Abstract
This dataset provides an in-depth analysis of rice yield and grain quality attributes in four successive four years across 27 diverse environments in Bangladesh. The analysis emphasizes assessing the performance of studied genotypes (GEN), environments (ENV), and their interrelations (GEI). The research aim is to detect a stable and adaptive rice cultivar that not only displays high yield, and better grain quality but also has molecular data to know favorable alleles and biotic and abiotic stress-related traits. The combined ANOVA revealed significant GEN, ENV, and GEI effects on grain yield (p ≤ 0.001). Cultivar BR9011-67-4-1 produced the highest mean yield at 4.77 t/ha, whereas BRRI dhan48 had the lowest yield at 4.66 t/ha. In the C1E2 environment, BR9011-67-4-1 exhibited the highest yield at 6.06 t/ha. The broad-sense heritability was found at 0.481 and selection accuracy was accounted (0.985). AMMI analysis displayed significant effects on yield from ENV, GEN, and GEI (<0.0001), with the first two principal components elucidating 100 % of the variance. BR9011-67-4-1 was found the most stable cultivar based on AMMI stability analysis, with the highest AMMI-based stability parameter (ASTAB), AMMI stability value (ASV), and AMMI stability index (ASI). Based on the GGE biplot, BR9011-67-4-1 performed better in eleven environments. In the grain quality analysis, BR9011-67-4-1 had the highest amylose (27.9 %) and protein content (9.5 %) compared to BR26 and BRRI dhan48. Head rice yield positively correlated with milled rice breadth and thousand seed weight. Based on single nucleotide polymorphism (SNP) markers, BR9011-67-4-1 possessed 9 significant QTLs having grain qualities traits (amylose content, chalkiness, and grain number), abiotic stress (salt and cold tolerance, drought), and biotic stress (blast, BLB, BPH) compare to other cultivars. Finally, the genotype BR9011-67-4-1 has been released as BRRI dhan98 in the year of 2020 by 103th National Seed Board committee and its outcome highlight the importance of selecting suitable cultivar for the commercial rice production throughout Bangladesh and accounting for environmental conditions in rice breeding programs.
Keywords: Multi-environment, Yield estimation, AMMI, Stability, GGE, Trait characterization, Grain quality, SNP
Specifications Table
| Subject | Agricultural and Biological Science |
| Specific subject area | Agronomy and Crop Science |
| Type of data | Table and Figures |
| Data collection | Grain yield was calculated over a 10.2 m2 area (excluding boundary rows) at a 14 % moisture content per plot. Then yield value was subsequently transformed to tons per hectare. For grain quality measurement, samples of rice were collected individually from each plot conducted over four successive growing seasons. The R software ‘metan’ package helped the derivation of tables and figures interpreting the data by calculating yield and grain quality traits. |
| Data source location | The study was conducted over four growing seasons across 27 environments in 13 locations (Sonagazi, Rangpur, Chuadanga, Chapainawabganj, Bogura, Kushtia, Habiganj, Jashore, Gazipur, Cumilla, Rajshahi, Chottogram, Barishal) throughout Bangladesh. The study area is located within Bangladesh's geographic coordinates, ranging from 22.53753° N latitude to 91.82884° E longitude, with elevations spanning from 10 m above sea level in the Coastal South to 105 m in the North. |
| Data accessibility |
Debsharma, Sanjoy Kumer (2024), “Yield Stability data”, Mendeley Data, V1, doi: 10.17632/5nwygjgn36.1 Repository name: https://data.mendeley.com/ Data identification number: (DOI:10.17632/5nwygjgn36.1) Direct URL to data: https://data.mendeley.com/datasets/5nwygjgn36/1 |
| Related research article | Research article will be submitted to reputed journal. |
1. Value of the Data
-
•
The dataset provides an in-depth understanding of genotype-environment interactions (GEI), an important aspect of creating stable and high-yield rice cultivars. Breeders can more effectively choose genotypes that function consistently in a range of situations by comprehending these interconnections.
-
•
The breeding program is seeking to improve rice varieties that can benefit greatly from the identification of stable genotypes such as BR9011-67-4-1, which exhibits high yield across the country and has excellent grain quality traits as well as the valuable effective QTLs exposed through 20 SNP trait genotyping.
-
•
Breeders and rice growers can optimize the best variety in the irrigated and partially-irrigated areas by using the dataset's useful information on how various genotypes perform in varied environmental conditions.
-
•
The results of the study on genetic characteristics, such as heritability, selection accuracy, and stability indexes, advance our knowledge of how genes regulate rice production and quality traits. This information is crucial for developing breeding techniques and furthering genetic studies as well as genetic gain estimation.
-
•
Correlation analyses between various quality attributes (such as amylose content, protein content, cooking time, and head rice yield) offer important insights for developing future breeding initiatives that specifically aim to increase quality characters.
-
•
SNP-based trait genotyping data gives the grain quality, biotic and abiotic stress related QTLs or genes information which is utilized to detect the insects and diseases of rice plants.
-
•
Access to processed data and statistical reports promotes reproducibility and openness in research, creating a cooperative atmosphere for further research and future insights.
2. Background
Rice (Oryza sativa L.) is a staple grain consumed by almost half of the world's population that contains essential elements such as dietary fibers, vitamins, minerals, and carbohydrates [1]. In Asia, where over ninety percent of the world's rice is produced and eaten. About 3 billion Asians eat between 35 and 75 percent of their calories from rice [2]. Bangladesh is the fourth-largest rice producer country in the world, where rice production will increase to 46.90, 54.09, and 60.85 MT by 2030, 2040, and 2050, correspondingly [3]. In Bangladesh as well as throughout the world, rice production is essential to sustaining food safety. To meet the food security of the country, breeders are always focusing their efforts on producing superior materials that are better than current ones based on yield performance and consumer preferences.
In Bangladesh, rice is grown during the three distinct seasons Aus, Aman, and Boro. Aus rice is planted in April, grows by summer rains (April and May) and is harvested in the month of July and August. Aman is grown during the rainy (monsoon) season, whereas Boro is grown during the dry season with irrigation. As a result, Transplanted Aus rice is also known as partially irrigated rice, which denotes that irrigation is only necessary during the initial crop establishment and land preparation stage in April, with adequate rainfall during the growing season (mid-June to mid-August) meeting the rainfall constraint [4].
Location-specific as well as climate-smart variety development is a time-demanding issue in modern plant breeding as well as crop production [5]. The genotype-environment (G-E) interaction is also influenced by location-specific meteorological variables, which makes it challenging to identify stable and the best genotypes [6]. Furthermore, high yield with stable and acceptable grain quality enriched as well as climate-smart rice variety development is the efficient way to overcome the negative consequences of climate conditions. GGE-biplot and AMMI models [7,8] both are available for assessing G-E interactions [9]. The AMMI model generates a ``which-won-where'' pattern and winning cultivar data and their adaptableness. It also separates environment and genotype significant impacts from GEI [10], providing information on GEI [9].
It is well known that Bangladeshis enjoy dishes containing a considerable amylose content and non-sticky rice. The term “grain quality” is nebulous and might signify different things to different individuals. The type of grain and how it will be used eventually choose its quality. It encompasses a variety of attributes that are characterized by inherent (starch, fat, hardness, and protein content), and physical (kernel size, moisture content) quality attributes. A grainʼs genetic characteristics, growing season, harvest date, equipment used for harvesting and handling grains, drying system, storage management techniques, and transportation methods all influence the grain's quality attributes [11]. Trait-based SNPs are used to characterize genotypes to investigate the prevalence of advantageous alleles for multiple factors connected with biotic and abiotic challenges as well as grain quality traits. The highly efficient SNP platform has enabled the opportunity to select genotypes utilizing cutting-edge breeding strategies, including marker-assisted breeding and genomic selection [6]. The objectives of this study were to identify stable and high-yield performer cultivars that can withstand a wide range of stresses as well as partially irrigated conditions by taking into account the eating habits and preferences of farmers and consumers.
3. Data Description
The dataset describes a comprehensive analysis of the selected cultivars on yield traits to show the stable and adaptable one across 27 various environments over four successive years from 2016 to 2019. Moreover, it contains an estimation of different quality traits, encompassing with trait correlations. The article displaying this dataset includes three figures and nine tables.
3.1. Combined analysis of variance (ANOVA)
The analysis of variance for twenty-seven environments, as well as combined data, showed significant (p ≤ 0.01) variation along with the three rice cultivars for grain yield across four years assessed (Table 1). The interaction variance between genotypes × environments was obtained to be significant for the selected traits, explaining the steady performance of the cultivars across the year with a significant aspect of environmental influences.
Table 1.
Combined ANOVA of studied three rice genotypes across 27 environments in four successive years.
| SV | Df | SS | MSS | F-value | Pr (>F) |
|---|---|---|---|---|---|
| ENV | 26 | 56.37 | 2.16 | 25.27 | <0.0001 |
| REP (ENV) | 54 | 5.64 | 0.10 | 1.22 | 0.0100 |
| GEN | 2 | 28.92 | 14.46 | 168.52 | <0.0001 |
| GEI | 43 | 21.31 | 0.49 | 5.78 | <0.0001 |
| Residuals | 90 | 7.72 | 0.08 | ||
| Overall mean | 4.48 | ||||
| CV (%) | 6.54 | ||||
source of variationsSV, environmentsENV, replicationsREP, R(E) replications within environmentsREP(ENV), genotypesGEN, genotype by environment interactionGEI, coefficient of variationCV, p-value connected with an effect's F-statistic or significant at the 0.01 probability levelPr (>F).
3.2. Yield estimation of studied cultivars
Table 2 shows the mean rice grain yield, trials, and environment name in four consecutive years. Among the cultivars, BR9011-67-4-1 showed the highest yield (4.77 tha−1), whereas the lowest one was recorded for BR26 at 4.01 tha−1. In the view of genotype-environment interactions, cultivar BR9011-67-4-1 with environment C1E2 produced the highest (6.06 tha−1) yield, superseded by cultivar BR9011-67-4-1 with environment C4E4 at 5.86 tha−1 and environment C4E9 at 5.68 tha−1.
Table 2.
Average grain yield, environment name, and code of three rice genotypes in twenty-seven testing environments.
| Trials name | Environmental Code | Environment name | Average yield (tha−1) |
||
|---|---|---|---|---|---|
| Name of genotypes | |||||
| BR9011-67-4-1 | BR26 | BRRI dhan48 | |||
| Preliminary Yield Trial (PYT), T. Aus 2016 | C1E1 | Gazipur | 4.97 | 3.50 | 4.12 |
| C1E2 | Rajshahi | 6.06 | 4.16 | 5.46 | |
| C1E3 | Rangpur | 4.77 | 3.22 | 3.93 | |
| C1E4 | Cumilla | 4.20 | 3.63 | 5.30 | |
| Regional Yield Trial (RYT), T. Aus 2017 | C2E1 | Gazipur | 4.17 | 4.43 | 4.91 |
| C2E2 | Rajshahi | 4.20 | 4.63 | 4.93 | |
| C2E3 | Rangpur | 5.40 | 4.15 | 4.90 | |
| C2E4 | Cumilla | 4.15 | 3.53 | 4.27 | |
| C2E5 | Sonagazi | 4.07 | 3.80 | 4.17 | |
| C2E6 | Kushtia | 3.60 | 4.37 | 4.93 | |
| C2E7 | Habiganj | 5.60 | 4.83 | 5.53 | |
| Advanced Lines Adaptive Research Trial (ALART), T. Aus 2018 | C3E1 | Gazipur | 5.27 | 4.12 | 4.47 |
| C3E2 | Chuadanga | 4.84 | 4.51 | 5.02 | |
| C3E3 | Chottogram | 4.21 | 3.75 | 4.14 | |
| C3E4 | Sonagazi | 4.87 | 3.51 | 4.14 | |
| C3E5 | Jashore | 4.15 | 3.48 | 4.84 | |
| C3E6 | Chapainawabganj | 4.78 | 3.23 | 4.63 | |
| C3E7 | Rangpur | 4.32 | 3.02 | 4.17 | |
| Proposed Variety Trial (PVT), T. Aus 2019 |
C4E1 | Bogura | 5.07 | 4.50 | – |
| C4E2 | Rangpur | 4.00 | 3.23 | – | |
| C4E3 | Sonagazi | 5.07 | 4.43 | – | |
| C4E4 | Cumilla | 5.86 | 4.90 | – | |
| C4E5 | Barishal | 4.49 | 3.72 | – | |
| C4E6 | Gazipur | 5.62 | 4.74 | – | |
| C4E7 | Jashore | 3.77 | 3.40 | – | |
| C4E8 | Habiganj | 5.62 | 4.71 | – | |
| C4E9 | Rajshahi | 5.68 | 4.69 | – | |
| Overall mean | 4.77 | 4.01 | 4.66 | ||
| CV | 6.54 | ||||
C denotes codes; E denotes environments.
3.3. Measurement of genetic parameters
The evaluation of genetic variability and genetic correlations among the yield-related traits and grain quality parameters enhances the existing breeding program which is greatly influenced by both genetic and environmental factors [12]. Different genetic parameters of three rice cultivars across 27 environments in four consecutive years are outlined through linear mixed model (LMM) in R software (Table 3). Among the variance estimates, genotypic variance revealed the highest percentage at 0.429 %, followed by genotypic variance at 0.200 %, and residual variance at 0.068 %. The broad-sense heritability was approximately medium at 0.481 %. The coefficient of determination for interaction effects (GEIr2) was measured at 0.319. Additionally, the heritability of means (h2 mg) stood at 0.971, representing high selection accuracy at 0.985. Furthermore, the genotype-environment correlation (rge) was detected to be high at 0.614, whereas the ratio of genotypic to the residual coefficient of variation was 1.55.
Table 3.
Genetic characteristics utilizing Linear mixed Models (LMM) of grain yield (tha−1) evaluated under 27 environments.
| Characteristics | Values estimated from grain yield data |
|---|---|
| Phenotypic variance | 0.429 |
| Genotypic variance | 0.200 |
| Residual variance | 0.068 |
| Heritability | 0.481 |
| Selection accuracy | 0.985 |
| h2 mg | 0.971 |
| GEIr2 | 0.319 |
| rge | 0.614 |
| CVg | 10.1 |
| CVr | 6.54 |
| CV ratio | 1.55 |
genotype by environment interactionGEI, GEI correlation of coefficientGEIr2, broad sense heritability of meanh2mg, genotype-environment correlationrge, genotypic coefficient of variationCVg, residual coefficient of variationCVr.
3.4. AMMI based variance analysis
Table 4 explains the AMMI analysis of variance, describing the effects of different factors on the percent total sum of squares (PTSS) exhibited through the grain yield dataset. The variance analysis revealed significant effects on grain yield from ENV, GEN, and GEI (<0.0001). The greater portion of the variation in the model was contributed to ENV (39.87 %), followed by GEN (20.46 %). Moreover, AMMI variance separated GEI into the first two principal components (PCs) along with a complement of 79.6 % and 20.4 % of the total percentages of GEI. The presence of a greater proportion of GEI necessitates a stable analysis of the rice cultivar in various environments (13).
Table 4.
AMMI ANOVA for rice grain yield for three genotypes studied across 27 environments.
| SV | DF | SS | MSS | PTSS | F-value | P-value (>F) | PC variance | AV of PC |
|---|---|---|---|---|---|---|---|---|
| ENV | 26 | 56.36 | 2.16 | 39.87 | 20.76 | <0.0001 | – | – |
| REP (ENV) | 54 | 5.63 | 0.1 | 3.99 | 1.21 | 0.0103 | – | – |
| GEN | 2 | 28.92 | 14.46 | 20.46 | 168.51 | <0.0001 | – | – |
| GEI | 43 | 21.31 | 0.49 | 15.08 | 5.77 | <0.0001 | – | – |
| PC1 | 27 | 17.02 | 0.63 | 12.05 | 7.34 | <0.0001 | 79.6 | 79.6 |
| PC2 | 25 | 4.37 | 0.17 | 3.09 | 2.04 | <0.0001 | 20.4 | 100 |
| Residuals | 90 | 7.72 | 0.08 | 5.46 | – | – | – | – |
| Total | 267 | 141.34 | 0.52 | 100.00 | – | – | – | – |
source of variationsSV, environmentsENV, replicationsREP, R(E) replications within environmentsREP(ENV), genotypesGEN, genotype by environment interactionGEI, principal componentPC, degrees of freedomDF, sum of squaresSS, mean sum of squaresMSS, Percentage total sum of the squarePTSS, Accumulated varianceAV, p-value connected with an effect's F-statistic or significant at the 0.01 probability level.
3.5. Stability statistics based on AMMI analysis
Table 5 illustrates the AMMI stability stats for grain yield on rice across 27 environments, concentrating on three selected cultivars. The ASTAB value was observed higher in cultivar BR9011-67-4-1 (1.56) and lower in BRRI dhan48 (0.88). BR9011-67-4-1 showed the highest ASI value (0.98) and ASV (4.81), whereas BRRI dhan48 exhibited the lowest one in both traits (ASI: 0.36 and ASV: 1.78). Cultivar BR9011-67-4-1 also shown the highest AVAMGE (7.47), DA (1.92), FA (3.67), MASI (0.98), MASV (4.81), ZA (0.67), and WAAS (1.01). The DZ and EV values remained constant for all cultivars at 0.81 and 0.33, respectively. Furthermore, the SIPC value was highest in BR26 (1.51) and lowest in BRRI dhan48 (1.25).
Table 5.
AMMI-based stability statistics of grain yield of the selected cultivars.
| GEN | ASTAB | ASI | ASV | AVAMGE | DA | DZ | EV | FA | MASI | MASV | SIPC | ZA | WAAS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BR9011-67-4-1 | 1.56 | 0.98 | 4.81 | 7.47 | 1.92 | 0.81 | 0.33 | 3.67 | 0.98 | 4.81 | 1.41 | 0.67 | 1.01 |
| BR26 | 1.15 | 0.67 | 3.32 | 6.09 | 1.48 | 0.81 | 0.33 | 2.20 | 0.67 | 3.32 | 1.51 | 0.55 | 0.80 |
| BRRI dhan48 | 0.88 | 0.36 | 1.78 | 3.97 | 1.12 | 0.81 | 0.33 | 1.26 | 0.36 | 1.78 | 1.25 | 0.36 | 0.49 |
AMMI based stability parameterASTAB, AMMI stability indexASI, AMMI stability valueASV, sum across environments of absolute value of GEI modelled by AMMIAVAMGE, annicchiarico's D parameter valuesDA, Zhang's D parameterDZ, sums of the averages of the squared eigenvector valuesEV, stability measure based on fitted AMMI modelFA, modified AMMI stability indexMASI, modified AMMI stability valueMASV, sums of the absolute value of the IPC scoresSIPC, absolute value of the relative contribution of IPCs to the interactionZA, weighted average of absolute scoresWAAS, genotypesGEN.
3.6. Multi-environment analysis based on GGE biplot
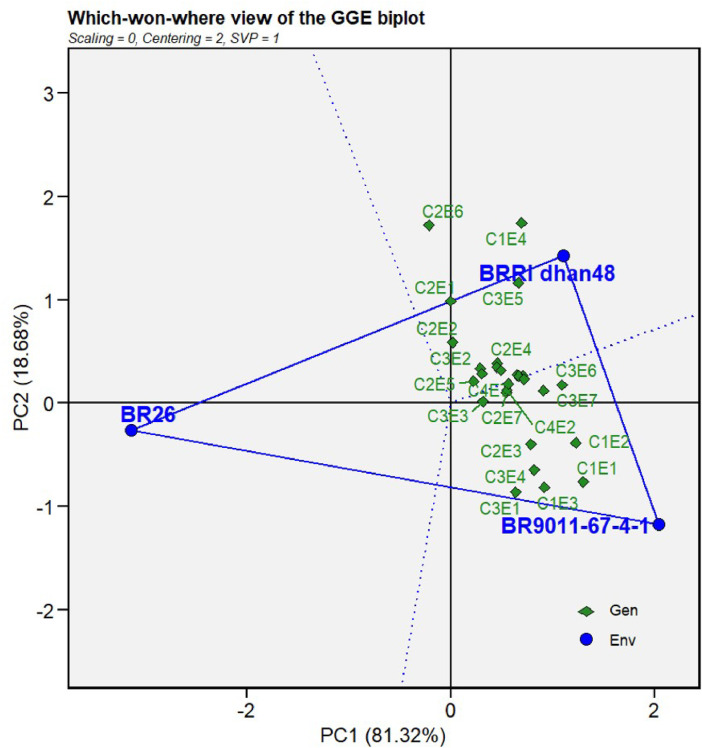
Fig. 1 illustrates the GGE biplot presenting the high-performing cultivars in their particular environments. In the biplot, a polygon is designed by linking the main genotypes (BR9011-67-4-1, BR26, and BRRI dhan48). A set of dotted lines that start at the plot's origin and extend perpendicular to the polygon's sides divides it into three sectors. Among the studied 27 environments, cultivar BR9011-67-4-1 was showed excellent performance in eleven environments (C1E1, C12E2, C1E3, C2E3, C2E7, C3E1, C3E3, C3E4, C3E6, C3E7 and C4E2). BRRI dhan48 exhibited well adaptability in eight environments (C1E4, C2E1, C2E2, C2E4, C2E5, C2E6, C3E2, and C3E5), whereas none of the environments showed superiority for BR26.
Fig. 1.
Which-won-where view for mega-environment analysis based on GGE biplot during four consecutive years under 27 environments.
3.7. Grain quality analysis among the studied traits
Table 6 describes the physio-chemical and milling properties of rice grain of the three studied cultivars. BR9011-67-4-1 and BR26 showed the MO at 66.4 % and 66.9 %, respectively. Cultivar BR9011-67-4-1 exhibited the HRY at 51.7 %. BR9011-67-4-1 had observed high values for MRL at 6.5 mm and BRRI dhan48 showed high for MRB at 2.3 mm. BR26 showed the highest value to LRB at 3.8. The TSW value was the highest for BRRI dhan48 at 23.3g. BR9011-67-4-1 had the highest ASPV at 4.0, followed by BR26 (3.3). BR9011-67-4-1 revealed the highest AMY and PRO at 27.9 % and 9.5 %, respectively with the lowest CT at 15.6 min. BR9011-67-4-1 exposed the lowest ER and IMR at 1.2 and 2.9, respectively.
Table 6.
Average values for studied grain quality parameters of the three cultivars.
| Cultivars | MO | HRY | MRL | MRB | LBR | TGW | ASPV | AMY | PRO | CT | ER | IMR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BR9011-67-4-1 | 66.4 | 51.7 | 6.5 | 2.0 | 3.3 | 22.6 | 4.0 | 27.9 | 9.5 | 15.6 | 1.2 | 2.9 |
| BR26 | 66.9 | 40.9 | 6.2 | 1.6 | 3.8 | 21.9 | 3.3 | 23.0 | 9.3 | 18.5 | 1.4 | 4.0 |
| BRRI dhan48 | 69.9 | 61.4 | 5.9 | 2.3 | 2.7 | 23.3 | 3.4 | 26.7 | 8.3 | 18.8 | 1.4 | 3.3 |
| RV | 5.82 | 1.39 | 0.03 | 0.01 | 0.00 | 0.12 | 0.08 | 0.04 | 0.21 | 0.04 | – | – |
| GM | 67.77 | 51.32 | 6.19 | 1.97 | 3.24 | 23.04 | 3.59 | 25.88 | 9.03 | 17.61 | – | – |
| LSD(0.05) | 5.47 | 2.68 | 0.38 | 0.19 | 0.15 | 0.79 | 0.63 | 0.44 | 1.03 | 0.43 | – | – |
| CV | 3.56 | 2.30 | 2.69 | 4.15 | 2.05 | 1.52 | 7.77 | 0.75 | 5.05 | 1.08 | – | – |
milling outturn(%)MO, head rice yield(%)HRY, milled rice length(mm)MRL, milled rice breadth(mm)MRB, length breath ratioLBR, thousand seed weight (g)TGW, alkali spreading valueASPV, amylose(%)AMY, protein(%)PR0, cooking time(min)CT, elongation ratioIR, imbibition ratioIMR, residual varianceRV, grand meanGM, least significant differenceLSD, coefficient of variationsCV.
3.8. Correlation among the studied traits based on grain quality data
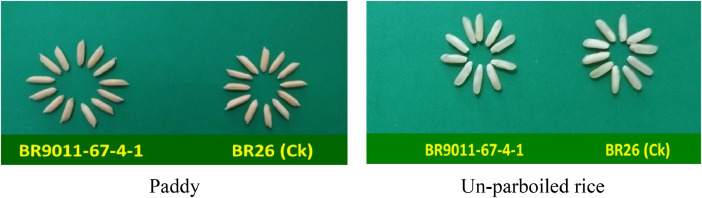
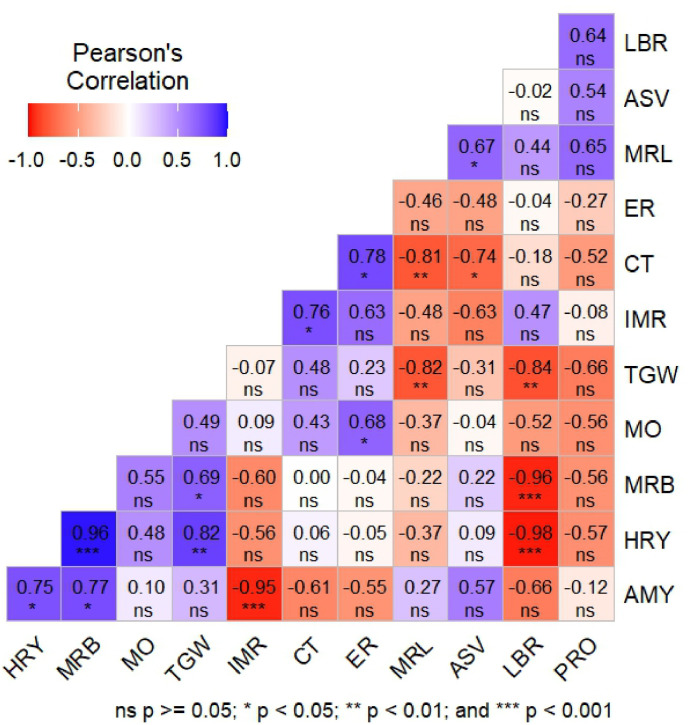
The original paddy and un-parbolied rice sample of the studied genotype BR9011-67-4-1 (BRRI dhan98) and the check variety BR26 are presented (Fig 3). For better understanding, the correlation analysis was done among the grain yield and yield-related twelve traits of the selected genotypes. In the Fig 2, AMY had a positive and significant relationship (p ≤ 0.05) with HRY and MRB, but had negative significant (p ≤ 0.001) association with IMR. HRY had a positively significant (p ≤ 0.001) correlation with MRB and TGW, whereas found to the negatively significant (p ≤ 0.001) with LBR. MRB also observed a negative significance (p ≤ 0.001) with LBR. TGW had a negative significance (p ≤ 0.01) with LBR and MRL. CT displayed a significantly negative association with MRL and ASV.
Fig. 3.
The pictorial view of grain quality parameters of the studied genotype and the check. The breeding lines BR9011-67-4-1 (BRRI dhan98) and BR26 (check) are represented in the figure as a paddy, whereas unparboiled rice was dehusked without boiling.
Fig. 2.
Visualization of correlation matrix utilizing Pearson's correlation coefficient, presents the association between different grain-related traits of the studied three cultivars across 27 environments (milling outturn(%)MO, head rice yield(%)HRY, milled rice length(mm)MRL, milled rice breadth(mm)MRB, length breath ratioLBR, thousand seed weight(g)TGW, alkali spreading valueASPV, amylose(%)AMY, protein(%)PR0, cooking time(min)CT, elongation ratioIR, imbibition ratioIMR).
3.9. Trait characterization of the studied three cultivars using QTLs-specific SNP markers
Table 7 shows three studied cultivars were genotyped and calculated twenty QTLs-specific SNP markers to the chosen traits by dint of grain quality [Wx-A_group {snpOS00445 (C)}, Wx-GBSSex10 {snpOS00038 (T)} for amylose content; chalk5_576 {snpOS00024 (G)} for chalkiness; Gn1a_1 {snpOS00396 (T)} for grain number; Saltol-Aus: snpOS0994 (T) confers salt tolerance [13]; qSCT1_4: snpOS0004 (C) confers cold tolerance; Pita: snpOS00006 (C) for rice blast; Xa4: snpOS00052 (A), xa13: snpOS00002 (G), Xa21_SKEP: snpOS00061 (C) for bacterial leaf blight; Bph17_1: snpOS00068 (T), BPH32: snpOS00442 (G), BPH3: snpOS00065 (T) for brown plant hopper] from the Intertek (https://www.intertek.com/agriculture/agritech/ (accessed on 18 February 2023) outsourcing provider. Based on SNP favorable data, BR9011-67-4-1 reveal 9 QTLs, BR26 concealed 5 QTLs and BRRI dhan48 bear 2 QTLs that circulates the trait of interest.
Table 7.
List of the QTL-based SNP markers with favorable allele utilized to illustrate three cultivars for the different traits of aspect.
| Traits | QTLs | QTL-based SNP with favorable allele | Cultivars |
||
|---|---|---|---|---|---|
| BR9011-67-4-1 | BR26 | BRRI dhan48 | |||
| Grain quality | |||||
| Amylose content | Wx-A_group | snpOS00445 (C) | C:C | T:T | C:C |
| Wx-GBSS-ex10 | snpOS00038 (T) | T:T | C:C | T:T | |
| Chalkiness | chalk5_576 | snpOS00024 (G) | G:G | G:G | A:A |
| Grain number | Gn1a_1 | snpOS00396 (T) | T:T | T:T | T:T |
| Abiotic stress | |||||
| Salt tolerance | Saltol-Aus | snpOS0994 (T) | T:T | – | G:G |
| Cold tolerance | qSCT1_4 | snpOS0004 (C) | C:C | – | T:T |
| Drought | DTY12-1_2 | IRRI_SNP1081 (A) | A:A | – | G:G |
| DTY3-2-IR64_1 | IRRI_SNP1016 (A) | A:A | – | A:A | |
| Biotic stress | |||||
| Blast | Pi-ta | snpOS00006 (C) | C:C | C:C | – |
| BLB | Xa4 | snpOS00052 (A) | A:A | A:A | – |
| Xa21_SKEP | snpOS00061 (C) | G:G | G:G | G:G | |
| Xa13 | snpOS00002 (G) | A:A | A:A | A:A | |
| BPH | Bph17_1 | snpOS00068 (T) | T:T | T:T | – |
| BPH32 | snpOS00442 (G) | G:G | – | – | |
| BPH3 | snpOS0065 (T) | T:T | C:C | – | |
| Number of desirable traits present in each genotype | 9 | 5 | 2 | ||
4. Experimental Design, Materials and Methods
4.1. Plant genetic materials
The investigation contained two rice cultivars (BR26, BRRI dhan48) and one elite breeding line BR9011-67-4-–1. After the four years yield evaluation, the National Seed Board committee has been approved this genotype BR9011-67-4-1 as a BRRI dhan98 in the year of 2020 for the commercial cultivation throughout the country of Bangladesh.
Table 8 shows information on the pedigree name, parentages, released year, salient characteristics of the considered rice genotypes.
Table 8.
Salient characteristics on the basis of distinctness, uniformity and stability (DUS) of studied three rice cultivars [14].
| Cultivars and pedigree name | Parentages | Year of release | Salient characteristics |
|---|---|---|---|
| BR9011-67-4-1 (BRRI dhan98) | MLT-145-2/ HR17512-11-2-3-1-4-2-3 | 2020 | Flag leaf (attitude of blade): Semi-erect Panicle exertion: Moderately exerted Grain length (without dehulling): Long Leaf senescence: Late & slow Polished grain (size of white core or Chalkiness): Small Endosperm (content of amylose): High |
| BR26 (IR44595-70-2-2-3) | IR18348-36-3 3/IR25863-61-3-2//IR58 | 1993 | Flag leaf (attitude of blade): Erect Panicle exertion: Well exerted Grain length (without dehulling): Very long Leaf senescence: Intermediate Polished grainsize of white (core or Chalkiness): Absent Endosperm (content of amylose): Intermediate |
| BRRI dhan48 (BR5563-3-3-4-1) | BR1543-9-2-1/IR13249-49-3-2-2 | 2008 | Flag leaf (attitude of blade): Erect Panicle exertion: Moderately exerted Grain length (without dehulling): Long Leaf senescence: Late & slow Polished grain (size of white core or Chalkiness): Absent Endosperm (content of amylose): High |
4.2. Tested environments
The different trials were conducted over four years from 2016 to 2019 in 13 locations (Sonagazi, Rangpur, Chuadanga, Chapainawabganj, Bogura, Kushtia, Habiganj, Jashore, Gazipur, Cumilla, Rajshahi, Chottogram, Barishal) across the country, involving four locations for the Preliminary Yield Trial (PYT), seven for the Regional Yield Trial (RYT), seven for the Advanced Lines Adaptive Research Trial (ALART), and nine for the Proposed Variety Trial (PVT) (Table 2). Table 9 displays average rainfall along with maximum and minimum temperatures (April to July month) of the four successive years from 2016 to 2019 across the different locations in Bangladesh. Fig. 4 exhibits geographical view of the study area over four successive growing seasons across Bangladesh.
Table 9.
Average rainfall, maximum and minimum temperatures (April to July month) data of the four successive years across the different locations in Bangladesh.
| Locations | Rainfall (mm) |
Max. Temperature (°C) |
Min. Temperature (°C) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2016 | 2017 | 2018 | 2019 | 2016 | 2017 | 2018 | 2019 | 2016 | 2017 | 2018 | 2019 | |
| Barishal | 28.7 | 30.8 | 37.2 | 46.8 | 29.4 | 29.8 | 30.4 | 30.6 | 16.8 | 17.0 | 17.7 | 17.2 |
| Bogura | 54.9 | 20.8 | 47.6 | 23.6 | 28.1 | 28.3 | 28.6 | 29.7 | 16.8 | 16.9 | 17.0 | 17.6 |
| Chottogram | 30.2 | 44.5 | 26.4 | 21.4 | 28.4 | 28.9 | 29.6 | 32.3 | 18.6 | 18.8 | 19.5 | 19.3 |
| Cumilla | 84.2 | 46.3 | 58.2 | 84.2 | 28.1 | 28.6 | 28.8 | 29.4 | 18.0 | 18.1 | 18.2 | 17.2 |
| Chapainawabganj | 38.2 | 28.5 | 24.1 | 56.3 | 28.7 | 29.1 | 29.9 | 29.9 | 16.0 | 15.9 | 16.0 | 16.1 |
| Chuadanga | 18.9 | 42.3 | 27.5 | 34.8 | 27.9 | 28.1 | 28.6 | 28.8 | 15.8 | 16.0 | 16.0 | 15.8 |
| Habiganj | 62.9 | 44.8 | 68.8 | 51.8 | 28.5 | 28.8 | 29.2 | 29.8 | 15.0 | 15.2 | 15.9 | 15.1 |
| Jashore | 24.3 | 18.9 | 50.2 | 87.2 | 29.4 | 29.9 | 31.0 | 30.7 | 16.2 | 16.4 | 16.9 | 17.4 |
| Gazipur | 77.8 | 50.8 | 83.1 | 91.7 | 28.9 | 29.2 | 29.8 | 30.2 | 18.2 | 18.6 | 18.9 | 19.2 |
| Sonagazi | 56.9 | 52.3 | 60.2 | 64.4 | 29.3 | 29.9 | 30.2 | 30.7 | 18.0 | 17.9 | 18.2 | 17.6 |
| Rangpur | 23.4 | 33.7 | 41.0 | 42.2 | 27.5 | 27.8 | 27.4 | 28.3 | 15.8 | 15.9 | 16.0 | 16.3 |
| Kushtia | 18.9 | 37.9 | 28.8 | 36.3 | 29.8 | 30.2 | 31.0 | 31.4 | 16.0 | 16.2 | 16.4 | 16.2 |
| Rajshahi | 25.6 | 36.5 | 23.3 | 19.2 | 29.9 | 30.1 | 30.9 | 31.2 | 16.0 | 16.1 | 16.3 | 16.5 |
Fig. 4.
Geographic map of the 13 studied locations over four successive growing seasons across Bangladesh. The pink colour designates the studied area where experimental trials were conducted.
4.3. Experimental layout
Eighteen to twenty-two-days aged seedlings were transplanted at a spacing of 20 cm × 15 cm, with two to three seedlings per hill. Each plot encompassed 10 rows, gauging 5.4 m in length, calculating in a total area of 10.8 m2. The layout of experiment applied a Randomized Complete Block Design (RCBD), replicated 3 times in the cultivated land. Each location and each research year, this experimental procedure was followed for the study.
4.4. Crop cultivation and management
The trials were done during the partially irrigated transplanted aus season over consecutive four years. The studied cultivars were soaked, sown, and transplanted at various experimental locations timely in the proper way. The sowing was done from mid-March to the first week of April, followed by transplanting from mid-April to the last week of April and harvesting through the mid-June to last week of June. The fertilizers were evenly distributed in the following ratios: 190:50:75:40:5 kg/ha of urea, TSP, MoP, gypsum, and ZnSO4 [15]. Except urea, which was handed out in three equal doses at final land preparation, 4–5 tillering stage, and 5–7 days before the panicle initiation stage after transplanting, other fertilizers were applied as basal. When necessary, weeding and irrigation were done following BRRI's suggested management techniques, and suitable pest control measures were put in place. Crop management tasks, such as weeding and watering, were carefully carried out under a schedule. An effective plan for managing diseases and pests was put into place [16].
4.5. Data collection and processing
The grain yield data that was assembled from measured 10.2 m2 plot size (excluding boundary rows). Every harvested plot had two consecutive rows along the borders removed to lessen the influence of border effects. Data on grain yield was collected per plot at a moisture content of 14 %. After that, the data was converted to tons ha−1. The data-collecting process followed the standard protocol described by Yoshida et al. [17].
4.6. Grain quality and chemical analysis of tested cultivars
Bangladesh Rice Research Instituteʼs Grain Quality and Nutrition (GQN) Division laboratory, situated in Gazipur, Bangladesh, served as the study's location. A set of rice samples was collected from studies conducted over three consecutive years. To evaluate the grain quality, the obtained sample was then divided into three segments (three replications). The grains in the collected samples were perfect, unbroken, and free of any disease, discoloration, insect infestation, or fractures. Important guidelines for conducting this investigation were provided by Dipti et al. [18]. Weight per thousand grains was determined from a total of one thousand unbroken grains, damaged or infested grains were excluded. The percentages of milled rice and head rice were calculated by manually and painstakingly sorting broken rice from whole grains. The length and width of the milled rice were measured using slide calipers, with millimeters serving as the unit of measurement. The length (L) was measured and divided by the breadth (B) to get the length-to-width ratio. The iodine-binding approach, which was recommended by Kongseree and Juliano [19], was used to determine the amylose content. According to Dipti et al. [18], the protein content was calculated by multiplying the nitrogen concentration by a factor of 5.95. Based on the point in time at which 90 % of the rice had undergone gelatinization, the cooking time was determined. The elongation ratio which is calculated by dividing the length of cooked rice by the length of uncooked rice, and the imbibition ratio which is derived from the water displacement technique, were both measured. The volume change that occurred when cooked rice was added to a graduated cylinder filled with water was measured in order to calculate the imbibition ratio [18].
4.7. Statistical analysis through R
The raw data from each replication was averaged to determine the replication mean. The average results for numerous traits were analyzed statistically and biometrically. Employing R software, the effects of genotypes (G), environment (E), and genotype-environment interactions (GEI) were investigated using a combined analysis of variance (ANOVA) and mean separation. The R “metan” package was used to carry out the genotype-genotype environment (GGE) biplot analysis, AMMI stability statistics, and AMMI ANOVA [20]. Utilizing the same package, Pearson's correlation coefficients were also computed.
Acknowledgments
Limitations
This dataset has some limitations. It was wise to include each agronomic character that contributes to yield in the dataset on grain yield, to observe the relationships between yield and its contributing yield qualities, which covers four growing years and 27 environments.
Ethics Statement
Our study satisfies the ethical guidelines for publication in Data in Brief, which have been read and followed by all authors. We don't conduct research on humans or animals in our work.
CRediT Author Statement
Sanjoy K. Debsharma: Conceptualization, Methodology, Investigation, Supervision, Software, Data curation, Formal analysis, Visualization, Writing - Original draft preparation, Reviewing and Editing; Mahmuda Khatun: Conceptualization, Methodology, Investigation, Supervision, Validation, Writing - Original draft preparation, Reviewing and Editing; Fahamida Akter, Md. Abu Syed, Mohammad Rafiqul Islam, Md. Rokebul Hasan, Md. Adil, Md. Ruhul Amin Sarker, Biswajit Karmakar: Investigation, Data curation, Validation, Writing - Reviewing and Editing.
Acknowledgments
The authors are grateful to Bangladesh Rice Research Institute for supplying planting materials for the research. No specific grant was taken for this research by public, private, or non-profit funding organizations.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
Data Availability
Mendeley DataYield Stability data (Original data).
References
- 1.FAO, FAOSTAT, Crops and livestock products, (2023).
- 2.Khush G.S. What it will take to feed 5.0 billion rice consumers in 2030. Pl. Mol. Biol. 2005;59:1–6. doi: 10.1007/s11103-005-2159-5. [DOI] [PubMed] [Google Scholar]
- 3.Kabir M.S., Salam M.U., Islam A.K., Sarkar M.A., Mamun M.A., Rahman M.C., Nessa B., Kabir M.J., Shozib H.B., Hossain M.B., Chowdhury A. Doubling rice productivity in Bangladesh: a way to achieving SDG 2 and moving forward. Bang. Rice J. 2020;24(2):1–47. [Google Scholar]
- 4.Khatun M., Ahmed M., Syed M., Akter F., Das S., Haq M., Dipti S. Identification of Ideal Trial Sites and Wide Adaptable T. Aus Rice Genotypes Suitable for Bangladesh. Bang. Rice J. 2020;23(2):77–85. doi: 10.3329/brj.v23i2.48250. [DOI] [Google Scholar]
- 5.Debsharma S.K., Roy P.R., Begum R.A., Iftekharuddaula K.M. Elucidation of genotype × environment interaction for identification of stable genotypes to grain yield of rice (Oryza sativa L.) varieties in Bangladesh rainfed condition. Bang. Rice J. 2020;24(1):59–71. doi: 10.3329/brj.v24i1.53240. [DOI] [Google Scholar]
- 6.Debsharma S.K., Rahman M.A., Khatun M., Disha R.F., Jahan N., Quddus M.R. Developing climate-resilient rice varieties (BRRI dhan97 and BRRI dhan99) suitable for salt-stress environments in Bangladesh. PLoS ONE. 2024;19(1) doi: 10.1371/journal.pone.0294573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zobel R.W., Wright M.G., Gauch H.G. Statistical analysis of a yield trial. Agron. J. 1988;80:388–393. [Google Scholar]
- 8.Crossa J., Gauch H.G., Zobel R.W. Additive main effects and multiplicative interaction analysis of two international maize cultivar trials. Crop Sci. 1990;30:493–500. [Google Scholar]
- 9.Yan W.K., Kang M.S., Ma B., Woods S., Cornelius P.L. GGE Biplot vs. AMMI analysis of genotype-by-environment data. Crop Sci. 2007;47:643–655. [Google Scholar]
- 10.Gauch H.G., Piepho H.P., Annicchiarico P. Statistical analysis of yield trials by AMMI and GGE: further considerations. Crop Sci. 2008;48:866–889. [Google Scholar]
- 11.Chen Y., Wang M., Ouwerkerk P.B. Molecular and environmental factors determining grain quality in rice. Food Energy Security. 2012;1(2):111–132. doi: 10.1002/fes3.11. [DOI] [Google Scholar]
- 12.Debsharma S.K., Syed M.A., Ali M.H., Maniruzzaman S., Roy P.R., Brestic M., Gaber A., Hossain A. Harnessing on genetic variability and diversity of rice (Oryza sativa L.) genotypes based on quantitative and qualitative traits for desirable crossing materials. Genes. 2023;14:10. doi: 10.3390/genes14010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.S.K. Debsharma, M.A. Rahman, M.R. Quddus, H. Khatun, R.F. Disha, P.R. Roy, S. Ahmed, M. El-Sharnouby, K.M. Iftekharuddaula, S. Aloufi, SNP based trait characterization detects genetically important and stable multiple stress tolerance rice genotypes in salt-stress environments, Plants 11 (2022) 1150, doi: 10.3390/plants11091150. [DOI] [PMC free article] [PubMed]
- 14.P.R. Roy, S.K. Debsharma, M.S. Islam, S.E. Jamal, F. Jenny, R. Parveen, M. Salahuddin, M.E. Rabbi., Distinctness, Uniformity and Stability (DUS) Test Characteristics of BRRI Developed 100 Rice Varieties (BR1 to Bangabandhu dhan100), Seed Certification Agency (SCA), Ministry of Agriculture, Gazipur-1701, Bangladesh (2022) 125. www.sca.gov.bd.
- 15.BARC, In: Fertilizer Recommendation Guide 2018, Bangladesh Agricultural Research Council (BARC), Farmgate, Dhaka (2018) 1215.
- 16.Bangladesh Rice Research Institute, Modern Rice Cultivation, 24th ed., Bangladesh Rice Research Institute, Gazipur, Bangladesh, 2022.
- 17.S. Yoshida, Routine procedures for growing rice plants in culture solution. Laboratory manual for physiological studies of rice, (1976).
- 18.Dipti S., Bari M.N., Kabir K.A. Grain quality characteristics of some Beruin rice varieties of Bangladesh. Pak. J. Nutr. 2003;2(4):242–245. [Google Scholar]
- 19.Kongseree N., Juliano B.O., Kongseree N. Physicochemical properties of rice grain and starch from lines differing amylose content and gelatinization temperature. J. Agric. Food Chem. 1972;20:714–718. doi: 10.1021/jf60181a020. [DOI] [Google Scholar]
- 20.R.Core Team, The R Project for Statistical Computing, Vienna, Austria, (2022), https://www.R-Project.Org/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Mendeley DataYield Stability data (Original data).