Abstract
Tuberculosis drug resistance contributes to the spread of tuberculosis. Immunotherapy is an effective strategy for treating tuberculosis, with the regulation of macrophage-mediated anti-tuberculosis immunity being crucial. Norcantharidin (NCTD), a drug used in tumor immunotherapy, has significant immunomodulatory effects. Thus, NCTD may have an anti-tuberculosis role by regulating immunity. Understanding how NCTD affects the proteome of Mtb-infected macrophages can provide valuable insights into potential treatments. This study aimed to investigate the impact of NCTD (10 μg/mL) on the proteome of macrophages infected with Mtb H37Ra using liquid chromatography-tandem mass spectrometry (LC–MS/MS) analysis. A total of 69 differentially regulated proteins (DRPs) were identified, with 28 up-regulated and 41 down-regulated in the NCTD-treated group. Validation of six DRPs (CLTCL1, VAV1, SP1, TRIM24, MYO1G, and WDR70) by Western blot analysis confirmed the accuracy of the LC–MS/MS method used in this study. NCTD modulates various protein expressions involved in chromatin-modifying enzymes, RHO GTPases activating PAKs, Fc gamma R-mediated phagocytosis, T cell receptor signaling pathway, and antigen processing and presentation. Overall, the research provides new insights into the effects of NCTD on the proteome of Mtb-infected macrophages. The identified changes highlight potential targets for future therapeutic interventions aimed at enhancing host immunity against Mtb infection or developing new anti-TB drugs based on these findings.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12953-024-00235-y.
Keywords: Norcantharidin, Mycobacterium tuberculosis, H37Ra, Macrophage, Proteome
Introduction
Tuberculosis is a serious public health problem that significantly threatens human well-being. Despite efforts to control its spread, the number of new cases remains alarmingly high. In 2021, an estimated 10.6 million people became ill with tuberculosis, and 1.6 million people died from it [1]. The infection rates have caused severe public health issues worldwide. The effectiveness of prevention and treatment for tuberculosis is inadequate due to several reasons: the prevalence of drug-resistant strains, the ineffectiveness of BCG (Bacillus Calmette-Guerin) vaccination in adults [2], difficulties in diagnosing latent infections, and poor treatment compliance. However, immunotherapy has gained attention in recent years as it offers a promising approach by modulating the host's immune status for treating tuberculosis.
Macrophages are host cells for Mycobacterium tuberculosis (Mtb) and serve as the first line of defense against this pathogen. However, pulmonary alveolar macrophages can also assist in the bacteria's escape from immune killing [3]. The autophagy function of macrophages is weakened, promoting the survival of intracellular mycobacteria [4]. The expression of MHC (major histocompatibility complex) molecules on the surface of macrophages is downregulated [5], leading to polarization towards an M2 phenotype [6]. Apoptosis induced by endoplasmic reticulum stress further promotes the survival of intracellular mycobacteria [5]. Therefore, modulating the immune function of macrophages is a suitable strategy for treating tuberculosis.
Norcantharidin, a derivative of cantharidin. It is mainly used in clinical practice for treating various cancers such as liver cancer, esophageal cancer, and gastric cancer, showing strong anti-tumor activity [7, 8] and unique immunomodulatory effects [9–11]. Currently, there are no reports on the use of Norcantharidin in tuberculosis treatment. However, recent studies have shown that Norcantharidin can enhance immune responses in LPS-induced macrophages by boosting AKT/p65 phosphorylation and NF-kB transcriptional activity. It also enhances macrophage secretion of IL-1β, IL-6, and TNF-α for indirect bactericidal effects [12, 13], regulates iron metabolism in macrophages through ferritin modulation [14, 15], induces autophagy and apoptosis in innate lymphoid cells via activation of the JNKs/c-Jun signaling pathway to promote their aggregation [16], and shows promising efficacy in improving disease progression by inhibiting IL-17 through STAT3 regulation in an acute lung injury model [17]. These findings collectively suggest potential immunomodulatory properties of Norcantharidin against MTB.
Currently, there is no published report about work reporting the characterization of Norcantharidin on the proteome of Mtb-infected macrophages; however, its regulatory influence on macrophages has not been fully elucidated. It has been hypothesized that Norcantharidin may affect the proteome of Mtb-infected macrophages, consequently influencing the production of numerous factors essential for anti-TB activity. Accordingly, in this work, using the tandem mass tag (TMT)-based quantitative proteomics method and liquid chromatography-tandem mass spectrometry (LC–MS/MS) analysis, we present, for the first time, a comprehensive proteomic analysis of macrophages infected with H37Ra.
Materials and methods
Main reagents and consumables
The primary reagents used in the LC–MS/MS analysis included Nanosep Centrifugal Devices 10 kD (Pall) and the iTRAQ 8 plex kit (SCIEX). For Western blot analysis, the following antibodies were used: anti-SP1 (ab231778; Abcam, USA), anti-TRIM24 (4208–1-AP; Proteintech, USA), anti-CLTCL1 (22,283–1-AP; Proteintech, USA), anti-VAV1 (AF6182; Affinity, USA), anti-WDR70 (PU451766S; Abmart, Shanghai, China), anti-MYO1G (LK43614S; Abmart, Shanghai, China), anti-Actin (Cell Signaling, USA), goat anti-rabbit IgG (SA00001-2; Proteintech, USA), and goat anti-mouse IgG (074–1806; KPL, USA). Norcantharidin was obtained from Shanghai Yuanye Bio-Technology Co., Ltd (5442–12-6).
Cell culture
THP-1 cells (Laboratory conservation) were maintained in RPMI-1640 (Solarbio) supplemented with 10% FBS (WISENT, Canada), 100 U/mL penicillin, and 100 μg/mL streptomycin at 37 °C and 5% CO2. All media components and reagents were endotoxin-free.
Macrophage phenotype induction and Mycobacterium tuberculosis H37Ra intracellular infection
THP-1 monocytes were cultured in a 10 cm diameter petri dish with 10^7 cells per well and induced to the macrophage phenotype by exposure to 1 μg/mL Phorbol 12-Myristate 13-Acetate (PMA, MultiSciences) for 24 h. Twenty-four hours after washing off PMA and incubating in complete medium (DMEM supplemented with 10% FBS), the cells were exposed to 10^8 colony forming units (CFU) of Mtb H37Ra (MOI = 10) in antibiotic-free medium for 6 h. The cells were then washed three times with PBS to remove extracellular bacteria and incubated in complete medium with Norcantharidin (5 μg/mL or 10 μg/mL) for 24 h. Afterwards, the cells were rinsed three times with PBS, 1 mL of fresh PBS was added, and the cells were scraped off, centrifuged, and the supernatant was discarded. The cell pellets were stored at −80 °C for subsequent use. Protein extraction using RIPA was performed for Western blot analysis.
Protein extraction
Protein extraction was performed by homogenizing 10^7 cells in 200 µL of lysis buffer (RIPA containing 1% PMSF) on ice for 30 min and then centrifuged (10,000 × g for 5 min at 4 °C). The supernatants were quantified using the Pierce BCA Protein Assay Kit. All analyses were performed in duplicate. Samples were stored at − 80 °C for further analysis.
Enzymatic digestion and iTRAQ labeling in protein sample solution
After pooling, the samples were evaporated by vacuum concentration to remove excess water, TEAB, and isopropanol. Aliquots of lysates were mixed with 200 μL of 8 M urea in Nanosep Centrifugal Devices (PALL). The device was centrifuged at 14,000 × g at 20 °C for 20 min. All following centrifugation steps were performed under the same conditions to ensure maximum concentration. The concentrate was diluted with 200 μL of 8 M urea in 0.1 M Tris–HCl, pH 8.5, and the device was centrifuged. Proteins were reduced with 10 mM DTT for 2 h at 56 °C. Subsequently, the samples were incubated in 5 mM iodoacetamide for 30 min in the dark to block reduced cysteine residues followed by centrifugation. The resulting concentrate was diluted with 200 μL of 8 M urea in 0.1 M Tris–HCl, pH 8.0, and concentrated again. This step was repeated twice, and the concentrate was subjected to proteolytic digestion overnight at 37 °C. The digests were collected by centrifugation. Each iTRAQ reagent (AB Sciex, USA) was dissolved in 70 μL of isopropanol and added to the respective peptide mixture for 120 min. The labeling reaction was quenched by the addition of 100 μL of Milli-Q water, and the seven labeled samples were then pooled into one sample according to the manufacturer’s instructions.
LC–MS/MS analysis
The lyophilized peptide fractions were re-suspended in ddH2O containing 0.1% formic acid, and 2 μL aliquots were loaded into a nanoViper C18 trap column (Acclaim PepMap 100, 75 μm × 2 cm). Chromatography separation was performed on the Easy nLC 1200 system (Thermo Fisher). The trapping and desalting procedures were carried out with 20 μL of 100% solvent A (0.1% formic acid). Then, an elution gradient of 5–38% solvent B (80% acetonitrile, 0.1% formic acid) over 60 min was used on an analytical column (Acclaim PepMap RSLC, 75 μm × 25 cm C18-2 μm 100 Å). Data-dependent acquisition (DDA) mass spectrometry techniques were used to acquire tandem MS data on a Thermo Fisher Q Exactive mass spectrometer fitted with a Nano Flex ion source. Data was acquired using an ion spray voltage of 1.9 kV and an interface heater temperature of 275℃. For a full mass spectrometry survey scan, the target value was 3 × 106, and the scan range was 350 to 2,000 m/z at a resolution of 70,000 with a maximum injection time of 100 ms. For the MS2 scan, only spectra with a charge state of 2–5 were selected for fragmentation by higher-energy collision dissociation with a normalized collision energy of 32. The MS2 spectra were acquired in the ion trap in rapid mode with an AGC target of 8,000 and a maximum injection time of 50 ms. Dynamic exclusion was set for 25 s.Peptides were analyzed using high-performance liquid chromatography tandem high-resolution mass spectrometry after labeling with iTRAQ reagent and subsequent enrichment, generating substantial mass spectrum data. PD software was used for protein identification in the samples under the following conditions: PSM FDR (false positive) < 0.01 and Protein FDR < 0.01. Protein identification was performed using the human Uniprot sequence database (20,399), with the database sequence file located at data/1.Identification/Uniprot-human.fasta.
The annotation of differentially functional proteins
Protein annotation involved using multiple functional databases, such as REACTOME and KEGG, to annotate the functions of identified proteins and reveal their functional classifications.
Network and functional analysis
To analyze the involvement of differentially regulated proteins in common biological processes, an enrichment ontology and pathway analysis was performed using g:Profiler software (version e104_eg51_p15_3922dba) based on REACTOME and KEGG databases (p-adjusted < 0.05).
Validation of LC–MS/MS results by Western blot
To validate the LC–MS/MS results, we performed a Western blot analysis for six chosen proteins (CLTCL1, VAV1, SP1, TRIM24, MYO1G, and WDR70) selected from the list of significantly changed proteins. The protein extract was used for Western blot analysis. Actin was used as a control for equal loading and to quantify the relative abundance of the examined proteins. Forty micrograms of total protein isolates were solubilized in a sample buffer (100 mM Tris–HCl, 4% SDS, 20% glycerol, 0.2% bromophenol blue, and 200 mM dithiothreitol, pH 6.8) and incubated for 7 min at 99.9℃. Samples were separated by SDS-PAGE electrophoresis and transferred onto a polyvinylidene fluoride (PVDF) membrane (Millipore, Ireland). A 5% BSA solution in TBST was used to block nonspecific binding. Next, the membranes were incubated at 4℃ overnight with specific primary antibodies: SP1 (1:1000; ab231778; Abcam, USA), TRIM24 (1:1000; 14,208–1-AP; Proteintech, USA), CLTCL1 (1:1000; 22,283–1-AP; Proteintech, USA), VAV1 (1:1000; AF6182; Affinity, USA), WDR70 (1:500; PU451766S; Abmart, Shanghai, China), MYO1G (1:500; LK43614S; Abmart, Shanghai, China), and actin (1:3000; Cell Signaling, USA). After washing, membranes were incubated with HRP-conjugated secondary antibodies (1.5 h, RT): goat anti-rabbit IgG (1:5000; SA00001-2, Proteintech, USA) for CLTCL1 (CHC22: clathrin heavy chain 22), VAV1, SP1, TRIM24, MYO1G, and WDR70, and goat anti-mouse IgG for actin (1:5000; 074–1806, KPL, USA). Visualization of immunocomplexes was carried out using chemiluminescence HRP substrate (Merck Millipore, Burlington, MA, USA) according to the manufacturer’s protocol and visualized with an AI 680 Imaging System (GE, USA). The results were quantified by optical density (OD) analysis of immunocomplexes with Image J (National Institutes of Health, USA). Data were presented as a ratio of examined protein relative to actin protein in arbitrary OD units. The normality of Western blot data distributions was confirmed using a Shapiro–Wilk test (p > 0.05), and the results were statistically checked by a Student’s t-test (p < 0.05) using Statistica software (GraphPad Prism 8, USA). Data were presented as mean ± SEM (n = 5).
Statistical analysis
A normality test was first performed to determine whether our dataset followed a normal distribution. Student’s t-test for two-sample and two-tailed comparisons was used with GraphPad Prism version 8.0 (GraphPad Software Inc., San Diego, CA, USA). In all cases, P < 0.05 was considered statistically significant.
Results
Quantitative analysis of differential protein expression
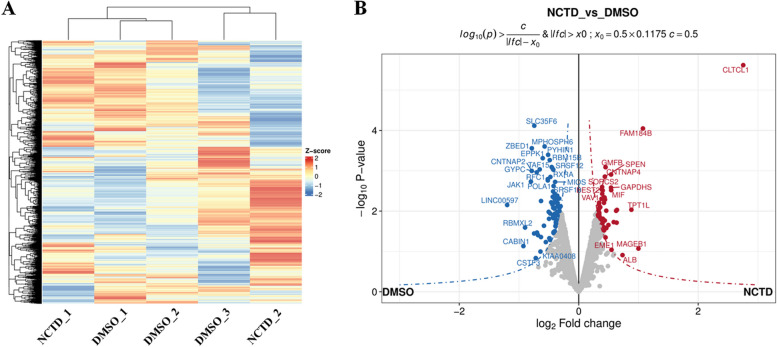
A total of 5599 proteins were identified in this study (Fig. 1A, Table S1). After relative quantification, filters (fold change cutoff 1.2 and p < 0.05) were applied to obtain the final list of 69 differentially regulated proteins (DRPs). Among all DRPs, 28 were up-regulated and 41 were down-regulated in the NCTD-treated group (Table S2). DRPs were visualized in a Volcano plot (Fig. 1B). In Volcano plots, the x-axis represents the logarithm base 2 of the ratio between comparison groups, ensuring a symmetrical distribution of ratios. A positive log2(ratio) indicates high protein expression while a negative log2(ratio) indicates low protein expression (Fig. 1B).
Fig. 1.
Volcano plot of proteins quantified by LC–MS/MS in NCTD vs. DMSO comparison. After intracellular infection with Mycobacterium tuberculosis H37Ra for 24 h, quantitative analysis of differential protein expression was performed using LC–MS/MS for NCTD (10 μg/mL, n = 2) vs. DMSO (n = 3) treatment. The heatmap illustrates the total number of identified proteins (A). The volcano plot displays the fold-change (x-axis) versus significance (y-axis) of the differentially regulated proteins (DRPs) under the influence of NCTD (B). The significance (non-adjusted p-value) and fold-change are converted to − Log10(p-value) and Log2(fold-change), respectively. The vertical and horizontal dotted lines indicate the cut-offs for fold-change (Log2 > 0) and p-value (0.05), respectively. The selected DRPs are labeled with their gene symbols
Differentially regulated proteins and functional annotations
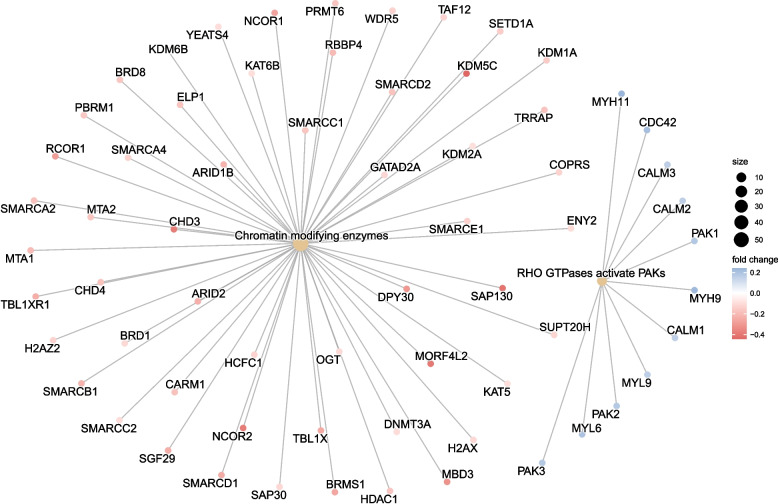
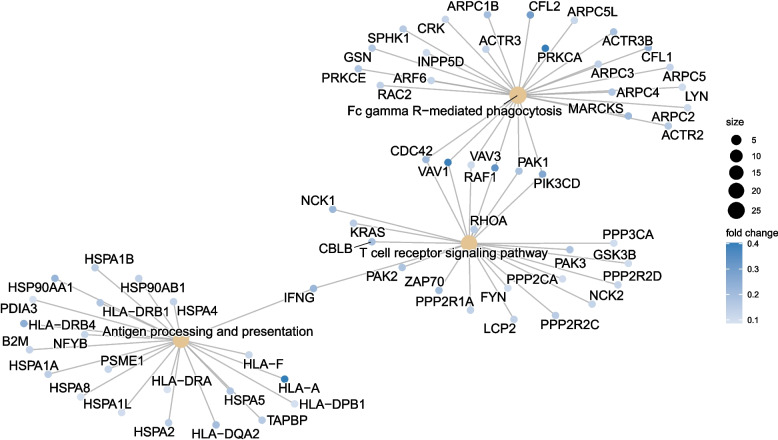
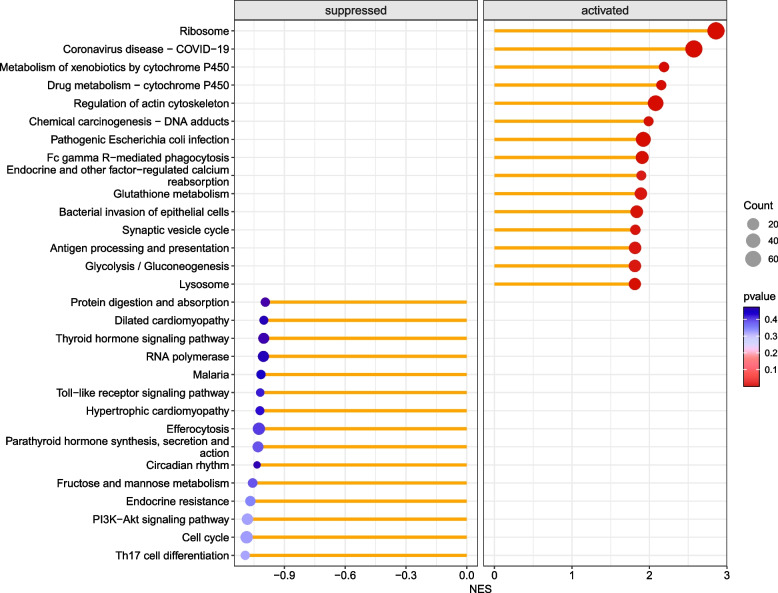
Through functional analysis of differential proteins, the identified proteins were subjected to enrichment or statistical analysis in various functional databases, including Cnetplot (REACTOME), Cnetplot (KEGG), GSEA (KEGG), and GSEA (REACTOME). Most of the DRPs were annotated to ‘Chromatin modifying enzymes’ (54 DRPs, Fig. 2), ‘RHO GTPases activate PAKs’ (11 DRPs, Fig. 2), ‘Fc gamma R-mediated phagocytosis’ (27 DRPs, Fig. 3), ‘T cell receptor signaling pathway’ (23 DRPs, Fig. 3), Antigen processing and presentation (23 DRPs, Fig. 3), Ribosome and Pathogenic Escherichia coli infection (Fig. 4), and Glycolysis/Gluconeogenesis (Fig. 4).
Fig. 2.
Cnetplot of enriched pathways of DRPs identified through REACTOME analysis. The REACTOME analysis revealed that the primary enriched pathways of DRPs included Chromatin modifying enzymes and RHO GTPases activating PAKs. The cnetplot visually depicts these enriched pathways
Fig. 3.
Cnetplot of enriched pathways of DRPs identified through KEGG analysis. The KEGG analysis revealed that the primary enriched pathways of DRPs included Fc gamma R-mediated phagocytosis, T cell receptor signaling pathway, and antigen processing and presentation
Fig. 4.
Enrichment of functional entries in genetic variations determined using KEGG annotation datasets. The genes upregulated and downregulated after NCTD treatment were annotated separately by KEGG. The color represents the negative log transformation of the adjusted p-value, with red being the most significant and blue being less significant. Term size is indicated by the size of the dot, while term names are indicated on the y-axis. The magnitude of NES reflects the extent to which gene sets are enriched on the x-axis
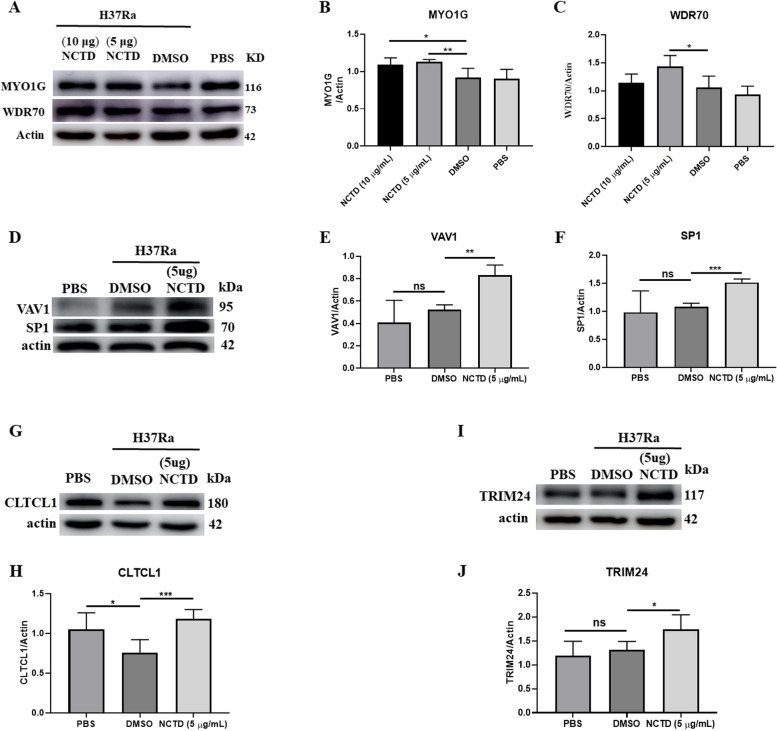
Revalidation of differentially expressed proteins through Western blot analysis
We used functional and differential expression analysis to identify genes involved in the onset and progression of Mtb, which were then validated by Western blot analysis. The results showed a significant upregulation in the expression of MYO1G (Fig. 5A,B), WDR70 (Fig. 5A,C), VAV1 (Fig. 5D,E), SP1 (Fig. 5D,F), CLTCL1 (Fig. 5G,H), and TRIM24 (Fig. 5I,J) in THP-1-derived macrophages following Norcantharidin treatment.
Fig. 5.
Validation of differentially expressed proteins through Western blot analysis. The identified differential proteins were validated through Western blot in THP-1 macrophages. Validation was performed for WDR70, MYO1G, VAV1, SP1, CLTCL1, and TRIM24 proteins with actin as the reference protein (P-value < 0.05). THP-1 macrophages were infected intracellularly with Mycobacterium H37Ra at MOI = 10 for 24 h. NCTD was added 6 h post-infection and continued until 24 h to eliminate extracellular bacteria. Representative Western blots demonstrate the alteration in protein expression of MYO1G (A), WDR70 (B), CLTCL1 (C), VAV1 (D), SP1 (E), and TRIM24 (F) in the NCTD-treated group (5 μg/mL and 10 μg/mL) compared to the control group (DMSO and PBS). Data were obtained from three independent experiments and are expressed as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control
Discussion
In this study, an LC–MS/MS analysis was performed for the first time to investigate differentially regulated proteins (DRPs) in THP-1-derived macrophages infected with Mtb and exposed to NCTD (10 μg/mL). After NCTD treatment, we identified 69 DRPs, with 28 proteins up-regulated and 41 proteins down-regulated compared to the control DMSO group. The differential expression of six proteins associated with tuberculosis immunity was confirmed by Western blotting. These findings strongly suggest that NCTD significantly influences the immune response against tuberculosis in macrophages and holds potential as an anti-tuberculosis therapeutic agent.
NCTD exhibits significant anti-tumor effects, clinically validated in cancer treatment. In addition to directly inhibiting tumor growth [18–20], NCTD demonstrates notable immunomodulatory effects. It can suppress immune-mediated pro-cancer effects induced by immune factors [21], regulate iron metabolism in macrophages [14]. However, in vitro studies on human peripheral blood immune cells have revealed that NCTD can inhibit the production of IL-2, IL-4, and IL-10 [22]. The immune functions regulated by NCTD are closely linked with anti-TB immunity, necessitating further exploration of their potential application in anti-TB immunotherapy. Additional investigation is required to elucidate its precise mechanisms of immunomodulation. This study reveals enrichment of differentially expressed proteins in the chromatin modifying enzymes and RHO GTPases activate PAKs signaling pathways. The DRPs were significantly enriched in key macrophage functions involved in anti-TB immunity, including Fc gamma R-mediated phagocytosis, T cell receptor signaling, antigen processing and presentation, lysosomal activity, and glycolysis. The findings imply that NCTD potentially exerts a pivotal influence on the regulation of macrophage differentiation and functionality.
The immune response of macrophages to Mtb plays a crucial role in the occurrence and progression of tuberculosis [3]. Targeting macrophages is an effective strategy for treating tuberculosis [23–25]. Macrophages are the primary target cells for M. tuberculosis infection, with the bacteria capable of persisting within them for extended periods. Understanding the mechanisms of immune evasion will help identify treatment targets for tuberculosis. The main strategies employed by M. tuberculosis to evade host immunity include disrupting the CD4 + T cell-macrophage immunological synapse [26], inhibiting DNA modification of host macrophages [27], and affecting lysosomal function [28]. Enhanced anti-tuberculosis immunity can be achieved through the activation of Fc gamma receptors [29] and the promotion of glycolysis [30]. Promoting macrophage autophagy can effectively eliminate tuberculosis bacteria [31, 32], making these important targets for treatment.
The THP-1 cell line, a human mononuclear cell line, serves as the primary cellular model for investigating the immune response of host macrophages to tuberculosis [33–38]. However, our study did not examine the impact of NCTD on TB bacterial growth in vitro and in vivo; this aspect will be the focus of future investigations. Furthermore, we utilized the Mtb H37Ra strain to establish a cell infection model. While H37Rv is a standard virulent strain, H37Ra exhibits reduced virulence. Infection with the virulent Mtb strain H37Rv elicits a more severe inflammatory immune response accompanied by increased tissue damage, providing insights into host–pathogen interaction dynamics and pathogenesis features across different Mtb strains [39, 40]. Nevertheless, due to less stringent environmental requirements, H37Ra-infected cells are commonly employed as cellular models [33, 41–44], which are subsequently validated using the virulent Mtb strain H37Rv.
Among the DRPs, we identified the most significantly expressed genes associated with infection: CLTCL1, VAV1, SP1, TRIM24, MYO1G, and WDR70. The CLTCL1 gene encodes the clathrin heavy chain 22 (CHC22) protein, which plays a crucial role in cellular glucose metabolism [45, 46] and autophagy [47]. We observed a significant up-regulation of CLTCL1 following NCTD treatment, suggesting that it may be a primary mechanism through which NCTD regulates macrophage glucose metabolism or cell autophagy to exert anti-tuberculosis immunity. VAV guanine nucleotide exchange factor 1 (VAV1) acts as a guanine nucleotide exchange factor, influencing foam macrophage formation [48] and macrophage chemotaxis [49]. It also plays a pivotal role in promoting NLRP3 inflammasome activation [50]. The cellular transcription factor Sp1 is involved in regulating macrophage polarization [51–53], pyroptosis [54], and CD8 + T cell exhaustion [55]. The E3 ligase Trim24 regulates M2 macrophage polarization through crosstalk with Stat6 [56]. Macrophage polarization, autophagy, and the immunomodulatory function of macrophages are closely associated with anti-tuberculosis immunity. Myo1G, a member of class I myosins, is specifically expressed in hematopoietic cells and is localized to the plasma membrane. It plays a crucial role in regulating cell elasticity through deformations of the actin network at the cell cortex [57]. Recent studies have shown that MYO1G is essential for B lymphocyte adhesion and migration [58], which are closely linked to macrophage-mediated anti-TB immune function [3, 59, 60]. Additionally, WDR70 is involved in DNA modification [61] and histone modification [62], contributing to epigenetic regulation [63]. Since TB exploits host DNA damage repair as its main strategy for immune evasion [27], this implies that NCTD's primary mechanism in targeting macrophages to exert its anti-TB effect may involve the regulation of these key proteins.
Supplementary Information
Supplementary Material 1: Figure S1. GSEA analysis of REACTOME for DRPs between the NCTD and DMSO groups. The Enrichment Score from the GSEA analysis of DRPs enriched in chromatin modifying enzymes was visualized through line plots and heat maps. Figure S2. GSEA analysis of KEGG for DRPs between the NCTD and DMSO groups. The Enrichment Score from the GSEA analysis of DRPs enriched in Fc gamma R-mediated phagocytosis, T cell receptor signaling pathway, and antigen processing and presentation was visualized through line plots and heat maps. Figure S3. Coomassie brilliant blue staining of gels. The integrity of protein samples was assessed using SDS-PAGE gel electrophoresis followed by Coomassie brilliant blue staining. Figure S4. The morphological characteristics show the viability of THP-1 cells treated with the NCTD. Following a 24-hour treatment of THP-1 cells with NCTD, the cellular morphology and density were examined under a microscope at various magnifications.
Acknowledgements
We thank the staffs of the Guangdong Second Provincial General Hospital and Guangdong Medical University for their help during this study, the Wininnovate Bio Company (Shenzhen, China) for data analysis.
Abbreviations
- NCTD
Norcantharidin
- LC–MS/MS
Liquid chromatography-tandem mass spectrometry
- Mtb
Mycobacterium tuberculosis
- DRPs
Differentially regulated proteins
- CLTCL1
Clathrin heavy chain like 1
- VAV1
VAV guanine nucleotide exchange factor 1
- SP1
Sp1 transcription factor
- TRIM24
E3 ligase Trim24
- MYO1G
Myosin IG
- WDR70
WD repeat domain 70
- BCG
Bacillus Calmette-Guerin
- MHC
major histocompatibility complex
- LPS
Lipopolysaccharide
- TMT
Tandem mass tag
- THP-1
Human acute monocytic leukemia cell line
Authors’ contributions
YLW, YTL, GBL, JLW and XRL performed the experiments, analyzed the data and drafted the manuscript. XXG, QDH, JL, JYO, YRD, JYW, YBL and YCG helped to analyze the data or revise the manuscript. XZC and JAZ were responsible for leading this work and revising the manuscript. All authors reviewed the manuscript.
Funding
This work was supported by the Guangdong Basic and Applied Basic Research Foundation (2021B1515140068, 2022A1515140190, 2019A1515110066), the Science and Technology Project of Dongguan (20211800904782, 20221800905862, 202050715005178), the Medical Science and Technology Research Foundation of Guangdong (A2023068, A2018434), the Science and Technology Program of Shenzhen (JCYJ20220530144402005), the Shenzhen Futian District Health Public Welfare Research Project (FTW2021057), the Guangdong Climbing Project Foundation (pdjh2022b0223), and the Science and Technology Innovation Fund of Guangdong Medical University (GDMU2021025, GDMU2022251, GDMU2023005).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
This study does not entail experiments involving human or animal subjects.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yi-Lin Wu, Yuan-Ting Li and Gan-Bin Liu contributed equally to this work.
Contributor Information
Xiao-Zhen Cai, Email: imcaixiaozhen@163.com.
Jun-Ai Zhang, Email: zhangjunai@gdmu.edu.cn.
References
- 1.Bagcchi S. Who’s global tuberculosis report 2022. Lancet Microb. 2023;4(1):e20. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez AH, Flores-Valdez MA. Can immunization with bacillus calmette-guerin be improved for prevention or therapy and elimination of chronic mycobacterium tuberculosis infection? Expert Rev Vaccines. 2019;18(12):1219–27. [DOI] [PubMed] [Google Scholar]
- 3.Lovey A, Verma S, Kaipilyawar V, et al. Early alveolar macrophage response and il-1r-dependent t cell priming determine transmissibility of mycobacterium tuberculosis strains. Nat Commun. 2022;13(1):884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paik S, Kim KT, Kim IS, et al. Mycobacterial acyl carrier protein suppresses tfeb activation and upregulates mir-155 to inhibit host defense. Front Immunol. 2022;13:946929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su H, Zhu S, Zhu L, et al. Recombinant lipoprotein rv1016c derived from mycobacterium tuberculosis is a tlr-2 ligand that induces macrophages apoptosis and inhibits mhc ii antigen processing. Front Cell Infect Microbiol. 2016;6:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang SA, Ali Y, Fedotova E, et al. Morphoproteomics identifies the foamy alveolar macrophage as an m2 phenotype with pd-l1 expression in the early lesion of post-primary tuberculosis: Implications for host immune surveillance and therapy. Ann Clin Lab Sci. 2020;50(4):429–38. [PubMed] [Google Scholar]
- 7.Wang L, Otkur W, Wang A, et al. Norcantharidin overcomes vemurafenib resistance in melanoma by inhibiting pentose phosphate pathway and lipogenesis via downregulating the mtor pathway. Front Pharmacol. 2022;13:906043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei M, Jiang Y, Sun R, et al. Self-assembly of a linear-dendritic polymer containing cisplatin and norcantharidin into raspberry-like multimicelle clusters for the efficient chemotherapy of liver cancer. ACS Appl Mater Interfaces. 2023;15(11):14664–77. [DOI] [PubMed]
- 9.Zhai BT, Sun J, Shi YJ, et al. Review targeted drug delivery systems for norcantharidin in cancer therapy. J Nanobiotechnology. 2022;20(1):509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao S, Wang Y, Ma W, et al. Intraperitoneal administration of thermosensitive hydrogel co-loaded with norcantharidin nanoparticles and oxaliplatin inhibits malignant ascites of hepatocellular carcinoma. Drug Delivery. 2022;29(1):2713–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou J, Ren Y, Tan L, et al. Norcantharidin: Research advances in pharmaceutical activities and derivatives in recent years. Biomed Pharmacother. 2020;131:110755. [DOI] [PubMed] [Google Scholar]
- 12.Zhao Q, Qian Y, Li R, et al. Norcantharidin facilitates lps-mediated immune responses by up-regulation of akt/nf-kappab signaling in macrophages. PLoS ONE. 2012;7(9):e44956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu XH, Blazsek I, Comisso M, et al. Effects of norcantharidin, a protein phosphatase type-2a inhibitor, on the growth of normal and malignant haemopoietic cells. Eur J Cancer. 1995;31A(6):953–63. [DOI] [PubMed] [Google Scholar]
- 14.Zheng J, Qian ZM, Sun YX, et al. Downregulation of hepcidin by norcantharidin in macrophage. Nat Prod Res. 2023;38(4):1–6. [DOI] [PubMed] [Google Scholar]
- 15.Zheng J, Wang JJ, Ma HM, et al. Norcantharidin down-regulates iron contents in the liver and spleen of lipopolysaccharide-treated mice. Redox Rep. 2022;27(1):119–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han Z, Li B, Wang J, et al. Norcantharidin inhibits sk-n-sh neuroblastoma cell growth by induction of autophagy and apoptosis [J]. Technol Cancer Res Treat. 2017;16(1):33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du LJ, Feng YX, He ZX, et al. Norcantharidin ameliorates the development of murine lupus via inhibiting the generation of il-17 producing cells. Acta Pharmacol Sin. 2022;43(6):1521–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu P, Wang S, Liu M, et al. Norcantharidin inhibits cell growth by suppressing the expression and phosphorylation of both egfr and c-met in human colon cancer cells. BMC Cancer. 2017;17(1):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z, Sun B, Lu J, et al. Norcantharidin inhibits the malignant progression of cervical cancer by inducing endoplasmic reticulum stress. Mol Med Rep. 2024;29(5):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li XP, Jing W, Sun JJ, et al. A potential small-molecule synthetic antilymphangiogenic agent norcantharidin inhibits tumor growth and lymphangiogenesis of human colonic adenocarcinomas through blocking vegf-a,-c,-d/vegfr-2,-3 “multi-points priming” mechanisms in vitro and in vivo. BMC Cancer. 2015;15:527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao Y, Li W, Liu R, et al. Norcantharidin inhibits il-6-induced epithelial-mesenchymal transition via the jak2/stat3/twist signaling pathway in hepatocellular carcinoma cells. Oncol Rep. 2017;38(2):1224–32. [DOI] [PubMed] [Google Scholar]
- 22.Chen YC, Chang SC, Wu MH, et al. Norcantharidin reduced cyclins and cytokines production in human peripheral blood mononuclear cells. Life Sci. 2009;84(7–8):218–26. [DOI] [PubMed] [Google Scholar]
- 23.Li B, Wang W, Zhao L, et al. Photothermal therapy of tuberculosis using targeting pre-activated macrophage membrane-coated nanoparticles. Nat Nanotechnol. 2024;19(6):834–45. [DOI] [PubMed] [Google Scholar]
- 24.Gupta A, Thirunavukkarasu S, Rangel-Moreno J, et al. Phospholipase c epsilon-1 (plce1) mediates macrophage activation and protection against tuberculosis. Infect Immun. 2024;92(4):e0049523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park JY, Kim HD, Abekura F, et al. A novel mycobacterium tuberculosis antigen, mtb48 enhances inflammatory response in lps-induced raw 264.7 macrophage immune cells. Mol Immunol. 2024;166:50–7. [DOI] [PubMed] [Google Scholar]
- 26.Mwebaza I, Shaw R, Li Q, et al. Impact of mycobacterium tuberculosis glycolipids on the cd4+ t cell-macrophage immunological synapse. J Immunol. 2023;211(9):1385–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ge B. Mycobacterium tuberculosis suppresses host DNA repair to boost its intracellular survival. Cell Host Microbe. 2023;2023(31):1–17. [DOI] [PubMed] [Google Scholar]
- 28.Bedard M, van der Niet S, Bernard EM, et al. A terpene nucleoside from m. Tuberculosis induces lysosomal lipid storage in foamy macrophages. J Clin Invest. 2023;133(6):e161944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maglione PJ, Xu J, Casadevall A, et al. Fc gamma receptors regulate immune activation and susceptibility during mycobacterium tuberculosis infection [J]. J Immunol. 2008;180(5):3329–38. [DOI] [PubMed] [Google Scholar]
- 30.Cox DJ, Coleman AM, Gogan KM, et al. Inhibiting histone deacetylases in human macrophages promotes glycolysis, il-1beta, and t helper cell responses to mycobacterium tuberculosis. Front Immunol. 2020;11:1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang S, Zhou X, Ou M, et al. Berbamine promotes macrophage autophagy to clear mycobacterium tuberculosis by regulating the ros/ca(2+) axis. mBio. 2023;14(4):e0027223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta PK, Jahagirdar P, Tripathi D, et al. Macrophage targeted polymeric curcumin nanoparticles limit intracellular survival of mycobacterium tuberculosis through induction of autophagy and augment anti-tb activity of isoniazid in raw 264.7 macrophages. Front Immunol. 2023;14:1233630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye Y, Liu J, Guo Y, et al. Ppargamma ameliorates mycobacterium tuberculosis h37ra-induced foamy macrophage formation via the abcg1-dependent cholesterol efflux pathway in thp-1 macrophages. Front Microbiol. 2022;13:829870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhengu KN, Singh R, Naidoo P, et al. Cytokine responses during mycobacterium tuberculosis h37rv and ascaris lumbricoides costimulation using human thp-1 and jurkat cells, and a pilot human tuberculosis and helminth coinfection study. Microorganisms. 2023;11(7):1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Priyanka, Medha, Bhatt P, et al. Late stage specific rv0109 (pe_pgrs1) protein of mycobacterium tuberculosis induces mitochondria mediated macrophage apoptosis. Microb Pathog. 2023,176:106021. [DOI] [PubMed]
- 36.Palcekova Z, De K, Angala SK, et al. Impact of methylthioxylose substituents on the biological activities of lipomannan and lipoarabinomannan in mycobacterium tuberculosis. ACS Infect Dis. 2024;10(4):1379–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Menezes YKT, Eto C, de Oliveira J, et al. The endogenous retinoic acid receptor pathway is exploited by mycobacterium tuberculosis during infection, both in vitro and in vivo. J Immunol. 2023;211(4):601–11. [DOI] [PubMed] [Google Scholar]
- 38.Yu X, Huang Y, Li Y, et al. Mycobacterium tuberculosis pe_pgrs1 promotes mycobacteria intracellular survival via reducing the concentration of intracellular free ca(2+) and suppressing endoplasmic reticulum stress. Mol Immunol. 2023;154:24–32. [DOI] [PubMed] [Google Scholar]
- 39.Pu W, Zhao C, Wazir J, et al. Comparative transcriptomic analysis of thp-1-derived macrophages infected with mycobacterium tuberculosis h37rv, h37ra and bcg. J Cell Mol Med. 2021;25(22):10504–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Cao W, Chen S, et al. Comparative interleukins and chemokines analysis of mice mesenchymal stromal cells infected with mycobacterium tuberculosis h37rv and h37ra. Arch Biochem Biophys. 2023;744:109673. [DOI] [PubMed] [Google Scholar]
- 41.Mendonca LE, Pernet E, Khan N, et al. Human alveolar macrophage metabolism is compromised during mycobacterium tuberculosis infection. Front Immunol. 2022;13:1044592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Wang S, Chen X, et al. Mutations in the promoter region of methionine transporter gene metm (rv3253c) confer para-aminosalicylic acid (pas) resistance in mycobacterium tuberculosis. mBio. 2024;15(2):e0207323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Son SH, Lee J, Cho SN, et al. Herp regulates intracellular survival of mycobacterium tuberculosis h37ra in macrophages by regulating reactive oxygen species-mediated autophagy. mBio. 2023;14(5):e0153523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Babele P, Midha MK, Rao KVS, et al. Temporal profiling of host proteome against different m. Tuberculosis strains reveals delayed epigenetic orchestration. Microorganisms. 2023;11(12):2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fumagalli M, Camus SM, Diekmann Y, et al. Genetic diversity of chc22 clathrin impacts its function in glucose metabolism. eLife. 2019;8:e41517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vassilopoulos S, Esk C, Hoshino S, et al. A role for the chc22 clathrin heavy-chain isoform in human glucose metabolism. Science. 2009;324(5931):1192–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Latomanski EA, Newton HJ. Interaction between autophagic vesicles and the coxiella-containing vacuole requires cltc (clathrin heavy chain). Autophagy. 2018;14(10):1710–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rahaman SO, Zhou G, Silverstein RL. Vav protein guanine nucleotide exchange factor regulates cd36 protein-mediated macrophage foam cell formation via calcium and dynamin-dependent processes. J Biol Chem. 2011;286(41):36011–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vedham V, Phee H, Coggeshall KM. Vav activation and function as a rac guanine nucleotide exchange factor in macrophage colony-stimulating factor-induced macrophage chemotaxis. Mol Cell Biol. 2005;25(10):4211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qiu J, Guo J, Liu L, et al. Vav1 promotes inflammation and neuronal apoptosis in cerebral ischemia/reperfusion injury by upregulating microglial and nlrp3 inflammasome activation. Neural Regen Res. 2023;18(11):2436–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu Q, Kong F, Zhao G, et al. Sp1 transcriptionally activates htr2b to aggravate traumatic spinal cord injury by shifting microglial m1/m2 polarization. J Orthop Surg Res. 2024;19(1):230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo H, Du M, Yang Y, et al. Sp1 regulates the m1 polarization of microglia through the hur/nf-kappab axis after spinal cord injury. Neuroscience. 2024;544:50–63. [DOI] [PubMed] [Google Scholar]
- 53.Zheng X, Sarode P, Weigert A, et al. The hdac2-sp1 axis orchestrates protumor macrophage polarization. Can Res. 2023;83(14):2345–57. [DOI] [PubMed] [Google Scholar]
- 54.Pan J, Li Y, Gao W, et al. Transcription factor sp1 transcriptionally enhances gsdme expression for pyroptosis. Cell Death Dis. 2024;15(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cao T, Zhang W, Wang Q, et al. Cancer slc6a6-mediated taurine uptake transactivates immune checkpoint genes and induces exhaustion in cd8(+) t cells. Cell. 2024;187(9):2288-2304 e2227. [DOI] [PubMed] [Google Scholar]
- 56.Yu T, Gan S, Zhu Q, et al. Modulation of m2 macrophage polarization by the crosstalk between stat6 and trim24. Nat Commun. 2019;10(1):4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olety B, Walte M, Honnert U, et al. Myosin 1g (myo1g) is a haematopoietic specific myosin that localises to the plasma membrane and regulates cell elasticity. FEBS Lett. 2010;584(3):493–9. [DOI] [PubMed] [Google Scholar]
- 58.Cruz-Zarate D, Lopez-Ortega O, Giron-Perez DA, et al. Myo1g is required for efficient adhesion and migration of activated b lymphocytes to inguinal lymph nodes. Sci Rep. 2021;11(1):7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patel NR, Bole M, Chen C, et al. Cell elasticity determines macrophage function. PLoS ONE. 2012;7(9):e41024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berg RD, Levitte S, O’Sullivan MP, et al. Lysosomal disorders drive susceptibility to tuberculosis by compromising macrophage migration. Cell. 2016;165(1):139–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mao X, Wu J, Zhang Q, et al. Requirement of wdr70 for pole3-mediated DNA double-strand breaks repair. Sci Adv. 2023;9(36):eadh2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeng M, Tang Z, Guo L, et al. Wdr70 regulates histone modification and genomic maintenance in fission yeast. Biochim Biophys Acta Mol Cell Res. 2020;1867(5):118665. [DOI] [PubMed] [Google Scholar]
- 63.Zeng M, Ren L, Mizuno K, et al. Crl4(wdr70) regulates h2b monoubiquitination and facilitates exo1-dependent resection. Nat Commun. 2016;7:11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Figure S1. GSEA analysis of REACTOME for DRPs between the NCTD and DMSO groups. The Enrichment Score from the GSEA analysis of DRPs enriched in chromatin modifying enzymes was visualized through line plots and heat maps. Figure S2. GSEA analysis of KEGG for DRPs between the NCTD and DMSO groups. The Enrichment Score from the GSEA analysis of DRPs enriched in Fc gamma R-mediated phagocytosis, T cell receptor signaling pathway, and antigen processing and presentation was visualized through line plots and heat maps. Figure S3. Coomassie brilliant blue staining of gels. The integrity of protein samples was assessed using SDS-PAGE gel electrophoresis followed by Coomassie brilliant blue staining. Figure S4. The morphological characteristics show the viability of THP-1 cells treated with the NCTD. Following a 24-hour treatment of THP-1 cells with NCTD, the cellular morphology and density were examined under a microscope at various magnifications.
Data Availability Statement
No datasets were generated or analysed during the current study.