Abstract
Total resection of glioblastoma (GB) tumors is nearly impossible, and systemic administration of temozolomide (TMZ) is often inadequate. This study presents a hybrid layered composite nanofiber mesh (LHN) designed for localized treatment in GB tumor bed. The LHN, consisting of polyvinyl alcohol and core-shell polylactic acid layers, was loaded with TMZ and rutin. In vitro analysis revealed that LHNTMZ and LHNrutin decelerated epithelial-mesenchymal transition and growth of stem-like cells, while the combination, LHNTMZ+rutin, significantly reduced sphere size compared to untreated and LHNTMZ-treated cells (P < 0.0001). In an orthotopic C6-induced GB rat model, LHNTMZ+rutin therapy demonstrated a more pronounced tumor-reducing effect than LHNTMZ alone. Tumor volume, assessed by magnetic resonance imaging, was significantly reduced in LHNTMZ+rutin-treated rats compared to untreated controls. Structural changes in tumor mitochondria, reduced membrane potential, and decreased PARP expression indicated the activation of apoptotic pathways in tumor cells, which was further confirmed by a reduction in PHH3, indicating decreased mitotic activity of tumor cells. Additionally, the local application of LHNs in the GB model mitigated aggressive tumor features without causing local tissue inflammation or adverse systemic effects. This was evidenced by a decrease in the angiogenesis marker CD31, the absence of inflammation or necrosis in H&E staining of the cerebellum, increased production of IFN-γ, decreased levels of interleukin-4 in splenic T cells, and lower serum AST levels. Our findings collectively indicate that LHNTMZ+rutin is a promising biocompatible model for the local treatment of GB.
Keywords: Glioblastoma, Local treatment, Temozolomide, Rutin, Hybrid layered composite nanofiber web
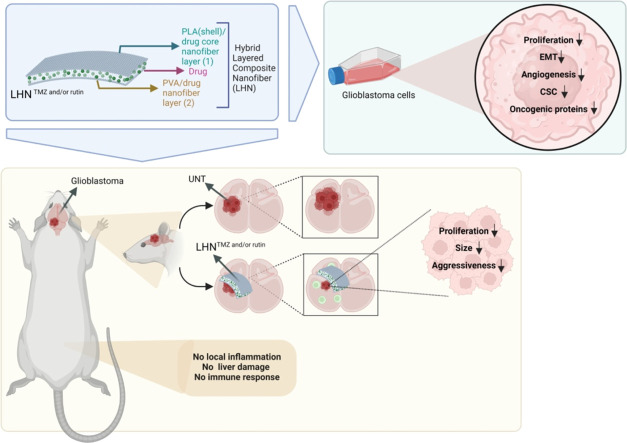
Graphical abstract
The developed hybrid layered composite nanofiber networks are promising as a treatment approach that can suppress the proliferation of tumor cells in the tumor area.
1. Introduction
Glioblastoma (GB) is the most aggressive and common form of brain tumor, accounting for 12 %−15 % of all brain tumors and 50 %−60 % of astrocytomas [1]. The overall survival time of patients is approximately 15 months with the current treatment, which mainly consists of a maximal surgical resection of the tumor, and adjuvant radiotherapy, and chemotherapy, which is temozolomide (TMZ) in most cases [2,3]. However, since complete resection of the tumor is not possible in most cases, recurrence is often encountered. Even though newly developed technologies improved the success of oncological therapy in a variety of cancer types, in the case of GB, systemic toxicity, limited penetration through the blood-brain barrier, inability to reach a sufficient concentration in the tumor bed, and short half-life of these therapies pose limitations to the success of approaches developed for GB therapy [[4], [5], [6], [7]]. Therefore, the need for new therapy approaches focusing on the local effect of anti-cancer agents at the tumor bed comes to the fore for GB treatment.
Pioneering studies have evidenced that local application of TMZ into GB tumor bed reduces the clinical side effects frequently observed in systemic therapy, by avoiding systemic circulation of TMZ and thus protecting normal tissues from the toxicity of the drug [8,9]. In addition, Gliadel® wafers, one of the pioneers of local therapy for intracranial tumors, consist of a biocompatible carrier vector to treat cancer with carmustine have opened a new era of biocompatible polymer technology for achieving a controlled release of different therapeutic agents at the tumor bed locally [[10], [11], [12]]. However, the reported “sink effect” of Gliadel® wafers, which refers to the loss of the drug in systemic circulation due to excessive diffusion, highlights the need for a controlled and long-term release method of the therapeutic agent from polymers [13]. Moreover, although carmustine could be used in treating many cancers, including glial tumors, current studies recommend using carmustine only as a supplement alongside TMZ for GB, and TMZ-based therapy is still the standard of care for GB patients [14]. Therefore, polymer-based controlled release systems using TMZ are one of the researchers' goals for the local treatment of GB in the tumor bed.
Among polymers, nanofibers produced by electrospinning have attracted much attention as implantable drug delivery systems in the tumor bed after surgical resection due to their indispensable properties, such as large surface area-to-volume ratio due to high loading capacity and high encapsulation efficiency [15]. Nanofibers enable controlled and long-term release of adjustable drug concentrations at the tumor site [16]. Nanofiber-based drug delivery systems loaded with TMZ, carmustine, doxorubicin and paclitaxel were reported to reduce systemic side effects while delivering a sufficient amount of these agents to the tumor site [[16], [17], [18], [19]]. Moreover, engineered nanofibers allow co-administration of more than one therapeutic agent to the tumor bed. For instance, nanofibers co-loaded with TMZ and paclitaxel were used to achieve long-term glioma cell inhibition in vitro [20]. Co-loading of multiple agents into a nanofiber can not only be used to deliver chemotherapeutic agents, but also opens the possibility of conducting preclinical trials for a large range of therapeutic candidates that cannot be used systemically. One of these anti-cancer therapeutic candidates is flavonoids [21,22]. So far, numerous studies have highlighted the antioxidant and cancer cell-killing effects of various flavonoids [23]. However, many of these flavonoids are recommended to be taken orally as supplements [22]. Ingestion of these flavonoids is beneficial to mitigate the signaling pathways that induce chronic inflammation [22]. However, the oral intake of high doses of flavonoids is inconvenient since they could act as mutagens and inhibitors of key enzymes in hormone metabolism [21,24,25]. Therefore, uncovering the effects of direct application of flavonoids, whose anti-cancer effects are well-defined in vitro, to the tumor bed, is worthy of research. One of those flavonoids, rutin, abundantly exists in herbs, fruits, vegetables, and plant-based beverages such as tea and wine [26]. Its inhibitory effect on oxidative stress, inflammation, cancer-cell proliferation and angiogenesis and promoting effect on apoptosis was evidenced in various cancers, including glioma, breast, liver, pancreas, and colon cancers in vitro [[27], [28], [29], [30], [31], [32]]. In particular, in our studies over the last decade, in which we mainly clarified the in vitro effects of olive leaf extract on GB cells, [[33], [34], [35], [36], [37], [38]] we evidenced that rutin is one of the essential components responsible for this effect [39]. In addition, rutin relieves a wide range of chemotherapy-related side effects and increases chemosensitivity [40]. Likewise, our previous studies showed more potent GB-cell inhibition by the concurrent use of TMZ and rutin than by TMZ-only, due to the additive effect of rutin [39]. Therefore, investigating the therapeutic efficiency of direct application of rutin and TMZ to the tumor bed with a vehicle to provide a long-term release of these agents may pave the way for a new treatment approach for GB patients.
Core-shell nanofibers can be one of the most convenient tools for loading TMZ and rutin, since they enable sufficient drug-release control by tailoring components in different layers that induce various polymer degradation rates and drug dissolution [41]. In one of our previous studies, polylactic acid (PLA) core-shell nanofiber networks enabled the controlled release of oleuropein, rutin, and TMZ. The release of oleuropein, rutin, and TMZ from PLA suppressed GB cell growth individually. However, a more potent GB cell-killing effect was observed when the cell lines were treated with oleuropein or rutin-loaded PLA [42]. Unlike the PLA single-layer nanofiber we used in our previous study, in this study, we designed a biocompatible hybrid layer composite nanofiber network (LHN) with different release characteristics in different sequential processes, which is more suitable for local treatment of GB in living systems. The first layer, polyvinyl alcohol (PVA) was designed for an immediate release of TMZ and rutin to kill the residual cancer cells after tumor resection. The second layer, a core-shell PLA nanofiber network, was designed for controlled long term release of these agents to prevent tumor growth. Additionally, the therapy was supported with the loaded TMZ and rutin between the layers. The designed TMZ and/ or rutin-loaded LHN (LHNTMZ, LHNrutin, LHNTMZ+rutin) were tested for their effects on the aggressiveness of GB cells by in vitro functional analysis, and the affected oncological processes and associated protein network were revealed. The impact of LHNTMZ, LHNrutin, and LHNTMZ+rutin on GB tumor progression and the risk of systemic side effects and effects on inflammatory response was determined using an orthotopic GB model.
2. Materials and methods
2.1. Materials
The PVA (purity 87.8 %, Mw ∼30,000 g/mol) was from ZAG Industrial Chemicals (Istanbul, Turkey). PLA Luminy LX 175 granules (Total Corbion) was provided from Kumrukimya (Istanbul, Turkey). N, N-dimethylformamide (DMF) (∼99.8 % purity) and acetone (∼99 % purity) were purchased from Honeywell Riedel-de-Haën (Germany) and Sigma-Aldrich Chemical Company (St. Louis, USA), respectively. Distilled water was used in the experiments, and all reagents were used without being purified. TMZ and rutin were provided from Sigma-Aldrich (St. Louis, USA). TMZ (10 mg/ml) was dissolved in dimethyl sulfoxide (DMSO), and rutin (6.25 mg/25 ml) was dissolved in distilled water as described previously [39]. LHN, encapsulated with IC50 doses of rutin (142.2 µM) (LHNrutin) and TMZ (1,000 µM) (LHNTMZ) [39] were used for all in vitro and in vivo analysis.
2.2. Nanofabrication and characterization
2.2.1. Preparation of the process
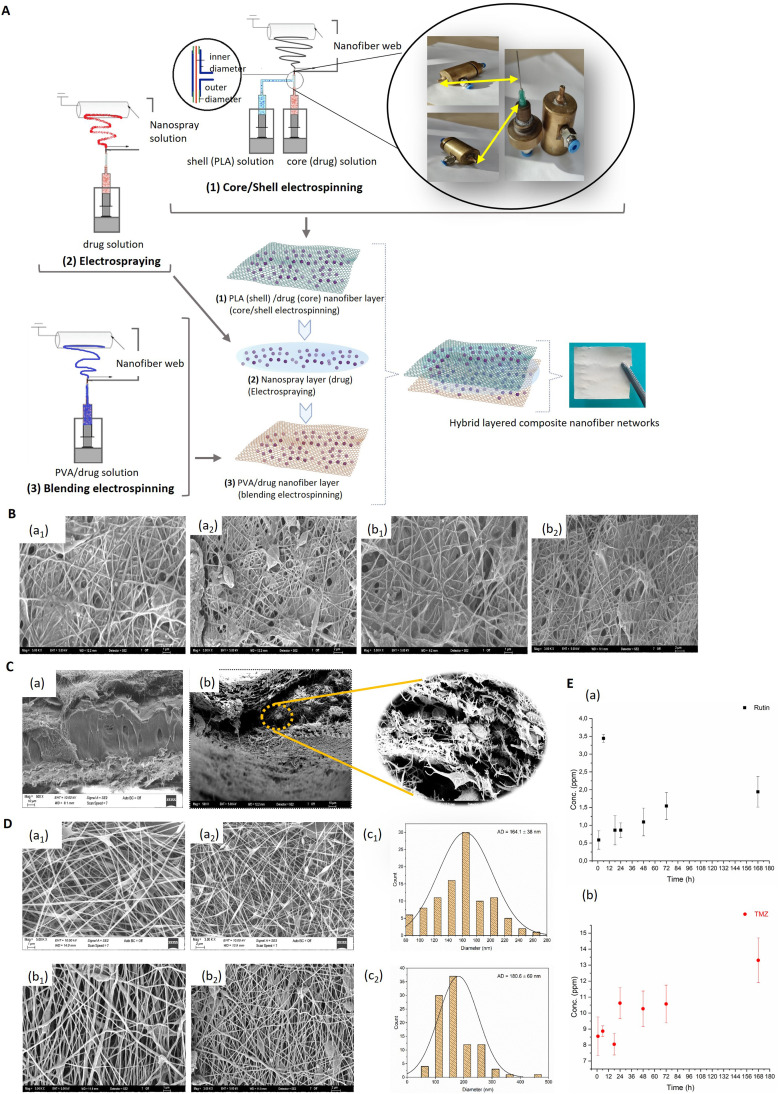
The drug-loaded PLA LHN webs were fabricated by electrospinning (INOVENSO NE300 Multinozzle Electrospinning Machine, Turkey). The LHN was produced in three layers: the first was composed of a core-shell nanofiber web, the second (inner layer) was formed as an active reagent deposited on the first layer, and the third was produced from PVA nanofiber, which contains active reagents. Firstly, PLA granules were dissolved in a binary-solvent system of DMF and acetone (4:1, v/v) to obtain a 10 % (w/v) PLA solution by stirring under room conditions overnight. A 10 ml PLA solution-filled plastic syringe was inserted into the syringe pump platform and connected to a needle. The needle has two components: the first consisted of an inner needle (a syringe needle with a diameter of 0.8 mm) and a core part, where the drug/agent solution of the nanofiber was spun from; the second component consisted of an outer needle, 2.11 mm in diameter, and the shell part, where the PLA solution of the nanofiber was spun from (Fig. 1A).
Fig. 1.
(A) Schematic illustration of the fabrication of LHN webs. (B) SEM micrographs of the first layer of the LHN webs: (a1, a2) LHNrutin and (b1, b2) LHNTMZ (Magnification: 5 kX, scale: 1 µm; 3 kX, scale: 2 µm). (C) SEM cross-sectional images of the LHN webs: (a) LHNrutin and (b) LHNTMZ (Magnification: 500 kX, scale: 10 µm). (D) SEM micrographs of the third LHN webs: (a1, a2) LHNrutin and (b1, b2) LHNTMZ (Magnification: 5 kX, scale: 1 µm; 3 kX, scale: 2 µm) and distribution of fiber sizes for (c1) LHNrutin and (c2) LHN™Z. (E) Drug release profiles of the LHN webs: (a) LHNrutin and (b) LHNTMZ. LHN: layered hybrid nanofiber,.
An electrospraying process formed the inner layer. In this process, rutin and TMZ (drug/agent solutions) were directly electrosprayed to a rotating drum on the first layer. The spraying was made from a syringe needle connected to the syringe by a tubing element. After preparing the homogeneous drug solution, it was filled in plastic syringes (10 ml). The last layer of the nanofiber webs was produced by blending electrospinning process. The PVA solution is prepared in distilled water (10 wt%), by stirring it at 90 °C for 3 h Then, rutin and TMZ were added separately in PVA (10 wt%) solutions, which was prepared by stirring at room conditions overnight. After preparing the homogeneous PVA-drug solution, it was filled in plastic syringes (10 ml) and connected to the syringe needle. LHN webs with PLA-rutin were labeled PPR, while those with PLA-TMZ were labeled PPT.
A high-voltage source or system, a spinneret and a conductive collector are the three essential components of conventional electrospinning [43]. The polymer solutions were exposed to a high 18–26 kV voltage. As the strength of the electrostatic forces increased, the polymer solution droplets at the needle's tip stretched and formed a cone shape. Polymer fibers were deposited as non-woven webs on the collector as the electrostatic field increased further, and the force exceeded the cone-shaped droplet's surface tension as described previously [44]. The electrospinning process parameters are shown in Table 1.
Table 1.
The parameters used in the production of LHN webs.
| Drugs/agents | First layer (Core - Shell) | Inner layer (Electrospraying) | Third layer (Blending) |
|---|---|---|---|
| TMZ | Inner part (TMZ): 1.5 ml | • TMZ: 0.9 ml | • TMZ: 0.6 ml |
| • Feeding rate: 0.2 ml/h | • Feeding rate: 0.3 ml/h | • PVA: 3.5 ml | |
| • Distance: 143 mm | • Distance: 143 mm | • Feeding rate: 0.4 ml/h | |
| • Voltage: 26 kV | • Voltage: 18 kV | • Distance: 110 mm | |
| • Rotation rate: 250 rpm | • Rotation rate: 250 rpm | • Voltage: 26 kV | |
| Outer part (PLA): 5 ml | • Rotation rate: 250 rpm | ||
| • Feeding rate: 0.85 ml/h | |||
| • Distance: 143 mm | |||
| • Voltage: 26 kV | |||
| Rotation rate: 250 rpm | |||
| Rutin | Inner part (Rutin): 5.22 ml | • Rutin: 3.14 ml | • Rutin: 2.1 ml |
| • Feeding rate: 0.3 ml/h | • Feeding rate: 1.5 ml/h | • PVA: 3.5 ml | |
| • Distance: 143 mm | • Distance: 143 mm | • Feeding rate: 0.6 ml/h | |
| • Voltage: 26 kV | • Voltage: 26 kV | • Distance: 112 mm | |
| • Rotation rate: 250 rpm | • Rotation rate: 250 rpm | • Voltage: 26 kV | |
| Outer part (PLA): 20.5 ml | • Rotation rate: 250 rpm | ||
| • Feeding rate: 2.0 ml/h | |||
| • Distance: 143 mm | |||
| • Voltage: 26 kV | |||
| • Rotation rate: 250 rpm |
2.2.2. Characterization
2.2.2.1. Scanning electron microscopy
A scanning electron microscope (SEM; Carl Zeiss/Gemini 300, ZEISS Ltd., Germany) visualized the surface morphologies of LHN webs at 10 kV. The samples were coated with gold/palladium around 20 min before analysis. The diameters of the nanofiber webs were measured using Image J software 1.520 V (National Institutes of Health, Bethesda, MD, USA) by randomly selecting 100 fibers for each sample.
2.2.2.2. In vitro drug release
The method for detecting in vitro release, tested using the total immersion method, was described in detail, and the calibration equation and calibration curve were shown in our previous study [42]. Briefly, the release of rutin or TMZ from nanofiber webs (1.5 cm × 1.5 cm) into a phosphate buffered saline (PBS) solution (5ml, pH: 7.4) was assessed in sealed glass tubes. The releasing pattern of each sample was individually observed for six different periods. Three replicates were analyzed for each release test. A UV–Vis–NIR spectrophotometer (UV-3600; Shimadzu Scientific Instruments, Japan) was used to measure the related absorbance value at the max wavelength (λmax), i.e. the distinctive peaks of rutin and TMZ. According to the calibration curve of the LHN web-released TMZ and rutin, the absorbance values of the rutin and TMZ were measured as 267 nm and 262 nm, respectively.
2.3. In vitro analyses to detect the effect of LHN webs on GB cells
2.3.1. Real-Time cell monitoring
Cell proliferation analysis was assessed using an xCELLigence System (ACEA Biosciences, Inc., San Diego, CA, USA). Briefly, 15 × 103 T98 G cells were seeded into each well of the E-plate containing the standard medium and placed into the cell analyzer in a 5 % CO2 humidified incubator at 37 °C. Proliferation pattern of the cells was monitored by measuring the cellular impedance in real-time every 15 min for 100 h after treatment with loaded LHN.
2.3.2. Mitochondrial membrane potential (Δψm) assay
Cell monolayers were treated with 200 nM tetramethylrhodamine ethyl ester (TMRE) (Cell Signaling Technology, Massachusetts, USA) at 37 °C for 20 min to stain active mitochondria. After removing the supernatant, the cell monolayer was washed thrice with PBS, and Δψm was visualized by a fluorescence microscope (EVOS M5000 Imaging System;Thermo Fisher Scientific, CA, USA). TMRE labeling of active mitochondria was detected at an excitation/emission wavelength of 550 nm/580 nm.
2.3.3. Scratch-Wound healing assay
2 × 105 T98 G cells were seeded into each well of a 6-well plate and incubated until 90 % confluency. The cell monolayer was rectangularly wounded using a 200-µl pipette tip, and a gentle PBS washing subsequently removed the cell debris. The change in the size of the wounded area was captured and measured before and 48 h after the incubation with differently loaded LHN groups. Experiments were performed in triplicate. The Image J measured the recovered wound area.
2.3.4. Microfluidic invasion assay
The IC (Invasion Chemotaxis) chip (Initio Biomedical Engineering, Izmır, Turkey) analyzed the invasion of T98 G cells. Briefly, a 1:1 diluted Matrigel (Corning, NY, USA) in a cold serum-free medium was loaded into the center of the IC—Chip. After 30 min polymerization of the Matrigel at 37 °C and 5 % CO2, 1 × 106 cells/ml serum-free medium was added from the top while the bottom of the IC—Chip was filled with 10 % serum-containing medium. The chips were incubated vertically for three days. The serum-containing medium was replaced daily with fresh medium. The fluorescence microscope visualized the cell invasion. Before the experiment, to make the cell movement visible on the IC—Chip, green fluorescent protein (GFP, Takara Bio, Shiga, Japan) plasmid was constructed to transfect T98 G cells using Lipofectamine™ 3000 (ThermoFisher Scientific, Waltham, MA, USA) following the manufacturer's instructions. The cell population, of whom 70 % expressed GFP, was applied to the IC—Chip.
2.3.5. HUVEC tube formation assay
The bottom of each well of a 96-well culture plate was coated with 50 µl Matrigel. 2 × 103 HUVEC cells/wells were seeded to each well in the continued medium of T98 G cells, which were treated with LHN variants. In addition, 4 ng/ml VEGF (Lonza, Basel, Switzerland.) was included in each well to stimulate the tube forming of endothelial cells. The formation of capillary-like structures were seen in the negative control samples in 3 h. All samples were followed for 24 h and captured to observe the effect of LHN-treated cell secretome on endothelial tube forming. The experiment was performed in three biological replicates.
2.3.6. Colony forming assay
T98 G cells were seeded into 6-well plates at a density of 1 × 105 cells and incubated for 24 h until the cells were ∼80 % confluent. Then, the cells were treated with different groups of loaded LHN and incubated for 48 h. The pre-treated cells were collected and re-seeded into 6-well plates at a density of 1 × 103 cells and allowed to form colonies in a standard growth medium for 10 d. The cell medium was renewed every 5 d. At the end of the incubation, the formed-colonies were stained according to the manufacturer's instructions using the CellMAX Colonogenic Assay Kit (BioPioneer, San Diego, CA, USA). Colonies were counted using the Image J with the following values: size: 0.01-infinity, circularity: 0.3–1000. The essay was performed in all groups simultaneously in three biological replicates.
2.3.7. Sphere-Forming assay
T98 G cells (80 cells/well) were seeded into an ultra-low attachment 96-well polystyrene microtiter plates (PerkinElmer Inc., Waltham, MA, USA) and placed in a humid incubator at 37 °C and 5 % CO2. The cells formed spherical shapes in 3 d and were captured under an inverted microscope. The spheroids were treated with different groups of loaded LHN for 96 h within a fresh medium. The spheroid was double stained with a mixture of 2 uM Calcein AM (Abcam, MA, USA) and 4.5 uM PI (Thermo Fisher Scientific, MA, USA) to detect the viability, and visualized by the fluorescence microscope. Calcein AM was detected at an excitation/emission wavelength of 488/520 nm, while PI was detected at 535 nm/615 nm.
2.3.8. Apoptosis assessment
In order to evaluate cell viability in various LHN-loaded groups, we used the Muse Annexin V & Dead Cell Assay Kit (Millipore; Burlington, MA, USA) according to the manufacturer's guidelines. Specifically, 15 × 104 cells were subjected to treatment with loaded LHN. Subsequently, the cells were collected and incubated with Annexin V and Dead Cell reagent in darkness for 20 min. The staining of cells with those markers was analyzed using a Muse Cell Analyzer (Millipore; Burlington, MA, USA). Each experiment was replicated three times for robustness.
2.3.9. Dual acridine orange/propidium iodide (AO/PI) fluorescent staining
Cells were seeded at a density of 5 × 104 cells per well in a 24-well plate and treated with variously loaded LHNs. Subsequently, the cells were exposed to a dual fluorescent staining solution (100 µl) containing 10 µg/ml AO and 5 µl PI (Sigma, St. Louis, MO, USA) for 5 min. The cell morphology was observed using the fluorescence microscope.
2.3.10. Real-Time quantitative PCR
The total RNA of the cells was extracted using a Zymo RNA Isolation Kit (Zymo Research, CA, USA). The quality and concentration of extracted RNA were measured at the ratio of 260:280 using a UV/Vis spectrophotometer (Beckman Coulter, Brea, Canada). A high-capacity cDNA Synthesis Kit (Thermo Fisher Scientific, USA) converted total RNA (100 ng) into cDNA. TaqMan Real-Time PCR Assays were used to detect RNA expression levels of TWIST (Hs01675818_s1), SNAIL (Hs_00195591_m1), ZEB1 (Hs01566408_m), N—Cadherin (Hs00983056_m1), E-Cadherin (Hs01013959_m1), CD133 (Hs01009259_m1), NANOG (Hs02387400_g1), SOX2 (Hs01053049_s1) and OCT4 (Hs04260367_gH) genes and long-non coding RNA levels of MALAT1 (Hs03453854_g1), HOTAIR (Hs03296661mH), LOXL1-AS1 (Hs04274784_m1), PVT1 (Hs01069023_m1) and H19 (Hs03663733_g1) in a StepOnePlus™ Real-Time PCR System according to the manufacturer's protocol.
2.3.11. Proteome analysis
2.3.11.1. Protein extraction
For liquid chromatography (LC)-mass spectrometry (MS)/MS analysis, the cells were grown in DMEM-F12 media. An equal number of cells (3 × 105) was seeded into T-75 flasks in triplicates. When the cell's confluency reached 70 %, the cells were washed with ample ice-cold PBS three times and removed from the plates by scraping. Excess PBS was decanted after 10 min centrifugation at 4 °C at 2,000× g, and the M-PER lysis buffer (Thermo Scientific) was added to each cell pellet. For further lysis of the cell pellets, they were homogenized in the lysis buffer using 0.1 mm stainless steel beads in a mechanical disruption device (Bullet Blender; Next Advance, Troy, NY). Homogenates were centrifuged at 15,000 g for 30 min at 4 °C to remove cell debris. Protein concentrations were determined by modified Bradford assay (BioRad, Hercules, CA), and protein extracts were aliquoted, snap-frozen in liquid nitrogen, and stored at −80 °C.
2.3.11.2. Peptide preparation for mass spectrometry
The protein extracts were reduced by adding 0.5 M DTT stock solution to a final concentration of 20 mM, followed by incubation at 95 °C for 5 min and cooling at room temperature for 10 min. Proteins were then alkylated in 40 mM of IAA for 45 min in darkness at room temperature. After reduction and alkylation, a filter-aided sample preparation (FASP) protocol was performed, and minor changes were made. Briefly, proteins were mixed with eight volumes of 8 M urea in 100 mM Tris–HCl pH 8. Microcon Ultracel 30 kDa (Millipore) units were prepared by briefly spinning 60 % methanol through the filter at 14,000× g. Protein samples were transferred to the filter units and spun at 14,000× g for 15 min. Detergent removal by buffer exchange was performed in two successive washes with 8 M urea in 100 mM Tris–HCl pH 8 with a 15 min spin at 14,000× g. One additional wash using 100 mM Tris–HCl pH 8, with a 15 min spin at 14,000× g, was included to remove excess urea. Protein digestion was achieved by adding 1 µg trypsin in 50 mM ammonium bicarbonate (ABC) and incubating at 37 °C overnight. Peptides were recovered and washed with 50 µl of 50 mM ABC, spinning at 14,000× g for 5 min. The mixture was then vacuumed and dried at 45 °C. Tryptic peptides were resuspended in 10 µl of 0.1 % formic acid.
2.3.11.3. Liquid chromatography-parameters
All peptide separations were carried out on a Thermo Scientific Ultimate 3000 Series RSLCnano pump coupled with a Thermo Scientific Ultimate 3000 Series TCC-3000RS column compartments and a Thermo Fisher Scientific UltiMate 3000 Series RS autosampler controlled by Xcalibur 4.0 Software (Thermo Fisher Scientific, US). For each analysis, the sample was loaded into a trapping column Acclaim PepMap 5 mm × 300 µm i.d., C18, 5 µm, 100 Å (Thermo Scientific, US) at 5 µl/min with an aqueous solution containing 0.05 % (v/v) trifluoroacetic acid and 1 % acetonitrile. After 3 min, the trapping column was put on-line with an analytical column Acclaim PepMap RSLC 15 cm × 75 µm i.d., C18, 2 µm, 100 Å (Thermo Scientific, US). Peptide elution was performed by applying a mixture of mobile phase A and B. Mobile phase A was HPLC grade water with 0.1 % (v/v) formic acid, and mobile phase B was HPLC grade acetonitrile with 0.1 % (v/v) formic acid. The cleaved samples were separated using a 130 min gradient elution at a flow rate of 0.30 µl/min, which was directly interfaced with a Thermo Q Exactive mass spectrometer. The applied gradient steps were as follows: 0–45 min linear increase from 6 % to 20 % B; 45–75 min linear increase to 40 % B; 75–90 in linear increase to 90 % B and hold 30 min; 120–125 decreasing to 6 % B; and 125–130 min 94 % A. The column temperature was set at 40 °C. Two microliter of each sample was injected. In positive mode, a heated electrospray ionization source (HESI) was used to ionize the target compounds. The parameters were set as follows: the ionization voltage was optimized at +2.3 kV; the capillary temperature was set at 300 °C; the vaporizer temperature was set to 300 °C; sheath gas and auxiliary gas flow were optimized at 50 and 30 arbitrary units, respectively. S lens RF level was set to 50.
2.3.11.4. Data-dependent acquisition
For data-dependent acquisitions using Q Exactive, the scan sequence began with an Orbitrap Full MS spectrum with the following parameters: resolution 70,000, scan range 400–2000 m/z, automatic gain control (AGC) target of 3 × 106, maximum injection time of 60 ms, and centroid spectrum data type. We selected the top 10 precursors for MS2 analysis, which consisted of high-energy collision dissociation (HCD) with the following parameters: resolution 17,500, AGC 1 × 105, maximum injection time 100 ms, isolation window 2 m/z, NCE 27, and acquired as a centroid spectrum data type. External calibration was performed by calibration solution (LTQ Velos ESI Positive Ion Calibration Solution 88323, Pierce, USA) in positive modes before each sample series. The underfill ratio was set at 9 %, which corresponds to a 1.5 × 105 intensity threshold. In addition, unassigned and singly charged species were excluded from MS2 analysis, and dynamic exclusion was set to automatic.
2.3.11.5. MS data analysis
The raw data generated from LC-MS/MS analysis and protein identification and quantification were processed using Proteome Discoverer 2.2 (Thermo Fisher Scientific, US). For protein identification, the following search parameters were used: peptide mass tolerance set to 10 ppm, MS/MS mass tolerance set to 0.02 Da, mass precision was set to 2 ppm, up to one missed cleavage allowed, minimal peptide length was set to six amino acids, carbamidomethylation of cysteine (+57.021464) was set as the fixed modifications, the oxidation of methionine (+15.994915) and deamidation of asparagine (0.984016) was set as the variable modification. For each protein, a few identified unique peptides were set to two. Input data were searched against the UniProtKB-SwissProt database (taxonomy: Homo sapiens, release 2017–08). Protein grouping was performed according to molecular function using the GeneOntology (GO) database available in the Proteome Discoverer 2.2 (Thermo Fisher Scientific, Bremen, US). Label-Free Quantification (LFQ) and validation were performed with the Proteome Discoverer 2.2 (Thermo Scientific, USA), the workflow used to generate the intensity-based quantitation data.
2.4. In vivo analyses to detect the effect of LHN webs on a GB model
2.4.1. Orthotopic GB model and LHN treatment
All animal experiments were approved by the Animal Care and Use Committee of Bursa Uludag University (Bursa, Turkey) (2021–02/10), and were conducted in concordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Sprague Dawley rats were inbred in the Experimental Animal Breeding and Research Unit of Bursa Uludag University, and they were acclimatized in a humidity and temperature-controlled facility for at least 3 d before surgery, under a 12-h light/dark cycle with ad libitum access to food and water. The C6 rat GB cell line was injected into 8–12 weeks rats (weight 280–330 g) to develop an orthotopic GB model as described previously [13]. In brief, rats were anesthetized with isoflurane (5 % for induction, 3 % for maintenance) and positioned on a stereotactic frame. The scalp was disinfected with 70 % ethanol, followed by a midline skin incision under a surgical microscope to expose the right frontal skull. Pericranial tissues were cleaned, and a 5 mm × 5 mm right frontal craniectomy was performed (1 mm anterior to the coronal suture and 1 mm lateral to the midline), followed by dura opening. Using stereotactic guidance, a volume of 10 µl containing 2.5 × 107 C6 cellsl PBS was slowly injected 2.5 mm deep from the cortical surface, 2 mm lateral and anterior to the bregma, using a Hamilton syringe (Hamilton Co, Reno, NV). The needle was kept in position for 5 min post-injection to prevent leakage before being withdrawn slowly over a period of 2 min. Rats were randomly divided into four groups (n = 8): (i) untreated control (UNT), (ii) LHNTMZ, (iii) LHNrutin and (iv) LHNTMZ+rutin. Nanofiber implantation was performed simultaneously with tumor implantation. After surgery, all animals were housed in wire cages in a temperature-controlled room with regularly alternating light and dark cycles. They were fed with a standard pellet diet and water ad libitum. Animal behaviors in each group were monitored daily for 11 d At the end of the experiment, the animals were sacrificed under deep anesthesia; tumor tissues, spleen and peripheral serum samples were collected for histological, molecular and biochemical analyses.
2.4.2. Magnetic resonance imaging
All magnetic resonance imaging (MRI) examinations were performed on a 3T MRI device (Philips Achieva dStream). MRI imaging consisted of T2 images (Repetition Time: 2,000 ms, Echo Time: 120 ms, Voxel Size: 0.2 × 0.2 × 2 mm3, no interslice gap, n = 6) acquired on axial and coronal planes. MRI images of rat brains were taken at the end of the 11th d after tumor transplantation and nano-treatment. Two blinded observers, one with 23 years and the other with 7 years of experience in neuroradiology, interpreted radiological images. Tumoral tissue was defined as a slight T2 hyperintense area that possibly corresponded to the infiltrative tumoral edema, whereas nanofiber was detected as a T2-marked hypointense structure with sharp angles. The long axis of the tumoral lesion and nanofiber were measured for each subject.
2.4.3. Immunohistochemistry (IHC) of target proteins
The tumor tissues resected from the brain were fixed in 4 % paraformaldehyde for 24 h and embedded into paraffin (FFPE). 3-µm serial tissue sections were processed for hematoxylin and eosin (H&E) staining and IHC for phospho-histone H3 (pHH3) and CD31. Primary antibodies were the monoclonal mouse CD31 and pHH3 antibodies (Dako, Carpinteria, California, USA). The sections were manually treated with primary antibodies for 30 min. After washing with PBS, the sections were incubated with a secondary antibody and the DAKO—HRP kit containing the avidin-biotin system for 20 min. Slides were washed with PBS, counterstained with hematoxylin, dehydrated, and closed with Entellam for examination under light microscopy (ECLIPSE Ci Series, Nikon, NY, USA). Two independent pathologists evaluated the immunolabelling, independently of other analyses. Strong nuclear immunoreactivity of PHH3 was considered as positive staining, while CD31, staining intensity was evaluated based on strong cytoplasmic staining [45,46].
2.4.4. Transmission electron microscopy (TEM)
The tumor tissue samples were fixed in a 2.5 % phosphate-buffered glutaraldehyde solution (Merck, Kenilworth, NJ, USA) at 4 °C for 24 h Tissue samples were then double-rinsed with PBS and fixed with 1 % OsO4 (Electron Microscopy Sciences; Hatfield, USA) for 2 h at room temperature, dehydrated using an ethanol series, and embedded in EPON (Electron Microscopy Sciences; Hatfield, USA). 700 nm sections of the tissues were collected using an ultramicrotome (Leica Ultracut RM; Wetzlar, Germany), stained with toluidine blue, and visualized in a light microscope to select suitable areas for TEM analysis. Ultrathin sections (60 nm) were contrasted with uranyl acetate-lead citrate and examined using a JEM-1220 microscope (JEOL; Tokyo, Japan). For the analysis of each sample, at least ten different areas of the grid were scanned, and evaluations were made in light of these common findings.
2.4.5. Western blotting
The M-PER lysis buffer extracted the cell-free proteins, and an SDS-PAGE separated the protein using 12 % acrylamide gels. Protein (30 µg per lane) was used for qualitative and quantitative analyses. Electrophoretic transfer of proteins onto positively charged nitrocellulose membranes was performed in a semi-dry electrophoretic transfer cell (Bio-Rad Laboratories, Inc.) for 20 min at 15 V, in a buffer containing 48 mM Tris (pH 9.2), 39 mM glycine, 20 % (v/v) methanol and 0.0375 g/l SDS. The membranes were blocked in TBS-T buffer (Tris–HCl 25 mM pH 7.2, NaCl 150 mM, and 0.1 % Tween 20) containing 5 % nonfat dry milk for 1 h at room temperature, and washed with TBS-T three times before incubation with primary antibodies diluted in TBS-T overnight at 4 °C. The membranes were then washed three times with TBS-T and incubated with HRP-conjugated secondary antibodies (goat anti-mouse; goat anti-rabbit, Bio-Rad) for 1 h at room temperature. Monoclonal anti-β-actin antibody (Santa Cruz Biotechnology, Inc.) and polyclonal anti-PARP (Cell Signaling) were used at the respective dilutions of 1:1,000 and 1:750. Following the subsequent three washes with TBST, protein bands were visualized with an enhanced chemiluminescence detection system (Thermo Fisher Scientific). Protein band intensities were analyzed using the Image J. β-actin was used as the internal normalization control for the band intensities.
2.4.6. Bio-safety evaluation
H&E staining of brain tissues of healthy rats treated with LHN determined the toxicity of LHN to healthy brain tissue. The aspartate aminotransferase (AST), alanine aminotrasnferase (ALT), creatine and urine levels of peripheral blood serum samples of GB rats determined the toxic effect of LHN on the liver and kidney. Burton Medical Analysis Laboratory, Bursa performed those biochemical tests. The IFN-γ and interleukin (IL)−4 production rate of splenocytes were used to assess the generation and regulation of immune responses.
2.4.6.1. The enzyme-linked immunosorbent spot (Elispot) assay
The PVDV membrane pre-coated 96-well plates (U-CyTech Bioscience, CM Utrecht, Netherlands) were coated with monoclonal rat anti-murine IL-4 and IFN-γ coating antibodies overnight at 4 °C, respectively. The plates were blocked for 1 h at 37 °C with 1 % BSA. Spleen cells were stimulated with phytohemagglutinin (PHA; 2.5 µg/ml), and 3 × 105 spleen cells were added into each well and incubated overnight (37 °C, 5 % CO2). The cells were removed by washing the plate with PBS-T, and the detection antibodies were added for 2 h, followed by AEC solution for 30 min at room temperature. When spots were visible, the color reaction was stopped by rinsing the plates with distilled water, and the spots were counted under 16 × magnification after drying. Spleen cells of the UNT incubated with 2.5 µg/ml PHA served as a positive control, while unstimulated cells were used as negative control.
2.5. Statistical analysis
Statistical analysis was done using the GraphPad Prism (8.0.1) software (GraphPad Software Inc.; San Diego, CA, USA). An independent sample T-test was utilized to evaluate the findings of cell proliferation, Δψm, scratch wound healing, microfluidic invasion, HUVEC Tube Formation in vitro and the tumor size, protein expression in immunohistochemistry, western blotting, biochemistry, ELISA and Elispot in vivo. The calculation detected the difference between LHNrutin-treated and UNT, LHNTMZ-treated and UNT, or LHNTMZ+rutin-treated and LHNTMZ-treated samples. The data are presented as mean ± SD. Significance was established at a value of P < 0.05.
3. Results and discussion
3.1. Morphological characterization of LHN webs
Several distinct factors largely control the result of the electrospinning process. The drug release profiles are significantly influenced by drug loading, fiber shape and different production processes [47]. Accordingly, SEM analysis was performed by imaging both cross-sections and surfaces of the samples. As seen in the layered structure in the cross-sectional images in Fig. 1B, the inner layer was structured in a filling form, while the upper and lower layers were structured in a fibrous form in both LHNTMZ and LHNrutin web samples. However, while the inner layers of LHNrutin were additive blocks, whereas in LHNTMZ it consisted of particles between the fibers (Fig. 1B). The average fiber diameter of LHNrutin webs is 164.1 ± 38 nm in the 89 – 253 nm range.
The first layer was produced and left to dry during the electrospinning process. Next, the drug/agent solution was directly sprayed onto this layer. The first layer had micropores due to the openness between nanofibers. So, the solution could transfer through the first layer to the backside during the spraying process. While the drug formed a film-like structure, the backside of the first layer was in the shape of a fiber-reinforced polymeric composite (Fig. 1C). Also, the drug deposition was formed between two nanofiber layers (Fig. 1B). The third layer was in a fiber structure. The third layer was produced after the electrospraying process and drying duration of the drug/agent solution. As the third layer was in a fiber structure, the drug/agent did not deposit on it (Fig. 1D). The TMZ fiber had bulges, while fibers that consisted rutin had a smooth form. These bulges on LHN™Z webs led to a non-uniform diameter ranging between 89 and 485 nm. Since the fibers were embedded in resin in the first layer, the diameter of the fiber, which consisted of rutin and TMZ, was measured from the last layer.
3.2. In vitro release of TMZ and rutin from LHN webs
DMF was used as the spinning solvent for rutin [48]. Electrospinning and electrospraying processes affected the amount of TMZ or rutin incorporated into the fiber. Thus, LHN surfaces with variable thickness at different sites could be formed, which led to a variable concentration of TMZ or rutin in different regions of the LHN surfaces. The areas of the LHN close to the edges were thinner than the average and consisted TMZ and/or rutin in a lower concentration. In contrast, the central parts of the LHN webs were the thickest regions and consisted sufficient concentration of TMZ and/or rutin. The results of cell assessment and controlled drug release could be affected by the variations in the thickness of LHN webs. In addition, the arrangement of the electrospinning device complicates optimizing the collector's reciprocating movement. Therefore, in vitro release analysis of TMZ and rutin from LHN webs was assessed from 3 different sides of the LHN surfaces to determine the releasing patterns.
The rutin-releasing of LHNrutin increased as a bursting release behavior for the first 6 h due to the dissolving of the PVA, and active reagent deposited layers. While the minimum rutin release was observed in the 1st h (0.586 ± 0.26 ppm), it released at a maximum level in the 6th h (3.44 ± 0.11 ppm). In contrast, the rutin release suddenly decreased in the 18th h (0.86 ± 0.41 ppm). Because the release of the active agent come from only core-shell layer. After this time, controlled release has started until 168th h, which has demonstrated in a constantly increasing release behavior. While the PVA and the deposit layer cause it to release in the form of bursting in the first 6 h due to its sudden dissolution in PBS, core-shell structured PLA exhibited the controlled release behavior in a way that constantly increases due to insolubility of the PLA structure in PBS. Actually, the occurrence of this condition coincides with the purpose of the study. In LHN application, high dose active agent release was targeted in the first stage, and then controlled release and low amount increments in the subsequent stages.
The TMZ release of LHNTMZ was gradually increased. The maximum TMZ release of LHNTMZ was detected at 168 h (0.66 ± 0.28 ppm), while the minimum release was at 18 h (8.05 ± 0.67 ppm) (Fig. 1E). The TMZ release at the 6th h increased compared to first h release. This increase did not occur in the form of bursting behavior as in the rutin release. Further, for the 18th h release behavior has decreased as in the rutin release. The reason for this decrease may be the result of the start of the release in the core-shell after the dissolution of the PVA and the deposit (active agent) layer. It was observed that there was a controlled release after 24 h, a similar release was made up to 72 h; however, it was observed that the amount of release increased in 168 h It has been seen that there is no release in the form of the expected bursting in the 1st h of TMZ release. But, it has been observed that controlled release occurs starting from 1 h.
3.3. TMZ and/or rutin-loaded LHN inhibits aggressive features of T98 G cells
The IC50 concentrations of TMZ and rutin [39] were encapsulated in LHN webs, covering an area of 0.5 cm2. LHN™Z reduced cell viability by 59.1 % ± 0.04 % compared to UNT cells in 24 h (P < 0.0001, Fig. 2A). In addition, LHNTMZ decreased T98 G cell proliferation by 50.4 % ± 0.24 % in 48 h (P < 0.0001, Fig. 2, Fig. 2), which was its maximum inhibitory effect. The proliferation rate of LHNTMZ-treated cells reached the rate of UNT cells from 48 to 96 h (Fig. 2A). LHNrutin decreased T98 G cell proliferation by 40.2 % ± 0.15 % (P < 0.0001) and 18.1 % ± 0.06 % (P < 0.0001) compared to UNT cells in 24 and 48 h, respectively (Fig. 2, Fig. 2). The proliferation rate of LHNrutin-treated cells remained stable after 48 h, but a decrease in growth rate became apparent over time. LHNTMZ+rutin suppressed T98 G cell proliferation by 30.7 % ± 0.152 % in 24 h (P < 0.0001). The maximum inhibitory effect of LHNTMZ+rutin was 37.5 % ± 0.20 % in 48 h (P < 0.0001). The cell inhibitory effect of LHNTMZ+rutin gradually increased compared to UNT cells until the end of 96 h The T98 G cell inhibitory effect of LHNTMZ+rutin was more substantial than the effect of LHNTMZ (P < 0.0001, Fig. 2A and B). In our previous study, the effects of the combination of rutin with TMZ on cells (additive, antagonistic or synergistic) was calculated using the R-package of the SynergyFinder calculation tool (https://bioconductor.org/packages/release)/bioc/html/synergyfinder.html). Accordingly, rutin's zero-interaction potency (ZIP) synergy score (T98GSZIP = −4.1) indicated an additive effect when used simultaneously with TMZ [39]. The effect of LHNTMZ, LHNrutin and LHNTMZ+rutin on the mitochondrial function of T98 G cells was assessed by measuring the mitochondrial membrane potential (Δψm) (Fig. 2C). The effect of LHNTMZ, LHNrutin, and LHNTMZ+rutin on the mitochondrial function of T98 G cells was assessed by measuring theΔψm (Fig. 2C) did not affect the Δψm compared to UNT cells. In contrast, LHNrutin decreased the Δψm compared to UNT T98 G cells (P = 0.0058, Fig. 2C). In addition, TMRE-labeling red fluorescence observed in LHNTMZ+rutin-treated cells was substantially lower compared to LHNTMZ treatment (P = 0.003, Fig. 2C).
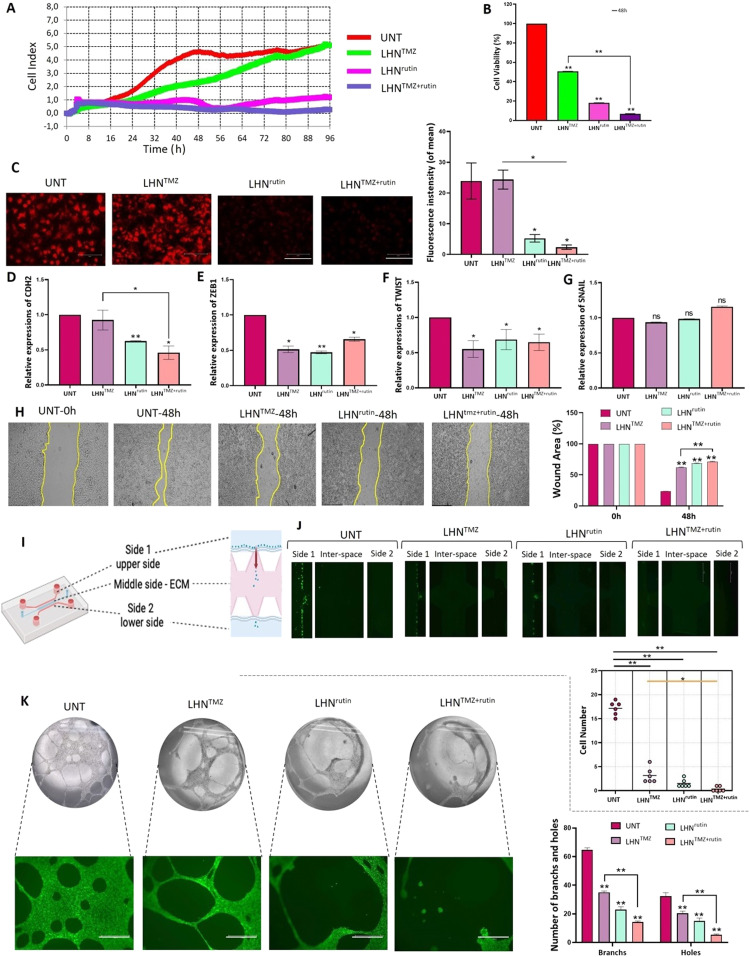
Fig. 2.
The effects of LHNTMZ, LHNrutin and LHNTMZ+rutin on T98 G cell growth and aggressiveness. T98 G cells were treated with 0.5 cm2 of LHNs encapsulated with IC50 doses of TMZ (1,000 µM), rutin (142.2 µM) and their combination. (A) The effects of LHNTMZ, LHNrutin, and LHNTMZ+rutin treatments on real-time cell proliferation of T98 G cells over 96 h (B) The T98 G cell viability percentage after 48-h treatment with LHNTMZ, LHNrutin and LHNTMZ+rutin (**P < 0.001). (C) The effect of LHNTMZ, LHNrutin and LHNTMZ+rutin on Δψm in T98 G cells. TMRE accumulation of cells (red color) is a sign of high membrane potential. Depolarized mitochondria result in reduced TMRE accumulation. (D) RNA expression of EMT-signaling gene, CDH2. (E-G) RNA expression of EMT regulator transcription factors ZEB1, TWIST and SNAIL. For D-G: Ct values were normalized to the Ct of GAPDH. (*P < 0.05, **P < 0.0001). (H) The representative images of the findings on the scratch wound healing assay. 90 % confluent cell monolayer scratched and recorded as the time 0. The change in the size of the wounded area was recorded after 48-h of treatment with LHNTMZ, LHNrutin or LHNTMZ+rutin. Scale bar: 595.2 µm. (I) The schema of the microfluidic invasion model on an IC—Chip. (J) The movement of GFP-expressing T98 G cells from side 1 (upper side) to side 2 (lower side) and their invasion to the EMC at the interspace was observed by fluorescent microscopy over 48 h 10 % FBS was used as the chemoattractant. Scale bar: 750 µm. *P < 0.05, **P < 0.0001. (K) The effect of the TMZ, rutin and TMZ+rutin release of LHN on tube forming of HUVEC cells. Calcein AM confirmed the viability of HUVEC cells of the capillary structure during decomposition by green color. Scale bar: 750 µm. **P < 0.0001 CDH-2: N-cadherin, ZEB-1: zinc finger E-box binding homeobox 1, TWIST-2: Twist-related protein, SNAIL: zinc finger protein SNAI1. The number of biological replicates was A-H:3, J:6 and K:5. All values were expressed as the mean ± SD (standard deviation) of the biological replicates. All P values were presented for comparison with the UNT samples.
The mesenchymal characteristics of neural cells contribute to a non-classical EMT-like process, or a glial-to-mesenchymal transition, which plays a role in the progression of GB, including invasion, migration and angiogenesis [49]. Therefore, the effects of LHNTMZ, LHNrutin, and LHNTMZ+rutin on RNA expression of EMT-involved proteins and transcription factors encoding genes was assessed. While LHNTMZ did not affect the expression of the CDH2, LHNrutin reduced it (P < 0.0001, Fig. 2D) compared to UNT cells. In addition, LHNTMZ+rutin significantly decreased the RNA expression of CDH2 compared to UNT (P = 0.0006) and LHNTMZ-treated cells (P = 0.020, Fig. 2D). Besides, LHNTMZ and LHNrutin reduced the RNA expression of the EMT-inducing transcription factors ZEB1 (LHNTMZ: P = 0.0003, LHNrutin: P < 0.0001) and TWIST (LHNTMZ:p = 0.0028, LHNrutin: P = 0.0255) than UNT cells (Fig. 2, Fig. 2). Also, LHNTMZ+rutin decreased ZEB1 and TWIST RNA expressions compared to UNT cells (P = 0.0002 and P = 0.0107, respectively). However, the involvement of rutin in LHNTMZ+rutin did not contribute to the ZEB1 and TWIST-reducing capacity of LHNTMZ. Besides, SNAIL was not affected by LHNTMZ, LHNrutin, and LHNTMZ+rutin (Fig. 2G).
EMT is followed by migration and invasion of GB cells [50]. In this study, a scratched wound assay was used to model cell migration. While UNT T98 G cells recovered 76.4 % ± 0.07 % of the scratched wound, LHNTMZ could repair 37.6 % ± 0.54 % of the scar in 48 h (P < 0.0001, Fig. 2H). The treatment of T98 G cells with LHNrutin resulted in a significantly slower rate of wound recovery compared to UNT cells (P < 0.0001) and LHNTMZ -treated cells (P < 0.0001), leading to the closure of only 32.1 % ± 0.46 % of the gap (Fig. 2H). In addition, LHNTMZ+rutin led to a sharp decrease in wound healing (29.4 % ± 0.48 % closure) compared to LHNTMZ (Fig. 2H).
To examine the effect of LHNTMZ LHNrutin, and LHNTMZ+rutin on the invasiveness of T98 G cell, an in vitro invasion microenvironment was simulated using an IC—Chip. The structure of the invasion model in IC—Chip is demonstrated in Fig. 2I. The movement of cells from side 1 to side 2 in the IC—Chip was visualized for 48 h to measure invasiveness. The results (Fig. 2J) indicated that the invasion of LHNTMZ-treated cells was reduced by 5.4 ± 2.6 fold compared to UNT T98 G cells (P < 0.0001). Additionally, LHNrutin treatment led to an 11.4 ± 1.5-fold decrease in invasion compared to UNT cells (P < 0.0001). Furthermore, LHNTMZ+rutin treatment resulted in a 51.5 ± 1.41-fold reduction in invasion in comparison to UNT cells (P < 0.0001), which is also 9.5 ± 0.98 fold lower than the invasion observed with LHNTMZ (P = 0.0021).
Capillary-like tube formation assay of HUVEC cells was used to study angiogenesis. As shown in Fig. 2K, the conditioned medium collected from LHNTMZ and LHNrutin-treated T98 G cells significantly reduced the formation of a network of capillary-like tube networks in HUVEC cells compared to UNT controls (P < 0.0001). In addition, the conditioned medium from LHNTMZ+rutin-treated T98 G cells resulted in a considerable reduction in capillary tube formation in HUVEC cells compared to UNT cells (P < 0.0001). Notably, the inhibitory effect of the conditioned medium from LHNTMZ+rutin-treated T98 G cells was significantly greater than that of the medium from LHNTMZ-treated cells (P < 0.0001).
3.4. LHNTMZ+rutin enhances the effect of LHNTMZ on the glioma stem cell phenotype of T98 G cells
The RNA expression of CD133, OCT4, NANOG and SOX2 defined the regulation of stem-like cell maintenance in T98 G cells. According to the findings, LHNTMZ and LHNrutin treatment reduced the RNA expression of CD133 compared to UNT T98 G cells (P = 0.001, P = 0.0023, respectively). Besides, LHNTMZ+rutin resulted in less CD133 expression in T98 G cells than UNT and LHNTMZ-treated cells (Fig. 3A). Similarly, RNA expression of OCT4 decreased with LHNTMZ, LHNrutin and LHNTMZ+rutin treatment compared to UNT cells (P < 0.0001). The reduction caused by LHNTMZ+rutin was significantly more than the effect of LHNTMZ (P = 0.0009; Fig. 3B). Moreover, while LHNTMZ treatment resulted in a significant reduction in the expression levels of NANOG (P = 0.0039) and SOX2 (P = 0.0039), these reductions were even more pronounced with by LHNrutin treatment compared to UNT T98 G cells (P < 0.0001; Fig. 3, Fig. 3). Although LHNTMZ+rutin significantly reduced NANOG and SOX2 expressions in comparison to UNT cells, this effect was negligible compared to LHNTMZ-treated cells. These findings indicated that the involvement of rutin in LHNTMZ+rutin could enhance the glioma stem cells (GSC) inhibitory effect of TMZ by reducing the expression of CD133 and OCT4.
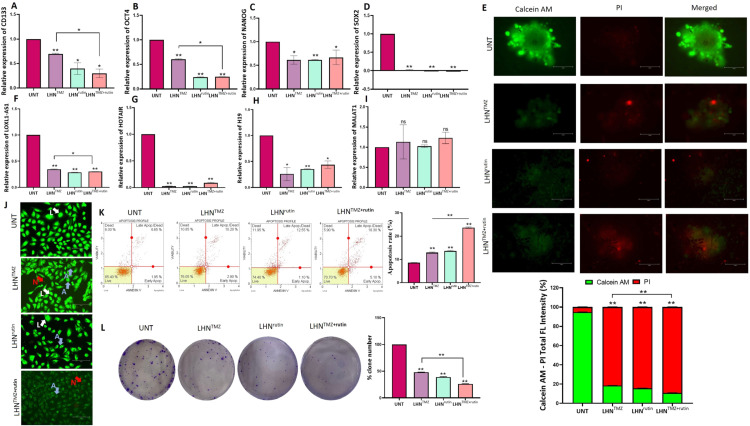
Fig. 3.
Effects of LHNTMZ, LHNrutin, and LHNTMZ+rutin on GSC phenotype of T98 G cells. The RNA expression of (A) CD133, (B) OCT4, (C) NANOG and (D) SOX2. (E) 3D-spheroid formation. T98 G cell spheres were formed in a 96-well, round-bottom, ultra-low attachment plate for 3 d Calcein-AM and PI staining identified the vitality of the spheres. Calcein-AM staining (green) indicates metabolically viable or proliferative regions, whereas PI staining (red) indicates the sites of necrosis or death. ImageJ software measured the size of captured tumor spheres and the ratio of the diameter of live/dead area. Scalebar: 150 µm. The RNA expression of (F) LOXL1-AS1, (G) HOTAIR, (H) H19 and (I) MALAT1. (J) Viability of T98 G cells after treatment with loaded LHNs by AO/PI staining. In morphological analysis, green cells with granular nuclei on one side showed apoptosis. In contrast, a circular nucleus evenly distributed in the center of the cell indicated that a cell is in interphase. Dark red cells with a vague rim indicated necrosis. Color-coded arrows indicate: live cells in white, early apoptosis in blue, late apoptosis in orange, and necrosis in red. (K) Analysis of apoptotic T98 G cells using Annexin V assay after loaded LHN's treatments. (L) The colony formation of T98 G cells after LHNTMZ, LHNrutin, and LHNTMZ+rutin treatments. All values represent mean ± SD (n = 3). *P < 0.05, **P < 0.0001. CSC: Cancer Stem Cell, CD133: Prominin-1, OCT4: POU5F1, NANOG: Nanog Homeobox, SOX2: SRY-box 2, L: alive cells, A: apoptosis, N: necrosis.
The sphere formation assay confirmed the effect of LHNTMZ, LHNrutin and LHNTMZ+rutin on GSC population residing in T98 G cells. The findings showed that the size of the LHNTMZ and LHNrutin-treated T98 G cell spheres was smaller than that of UNT cell. In addition, LHNTMZ+rutin significantly reduced the sphere size compared to UNT and LHNTMZ-treated cells. Upon Calcein-AM and PI staining, the intensity of Calcein-AM staining (green), a sign of vitality, decreased. At the same time, LHNTMZ and LHNrutin treatments increased the intensity of PI staining (red) compared to UNT T98 G cell spheres (Fig. 3E). In addition, after LHNTMZ+rutin, the pallor in Calcein AM staining became more evident. Notably, while PI-dependent red fluorescence was focally observed in the core region of UNT cell spheres, after treatment, there was no PI staining, which was focal in the core region. However, an irregularly distributed red fluorescence was observed throughout the sphere (Fig. 3E). This finding is suggested to be a disruption of the integrity of the sphere due to the loss of cell viability rather than the presence of hypoxia. Our previous study showed that Olea europaea leaf extract decreases the GB tumor sphere size by inhibiting the expression of LncRNA LOXL1-AS1, HOTAIR, H19 and MALAT1 [51]. Similarly, in this study, the expression of LOXL1-AS1, HOTAIR and H19 LncRNA was reduced by LHNTMZ and LHNrutin (Figs. 3F-H). In contrast, MALAT1 was not affected by LHNTMZ, LHNrutin and LHNTMZ+rutin treatments (Fig. 3I).
Morphological analysis of T98 G cells treated with LHNTMZ, LHNrutin and LHNTMZ+rutin was performed using the fluorescent dye staining method AO/PI. UNT T98 G cells had a uniform circular nucleus at the center, indicating viability. In contrast, cell nuclei treated with LHNTMZ, LHNrutin and LHNTMZ+rutin showed a horseshoe-shaped configuration, a sign of nuclear fragmentation seen during apoptosis (Fig. 3J). In addition, according to annexin V analysis, the apoptosis effect of LHNrutin was greater compared to UNT cells (P < 0.0001). The highest rate of apoptosis was observed in cells treated with LHNTMZ+rutin (P < 0.0001, Fig. 3K).
A clonogenic assay was used to test the ability of T98 G cells to retain proliferative capacity, survival, and expansion into a clonal population over time. The number of colonies formed by LHNTMZ and LHNrutin-treated T98 G cells was reduced by 2.1-fold and 2.6-fold, respectively, compared to UNT T98 G cells, (P < 0.0001). Moreover, the number of colonies formed by LHNTMZ+rutin-treated T98 G cells was 3.8-fold lower than that of UNT cells (P < 0.0001), and 1.8-fold lower than that of LHNTMZ-treated cells (P < 0.0001) (Fig. 3L).
3.5. LHNrutin and LHN™Z+rutin affected proteins that are involved in apoptosis, energy metabolism and protein transport
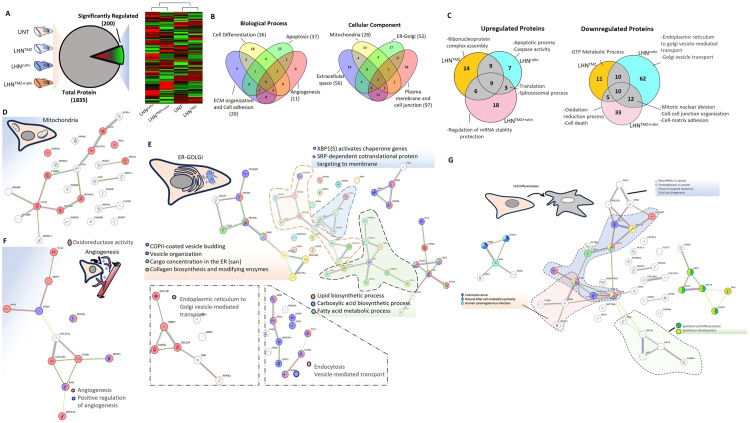
Following LFQ analyses data, a total of 1835 proteins were identified, with 200 proteins showing significant regulation between groups. Heatmap clustering analysis was conducted to assess group similarities. The hierarchical clustering of protein expression revealed two distinct trends. The control group and LHNTMZ group formed one cluster, while the LHNrutin and LHNTMZ+rutin groups formed the other (Fig. 4A). Seventy-two regulated proteins are involved in biological processes such as cell differentiation, apoptosis, angiogenesis, extracellular matrix organization, and cell-cell adhesion. Furthermore, most of the 163 proteins were observed to be located in the plasma membrane, cell junctions, ER-Golgi, mitochondria and the extracellular space component of the cell (Fig. 4B). Upon analysis of the upregulated proteins affected by LHNTMZ, LHNrutin and combined treatment (Fig. S1), we identified associations with apoptotic and spliceosomal processes, translation and nucleic acid metabolism. Meanwhile, the downregulated proteins were linked to energy metabolism, oxidoreductase activity, endoplasmic reticulum (ER)-Golgi transport and regulation, and cell-to-cell and cell-matrix regulation (Fig. 4C).
Fig. 4.
(A) A heatmap depicting differentially expressed proteins resulting from the label-free proteomics analysis of treatment groups. The clustering was generated using the average-linkage method along with Euclidean distance, utilizing the Heatmapper online tool (http://www.heatmapper.ca/expression/). (B) Venn diagram of biological process and cellular localization of the differentially up- and down-regulated proteins. (C) Venn diagram of up- and down-regulated proteins and their biological process. Analysis of the down/up-regulated mitochondria-associated proteins (D), ER-Golgi-associated proteins (E), angiogenesis-associated proteins (F), and cell differentiation-associated proteins (G) interactions created using the STRING analysis tool. GTP: Guanosine-5′-triphosphate.
Mitochondria, the powerhouse of the cell, represents one of the most crucial hubs in cancer metabolism, playing a vital role in cancer cell proliferation, survival, and metastasis [52]. Consequently, mitochondria become a significant target in cancer treatment. The first protein interaction analysis of mitochondria-related proteins pointed out that oxidoreductase activities were influenced (Fig. 4D). However, two major upregulated protein groups associated with mitochondrial activity were observed when further analysis was performed. The first group includes ATP5ME, PDZD8, HSDL2, NUDT1 and ALDH7A1 proteins, which play a crucial role in maintaining the structural and functional integrity of mitochondria. These proteins exhibit high expression levels in various cancer cells and are involved in processes related to ATP synthesis, maintenance of mitochondrial membrane lipid bilayer, oxidative stress and lipid peroxidation [[53], [54], [55], [56]]. These proteins have a protective effect on mitochondria damage as a response to the treatment. However, besides the induction of protective protein expression, an increase in protein levels that stimulate the initiation of mitochondria-associated cell death, such as CYC3 and CYB5R3, was observed. On the other hand, protein levels of CHCHD2P9 and TMX1, which are upregulated in various cancers and crucial for maintaining mitochondria homeostasis and chemotherapy resistance by controlling the flux of Ca2+ between the ER and mitochondria, were decreased in the treated group [[57], [58], [59], [60], [61]]. Additionally, anti-apoptotic proteins CIAPIN1 and WDR26 were downregulated compared to the control groups [[62], [63], [64]]. These results illustrate the battle between the apoptotic effect of chemotherapy on mitochondria and the response of protective cellular mechanisms. Moreover, the LHNrutin application exhibited a more significant effect on DNA and RNA metabolism, whereas the LHNTMZ group showed a more notable impact on lipid metabolism. It has been noticed that the combined application of LHNTMZ+rutin affects the main processes of ATP synthesis.
Analysis of ER and Golgi-associated proteins exhibiting increased expression levels following treatment in the ER-Golgi proteome revealed heightened signals, including ALIX, associated with cancer migration and invasion; SCD1, implicated in angiogenesis and immune regulation; PPT1, functional in drug resistance; and B2 M, involved in cancer cell evasion from the immune system and the provision of survival signals [[65], [66], [67], [68], [69], [70]]. However, some proteins, like ANXA7, known as a tumor suppressor in GB, were upregulated due to the treatments [71].When assessing the downregulated proteins in ER-Golgi, a significant decrease was observed in the levels of TMEM67, UBXN1, and PDCD6 proteins within the LHN™Z group. These proteins play crucial roles in cell proliferation and development [[72], [73], [74]]. Compared to LHNTMZ, LHNrutin treatment had a significant effect on protein downregulation. Most downregulated proteins in the LHNrutin treatment group were associated with ER-ERGIC-Golgi transport. In cancer cells, the transport between the ER and Golgi increases because of increased protein synthesis, ER stress, EMT, and active synthesis of secretory proteins such as matrix metalloproteases (Fig. 4E) [75]. Thus, the expression of proteins involved in ER-ERGIC-Golgi transport was expected to decrease during LHNrutin treatment. However, unlike LHNrutin treatment, this effect was not observed in the LHNTMZ-treated group. Angiogenesis is another crucial process in the development and metastasis of cancer. Results indicate that applying LHNTMZ increased the levels of COL15A1 and COL27A1, collagen proteins associated with angiogenesis (Fig. 4F). These collagens are released into the extracellular region and contribute to various processes such as cell differentiation, cellular development and differentiation regulation, and tumor suppression. COL15A1 is essential in regulating the extracellular matrix (ECM) in cancer cells and suppressing receptor-dependent metastasis, but COL27A1 is not well-characterized [76,77]. The findings indicated that both LHNTMZ and LHNrutin treatments independently influenced angiogenesis. However, the combined treatment appeared to have a greater effect. Specifically, the combination treatment resulted in a significant increase in proteins involved in angiogenesis. It upregulated the levels of essential proteins such as ENG, ERAP1, TGFBI, MTDH and GLUL, which play a crucial role in angiogenesis, metastasis and drug resistance [[78], [79], [80], [81], [82], [83], [84]]. These results showed that combining LHNTMZ and LHNrutin treatment proved more effective in suppressing angiogenesis in GB cells.
One of the main goals of cancer treatment is to selectively induce apoptosis in cancer cells. As a result of treatment, there was an increase in the levels of essential proteins that play a role in apoptotic processes, such as CASP3, CYCS and PDCD6IP, in all three groups [[85], [86], [87]]. Compared to LHNTMZ treatment, LHNrutin affected more proteins. Significant decreases in protein levels were associated with ER-Golgi-related processes and some of these proteins, like PGRMC1. ACSL4, GLUL, APEX1, CCAR2 and TMX1 especially play an essential role in the mitochondria-ER relationship. These proteins are observed to be effective in regulating calcium balance between ER and mitochondria, lipid exchange, and ER-mitochondria contact [60,[88], [89], [90], [91]]. It is believed that these changes may disrupt the relationship between ER and mitochondria, leading to mitochondrial degradation and the induction of apoptotic processes.
Cellular differentiation processes are another important mechanism for cancer development and metastases. During the treatment, there was a significant change in the levels of 36 proteins, which were mainly associated with epithelial cell differentiation, cancer development, and extracellular matrix organization (Fig. 4G). There was a decrease in the levels of essential proteins such as CDK5, DDX3X and CIAPIN1, which are involved in cellular differentiation and apoptosis processes in LHNrutin treatment [[92], [93], [94]]. Similar results were observed in the LHNTMZ application. Proteins such as ANXA7, which plays a pro-apoptotic role in GB cells, were increased, while anti-apoptotic proteins like CIAPIN1 were downregulated in the LHNTMZ group [95]. However, the application of LHNTMZ-only exhibits a comparatively slight effect on cancer cells compared to LHNrutin-only treatment.
3.6. Though local application of LHNTMZ reduced the tumor growth in GB cells
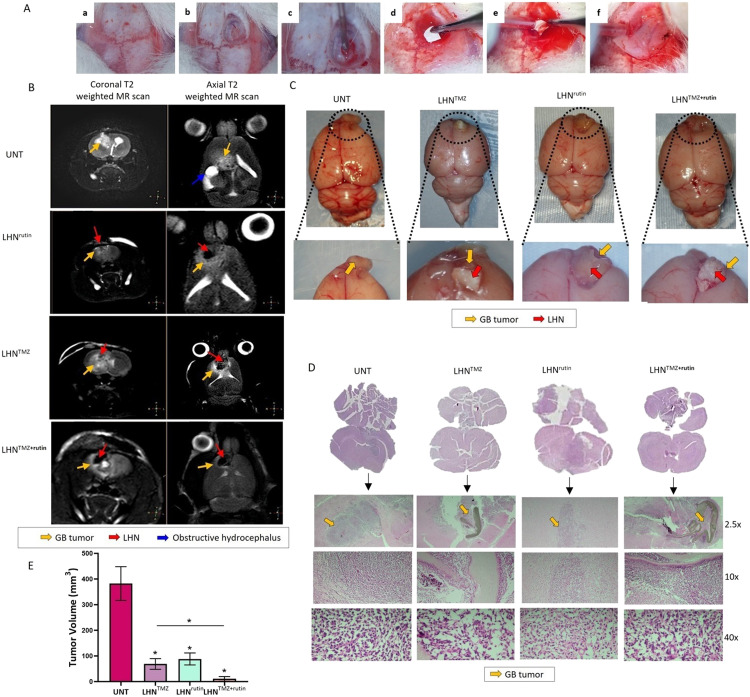
The right frontal skull of the rat brains was exposed, and the dura was opened by performing a right frontal craniectomy. Rat GB cells, C6 (2.5 × 106 cells in a 10 µl medium), were slowly injected to a 2.5 mm depth from the cortical surface with stereotactic guidance (Fig. 5A). To mimic a GB model where the tumor is inaccessible or unresectable by surgery, LHNTMZ, LHNrutin, or LHNTMZ+rutin webs were incorporated into the cavity area, following the cell seeding (Fig. 5A). The behaviors of the rats were monitored, and the presence of the tumor was confirmed by MRI on Day 11 (Fig. 5B). Untreated rats exhibited tumor extension to other regions of the brain, along with infiltrative edema in the deep white matter of the right frontal lobe (Fig. 5B).
Fig. 5.
The effect of LHNTMZ, LHNrutin and LHNTMZ+rutin on GB tumor growth. (A) Generation of an allograft GB model and insertion of LHN webs. (a) A midline skin incision exposed the right frontal intracranial surfaces. (b) The dura was opened following a 5 mm x 5 mm right frontal craniectomy. (c) Injected 2.5 × 106/10 µl of C6 cell into 2 mm lateral and anterior to bregma. (d, e) LHN webs were inserted into the resected area. (f) A bone wax was used to cover the resection area. (B) Coronal T2 weighted and axial T2 weighted MR scans on Day 11. UNT GB rats: Tumor lesion causing infiltrative edema in the right frontal deep white matter (yellow arrow) and hydrocephalus secondary to compression (blue arrow). After LHNTMZ: a slightly hyperintense infiltrative tumoral lesion (yellow arrow) in the right frontal region extends transcallosally from the anterior part of the corpus callosum, with the LHN network observed at its center (red arrow). After LHNrutin: In the deep white matter of the right frontal lobe, tumor lesion (yellow arrow) and a prominent hypointense large-sized nanofiber (red arrow) are observed. The lesion extends transcallosally to the opposite hemisphere, exerting pressure on the LHN network. After LHNTMZ+rutin: Hypointense LHN (red arrow) in the right frontal area and a small tumoral lesion of a millimetric thickness (yellow arrow) surrounding it. (C) The appearance of the resected brain and the tumor at the end of the experiment. (D) H&E staining of brain sections on Day 11. (E) The volume of the GB tumors. *P < 0.05.
In LHNTMZ and LHNrutin-treated rats, the tumor size was smaller than that of UNT rats, and infiltrative edema was not observed (Fig. 5B). The images of the resected brain tissue and the tumor volume at the end of the experiment, is shown in Fig. 5C. Notably, LHNTMZ, LHNrutin, and LHNTMZ+rutin webs remained present on Day 11 when the tumors were resected (Fig. 5C). A H&E staining of coronal sections of brain tissue confirmed the site of tumor growth (Fig. 5D). Based on tumor volumes determined by H&E staining, LHNTMZ and LHNrutin reduced tumor size compared to UNT GB rats (P = 0.0037 and P = 0.0018, respectively, Fig. 5E). The tumor size in UNT GB rats was 382 ± 65.7 mm3, 69 ± 21 mm3 after LHNTMZ, and 88.5 ± 23.33 mm3 after LHNrutin. In addition, LHNTMZ+rutin treatment led to a significant decrease in the tumor size (11 ± 9 mm3) compared to LHNTMZ-treated rats with GB (P = 0.011). These findings supported the promising additive effect of rutin in increasing the success of TMZ on GB, which we initially demonstrated in vitro experimental models.
3.7. LHNrutin and LHNTMZ+rutin decreased the proliferation capacity and mitochondrial function of GB tumors
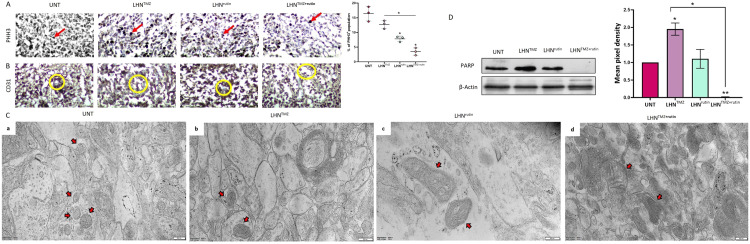
PHH3, a marker for cell proliferation, specific for GB cells in mitosis, was expressed at the highest levels in the tumor tissue of UNT GB rats (12.75 % ± 2.51 %, Fig. 6A). LHNrutin treatment resulted in a significant reduction in PHH3 expression of GB tumors compared to the UNT group (LHNrutin: 7.83 % ± 0.94 %, P = 0.0052). In addition, the treatment with LHNTMZ+rutin resulted in the lowest PHH3 expression (3.5 % ± 1.25 %), which was significantly lower than the tumors of UNT (P = 0.0013) and LHNTMZ-treated rats (P = 0.0010; Fig. 6A). In addition, the intratumoral microvessel density was analyzed by CD31 expression to analyze the angiogenesis of the tumors. According to the findings, the CD31 inhibitory effect of LHNTMZ, LHNrutin and LHNTMZ+rutin was negligible compared to UNT GB rats (Fig. 6B).
Fig. 6.
The effects of LHNTMZ, LHNrutin and LHNTMZ+rutin on GB tumor in pathological examination (A) IHC staining of PHH3 indicates the mitotic and proliferation capacity (staining areas are shown with red arrows). (B) IHC staining of CD31 shows the intratumoral microvessel density (staining areas are shown with yellow circles). (C) TEM images of GB tumor cells focusing on the structure of mitochondria (red arrows). (D) Immunoblotting of PARP protein expression in GB tumors. All values were expressed as the biological replicates' mean± SD. *P < 0.05, **P < 0.0001.
Tumor sections were imaged with TEM to evaluate the tumor at the cellular level (Fig. 6C). In the tumors of UNT and LHNTMZ treated-GB rats, the outer membrane and cristae of the mitochondria were transparent, and the matrix had an average electron density (Fig. 6C). In contrast, in tumors of LHNrutin-treated GB rats, the size of the mitochondria was enlarged, and the crista was irregular and could be poorly defined in some parts. In addition, the matrix was less condensed than that in UNT and LHNTMZ-treated rats (Fig. 6C). This could be a sign of suppression of ATP production and mitochondrial function due to damage and swelling of mitochondria after LHNrutin treatment [96,97]. In addition, in tumors of LHNTMZ+rutin-treated GB rats, the size of the mitochondria was more significant than that of UNT GB rats and those treated with LHNTMZ. The matrix of these tumors' mitochondria was more electron-dense than that of UNT rats and less condensed than that of LHNTMZ-treated GB rats. Moreover, similar to LHNrutin-treated GB rats, the crista of LHNTMZ+rutin-treated GB rats' mitochondria was irregular and barely distinguishable compared to UNT and LHNTMZ-treated GB rats (Fig. 6C). The TEM images of the mitochondria of GB-rats suggest that LHNrutin reduced the mitochondrial activity of GB tumors and could also decrease the LHNTMZ-mediated mitochondrial function. In line with this finding, while the tumors of LHNTMZ-treated GB rats showed an induced PARP expression, it was reduced by LHNrutin and LHNTMZ+rutin treatment (Fig. 6D). These findings suggested that LHNrutin and LHNTMZ+rutin mediated inhibition of the mitochondrial activity resulted in the suppression of PARP, which depends on the mitochondrial activity to use NAD+ as a substrate to regulate its cellular functions in GB tumors.
3.8. Local application of LHNrutin and LHNTMZ+rutin into GB tumors is biologically safe
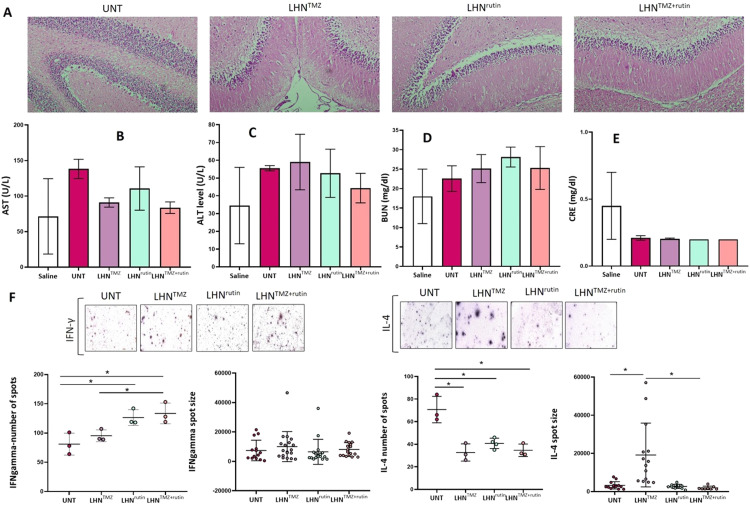
LHNTMZ, LHNrutin and LHNTMZ+rutin were inserted into the brains of rats with tumors for 11 d and the brains were collected to analyze the potential LHN-mediated tissue damage and inflammation. The H&E staining of the cerebellum did not show a sign of LHNTMZ, LHNrutin and LHNTMZ+rutin-induced inflammation or necrosis (Fig. 7A). The serum AST levels were reduced by LHNTMZ, LHNrutin and LHNTMZ+rutin compared to UNT. While LHNrutin reduced the release of ALT into the peripheral blood, LHNTMZ was not affected. In addition, LHNTMZ+rutin reduced the LHNTMZ-mediated leak of ALT (P = 0.2246) (Fig. 7, Fig. 7). These findings provided evidence that LHNrutin and LHNTMZ+rutin protected rats from liver inflammation [98]. In contrast, LHNrutin induced the serum urea level compared to UNT and LHNTMZ-treated rats (P > 0.05, Fig. 7D). However, LHNTMZ+rutin reduced the LHNrutin-mediated level of urea to the level of urea caused by LHNTMZ. Notably, the maximum urea level observed after LHNrutin treatment was between the reference intervals of the acceptable urea levels for rats [99]. Besides, the serum creatinine level was unaffected by LHNTMZ, LHNrutin, and LHNTMZ+rutin compared to that of UNT GB rats. These findings provided evidence that LHNTMZ, LHNrutin, and LHNTMZ+rutin did not cause any adverse effects on the renal system of the rats [100].
Fig. 7.
The biocompatibility of LHNTMZ, LHNrutin and LHNTMZ+rutin. (A) H&E staining of the cerebellum after LHNTMZ, LHNrutin and LHNTMZ+rutin application. The peripheral blood serum (B) AST, (C) ALT, (D) BUN and (E) CRE. (F) Analysis of IFN-γ and IL-4 levels at splenic cells by ELISPOT assays. Spot numbers were normalized according to the background seen in PBS. The experiments were performed in three biological replicates. All data are presented as mean ± SE; *P < 0.05.
IFN-γ and IL-4 production of splenic cells were analyzed to observe the changes in the inflammatory response of rats. ELISPOT analysis showed that LHNTMZ did not affect the number of splenic T cells that produce IFN-γ and the production level (Fig. 7F). In addition, while LHNTMZ reduced the number of splenic cells that produce IL-4 (P = 0.0093), the level of production was increased for each cell compared to UNT rats (P = 0.0016). In contrast, LHNrutin treatment increased the number of splenic cells that produce IFN-γ (P = 0.0273), while decreasing the number of splenic cells that produce IL-4- (P = 0.005) when compared to that of UNT rats. However, the level of IFN-γ and IL-4 production of the splenic cells was not affected by LHNrutin (Fig. 7F). LHNTMZ+rutin treatment increased the number of cells that produce IFN-γ (P = 0.0242) and decreased the number of cells that produce IL-4 (P = 0.0086) compared to UNT (Fig. 7F). Moreover, the number of splenic cells that produce IFN-γ was higher in LHNTMZ+rutin-treated rats compared to LHNTMZ-treated (P = 0.0317; Fig. 7F). However, LHNTMZ+rutin did not affect the production level of IFN-γ and IL-4. Collectively, these findings suggest that LHNTMZ, LHNrutin and LHNTMZ+rutin treatments induce production level of Thelper1 (Th1) cells, which promotes cellular immunity, while suppressing production level of Th2 cells which exerts adverse immunoregulatory effects on cellular immunity, but upregulating humoral immunity. However, they did not affect the functionality of those cells.
3.9. Discussion
TMZ, a key component in GB treatment, is administered orally or intravenously [101]. Although it is an effective agent, it causes serious side effects [102]. While TMZ is effective against some GB cell lines like A172, U87, U251 and U373, others like T98 G, U138 and LN-18 show resistance due to dysfunctional DNA repair systems or GSCs [103]. Flavonoids like rutin, found in olives, enhance chemotherapy response and have anti-cancer properties [26,[104], [105], [106], [107]]. Our previous study showed that rutin enhances TMZ-induced GB cell death by suppressing ROS and arresting the cell cycle [39]. However, rutin's limited oral bioavailability due to its lipophilic nature, poor water solubility, and rapid biodegradability pose challenges [108]. We previously demonstrated that rutin released from PLA induced apoptosis and reduced the size of cell spheres in GB cells [42]. Combining PLArutin with PLATMZ further reduced GB cell aggressiveness in vitro [42], highlighting the potential of such combinations in cancer treatment. However, the rapid release rate of PLA necessitates improvements for achieving sustained release in vivo [42]. To address this, a biocompatible three-layer hybrid composite nanofiber was developed in this study to enable sustained TMZ and/or rutin release. LHNTMZ+rutin, an advancement over PLATMZ+rutin, offers a superior controlled and sustained release of TMZ and rutin due to different dissolution rates in each layer. LHN's core-shell structure and multiple layers also provide a higher surface area, enhancing drug-loading capacity. This is particularly advantageous for surgical applications, where mechanical strength is crucial for effective use. Therefore, the integrated combination of materials in LHN surpasses PLA in terms of mechanical durability. Unlike previous single-layer PLA core-shell nanofibers, this design integrates two polymers with LHN, enhancing release capability and cellular impact, making it suitable for in vivo applications.
TMZ from LHN exhibited gradual release, while rutin showed an initial burst before stabilizing due to polymer interaction [109]. Despite rutin's insolubility in water, its glycosidic structure suggests potential polymer interaction forming hydrogen bonds [110]. Rutin's release behavior has been previously observed in other nanofibers [[111], [112], [113]]. This study's in vitro analyses confirmed that LHN maintained TMZ and rutin's effects on GB cells [39]. LHNTMZ reduced EMT-inducing factors ZEB1 and TWIST but not mitochondrial activity, allowing ROS-mediated EMT signaling [[114], [115], [116]]. LHNrutin effectively interrupted EMT and reduced mitochondrial activity, migration and angiogenesis. Combined LHNTMZ+rutin treatment significantly reduced CDH2 expression and mitochondrial activity, enhancing treatment efficacy.
Hypoxia and EMT sustain GSCs [117]. LHNrutin and LHNTMZ+rutin treatment decreased GSC marker expression, with greater reductions in LHNTMZ-treated T98 G cells. Silencing CD133 suppressed MDR1 via the AKT/NF-κB pathway, enhancing the efficacy of chemotherapy [118]. NF-κB inhibition also reduced stem-like cancer cells [119]. Our study found decreased lncRNA LOXL1-AS1, HOTAIR, and H19 after treatment. LOXL1-AS1 suppression reduces mesenchymal traits in GB cells [120,121], and H19 regulates GSC maintenance and GB tumor invasion [122,123]. Thus, LHNTMZ and LHNrutin likely reduce the stem cell-like properties of T98 G cell by affecting LOXL1-AS1 and H19 regulation. Sphere formation assays confirmed this effect, with treated spheres shrinking and losing viability. The hypoxic core regions weakened and decreased HOTAIR expression supported reduced sphere growth [124]. LHNrutin had a more pronounced effect, significantly inhibiting CD133 and OCT4, resulting in smaller spheres with non-apparent hypoxic cores. Colony-forming assays showed reduced T98 G cell survival over time with LHNrutin, highlighting its potent role in reducing GSC maintenance and cell renewal compared to LHNTMZ alone.
In vitro experiments demonstrated that TMZ and rutin retain their GB cell mitigating effects when loaded in LHN. Supporting this, LHNTMZ reduced GB tumor size in rats, while a more substantial reduction was observed with LHNTMZ+rutin. These treatments also reduced tumor mitotic activity, suggesting decreased proliferative capacity. However, they did not affect intratumor vascularization, as indicated by unchanged CD31 expression and MRI images [125].
Mitochondrial structural changes suggest metabolic alterations in GB tumors. LHNrutin and LHNTMZ+rutin led to the unraveling of mitochondrial cristae, indicating a decline in Δψm in GB tumors [126]. Enlarged mitochondria with reduced electron density suggested swelling and osmotic imbalance, impairing ion homeostasis [127]. These changes can trigger mitochondria-mediated cell death or necrosis, influenced by PARP activation [128,129]. Excessive PARP activation causes energy failure and cell death, whereas PARP inhibition preserves ATP for apoptosis [128,129]. The significant decrease in PARP expression after LHNTMZ+rutin suggests these mitochondrial changes likely promote apoptosis over necrosis, aligning with previous findings on rutin's apoptosis-promoting effect [39]. Notably, we emphasize the need for fundamental analysis to understand the cellular mechanism by which LHNTMZ+rutin suppress PARP expression. Considering the anti-tumoral effect of PARP inhibition, this finding correlates with observed changes in tumor aggressiveness, such as the label-free proteomic analysis indicating significant changes in mitochondria, ER and Golgi proteins and alterations in the extracellular environment. These changes counteract carcinogenesis by affecting collagen and cadherin content, vesicle transport, immune cell communication, and cell migration suppression. Proteins involved in nucleic acid protection and regulation were also affected, supporting anticarcinogenic and apoptotic processes. Notably, LHN™Z alone had a lower impact on the cell proteome than LHNrutin and LHNTMZ+rutin, suggesting that combination treatment is more effective.
The primary goal of using LHNTMZ, LHNrutin and LHNTMZ+rutin for GB tumors is to target residual tumor cells post-resection or treat inoperable tumors, maintaining local treatment after surgery. These drug-loaded nanomaterials aim to suppress the proliferation of remaining tumor cells. The biocompatibility of these treatments is crucial for their retention at the tumor site, as evidenced by the absence of inflammation in the cerebellum of healthy rats and a reduction in serum AST and ALT levels, indicating liver safety [130]. While TMZ can cause liver injury [98], our findings suggest that local application of LHNTMZ is safer. Additionally, these treatments did not have a negative effect on kidney functions [100].
We also assessed the immune response, focusing on the differentiation of splenic cells into Th1 and Th2 cells. Th1 cells enhance anti-tumor response, while Th2 cells support a pro-tumoral response [131,132]. Our results showed that these treatments did not alter the production of IFN-γ (Th1 cytokine) and IL-4 (Th2 cytokine) in splenic cells [133]. However, a shift towards Th1 cell transformation was observed, indicating a favorable immune response [133,134]. Thus, LHNTMZ, LHNrutin and LHNTMZ+rutin are effective and safe for GB treatment, with promising immune response outcomes.
4. Conclusion
In this study, the anti-GB effects of LHNTMZ, LHNrutin and LHNTMZ+rutin were likely due to their individual effects, as shown previously. Therefore, this study demonstrated that rutin retains its anti-GB effects when loaded into LHN. Additionally, our findings emphasized the mitigating effect of LHNTMZ and LHNrutin on aggressive features of GB tumors in the GB rat model when locally applied at the tumor site. Notably, the tumor-reducing effect of LHNTMZ+rutin was noteworthy when compared with LHN™Z. Therefore, our findings are promising for LHNTMZ+rutin as a treatment approach that can suppress the proliferation of tumor cells at the tumor site. The fact that LHNTMZ, LHNrutin and LHNTMZ+rutin do not cause local tissue inflammation and that AST, ALT, creatinine and urea levels, which are indicators of adverse effects on liver and kidney, are not affected, supports that the produced material can be used. Therefore, applying LHNTMZ, LHNrutin and LHNTMZ+rutin to the GB tumor site was considered biocompatible. However, it should be noted that translating LHNTMZ+rutin into clinical use as a GB treatment will only be possible by investigating it in a manner that reflects the heterogeneity and tumor microenvironment in GB tumors with different pathological features. Additionally, its biocompatibility and side effects need to be elucidated in more detail, and its impact on disease-free survival and quality of life should be observed over extended follow-up periods.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
This study represents a part of Melis Erçelik's doctoral thesis research at Bursa Uludag University, Department of Medical Biology. The author Melis Ercelik is granted by 100/2000 The Council of Higher Education (CoHE) as a PhD Scholar in Molecular Biology and Genetics (Gene Therapy and Genome Studies). We greatly acknowledge to the Scientific and Technological Research Council of Turkey (TÜBİTAK) for supporting author Melis Ercelik within the scope of the 2211-C priority areas doctoral program scholarship. We greatly acknowledge to the Scientific and Technological Research Council of Turkey (TÜBİTAK) for supporting author Cagla Tekin within the scope of the 2211-A doctoral program scholarship. Special thanks to Dr. Tugba Bagci Onder from Koc University, for kindly providing the GB cell line. We thanks to Prof. Dr. Şükrü Yıldırım from Istanbul Maltepe University for his support in IHC analysis and Prof. Dr. Tülin Alkan from Bursa Uludağ University for sharing the supplementary material in vivo experiments with us. We would also like to thanks to MRI technician Senem Elber (magnetic resonance imaging technologist) for special assistance with in vivo imaging. This study was supported by Scientific and Technological Research Council of Turkey (TUBITAK) under the Grant Number 121S624. The authors thank to TUBITAK for their supports.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ajps.2024.100971. The figures with “S” before the serial number are included in the Supplementary material.
Appendix. Supplementary materials
References
- 1.Iacob G., Dinca E.B. Current data and strategy in glioblastoma multiforme. J Med Life. 2009;2:386–393. [PMC free article] [PubMed] [Google Scholar]
- 2.Templeton A., Hofer S., Töpfer M., Sommacal A., Fretz C., Cerny T., et al. Extraneural spread of glioblastoma–report of two cases. Onkologie. 2008;31(4):192–194. doi: 10.1159/000118627. [DOI] [PubMed] [Google Scholar]
- 3.Castañeda C.A., Casavilca S., Orrego E., García-Corrochano P., Deza P., Heinike H., et al. Glioblastoma: análisis molecular y sus implicancias clínicas [Glioblastoma: molecular analysis and its clinical implications] Rev Peru Med Exp Salud Publica. 2015;32(2):316–325. [PubMed] [Google Scholar]
- 4.Chacko A.M., Li C., Pryma D.A., Brem S., Coukos G., Muzykantov V. Targeted delivery of antibody-based therapeutic and imaging agents to CNS tumors: crossing the blood-brain barrier divide. Expert Opin Drug Deliv. 2013;10(7):907–926. doi: 10.1517/17425247.2013.808184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Juillerat-Jeanneret L. The targeted delivery of cancer drugs across the blood-brain barrier: chemical modifications of drugs or drug-nanoparticles? Drug Discov Today. 2008;13:1099–1106. doi: 10.1016/j.drudis.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Löscher W., Potschka H. Drug resistance in brain diseases and the role of drug efflux transporters. Nat Rev Neurosci. 2005;6:591–602. doi: 10.1038/nrn1728. [DOI] [PubMed] [Google Scholar]
- 7.Siegal T. Which drug or drug delivery system can change clinical practice for brain tumor therapy? Neuro Oncol. 2013;15:656–669. doi: 10.1093/neuonc/not016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong L.T.A., Kim Y.M., Park H.H., Hwang D.H., Cui Y., Lee E.M., et al. An injectable hydrogel enhances tissue repair after spinal cord injury by promoting extracellular matrix remodeling. Nat Commun. 2017;8(1):533. doi: 10.1038/s41467-017-00583-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bastiancich C., Vanvarenberg K., Ucakar B., Pitorre M., Bastiat G., Lagarce F., et al. Lauroyl-gemcitabine-loaded lipid nanocapsule hydrogel for the treatment of glioblastoma. J Control Release. 2016;225:283–293. doi: 10.1016/j.jconrel.2016.01.054. [DOI] [PubMed] [Google Scholar]
- 10.Westphal M., Hilt D.C., Bortey E., Delavault P., Olivares R., Warnke P.C., et al. A phase 3 trial of local chemotherapy with biodegradable carmustine (BCNU) wafers (Gliadel wafers) in patients with primary malignant glioma. Neuro Oncol. 2003;5(2):79–88. doi: 10.1215/S1522-8517-02-00023-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bota D.A., Desjardins A., Quinn J.A., Affronti M.L., Friedman H.S. Interstitial chemotherapy with biodegradable BCNU (Gliadel) wafers in the treatment of malignant gliomas. Ther Clin Risk Manag. 2007;3(5):707–715. [PMC free article] [PubMed] [Google Scholar]
- 12.Giese A., Kucinski T., Knopp U., Goldbrunner R., Hamel W., Mehdorn H.M., et al. Pattern of recurrence following local chemotherapy with biodegradable carmustine (BCNU) implants in patients with glioblastoma. J Neurooncol. 2004;66(3):351–360. doi: 10.1023/b:neon.0000014539.90077.db. [DOI] [PubMed] [Google Scholar]
- 13.Ramachandran R., Junnuthula V.R., Gowd G.S., Ashokan A., Thomas J., Peethambaran R., et al. Theranostic 3-dimensional nano brain-implant for prolonged and localized treatment of recurrent glioma. Sci Rep. 2017;7:43271. doi: 10.1038/srep43271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao Z.Z., Wang Z.F., Lan T., Huang W.H., Zhao Y.H., Ma C., et al. Carmustine as a supplementary therapeutic option for glioblastoma: a systematic review and meta-analysis. Front Neurol. 2020;11:1036. doi: 10.3389/fneur.2020.01036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norouzi M., Boroujeni S.M., Omidvarkordshouli N., Soleimani M. Advances in skin regeneration: application of electrospun scaffolds. Adv Healthc Mater. 2015;4:1114–1133. doi: 10.1002/adhm.201500001. [DOI] [PubMed] [Google Scholar]
- 16.Qiu K., He C., Feng W., Wang W., Zhou X., Yin Z., et al. Doxorubicin-loaded electrospun poly(l-lactic acid)/mesoporous silica nanoparticles composite nanofibers for potential postsurgical cancer treatment. J Mater Chem B. 2013;1(36):4601–4611. doi: 10.1039/c3tb20636j. [DOI] [PubMed] [Google Scholar]
- 17.Ranganath S.H., Wang C.H. Biodegradable microfiber implants delivering paclitaxel for post-surgical chemotherapy against malignant glioma. Biomaterials. 2008;29:2996–3003. doi: 10.1016/j.biomaterials.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 18.Brem S., Tyler B., Li K., Pradilla G., Legnani F., Caplan J., et al. Local delivery of temozolomide by biodegradable polymers is superior to oral administration in a rodent glioma model. Cancer Chemother Pharmacol. 2007;60(5):643–650. doi: 10.1007/s00280-006-0407-2. [DOI] [PubMed] [Google Scholar]
- 19.Tseng Y.Y., Liao J.Y., Chen W.A., Kao Y.C., Liu S.J. Sustainable release of carmustine from biodegradable poly [(d,L)-lactide-co-glycolide] nanofibrous membranes in the cerebral cavity: in vitro and in vivo studies. Expert Opin Drug Deliv. 2013;10:879–888. doi: 10.1517/17425247.2013.758102. [DOI] [PubMed] [Google Scholar]
- 20.Ni S., Fan X., Wang J., Qi H., Li X. Biodegradable implants efficiently deliver combination of paclitaxel and temozolomide to glioma C6 cancer cells in vitro. Ann Biomed Eng. 2014;42(1):214–221. doi: 10.1007/s10439-013-0903-6. [DOI] [PubMed] [Google Scholar]
- 21.Kuo S.M. Dietary flavonoid and cancer prevention: evidence and potential mechanism. Crit Rev Oncog. 1997;8(1):47–69. doi: 10.1615/critrevoncog.v8.i1.30. [DOI] [PubMed] [Google Scholar]
- 22.Kopustinskiene D.M., Jakstas V., Savickas A., Bernatoniene J. Flavonoids as anticancer agents. Nutrients. 2020;12(2):457. doi: 10.3390/nu12020457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slika H., Mansour H., Wehbe N., Nasser S.A., Iratni R., Nasrallah G., et al. Therapeutic potential of flavonoids in cancer: rOS-mediated mechanisms. Biomed Pharmacother. 2022;146 doi: 10.1016/j.biopha.2021.112442. [DOI] [PubMed] [Google Scholar]
- 24.Galati G., O'Brien P.J. Potential toxicity of flavonoids and other dietary phenolics: significance for their chemopreventive and anticancer properties. Free Radic Biol Med. 2004;37(3):287–303. doi: 10.1016/j.freeradbiomed.2004.04.034. [DOI] [PubMed] [Google Scholar]
- 25.Skibola C.F., Smith M.T. Potential health impacts of excessive flavonoid intake. Free Radic Biol Med. 2000;29(3–4):375–383. doi: 10.1016/s0891-5849(00)00304-x. [DOI] [PubMed] [Google Scholar]
- 26.Pandey P., Khan F., Qari H.A., Oves M. Rutin (bioflavonoid) as cell signaling pathway modulator: prospects in treatment and chemoprevention. Pharmaceuticals (Basel) 2021;22(11):1069. doi: 10.3390/ph14111069. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganeshpurkar A., Saluja A.K. The pharmacological potential of rutin. Saudi Pharmaceutical J. 2017;25(2):149–164. doi: 10.1016/j.jsps.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perk A.A., Shatynska-Mytsyk I., Gerçek Y.C., Boztaş K., Yazgan M., Fayyaz S., Farooqi A.A. Rutin mediated targeting of signaling machinery in cancer cells. Cancer Cell Int. 2014;14(1):124. doi: 10.1186/s12935-014-0124-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang C., Zhang L., Miao Y., Fang B., Yang Z. Anticancer and apoptotic-inducing effects of rutin-chitosan nanoconjugates in triple negative breast cancer cells. J Clust Sci. 2021;32:331–340. [Google Scholar]
- 30.Salam H.S., Tawfik M.M., Elnagar M.R., Mohammed H.A., Zarka M.A., Awad N.S. Potential apoptotic activities of hylocereus undatus peel and pulp extracts in MCF-7 and Caco-2 cancer cell lines. Plants (Basel) 2022;11(17):2192. doi: 10.3390/plants11172192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ben Sghaier M., Pagano A., Mousslim M., Ammari Y., Kovacic H., Luis J. Rutin inhibits proliferation, attenuates superoxide production and decreases adhesion and migration of human cancerous cells. Biomed Pharmacother. 2016;84:1972–1978. doi: 10.1016/j.biopha.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 32.Yan X., Hao Y., Chen S., Jia G., Guo Y., Zhang G., et al. Rutin induces apoptosis via P53 up-regulation in human glioma CHME cells. Transl Cancer Res. 2019;8(5):2005–2013. doi: 10.21037/tcr.2019.09.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tunca B., Tezcan G., Cecener G., Egeli U., Ak S., Malyer H., et al. Olea europaea leaf extract alters microRNA expression in human glioblastoma cells. J Cancer Res Clin Oncol. 2012;138(11):1831–1844. doi: 10.1007/s00432-012-1261-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tezcan G., Tunca B., Bekar A., Budak F., Sahin S., Cecener G., et al. Olea europaea leaf extract improves the treatment response of GBM stem cells by modulating miRNA expression. Am J Cancer Res. 2014;4(5):572–590. [PMC free article] [PubMed] [Google Scholar]
- 35.Tezcan G., Taskapilioglu M.O., Tunca B., Bekar A., Demirci H., Kocaeli H., et al. Olea europaea leaf extract and bevacizumab synergistically exhibit beneficial efficacy upon human glioblastoma cancer stem cells through reducing angiogenesis and invasion in vitro. Biomed Pharmacother. 2017;90:713–723. doi: 10.1016/j.biopha.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 36.Tezcan G., Tunca B., Demirci H., Bekar A., Taskapilioglu M.O., Kocaeli H., et al. Olea europaea leaf extract improves the efficacy of temozolomide therapy by inducing MGMT methylation and reducing P53 expression in glioblastoma. Nutr Cancer. 2017;69(6):873–880. doi: 10.1080/01635581.2017.1339810. [DOI] [PubMed] [Google Scholar]
- 37.Mutlu M., Tunca B., Ak Aksoy S., Tekin C., Egeli U., Cecener G. Inhibitory effects of olea europaea leaf extract on mesenchymal transition mechanism in glioblastoma cells. Nutr Cancer. 2021;73(4):713–720. doi: 10.1080/01635581.2020.1765260. [DOI] [PubMed] [Google Scholar]
- 38.Erçelik M., Tunca B., S Ak Aksoy, Ç Tekin, Tezcan G. Olea europaea L. Leaf extract attenuates temozolomide-induced Ssenescence-associated secretion phenotype in glioblastoma. Turk J Pharm Sci. 2023;20(2):68–77. doi: 10.4274/tjps.galenos.2022.57639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ercelik M., Tekin C., Tezcan G., Ak Aksoy S., Bekar A., Kocaeli H., et al. Olea europaea leaf phenolics oleuropein, hydroxytyrosol, tyrosol, and rutin induce apoptosis and additionally affect temozolomide against glioblastoma: in particular, oleuropein inhibits spheroid growth by attenuating stem-like cell phenotype. Life (Basel) 2023;13(2):470. doi: 10.3390/life13020470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang P., Sun S., Li N., Ho A.S.W., Kiang K.M.Y., Zhang X., et al. Rutin increases the cytotoxicity of temozolomide in glioblastoma via autophagy inhibition. J Neurooncol. 2017;132(3):393–400. doi: 10.1007/s11060-017-2387-y. [DOI] [PubMed] [Google Scholar]
- 41.Son Y.J., Kim H.S., Choi D.H., Yoo H.S. Multilayered electrospun fibrous meshes for restenosis-suppressing metallic stents. J Biomed Mater Res B Appl Biomater. 2017;105(3):628–635. doi: 10.1002/jbm.b.33583. [DOI] [PubMed] [Google Scholar]
- 42.Ercelik M., Tekin C., Parin F.N., Mutlu B., Dogan H.Y., Tezcan G., et al. Co-loading of temozolomide with oleuropein or rutin into polylactic acid core-shell nanofiber webs inhibit glioblastoma cell by controlled release. Int J Biol Macromol. 2023;253(Pt 2) doi: 10.1016/j.ijbiomac.2023.126722. [DOI] [PubMed] [Google Scholar]
- 43.Sabzehmeidani M.M., Ghaedi M., Karimi H. Photocatalytic activity based on electrospun nanofibers. Interface Sci Technol. 2021;32:625–672. [Google Scholar]
- 44.Parin F.N., Terzioğlu P., Sicak Y., Yildirim K., Öztürk M. Pine honey–loaded electrospun poly (vinyl alcohol)/gelatin nanofibers with antioxidant properties. J Textile Institute. 2021;112(4):628–635. [Google Scholar]
- 45.Tracht J., Zhang K., Peker D. Grading and prognostication of neuroendocrine tumors of the pancreas: a comparison study of Ki67 and PHH3. J Histochem Cytochem. 2017;65(7):399–405. doi: 10.1369/0022155417708186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newman P.J., Newman D.K. Signal transduction pathways mediated by PECAM-1: new roles for an old molecule in platelet and vascular cell biology. Arterioscler. Thromb. Vasc. Biol. 2003;23(6):953–964. doi: 10.1161/01.ATV.0000071347.69358.D9. [DOI] [PubMed] [Google Scholar]
- 47.Luraghi A., Peri F., Moroni L. Electrospinning for drug delivery applications: a review. J Control release. 2021;334:463–484. doi: 10.1016/j.jconrel.2021.03.033. [DOI] [PubMed] [Google Scholar]
- 48.Stoyanova N., Spasova M., Manolova N., Rashkov I., Georgieva A., Toshkova R. Quercetin- and rutin-Containing electrospun cellulose acetate and polyethylene glycol fibers with antioxidant and anticancer properties. Polymers (Basel) 2022;14(24):5380. doi: 10.3390/polym14245380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahabir R., Tanino M., Elmansuri A., Wang L., Kimura T., Itoh T., et al. Sustained elevation of Snail promotes glial-mesenchymal transition after irradiation in malignant glioma. Neuro Oncol. 2014;16(5):671–685. doi: 10.1093/neuonc/not239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iwadate Y. Epithelial-mesenchymal transition in glioblastoma progression. Oncol Lett. 2016;11(3):1615–1620. doi: 10.3892/ol.2016.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mutlu M., Tunca B., Aksoy S.A., Tekin C., Cecener G., Egeli U. Olea europaea leaf extract decreases tumour size by affecting the LncRNA expression status in glioblastoma 3D cell cultures. Eur J Integr Med. 2021;45:1876–3820. [Google Scholar]
- 52.Vasan K., Werner M., Chandel N.S. Mitochondrial metabolism as a target for cancer therapy. Cell Metab. 2020;32(3):341–352. doi: 10.1016/j.cmet.2020.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee J.S., Lee H., Woo S.M., Jang H., Jeon Y., Kim H.Y., et al. Overall survival of pancreatic ductal adenocarcinoma is doubled by Aldh7a1 deletion in the KPC mouse. Theranostics. 2021;11(7):3472–3488. doi: 10.7150/thno.53935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin Y., Zhang F., Jin Y., Zhong Q., Tan W., Liu J., et al. NUDT1 could be a prognostic biomarker and correlated with immune infiltration in clear cell renal cell carcinoma. Appl Bionics Biomech. 2022;2022 doi: 10.1155/2022/3669296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu T., Wang L., Chen G., Tong L., Ye X., Yang H., et al. PDZD8-mediated endoplasmic reticulum-mitochondria associations regulate sympathetic drive and blood pressure through the intervention of neuronal mitochondrial homeostasis in stress-induced hypertension. Neurobiol Dis. 2023;183 doi: 10.1016/j.nbd.2023.106173. [DOI] [PubMed] [Google Scholar]
- 56.Ruokun C., Yake X., Fengdong Y., Xinting W., Laijun S., Xianzhi L. Lentivirus-mediated silencing of HSDL2 suppresses cell proliferation in human gliomas. Tumour Biol. 2016;37(11):15065–15077. doi: 10.1007/s13277-016-5402-6. [DOI] [PubMed] [Google Scholar]
- 57.Lamb R., Bonuccelli G., Ozsvári B., Peiris-Pagès M., Fiorillo M., Smith D.L., et al. Mitochondrial mass, a new metabolic biomarker for stem-like cancer cells: understanding WNT/FGF-driven anabolic signaling. Oncotarget. 2015;6(31):30453–30471. doi: 10.18632/oncotarget.5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Im J.Y., Kim S.J., Park J.L., Han T.H., Kim W.I., Kim I., et al. CYB5R3 functions as a tumor suppressor by inducing ER stress-mediated apoptosis in lung cancer cells via the PERK-ATF4 and IRE1α-JNK pathways. Exp Mol Med. 2024;56(1):235–249. doi: 10.1038/s12276-024-01155-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gundamaraju R., Lu W., Manikam R. CHCHD2: the power house's potential prognostic factor for cancer? Front Cell Dev Biol. 2020;8 doi: 10.3389/fcell.2020.620816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raturi A., Gutiérrez T., Ortiz-Sandoval C., Ruangkittisakul A., Herrera-Cruz M.S., Rockley J.P., et al. TMX1 determines cancer cell metabolism as a thiol-based modulator of ER-mitochondria Ca2+ flux. J Cell Biol. 2016;214(4):433–444. doi: 10.1083/jcb.201512077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Z., Zhao X., Zhang L., Pei B. Cytochrome C inhibits tumor growth and predicts favorable prognosis in clear cell renal cell carcinoma. Oncol Lett. 2019;18(6):6026–6032. doi: 10.3892/ol.2019.10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Han J.H., Heo K.S., Myung C.S. CIAPIN1 accelerates vascular remodelling via p53 and JAK2-STAT3 regulation in vascular smooth muscle cells. Br J Pharmacol. 2021;178:4533–4551. doi: 10.1111/bph.15631. [DOI] [PubMed] [Google Scholar]
- 63.Huffman N., Palmieri D., Coppola V. The CTLH complex in cancer cell plasticity. J Oncol. 2019 doi: 10.1155/2019/4216750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feng Y., Zhao J., Hou H., Zhang H., Jiao Y., Wang J., et al. WDR26 promotes mitophagy of cardiomyocytes induced by hypoxia through Parkin translocation. Acta Bziochim Biophys Sin (Shanghai) 2016;48(12):1075–1084. doi: 10.1093/abbs/gmw104. [DOI] [PubMed] [Google Scholar]
- 65.Xie Q.H., Wang W.M., Yang J.G., Xia H.F., Xiao B.L., Chen G.H., Huang J., et al. ALIX promotes cell migration and invasion of head and neck squamous cell carcinoma by regulating the expression of MMP9, MMP14, VEGF-C. Arch Oral Biol. 2023;151 doi: 10.1016/j.archoralbio.2023.105696. [DOI] [PubMed] [Google Scholar]
- 66.Sen U., Coleman C., Sen T. Stearoyl coenzyme A desaturase-1: multitasker in cancer, metabolism, and ferroptosis. Trends Cancer. 2023;9(6):480–489. doi: 10.1016/j.trecan.2023.03.003. [DOI] [PubMed] [Google Scholar]
- 67.Tracz-Gaszewska Z., Dobrzyn P. Stearoyl-CoA Desaturase 1 as a therapeutic target for the treatment of cancer. Cancers (Basel) 2019;11(7):948. doi: 10.3390/cancers11070948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sharma G., Ojha R., Noguera-Ortega E., Rebecca V.W., Attanasio J., Liu S., et al. PPT1 inhibition enhances the antitumor activity of anti-PD-1 antibody in melanoma. JCI Insight. 2022;7(20) doi: 10.1172/jci.insight.165688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rebecca V.W., Nicastri M.C., Fennelly C., Chude C.I., Barber-Rotenberg J.S., Ronghe A., et al. PPT1 promotes tumor growth and is the molecular target of chloroquine derivatives in cancer. Cancer Discov. 2019;9(2):220–229. doi: 10.1158/2159-8290.CD-18-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang H., Liu B., Wei J. Beta2-microglobulin(B2M) in cancer immunotherapies: biological function, resistance and remedy. Cancer Lett. 2021;517:96–104. doi: 10.1016/j.canlet.2021.06.008. [DOI] [PubMed] [Google Scholar]
- 71.Guo C., Liu S., Greenaway F., Sun M.Z. Potential role of annexin A7 in cancers. Clin Chim Acta. 2013;423:83–89. doi: 10.1016/j.cca.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 72.Rezvani K. UBXD proteins: a family of proteins with diverse functions in cancer. Int J Mol Sci. 2016;17(10):1724. doi: 10.3390/ijms17101724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang X., Wu F., Wang H., Duan X., Huang R., Tuersuntuoheti A., et al. PDCD6 cooperates with C-Raf to facilitate colorectal cancer progression via Raf/MEK/ERK activation. J Exp Clin Cancer Res. 2020;39(1):147. doi: 10.1186/s13046-020-01632-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao Y., Xu L., Wang X., Niu S., Chen H., Li C. A novel prognostic mRNA/miRNA signature for esophageal cancer and its immune landscape in cancer progression. Mol Oncol. 2021;15(4):1088–1109. doi: 10.1002/1878-0261.12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hong S.H., Chang S.H., Cho K.C., Kim S., Park S., Lee A.Y., et al. Endoplasmic reticulum-Golgi intermediate compartment protein 3 knockdown suppresses lung cancer through endoplasmic reticulum stress-induced autophagy. Oncotarget. 2016;7(40):65335–65347. doi: 10.18632/oncotarget.11678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cui Y., Miao C., Liu S., Tang J., Zhang J., Bu H., et al. Clusterin suppresses invasion and metastasis of testicular seminoma by upregulating COL15a1. Mol Ther Nucleic Acids. 2021;26:1336–1350. doi: 10.1016/j.omtn.2021.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martínez-Nieto G., Heljasvaara R., Heikkinen A., Kaski H.K., Devarajan R., Rinne O., et al. Deletion of Col15a1 modulates the tumour extracellular matrix and leads to increased tumour growth in the MMTV-PyMT mouse mammary carcinoma model. Int J Mol Sci. 2021;22(18):9978. doi: 10.3390/ijms22189978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Eelen G., Dubois C., Cantelmo A.R., Goveia J., Brüning U., DeRan M., et al. Role of glutamine synthetase in angiogenesis beyond glutamine synthesis. Nature. 2018;561(7721):63–69. doi: 10.1038/s41586-018-0466-7. [DOI] [PubMed] [Google Scholar]
- 79.Dhiman G., Srivastava N., Goyal M., Rakha E., Lothion-Roy J., Mongan N.P., et al. Metadherin: a therapeutic target in multiple cancers. Front Oncol. 2019;9:349. doi: 10.3389/fonc.2019.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ferrari G., Cook B.D., Terushkin V., Pintucci G., Mignatti P. Transforming growth factor-beta 1 (TGF-beta1) induces angiogenesis through vascular endothelial growth factor (VEGF)-mediated apoptosis. J Cell Physiol. 2009;219(2):449–458. doi: 10.1002/jcp.21706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zonneville J., Safina A., Truskinovsky A.M., Arteaga C.L., Bakin A.V. TGF-β signaling promotes tumor vasculature by enhancing the pericyte-endothelium association. BMC Cancer. 2018;18(1):670. doi: 10.1186/s12885-018-4587-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cifaldi L., Romania P., Lorenzi S., Locatelli F., Fruci D. Role of endoplasmic reticulum aminopeptidases in health and disease: from infection to cancer. Int J Mol Sci. 2012;13(7):8338–8352. doi: 10.3390/ijms13078338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rossi E., Bernabeu C., Smadja D.M. Endoglin as an adhesion molecule in mature and progenitor endothelial cells: a function beyond TGF-β. Front Med (Lausanne) 2019;6:10. doi: 10.3389/fmed.2019.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sánchez F.S.L., Martínez J.A.C., Méndez-García L., García-Cenador M.B., Pericacho M. Endoglin and other angiogenesis markers in recurrent varicose veins. J Pers Med. 2022;12(4):528. doi: 10.3390/jpm12040528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hashemi M., Yousefi J., Hashemi S.M., Amininia S., Ebrahimi M., Taheri M., et al. Association between programmed cell death 6 interacting protein insertion/deletion polymorphism and the risk of breast cancer in a sample of Iranian population. Dis Markers. 2015;2015 doi: 10.1155/2015/854621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kulikov A.V., Shilov E.S., Mufazalov I.A., Gogvadze V., Nedospasov S.A., Zhivotovsky B. Cytochrome c: the Achilles' heel in apoptosis. Cell Mol Life Sci. 2012;69(11):1787–1797. doi: 10.1007/s00018-011-0895-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Asadi M., Taghizadeh S., Kaviani E., Vakili O., Taheri-Anganeh M., Tahamtan M., et al. Caspase-3: structure, function, and biotechnological aspects. Biotechnol Appl Biochem. 2022;69(4):1633–1645. doi: 10.1002/bab.2233. [DOI] [PubMed] [Google Scholar]
- 88.Restelli M., Magni M., Ruscica V., Pinciroli P., De Cecco L., Buscemi G., et al. A novel crosstalk between CCAR2 and AKT pathway in the regulation of cancer cell proliferation. Cell Death Dis. 2016;7(11):e2453. doi: 10.1038/cddis.2016.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peng L., Liu Y., Chen J., Cheng M., Wu Y., Chen M., et al. APEX1 regulates alternative splicing of key tumorigenesis genes in non-small-cell lung cancer. BMC Med Genomics. 2022;15(1):147. doi: 10.1186/s12920-022-01290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen F., Kang R., Liu J., Tang D. The ACSL4 network regulates cell death and autophagy in diseases. Biology (Basel) 2023;12(6):864. doi: 10.3390/biology12060864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peluso J.J., Liu X., Gawkowska A., Lodde V., W C.A. Progesterone inhibits apoptosis in part by PGRMC1-regulated gene expression. Mol. Cell Endocrinol. 2010;320(1–2):153–161. doi: 10.1016/j.mce.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Luo Z., Wang Y., Bi X., Ismtula D., Wang H., Guo C. Cytokine-induced apoptosis inhibitor 1: a comprehensive analysis of potential diagnostic, prognosis, and immune biomarkers in invasive breast cancer. Transl Cancer Res. 2023;12(7):1765–1786. doi: 10.21037/tcr-23-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gadek M., Sherr E.H., Floor S.N. The variant landscape and function of DDX3X in cancer and neurodevelopmental disorders. Trends Mol Med. 2023;29(9):726–739. doi: 10.1016/j.molmed.2023.06.003. [DOI] [PubMed] [Google Scholar]
- 94.Pozo K., Bibb J.A. The emerging role of Cdk5 in cancer. Trends Cancer. 2016;2(10):606–618. doi: 10.1016/j.trecan.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu H., Guo D., Sha Y., Zhang C., Jiang Y., Hong L., et al. ANXA7 promotes the cell cycle, proliferation and cell adhesion-mediated drug resistance of multiple myeloma cells by up-regulating CDC5L. Aging (Albany NY) 2020;12(11):11100–11115. doi: 10.18632/aging.103326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Strubbe-Rivera J.O., Schrad J.R., Pavlov E.V., Conway J.F., Parent K.N., Bazil J.N. The mitochondrial permeability transition phenomenon elucidated by cryo-EM reveals the genuine impact of calcium overload on mitochondrial structure and function. Sci Rep. 2021;11(1):1037. doi: 10.1038/s41598-020-80398-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Burke PJ. Mitochondria, bioenergetics and apoptosis in cancer. Trends Cancer. 2017;3(12):857–870. doi: 10.1016/j.trecan.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Aygun C., Altınok A.Y., Çakır A., Agan A.F., Balaban Y. Acute temozolomide induced liver injury: mixed type hepatocellular and cholestatic toxicity. Acta Gastroenterol Belg. 2016;79(2):363–365. [PubMed] [Google Scholar]
- 99.Houtmeyers A., Duchateau L., Grünewald B., Hermans K. Reference intervals for biochemical blood variables, packed cell volume, and body temperature in pet rats (Rattus norvegicus) using point-of-care testing. Vet Clin Pathol. 2016;45(4):669–679. doi: 10.1111/vcp.12419. [DOI] [PubMed] [Google Scholar]
- 100.Salazar J.H. Overview of urea and creatinine. Lab Med. 2014;45:e19–e20. [Google Scholar]
- 101.Wesolowski J.R., Rajdev P., Mukherji S.K. Temozolomide (Temodar) AJNR Am J Neuroradiol. 2010;31(8):1383–1384. doi: 10.3174/ajnr.A2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.CTEP. Possible side effects of temozolomide, 2024. Availlable from: https://ctep.cancer.gov/protocolDevelopment/docs/sideeffects/SideEffects-Temozolomide.docx. (Accessed April 25, 2024).
- 103.Lee S.Y. Temozolomide resistance in glioblastoma multiforme. Genes Dis. 2016;3(3):198–210. doi: 10.1016/j.gendis.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Meerson A., Khatib S., Mahajna J. Natural products targeting cancer stem cells for augmenting cancer therapeutics. Int J Mol Sci. 2021;2(23):13044. doi: 10.3390/ijms222313044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sharma S., Ali A., Ali J., Sahni J.K., Rutin Baboota S. Therapeutic po- tential and recent advances in drug delivery. Expert Opin Investig Drugs. 2013;22:1063–1079. doi: 10.1517/13543784.2013.805744. [DOI] [PubMed] [Google Scholar]
- 106.Lukšič L., Bonafaccia G., Timoracka M., Vollmannova A., Trček J., Nyambe T.K., et al. Rutin and quercetin transformation during preparation of buckwheat sourdough bread. J Cereal Sci. 2016;69:71–76. [Google Scholar]
- 107.Nasiri F., Kismali G., Alpay M., Kosova F., Çakir D.U., Sel T. Rutin enhances the antiproliferative effect of 5-FU and oxaliplatin in colon cancer cells. Cancer Res. 2016;76:2177. [Google Scholar]
- 108.Costa R., Costa Lima S.A., Gameiro P., Reis S. On the development of a cutaneous flavonoid delivery system: advances and limitations. Antioxidants (Basel) 2021;10(9):1376. doi: 10.3390/antiox10091376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ishak R.A., Mostafa N.M., Kamel A.O. Stealth lipid polymer hybrid nanoparticles loaded with rutin for effective brain delivery–comparative study with the gold standard (Tween 80): optimization, characterization and biodistribution. Drug Deliv. 2017;24(1):1874–1890. doi: 10.1080/10717544.2017.1410263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ahmad H., Arya A., Agrawal S., Mall P., Samuel S.S., Sharma K., Singh P.K., et al. Rutin phospholipid complexes confer neuro-protection in ischemic-stroke rats. RSC Adv. 2016;6:96445–96454. [Google Scholar]
- 111.Zhou L., Cai L., Ruan H., Zhang L., Wang J., Jiang H., et al. Electrospun chitosan oligosaccharide/polycaprolactone nanofibers loaded with wound-healing compounds of Rutin and Quercetin as antibacterial dressings. Int J Biol Macromol. 2021;183:1145–1154. doi: 10.1016/j.ijbiomac.2021.05.031. [DOI] [PubMed] [Google Scholar]
- 112.Zhao X., Liang Y., Guo B., Yin Z., Zhu D., Han Y. Injectable dry cryogels with excellent blood-sucking expansion and blood clotting to cease hemorrhage for lethal deep-wounds, coagulopathy and tissue regeneration. Chemical Engineering J. 2021;403 [Google Scholar]
- 113.Li B., Yang X. Rutin-loaded cellulose acetate/poly(ethylene oxide) fiber membrane fabricated by electrospinning: a bioactive material. Mater Sci Eng C Mater Biol App. 2020;109 doi: 10.1016/j.msec.2019.110601. [DOI] [PubMed] [Google Scholar]
- 114.Vander Heiden M.G., Plas D.R., Rathmell J.C., Fox C.J., Harris M.H., Thompson C.B. Growth factors can influence cell growth and survival through effects on glucose metabolism. Mol Cell Biol. 2001;21(17):5899–5912. doi: 10.1128/MCB.21.17.5899-5912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Starkov A.A., Fiskum G. Regulation of brain mitochondrial H2O2 production by membrane potential and NAD(P)H redox state. J. Neurochem. 2003;86(5):1101–1107. doi: 10.1046/j.1471-4159.2003.01908.x. [DOI] [PubMed] [Google Scholar]
- 116.Lee S.Y., Ju M.K., Jeon H.M., Lee Y.J., Kim C.H., Park H.G., et al. Reactive oxygen species induce epithelialmesenchymal transition, glycolytic switch, and mitochondrial repression through the Dlx2/Snail signaling pathways in MCF7 cells. Mol Med Rep. 2019;20(3):2339–2346. doi: 10.3892/mmr.2019.10466. [DOI] [PubMed] [Google Scholar]
- 117.Boyd N.H., Tran A.N., Bernstock J.D., Etminan T., Jones A.B., Gillespie G.Y., et al. Glioma stem cells and their roles within the hypoxic tumor microenvironment. Theranostics. 2021;11(2):665–683. doi: 10.7150/thno.41692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yuan Z., Liang X., Zhan Y., Wang Z., Xu J., Qiu Y., et al. Targeting CD133 reverses drug-resistance via the AKT/NF-κB/MDR1 pathway in colorectal cancer. Br J Cancer. 2020;122(9):1342–1353. doi: 10.1038/s41416-020-0783-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zakaria N., Mohd Yusoff N., Zakaria Z., Widera D., Yahaya B.H. Inhibition of NF-κB signaling reduces the stemness characteristics of lung cancer stem cells. Front Oncol. 2018;8:166. doi: 10.3389/fonc.2018.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kabacaoglu D., Ruess D.A., Ai J., Algül H. NF-κB/Rel transcription factors in pancreatic cancer: focusing on RelA, c-Rel, and RelB. Cancers (Basel) 2019;11(7):937. doi: 10.3390/cancers11070937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang H., Li L., Yin L. Silencing LncRNA LOXL1-AS1 attenuates mesenchymal characteristics of glioblastoma via NF-κB pathway. Biochem. Biophys. Res. Commun. 2018;500(2):518–524. doi: 10.1016/j.bbrc.2018.04.133. [DOI] [PubMed] [Google Scholar]
- 122.Jiang X., Yan Y., Hu M., Chen X., Wang Y., Dai Y., et al. Increased level of H19 long noncoding RNA promotes invasion, angiogenesis, and stemness of glioblastoma cells. J Neurosurg. 2016;124(1):129–136. doi: 10.3171/2014.12.JNS1426. [DOI] [PubMed] [Google Scholar]
- 123.Li W., Jiang P., Sun X., Xu S., Ma X., Zhan R. Suppressing H19 Modulates tumorigenicity and stemness in U251 and U87MG glioma cells. Cell Mol Neurobiol. 2016;36(8):1219–1227. doi: 10.1007/s10571-015-0320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Huang K., Sun J., Yang C., Wang Y., Zhou B., Kang C., et al. HOTAIR upregulates an 18-gene cell cycle-related mRNA network in glioma. Int J Oncol. 2017;50(4):1271–1278. doi: 10.3892/ijo.2017.3901. [DOI] [PubMed] [Google Scholar]
- 125.Hlatky L., Hahnfeldt P., Folkman J. Clinical application of antiangiogenic therapy: microvessel density, what it does and doesn't tell us. J. Natl. Cancer Inst. 2002;94(12):883–893. doi: 10.1093/jnci/94.12.883. [DOI] [PubMed] [Google Scholar]
- 126.Gottlieb E., Armour S.M., Harris M.H., Thompson C.B. Mitochondrial membrane potential regulates matrix configuration and cytochrome c release during apoptosis. Cell Death Differ. 2003;10(6):709–717. doi: 10.1038/sj.cdd.4401231. [DOI] [PubMed] [Google Scholar]
- 127.Javadov S., Chapa-Dubocq X., Makarov V. Different approaches to modeling analysis of mitochondrial swelling. Mitochondrion. 2018;38:58–70. doi: 10.1016/j.mito.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Boulares A.H., Yakovlev A.G., Ivanova V., Stoica B.A., Wang G., Iyer S., et al. Role of poly(ADP-ribose) polymerase (PARP) cleavage in apoptosis. Caspase 3-resistant PARP mutant increases rates of apoptosis in transfected cells. J Biol Chem. 1999;274(33):22932–22940. doi: 10.1074/jbc.274.33.22932. [DOI] [PubMed] [Google Scholar]
- 129.FJr Gallyas, Sumegi B. Mitochondrial protection by PARP inhibition. Int J Mol Sci. 2020;21(8):2767. doi: 10.3390/ijms21082767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kalas M.A., Chavez L., Leon M., Taweesedt P.T., Surani S. Abnormal liver enzymes: a review for clinicians. World J Hepatol. 2021;13(11):1688–1698. doi: 10.4254/wjh.v13.i11.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Thorsson V., Gibbs D.L., Brown S.D., Wolf D., Bortone D.S., Ou Yang T.H., et al. The immune landscape of cancer. immunity. 2018;48(4):812–830. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ellyard J.I., Simson L., Parish C.R. Th2-mediated anti-tumour immunity: friend or foe? Tissue antigens. 2007;70(1):1–11. doi: 10.1111/j.1399-0039.2007.00869.x. [DOI] [PubMed] [Google Scholar]
- 133.Paludan S.R. Interleukin-4 and interferon-gamma: the quintessence of a mutual antagonistic relationship. Scand J Immunol. 1998;48(5):459–468. doi: 10.1046/j.1365-3083.1998.00435.x. [DOI] [PubMed] [Google Scholar]
- 134.Sibley L.S., White A.D., Marriott A., Dennis M.J., Williams A., Marsh P.D., et al. ELISPOT refinement using spot morphology for assessing host responses to tuberculosis. Cells. 2012;1(1):5–14. doi: 10.3390/cells1010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.