ABSTRACT
Acinetobacter baumannii is a leading cause of multidrug-resistant bacterial infections worldwide, and the capsular polysaccharide (CPS) is a major virulence determinant. A previous study of A. baumannii from intubated and asymptomatic patients admitted to the intensive care unit (ICU) at the Hospital for Tropical Diseases in Ho Chi Minh City in Vietnam revealed multiple lineages with diverse antibiotic resistance profiles and CPS biosynthesis loci. Here, we show that 48_n, an asymptomatic nasal carriage isolate belonging to ST142, is extensively antibiotic resistant and carries acquired resistance determinants accounting for the resistance profile. 48_n carries the novel KL71 CPS biosynthesis locus in the chromosome. The structure of the CPS produced by 48_n was established using 1H and 13C nuclear magnetic resonance spectroscopy, including two-dimensional 1Н,1Н COSY, 1Н,1Н TOCSY, 1Н,1Н ROESY, 1Н,13C HSQC, and 1Н,13C HMBC experiments, and confirmed by Smith degradation. Consistent with the genetic content of KL71, the K71 CPS was found to be made up of octasaccharide K units containing six l-rhamnose residues and one residue each of N-acetyl-d-glucosamine and d-glucuronic acid. K71 CPS was branched and closely related to the K74 CPS produced by BAL_309, an antibiotic susceptible ST142 isolate recovered from an intubated patient in the same ICU 7 years later. K71 and K74 differ only in the linkage between K units, and this is due to the replacement of a single gene at the K locus that codes for the Wzy polymerase.
IMPORTANCE
The majority of Acinetobacter baumannii genomes sequenced and analyzed to develop an understanding of extensively drug-resistant (XDR) isolates belong to the globally disseminated CC2 clonal complex. While XDR isolates belonging to rarer lineages are often unexplored, detailed analyses could provide novel insights into the spread of resistance, as well as cell surface features such as the CPS that determine the specificity of non-antibiotic therapeutics required to treat XDR infections that resist antimicrobial chemotherapy. Here, we describe the properties of an XDR asymptomatic nasal carriage isolate recovered in Vietnam that belongs to ST142, a rarely encountered sequence type. We report the resistance profile and correlate this with detected resistance determinants. We also solve the structure of the CPS and reveal its relationship with CPS produced by other A. baumannii isolates.
KEYWORDS: Acinetobacter baumannii, capsular polysaccharide, K locus, K71, rhamnose
INTRODUCTION
Acinetobacter baumannii is an opportunistic Gram-negative bacterial pathogen that is listed among the leading etiological agents of mortality attributed to multidrug resistance worldwide (1). Alternate therapeutic interventions being investigated for clinical use include monoclonal antibodies (2) and bacteriophage (3) that target the capsular polysaccharide (CPS) on the cell surface. The specific structure of the CPS produced by individual isolates can vary greatly in carbohydrate composition, the number and types of glycosidic linkages, and non-carbohydrate additions. Hence, elucidation of the structures of CPS produced by multiple, extensive, and pan antibiotic-resistant isolates is important to provide an understanding of target specificity that underpins the design and development of these therapies.
Previously, a study reported the short read data for 159 antibiotic-resistant A. baumannii isolates recovered from patients admitted to the intensive care unit (ICU) at the Hospital for Tropical Diseases (HTD) in Ho Chi Minh City, Vietnam (4). These isolates were obtained from specimens taken from patients assessed for asymptomatic colonization (carriage) following admission to the ICU during 2003–2007, or from intubated patients with ventilator-associated pneumonia (VAP) recovered during an outbreak that occurred between 2008 and 2012. Among this collection, several strains harbored novel CPS loci, and we have since reported the CPS structures for four of them (5–7).
While most isolates belonged to the major globally disseminated clonal complex, CC2 (also known as global or international clone 2), two were reported to belong to sequence type (ST) 142 and carry different sequences at the CPS biosynthesis K locus (KL). One of these isolates, BAL_309, was recovered in 2011 from a VAP patient during the ICU outbreak and was reported to be susceptible to antibiotics and carry the KL74 CPS locus (4). We previously determined the structure of the K74 CPS produced by BAL_309 and showed that it was composed of octasaccharide units consisting of l-rhamnose (l-Rhap), d-glucuronic acid (d-GlcpA), and N-acetyl-d-glucosamine (d-GlcpNAc) (6).
The second ST142 isolate, 48_n, was recovered in 2004, 7 years earlier than BAL_309, from a patient assessed for asymptomatic nasal carriage and was reported to carry a different set of genes, designated KL71, at the CPS biosynthesis locus (4). It was recorded as being resistant to amikacin, ceftazidime, ceftriaxone, and cefalexin, as well as having intermediate resistance to piperacillin/tazobactam, but sensitive to imipenem, ciprofloxacin, and colistin. In this study, we report an extended resistance profile and the genes found in the 48_n genome that account for its antibiotic resistance profile. We also describe the gene content of KL71 and its relationship to KL74. We determined the structure of the K71 CPS produced by 48_n and correlate this to the genetic content of the KL71 locus. We also describe the relationship between isolates 48_n and BAL_309.
RESULTS
A. baumannii 48_n is an extensively antibiotic-resistant isolate
The 48_n isolate was recorded as resistant to beta-lactams ceftriaxone and ceftazidime but susceptible to imipenem. It was also resistant to aminoglycosides amikacin and gentamicin and susceptible to ciprofloxacin and colistin (4). Using a more extensive group of antibiotics, here, resistance to the carbapenems meropenem and doripenem was detected as well as resistance to additional aminoglycosides, tobramycin and netilmicin, kanamycin and neomycin, and streptomycin and spectinomycin. 48_n was also resistant to sulfonamides, tetracycline, doxycycline, and trimethoprim but susceptible to fluoroquinolones. Hence, this sporadic carriage isolate was extensively antibiotic resistant.
The draft genome sequence of A. baumannii 48_n was assembled from the available short read data [Sequence Read Archive (SRA) accession number ERR197591] and was submitted to National Center for Biotechnology Information (NCBI) under accession number JAWXYJ000000000 (BioProject number PRJNA893697 and BioSample number SAMN38236054). The genome was found to belong to ST142IP in the Institut Pasteur (IP) scheme and ST3245OX in the Oxford (OX) scheme and to carry the oxaAb gene variant encoding OXA-510. Genes accounting for the resistance profile were detected; aminoglycoside resistance genes aacC2d, aadB, aphA6, and aadA1 account for the observed aminoglycoside resistance pattern, sul1 and sul2 for sulfonamide resistance, tet(X3) for tetracycline resistance, and dfrA41 for trimethoprim resistance. Three genes encoding beta-lactamases were present (oxa10, blaCARB-49, and blaVEB-7), and the oxa10 gene likely accounts for the observed carbapenem resistance (8). The cmlA1, floR, and arr2 genes and the msrE-mphE gene pair were also detected. The array of cassette-associated genes, aadB:arr2:cmlA1:oxa10:aadA1, was found together with sul1 on a single contig, but the location of these and other resistance genes could not be determined from the draft genome. Some may be on the r3-T13 and r3-T21 plasmids detected.
The KL71 CPS biosynthesis locus
The sequence at the K locus in the 48_n genome was previously identified as KL71 (4), though the genetic content and arrangement of KL71 was not described. KL71 (Fig. 1A, annotated sequence released under GenBank accession number PP621479.1) includes a module of CPS export genes (wza-wzb-wzc) and a module of genes for simple nucleotide-linked sugars and precursors (galU-pgm) (9) on either side of a central region that includes rmlBDAC genes for synthesis of l-Rhap (10), a ugd4 gene for conversion of UDP-d-Glcp to UDP-d-GlcpA (6, 11, 12), five glycosyltransferase genes (gtr#), a wzx translocase gene, and a wzy polymerase gene. It also includes a gene for the ItrA3 initiating transferase that begins K-unit synthesis on the inner membrane via transfer of d-GlcpNAc-1P to the undecaprenol phosphate lipid carrier (10).
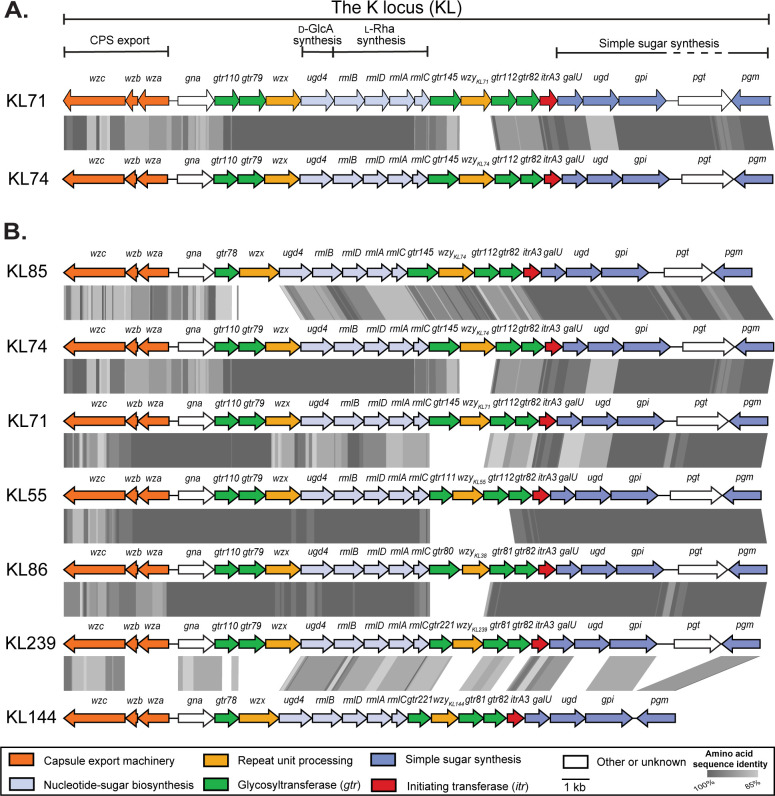
Fig 1.
Organization of the KL71 locus and related K loci. Genes drawn as arrows are colored according to predicted functions of gene products as indicated by the scheme below. Gray shading indicates regions of >85% amino acid sequence identity between predicted proteins determined by tBLASTx using EasyFig v.2.2.2 (13). (A) Comparison of KL71 with KL74. (B) Comparison of KL sequences from isolates producing CPS related to K71. Figure annotated in Adobe Illustrator. Figure drawn to scale using sequences available in the A. baumannii KL reference sequence database (14).
A pairwise sequence comparison of KL71 and the KL74 sequence in the BAL_309 chromosome (GenBank accession number MN148383.1) revealed that the two sequences share 96.97% nucleotide sequence identity over 95% of the locus. KL71 and KL74 (Fig. 1A) share 21 of 22 genes, differing only in the sequence of the gene for the Wzy polymerase responsible for polymerizing K units to form the long-chain CPS. In the K74 CPS, WzyKL74 forms a β-d-GlcpNAc-(1→3)-d-GlcpA linkage between octasaccharide units (6). Although WzyKL71 and WzyKL74 polymerases (GenPept accession numbers WZD86369 and QHE90354.1, respectively) both belong to the EpsG protein family (IPR049458/PF14897), the translated protein sequences do not share significant sequence identity. Hence, KL71 is predicted to produce the same octasaccharide K unit as KL74, but the K71 and K74 CPS structures are likely to have a different linkage between K units.
Structure of the K71 CPS produced by A. baumannii 48_n
A CPS sample was isolated from cells of A. baumannii 48_n by phenol–water extraction (15). Sugar analysis of the CPS revealed rhamnose (Rha) and glucosamine in the ratio 2.9:1.0 (GLC detector response). The l configuration of rhamnose was established by GLC of the acetylated (S)−2-octyl rhamnosides (16). The d configuration of GlcNAc was established by the 13C nuclear magnetic resonance (NMR) data of the CPS using known regularities in glycosylation effects (17).
In the 1H and 13C NMR spectra and a two-dimensional 1Н,13C HSQC spectrum (Fig. 2) of the CPS, there were eight signals for anomeric atoms, including those for six deoxyhexose (Rhap) residues. The presence of signals for an NAc group at δH 2.07 in the 1H NMR spectrum and δC 24.0 (Me) and 175.6 (CO) in the 13C NMR spectrum as well as a signal at δC 56.9 for C-2 in the 13C NMR spectrum showed that one GlcpNAc residue was present. A correlation between a CO2H group (δC 175.1) and H-5 at δH 3.90 in the two-dimensional 1H,13С HMBC spectrum showed that a glucuronic acid (GlcpA) residue was present. Therefore, the CPS has an octasaccharide repeating unit containing six Rhap residues and one residue each of GlcpNAc and GlcpA.
Fig 2.
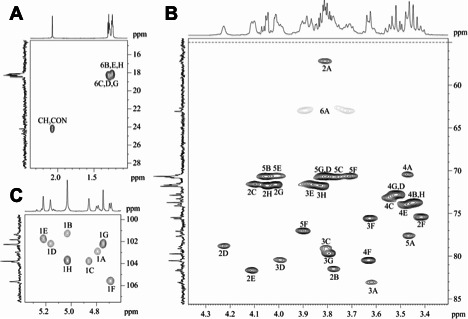
Expansion of 1H,13C HSQC spectrum of K71 OPS. (A) Methyl region. (B) Ring region. (C) Anomeric region. Arabic numerals refer to the carbon atoms in the residues as designated in Table 1.
The 1Н and 13С NMR spectra of the CPS were assigned using two-dimensional 1Н,1Н COSY, 1Н,1Н TOCSY, 1Н,1Н ROESY, and 1Н,13C HSQC experiments (Table 1). The configurations of the glycosidic linkages were established by 13C NMR chemical shifts of C-5 compared with published data of the corresponding α- and β-pyranosides (18, 19). The β configuration of the GlcpNAc (A) and GlcpA (F) residues was confirmed by relatively large coupling constants J1,2 8.5 and 7.8 Hz, respectively, and by H-1–H-3 and H-1–H-5 correlations in the 1H,1H ROESY spectrum of the CPS. The α configuration of the Rha residues (B, C, D, E, G, and H) was confirmed by H-1–H-2 correlations in the 1H,1H ROESY spectrum with no H-1–H-3 and H-1–H-5 correlations.
TABLE 1.
| Monosaccharide residuec | C1 | C2 | C3 | C4 | C5 | C6 | |
|---|---|---|---|---|---|---|---|
| H1 | H2 | H3 | H4 | H5 | H6 (6a,6b) | ||
| CPS | |||||||
| →3)-β-d-GlcpNAc-(1→ | A | 102.7 4.79 |
56.9 3.81 |
82.9 3.62 |
70.2 3.47 |
77.4 3.47 |
62.8 3.71, 3.89 |
| →2)-α-l-Rhap-(1→ | B | 101.1 5.03 |
81.3 3.78 |
71.5 3.85 |
73.5 3.46 |
70.4 3.78 |
18.1 1.28 |
| →3)-α-l-Rhap-(1→ | C | 103.6 4.86 |
71.4 4.11 |
78.9 3.81 |
73.1 3.55 |
4.05 70.4 |
17.9 1.25 |
| →2,3)-α-l-Rhap-(1→ | D | 102.0 5.17 |
78.6 4.23 |
80.3 4.00 |
72.6 3.52 |
70.4 3.82 |
18.1 1.30 |
| →2)-α-l-Rhap-(1→ | E | 101.6 5.22 |
81.4 4.11 |
71.4 3.86 |
73.4 3.48 |
70.4 3.79 |
18.1 1.29 |
| →4)-β-d-GlcpA-(1→ | F | 105.4 4.69 |
75.2 3.42 |
75.4 3.63 |
80.3 3.63 |
76.8 3.90 |
175.1 |
| →3)-α-l-Rhap-(1→ | G | 102.0 4.75 |
71.4 4.02 |
79.5 3.80 |
72.6 3.52 |
70.4 4.00 |
17.9 1.24 |
| α-l-Rhap-(1→ | H | 103.6 5.09 |
71.4 4.05 |
71.7 3.82 |
73.9 3.46 |
70.4 3.72 |
17.9 1.25 |
| OSd | |||||||
| β-d-GlcpNAc-(1→ | A'e | 104.1 4.74 |
57.5 3.75 |
75.3 3.57 |
71.5 3.46 |
77.3 3.45 |
62.3 3.76, 3.92 |
| →3)-α-l-Rhap-(1→ | D' | 103.4 5.04 |
71.4 4.27 |
75.3 3.57 |
73.0 3.55 |
70.7 3.96 |
18.2 1.28 |
| →3)-α-l-Rhap-(1→ | C' | 101.0 4.97 |
71.8 4.08 |
80.0 3.87 |
72.6 3.52 |
70.8 3.83 |
18.2 1.28 |
| →2)-R | B' | 90.9 5.11 |
81.6 3.64 |
61.2 3.74; 3.86 |
|||
Structures of the K71 CPS and OS are shown in Fig. 3. 1H NMR chemical shifts are shown in italics.
R indicates glyceraldehyde in the hydrated form HOCH2CH(O–)CH(OH)2.
Signals for the N-acetyl group of GlcpNAc are at δС 24.0 (Me) and 175.6 (CO), δH 2.07 in the CPS, δС 23.8 (Me) and 176.2 ppm (CO), δH 2.04 in the OS.
OS, oligosaccharide.
Bold face text refers to the residues detected and their order
The glycosylation pattern of the monosaccharides was established by low-field positions of the linkage carbons: C-3 of units A, C, and G, C-2 of units B and E, C-2 and C-3 of unit D, and C-4 of unit F at δ 78.6–82.9 in the 13C NMR spectrum of the CPS, as compared with their positions at δ 71–75 in the corresponding non-substituted monosaccharides (18, 19). The observed downfield shifts were due to glycosylation effects on the linkage carbons (18).
Linkage and sequence analyses of the CPS were performed using a two-dimensional 1H,13C HMBC experiment (Fig. 4). The HMBC spectrum showed inter alia the following inter-residue 1H/13C correlation peaks A H-1/D C-3, B H-1/A C-3, C H-1/B C-2, D H-1/C C-3, E H-1/D C-2, F H-1/E C-2, G H-1/F C-4, and H H-1/G C-3. Accordingly, the following inter-residue correlations between the anomeric protons and linkage carbons were observed in the HSQC–NOESY spectrum (Fig. 5): A H-1/D C-3, D H-1/C C-3, C H-1/B C-2, B H-1/A C-3, E H-1/D C-2, F H-1/E C-2, G H-1/F C-4, and H H-1/G C-3. Hence, K71 includes branched octasaccharide K units with one GlcNAc and three Rha residues in the main chain and one GlcA and three Rha residues in the side chain (Fig. 3A).
Fig 4.
Anomeric region of a 1H,13C of HMBC of the K71 CPS. Arabic numerals before slash refer to protons and after slash to carbons of sugar residues as designated in Table 1.
Fig 5.
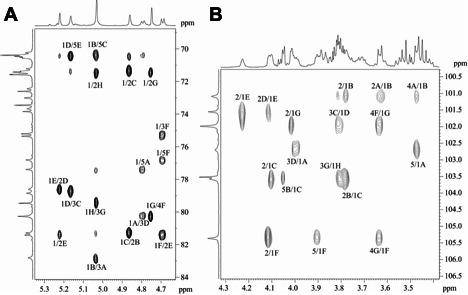
Expansion of 1H, 13C HSQC–NOESY spectrum of K71 OPS. (A) Anomeric region. (B) Ring region. Arabic numerals before slash refer to the protons, and those after slash refer to carbon atoms in the corresponding residues.
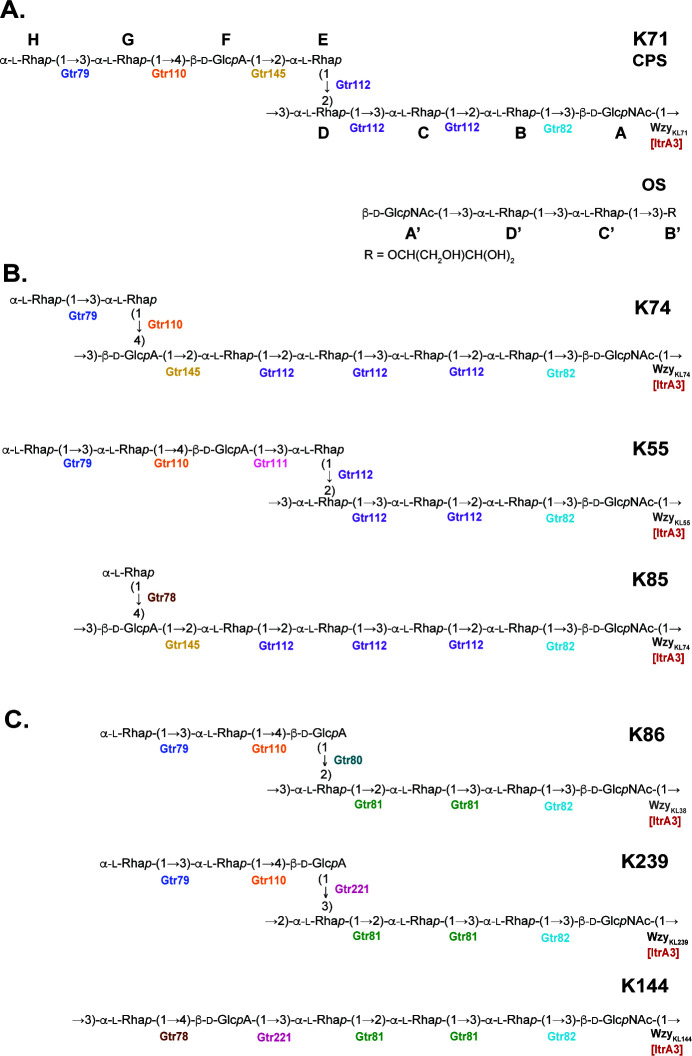
Fig 3.
(A) Structures of the K71 CPS and OS. (B and C) Structures of A. baumannii CPS types related to K71. Individual glycosyltransferases (Gtrs), Wzy polymerases, and initiating transferases (Itrs) are colored and indicated next to the linkages they are predicted to form.
This structure was confirmed by Smith degradation of the CPS, which resulted in an oligosaccharide (OS) (Fig. 3A) as a result of cleavage of GlcA and four of the Rha residues. The OS contained two Rha residues, one GlcNAc residue, and a glyceraldehyde aglycon derived from a 2-substituted Rha residue. The structure of the OS was established by NMR spectroscopy as described for the CPS (for the assigned 1Н and 13C NMR chemical shifts, see Table 1). The OS structure was confirmed by determination of its molecular mass by negative ion mode electrospray ionization mass spectrum, which showed peaks of the [M + Na]+ ion at m/z 626.2263 and [M + K]+ ion at m/z 642.2017 against the calculated values m/z 626.2267 and 642.2006, respectively.
Configuration of the K71 unit and assignment of the WzyKL71 polymerase
As the K71 octasaccharide unit includes a single d-GlcpNAc, this residue was assigned as the first sugar due to the presence of itrA3 in KL71 that codes for an ItrA3 d-GlcpNAc-1P initiating transferase. Hence, in the K71 CPS, there is a tetrasaccharide main chain of →3)-α-l-Rhap-(1→3)-α-l-Rhap-(1→2)−α-l-Rhap-(1→3)-β-d-GlcpNAc-(1→, and an α-l-Rhap-(1→3)-α-l-Rhap-(1→4)-β-d-GlcpA-(1→2)-l-Rhap side branch joined α-(1→4) to the terminal l-Rhap residue in the main chain (Fig. 3A). The bond between individual K units is therefore a β-d-GlcpNAc-(1→3)-α-l-Rhap linkage. As the bond between K units is the only linkage that differs between K71 and K74, which has a six sugar main chain and a disaccharide side chain, and the linkages formed by the encoded glycosyltransferases were previously assigned for K74 [as shown in Fig. 3B (6)], the WzyKL71 polymerase encoded by KL71 could be assigned to the β-d-GlcpNAc-(1→3)-α-l-Rhap linkage. This assignment is supported by WzyKL71 sharing 48.73% amino acid sequence identity with WzyKL55 reported to form a β-d-GlcpNAc-(1→3)-α-l-Rhap linkage between units in the A. baumannii K55 CPS (6) (see Fig. 3).
Distribution of KL71 and KL74 in A. baumannii genomes
A total of 27,079 A. baumannii genome assemblies available in NCBI (as of 21 May 2024) were screened for the presence of the KL71 CPS biosynthesis gene cluster to assess the distribution of the K71 CPS in A. baumannii. The KL71 sequence was found in only two further genomes from isolates PUMA0075 and IK20 (NCBI assembly accession numbers GCA_030552105 and GCA_032941345, respectively). Both are environmental isolates recovered in Southeast Asia and belong to ST142. Similarly, KL74 was only otherwise identified in two genomes that also belong to ST142. These are isolate A156 (NCBI assembly accession number GCA_022937765) from a respiratory specimen recovered in China in 2018 and isolate ARLG-1813 (NCBI assembly accession number GCA_002136815) recovered in the United States of America. The average nucleotide identity of these six genomes was >99.8%, indicating that they are highly related. This suggests that both KL71 and KL74 are rarely encountered CPS types in A. baumannii and are currently restricted to the ST142 lineage.
DISCUSSION
The majority of extensively resistant A. baumannii that warrant alternate therapeutic inventions belong to the globally disseminated clonal complex, CC2. ST142 is a relatively uncommon lineage with only four ST142 isolates reported in the literature to date. These include one isolate recovered from the cerebrospinal fluid of a hospitalized patient in China (20); an environmental isolate recovered from tank milk in Bogor, Indonesia (described above) (21); and BAL_309 and 48_n isolates recovered from the same ICU at the HTD in Ho Chi Minh City, Vietnam (4). In this study, we report that isolate 48_n is extensively antibiotic resistant and carries a large suite of acquired resistance genes that account for the observed resistance profile. However, while BAL_309 was recovered during an outbreak of VAP in the ICU, 7 years after 48_n, this isolate is susceptible to antibiotics. The oxa10 gene does not confer resistance to carbapenems in Escherichia coli, where it is usually studied, and its ability to confer resistance to some carbapenem antibiotics in A. baumannii (8) is not widely appreciated. Here, the carbapenem resistance phenotype of 48_n, namely, susceptible to imipenem but resistant to meropenem and doripenem, is consistent with the phenotype reported earlier (8) for oxa10 in A. baumannii.
Structural resolution of the CPS produced by 48_n confirmed that the sequence of the K71 unit is identical to that of K74 produced by BAL_309 (6) and that the two CPSs differ only in the nature of the linkage between K units that is formed by the Wzy polymerase. This difference arises due to the replacement of the wzy gene at the K locus, for which the encoded products share little sequence homology and select different donor sugars to participate in the bond (l-Rhap for WzyKL71 and d-GlcpA for WzyKL74), resulting in a change to the overall topology where different numbers of monosaccharides are present in the main chain and side branch of the CPS (Fig. 3B; K71 = four main-chain and four side-branch sugars; K74 = six main-chain and two side-branch sugars).
These two structures also share the same monosaccharide constituents as five other CPS types previously described for A. baumannii (Fig. 3). These types (K55, K85, K86, K144, and K239) are all initiated by d-GlcpNAc followed by an l-Rhap sugar α-(1→3) linked to this residue by a Gtr82 glycosyltransferase. This group of related CPS can be further split into two subgroups based on the sequence of the 2-substitued and 3-substituted Rha residues in the main chain, and this is determined by the activity of either the multifunctional Gtr112 glycosyltransferase that forms three linkages (Fig. 3B) or bifunctional Gtr81 glycosyltransferase that forms two linkages (Fig. 3C) as described previously (6). Consistent with this, the corresponding gene clusters for these CPSs share a central ugd4-rmlBDAC gene module (Fig. 1B) but differ in the number and types of glycosyltransferase genes and/or the sequence for the Wzy polymerase. While four of these CPSs share a β-d-GlcpNAc-(1→3)-α-l-Rhap linkage between their K units, the encoded Wzy polymerases share little sequence identity to each other despite belonging to the same protein family (Table 2). Hence, the specificity of Wzy polymerases is complex and likely involves multiple structural epitopes present in the A. baumannii CPS.
TABLE 2.
Comparison of Wzy polymerases showing amino acid sequence identity matrix on the right
| Wzy | Protein family | Length (aa) | No. of TMS | Unit topologya | Linkage | WzyKL71 | WzyKL55 | WzyKL144 | WzyKL86 | WzyKL239 | WzyKL74/ WzyKL85 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| WzyKL71 | EpsG | 359 | 9 | 4 + 4 | β-d-GlcpNAc-(1→3)-l-Rhap | 100 | 48.73 | 23.91 | 21.17 | 22.46 | 19.62 |
| WzyKL55 | EpsG | 365 | 10 | 4 + 4 | β-d-GlcpNAc-(1→3)-l-Rhap | 48.73 | 100 | 19.32 | 19.47 | 22.46 | 17.57 |
| WzyKL144 | EpsG | 321 | 9 | 6 + 0 | β-d-GlcpNAc-(1→3)-l-Rhap | 23.91 | 19.32 | 100 | 27.56 | 20.00 | 23.21 |
| WzyKL86 | EpsG | 327 | 9 | 4 + 3 | β-d-GlcpNAc-(1→3)-l-Rhap | 21.17 | 19.47 | 27.56 | 100 | 16.91 | 19.80 |
| WzyKL239 | –b | 366 | 9 | 4 + 3 | β-d-GlcpNAc-(1→2)-l-Rhap | 22.46 | 22.46 | 20.00 | 16.91 | 100 | 19.92 |
| WzyKL74/KL85cc | EpsG | 412 | 9 | 6 + 2/1d | β-d-GlcpNAc-(1→3)-d-GlcpA | 19.62 | 17.57 | 23.21 | 19.80 | 19.92 | 100 |
Number of residues in the K-unit main chain versus number of residues in the K-unit side branch.
No detected Pfam.
WzyKL74 from KL74 and KL85 are 98.8% identical.
K74 has a disaccharide side branch, whereas K85 has a monosaccharide side branch.
TMS = transmembrane segments.
MATERIALS AND METHODS
Isolate, cultivation, and resistance profiling
A. baumannii isolate 48_n was obtained in 2004 from a patient nasal carriage sample at the HTD in Ho Chi Minh City, Vietnam (4). The antibiotic resistance profile of 48_n was determined as described previously (22). For CPS structural analysis, cells of 48_n were cultivated in 2TY medium for 16 h, harvested by centrifugation (10,000 × g, 20 min), washed with phosphate buffered saline, then suspended in an acetone–water mixture [7:3 (vol/vol)], precipitated by centrifugation, and dried on air.
Whole-genome assembly and bioinformatics analyses
Short read data for A. baumannii 48_n (SRA accession number SRR26815235) was quality checked then de novo assembled using SPAdes v.3.15.5 (23). The assembled genome consisted of 277 contigs with a total length of 4,182,363 bases, an N50 of 60.7 kb, and a read depth of 80×. The assembly was annotated using Prokka (https://github.com/tseemann/prokka) and uploaded to NCBI under accession number JAWXYJ000000000 (BioProject number PRJNA893697 and BioSample number SAMN38236054). OrthoANI values were calculated with the OAT tool (24).
Multilocus sequence typing was performed using the A. baumannii IP and OX schemes available at https://pubmlst.org/bigsdb?db=pubmlst_abaumannii_seqdef. Resistance genes were identified using NCBI AMRFinder (https://www.ncbi.nlm.nih.gov/pathogens/antimicrobial-resistance/AMRFinder/). Kaptive v.2.0.7 (https://github.com/klebgenomics/Kaptive) was used to identify the sequence at the K locus using the latest A. baumannii KL reference sequence database that includes 241 KL (14). The KL71 sequence in the A. baumannii 48_n genome was extracted, manually annotated according to the established nomenclature system (9), and submitted to NCBI GenBank under accession number PP621479.1.
Isolation of CPS
Cell mass (3.05 g) was extracted with 45% aqueous phenol (70°C, 1 h) (15). The extract was dialyzed without layer separation and freed from insoluble contaminations by centrifugation. The resultant solution was concentrated and treated with cold aqueous 50% CCl3CO2H at 0°C for 1 h; after centrifugation, the supernatant was dialyzed against distilled water. A crude CPS sample (100 mg) was hydrolyzed with 2% CH3CO2H (100°C, 2 h). Fractionation of the products by gel-permeation chromatography on a column (56 × 2.5 cm) of Sephadex G-50 Superfine (Healthcare) in 0.05-M pyridinium acetate (pH 4.5) as eluent gave a purified CPS sample (56 mg).
Chemical analyses
A CPS sample (1 mg) was hydrolyzed with 2-M CF3CO2H (120°C, 2 h) then blown in air current with MeOH. The products were reduced with NaBH4 in 1-M NH4OH (0.5 mL, 10 mg, 20°C, 1 h) and after blowing off in AcOH 2% in MeOH acetylated by a 1:1 (vol/vol) mixture of pyridine and Ac2O (120°C, 2 h). The alditol acetates obtained were analyzed by GLC on a Maestro chromatograph (Agilent 7820; Interlab, Russia) equipped with an HP-5 column (0.32 mm × 30 m) using a temperature program of 160°C (1 min) to 290°C at 7 °C/min.
To determine the absolute configuration of the monosaccharides, a CPS sample (1 mg) was hydrolyzed with 2-M CF3CO2H (120°C, 2 h) then blown in air current with MeOH. Following this, (+)−2-octanol (0.1 mL) and CF3CO2H (15 µL) were added (16 h, 120°C.) After blowing off with MeOH, the products were acetylated with a 1:1 (vol/vol) mixture of pyridine and Ac2O (120°C, 2 h). The acetylated 2-octyl glycosides were analyzed by GLC as indicated above.
Smith degradation
A CPS sample (21 mg) was oxidized with aqueous NaIO4 (25 mg in 3.2 mL of H2O) at 20°C for 40 h in the dark and reduced with NaBH4 (25 mg) at 20°C for 16 h. The excess of NaBH4 was destroyed with concentrated HOAc; the solution was evaporated; then methanol was added to the residue (3 × 1 mL) and evaporated. The residue was dissolved in 0.3-mL water and applied to a column (80 × 1.6 cm) of TSK HW-40 (S) in 1% HOAc. A modified polysaccharide was eluted with aqueous 0.1% HOAc and hydrolyzed with 2% CF3CO2H (100°C, 2 h). Fractionation of the products by gel-permeation chromatography on a column (80 × 1.6 cm) of TSK HW-40 (S) in 1% HOAc gave an OS sample (7.4 mg).
NMR spectroscopy
Samples were deuterium-exchanged by freeze-drying from 99.9% D2O and then examined as solutions in 99.95% D2O. NMR spectra were recorded on a Bruker Avance II 600 MHz spectrometer (Germany) at 60°C. Sodium 3-trimethylsilylpropanoate-2,2,3,3-d4 (δH 0, δC −1.6) was used as internal reference for calibration. Two-dimensional NMR spectra were obtained using standard Bruker software, and Bruker TopSpin v.2.1 program was used to acquire and process the NMR data. A 60-ms MLEV-17 mixing time and a 200-ms spin-lock time were used in 1H,1H TOCSY and ROESY experiments, respectively. A 60-ms delay was used for evolution of long-range couplings to optimize a 1H,13C HMBC experiment for the coupling constant of JH,C 8 Hz. A mixing time of 0.3 s was used in a HSQC–NOESY experiment.
Mass spectrometry
High-resolution electrospray ionization mass spectrometry was performed in the negative ion mode using a micrOTOF II instrument (Bruker Daltonics). An oligosaccharide sample (~50 ng/L) was dissolved in a 1:1 (vol/vol) water–acetonitrile mixture and injected with a syringe at a flow rate of 3 µL/min. Capillary entrance voltage was set at 3,200 V, and the interface temperature was set at 180°C. Nitrogen was used as the drying gas. Mass range was from m/z 50 to 3,500. Internal calibration was done with ESI Calibrant Solution (Agilent).
ACKNOWLEDGMENTS
We thank Stephen Baker for providing A. baumannii isolate 48_n.
This work was supported by an Australian Research Council Future Fellowship (FT230100400, to J.J.K.), a National Health and Medical Research Council Australia Investigator Fellowship (GNT1194978, to R.M.H.), and the Russian Science Foundation (24-14-00236 to Y.A.K.).
Note
TMS = transmembrane segment
Contributor Information
Johanna J. Kenyon, Email: j.kenyon@griffith.edu.au.
Olaya Rendueles, Centre de Biologie Integrative, Toulouse, France.
REFERENCES
- 1. Murray CJL, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, Han C, Bisignano C, Rao P, Wool E, et al. 2022. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet 399:629–655. doi: 10.1016/S0140-6736(21)02724-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nielsen TB, Yan J, Slarve M, Lu P, Li R, Ruiz J, Lee B, Burk E, Talyansky Y, Oelschlaeger P, Hurth K, Win W, Luna BM, Bonomo RA, Spellberg B. 2021. Monoclonal antibody therapy against Acinetobacter baumannii Infect Immun 89:e0016221. doi: 10.1128/IAI.00162-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu M, Hernandez-Morales A, Clark J, Le T, Biswas B, Bishop-Lilly KA, Henry M, Quinones J, Voegtly LJ, Cer RZ, Hamilton T, Schooley RT, Salka S, Young R, Gill JJ. 2022. Comparative genomics of Acinetobacter baumannii and therapeutic bacteriophages from a patient undergoing phage therapy. Nat Commun 13:3776. doi: 10.1038/s41467-022-31455-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schultz MB, Pham Thanh D, Tran Do Hoan N, Wick RR, Ingle DJ, Hawkey J, Edwards DJ, Kenyon JJ, Phu Huong Lan N, Campbell JI, Thwaites G, Thi Khanh Nhu N, Hall RM, Fournier-Level A, Baker S, Holt KE. 2016. Repeated local emergence of carbapenem-resistant Acinetobacter baumannii in a single hospital ward. Microb Genom 2:e000050. doi: 10.1099/mgen.0.000050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arbatsky NP, Kenyon JJ, Kasimova AA, Shashkov AS, Shneider MM, Popova AV, Knirel YA, Hall RM. 2019. K units of the K8 and K54 capsular polysaccharides produced by Acinetobacter baumannii BAL 097 and RCH52 have the same structure but contain different di-N-acyl derivatives of legionaminic acid and are linked differently. Carbohydr Res 483:107745. doi: 10.1016/j.carres.2019.107745 [DOI] [PubMed] [Google Scholar]
- 6. Kenyon JJ, Arbatsky NP, Sweeney EL, Zhang Y, Senchenkova SN, Popova AV, Shneider MM, Shashkov AS, Liu B, Hall RM, Knirel YA. 2021. Involvement of a multifunctional rhamnosyltransferase in the synthesis of three related Acinetobacter baumannii capsular polysaccharides, K55, K74 and K85. Int J Biol Macromol 166:1230–1237. doi: 10.1016/j.ijbiomac.2020.11.005 [DOI] [PubMed] [Google Scholar]
- 7. Kenyon J.J, Kasimova AA, Shashkov AS, Hall RM, Knirel YA. 2018. Acinetobacter baumannii isolate BAL_212 from Vietnam produces the K57 capsular polysaccharide containing a rarely occurring amino sugar N-acetylviosamine. Microbiology (Reading) 164:217–220. doi: 10.1099/mic.0.000598 [DOI] [PubMed] [Google Scholar]
- 8. Antunes NT, Lamoureaux TL, Toth M, Stewart NK, Frase H, Vakulenko SB. 2014. Class D β-lactamases: are they all carbapenemases? Antimicrob Agents Chemother 58:2119–2125. doi: 10.1128/AAC.02522-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kenyon JJ, Hall RM. 2013. Variation in the complex carbohydrate biosynthesis loci of Acinetobacter baumannii genomes. PLoS ONE 8:e62160. doi: 10.1371/journal.pone.0062160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kenyon J.J, Shashkov AS, Senchenkova SN, Shneider MM, Liu B, Popova AV, Arbatsky NP, Miroshnikov KA, Wang L, Knirel YA, Hall RM. 2017. Acinetobacter baumannii K11 and K83 capsular polysaccharides have the same 6-deoxy-L-talose-containing pentasaccharide K units but different linkages between the K units. Int J Biol Macromol 103:648–655. doi: 10.1016/j.ijbiomac.2017.05.082 [DOI] [PubMed] [Google Scholar]
- 11. Kenyon JJ, Kasimova AA, Sviridova AN, Shpirt AM, Shneider MM, Mikhaylova YV, Shelenkov AA, Popova AV, Perepelov AV, Shashkov AS, Dmitrenok AS, Chizov AO, Knirel YA. 2021. Correlation of Acinetobacter baumannii K144 and K86 capsular polysaccharide structures with genes at the K locus reveals the involvement of a novel multifunctional rhamnosyltransferase for structural synthesis. Int J Biol Macromol 193:1294–1300. doi: 10.1016/j.ijbiomac.2021.10.178 [DOI] [PubMed] [Google Scholar]
- 12. Arbatsky NP, Popova AV, Shneider MM, Shashkov AS, Hall RM, Kenyon JJ, Knirel YA. 2021. Structure of the K87 capsular polysaccharide and KL87 gene cluster of Acinetobacter baumannii LUH5547 reveals a heptasaccharide repeating unit. Carbohydr Res 509:108439. doi: 10.1016/j.carres.2021.108439 [DOI] [PubMed] [Google Scholar]
- 13. Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cahill SM, Hall RM, Kenyon JJ. 2022. An update to the database for Acinetobacter baumannii capsular polysaccharide locus typing extends the extensive and diverse repertoire of genes found at and outside the K locus. Microb Genom 8:e000878. doi: 10.1099/mgen.0.000878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Westphal O, Jann K. 1965. Bacterial lipopolysaccharides: extraction with phenol-water and further applications of the procedure. methods in carbohydrate chemistry, p 83–91. Academic press, New York. [Google Scholar]
- 16. Leontein K, Lönngren J. 1993. Determination of the absolute configuration of sugars by gas–liquid chromatography of their acetylated 2-octyl glycosides. Methods Carbohydr Chem 9:87–89. doi: 10.1016/s0021-9673(96)00987-9 [DOI] [Google Scholar]
- 17. Shashkov AS, Lipkind GM, Knirel YA, Kochetkov NK. 1988. Stereochemical factors determining the effects of glycosylation on the 13 C chemical shifts in carbohydrates . Mag Reson Chem 26:735–747. doi: 10.1002/mrc.1260260904 [DOI] [Google Scholar]
- 18. Lipkind GM, Shashkov AS, Knirel YA, Vinogradov EV, Kochetkov NK. 1988. A computer-assisted structural analysis of regular polysaccharides on the basis of 13C-n.m.r. data. Carbohydr Res 175:59–75. doi: 10.1016/0008-6215(88)80156-3 [DOI] [PubMed] [Google Scholar]
- 19. Jansson P-E, Kenne L, Schweda E. 1987. Nuclear magnetic resonance and conformational studies on monoacetylated methyl D-gluco- and D-galacto-pyranosides. J Chem Soc Perkin Trans 1:377–383. doi: 10.1039/p19870000377 [DOI] [Google Scholar]
- 20. Chen T, Fu Y, Hua X, Xu Q, Lan P, Jiang Y, Yu Y, Zhou Z. 2021. Acinetobacter baumannii strains isolated from cerebrospinal fluid (CSF) and bloodstream analysed by cgMLST: the dominance of clonal complex CC92 in CSF infections. Int J Antimicrob Agents 58:106404. doi: 10.1016/j.ijantimicag.2021.106404 [DOI] [PubMed] [Google Scholar]
- 21. Sykes EME, Mateo-Estrada V, Engelberg R, Muzaleva A, Zhanel G, Dettman J, Chapados J, Gerdis S, Akineden Ö, Khan IUH, Castillo-Ramírez S, Kumar A. 2024. Phylogenomic and phenotypic analyses highlight the diversity of antibiotic resistance and virulence in both human and non-human Acinetobacter baumannii mSphere 9:e0074123. doi: 10.1128/msphere.00741-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Holt KE, Kenyon JJ, Hamidian M, Schultz MB, Pickard DJ, Dougan G, Hall RM. 2016. Five decades of genome evolution in the globally distributed, extensively antibiotic-resistant Acinetobacter baumannii global clone 1. Microb Genom 2:e000052. doi: 10.1099/mgen.0.000052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee I, Ouk Kim Y, Park S-C, Chun J. 2016. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103. doi: 10.1099/ijsem.0.000760 [DOI] [PubMed] [Google Scholar]