Summary
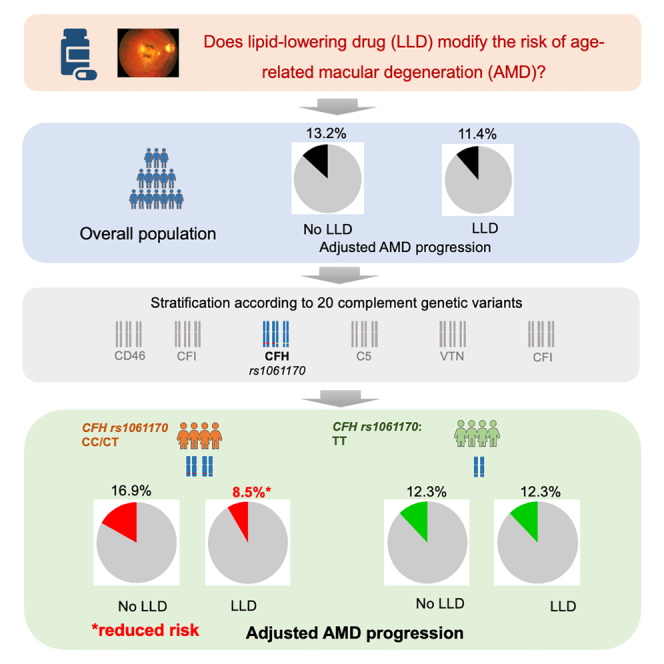
We investigated whether the effect of lipid-lowering drugs (LLDs) on age-related macular degeneration (AMD) differs according to the main complement genetic variants in Singapore Epidemiology of Eye Diseases (SEED) (n = 5,579) and UK Biobank studies (n = 445,727). The effect of LLD was determined for each stratum of 20 complement genetic variants. In SEED, 484 individuals developed AMD and 216 showed progression over 6 years. In the UK Biobank, 913 participants developed AMD over 11 years. rs1061170 variant (complement factor H gene) was the only variant for which we found a protective effect in both populations. This effect was found in individuals carrying at least one C allele in SEED (odds ratio [OR] = 0.41; 95% confidence interval [CI], 0.19–0.87) and in individuals carrying two C alleles in UK Biobank (hazard ratio [HR] = 0.65; 95% CI, 0.45–0.93). These effects corresponded to a 50% and 35% decrease in AMD risk, respectively. Our study highlights the potential for personalized therapy for AMD based on complement genotyping.
Subject areas: Health sciences, Medicine, Medical specialty, Internal medicine, Cardiovascular medicine, Pharmacology
Graphical abstract
Highlights
-
•
Lipid-lowering drugs protect against age-related macular degeneration progression
-
•
This effect is specific to individuals with C allele of the rs1061170 variant in CFH gene
Health sciences; Medicine; Medical specialty; Internal medicine; Cardiovascular medicine; Pharmacology
Introduction
Age-related macular degeneration (AMD) is a chronic, progressive disease causing severe and irreversible vision loss in the elderly.1 It impacts nearly 200 million individuals globally,2 accounting for 15%–20% of irreversible vision loss cases in individuals aged 50 and over in Europe and North America.3 The burden of AMD will increase further with the aging population. Current treatments, i.e., the anti-vascular endothelial growth factor agents, primarily stabilize vision in wet AMD,1 while treatment options to prevent progression from the early or intermediate to the sight-threatening late stage of AMD remain limited.
Numerous studies have suggested that lipid metabolism plays a significant role in the pathophysiology of AMD,4,5,6,7 indicating that lipid-lowering drug (LLD) may be a potential treatment strategy for AMD. However, both clinical and non-clinical studies, including meta-analyses, have reported inconsistent findings regarding the association between LLD and AMD, with some showing beneficial effect of LLD,8,9,10,11 while others showing no effect.12,13,14,15,16,17 Clarifying the effect of LLD on AMD is important for improving treatment strategies for AMD.
We hypothesized that the complement system activity, driven by genotype polymorphism, might modify the effect of LLD on AMD and thus explain the discrepancy in current evidence. Firstly, complement system can regulate lipid metabolism by modulating the inflammatory properties of lipoproteins.18 Secondly, strong associations between measurements made on systemic complement activation with lipoprotein sub-fractions have been observed.19,20 Therefore, in this prospective study, including participants from Singapore Epidemiology of Eye Diseases (SEED) and the UK Biobank studies, we aimed to determine whether the effect of LLD on AMD incidence and progression differs according to the main complement genetic variants and to explore how lipoprotein sub-fractions could mediate this effect.
Results
Characteristics of participants included in the analysis
In SEED, among the 10,033 participants recruited at baseline, 6,762 had follow-up visits. We excluded 1,154 participants without AMD grading at baseline and/or follow-up (no or ungradable fundus images). Among the remaining 5,608 participants, 1,251 (22.3%) took LLD at baseline (29 did not have this information available). The medications were mainly statin (n = 1,136; 90.8%) and fibrates (n = 102; 8.2%), the remaining were either labeled other LLD (n = 29; 2.3%) or medication not specified (n = 2; 0.2%). The characteristics of the individuals taking LLD showed significant difference with individuals not taking these medications (Table 1). Individuals taking LLD were older and more likely to have hypertension, diabetes, and cardiovascular disease (CVD). The absolute standardized difference (ASD) for these four variables were higher than 0.50 (Table 1), suggesting that classical regression adjustment might not be trustworthy, and thus advocating for the use of a propensity score approach.21
Table 1.
Characteristics of the Singapore Epidemiology of Eye Diseases study participants according to lipid-lowering drug
| No LLD |
LLD |
p value | ASD | |
|---|---|---|---|---|
| n = 4,328 | n = 1,251 | |||
| Age, years, median (IQR) | 52.8 (48.0, 60.2) | 61.1 (54.3, 66.9) | <0.001 | 0.73 |
| Female, n (%) | 2,260 (52.2) | 622 (49.7) | 0.119 | 0.05 |
| Ethnicity, n (%) | <0.001 | 0.31 | ||
| Chinese | 1,704 (39.4) | 490 (39.2) | ||
| Indian | 1,292 (29.9) | 523 (41.8) | ||
| Malay | 1,332 (30.8) | 238 (19.0) | ||
| Hypertension, n (%) | 2,131 (49.4) | 1,018 (81.4) | <0.001 | 0.72 |
| Diabetes, n (%) | 727 (16.8) | 595 (47.6) | <0.001 | 0.70 |
| BMI, kg/m2, median (IQR) | 24.6 (22.0, 27.6) | 25.5 (23.2, 28.5) | <0.001 | 0.24 |
| Smoking status, n (%) | <0.001 | 0.13 | ||
| Never smoked | 3,094 (71.5) | 925 (74.0) | ||
| Current smoker | 689 (15.9) | 147 (11.8) | ||
| Past smoker | 542 (12.5) | 178 (14.2) | ||
| CVD, n (%) | 156 (3.6) | 262 (20.9) | <0.001 | 0.55 |
| Education level, n (%) | <0.001 | 0.16 | ||
| No or primary | 2,205 (51.0) | 736 (59.0) | ||
| Higher than primary | 2,118 (49.0) | 512 (41.0) |
LLD, lipid-lowering drug; IQR, interquartile range; ASD, absolute standardized difference; BMI, body mass index; CVD, cardiovascular disease.
To build our incidence outcome in SEED, among the 5,608 individuals having a gradable fundus at both visits, we excluded 2,159 prevalent AMD cases at baseline. We further excluded 44 individuals with incomplete medical records. Among the remaining 3,405 individuals, 2,586 (75.9%) had genotyping available. In this population, 484 (18.7%) developed AMD at the follow-up visit (317 early, 163 intermediate, and 4 late AMD). Compared to individuals who remained free of any AMD between the two visits, individuals that developed AMD at the follow-up visit were older and were more likely to be male, to be Chinese or Malay, to have hypertension, to be past smokers, and to have no primary education level (Table S1).
We then built our progression outcome. Among the 5,608 individuals having a gradable fundus at both visits, 2,133 had early or intermediate AMD at baseline. We excluded 31 individuals due to incomplete medical records. Among the remaining 2,102 individuals, 1,620 (77.1%) had genotyping available. Out of them, 216 participants progressed between the 2 visits (early to intermediate, n = 199; early to late, n = 3, and intermediate to late AMD, n = 14). Compared to individuals that remained within their baseline AMD categories between the two visits, individuals that progressed to a more severe stage were more likely to be Indian or Malay and had higher body mass index (BMI) (Table S1).
In UK Biobank, among the 449,297 participants included with complete medical records, 77,864 (17.3%) took LLD at baseline. The prevalent cases at baseline were removed (n = 3,984). Among the remaining individuals, 913 developed AMD during the follow-up at a mean follow-up time of 6.9 ± 2.8 years. Compared to individuals who remained free of AMD during the follow-up, individuals who developed AMD were older and were more likely to have diabetes and CVD, and to be past smokers (Table S2).
Association of LLD with AMD incidence and progression
Overall, in SEED, the effect of LLD was not associated with AMD incidence (inverse treatment probability weights [ITPW] method: odds ratio [OR], 0.93; 95% confidence interval [CI], 0.68, 1.27; overlap weights [OW] method: OR, 1.08; 95% CI, 0.83, 1.41). However, when stratifying for complement genotypes, LLD was associated with AMD according to rs7523273 (CD46) and rs10033900 (CFI) with decreased risks of AMD and according to rs41347947 (CD93) with increased risk of AMD (Table S3). We used UK Biobank to confirm these possible associations by estimating the effect of LLD according to the same genetic variants. None of these associations were found in UK Biobank (Table S4; Figure S1).
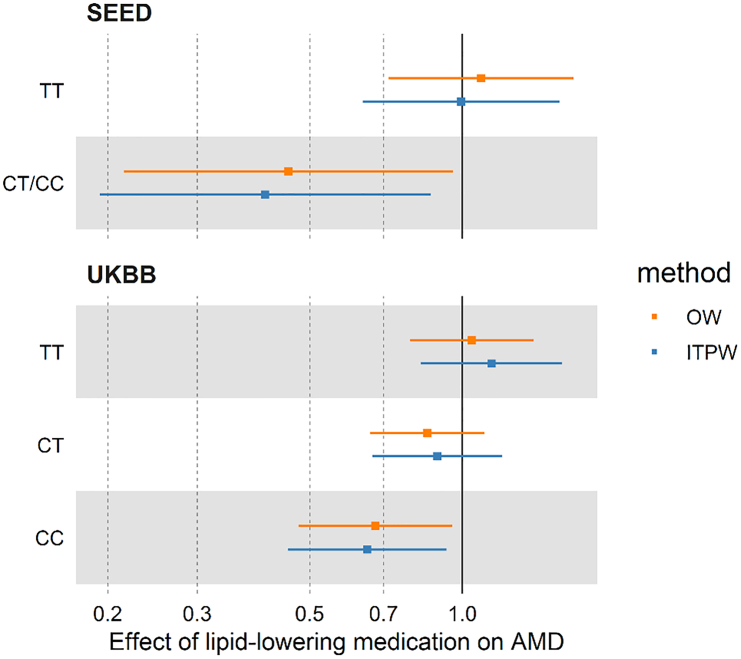
Overall, in SEED, the effect of LLD was not associated with progression (ITPW method: OR, 0.83; 95% CI, 0.56, 1.22; OW method: OR, 0.93; 95% CI, 0.64, 1.34). However, when stratifying for the complement genotype, LLD was associated with a decreased risk of AMD progression according to rs1061170 (complement factor H [CFH]) for individuals with CT or CC genotype, and rs11080055 (VTN) for individuals with CC genotype (Table 2; Table S3). These associations were tested in UK Biobank. LLD was only associated with a decreased risk of AMD incidence in individuals with the CC genotype of rs1061170 (CFH) (ITPW method: hazards ratio [HR], 0.65; 95% CI, 0.45, 0.93; OW method: HR, 0.67; 95% CI, 0.47, 0.96) (Table 2; Figure 1; Table S4; Figure S1).Compared to SEED study, the larger sample size of UK Biobank allowed us to detect that protective effect specifically for individuals with two C alleles (CC genotype). The reduction in the ASD when using the propensity score methods (Figures S2 and S3) showed a drastic reduction of bias due to differences between individual taking and not taking LLD, which confirms the advantage of using a propensity score approach.
Table 2.
Effect of lipid-lowering drug on the risk of AMD in Singapore Epidemiology of Eye Diseases study and in UK Biobank according to the CFH rs1061170 genetic variant
| Cohort | allele | n | Method | Effect sizea | p value |
|---|---|---|---|---|---|
|
SEED AMD progression |
OR (95% CI) | ||||
| CFH rs1061170 | TT | 1,281 | ITPW | 1.00 (0.64, 1.55) | 0.983 |
| TT | 1,281 | OW | 1.09 (0.72, 1.66) | 0.688 | |
| CT/CC | 339 | ITPW | 0.41 (0.19, 0.87) | 0.020 | |
| CT/CC | 339 | OW | 0.45 (0.22, 0.96) | 0.039 | |
|
UK Biobank AMD incidence |
HR (95% CI) | ||||
| CFH rs1061170 | TT | 171,893 | ITPW | 1.14 (0.83, 1.57) | 0.416 |
| TT | 171,893 | OW | 1.04 (0.79, 1.38) | 0.760 | |
| CT | 209,130 | ITPW | 0.89 (0.67, 1.20) | 0.456 | |
| CT | 209,130 | OW | 0.85 (0.66, 1.11) | 0.230 | |
| CC | 64,704 | ITPW | 0.65 (0.45, 0.93) | 0.019 | |
| CC | 64,704 | OW | 0.67 (0.47, 0.96) | 0.027 | |
SEED, Singapore Epidemiology of Eye Diseases; ITPW, inverse treatment probability weighting; OW, overlap weights.
The effects are expressed as odds ratios (OR) or hazard ratios (HRs) with their 95% confidence intervals (CIs).
Figure 1.
Effect of lipid-lowering drug on the risk of AMD in Singapore Epidemiology of Eye Diseases and UK Biobank according to the rs1061170 genetic variant
The squares represent the odds ratios (Singapore Epidemiology of Eye Diseases [SEED]) or hazard ratios (UK Biobank [UKBB]), with the horizontal bars indicating the 95% confidence intervals. The outcome is AMD progression in SEED study and AMD incidence in UKBB study. ITPW, inverse treatment probability weighting; OW, overlap weights.
In SEED, we performed a subgroup analysis by considering only individuals taking statin (instead of LLD as a whole) and found similar trends (ITPW method: OR, 0.46; 95% CI, 0.21, 1.01; p = 0.052; OW method: OR, 0.52; 95% CI, 0.24, 1.11; p = 0.091) for individuals with at least one C allele (CC/CT genotype) in rs1061170 genetic variant (CFH).
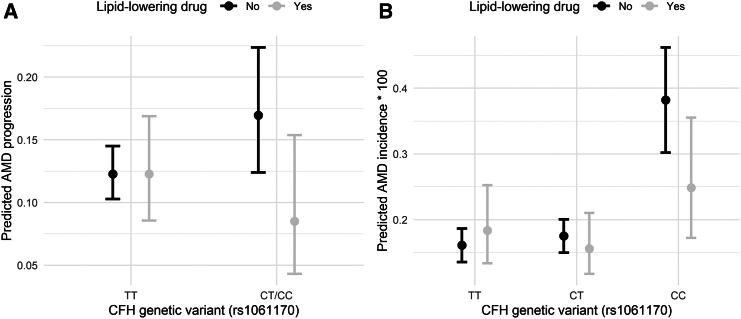
Furthermore, we predicted AMD progression and incidence according to LLD intake and rs1061170 polymorphism in SEED and UK Biobank (Figure 2). A large reduction was found for individuals taking LLD with at least one C allele in SEED (genotypes CT/CC), from 16.9% to 8.5%, compared to a stable rate of 12.3% in individuals homozygous for the reference allele (genotype TT) (Figure 2A). A similar reduction associated with LLD intake was found in UK Biobank for individuals homozygous for the C allele (genotype CC), with a reduction from 0.38% to 0.25% (Figure 2B).
Figure 2.
Predicted AMD progression and incidence according to the lipid-lowering drug and the CFH rs1061170 genotype
(A) Predicted AMD progression in Singapore Epidemiology of Eye Diseases (SEED) study according to the lipid-lowering drug (LLD) and the CFH rs1061170 genotype.
(B) Predicted AMD incidence in UK Biobank populations according to the LLD and the CFH rs1061170 genotype. The points represent the mean predicted progression rate (SEED) or incidence rate (UKBB), with the vertical bars indicating their 95% confidence intervals. Predictions from the UK Biobank were multiplied by 100 to facilitate the reading. These predictions were calculated using the LLD effects estimated from the multivariable models (as shown in Figure 1) using the inverse treatment probability weighting approach, and applied to the untreated population (see STAR Methods for details).
Lipoprotein sub-fractions
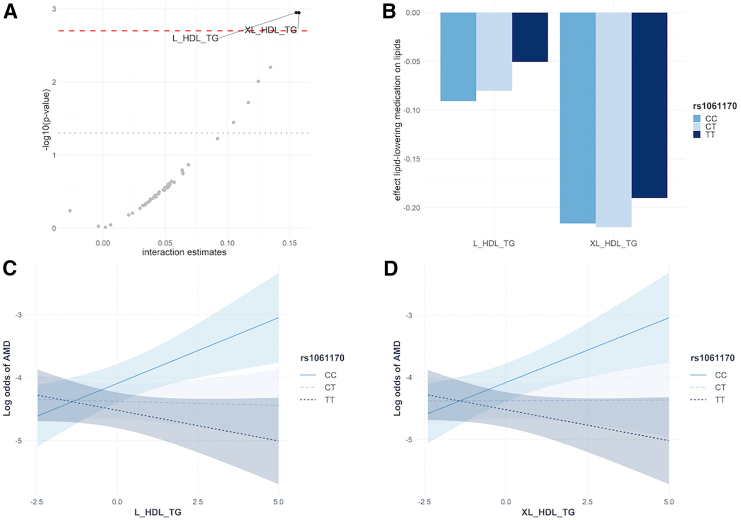
An interaction between two lipoprotein sub-fractions and rs1061170 genetic variant was observed in relation to AMD after correction for multiple testing: triglycerides in large high-density lipoprotein (HDL) and triglycerides in very large HDL (Figure 3A; Table S5). While the direction of effect of triglycerides in large HDL and triglycerides in very large HDL on AMD was negative for individuals with TT genotype, the association was positive for individuals with the CC genotype (Figures 3C and 3D; Table S5A). Furthermore, taking LLD was associated with decreased levels of triglycerides in very large HDL regardless of the rs1061170 genotype, and associated with decreased levels of triglycerides in large HDL for individuals with CC and CT genotypes (Figures 3B and Table S5B).
Figure 3.
Effect of lipoprotein sub-fractions and lipid-lowering drug according to CFH rs1061170 genotype
(A) Estimates corresponding to the interactions between 49 low-density lipoprotein and high-density lipoprotein (HDL) sub-fractions and CFH genetic variant (rs1061170) on AMD. The gray dotted and red dashed lines corresponded to 5% significance threshold before and after FDR correction, respectively. The two HDL sub-fractions with FDR corrected p values < 5% (above the red dashed line) were considered for the analyses (B), (C), and (D).
(B) Effect of lipid-lowering drug on triglycerides sub-fractions in large and very large HDL according to rs1061170 genetic variant.
(C) Effects of triglycerides in large HDL (L_HDL_TG) on AMD according to rs1061170 genetic variant. The shaded areas around the solid and dotted lines corresponded to the 95% confidence intervals.
(D) Effects of triglycerides in very large HDL (XL_HDL_TG) on AMD according to rs1061170 genetic variant. The shaded areas around the solid and dotted lines corresponded to the 95% confidence intervals.
Discussion
We have shown in two large prospective cohorts that LLD had a protective effect on AMD according to the rs1061170 genetic variant located in the CFH gene. In SEED, this decreased risk was evidenced for individuals with at least one C allele, and in UK Biobank, due to its bigger sample size, this effect was evidenced for individuals with the CC genotype. In SEED, the protective effect of LLD corresponded to a 50% decrease in progression rate. A similar protective effect with 35% decreased risk was found in the UK Biobank for individuals with two CC alleles. Further analyses showed that this protective effect could be mediated by a decrease in the triglycerides sub-fraction in very large HDL.
Because lipid metabolism is involved in the AMD pathophysiology,4,5 LLDs have previously been investigated as a potential therapeutic option; however, the results have been inconsistent. Numerous studies, including meta-analyses, were insufficient to establish a role for LLD as a preventive strategy for delaying the onset or progression of AMD.12,13,14,15,16,17 Conversely, a recent meta-analysis of 38,694 participants from 14 European populations revealed a 15% reduction in risk of any AMD in patients who were on LLD.11 While this is the largest association study to date on LLD and AMD, the main limitation was its cross-sectional nature. Several small clinical trials have also attempted to compare the effects of statins on AMD progression. In a small trial of 26 patients with large drusenoid deposits, the group treated with high-dose atrovastin had regression of drusen.22 A double-blind randomized controlled trial showed that simvastatin was associated with a significant 2-fold decrease in the risk of progression. We suggest that these inconsistencies in prior literature were due to the complement genotype polymorphism. In our study, we found a protective effect of LLD for people with one or two C alleles in the rs1061170 genetic variant in SEED and UK Biobank, respectively. The magnitudes of the effect were strong with 50% and 35% decreases in the progression rates in SEED and UK Biobank, respectively. In SEED, unfortunately, the number of individuals with a CC genotype for the rs1061170 genetic variant was too limited to determine the effect of LLD in this group. Consistent with our findings, Guymer et al. found that the most prominent effect of simvastatin was observed among those homozygous for the at-risk allele C of this genetic variant.23
The protective effect of LLD in a sub-group of individuals according to their complement genotype may be due to the complex interplay between complement system and lipid metabolism in the retina. HDL lipoprotein particles contain various complement components18,24,25 and this may explain the interactions between these two biological systems.20 It has been shown that increased CFH concentration can trigger the anti-inflammatory properties of large HDL particles.18 In our analyses, we showed that LLD was associated with an overall decrease in the level of the triglyceride sub-fraction in very large HDL particles. We also showed that the effect of this sub-fraction on AMD was modified by rs1061170 polymorphism, with the opposite direction of effects for individuals with CC and TT alleles. Interestingly, lower level of this sub-fraction was associated with a decreased risk of AMD only in individuals with CC genotype. Therefore, the protective effect of LLD for AMD progression for these individuals may be due to decreased inflammation levels associated with a lower concentration of triglycerides sub-fraction in very large HDL particles. Proper biological studies are needed to determine the functional effect of rs1061170 genetic variant polymorphism.
Our findings suggest that LLD could be used for AMD in a personalized medicine framework. Despite the variation of the allele frequency of the rs1061170 genetic variant between populations, the high magnitude of effect of LLD reported here has the potential for a large public health impact on the disease burden of the 200 million individuals estimated to have AMD. Here, we reported between 50% (SEED) and 35% (UK Biobank) decreased risk in individuals on LLD with one or two C alleles for the genetic variant rs1061170. In contrast, the Age-Related Eye Disease Study supplement formulary had a moderate effect (25%) in individuals with high-risk features and no effect at earlier stages.26 An approach could be to genotype for CFH genetic variant of interest in patients with early AMD, and appropriate LLD commenced if a risk allele is detected. These drugs are well known, safe, and are widely used for common systemic conditions and would require only a change in indication if proven to prevent AMD progression.
The key strength of this study was the utilization of longitudinal data from two independent cohorts. We believe this is an important strength compared to prior cross-sectional observational studies. Moreover, the validation of our findings in the large UK Biobank cohort demonstrated the robustness of our findings. Furthermore, the severity of AMD in SEED was performed using Beckman classification system,27 graded by a reading center, ensuring that the progression of disease was unequivocal as each increase in severity grade required a mark change in clinical features. Finally, we used appropriate statistical methods based on the estimation of propensity scores to account for the important differences in the characteristics of the participants according to the use of LLD.
Overall, our study conducted in two large independent cohorts sheds new light on the use of LLD for AMD therapeutics. We show that, while LLD may not be beneficial overall, individuals with a C allele for the genetic variant rs1061170 in the CFH gene have a substantially decreased risk of AMD. This indicates a possible avenue for the development of a targeted, personalized therapy based on the complement genotyping for patients with early form of the disease and thus at risk of progressing.
Limitations of the study
Some limitations must also be acknowledged. Firstly, no information was available on the dose of the LLD taken; we could not thus explore a possible dose-relationship effect of these medications on AMD progression. Secondly, in the AMD incidence analysis in UK Biobank, the AMD status was based on self-reported information and hospital consultations. However, we manually graded the images of all the AMD cases and found that 45.3%, 41.7%, and 3.9% of them had signs of early, intermediate, and late AMD, respectively, while only 9.1% showed no AMD in fundus image. Although the proportion of false-negative cases is unknown, this indicates a low proportion of false-positive cases.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Simon Nusinovici (simon65@nus.edu.sg).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Regarding the Singapore Epidemiology of Eye Disease data, as the study involves human participants, the data cannot be made freely available in the manuscript, the supplemental files, or a public repository due to ethical restrictions. Nevertheless, the data are available from the Singapore Eye Research Institutional Ethics Committee for researchers who meet the criteria for access to confidential data. Interested researchers can send data access requests to the Singapore Eye Research Institute using the following email address: seri@seri.com.sg.
The UK Biobank data were obtained from UK Biobank (application number 45925). Data cannot be shared publicly due to the violation of patient privacy and lack of informed consent for data sharing.
The code used for the analyses is available from the corresponding author upon reasonable request (simon65@nus.edu.sg).
Acknowledgments
This study was supported by the National Medical Research Council (NMRC), Singapore (grant numbers: NMRC/CIRG/1417/2015, NMRC/CIRG/1488/2018, and NMRC/OFLCG/004a/2018).
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Author contributions
Conception or design of the study, Y.C.T., S.T., U.C., C.-Y.C., and S.N.; acquisition of data and analysis of the data, H.L. and S.N.; interpretation of the results, C.C.X., K.Y.C.T., Y.C.T., H.L., S.T., C.S., Q.F., D.L.S., X.W., U.C., C.M.G.C., T.Y.W., C.-Y.C., and S.N.; drafted the manuscript, C.C.X., K.Y.C.T., and S.N.; revision of the manuscript, C.C.X., K.Y.C.T., Y.C.T., H.L., S.T., C.S., Q.F., D.L.S., X.W., U.C., C.M.G.C., T.Y.W., C.-Y.C., and S.N.; approval and agreement of the submitted version, C.C.X., K.Y.C.T., Y.C.T., H.L., S.T., C.S., Q.F., D.L.S., X.W., U.C., C.M.G.C., T.Y.W., C.-Y.C., and S.N. All authors confirm that they had full access to all the data in the study and accept responsibility to submit for publication.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Software and algorithms | ||
| R Project | RStudio | https://posit.co/products/open-source/rstudio/ |
| Other | ||
| Singapore Epidemiology of Eye Disease (SEED) data | Singapore Epidemiology of Eye Disease (SEED) study | https://www.snec.com.sg/research-innovation/key-programme-singapore-epidemiology-of-eye-diseases |
| UK Biobank data | UK Biobank | https://www.ukbiobank.ac.uk/ |
Experimental model and study participant details
This is a multi-ethnic prospective study. Participants were from the Singapore Epidemiology of Eye Diseases (SEED) and the UK Biobank studies. SEED is a prospective population-based study of 10,033 subjects aged 40 years and over from the three main ethnic groups in Singapore: Chinese Indian, and Malay, recruited at 2004 and followed up 6 years later, a standardised interview, laboratory and ophthalmic investigations at both visits.28 The UK Biobank study is a prospective cohort in the UK with over 500,000 participants aged 37–73 years recruited during 2006–2010.29 The study has collected extensive phenotypic detail, including data from questionnaires, physical measures, and sample assays and longitudinal follow-up for a wide range of health-related outcomes. Age, sex, ethnicity and other relevant demographics and clinical information for SEED participants and UK Biobank participants included in this study are presented in Tables S1 and S2.
Ethics
Informed, written consent was obtained from all the participants of the SEED study, with ethical approval obtained from the Institutional Review Board of SingHealth. Similarly, written informed consent was obtained from all the participants of the UK Biobank study, which was approved by the North West Center for Research Ethics Committee.
Method details
AMD grading
In SEED, the presence and severity of AMD was determined by the Singapore National Eye Center Ocular reading center, based on fundus photographs, according to the Beckman classification system.27 Early AMD was defined as the presence of medium (63–125 μm) drusen and without any pigmentary abnormalities. Intermediate AMD was defined as the presence of large drusen (larger than 125μm) and/or the presence of pigmentary abnormalities with medium drusen. Late AMD was defined as the presence of features of wet AMD or geographic atrophy. The analyses were performed at the individual level with the more severe eye considered for each individual (Table S6).
We identified AMD incidence and progression based on the grading at baseline and 6-year follow-up. Individuals who were free of AMD at baseline but had any form of AMD at the follow-up visit were defined as having incident AMD, with the corresponding controls being those free of AMD at both visits. Individuals with AMD progression was defined as those who progressed from early AMD to intermediate/late AMD, or from intermediate to late AMD. The controls were individuals whose AMD severity remained unchanged between the two visits.
LLD and participant’s characteristics
We obtained the patient reported use of LLD (statins, fibrates and unspecified) at the baseline visit. No information was available on the dosage. In the main analysis, LLD was used as a whole. In a sensitivity analysis, only statins were considered. To account for potential confounders, we considered the following participant’s characteristics: age, sex, ethnicity, hypertension, cardiovascular disease, diabetes status, BMI, smoking status and education level.
Complement system genotype
We considered the single nucleotide polymorphisms (SNPs) associated with AMD in the latest large international genome-wide association studies conducted on 16,144 patients with AMD and 17,832 controls.30 Among these SNPs, we selected those flanked 250kb of each complement system genes. For each gene, we used the lead SNP with the smallest p-value, which led us to consider 36 SNPs. These SNPs were extracted in SEED, and for the SNPs not available, we used the next available lead SNP instead. Finally, we excluded those with mean R2 imputation quality <0.60 and thus kept a total of 20 SNPs (Table S7).
To determine whether the effect of LLD differ according to the complement system genotype, we stratified the analyses according to these SNPs, i.e., one analysis was run in each genetic stratum: homozygous for the reference allele, heterozygous, and homozygous for the risk allele. If the number of individuals in the latest group was too small, then two groups were considered: homozygous for the reference allele and individuals with at least one risk allele.
Replication of analysis in UK biobank
In UK Biobank, AMD was defined based on a combination of the following information: 1) in-patient and mortality data using predefined International Classification of Diseases (ICD) 10 (code: H35.3) 2) self-reported information (data field: 20002; code: 1528 and date field: 6148). Prevalent AMD cases were defined as individuals who received an initial diagnosis of AMD at or before recruitment. Incident AMD cases were defined as individuals who initially received a diagnosis of AMD between recruitment and during the 11 years of follow up. The use of LLD at the baseline visit were collected. All the 20 complement genotype SNPs used for the analyses in SEED were available in UK Biobank and all had R2 imputation values ≥ 0.90 (Table S7).
Lipoprotein sub-fractions
We used nuclear magnetic resonance (NMR) metabolomics data available in UK biobank at the baseline visit. Among the 170 blood metabolites quantified, we included 49 HDL and low-density lipoprotein (LDL) sub-fractions. HDL and LDL were classified as very large (XL), large (L), medium (M), and small (S); and L, M and S, respectively. For each lipoprotein subclass, the concentrations of lipids, triglycerides, cholesterol esters, free cholesterol, and phospholipids were considered. The list of the lipid-related metabolites considered is shown in Table S8.
Quantification and statistical analysis
The effect of LLD on AMD was determine according to each complement system genetic variant. To account for the difference in characteristics between individuals taking or not LLD, we used a propensity score approach.31 This method is very efficient to determine the effect of a treatment in an observational study because it allows to appropriately removes confusion effects.32 It has been indeed shown that propensity score studies produce results generally consistent with the findings of randomized clinical trials.33 First, we estimated the probability of the participants to be taking LLD using a logistic regression model with the following covariates: age, sex, ethnicity (Chinese, Malay or Indian), hypertension (yes/no), diabetes status (yes/no), BMI (continuous), smoking status (never smoked, past smoker, current smoker), self-reported history of cardio-vascular disease (yes/no), and education level (no formal education or primary education, O/N levels, A levels or university education). These probabilities (predicted scores) were then used to calculate weights which were used in a logistic regression model (SEED) and Cox proportional hazards model (UK Biobank) with AMD outcomes as the dependent variables, and LLD as the exposure variable. Two different weights were used to determine the robustness of the results: IPTW34 and OW.35 The effect of LLD on AMD were expressed in odds-ratios (OR) in SEED and in hazard ratios (HR) in UK Biobank. More details regarding these methods are presented in the Methods S1.
Furthermore, we calculated the adjusted predicted AMD progression and incidence according to LLD and the CFH rs1061170 genotype. In SEED, we first converted the OR (estimated using the logistic multivariable model) into relative risks (RR) using the formula: RR = OR/((1-P0)+(P0∗OR)),36 with P0 being the AMD probability in individuals not taking LLD. In UK Biobank, we converted the HR (estimated using the multivariable Cox model) into RR using the following formula: RR=(1-exp(HR∗log(1-P0)))/P0. The adjusted AMD predictions were then obtained using the formula: adjusted AMD predictions = RR∗P0/(1-P0+(RR∗P0)). In SEED, P0 with their 95% CI were calculated using a binomial distribution and in UK Biobank as 1 – survival probability of a Cox model with only the intercept.
Finally, to explore whether lipoprotein sub-fractions mediated the effect of LLD on AMD, we performed the following cross-sectional analyses in UK Biobank using the prevalent AMD cases at baseline. Firstly, we determined whether the effect of the HDL and LDL sub-fractions on AMD varied according to rs1061170 genetic variant by testing the interaction effect between these sub-fractions and rs1061170 coded as continuous variable. The same set of covariates as the one used in the main analyses was used to correct for possible confounders. Then, for the significant interactions after false discovery rate correction, we quantified the variations of lipoprotein sub-fractions associated with LLD using the same propensity score approach as the one used for the main analyses. For these analyses, we standardized the lipoprotein sub-fractions distributions with Z score normalizations.
Published: November 8, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.111344.
Supplemental information
References
- 1.Mitchell P., Liew G., Gopinath B., Wong T.Y. Age-related macular degeneration. Lancet. 2018;392:1147–1159. doi: 10.1016/S0140-6736(18)31550-2. [DOI] [PubMed] [Google Scholar]
- 2.Wong W.L., Su X., Li X., Cheung C.M.G., Klein R., Cheng C.Y., Wong T.Y. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet. Glob. Health. 2014;2:e106–e116. doi: 10.1016/S2214-109X(13)70145-1. [DOI] [PubMed] [Google Scholar]
- 3.Flaxman S.R., Bourne R.R.A., Resnikoff S., Ackland P., Braithwaite T., Cicinelli M.V., Das A., Jonas J.B., Keeffe J., Kempen J.H., et al. Global causes of blindness and distance vision impairment 1990–2020: a systematic review and meta-analysis. Lancet. Glob. Health. 2017;5:e1221–e1234. doi: 10.1016/S2214-109X(17)30393-5. [DOI] [PubMed] [Google Scholar]
- 4.Colijn J.M., den Hollander A.I., Demirkan A., Cougnard-Grégoire A., Verzijden T., Kersten E., Meester-Smoor M.A., Merle B.M.J., Papageorgiou G., Ahmad S., et al. Increased High-Density Lipoprotein Levels Associated with Age-Related Macular Degeneration. Ophthalmology. 2019;126:393–406. doi: 10.1016/j.ophtha.2018.09.045. [DOI] [PubMed] [Google Scholar]
- 5.Fan Q., Maranville J.C., Fritsche L., Sim X., Cheung C.M.G., Chen L.J., Gorski M., Yamashiro K., Ahn J., Laude A., et al. HDL-cholesterol levels and risk of age-related macular degeneration: a multiethnic genetic study using Mendelian randomization. Int. J. Epidemiol. 2017;46:1891–1902. doi: 10.1093/ije/dyx189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nusinovici S., Zhou L., Wang X., Tham Y.C., Wang X., Wong T.Y., Chakravarthy U., Cheng C.Y. Contributions of Lipid-Related Metabolites and Complement Proteins to Early and Intermediate Age-Related Macular Degeneration. Ophthalmol. Sci. 2024;4:100538. doi: 10.1016/j.xops.2024.100538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sim R.Z.H., Tham Y.C., Betzler B.K., Zhou L., Wang X., Sabanayagam C., Cheung G.C.M., Wong T.Y., Cheng C.Y., Nusinovici S. Relationships between Lipid-Related Metabolites and Age-Related Macular Degeneration Vary with Complement Genotype. Ophthalmol. Sci. 2022;2:100211. doi: 10.1016/j.xops.2022.100211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbosa D.T.Q., Mendes T.S., Cíntron-Colon H.R., Wang S.Y., Bhisitkul R.B., Singh K., Lin S.C. Age-related macular degeneration and protective effect of HMG Co-A reductase inhibitors (statins): results from the National Health and Nutrition Examination Survey 2005–2008. Eye. 2014;28:472–480. doi: 10.1038/eye.2014.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein R., Klein B.E.K., Tomany S.C., Danforth L.G., Cruickshanks K.J. Relation of Statin Use to the 5-Year Incidence and Progression of Age-Related Maculopathy. Arch. Ophthalmol. 2003;121:1151–1155. doi: 10.1001/archopht.121.8.1151. [DOI] [PubMed] [Google Scholar]
- 10.Fong D.S., Contreras R. Recent Statin Use and 1-Year Incidence of Exudative Age-Related Macular Degeneration. Am. J. Ophthalmol. 2010;149:955–958.e1. doi: 10.1016/j.ajo.2009.12.037. [DOI] [PubMed] [Google Scholar]
- 11.Mauschitz M.M., Verzijden T., Schuster A.K., Elbaz H., Pfeiffer N., Khawaja A., Luben R.N., Foster P.J., Rauscher F.G., Wirkner K., et al. Association of lipid-lowering drugs and antidiabetic drugs with age-related macular degeneration: a meta-analysis in Europeans. Br. J. Ophthalmol. 2023;107:1880–1886. doi: 10.1136/bjo-2022-321985. [DOI] [PubMed] [Google Scholar]
- 12.Al-Holou S.N., Tucker W.R., Agrón E., Clemons T.E., Cukras C., Ferris F.L., III, Chew E.Y. The Association of Statin Use with Age-Related Macular Degeneration Progression. Ophthalmology. 2015;122:2490–2496. doi: 10.1016/j.ophtha.2015.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maguire M.G., Ying G.s., McCannel C.A., Liu C., Dai Y., Complications of Age-related Macular Degeneration Prevention Trial CAPT Research Group Statin Use and the Incidence of Advanced Age-related Macular Degeneration in the Complications of Age-related Macular Degeneration Prevention Trial. Ophthalmology. 2009;116:2381–2385. doi: 10.1016/j.ophtha.2009.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shalev V., Sror M., Goldshtein I., Kokia E., Chodick G. Statin Use and the Risk of Age Related Macular Degeneration in a Large Health Organization in Israel. Ophthalmic Epidemiol. 2011;18:83–90. doi: 10.3109/09286586.2011.560746. [DOI] [PubMed] [Google Scholar]
- 15.Roizenblatt M., Naranjit N., Maia M., Gehlbach P.L. The Question of a Role for Statins in Age-Related Macular Degeneration. IJMS. 2018;19:3688. doi: 10.3390/ijms19113688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eshtiaghi A., Popovic M.M., Sothivannan A., Muni R.H., Kertes P.J. STATIN USE AND THE INCIDENCE OF AGE-RELATED MACULAR DEGENERATION: A Meta-Analysis. Retina. 2022;42:643–652. doi: 10.1097/IAE.0000000000003398. [DOI] [PubMed] [Google Scholar]
- 17.Gehlbach P., Li T., Hatef E. Statins for age-related macular degeneration. Cochrane Database Syst. Rev. 2016;2016 doi: 10.1002/14651858.CD006927.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y., Gordon S.M., Xi H., Choi S., Paz M.A., Sun R., Yang W., Saredy J., Khan M., Remaley A.T., et al. HDL subclass proteomic analysis and functional implication of protein dynamic change during HDL maturation. Redox Biol. 2019;24 doi: 10.1016/j.redox.2019.101222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Acar İ.E., Lores-Motta L., Colijn J.M., Meester-Smoor M.A., Verzijden T., Cougnard-Gregoire A., Ajana S., Merle B.M.J., de Breuk A., Heesterbeek T.J., et al. Integrating Metabolomics, Genomics, and Disease Pathways in Age-Related Macular Degeneration. Ophthalmology. 2020;127:1693–1709. doi: 10.1016/j.ophtha.2020.06.020. [DOI] [PubMed] [Google Scholar]
- 20.Nusinovici S., Zhou L., Raghavan L., Tham Y.C., Li H., Cheung D., Wang X., Gemmy Cheung C.M., Wong T.Y., Chakravarthy U., et al. Interplay between lipids and complement proteins – How multi-omics data integration can help unravel age-related macular degeneration pathophysiology, a proof-of-concept study. Ophthalmol. Sci. 2024;5 doi: 10.1016/j.xops.2024.100629. Published online October 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.RUBIN D.B. Using Propensity Scores to Help Design Observational Studies: Application to the Tobacco Litigation. Health Serv. Outcome Res. Methodol. 2001;2:169–188. https://link.springer.com/content/pdf/10.1023%2FA%3A1020363010465.pdf [Google Scholar]
- 22.Vavvas D.G., Daniels A.B., Kapsala Z.G., Goldfarb J.W., Ganotakis E., Loewenstein J.I., Young L.H., Gragoudas E.S., Eliott D., Kim I.K., et al. Regression of Some High-risk Features of Age-related Macular Degeneration (AMD) in Patients Receiving Intensive Statin Treatment. EBioMedicine. 2016;5:198–203. doi: 10.1016/j.ebiom.2016.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guymer R.H., Baird P.N., Varsamidis M., Busija L., Dimitrov P.N., Aung K.Z., Makeyeva G.A., Richardson A.J., Lim L., Robman L.D. Proof of Concept, Randomized, Placebo-Controlled Study of the Effect of Simvastatin on the Course of Age-Related Macular Degeneration. PLoS One. 2013;8 doi: 10.1371/journal.pone.0083759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaisar T., Pennathur S., Green P.S., Gharib S.A., Hoofnagle A.N., Cheung M.C., Byun J., Vuletic S., Kassim S., Singh P., et al. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Invest. 2007;117:746–756. doi: 10.1172/JCI26206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barbu A., Hamad O.A., Lind L., Ekdahl K.N., Nilsson B. The role of complement factor C3 in lipid metabolism. Mol. Immunol. 2015;67:101–107. doi: 10.1016/j.molimm.2015.02.027. [DOI] [PubMed] [Google Scholar]
- 26.Evans J.R., Lawrenson J.G. Antioxidant vitamin and mineral supplements for slowing the progression of age-related macular degeneration. Cochrane Eyes and Vision Group. Cochrane Database Syst. Rev. 2023;2023 doi: 10.1002/14651858.CD000254.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferris F.L., Wilkinson C.P., Bird A., Chakravarthy U., Chew E., Csaky K., Sadda S.R., Beckman Initiative for Macular Research Classification Committee Clinical Classification of Age-related Macular Degeneration. Ophthalmology. 2013;120:844–851. doi: 10.1016/j.ophtha.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Majithia S., Tham Y.C., Chee M.L., Nusinovici S., Teo C.L., Chee M.L., Thakur S., Soh Z.D., Kumari N., Lamoureux E., et al. Cohort profile: The Singapore Epidemiology of Eye Diseases study (SEED) Int. J. Epidemiol. 2021;50:41–52. doi: 10.1093/ije/dyaa238. Published online January 4, 2021. [DOI] [PubMed] [Google Scholar]
- 29.Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., Downey P., Elliott P., Green J., Landray M., et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLoS Med. 2015;12 doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fritsche L.G., Igl W., Bailey J.N.C., Grassmann F., Sengupta S., Bragg-Gresham J.L., Burdon K.P., Hebbring S.J., Wen C., Gorski M., et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 2016;48:134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Austin P.C. An Introduction to Propensity Score Methods for Reducing the Effects of Confounding in Observational Studies. Multivariate Behav. Res. 2011;46:399–424. doi: 10.1080/00273171.2011.568786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D’Agostino R.B. Propensity score methods for bias reduction in the comparison of a treatment to a non-randomized control group. Stat. Med. 1998;17:2265–2281. doi: 10.1002/(sici)1097-0258(19981015)17:19<2265::aid-sim918>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 33.Kitsios G.D., Dahabreh I.J., Callahan S., Paulus J.K., Campagna A.C., Dargin J.M. Can We Trust Observational Studies Using Propensity Scores in the Critical Care Literature? A Systematic Comparison With Randomized Clinical Trials. Crit. Care Med. 2015;43:1870–1879. doi: 10.1097/CCM.0000000000001135. [DOI] [PubMed] [Google Scholar]
- 34.Austin P.C., Stuart E.A. Moving towards best practice when using inverse probability of treatment weighting (IPTW) using the propensity score to estimate causal treatment effects in observational studies. Stat. Med. 2015;34:3661–3679. doi: 10.1002/sim.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas L.E., Li F., Pencina M.J. Overlap Weighting: A Propensity Score Method That Mimics Attributes of a Randomized Clinical Trial. JAMA. 2020;323:2417–2418. doi: 10.1001/jama.2020.7819. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J., Yu K.F. What’s the Relative Risk?: A Method of Correcting the Odds Ratio in Cohort Studies of Common Outcomes. JAMA. 1998;280:1690–1691. doi: 10.1001/jama.280.19.1690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Regarding the Singapore Epidemiology of Eye Disease data, as the study involves human participants, the data cannot be made freely available in the manuscript, the supplemental files, or a public repository due to ethical restrictions. Nevertheless, the data are available from the Singapore Eye Research Institutional Ethics Committee for researchers who meet the criteria for access to confidential data. Interested researchers can send data access requests to the Singapore Eye Research Institute using the following email address: seri@seri.com.sg.
The UK Biobank data were obtained from UK Biobank (application number 45925). Data cannot be shared publicly due to the violation of patient privacy and lack of informed consent for data sharing.
The code used for the analyses is available from the corresponding author upon reasonable request (simon65@nus.edu.sg).