Abstract
Whole genome sequencing-based methodologies have become extremely relevant for the molecular surveillance of human pathogens and are being increasingly introduced into national reference laboratory services. In this study, we describe the validation and implementation of core-genome Multi-Locus Sequence Typing (cgMLST) and whole genome single-nucleotide polymorphism (wgSNP) analysis at the Irish Mycobacteria Reference Laboratory, as a replacement for Mycobacterial Interspersed Repetitive Unit-Variable Number Tandem Repeat (MIRU-VNTR) typing. Concordance of clustering, discriminatory power, and ease-of-use of both WGS analytical methods were evaluated. Although wgSNP analysis (MTBseq) was the most discriminatory method (p < 0,001), we recommend cgMLST (SeqSphere+), as the first-line approach for molecular typing of Mycobacterium tuberculosis isolates in the context of routine surveillance work due to its ease of use and decreased turnaround time, while reserving wgSNP-analysis for a more in-depth cluster analysis of new isolates that show a distance of ≤12 alleles to any other isolate(s) in the cgMLST database.
Keywords: Tuberculosis, MTBC, WGS, cgMLST, wgSNP, Molecular surveillance
1. Introduction
Although the number of new cases of tuberculosis (TB) infection has been in a declining trend in Ireland in recent years, TB is still a considerable burden to human health worldwide. TB is a leading cause of death caused by a single infectious agent globally, particularly impacting vulnerable populations such as migrants, prison inmates and/or immunocompromised individuals (e.g., people co-infected with HIV) [[1], [2], [3]].
The WHO estimates that approximately 10.6 million people fell ill with TB around the globe in 2021, from which the WHO European Region accounted for 2.5 % of all cases with an incidence of 8.4 cases per 100,000 population within the EU/EEA region in 2021 [2,3]. According to the Irish Health Protection Surveillance Centre, a total of 222 TB cases were notified in Ireland for 2022, representing a national crude incidence rate of 4.7 cases per 100,000 population [4], which is below the EU/EEA average, but still far from the target established by WHO in their END TB strategy of 2.4 per 100,000 population TB incidence by the end of 2030 [5].
One of the goals set by WHO's END TB strategy [5] is to control and end TB transmission in the European Union by 2030. To achieve this goal, one fundamental aspect is the timely reporting and accurate genotyping of Mycobacterium tuberculosis complex (MTBC isolates), in an effort to identify, keep track of, and ultimately break transmission chains.
Mycobacterial Interspersed Repetitive Unit - Variable Number Tandem Repeat (MIRU-VNTR) genotyping, a reference method previously adopted worldwide since the 2000s, including at the Irish Mycobacteria Reference Laboratory (IMRL), clustered cultured isolates based on their variable number of tandem repeats at 24 informative loci across the genome [6,7]. As results were expressed as numerical codes (24-digit code), comparison and data exchange between different institutions worldwide was relatively straightforward [[8], [9], [10]]. However, MIRU-VNTR presented limitations, as it did not have sufficient discriminatory power in some cases to correctly establish routes of transmission since it covered less than 1 % of the mycobacterial genome and did not provide valuable information on antibiotic resistance or virulence gene determinants.
Core genome multi-locus sequence typing (cgMLST) is a typing methodology based on the genome-wide gene-by-gene allele calling of hundreds or thousands of genes belonging to the “core genome” of a certain bacterial species, representing an extension of the traditional MLST concept [11,12]. Different commercially available platforms, such as Ridom Bioinformatics' SeqSphere+ and Applied Maths’ BioNumerics, have been developed to be used both in research and routine surveillance in public health institutions.
Whole genome single nucleotide polymorphism (wgSNP) analysis differs from cgMLST as it is based on the calling of single-nucleotide variants in the bacterial genome, including non-coding regions, adding a further level of resolution to the analysis. MTBseq pipeline [13] was specifically developed for wgSNP analysis of MTBC isolates, which employs a reference mapping based workflow, reports detected single nucleotide polymorphisms (SNPs) with known association with antibiotic resistance and performs a lineage classification based on phylogenetic SNPs [14,15].
Following the trend of many EU/EEA countries and after participating in a successful European pilot study led by the European Centre for Disease Prevention and Control (ECDC) [[16], [17], [18], [19]] on the use of WGS analytical methods for molecular surveillance of multidrug resistant TB strains, the IMRL started its transition to routine WGS surveillance in 2019 and validated the use of cgMLST and wgSNP analysis for identification, resistance prediction and epidemiological analysis of MTBC isolates.
The aim of this work is to describe the process of validation and implementation of the two methods at the IMRL, and offer some guidance to fellow colleagues when it comes to the choice of WGS analytical platforms that will fulfil their institute needs and resources.
2. Results
2.1. Ease of use
Ridom SeqSphere+ provides a graphical user interface (GUI) that offers the user an intuitive experience when using the platform for the first time. Free tutorials are available online on Ridom's website that provide a helpful introduction to the software and cover the main functionalities of the platform. A specific tutorial for TB molecular typing is also available on the webpage.
The software works directly with the raw short-read FASTQ files produced by a next-generation sequencer and the pipeline mode allows importation, assembly, and analysis of hundreds of read files in an automated manner just with the click of few buttons. It is also possible to manually input additional epidemiological information for an individual sample or to batch import this information through a metadata excel file. A comparison table is built with the selected samples containing several fields of information, namely QC parameters, the complex type attributed to the sample, any epidemiological data associated with the sample (if this data was provided), cgMLST alleles present/absent. Additionally, a minimum spanning tree can be generated and directly visualised in SeqSphere+ platform and exported in different graphical formats.
In relation to MTBseq, this is an open-source pipeline composed of different modules that can be run independently from each other, allowing the user maximum flexibility and customisation of the workflow. However, to be able to run this pipeline, some previous knowledge of command line is required since no GUI is available, and it runs from a terminal window only.
A script (TBfull.sh) is run to retrieve the read files from their storage folder and to feed them into the pipeline. The main outputs of the pipeline are a statistics file of mapping quality, a joint list of variants, a FASTA alignment of SNP positions, and a lineage classification. The genome alignment FASTA file can then be used as an input to generate a phylogenetic tree using other relevant software programs.
One important aspect to consider when running MTBseq pipeline is the requirement that genomes have Illumina's output format, which can limit the use of the platform if different sequencing technologies are used in the laboratory. Table 1 summarises the main characteristics of SeqSphere+ and MTBSeq pipelines.
Table 1.
– Comparison of features of SeqSphere+ platform and MTBseq pipeline.
| SEQSPHERE+ | MTBSEQ | |
|---|---|---|
| LICENSE COST | 1/3/5-years license | Free, Open-source |
| COST PER ANALYSIS | Unlimited analysis | Unlimited analysis |
| GUI | Yes | No |
| OPERATING SYSTEM | MS Windows/Linux | Linux |
| ASSEMBLY | Reference/de novo | Reference |
| VIRULENCE ANALYSIS | Yes | No |
| AMRANALYSISa | Nob | Yes |
Not assessed as it was not within the scope of this study.
Not automatically available for M. tuberculosis but a database can be manually curated to include the mutations most frequently found in the population.
2.2. Concordance of clustering analysis methods
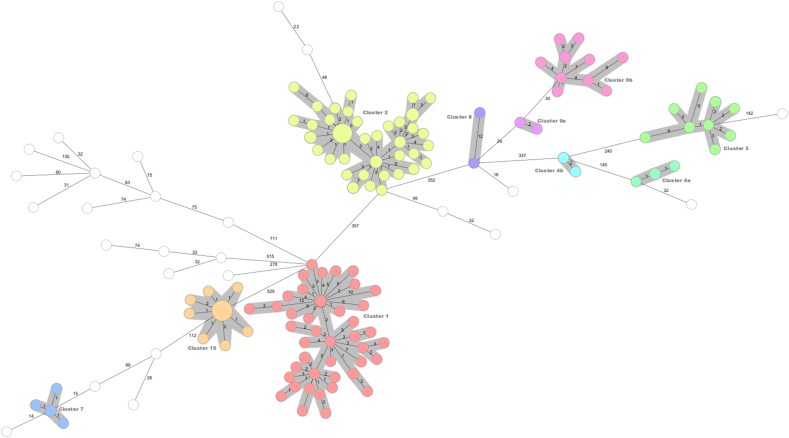
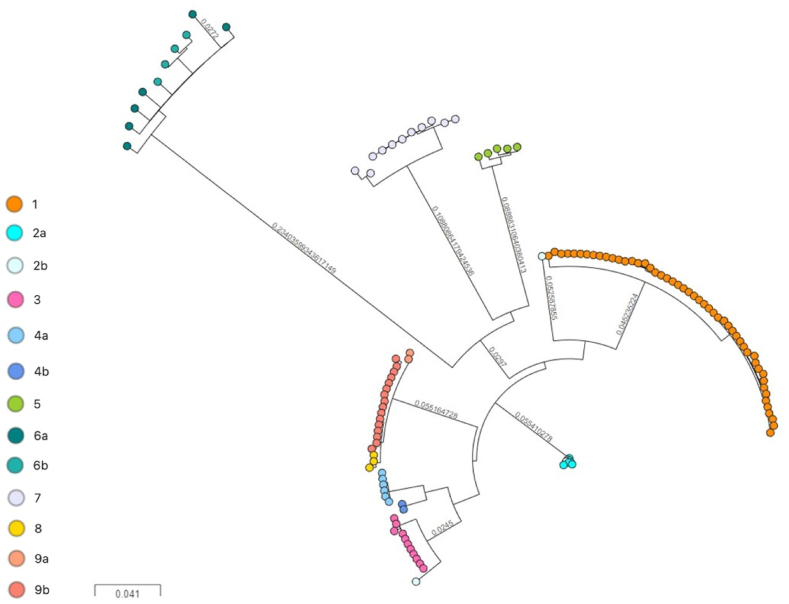
The isolates selected for this study were previously characterised by MIRU-VNTR genotyping and were representative of 14 clusters of M. tuberculosis present in Ireland. Analysis by cgMLST using SeqSphere + default settings of 12 allelic differences generated 10 distinct clusters and 23 unique strains (Fig. 1 and Table 2, supplementary data), herein defined as isolates that did not cluster with any other isolate in the dataset. On the other hand, wgSNP-analysis, using the threshold of ≤5 SNPs to define a cluster [20], resolved the dataset into 17 sub-clusters and 28 unique strains (see Fig. 2,Table 2 and supplementary data). If a threshold of ≤12 SNPs was used, 14 clusters and 13 unique strains were found. If a threshold of ≤20 SNPs was employed, 13 clusters and 7 unique strains were detected. The largest distance between isolates in each cluster ranged from 0 to 116 SNPs.
Fig. 1.
cgMLST analysis of the 223 M. tuberculosis complex isolates based on a scheme of 2891 core genes, pairwise ignoring missing values, with clusters defined as group of isolates with a maximum of 12 allelic differences of distance and with a minimum of 2 isolates (isolates belonging to the cluster are shaded in grey).
Table 2.
–Molecular typing of the isolates previously clustered by MIRU-VNTR and analysed by cgMLST (n = 223) and wgSNP analysis (n = 199). A total of 218 isolates were analysed by both cgMLST and wgSNP analysis.
| MIRU-VNTR Cluster | MIRU-VNTR Genotype | cgMLST cluster |
wgSNP cluster (≤5 SNP) | wgSNP cluster (≤12 SNP) | wgSNP cluster (<20 SNP) | Lineage | Sub-lineage | Max. distance (SNP) |
|---|---|---|---|---|---|---|---|---|
| IMRL 1 (50) | 22322534233442514332332 | 1 (50) | 1a (28) | 1a (44) | 1 (50) | Euro-American | X type | 28 |
| 1b (7) | 1b (3) | 4.1.1.2 | ||||||
| 1c (3) | 1c (2) | |||||||
| 1d (2) | unique (1) | |||||||
| 1e (2) | ||||||||
| unique (8) | ||||||||
| IMRL 2a (40) | 142244332224126143322622 | 2 (38) | 2a (26) | 2a (39) | 2a (39) | Euro-American | LAM | 13 |
| unique (2) | 2b (11) | n.d. (1) | n.d. (1) | 4.3.3 | ||||
| 2c (2) | ||||||||
| n.d. (1) | ||||||||
| IMRL 2b (40) | 142244332224126153322622 | 2 (36) | 2b (37) | 2a (39) | 2a (39) | Euro-American | LAM | 16 |
| unique (4) | 2d (2) | n.d. (1) | n.d. (1) | 4.3.3 | ||||
| n.d. (1) | ||||||||
| IMRL 3 (11) | 245243122334225143335522 | 3 (11) | 3 (9) | 3 (9) | 3 (9) | Euro-American | Mainly T | 16 |
| unique (2) | unique (2) | unique (2) | 4.8 | |||||
| IMRL 4a (6) | 224213322534226153332422 | 4a (5) | 4a (5) | 4a (5) | 4a (5) | Euro-American | H37Rv | 55 |
| unique (1) | unique (1) | unique (1) | unique (1) | 4.9 | ||||
| IMRL 4b (3) | 224213322534226153335422 | 4b (3) | 4b (2) | 4b (2) | 4b (2) | Euro-American | H37Rv | 0 |
| n.d. (1) | n.d. (1) | n.d. (1) | 4.9 | |||||
| IMRL 5 (4) | 242247432244225113342543 | unique (4) | 5 (2) | 5 (4) | 5 (4) | East African-Indian | Delhi - CAS | 12 |
| unique (2) | 3.1.1 | |||||||
| IMRL 6a (5) | 2145243a2843266223342713 | unique (5) | unique (5) | 6a (2) | 6a (4) | Indo-Oceanic | EAI Manila | 31 |
| unique (3) | unique (1) | 1.2.1 | ||||||
| IMRL 6b (4) | 2145243a2943266223342713 | unique (4) | unique (4) | 6b (3) | 6b (3) | Indo-Oceanic | EAI Manila | 24 |
| unique (1) | unique (1) | 1.2.1 | ||||||
| IMRL 7 (10) | 244233352644425153353623 | 7 (6) | 7 (6) | 7 (7) | 7 (8) | East-Asian | Beijing | 116 |
| unique (4) | unique (4) | unique (3) | unique (2) | 2.2.1 | ||||
| IMRL 8 (3) | 22421333154422515333_522 | 8 (2) | unique (2) | unique (2) | 8 (2) | Euro-American | Cameroon | 13 |
| unique (1) | n.d. (1) | n.d. (1) | n.d. (1) | 4.6.2.2 | ||||
| IMRL 9a (2) | 224213331644225153332522 | 9a (2) | 9a (2) | 9a (2) | 9a (2) | Euro-American | Cameroon | 9 |
| 4.6.2.2 | ||||||||
| IMRL 9b (13) | 22421433164422515333_522 | 9b (13) | 9b (13) | 9b (13) | 9b (13) | Euro-American | Cameroon | 3 |
| 4.6.2.2 | ||||||||
| IMRL 10 (32) | 244233352644424173353823 | 10 (32) | 10 (32) | 10 (32) | 10 (32) | East-Asian | Beijing | 5 |
| 2.2.1 |
Legend: n.a – not assigned; n.d. – not determined. (n) are the total numbers of isolates.
Fig. 2.
wgSNP-based phylogeny of 199 M. tuberculosis complex isolates using a threshold of 5 SNPs to define clusters [excluding Cluster 10 (n = 32) published in Ref. [31]; and 9 other isolates from clusters 1 (n = 5), 2a (n = 2), 2b (n = 1) and 4b (n = 1)]. The final SNP tree was generated by RAxML and visualised with MicroReact. Regions annotated as repetitive elements, InDels, multiple consecutive SNPs in a 12-bp window, and 92 genes implicated in antibiotic resistance were excluded for the phylogenetic reconstruction. Isolates are coloured according to their MIRU-VNTR cluster.
Both WGS-based analyses disassembled three of the MIRU-VNTR clusters: cluster 5 and clusters 6a and 6b. All isolates within these clusters were shown to be unique by WGS, rejecting the possibility of recent transmission between cases within each MIRU-VNTR cluster [20]. MIRU-VNTR clusters 2a and 2b, which differ by a single repeat at the MIRU-VNTR locus 2996, were merged into a single cluster by cgMLST. In contrast, wgSNP analysis (≤5 SNP threshold) further divided the two MIRU-VNTR clusters into 3 sub-clusters containing a mixture of isolates from both MIRU-VNTR clusters 2a and 2b.
Cluster 8 comprised two isolates that were shown to differ by 12 alleles by cgMLST, however, by wgSNP-analysis these two isolates showed less than 5 SNPs difference. Even though the cluster was concordant between the two methods, wgSNP analysis revealed much less diversity than might be expected from the cgMLST results.
In terms of overall concordance of clustering analysis, cgMLST and wgSNP analysis showed 88 % and 76 % concordance with MIRU-VNTR genotyping, respectively, and 88 % concordance of results with each other. However, if a looser wgSNP threshold was applied (i.e. cluster = ≤12 SNPs rather than 5), the agreement between cgMLST and wgSNP-analysis would increase to 96 % (i.e. 14 clusters and 13 unique strains).
2.3. Discriminatory power
Discriminatory power was measured by Simpson's index of diversity (Table 3) [21,22]. wgSNP-analysis showed the highest discriminatory power of all three-methods [SID: 0.895 vs. 0.807 (cgMLST) and 0.856 (MIRU-VNTR), p < 0,001). Even though a small overlap between the 95 % confidence intervals of MIRU-VNTR and wgSNP analysis could be observed, the p-value between the two SIDs was <0.001, which makes the difference in discriminatory power statistically significant. Similarly, MIRU-VNTR appears to have a higher discriminatory power when compared to cgMLST (0.856 vs 0.807), even though cgMLST was able to divide the dataset into more partitions and had a higher level of concordance with wgSNP analysis than MIRU-VNTR (88 % vs 76 %).
Table 3.
Assessment of discriminatory power of MIRU-VNTR, cgMLST and wgSNP analysis by calculation of the Simpson's Index of Diversity and corresponding 95 % confidence intervals. Calculations were performed using the freely available tools at the website Comparing Partitions (http://www.comparingpartitions.info/index.php?link=Home).
| NAME | PARTITIONS | SIMPSON’S ID | CI (95 %) |
|---|---|---|---|
| wgSNP (≤5 SNPs) | 45 | 0.895 | 0.874–0.916 |
| cgMLST | 33 | 0.807 | 0.775–0.839 |
| MIRU-VNTR | 14 | 0.856 | 0.836–0.875 |
3. Discussion
This study as part of a larger validation to introduce WGS-based methods at the IMRL as part of the routine diagnostic algorithm, which is accredited to ISO 15189 standards (Fig. S1, supplementary data). The main aim was to evaluate the performance of two WGS analytical methods, more specifically cgMLST (Ridom SeqSphere+) and wgSNP analysis (MTBSeq pipeline), and to define the best strategy to implement these into the molecular surveillance and relatedness of M. tuberculosis isolates in Ireland. Both WGS analysis methods were evaluated in terms of ease of use of the respective platforms, concordance of clustering results with MIRU-VNTR (and with each other), and discriminatory power.
In terms of overall experience and ease of use, SeqSphere+ platform (cgMLST) Ridom Bioinformatics is an intuitive graphical user interface that constitutes a unified platform for clustering analysis and computation of minimum spanning trees based on cgMLST clustering, allowing its direct visualisation in the GUI. On the other hand, even though MTBseq pipeline offers a high level of flexibility to adapt to the user's needs and allows deeper investigation of phylogenetic relatedness between isolates, additional software is required to generate/visualise the phylogenetic tree and no GUI is provided to perform the analysis. Instead, the analysis needs to be run using the command line, resulting in a less intuitive and user-friendly experience when compared to SeqSphere+ platform (cgMLST).
Although the cost of analysis was not set as a main criterion for the evaluation of WGS methods in this study, this is a factor that imperatively comes into the balance when deciding on which method to implement. In this sense, cgMLST would represent a more expensive choice, since it requires a paid license to carry out the analysis for the contracted time, as opposed to MTBseq pipeline that is freely available for download and installation on GitHub (https://github.com/ngs-fzb/MTBseq_source) (Table 1). However, for laboratories who may not have access to command line languages and bioinformatics expertise, SeqSphere+ appears to be a sensible choice to implement in routine and real-time surveillance of MTBC.
According to our analysis and using the 12-allele threshold to define a cluster in cgMLST [23] and ≤5 SNPs for wgSNP analysis [20], the two methods showed 88 % concordance between each other, and 88 % and 76 % concordance with MIRU-VNTR, respectively. However, full concordance was not to be expected, considering that the discriminatory power of wgSNP analysis is known to be superior, which was confirmed by our results.
Cluster 8 comprised two isolates that were shown to differ by 12 alleles by cgMLST, however, by wgSNP-analysis these two isolates showed less than 5 SNPs difference. Even though the cluster was concordant between the two methods, wgSNP analysis revealed much less diversity than might be expected from the cgMLST results. This is most likely to do with the differences between the methodologies here (i.e. core genes used for cgMLST may not all be used for wgSNP analysis following filtering), and this is the most extreme example within the cohort. However, the algorithm proposed (i.e. cgMLST followed by wgSNP analysis where a cluster is detected) has proven to be robust enough to detect the cluster which gives us further confidence (Fig. S1, supplementary data).
Moreover, a small overlap of confidence intervals was observed when comparing both WGS methods to MIRU-VNTR. This small overlap in confidence intervals indicates that MIRU-VNTR is indeed a very good classical typing method for MTBC, particularly useful when excluding unrelated isolates from clusters. The difficulty arises when differentiating isolates that share the same MIRU-VNTR profile (24/24 loci match) but do not share an apparent epidemiological link. This becomes particularly relevant in epidemiological investigations of TB clusters in low TB incidence countries, where clusters may have a predominance of isolates from immigrant populations from high TB incidence countries with the same MIRU-VNTR profile [[23], [24], [25], [26]]. In this study, WGS-based analysis showed that MIRU-VNTR had overestimated the size of nine of the initial 14 clusters and that clusters 5, 6a and 6b were, in fact, artefacts resulting from over-clustering of isolates that were genetically distant (>30 alleles/12–31 SNPs). This highlights the importance of good quality epidemiological data when interpreting results and how a case-by-case approach should always be considered.
Considering the strong concordance between the two WGS methods documented in this study and since wgSNP analysis on every isolate could prove to be too much of a computational burden in a routine laboratory setting, it justified a strategy to use cgMLST as a first-line method, followed by wgSNP analysis on any cluster detected (see WGS pipeline algorithm in supplementary data, Fig. S1). So, when an isolate is sequenced, it is first compared to every strain already sequenced at the IMRL using cgMLST. If the isolate clusters with any other strain in the database, all isolates within that cluster are analysed using wgSNP in order to confirm whether there is, indeed, a close relationship found, indicating possible recent transmission, or not. The default threshold for the MTBC cgMLST scheme in SeqSphere is 12 alleles, therefore it was decided to set the threshold there, in order to catch all possible related strains prior to wgSNP analysis.
cgMLST can be performed on each new isolate received in order to compare it to all isolates in the database. In practice, this takes approximately 10 min, compared to a much longer time if wgSNP analysis was used to compare a new isolate to every other strain in the database. Of course, this depends on the computational capacity available, analysis pipeline in place, and the technical specifications of the PC being used. Decreasing turnaround times is always a priority in the laboratory, which makes this algorithm a good proposal in our view. Because Seqsphere+ retains the mapped files for each isolate, the comparison also requires less computational power and can be performed on a regular PC; another advantage in a resource-constrained environment.
The commonly used threshold for wgSNP analysis of MTBC is ≤ 5 SNPs and represents possible recent transmission. However, based on what is described in the literature, the estimated mutation rate per year for MTBC is 0.5 SNPs per genome [27], meaning that while isolates showing up to 5 SNPs difference can be considered linked by a transmission event, more than 5 but lower or equal to 12 (>5 - ≤ 12) SNPs could still be considered possible transmission. In fact, if a broader cluster definition is considered and a distance ≤12 SNPs (indicating recent and possible transmission) between isolates is used to define a wgSNP cluster, the concordance of results between cgMLST and wgSNP-analysis would actually increase to 96 %, making the two methods more comparable (see Table 2). Ireland is a low-incidence, relatively high-income country, where a threshold of ≤5 SNPs is appropriate to focus public health contact tracing resources, but this may differ for other settings.
Clusters 2a and 2b were originally differentiated by MIRU-VNTR based on one repeat difference in a single locus out of the 24 targeted loci (Table 2). Cluster 2a contained 4 repeats at the 2996 locus, while cluster 2b contained 5 repeats. However, cgMLST aggregated these two clusters into a single cluster, while wgSNP analysis resolved it into three sub-clusters. Because of this discrepancy in clustering and the fact that Cluster 2a and 2b account for 33 % of the dataset, the Simpsons’ ID of cgMLST was inappropriately decreased compared to MIRU-VNTR, even though it was able to resolve the dataset into more partitions (10 clusters and 23 unique strains vs 14 clusters).
It is important to note that this study comes with some limitations. One is the criterion used for the selection of our dataset, since only isolates that clustered together by MIRU-VNTR were selected. This introduced a selection bias right at the starting point as our dataset is not representative of the whole population diversity of MTBC. In order to resolve this, 2019 and 2020 prospective datasets (all isolates received in both years) were added to the overall validation and the concordance was confirmed (unpublished data). Another limitation would be that we are not comparing like with like, i.e. MIRU-VNTR genotyping is based on the fragment analysis of repeat regions that are masked by the whole genome analysis methods such as cgMLST and wgSNP analysis, which could account for some differences. For instance, double alleles found using MIRU-VNTR genotyping have long been a source of confusion for the TB community and have been associated with either a sub-population, mixture, or genetic drift [28]. Similarly, cgMLST and wgSNP analysis are not exactly ‘like’ for ‘like’ since cgMLST compares alleles across 2891 core genes and wgSNP analysis using MTBseq uses a set of filters to remove resistance genes and repetitive regions during analysis. The final limitation of this methodology in its current form is the fact that mixtures of closely related strains cannot be distinguished readily at present, highlighting the importance of good quality epidemiological data for appropriate interpretation of molecular data. This is a challenge for the TB community as a whole and the IMRL is monitoring the literature closely in order to update our methods if and when this limitation has been resolved.
4. Conclusion
The adoption of WGS-based analytical methods for real-time surveillance of MTBC isolates is of paramount importance for improving the capacity of national reference laboratories and public health institutions to help inform public health action and design policies to contain the spread of TB around the globe.
However, the choice of methods to implement into the public health setting for clinical and routine surveillance work is not always easy and will naturally need to be tailored to each institution's needs, as the capacities and weight of several parameters need to be considered (i.e. ease of use, staff expertise, turn-around time, maintenance, customer-support, etc.).
Based on our study results although not quite as discriminatory as wgSNP-analysis, cgMLST appears to be a good alternative to MIRU-VNTR for a first-line genotyping method as it provides sufficient resolution for routine surveillance work, conveniently supported by epidemiological data and its platform, SeqSphere+, is relatively easy to use and to implement. WgSNP analysis using MTBseq pipeline would be recommended as a second-line analysis on any cgMLST clusters found, providing a more in-depth analysis, namely for outbreak investigations where a higher resolution is needed to ascertain chains of transmission. With the utility of WGS-based genotyping, the IMRL hopes to inform public health action and to ultimately contain the spread of TB in Ireland, and consequently in the EU/EEA region.
5. Materials and methods
5.1. Sample selection
Clusters of M. tuberculosis isolates (24/24 or 23/24 MIRU-VNTR identical) were chosen in order to include the largest circulating clusters as well as a representative selection of different lineages. A collection of 240 M. tuberculosis clinical isolates (n = 14 clusters, sizes ranging from 2 to 84) submitted to the IMRL for investigation, between 1998 and 2019, were included. All isolates had been previously typed by MIRU-VNTR as part of routine surveillance, and whole-genome sequencing was performed using Illumina's MiniSeq platform (Illumina Inc., San Diego, CA, USA) as part of a larger molecular epidemiological study (unpublished work).
All data was handled blindly and analysed using the two WGS-based typing approaches: cgMLST and wgSNP analysis, using the workflow proposed by Tagliani et al. as reference [19]. The same fastq files were used for both pipelines (please refer to WGS pipeline, supplementary data).
5.2. Core-genome MLST
Raw sequence data of 230 isolates was uploaded to the Ridom SeqSphere+ platform (version 7; Ridom GmbH, Munich, Germany) using the ‘pipeline starter mode’ (please refer to supplementary data for more information). Sequence data was mapped to the Mycobacterium tuberculosis H37Rv strain genome (GenBank ID: NC_000962.3) and uploaded into the study database using pre-defined quality criteria (≥90 % cgMLST targets and mean read coverage ≥30x). Seven samples did not meet the QC parameters and failed cgMLST genotyping, and were therefore excluded from the analysis, bringing the dataset number to 223 isolates (from 221 patients).
cgMLST analysis was carried out for the remaining 223 isolates using a specific cgMLST scheme for M. tuberculosis complex typing published in 2018 [27], based on 2891 core genes to assign clusters (single linkage clustering). Clusters were defined by using a maximum threshold of 12 allelic differences, which corresponds to the default settings established for MTBC by SeqSphere+ [27]. cgMLST allele numbers were automatically retrieved from and stored at cgMLST.org, a web-based nomenclature server.
5.3. wgSNP-based phylogenetic analysis
In total, raw sequence data of 231 MTBC isolates (from 226 patients) underwent wgSNP analysis using the MTBseq pipeline (v1.0.4) (please refer to supplementary data for more information) [13]. Briefly, reads were mapped to the H37Rv genome (GenBank ID: NC_000962.3) with BWA and alignments were then refined with the GATK and Samtools toolkits for base quality recalibration and alignment corrections for possible PCR and InDel artefacts. The TBfull script was used with default settings and the --distance 5 command. From the concatenated sequence alignments, isolates were grouped by agglomerative clustering with a maximum distance threshold of ≤5 SNPs to the nearest isolate in the same group [20]. Regions annotated as repetitive elements, InDels, multiple consecutive SNPs in a 12-bp window, and 92 genes implicated in antibiotic resistance were excluded for the phylogenetic reconstruction.
Subsequently, TBjoin was used with default settings and the --distance 5 --continue command for phylogenetic analysis. The FASTA alignment of SNP positions obtained through MTBseq pipeline was used as input for RaxML GUI 2.0 [29] to generate the final SNP tree and visualised using the freely available online platform Microreact [30].
5.4. Evaluation of the performance of WGS-based methods
The two WGS analytical methods were evaluated using three parameters: overall ease of use, concordance of clustering results and discriminatory power.
Ease of use was evaluated in terms of required knowledge to use the software for cgMLST and wgSNP analysis.
The percentage of concordance of results obtained by cgMLST or wgSNP-analysis versus MIRU-VNTR results was calculated based on the number of matching classifications divided by the total number of isolates analysed.
Additionally, discriminatory power was assessed by calculating the Simpson's Index of Diversity (SID) [21] using the freely available online tools at the Comparing Partitions website (http://www.comparingpartitions.info/index.php?link=Home) [22]. To determine if the typing methods differed in their discriminatory power, the 95 % Confidence intervals (CI) and the p-values between the two coefficients were both considered.
CRediT authorship contribution statement
Diana Espadinha: Writing – review & editing, Writing – original draft, Visualization, Methodology, Formal analysis, Conceptualization. Emma Roycroft: Writing – review & editing, Supervision, Methodology, Conceptualization. Peter R. Flanagan: Writing – review & editing, Software. Simone Mok: Writing – review & editing, Software. Eleanor McNamara: Writing – review & editing, Supervision, Conceptualization. Thomas R. Rogers: Writing – review & editing, Supervision, Conceptualization. Margaret M. Fitzgibbon: Writing – review & editing, Supervision, Methodology, Conceptualization.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Silvia Herrera Leon, EUPHEM coordinator for her help in reviewing this manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e40279.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Lawn S.D., Zumla A.I. Tuberculosis. Lancet. 2011;378:57–72. doi: 10.1016/S0140-6736(10)62173-3. [DOI] [PubMed] [Google Scholar]
- 2.European Centre for Disease Prevention and Control, World Health Organization Regional Office for Europe Tuberculosis surveillance and monitoring in Europe 2021 - 2019 data. 2021. https://www.ecdc.europa.eu/en/publications-data/tuberculosis-annual-epidemiological-report-2019 Copenhagen.
- 3.World Health Organization . World Health Organization; Geneva: 2021. Global Tuberculosis Report 2021.https://apps.who.int/iris/rest/bitstreams/1379788/retrieve [Google Scholar]
- 4.Health Protection Surveillance Centre (HPSC) Tuberculosis in Ireland: provisional trends in surveillance data. Dublin. 2023 [Google Scholar]
- 5.World Health Organization . World Health Organization; 2015. Health in 2015: from MDGs, Millennium Development Goals to SDGs, Sustainable Development Goals. [Google Scholar]
- 6.Allix-Béguec C., Fauville-Dufaux M., Supply P. Three-year population-based evaluation of standardized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 2008;46:1398–1406. doi: 10.1128/JCM.02089-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Supply P., Mazars E., Lesjean S., Vincent V., Gicquel B., Locht C. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 2000;36:762–771. doi: 10.1046/j.1365-2958.2000.01905.x. [DOI] [PubMed] [Google Scholar]
- 8.Evans J.T., Hawkey P.M., Smith E.G., Boese K.A., Warren R.E., Hong G. Automated high-throughput mycobacterial interspersed repetitive unit typing of Mycobacterium tuberculosis strains by a combination of PCR and nondenaturing high-performance liquid chromatography. J. Clin. Microbiol. 2004;42:4175–4180. doi: 10.1128/JCM.42.9.4175-4180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frothingham R., Meeker-O’Connell W.A. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology. 1998;144:1189–1196. doi: 10.1099/00221287-144-5-1189. [DOI] [PubMed] [Google Scholar]
- 10.Supply P., Allix C., Lesjean S., Cardoso-Oelemann M., Rüsch-Gerdes S., Willery E., et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 2006;44:4498–4510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mellmann A., Harmsen D., Cummings C.A., Zentz E.B., Leopold S.R., Rico A., et al. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One. 2011;6 doi: 10.1371/journal.pone.0022751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maiden M.C.J., Van Rensburg M.J.J., Bray J.E., Earle S.G., Ford S.A., Jolley K.A., et al. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat. Rev. Microbiol. 2013;11:728–736. doi: 10.1038/nrmicro3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohl T.A., Utpatel C., Schleusener V., De Filippo M.R., Beckert P., Cirillo D.M., et al. MTBseq: a comprehensive pipeline for whole genome sequence analysis of Mycobacterium tuberculosis complex isolates. PeerJ. 2018;2018 doi: 10.7717/peerj.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Homolka S., Projahn M., Feuerriegel S., Ubben T., Diel R., Nübel U., et al. High resolution discrimination of clinical mycobacterium tuberculosis complex strains based on single nucleotide polymorphisms. PLoS One. 2012;7 doi: 10.1371/journal.pone.0039855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coll F., McNerney R., Guerra-Assunção J.A., Glynn J.R., Perdigão J., Viveiros M., et al. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat. Commun. 2014;5 doi: 10.1038/ncomms5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roetzer A., Diel R., Kohl T.A., Rückert C., Nübel U., Blom J., et al. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med. 2013;10 doi: 10.1371/journal.pmed.1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mellmann A., Bletz S., Böking T., Kipp F., Becker K., Schultes A., et al. Real-time genome sequencing of resistant bacteria provides precision infection control in an institutional setting. J. Clin. Microbiol. 2016;54:2874–2881. doi: 10.1128/JCM.00790-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyllie D.H., Davidson J.A., Grace Smith E., Rathod P., Crook D.W., Peto T.E.A., et al. A quantitative evaluation of MIRU-VNTR typing against whole-genome sequencing for identifying Mycobacterium tuberculosis transmission: a prospective observational cohort study. EBioMedicine. 2018;34:122–130. doi: 10.1016/j.ebiom.2018.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tagliani E., Anthony R., Kohl T.A., De Neeling A., Nikolayevskyy V., Ködmön C., et al. Use of a whole genome sequencingbased approach for Mycobacterium tuberculosis surveillance in Europe in 2017-2019: an ECDC pilot study. Eur. Respir. J. 2021;57 doi: 10.1183/13993003.02272-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker T.M., Ip C.L.C., Harrell R.H., Evans J.T., Kapatai G., Dedicoat M.J., et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect. Dis. 2013;13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunter P.R., Gaston M.A. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carriço J.A., Silva-Costa C., Melo-Cristino J., Pinto F.R., De Lencastre H., Almeida J.S., et al. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J. Clin. Microbiol. 2006;44:2524–2532. doi: 10.1128/JCM.02536-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stucki D., Ballif M., Egger M., Furrer H., Altpeter E., Battegay M., et al. Standard genotyping overestimates transmission of mycobacterium tuberculosis among immigrants in a low-incidence country. J. Clin. Microbiol. 2016;54:1862–1870. doi: 10.1128/JCM.00126-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fenner L., Gagneux S., Helbling P., Battegay M., Rieder H.L., Pfyffer G.E., et al. Mycobacterium tuberculosis transmission in a country with low tuberculosis incidence: role of immigration and HIV infection. J. Clin. Microbiol. 2012;50:388–395. doi: 10.1128/JCM.05392-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munang M.L., Browne C., Khanom S., Evans J.T., Grace Smith E., Hawkey P.M., et al. Tuberculosis microepidemics among dispersed migrants, Birmingham, UK, 2004–2013. Emerg. Infect. Dis. 2015;21:524–527. doi: 10.3201/eid2103.140209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sloot R., Borgdorff M.W., De Beer J.L., Van Ingen J., Supply P., Van Soolingen D. Clustering of tuberculosis cases based on variable-number tandem-repeat typing in relation to the population structure of Mycobacterium tuberculosis in The Netherlands. J. Clin. Microbiol. 2013;51:2427–2431. doi: 10.1128/JCM.00489-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohl T.A., Harmsen D., Rothgänger J., Walker T., Diel R., Niemann S. Harmonized genome wide typing of tubercle Bacilli using a web-based gene-by-gene nomenclature system. EBioMedicine. 2018;34:131–138. doi: 10.1016/j.ebiom.2018.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jajou R., Kamst M., van Hunen R., de Zwaan C.C., Mulder A., Supply P., et al. Occurrence and nature of double alleles in variable-number tandem-repeat patterns of more than 8,000 Mycobacterium tuberculosis complex isolates in The Netherlands. J. Clin. Microbiol. 2018;56 doi: 10.1128/JCM.00761-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edler D., Klein J., Antonelli A., Silvestro D. raxmlGUI 2.0: a graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol. Evol. 2021;12:373–377. doi: 10.1111/2041-210X.13512. [DOI] [Google Scholar]
- 30.Argimón S., Abudahab K., Goater R.J.E., Fedosejev A., Bhai J., Glasner C., et al. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb. Genomics. 2016;2 doi: 10.1099/MGEN.0.000093/CITE/REFWORKS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roycroft E., Fitzgibbon M.M., Kelly D.M., Scully M., McLaughlin A.M., Flanagan P.R., et al. The largest prison outbreak of TB in Western Europe investigated using whole-genome sequencing. Int. J. Tuberc. Lung Dis. 2021;25:491–497. doi: 10.5588/ijtld.21.0033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.