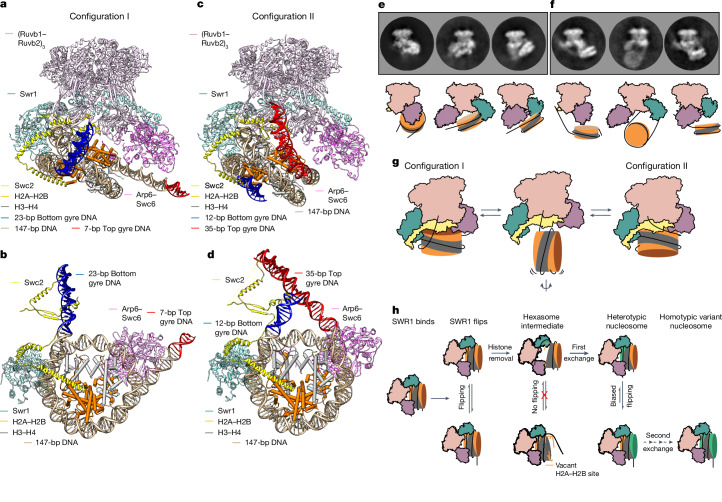
Fig. 5. Structural basis of SWR1-mediated nucleosome flipping.
a, The built-in coordinates of SWR1 in complex with a canonical nucleosome at 3.8 Å resolution in configuration I. Note that the DNA emanating from the lower gyre of the nucleosome (highlighted in blue) is bent up and binding across the surface of SWR1. b, A bottom view of the SWR1–nucleosome structure in configuration I. For clarity, only Swr1 (HD1 and HD2), Swc2 and the Arp6–Swc6 complex of SWR1 are shown. c, The built-in coordinates of SWR1 in complex with a canonical nucleosome at 4.7 Å resolution in configuration II. Note that the DNA emanating from the upper gyre of the nucleosome (highlighted in red) is binding across the surface of SWR1. d, A bottom view of the SWR1–nucleosome structure in configuration II. For clarity, only Swr1 (HD1 and HD2), Swc2 and the Arp6–Swc6 complex of SWR1 are shown. e, Three representative 2D class averages of SWR1–nucleosome in the canonical conformation. A cartoon representation of each 2D class is shown beneath. f, Three representative 2D class averages of SWR1-mediated nucleosome flipping with SWR1 orientated as in panel e. A cartoon representation of each 2D class is shown beneath. g, Cartoon summary of SWR1-mediated nucleosome flipping. h, Cartoon summary of kinetic proofreading and processivity of histone exchange by the SWR1 remodeller.