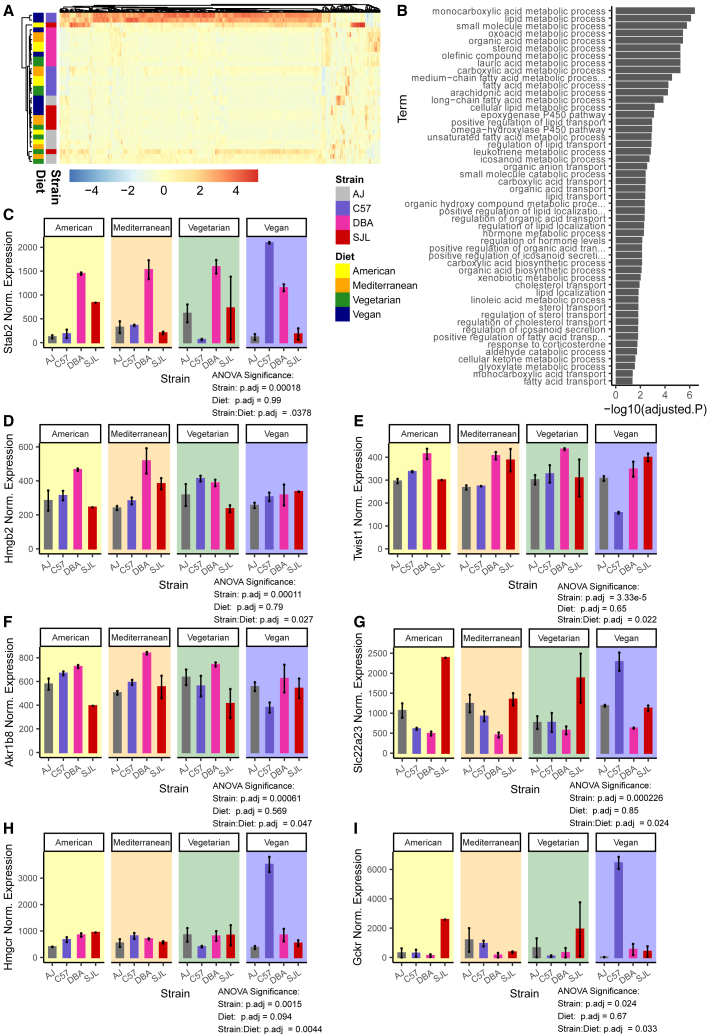
Figure 4.
Gene expression response driven by strain:diet interaction effects in visceral adipose tissue correlates with metabolic responses
(A) Heatmap of 421 visceral adipose tissue genes that exhibit significant strain:diet-dependent effects. Color represents the normalized expression value across samples, normalization performed with DESeq2.
(B) Pathway analysis of 421 visceral adipose tissue genes that exhibit significant strain:diet-dependent effects. Significantly enriched pathways from the Gene Ontology-biological processes database. P-values adjusted using FDR correction for multiple tests.
(C) Expression of gene Stab2 in visceral adipose tissue, with significant strain:diet-dependent effects (n = 2, 18 weeks old, male).
(D) Expression of gene Hmgb2 in visceral adipose tissue, with significant strain:diet-dependent effects (n = 2, 18 weeks old, male).
(E) Expression of gene Twist1 in visceral adipose tissue, with significant strain:diet-dependent effects (n = 2, 18 weeks old, male).
(F) Expression of gene Akr1b8 in visceral adipose tissue, with significant strain:diet-dependent effects (n = 2, 18 weeks old, male).
(G) Expression of gene Slc22a23 in visceral adipose tissue, with significant strain:diet-dependent effects (n = 2, 18 weeks old, male).
(H) Expression of gene Hmgcr in visceral adipose tissue, with significant strain:diet-dependent effects (n = 2, 18 weeks old, male).
(I) Expression of gene Gckr in visceral adipose tissue, with significant strain:diet-dependent effects (n = 2, 18 weeks old, male).
Error bars represent mean ± SEM, p-values from two-way ANOVA performed considering the effects of Strain, Diet, and interaction effects, p-values adjusted using FDR correction (n = ∼31,000) (Table S4). Gene expression plots show normalized counts.
See also Figures S12–S20.