Abstract
Transition metals like Zn are essential for all organisms including bacteria, but fluctuations of their concentrations in the cell can be lethal. Organisms have thus evolved complex mechanisms for cellular metal homeostasis. One mechanistic paradigm involves pairs of transcription regulators sensing intracellular metal concentrations to regulate metal uptake and efflux. Here we report that Zur and ZntR, a prototypical pair of regulators for Zn uptake and efflux in E. coli, respectively, can coordinate their regulation through DNA, besides sensing cellular Zn2+ concentrations. Using a combination of live-cell single-molecule tracking and in vitro single-molecule FRET measurements, we show that unmetallated ZntR can enhance the unbinding kinetics of Zur from DNA by directly acting on Zur-DNA complexes, possibly through forming heteromeric ternary and quaternary complexes that involve both protein-DNA and protein-protein interactions. This ‘through-DNA’ mechanism may functionally facilitate the switching in Zn-uptake regulation when bacteria encounter changing Zn environments, such as facilitating derepression of Zn-uptake genes upon Zn depletion; it could also be relevant for regulating the uptake-vs.-efflux of various metals across different bacterial species and yeast.
Subject terms: Single-molecule biophysics, Bacterial transcription, Cellular microbiology
Proteins Zur and ZntR sense intracellular zinc concentrations and regulate zinc transport, respectively, in E. coli. Here the authors show that the proteins coordinate their regulation through DNA, by forming heteromeric complexes that involve protein-DNA and protein-protein interactions.
Introduction
For all life forms including bacteria, transition metals like Zn are essential but their excess is also detrimental1–15. Host cells can sequester metals to curb bacterial proliferation during infection, while metal stress can also be effective bactericidal treatments7–10,16. For growth and survival, bacteria have evolved exquisite mechanisms to regulate metal uptake and efflux5,10,11,13–24. Studying bacteria has thus produced mechanistic paradigms not only for understanding metal homeostasis in general but also for developing antibiotic treatments6,25–27.
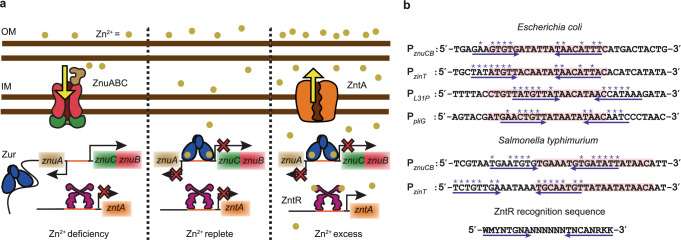
One such paradigm is the “set-point’’ mechanism that bacteria use to regulate cellular concentrations of a variety of transition metals (e.g., Zn2+, Fe2+, Ni2+, etc.)1–5,10. Here, the cellular free metal concentration [Mn+]free is bound by the metal-binding affinities of the respective uptake and efflux regulators. In E. coli, the Fur-family metalloregulator Zur28 and the MerR-family metalloregulator ZntR29,30 are the major Zn-uptake and efflux regulators that control the free or bio-available Zn2+ concentration in the cell (i.e., [Zn2+]free) to a range set by their respective Zn2+ binding affinities (Fig. 1a). Under Zn deficiency ([Zn2+]free < 0.2 fM), Zur has vacant regulatory Zn-binding sites and is a non-repressor that binds to non-consensus DNA sites31; here Zn-uptake genes (e.g., znuABC) are actively transcribed, while ZntR is at its apo state (i.e., ZntRapo) and binds its cognate promoter tightly, repressing Zn-efflux genes (e.g., zntA). Under Zn-replete conditions where [Zn2+]free exceeds 0.2 fM, Zur becomes fully metallated (i.e., ZurZn) and binds to its cognate promoter sites tightly, repressing Zn uptake. When [Zn2+]free rises further above 1.1 fM (i.e., Zn excess), ZntR is metallated (i.e., ZntRZn) to become an activator at its cognate promoters, activating Zn efflux. (It is worth noting that there are also other Zn-uptake and efflux systems4,32, such as ZupT33 and ZitB34 that are not regulated by Zur and ZntR in E. coli; see also Supplementary Notes 1 on discussion of other Zn-regulatory systems.)
Fig. 1. Sequences partially similar to ZntR recognition motifs are located near Zur-binding sites in 10.1038/s41467-024-55017-z Zur-regulon promoters.
a Present functional paradigm of Zur and ZntR in bacteria. Left: Under Zn2+ deficiency, Zur, without metallation at its regulatory sites, is a non-repressor that binds to non-consensus DNA sites but not to its recognition sequence (i.e., Zur box) in the promoter regions of its regulons (e.g., the divergent znuABC operon); here Zn2+ uptake genes are derepressed, while the non-metallated ZntR binds to its recognition sequence in the promoter regions of its regulons (e.g., zntA), repressing Zn2+ efflux. Center: Under Zn2+ replete conditions, Zur starts to be fully metallated (ZurZn) and binds to Zur box, repressing Zn2+ uptake, while ZntR is still predominantly in its apo form, repressing Zn2+ efflux. Right: Under Zn2+ excess, fully metallated ZurZn keeps repressing Zn2+ uptake, while the metallated ZntRZn at its cognate promoters distorts the DNA to result in activation of Zn2+ efflux genes. Both Zur and ZntR also have a freely diffusing population in the cell (not shown). IM inner membrane, OM outer membrane. b Promoter region sequences of Zur regulons in two different bacteria. Pink shades: Zur boxes. Double blue arrows: possible dyad symmetric sequences recognized by ZntR, whose consensus recognition sequence is shown at the bottom. Asterisk (*) denotes matches with the consensus sequence. Analysis of other species in Supplementary Notes 1.
While the binding of Zur and ZntR to their cognate promoters leads to transcription repression or activation of Zn-uptake or efflux genes, their unbinding from DNA is key to resetting regulation status when environmental and cellular Zn levels change. We have recently uncovered an unusual, facilitated unbinding mechanism for Zur and ZntR from DNA31,35,36. There, a homotypic freely diffusing protein can either assist the dissociation of the incumbent protein on DNA or directly substitute it, likely through an intermediate ternary protein2-DNA complex enabled by the multivalent contact between the protein and DNA, leading to protein-concentration−enhanced unbinding kinetics. Such facilitated unbinding allows for more facile switching between repression and derepression of Zn-uptake genes or between activation and deactivation of Zn-efflux genes; it was also observed for other types of DNA-binding proteins36–43. Additionally, Zur shows an impeded unbinding from its oligomerization-rendered stabilization on DNA31, which allows for storing the non-repressor form of Zur longer at non-consensus chromosomal sites to not interfere with its repressor form at cognate promoter sites. Overall, the apparent first-order unbinding rate constant k–1 of Zur and ZntR from tight binding sites on DNA follows31,35:
| 1 |
Here is a first-order intrinsic unbinding rate constant. The second term only applies to Zur and accounts for its impeded unbinding pathway, where kr is a first-order rate constant; Km is an effective dissociation constant of protein oligomer on DNA; and [P] is the free protein concentration. The third term is for the facilitated unbinding pathway for both Zur and ZntR with kf being a second-order rate constant.
As both Zur and ZntR can act on themselves on DNA, we wondered whether they could also act on each other, leading to cross-communication directly on DNA between the two regulatory systems. Here, we report the discovery of partial ZntR recognition sequences that overlap with Zur binding boxes in the promoters of Zur regulons. Using single-molecule tracking and single-cell protein quantitation, we show that in live E. coli cells, the unmetallated ZntRapo can enhance the unbinding kinetics of both repressor and non-repressor forms of Zur from DNA, whereas metallated ZntRZn cannot. We further show, through in vitro single-molecule FRET measurements, that ZntRapo directly acts on Zur-DNA complexes, possibly through forming heteromeric ternary and quaternary complexes that involve both protein-DNA and ZntR-Zur protein-protein interactions; this direct action gives rise to a ‘through-DNA’ mechanism for their cross-actions in regulating Zn homeostasis. Moreover, this mechanism is likely functionally significant in facilitating the switching in Zn-uptake regulation when an E. coli cell encounters changing Zn environments, for example in an observed facilitated derepression of a Zn-uptake gene upon Zn depletion; it may even be broadly relevant for regulating uptake-vs.-efflux of Zn and other metals for different bacterial species and yeast. (Experimental details are described in Methods, Supplementary Methods, Supplementary Fig. 1–8, and Supplementary Table 1–4.)
Results
Zur cognate promoters contain partial ZntR recognition sequences
We examined the DNA sequences around Zur’s and ZntR’s operator sites in E. coli’s genome (Supplementary Notes 1; Supplementary Figs. 9 and 10). Strikingly, at the promoters of the four known Zur regulons [i.e., znuABC, zinT (a periplasmic Zn chaperone), l31p and l36p (a pair of ribosomal proteins), and pliG (a periplasmic lysozyme inhibitor)]28,32,44–46, the Zur box overlaps with sequences that match significantly with ZntR’s recognition sequence (Fig. 1b), suggesting possible direct involvement of ZntR in Zur-DNA interactions. At promoters of known ZntR regulons (e.g., zntA), we did not identify clear Zur binding sites.
ZntRapo enhances unbinding of repressor ZurZn from DNA in cells
The discovery of partial ZntR recognition sequences around Zur boxes prompted us to examine whether ZntR can affect Zur unbinding from DNA. We first examined how ZntRapo may affect ZurZn unbinding, as ZntRapo and ZurZn coexist in the cell under the normal Zn-replete conditions (Fig. 1a, center).
To visualize ZurZn in the cell, we tagged Zur at its C-terminus with the photoconvertible fluorescent protein mEos3.2 (i.e., ZurmE), either at its chromosomal locus or additionally on an inducible plasmid to access a broader range of cellular protein concentrations, as previously reported (Methods; Supplementary Methods 1.1; Supplementary Notes 2; Supplementary Figs. 11, 12)31. We cultured and imaged cells in the presence of 20 μM Zn2+, under which cellular Zur is known to be dominantly in its metallated repressor form 31. The unbinding kinetics of from DNA in the cell was measured using single-molecule tracking (SMT), as we reported (Supplementary Fig. 1; Supplementary Methods 1.2)31. Briefly, we used controlled photoconversion coupled with time-lapse stroboscopic imaging to track single molecules until the fluorescent tag photobleached (Fig. 2a). The displacement length r distribution obtained from SMT can resolve its three diffusion states: freely diffusing in the cytosol (FD), non-specifically bound to DNA (NB), and tightly bound to DNA (TB), including their effective diffusion constants and fractional populations (Fig. 2b; Supplementary Notes 3; Supplementary Figs. 13–16; Supplementary Tables 5 and 6). Thresholding the displacement-vs.-time trajectories allowed us to extract Zur’s microscopic residence times τ on DNA that are dominated by protein residing at tight binding sites (e.g., operator sites) (Supplementary Fig. 17). Using a three-state kinetic model previously validated (Fig. 2c)31, we can analyze the distribution of τ to extract the apparent unbinding rate constant k−1 (Fig. 2d; Supplementary Notes 4; Supplementary Figs. 17–19). This SMT, along with single-cell total fluorescence quantitation, also enabled us to quantify the protein copy number, from which protein concentration in the cell can be determined (Supplementary Methods 1.2.3).
Fig. 2. Single-molecule tracking shows that ZntRapo enhances the kinetics of ZurZn unbinding from DNA in cells.
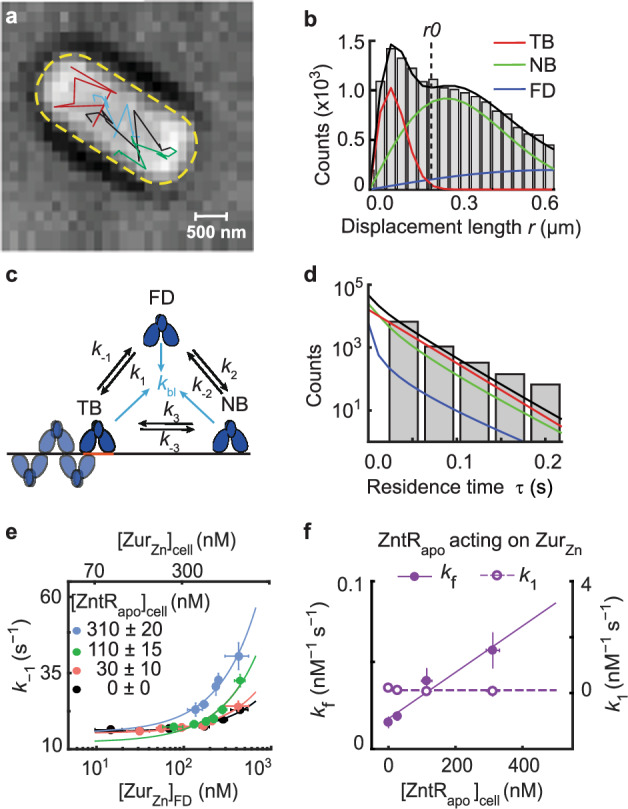
a Exemplary single-molecule tracking trajectories (colored lines) overlaid on the bright-field transmission image of a live E. coli cell from a strain expressing both ZurmE and from plasmids grown in the presence of 20 µM Zn2+. 16 biological replicates were conducted and in total 211,838 tracking trajectories were imaged in 4267 cells. Yellow dashed line: cell contour. b Exemplary distribution of 20454 displacement lengths r per time lapse (40 ms) for in cells having ~250 nM ZurmE and ~30 nM from a strain expressing both ZurmE and from plasmids grown in the presence of 20 µM Zn2+. Solid lines: the overall fit (black), and the resolved TB, NB, and FD diffusion states (Supplementary Notes 3). Vertical dashed line: the displacement threshold r0 below which >99.5% TB state is included. Bin width is 0.034 µm. c 3-state kinetic model for Zur-DNA interactions in cell. k’s: rate constants. d Histogram of 8435 microscopic residence time τ of in cells having ~250 nM ZurmE and ~30 nM . Black line: fit with Supplementary Eq. S12; the deconvoluted contributions from the three diffusion states are in red, green, and blue as color-coded in b. Bin width is 0.044 s. e Dependence of the apparent unbinding rate constant k−1 of on its own concentration and at different [] in the cell (black: n = 1379 cells; orange: n = 1152 cells; green: n = 978 cells; blue: n = 505 cells). Lines: fits with Eq. (1) including 1st and 3rd terms only. f The facilitated unbinding rate constant kf (solid circle) and the binding rate constant k1 (open circle) of vs. cellular [] (n = 4267 cells). Solid line: linear fit; dashed line: horizontal line fit. Error bars in (e–f) are SEM. Source data are provided as a Source Data file and also available in Supplementary Table 7.
For ZntRapo, because quantifying the metallated state of ZntR in vivo is challenging, we used the ZntRC115S mutant as a mimic. This C115S mutation at ZntR’s binuclear Zn-binding site abolishes its ability to activate Zn-efflux genes under Zn stress47, and the protein can still bind DNA tightly inside cells35; both are characteristic of ZntRapo. In vitro Zn-binding assay further confirmed that ZntRC115S’s Zn-binding affinity is severely diminished, weaker than 0.1 μM, 108 times weaker than wild-type ZntR (Supplementary Notes 5; Supplementary Fig. 20). Therefore, ZntRC115S behaves effectively as ZntRapo and is referred as so below. We further tagged ZntRapo with super-folder GFP (i.e., ) in an inducible plasmid on top of ΔzntR in the chromosome. This GFP tagging maintains ZntR’s function (Supplementary Notes 2) and allows for spectral separation from the red fluorescent form of and for quantification in the cell.
By sorting individual cells into groups of similar cellular and concentrations and analyzing each group separately (Supplementary Fig. 13), we overcame large cell-to-cell protein expression heterogeneity and determined the apparent unbinding rate constant k−1 from DNA for as a function of its cellular concentration and at different cellular [] (Fig. 2e). In the ΔzntR strain with no present, k−1 of increases with increasing free (and total) [] in the cell (Fig. 2e, black), reflecting its facilitated unbinding, as we reported previously31 where the slope is the facilitated unbinding rate constant kf (Eq. 1). The impeded unbinding for ZurZn occurs at lower than accessible concentrations in the cell and is unobservable here31. Strikingly, with increasing [], k−1 of progressively increases (Fig. 2e), and its slope, , increases linearly with [] (Fig. 2f, solid symbols), suggesting that ZntRapo can enhance the facilitated unbinding of .
The kinetic model in Fig. 2c also allowed for analyzing the fractional populations of Zur’s three states, leading to extraction of other kinetic parameters (Supplementary Table 7; Supplementary Notes 4.2)31, including ’s binding rate constant k1 to tight binding sites. Interestingly, k1 shows no dependence on [] (Fig. 2f, open symbols). This independence indicates that ZntRapo does not block ZurZn binding to operator sites and suggests that ZntRapo does not bind on its own to the partial recognition sequences that overlap with Zur boxes on DNA, but instead requires Zur to be already bound on DNA, possibly involving both ZntR-Zur interaction on DNA and the partial ZntR-DNA interaction.
ZurZn-DNA interactions are dynamic in vitro
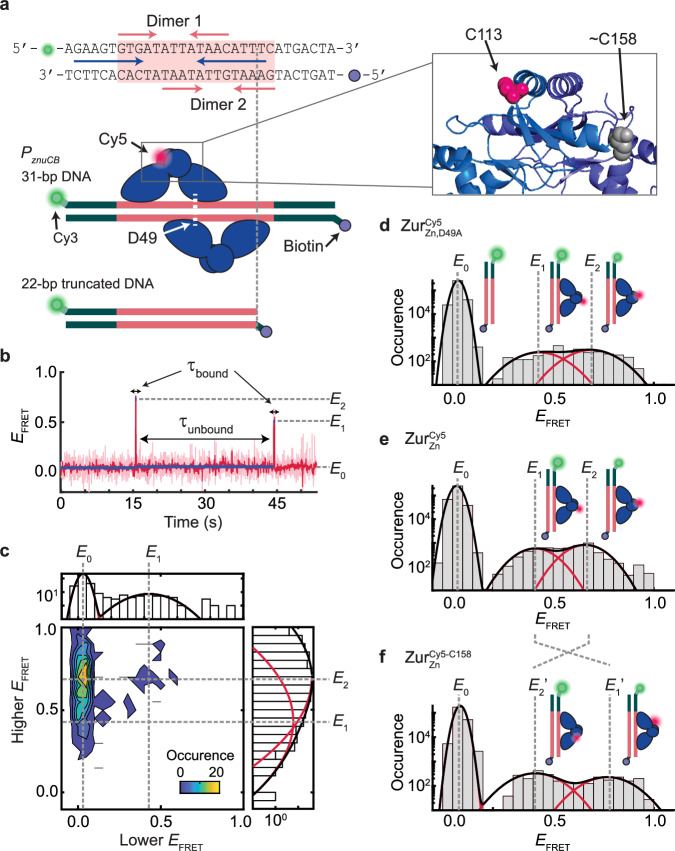
The above results show that the unbinding of the Zn-uptake repressor ZurZn from DNA can be enhanced by the apo-repressor form of the Zn-efflux regulator ZntRapo in the cell. Given the complexity of cellular environments, this enhancement may or may not be from direct actions of ZntRapo on DNA-bound ZurZn. To remove cellular complexity, we examined whether and how ZntRapo can directly affect ZurZn unbinding from its cognate DNA in vitro. Here we labeled a 31-bp DNA at one end with the FRET donor Cy3, immobilized it on a surface via biotin-neutravidin linkage at the other end, and examined its interaction with Cy5-labeled ZurZn (a homodimeric protein) in the surrounding solution using single-molecule FRET (smFRET) measurements (Fig. 3a; Supplementary Fig. 7; Supplementary Methods 1.3; Supplementary Notes 6–9; Supplementary Figs. 21–25; Supplementary Tables 8, 9). The DNA sequence was from the bidirectional promoter of E. coli’s znuCB/znuA genes, encompassing the two-dyad Zur binding box and a potential partial ZntR recognition sequence (Fig. 3a, top). We also constructed a 22-bp DNA, in which one dyad is truncated to allow only one Zur dimer to bind (Fig. 3a, lower).
Fig. 3. SmFRET measurements show dynamic ZurZn-DNA interactions in vitro.
a Top: the DNA oligomer sequence from E. coli znuCB promoter and Cy3/biotin label positions for smFRET measurements. Pink shade: Zur box; pink arrows: two dyads for binding two ZurZn dimers; blue arrows: potential ZntR recognition dyad (Fig. 1b). Bottom: Two different-length DNA constructs. A single Cy5 is labeled at C113 or C158 on Zur. Zoom-in box: C113 and approximate C158 positions (shown on different monomers for clarity) in the dimeric ZurZn’s crystal structure (PDB: 4MTD44; residues 153-171 are unresolved). b Representative single-molecule EFRET trajectory of an immobilized 22-bp truncated DNACy3 interacting with (Cy5 at C113) (4 nM). Pink line: raw data; red line: after non-linear filtering; blue line: mean value of each EFRET state. Three EFRET states (E0, E1, E2) are denoted. Dwell times on E0 state are designated as τunbound and those on higher states (E1 and E2) as τbound. c Two-dimensional histogram of the lower vs. higher EFRET state values from single-molecule EFRET trajectories, as in (b). Top/right: corresponding one-dimensional projections; red (black) lines: Gaussian resolved (overall) fits. d Histogram of EFRET trajectories, as in (b). e Same as (d), but with (Cy5 at C113) (4 nM). f Same as (d), but with (4 nM). Cartoons in (d–f) show free DNA and DNA-bound ZurZn in two binding orientations differentiated by the Cy5-label. The FRET donor (green sphere) and acceptor (red sphere) are drawn on DNA and Zur at their approximate locations. Histograms for (d–f) are compiled from 305, 389, and 238 EFRET trajectories, respectively; bin size = 0.05. Source data are provided as a Source Data file.
First, we studied this truncated 22-bp DNACy3 interacting with the mutant ; this D49A mutation eliminates the key inter-dimer salt-bridge interactions, resulting in dominantly single dimer-DNA interactions at up to 10 nM ZurZn concentrations44. The single Cy5 label is at C113 of one monomer of the dimeric protein (unless otherwise noted), and C113 is a non-conserved surface-exposed natural cysteine distant from Zur’s DNA-binding domains (Fig. 3a, inset; Supplementary Fig. 21; Supplementary Notes 6.1). At 4 nM , single-molecule EFRET trajectories show distinct states and their dynamic transitions: some show three EFRET states (Fig. 3b), but most show two states during the limited observation time before label photobleaching (Supplementary Fig. 31). We pooled hundreds of such trajectories and examined the two-dimensional histogram of lower vs. higher EFRET values: three states are clearly resolved at EFRET ~ 0.03, 0.43, and 0.69, denoted as E0, E1, and E2, respectively (Fig. 3c). The same three states also fit the one-dimensional EFRET histogram (Fig. 3d): the lowest EFRET state (E0) is assigned as the free DNA state, which dominates the distribution expectedly. The E1 and E2 states can be assigned as the two orientations of a single dimer bound to the truncated DNACy3, where the single Cy5-label breaks the symmetry (Fig. 3d, cartoons). Consistently, their relative populations are almost equal (slight difference could be due to the single Cy5 labeling).
We then studied , which contains the natural D49 residue for inter-dimer salt-bridge interactions, interacting with the truncated DNACy3. Expectedly, behaves similarly as , showing the same EFRET states (Fig. 3e), because the truncated DNACy3 only allows for one Zur dimer binding regardless whether or not Zur can form inter-dimer interactions. To further improve data analysis reliability, we globally fitted the results of and interacting with the truncated DNACy3 to obtain E1 and E2 values at ~0.44 and ~0.65, respectively (Supplementary Notes 7), which agree with predictions from the ZurZn-DNA complex structure (Supplementary Notes 6.2; Supplementary Table 8).
To further confirm the assignment of E1 and E2 states, we moved the Cy5 from C113 to C158, another non-conserved surface-exposed natural cysteine (; Fig. 3a, inset). Based on the ZurZn-DNA complex structure, this C158 position is closer to the FRET donor in the complex for the E1 orientation but further for the E2 orientation (Supplementary Fig. 22a–c; Supplementary Notes 6.2). Indeed, E1 increases to ~0.77 (E1′ state) and E2 decreases to ~0.41 (E2′ state), while maintaining the same population ratio (Fig. 3e, f).
ZntRapo enhances facilitated unbinding of ZurZn from DNA in vitro: a 'through-DNA' mechanism
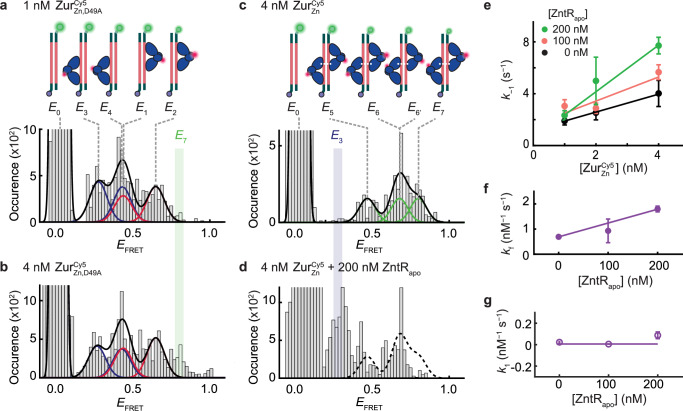
Next, we used the 31-bp DNACy3, which contains the full two-dyad Zur box to maximally bind two ZurZn dimers and also encodes the potential ZntR recognition sequence (Fig. 3a). For , the salt-bridge mutant that has weakened inter-dimer interactions, the EFRET histogram can be fitted by four protein-bound states (Supplementary Notes 7), with approximately equal populations, besides the free DNA state E0 at 1 nM protein concentration (Fig. 4a). Two of them, E1 and E2, are the same as those in ’s interaction with the truncated DNACy3 (Fig. 3d) and are thus assigned similarly as the two orientations of a single dimer bound at the dyad proximal to Cy3. The other two (E3 and E4) have lower EFRET values (~0.27 and ~0.43) and can be assigned as the two orientations of a single dimer bound at the dyad distal to Cy3 (Fig. 4a, cartoons). All EFRET values agree with predictions from the ZurZn-DNA structure (Supplementary Notes 6.2; Supplementary Table 8). Upon increasing [] to 4 nM, a new EFRET state appeared at ~0.8 (E7; Fig. 4b), attributable to two dimers concurrently bound to the 31-bp DNA, now possible due to increased protein concentration. Consistently, when we swapped out 75% of the 4 nM to its unlabeled form, the EFRET ~ 0.8 peak disappeared (Supplementary Fig. 33).
Fig. 4. ZntRapo enhances facilitated unbinding of ZurZn from DNA in vitro.
a Histogram of EFRET trajectories of immobilized 31-bp DNACy3 interacting with (1 nM). Colored lines: Gaussian resolved fits, where each color corresponds to one dimer at one of the two dyads of Zur box in two orientations, as shown by the inset cartoons. Black line: overall fit. Bin size = 0.02. b Same as (a), but with at 4 nM. E7 state (green-shaded) only appears when two dimers are bound to DNA. c Same as (a), but with (4 nM). Cartoons show DNA-bound with four different combinations of two dimer-bound form. d Same as (c) but with an introduction of unlabeled ZntRapo (200 nM). E3 state (blue-shaded) only appears when a single ZurZn dimer is bound to DNA. Histograms for (a–d) are compiled from 405, 433, 255, and 168 EFRET trajectories, respectively. e Apparent unbinding rate constant k−1 of on the 31-bp DNA vs. its own concentration at 0 (black), 100 (orange), 200 nM (green) of ZntRapo. Lines: linear fits; the slope is the facilitated unbinding rate constant kf for . kf are extracted from total 3238 dwell times. f ZntRapo-concentration-dependent kf for extracted from (e). kf are extracted from 1005, 1339, 894 dwell times for 0, 100, 200 nM of ZntRapo, respectively. Line: linear fit. g The binding rate constant k1 for to 31-bps DNA shows no significant dependence on [ZntRapo]. k1 are extracted from 249, 569, 493 dwell times for 0, 100, 200 nM of ZntRapo, respectively. Line: horizontal line fit. The binding and unbinding rate constants are extracted from single exponential fits (y = A*exp(-k*τ)) of the distributions of dwell time (Supplementary Fig. 34, 35), and their error bars are 90% confidence intervals from fits. Source data are provided as a Source Data file.
Then, we studied the 31-bp DNACy3 in interacting with . At 4 nM, is expected to bind as two dimers because of inter-dimer salt-bridge interactions44. Consistently, at EFRET ~ 0.27, the lowest EFRET state for a single dimer-bound form (i.e., E3 in Fig. 4a, b) and where the two-dimer-bound form should not have a FRET signal (Supplementary Notes 6.2; Supplementary Table 8), no population was observed (Fig. 4c). The higher EFRET states can be resolved into three states with a 1:2:1 population ratio at EFRET ~ 0.47, 0.68, and 0.80, denoted as E5, E6 (E6′), and E7, respectively, and assigned as the four different combinations of two-dimer-bound form, each dimer carrying a single Cy5 label (Fig. 4c, cartoons). Note that the E6 and E6′ states are unresolved from each other in the EFRET histogram. All these EFRET values also agree with the predictions from ZurZn-DNA complex structure (Supplementary Notes 6.2; Supplementary Table 8). Moreover, the E7 state is at the same position as the additional EFRET ~ 0.8 peak in Fig. 4b, when the salt-bridge mutant is at a higher concentration, supporting it being from the two-dimer-bound form.
We next added ZntRapo (i.e., ZntRC115S mutant) to probe whether it can enhance unbinding from the 31-bp DNACy3. Strikingly, E7, a two-dimer-bound state that does not overlap with any of the single-dimer-bound states, is substantially depopulated, while the E3 state, unique to single-dimer-bound form, appears (Fig. 4d; Supplementary Fig. 25g–l). Both observations indicate that ZntRapo disrupts ZurZn-DNA interactions. Moreover, the states at higher EFRET values (i.e., EFRET ~ 0.5–0.7) are more depopulated than those at lower EFRET values (i.e., EFRET ~ 0.3–0.5), which is even clearer at lower (Supplementary Notes 8). Therefore, the dimer bound at the proximal dyad to Cy3 is preferentially disrupted by ZntRapo, consistent with the fact that the partial ZntRapo recognition sequence on the 31-bp DNACy3 is proximal to Cy3 (Fig. 3a).
We further analyzed the residence times τbound and τunbound from the EFRET trajectories to extract ZurCy5-DNACy3 interaction kinetics (Fig. 3b; Supplementary Table 9 and Supplementary Figs. 34, 35). The unbinding rate constant k−1 for on the 31-bp DNA clearly shows a linear dependence on its own concentration (Fig. 4e, black), reflecting its facilitated unbinding that was observed in cells (Fig. 2e), where the slope is the facilitated unbinding rate constant kf31. Moreover, with increasing [ZntRapo], kf increases (Fig. 4e, f), as observed in cells (Fig. 2f, solid symbols), indicating that ZntRapo directly enhances the facilitated unbinding of ZurZn from DNA. Meanwhile, the binding rate constant k1 of is independent of ZntRapo (Fig. 4g), suggesting that ZntRapo alone does not bind directly to this 31-bp DNA, but needs incumbent ZurZn on DNA, similarly as observed in cells (Fig. 2f, open symbols). These results further support that possible ZntR-Zur interactions on DNA are important for ZntRapo-enhanced ZurZn unbinding from DNA.
Altogether, the in vitro experiments demonstrate that ZntRapo enhances Zur’s facilitated unbinding from DNA through its direct actions on Zur-DNA complex, enabled by the overlapping Zur and ZntR recognition sequences, where both protein-DNA and protein-protein interactions are important. We postulate that this ‘through-DNA’ mechanism possibly occurs via ZntRapo acting directly on the Zur-DNA ternary complex to form a heteromeric ZntR-Zur-DNA quaternary complex (Fig. 5f, step 5).
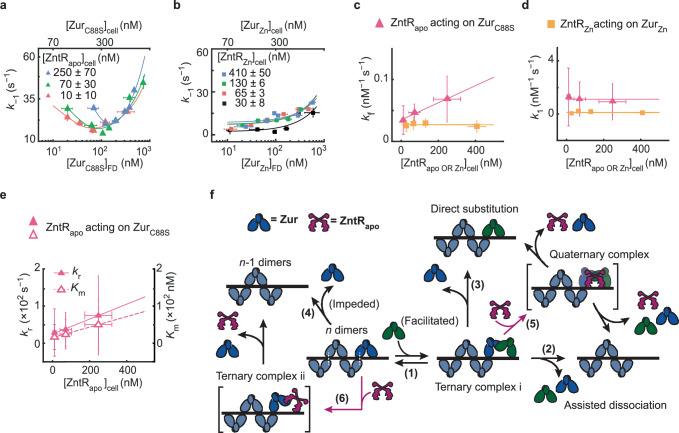
Fig. 5. ZntR effects on Zur-DNA interactions.
a Dependence of the apparent unbinding rate constant k−1 of on its own concentration and at different [] in the cell (orange: n = 514 cells; green: n = 642 cells; blue: n = 532 cells). Lines: fits with Eq. (1). b Same as (a), but for k−1 of and at different [] in the cell (black: n = 611 cells; orange: n = 287 cells; green: n = 494 cells; blue: n = 623 cells). Lines: fits with Eq. (1) including 1st and 3rd terms only. c The facilitated unbinding rate constant kf of vs. cellular [] (magenta triangle) and of vs. the cellular [] (yellow square) (n = 1978 cells). Lines: linear (magenta) and horizontal line (yellow) fits. d Same as (c) but for the binding rate constant k1. Lines: horizontal line fits (n = 2039 cells). e The impeded unbinding rate constant kr (solid triangle) and the effective oligomer dissociation constant Km (open triangle) of vs. cellular [] (n = 2039 cells). Lines: linear fits. Error bars in (a–e) are SEM. f Mechanistic model for ZntRapo-dependent Zur unbinding kinetics. Starting with oligomerized Zur (dark and light blue) at a tight-binding site on DNA, the unbinding of an incumbent Zur protein (dark blue) can be facilitated by a freely diffusing Zur (dark green) through the formation of a ternary complex i (step 1), leading to assisted dissociation (step 2) or direct substitution (step 3); this facilitated unbinding of Zur can be enhanced by ZntRapo through the formation of a heteromeric quaternary complex (step 5). The oligomer-induced impedance of Zur unbinding (step 4) can be weakened by ZntRapo through the formation of a heteromeric ternary complex ii (step 6), leading to faster Zur unbinding as well. White dashed lines on the ‘n dimers’ denote salt bridge interactions between Zur dimers. Source data are provided as a Source Data file and also available in Supplementary Table 7.
ZntRapo enhances unbinding of Zur non-repressor form as well
Having shown that ZntRapo directly acts on ZurZn-DNA complex to enhance ZurZn unbinding from its recognition sites, we continued to examine whether ZntRapo can act on the DNA-bound non-repressor form of Zur, as these two coexist in the cell under Zn-deficient (and replete) conditions (Fig. 1a). To ensure Zur being in the non-repressor form in the cell, we examined the ZurC88S mutant whose Cys88 at the regulatory Zn-binding site was mutated to make it a constitutive non-repressor1,44 and which binds tightly to DNA but at unidentified sites distinct from Zur boxes at its regulon promoters31. We again sorted individual cells into groups of similar [] and [] and analyze the groups separately.
At any cellular [], the apparent unbinding rate constant k−1 of shows the biphasic unbinding behavior (Fig. 5a): initially decreases with increasing cellular [] (i.e., impeded unbinding), reaches a minimum, and then increases toward higher concentrations (i.e., facilitated unbinding), as we previously discovered and described by Eq. 131. Strikingly, with increasing [], the biphasic behavior of k−1 shifts toward the upper-right of the plot (Fig. 5a). The extracted facilitated unbinding rate constant kf of increases (Fig. 5c, magenta), similarly as observed for ZurZn above, and consistent with ZntRapo acting on DNA-bound Zur non-repressor form, possibly through the same heteromeric quaternary complex (Fig. 5f, step 5).
The impeded unbinding rate constant kr of also increases with increasing [] (Fig. 5e, solid symbols); so does Km, the effective dissociation constant of Zur oligomerization on DNA (Fig. 5e, open symbols), which correlates with the minimum position of k−1 in Fig. 5a. Both changes of kr and Km indicate that ZntRapo can act on oligomerized ZurC88S on DNA, weakening its oligomerization and diminishing its impedance on unbinding. We attribute this weakening to ZntRapo directly interacting with oligomerized ZurC88S, for example via a possible heteromeric ternary complex (Fig. 5f, step 6). Since the non-repressor ZurC88S binds to sequences outside Zur’s regulon promoters, ZntRapo’s effects on ZurC88S-DNA interaction suggest that ZntR recognition sites must exist at other places on the chromosome as well. Indeed, we discovered previously >80 potential ZntR recognition sites across the E. coli chromosome35.
The binding rate constant k1 of shows no dependence on [] (Fig. 5d, magenta), suggesting that ZntRapo does not block binding to DNA and corroborating that ZntRapo’s interaction with the Zur-DNA complex involves ZntR-Zur interactions besides ZntR-DNA interactions (Fig. 5f, steps 5 and 6).
Combining the results on ZurZn and ZurC88S, ZntRapo enhances the unbinding of Zur from DNA, regardless of whether Zur binds tightly to Zur boxes or other sequences, in a ‘through-DNA’ mechanism. Using the kinetic model in Fig. 5f, we derived the relation between Zur’s unbinding rate constant k−1 and the concentrations of Zur and ZntRapo (Supplementary Eq. S43; Supplementary Notes 10). Supplementary Eq. S43 satisfactorily describes the experimental data, further supporting the ‘through-DNA’ mechanism for ZntRapo acting on DNA-bound Zur.
ZntRZn has no effect on repressor ZurZn unbinding from DNA
Having shown that ZntRapo can enhance the unbinding of both the repressor and non-repressor forms of Zur from DNA, we pondered about the metallated ZntRZn, which is the activator for Zn efflux. As ZntRZn and Zurnon-repressor do not coexist in the cell under physiological conditions (Fig. 1a), we examined ZntRZn’s effect on the unbinding of ZurZn, which coexist in the cell under Zn-excess conditions (i.e., in the presence of 100 μM Zn2+ in the media31,47; Fig. 1a, right). At a given [] in the cell, the apparent unbinding rate constant k−1 of expectedly shows facilitated unbinding (Fig. 5b). More importantly, changing ZntRZn’s cellular concentration has no discernible effect on the facilitated unbinding rate constant kf of , in contrast to that of ZntRapo (Fig. 5c, yellow vs. magenta). Therefore, ZntR’s interaction with Zur-DNA complex only applies to ZntR’s apo-repressor form and not its holo-activator form. This difference could come from that apo and holo ZntR have different conformations30, which may lead to their different interactions with Zur-DNA complex. Consistently, the binding rate constant k1 of shows no dependence on [], either (Fig. 5d, yellow).
Functional consequence of ZntRapo on the expression of Zur regulon
In bacteria’s natural habitats, the availability of essential micronutrients like Zn2+ can fluctuate substantially. The ability of the efflux regulator ZntRapo in enhancing the unbinding of both the repressor and non-repressor forms of the uptake regulator Zur from DNA could have functional significance in facilitating the switching in Zn-uptake regulation when an E. coli cell encounters changing Zn environments. Starting from Zn-replete conditions where ZurZn repressor and ZntRapo coexist in the cell (Fig. 1a, center), if the cell encounters a Zn deficient environment, the enhancement of ZurZn’s unbinding from its regulon promoters by free ZntRapo would facilitate the derepression of Zn-uptake genes. On the other hand, starting from Zn deficient conditions where Zur’s non-repressor form coexists with ZntRapo in the cell (Fig. 1a, left), if the cell moves into Zn-replete or excess conditions, ZntRapo-enhanced Zur unbinding from non-operator sites would facilitate Zur release from DNA for binding to operator sites, upon Zn-metallation, to repress Zn uptake.
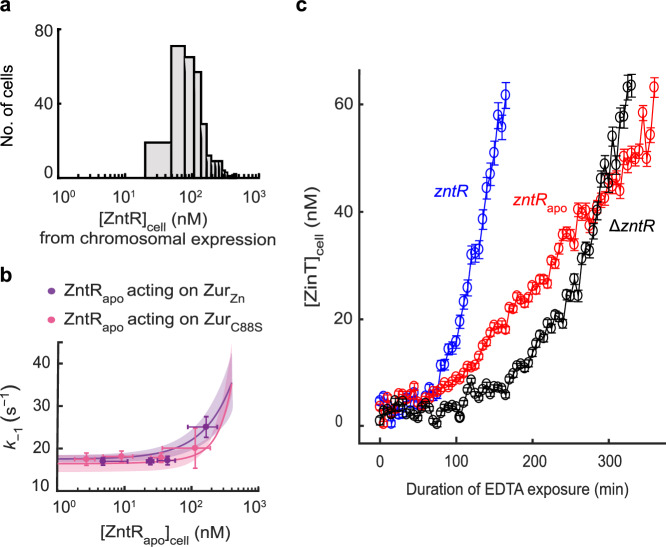
Using chromosomal tagging only, we determined ZntR’s physiological concentration in the cell to range from ~30 to ~400 nM (Fig. 6a). Under physiological expression from chromosomal locus, the cellular concentration of Zur is expected to be similar to that of ZntR9,48, and we previously determined that it ranged from ~50 to ~250 nM or from ~60 to ~300 nM when E. coli cells were grown in minimal or Zn2+-supplemented media28. Using the experimentally determined kinetic parameters of Zur-DNA interactions in the cell in the presence of ZntRapo (Supplementary Table 7), we simulated the dependences of the apparent unbinding rate constant k−1 of ZurZn (repressor) and ZurC88S (non-repressor) on [ZntRapo] in these physiological concentration ranges (Fig. 6b, lines and bands). ZntRapo can enhance k−1 of ZurZn and ZurC88S from ~16 to ~35 s−1, by a factor of ~2. We then sorted out the individual cells that possessed the physiological [Zur] range, and extracted the apparent Zur unbinding rate constant, k−1, for different [ZntRapo] concentration groups within ZntR’s physiological concentration range; the results expectedly fall within simulation predictions (Fig. 6b, symbols). We further simulated such dependences using kinetic parameters from in vitro smFRET measurements (Supplementary Table 9); again, ZntRapo can enhance k−1 of ZurZn by a factor of ~4 in the physiological protein concentration ranges (Supplementary Fig. 36). Altogether, these results suggest that facilitation by ZntRapo could have significant kinetic effects in transcription regulation by Zur.
Fig. 6. ZntRapo-induced enhancement of Zur unbinding from DNA is correlated with more facile derepression of zur regulon in cells under physiological expression.
a Distribution of [ZntRmE] in the cell, when expressed solely from its chromosomal locus under our imaging condition in regular M9 media. b Dependence of the apparent unbinding rate constant of (n = 2888 cells) and of (n = 1978 cells) on [] in the cell (symbols), with both proteins in the range of their respective physiological concentrations. x, y axes both in logscale. Error bars are SEM. Lines: Simulated dependences of k−1 of ZurZn and ZurC88S on [ZntRapo] in the cell in physiological concentration ranges using Eq. S43 and experimentally determined rate constants (Supplementary Table 7); colored bands: simulated upper/lower bounds in the ranges of 119 ± 33 nM (average ± s.d.) for ZurC88S (pink) and 150 ± 48 nM for ZurZn (purple) that correspond to the physiological Zur concentrations in E. coli grown in minimal or Zn2+-supplemented media, respectively31. c Time profiles of cellular protein expression of zinTG, a Zur regulated gene, upon 2 mM EDTA treatment in chromosomal ΔzntR, zntRapo, and wild-type zntR strains. Error bars are SEM from imaging >100 cells in three independent replicates (n = 223 cells) (zntRapo, red); 220 cells (ΔzntR, black); 156 cells (wild-type zntR, blue). Source data are provided as a Source Data file.
To probe whether ZntRapo can indeed influence transcription regulation by Zur, we examined the derepression in the cell of chromosomal zinT gene (tagged with the fast-maturing sfGFP for quantitation), a periplasmic zinc chaperone whose transcription is repressed by ZurZn (Fig. 1b)45. Here we added EDTA to deplete extracellular Zn2+ to induce derepression (Supplementary Methods 1.2.4). In the ΔzntR strain, the derepression of ZinT is reflected by an increase of [ZinT] in the cell after ~100 min EDTA exposure (Fig. 6c, black). (Note that the timescale here is an overall timescale that encompasses many underlying processes, including ZurZn unbinding from regulon promoter as well as the lowering of intracellular Zn2+ concentration that could be a slow process.) Strikingly, in the zntRapo strain whose chromosomal zntR gene carries the C115S mutation, such ZinT derepression appears much earlier at ~70 min EDTA exposure (Fig. 6c, red), supporting that the presence of ZntRapo can indeed facilitate the derepression of Zn uptake genes when E. coli encounters changing Zn environment (i.e., Zn depletion here). Moreover, when the chromosomal zntR gene is unmodified and cellular ZntR exists initially in a mixture of apo and holo forms, the derepression of ZinT occurs even faster (Fig. 6c, blue), which we attribute to that ZntRapo can facilitate the derepression as well as that the cell initially had higher levels of Zn-efflux pumps (activated by ZntRZn) and thus faster response to EDTA-induced Zn depletion.
Discussion
The through-DNA actions of an efflux regulator on its corresponding uptake regulator, or vice versa, likely extend beyond E. coli to other bacterial species and other metal ion homeostasis (Supplementary Notes 1). We find that partial recognition sequences of ZntR or its homolog also exist around Zur boxes in other bacteria, such as in S. typhimurium and P. aeruginosa (Fig. 1b and Supplementary Fig. 9). Such sequence overlaps also occur to other Zn efflux-uptake regulator pairs, including the CzrA-Zur pair in B. subtilis and the SczA-AdcR pair in P. aeruginosa (Supplementary Fig. 10a, b), as well as to bacterial efflux-uptake regulator pairs for other metals like Fe and Ni (Supplementary Fig. 10c, d). Moreover, opposite to the pattern of partial efflux regulator recognition sequences near uptake regulator boxes, Helmann et al. identified two potential Fur (Fe uptake regulator) recognition sites around a known PerR (Fe efflux regulator) binding box49. Similar through-DNA actions between regulator pairs could even exist in yeast, a higher organism, as partial efflux regulator recognition sequences can be found near promoter binding boxes of uptake regulators for Fe homeostasis (Supplementary Fig. 10e). Altogether, these observations of sequence overlaps on DNA suggest a broad relevance of the through-DNA mechanism between uptake and efflux regulators of many metals, across a range of regulatory proteins, and through different levels of organisms, which may constitute another mechanistic paradigm for metal regulation in biology.
Methods
Construction of strains for in vivo and in vitro experiments
For live cell single-molecule imaging and tracking of Zur as a function of Zur and ZntR concentrations in E. coli BW25113 cells, the proteins Zur and ZntR were tagged genetically with photo-convertible protein mEos3.2 and sfGFP, respectively, to separate them spectrally. The zur and zntR genes were either tagged chromosomally or the tagged genes were encoded into a plasmid rather than in the chromosome, to access a broad range of protein concentrations.
For in vitro smFRET measurement, site-directed mutagenesis was used to make Zur variants that contain a uniquely labelable cysteine in each monomer to label Zur with the FRET acceptor Cy5. All Zur variants and ZntR(C115S) mutant were cloned in a pET3a vector, and the proteins were expressed in E. coli (BL21 DE3) cells. See details in Supplementary Methods.
Protein purification and DNA labeling for in vitro smFRET
All Zur variants were purified as previously described44. Briefly, the proteins are overexpressed in cells by isopropyl-beta-D-thiogalactopyranoside (IPTG) and then lysed with lysozyme in lysis buffer, followed by freeze and thaw cycle and sonication. The proteins were collected by centrifugation, then the protein was purified by a series of columns. Protein purity was confirmed by SDS-PAGE, quantified using UV measurement at 280 nm, and protein identity was confirmed by mass spectrometry. After then, Cy5 FRET acceptor was labeled at the targeted cysteine in protein via maleimide chemistry. The mono-labeled fraction was purified using an anion exchange column. For ZntR(C115S) mutant, the protein is expressed in the same way as Zur using IPTG, and purified as previously described30,47. Briefly, the supernatant was collected after centrifugation and the proteins were precipitated out with 45% saturated (NH4)2SO4 overnight. The precipitated proteins were resuspended in Tris buffer and purified via a series of columns. Protein purity was confirmed by SDS-PAGE, quantified using Bradford assay. Protein identity was confirmed by mass spectrometry. For Cy3 labeling to DNA, the Cy3 and biotin-tagged DNA oligomeric strands were purchased from Integrated DNA Technologies (IDT, Coralville, IA) and dissolved in Buffer, and annealed together. Two types of double-strand DNA (dsDNA) constructs were used. The sequences of both constructs were from the znuCB gene promoter and contain the specific two-dyad sequence recognized by two Zur dimers and the complementary. The other construct is truncated DNA that has only one dyad sequence and its complementary. See details in Supplementary Methods 1.3.
Sample preparation, experimental procedure, and data processing for live cell single-molecule tracking and protein quantification
The E. coli cells were grown in LB medium overnight and later diluted in minimal media with vitamins, amino acids and glucose. Cells were grown to an OD600 of 0.3 and l-arabinose was used to induce plasmid expression when applicable. Zn2+ was used for Zinc stress to a final concentration of 20uM or 100uM. The cells were then washed with the same minimal media, pelleted, and added onto an agarose gel pad in a glass slide which was then sandwiched by a glass coverslip pretreated with gold nano-particles as position markers, and sealed with epoxy-glue.
An Olympus IX71 inverted microscope with a TIRF oil immersion objective (Olympus PlanApo N 60× oil 1.45) and an EMCCD camera (Andor Technology, DU-897E-CSO-#BV, pixel size 16 × 16 μm2) was used to perform the imaging. For single-molecule tracking and protein quantification for mEos3.2-tagged Zur was done as reported previously (Supplementary Fig. 1)31,35. First, a 405 nm (1–100 W/cm2) (CrystaLaser, DL405-100) laser was used to photo-convert a single mEos3.2 tagged Zur proteins from their green to red emissive forms. The photo-converted protein was then tracked with 561 nm laser (21 kW/cm2) (Coherent, Sapphire 561-200CW), with 4 ms exposure time and a time-lapse of 40 ms; the fluorescence was collected through a dichroic filter (Chroma, Z408/488/561 rpc) inside the filter cube and a red band pass emission filter (Semrock, FF01-617/73). This was done for several cycles, to obtain a single molecule tracking movie. To quantify the total Zur concentration in the cells, the 405 nm laser was used to photo-convert all the mEos3.2 proteins to their red fluorescent form. The 561 nm laser was then used to obtain the total red intensity of the cell. This step was repeated multiple times to photo-bleach all red mEos3.2 proteins. After this step, the total ZntR concentration in the cell was quantified using the 488 nm (7 kW/cm2) laser (CrystaLaser, DL488-050) to obtain the intensity of all the green sfGFP protein tagged ZntR in the cell. The fluorescence was collected through the dichroic filter and a green emission filter (Chroma, ET525/50 M).
We used custom MATLAB codes, iQPALM35, to analyze the single molecule tracking movies. Here, the candidate single-molecule spots were selected within defined cell boundaries using a two-dimensional Gaussian fitting and for each determined spot, the intensity of the single mEos3.2 was extracted. The total Zur copy number in the cells was obtained by dividing the whole cell mEos3.2 red intensity by the single molecule intensity. The total cellular ZntR concentration was also determined in a similar way from the whole cell sfGFP green intensity and the single sfGFP intensity determined from separate experiments. See Supplementary Methods 1.2 for details.
Resolution and extraction of effective Zur diffusion states in the cells
By determining the centroid position of each single molecule spot using the two-dimensional Gaussian fitting as described above and in Supplementary Methods 1.2, we were able to extract the position trajectory of the single molecules, as done previously31,35. From these trajectories, we could also obtain the displacement lengths, r, of individual mE-tagged Zur proteins.
Further the cells were sorted by their Zur and ZntR concentrations, into similar concentration ranges to overcome large cell-to-cell heterogeneity, and each individual concentration group was analyzed.
The distributions of the displacement lengths at different Zur and ZntR concentrations, were globally fitted with a probability distribution function, using linear combinations of three Brownian diffusion states, assuming a quasi-static approximation31,35. From the fitting results, we could extract the effective diffusion coefficients (D’s) and their corresponding fractional population (A’s). The assignment of the three diffusive states, corresponding to three Zur populations, cytoplasmic diffusion (FD), Tight binding to DNA (TB) or non-specific binding to DNA (NB) were previously reported and rationalized31. See Supplementary Notes 3 for details.
Sample preparation, imaging, and data analysis for in vitro smFRET studies
To immobilize DNA for in vitro studies, quartz slides were first amine-functionalized, followed by coating with biotinylated-polyethylene glycol (PEG) polymers. The biotinylated terminal group forms biotin-neutravidin linkages for immobilizing biotinylated DNA molecules (Supplementary Fig. 7)50,51. Coverslips were also amine-functionalized and coated with PEG polymers. A microfluidic channel was formed by double-sided tape sandwiched between a quartz slide and a borosilicate cover slip. After then, neutravidin, Cy3-labeled biotinylated DNA solution flowed through the channel for immobilization. Then, the Cy5-labeled Zur solution containing an oxygen scavenging system52 in the same buffer, and if applicable, containing ZntRapo, was flowed for fluorescence imaging.
The single-molecule fluorescence experiments were performed using a prism-type total internal reflection microscope based on an Olympus IX71 inverted microscope, similarly as we previously reported51,53,54. The immobilized Cy3-labeled DNA was excited by a continuous-wave circularly polarized 532-nm laser (CrystaLaser, GCL-025-L-0.5%) on the sample. The fluorescence of both Cy3 and Cy5 was collected by a 60× NA 1.2 water-immersion objective and split by a dichroic mirror into two channels using a Dual-View system (Optical Insights). Each channel of fluorescence was further filtered (Chroma, HQ580-60m or HQ660LP) and projected onto one-half of the imaging area of an EMCCD camera (Andor Ixon DV887) controlled by Andor IQ software. All image analysis was done by custom-written codes in MATLAB (compatible with MATLAB R2019b, Supplementary Software S1). Individual Cy3 and Cy5 fluorescence intensity trajectories were extracted and the FRET efficiency (EFRET) was computed as an approximation using the relationship: ICy5/(ICy5 + ICy3), where ICy3 and ICy5 are the fluorescence intensities. In order to obtain higher resolution EFRET histograms, a forward-backward non-linear (fnbl) filter was used to reduce the noise in the fluorescence trajectories (Supplementary Fig. 8)55,56 and thresholded to distinguish EFRET states. EFRET value of each state was taken from the original EFRET trajectories to avoid value changes by fbnl filtering. See details in Supplementary Methods 1.3.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
This research is supported by NIH Grant GM109993. We thank Dr. Bing Fu of Cornell University for discussions.
Author contributions
U.K.C. designed and performed live-cell imaging experiments, constructed strains, expressed and purified proteins, performed biochemical experiments, wrote MATLAB codes, analyzed data, and wrote the manuscript; Y.P. designed and performed in vitro smFRET experiments, expressed and purified proteins, performed biochemical experiments, wrote MATLAB codes, analyzed data, and wrote the manuscript; K.S. purified and characterized Zur proteins, performed early smFRET and biochemical experiments; W.J. wrote MATLAB codes for smFRET image analysis; C.P.J. constructed Zur expression plasmids, designed mutant Zur variants, and purified Zur proteins; D.H.F. contributed to plasmid constructions; P.C. directed research and wrote the manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
All data are available in the main text, the Supplementary Information, or Source Data. Raw data supporting the findings of this study are available upon request due to their substantial volume. Source data are provided with this paper.
Code availability
MATLAB codes and detailed instructions are included in Supplementary Software 1.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Udit Kumar Chakraborty, Youngchan Park.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-55017-z.
References
- 1.Outten, C. E. & O’Halloran, T. V. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science292, 2488–2492 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Foster, A. W., Osman, D. & Robinson, N. J. Metal preferences and metallation. J. Biol. Chem.289, 28095–28103 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reyes-Caballero, H., Campanello, G. C. & Giedroc, D. P. Metalloregulatory proteins: metal selectivity and allosteric switching. Biophys. Chem.156, 103–114 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Capdevila, D. A., Wang, J. & Giedroc, D. P. Bacterial strategies to maintain zinc metallostasis at the host-pathogen interface. J. Biol. Chem.291, 20858–20868 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murdoch, C. C. & Skaar, E. P. Nutritional immunity: the battle for nutrient metals at the host–pathogen interface. Nat. Rev. Microbiol.20, 657–670 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brophy, M. B. & Nolan, E. M. Manganese and microbial pathogenesis: sequestration by the mammalian immune system and utilization by microorganisms. ACS Chem. Biol.10, 641–651 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodgkinson, V. & Petris, M. J. Copper homeostasis at the host-pathogen interface. J. Biol. Chem.287, 13549–13555 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Djoko, K. Y., Ong, C. Y., Walker, M. J. & McEwan, A. G. The role of copper and zinc toxicity in innate immune defense against bacterial pathogens. J. Biol. Chem.290, 18954–18961 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osman, D. et al. Bacterial sensors define intracellular free energies for correct enzyme metalation. Nat. Chem. Biol.15, 241–249 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waldron, K. J., Rutherford, J. C., Ford, D. & Robinson, N. J. Metalloproteins and metal sensing. Nature460, 823–830 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Coleman, J. E. Zinc enzymes. Curr. Opin. Chem. Biol.2, 222–234 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Yu, Y. et al. A brief history of metal recruitment in protozoan predation. Trends Microbiol.32, 465–476 (2024). [DOI] [PubMed] [Google Scholar]
- 13.Hu, Y.-M., Boehm, D. M., Chung, H., Wilson, S. & Bird, A. J. Zinc-dependent activation of the Pho8 alkaline phosphatase in Schizosaccharomyces pombe. J. Biol. Chem.294, 12392–12404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dosanjh, N. S. & Michel, S. L. Microbial nickel metalloregulation: NikRs for nickel ions. Curr. Opin. Chem. Biol.10, 123–130 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Graham, A. I. et al. Severe zinc depletion of Escherichia coli. J. Biol. Chem.284, 18377–18389 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lonergan, Z. R. & Skaar, E. P. Nutrient zinc at the host–pathogen interface. Trends Biochem. Sci.44, 1041–1056 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osman, D. et al. Fine control of metal concentrations is necessary for cells to discern zinc from cobalt. Nat. Commun.8, 1884 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tottey, S., Harvie, D. R. & Robinson, N. J. Understanding how cells allocate metals using metal sensors and metallochaperones. Acc. Chem. Res.38, 775–783 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Waldron, K. J. & Robinson, N. J. How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Microbiol.7, 25–35 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Argüello, J. M., Raimunda, D. & Padilla-Benavides, T. Mechanisms of copper homeostasis in bacteria. Front. Cell. Infect. Microbiol. 3, 73 (2013). [DOI] [PMC free article] [PubMed]
- 21.Huat Lu, Z., Dameron, C. T. & Solioz, M. The Enterococcus hirae paradigm of copper homeostasis: copper chaperone turnover, interactions, and transactions. Biometals16, 137–143 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Bütof, L., Große, C., Lilie, H., Herzberg, M. & Nies, D. H. Interplay between the Zur regulon components and metal resistance in Cupriavidus metallidurans. J. Bacteriol. 201, e00192–19 (2019). [DOI] [PMC free article] [PubMed]
- 23.Shafer, C. M., Tseng, A., Allard, P. & McEvoy, M. M. Strength of Cu-efflux response in Escherichia coli coordinates metal resistance in Caenorhabditis elegans and contributes to the severity of environmental toxicity. J. Biol. Chem.297, 101060 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Summers, A. O. Damage control: regulating defenses against toxic metals and metalloids. Curr. Opin. Microbiol.12, 138–144 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Helsel, M. E. & Franz, K. J. Pharmacological activity of metal binding agents that alter copper bioavailability. Dalt. Trans.44, 8760–8770 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunsaker, E. W. & Franz, K. J. Emerging opportunities to manipulate metal trafficking for therapeutic benefit. Inorg. Chem.58, 13528–13545 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frei, A., Verderosa, A. D., Elliott, A. G., Zuegg, J. & Blaskovich, M. A. T. Metals to combat antimicrobial resistance. Nat. Rev. Chem.7, 202–224 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patzer, S. I. & Hantke, K. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol. Microbiol.28, 1199–1210 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Brocklehurst, K. R. et al. ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol. Microbiol.31, 893–902 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Outten, C. E., Outten, F. W. & O’Halloran, T. V. DNA distortion mechanism for transcriptional activation by ZntR, a Zn(II)-responsive MerR homologue in Escherichia coli. J. Biol. Chem.274, 37517–37524 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Jung, W., Sengupta, K., Wendel, B. M., Helmann, J. D. & Chen, P. Biphasic unbinding of a metalloregulator from DNA for transcription (de)repression in live bacteria. Nucleic Acids Res.48, 2199–2208 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hantke, K. Bacterial zinc uptake and regulators. Curr. Opin. Microbiol.8, 196–202 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Grass, G., Wong, M. D., Rosen, B. P., Smith, R. L. & Rensing, C. ZupT is a Zn(II) uptake system in Escherichia coli. J. Bacteriol.184, 864–866 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grass, G. et al. ZitB (YbgR), a member of the cation diffusion facilitator family, is an additional zinc transporter in Escherichia coli. J. Bacteriol.183, 4664–4667 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen, T.-Y. et al. Concentration- and chromosome-organization-dependent regulator unbinding from DNA for transcription regulation in living cells. Nat. Commun.6, 7445 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen, T. Y., Cheng, Y. S., Huang, P. S. & Chen, P. Facilitated unbinding via multivalency-enabled ternary complexes: new paradigm for protein-DNA interactions. Acc. Chem. Res.51, 860–868 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamar, R. I. et al. Facilitated dissociation of transcription factors from single DNA binding sites. Proc. Natl. Acad. Sci. USA114, E3251–E3257 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erbaş, A. & Marko, J. F. How do DNA-bound proteins leave their binding sites? The role of facilitated dissociation. Curr. Opin. Chem. Biol.53, 118–124 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spinks, R. R. et al. DnaB helicase dynamics in bacterial DNA replication resolved by single-molecule studies. Nucleic Acids Res.49, 6804–6816 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson, A. & van Oijen, A. M. Bacterial replication, transcription and translation: mechanistic insights from single-molecule biochemical studies. Nat. Rev. Microbiol.11, 303–315 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Ma, C. J., Gibb, B., Kwon, Y., Sung, P. & Greene, E. C. Protein dynamics of human RPA and RAD51 on ssDNA during assembly and disassembly of the RAD51 filament. Nucleic Acids Res.45, 749–761 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duzdevich, D., Redding, S. & Greene, E. C. DNA dynamics and single-molecule biology. Chem. Rev.114, 3072–3086 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaniecki, K., De Tullio, L. & Greene, E. C. A change of view: homologous recombination at single-molecule resolution. Nat. Rev. Genet.19, 191–207 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gilston, B. A. et al. Structural and mechanistic basis of zinc regulation across the E. coli Zur regulon. PLoS Biol.12, e1001987 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panina, E. M., Mironov, A. A. & Gelfand, M. S. Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc. Natl. Acad. Sci. USA100, 9912–9917 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hemm, M. R. et al. Small stress response proteins in Escherichia coli: proteins missed by classical proteomic studies. J. Bacteriol.192, 46–58 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khan, S., Brocklehurst, K. R., Jones, G. W. & Morby, A. P. The functional analysis of directed amino-acid alterations in ZntR from Escherichia coli. Biochem. Biophys. Res. Commun.299, 438–445 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Young, T. R. et al. Calculating metalation in cells reveals CobW acquires CoII for vitamin B12 biosynthesis while related proteins prefer ZnII. Nat. Commun.12, 1195 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pinochet-Barros, A. & Helmann, J. D. Bacillus subtilis Fur is a transcriptional activator for the PerR-repressed pfeT gene, encoding an iron efflux pump. J. Bacteriol.202, e00697–19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roy, R., Hohng, S. & Ha, T. A practical guide to single-molecule FRET. Nat. Methods5, 507–516 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Joshi, C. P. et al. Direct substitution and assisted dissociation pathways for turning off transcription by a MerR-family metalloregulator. Proc. Natl. Acad. Sci. USA109, 15121–15126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aitken, C. E., Marshall, R. A. & Puglisi, J. D. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J.94, 1826–1835 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martell, D. J. et al. Metalloregulator CueR biases RNA polymerase’s kinetic sampling of dead-end or open complex to repress or activate transcription. Proc. Natl. Acad. Sci. USA112, 13467–13472 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keller, A. M. et al. Dynamic multibody protein interactions suggest versatile pathways for copper trafficking. J. Am. Chem. Soc.134, 8934–8943 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chung, S. H. & Kennedy, R. A. Forward-backward non-linear filtering technique for extracting small biological signals from noise. J. Neurosci. Methods40, 71–86 (1991). [DOI] [PubMed] [Google Scholar]
- 56.Haran, G. Noise reduction in single-molecule fluorescence trajectories of folding proteins. Chem. Phys.307, 137–145 (2004). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All data are available in the main text, the Supplementary Information, or Source Data. Raw data supporting the findings of this study are available upon request due to their substantial volume. Source data are provided with this paper.
MATLAB codes and detailed instructions are included in Supplementary Software 1.