-responsive genes to important metabolic parameters. Our data suggest modulating effects of macronutrient sources.
The American diet, with the highest content of animal sourced macronutrients when compared to mostly or only plant-based vegetarian and vegan diets, led to higher weight gains in DBA/2J, C57BL/6J, and A/J mice and, at least in part, higher triglyceride and insulin levels. Although larger mouse studies will be required to further corroborate these results, the observation is consistent with human studies. Plant-based diets in humans consistently outperform American diets in beneficial effects on metabolic parameters when the general population is considered.2,3 In further support of differential responses to plant-based diets, we find that the PLSR model cleanly divides between animal and plant sources. Also, when compared to the American diet, the three more plant-based diets showed dramatically different gene expression responses in VAT; the Mediterranean diet upregulated genes involved in immune responses, the vegetarian diet promoted muscle-like expression patterns, and the vegan diet modified gene expression in multiple metabolic pathways. The vegan diet seemed to be the most different from the American diet, resulting in the largest number of differentially expressed genes.
Although the Mediterranean diet affected immune response gene expression in VAT, we did not observe large differences in immune cell proportions due to diet or strain:diet interactions. Most calculated cell type proportions only differed between strains. SJL mice are known to have elevated T cell levels80 which we confirm; we observe the highest T cell proportions in SJL mice on plant-based diets.
Our analysis identified ∼400 genes in VAT for which expression is altered by strain:diet interactions. Several of these were previously shown to modify the effect of diet on metabolic parameters in humans. This includes genetic variation in HMGCR48 and GCKR34,35; we show that expression in mouse visceral adipose tissue is responsive to strain:diet interactions. We identified Ucp2 as strain and diet responsive but did not show a significant interaction effect; previous studies found interactions between dietary energy intake and SNPs in UCP2 on metabolic parameters in humans.51 We detected many genes known to control adipose tissue expansion and homeostasis, including Hmgb2, Akr1b8, Slc22a23, Kcnk10,81 Twist1 and Acp1. An SNP in Ncan was previously found to be associated with fatty liver disease in humans.82 Acp1 catalyzes riboflavin production, and promotes adipogenesis and tissue expansion.83,84 Human ACP1 genetic variants are associated with glycemia, dyslipidemia, and obesity.85,86 Interestingly, in our study, Acp1-containing module 1 aggregate expression was inversely correlated with adipose tissue weights, and triglyceride and insulin levels. Based on correlations we identified, we predict that Hmgb2, Akr1b8, and Twist1 expression influence fat storage phenotypes, as most evident in DBA/2J mice. We also identified genes that regulate fatty acid and sterol metabolism and had previously been studied in the liver (Agxt, Agxt2, Bdh1, Slc40a1, Hmgcr, Adck2, and Gckr).87,88,89,90,91,92 These genes had highest expression in C57L/6J-vegan fed mice specifically.
Slc transporter genes with highest expression specifically in C57BL/6J vegan fed mice were identified as potentially important for sugar transport in the PLSR model based on their proximity to the dietary nutrients fructose and dextrose. Fructose and dextrose levels are lower in the vegan and vegetarian diets, and many mice on vegan and vegetarian diets are plotted near these sugars as well. Importantly, Slc2a2, a well-known glucose transporter and glucose sensor,93 is found near the sugars in the plot, and could be responding to lower amounts of one or both sugars. Slc39a2 is also found near the sugars in the RNA-PLSR plot while Slc30a1 is found opposite. Both transport zinc, and in response to zinc deficiency, zinc importer Slc39a2 is up-regulated94 while zinc exporter Slc30a1 is downregulated.95 While we did not measure the zinc content of our diets, red meat and poultry sources naturally contain zinc. Dietary zinc content may thus have contributed to the observed response. Slc40a1 is an iron exporter that is important for macrophage metabolism but may not be necessary for adipocyte function.96,97 Slc15a5 and Slc22a23 are not well studied, though they may also be important to the sugar and zinc transport responses of visceral adipose tissue for C57BL/6J fed a vegan diet based on their similar location in the plot. Both genome-wide ANOVA and co-expression network analyses also found that carnitine/carnosine transporter Slc22a15 was regulated by strain and diet.98 Tmem144 is a novel gene hypothesized to be a carbohydrate transporter. Based on correlations with phenotypes, Slc22a15 and Tmem144 may regulate adipose tissue size and insulin levels, and based on the identity of co-expressed genes, they may regulate the Hippo signaling pathway.
In conclusion, we identified known and novel candidate genes that respond to macronutrient source uniquely by genetic background. Mechanistic studies will be required to determine if and how these genes respond to diet components and/or genetic background. Future studies in C57BL/6J mice that showed the most pronounced effects on a vegan diet (highest number of differentially expressed gene in VAT and simultaneously lowest insulin levels compared to C57BL/6J mice on the other 3 diets) might uncover genetic markers that could identify individuals that would most benefit from a vegan diet. Commonalities between our study and the earlier human studies support the concept that results from mice may be translatable to humans. Our study further illustrates that inclusion of relatively few mice in well controlled studies can yield relevant effects that can then be tested in humans at a limited scale. Thus, our study provides a novel resource for precision nutrition; to apply diets to populations in which they would be most beneficial99 and prevent or ameliorate disease progression based on genetic risk, among other factors.
Limitations of the study
Due to limited resources, we used only four mice per group in study 2 making it underpowered to detect smaller differences. Similarly, with n = 2 mice per group, RNA-seq did not have enough power to identify the full spectrum of strain:diet responsive genes; only large magnitude changes in gene expression were identified as significant. We only used male mice which limits our conclusions to this population.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Susanna R. Keller (srk4b@virginia.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
RNA-seq data (raw FASTQ files and processed counts) are deposited at GEO and are publicly available with the accession code GSE260764.
-
•
All original code has been deposited on Github at https://github.com/jnr3hh/Reed_Civelek_Keller_2024_Manuscript and is publicly available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in this article is available from the lead contact upon request.
-
•
Counts, TPMs and fastq files from the RNA-seq data generated in this study are publicly available on GEO (accession code GSE260764). Codes to perform the analyses in this study are available on https://github.com/jnr3hh/Reed_Civelek_Keller_2024_Manuscript.
Acknowledgments
We thank Sara Sturgess MS, RDN at Research Diets Inc. for her thoughtful and careful work in translating the human diets into mouse chow, Alice C. Innis for her help with animal work for study 2, Jie Li, Soumen Paul, and Jack Roy for their assistance in FDG PET and MRI imaging. Imaging data was acquired through the University of Virginia Molecular Imaging Core Laboratory under the former and current direction of Drs. Stuart Berr and Maurits Jensen, respectively, with NIH S10OD021672 funding for the Albira Si trimodal scanner and NIH S10OD025024 funding for the Bruker 9.4T MRI scanner. The study was made possible through a University of Virginia 3Cavalier Award (to SRK, MC, SK), University of Virginia Engineering in Medicine Seed Grant (to MC, BKK, SRK, SK, HAF) and a University of Virginia Double Hoos Award (to DB and JNR).
Author contributions
SRK, MC, SK, BKK, and HAF designed the study. FH and SK designed the human diet patterns and translated them into mouse diets. JNR, AK, DB, JH, PS, NS, SCS, SEN, BKK, and SRK conducted experiments and analyzed data. JNR and SRK wrote the article with input from all authors.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| 2-[18F] fluoro-2-deoxy-D-glucose | PETNET, subsidiary of Siemens Medical Solutions | |
| TRIzol | ThermoFisher Scientific | Cat#15596026 |
| Critical commercial assays | ||
| Blood Glucose Testing System AimStrip Plus | Germaine Laboratories | Cat#SKU37350 |
| HR Series NEFA-HR(2) assay | FUJIFILM Wako Diagnostics USA Corporation | Color Reagent A Cat#999-34691, Solvent A Cat#995-34791, Color Reagent B Cat#991-34891, Solvent B Cat#993-35191, NEFA Standard Solution Cat#276-76491 |
| Insulin Rodent Chemiluminescence ELISA | ALPCO | Cat#80-INSMR |
| L-Type Triglyceride M Assay | FUJIFILM Wako Diagnostics USA Corporation | Triglyceride M Color A Cat#994-02891, Triglyceride M Color B Cat#990-02991, Multi-Lipid Calibrator Cat#464-01601 |
| RNA Broad Range Assay Kit | ThermoFisher Scientific | Cat#Q10210 |
| RNAse-free DNAse kit | Qiagen | Cat#79254 |
| RNeasy Micro Kit | Qiagen | Cat#74004 |
| TapeStation RNA Broad Range Assay | Agilent | RNA Screen Tape Cat#5067-5576, RNA ScreenTape Sample Buffer Cat#5067-5577, RNA Screen Tape Ladder Cat#5067-5578 |
| Deposited data | ||
| Raw Fastq Files and processed counts | This paper | GEO: GSE260764 |
| Mouse Reference Genome GRCm39 | Gencode | Gencode: GRCm39; https://www.gencodegenes.org/mouse/ |
| Experimental models: organisms/strains | ||
| A/J | Jackson Laboratories | Stock#000646; RRID:IMSR_JAX:000646 |
| C57BL6/J | Jackson Laboratories | Stock#000664; RRID:IMSR_JAX:000664 |
| DBA/2J | Jackson Laboratories | Stock#000671; RRID:IMSR_JAX:000671 |
| SJL/J | Jackson Laboratories | Stock#000686; RRID:IMSR_JAX:000686 |
| SJL/JCrNTac | Taconic | Model#SJL-M; RRID:IMSR_TAC:SJL |
| Software and algorithms | ||
| Original Code | This paper | Github: https://github.com/jnr3hh/Reed_Civelek_Keller_2024_Manuscript |
| Base R functions: pca(), aov() | R Core Team100 | R Core Team100 |
| R package ggplot2 | Wickham101 | Wickham101 |
| R package DEseq2 | Love et al.102 | Love et al.102 |
| R package mixOmics | Kim-Anh Le Cao, Florian Rohart, Ignacio Gonzalez, Sebastien Dejean with key contributors Benoit Gautier, Francois Bartolo, contributions from Monget et al.103 |
Monget et al.103 |
| Python package iterativeWGCNA | Greenfest-Allen et al. 79 | Greenfest-Allen et al.79; https://github.com/cstoeckert/iterativeWGCNA |
| fastQC | Andrews104 | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| trimGalore | Krueger105 | https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
| hisat2 | Kim et al.106 | Kim et al.106 |
| htseq | Putri et al.107 | Putri et al.107 |
| samtools | Danecek et al.108 | Danecek et al.108 |
| CIBERSORTx | Newman et al.109 | Newman et al.109; https://cibersortx.stanford.edu/ |
| PMOD version 3.9 | PMOD Technologies | |
| Other | ||
| American Diet | Research Diets, Inc. | D19062002 |
| Mediterranean Diet | Research Diets, Inc. | D19062003 |
| Vegetarian Diet | Research Diets, Inc. | D19062004 |
| Vegan Diet | Research Diets, Inc. | D19062005 |
Experimental model and study participant details
Animals: Male A/J (Stock #000646), C57BL/6J (Stock #000664), DBA/2J (Stock #000671) mice were purchased at 9 weeks of age from Jackson Laboratories. For the first study SJL/JCrNTac (Model # SJL-M) were obtained from Taconic Biosciences at 7-9 weeks of age. For the second study SJL/J mice were acquired from Jackson Laboratories (SJL/J, Stock #000686) at 5 weeks of age (not available for purchase at a later age). Mice were housed under temperature- (22oC) and humidity-controlled conditions and a constant light-dark cycle with free access to water and chow. They were fed a standard chow diet (cat. no. 7912 Teklad LM-485; Harlan Laboratories) until age 14-15 weeks (study 1) and 12-13 weeks (study 2). At this time mice were placed on one of four humanized diets (American, Mediterranean, vegetarian or vegan (described below)) until euthanasia with carbon dioxide asphyxia (after 16 weeks on diets in study 1 and 6 weeks in study 2 (Figure S1F)). Before starting the diets, mice were weighed and assigned to diets such that the average body weight of mice on each diet was similar for each individual strain. Mice were housed four per cage with the exception of SJL mice; they required single housing due to aggressive behavior against cage mates. Body weights and food intake were determined weekly, up to 10 weeks on diets and at euthanasia in study 1 (body weight), and until euthanasia in study 2. Blood collections for measurements of glucose, insulin, triglycerides, and non-esterified fatty acids (NEFA) were performed before starting the diets and at 4, 8, and 16 weeks on diets in study 1 and at the start of the diets and at euthanasia in study 2. In study 2, glucose uptake in muscle and fat tissues was measured in live mice after 4 weeks on diets using 2-[18F] fluoro-2-deoxy-D-glucose positron emission tomography (FDG-PET), and magnetic resonance imaging (MRI) was performed at 5 weeks on diets. At the beginning of studies, the number of mice for each strain and diet was n=7-8 (study 1) and n=4 (study 2). There were a few deaths (for unknown reasons) during study 1 (1 C57BL/6J on Mediterranean and vegetarian diets, 1 DBA/2J on Mediterranean and vegetarian diets) and in study 2 (1 A/J on American diet). After euthanasia, blood was collected by cardiac puncture and tissues were dissected. Tissues were frozen in liquid nitrogen and stored at -80oC until further processing. All animal procedures were approved by the University of Virginia Institutional Animal Care and Use Committee.
Method details
Design of humanized mouse diets
We designed mouse versions of four common human diets; American, Mediterranean, vegetarian, and vegan. Macronutrient composition was the same for all diets (35% fat, 50% carbohydrates, 15% protein) and was based on the average/mean macronutrient intake among adults in the US68 (33-34% kcal from fat, 49-51% kcal from carbohydrates, 15-16% kcal from protein). Meal plans containing the desired macronutrient composition were designed as following. Dietary intake patterns for the four diets were generated by developing 7-day meal plans for breakfast, lunch, dinner and two snacks per day for humans (examples shown in Figures S1A–S1D). The meal plans followed the NHANES-based pattern for typical American and vegetarian diets, the Food Pyramid for the Mediterranean diet, and lastly, to generate the pattern for the vegan diet, the vegetarian diet pattern was modified to remove all animal sources. All diet patterns were based on an average intake of 2,000 kcal per day and matched for macronutrient content. The Nutrient Database for Research (NDS-R, Minnesota) version 2018 was used for all macro- and micronutrient analysis. The diet patterns of all four human diets were translated into mouse chow replica in collaboration with Research Diets, Inc. (New Brunswick, NJ, USA). The food sources and nutrient profiles of the mouse chows reflected that of the human diet patterns and supplied adequate nutritional requirements according to NRC nutrient guidelines for mice. All four mouse diets were designed to have similar kcal/g food. Diet ingredients and compositions are shown in Tables S1 and S2, and Figure S1E.
Food intake measurements
Food was weighed before addition to mouse cages. A week later remaining food was weighed. To obtain food intake/cage during the week, food weight at the end of the week was subtracted from food weight at the beginning of the week.
Blood collections
Blood was collected between 8-9 AM from nicked tail veins of mice fed ad libitum. The first drop was used for blood glucose measurements using a glucometer (AimStrip Plus Blood Glucose Testing System (Germaine Laboratories, Inc, San Antonio TX, USA). Additional blood was collected into heparinized (for preparation of plasma for insulin and triglyceride measurements at 0, 4 and 8 weeks on diets) and non-heparinized capillary tubes (for preparation of serum for non-esterified fatty acid (NEFA) measurements). Blood was transferred to microfuge tubes and kept on ice before centrifugation at 1,600 g for 30 min at 4oC. Plasma or serum was collected and stored at -20oC until further analysis. At euthanasia blood glucose measurements were performed before animals were euthanized and additional blood (non-heparinized) was collected immediately after CO2 asphyxia by cardiac puncture with a 1 ml syringe connected to a 25G-5/8 needle, transferred to a 1.5 ml microcentrifuge tube, allowed to clot for 30 min on ice, and then centrifuged at 1,600 g for 30 min at 4oC. Serum was stored at -20°C (short-term storage) and -80oC (long-term storage) and used for insulin, triglyceride, and NEFA measurements.
Measurements of plasma and serum insulin, triglycerides, and non-esterified fatty acids
Insulin and triglycerides were measured in plasma or serum (terminal bleed only) using an Insulin Rodent Chemiluminescence ELISA (cat. no. 80-INSMR from ALPCO, Salem NH, USA) and the L-Type Triglyceride M Assay (Triglyceride M Color A 994-02891, Triglyceride M Color B cat. no. 990-02991 and Multi-Lipid Calibrator cat. no. 464-01601 from FUJIFILM Wako Diagnostics USA Corporation, Richmond, VA, USA). Non-esterified fatty acid levels (NEFA) were determined in serum using the HR Series NEFA-HR(2) assay (Color Reagent A cat. no. 999-34691, Solvent A cat. no. 995-34791, Color Reagent B cat. no. 991-34891, Solvent B cat. no. 993-35191, NEFA Standard Solution cat. no. 276-76491 from FUJIFILM Wako Diagnostics USA Corporation, Richmond, VA, USA).
2-[18F] Fluoro-2-deoxy-D-glucose positron emission tomography (FDG-PET) imaging in vivo
Mice fed ad libitum were injected intraperitoneally with 2-[18F] fluoro-2-deoxy-D-glucose (100-200 μCi) between 8-9 AM. After 30 min, during which mice were allowed to move freely in cages, they were anesthetized with isoflurane (3% in oxygen/air (50/50, 1 L/min) and imaged for 10 min in a Bruker Albira Si PET/SPECT/CT trimodal rodent scanner (Bruker, Billerica, MA) to acquire a single static PET image. This was followed by a 5 min computed tomography (CT) scan for attenuation correction and anatomical co-registration. Anesthesia was maintained during scanning using 1.5-2% isoflurane. The images were automatically reconstructed and registered using the Albira reconstruction toolbox as both PET and CT images are in the same image space. Visualization and quantification of PET/CT data were performed using Pixel wise MODeling (PMOD)’s image View tool (version 3.9, PMOD Technologies Ltd., Zurich, Switzerland). Briefly, the whole-body PET and CT images were read using the View tool and volumes of interest (VOI) drawn on CT images for regions corresponding to epididymal white adipose tissue (visceral adipose tissue, VAT), interscapular brown adipose tissue (BAT), and quadriceps skeletal muscle (Quad). The same VOI were then automatically drawn on the PET images. The Contour tool with appropriate thresholding was applied for accurate delineation of the boundaries of VAT, BAT, and Quad. Standardized uptake values (SUV) were computed for the VOI normalizing for the injected dose per gram body weight. SUV computations were performed using PMOD.
Magnetic resonance imaging (MRI) in vivo
MRI was performed using a 9.4T/20 (bore diameter 20 cm) horizontal bore BioSpec AVANCE neo preclinical imaging system equipped with a 116 mm bore gradient insert (Bruker BioSpin GmbH, Germany, maximum gradient strength 660 mT/m). Between 8-10 AM mice fed ad libitum were anaesthetized with 3% isoflurane and secured in an MRI compatible cradle. Anesthesia was maintained during scanning using 1.5-2% isoflurane in oxygen/air (50/50, 1 L/min). Respiration rate and rectal temperature were monitored throughout and body temperature maintained at 37°C (SA Instruments Inc, Stony Brook, NY, USA). A 40-mm inner diameter quadrature radiofrequency coil was used for signal transmission and reception. Scout images were taken to confirm correct positioning and orientation of subsequent scans. A 2-dimensional 28-slice T2-weighted (RARE) 3-point DIXON scan was performed for water-fat separation (1 mm slice thickness, image matrix = 256 x 128, field of view = 50 x 35 mm, repetition time = 1404 ms, echo time = 18 ms, signal averages = 2, RARE-factor = 4). The fat fraction (FF) map was calculated from the separated fat images. Quantification of fat volumes (from FF maps) were performed using PMOD’s VOI and contour tools (with appropriate thresholding) as described above. Volumes were normalized to mouse body weights.
Isolation and sequencing of mature RNA species
Homogenization of visceral (VAT) and subcutaneous (SAT) adipose tissues, quadriceps muscle and liver
Since some of the adipose tissues, quadriceps muscle and livers were large and/or dense, we homogenized tissues first at liquid nitrogen (LN2) temperatures using the Cell Crusher Tissue Pulverizer (from CELLCRUSHER, Portland Oregon, USA). Briefly, tissues were kept on dry ice until use, and all devices and tools were cooled in LN2. The whole tissue was inserted into the cell crusher device and crushed, then the powdered tissue was collected in 1.5 ml tubes in LN2. Samples were immediately used for RNA isolation or frozen at -70oC until use.
RNA isolation
All tissues were weighed and kept on dry ice before powdering or addition of TRIzol (cat. no. 15596026 from ThermoFisher Scientific, Waltham, MA, USA). For BAT and small SAT tissues, the whole tissue was added to 1 mL TRIzol. For powdered tissues (VAT, liver, quadriceps muscle, large SAT), ∼0.3 g of powdered tissue (not weighed to keep samples cold) was added to 1 mL TRIzol. Samples were spun at 4oC at 15,000 g for 15 minutes to remove lipid and cell debris. RNA was then extracted using the RNeasy Micro Kit (cat. no. 74004 from Qiagen, Germantown, MD, USA) following manufacturer’s protocol. DNA on Qiagen spin column was digested using the RNAse-free DNAse kit (cat. no. 79254 from Qiagen, Germantown, MD, USA) following manufacturer’s protocol. The isolated RNA was quantified using the Qubit Fluorometric Quantification system with the RNA Broad Range assay kit (cat. no. Q10210 from ThermoFisher Scientific, Waltham, Massachusetts, USA) following manufacturer’s protocol. RNA quality was assayed using the TapeStation RNA Broad Range assay (Agilent, Santa Clara California, USA). RNA species with RNA Integrity Number (RIN) > 7.5 were used in RNA-seq analyses.
Sequencing and quantification
RNA species were sequenced by Psomagen, Inc (Rockville, MD, USA). Raw FASTQ files are available on GEO (accession code GSE260764). Using fastQC, the quality of the sequenced reads was assessed, and low quality reads (Phred < 28) were removed with trimGalore. We aligned our sequenced reads to the mouse genome GRCm39 using hisat2 and quantified the aligned reads using htseq. Quantified transcripts per sample and transcripts per million (TPMs) are available on GEO (accession code GSE260764).
Quantification and statistical analysis
All data are presented as mean ± SEM. All gene expression plots show normalized counts.
Analysis of variance
Gene expression was normalized using R package DESeq2 with a design matrix Strain∗Diet. Genes and traits with zero values in more than 75% of the samples were removed. Statistical analyses were performed using R, ANOVA tests were performed using the base R function aov(). [2-way: trait or gene expression ∼ Strain∗Diet, 3-way: trait ∼ Strain∗Diet∗Time, 3-way removing batch effects: trait ∼ Strain∗Diet∗Time+batch]. Batch effects refer to sample effects due to differences in the two RNA-sequencing runs. P-values were adjusted using false discovery rate (FDR) correction for the number of traits or genes tested.
Principal component analysis
Principal component analysis was performed using base R function pca() on transcripts per million (TPM) normalized gene expression data or phenotypic traits. We removed genes with less than 0.1 TPM in 75% of the samples. For phenotypic data, we removed traits with many zeros (>50%), then removed samples with any zeros. Outliers were removed based on the results of the principal component analysis (Figure S21).
Differential expression analysis
To characterize the effects of diet, we performed differential gene expression analysis for visceral adipose tissue comparing RNA-seq data for the American diet to data for the other three diets. We used the DESeq2 R package with a design matrix Strain∗Diet+batch accounting for the strain, diet, and RNA sequencing batch. Volcano plots were made using package EnchancedVolcano.
Immune cell-type deconvolution
To determine which immune cell types were present in the bulk tissue RNA-seq data, we used CIBERSORTx.109 This program employs a machine learning algorithm that compares purified cells or single-cell gene signatures to infer the fraction of that cell type present in the bulk tissue RNA-seq data. We used the previously validated gene signature set ImmuneCC. It contains 511 reference genes corresponding to 25 unique immune cell types found in normal mouse immune cells, non-hematopoietic mouse tissues, and mouse tumor tissues.110 Briefly, to generate the ImmuneCC data set, PCA was used to cluster 115 publicly available datasets into 25 cell types. Genes highly expressed in non-hematopoietic cell types were removed, then differential expression analysis identified the top expressed genes in each cell type compared to the other 24 cell types. Using the ImmuneCC data set, we ran CIBERSORTx on the javabased web platform in relative mode using 100 iterations for each adipose tissue. For simplicity we averaged similar cell types and ran the program using 16 cell types.
Enrichment analysis
To determine if a set of genes was overrepresented within known pathways, we compared them to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) murine reference pathways. We performed these analyses on genes differentially expressed between diets (Figure S10) and the 421 strain:diet dependent genes (Table S4). In brief, the number of KEGG or GO pathway genes in the set of genes of interest was compared to the number of pathway genes in the ∼30,000 background genes expressed in VAT. Significance of enrichment was determined using Fisher’s exact test, performed in R using the packages org.Mm.eg.db, topGO, KEGGREST, and bc3net. P-values were adjusted using false discovery rate (FDR) correction for the number of pathways tested.
Partial least squares regression
Partial least squares regression is a dimensionality reduction technique used to determine how the variance in a set of X variables is related to the variance in a set of Y outcome variables.111,112,113,114 Like PCA, PLSR uses linear combinations of input variables to represent the multidimensional data in 2D. In PCA, the goal is to find principal component vectors such that the variance in one dataset is maximized. In PLSR, the goal is to find component vectors that represent axes through which the covariance of X and Y is maximized. Let X be a matrix of dimension N x K and let Y be a matrix N x M, where N is the number of samples (31 mice) and K is the number of diet sources (38 sources), and M is the number of genes (421 genes) (Figure S22). X and Y were centered and scaled. Both X and Y can be decomposed into latent variable vectors and loading vectors, such that ;
where t and u are latent variables and a and b are loading vectors. Loading vectors a and b are vectors of length K and M, respectively, that represent the direction of change in component space, while latent variables t and u are vectors of length n that represent the magnitude of change in component space. To obtain the values of a, t, b, and u that will maximize the covariance between X and Y in the new component space, we solve formula:
We used the package mixOmics115 function pls() to solve this function. In practice, solving the above function requires initializing a and b to any value, then iteratively computing a, t, b, and u as
until a converges. When a converges, the matrices X and Y are updated to remove the information of this component, where
The process of finding new a, t, b, and u values begins again with the residual X and Y matrix and continues for as many components will define the new space. The number of components included in the model is determined empirically using cross validation; we found that 3 components was optimal for this application. Loading vectors a and b now define the position of X input variables (diet sources) and Y output variables (genes) in the new component space, which were scaled and plotted together (Figure 5). Latent variables t and u now define the position of each sample in component space; in this study they define the position of each mouse (by strain and diet). These are projections of the samples into X and Y space, respectively. To plot both t and u information in the shared space, we averaged the X and Y projections. In this shared space, the covariance between diet sources and gene expression is related to the cosine of the angle between them; items nearby each other are strongly related, items opposite each other along a component axis are negatively related, and items at 90-degree angles apart are not related to each other. Because local regression is used to find a, t, b, and u, PLS is able to handle noisy or incomplete data, and particularly data with multiple correlated variables.
Co-expression network analysis
Co-expression networks identify groups of highly correlated, likely co-regulated genes, within a large gene set in an unbiased way. We used the iterativeWGCNA79 package to identify these groups, or modules, of related genes in the visceral adipose tissue gene expression. We used transcripts per million format, removed low-expressed genes (expression < 0.1 in > 75% of samples), and performed log normalization. The iterativeWGCNA program then computes the correlation between all genes, which is contained in a matrix. By clustering the correlation matrix using hierarchical clustering, the program identifies groups of genes with high correlations and thus similar patterns of expression, termed modules. iterativeWGCNA reassesses the strength of each gene’s membership in the module, and iteratively removes and reassigns genes to obtain more homogenous modules.
We defined the aggregate expression value of each module as the eigengene, which is the eigenvector or first principal component through the expression data for each module. We determined which module eigengenes were dependent on strain and diet using a 2-way ANOVA and which were correlated with metabolic traits using Pearson correlation. Both were corrected for the number of module eigengenes tested using FDR multiple test correction.
Published: November 4, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.111323.
Contributor Information
Heather A. Ferris, Email: hf4f@uvahealth.org.
Bijoy K. Kundu, Email: bkk5a@virginia.edu.
Sibylle Kranz, Email: sibylle.kranz@virginia.edu.
Mete Civelek, Email: mete@virginia.edu.
Susanna R. Keller, Email: srk4b@virginia.edu.
Supplemental information
References
- 1.Hales C.M., Carroll M.D., Fryar C.D., Ogden C.L. NCHS Data Brief; 2020. Prevalence of Obesity and Severe Obesity Among Adults: United States, 2017-2018; pp. 1–8. [PubMed] [Google Scholar]
- 2.Gardner C.D., Vadiveloo M.K., Petersen K.S., Anderson C.A.M., Springfield S., Van Horn L., Khera A., Lamendola C., Mayo S.M., Joseph J.J., American Heart Association Council on Lifestyle and Cardiometabolic Health Popular Dietary Patterns: Alignment With American Heart Association 2021 Dietary Guidance: A Scientific Statement From the American Heart Association. Circulation. 2023;147:1715–1730. doi: 10.1161/CIR.0000000000001146. [DOI] [PubMed] [Google Scholar]
- 3.Evert A.B., Dennison M., Gardner C.D., Garvey W.T., Lau K.H.K., MacLeod J., Mitri J., Pereira R.F., Rawlings K., Robinson S., et al. Nutrition Therapy for Adults With Diabetes or Prediabetes: A Consensus Report. Diabetes Care. 2019;42:731–754. doi: 10.2337/dci19-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guasch-Ferré M., Willett W.C. The Mediterranean diet and health: a comprehensive overview. J. Intern. Med. 2021;290:549–566. doi: 10.1111/joim.13333. [DOI] [PubMed] [Google Scholar]
- 5.Estruch R., Ros E. The role of the Mediterranean diet on weight loss and obesity-related diseases. Rev. Endocr. Metab. Disord. 2020;21:315–327. doi: 10.1007/s11154-020-09579-0. [DOI] [PubMed] [Google Scholar]
- 6.Kahleova H., Petersen K.F., Shulman G.I., Alwarith J., Rembert E., Tura A., Hill M., Holubkov R., Barnard N.D. Effect of a Low-Fat Vegan Diet on Body Weight, Insulin Sensitivity, Postprandial Metabolism, and Intramyocellular and Hepatocellular Lipid Levels in Overweight Adults: A Randomized Clinical Trial. JAMA Netw. Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.25454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Termannsen A.-D., Clemmensen K.K.B., Thomsen J.M., Nørgaard O., Díaz L.J., Torekov S.S., Quist J.S., Faerch K. Effects of vegan diets on cardiometabolic health: A systematic review and meta-analysis of randomized controlled trials. Obes. Rev. 2022;23 doi: 10.1111/obr.13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yokoyama Y., Levin S.M., Barnard N.D. Association between plant-based diets and plasma lipids: a systematic review and meta-analysis. Nutr. Rev. 2017;75:683–698. doi: 10.1093/nutrit/nux030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pilis W., Stec K., Zych M., Pilis A. Health benefits and risk associated with adopting a vegetarian diet. Rocz. Panstw. Zakl. Hig. 2014;65:9–14. [PubMed] [Google Scholar]
- 10.Gardner C.D., Trepanowski J.F., Del Gobbo L.C., Hauser M.E., Rigdon J., Ioannidis J.P.A., Desai M., King A.C. Effect of Low-Fat vs Low-Carbohydrate Diet on 12-Month Weight Loss in Overweight Adults and the Association With Genotype Pattern or Insulin Secretion: The DIETFITS Randomized Clinical Trial. JAMA. 2018;319:667–679. doi: 10.1001/jama.2018.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paoli A., Rubini A., Volek J.S., Grimaldi K.A. Beyond weight loss: a review of the therapeutic uses of very-low-carbohydrate (ketogenic) diets. Eur. J. Clin. Nutr. 2013;67:789–796. doi: 10.1038/ejcn.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kosinski C., Jornayvaz F.R. Effects of Ketogenic Diets on Cardiovascular Risk Factors: Evidence from Animal and Human Studies. Nutrients. 2017;9:517. doi: 10.3390/nu9050517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li M., Yuan J. Effects of very low-carbohydrate ketogenic diet on lipid metabolism in patients with type II diabetes mellitus: a meta-analysis. Nutr. Hosp. 2022;39:916–923. doi: 10.20960/nh.03987. [DOI] [PubMed] [Google Scholar]
- 14.Rosenbaum M., Hall K.D., Guo J., Ravussin E., Mayer L.S., Reitman M.L., Smith S.R., Walsh B.T., Leibel R.L. Glucose and Lipid Homeostasis and Inflammation in Humans Following an Isocaloric Ketogenic Diet. Obesity. 2019;27:971–981. doi: 10.1002/oby.22468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiu S., Bergeron N., Williams P.T., Bray G.A., Sutherland B., Krauss R.M. Comparison of the DASH (Dietary Approaches to Stop Hypertension) diet and a higher-fat DASH diet on blood pressure and lipids and lipoproteins: a randomized controlled trial. Am. J. Clin. Nutr. 2016;103:341–347. doi: 10.3945/ajcn.115.123281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghaedi E., Mohammadi M., Mohammadi H., Ramezani-Jolfaie N., Malekzadeh J., Hosseinzadeh M., Salehi-Abargouei A. Effects of a Paleolithic Diet on Cardiovascular Disease Risk Factors: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Adv. Nutr. 2019;10:634–646. doi: 10.1093/advances/nmz007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adeva-Andany M.M., González-Lucán M., Fernández-Fernández C., Carneiro-Freire N., Seco-Filgueira M., Pedre-Piñeiro A.M. Effect of diet composition on insulin sensitivity in humans. Clin. Nutr. ESPEN. 2019;33:29–38. doi: 10.1016/j.clnesp.2019.05.014. [DOI] [PubMed] [Google Scholar]
- 18.Martín-Peláez S., Fito M., Castaner O. Mediterranean Diet Effects on Type 2 Diabetes Prevention, Disease Progression, and Related Mechanisms. A Review. Nutrients. 2020;12:2236. doi: 10.3390/nu12082236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sofi F., Dinu M., Pagliai G., Cesari F., Gori A.M., Sereni A., Becatti M., Fiorillo C., Marcucci R., Casini A. Low-Calorie Vegetarian Versus Mediterranean Diets for Reducing Body Weight and Improving Cardiovascular Risk Profile: CARDIVEG Study (Cardiovascular Prevention With Vegetarian Diet) Circulation. 2018;137:1103–1113. doi: 10.1161/CIRCULATIONAHA.117.030088. [DOI] [PubMed] [Google Scholar]
- 20.Golzarand M., Mirmiran P., Azizi F. Adherence to the MIND diet and the risk of cardiovascular disease in adults: a cohort study. Food Funct. 2022;13:1651–1658. doi: 10.1039/d1fo02069b. [DOI] [PubMed] [Google Scholar]
- 21.Glenn A.J., Lo K., Jenkins D.J.A., Boucher B.A., Hanley A.J., Kendall C.W.C., Manson J.E., Vitolins M.Z., Snetselaar L.G., Liu S., Sievenpiper J.L. Relationship Between a Plant-Based Dietary Portfolio and Risk of Cardiovascular Disease: Findings From the Women’s Health Initiative Prospective Cohort Study. J. Am. Heart Assoc. 2021;10 doi: 10.1161/JAHA.121.021515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kahleova H., Salas-Salvadó J., Rahelić D., Kendall C.W., Rembert E., Sievenpiper J.L. Dietary Patterns and Cardiometabolic Outcomes in Diabetes: A Summary of Systematic Reviews and Meta-Analyses. Nutrients. 2019;11:2209. doi: 10.3390/nu11092209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo Y., Wang J., Sun L., Gu W., Zong G., Song B., Shen C., Zhou P., Chen Y., Wu Y., et al. Isocaloric-restricted Mediterranean Diet and Chinese Diets High or Low in Plants in Adults With Prediabetes. J. Clin. Endocrinol. Metab. 2022;107:2216–2227. doi: 10.1210/clinem/dgac303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sotos-Prieto M., Smith C.E., Lai C.-Q., Tucker K.L., Ordovas J.M., Mattei J. Mediterranean Diet Adherence Modulates Anthropometric Measures by TCF7L2 Genotypes among Puerto Rican Adults. J. Nutr. 2020;150:167–175. doi: 10.1093/jn/nxz210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mattei J., Qi Q., Hu F.B., Sacks F.M., Qi L. TCF7L2 genetic variants modulate the effect of dietary fat intake on changes in body composition during a weight-loss intervention. Am. J. Clin. Nutr. 2012;96:1129–1136. doi: 10.3945/ajcn.112.038125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bodhini D., Gaal S., Shatwan I., Ramya K., Ellahi B., Surendran S., Sudha V., Anjana M.R., Mohan V., Lovegrove J.A., et al. Interaction between TCF7L2 polymorphism and dietary fat intake on high density lipoprotein cholesterol. PLoS One. 2017;12 doi: 10.1371/journal.pone.0188382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grau K., Cauchi S., Holst C., Astrup A., Martinez J.A., Saris W.H.M., Blaak E.E., Oppert J.-M., Arner P., Rössner S., et al. TCF7L2 rs7903146-macronutrient interaction in obese individuals’ responses to a 10-wk randomized hypoenergetic diet. Am. J. Clin. Nutr. 2010;91:472–479. doi: 10.3945/ajcn.2009.27947. [DOI] [PubMed] [Google Scholar]
- 28.Gomez-Delgado F., Alcala-Diaz J.F., Leon-Acuña A., Lopez-Moreno J., Delgado-Lista J., Gomez-Marin B., Roncero-Ramos I., Yubero-Serrano E.M., Rangel-Zuñiga O.A., Vals-Delgado C., et al. Apolipoprotein E genetic variants interact with Mediterranean diet to modulate postprandial hypertriglyceridemia in coronary heart disease patients: CORDIOPREV study. Eur. J. Clin. Invest. 2019;49 doi: 10.1111/eci.13146. [DOI] [PubMed] [Google Scholar]
- 29.Campos-Perez W., Perez-Robles M., Torres-Castillo N., Rodríguez-Reyes S.C., De la Cerda Trujillo L.F., Navarro-Muñiz E., Lopez-Lizárraga C.R., Llamas-Covarrubias I.M., Martinez-Lopez E. Physical inactivity and excessive sucrose consumption are associated with higher serum lipids in subjects with Taq1B CETP polymorphism. J. Hum. Nutr. Diet. 2020;33:299–307. doi: 10.1111/jhn.12747. [DOI] [PubMed] [Google Scholar]
- 30.Wuni R., Kuhnle G.G.C., Wynn-Jones A.A., Vimaleswaran K.S. A Nutrigenetic Update on CETP Gene-Diet Interactions on Lipid-Related Outcomes. Curr. Atheroscler. Rep. 2022;24:119–132. doi: 10.1007/s11883-022-00987-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khatibi N., Mirzababaei A., Shiraseb F., Abaj F., Koohdani F., Mirzaei K. Interactions between caveolin 1 polymorphism and the Mediterranean and Mediterranean-DASH Intervention for Neurodegenerative Delay diet (MIND) diet on metabolic dyslipidemia in overweight and obese adult women: a cross-sectional study. BMC Res. Notes. 2021;14:364. doi: 10.1186/s13104-021-05777-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abaj F., Koohdani F., Rafiee M., Alvandi E., Yekaninejad M.S., Mirzaei K. Interactions between Caveolin-1 (rs3807992) polymorphism and major dietary patterns on cardio-metabolic risk factors among obese and overweight women. BMC Endocr. Disord. 2021;21:138. doi: 10.1186/s12902-021-00800-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Senftleber N., Jørgensen M.E., Jørsboe E., Imamura F., Forouhi N.G., Larsen C.L., Bjerregaard P., Hansen T., Albrechtsen A. Genetic study of the Arctic CPT1A variant suggests that its effect on fatty acid levels is modulated by traditional Inuit diet. Eur. J. Hum. Genet. 2020;28:1592–1601. doi: 10.1038/s41431-020-0674-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tam C.H.T., Wang Y., Lee H.M., Luk A.O.Y., Tong P.C.Y., Chan M.H.M., Ozaki R., Kong A.P.S., So W.Y., Chan J.C.N., Ma R.C.W. Early gene-diet interaction between glucokinase regulatory protein (GCKR) polymorphism, vegetable and fish intakes in modulating triglyceride levels in healthy adolescents. Nutr. Metab. Cardiovasc. Dis. 2015;25:951–958. doi: 10.1016/j.numecd.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 35.Fernandes Silva L., Vangipurapu J., Kuulasmaa T., Laakso M. An intronic variant in the GCKR gene is associated with multiple lipids. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-46750-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X., Qi Q., Zhang C., Smith S.R., Hu F.B., Sacks F.M., Bray G.A., Qi L. FTO genotype and 2-year change in body composition and fat distribution in response to weight-loss diets: the POUNDS LOST Trial. Diabetes. 2012;61:3005–3011. doi: 10.2337/db11-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin X., Qi Q., Zheng Y., Huang T., Lathrop M., Zelenika D., Bray G.A., Sacks F.M., Liang L., Qi L. Neuropeptide Y genotype, central obesity, and abdominal fat distribution: the POUNDS LOST trial. Am. J. Clin. Nutr. 2015;102:514–519. doi: 10.3945/ajcn.115.107276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corella D., Qi L., Sorlí J.V., Godoy D., Portolés O., Coltell O., Greenberg A.S., Ordovas J.M. Obese subjects carrying the 11482G>A polymorphism at the perilipin locus are resistant to weight loss after dietary energy restriction. J. Clin. Endocrinol. Metab. 2005;90:5121–5126. doi: 10.1210/jc.2005-0576. [DOI] [PubMed] [Google Scholar]
- 39.Smith C.E., Tucker K.L., Yiannakouris N., Garcia-Bailo B., Mattei J., Lai C.-Q., Parnell L.D., Ordovás J.M. Perilipin polymorphism interacts with dietary carbohydrates to modulate anthropometric traits in hispanics of Caribbean origin. J. Nutr. 2008;138:1852–1858. doi: 10.1093/jn/138.10.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corella D., Qi L., Tai E.S., Deurenberg-Yap M., Tan C.E., Chew S.K., Ordovas J.M. Perilipin gene variation determines higher susceptibility to insulin resistance in Asian women when consuming a high-saturated fat, low-carbohydrate diet. Diabetes Care. 2006;29:1313–1319. doi: 10.2337/dc06-0045. [DOI] [PubMed] [Google Scholar]
- 41.Heianza Y., Ma W., Huang T., Wang T., Zheng Y., Smith S.R., Bray G.A., Sacks F.M., Qi L. Macronutrient Intake-Associated FGF21 Genotype Modifies Effects of Weight-Loss Diets on 2-Year Changes of Central Adiposity and Body Composition: The POUNDS Lost Trial. Diabetes Care. 2016;39:1909–1914. doi: 10.2337/dc16-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Luis D.A., Primo D., Izaola O., Aller R. Adiponectin Gene Variant rs266729 Interacts with Different Macronutrient Distribution of Two Different Hypocaloric Diets. Lifestyle Genom. 2020;13:20–27. doi: 10.1159/000503863. [DOI] [PubMed] [Google Scholar]
- 43.Goni L., Sun D., Heianza Y., Wang T., Huang T., Martínez J.A., Shang X., Bray G.A., Smith S.R., Sacks F.M., Qi L. A circadian rhythm-related MTNR1B genetic variant modulates the effect of weight-loss diets on changes in adiposity and body composition: the POUNDS Lost trial. Eur. J. Nutr. 2019;58:1381–1389. doi: 10.1007/s00394-018-1660-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahmoudinezhad M., Khosravaniardakani S., Saljoughi Badelou L., Fayyazishishavan E., Kahroba H., Farhangi M.A. The integrative panel of fatty acid desaturase-2 (FADS2) rs174583 gene polymorphism and dietary indices (DQI-I and HEI) affects cardiovascular risk factors among obese individuals. BMC Endocr. Disord. 2023;23:41. doi: 10.1186/s12902-023-01289-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khodarahmi M., Javidzade P., Farhangi M.A., Hashemzehi A., Kahroba H. Interplay between fatty acid desaturase2 (FADS2) rs174583 genetic variant and dietary antioxidant capacity: cardio-metabolic risk factors in obese individuals. BMC Endocr. Disord. 2022;22:167. doi: 10.1186/s12902-022-01075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang T., Wang T., Heianza Y., Wiggs J., Sun D., Choi H.-K., Chai J.F., Sim X., Khor C.C., Friedlander Y., et al. Fish and marine fatty acids intakes, the FADS genotypes and long-term weight gain: a prospective cohort study. BMJ Open. 2019;9 doi: 10.1136/bmjopen-2018-022877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang T., Wang T., Heianza Y., Sun D., Ivey K., Durst R., Schwarzfuchs D., Stampfer M.J., Bray G.A., Sacks F.M., et al. HNF1A variant, energy-reduced diets and insulin resistance improvement during weight loss: The POUNDS Lost trial and DIRECT. Diabetes Obes. Metab. 2018;20:1445–1452. doi: 10.1111/dom.13250. [DOI] [PubMed] [Google Scholar]
- 48.Freitas R.N., Khaw K.-T., Wu K., Bowman R., Jeffery H., Luben R., Wareham N.J., Bingham S.A. A single nucleotide polymorphism in the 3-hydroxy-3-methylglutaryl-coenzyme A reductase gene (HMGCR) influences the serum triacylglycerol relationship with dietary fat and fibre in the European Prospective Investigation into Cancer and Nutrition in Norfolk (EPIC-Norfolk) study. Br. J. Nutr. 2010;104:765–772. doi: 10.1017/S0007114510001145. [DOI] [PubMed] [Google Scholar]
- 49.Corella D., Peloso G., Arnett D.K., Demissie S., Cupples L.A., Tucker K., Lai C.-Q., Parnell L.D., Coltell O., Lee Y.-C., Ordovas J.M. APOA2, dietary fat, and body mass index: replication of a gene-diet interaction in 3 independent populations. Arch. Intern. Med. 2009;169:1897–1906. doi: 10.1001/archinternmed.2009.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qi Q., Bray G.A., Smith S.R., Hu F.B., Sacks F.M., Qi L. Insulin receptor substrate 1 gene variation modifies insulin resistance response to weight-loss diets in a 2-year randomized trial: the Preventing Overweight Using Novel Dietary Strategies (POUNDS LOST) trial. Circulation. 2011;124:563–571. doi: 10.1161/CIRCULATIONAHA.111.025767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muhammad H.F.L., Sulistyoningrum D.C., Huriyati E., Lee Y.Y., Muda W.A.M.W. The interaction between energy intake, physical activity and UCP2 -866G/A gene variation on weight gain and changes in adiposity: an Indonesian Nutrigenetic Cohort (INDOGENIC) Br. J. Nutr. 2021;125:611–617. doi: 10.1017/S0007114520003104. [DOI] [PubMed] [Google Scholar]
- 52.de Luis D.A., Izaola O., Primo D., Gómez J.J.L. Role of beta-2 adrenergic receptor polymorphism (rs1042714) on body weight and glucose metabolism response to a meal-replacement hypocaloric diet. Nutrition. 2023;116 doi: 10.1016/j.nut.2023.112170. [DOI] [PubMed] [Google Scholar]
- 53.Francis M., Li C., Sun Y., Zhou J., Li X., Brenna J.T., Ye K. Genome-wide association study of fish oil supplementation on lipid traits in 81,246 individuals reveals new gene-diet interaction loci. PLoS Genet. 2021;17 doi: 10.1371/journal.pgen.1009431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roa-Díaz Z.M., Teuscher J., Gamba M., Bundo M., Grisotto G., Wehrli F., Gamboa E., Rojas L.Z., Gómez-Ochoa S.A., Verhoog S., et al. Gene-diet interactions and cardiovascular diseases: a systematic review of observational and clinical trials. BMC Cardiovasc. Disord. 2022;22:377. doi: 10.1186/s12872-022-02808-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giovanella J., Wollinger L.M., Capra L., Dresch F., Genro J.P., Contini V. Diet-gene interaction: effects of polymorphisms in the ACE, AGT and BDKRB2 genes and the consumption of sodium, potassium, calcium, and magnesium on blood pressure of normotensive adult individuals. Mol. Cell. Biochem. 2021;476:1211–1219. doi: 10.1007/s11010-020-03983-5. [DOI] [PubMed] [Google Scholar]
- 56.Baratta F., Angelico F., Del Ben M. Challenges in Improving Adherence to Diet and Drug Treatment in Hypercholesterolemia Patients. Int. J. Environ. Res. Public Health. 2023;20:5878. doi: 10.3390/ijerph20105878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maston G., Franklin J., Hocking S., Swinbourne J., Gibson A., Manson E., Sainsbury A., Markovic T. Dietary adherence and program attrition during a severely energy-restricted diet among people with complex class III obesity: A qualitative exploration. PLoS One. 2021;16 doi: 10.1371/journal.pone.0253127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vitolins M.Z., Case T.L. What Makes Nutrition Research So Difficult to Conduct and Interpret? Diabetes Spectr. 2020;33:113–117. doi: 10.2337/ds19-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lusis A.J., Seldin M.M., Allayee H., Bennett B.J., Civelek M., Davis R.C., Eskin E., Farber C.R., Hui S., Mehrabian M., et al. The Hybrid Mouse Diversity Panel: a resource for systems genetics analyses of metabolic and cardiovascular traits. J. Lipid Res. 2016;57:925–942. doi: 10.1194/jlr.R066944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Griffin L.E., Essenmacher L., Racine K.C., Iglesias-Carres L., Tessem J.S., Smith S.M., Neilson A.P. Diet-induced obesity in genetically diverse collaborative cross mouse founder strains reveals diverse phenotype response and amelioration by quercetin treatment in 129S1/SvImJ, PWK/EiJ, CAST/PhJ, and WSB/EiJ mice. J. Nutr. Biochem. 2021;87 doi: 10.1016/j.jnutbio.2020.108521. [DOI] [PubMed] [Google Scholar]
- 61.Yam P., Albright J., VerHague M., Gertz E.R., Pardo-Manuel de Villena F., Bennett B.J. Genetic Background Shapes Phenotypic Response to Diet for Adiposity in the Collaborative Cross. Front. Genet. 2020;11 doi: 10.3389/fgene.2020.615012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bennett B.J., Davis R.C., Civelek M., Orozco L., Wu J., Qi H., Pan C., Packard R.R.S., Eskin E., Yan M., et al. Genetic Architecture of Atherosclerosis in Mice: A Systems Genetics Analysis of Common Inbred Strains. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parks B.W., Sallam T., Mehrabian M., Psychogios N., Hui S.T., Norheim F., Castellani L.W., Rau C.D., Pan C., Phun J., et al. Genetic architecture of insulin resistance in the mouse. Cell Metab. 2015;21:334–347. doi: 10.1016/j.cmet.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nelson M.E., Madsen S., Cooke K.C., Fritzen A.M., Thorius I.H., Masson S.W.C., Carroll L., Weiss F.C., Seldin M.M., Potter M., et al. Systems-level analysis of insulin action in mouse strains provides insight into tissue- and pathway-specific interactions that drive insulin resistance. Cell Metab. 2022;34:227–239.e6. doi: 10.1016/j.cmet.2021.12.013. [DOI] [PubMed] [Google Scholar]
- 65.Parks B.W., Nam E., Org E., Kostem E., Norheim F., Hui S.T., Pan C., Civelek M., Rau C.D., Bennett B.J., et al. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 2013;17:141–152. doi: 10.1016/j.cmet.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Börgeson E., Boucher J., Hagberg C.E. Of mice and men: Pinpointing species differences in adipose tissue biology. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barrington W.T., Wulfridge P., Wells A.E., Rojas C.M., Howe S.Y.F., Perry A., Hua K., Pellizzon M.A., Hansen K.D., Voy B.H., et al. Improving Metabolic Health Through Precision Dietetics in Mice. Genetics. 2018;208:399–417. doi: 10.1534/genetics.117.300536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Table 056. Hyattsville, MD. 2017. https://www.cdc.gov/nchs/hus/data-finder.htm
- 69.Kirby A., Kang H.M., Wade C.M., Cotsapas C., Kostem E., Han B., Furlotte N., Kang E.Y., Rivas M., Bogue M.A., et al. Fine mapping in 94 inbred mouse strains using a high-density haplotype resource. Genetics. 2010;185:1081–1095. doi: 10.1534/genetics.110.115014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blencowe M., Chen X., Zhao Y., Itoh Y., McQuillen C.N., Han Y., Shou B.L., McClusky R., Reue K., Arnold A.P., Yang X. Relative contributions of sex hormones, sex chromosomes, and gonads to sex differences in tissue gene regulation. Genome Res. 2022;32:807–824. doi: 10.1101/gr.275965.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ghigliotti G., Barisione C., Garibaldi S., Fabbi P., Brunelli C., Spallarossa P., Altieri P., Rosa G., Spinella G., Palombo D., et al. Adipose Tissue Immune Response: Novel Triggers and Consequences for Chronic Inflammatory Conditions. Inflammation. 2014;37:1337–1353. doi: 10.1007/s10753-014-9914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maeda-Smithies N., Hiller S., Dong S., Kim H.-S., Bennett B.J., Kayashima Y. Ectopic expression of the Stabilin2 gene triggered by an intracisternal A particle (IAP) element in DBA/2J strain of mice. Mamm. Genome. 2020;31:2–16. doi: 10.1007/s00335-019-09824-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen K., Zhang J., Liang F., Zhu Q., Cai S., Tong X., He Z., Liu X., Chen Y., Mo D. HMGB2 orchestrates mitotic clonal expansion by binding to the promoter of C/EBPβ to facilitate adipogenesis. Cell Death Dis. 2021;12:666. doi: 10.1038/s41419-021-03959-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun L., Ji S., Xie X., Si L., Liu S., Lin Y., Wang Y., Song Z., Fang N., An Y., Yang J. Deciphering the interaction between Twist1 and PPARγ during adipocyte differentiation. Cell Death Dis. 2023;14:764. doi: 10.1038/s41419-023-06283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hunyenyiwa T., Hendee K., Matus K., Kyi P., Mammoto T., Mammoto A. Obesity Inhibits Angiogenesis Through TWIST1-SLIT2 Signaling. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.693410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pastel E., Pointud J.-C., Volat F., Martinez A., Lefrançois-Martinez A.-M. Aldo-Keto Reductases 1B in Endocrinology and Metabolism. Front. Pharmacol. 2012;3:148. doi: 10.3389/fphar.2012.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhong L., Liu Z., Yan R., Johnson S., Zhao Y., Fang X., Cao D. Aldo-keto reductase family 1 B10 protein detoxifies dietary and lipid-derived alpha, beta-unsaturated carbonyls at physiological levels. Biochem. Biophys. Res. Commun. 2009;387:245–250. doi: 10.1016/j.bbrc.2009.06.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song W., Luo Q., Zhang Y., Zhou L., Liu Y., Ma Z., Guo J., Huang Y., Cheng L., Meng Z., et al. Organic cation transporter 3 (Oct3) is a distinct catecholamines clearance route in adipocytes mediating the beiging of white adipose tissue. PLoS Biol. 2019;17 doi: 10.1371/journal.pbio.2006571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Greenfest-Allen E., Cartailler J.-P., Magnuson M.A., Stoeckert C.J. iterativeWGCNA: iterative refinement to improve module detection from WGCNA co-expression networks. bioRxiv. 2017 doi: 10.1101/234062. Preprint at. [DOI] [Google Scholar]
- 80.Hussain S., Kirwin S.J., Stohlman S.A. Increased T regulatory cells lead to development of Th2 immune response in male SJL mice. Autoimmunity. 2011;44:219–228. doi: 10.3109/08916934.2010.519746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nishizuka M., Hayashi T., Asano M., Osada S., Imagawa M. KCNK10, a Tandem Pore Domain Potassium Channel, Is a Regulator of Mitotic Clonal Expansion during the Early Stage of Adipocyte Differentiation. Int. J. Mol. Sci. 2014;15:22743–22756. doi: 10.3390/ijms151222743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gorden A., Yang R., Yerges-Armstrong L.M., Ryan K.A., Speliotes E., Borecki I.B., Harris T.B., Chu X., Wood G.C., Still C.D., et al. Genetic Variation at NCAN Locus is Associated with Inflammation and Fibrosis in Non-alcoholic Fatty Liver Disease in Morbid Obesity. Hum. Hered. 2013;75:34–43. doi: 10.1159/000346195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stanford S.M., Collins M., Diaz M.A., Holmes Z.J., Gries P., Bliss M.R., Lodi A., Zhang V., Tiziani S., Bottini N. The low molecular weight protein tyrosine phosphatase promotes adipogenesis and subcutaneous adipocyte hypertrophy. J. Cell. Physiol. 2021;236:6630–6642. doi: 10.1002/jcp.30307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pandey S.K., Yu X.X., Watts L.M., Michael M.D., Sloop K.W., Rivard A.R., Leedom T.A., Manchem V.P., Samadzadeh L., McKay R.A., et al. Reduction of low molecular weight protein-tyrosine phosphatase expression improves hyperglycemia and insulin sensitivity in obese mice. J. Biol. Chem. 2007;282:14291–14299. doi: 10.1074/jbc.M609626200. [DOI] [PubMed] [Google Scholar]
- 85.De Lorenzo A., Di Renzo L., Puja A., Saccucci P., Gloria-Bottini F., Bottini E. A study of acid phosphatase locus 1 in women with high fat content and normal body mass index. Metabolism. 2009;58:351–354. doi: 10.1016/j.metabol.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 86.Lucarini N., Antonacci E., Bottini N., Gloria Bottini F. Low-molecular-weight acid phosphatase (ACP1), obesity, and blood lipid levels in subjects with non-insulin-dependent diabetes mellitus. Hum. Biol. 1997;69:509–515. [PubMed] [Google Scholar]
- 87.Xu B.T., Teng F.Y., Wu Q., Wan S.R., Li X.Y., Tan X.Z., Xu Y., Jiang Z.Z. Bdh1 overexpression ameliorates hepatic injury by activation of Nrf2 in a MAFLD mouse model. Cell Death Discov. 2022;8:49. doi: 10.1038/s41420-022-00840-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fernández-Real J.M., McClain D., Manco M. Mechanisms Linking Glucose Homeostasis and Iron Metabolism Toward the Onset and Progression of Type 2 Diabetes. Diabetes Care. 2015;38:2169–2176. doi: 10.2337/dc14-3082. [DOI] [PubMed] [Google Scholar]
- 89.Gianmoena K., Gasparoni N., Jashari A., Gabrys P., Grgas K., Ghallab A., Nordström K., Gasparoni G., Reinders J., Edlund K., et al. Epigenomic and transcriptional profiling identifies impaired glyoxylate detoxification in NAFLD as a risk factor for hyperoxaluria. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109526. [DOI] [PubMed] [Google Scholar]
- 90.Barbara M., Scott A., Alkhouri N. New insights into genetic predisposition and novel therapeutic targets for nonalcoholic fatty liver disease. Hepatobiliary Surg. Nutr. 2018;7:372–381. doi: 10.21037/hbsn.2018.08.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mooli R.G.R., Ramakrishnan S.K. Emerging Role of Hepatic Ketogenesis in Fatty Liver Disease. Front. Physiol. 2022;13 doi: 10.3389/fphys.2022.946474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nagashima S., Yagyu H., Ohashi K., Tazoe F., Takahashi M., Ohshiro T., Bayasgalan T., Okada K., Sekiya M., Osuga J.i., Ishibashi S. Liver-specific deletion of 3-hydroxy-3-methylglutaryl coenzyme A reductase causes hepatic steatosis and death. Arterioscler. Thromb. Vasc. Biol. 2012;32:1824–1831. doi: 10.1161/ATVBAHA.111.240754. [DOI] [PubMed] [Google Scholar]
- 93.Zhou K., Yee S.W., Seiser E.L., van Leeuwen N., Tavendale R., Bennett A.J., Groves C.J., Coleman R.L., van der Heijden A.A., Beulens J.W., et al. Variation in the glucose transporter gene SLC2A2 is associated with glycemic response to metformin. Nat. Genet. 2016;48:1055–1059. doi: 10.1038/ng.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Peters J.L., Dufner-Beattie J., Xu W., Geiser J., Lahner B., Salt D.E., Andrews G.K. Targeting of the mouse Slc39a2 (Zip2) gene reveals highly cell-specific patterns of expression, and unique functions in zinc, iron, and calcium homeostasis. Genesis. 2007;45:339–352. doi: 10.1002/dvg.20297. [DOI] [PubMed] [Google Scholar]
- 95.Ryu M.-S., Lichten L.A., Liuzzi J.P., Cousins R.J. Zinc transporters ZnT1 (Slc30a1), Zip8 (Slc39a8), and Zip10 (Slc39a10) in mouse red blood cells are differentially regulated during erythroid development and by dietary zinc deficiency. J. Nutr. 2008;138:2076–2083. doi: 10.3945/jn.108.093575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Britton L., Jaskowski L.-A., Bridle K., Secondes E., Wallace D., Santrampurwala N., Reiling J., Miller G., Mangiafico S., Andrikopoulos S., et al. Ferroportin Expression in Adipocytes Does Not Contribute to Iron Homeostasis or Metabolic Responses to a High Calorie Diet. Cell. Mol. Gastroenterol. Hepatol. 2018;5:319–331. doi: 10.1016/j.jcmgh.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Winn N.C., Wolf E.M., Cottam M.A., Bhanot M., Hasty A.H. Myeloid-specific deletion of ferroportin impairs macrophage bioenergetics but is disconnected from systemic insulin action in adult mice. Am. J. Physiol. Endocrinol. Metab. 2021;321:E376–E391. doi: 10.1152/ajpendo.00116.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yee S.W., Buitrago D., Stecula A., Ngo H.X., Chien H.-C., Zou L., Koleske M.L., Giacomini K.M. Deorphaning a solute carrier 22 family member, SLC22A15, through functional genomic studies. FASEB J. 2020;34:15734–15752. doi: 10.1096/fj.202001497R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Voruganti V.S. Precision Nutrition: Recent Advances in Obesity. Physiology. 2023;38:1. doi: 10.1152/physiol.00014.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.R Core Team . R Foundation for Statistical Computing; 2023. R: A Language and Environment for Statistical Computing.https://www.R-project.org/ [Google Scholar]
- 101.Wickham H. Springer-Verlag; 2016. ggplot2: Elegant Graphics for Data Analysis.https://ggplot2.tidyverse.org [Google Scholar]
- 102.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Monget P., Coquery J., Yao F.Z., Liquet B. Data Integration Project; 2016. mixOmics: Omics.https://CRAN.R-project.org/package=mixOmics R package version 6.1.1. [Google Scholar]
- 104.Andrews S. Babraham Bioinformatics. 2010. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 105.Krueger F. Babraham Bioinformatics. 2012. https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
- 106.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Putri G., Anders S., Pyl P.T., Pimanda J.E., Zanini F. Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics. 2022;38:2943–2945. doi: 10.1093/bioinformatics/btac166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M., Li H. Twelve years of SAMtools and BCFtools. GigaScience. 2021;10 doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Newman A.M., Steen C.B., Liu C.L., Gentles A.J., Chaudhuri A.A., Scherer F., Khodadoust M.S., Esfahani M.S., Luca B.A., Steiner D., et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen Z., Huang A., Sun J., Jiang T., Qin F.X.-F., Wu A. Inference of immune cell composition on the expression profiles of mouse tissue. Sci. Rep. 2017;7 doi: 10.1038/srep40508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wold H. Soft Modelling by Latent Variables: The Non-Linear Iterative Partial Least Squares (NIPALS) Approach. J. Appl. Probab. 1975;12:117–142. doi: 10.1017/S0021900200047604. [DOI] [Google Scholar]
- 112.Wold S., Sjöström M., Eriksson L. PLS-regression: a basic tool of chemometrics. Chemometr. Intell. Lab. Syst. 2001;58:109–130. doi: 10.1016/S0169-7439(01)00155-1. [DOI] [Google Scholar]
- 113.Lê Cao K.-A., Martin P.G.P., Robert-Granié C., Besse P. Sparse canonical methods for biological data integration: application to a cross-platform study. BMC Bioinf. 2009;10:34. doi: 10.1186/1471-2105-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lê Cao K.-A., Rossouw D., Robert-Granié C., Besse P. A Sparse PLS for Variable Selection when Integrating Omics data. Stat. Appl. Genet. Mol. Biol. 2008;7:35. doi: 10.2202/1544-6115.1390. [DOI] [PubMed] [Google Scholar]
- 115.Rohart F., Gautier B., Singh A., Lê Cao K.A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017;13 doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
RNA-seq data (raw FASTQ files and processed counts) are deposited at GEO and are publicly available with the accession code GSE260764.
-
•
All original code has been deposited on Github at https://github.com/jnr3hh/Reed_Civelek_Keller_2024_Manuscript and is publicly available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in this article is available from the lead contact upon request.
-
•
Counts, TPMs and fastq files from the RNA-seq data generated in this study are publicly available on GEO (accession code GSE260764). Codes to perform the analyses in this study are available on https://github.com/jnr3hh/Reed_Civelek_Keller_2024_Manuscript.