Abstract
1. The pyruvate dehydrogenase complex was purified from Bacillus stearothermophilus in high yield. The specific activity (about 40nkat/mg of protein) was substantially lower than that of the pyruvate dehydrogenase complex from Escherchia coli (about 570nkat/mg of protein) measured at 30 degrees C under the same conditions. 2. The relative molecular masses of the four types of polypeptide chain i the complex were estimated by means of sodium dodecyl sulphate/polyacrylamide-gel electrophoresis to be 57 000, 54 000, 42 000 and 36 000 respectively. These polypetide chains showed no evidence of seriously anomalous behavior during tests of electrophoretic mobility. 3. The enzyme complex was resolved into its constituent proteins by means of gelfiltration on Sepharose CL-6B in the presence of 2M-KI, followed by chromatography on hydroxyapatite in the presence of 8M-urea. These harsh conditions were necessary to cause suitable dissociation of the enzyme complex. 4. The amino-acid compositions of the four constituent proteins after resolution were determined and their chain ratios were measured for several preparations of the complex. Some variability was noted between preparations but all samples contained a significant molar excess of the chains thought to contribute the pyruvate decarboxylase (EC 1.2.4.1) activity. 5. From the relative molecular masses and chain ratios of the four constituent proteins, it was calculated that the empirical unit must be repeated at least 50 times to make up the assembled complex. This conclusion is fully consistent with the demonstration by means of electron microscopy of apparent icosahedral symmetry for the Bacillus stearothermophilus complex, implying a 60-fold repeat. The structure stands in sharp contrast with the octahedral symmetry (24-fold repeat) of the Escherichia coli enzyme.
Full text
PDF


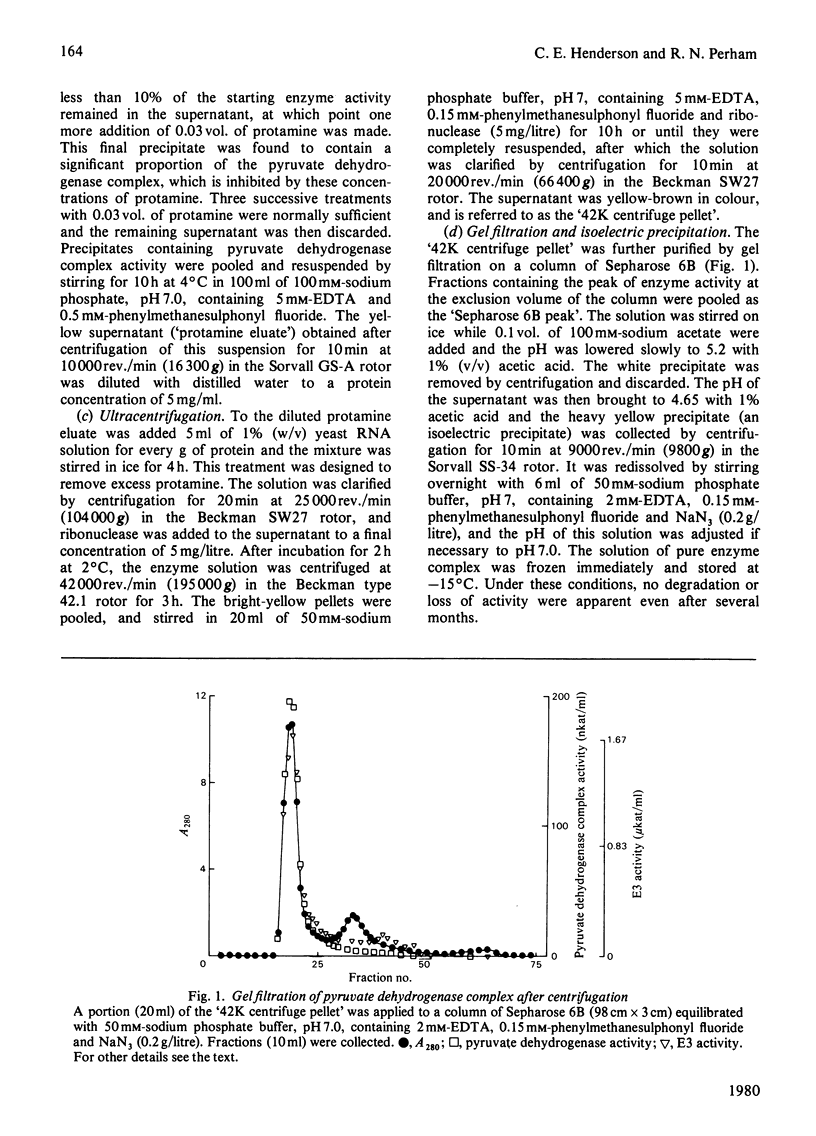

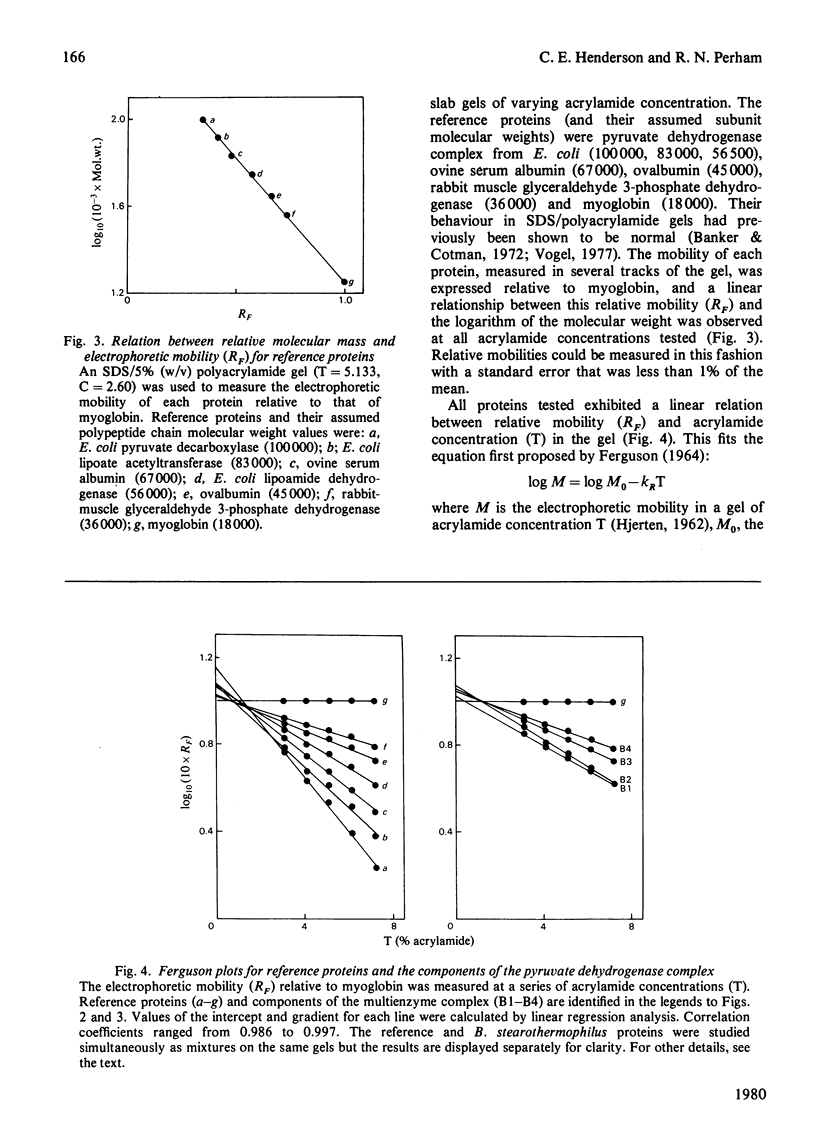
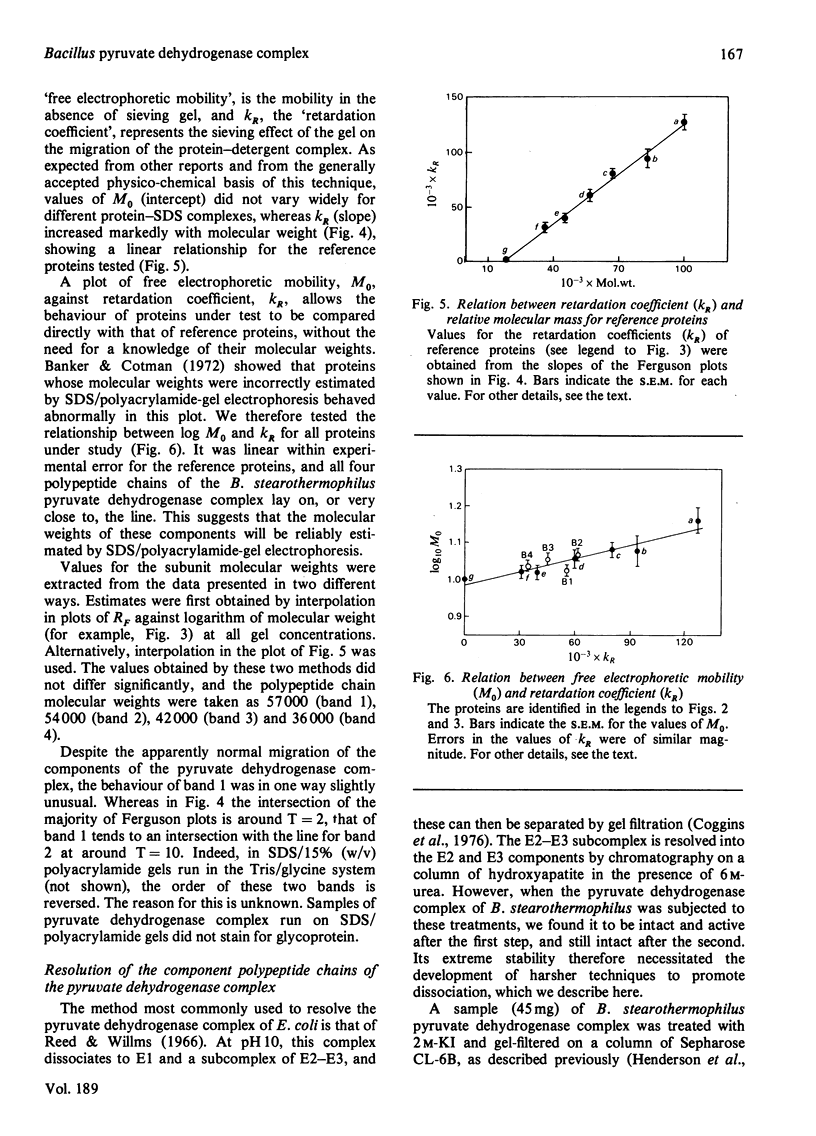
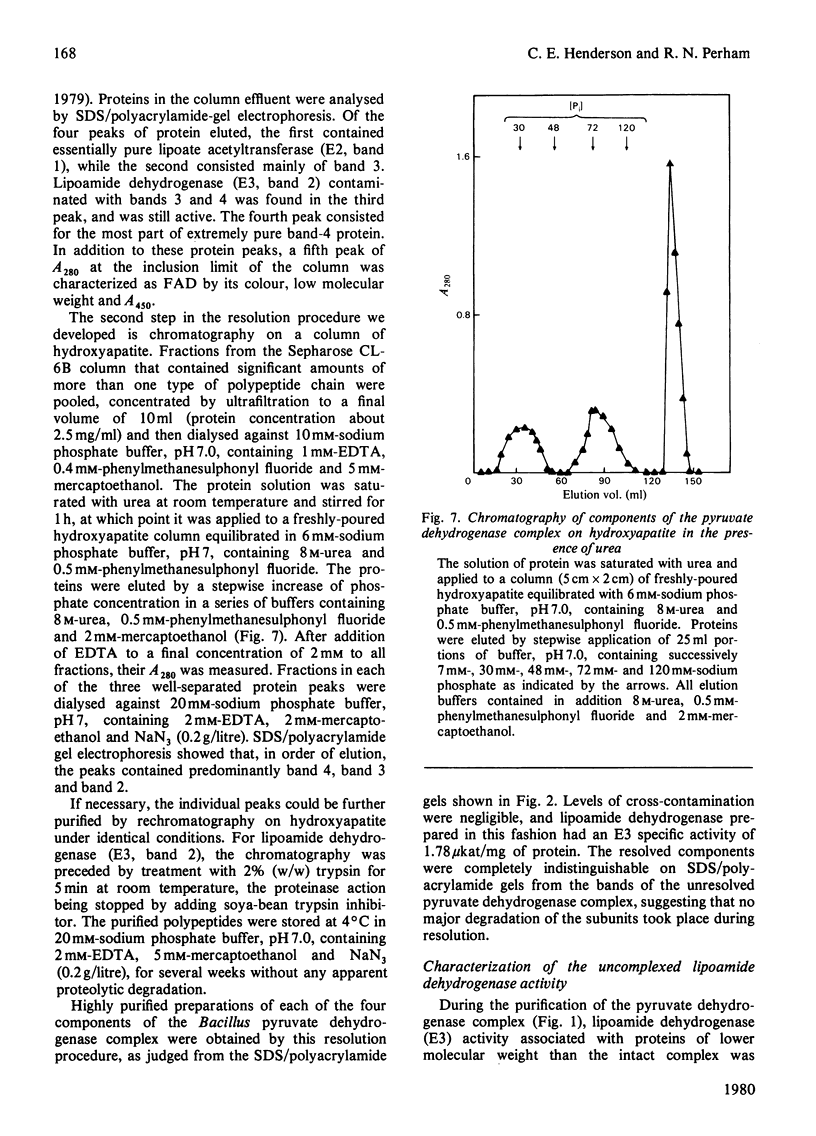




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banker G. A., Cotman C. W. Measurement of free electrophoretic mobility and retardation coefficient of protein-sodium dodecyl sulfate complexes by gel electrophoresis. A method to validate molecular weight estimates. J Biol Chem. 1972 Sep 25;247(18):5856–5861. [PubMed] [Google Scholar]
- Barrera C. R., Namihira G., Hamilton L., Munk P., Eley M. H., Linn T. C., Reed L. J. -Keto acid dehydrogenase complexes. XVI. Studies on the subunit structure of the pyruvate dehydrogenase complexes from bovine kidney and heart. Arch Biochem Biophys. 1972 Feb;148(2):343–358. doi: 10.1016/0003-9861(72)90152-x. [DOI] [PubMed] [Google Scholar]
- Bates D. L., Danson M. J., Hale G., Hooper E. A., Perham R. N. Self-assembly and catalytic activity of the pyruvate dehydrogenase multienzyme complex of Escherichia coli. Nature. 1977 Jul 28;268(5618):313–316. doi: 10.1038/268313a0. [DOI] [PubMed] [Google Scholar]
- Bates D. L., Harrison R. A., Perham R. N. The stoichiometry of polypeptide chains in the pyruvate dehydrogenase multienzyme complex of E. coli determined by a simple novel method. FEBS Lett. 1975 Dec 15;60(2):427–430. doi: 10.1016/0014-5793(75)80764-2. [DOI] [PubMed] [Google Scholar]
- Bretscher M. S. Major human erythrocyte glycoprotein spans the cell membrane. Nat New Biol. 1971 Jun 23;231(25):229–232. doi: 10.1038/newbio231229a0. [DOI] [PubMed] [Google Scholar]
- Coggins J. R., Hooper E. A., Perham R. N. Use of dimethyl suberimidate and novel periodate-cleavable bis(imido esters) to study the quaternary structure of the pyruvate dehydrogenase multienzyme complex of Escherichia coli. Biochemistry. 1976 Jun 15;15(12):2527–2533. doi: 10.1021/bi00657a006. [DOI] [PubMed] [Google Scholar]
- Danson M. J., Hale G., Johnson P., Perham R. N., Smith J., Spragg P. Molecular weight and symmetry of the pyruvate dehydrogenase multienzyme complex of Escherichia coli. J Mol Biol. 1979 Apr 25;129(4):603–617. doi: 10.1016/0022-2836(79)90471-6. [DOI] [PubMed] [Google Scholar]
- Danson M. J., Hooper E. A., Perham R. N. Intramolecular coupling of active sites in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. Biochem J. 1978 Oct 1;175(1):193–198. doi: 10.1042/bj1750193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danson M. J., Perham R. N. Evidence for two lipoic acid residues per lipoate acetyltransferase chain in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. Biochem J. 1976 Dec 1;159(3):677–682. doi: 10.1042/bj1590677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eley M. H., Namihira G., Hamilton L., Munk P., Reed L. J. -Keto acid dehydrogenase complexes. 18. Subunit composition of the Escherichia coli pyruvate dehydrogenase complex. Arch Biochem Biophys. 1972 Oct;152(2):655–669. doi: 10.1016/0003-9861(72)90262-7. [DOI] [PubMed] [Google Scholar]
- FERGUSON K. A. STARCH-GEL ELECTROPHORESIS--APPLICATION TO THE CLASSIFICATION OF PITUITARY PROTEINS AND POLYPEPTIDES. Metabolism. 1964 Oct;13:SUPPL–SUPPL1002. doi: 10.1016/s0026-0495(64)80018-4. [DOI] [PubMed] [Google Scholar]
- Furthmayr H., Timpl R. Characterization of collagen peptides by sodium dodecylsulfate-polyacrylamide electrophoresis. Anal Biochem. 1971 Jun;41(2):510–516. doi: 10.1016/0003-2697(71)90173-4. [DOI] [PubMed] [Google Scholar]
- Gibbons I., Perham R. N. The reaction of aldolase with 2-methylmaleic anhydride. Biochem J. 1970 Mar;116(5):843–849. doi: 10.1042/bj1160843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale G., Hooper E. A., Perham R. N. Amidination of pyruvate dehydrogenase complex of Escherichia coli under denaturing conditions. Biochem J. 1979 Jan 1;177(1):136–137. doi: 10.1042/bj1770136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale G., Perham R. N. Polypeptide-chain stoicheiometry and lipoic acid content of the pyruvate dehydrogenase complex of Escherichia coli. Biochem J. 1979 Jan 1;177(1):129–136. doi: 10.1042/bj1770129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada M., Otsuka K. I., Tanaka N., Ogasahara K., Koike K. Purification properties and subunit composition of pig heart lipoate acetyltransferase. J Biochem. 1975 Jul;78(1):187–197. [PubMed] [Google Scholar]
- Henderson C. E., Perham R. N., Finch J. T. Structure and symmetry of B. stearothermophilus pyruvate dehydrogenase multienzyme complex and implications for eucaryote evolution. Cell. 1979 May;17(1):85–93. doi: 10.1016/0092-8674(79)90297-6. [DOI] [PubMed] [Google Scholar]
- James G. T. Inactivation of the protease inhibitor phenylmethylsulfonyl fluoride in buffers. Anal Biochem. 1978 Jun 1;86(2):574–579. doi: 10.1016/0003-2697(78)90784-4. [DOI] [PubMed] [Google Scholar]
- Koike M., Koike K. Structure, assembly and function of mammalian alpha-keto acid dehydrogenase complexes. Adv Biophys. 1976:187–227. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lehtovaara P. Anomalous migration of leghaemoglobin on sodium dodecyl sulphate/polyacrylamide-gel electrophoresis. Biochem J. 1978 Jan 1;169(1):251–253. doi: 10.1042/bj1690251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panyim S., Chalkley R. The molecular weights of vertebrate histones exploiting a modified sodium dodecyl sulfate electrophoretic method. J Biol Chem. 1971 Dec 25;246(24):7557–7560. [PubMed] [Google Scholar]
- Perham R. N. Self-assembly of biological macromolecules. Philos Trans R Soc Lond B Biol Sci. 1975 Nov 6;272(915):123–136. doi: 10.1098/rstb.1975.0075. [DOI] [PubMed] [Google Scholar]
- Reed L. J., Oliver R. M. The multienzyme alpha-keto acid dehydrogenase complexes. Brookhaven Symp Biol. 1968 Jun;21(2):397–412. [PubMed] [Google Scholar]
- Shapiro A. L., Viñuela E., Maizel J. V., Jr Molecular weight estimation of polypeptide chains by electrophoresis in SDS-polyacrylamide gels. Biochem Biophys Res Commun. 1967 Sep 7;28(5):815–820. doi: 10.1016/0006-291x(67)90391-9. [DOI] [PubMed] [Google Scholar]
- Sugden P. H., Randle P. J. Regulation of pig heart pyruvate dehydrogenase by phosphorylation. Studies on the subunit and phosphorylation stoicheiometries. Biochem J. 1978 Aug 1;173(2):659–668. doi: 10.1042/bj1730659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung J. S., Knight C. A. Effect of charge on the determination of molecular weight of proteins by gel electrophoresis in SDS. Biochem Biophys Res Commun. 1971 Mar 19;42(6):1117–1121. doi: 10.1016/0006-291x(71)90020-9. [DOI] [PubMed] [Google Scholar]
- Vogel O., Hoehn B., Henning U. Molecular structure of the pyruvate dehydrogenase complex from Escherichia coli K-12. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1615–1619. doi: 10.1073/pnas.69.6.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel O. Redetermination of the molecular weights of the components of the pyruvate dehydrogenase complex from E. coli Kl2+. Biochem Biophys Res Commun. 1977 Feb 7;74(3):1235–1241. doi: 10.1016/0006-291x(77)91650-3. [DOI] [PubMed] [Google Scholar]
- Zacharius R. M., Zell T. E., Morrison J. H., Woodlock J. J. Glycoprotein staining following electrophoresis on acrylamide gels. Anal Biochem. 1969 Jul;30(1):148–152. doi: 10.1016/0003-2697(69)90383-2. [DOI] [PubMed] [Google Scholar]



