Highlights
-
•
Huang et al. performed genome-wide expression profiling microarray in the tissues of 81 rectal cancer patients before neoadjuvant therapy to analyze and discover the differentially expressed genes related to chemoradiotherapy resistance, and the results of the above analyses were verified in 45 rectal cancer patients, and finally a 20-gene signature was proposed to be a predictor of chemoradiotherapy response. Identifying a signature with group of genes can help predict the response to chemoradiotherapy of rectal cancer patients.
-
•
Among the 20 genes identified above, zinc finger protein 37A (ZNF37A) was the most significantly differentially expressed gene in rectal cancer tissues with different responses. Molecular biology experiments revealed that ZNF37A bound to the tumor necrosis factor receptor superfamily member 6B (TNFRSF6B) promoter region, thereby suppressing its transcriptional activity, thus inducing chemotherapy resistance by inhibiting apoptosis in rectal cancer.
-
•
This study established a multigene expression model for predicting the sensitivity of rectal cancer to chemoradiotherapy and elucidated the mechanism of resistance to synchronized chemoradiotherapy. These findings will serve as valuable clues for the accurate assessment of the sensitivity of chemoradiotherapy in rectal cancer patients and the development of drugs related to overcoming therapy resistance.
Keywords: Rectal cancer, Gene expression, Chemoradiotherapy resistance, ZNF37A, TNFRSF6B
Abstract
The identification a signature comprising a group of genes as markers of cancer response to chemoradiotherapy would be more appropriate and effective for predicting chemoradiotherapy efficacy. This study investigated the differentially expressed genes (DEGs) related to chemoradiotherapy resistance and established a multigene expression model for predicting the sensitivity of rectal cancer to chemoradiotherapy in rectal cancer patients, elucidated the mechanism of resistance to synchronized chemoradiotherapy. The genome-wide expression profiling microarray were performed in the tissues of 81 rectal cancer patients before neoadjuvant therapy to analyze and discover DEGs related to chemoradiotherapy resistance, and the results were verified in 45 rectal cancer patients, and finally a 20-gene signature was proposed to be a predictor of chemoradiotherapy response. Molecular biology experiments revealed that zinc finger protein 37A (ZNF37A) downregulation leads to therapeutic resistance. This study identified a 20-gene signature with group of genes can help predict the response to chemoradiotherapy of rectal cancer patients. ZNF37A demonstrated a statistically significant correlation with sensitivity to chemoradiotherapy and survival in patients with LARC who underwent chemoradiotherapy. The findings revealed that ZNF37A bound to the tumor necrosis factor receptor superfamily member 6B (TNFRSF6B) promoter region, thereby suppressing its transcriptional activity. Reduced expression of ZNF37A induces chemoradiation resistance by inhibiting apoptosis in colorectal cancer (CRC) cells. TNFRSF6B Knockdown restored the sensitivity of CRC to chemoradiotherapy. ZNF37A is an effective modulator of chemoradiotherapy response in rectal cancer. These findings elucidate the molecular mechanism underlying chemoradiotherapy resistance and provide potential applications for individualized clinical therapy.
Introduction
Colorectal cancer (CRC) is the third most common cancer among males and females globally [1]. Approximately 30 % of all CRC cases are rectal carcinomas [2]. Currently, neoadjuvant radical surgery is the standard treatment for locally advanced rectal cancer (LARC) [3]. Approximately 10–30 % of patients who undergo surgical resection after neoadjuvant therapy achieve complete pathological remission, and the 5-year survival rate can be as high as 90 % [4,5]. However, approximately 50 % of patients with rectal cancer do not experience tumor downstaging or even disease progression during neoadjuvant therapy. This group of patients has significantly worse disease-free and overall survival than those who are effectively treated and also experience toxic side effects associated with the therapy [6,7]. Therefore, pre-treatment screening of patients with rectal cancer who are sensitive to chemoradiotherapy can help increase the rate of complete pathological remission, reduce treatment-related injury and risk, substantially prolong patient survival and improve the life quality.
In this study, a genome-wide expression profiling microarray was employed to investigate differentially expressed genes (DEGs) related to chemoradiotherapy resistance in the tissues of 81 patients with rectal cancer before neoadjuvant therapy. The results of the aforementioned analysis were verified in 45 patients with rectal cancer, and a 20-gene signature was proposed as a predictor of chemoradiotherapy response.
Among these, Zinc-finger protein 37A (ZNF37A) exhibited the most significant differential expression in rectal cancer tissues with varying responses. ZNF37A is a member of the Kupple-C2H2 zinc-finger protein family and is a principal mediator of transcriptional inhibition. A previous study indicated that ZNF37A was highly expressed in poorly differentiated CRC and promoted tumor metastasis via transcriptional regulation of the THSD4/TGF-β axis [8]. Gauthier et al. found that ZNF37A acted as a transcriptional suppressor that inhibited interleukin-13 expression in type I myotonic dystrophy [9]. However, the Kupple-C2H2 zinc finger protein family is the largest family of transcriptional regulators in higher vertebrates. They are characterized by an N-terminal KRAB structural domain and a C-terminal DNA-binding zinc finger array that, together with the cofactor KAP1, is involved in repressing transposable element sequences [10]. Nowadays, more and more studies have also confirmed the key role of KRAB-ZFPs members in regulating tumor growth, progression and drug resistance [[11], [12], [13]]. Therefore, further studies are needed to explore the molecular mechanism of ZNF37A's involvement in regulating the sensitivity to radiotherapy in intestinal cancer.
Tumor necrosis factor receptor superfamily member 6B (TNFRSF6B), also known as decoy receptor 3 (DcR3), an analog of a cell surface receptor belonging to the tumor necrosis factor superfamily, cannot activate downstream signaling pathways after ligand binding due to the absence of peptides in the cell membrane. TNFRSF6B can inhibit apoptosis by interfering with the activation of multiple signaling pathways mediated by TLIA−DR3, LIGHT−LTβR, LIGHT−HVEM, and FasL−Fas [[14], [15], [16], [17], [18]], and also can be an endogenous immunomodulator in cancer growth and inflammatory reactions [19,20]. In our study, further experimental research revealed that ZNF37A bound to the promoter region of TNFRSF6B and inhibited its transcriptional activity, thereby inducing chemotherapy resistance by inhibiting apoptosis in rectal cancer.
Materials and methods
Patients and tumor tissues
As described in our previous study, the study group comprised 81 patients with LARC in the discovery stage and 45 patients in the validation stage who were enrolled from January 2006 to June 2013 at the Cancer Hospital, Chinese Academy of Medical Sciences, Beijing [21]. Pre-treatment endoscopic biopsy tissue samples were promptly frozen in liquid nitrogen and stored at –80 °C until further processing. The study population consisted of patients who met the following criteria: histologically confirmed rectal adenocarcinoma; primary and locally advanced tumors without distant spread; Karnofsky performance score (KPS) ≥ 70; life expectancy ≥ 6 months; and adequate organ function. All patients received a total radiation dose of 50 Gy administered in 25 fractions over 5 weeks, along with twice daily doses of 825 mg/m2/d Capecitabine and 50 mg/m2/week Oxaliplatin for 2 weeks, with 1 week of rest in every 21-day cycle (days 1–14 and days 21–35). Standardized surgery was performed 4–6 weeks after chemoradiotherapy. The histological response to chemoradiotherapy was evaluated accordance with the tumor regression classification of Mandard et al. [22] Tissue samples with < 10 % of residual tumor cells were designated as good responders (tumor regression grade TRG 1 and TRG 2); tissue samples with 10–50 % residual tumor cells were defined as intermediate responders (TRG 3); and tissue samples with > 50 % of residual tumor cells were considered as non-responders (TRG 4 and 5) (Fig. S1A). Disease-free survival (DFS) was measured from the date of the surgical procedure until tumor progression, death, or last follow-up. The 81 and 45 patients in discovery cohort and validation cohort were followed up until October 31, 2021, with median follow-up durations of 125 and 94 months, respectively. No participants were lost to follow-up. This study was approved by the Institutional Review Board of the Chinese Academy of Medical Sciences Cancer Institute (IRB No. 23/088–3827), and the recruitment process was conducted with the informed consent of each participant.
RNA extraction and gene expression profiling
RNA was extracted from pre-treatment endoscopic biopsy speciments containing > 60 % tumor tissue using phenol/chloroform (TRIzol; Invitrogen) and purified using column chromatography (RNeasy Mini kit; Qiagen) according to the manufacturer's guide. Subsequently, RNA integrity was checked using a bioanalyzer (Agilent 2100, Agilent Technologies); all samples showed good 18S and 28S ribosomal integrity (RIN > 7). Labeled and fragmented RNA targets were prepared, hybridized, and scanned as described by the manufacturer (Agilent Technologies). All 81 processed samples passed quality control. The expression data of each sample were extracted using Feature Extraction Software.
Quantitative real-time PCR analysis
Total RNA was isolated from tissue samples using TRIzol (Invitrogen). The relative gene expression of selected genes was quantified using the ABI Prism 7900 Sequence Detection System (Applied Biosystems) with the SYBR Green method. The primer sets used to amplify the selected genes were presented in Table S1.
Cell lines and cell culture
HT29, HCT8, SW480, and SW620 cells were cultured in RPMI-1640 medium with 10 % fetal bovine serum (FBS), while sw1116, RKO, LoVo, HCT116, and HEK293T cells were cultured in DMEM with 10 % FBS. All cells were maintained at 37 °C in a 5 % CO2 humidified incubator.
Establishment of CRC cell lines with ZNF37A overexpression or knockdown
The vector used to construct the ZNF37A protein-coding region was PLVX-IRES-Neo, purchased from Clontech. It was selected using neomycin so that the target gene could be stably expressed in mammalian cells. The full length DNA sequence of the ZNF37A protein-coding region was cloned into the PLVX-IRES-Neo Vector. The vector used to construct stable ZNF37A knockdown was pSIH−H1 (Invitrogen, Carlsbad, CA), which was selected with puromycin. Small interfering RNAs (Control siRNA: 5′−UUC UCC GAA CGU GUC ACG UTT−3′; siRNA-1: 5′−CGA GGA GCC AUG GAU AUU ATT−3′ and siRNA-2: 5′−CCC ACU CAA UUA ACA AUA UTT−3′) of ZNF37A and their antisense chains were synthesized by SinoGenoMax, and the synthesized fragments were attached to the pSIH−H1 vector. The plasmids were transfected into HEK293T packaging cells using Lipofectamine 2000 (Invitrogen) for lentivirus collection. LoVo, HCT8, and HCT116 cells were infected and selected with neomycin and puromycin, followed by validation using western blot and qPCR assays.
RNA-sequencing analysis
Total RNA was extracted from HCT8 and HCT116 cell lines overexpressing or knocking down ZNF37A and sequenced. RNA-sequencing data were mapped to the GRCh38 human genome by HISAT2, normalized, and log2 transformed. The DESeq2 R package was used for differential expression analysis. Significant DEGs were determined using log2 (fold change) and corrected P value. Gene enrichment analysis of normalized RNA-sequencing data was conducted using the Metascape database (http://metascape.org).
Colony formation assays
Cells were seeded in six-well plates (2000 cells/well) with or without chemotherapy (5 μg/ml 5-fluorouracil and 1 μg/ml oxaliplatin) and radiotherapy (2 Gy) treatment. After approximately 1 week, the colonies were fixed with methanol and stained with 0.5 % crystal violet. Colony number was quantified with ImageJ software.
Western blot analysis
Cells were lysed with RIPA lysis buffer (Solarbio, R0020). Protein concentration was determined by a BCA Protein Assay Kit (Thermo Fisher Scientific). Lysate containing 20 μg protein was separated by SDS-PAGE and transferred to a PVDF membranes (Millipore). The antibodies used in this study were: ZNF37A (PA5-30254, Invitrogen), TNFRSF6B (ab8405, Abcam), caspase-3 (#9662, Cell Signaling Technology), cleaved caspase-3 (#9661, Cell Signaling Technology), PARP (#9542, Cell Signaling Technology) and cleaved PARP (#5625, Cell Signaling Technology), and β-ACTIN (sc-47778, Santa). The signal was detected with ECL western blotting substrate (Thermo Fisher Scientific) on Amersham Imager 600. Quantification of the protein bands was performed by grayscale scanning using ImageJ software.
Immunofluorescence flow cytometry
An Annexin V/PI double-staining apoptosis detection kit (KeyGEN) was used. Cells from different treatment groups were digested, suspended, collected by centrifuging at 1000 rpm for 5 min, incubated with PI and Annexin V reagent at room temperature for 15 min, and analyzed using a BD Flow cytometry (LSRII).
Dual-luciferase reporter assays
The Nano-Glo Dual-Luciferase Reporter (NanoDLR™) System was used for dual-luciferase reporter assays according to the manufacturer's instructions (N1620; Promega). After cell adhesion in 48-well plates (6 × 104 cells/well), reporter plasmids were transfected into cells with Lipofectamine 2000 (Invitrogen) and incubated for 24 h. The Firefly and Renilla luciferase activities were measured using GENE5 software.
Chromatin immunoprecipitation assay
The EZ-ChIP™ Chromatin Immunoprecipitation Kit (Millipore, USA) was used. For crosslinking, 37 % formaldehyde was added to the complete growth medium, and the reaction was terminated 10 min later using glycine (Sigma). The cells were lysed and treated with ultrasound to fragment the chromatin. Chromatin fragments were incubated with corresponding antibodies against the target protein and Protein G magnetic beads for immunoprecipitation, and then they were eluted from the antibody/protein G microbeads and de-cross-linked. Purified DNA was analyzed using quantitative PCR.
Animal experiments
Male BALB/c mice aged 4–6 weeks (Beijing IDMO Co.) were injected subcutaneously with HCT8−Vector, HCT8−ZNF37A, HCT116−shCtrl, HCT116−sh1, and HCT116−sh2 CRC cells (2 × 106) and housed for 30 days. The mice were randomly categorized into vehicle and CT/RT groups. Mice in the CT/RT group were treated with radiotherapy (2 Gy) and chemotherapy (20 mg/kg 5-fluorouracil and 5 mg/kg oxaliplatin, i.p) for 2 weeks, whereas those in the control group received the same amount of normal saline without chemoradiotherapy treatment.
Statistical analysis
Data were processed and analyzed using the statistical software package R and SAS software version 9.3 (SAS Institute). First, the effects of risk factors, including age, sex, clinical stage, and KPS score, on response status and DFS were examined using the χ2 test and log-rank test. The microarray data were normalized using the “limma” package. Differential expression between responders (TRG1 and TRG2) and non-responders (TRG4 and TRG5) was analyzed using Student's t-test. Genes with P < 0.05 (after Bonferroni correction) on the t-test and fold change ≥ 2.0 or ≤ 0.5 were selected to generate a list of significant DEGs. Patients’ subsets grouped according to their gene expression levels were compared for DFS time using Kaplan–Meier curves and Cox proportional hazards model, and adjusted for gender, age, tumor stage, tumor grade, KPS, surgical procedure and tumor location as covariates. Differences in residual cancer cell proportions after chemoradiotherapy were analyzed using the Mann-Whitney U test. A receiver operating characteristic (ROC) curve was constructed to estimate the predictive value of the 20-gene signature in the validation cohort. All statistical tests were two-sided, with statistical significance set at P < 0.05.
Results
A 20-gene signature can predict treatment response in patients with rectal cancer
In a discovery cohort, 30 patients (37.0 %) were responders (TRG1, n = 7; TRG2, n = 23), 14 (17.3 %) were non-responders (TRG4, n = 12; TRG5, n = 2), and 37 (45.7 %) achieved an intermediate response (TRG3, n = 37), according to Mandard et al. [22], all of whom met the enrollment criteria. Examination of the clinical factors in this study showed no association with the clinical response. In addition, DFS was significantly associated with response status (P = 0.0028), making it a measure of response (Table 1). In a validation cohort of 45 samples, which included 27 (60.0 %) responders (TRG1, n = 10; TRG2, n = 17), 12 (26.7 %) intermediate responders (TRG3, n = 12), and 6 (13.3 %) non-responders (TRG4, n = 6), the relationships between sex, age, clinical stage, KPS, and response to chemoradiotherapy were not statistically significant, and DFS had a significant association with response status (P = 0.0527) (Table 1).
Table 1.
Characteristics of rectal cancer samples in discovery and validation stage and correlation with concurrent chemoradiotherapy response.
| Characteristic | Discovery cohort (N = 81) |
Validation cohort (N = 45) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total (%) | Responder (%) | Intermediate | Non-responder (%) | P † | Total (%) | Responder (%) | Intermediate responder (%) | Non-responder (%) | P † | |
| Responder (%) | ||||||||||
| No. of Patients | 81 (100.0) | 30 (37.0) | 37 (45.7) | 14 (17.3) | 45 (100.0) | 27 (60.0) | 12 (26.7) | 6 (13.3) | ||
| Age, years | 0.7325 | 0.1342 | ||||||||
| < 54 | 40 (49.4) | 16 (53.3) | 17 (45.9) | 7 (50.0) | 22 (48.9) | 13 (48.1) | 4 (33.3) | 5 (83.3) | ||
| ≥ 54 | 41 (50.6) | 14 (46.7) | 20 (54.1) | 7 (50.0) | 23 (51.1) | 14 (51.9) | 8 (66.7) | 1 (16.7) | ||
| Sex | 0.9124 | 0.5391 | ||||||||
| Male | 54 (66.7) | 20 (66.7) | 25 (67.6) | 9 (64.3) | 31 (68.9) | 19 (70.4) | 9 (75.0) | 3 (50.0) | ||
| Female | 27 (33.3) | 10 (33.3) | 12 (32.4) | 5 (35.7) | 14 (31.1) | 8 (29.6) | 3 (25.0) | 3 (50.0) | ||
| Clinical stage | 0.4096 | 0.4644 | ||||||||
| II | 19 (23.5) | 8 (26.7) | 9 (24.3) | 2 (14.3) | 12 (26.7) | 9 (33.3) | 2 (16.7) | 1 (16.7) | ||
| III | 62 (76.5) | 22 (73.3) | 28 (75.7) | 12 (85.7) | 33 (73.3) | 18 (66.7) | 10 (83.3) | 5 (83.3) | ||
| KPS | 0.1770 | 0.1642 | ||||||||
| 80–89 | 34 (42.0) | 16 (53.3) | 13 (35.1) | 5 (35.7) | 17 (37.8) | 9 (33.3) | 6 (50.0) | 2 (33.3) | ||
| 90–100 | 47 (58.0) | 14 (46.7) | 24 (64.9) | 9 (64.3) | 28 (62.2) | 18 (66.7) | 6 (50.0) | 4 (66.7) | ||
| DFS events | 0.0028 | 0.0527 | ||||||||
| Alive | 42 (51.9) | 23 (76.7) | 14 (37.8) | 5 (35.7) | 29 (64.4) | 20 (74.1) | 7 (58.3) | 2 (33.3) | ||
| Dead | 39 (48.1) | 7 (23.3) | 23 (62.2) | 9 (64.3) | 16 (35.6) | 7 (25.9) | 5 (41.7) | 4 (66.7) | ||
KPS, Karnofsky performance score; DFS, Disease-free survival. † On the basis of χ2 test or Fisher exact test.
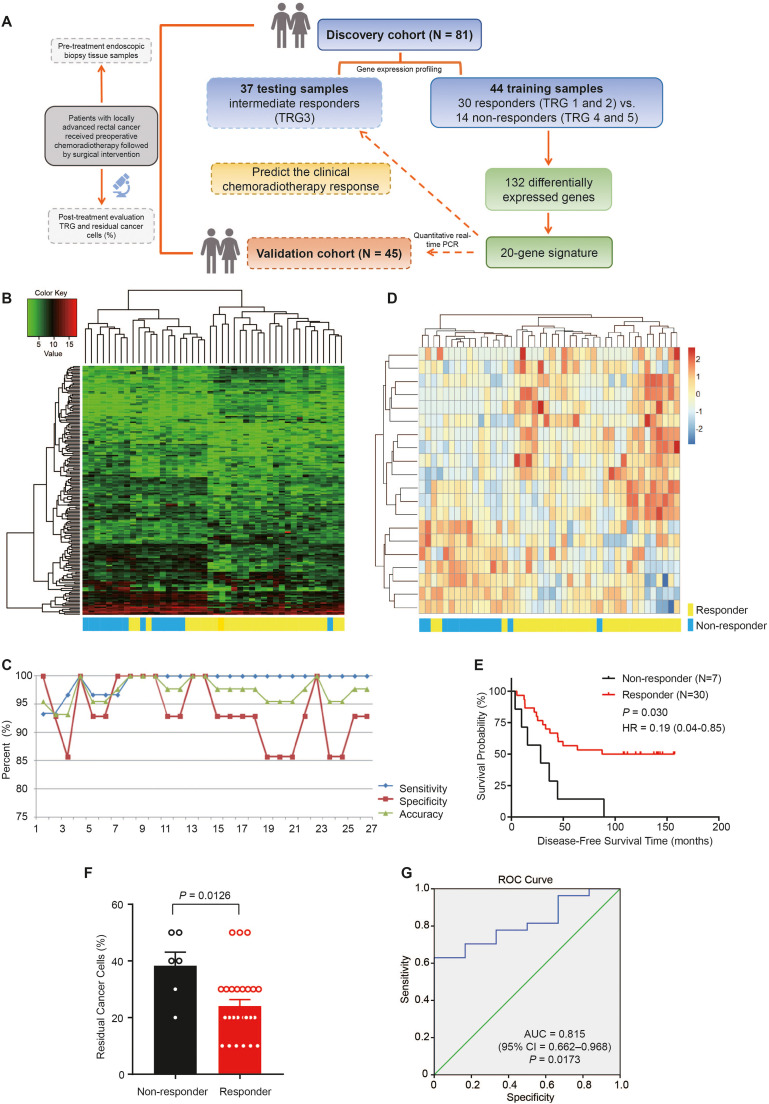
To identify the molecular signatures of tumor response to chemoradiotherapy, we performed gene expression profiling using a DNA array in all samples consisting of training and test samples in the discovery cohort. Among the 44 training samples, we identified a list of 179 probes representing 132 DEGs (P < 0.05 and Fold Change ≥ 2.0 or ≤ 0.5) between responders and non-responders (Fig. 1A and B). Responders and non-responders were grouped into two distinct groups, with 13 of 14 non-responders on the left side of the cluster and 23 of 30 responders on the right side. The clustering results of the 179 probe sets were highly consistent with the clinical pathology assessment (81.8 %). In addition, we used these 179 discriminating probes to generate a two-dimensional plot, in which the responders (red) and non-responders (green) were categorized into two distinct gene expression data spaces (Fig. S1B). Moreover, by analyzing the enrichment of 132 DEGs, we found that these genes are mainly related to metabolic process and various biological regulatory processes (Fig. S1C).
Fig. 1.
A 20-gene signature can predict treatment response in rectal cancer patients. A. Schematic of the strategy for cohorts, generation, and validation of an optimum predictive classifier. B. Cluster analysis of 179 differentially expressed genes in 44 training samples. C. Sensitivity, specificity and accuracy of different gene groups in predicting chemoradiotherapy efficacy. D. Heat map of 20 genes with the most significant differences in 44 training samples. E. Kaplan-Meier curve for disease-free survival time in testing samples (n = 37) of rectal cancer patients with different sensitivity prediction subgroups. F. Proportion of residual cancer cells in testing samples (n = 33, 4 samples miss data) of rectal cancer patients with different sensitivity prediction subgroups. P value was calculated by the Mann-Whitney U test. G. ROC curves of 20 gene prediction models in the validation cohort (27 responders and 6 non-responders, n = 33) of patients with rectal cancer.
To generate an optimum predictive classifier from this cohort, we first sorted the 179 discriminating probes (132 genes) by P value in ascending order. The top five genes constituted the first predictive classifier, and additional genes were gradually added in increments of five. We calculated the sensitivity, specificity, and accuracy of the predictive values in the 44 samples for each group of genes (Fig. 1C). Using the top 20 genes, we obtained the best prediction results. Clustering of the 20-gene sets was consistent with the clinical pathology assessment (90.9 %). Thirteen of the 14 non-responders were in the left cluster, and 27 of the 30 responders congregated on the right side (Fig. 1D). The molecular signature with the top 20 genes effectively separated responders and non-responders into two groups, better than 179 probes based on principal component analysis (Fig. S1D). The top 20 genes identified are presented in Table S2.
We used a prediction model with a 20-gene signature to predict the clinical response to chemoradiotherapy in 37 intermediate responders, which were categorized 30 responders and 7 non-responders. Notably, DFS and the proportion of residual cancer cells after chemoradiotherapy were distinctly different between the two groups (P = 0.030 and P = 0.0126, respectively; Fig. 1E and F). Furthermore, we conducted an ROC analysis of the predictions by the 20-gene model in 27 responders and 6 non-responders in the validation cohort, which showed a highly significant ability to discriminate responders from non-responders in the cohort (AUC = 0.815, 95 % CI = 0.662–0.968; P = 0.0173; Fig. 1G).
ZNF37A influences the sensitivity of CRC cells to chemoradiotherapy
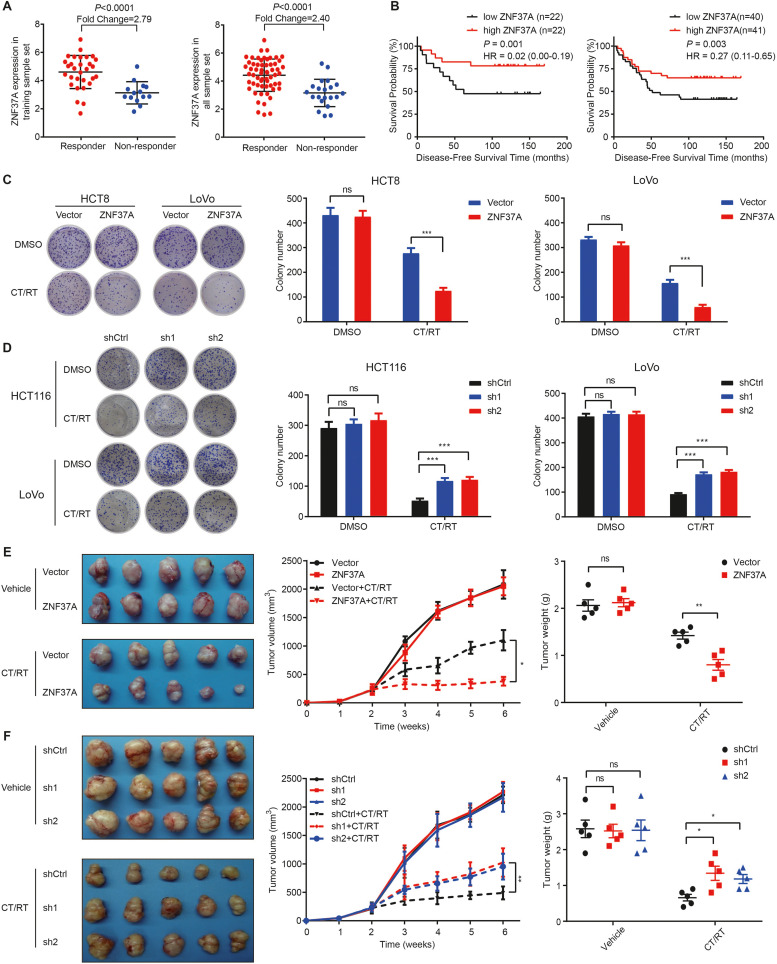
To explore the specific functions of the top 20 genes in our prediction model, we knocked down the top 10 genes in HCT8 CRC cells and conducted a colony formation assay. Our findings revealed that the experimental results for the six genes were consistent with the expression profiling results, with ZNF37A showing the most significant differences (Fig. S2). In addition, patients with high ZNF37A expression responded better to preoperative chemoradiotherapy and had prolonged DFS in 44 training sample set and 81 all-sample set (Fig. 2A and B). Therefore, we hypothesized that ZNF37A might be a key gene influencing the sensitivity of CRC to chemoradiotherapy. To confirm this hypothesis, we searched the TCGA database and found that across various tumor types, ZNF37A expressed higher in tumors than in normal tissues (Fig. S3A). Although ZNF37A was highly expressed in colon adenocarcinoma (COAD), no significant differences were noted among tumor progression stages (Fig. S3B). Survival analysis based on data from The Cancer Genome Atlas Rectal Adenocarcinoma Program (TCGA-READ) indicated that patients with high ZNF37A expression had a trend toward longer survival outcomes (P = 0.089, Fig. S3C). We further detected ZNF37A mRNA and protein expression in the eight CRC cell lines (Fig. S4A). Higher expression levels of ZNF37A in cells correlated with a lower percentage of colony formation after chemoradiotherapy compared with that in the untreated group, indicating higher sensitivity of the cells to chemoradiotherapy (Fig. S4B and C).
Fig. 2.
ZNF37A increases the sensitivity of colorectal cancer cells to chemoradiotherapy. A. Expression level of ZNF37A between responders and non-responders in 44 training sample set and 81 all sample set. Data were mean±SEM. B. Kaplan-Meier curves of disease-free survival according to ZNF37A expression in 44 training sample set and 81 all sample set. Low and high ZNF37A groups were separated according to the median of the chip detection values. C. Colony formation assays of HCT8-Vector, HCT8-ZNF37A, LoVo-Vector and LoVo-ZNF37A cells. DMSO, control group treated with Dimethyl sulfoxide. CT/RT, the treatment group treated with radiotherapy (2 Gy) and chemotherapy (5 μg/ml 5-Fu and 1 μg/ml Oxaliplatin). D. Colony formation assays of HCT116-shCtrl, HCT116-ZNF37A sh1, HCT116-ZNF37A sh2, LoVo-shCtrl, LoVo-ZNF37A sh1 and LoVo-ZNF37A sh2 cells. Right panels represented data (mean±SEM) from three independent experiments. E. Images of subcutaneous tumors at the end of the treatment period, proliferation curves, and tumor weights for HCT8-Vector and HCT8-ZNF37A tumors transplanted in nude mice with or without chemoradiotherapy treatment. Data were presented as mean±SEM of tumor volumes from five mice at each time point. F. Images of subcutaneous tumors at the end of the treatment period, proliferation curves, and tumor weights for HCT116-shCtrl, HCT116-ZNF37A sh1, and HCT116-ZNF37A sh2 tumors transplanted in nude mice with or without chemoradiotherapy treatment. Data were presented as mean±SEM of tumor volumes from five mice at each time point. *, P < 0.05; **, P < 0.01; ***, P < 0.001 and ns, not significant of Student's t-test (C-E). *, P < 0.05; **, P < 0.01 and ns, not significant of one-way ANOVA (F).
Subsequently, we performed gene expression analysis in the CRC cell lines HCT8, HCT116, and LoVo to confirm that the expression of ZNF37A may influence the sensitivity of CRC to chemoradiotherapy. In the absence of chemoradiotherapy, ZNF37A overexpression (ZNF37A-OE) in CRC cells or knockdown of ZNF37A (ZNF37A-KD) did not affect colony formation. However, colony formation survival was greatly suppressed in ZNF37A overexpressing cells in comparison with the control cells when treated with chemoradiotherapy. Conversely, ZNF37A knockdown had the opposite effect (Figs. 2C and D and S4D and E). Therefore, ZNF37A expression had an influence on the sensitivity of CRC to chemoradiotherapy.
Next, we conducted in vivo experiments to compare differences in proliferative ability and sensitivity to chemoradiotherapy. We transplanted ZNF37A-OE HCT8 cells and control cells into nude mice; when not treated with chemoradiotherapy, ZNF37A overexpression did not influence the progression of xenograft tumors versus control. However, after chemoradiotherapy, ZNF37A overexpression significantly suppressed growth of tumor versus control (Fig. 2E). Furthermore, when ZNF37A-KD and control HCT116 cells were transplanted into nude mice, tumor growth was similar between the ZNF37A-KD and control group in the absence of chemoradiotherapy. After chemoradiotherapy, ZNF37A knockdown enhanced the growth of xenograft tumors compared with control (Fig. 2F).
ZNF37A enhances sensitivity to chemoradiotherapy by promoting apoptosis in CRC cells
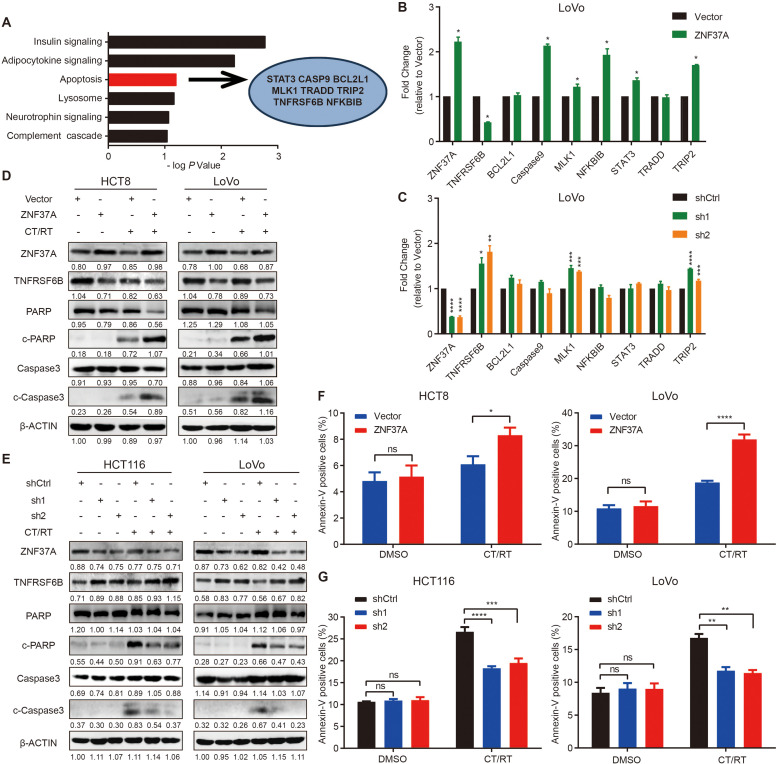
RNA-sequencing analyses were conducted on HCT8 and HCT116 cell lines to investigate the mechanism underlying ZNF37A regulation in the sensitivity of CRC to chemoradiotherapy. Our findings indicated that the upregulated genes after ZNF37A overexpression (Fig. S5A and B) and the downregulated genes after ZNF37A knockdown (Fig. S5C and D) all correlated with positive regulation of the cell death pathway. Given that ZNF37A belongs to the Kupple-C2H2 family of zinc-finger proteins, which play a significant role in transcriptional inhibition, we further analyzed genome-wide expression profiling data and found that 372 genes showed a significant negative correlation with ZNF37A. Subsequent KEGG pathway enrichment analysis revealed that these 372 genes were accumulated in the apoptosis signaling pathway, which is closely related to the response to radiotherapy and chemotherapy (Fig. 3A). Therefore, we hypothesized that ZNF37A regulates the sensitivity of CRC to chemoradiotherapy via transcriptional modulation of apoptosis-related gene expression. Accordingly, we identified eight candidate apoptosis-related genes among the 372 genes and investigated their correlation with ZNF37A expression (Fig. 3A). RT-PCR analysis revealed that TNFRSF6B expression was diminished in ZNF37A-OE cells relative to the control group (Figs. 3B and S5E, Table S1) and increased in ZNF37A-KD cells (Figs. 3C and S5F, Table S1). Conversely, the expression levels of the other seven candidate genes showed no significant association with the expression of ZNF37A.
Fig. 3.
ZNF37A enhances sensitivity to chemoradiotherapy by promoting apoptosis in CRC cells. A. Analysis of the expression profiling data revealed that there were 372 genes significantly negatively correlated with ZNF37A. KEGG pathway enrichment result of 372 genes negatively related to the expression of ZNF37A and 8 candidate apoptosis-related genes were selected from the 372 genes. B. The mRNA expression levels of ZNF37A and eight apoptosis related genes were detected by real-time quantitative PCR in LoVo-Vector and LoVo-ZNF37A cells. C. The mRNA expression levels of ZNF37A and eight apoptosis related genes were detected by real-time quantitative PCR in LoVo-shCtrl, LoVo-ZNF37A sh1 and LoVo-ZNF37A sh2cells. D-E. Western blot analysis of ZNF37A, TNFRSF6B and several apoptosis-related protein levels in chemoradiotherapy-treated and untreated groups of ZNF37A-OE HCT8 and LoVo cells (D), ZNF37A-KD HCT116 and LoVo cells (E). F-G. Proportion of apoptotic cells in the chemoradiotherapy-treated and untreated groups of ZNF37A-OE HCT8 and LoVo cells (F), ZNF37A-KD HCT116 and LoVo cells (G). Data were shown as mean±SEM from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 and ns, not significant of Student's t-test (B, F). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 and ns, not significant of one-way ANOVA (C, G).
The detection of protein levels revealed that TNFRSF6B expression decreased in ZNF37A-OE cells. Moreover, when no chemoradiotherapy was administered, neither cleaved caspase-3 nor cleaved PARP expression was detected in the control or ZNF37A-OE cells. After chemoradiotherapy, the cleaved caspase-3 and cleaved PARP levels was markedly elevated in ZNF37A-OE cells (Fig. 3D). Conversely, TNFRSF6B expression was up-regulated in ZNF37A-KD cells. Additionally, neither cleaved caspase-3 nor cleaved PARP was detected in either control or ZNF37A-KD cells without chemoradiotherapy. However, after chemoradiotherapy, a notable decline in cleaved caspase-3 and cleaved PARP expression was observed in ZNF37A low-expressing cells relative to the control group (Fig. 3E).
Flow cytometry confirmed that when not treated with chemoradiotherapy, neither ZNF37A overexpression nor knockdown altered the proportion of apoptotic cells. However, after treatment with chemoradiotherapy, the proportion of apoptotic ZNF37A-OE cells was markedly elevated compared to the control group (Fig. 3F), and a substantial attenuation in the proportion of apoptotic ZNF37A-KD cells was detected (Fig. 3G).
ZNF37A binds to the TNFRSF6B promoter region and represses its transcriptional activity
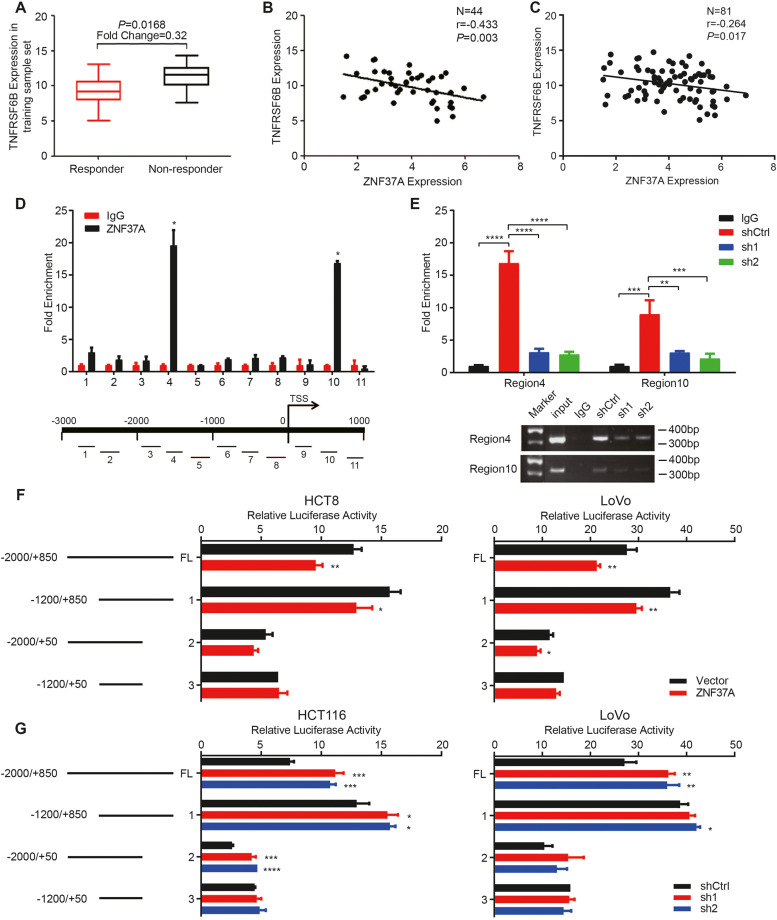
We explored the ZNF37A and TNFRSF6B expression levels in the gene expression profiles of 44 training samples. Patients with high TNFRSF6B expression were insensitive to preoperative chemoradiotherapy (fold change = 0.32, P = 0.0168; Fig. 4A). The TNFRSF6B expression levels in 44 training samples and 81 all samples were negatively correlated with the ZNF37A expression level (P = 0.003 and P = 0.017, Fig. 4B and C).
Fig. 4.
ZNF37A binds to the TNFRSF6B promoter region and represses its transcriptional activity. A. Differences of TNFRSF6B Expression between responders and non-responders in the 44 training sample set. The values of different groups were the median and extreme values of chip detection values. B-C. Negative correlation between ZNF37A and TNFRSF6B expression in the 44 training sample set and 81 all sample set. D. Eleven pairs of primers designed for continuous segments covering 3000 bp upstream and 1000 bp downstream of the transcription start site (TSS) of TNFRSF6B and the ChIP-qPCR results showed significant enrichment of ZNF37A in the fourth and tenth primer regions. E. ChIP-qPCR results and gel diagram of ChIP-qPCR products for the fourth and tenth primer regions mentioned above in the LoVo-shCtrl, LoVo-sh1 and LoVo-sh2 cell lines. F. Luciferase activity of transfection of full-length and different truncated recombinant plasmid of the TNFRSF6B promoter region in HCT8-Vector, HCT8-ZNF37A, LoVo-Vector and LoVo-ZNF37A cells. G. Luciferase activity of transfection of full-length and different truncated recombinant plasmid of the TNFRSF6B promoter region in HCT116-shCtrl, HCT116-ZNF37A sh1, HCT116-ZNF37A sh2, LoVo-shCtrl, LoVo-ZNF37A sh1 and LoVo-ZNF37A sh2 cells. Data were shown as mean±SEM from three independent experiments. *, P < 0.05; **, P < 0.01 of Student's t-test (D, F). *, P < 0.05; **, P < 0.01; ***, P < 0.001 and ****, P < 0.0001 of one-way ANOVA (E, G).
In the validation cohort of 45 patients, quantitatively analyses through RT-PCR revealed that the ZNF37A expression in responders was highly increased compared with that in non-responders (P = 0.007, Fig. S6A), whereas TNFRSF6B expression was markedly diminished (P = 0.044, Fig. S6B). A notable negative correlation was identified between TNFRSF6B and ZNF37A expression levels in these 45 samples (P = 0.0279, Fig. S6C). This inverse relationship between TNFRSF6B and ZNF37A expression was also detected in the TCGA-COAD and TCGA-READ datasets (Fig. S6D and E).
Furthermore, we conducted chromatin immunoprecipitation (ChIP)-qPCR experiments to ascertain the potential binging of ZNF37A to the promoter region of TNFRSF6B and its capacity to regulate transcription. First, we performed ChIP to enrich the DNA that binds to the ZNF37A protein and designed 11 pairs of primers for continuous segments covering 3000 bp upstream and 1000 bp downstream of the transcription start site (TSS) of TNFRSF6B (Table S3). The RT-PCR results indicated significant enrichment of ZNF37A binding in the region from –1880 bp to –1550 bp upstream of TSS and from +330 bp to +700 bp downstream of TSS (Fig. 4D). Furthermore, the ChIP-qPCR results showed that ZNF37A-KD cells exhibited less enrichment in these DNA regions than the control cells (Fig. 4E). Based on these results, we constructed reporter gene vectors containing the full-length TNFRSF6B promoter region, as well as vectors lacking either one or both of the identified ZNF37A binding regions. Dual luciferase reporter assays indicated that transfection with the full-length TNFRSF6B promoter resulted in decreased luciferase activity in cells stably overexpressing ZNF37A and increased luciferase activity in cells with ZNF37A knockdown compared with control cells, suggesting that ZNF37A inhibited the transcriptional activity in the TNFRSF6B promoter region. Similar observations were made when transfecting vectors lacking either of the two identified ZNF37A binding regions; nevertheless, no obvious difference in luciferase activity was observed between control cells and those stably up- or downregulated by ZNF37A when transfected with a vector lacking both binding regions simultaneously (Fig. 4F and G). This suggests that the two ZNF37A-binding regions identified by ChIP-qPCR are the key regions for ZNF37A in regulating the transcriptional activity of TNFRSF6B.
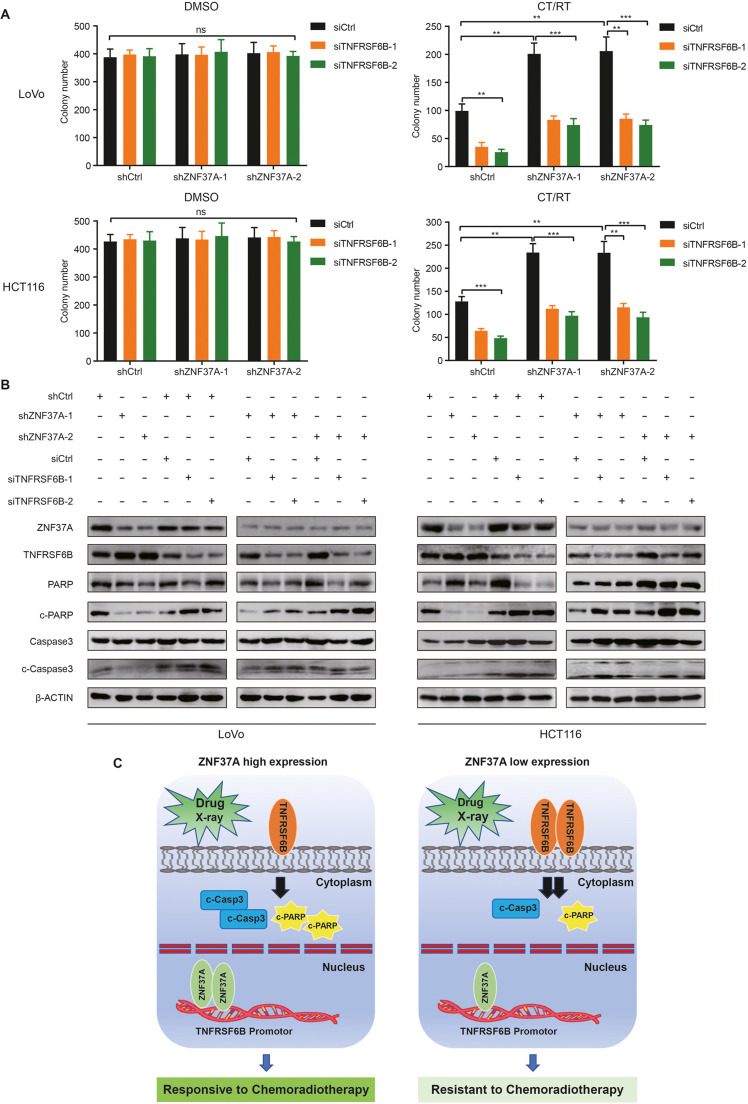
Knocking down TNFRSF6B restores sensitivity of CRC to chemoradiotherapy
Subsequently, TNFRSF6B expression was silenced in ZNF37A-KD cells. Colony formation assays revealed that TNFRSF6B low-expressing cells exhibited growth potential comparable to that of control cells in the absence of chemoradiotherapy. However, a significant decline in colony formation ability was observed after reducing TNFRSF6B expression in the chemoradiotherapy-treated group versus control (Figs. 5A and S7). This indicates that lowering TNFRSF6B expression in ZNF37A-KD cell lines restored sensitivity to chemotherapy. Moreover, we examined the expression levels of apoptotic proteins following TNFRSF6B knockdown in ZNF37A-KD cell lines. Our findings revealed that after chemoradiotherapy, a reduction in TNFRSF6B expression led to an increase in the levels of cleaved caspase3 and cleaved PARP when compared with the control group (Fig. 5B). Chemotherapy drugs and radiation stimulation can induce cell apoptosis. When the expression of ZNF37A is high in colorectal cancer, the transcriptional activity of TNFRSF6B is low. Consequently, tumor cell apoptosis is increased, which shows that it is sensitive to chemoradiotherapy. Conversely, when the expression of ZNF37A in CRC is low, the transcriptional activity of TNFRSF6B is high; thus, apoptosis is reduced, and the tumor cells are not sensitive to radiotherapy and chemotherapy (Fig. 5C).
Fig. 5.
Knocking down TNFRSF6B can restore the sensitivity of colorectal cancer to chemoradiotherapy. A. Colony formation assays of Knocking down TNFRSF6B in LoVo-shCtrl, LoVo-ZNF37A sh1 and LoVo-ZNF37A sh2, HCT116-shCtrl, HCT116-ZNF37A sh1, HCT116-ZNF37A sh2 cells. DMSO, control group treated with dimethyl sulfoxide. CT/RT, the treatment group treated with radiotherapy (2 Gy) and chemotherapy (5-Fu and Oxaliplatin). Data were shown as mean±SEM from three independent experiments. **, P < 0.01; ***, P < 0.001 and ns, not significant of Student's t-test. B. Detection of apoptosis-related proteins by western blot after chemoradiotherapy when knocking down TNFRSF6B in LoVo-shCtrl, LoVo-ZNF37A sh1 and LoVo-ZNF37A sh2, HCT116-shCtrl, HCT116-ZNF37A sh1, HCT116-ZNF37A sh2 cells. C. Scatter plots showed the effect of ZNF37A expression level on chemoradiotherapy sensitivity of CRC.
Discussion
The predictive role of monomolecules in the efficacy of preoperative chemoradiotherapy in patients with rectal cancer has been extensively studied. However, the response of LARC to neoadjuvant therapy results from a combination of clinical factors and molecular biological features of tumors, involving multiple genes and biological pathways [[23], [24], [25]]. Researchers have reached a consensus that dysregulated gene expression is a major contributor to rectal cancer development and differences in efficacy and survival time [[26], [27], [28]]. Therefore, identifying a signature comprising a group of genes as markers of cancer response to chemoradiotherapy would be more appropriate and effective. Gene and protein signatures and expression modules related to sensitivity to chemoradiotherapy have been identified with the rapid development of multi-omics sequencing [[29], [30], [31], [32]]. In our previous study of these 126 patients in total, whole-exome sequencing was performed on 28 paired tumors collected before and after chemoradiotherapy from the same patients who did not respond to chemoradiotherapy treatment. The analysis demonstrated that recurrent mutations in CTDSP2, APC, KRAS, TP53 and NFKBIZ conferred a selective advantage to cancer cells, making them resistant to chemoradiotherapy treatment. This study revealed genome landscapes in rectal cancer before and after chemoradiotherapy and tumors evolution under chemoradiotherapy stress [21]. A recent analysis of gene expression profiles predicting sensitivity to chemoradiation (capecitabine plus radiation) for LARC has identified 213 DEGs [33]. Although our study includes patients who underwent different chemoradiotherapy regimens, we identified four overlapping DEGs (namely, ARL11, CYP4F2, PROS1, and NOTUM) among the 132 DEGs and 213 DEGs mentioned above, all reported to be involved in apoptosis regulation [[34], [35], [36], [37]]. This indicates the relationship between apoptosis and sensitivity to chemoradiotherapy. In this study, we examined genome-wide gene expression in 81 patients with rectal cancer and established a set of 20 genes to predict treatment response to neoadjuvant chemoradiotherapy.
Among the 20 genes identified, the ZNF37A expression level exhibited the most significant correlation with preoperative sensitivity to chemoradiotherapy in rectal cancer. Additional examination of the clinical data revealed that patients with CRC who displayed high ZNF37A expression exhibited increased sensitivity to neoadjuvant chemoradiotherapy and had prolonged DFS. Subsequently, we confirmed that low ZNF37A expression decreased the sensitivity of rectal cancer cells to chemoradiotherapy both in vivo and in vitro. Despite limited research on ZNF37A, the Kupple-C2H2 zinc finger protein family has been recognized to play an important role in transcriptional inhibition by binding scaffold protein KAP1 to recruit various transcriptional suppressor molecules. Furthermore, this family plays a pivotal role in many crucial biological activities, ranging from embryonic development and cell differentiation to apoptosis and tumor progression [[38], [39], [40]]. This study is the first to identify a potential correlation between ZNF37A, a Kupple-C2H2 zinc-finger protein member, and the sensitivity of rectal cancer to chemoradiotherapy.
A notable finding of this study is the association of ZNF37A with the response to chemoradiotherapy in rectal cancer, potentially mediated by apoptotic signaling pathways modulation. Gene enrichment analysis revealed that genes negatively correlated with ZNF37A expression were significantly enriched in the apoptotic signaling pathway. Further experimental studies demonstrated that the ZNF37A expression influenced the level of apoptosis in CRC cells following chemoradiotherapy. Apoptosis is a type of programmed cell death that is regulated by a dedicated balance between multiple signaling pathways that are pro-survival and pro-apoptotic [[41], [42], [43]]. Apoptosis is a hallmark of cancer, and tumorigenesis occurs when this balance favors cell survival. Drugs targeting apoptotic pathway molecules can directly activate apoptotic machinery in malignant cells [[44], [45], [46]]. Therefore, the early identification and characterization of precise preclinical disease models and apoptosis-associated resistance-regulating molecules are essential for improving the long-term treatment response rate of patients with tumors. Additionally, the study findings revealed that rectal cancer tumor cells with abnormally low ZNF37A expression could tolerate chemoradiotherapy-induced apoptosis, contributing to the long-term survival of tumor cells. Our findings revealed that patients with CRC and high ZNF37A expression exhibited a longer disease-free survival.
Furthermore, our findings have revealed a previously unreported binding interaction between ZNF37A and the promoter region of TNFRSF6B, promoting the apoptosis of CRC cells after chemoradiotherapy by inhibiting TNFRSF6B transcription. Knocking down TNFRSF6B expression in ZNF37A-KD cells increased sensitivity to chemoradiotherapy. Given the role of TNFRSF6B in regulating apoptosis in tumor cells, increased transcript levels of TNFRSF6B due to low ZNF37A expression in our cohort of patients with rectal cancer may significantly contribute to chemoradiotherapy in rectal cancer.
However, our study had some limitations. Our study primarily focused on the regulation of the apoptosis signaling pathway by ZNF37A; however, other signaling pathways were also involved in the pathway enrichment analysis, indicating its potential involvement in the regulation of multiple pathways. Second, in our study, ZNF37A, a transcriptional repressor, binds to the promoter region of TNFRSF6B, potentially regulated by other transcription factors. Further research is necessary to investigate the regulatory mechanisms of TNFRSF6B at the transcriptional level. Multiple signaling pathways, including JAK-STAT [47], TGF-β3/SMAD [48], PI3K/Akt [49] and NF-κB [50], are activated downstream of TNFRSF6B in human tumors. However, the potential cross-talk between ZNF37A/TNFRSF6B and other key signaling pathways needs to be explored by further studies. Third, the clinical prediction model proposed in this study was developed using retrospective data and a limited sample size. Consequently, the results require further validation in a larger prospective cohort. Fourth, although we analyzed the effects of age, gender, and clinical stage on the proportion of residual cancer cells after chemotherapy in the enrolled samples, we did not adequately consider the effects of other variables, such as genetic heterogeneity or treatment adherence, which need to be improved by more rigorous and in-depth studies.
Therefore, our study established a multigene expression model for predicting the sensitivity of rectal cancer to chemoradiotherapy and elucidated the mechanism underlying resistance to preoperative synchronized chemoradiotherapy in rectal cancer. Our findings offer a novel perspective on accurately assessing sensitivity to chemoradiotherapy in patients with rectal cancer and developing drugs to overcome therapy resistance.
Conclusions
ZNF37A is an effective modulator of chemoradiotherapy response in rectal cancer. ZNF37A enhances the sensitivity of CRC to chemoradiotherapy by downregulating its transcription level after binding to the promoter region of TNFRSF6B, resulting in an elevated proportion of apoptotic rectal cancer cells after chemoradiotherapy. These findings have significant theoretical implications and potential clinical application for elucidating the molecular mechanism of chemoradiotherapy resistance in rectal cancer and for providing individualized clinical therapy.
CRediT authorship contribution statement
Ying Huang: Writing – original draft, Methodology, Investigation, Data curation. Jing Jin: Writing – review & editing, Resources, Methodology, Data curation. Ningxin Ren: Writing – original draft, Methodology, Investigation, Formal analysis, Data curation. Hongxia Chen: Project administration, Investigation, Data curation. Yan Qiao: Methodology, Formal analysis, Data curation. Shuangmei Zou: Resources, Methodology, Data curation. Xin Wang: Resources, Methodology, Data curation. Linlin Zheng: Writing – original draft, Software, Methodology, Data curation. Ye-Xiong Li: Writing – review & editing, Supervision, Resources, Data curation, Conceptualization. Wen Tan: Writing – review & editing, Supervision, Project administration, Funding acquisition, Conceptualization. Dongxin Lin: Writing – review & editing, Supervision, Project administration, Funding acquisition, Conceptualization.
Declaration of competing interest
Ying Huang, Jing Jin, Ningxin Ren, Hongxia Chen, Yan Qiao, Shuangmei Zou, Xin Wang, Linlin Zheng, Ye-Xiong Li, Wen Tan and Dongxin Lin are the authors of manuscript entitled “ZNF37A downregulation promotes TNFRSF6B expression and leads to therapeutic resistance to concurrent chemoradiotherapy in rectal cancer patients”. These authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work is supported by grants from National Natural Science Foundation (grant no. 81972859 to W.T.), Beijing Municipal Science & Technology Commission Grant (grant no. D0905001040531 to D.L.), and State Key Laboratory of Molecular Oncology Grant (grant no. SKLMO-KF2023-03 to D.L.). We would like to express our sincere gratitude to the patients and clinicians who participated in this study. Special thanks to Kaitai Zhang and Xuebin Di for providing the genome-wide expression profiling data services.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2024.102203.
Contributor Information
Ye-Xiong Li, Email: yexiong12@163.com.
Wen Tan, Email: tanwen@cicams.ac.cn.
Appendix. Supplementary materials
Data availability
The genome-wide expression profiling data of 81 rectal cancer patients with neoadjuvant chemoradiotherapy in this study have been deposited in the National Genomic Data Center database (https://ngdc.cncb.ac.cn/gsub/) and can be obtained under the reasonable request to the corresponding author (Project ID: PRJCA027384). Public datasets used in this work can download from The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/).
References
- 1.Siegel R.L., Miller K.D., Wagle N.S., Jemal A. Cancer statistics, 2023. CA Cancer J. Clin. 2023;73(1):17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Wagle N.S., Cercek A., Smith R.A., Jemal A. Colorectal cancer statistics, 2023. CA Cancer J. Clin. 2023;73(3):233–254. doi: 10.3322/caac.21772. [DOI] [PubMed] [Google Scholar]
- 3.van Gijn W., Marijnen C.A., Nagtegaal I.D., Kranenbarg E.M., Putter H., Wiggers T., Rutten H.J., Påhlman L., Glimelius B., van de Velde C.J., et al. Preoperative radiotherapy combined with total mesorectal excision for resectable rectal cancer: 12-year follow-up of the multicentre, randomised controlled TME trial. Lancet. Oncol. 2011;12(6):575–582. doi: 10.1016/S1470-2045(11)70097-3. [DOI] [PubMed] [Google Scholar]
- 4.Maas M., Nelemans P.J., Valentini V., Das P., Rödel C., Kuo L.J., Calvo F.A., García-Aguilar J., Glynne-Jones R., Haustermans K., et al. Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: a pooled analysis of individual patient data. Lancet. Oncol. 2010;11(9):835–844. doi: 10.1016/S1470-2045(10)70172-8. [DOI] [PubMed] [Google Scholar]
- 5.Martin S.T., Heneghan H.M., Winter D.C. Systematic review and meta-analysis of outcomes following pathological complete response to neoadjuvant chemoradiotherapy for rectal cancer. Br. J. Surg. 2012;99(7):918–928. doi: 10.1002/bjs.8702. [DOI] [PubMed] [Google Scholar]
- 6.Zhu J., Liu A., Sun X., Liu L., Zhu Y., Zhang T., Jia J., Tan S., Wu J., Wang X., et al. Multicenter, randomized, phase III trial of neoadjuvant chemoradiation with capecitabine and irinotecan guided by UGT1A1 status in patients with locally advanced rectal cancer. J. Clin. Oncol. 2020;38(36):4231–4239. doi: 10.1200/JCO.20.01932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conroy T., Bosset J.F., Etienne P.L., Rio E., François É., Mesgouez-Nebout N., Vendrely V., Artignan X., Bouché O., Gargot D., et al. Neoadjuvant chemotherapy with FOLFIRINOX and preoperative chemoradiotherapy for patients with locally advanced rectal cancer (UNICANCER⁃PRODIGE 23): a multicentre, randomised, open⁃label, phase 3 trial. Lancet. Oncol. 2021;22(5):702–715. doi: 10.1016/S1470-2045(21)00079-6. [DOI] [PubMed] [Google Scholar]
- 8.Liu J., Huang Z., Chen H.N., Qin S., Chen Y., Jiang J., Zhang Z., Luo M., Ye Q., Xie N., et al. ZNF37A promotes tumor metastasis through transcriptional control of THSD4/TGF-β axis in colorectal cancer. Oncogene. 2021;40(19):3394–3407. doi: 10.1038/s41388-021-01713-9. [DOI] [PubMed] [Google Scholar]
- 9.Gauthier M., Marteyn A., Denis J.A., Cailleret M., Giraud-Triboult K., Aubert S., Lecuyer C., Marie J., Furling D., Vernet R., et al. A defective Krab-domain zinc-finger transcription factor contributes to altered myogenesis in myotonic dystrophy type 1. Hum. Mol. Genet. 2013;22(25):5188–5198. doi: 10.1093/hmg/ddt373. [DOI] [PubMed] [Google Scholar]
- 10.Ecco G., Imbeault M., Trono D. KRAB zinc finger proteins. Development. 2017;144:2719–2729. doi: 10.1242/dev.132605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye Q., Liu J., Xie K. Zinc finger proteins and regulation of the hallmarks of cancer. Histol. Histopathol. 2019;34(10):1097–1109. doi: 10.14670/HH-18-121. [DOI] [PubMed] [Google Scholar]
- 12.Chen G., Chen J., Qiao Y., Shi Y., Liu W., Zeng Q., Xie H., Shi X., Sun Y., Liu X., et al. ZNF830 mediates cancer chemoresistance through promoting homologous recombination repair. Nucl. Acid. Res. 2017;46:1266–1279. doi: 10.1093/nar/gkx1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang W., Zhangyuan G., Wang F., Jin K., Shen H., Zhang L., Yuan X., Wang J., Zhang H., Yu W., et al. The zinc finger protein Miz1 suppresses liver tumorigenesis by restricting hepatocyte-driven macrophage activation and inflammation. Immunity. 2021;54(6):1168–1185. doi: 10.1016/j.immuni.2021.04.027. e8. [DOI] [PubMed] [Google Scholar]
- 14.Pitti R.M., Marsters S.A., Lawrence D.A., Roy M., Kischkel F.C., Dowd P., Huang A., Donahue C.J., Sherwood S.W., Baldwin D.T., et al. Genomic amplification of a decoy receptor for Fas ligand in lung and colon cancer. Nature. 1998;396(6712):699–703. doi: 10.1038/25387. [DOI] [PubMed] [Google Scholar]
- 15.Yu K.Y., Kwon B., Ni J., Zhai Y., Ebner R., Kwon B.S. A newly identified member of tumor necrosis factor receptor superfamily (TR6) suppresses LIGHT-mediated apoptosis. J. Biol. Chem. 1999;274(20):13733–13736. doi: 10.1074/jbc.274.20.13733. [DOI] [PubMed] [Google Scholar]
- 16.Migone T.S., Zhang J., Luo X., Zhuang L., Chen C., Hu B., Hong J.S., Perry J.W., Chen S.F., Zhou J.X., et al. TL1A is a TNF-like ligand for DR3 and TR6/DcR3 and functions as a T cell costimulator. Immunity. 2002;16(3):479–492. doi: 10.1016/s1074-7613(02)00283-2. [DOI] [PubMed] [Google Scholar]
- 17.Zhou J., Song S., He S., Wang Z., Zhang B., Li D., Zhu D. Silencing of decoy receptor 3 (DcR3) expression by siRNA in pancreatic carcinoma cells induces Fas ligand-mediated apoptosis in vitro and in vivo. Int. J. Mol. Med. 2013;32(3):653–660. doi: 10.3892/ijmm.2013.1437. [DOI] [PubMed] [Google Scholar]
- 18.Roth W., Isenmann S., Nakamura M., Platten M., Wick W., Kleihues P., Bähr M., Ohgaki H., Ashkenazi A., Weller M. Soluble decoy receptor 3 is expressed by malignant gliomas and suppresses CD95 ligand-induced apoptosis and chemotaxis. Cancer. Res. 2001;61(6):2759–2765. [PubMed] [Google Scholar]
- 19.Hsieh S.L., Lin W.W. Decoy receptor 3: an endogenous immunomodulator in cancer growth and inflammatory reactions. J. Biomed. Sci. 2017;24(1):39. doi: 10.1186/s12929-017-0347-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang C., Li H., Huang Y., Tang Y., Wang J., Cheng Y., Wei Y., Zhu D., Cao Z., Zhou J. Integrative analysis of TNFRSF6B as a potential therapeutic target for pancreatic cancer. J. Gastrointest. Oncol. 2021;12(4):1673–1690. doi: 10.21037/jgo-21-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J., Lin Y., Huang Y., Jin J., Zou S., Zhang X., Li H., Feng T., Chen J., Zuo Z., et al. Genome landscapes of rectal cancer before and after preoperative chemoradiotherapy. Theranostics. 2019;9(23):6856–6866. doi: 10.7150/thno.37794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandard A.M., Dalibard F., Mandard J.C., Marnay J., Henry-Amar M., Petiot J.F., Roussel A., Jacob J.H., Segol P., Samama G., et al. Pathologic assessment of tumor regression after preoperative chemoradiotherapy of esophageal carcinoma. Clinicopathologic correlations. Cancer. 1994;73(11):2680–2686. doi: 10.1002/1097-0142(19940601)73:11<2680::aid-cncr2820731105>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 23.Holohan C., Van Schaeybroeck S., Longley D.B., Johnston P.G. Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer. 2013;13(10):714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 24.Foersch S., Glasner C., Woerl A.C., Eckstein M., Wagner D.C., Schulz S., Kellers F., Fernandez A., Tserea K., Kloth M., et al. Multistain deep learning for prediction of prognosis and therapy response in colorectal cancer. Nat. Med. 2023;29(2):430–439. doi: 10.1038/s41591-022-02134-1. [DOI] [PubMed] [Google Scholar]
- 25.Wang H., Jia H., Gao Y., Zhang H., Fan J., Zhang L., Ren F., Yin Y., Cai Y., Zhu J., et al. Serum metabolic traits reveal therapeutic toxicities and responses of neoadjuvant chemoradiotherapy in patients with rectal cancer. Nat. Commun. 2022;13(1):7802. doi: 10.1038/s41467-022-35511-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ried T., Meijer G.A., Harrison D.J., Grech G., Franch-Expósito S., Briffa R., Carvalho B., Camps J. The landscape of genomic copy number alterations in colorectal cancer and their consequences on gene expression levels and disease outcome. Mol. Aspect. Med. 2019;69(10):48–61. doi: 10.1016/j.mam.2019.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Ahluwalia P.J., Kolhe R., Gahlay G.K. The clinical relevance of gene expression based prognostic signatures in colorectal cancer. Biochim. Biophys. Acta. Rev. Cancer. 2021;1875(2) doi: 10.1016/j.bbcan.2021.188513. [DOI] [PubMed] [Google Scholar]
- 28.Battisti N.M.L., De Glas N., Soto-Perez-de-Celis E., Liposits G., Bringuier M., Walko C., Lichtman S.M., Aapro M., Cheung K.L., Biganzoli L., et al. Chemotherapy and gene expression profiling in older early luminal breast cancer patients: an international society of geriatric oncology systematic review. Eur. J. Cancer. 2022;172(9):158–170. doi: 10.1016/j.ejca.2022.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park I.J., Yu Y.S., Mustafa B., Park J.Y., Seo Y.B., Kim G.D., Kim J., Kim C.M., Noh H.D., Hong S.M., et al. A nine-gene signature for predicting the response to preoperative chemoradiotherapy in patients with locally advanced rectal cancer. Cancers (Basel) 2020;12:800. doi: 10.3390/cancers12040800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emons G., Auslander N., Jo P., Kitz J., Azizian A., Hu Y., Hess C.F., Roedel C., Sax U., Salinas G., et al. Gene-expression profiles of pretreatment biopsies predict complete response of rectal cancer patients to preoperative chemoradiotherapy. Br. J. Cancer. 2022;127:766–775. doi: 10.1038/s41416-022-01842-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Islam Khan M.Z., Tam S.Y., Azam Z., Law H.K.W. Proteomic profiling of metabolic proteins as potential biomarkers of radioresponsiveness for colorectal cancer. J. Proteom. 2022;262 doi: 10.1016/j.jprot.2022.104600. [DOI] [PubMed] [Google Scholar]
- 32.Li Y., Wang B., Ma F., Jiang D., Wang Y., Li K., Tan S., Feng J., Wang Y., Qin Z., et al. Proteomic characterization of the colorectal cancer response to chemoradiation and targeted therapies reveals potential therapeutic strategies. Cell. Rep. Med. 2023;4(12) doi: 10.1016/j.xcrm.2023.101311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y., Shi J., Liu W., Tang Y., Shu X., Wang R., Chen Y., Shi X., Jin J., Li D. A deep neural network predictor to predict the sensitivity of neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Cancer Lett. 2024;589 doi: 10.1016/j.canlet.2024.216641. [DOI] [PubMed] [Google Scholar]
- 34.Yendamuri S., Trapasso F., Calin G.A. ARLTS1 - a novel tumor suppressor gene. Cancer Lett. 2008;264(1):11–20. doi: 10.1016/j.canlet.2008.02.021. [DOI] [PubMed] [Google Scholar]
- 35.Wan S., Pan Q., Yang G., Kuang J., Luo S. Role of CYP4F2 as a novel biomarker regulating malignant phenotypes of liver cancer cells via the Nrf2 signaling axis. Oncol. Lett. 2020;20(4):13. doi: 10.3892/ol.2020.11874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Che Mat M.F., Abdul Murad N.A., Ibrahim K., Mohd Mokhtar N., Wan Ngah W.Z., Harun R., Jamal R. Silencing of PROS1 induces apoptosis and inhibits migration and invasion of glioblastoma multiforme cells. Int. J. Oncol. 2016;49(6):2359–2366. doi: 10.3892/ijo.2016.3755. [DOI] [PubMed] [Google Scholar]
- 37.Gong H., Niu Q., Zhou Y., Wang Y.X., Xu X.F., Hou K.Z. Notum palmitoleoyl-protein carboxylesterase regulates Fas cell surface death receptor-mediated apoptosis via the Wnt signaling pathway in colon adenocarcinoma. Bioengineered. 2021;12(1):5241–5252. doi: 10.1080/21655979.2021.1961657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lupo A., Cesaro E., Montano G., Zurlo D., Izzo P., Costanzo P. KRAB-zinc finger proteins: a repressor family displaying multiple biological functions. Curr. Genom. 2013;14(4):268–278. doi: 10.2174/13892029113149990002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Groner A.C., Meylan S., Ciuffi A., Zangger N., Ambrosini G., Dénervaud N., Bucher P., Trono D. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet. 2010;6(3) doi: 10.1371/journal.pgen.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Najafabadi H.S., Mnaimneh S., Schmitges F.W., Garton M., Lam K.N., Yang A., Albu M., Weirauch M.T., Radovani E., Kim P.M., et al. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotechnol. 2015;33(5):555–562. doi: 10.1038/nbt.3128. [DOI] [PubMed] [Google Scholar]
- 41.Morana O., Wood W., Gregory C.D. The apoptosis paradox in cancer. Int. J. Mol. Sci. 2022;23(3):1328. doi: 10.3390/ijms23031328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warren C., Wong-Brown M.W., Bowden N.A. BCL-2 family isoforms in apoptosis and cancer. Cell Death Dis. 2019;10(3):177. doi: 10.1038/s41419-019-1407-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gourisankar S., Krokhotin A., Ji W. Rewiring cancer drivers to activate apoptosis. Nature. 2023;620(7973):417–425. doi: 10.1038/s41586-023-06348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le Tourneau C., Tao Y., Gomez-Roca C., Cristina V., Borcoman E., Deutsch E., Bahleda R., Calugaru V., Modesto A., Rouits E., et al. Phase I trial of Debio 1143, an antagonist of inhibitor of apoptosis proteins, combined with cisplatin chemoradiotherapy in patients with locally advanced squamous cell carcinoma of the head and neck. Clin. Cancer Res. 2020;26(24):6429–6436. doi: 10.1158/1078-0432.CCR-20-0425. [DOI] [PubMed] [Google Scholar]
- 45.Ferris R.L., Harrington K., Schoenfeld J.D., Tahara M., Esdar C., Salmio S., Schroeder A., Bourhis J. Inhibiting the inhibitors: development of the IAP inhibitor xevinapant for the treatment of locally advanced squamous cell carcinoma of the head and neck. Cancer Treat. Rev. 2023;113(2) doi: 10.1016/j.ctrv.2022.102492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carneiro B.A., El-Deiry W.S. Targeting apoptosis in cancer therapy. Nat. Rev. Clin. Oncol. 2020;17(7):395–417. doi: 10.1038/s41571-020-0341-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei Y., Chen X., Yang J., Yao J., Yin N., Zhang Z., Li D., Zhu D., Zhou J. DcR3 promotes proliferation and invasion of pancreatic cancer via a DcR3/STAT1/IRF1 feedback loop. Am. J. Cancer Res. 2019;9(12):2618–2633. [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y.P., Zhu H.F., Liu D.L., Hu Z.Y., Li S.N., Kan H.P., Wang X.Y., Li Z.G. DcR3 induces epithelial-mesenchymal transition through activation of the TGF-beta3/SMAD signaling pathway in CRC. Oncotarget. 2016;7:77306–77318. doi: 10.18632/oncotarget.12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ge H., Liang C., Li Z., An D., Ren S., Yue C., Wu J. DcR3 induces proliferation, migration, invasion, and EMT in gastric cancer cells via the PI3K/AKT/GSK-3beta/beta-catenin signaling pathway. Onco. Target. Ther. 2018;11:4177–4187. doi: 10.2147/OTT.S172713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin Z., Liu S., Zhan Q., Shao X., Ma J., Pan L. Decoy receptor 3 alleviates hepatic fibrosis through suppressing inflammation activated by NF-kappaB signaling pathway. Adv. Clin. Exp. Med. 2018;27:441–447. doi: 10.17219/acem/68387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome-wide expression profiling data of 81 rectal cancer patients with neoadjuvant chemoradiotherapy in this study have been deposited in the National Genomic Data Center database (https://ngdc.cncb.ac.cn/gsub/) and can be obtained under the reasonable request to the corresponding author (Project ID: PRJCA027384). Public datasets used in this work can download from The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/).